Significance
Analyzing complex microbial communities is the milestone of modern microbiology, calling for “deep functional profiling” techniques. While next generation sequencing revolutionized our understanding of microbiota communities, we still lack high-throughput technologies to precisely determine their functionality. Here we show how cultivation of individual bacteria inside droplets of microfluidic double water-in-oil-in-water emulsion enables us to isolate the clones with a desired activity. This approach allows us not only to select the potent antibiotic producer but also to discover a distinct mechanism of self-resistance as well as assess its efficiency on entire microbiomes. The outcome of this methodology shows that it could be effectively transferred to numerous applications in microbiology and biotechnology.
Keywords: microbiome, microfluidics, deep functional profiling, antibiotic resistance, single-cell cultivation
Abstract
Microbiome spectra serve as critical clues to elucidate the evolutionary biology pathways, potential pathologies, and even behavioral patterns of the host organisms. Furthermore, exotic sources of microbiota represent an unexplored niche to discover microbial secondary metabolites. However, establishing the bacterial functionality is complicated by an intricate web of interactions inside the microbiome. Here we apply an ultrahigh-throughput (uHT) microfluidic droplet platform for activity profiling of the entire oral microbial community of the Siberian bear to isolate Bacillus strains demonstrating antimicrobial activity against Staphylococcus aureus. Genome mining allowed us to identify antibiotic amicoumacin A (Ami) as responsible for inhibiting the growth of S. aureus. Proteomics and metabolomics revealed a unique mechanism of Bacillus self-resistance to Ami, based on a subtle equilibrium of its deactivation and activation by kinase AmiN and phosphatase AmiO, respectively. We developed uHT quantitative single-cell analysis to estimate antibiotic efficacy toward different microbiomes and used it to determine the activity spectra of Ami toward human and Siberian bear microbiota. Thus, uHT microfluidic droplet platform activity profiling is a powerful tool for discovering antibiotics and quantifying external influences on a microbiome.
The latest insights into microbiome revealed close links between the spectra of coexisting bacteria and progression of several pathologies in human hosts (1–3). Microbiome spectra turned out to be a viable marker of behavior and habits of Homo sapiens and Neanderthals, as well as the environmental and historical conditions they lived in (4). The bactericide properties of microbiome spectra and bacterial coexistence are becoming a hallmark of present-day biomedical investigations in humans (5, 6). Scrutinizing the microbiota of a plethora of organisms is becoming a mainstream of modern microbiology. The microbiota of wild, captive, and domesticated animals as well as birds, reptiles, and gene-modified animal models (6–10) were analyzed, offering valuable answers to the long-standing issues of biology. Microbiomes have also come into the limelight of evolutionary studies (11).
The revolution in screening technologies complemented by that of functional and structural analyses of large arrays of microbiota species on a single-cell level (12) allows us to isolate and characterize clones with different activities, such as microbial killers, antifungi and antiparasite drugs, as well as probiotic bacterial strains. Microbiota of wild animals is an underestimated resource for this type of screenings. The ability of wild animals to thrive while surrounded by aggressive microorganisms may be partially mediated by their microbiota, making this kind of microbiota a potentially attractive niche for a targeted screening of antibiotics and prospective probiotic strains. In this work, we adjusted our ultrahigh-throughput (uHT) microfluidic droplet platform (13) to perform functional screening of wild-animal microbiota. Our platform allows for packaging the individual bacterial clones in double emulsion droplets and screening these microcompartments by FACS. This technique enables functional screening of the cell libraries with an enormous biodiversity displaying productivity reaching 108 variants per hour. The critical advantage of the system lies in the possibility of microbiome functional profiling on a single-cell level.
Here we present the analyses of the microbiome collected from East Siberian brown bear (Ursus arctos collaris) obtained immediately after capture in the taiga. We aimed to screen this microbiome resource and search for probiotics and physiologically active compounds. A Bacillus pumilus strain producing an unstable antibiotic amicoumacin A (Ami) was isolated, enabling us to identify the B. pumilus Ami biosynthetic gene cluster and discover a crucial role of Ami kinase/phosphatase in the regulation of Ami production. We than applied uHT screening to quantify the external influence on the microbiome diversity, thus obtaining a detailed description of Ami activity spectra toward human and Siberian bear microbiota.
