Abstract
Aggregation of mutant superoxide dismutase 1 (SOD1) is a pathological hallmark of a subset of familial ALS patients. However, the possible role of misfolded wild type SOD1 in human ALS is highly debated. To ascertain whether or not misfolded SOD1 is a common pathological feature in non-SOD1 ALS, we performed a blinded histological and biochemical analysis of post mortem brain and spinal cord tissues from 19 sporadic ALS, compared with a SOD1 A4V patient as well as Alzheimer’s disease (AD) and non-neurological controls. Multiple conformation- or misfolded-specific antibodies for human SOD1 were compared. These were generated independently by different research groups and were compared using standardized conditions. Five different misSOD1 staining patterns were found consistently in tissue sections from SALS cases and the SOD1 A4V patient, but were essentially absent in AD and non-neurological controls. We have established clear experimental protocols and provide specific guidelines for working, with conformational/misfolded SOD1-specific antibodies. Adherence to these guidelines will aid in the comparison of the results of future studies and better interpretation of staining patterns. This blinded, standardized and unbiased approach provides further support for a possible pathological role of misSOD1 in SALS.
Introduction
Amyotrophic Lateral Sclerosis (ALS) is a heterogeneous neurodegenerative syndrome characterized by adult-onset progressive loss of primarily motor neurons in the cerebral cortex, brain stem, and spinal cord. ALS leads to progressive muscle weakness, atrophy and typically death 3 to 5 years after symptom onset1,2. About 5–10% of ALS cases are hereditary (familial; FALS)3,4. The remaining ≈90% of cases lack an overt familial history and are referred to as sporadic (SALS)3–5. Repeat DNA expansion in C9ORF72 as well as mutations in approximately 40 other genes have been associated with ALS; most frequently SOD1, TARDBP (TDP-43) and FUS/TLS (for review see6).
Neurodegeneration in ALS is associated with protein misfolding and aggregation and the formation of inclusions in regions of the central nervous system (CNS) affected by the disease7. For example, cytoplasmic inclusions of TDP-43 are frequently observed in motor neurons in a majority of non-SOD1 FALS cases, with or without TARDBP mutations, and in many SALS cases8–17. Inclusions of human mutant or wild type SOD1 are found in transgenic mouse models overexpressing the proteins18–24 and in patient material at autopsy8,19,25–28. The findings that SOD1 mutant proteins with biophysical properties similar to wild type SOD1 (wtSOD1); including D90A29 and L117V30, cause ALS, as well that overexpression of wtSOD1 causes an ALS-like disease in transgenic mice24, supports an emerging hypothesis that wtSOD1 contributes to ALS. Interestingly, it has been also reported that wtSOD1 can acquire an aberrant conformation, implying a possible shared pathological pathway between mutant SOD1-linked FALS and SALS27. Other studies have also demonstrated that wild type human SOD1 acquires toxic properties upon oxidative damage31–33. It also lead to an exacerbation of disease phenotype in transgenic mice expressing different SOD1 mutants34,35. Hence, it is possible that wtSOD1 may be a contributor to disease pathogenesis in sporadic ALS.
Conformational- and misfolded-specific antibodies, recognizing either naturally buried epitopes or completely misfolded forms of SOD1 have emerged as valuable tools with which to distinguish unfolded or misfolded SOD1 species from the natively folded protein20,26–28,36,37. These antibodies have been used to detect misSOD1 in spinal motor neurons of SALS patients without SOD1 mutations19,26–28. However, several other studies have concluded that misSOD1 is not present in SALS38–42. Since this discrepancy could depend on differences in the methodology used for immunohistochemistry, we aimed to address this in a range of SALS patient material. Using a panel of different antibodies that bind specifically to unfolded/misfolded conformations of SOD1 and following standardized experimental protocols, we have undertaken a blinded histopathological study and also biochemical analysis of misSOD1. The results provide supporting evidence that strongly suggest a role for misSOD1 in non-SOD1 SALS.
Results
Specific staining of misSOD1 in FALS
Four different polyclonal, as well as one monoclonal antibody, covering epitopes across the entire SOD1 protein (N-terminus, central region and C-terminus) were used (Fig. 1). All antibodies chosen have previously been shown to be highly specific for human misSOD126–28. First, immunohistochemical analyses were performed in post-mortem spinal cord tissues from a FALS patient heterozygous for the A4V SOD1 mutation (positive control; Fig. 2) and compared with 2 non-neurological controls (negative controls), in order to standardize the immunostaining protocol.
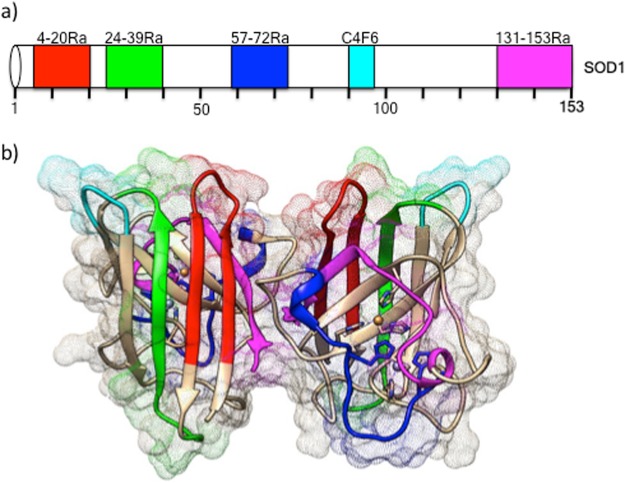
Figure 1.
Human SOD1 protein schematic representation with highlighted misSOD1 epitope mapping regions (a) Linear representation of the 5 tested misfolded/conformational SOD1 specific antibody’s binding regions. (b) 3D protein structure representation with highlighted misfolded/conformational SOD1 antibody’s bindging regions. Ra: Rabbit polyclonal. C4F6: mouse monoclonal.
Figure 2.
