Achlorophyllous unicellular microalgae of the genus Prototheca (Trebouxiophyceae, Chlorophyta) are the only known plants that cause infections in both humans and animals, collectively referred to as protothecosis. Human protothecosis, most commonly manifested as cutaneous, articular, and disseminated disease, is primarily caused by Prototheca wickerhamii, followed by Prototheca zopfii and, sporadically, by Prototheca cutis and Prototheca miyajii.
KEYWORDS: cytochrome b, Prototheca, algae, cytb, genotypic identification
ABSTRACT
Achlorophyllous unicellular microalgae of the genus Prototheca (Trebouxiophyceae, Chlorophyta) are the only known plants that cause infections in both humans and animals, collectively referred to as protothecosis. Human protothecosis, most commonly manifested as cutaneous, articular, and disseminated disease, is primarily caused by Prototheca wickerhamii, followed by Prototheca zopfii and, sporadically, by Prototheca cutis and Prototheca miyajii. In veterinary medicine, however, P. zopfii is a major pathogen responsible for bovine mastitis, which is a predominant form of protothecal disease in animals. Historically, identification of Prototheca spp. has relied upon phenotypic criteria; these were later replaced by molecular typing schemes, including DNA sequencing. However, the molecular markers interrogated so far, mostly located in the ribosomal DNA (rDNA) cluster, do not provide sufficient discriminatory power to distinguish among all Prototheca spp. currently recognized. Our study is the first attempt to develop a fast, reliable, and specific molecular method allowing identification of all Prototheca spp. We propose the mitochondrial cytb gene as a new and robust marker for diagnostics and phylogenetic studies of the Prototheca algae. The cytb gene displayed important advantages over the rDNA markers. Not only did the cytb gene have the highest discriminatory capacity for resolving all Prototheca species, but it also performed best in terms of technical feasibility, understood as ease of amplification, sequencing, and multiple alignment analysis. Based on the species-specific polymorphisms in the partial cytb gene, we developed a fast and straightforward PCR-restriction fragment length polymorphism (RFLP) assay for identification and differentiation of all Prototheca species described so far. The newly proposed method is advocated to be a new gold standard in diagnostics of protothecal infections in human and animal populations.
INTRODUCTION
The genus Prototheca (Trebouxiophyceae, Chlorophyta), originally established by Krüger in 1894, includes unicellular, achlorophyllous microalgae, phylogenetically related to Chlorella spp. (1). So far, eight species have been accommodated within the genus, namely, P. wickerhamii, P. zopfii (classified into genotypes 1 and 2), P. blaschkeae, P. cutis, P. miyajii, P. stagnora, P. ulmea, and, very recently, P. tumulicola (2–6); all but the last three are implicated in human and animal pathologies, collectively referred to as protothecosis. Human protothecosis, most commonly manifested as cutaneous, articular, and disseminated disease, is primarily caused by P. wickerhamii, followed by P. zopfii and, sporadically, by P. cutis, and P. miyajii (7), whereas in veterinary medicine P. zopfii is a major pathogen and responsible for a disproportionate number of cases of bovine mastitis, which is a predominant form of protothecal disease in animals. Occasionally, P. blaschkeae and P. wickerhamii have been involved in bovine mammary protothecosis (8, 9). The latter species has also been the main cause of infections, very rarely reported, in small animals, such as dogs, cats, goats, and fish (10–13). Prototheca spp. are emerging pathogens whose incidence has been on the rise worldwide. For instance, until 2012, the total number of cases of human protothecosis was 160, up from 76 before 1996 and 22 before 1980 (7). This accelerating trend is a product of an expanding population of senile and immunocompromised patients, as well as of increasing clinical awareness and technological improvements in diagnostic instrumentation.
Historically, identification of Prototheca spp. has relied upon phenotypic criteria, including gross colonial morphology, micromorphology in histopathological sections or culture, and biochemical activity, typically assessed by auxanographic carbohydrate assimilation assays. However, these conventional, phenotype-based methods are laborious, time-consuming, and expertise demanding, and, above all, the results delivered are often ambiguous and poorly reproducible. The phenotypic typing methods, although still in use, are now increasingly being superseded by molecular, DNA-based modalities, a large battery of which has been developed over the last 2 decades, rendering the identification of Prototheca spp. faster and more accurate (3, 14–25). Yet none of these methods are exempt from limitations. One of the earliest and most popular typing systems for Prototheca isolates has been genotype-specific PCR and restriction fragment length polymorphism (RFLP) analysis targeted on a 450-bp region of the 18S rRNA gene (3, 25). Both of these approaches have successfully been used to identify bovine mastitis-related Prototheca pathogens, that is, P. zopfii genotypes 1 and 2 and P. blaschkeae (25–32). No other species can be differentiated with these methods. Moreover, to achieve a definite result, sometimes three PCR or PCR-RFLP assays need to be performed. In an effort to streamline and simplify the molecular identification of the Prototheca algae, several other PCR-based protocols have been developed, with some aimed at improving specifically the diagnosis of protothecal mastitis and others aimed at covering the widest possible spectrum of Prototheca spp. Among the former are nested-PCR (22) and duplex PCR (14), both targeting the 18S ribosomal RNA (rDNA) region, applicable directly in milk samples, and allowing identification of P. zopfii and P. zopfii genotype 2, respectively. With the same molecular targets, two real-time PCR assays were designed to detect P. zopfii, with discrimination between genotypes 1 and 2 (20, 21). A combination of two assays based on PCR amplification of the ribosomal internal transcribed spacer (ITS) region, encompassing ITS1, the 5.8S rRNA gene, and ITS2, was proposed to detect and identify P. zopfii genotype 2 and P. blaschkeae in bovine mastitic milk (19).
Of the methods conceived as pan-generic molecular identification schemes, worthy of note are the following three: first, a two-step, 18S rDNA-based reverse transcription-PCR (RT-PCR) followed by DNA resolution melting analysis (RMA), providing differentiation between P. zopfii genotypes 1 and 2 and P. blaschkeae in the first assay and between P. wickerhamii, P. stagnora, and P. ulmea in the second assay (15); second, a two-step PCR single-strand conformation polymorphism (SSCP) analysis directed toward two regions of the 18S rDNA, distinguishing P. zopfii genotypes 1 and 2 and P. blaschkeae, as well as P. stagnora and P. ulmea (16); third, a single multiplex PCR, with primers amplifying extranuclear 18S and 23S rRNA-coding partial sequences, separating P. zopfii genotype 2, P. blaschkeae, P. wickerhamii, and, as a group, P. zopfii genotype 1, P. stagnora, and P. ulmea (17). From this brief overview, it is clear that none of the methods allow identification of all of the currently accepted Prototheca species. They all lack P. cutis, P. miyajii, and P. tumulicola, albeit the last two could not be included due to the recency of their discoveries. Moreover, the two most discriminatory techniques, that is, quantitative PCR/high-resolution melting (qPCR/HRM) and PCR-SSCP, suffer from certain disadvantages. The success of qPCR/HRM depends largely on not only the quality of the obtained amplimer but also the dye chemistry, instrument resolution, and data analysis or software used (33). Each of these elements may affect reproducibility and reliability of the results. An important limitation of PCR-SSCP is a tendency of single-stranded DNA to adopt several conformational forms under different physical conditions, such as temperature and ionic environment. Furthermore, the mobility of single-stranded DNA conformers may vary considerably in the context of the applied electrophoretic parameters (34). All of this may alter the PCR-SSCP patterns, posing interpretative difficulties or even precluding identification.
For the identification of Prototheca spp., an alternative to all methods mentioned above is DNA sequencing, most commonly performed within the ribosomal operon. Indeed, PCR amplification followed by sequencing of the small-subunit (SSU) rRNA gene (18S rRNA), ITS region, and/or large-subunit (LSU) rRNA gene (28S rRNA) portions has been a common strategy for species delineation also in the clinical setting (4, 35–41) and for inference of phylogenetic and evolutionary relationships within and beyond the Prototheca genus (18, 19, 23, 24, 42, 43). What hampers this approach from becoming a routine diagnostic tool are mainly time and financial constraints, especially for large-scale investigations.
Sequencing procedures, along with preparatory activities and postsequencing data analyses, cannot be performed in less than 24 h. Moreover, the intragenomic sequence divergences in the protothecal rRNA genes and ITS loci (23, 43) necessitate cloning the PCR-amplified DNA fragment prior to sequencing to avoid potential chimeric sequences from direct PCR product, which further prolongs the identification process.
Finally, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has recently been adapted for the identification of Prototheca spp. (44). Apart from several experimental variables influencing the quality and reproducibility of the MALDI-TOF profiling results, a key limitation of this technology is that it requires a high degree of technical training and proprietary equipment, which are hardly available in most molecular diagnostic laboratories.
Overall, a fast, reliable, easy-to-perform, and cost-effective method for detection and species delimitation of Prototheca algae is still needed.
In this study, we propose a new typing system for Prototheca spp. based on the mitochondrion-encoded cytochrome b (cytb) gene partial sequence. The resolving power of the new marker was compared with that of SSU, partial LSU, and ITS markers.
MATERIALS AND METHODS
Strains.