Results
Microfluidic Selection of Antibiotic Activity.
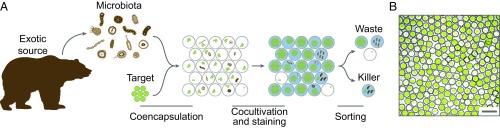
The saliva samples collected from an oral cavity of the Siberian brown bear were screened for bacteria inhibiting the growth of pathogenic Staphylococcus aureus cells using a microfluidic platform (Fig. 1). It is based on cocultivation of oral microbiota members with the target S. aureus strain producing GFP reporter in droplets of microfluidic double water-in-oil-in-water emulsion (MDE).
Fig. 1.
(A) Principal scheme of bacteria isolation demonstrating antimicrobial activity against the target S. aureus from oral microbiota of the Siberian bear using the uHT microfluidic droplet platform. The isolated microbiota was coencapsulated with the target pathogen producing a GFP reporter. After in droplet cocultivation, the bacteria were stained and sorted for isolating the droplets with a high initial target load, low GFP level, and high metabolic activity. (B) MDE droplets with coencapsulated target S. aureus cells and microbiota species after in droplet cocultivation. (Scale bar: 50 μm.)
The combination of three independent fluorescent signals was used to isolate MDE droplets by FACS. The isolated droplets had to conform to the following criteria simultaneously; they exhibited a high initial S. aureus load and a low S. aureus count after in droplet cocultivation accompanied by the presence of live, metabolically active cells. The overall throughput of this platform was estimated to embrace 30,000 droplets per second, which enabled deep probing of microbial community based on anti-S. aureus activity. Several bacterial clones with anti-S. aureus activity were selected and identified as Enterococcus casseliflavus, Weissella confusa, and B. pumilus. These strains were not selected during the preliminary standard testing of bear’s microbiota. This indicates a substantial enrichment of bacteria displaying antagonistic properties against S. aureus after uHT screening in MDE droplets. In what follows, we focus on the isolated B. pumilus, as it was the most efficient inhibitor of S. aureus growth in a plate overlay assay.
Antibacterial Activity of B. pumilus Is Mediated by the Production of Amicoumacin A.
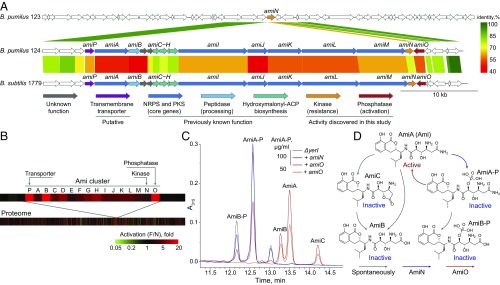
The isolated B. pumilus 124 strain displaying antibacterial activity against S. aureus was analyzed for the biologically active metabolites and found to produce Ami, an antibiotic previously discovered for Bacillus (14–16) and Xenorhabdus bovienii (17). Whole-genome sequencing with a subsequent mining of the B. pumilus 124 genome identified a hybrid PKS–NRPS cluster of Ami biosynthetic genes. The identified cluster had a similar architecture, but limited similarity to the previously described Ami cluster (18) from Bacillus subtilis (Fig. 2A).
Fig. 2.
A multiomics approach applied to discover the regulation of Ami production. (A) NGS and genome mining were used to identify Ami biosynthetic gene clusters. Comparison of Ami biosynthesis gene clusters from producing the B. pumilus 124 strain (NCBI QENN00000000), nonproducing B. pumilus 123 strain (NCBI QENO00000000), and the previously reported B. subtilis 1779 strain (18). Scale bar indicates protein identities. (B) Proteomics was used to determine the borders of the Ami cluster. Differential profile of protein level in activated (F) and inactivated (N) conditions on a proteome level and Ami biosynthetic cluster. Scale bar indicates a fold difference between F and N. (C) Metabolomics was used to confirm the biological functions of AmiN and AmiO. Ami activation via dephosphorylation was observed after amiN knockout and amiO heterologous expression in B. subtilis. (D) General scheme illustrating Ami interconversion by AmiN and AmiO and its spontaneous inactivation by deamidation. AmiA (Ami) denotes amicoumacin A; AmiB and AmiC, amicoumacin B and C; and AmiA-P and AmiB-P, phosphorylated AmiA and AmiB, respectively.