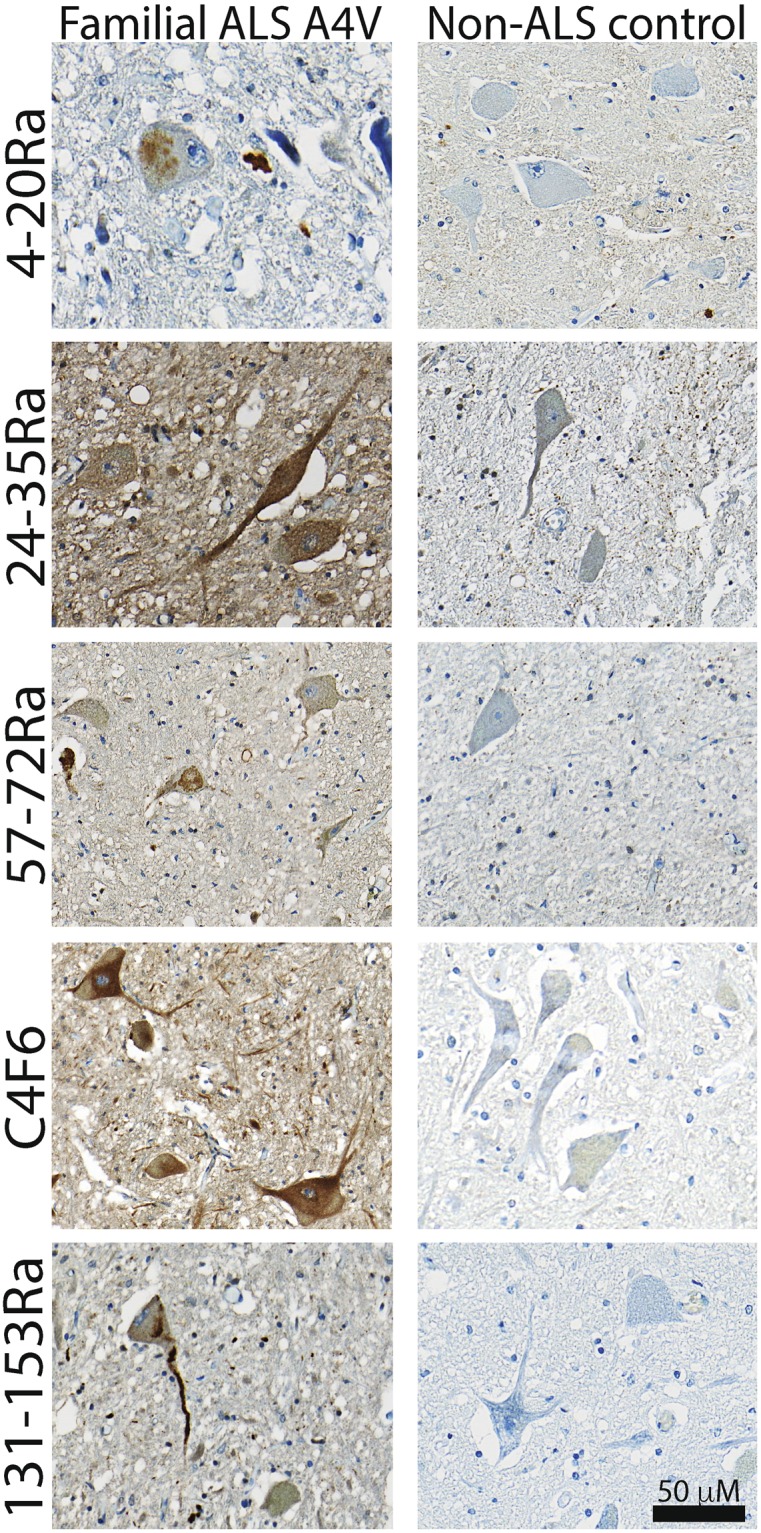
Immunodetection of misSOD1 accumulation in spinal cord of A4V SOD1 FALS patient Lumbar spinal cord sections from A4V-SOD1 FALS autopsied patient (positive control) and non-neurological control individual (negative control) immunostained with 5 misSOD1 conformational antibodies (4–20Ra, 24–39Ra, 57–72Ra, C4F6 and 131–153 R). Characteristic misSOD1-positive cytoplasmic accumulation in spinal motor neurons can be observed with all tested antibodies in the A4V-SOD1 FALS patient, whereas no immunostaining is detected in the non-neurological control. Scale bar: 50 mm.
All 5 antibodies revealed the presence of cytoplasmic misSOD1 accumulation within spinal motor neurons of the grey matter in the FALS patient harbouring the A4V SOD1 mutation. Strong axonal and dendritic misSOD1 accumulation, as well as intense punctate immunostaining in the neuropil were also observed. In agreement with previous observations26,28, no immunoreactive signal was detected in the control spinal cord sections, confirming the high specificity of all of the tested misSOD1 antibodies.
Widespread staining of misSOD1 in SALS
Having validated the panel of SOD1 antibodies in the FALS vs control material, we next analysed a cohort of 13 French Canadian SALS cases, 2 neurological AD cases and 2 non-neurological controls (Table 1). Material from cortical (frontal, temporal and occipital lobes as well as entorhinal sections) and spinal cord (lumbar, cervical, thoracic and sacral spinal sections) regions were examined in a blinded manner. Positive misSOD1 immunoreactive signals were detected in all of the 13 SALS cases using the 5 misSOD1 antibodies (Fig. 3, Supplementary Table 1 and Supplementary Figures 1–5). Five characteristic misSOD1-positive immunostaining patterns were observed and can be summarized as; (1) diffuse misSOD1 staining in the cytoplasm of motor neurons, (2) misSOD1-positive deposits in the cytoplasm of motor neurons, (3) dendritic and axonal misSOD1 accumulation, (4) misSOD1-positive perivacuolar ring-like structures, and (5) misSOD1 nuclear accumulation (Fig. 3). We determined a cut-off, whereby to be considered as misSOD1positive, a case should display more than one of the above immunostaining patterns in two or more regions of the CNS. Interestingly, not all tested antibodies detected the misSOD1 immunostaining patterns with the same intensity in each of the patients (Supplementary Figures 1–5). Based on these results, more than one conformational- and misSOD1-specific antibody was consequently used to detect the presence of misSOD1 species in at least two different spinal cord regions for each of the tested SALS and control individuals.
Table 1.
Summary of the studied ALS patients and control individuals.
