Significance
Antibodies binding to their cognate cellular receptors can trigger important downstream immune responses. We mapped out critical amino acids on the IgA1 antibody that govern binding to its specific receptor, FcαRI. We found that two of the most important amino acids were located on a different IgA1 domain than the rest of the binding site, separated by a flexible linker. To better understand the interplay between receptor binding and dynamic motions in IgA1, we conducted dynamics simulations on the system. The results indicate that receptor binding perturbs IgA1 conformational dynamics over long distances and can link the receptor binding site to the hinge region of IgA1. We validate this finding experimentally, which has implications for the kidney disease IgA nephropathy.
Keywords: IgA1 antibody, binding energetics, molecular-dynamics simulations, surface plasmon resonance, principal-component analysis
Abstract
IgA effector functions include proinflammatory immune responses triggered upon clustering of the IgA-specific receptor, FcαRI, by IgA immune complexes. FcαRI binds to the IgA1–Fc domain (Fcα) at the CH2–CH3 junction and, except for CH2 L257 and L258, all side-chain contacts are contributed by the CH3 domain. In this study, we used experimental and computational approaches to elucidate energetic and conformational aspects of FcαRI binding to IgA. The energetic contribution of each IgA residue in the binding interface was assessed by alanine-scanning mutagenesis and equilibrium surface plasmon resonance (SPR). As expected, hydrophobic residues central to the binding site have strong energetic contributions to the FcαRI:Fcα interaction. Surprisingly, individual mutation of CH2 residues L257 and L258, found at the periphery of the FcαRI binding site, dramatically reduced binding affinity. Comparison of antibody:receptor complexes involving IgA or its precursor IgY revealed a conserved receptor binding site at the CH2–CH3 junction (or its equivalent). Given the importance of residues near the CH2–CH3 junction, we used coarse-grained Langevin dynamics simulations to understand the functional dynamics in Fcα. Our simulations indicate that FcαRI binding, either in an asymmetric (1:1) or symmetric (2:1) complex with Fcα, propagated long-range conformational changes across the Fc domains, potentially impacting the hinge and Fab regions. Subsequent SPR experiments confirmed that FcαRI binding to the Fcα CH2–CH3 junction altered the kinetics of HAA lectin binding at the IgA1 hinge. Receptor-induced long-distance conformational transitions have important implications for the interaction of aberrantly glycosylated IgA1 with anti-glycan autoantibodies in IgA nephropathy.
IgA is the second most prevalent antibody isotype in serum and the most abundant isotype in mucosal secretions (1, 2); it performs an important role in preventing and countering pathogenic challenge to the immune system. IgA can be divided into IgA1 and IgA2 subclasses, which differ in the number of glycosylation sites and the length of the hinge region. The IgA1 subclass features a heavily O-glycosylated hinge region, with up to six potential O-glycans. Aberrant O-glycosylation of the IgA1 hinge is a key feature seen in patients with the autoimmune disease IgA nephropathy (IgAN), as it forms a neoepitope for anti-glycan autoantibodies and leads to deposition of immune complexes in the glomerular mesangium (3).
In the presence of multivalent antigen, IgA initiates signaling via the IgA-specific receptor FcαRI on immune cells, triggering a range of proinflammatory responses (2, 4, 5). The FcαRI ectodomain consists of two orthogonal Ig-like domains, D1 and D2 (6, 7). The N-terminal region of FcαRI D1 contacts IgA at the CH2–CH3 (Cα2–Cα3) domain interface (Fig. 1A) (6, 8–13). Analytical ultracentrifugation, biosensor, and crystallographic studies have shown that two FcαRI molecules can bind a single IgA antibody (6, 13, 14). The FcαRI ectodomain can be shed upon activation by the action of ADAM10 and ADAM17, resulting in soluble FcαRI in serum (15); this soluble receptor form has been implicated in the progression of IgAN (16, 17). However, the role played by soluble FcαRI in IgAN remains unclear.
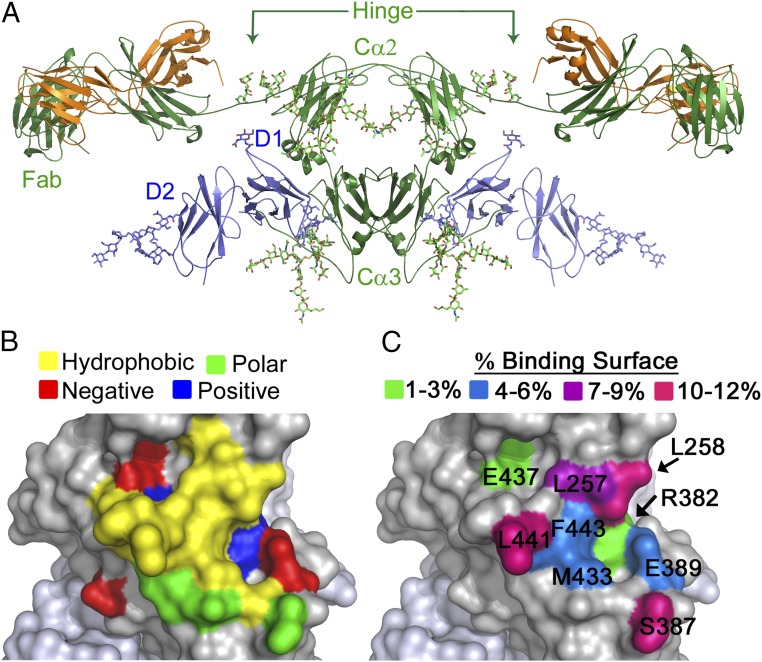
Fig. 1.
FcαRI binds IgA1 at a hydrophobic region of the Cα2–Cα3 junction. (A) Model of the 2:1 complex between FcαRI (blue) and IgA1 (green/orange) (3, 6) based on the crystal structure of the FcαRI:Fcα complex (PDB ID code 1OW0) and the solution structure of full-length IgA1 (PDB ID code 1IGA). (B and C) Characteristics of FcαRI binding site on Fcα: amino acid properties (hydrophobic, yellow; positively charged, blue; negatively charged, red; polar, green) (B); percent contribution of each Fcα residue with side chain contacts to the binding surface (C).
Here, we report binding data and computational analyses, providing information on energetic and dynamic aspects of the FcαRI:IgA1 interaction. We combined alanine-scanning mutagenesis and equilibrium biosensor experiments to complete the first systematic analysis of the energetic contributions of individual IgA residues whose side chains contact FcαRI, identifying the Fcα energetic hot-spot residues in the binding interface. Comparing these results to other related antibody:receptor pairs revealed a common mode of binding. Using the FcαRI:Fcα crystal structure, we performed coarse-grained molecular-dynamics (MD) simulations and principal-component analysis (PCA) to elucidate the role of IgA1–Fc domain motion in receptor binding. We discerned functionally relevant hinge-based dynamics of the IgA1 Cα2 and Cα3 domains based on the compatibility of corresponding principal eigenvectors with the conformational change induced by receptor binding. The analysis predicted that receptor binding at the Cα2–Cα3 interface would induce long-range conformational changes propagating up to the O-glycosylated hinge, which was confirmed using biosensor experiments with the hinge-binding lectin HAA. Thus, we propose that long-distance communication in IgA1 is mediated by extensive allosteric networks that couple antigen-binding (Fab) regions and receptor-binding (Fc) regions (18–20). Our results are consistent with experimental and computational studies of widely diverse classes of proteins that revealed that allosteric mechanisms are responsible for inducing large-scale conformational changes (21–24), promoting dynamic coupling (25, 26), or selecting functionally relevant folding pathways (27, 28).
Results
Energetic Analysis of FcαRI Binding to IgA1.