A total of 21 strains of Prototheca spp. were included in the study (Table 1). Within this number were nine Prototheca type strains representing eight species and genotypes (Table 1) and 12 additional strains retrieved from an in-house strain depository, purchased from international culture collections or kindly provided by collaborating laboratories (Table 1). The strains were stored in Viabank cryopreservation vials (Medical Wire and Equipment Co., Ltd., Corsham, United Kingdom) at −70°C and were revived by streaking a loopful (10 μl) of the frozen culture onto yeast-peptone-dextrose (YPD) (Difco, Franklin Lakes, NJ) agar plates and incubation at either 30°C (P. stagnora, P. ulmea, and P. tumulicola) or 37°C (all remaining species) aerobically for 72 h. Subcultures were maintained on the same medium and under the same conditions as described above.
TABLE 1.
Prototheca sp. strains used in this study
| Strain group and Species/genotypeg | Straina | Source | GenBank accession no(s). (clone no.)f |
Reference | |||
|---|---|---|---|---|---|---|---|
| SSU | ITS | LSU | cytb | ||||
| Type strains | |||||||
| P. zopfii genotype 1 | SAG 2063b | Swine manure, Germany | MF163506 (1), MF163507 (2) | MF163488 (1), MF163489 (2), MF163490 (3), MF163491 (4) | KY654530† | MF163464 | 3 |
| P. zopfii genotype 2 | SAG 2021b | Bovine mastitis, Germany | MF163508 (1), MF163509 (2), MF163510 (3) | MF163492 (1), MF163493 (2), MF163494 (3) | MH374907 | MF163469 | |
| P. blaschkeae | SAG 2064b | Human onychomycosis, Germany | MF163511 (1), MF163512 (2), MF163513 (3) | MF163498 (1), MF163499 (2), MF163500 (3) | KY654532† | MF163449 | |
| P. cutis | ATCC PRA-338c | Human dermatitis, Japan | MF163514 (1), MF163515 (2) | MF163476 (1), MF163477 (2), MF163478 (3), MF163479 (4) | AB470469† | MF163453 | 4 |
| P. ulmea | ATCC 50112c | Sap from wounded elm (Ulmus amercicana), USA | MF163516 (1), MF163517 (2), MF163518 (3) | MF163495 (1), MF163496 (2), MF163497 (3) | AB097094† | MF163456 | |
| P. stagnora | ATCC 16528c | Digested sludge, USA | MF163519 (1), MF163520 (2), MF163521 (3) | MF163485 (1), MF163486 (2), MF163487 (3) | AB097091† | MF163455 | |
| P. wickerhamii | ATCC 16529c | Household plumbing, USA | MF163504 (1), MF163505 (2) | MF163474 (1), MF163475 (2), MF163483 (3), MF163484 (4) | AB183198† | MF163459 | |
| P. miyajii | IFM 53848d | Human dermatitis, Japan | MF163522 (1), MF163523 (2), MF163524 (3), MF163525 (4) | MF163480 (1), MF163481 (2), MF163482 (3) | KP898385† | MF163454 | 5 |
| P. tumulicola | JCM 31123e | Soil from a stone chamber, Japan | MG906573 (1), MH130092 (2), MH130093 (3) | MG910494 (1), MH130090 (2), MH130091 (3) | LC090551† | MG911719 | 6 |
| Confirmed strains | |||||||
| P. zopfii genotype 1 | 18/IV* | Rectal swab, cow, Poland | ND | ND | MH374897 | MF163465 | |
| 48/IV* | Cow's water bowl, Poland | MH374898 | MF163466 | ||||
| 70/IV* | Cow manure, Poland | MH374899 | MF163467 | ||||
| P. zopfii genotype 2 | P535** | Bovine mastitis, Germany | MH374900 | MF163471 | |||
| P232** | Canine protothecosis | MH374901 | MF163470 | ||||
| POL1* | Bovine mastitis, Poland | MH374902 | MF163472 | ||||
| P. blaschkeae | BEL-2*** | Bovine mastitis, Belgium | MH374903 | MF163451 | |||
| BEL-7*** | Bovine mastitis, Belgium | MH374904 | MF163452 | ||||
| 49/IV* | Cow's water bowl, Poland | MH374905 | MF163450 | ||||
| P. wickerhamii | PW-A* | Skin lesion (Cyprinus carpio), Poland | MF163526 (1) | MF163501 (1), MF163502 (2), MF163503 (3) | KX828712† | ND | 13 |
| PWPL1* | Human neuroinfection, Poland | ND | ND | JX256253† | MF163461 | 35 | |
| 201/09**** | Human disseminated algaemia, Malaysia | MH374906 | MF163460 | 75 | |||
Sources of confirmed strains are indicated as follows: *, in-house collection; **, Uwe Roesler, Institute of Animal Hygiene and Environmental Health, Centre for Infectious Medicine, Freie Universität Berlin, Germany; ***, Paul-Émile Lagneau, Regional Association for Health and Animal Identification, Ciney, Belgium; ****, Ratna Mohd Tap, Bacteriology Unit, Institute for Medical Research, Jalan Pahang, Kuala Lumpur, Malaysia.
Culture Collection of Algae at the University of Göttingen (SAG), Göttingen, Germany.
American Type Culture Collection (ATCC), Manassas, VA, USA.
Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University, Chiba, Japan.
Japan Collection of Microorganisms (JCM) RIKEN Hirosawa, Wako-shi, Saitama, Japan.
Numbers in parentheses indicate different clones derived from a single Prototheca strain. A number of the LSU sequences have been previously published, as indicated (†). ND, not determined.
See the text for the distinctions between the two groups of strains.
Apart from the well-described strains, a panel of 70 Prototheca isolates cultured from mastitis milk and environmental samples were used for validation purposes (see Table S1 in the supplemental material). For the species- or genotype-level identification of Prototheca isolates, genotype-specific PCR (3) was used as a reference method.
DNA extraction.
A loopful of Prototheca sp. cells from a single colony grown on YPD agar was used for a DNA extraction procedure. This was performed with a GeneMATRIX Environmental DNA & RNA purification kit (EURx, Gdańsk, Poland) and involved mechanical cell disruption by vigorous shaking with glass beads in a detergent-rich environment and combined action of lysozyme and proteinase K. All steps, including additional treatment with lyticase (100 μg/ml) (Sigma, Saint Louis, MO, USA) and β-mercaptoethanol (1 μl/ml) (Sigma, Saint Louis, MO, USA) were performed strictly according to the manufacturer's instructions. The purified DNA, dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), was quantified with a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, USA) and stored at −20°C until used.
In silico analysis.
The whole-genome sequences (WGS) of nine reference Prototheca sp. strains, generated with the MiSeq platform (Illumina, San Diego, CA, USA) (data not published) were searched using blastn software (www.ncbi.nlm.nih.gov/BLAST) (45) for the presence of six single-locus mitochondrial genes involved in the respiratory functions of the mitochondrion, i.e., atp6, coding for subunit 6 of the mitochondrial ATPase complex, cox1 to cox3, encoding subunits 1, 2, and 3 of the cytochrome oxidase, nad1, encoding subunit 1 of the NADH dehydrogenase, and cytB, which encodes apocytochrome B. The corresponding genes from the mitochondrial genome sequence of the Prototheca wickerhamii strain (SAG 263-11) published by Wolff et al. (46) and available under GenBank accession number NC_001613.1 served as a reference for all BLAST searches. Once mapped, nucleotide sequences of the six target genes were extracted from the WGS data set and subjected to multiple alignments performed in AliView (47) using the Muscle algorithm (48). Each single gene alignment was analyzed individually and was composed of DNA sequences representing nine Prototheca species (genotypes). Based on different conserved regions identified upon sequence cross-matching, primer design was attempted with primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (49) with the prerequisite that the primers amplify a product ranging in size from 300 to 700 bp, with a GC content of 45 to 65% and devoid of long (>9 bp) homopolymer tracts or tandem repeats.
PCR amplification and sequencing.
The cytb partial gene was amplified in 20-μl reaction mixtures containing 2 μl (ca. 10 ng) of template DNA, 0.1 μl (5 U/μl) of OptiTaq DNA polymerase (Pol) (EURx, Gdańsk Poland), 2 μl of 10× Pol buffer C with MgCl2 (1.5 mM), 0.8 μl of deoxynucleoside triphosphates (dNTPs) (0.2 mM each), and 0.4 μl (0.2 μM) of primer cytb-F1 and cytb-R1 (Table S2). The PCR conditions were the following: 3 min of initial denaturation at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C, with a final extension period of 5 min at 72°C. The amplified products were visualized by agarose gel electrophoresis (1%, wt/vol) and ethidium bromide staining. The amplicons were purified using an EPPiC Fast kit (A&A Biotechnology, Gdańsk, Poland) and directly sequenced with the same primers used for PCR amplification.