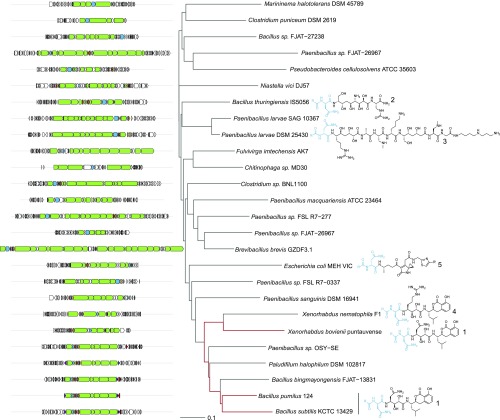
Using genome mining, we found the biosynthetic clusters related to Ami cluster (SI Appendix, Fig. S1), some of which were previously described as responsible for producing the following antibiotics: zwittermicin (19), paenilamicin (20), xenocoumacin (21), and colibactin (22, 23) (Fig. 3).
Fig. 3.
Representative Ami homologous gene clusters that were identified through genome mining. The core biosynthetic genes are green, the activating peptidases, blue. A neighbor joining tree of different clusters was built based on a reciprocal blast score. The known products of biosynthetic clusters are represented as prodrugs that are further proteolytically processed up to their active form by N-acyl-d-Asn (blue; R, fatty acid residue) cleavage. Preantibiotic forms of 1, amicoumacin A; 2, zwittermicin A; 3, paenilamicin B1; 4, xenocoumacin 1; and 5, proposed colibactin.
The notable peculiarity of these clusters is the presence of a gene coding the specific peptidase, activating cognate preantibiotics during the export process. There is, however, an additional gene encoding N-acetyltransferase AmiS inactivating Ami in the case of Ami-like gene cluster from X. bovienii (17). The fact that amiS homolog is absent in both B. pumilus 124 and B. subtilis 1779 strains gave rise to an idea that Bacilli should have an alternative mechanism of self-resistance. Hashimoto et al. (24) have reported the presence of a phosphorylated Ami, produced by B. pumilus, and demonstrated that the phosphorylated Ami is inactive. We found two genes, amiN and amiO, encoding a putative kinase and an alkaline phosphatase, respectively, adjacent to the core biosynthetic genes of the Ami cluster and hypothesized that their products may contribute to both self-resistance and Ami biosynthesis. The tandem-mass spectral analysis was used to find molecular weight of Ami derivates, namely the phosphorylated AmiA-P and AmiB-P (Fig. 2D). It was shown that these compounds have exactly the same mass value as described by Hashimoto et al. (24), where the site of Ami phosphorylation by NMR was determined.
The Regulation of Ami Production in B. pumilus.
The essential role of genes amiA–amiM in Ami biosynthesis was previously demonstrated through a homologous B. subtilis 1779 cluster (18); yet the mechanism was not elucidated. Here we observe Ami production to be inducible and regulated at several additional levels in B. pumilus. Ami production was induced while cultivated in a thin layer without shaking. Cultivation with limited aeration and shaking resulted in more than a 20-fold decrease in Ami production, which was insufficient to inhibit Staphylococcus (SI Appendix, Fig. S2). Metabolomic analysis of B. pumilus 124 cultivated in “activated” (F) and “inactivated” (N) conditions revealed that the inactive phosphorylated Ami derivatives were present exclusively inside the bacterial cells, and absent in the culture medium. A dramatic difference in Ami phosphorylation balance between F and N states was also manifested, suggesting that Ami was efficiently inactivated via phosphorylation in N conditions.
Ami production in F conditions entailed the formation of a slimy film that was essential for inhibiting S. aureus during cocultivation (SI Appendix, Fig. S3). This biofilm enabled B. pumilus to entrap S. aureus and restrict its growth and was further efficiently attacked by DNase treatment. While biofilm production was observed both for Ami-producing B. pumilus 124 and B. pumilus 123 without the Ami cluster, only B. pumilus 124 inhibited S. aureus growth (SI Appendix, Fig. S3).