Using the crystal structure of the complex, Fcα residues involved in the FcαRI:Fcα interaction were identified (6). Nineteen residues, located at the Cα2–Cα3 interface, contact FcαRI. Ten of these residues have side chains contacting FcαRI, defined as being within 4 Å of FcαRI (L257, L258, R382, S387, E389, M433, E437, L441, A442, and F443) (6). Except for L257 and L258, which are located on the AB helix/loop of the Cα2 domain of IgA, all residues are in the Cα3 domain. The FcαRI binding site on IgA is composed of a central hydrophobic region surrounded by polar and charged residues, an arrangement typical of many protein–protein binding interfaces (Fig. 1B) (6, 29–31). The contribution of each side chain to the buried surface area in the FcαRI:Fcα complex is shown in Fig. 1C.
Each Fcα residue whose side chain contacts FcαRI was individually mutated to alanine (with the exception of A442). Binding of Fcα variants to FcαRI was measured by surface plasmon resonance (SPR) (SI Appendix, Fig. S1). Since two FcαRI molecules can bind a single Fcα protein, equilibrium binding data were fitted to a two-site binding model to determine KD1 and KD2, equilibrium binding constants corresponding to the first and second binding events (6, 13, 14). Similar to previously published values, wild-type Fcα bound FcαRI with KD1 and KD2 values of 46.2 and 223 nM, respectively (Table 1) (13, 14). KD1 values were used to compute ΔΔG values for the various mutations. All Fcα mutants had positive ΔΔG values, indicating that each mutation resulted in a less favorable interaction with FcαRI compared with wild type (Fig. 2A and Table 1). Far-UV circular dichroism experiments demonstrated that all mutant and wild-type Fcα proteins had similar secondary structure content, indicating that decreases in affinity were not due to global structural changes (SI Appendix, Fig. S2 and Table S1).
Table 1.
Equilibrium parameters for FcαRI binding to Fcα wild type and mutants
| Fcα ligand | KD1, nM | KD2, nM | ΔG, kcal/mol | ΔΔG, kcal/mol |
| Wild type | 46 ± 2 | 220 ± 10 | −10.00 | 0.00 |
| R382A | 63 ± 3 | 520 ± 40 | −9.82 | +0.18 |
| E437A | 64 ± 2 | 450 ± 30 | −9.81 | +0.19 |
| S387A | 73 ± 2 | 620 ± 30 | −9.73 | +0.27 |
| E389A | 292 ± 6 | 2,900 ± 100 | −8.91 | +1.09 |
| L441A | 360 ± 10 | 2,600 ± 100 | −8.79 | +1.21 |
| L257A | 2,390 ± 70 | 62,000 ± 6,000 | −7.67 | +2.34 |
| M433A | 3,000 ± 2,000 | ∼29,000 | −7.52 | +2.49 |
| F443A | ∼6,000 | ∼45,000 | −7.14 | +2.93 |
| L258A | ∼15,000 | ∼78,000 | −6.56 | +3.44 |
Equilibrium parameters for Fcα proteins were derived from analyses with at least nine different concentrations of injected FcαRI. KD1 values for Fcα F443A and L258A, and KD2 values for M433A, F443A, and L258A could not be determined with a high degree of confidence under the experimental conditions. Each binding experiment was carried out in duplicate.
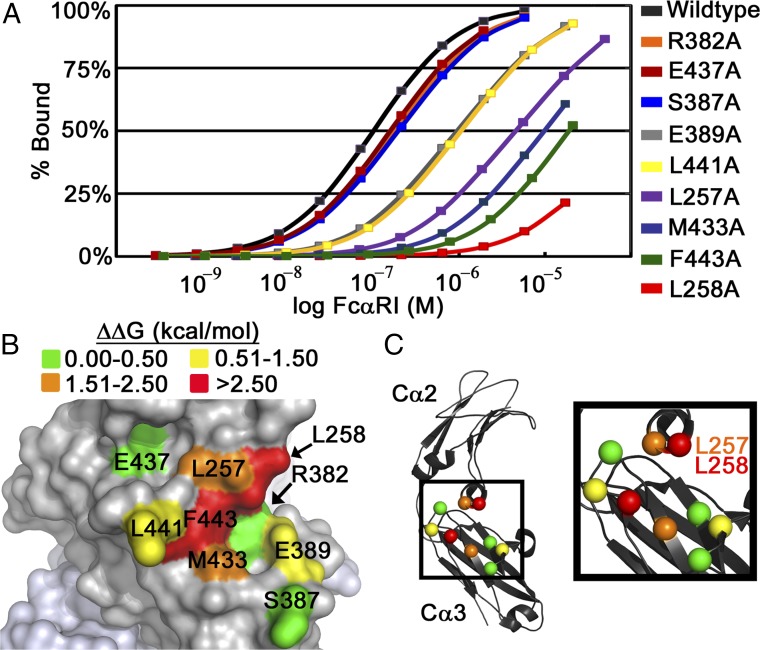
Fig. 2.
SPR analysis of FcαRI binding to Fcα variants identifies energetic hot-spot residues. (A) Coplotted SPR binding isotherms of wild-type Fcα and mutants binding to FcαRI. Fcα variants were coupled to the SPR chip and soluble FcαRI was flowed over. Fcα L257A, M433A, F443A, and L258A mutations resulted in the largest decreases in binding affinity. (B) Plotting of the experimentally determined ΔΔG values on Fcα. (C) Location of the Cα of all mutated residues, colored according to the ΔΔG values for each alanine mutant.
Mutation of charged or polar Fcα residues located at the periphery of the binding site (R382, S387, and E437) had only mild effects on binding affinity, with ΔΔG values of less than +0.3 kcal/mol compared with wild type (Fig. 2B and Table 1). Two additional mutations, E389A and L441A, had an intermediate effect on binding affinity, with ΔΔG values between +1.0 and +1.2 kcal/mol. In the complex, E389 is sandwiched between FcαRI residues R52 and R53 and Fcα residue R382, so the E389A mutation likely results in electrostatic repulsion between the arginine side chains. L441 is a hydrophobic residue located in close proximity to the center of the FcαRI binding site on Fcα. The loss of the hydrophobic side chain in the L441A variant results in a loss of van der Waals interactions with FcαRI residues Y35, F56, and H85.
Mutation of hydrophobic Fcα residues L257, L258, M433, or F443 resulted in the largest decreases in FcαRI affinity (ΔΔG values between +2.34 and +3.44 kcal/mol). The side chain of each of these hot-spot residues has a high percentage of its accessible surface area buried in the FcαRI:Fcα complex (between 90% and 100%; SI Appendix, Table S2). However, buried surface area alone is not a reliable indicator of a mutation’s effect. In the case of S387, whose surface area is 70% buried in the interface, mutation to alanine resulted in a much smaller ΔΔG value of +0.27 kcal/mol. Experimental ΔΔG values also did not have a strong correlation with the Fcα residues’ individual surface area contributions to the total binding interface (Figs. 1C and 2B and SI Appendix, Table S2) (32).
The importance of M433A and F443A is expected due to the residues’ location in the central hydrophobic region of the protein binding interface (Fig. 1B), which typically contributes significantly to the energetics of complex formation (29, 33–36). Mutation of these hydrophobic residues to alanine would result in a loss of significant van der Waals interactions by disrupting the tight packing of FcαRI residues Y35, L54, F56, G84, and H85 against the central hydrophobic region of Fcα, with a negative impact on occlusion of bulk solvent at the binding interface.