In addition to the cytb gene, three loci from the rDNA cluster were analyzed, i.e., the 18S small-subunit (SSU) rRNA gene, the D1/D2 region of the 28S large-subunit (LSU) rRNA gene, and the internal transcribed spacer (ITS) locus. The complete ITS region, including the two spacers (ITS1 and ITS2) and the 5.8S rRNA gene, was amplified with the universal primers ITS4 and ITS5, anchoring at the 5′ end of the 28S rDNA and 3′ end of the 18S rDNA, respectively. PCRs were carried out in 50-μl volumes containing 2 μl (ca. 10 ng) of template DNA, 0.25 μl (5 U/μl) of OptiTaq DNA polymerase (EURx, Gdańsk Poland), 5 μl of 10× Pol buffer B with MgCl2 (1.5 mM), 2 μl of dNTPs (0.2 mM each), 3% (vol/vol) dimethyl sulfoxide (DMSO), and 1 μl (0.2 μM) of each primer. The thermal cycling profile was 96°C for 3 min, 30 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 1 min, with a final step at 72°C for 7 min. The full-length SSU gene and the D1/D2 domain of the LSU rRNA gene were amplified in 50-μl mixtures whose composition was identical to that for ITS amplification, except that primers SSU-F1 and SSU-F2 (SSU) or NL1 and NL4 (LSU) were used (Table S2). The thermocycling conditions for SSU were as previously described (50). The thermal profile for LSU was 95°C for 5 min, followed by 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s and a final step at 72°C for 7 min. The amplicons of SSU and ITS were electrophoresed in 1% (wt/vol) agarose gels, purified with an ExtractMe Gel-Out kit (Blirt, Gdańsk, Poland), and cloned into a plasmid vector using a pCR-Script Amp SK(+) cloning kit (Promega, Madison, WI, USA), according to the vendor's protocol. At least two positive clones from each cloning experiment were selected for sequencing with primers listed in Table S2. For LSU amplicons, direct sequencing was performed.
All PCR assays were carried out on an ABI 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA). All sequencing reactions were run on an Applied Biosystems 3730xl genetic analyzer, using BigDye, version 3.1, chemistry (Applied Biosystems, Foster City, CA, USA). GC-rich PCR templates were sequenced with addition of 1 M betaine (Sigma, Saint Louis, MO, USA) and dGTP BigDye, version 3.0, chemistry (Applied Biosystems, Foster City, CA USA). Sequence data were analyzed using FinchTV, version 1.4.0 (Geospiza, Akron, OH, USA). Consensus sequences were obtained with Seqman Pro, version 9.1, software (DNAStar, Madison, WI, USA).
Multiple sequence alignments to visualize locations of primers used to amplify the SSU, LSU, and ITS loci were performed in MAFFT, version 7310. The SSU, LSU, or ITS sequence representative for a single species was sequenced from different clones, merged into consensus sequences in AliView (47), and visualized with the help of ESPript (http://espript.ibcp.fr) (51).
Phylogenetic analyses.
The Prototheca sp. sequences from four genetic loci (SSU, LSU, ITS, and cytB) were aligned with respective sequences from other Chlorophyta genera (Auxenochlorella, Chlamydomonas, Chlorella, and Helicosporidium), retrieved from the GenBank database, in MAFFT, version 7.310 (52), with default settings. Poorly aligned regions were automatically removed with trimAl, version 1.3 (53), using a gap threshold of 0.3 (settings: –gt 0.3 –st 0.001). Phylogenetic trees were inferred through the maximum likelihood (ML) analysis using randomized accelerated maximum likelihood (RAxML, version 8.2.9) software (54), under the general time-reversible categorical (GTRCAT) model of evolution. The bootstrap option was used with 100 replicates to infer statistical support of branching patterns. Pairwise identity matrices were generated in MEGA, version 7.0.26, based on a simple number of differences model, based on alignments with poorly aligned regions removed (55).
PCR-RFLP.
To develop a PCR-RFLP assay for species- or genotype-specific identification of Prototheca algae, single nucleotide polymorphisms (SNPs) were detected upon alignment of the cytb gene partial sequences generated from each Prototheca species (genotype), as described above. To check if the SNPs located within the restriction enzyme recognition sites that would produce an RFLP, the sequences were screened with Clone Manager software, version 9.0 (Sci-Ed Software, Denver, CO, USA). In silico restriction digestions were carried out, and predicted restriction patterns were determined for each species (genotype). Of several enzyme systems designed, only those yielding distinct, easily resolved patterns for each species (genotype) and recruiting the fewest, readily available, and inexpensive enzymes were sought. A combination of RsaI and TaiI restriction enzymes was selected for PCR-RFLP assays. Experimentally, 644-bp amplimers of the partial cytb gene, produced with the PCR mixture and cycling conditions identical to those described above, were doubly digested with FastDigest RsaI and TaiI (Thermo Fisher Scientific, Waltham, MA, USA). Restriction reaction mixtures consisted of 1 μl of each enzyme, 3 μl of 10× restriction enzyme buffer, 10 μl (ca. 0.2 μg) of PCR product, and 17 μl of Milli-Q water to make a final volume of 30 μl. Digestions were performed at 37°C for 5 min (RsaI) followed by 5 min at 65°C (TaiI), as recommended by the supplier. The restriction products were fractionated on 4% agarose gels and visualized by ethidium bromide staining and exposure to UV light. Analysis of the electropherograms was done with a UVP BioDoc-IT imaging system (Analityk Jena, Jena, Germany).
To further distinguish species (P. miyajii versus P. tumulicola) that yielded identical RsaI and TaiI double-digestion patterns, the 644-bp cytb PCR fragment was restricted with MboI FastDigest enzyme (Thermo Fisher Scientific, Waltham, MA, USA) under conditions recommended by the manufacturer using 1 μl of enzyme, 3 μl of 10× restriction enzyme buffer, 10 μl (ca. 0.2 μg) of PCR product, and 17 μl of Milli-Q water to a final volume of 30 μl. DNA fragments were electrophoresed on 2% agarose gels and visualized as described above.
Accession number(s).
The SSU, LSU, ITS, and cytb gene sequences obtained in this study were deposited in the GenBank database under the accession numbers provided in Table 1.
RESULTS
Of the six mitochondrial genes (atp6, cox1, cox2, cox3, ndh1, and cytb), primer design was feasible only for atp6 and cytb. Failure in the development of the primers for the cox and ndh genes was due to either lack of conserved primer binding sites, inadequate sequence length, sequence variability, or a combination of these factors. The optimal primers for amplifying partial atp6 and cytb genes were selected among a set of candidate primer pairs through a series of in silico PCR simulations. Primers atp6_F2 and atp6_R2 were designed to generate a product of 527 bp, corresponding to positions 802 to 1328 in the P. wickerhamii SAG 263-11 ATPase subunit 6 coding sequence (GenBank accession no. U02970), whereas primers cytb_F1 and cytb_R2 were designed to amplify a 644-bp product spanning coordinates 197 and 840 of the cytb gene in the P. wickerhamii SAG 263-11 mitochondrial genome (GenBank accession no. U02970) (see Table S2 in the supplemental material).
For each Prototheca species (genotype) reference strain, primers cytb_F1 and cytb_R2 resulted in amplification products of the predicted size (Fig. 1). This was achieved by applying an optimized PCR protocol, as described in Materials and Methods. In contrast, the PCR results for the atp6 partial gene were consistently negative for all Prototheca species (genotype) reference strains. Despite several optimization attempts, amplification of the atp6 gene segment was unsuccessful, and consequently further exploration of this gene was abandoned.
FIG 1.
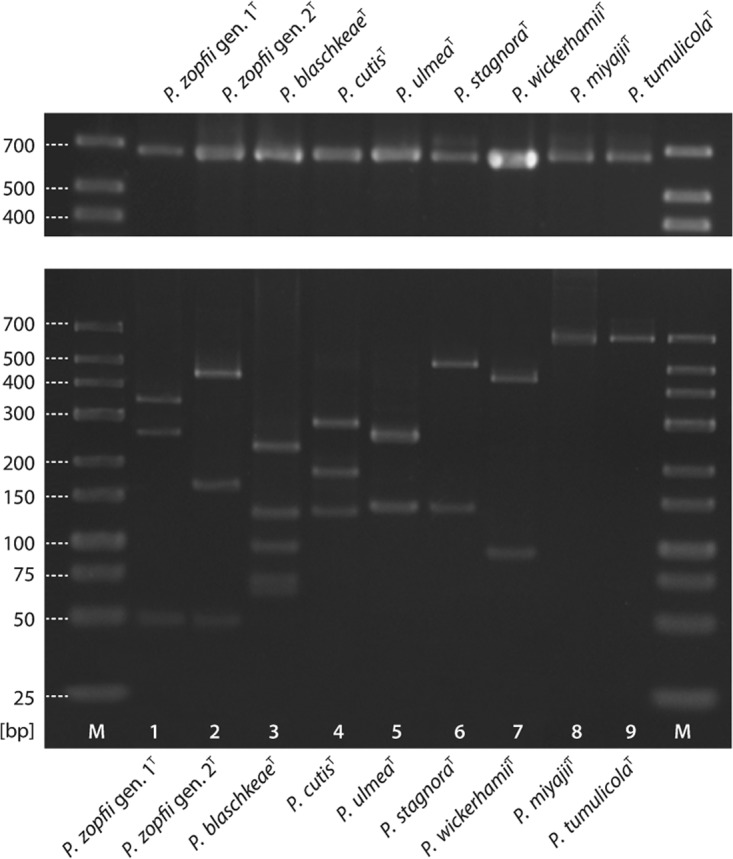
PCR products of the partial cytb gene (above) and restriction patterns of these products (below) produced upon RsaI/TaiI double digestion for nine Prototheca species (genotypes) type strains. Lane 1, P. zopfii genotype 1; lane 2, P. zopfii genotype 2; lane 3, P. blaschkeae; lane 4, P. cutis; lane 5, P. ulmea; lane 6, P. stagnora; lane 7, P. wickerhamii; lane 8, P. miyajii; lane 9, P. tumulicola; lanes M, size marker (Gene Ruler Low Range; Thermo Fisher Scientific, Waltham, MA, USA).