| Patient | Genotype | Age of onset | Age of death | Time to autopsy (Hours) | Gender | Pathological TDP-43 status | Origin |
|---|---|---|---|---|---|---|---|
| 1 | FALS SOD1/A4V | 72 | 73 | 22 | F | Nuclear | Umeå, Sweden |
| 2 | SALS | 53 | 63 | 16 | F | Cytoplasmic | Québec, Canada |
| 3 | SALS | 68 | 78 | 18 | M | Cytoplasmic | Québec, Canada |
| 4 | SALS | 53 | 63 | 15 | F | Cytoplasmic | Québec, Canada |
| 5 | SALS | 79 | 81 | 22 | M | Cytoplasmic | Québec, Canada |
| 6 | SALS | 52 | 54 | Unavailable | M | Cytoplasmic | Québec, Canada |
| 7 | SALS | Unavailable | 73 | Unavailable | M | Cytoplasmic | Québec, Canada |
| 8 | SALS | 60 | 67 | Unavailable | F | Cytoplasmic | Québec, Canada |
| 9 | SALS | 72 | 74 | Unavailable | M | Cytoplasmic | Québec, Canada |
| 10 | SALS | 72 | 74 | 23 | M | Cytoplasmic | Québec, Canada |
| 11 | SALS | 49 | 53 | 20 | F | Cytoplasmic | Québec, Canada |
| 12 | SALS | Unavailable | 61 | 15 | M | Cytoplasmic | Québec, Canada |
| 13 | SALS | Unavailable | 78 | 24 | F | Cytoplasmic | Québec, Canada |
| 14 | SALS | 67 | 69 | 24 | M | Cytoplasmic | Québec, Canada |
| 15 | SALS | 68 | 70 | 21 | M | Cytoplasmic | Umeå, Sweden |
| 16 | SALS | 60 | 61 | 23 | F | Cytoplasmic | Umeå, Sweden |
| 17 | SALS | 64 | 65 | 18 | M | Cytoplasmic | Umeå, Sweden |
| 18 | SALS | 62 | 64 | 20 | M | Cytoplasmic | Umeå, Sweden |
| 19 | SALS | 64 | 69 | Unavailable | F | Cytoplasmic | Umeå, Sweden |
| 20 | SALS | 64 | 67 | Unavailable | F | Cytoplasmic | Umeå, Sweden |
| 21 | Non-neurological Control | Not applicable | 65 | 15 | M | Nuclear | NIH Neurobiobank, USA |
| 22 | Non-neurological Control | Not applicable | 73 | 22 | M | Nuclear | Umeå, Sweden |
| 23 | Neurological Control (Alzheimer) | Not applicable | 65 | 23 | F | Nuclear | Québec, Canada |
| 24 | Neurological control (Alzheimer) | Not applicable | 58 | 20 | M | Nuclear | Québec, Canada |
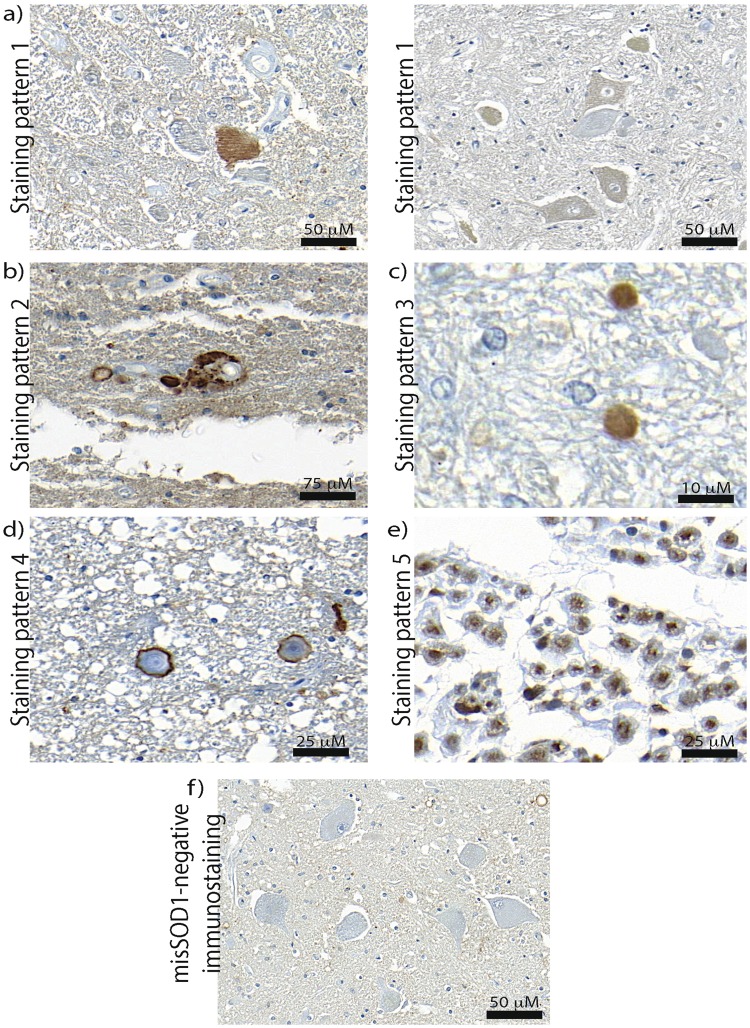
Figure 3.
Detected misSOD1 immunostaining patterns MisSOD1 immunostaining patterns detected in SALS individuals were (a) Diffuse misSOD1 staining in the cytoplasm of motor neurons (immunostaining pattern 1). Note that different misSOD1 immunosignal intensity levels were detected. As misSOD1-positive motor neurons can be detected beside misSOD1-negative motor neurons on the same tissue section, it is unlikely that the observed misSOD1 accumulation represents a false-positive signal (b) misSOD1-positive deposits detecetd in the cytoplasm of motor neurons (immunostaining pattern 2) (c) Dendritic and axonal misSOD1 accumulation (immunostaining pattern 3) (d) MisSOD1-positive perivacuolar ring-like structures (immunostaining pattern 4) (e) MisSOD1 nuclear accumulation (immunostaining pattern 5) and (f) misSDO1-negative immunostaining observed in non-neurological control. Scale bars are presented in the lower right corner for each detected immunostaining patterns.
Distinct patterns of misSOD1 in SALS
Immunohistopathological analyses using the 4–20Ra misSOD1 antibody, targeting the N-terminal region of the human SOD1 protein, showed diffuse cytoplasmic misSOD1 accumulation in spinal motor neurons of all SALS cases (Supplementary Figure 1, and Supplementary Table 1). Axonal segments in the ventral horn, perivacuolar ring-like structure, as well as nuclear misSOD1 accumulation were identified SALS cases (Supplementary Figure 1, and Supplementary Table 1). Immunostaining, using both the 24–39Ra and the 57–72Ra antibodies, revealed diffuse misSOD1 accumulation as well as misSOD1-positive deposits in the cytoplasm of motor neurons in almost all of the SALS cases studied (Supplementary Figures 2,3, and Supplementary Table 1). Axonal, perivacuolar and nuclear misSOD1 positive immunostainings were also observed in the vast majority of SALS spinal cord sections (Supplementary Table 1). MisSOD1 species in the cytoplasm of motor neurons were also detected in SALS patients, using both C4F6 and 131–153Ra antibodies, targeting the C-terminal part of the SOD1 protein (Supplementary Figures 4, 5, and Supplementary Table 1). MisSOD1-positive immunostaining was also frequently observed in large myelinated fibers in the lumbar ventral horn for the vast majority of SALS patients using all of the tested antibodies (Fig. 3 and Supplementary Table 1).
Interestingly, misSOD1-positive ring-like immunostainings, surrounding corpora amylacea (CA)-like structures, were densely located within the grey matter of spinal anterior horns, whereas misSOD1-negative CA-like structures were rather observed at the spinal cord periphery (Fig. 4).
Figure 4.