The dramatic effect of mutating L257 or L258, the only two Cα2 residues with side-chain contacts, indicates the Cα2 domain also plays a very important role in the stability of the FcαRI:Fcα complex (Fig. 2 B and C). In the complex, the side chain of L257 interacts with FcαRI residues Y35 and R82 and forms part of a hydrophobic pocket into which the side chain of residue H75 packs; this pocket is responsible for the pH dependence of the FcαRI:Fcα interaction (14). The L258 side chain interacts with FcαRI residues Y35, R52, and R53. The importance of L257 and L258 for stable complex formation is further supported by the observation that the IgA1 Cα3 domain alone (expressed in bacteria and refolded from inclusion bodies) showed nearly undetectable binding to FcαRI by SPR (SI Appendix, Fig. S3), confirming that Cα3 alone does not confer stable binding to FcαRI, despite contributing 80% of the residues whose side chains contact FcαRI.
Importance of the CH2 Domain in Related Antibody:Receptor Interactions.
Given the importance of the Cα2 L257 and L258 residues in the FcαRI:Fcα interaction, we compared this complex to a related Ig:receptor pair to ascertain the role of analogous residues. IgY, the predominant serum antibody of lower vertebrates including reptiles, amphibians, and birds (37), is believed to be an ancestor of human IgA (38). The two C-terminal domains of human IgA and chicken IgY share 34% amino acid identity, and the interaction between chicken IgY and the chicken Ig-like receptor, CHIR–AB1, closely resembles the FcαRI:Fcα interaction (39). Similar to FcαRI, the gene encoding the CHIR–AB1 receptor is found within the leukocyte receptor cluster and signaling requires the associated FcRγ coreceptor. Furthermore, CHIR–AB1 binds the Fc domain of chicken IgY (Fcυ) to form a 2:1 complex (40–43). Mutational analyses mapped the IgY contact residues to the CH3–CH4 (Cυ3–Cυ4) junction, which is analogous to the IgA Cα2–Cα3 junction (39, 44). Individual mutation of Cυ3 residues 362–364 (LYI), analogous to Cα2 residues 256–258 (LLL), as well as mutation of Cυ4 Pro and Arg residues within the PMRF motif (residues 554–557; analogous to Cα3 PLAF residues 440–443), resulted in the greatest decrease in binding of IgY to the CHIR–AB1 receptor. Thus, accessible residues in the Cα2/Cα3 (or equivalent Cυ3/Cυ4) interfaces involved in receptor interactions are conserved between IgA and IgY (39).
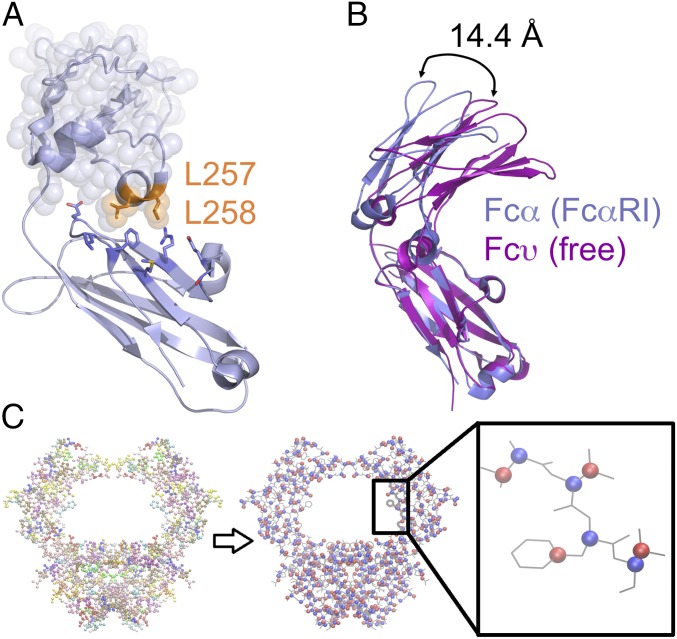
The critical contribution of the Cα2 L257 and L258 residues, and analogous Cυ3 residues, to interactions with their respective receptors is reflective of a conserved binding mode, despite the fact that these residues are located across from the Cα3 domain boundary where most of the receptor contacts occur (Fig. 3A). A flexible linker exists between the Cα2 and Cα3 domains in Fcα, which could act as a major hinge point between these two domains. Backbone alignment of Fcα (from the FcαRI:Fcα structure) with the unbound IgY–Fc fragment (42) showed a 14.4-Å difference in the position of the upper domains (Fig. 3B). Unlike IgA, IgY does not have a disulfide bond linking its Cυ3 domains (analogous to IgA Cα2 domains), which may account for the observed degree of variation in the position of the upper domains. The variability of the position of Cα2 or Cυ3 in these structural alignments indicates a substantial degree of flexibility at the Cα2–Cα3 (Cυ3–Cυ4) junction.
Fig. 3.
Comparison of Fcα and Fcυ crystal structures reveals variability in the CH2 domain position, indicating the Cα2–Cα3 junction acts as a hinge point. (A) Structure of the Fcα heavy chain, showing the location of L257 and L258 at the bottom of the Cα2 domain (highlighted with transparent spheres). (B) Overlay of Fcα from the FcαRI-bound complex (blue) with unbound IgY Cυ3–Cυ4 (magenta) revealed a 14.4-Å shift between the top of the Cα2 and Cυ3 domains. (C) The coarse-grained model of Fcα is shown in a bead representation, with each amino acid represented using two beads. First bead (blue), representing the backbone, is located at Cα position, and the second bead (red), representing the side chain, is located at the center of mass of the amino acid’s side chain.
FcαRI Binding Dampens Intradomain Motions of Fcα.
To elucidate the effect of FcαRI binding on Fcα flexibility, we performed Langevin dynamics (LD) simulations of a coarse-grained model of the 2:1 FcαRI:Fcα complex, a 1:1 FcαRI:Fcα complex (through removal of the trans FcαRI receptor), and the unliganded Fcα (through removal of both cis and trans FcαRI receptors). The initial configurations of these systems were obtained from the crystal structure of the 2:1 FcαRI:Fcα complex (PDB ID code 1OW0) (6). As indicated in Materials and Methods, the coarse-grained procedure describes amino acids by using two virtual particles, Cα and side chain (Cα–SC), that represent backbone and side-chain atoms (Fig. 3C). Comparison of the B-factor profile of Fcα heavy chains in the 2:1 FcαRI:Fcα complex with the corresponding experimental values in the crystal structure (SI Appendix, Fig. S4) indicates a similar pattern of structural flexibility, which supports the validity of our LD simulation protocol.
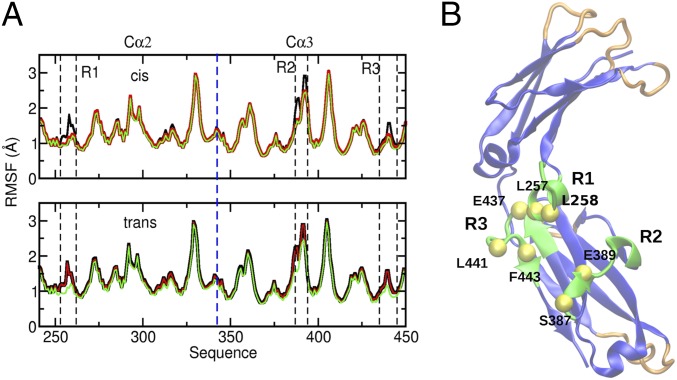
We characterized quantitatively the degree of Fcα flexibility (Fig. 4) in these three systems by computing root-mean-square fluctuations (RMSFs) of protein amino acids in LD trajectories (Materials and Methods). Consistent with the stoichiometry of receptor binding, residue fluctuations (Fig. 4A) in the two Fcα chains are symmetric in Fcα and 2:1 FcαRI–Fcα conformations and asymmetric in 1:1 FcαRI–Fcα conformations. We note that large flexibility is present in the intersubunit loop regions (Fig. 4); however, subunit structure is largely preserved in our simulations. We propose that the combination of structural stability in the receptor-binding region and conformational flexibility in loop regions at the intersubunit interfaces is important to mediate functional allosteric communication between subunits. As shown in Fig. 4A, comparison of RMSF profiles indicates that the major effect of receptor binding is to reduce flexibility of three Fcα regions, R1 (amino acids 255–260), R2 (380–390), and R3 (430–445). This dampening effect is the direct result of formation of the FcαRI–Fcα interface as indicated by the nearly identical RMSF differences in these regions in the cis Fcα chain in the asymmetric 1:1 and in the symmetric 2:1 complexes compared with the unliganded Fcα (Fig. 4A). As discussed above, these regions contribute to the FcαRI:Fcα interface and include the hinge formed by CH2 and CH3 domains. The largest changes in RMSF values in these regions correspond to amino acids L257, L258, G259, S260, S387, Q388, E389, R392, E393, P440, L441, A442, and F443 (Fig. 4). We note that this set includes L257, L258, S387, E389, L441, A442, and F443, which are highlighted as important for FcαRI binding in our mutagenesis studies.