The in silico analysis split the analyzed Prototheca cytb sequences into eight distinct patterns on the basis of double digestion with RsaI and TaiI, as shown in Table 2. The RFLPs obtained for the Prototheca sp. type strains showed perfect matches with those expected to be seen upon gel electrophoresis, i.e., directly derived from in silico predictions and corrected for low-molecular-weight bands (≤25 bp) or comigrating bands (e.g., 247- and 257-bp fragments for P. stagnora), precluding their detection on standard-resolution agarose gels (Fig. 1).
TABLE 2.
Molecular differentiation of Prototheca species/genotypes by PCR-RFLP analysis with RsaI and TaiI and with MboI of the cytb gene fragment
| Enzyme(s) and species/genotype | Length of fragment(s) (bp)a |
|
|---|---|---|
| Determined in silico | Expected upon gel electrophoresis | |
| RsaI/TaiI | ||
| P. zopfii genotype 1 | 25, 47, 253, 319 | 50, 250, 320 |
| P. zopfii genotype 2 | 25, 47, 161, 411 | 50, 160, 410 |
| P. blaschkeae | 22, 25, 25, 61, 68, 93, 128, 222 | 60, 70, 90, 130, 220 |
| P. cutis | 25, 25, 129, 183, 282 | 130, 180, 280 |
| P. ulmea | 140, 247, 257 | 140, 250 |
| P. stagnora | 25, 140, 479 | 140, 480 |
| P. wickerhamii | 25, 25, 89, 94, 411 | 90, 400 |
| P. miyajii | 25, 619 | 620 |
| P. tumulicola | 15, 629 | 620 |
| MboI | ||
| P. miyajii | 151, 221, 272 | 150, 220, 270 |
| P. tumulicola | 644 | 640 |
The sizes were rounded to the nearest 10. Fragments of ca. 20 bp in length were excluded since they are hardly visible even in high-percentage (4%) agarose gels.
All species (genotypes) were clearly identified upon the digestion, except that P. miyajii and P. tumulicola could not be distinguished from each other as they yielded a single band of the same size on a gel. According to in silico analysis, both of these species produce nearly identical restriction patterns (Table 2). The two species, however, could be easily separated when the cytb PCR fragment was restricted with MboI enzyme. Expectedly, three fragments were shown for P. miyajii, while the amplicon of P. tumulicola was not digested (Fig. 2).
FIG 2.

Restriction patterns of the partial cytb gene produced upon MboI digestion for two Prototheca species indistinguishable by RsaI/TaiI double digestion. Lane 1, P. miyajii; lane 2, P. tumulicola; lane M, size marker (Gene Ruler Low Range; Thermo Fisher Scientific, Waltham, MA, USA).
The cytb PCR-RFLP assay was evaluated on 12 strains of confirmed species (genotype) identity (Table 1). All strains belonging to the same species (genotype), i.e., either P. zopfii genotypes 1 and 2, P. blaschkeae, or P. wickerhamii, showed identical patterns easily distinguishable from those of other species (genotypes) (Fig. 3). The assay was further used for species (genotype) identification of 70 Prototheca isolates cultured from mastitis milk and environmental samples, yielding 2 profiles specific for P. zopfii genotype 1, 65 profiles specific for P. zopfii genotype 2, and 3 profiles specific for P. blaschkeae (Table S1). As all of these isolates were also identified to the species (genotype) level with the genotype-specific PCR (3), a full agreement between the two analyses was observed.
FIG 3.
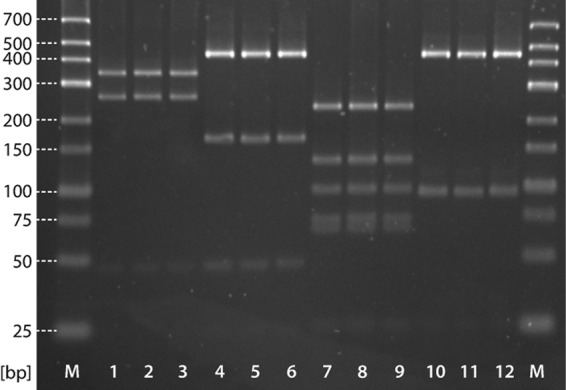
Evaluation of the cytb PCR restriction enzyme analysis profiling for selected Prototheca sp. strains. Lanes 1 to 3, P. zopfii genotype 1; lanes 4 to 6, P. zopfii genotype 2; lanes 7 to 9, P. blaschkeae; lanes 10 to 12 – P. wickerhamii; lanes M, size marker (Gene Ruler Low Range; Thermo Fisher Scientific, Waltham, MA, USA).
Sequencing of the partial cytb gene was performed for all 21 Prototheca strains under study, and the resulting sequences were submitted to the GenBank under the accession numbers provided in Table 1. A total of 335 variable nucleotide sites were detected, which comprised 55.9% (335/599 bp) of the length of a multiple, 21-sequence alignment. For species (genotypes) represented by more than two strains, the cytb gene sequences differed at most in 14 (P. zopfii genotype 1), 3 (P. blaschkeae and P. wickerhamii), or 1 (P. zopfii genotype 2) nucleotide position, translating into 97.7%, 99.5%, and 99.8% sequence similarity, respectively. Pairwise sequence comparison showed that individual Prototheca species (genotypes) shared no more than 94.5% identity (range, 78.6% to 94.5%) (Table S3).
The reliability of cytb as a molecular marker for Prototheca species (genotype) identification was compared with that of SSU, LSU, and ITS. For this purpose, sequencing of the three loci for nine Prototheca sp. type strains was performed. Since previous observations clearly indicated the intragenomic heterogeneity of the rDNA repeat units in Prototheca spp., a strategy of cloning the SSU and ITS PCR products was employed with sequence determination for individual (at least two) clones. The assembled sequences were deposited in the GenBank under the accession numbers provided in Table 1. Altogether, 26 SSU and 33 ITS clone-specific sequences were available for comparative analyses. Given the intrastrain homogeneity of the LSU sequences, one LSU sequence per strain was analyzed. A total of 463 variable nucleotide sites were detected for the SSU, equivalent to 28.8% (463/1,610 bp) of the alignment length. The clonal (i.e., derived from a single strain) SSU sequences were either identical (P. cutis, P. tumulicola, and P. ulmea) or differed at most in 71 (P. wickerhamii), 5 (P. miyajii), 3 (P. zopfii genotype 1), or 2 (P. zopfii genotype 2, P. blaschkeae, and P. stagnora) nucleotide positions, translating into 95.6%, 99.7%, 99.8%, and 99.9% sequence similarity, respectively. Pairwise sequence comparison showed an interspecies identity range of 77.8% to 99.8% (Table S4). The highest sequence identity was observed between P. zopfii genotypes 1 and 2 (up to 99.8%) and between P. cutis and P. miyajii (up to 98.6%).
A total of 317 variable nucleotide sites were identified among the ITS sequences, which comprised 61.2% (317/518 bp) of the alignment length. The intrastrain variation between the sequences was observed for P. wickerhamii, P. cutis, and P. blaschkeae, with a maximum number of nucleotide differences of 61 (sequence similarity, 87.6%), 5 (99.1%), and 2 (99.5%), respectively. There were no two Prototheca species (or genotypes) that shared more than 96.8% sequence similarity in their ITS loci (range, 69.6 to 96.8%). For P. wickerhamii, however, the sequence similarity spanned a wide range of 87.6 to 100% (Table S5).
A total of 428 variable nucleotide sites were identified among the LSU sequences, comprising 59.8% (428/716 bp) of the alignment length. Pairwise sequence comparison showed an interspecies identity range of 79.7% to 99.2% (Table S6). The highest sequence identity was observed between P. zopfii genotypes 1 and 2 (up to 99.16%)
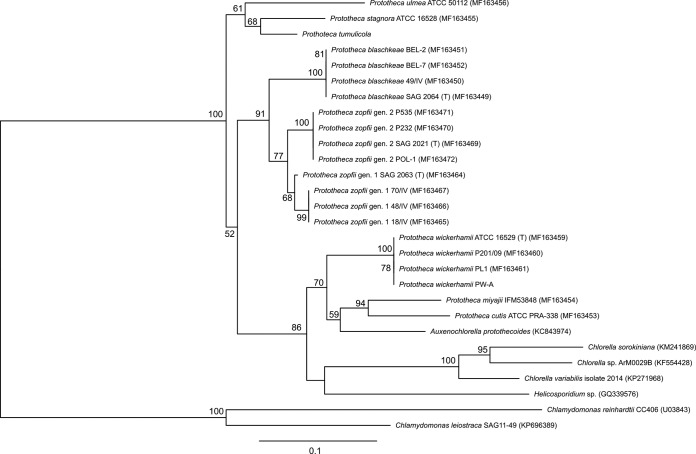
A phylogenetic tree constructed based on the partial cytb gene sequences (599 bp) clearly separated all Prototheca species (genotypes) (Fig. 4). All strains belonging to the same species (genotype) formed distinct clusters, supported by high bootstrap values (up to 100%). More specifically, all Prototheca species grouped in three separate clades. P. miyajii and P. cutis grouped together with Auxenochlorella, with low bootstrap support (59%), as a sister group to P. wickerhamii (bootstrap value, 70%). P. blaschkeae and P. zopfii of both genotypes formed a second well-supported clade (bootstrap value, 91%). The last three species, namely, P. ulmea, P. stagnora, and P. tumulicola, represented by single strains, formed the third monophyletic group (bootstrap value, 61%). At a more general level, P. miyajii, P. cutis, and P. wickerhamii grouped together with the genera Auxenochlorella, Chlorella, and Helicosporidium (bootstrap value, 86%), indicating paraphyly of the genus Prototheca.