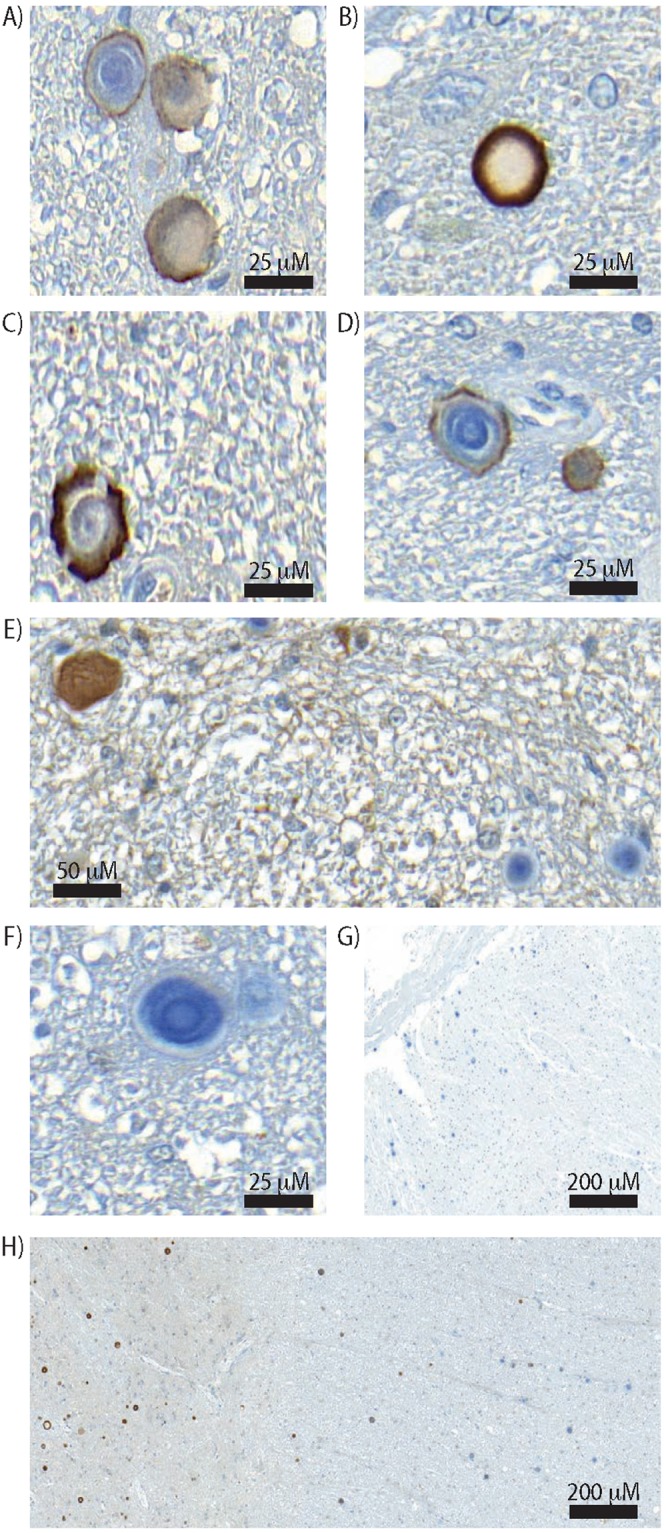
Detection of misSOD1-positive ring-like structure in the SALS and control individuals Representative images of misSOD1-positive ring-like structures in the extracellular space. (A–D) misSOD1-positive serpiginous structures detected in SALS cases detected in spinal horn grey matter. (E–G) misSOD1-negative CA-like structures detected in SALS cases located in the spinal cord tissue section periphery. (H) Spinal ventral horn grey matter accumulation of misSOD1-positive CA-like structures and, peripheral white matter negative misSOD1 CA-like deposits within the spinal cord.
MisSOD1 accumulation was also detected with at least 2 or more of the above-described patterns of staining, for all of the tested conformation specific misSOD1 antibodies, in post-mortem brain tissue sections (frontal, temporal and occipital lobes as well as enthorinal sections) from SALS patients (Fig. 5). MisSOD1 immunoreactivity was negligable in the 2 individuals with Alzheimer’s disease and in the non-neurological controls. Furthermore, no misSOD1-positive staining of CA-like structures was observed in control material.
Figure 5.
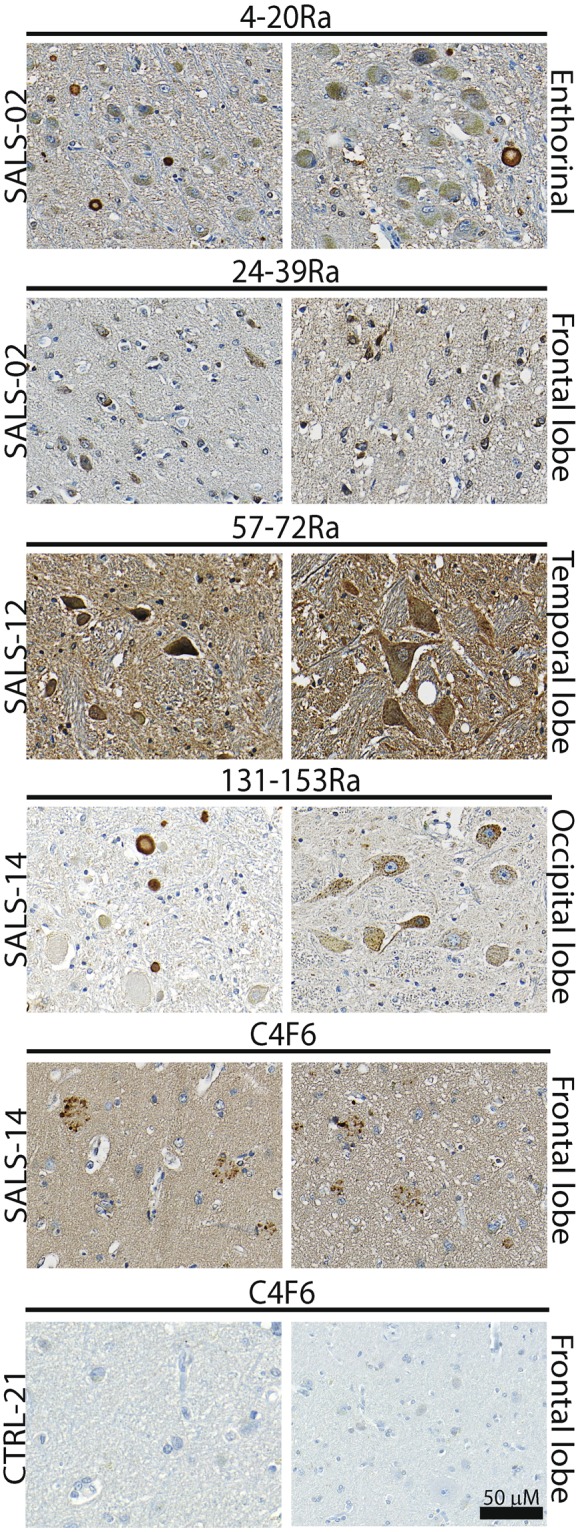
Immunodetection of misfolded SOD1 aggregates detected in brain tissue sections of SALS patients by immunohistochimestry using the misfolded SOD1/conformational-specific 4–20Ra, 24–39Ra, 57–72Ra, C4F6 and 131–153Ra antibodies Brain tissue sections from SALS patients and non-ALS control individuals immunostained using 5 different misSOD1/conformational-specific antibodies. Two representative pictures per SALS patients and control are showed. Different brain areas were analysed and misSOD1-positive accumulation can be observed.
Immunocapture of misSOD1 from SALS spinal cord
To validate the presence of misSOD1 in SALS spinal cord protein extracts, we used an independent method to immunocapture misSOD1 from whole tissue extracts with the C4F6 and 24–39Ra antibodies. Although all misfolded/conformational SOD1 antibodies used in this study worked in immunohistochemistry, the C4F6 and 24–39Ra misfolded/conformational SOD1 antibodies were the best working antibodies for this specific experiment. Available frozen and unfixed spinal cords from SALS patients were used. As positive controls we also included spinal cord tissue from an A4V SOD1 mutation carrier and from an end-stage G93ASOD1 mouse. Spinal cord protein extract, collected from one non-neurological control, was used as a negative control. At a similar level of SOD1 in the input sample, significant amounts of misSOD1 were immunocaptured from SALS and A4V SOD1 spinal cords (Fig. 6). However, no misSOD1 was detected above background threshold in the non-neurological control lysate.