Fig. 4.
Backbone flexibility in distinct Fcα complexes. (A) The root-mean-square fluctuations (RMSFs) of Cα atoms of Fcα amino acids in unliganded Fcα (black); 1:1 FcαRI–Fcα (red) and 2:1 FcαRI–Fcα (green) complexes are shown for the cis (trans) heavy chain in the Upper (Lower) panel. In the asymmetric 1:1 FcαRI–Fcα complex, the receptor is bound to the cis Fcα heavy chain. (B) Amino acids (green) corresponding to the R1, R2, and R3 regions in A, located primarily near the linker between the Cα2 and Cα3 domains, experience the strongest RMSF dampening upon receptor binding. The yellow spheres with labels indicate the amino acid positions in this set, which are highlighted as important for receptor binding in mutagenesis studies. Six structural regions (orange and green loop in the Cα3 domain) with the largest flexibility (RMSF > 1.5 Å) primarily include loops involved in the intersubunit interface.
FcαRI Binding Results in Weak Perturbation of Fundamental Motions of Fcα.
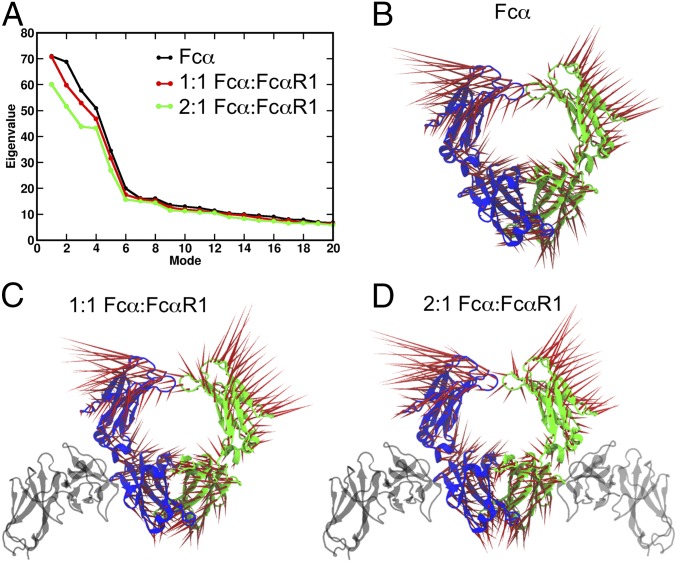
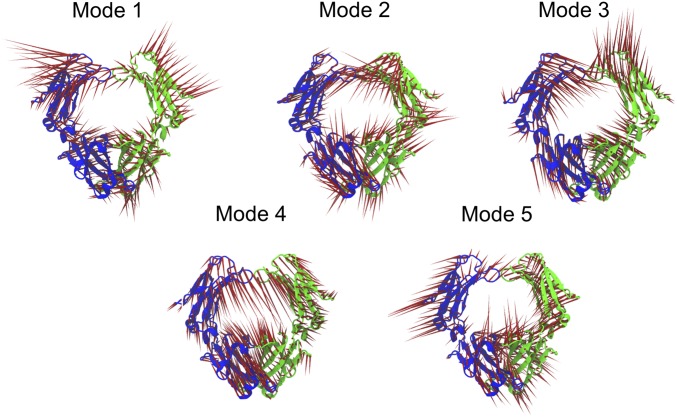
Receptor-induced conformational changes in proteins are mediated by allosteric networks that span long distances and may include interdomain and intersubunit interactions. To reveal long-range communication within the Fcα structure activated upon FcαRI binding, we use PCA, which probes collective motions of distinct FcαRI:Fcα complexes (Materials and Methods). In this approach, the covariance matrix is diagonalized to yield the set of eigenvectors that characterize the direction of motion in independent modes and eigenvalues that determine the amplitudes of motions (45). Zero eigenvalues, which correspond to rotations and translations of the entire structure, are excluded from the analysis and nonzero eigenvalues are ranked in order of decreasing value (Fig. 5A). Generally, it is found that collective motions with the largest contribution to the RMSF correspond to a small number of principal component (PC) modes with the largest eigenvalues. In each of the systems studied, we find that the five highest-ranked PC modes have significantly larger eigenvalues than all other modes (Fig. 5A), which indicates that these are the most relevant modes in describing the functional dynamics of the Fcα structure. The common aspect of the eigenvalue profiles for the three systems is consistent with the similar overall pattern of RMSFs. Smaller eigenvalues corresponding to 1:1 and 2:1 FcαRI–Fcα complexes reflect the dampening effect of FcαRI binding on Fcα motions. The smaller difference between the Fcα and 1:1 FcαRI–Fcα profiles indicates that single receptor binding results in a weak perturbation of Fcα motion, while the larger change in eigenvalues upon binding of the second receptor indicates a stronger perturbation in the symmetric complex. To characterize, at the amino acid level, the five significant PC modes that contribute to conformational changes in Fcα, we examined in detail the associated motions and directional correlations of amino acid pairs (Materials and Methods). Fcα motions associated with the five highest-ranked PC modes involve primarily rigid-body domain motions of the Cα2 and Cα3 domains around their flexible common joints (Fig. 6, SI Appendix, Fig. S5, and Movies S1–S15). These movements satisfy constraints imposed by intersubunit interfaces (Cα2–Cα2 and Cα3–Cα3), in addition to those resulting from intrasubunit hinges (Cα2–Cα3). Extensive contacts between Cα3 domains of the two chains strongly constrain the relative mobility of these two domains, so that, in all five PC modes, their motions consist largely of rigid-body rotations around the common interface (SI Appendix, Fig. S5 and Movies S1–S15). Distinct motions of the five PC modes arise primarily from the more flexible Cα2–Cα2 interface, which is dominated by disulfide bonds (Movies S1–S15). In a given PC mode, associated Fcα motions are similar for all three complexes. For example, mode 1 (Fig. 5B and Movies S1–S3) corresponds to torsional motions of the Cα3 domains and swing-like motions of Cα2 domains. Comparative study of motions and correlations of amino acid pairs further confirms that Cα2 and Cα3 domains have a higher flexibility to move around their hinge in the unliganded Fcα compared with more restricted movements of these domains in 1:1 and 2:1. Overall, our analysis highlights the importance of the intersubunit (Cα2–Cα2) disulfide bond region for effecting Fcα conformational changes. This indicates that perturbations at the intrasubunit junctions, such as those effected by receptor binding, are transmitted primarily to the Cα2–Cα2 interface and, therefore, are likely to influence conformational fluctuations at the IgA1 hinge. In addition, the common fundamental motions of the three systems lead us to conclude that FcαRI binding yields tighter coupling of sites near the Fcα Cα2–Cα3 intrasubunit junction without significantly distorting global Fcα motions.