FIG 4.
Phylogenetic tree constructed through maximum likelihood analysis based on cytb sequences. The bootstrap values obtained by the analysis are marked at the nodes.
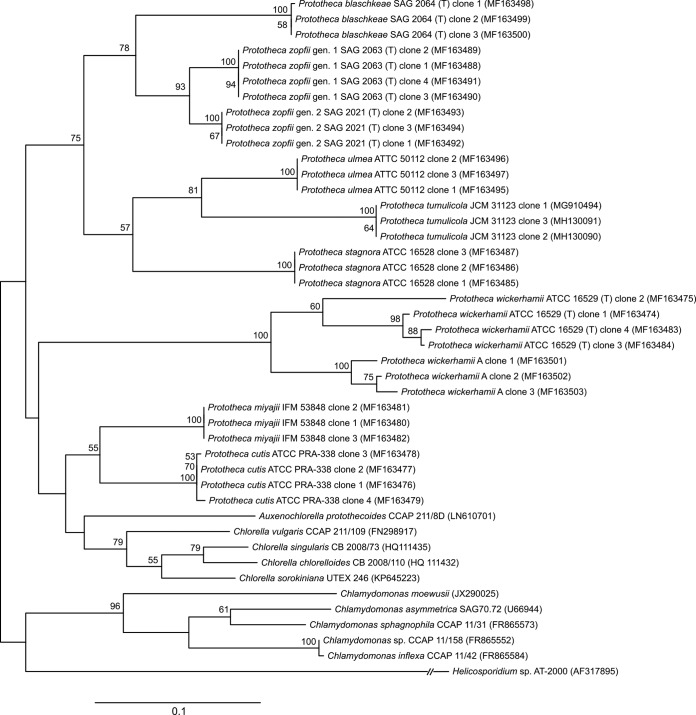
Phylogenetic analysis of Prototheca sp. type strains, inferred from the SSU sequences (1,610 bp), positioned P. zopfii genotypes 1 and 2 in the same cluster, as was the case for P. cutis and P. miyajii (Fig. 5). Both of these topologies were highly supported (bootstrap values of ≥97%).
FIG 5.
Phylogenetic tree constructed through maximum likelihood analysis based on SSU rDNA sequences. The bootstrap values obtained by the analysis are marked at the nodes.
All Prototheca species (genotypes) could be discriminated in the phylogenetic tree generated from alignment of the ITS sequences (518 bp), yet P. wickerhamii was spread into two clusters, one specific for P. wickerhamii strain ATCC 16529 and the other specific for P. wickerhamii strain A (Fig. 6). These two groupings had different statistical confidence levels (bootstrap values of 60% and 100%, respectively).
FIG 6.
Phylogenetic tree constructed through maximum likelihood analysis based on ITS sequences. The bootstrap values obtained by the analysis are marked at the nodes.
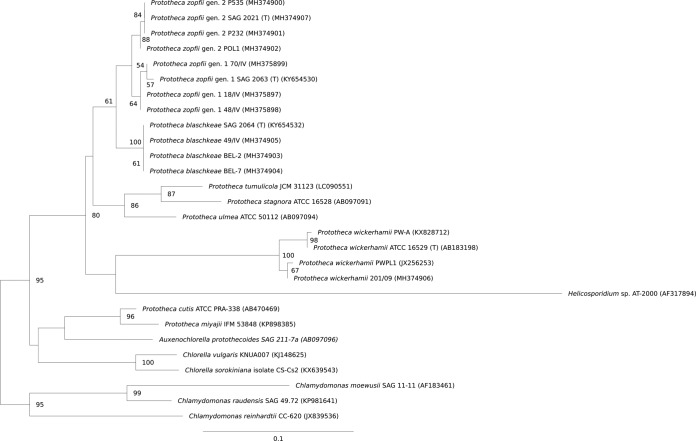
Based on the LSU-derived phylogram, clustering of the Prototheca species (genotypes) was quite similar to that developed from the cytb gene. Still, LSU sequences of the P. wickerhamii strains were more diverse, classified as two subclusters. Also, LSU sequence clusters of P. zopfii genotypes 1 and 2 were less clearly separated than the respective cytb gene clusters (Fig. 7).
FIG 7.
Phylogenetic tree constructed through maximum likelihood analysis based on D1/D2 LSU sequences. The bootstrap values obtained by the analysis are marked at the nodes.
DISCUSSION
The nuclear rRNA gene (rDNA) cluster has been the most extensively scrutinized region for taxonomic and phylogenetic studies across the entire tree of life. The small-subunit (SSU; 16S or 18S) and large-subunit (LSU; 23S, 26S, or 28S) rRNA genes and the internal transcribed spacer (ITS) regions 1 and 2, including the 5.8S rRNA gene, are the backbone of bacterial, fungal, and plant identification and systematics (56–58). While the ITS has recently been declared the primary barcode for fungi, it has not received such status in plants. Here, a multilocus marker system, including the ITS along with plastid-encoded maturase K (matK) and ribulose-1,5-bisphosphate carboxylase (rbcL) genes, has been recommended for species delineation and phylogenetic analyses (57, 58). So far, studies investigating the phylogeny and taxonomy of the Prototheca algae have relied exclusively on rDNA sequence data. Consequently, any identification or typing schemes developed for Prototheca spp. target sequences from the rDNA cluster (14, 15, 19–22, 24). (Primers used in all PCR-based assays so far developed for the identification of Prototheca spp. are shown in Fig. 8, 9, and 10 and are listed in Table S7 in the supplemental material). However, as evidenced in this study, none of the three rDNA (SSU, LSU, and ITS) loci provided adequate resolution to define all Prototheca species (genotypes). For instance, sequence divergence for the SSU locus between P. cutis and P. miyajii ranged from 1.4% to 1.7%, which is well below a 3% threshold, commonly used for species delimitation (59). Even lower (0.8 to 1.4%) was the minimum genetic distance between P. zopfii genotypes 1 and 2 at the D1/D2 LSU locus. Still, both genotypes could be easily separated from each other. Conversely, the genetic distance was much too high to link all P. wickerhamii ITS clones with the same species category.
FIG 8.
Multiple alignment of the SSU sequences of the Prototheca sp. type strains. Nucleotides identical across all displayed species are shaded in red, and those present in at least six sequences are boxed in blue. Blue and orange arrows indicate forward and reverse primers, respectively. Alignment positions boxed in gray are nucleotide coordinates for the adjacent alignment block. Additional black lines indicate sequences based on which species-specific primers were designed. Primer names are given above the arrows, and numbers in brackets correspond to numbers provided in Table S7 in the supplemental material. Dots in the alignment represent the intersequence gaps, while an ellipsis beneath the proto18S-4r-1 (#8) primer in the P. miyajii IFM 53848 SSU sequence indicates a large insert in that region.
FIG 9.
Multiple alignment of the ITS sequences of the Prototheca sp. type strains. Nucleotides identical across all displayed species are shaded in red, and those present in at least six sequences are boxed in blue. Blue and orange arrows indicate forward and reverse primers, respectively. Alignment positions boxed in gray are nucleotide coordinates for the adjacent alignment block. Additional black lines indicate specific strains for which identification primers were designed. Primer names are given above the arrows, and numbers in brackets correspond to numbers provided in Table S7 in the supplemental material. Dots in the alignment represent the intersequence gaps.
FIG 10.
Multiple alignment of the D1/D2 LSU sequences of the Prototheca spp. type strains. Nucleotides identical across all displayed species are shaded in red, and those present in at least six sequences are boxed in blue. Blue and orange arrows indicate forward and reverse primers, respectively. Alignment positions boxed in gray are nucleotide coordinates for the adjacent alignment block. Primer names are given above the arrows, and numbers in brackets correspond to numbers provided in Table S7 in the supplemental material. Dots in the alignment represent the intersequence gaps.
Interestingly, in the case of SSU and ITS loci, this conspecificity between certain Prototheca species might have been overlooked if the multiclone sequencing strategy had not been undertaken. In fact, this strategy was necessitated by a number of ambiguities produced upon direct sequencing of the PCR-amplified rDNA products. This relates to the phenomenon of intragenomic, also referred to as intrastrain or intraindividual, variability of the rDNA units.