We analyzed the proteome of B. pumilus 124 in cells cultivated under the activated and inactivated conditions to reveal the mechanism by which Ami synthesis is activated and regulated (Fig. 2B). We observed the increased levels of enzymes involved in de novo inosine monophosphate (IMP) biosynthesis, peptidoglycan synthesis and remodeling, as well as flagellum organization. We speculate that the enzymes involved in peptidoglycan synthesis and remodeling influence the process of biofilm formation, while the improved flagella biosynthesis helps overcome an increased viscosity and contribute to S. aureus entrapment. The protein levels of AmiA–AmiM as well as AmiN, AmiO, and putative MFS family major facilitator transporter AmiP were significantly higher upon activation. Moreover, a drastic difference in the protein level was documented in case of a putative Ami transporter AmiP, activating peptidase AmiB and phosphatase AmiO (Fig. 2B).
Recombinant AmiN and AmiO were produced in Escherichia coli (SI Appendix, Fig. S4). We observed a rapid phosphorylation of Ami by AmiN in vitro using purified AmiN, while purified AmiO dephosphorylated phosphorylated Ami (Ami-P). As expected, the major difference between the minimum inhibitory concentration (MIC) of Ami and Ami-P (0.25 and >100 µg/mL) was observed using S. aureus reporter, indicating that Ami was inactivated via phosphorylation and further reactivated by dephosphorylation.
The biological function of AmiN was elucidated using E. coli BL21(DE3) and B. subtilis 168 surrogate hosts (SI Appendix, Table S1). Transformation of E. coli with a plasmid encoding AmiN conferred the resistance to Ami. B. subtilis 168 has an amiN homolog (yerI) that also phosphorylates Ami, mediating natural B. subtilis resistance to the drug. A knockout of yerI made B. subtilis susceptible to Ami (MIC 0.8 µg/mL) but its resistance was restored after transformation with a plasmid expressing amiN.
Production of AmiO by B. subtilis 168 ΔyerI resulted in Ami-P dephosphorylation (Fig. 2C). We did not observe any background phosphatase activity in B. subtilis even though it encodes two homologous alkaline phosphatases 3 and 4, pointing at AmiO specificity toward Ami-P.
Technique for uHT Profiling of Ami Activity in Different Microbiota Sources.
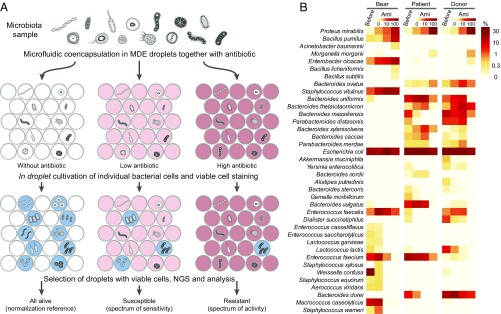
Differential levels of antibiotic resistance, growth rate, and intricate interactions between members hamper analysis of microbial communities. To devise a simple technique for uHT profiling of external influence on microbiome, we developed a method grounded in the MDE platform (Fig. 4A).
Fig. 4.
(A) The principal scheme of the activity/sensitivity spectrum assessment using highly heterogeneous bacteria population (microbiota) and MDE screening platform. This figure is instructional and virtually identical to figure 3 of ref. 13. (B) Heat map indicating the portion of bacteria and influence of Ami on different microbiota samples: oral microbiota of Siberian bear (bear), human fecal microbiota from a patient with colitis (patient), and a healthy human donor (donor). The data were obtained before (before) and after applying MDE screening platform, i.e., single-cell cultivation in droplets with various Ami concentrations (0, 10, and 100 µg/mL), selection of metabolically active population of cells encapsulated in droplets with subsequent metagenomic sequencing and bioinformatic analysis.
The main idea of this approach is based on cultivating the individual cells from microbiota samples together with antibiotic inside MDE droplets. Subsequently, the droplets are stained for metabolic activity, selected using FACS, and analyzed by NGS and bioinformatic analysis. The single-cell in droplet cultivation abolishes any effect of proximal bacterial clones and allows the use of uHT of broad microbial biodiversity. To validate the procedure, we determined the effects of Ami administration on the oral microbiota from the Siberian bear and human fecal microbiota from a patient with colitis and a healthy donor (Fig. 4B).