Fig. 5.
Principal-component analysis (PCA) of MD trajectories of distinct Fcα complexes. (A) The largest 20 eigenvalues of the PC modes for different Fcα complexes: Fcα (black), 1:1 FcαRI–Fcα (red), and 2:1 FcαRI–Fcα (green). (B–D) Fcα motions associated with mode 1 (largest eigenvalue) in the three systems studied. The red vectors illustrate the amplitude and the direction of residue motion for (B) Fcα alone, (C) 1:1 FcαRI–Fcα, and (D) 2:1 FcαRI–Fcα.
Fig. 6.
Fcα motions associated with the highest ranked PC modes. The five panels indicate the mode motions for modes 1–5 for unliganded Fcα. The red vectors indicate the direction of residue motion, and the vector length indicates the relative amplitude of the residue motion in each mode.
Receptor Binding Activates a Long-Range Allosteric Network in Fcα.
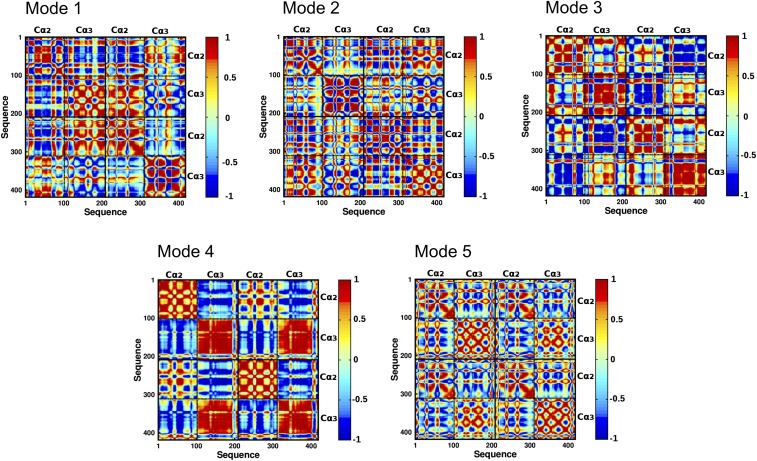
To characterize long-range communication between Fcα regions, we computed cross-correlation maps of residue fluctuations along the five highest ranked PC modes for the three distinct Fcα systems (Materials and Methods). As illustrated in Fig. 7 and SI Appendix, Fig. S5, in each of the five modes, intradomain motions of liganded or unliganded Fcα are strongly correlated, consistent with the rigid-body domain motions noted above. In addition, we find strong correlation between motions of regions of distinct domains, which supports the existence of long-range interactions and coordinated domain movements (Fig. 7). For example, the cross-correlation map of PC modes 1 and 2 indicates strong coupling involving regions of the Cα2 and Cα3 domains of distinct subunits. In modes 3–5, intersubunit coupling is primarily mediated by strong correlations involving the Cα2–Cα2 and Cα3–Cα3 interfaces. In addition, strong intrasubunit anticorrelation between Cα2 and Cα3 domains, observed in modes 1 and 4, highlights hinge-based motions of IgA1. Overall, we surmise that the collective motions of the Fcα molecule are largely determined by strong correlation of intersubunit motions and anticorrelation of intrasubunit motions. These results suggested that allosteric interdomain communications control the global motions of Fcα. We also note that the pattern of interdomain coupling in 1:1 and 2:1 complexes is similar to that identified in the unliganded Fcα (SI Appendix, Fig. S5), which is consistent with the common fundamental motions of Fcα in the three systems. Nevertheless, the cross-correlation maps of the 1:1 complex reveal the allosteric effects of receptor binding to the cis subunit on the dynamics of the trans subunit.
Fig. 7.
Maps of directional correlation coefficients of all amino acid pairs in the unliganded Fcα for the five highest ranked PC modes. Correlation of amino acid pairs in modes 1–5. Strong correlation of a given pair of residues is indicated in red, and strong anticorrelation is shown in blue.
Fcα Residue Network Mediates Receptor-Induced Intersubunit Communication.
To pinpoint the effect of receptor binding on the Fcα allosteric network, we highlight pairs of amino acids within distinct subunits that have significantly modified correlation properties within 1:1 and 2:1 complexes compared with the unliganded Fcα. To this end, we consider residue pairs with weakly correlated motions in the unliganded Fcα and strongly correlated/anticorrelated motions in 1:1 and 2:1, as well as pairs that switch from strongly correlated (anticorrelated) motions in the unliganded Fcα to strongly anticorrelated (correlated) motions in FcαRI–Fcα complexes (Materials and Methods). Residue pairs identified according to our criteria for long-range communication are indicated in SI Appendix, Tables S3 and S4 and their structural location is illustrated in SI Appendix, Fig. S6 for the five highest ranked PC modes. We find that modes 2 and 3 include the largest populations of these pairs, which suggests that intersubunit coupling associated with modes 2 and 3 provides a large contribution to the propagation of receptor-induced perturbation over long distances. In modes 1, 4, and 5, intersubunit residue correlations are less affected by receptor binding (SI Appendix, Fig. S5), indicating that these mode motions are primarily responsible for regulating the other biological roles of Fcα molecule.
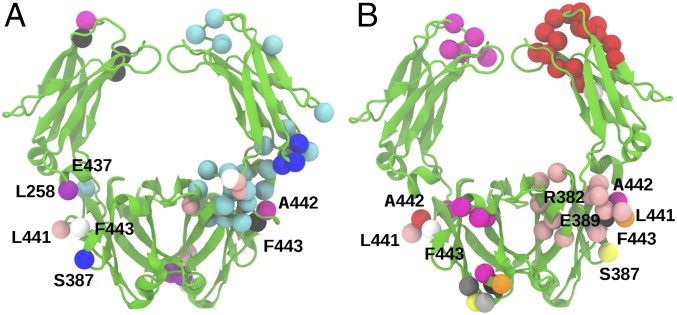
We examined in detail the long-distance coupling of each of the nine residues that comprise the receptor site (L257, L258, R382, S387, E389, M433, E437, L441, and F443), which were highlighted by our binding experiments. As shown in Fig. 8 and SI Appendix, Tables S3 and S4, long-distance communication involving these nine residues is significantly perturbed by receptor binding to Fcα. We find that FcαRI binding results in the strongest perturbation of directional correlation of residues L441 and F443 with distant residues. Two regions include large clusters of residues (Fig. 8) that are involved in long-distance communication with the FcαRI binding site. One of these regions is the Cα2–Cα3 junction, consistent with the signaling between the cis and trans receptor-binding sites. The second region is located at the Cα2–Cα2 junction near the IgA1 hinge, which suggests that receptor binding could induce long-range conformational changes in the hinge and Fab regions of IgA1. Notably, our results reveal that asymmetric binding of FcαRI to the cis Fcα subunit, as illustrated by the 1:1 complex, elicits strong long-distance response in the trans Fcα subunit (Fig. 8). Overall, we conclude that FcαRI binding induces tighter coupling of Fcα subunits by altering the underlying allosteric network without strongly perturbing the global fundamental motions.
Fig. 8.
Long-distance allosteric interactions between intersubunit pairs involving FcαRI binding sites. (A) Intersubunit residue pairs that switch from weak directional correlation in unliganded Fcα to strong correlation or anticorrelation upon receptor binding. Amino acid pairs, highlighted by distinct colors, include experimentally identified FcαRI-binding sites L258 (purple, cis), S387 (blue, cis), E437 (cyan, cis), L441 (pink, cis), A442 (magenta, trans), and F443 (white, cis; black, trans). (B) Intersubunit residue pairs that switch from strongly correlated (anticorrelated) motions in the unliganded Fcα to strongly anticorrelated (correlated) motions in FcαRI:Fcα complexes. Shown are pairs that include FcαRI binding sites L441 (pink, cis), A442 (red, cis), F443 (white, cis), R382 (silver, trans), S387 (yellow, trans), E389 (gray, trans), L441 (orange, trans), L442 (magenta, trans), and F443 (black, trans).