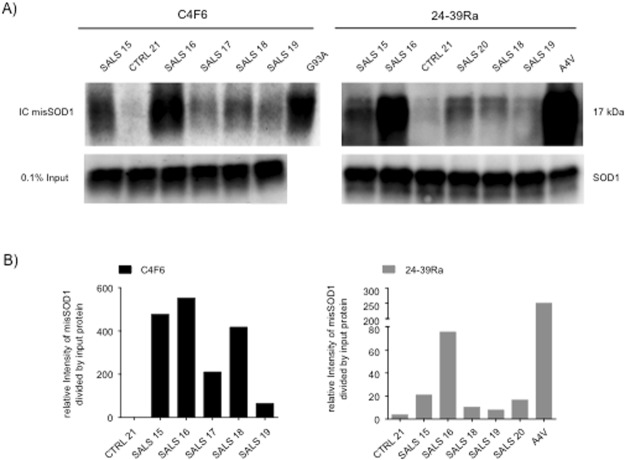
Figure 6.
Immunocapture of misSOD1 in SALS spinal cord tissue samples using the C4F6 and 24–39Ra misSDO1 specific antibodies (A) Immunocapture of misSOD1 in total spinal cord protein extracts, obtained from SALS patients, A4V-SOD1 FALS patient, non-ALS control individual and transgenic mice over-expressing G93ASOD1 mutant protein is illustrated. (B) Quantification data of the immunocaptured misSOD1 protein shown in panel A. Immunodetection of the transferred immunucaptured products, using the 24–39Ra and the C4F6 misSOD1/conformational specific antibodies, revealed that both antibodies yielded clear and positive immunoreactivity signals and a high degree of specificity whilst only very low levels were detected in spinal cord extracts collected from the non-ALS control individual. A fraction of the total amount of proteins used (0.1%) was also run on a SDS–PAGE to validate that an equal amount of protein was loaded from each sample. Note that, due to the availability of patient materials, SALS17 and SALS20 spinal cord extracts were only immunocaptured using either the C4F6 or 24–39Ra antibodies.
Discussion
To ascertain the potential importance of misfolded SOD1 in the pathology in SALS, we conducted blinded, controlled, histopathological and biochemical analyses of misSOD1 in brain and spinal cord samples from a FALS A4V-SOD1 patient and 19 SALS patients (13 analyzed by IHC and 6 by IC/Western blotting). Non-neurological control as well as non-ALS neurological control individuals showed no specific misSOD1 positive immunostaining patterns (Supplementary Table 1). In SALS individuals, variability in misSOD1 staining patterns was observed, which were found to be highly specific to SALS, whereas no misSOD1 immunoreactivity was detected in all tested control individuals including neurological Alzheimer patients. Detection of misSOD1 in available SALS spinal cords was also confirmed by immunocapture analysis. Hence, differential staining patterns may correspond either to a pathological – clinical correlation, or reflect the presence of different misSOD1 species.
An evolving hypothesis has proposed a common final neurodegenerative pathway shared by at least some FALS and SALS cases that involves cytotoxic, non-native conformers of SOD1. This is supported by several studies, which have demonstrated that: (1) wtSOD1 acquires properties of ALS-linked mutant human SOD1 species27,32; (2) Mice that overexpress wild-type human SOD1 at high rate develop both SOD1 aggregation and a fatal ALS-like disease24; (3) Expression of human wtSOD1 exacerbates disease in transgenic mice expressing different mutant SOD1s34,35; (4) Aberrant cytoplasmic TDP-43 or FUS accumulation can trigger misfolding of the human wtSOD1 protein8; and (5) Misfolded forms of human wtSOD1 have been detected in post-mortem CNS tissue of both SALS and FALS without SOD1 mutations8,19,26,28. However, the presence of misSOD1 in SALS is disputed (Table 3). Variability in the fixation and storage of autopsy material and in the standardization of immunostaining protocols may be one of the reasons for such divergent interpretations. Regardless, the difficulty to detect misSOD1 species in SALS or the presence of misSOD1 immunostaining in control individuals does not formally rule out its role as a pathological component in SALS. Lack of compliance with the published instructions for the staining of misSOD1 aggregates and with the actual proposed standardized immunodetection guidelines (Supplementary Table 2), which may lead to false-negative or false-positive results, are most likely the reasons for the divergent interpretations.
Table 3.
Studies describing the presence or absence of misSOD1 in human SALS post-mortem tissues.
| misSOD1 | # of SALS cases | |
|---|---|---|
| Bosco et al., 2010, Nat. Neuro27. | Yes | 9 |
| Forsberg et al., 2010, PLoS ONE26 | Yes | 29 |
| Forsberg et al., 2011, Acta Neuropatho28. | Yes | 51 |
| Pokrishevsky et al., 2012, PLoS ONE8 | Yes | 3 |
| Grad et al., 2014, PNAS19 | Yes | 20 |
| Ayers et al., 2014, Acta Neuropatho. Comm41. | Yes but no difference with controls | 25 |
| Brotherton et al., 2012, PNAS39 | No | 25 |
| Liu et al., 2009, Ann. Neuro38. | No | 13 |
| Kerman et al., 2010, Acta Neuropatho40. | No | 10 |
| Da Cruz et al., 2017, Acta Neuropatho42. | Yes but no difference with controls by IHC and no misSOD1 detection in SALS by IF | 30 |
A common denominator between all published studies evaluating the presence of misSOD1 species in SALS is the lack of standardization in IHC experimental protocols. This may lead to inconsistent results and so far standardized protocols for the evaluation of SOD1 staining patterns has been limited by a lack of standard criteria defining misSOD1 immunostaining. This has made it difficult to compare results of retrospective or prospective studies. To overcome this drawback, we have sought to establish clear experimental methods and guidelines for the comparison of misfolded SOD1- and conformational-specific antibodies (summarized in Table 4 and fully described in the Supplementary Table 2). We suggest that the adherence to these guidelines will aid in the interpretation of staining patterns with misSOD1/conformational antibodies and propose that the same guidelines might also be applicable to other misfolded-specific and conformational SOD1 antibodies.
Table 4.
Summary of the proposed guidelines.
| Guidelines | |
|---|---|
| 1 | To use more than one misfolded SOD1/conformational-specific antibody before concluding any results |
| 2 | To test and use optimal antibodies working concentrations |
| 3 | To optimize antigen retrieval time for each antibody |
| 4 | To use citrate-based instead of TRIS/EDTA-based buffers |
| 5 | To test different central nervous system regions (cervical, lumbar, thoracic spinal cord sections and other brain regions) |
| 6 | To perform, in parallel, Hematoxylin/Eosin coloration on adjacent sections |
In contrast to our results, Da Cruz and colleagues firmly excludes the possibility that misfolded SOD1 is a common feature of sporadic ALS42. The authors found no difference between SALS and controls by IHC, and an absence of misSOD1 immunoreactive signal by IF in any of the tested cases. It is important to highlight here that this discrepancy with our actual data depends on differences in the methodology used for both IHC and IF protocols. As a matter of fact, Da Cruz and colleagues used Tris/EDTA based-buffers to retrieve antigens, which most likely led to false-positive misSOD1 staining in their controls. Indeed, EDTA is known to be a chelator of bivalent metals that may interfere with the proper folding of metalloproteins such as SOD1, and may therefore explain the presence of false-positive misSOD1 signals20,43–45. The use of the reducing agent DTT, preventing intra-molecular disulfide bond formation necessary for the proper folding of the protein, could also explain the false-positive detection of misSOD1 by immunocapture obtained by Da Cruz and colleagues in non-ALS controls. Moreover, the antigen retrieval step, shown to be crucial when working with misSOD1 antibodies, was omitted during the IF protocol used by Da Cruz and colleagues, as well as questionable antibody working dilutions (e.g. 1/60,000) for the detection of misSOD1, could also explain the lack of misSOD1 immunoreactive signal.