The composition of bear oral microbiota dramatically differs from human oral microbiota and has much more in common with human fecal microbiota (SI Appendix, Fig. S5). We observed that 83% of bacteria were culturable in droplets (SI Appendix, Fig. S6). Cultivation of fast-growing species in droplets restricted their propagation to MDE compartments, thus allowing assessment of minor microbiota species that represent less than 0.1% of the initial population.
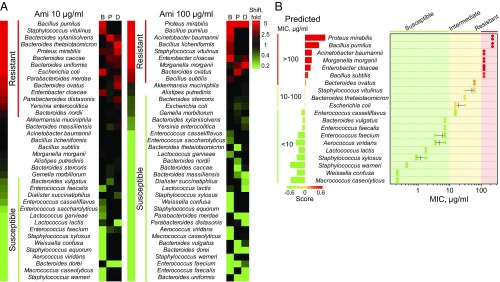
uHT screening of Ami activity brought about an increase in frequency of some bacteria and a decrease in frequency of others (Fig. 5A), which enabled us to estimate susceptibility/resistance to Ami for each particular bacterium in every microbiota sample (Fig. 5B). Our estimations were confirmed using clinical isolates. We found that the MIC ranges obtained using microbiota composition shifts after a single-cell cultivation in MDE compartments were in agreement with MIC of clinical isolates measured using the standard assay (Fig. 5B). Furthermore, we observed a strong correlation between the values of composition shift (scores) and MIC measured in vitro. This correlation analysis allowed us to predict the ranges of MIC for each investigated bacterium (SI Appendix, Fig. S7).
Fig. 5.
(A) Heat map indicating a shift of microbiota composition after the single-cell cultivation in MDE compartments in the presence of 10 and 100 µg/mL Ami, relative to the respective cultivation without an antibiotic. Indication is represented as follows: bear oral microbiota (B), human fecal microbiota from a patient with colitis (P), a healthy human donor (D). (B) Comparison between MIC predicted using single-cell cultivation in MDE in the presence of Ami (Left, scale with bars) and MIC of clinical isolates measured in vitro (Right, scatterplot). Scale bars (Left) indicate the values of scores obtained from the shift of microbiota composition after single-cell cultivation in MDE compartments in the presence of Ami. The scores enabled us to range bacteria according to their resistance to Ami and subdivide them into groups according to the values of their predicted MIC (<10, 10–100, >100 µg/mL). Each point of the scatterplot (Right) represents MIC measured in vitro for a particular bacterial strain, the horizontal lane represents the range, and the vertical lane indicates the mean.
Gram-negative Proteus mirabilis, Acinetobacter baumannii, Morganella morganii, and Enterobacter cloacae were resistant, while Gram-positive Macrococcus caseolyticus, Staphylococcus (except Staphylococcus vitulinus), Aerococcus viridans, and W. confusa were sensitive to Ami. Bacilli (B. subtilis, Bacillus licheniformis, and B. pumilus) were also resistant, likely due to the presence of AmiN or its homologs. The major components which were overrepresented in the patient microbiota over the donor one (SI Appendix, Fig. S8), i.e., Enterococcus faecium and Bacteroides vulgatus, were also susceptible to Ami. Hence, Ami-producing B. pumilus 124 may hold promise as a probiotic for controlled remodeling of human gut microbiota.
Discussion
Exploration of uncommon microbiota sources appears to be a promising approach to search for and select both antibiotics and probiotics from wild nature. Classical microbiology, however, is inefficient for deep functional profiling of entire microbiomes. Here we show how microfluidic uHT screening technologies could be applied for classical microbiological problems, i.e., antibiotic/probiotic selection and susceptibility/resistance testing. The technological advantage of MDE encapsulation provides an opportunity of a single-cell manipulation and detailed ultrahigh-throughput functional characterization of microbial communities.