FcαRI Binding at the Cα2–Cα3 Junction Affects HAA Binding at IgA1 Hinge.
We have shown that binding of FcαRI occurs at a hot spot for dynamic transitions and that FcαRI binding dampens IgA1 domain motions. Furthermore, the negative cooperativity seen in this study and previous (13, 14) SPR experiments (i.e., KD2 is 4.8-fold weaker than KD1) and the PCA analysis together demonstrate the existence of long-range conformational effects across the Fcα dimer, suggesting that receptor binding at the Cα2–Cα3 interface could influence dynamics near the Cα2–Cα2 interface and the IgA1 hinge regions. Thus, we conducted SPR binding experiments to determine whether FcαRI binding can affect recognition events at the IgA1 hinge. Each IgA1 hinge region contains six potential O-linked glycosylation sites (46–52). The O-glycans consist of a core N-acetylgalactosamine (GalNAc) linked to a galactose, both of which may be sialylated (48–52). This heavy O-glycosylation of the IgA1 hinge causes it to be less flexible than IgG hinges and therefore potentially more likely to transmit long-range conformational changes.
Patients with IgAN, a kidney disease characterized by glomerular deposition of IgA1 immune complexes, have aberrantly glycosylated IgA1 that is undergalactosylated compared with control samples (53), resulting in the exposure of GalNAc moieties (54). HAA, a snail lectin, is a hexamer (a dimer of trimers) that specifically recognizes terminal GalNAc residues (55, 56). We have previously shown that HAA is functionally bivalent when binding IgA1 and can simultaneously bind two GalNAc residues on a single IgA1 antibody, presumably one from each hinge (56). If FcαRI binding at the Cα2–Cα3 junction induces long-range conformational changes that propagate to the IgA1 hinge, this could result in altered recognition of the O-glycans on the hinge by HAA due to changes in relative orientation of the GalNAc residues. Therefore, we measured binding of HAA to a myeloma IgA1 protein (IgA1κ) in the presence or absence of saturating concentrations of FcαRI to assess any differences in the affinity or kinetics of HAA binding (13).
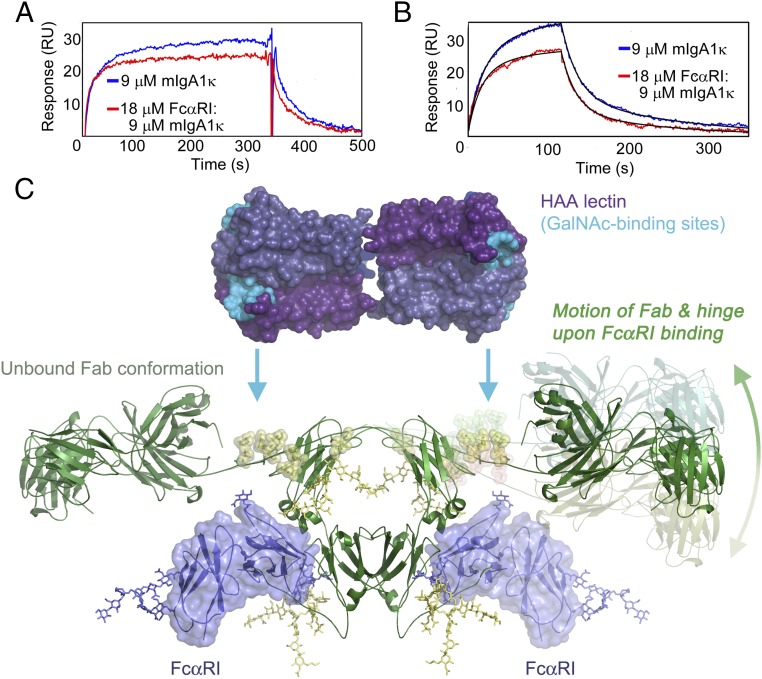
Equilibrium binding experiments were conducted to evaluate whether the affinity of IgA1κ for immobilized HAA was affected by the binding of FcαRI. Due to the low affinity of the IgA1–HAA interaction, full equilibrium binding curves could not be measured, but inspection of SPR sensorgrams from a low-flow rate experiment indicated a potential difference in the kinetics of binding (Fig. 9A). To verify this observation, we repeated binding experiments at 30 μL/min for kinetic analysis and fitted the data to a bivalent analyte model. The affinity of free IgA1κ for HAA (222 μM KD1; 4.4 μM KD2) was weaker compared with the FcαRI:IgA1κ complex (170 μM KD1; 2.8 μM KD2) (Fig. 9B and Table 2). The higher-affinity interaction of HAA with the FcαRI:IgA1 complex is solely due to faster association rates for both binding events (30% faster k1,on; 54% faster k2,on). These faster on-rates are the opposite trend of what would be expected based simply on a diffusion-limited binding event, since the FcαRI:IgA1κ complex has ∼40% larger mass than IgA1κ alone. Thus, binding of FcαRI to the Cα2–Cα3 interface of IgA1 induces conformational changes substantial enough to alter the kinetics of HAA binding to a distal site at the IgA1 hinge.
Fig. 9.
FcαRI binding influences HAA binding at the IgA1 hinge. (A) Comparison of SPR curves under steady-state conditions for mIgA1κ binding to the lectin HAA in the presence or absence of FcαRI indicates differences in kinetics of binding. (B) Kinetic binding data comparing HAA binding to mIgA1κ in the presence and absence of FcαRI binding revealed faster on rates of binding in the presence of FcαRI. Kinetic data and fits are shown in detail in SI Appendix, Fig. S7. (C) Model for scissor-like action of Fcα and hinge when FcαRI binds. FcαRI binding at the Cα2–Cα3 junction induces long-range conformational changes that are transmitted up into the hinge and Fab regions. The change in relative proximity of hinge O-glycans increases the rate of binding by the lectin HAA. For clarity, the conformation of the hinge and Fab in the unbound form are shown on the left side, and the proposed conformational change is illustrated on the right side.
Table 2.
Kinetic parameters for HAA binding to IgA1κ in the presence or absence of FcαRI
| Analyte | k1,on,* (M⋅s)−1 | k1,off, s−1 | k2,on, (M⋅s)−1 | k2,off, s−1 | KD1,† μM | KD2,† μM |
| mIgA1κ alone | 460 ± 10 | 0.051 ± 0.001 | 500 ± 20 | 0.0044 ± 0.0001 | 222 ± 6 | 4.4 ± 0.2 |
| 2:1 FcαRI:mIgA1κ | 600 ± 20 | 0.051 ± 0.001 | 780 ± 40 | 0.0044 ± 0.0002 | 170 ± 7 | 2.8 ± 0.2 |
Kinetic parameters were determined using the bivalent analyte model in the program BIAevaluation.
KD1 and KD2 were corrected by statistical factors, as described in Materials and Methods.
Discussion
We have conducted a systematic analysis of the relative energetic contribution of IgA1 side-chain contacts to the FcαRI:IgA1 complex, which complements previous mutational studies (10, 11, 57). Mutation of IgA1 hydrophobic residues central to the binding interface greatly reduced binding to FcαRI, as expected (10, 11). For example, mutation of L441, M433, or F443 at the center of the binding site results in 12%, 25%, or 29% reductions in binding free energy, respectively. Mutation of charged and polar residues forming the outer periphery of the binding site (R382, E437, and S387) had a much milder effect, with reductions in ΔG of only 1.8–2.7%. Mutation of E389 is more deleterious (11% reduction in ΔG), presumably due to its role in mitigating electrostatic repulsion between a cluster of basic residues. It was interesting that mutation of Cα2 AB helix/loop residues L257 or L258 also resulted in a dramatic loss in binding affinity, causing a 23% or 34% loss in binding free energy, respectively. The very low affinity of FcαRI for a dimeric Cα3-only construct of IgA1 highlights the critical role of the Cα2 domain (SI Appendix, Fig. S3). The importance of these residues in the Cα3 energetic hot spot and the Cα2 domain are underlined by their conservation in the related CHIR–AB1:IgY complex. Combined, these data indicate a similar mode of binding and we hypothesize that the Cα2 domain plays a key role in complex formation.