Different patterns of misSOD1 immunostaining were observed in this study, including nuclear misSOD1 accumulation and perivacuolar misSOD1-positive ring-like structures in the extracellular space (Figs 3 and 4). Although nuclear wtSOD1 as well as misSOD1 nuclear expression as been previously reported28,46, clear evidence of a nuclear function of SOD1 protein, either native or misfolded, has not been ascertained. Taking into account the serpiginous aspect and the size of the detected misSOD1 positive ring-like structures, it is likely that these could be corpora amylacea (CA), which are small hyaline masses of unknown origin. CA are round extracellular structures of 10 to 50 μm in diameter and are frequently found beneath the pia matter within the normal aging brain as well as in a variety of neurological conditions including AD, multiple sclerosis, hippocampal sclerosis and epilepsy47–53. Their composition includes ubiquitin, heat-shock proteins, myelin basic protein, NeuN, S100 proteins, alpha-synuclein (for review see54) which suggests protein contents probably originates from neuron and/or neuropil degradation products. Their ability to calcify over time is an additional argument in favour of this hypothesis. CA were shown to be less abundant in ALS or in Parkinson patients as compared to CNS samples from AD patients51. In contrast, misSOD1-positive CA-like structures were frequently observed in spinal cord CNS samples from SALS patients in the present study (Supplementary Table 1). On the other hand, we were not able to detect these misSOD1-positive structures in the neurological and non-neurological controls, indicating that the detected misSOD1 immunoreactive signals may be specific to SALS. They can be witnesses of the evolution of the disease but can also have an alternative protective function by sequestering aggregated proteins. The precise role or relevance of misSOD1-positive CA-like structures in our study remain elusive. Interestingly, misSOD1 CA-like structures were also densely located within the grey matter of spinal anterior horns in SALS patients while negative CA were often located at the spinal cord periphery (Fig. 4). Our data support the idea that different CA counterparts, arising from different origin, may exist55 (Fig. 4 and Supplementary Figure 6). This study interestingly showed that CA lying beneath the pia matter, often larger in size, are unaffected by changes in the neuronal population, whereas CA lying in the grey matter may be more responsive to neuronal loss. To date, our data are in favour of their specific character in SALS and encourages to consider them as positive marker of the disease (pattern 4 of the reading grid) even if further studies are necessary in order to clearly delineate their role and their specificity in ALS.
Material and Methods
Animals
Transgenic mice overexpressing G93A mutant SOD1 (www.jax.org/strain/002726) were maintained and cared for according to the guidelines of the Animal Care and Use Committee of Umeå University, Sweden. The Regional Ethical Committee for Animal Experiments approved experimental protocols involving animals beforehand, under a project with reference A49–14.
Human samples and neuropathological examination
This study was performed in accordance with the Declaration of Helsinki (WMA, 1964) and approved by the appropriate national ethical review boards in Canada (Ethical research board of the CHU de Québec. Protocol number: 2012-1316. For more information, please contact gurecherche@chuq.qc.ca) or Sweden (Ethical Review Board Ref. No. 14-17-31 M). Human autopsies were performed as described56. Informed consent for study participation has been obtained from each participant.
CNS tissue was obtained from autopsy material from 19 SALS and one A4V-SOD1 FALS patient, as well as 2 non-neurological control individuals and 2 neurological (Alzheimer’s Disease) controls (see Table 1 for clinical information). The ALS patients met the El Escorial criteria for clinically definite, probable or laboratory supported ALS57. All died from respiratory failure, some with concomitant pneumonia. None had received treatment with invasive ventilation through a tracheotomy, or experimental therapy apart from treatment with riluzole. A diagnosis of FALS or SALS was deduced after collecting genealogical and medical data from at least three prior generations in accordance with Byrne et al.58. All ALS patients were also screened for mutations in a panel of known ALS-causing genes including SOD1, TARDBP, FUS and a hexanucleotide-repeat expansion in C9ORF72 (for details see59–63). Analyses were performed on genomic DNA extracted from peripheral blood leucocytes. Post-mortem tissue samples from one non-neurological individual and from 2 Alzheimer’s disease patients were obtained from the NIH Neurobiobank at the University of Maryland, Baltimore, MD. Post-mortem CNS tissues were fixed by immersion in 20% buffered formalin, or stored unfixed in a −80 °C freezer until use. Paraffin-embedded material was used for immunohistochemical analyses. Sectioning of SALS and control post-mortem tissues (10 mm thickness) was performed at the CHU de Quebec. All slides were encoded and sent to Umea University for immunohistochemical analyses.
Immunohistochemistry
Sections were stained using the automated BenchMark ULTRA (Ventana Tucson, AZ, USA). Deparaffinization of tissue sections was performed by heating at 72 °C for 12 minutes in Ventana EZ solution. Antigen retrieval was done using a sodium citrate solution (CC2, Ventana Tucson, AZ, USA) at 91 °C for 44 minutes (24 minutes for 57–72Ra antibody) to obtain the optimal signal to noise ratio. Primary antibodies were incubated for 32 minutes at 37 °C. Sections were then stained using the ultraView Universal DAB detection kit and counterstained with hematoxylin and lithium carbonate (Bluing reagent). Slides were dehydrated with alcohol and xylene before mounting. Imaging was performed using a digital slide scanner. Antibodies, dilutions and antigen retrieval incubation times are listed in Table 2.
Table 2.
Immunohistochemistry conditions using the Ventana BenchMark ULTRA.
| Antibody | Antigen retrieval incubation time | Working dilution |
|---|---|---|
| 4–20Ra | 44 minutes | 1/350 |
| 24–39Ra | 44 minutes | 1/1,000 |
| 57–72Ra | 24 minutes | 1/3,000 |
| C4F6 (Non-purified) | 44 minutes | 1/2 |
| 131–153Ra | 44 minutes | 1/1,000 |
Immunocapture
End-stage mutant G93ASOD1 transgenic mice were sacrificed by intraperitoneal injection of pentobarbital. The spinal cord was isolated by flushing with saline and homogenized immediately in 25 volumes of ice-cold immunocapture (IC) buffer (PBS, 40 mM iodoacetamide (IAM, Thermo Scientific, Rockford, IL, USA), Complete EDTA-Free Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) and 0.5% Nonidet P-40 using a tissue homogenizer followed by sonication. Lysates were centrifuged at 25,000 g for 30 min. at 4 °C and supernatants were stored at −80 °C until analysis.