Our technique enabled an efficient and straightforward single-step isolation of bacteria displaying the desirable anti-S. aureus activity from an exotic microbiota source. Subsequent metabolomic analysis, sequencing, and genome mining revealed the main active compound (Ami) and its biosynthetic cluster in the genome of a promising probiotic candidate B. pumilus. When we were analyzing the metagenomes of the bacteria selected after uHT screening, we observed the amplification of numerous bacteria that were unculturable under the specific conditions of the experiment. Although B. pumilus was selected by uHT screening as a culturable component of bear microbiome, we believe that this principle is particularly interesting as a source of data for deep metagenome mining and selection of unexplored biosynthetic clusters as well as for uHT analysis of unculturable species.
The selected Ami cluster from B. pumilus was characterized using proteomics and heterologous expression, which revealed a specific mechanism of Ami inactivation/activation via kinase AmiN and phosphatase AmiO activity. We found that the Ami cluster is present only in a few B. pumilus reference genomes; the presence of amiN, however, is almost ubiquitous (SI Appendix, Fig. S9A). B. pumilus have amiN and the Ami cluster in the same neighborhood (Fig. 2A), while strains without the Ami cluster have a stand-alone amiN homolog (SI Appendix, Fig. S9B). We hypothesize that stand-alone AmiN-like kinases have a promiscuous specificity toward Ami analogs (or related antibiotics like zwittermycin, Fig. 3), while the bona fide AmiN is a dedicated enzyme that evolved for efficient inactivation of and self-resistance to Ami.
AmiN homologs are found in numerous Bacillus species that stress their functional significance (SI Appendix, Fig. S10). Moreover, we found it in thermoactinomycete Paludifilum halophilum that also contains a homologous Ami cluster (Fig. 3). To probe the effectiveness of a particular external influence on the entire microbial community, we developed the uHT technique based on a single-cell in droplet cultivation. The only limitations of this method lie in in droplet cultivability and accuracy of bioinformatic analysis. We observed, however, that the overwhelming majority of bacterial species in our samples were culturable in droplets and lending themselves well to the freely available algorithms such as MetaPhlAn2. The spectrum of Ami activity was analyzed using different microbiomes and quantitative estimations of Ami activity on bacteria were made. We found Ami to be especially active against Gram-positive bacteria. Furthermore, it is active against some bacteria relevant to dysbiosis, and we suppose that Ami-producing strains could be used for controlled microbiota remodeling. Further investigations of this approach, however, must be done in terms of its safety and efficiency in vivo. Finally, we assume that the demonstrated approach is not limited to bacterial communities and could be efficiently expanded to the deep functional profiling of eukaryotic cells in applications such as biomarker probing and chemotherapy resistance/efficiency screening.
Materials and Methods
Selection of Bacteria Displaying Anti-S. aureus Activity.
The detailed selection procedure was previously described in ref. 13 and is specified in SI Appendix, SI Materials and Methods.
Microbiota Collection and Storage.
The study was approved by the Local Ethics Committee of the Federal Research and Clinical Centre of Physical-Chemical Medicine (FRCC PCM), conclusion No. 2017/02 from 13.04.2017. All donors provided written informed consent.
NGS Sequencing.
The selected MDE droplets were freeze dried and total DNA was isolated using the QIAamp DNA Investigator Kit (Qiagen). Whole-genome amplification was performed using the REPLI-g Single Cell Kit (Qiagen). For individual strains, genomic DNA (100 ng for each sample) was disrupted into 400- to 550-bp fragments using the Covaris S220 System (Covaris). Sequencing of libraries was performed using the genetic analyzer HiSeq2500, the HiSeq PE Cluster Kit v4 cBot and the HiSeq SBS Kit v4 (250 cycles) (Illumina) according to the manufacturer’s instructions. For detailed information please refer to SI Appendix, SI Materials and Methods.
Deep Profiling of Ami Activity.