PCA of Fcα motions determined in MD simulations suggests that, despite disulfides tethering the Cα2 domains, free IgA1 exhibits a significant degree of motion of the Cα2 domains relative to the Cα3 domains. Binding of FcαRI causes a loss in Fcα intradomain and interdomain flexibility. The PC analysis reveals that binding of FcαRI to only one side of Fcα (forming a 1:1 complex) induces conformational changes across the dimer interface to the opposite heavy chain, explaining the consistent observation of negative cooperativity in SPR binding data for the FcαRI:Fcα interaction (13, 14). Analysis of RMSFs of Fcα showed that there are three major sites containing dynamically relevant residues, all of which are located at the Cα2–Cα3 junction, including the energetic hot-spot residues. The concentration of the dynamic and energetic hot-spot residues at the Cα2–Cα3 junction accounts for the common mode of binding seen in the interactions of receptors and bacterial immune evasion proteins such as staphylococcal SSL7 (58, 59) with IgA or IgY.
An interesting and exciting prediction from this PC analysis is that FcαRI binding can induce long-range conformational changes in IgA1. A consensus model of intact IgA1 bound to FcαRI based on both crystal and solution structures (6, 60) shows that the FcαRI-binding site is distal from the hinges and Fab regions (Fig. 1A) (3). However, the solution structures of monomeric, dimeric, and secretory IgA1 (60–62) indicate that the C-terminal portion of each hinge comes into close contact with the opposite Fcα heavy chain. The swing-like motions of Cα2 of the Fcα fragment that can occur upon FcαRI binding as described by the PC analysis (Fig. 5B) might therefore be propagated from the Fc region to the hinge. We confirmed this hypothesis by demonstrating that IgA1 binds with a significantly faster on rate to the GalNAc-specific lectin HAA when FcαRI is prebound at the IgA1 Cα2–Cα3 interface, indicating that FcαRI binding induces long-range conformational effects at the IgA1 hinge. HAA is useful as a diagnostic tool for identifying patients with IgAN, since it binds to a similar epitope to that recognized by anti-glycan autoantibodies in IgAN patients (47, 55, 63).
Soluble FcαRI has been implicated in the pathogenesis of IgAN (16, 17, 64), although its role has been puzzling. We previously showed that alterations in IgA1 N-glycans have no effect on FcαRI binding (13), and the FcαRI binding site is distal from the O-glycosylated hinges (Fig. 1A) (3, 6). The long-range conformational change described here that occurs in IgA1 upon receptor binding can finally provide a potential mechanism for the role of FcαRI in IgAN pathogenesis. In particular, it suggests that the binding of soluble FcαRI to IgA1 might alter its affinity for important IgA1 hinge-targeting anti-glycan autoantibodies (63, 65) or mesangial cell-expressed transferrin receptor (66, 67). Thus, the presence of soluble FcαRI in serum could very well increase the likelihood of IgA1 immune complexes being deposited within the mesangial region of the glomerulus.
Finally, because the heavily O-glycosylated mucin-like IgA1 hinge is more rigid than a typical IgG hinge, long-range dynamic motions induced upon FcαRI binding could propagate all of the way through to the Fab regions. Such long-range communication between Fab and Fc regions of antibodies has precedent; for example, binding of either streptococcal protein A (SpA) to the Cγ2–Cγ3 junction or protein G (SpG) to the Cγ1 and Cγ2–Cγ3 domains of IgG2a is inhibited in the presence of hapten (68, 69). IgA antibodies have been shown to possess excellent properties for antitumor immunotherapy (70–72). Such long-range conformational effects upon ligand or receptor binding at the Cα2–Cα3 junction could have important implications for IgA antibody design in biotechnology, biomaterial engineering, and therapeutics.
Materials and Methods
Fcα and FcαRI Cloning, Expression, and Purification.
Fcα (residues C242–K450) lacking the hinge and tail-piece regions was cloned into pcDNA 3.0 (13, 14). Site-directed mutagenesis was performed using the QuikChange II XL kit (Stratagene) and verified by DNA sequencing. COS-7 cells were transiently transfected with 5 μg of Fcα using Lipofectamine 2000 (Invitrogen) and cultured as previously described (13). Secreted Fcα was dialyzed into 20 mM Tris⋅HCl, pH 7.4, and 300 mM NaCl, and purified as described (13).
The FcαRI construct encoding the 195-residue soluble FcαRI ectodomain (Q22–T216) was previously cloned into the baculovirus expression vector pAcGP67 (BD Pharmingen) using the upstream EcoRI and downstream HindIII sites (14). Viral amplification was performed in Sf9 cells cultured in Gibco Sf900 II media. FcαRI protein was expressed in High Five cells cultured in HyClone SFX media and purified as described (14). All proteins were determined to be >95% pure by SDS/PAGE. Protein concentrations were determined using extinction coefficients at 280 nm of 64,940 M−1⋅cm−1 for Fcα and 33,140 M−1⋅cm−1 for FcαRI (14).
Biosensor Analyses.
SPR assays were carried out on a BIAcore 3000 instrument at 25 °C. Fcα variants were immobilized on BIAcore CM5 chips to ∼200 response units (RU) by standard random-amine chemistry. For equilibrium binding experiments, 20-μL aliquots of threefold serial dilutions of FcαRI (292 pM to 50 μM) in degassed TBS-P (0.02 M Tris⋅HCl, pH 7.4, 0.15 M NaCl, 0.005% Surfactant P20) were injected at 5 μL/min. Equilibrium data were fitted globally in the program Scientist 3.0 (Micromath) to a single-site or a bivalent ligand binding model to determine KD1 and KD2, the binding affinity of the first and second binding events. For comparison of FcαRI binding to Fcα variants, ΔG values were calculated based on KD1 values, according to Eq. 1:
| [1] |
where T is temperature in kelvin, R is the gas constant (1.985 cal⋅K−1⋅mol−1), and Ka = 1/Kd. This allowed calculation of ΔΔG, the difference between the change in free energy of FcαRI binding to wild-type and mutant Fcα proteins. Reported binding parameters are averaged from two different experiments.
Snail lectin HAA (lot #101H3871; Sigma-Aldrich) and myeloma patient-derived IgA1κ (lot #14C06810; Meridian Biosciences) were prepared as described (56). A CM5 chip was immobilized with HAA via random-amine chemistry to final densities of 200, 300, or 400 RU. For equilibrium analysis, 10 μL of serial threefold dilutions of 9 μM IgA1κ in the presence or absence of 18 μM FcαRI were injected at 5 μL/min in degassed TBS-P. For kinetic analysis, threefold serial dilutions of IgA1κ alone, FcαRI alone, or the mixtures of FcαRI:IgA1κ were injected at 30 μL/min. Injection of 9 μM IgA1κ alone or 18 μM FcαRI:9 μM IgA1κ mixtures at 30, 50, 75, and 100 μL/min yielded superimposable binding curves, demonstrating that binding to HAA was not mass transport limited. Data were fitted to a kinetic bivalent analyte model without allowing for bulk refractive index shift. The on rate for the second binding event was converted to molar units using the formula: k2,on [(M⋅s)−1] = k2,on [(RU⋅s)−1] × 100 × analyte molecular weight in daltons (150,000 Da for IgA1κ alone; 212,000 for 2:1 FcαRI:IgA1κ complex). KD values were calculated as KD1 = 2k1,off/k1,on and KD2 = k2,off/2k2,on, where the factors of 2 are statistical correction terms relating the apparent and intrinsic rate constants (14, 73). Additional detailed SPR methods are included in SI Appendix.