It is generally assumed that all copies of rRNA genes within an organism are identical or nearly identical in their nucleotide sequences. The homogeneity of rRNA gene copies has been explained by a concerted evolution model, under which the repeated genes are subject to sequence homogenization through either unequal crossing over or gene conversion (60). However, there have been a number of reports describing considerable differences in nucleotide sequences between copies of rRNA genes in a single organism. This phenomenon has been documented in both prokaryotes and eukaryotes, including fungi, animals, and plants (61–64). The origins of intragenomic rRNA gene polymorphisms are poorly understood. In prokaryotes, the variation of rRNA gene copy numbers has been attributed to horizontal gene transfer (65), whereas in eukaryotes, it has been speculated to occur via birth-and-death evolution, which involves repeated genetic duplications with strong purifying selection. The intragenomic ITS sequence polymorphisms in a strain of the yeast species Pichia membranifaciens have been shown to be a product of intergenomic rDNA recombination of different strains harboring significantly different ITS sequences. Intragenomic recombinations between the polymorphic ITS repeats were also demonstrated (62). Moreover, a defect in the gene conversion mechanisms required for concerted evolution of rRNA genes has been proposed for explaining the maintenance of the polymorphic repeats (62).
Prototheca algae seem to be particularly notorious for displaying high levels of intrastrain rDNA polymorphism. Previous studies, similar to the present one, revealed an important degree of sequence heterogeneity between different copies of the SSU rRNA gene and the ITS locus within a single Prototheca strain (19, 43, 66). Although not seen in this study, the intrastrain sequence heterogeneity had also been reported among copies of the protothecal LSU rRNA gene (23, 66).
Even though the amount of intragenomic variation for each of the rDNA loci has not been well quantified in Prototheca spp., it might be as high as in P. wickerhamii, with 17 different SSU haplotypes demonstrated in a single strain (43). Given that the SSU is the slowest evolving rDNA marker, the variations in the LSU and ITS loci are expected to be much higher.
Mitochondrial genes were among the first markers used for molecular phylogenetic investigations. Compared with nuclear DNA, mitochondrial DNA offers certain advantageous characteristics, including a high rate of evolution, limited proneness to recombination, haploidy (single-locus genes), and high copy number per cell (59, 67). Among many different mitochondrion-based markers, the cytb gene, coding for cytochrome b, a transmembrane protein forming the core of the mitochondrial cytochrome bc1 complex of the respiratory chain, has been one of the most widely exploited and has been successfully used in resolving phylogenetic relationships across the broad spectrum of eukaryotic lineages at a variety of taxonomic levels (68–71). This is because the cytb gene is variable enough to allow discrimination between even very closely related species and conservative enough to define relationships above the species level.
Despite these attractive features, the cytb gene has very rarely been employed in studies on the phylogeny of microalgae. This reluctance may be attributed to the prevailing notion that plant mitochondrial genes have low mutation rates, which translates into their low intra- and interspecies discriminatory powers, precluding their use as plant barcodes (72). However, there has been a growing amount of evidence that the low polymorphism of the mitochondrial genome does not apply across all plant taxa (72, 73). Nevertheless, the potential of the cytb gene for identification and phylogenetic sorting of the Prototheca microalgae had never been explored. To our knowledge, this is the first study in which the sequences of the partial cytb gene were used to investigate the geno-taxonomic relations within the Prototheca genus. The study is also the first attempt to develop a fast, reliable, and specific molecular method to identify all Prototheca spp., based on polymorphisms in the cytb gene.
The intraspecies (intragenotype) sequence similarities calculated for the SSU, ITS, LSU, and cytb loci fell within the ranges of 95.6 to 100%, 87.6 to 100%, 97.5 to 100%, and 97.7 to 100%, respectively, while the interspecies (intergenotype) similarities for the same loci were of 77.9 to 99.8%, 69.6 to 96.8%, 79.7 to 99.2%, and 78.6 to 94.5%, respectively. These values show that the cytb gene provides higher taxonomic resolution than the three other markers. The SSU marker could not accurately discriminate between P. zopfii genotypes 1 and 2 or between P. cutis and P. miyajii, whereas the ITS failed to maintain P. wickerhamii as an integral species. Inasmuch as the high level of the SSU sequence identity (98.3 to 98.6%) between P. cutis and P. miyajii leads to obliteration of their species-level differences, the high intraspecies variation of the ITS sequences (87.6 to 100%) resulted in a breakdown of species boundaries for P. wickerhamii. Interestingly, unlike all other species, but most notably P. miyajii and P. wickerhamii, nonpathogenic Prototheca species (i.e., P. stagnora, P. tumulicola, and P. ulmea) showed, at both rDNA loci, no intraspecific variation at all. It may be speculated that whereas saprotrophy as the sole lifestyle strategy favors sequence homogenization, the alternative trophic mode and pathogenic specialization, forcing an interplay with the animal host, may trigger accelerations of rDNA evolution.
The increased accumulation of nucleotide substitutions in the ribosomal genes in certain heterotrophic (parasitic) plant species compared to that in their autotrophic relatives has been well documented in the literature (74).
The partial cytb gene displayed important advantages over the rDNA markers. Not only did it show the highest discriminatory power resolving all Prototheca species with strong statistical support, but it also performed best in terms of technical feasibility, understood as ease of amplification, sequencing, and multialignment analysis (Fig. 11). Based on the species-specific polymorphisms in the partial cytb gene, we developed a fast and simple PCR-RFLP method for identification and differentiation of all nine currently recognized Prototheca species (genotypes). The method involves two RFLP assays on the same, 644-bp-long PCR product: first, a double enzyme digestion, producing seven species- or genotype-specific profiles and one shared by P. miyajii and P. tumulicola and, second, a one-enzyme reaction separating these two species. The method was evaluated by analyzing 12 strains of confirmed species (genotype) identity and an additional 70 Prototheca isolates cultured from mastitis milk and environmental samples, in every case providing a positive and unambiguous species (genotype) assignment.
FIG 11.
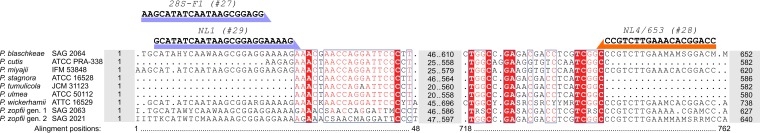
Multiple alignment of the partial sequences of the cytb gene of the Prototheca sp. type strains. Nucleotides identical across all displayed species are shaded in red, and positions with a maximum of two different nucleotides are boxed in blue. The blue-, green-, and black-shaded nucleotides indicate recognition sites for RsaI (GT∧AC), TaiI (ACGT∧), and MboI (∧GATC) restriction enzymes, respectively. The caret represents where the enzyme cuts the sequence.
The reagent costs for the basic PCR-RFLP assay, distinguishing all Prototheca spp. except P. miyajii and P. tumulicola, were estimated at $3.00 per sample. The cost of the complete, two-assay algorithm, which differentiates between P. miyajii and P. tumulicola, was calculated to be $5.00 per sample. Although the overall cost for the commonly used genotype-specific PCR is lower, it allows identification of only two Prototheca species (P. zopfii genotypes 1 and 2 and P. blaschkeae).
Altogether, the system proposed allows accurate and robust identification of Prototheca spp. in a short time (<3 h), with low costs and technical requirements. The limitation of the method is its culture dependency, which extends the total time of analysis by ca. 48 to 72 h. The potential of the method to be applied on clinical material directly is now under investigation. Still, the PCR-RFLP analysis described herein, along with PCR sequencing, a more lengthy and more expensive option, is the only approach currently available capable of identifying all known Prototheca species (genotypes).
Certain attention has to be given to the general phylogeny of the Prototheca genus, as inferred from the partial cytb gene analysis. Upon inspection of the phylogram, four major observations were made. First, the genus Prototheca appeared to be paraphyletic with not only Auxenochlorella and Helicosporidium but also Chlorella, thus expanding the previously proposed AHP (for Auxenochlorella, Helicosporidium, and Prototheca) lineage (2, 41). Second, P. zopfii genotype 1, P. zopfii genotype 2, and P. blaschkeae were clearly monophyletic, with P. zopfii genotypes 1 and 2 sharing a particularly close relationship, supporting their conspecificity (3, 28). Third, P. wickerhamii, but more pronouncedly P. cutis and P. miyajii, were more closely related to Auxenochlorella protothecoides than to other Prototheca species. The monophyly of P. wickerhamii and A. protothecoides had been suggested earlier, according to SSU-based phylogenies (2). Fourth, the strictly saprotrophic (nonpathogenic) species of P. stagnora, P. ulmea, and the very recently described P. tumulicola formed a group of sister lineages separated from other Prototheca species much more distinctly, as with the SSU- and ITS-derived phylogenies from this and past studies (2, 41).
In conclusion, this is the first report to investigate the mitochondrial cytb gene as a molecular marker for identification and phylogenetic analysis of the Prototheca microalgae. The 644-bp fragment of the cytb gene examined in this study has proved effective for discrimination and phylogenetic studies of Prototheca spp. The PCR-RFLP assay targeting the partial cytb gene was developed and, unlike any other method, allowed fast and reliable identification of all Prototheca species described so far. We would advocate the use of this technique and suggest that it could replace ribotyping as the gold standard for identification and taxonomic classification of the Prototheca algae. Since only one available strain for each of five Prototheca species (P. cutis, P. miyajii, P. stagnora, P. ulmea, and P. tumulicola) was examined in this study, the method will require further validation with the recovery of other strains representing these species.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Marcin Świstak for technical assistance.