Segments of un-fixed frozen lumbar spinal cord from patients were used for immunocapture. Weighed spinal cord sections were homogenized in 10 volumes of IC buffer as described above, centrifuged at 1,000 g for 10 min. at 4 °C and the resulting supernatant was analyzed by immunocapture.
Anti-human SOD1 antibodies (24–39Ra or C4F6) were cross-linked to Dynabeads M-270 Epoxy with the Dynabeads Antibody Coupling Kit (Invitrogen, California, USA). Beads were isolated with a magnet, washed to remove unbound antibodies and equilibrated with IC buffer. Antibody-coated beads were incubated with equal volumes of spinal cord extracts for 1 hour at 23 °C. The beads were washed 5 times with IC buffer and samples were eluted by boiling in 1x sample buffer containing 40 mM IAM. Immunocaptured proteins and the spinal cord extracts were analyzed using non-reduced SDS-PAGE and western blotting as previously described64–66.
The different binding regions, recognized by the misfolded/conformational SOD1 antibodies were visualized using the SOD1 1AZV protein data bank entry67 with the Chimera protein structure visualization system68. Note that the minimal binding epitope for C4F6 contains, but not necessarily restricted to, amino acids 90–9341.
Electronic supplementary material
Acknowledgements
We would like to thank Lily-Ann Franche, Lydia Touzel Deschênes, Manon Labrecque, Matthew Marklund, Helena Alstermark, Eva Jonsson, Ann-Charloth Nilsson, Karin Hjertkvist, Anna-Lena Bolender and Ulla-Stina Spetz for skillful technical assistance. This work was supported by the Canadian Institutes for Health Research, the W. Garfield Weston Foundation through the Weston Brain Institute, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the Bertil Hållsten Foundation, the Torsten and Ragnar Söderberg Foundation, the Swedish Brain Fund, the Stratneuro Initiative, Västerbotten County Council and the Kempe Foundations. FGL is the recipient of a tier 2 Canada research Chair. BP is the recipient of a Fond de recherche du Québec en santé (FRQS) and an ALS Canada Doctoral Research award.
Author Contributions
Experimental and technical inputs: B.P., M.L., U.N., J.G., T.B. and F.G.L. Study design: B.P., M.L., T.B., S.M., P.M.A. and F.G.L. Recruitment and clinical assessment of patients: M.B., N.D., P.G., T.B. and P.M.A. Neuropathological examination: S.S., K.F., P.G. and T.B. Writing of the article: B.P., M.L., J.G., M.B., U.N., J.-P.J., N.C., T.B., S.M., P.M.A., K.F., S.S., N.D., P.G., N.R.C. and F.G.L. Operational grant money: J.G., T.B., P.M.A., S.M. and F.G.L.
Data Availability
All data generated or analysed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31773-z.
References
- 1.Mulder DW, Kurland LT, Offord KP, Beard CM. Familial adult motor neuron disease: amyotrophic lateral sclerosis. Neurology. 1986;36:511–517. doi: 10.1212/WNL.36.4.511. [DOI] [PubMed] [Google Scholar]
- 2.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 2013;14:248–64. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Sayana P, Zhang X, Le W. Genetics of amyotrophic lateral sclerosis: an update. Mol. Neurodegener. 2013;8:28. doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim. Biophys. Acta. 2006;1762:956–72. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat. Rev. Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 6.Ghasemi Mehdi, Brown Robert H. Genetics of Amyotrophic Lateral Sclerosis. Cold Spring Harbor Perspectives in Medicine. 2017;8(5):a024125. doi: 10.1101/cshperspect.a024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blokhuis AM, Groen EJN, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–94. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pokrishevsky E, et al. Aberrant Localization of FUS and TDP43 Is Associated with Misfolding of SOD1 in Amyotrophic Lateral Sclerosis. Plos One. 2012;7:e35050. doi: 10.1371/journal.pone.0035050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brettschneider J, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathologica. 2012;123(6):825–839. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper-Knock J, et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135(3):751–764. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marco GD, et al. Cytoplasmic accumulation of TDP-43 in circulating lymphomonocytes of ALS patients with and without TARDBP mutations. Acta Neuropathologica. 2011;121(5):611–622. doi: 10.1007/s00401-010-0786-7. [DOI] [PubMed] [Google Scholar]
- 12.Hart MP, Brettschneider J, Lee VMY, Trojanowski JQ, Gitler AD. Distinct TDP-43 pathology in ALS patients with ataxin 2 intermediate-length polyQ expansions. Acta Neuropathologica. 2012;124(2):221–230. doi: 10.1007/s00401-012-0985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayaki, T. et al. Immunoreactivity of valosin-containing protein in sporadic amyotrophic lateral sclerosis and in a case of its novel mutant. Acta Neuropathologica Communications2(1) (2014). [DOI] [PMC free article] [PubMed]
- 14.Neumann M, et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto Y, et al. An autopsy case of SOD1-related ALS with TDP-43 positive inclusions. Neurology. 2011;77(22):1993–1995. doi: 10.1212/WNL.0b013e31823a0cfc. [DOI] [PubMed] [Google Scholar]
- 16.Stewart H, et al. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathologica. 2012;123(3):409–417. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan C-F, et al. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathologica. 2007;113(5):535–542. doi: 10.1007/s00401-007-0206-9. [DOI] [PubMed] [Google Scholar]
- 18.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat. Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 19.Grad LI, et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA. 2014;111:3620–5. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gros-Louis F, Soucy G, Larivière R, Julien JP. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J. Neurochem. 2010;113:1188–1199. doi: 10.1111/j.1471-4159.2010.06683.x. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson PA, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain. 2004;127:73–88. doi: 10.1093/brain/awh005. [DOI] [PubMed] [Google Scholar]
- 22.Tokuda E, et al. Immunochemical characterization on pathological oligomers of mutant Cu/Zn-superoxide dismutase in amyotrophic lateral sclerosis. Mol. Neurodegener. 2017;12:2. doi: 10.1186/s13024-016-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergemalm D, et al. Superoxide dismutase-1 and other proteins in inclusions from transgenic amyotrophic lateral sclerosis model mice. J. Neurochem. 2010;114:408–418. doi: 10.1111/j.1471-4159.2010.06753.x. [DOI] [PubMed] [Google Scholar]
- 24.Graffmo KS, et al. Expression of wild-type human superoxide dismutase-1 in mice causes amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:51–60. doi: 10.1093/hmg/dds399. [DOI] [PubMed] [Google Scholar]
- 25.Rotunno, M. S. & Bosco, D. A. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. 7, 1–16 (2013). [DOI] [PMC free article] [PubMed]
- 26.Forsberg K, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. Plos One. 2010;5:1–9. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosco Da, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forsberg K, Andersen PM, Marklund SL, Brännström T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen PM, et al. Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat. Genet. 1995;10:61–6. doi: 10.1038/ng0595-61. [DOI] [PubMed] [Google Scholar]
- 30.Synofzik M, et al. Mutant superoxide dismutase-1 indistinguishable from wild-type causes ALS. Hum. Mol. Genet. 2012;21:3568–3574. doi: 10.1093/hmg/dds188. [DOI] [PubMed] [Google Scholar]