Different microbiota samples were serially coencapsulated with Ami to the final concentrations of 0, 10, and 100 µg/mL Ami inside MDE droplets. Calcein Violet AM (Thermo Fisher Scientific) was added to the droplet emulsion to the final concentration of 10 μM for metabolic activity staining after 10 h of incubation at 35 °C. Following the incubation for 30 min at room temperature, the droplets with Calcein Violethigh fluorescence (450/50-nm filter) were sorted using a FACSAria III cell sorter (BD Biosciences). The collected droplets were frozen in liquid nitrogen, freeze dried, and analyzed by whole-genome sequencing, and bioinformatics resulted in quantification of bacteria in samples (Fig. 5B). Bacteria distributions in samples with Ami were normalized to the respective distribution in samples without Ami, resulting in shift fold calculated for each bacterium in the sample (Fig. 5A). The natural logarithm of the shift fold was defined as a score. Bacteria that have negative scores (depleted) at 10 µg/mL Ami concentration were defined as susceptible (MIC < 10 µg/mL), positive scores (increased) at 100 µg/mL as resistant (MIC > 100 µg/mL), and positive scores at 10 µg/mL and negative scores at 100 µg/mL as intermediate (MIC = 10–100 µg/mL), respectively. Confidential intervals of MICs were predicted using a linear regression between the scores and the mean values of MIC measured in vitro for particular bacterial strains using bacterial collection.
Supplementary Material
Acknowledgments
This work was supported by Grant RFMEFI60716X0145 from the Ministry of Education and Science of Russia. Experiments were partially carried out using the equipment provided by the Institute of Bioorganic Chemistry core facility (CKP IBCH).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the GenBank database, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. QENN00000000 and QENO00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811250115/-/DCSupplemental.
References
- 1.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 3.Halfvarson J, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weyrich LS, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. doi: 10.1038/nature21674. [DOI] [PubMed] [Google Scholar]
- 5.Miller AW, Dale C, Dearing MD. The induction of oxalate metabolism in vivo is more effective with functional microbial communities than with functional microbial species. mSystems. 2017;2:e00088-17. doi: 10.1128/mSystems.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sassone-Corsi M, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosshart SP, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang HY, et al. Diets alter the gut microbiome of crocodile lizards. Front Microbiol. 2017;8:2073. doi: 10.3389/fmicb.2017.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kropáčková L, et al. Variation between the oral and faecal microbiota in a free-living passerine bird, the great tit (Parus major) PLoS One. 2017;12:e0179945. doi: 10.1371/journal.pone.0179945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Front Microbiol. 2017;8:1705. doi: 10.3389/fmicb.2017.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hird SM. Evolutionary biology needs wild microbiomes. Front Microbiol. 2017;8:725. doi: 10.3389/fmicb.2017.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terekhov SS, et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc Natl Acad Sci USA. 2017;114:2550–2555. doi: 10.1073/pnas.1621226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh J, Omoto S, Nishizawa N, Kodama Y, Inouye S. Chemical structures of amicoumacins produced by Bacillus pumilus. Agric Biol Chem. 1982;46:2659–2665. [Google Scholar]
- 15.Pinchuk IV, Bressollier P, Sorokulova IB, Verneuil B, Urdaci MC. Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res Microbiol. 2002;153:269–276. doi: 10.1016/s0923-2508(02)01320-7. [DOI] [PubMed] [Google Scholar]
- 16.Polikanov YS, et al. Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome. Mol Cell. 2014;56:531–540. doi: 10.1016/j.molcel.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park HB, Perez CE, Perry EK, Crawford JM. Activating and attenuating the amicoumacin antibiotics. Molecules. 2016;21:824. doi: 10.3390/molecules21070824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Sci Rep. 2015;5:9383. doi: 10.1038/srep09383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kevany BM, Rasko DA, Thomas MG. Characterization of the complete zwittermicin A biosynthesis gene cluster from Bacillus cereus. Appl Environ Microbiol. 2009;75:1144–1155. doi: 10.1128/AEM.02518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller S, et al. Paenilamicin: Structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew Chem Int Ed Engl. 2014;53:10821–10825. doi: 10.1002/anie.201404572. [DOI] [PubMed] [Google Scholar]
- 21.Reimer D, Pos KM, Thines M, Grün P, Bode HB. A natural prodrug activation mechanism in nonribosomal peptide synthesis. Nat Chem Biol. 2011;7:888–890. doi: 10.1038/nchembio.688. [DOI] [PubMed] [Google Scholar]
- 22.Nougayrède J-P, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 23.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto M, et al. Isolation of 8′-phosphate ester derivatives of amicoumacins: Structure-activity relationship of hydroxy amino acid moiety. J Antibiot (Tokyo) 2007;60:752–756. doi: 10.1038/ja.2007.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.