This work was supported by the National Science Centre within the SONATA funding scheme (contract no. 2014/15/D/NZ7/01797). Additionally, Anna Karnkowska and Kacper Maciszewski were supported by a National Science Centre grant (SONATA 2016/21/D/NZ8/01288).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00584-18.
REFERENCES
- 1.Jagielski T, Lagneau PE. 2007. Protothecosis. A pseudofungal infection. J Mycol Med 17:261–270. doi: 10.1016/j.mycmed.2007.08.003. [DOI] [Google Scholar]
- 2.Ueno R, Urano N, Suzuki M. 2003. Phylogeny of the non-photosynthetic green micro-algal genus Prototheca (Trebouxiophyceae, Chlorophyta) and related taxa inferred from SSU and LSU ribosomal DNA partial sequence data. FEMS Microbiol Lett 223:275–280. doi: 10.1016/S0378-1097(03)00394-X. [DOI] [PubMed] [Google Scholar]
- 3.Roesler U, Möller A, Hensel A, Baumann D, Truyen U. 2006. Diversity within the current algal species Prototheca zopfii: a proposal for two Prototheca zopfii genotypes and description of a novel species, Prototheca blaschkeae sp. nov. Int J Syst Evol Microbiol 56:1–7. doi: 10.1099/ijs.0.64188-0. [DOI] [PubMed] [Google Scholar]
- 4.Satoh K, Ooe K, Nagayama H, Makimura K. 2010. Prototheca cutis sp. nov., a newly discovered pathogen of protothecosis isolated from inflamed human skin. Int J Syst Evol Microbiol 60:1236–1240. doi: 10.1099/ijs.0.016402-0. [DOI] [PubMed] [Google Scholar]
- 5.Masuda M, Hirose N, Ishikawa T. 2016. Prototheca miyajii sp. nov., isolated from a patient with systemic protothecosis. Int J Syst Evol Microbiol 66:1510–1520. doi: 10.1099/ijsem.0.000911. [DOI] [PubMed] [Google Scholar]
- 6.Nagatsuka Y, Kiyuna T, Kigawa R, Sano C, Sugiyama J. 2016. Prototheca tumulicola sp. nov., a novel achlorophyllous yeast-like microalgal isolated from the stone chamber interior of the Takamatsuzuka Tumulus. Mycoscience 58:53–59. doi: 10.1016/j.myc.2016.09.005. [DOI] [Google Scholar]
- 7.Todd JR, King JW, Oberle A, Matsumoto T, Odaka Y, Fowler M, Pore RS, Shahan TA, Yin L, Sanusi ID. 2012. Protothecosis: report of a case with 20-year follow-up, and review of previously published cases. Med Mycol 50:673–689. doi: 10.3109/13693786.2012.677862. [DOI] [PubMed] [Google Scholar]
- 8.Marques S, Silva E, Carvalheira J, Thompson G. 2006. Short communication: in vitro antimicrobial susceptibility of Prototheca wickerhamii and Prototheca zopfii isolated from bovine mastitis. J Dairy Sci 89:4202–4204. doi: 10.3168/jds.S0022-0302(06)72465-1. [DOI] [PubMed] [Google Scholar]
- 9.Jagielski T, Lassa H, Ahrholdt J, Roesler U, Malinowski E. 2010. Molecular characterization of Polish Prototheca zopfii mastitis isolates and first isolation of Prototheca blaschkeae in Poland. Pol J Vet Sci 13:725–729. doi: 10.2478/v10181-010-0025-0. [DOI] [PubMed] [Google Scholar]
- 10.Tsuji H, Kano R, Hirai A, Murakami M, Yanai T, Namihira Y, Chiba J, Hasegawa A. 2006. An isolate of Prototheca wickerhamii from systemic canine protothecosis. Vet Microbiol 118:305–311. doi: 10.1016/j.vetmic.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Endo S, Sekiguchi M, Kishimoto Y, Kano R, Aoki S, Sichinohe T, Hasegawa A. 2010. The first case of feline Prototheca wickerhamii infection in Japan. J Vet Med Sci 72:1351–1353. doi: 10.1292/jvms.09-0504. [DOI] [PubMed] [Google Scholar]
- 12.Camboim EK, Garino FJ, Dantas AF, Simões SV, Melo MA, Azevedo EO, Mota RA, Riet-Correa F. 2011. Protothecosis by Prototheca wickerhamii in goats. Mycoses 54:e196–e200. doi: 10.1111/j.1439-0507.2010.01864.x. [DOI] [PubMed] [Google Scholar]
- 13.Jagielski T, Dyląg M, Roesler U, Murugaiyan J. 2017. Isolation of infectious microalga Prototheca wickerhamii from a carp (Cyprinus carpio)—a first confirmed case report of protothecosis in a fish. J Fish Dis 40:1417–1421. doi: 10.1111/jfd.12614. [DOI] [PubMed] [Google Scholar]
- 14.Gao J, Hou RG, Zhang HQ, He JZ, Li SM, Su JL, Han B. 2011. A novel DNA extraction and duplex polymerase chain reaction assay for the rapid detection of Prototheca zopfii genotype 2 in milk. Lett Appl Microbiol 53:278–282. doi: 10.1111/j.1472-765X.2011.03102.x. [DOI] [PubMed] [Google Scholar]
- 15.Ricchi M, Cammi G, Garbarino CA, Buzzini P, Belletti GL, Arrigoni N. 2011. A rapid real-time PCR/DNA resolution melting method to identify Prototheca species. J Appl Microbiol 110:27–34. doi: 10.1111/j.1365-2672.2010.04861.x. [DOI] [PubMed] [Google Scholar]
- 16.Cremonesi P, Pozzi F, Ricchi M, Castiglioni B, Luini M, Chessa S. 2012. Technical note: identification of Prototheca species from bovine milk samples by PCR-single strand conformation polymorphism. J Dairy Sci 95:6963–6968. doi: 10.3168/jds.2012-5785. [DOI] [PubMed] [Google Scholar]
- 17.Capra E, Cremonesi P, Cortimiglia C, Bignoli G, Ricchi M, Moroni P, Pesce A, Luini M, Castiglioni B. 2014. Simultaneous identification by multiplex PCR of major Prototheca spp. isolated from bovine and buffalo intramammary infection and bulk tank. Lett Appl Microbiol 59:642–647. doi: 10.1111/lam.12326. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Fu YF, Wang RY, Li L, Cao YH, Chen YQ, Zhao HZ, Zhang QQ, Wu JQ, Weng XH, Cheng XJ, Zhu LP. 2014. Identification of clinically relevant fungi and Prototheca species by rRNA gene sequencing and multilocus PCR coupled with electrospray ionization mass spectrometry. PLoS One 9:e98110. doi: 10.1371/journal.pone.0098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques S, Huss VA, Pfisterer K, Grosse C, Thompson G. 2015. Internal transcribed spacer sequence-based rapid molecular identification of Prototheca zopfii and Prototheca blaschkeae directly from milk of infected cows. J Dairy Sci 98:3001–3009. doi: 10.3168/jds.2014-9271. [DOI] [PubMed] [Google Scholar]
- 20.Sobukawa H, Ibaraki M, Kano R, Ito T, Suzuki K, Kamata H, Hasegawa A. 2013. Rapid molecular typing of Prototheca zopfii by high resolution melting real-time PCR (PCR-HRM). Med Mycol J 54:341–344. doi: 10.3314/mmj.54.341. [DOI] [PubMed] [Google Scholar]
- 21.Onozaki M, Makimura K, Satoh K, Hasegawa A. 2013. Detection and identification of genotypes of Prototheca zopfii in clinical samples by quantitative PCR analysis. Jpn J Infect Dis 66:383–390. doi: 10.7883/yoken.66.383. [DOI] [PubMed] [Google Scholar]
- 22.Onozaki M, Makimura K, Hasegawa A. 2009. Rapid identification of Prototheca zopfii by nested polymerase chain reaction based on the nuclear small subunit ribosomal DNA. J Dermatol Sci 54:56–59. doi: 10.1016/j.jdermsci.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Hirose N, Nishimura K, Inoue-Sakamoto M, Masuda M. 2013. Ribosomal internal transcribed spacer of Prototheca wickerhamii has characteristic structure useful for identification and genotyping. PLoS One 8:e81223. doi: 10.1371/journal.pone.0081223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishimoto Y, Kano R, Maruyama H, Onozaki M, Makimura K, Ito T, Matsubara K, Hasegawa A, Kamata H. 2010. 26S rDNA-based phylogenetic investigation of Japanese cattle-associated Prototheca zopfii isolates. J Vet Med Sci 72:123–126. doi: 10.1292/jvms.09-0115. [DOI] [PubMed] [Google Scholar]
- 25.Möller A, Truyen U, Roesler U. 2007. Prototheca zopfii genotype 2: the causative agent of bovine protothecal mastitis? Vet Microbiol 120:370–374. doi: 10.1016/j.vetmic.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 26.Zaini F, Kanani A, Falahati M, Fateh R, Salimi-Asl M, Saemi N, Farahyar SH, Kheirabad AK, Nazeri M. 2012. Identification of Prototheca zopfii from bovine mastitis. Iran J Public Health 41:84–88. [PMC free article] [PubMed] [Google Scholar]
- 27.Jagielski T, Lassa H, Ahrholdt J, Malinowski E, Roesler U. 2011. Genotyping of bovine Prototheca mastitis isolates from Poland. Vet Microbiol 149:283–287. doi: 10.1016/j.vetmic.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 28.Shahid M, Ali T, Zhang L, Hou R, Zhang S, Ding L, Han D, Deng Z, Rahman A, Han B. 2016. Characterization of Prototheca zopfii genotypes isolated from cases of bovine mastitis and cow barns in China. Mycopathologia 181:185–195. doi: 10.1007/s11046-015-9951-9. [DOI] [PubMed] [Google Scholar]
- 29.Ricchi M, Goretti M, Branda E, Cammi G, Garbarino CA, Turchetti B, Moroni P, Arrigoni N, Buzzini P. 2010. Molecular characterization of Prototheca strains isolated from Italian dairy herds. J Dairy Sci 93:4625–4631. doi: 10.3168/jds.2010-3178. [DOI] [PubMed] [Google Scholar]