- 31.Rakhit R, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 32.Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J. Neurochem. 2007;102:170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 33.Guareschi S, et al. An over-oxidized form of superoxide dismutase found in sporadic amyotrophic lateral sclerosis with bulbar onset shares a toxic mechanism with mutant SOD1. Proc. Natl. Acad. Sci. 2012;109:5074–5079. doi: 10.1073/pnas.1115402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, L. et al. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. 18 1642–1651 (2009). [DOI] [PMC free article] [PubMed]
- 35.Deng H-X, et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl. Acad. Sci. USA. 2006;103:7142–7. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayers JI, Fromholt SE, O’Neal VM, Diamond JH, Borchelt DR. Prion-like propagation of mutant SOD1 misfolding and motor neuron disease spread along neuroanatomical pathways. Acta Neuropathol. 2016;131:103–114. doi: 10.1007/s00401-015-1514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urushitani M, Ezzi SA, Julien J-P. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2007;104:2495–2500. doi: 10.1073/pnas.0606201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu HN, et al. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Ann. Neurol. 2009;66:75–80. doi: 10.1002/ana.21704. [DOI] [PubMed] [Google Scholar]
- 39.Brotherton TE, et al. Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proc. Natl. Acad. Sci. USA. 2012;109:5505–10. doi: 10.1073/pnas.1115009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerman A, et al. Amyotrophic lateral sclerosis is a non-amyloid disease in which extensive misfolding of SOD1 is unique to the familial form. Acta Neuropathol. 2010;119:335–344. doi: 10.1007/s00401-010-0646-5. [DOI] [PubMed] [Google Scholar]
- 41.Ayers JI, et al. Conformational specificity of the C4F6 SOD1 antibody; low frequency of reactivity in sporadic ALS cases. Acta Neuropathol. Commun. 2014;2:55. doi: 10.1186/2051-5960-2-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Da Cruz Sandrine, Bui Anh, Saberi Shahram, Lee Sandra K., Stauffer Jennifer, McAlonis-Downes Melissa, Schulte Derek, Pizzo Donald P., Parone Philippe A., Cleveland Don W., Ravits John. Misfolded SOD1 is not a primary component of sporadic ALS. Acta Neuropathologica. 2017;134(1):97–111. doi: 10.1007/s00401-017-1688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strange RW, et al. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 2003;328:877–891. doi: 10.1016/S0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 44.Johansson A-S, et al. Cytotoxicity of superoxide dismutase 1 in cultured cells is linked to Zn2+ chelation. Plos One. 2012;7:e36104. doi: 10.1371/journal.pone.0036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strange RW, Yong CW, Smith W, Hasnain SS. Molecular dynamics using atomic-resolution structure reveal structural fluctuations that may lead to polymerization of human Cu-Zn superoxide dismutase. Proc. Natl. Acad. Sci. USA. 2007;104:10040–4. doi: 10.1073/pnas.0703857104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoue E, et al. SOD1 Is Essential for the Viability of DT40 Cells and Nuclear SOD1 Functions as a Guardian of GenomicDNA. J. Nucleic Acids. 2010;2010:1–11. doi: 10.4061/2010/795946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller JN. Age-related neuropathology, cognitive decline, and Alzheimer’s disease. Ageing Res. Rev. 2006;5:1–13. doi: 10.1016/j.arr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 48.Nishio, S. et al. Corpora Amylacea Replace the Hippocampal Pyramidal Cell Layer in a Patient with Temporal Lobe Epilepsy. 42 960–962 (2001). [DOI] [PubMed]
- 49.Song W, et al. Astroglial Heme Oxygenase-1 and the Origin of Corpora Amylacea in Aging and Degenerating Neural Tissues. Exp Neurol. 2014;254:78–89. doi: 10.1016/j.expneurol.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Mrak R, T. Griffin WS, I. Graham D. Aging-associated Changes in Human Brain. J. Neuropathol. Exp. Neurol. 1997;56:1269–1275. doi: 10.1097/00005072-199712000-00001. [DOI] [PubMed] [Google Scholar]
- 51.Pisa D, Alonso R, Rábano A, Carrasco L. Corpora Amylacea of Brain Tissue from Neurodegenerative Diseases Are Stained with Specific Antifungal Antibodies. Front. Neurosci. 2016;10:1–12. doi: 10.3389/fnins.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selmaj K, et al. Corpora amylacea from multiple sclerosis brain tissue consists of aggregated neuronal cells. Acta Biochim. Pol. 2008;55:43–49. [PubMed] [Google Scholar]
- 53.Paesschen WV, Revesz T, Duncan JS. Corpora amylacea in hippocampal sclerosis. J. Neurol. Neurosurg. Psychiatry. 1997;63:513–515. doi: 10.1136/jnnp.63.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohn TT. Corpora Amylacea in Neurodegenerative Diseases: Cause or. Int J Neurol Neurother. 2015;2:1–10. doi: 10.23937/2378-3001/2/2/1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavanagh JB. Spinal corpora amylacea and motor neuron disease: a quantitative study. J. Neurol. Neurosurg. Psychiatry. 1998;65:488–491. doi: 10.1136/jnnp.65.4.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paré B, et al. Early detection of structural abnormalities and cytoplasmic accumulation of TDP-43 in tissueengineered skins derived from ALS patients. Acta Neuropathol. Commun. 2015;3:5. doi: 10.1186/s40478-014-0181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 58.Byrne S, Elamin M, Bede P, Hardiman O. Absence of consensus in diagnostic criteria for familial neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry. 2012;83:365–367. doi: 10.1136/jnnp-2011-301530. [DOI] [PubMed] [Google Scholar]
- 59.Leblond CS, et al. Replication study of MATR3 in familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2016;37:209.e17–209.e21. doi: 10.1016/j.neurobiolaging.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 60.Gros-Louis F, et al. Chromogranin B P413L variant as risk factor and modifier of disease onset for amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2009;106:21777–21782. doi: 10.1073/pnas.0902174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daoud H, et al. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol. Aging. 2012;33:839.e5–839.e9. doi: 10.1016/j.neurobiolaging.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 62.Belzil VV, et al. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohta Y, et al. Sex-dependent effects of chromogranin B P413L allelic variant as disease modifier in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2016;25:4771–4786. doi: 10.1093/hmg/ddw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jonsson PA, et al. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain. 2006;129:451–464. doi: 10.1093/brain/awh704. [DOI] [PubMed] [Google Scholar]
- 65.Zetterstrom P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proceedings of the National Academy of Sciences. 2007;104(35):14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zetterström P, Graffmo KS, Andersen PM, Brännström T, Marklund SL. Composition of Soluble Misfolded Superoxide Dismutase-1 in Murine Models of Amyotrophic Lateral Sclerosis. NeuroMolecular Medicine. 2013;15(1):147–158. doi: 10.1007/s12017-012-8204-z. [DOI] [PubMed] [Google Scholar]
- 67.Eisenberg D, et al. Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis. Protein Sci. 1998;7:545–555. doi: 10.1002/pro.5560070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article.