- 30.Osumi T, Kishimoto Y, Kano R, Maruyama H, Onozaki M, Makimura K, Ito T, Matsubara K, Hasegawa A. 2008. Prototheca zopfii genotypes isolated from cow barns and bovine mastitis in Japan. Vet Microbiol 131:419–423. doi: 10.1016/j.vetmic.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Aouay A, Coppée F, Cloet S, Cuvelier P, Belayew A, Lagneau P, Mullender C. 2008. Molecular characterization of Prototheca strains isolated from bovine mastitis. Med Mycol 18:224–227. doi: 10.1016/j.mycmed.2008.10.002. [DOI] [Google Scholar]
- 32.Gao J, Zhang HQ, He JZ, He YH, Li SM, Hou RG, Wu QX, Gao Y, Han B. 2012. Characterization of Prototheca zopfii associated with outbreak of bovine clinical mastitis in herd of Beijing, China. Mycopathologia 173:275–281. doi: 10.1007/s11046-011-9510-y. [DOI] [PubMed] [Google Scholar]
- 33.Erali M, Wittwer CT. 2010. High resolution melting analysis for gene scanning. Methods 50:250–261. doi: 10.1016/j.ymeth.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menounos PG, Patrinos GP. 2010. Mutation detection by single-strand conformation polymorphism and heteroduplex analysis, p 45–48. In Patrinos GP, Ansorge W (ed) Molecular diagnostics, 2nd ed Elsevier Academic Press, Boston, MA. [Google Scholar]
- 35.Żak I, Jagielski Kwiatkowski T, Bielecki S, J. 2012. Prototheca wickerhamii as a cause of neuroinfection in a child with congenital hydrocephalus. First case of human protothecosis in Poland. Diagn Microbiol Infect Dis 74:186–189. doi: 10.1016/j.diagmicrobio.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 36.McMullan B, Muthiah K, Stark D, Lee L, Marriott D. 2011. Prototheca wickerhamii mimicking yeast: a cautionary tale. J Clin Microbiol 49:3078–3081. doi: 10.1128/JCM.00487-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takano M, Hoshi S, Nagai K, Ishidaira H, Onozaki M, Satoh K, Makimura K. 2014. The first case of human protothecosis caused by Prototheca zopfii in Japan. J Infect Chemother 20:647–649. doi: 10.1016/j.jiac.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Lanotte P, Baty G, Senecal D, Dartigeas C, Bailly E, Duong TH, Chandenier J, Goudeau A. 2009. Fatal algaemia in patient with chronic lymphocytic leukemia. Emerg Infect Dis 15:1129–1131. doi: 10.3201/eid1507.090373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang QQ, Li L, Zhu LP, Zhao Y, Wang YR, Zhu JH, Zhu M. 2012. Cutaneous protothecosis in patient with diabetes mellitus and review of published case reports. Mycopathologia 173:163–171. doi: 10.1007/s11046-011-9480-0. [DOI] [PubMed] [Google Scholar]
- 40.Yun CH, Jeong JH, Ryu HR, Kim JH, Baek JO, Lee JR, Seo YH, Roh JY. 2016. Cutaneous protothecosis responds rapidly to voriconazole. Int J Dermatol 55:1373–1377. doi: 10.1111/ijd.13160. [DOI] [PubMed] [Google Scholar]
- 41.Marques S, Silva E, Kraft C, Carvalheira J, Videira A, Huss VA, Thompson G. 2008. Bovine mastitis associated with Prototheca blaschkeae. J Clin Microbiol 46:1941–1945. doi: 10.1128/JCM.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueno R, Hanagata N, Urano N, Suzuki M. 2005. Molecular phylogeny and phenotypic variation in the heterotrophic green algal genus Prototheca. J Phycol 41:1268–1280. doi: 10.1111/j.1529-8817.2005.00142.x. [DOI] [Google Scholar]
- 43.Ueno R, Huss VA, Urano N, Watabe S. 2007. Direct evidence for redundant segmental replacement between multiple 18S rRNA genes in a single Prototheca strain. Microbiology 153:3879–3893. doi: 10.1099/mic.0.2007/009365-0. [DOI] [PubMed] [Google Scholar]
- 44.Murugaiyan J, Ahrholdt J, Kowbel V, Roesler U. 2012. Establishment of a matrix-assisted laser desorption ionization time-of-flight mass spectrometry database for rapid identification of infectious achlorophyllous green micro-algae of the genus Prototheca. Clin Microbiol Infect 18:461–467. doi: 10.1111/j.1469-0691.2011.03593.x. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Wolff G, Plante I, Lang BF, Kuck U, Burger G. 1994. Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii. Gene content and genome organization. J Mol Biol 237:75–86. [DOI] [PubMed] [Google Scholar]
- 47.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao LH, Escalante L, Yang CF, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–380. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 57.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc 90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 59.Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci 270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganley ARD, Kobayashi T. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res 17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vĕtrovský T, Baldrian P. 2013. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu ZW, Wang QM, Liu XZ, Bai FY. 2016. Intragenomic polymorphism and intergenomic recombination in the ribosomal RNA genes of strains belonging to a yeast species Pichia membranifaciens. Mycology 7:102–111. doi: 10.1080/21501203.2016.1204369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song J, Shi L, Li D, Sun Y, Niu Y, Chen Z, Luo H, Pang X, Sun Z, Liu C, Lv A, Deng Y, Larson-Rabin Z, Wilkinson M, Chen S. 2012. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS One 7:e43971. doi: 10.1371/journal.pone.0043971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stage DE, Eickbush DG. 2007. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res 17:1888–1897. doi: 10.1101/gr.6376807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yap WH, Zhang Z, Wang Y. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J Bacteriol 181:5201–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirose N, Hua Z, Kato Y, Zhang Q, Li R, Nishimura K, Masuda M. 2018. Molecular characterization of Prototheca strains isolated in China revealed the first cases of protothecosis associated with Prototheca zopfii genotype 1. Med Mycol 56:279–287. doi: 10.1093/mmy/myx039. [DOI] [PubMed] [Google Scholar]
- 67.Waugh J. 2007. DNA barcoding in animal species: progress, potential and pitfalls. Bioessays 29:188–197. doi: 10.1002/bies.20529. [DOI] [PubMed] [Google Scholar]
- 68.Biswas SK, Wang L, Yokoyama K. 2005. Molecular phylogenetics of the genus Trichosporon inferred from mitochondrial cytochrome b gene sequences. J Clin Microbiol 43:5171–5178. doi: 10.1128/JCM.43.10.5171-5178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giresse X, Ahmed S, Richard-Cervera S, Delmotte F. 2009. Development of new Oomycete taxon-specific mitochondrial cytochrome b region primers for use in phylogenetic and phylogeographic studies. J Phytopathol 158:321–327. doi: 10.1111/j.1439-0434.2009.01619.x. [DOI] [Google Scholar]
- 70.Tobe SS, Kitchener AC, Linacre AM. 2010. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome B and cytochrome oxidase subunit I mitochondrial genes. PLoS One 5:e14156. doi: 10.1371/journal.pone.0014156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arenas M, Campos R, Coronado X, Ortiz S, Solari A. 2012. Trypanosoma cruzi genotypes of insect vectors and patients with Chagas of Chile studied by means of cytochrome b gene sequencing, minicircle hybridization, and nuclear gene polymorphisms. Vector Borne Zoonotic Dis 12:196–205. doi: 10.1089/vbz.2011.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. 2007. Extensive variation in synonymous substitution rates in mitochondrial gene of seed plants. BMC Evol Biol 7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sloan DB, Oxelman B, Rautenberg A, Taylor DR. 2009. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol 9:260. doi: 10.1186/1471-2148-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lemaire B, Huysmans S, Smets E, Merckx V. 2011. Rate accelerations in nuclear 18S rDNA of mycoheterotrophic and parasitic angiosperms. J Plant Res 124:561–576. doi: 10.1007/s10265-010-0395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mohd Tap R, Sabaratnam P, Salleh MA, Abd Razak MF, Ahmad N. 2012. Characterization of Prototheca wickerhamii isolated from disseminated algaemia of kidney transplant patient from Malaysia. Mycopathologia 173:173–178. doi: 10.1007/s11046-011-9469-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.