MD Simulations.
Coarse-grained model of distinct Fcα complexes.
Our MD simulations used coarse-grained descriptions of the 2:1 FcαRI:Fcα complex, the 1:1 FcαRI:Fcα complex, and the unliganded Fcα dimer of heavy chains. The coarse-graining procedure was performed by representing each amino acid using two virtual particles. One particle, representing the backbone, is located at the Cα position and the other, representing the side chain, is located at the center of mass of the amino acid side chain (SC). The crystal structure (PDB ID code 1OW0) (6) of the 2:1 FcαRI:Fcα complex was used to obtain the initial configuration of coarse-grained models. The potential energy of the protein is represented by the following equation:
| [2] |
where VBL is the bond length potential, and VBA and VDA are the bond angle and the dihedral angle potentials, respectively (74). The nonbonded potential (VNB) is calculated using the Lennard–Jones potential:
| [3] |
where σij is the hard-core radius and εij is the well depth of the interaction between two virtual particles i, j separated by a distance rij. In this Gō-type model, native contacts, identified as amino acid pairs found within a cutoff distance of 8 Å in the crystal structure, were assigned εij = –1.25 kcal/mol for Cα–Cα as well as for the Cα–SC pairs. Native SC–SC pairs were assigned εij coefficients based on statistical potentials obtained from table 3 of Kolinski et al. (75). For all native pairs, σij is chosen based on the native distance obtained from the crystal structure. Repulsive nonbonded interactions corresponding to nonnative contacts were described by using a Lennard–Jones potential with σij = 45.42 Å and εij = –10−12 kcal/mol.
Langevin dynamics simulations.
The CHARMM program (76) was used to perform Langevin dynamics simulations for the three distinct Fcα configurations at the temperature of 300 K. We used a friction coefficient of 10 ps−1 and a time step of 5 fs in our MD simulations. For each Fcα configuration, we obtained 80 trajectories consisting of 2 × 107 steps (representing 100 ns) for a total simulation time of 8 μs. The effective timescales probed in our simulations are longer by up to several orders of magnitude due to the coarse-grained description of protein amino acids and the absence of explicit solvent representation in our model. Estimates of the real timescales can be obtained by using reduced units (77). The natural unit of time is τ = (mσ2/ε)1/2 = 3 ps, where m = 5 × 10−22 g is the average mass of an amino acid, σ = 3.8 Å is the length of the virtual Cα–Cα bond, and the energy ε = 1.25 kcal/mol. Thus, the time step is 0.002τ, the friction coefficient is 30/τ, and the total simulation time is 3.2 × 10^6τ.
MD Analysis.
PCA.
We probed the functional dynamics of IgA1 upon receptor binding by comparing the principal collective motions of the Fcα dimer in MD trajectories of the three systems studied. Collective motions in dynamic trajectories were characterized by performing PCA, which consists of diagonalizing the covariance matrix, to determine the set of eigenvectors and eigenvalues for each simulation type. represents the position vector of particle i at a given time, and is the ensemble average over all of the frames of a trajectory type. In this analysis, amino acid positions were described using the Cα virtual particles. For the PCA, we used 2,000 time frames, separated by 50 ps (16.7τ), within each trajectory. Rigid-body translations and rotations of the Fcα dimer were removed by aligning conformations corresponding to each frame with the crystal structure. The PCA calculations were performed using the CARMA MD simulation analysis package by Glykos (78).
RMSFs and B factors.
Changes in Fcα residue flexibility upon binding to the receptor were probed by computing the RMSFs in each distinct MD simulation. RMSF values were obtained by taking the square root of diagonal elements of the covariance matrix:
| [4] |
To validate the MD simulation protocol, we compared the computational values of B factors, , with the corresponding experimental values.
Directional correlation maps.
The directional correlation coefficient of a residue pair (i, j) in a given PC mode M was calculated by using , where is the unit vector in the direction of the displacement of the ith residue in mode M. These coefficients evaluate the directional similarity of motions of pairs of amino acids. We were able to probe short- and long-range coupling of IgA1 regions using maps of directional correlation coefficients of all amino acid pairs.
Long-range structural perturbation.
Residue pairs that mediate long-range structural perturbation across the two Fcα heavy chains were identified based on their weak coupling in the unliganded Fcα dimer and strong coupling in response to FcαRI binding or switching from strongly correlated (anticorrelated) motions to strongly anticorrelated (correlated) motions. To this end, we determined all residue pairs (i, j), where i and j belong to distinct Fcα heavy chains, which are separated by a minimum distance Å in the crystal structure. Changes from weak to strong coupling were evaluated by identifying pairs with in the 2:1 FcαRI:Fcα complex, in the 1:1 FcαRI:Fcα complex, and in the unliganded Fcα dimer. Changes from strong correlation to strong anticorrelation or vice versa were evaluated by in the 2:1 FcαRI:Fcα complex and in the unliganded Fcα, with .
Supplementary Material
Acknowledgments
We thank Bryan W. Poulsen for assistance in Cα3 refolding, Dr. Michelle Gomes for advice with SPR, and Dr. Sohaib Khan (Department of Cancer and Cell Biology, University of Cincinnati) for the use of the BIAcore 3000 instrument. FcαRI baculovirus stock was obtained from Caltech Protein Expression Center. In-gel trypsin digestion and MALDI-TOF/TOF sequencing of the soluble FcαRI ectodomain was carried out at the University of Cincinnati Proteomics Laboratory under the direction of Dr. Kenneth Greis. This work was supported by funds from the State of Ohio Eminent Scholar Program and grants from NIH/National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK071802), the V Foundation for Cancer Research, and the Leukemia Research Foundation (to A.B.H.), and from the National Science Foundation [Faculty Early Career Development Grant MCB-0952082 and Grant MCB-1516918 (to G.S.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807478115/-/DCSupplemental.
References
- 1.van Egmond M, et al. IgA and the IgA Fc receptor. Trends Immunol. 2001;22:205–211. doi: 10.1016/s1471-4906(01)01873-7. [DOI] [PubMed] [Google Scholar]
- 2.Monteiro RC, Van De Winkel JG. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H, et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol. 2011;22:1795–1803. doi: 10.1681/ASN.2011050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian K, et al. Functional expression of IgA receptor FcalphaRI on human platelets. J Leukoc Biol. 2008;84:1492–1500. doi: 10.1189/jlb.0508327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otten MA, van Egmond M. The Fc receptor for IgA (FcalphaRI, CD89) Immunol Lett. 2004;92:23–31. doi: 10.1016/j.imlet.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 6.Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- 7.Ding Y, et al. Crystal structure of the ectodomain of human FcalphaRI. J Biol Chem. 2003;278:27966–27970. doi: 10.1074/jbc.C300223200. [DOI] [PubMed] [Google Scholar]
- 8.Wines BD, Sardjono CT, Trist HH, Lay CS, Hogarth PM. The interaction of Fc alpha RI with IgA and its implications for ligand binding by immunoreceptors of the leukocyte receptor cluster. J Immunol. 2001;166:1781–1789. doi: 10.4049/jimmunol.166.3.1781. [DOI] [PubMed] [Google Scholar]
- 9.Wines BD, et al. Identification of residues in the first domain of human Fc alpha receptor essential for interaction with IgA. J Immunol. 1999;162:2146–2153. [PubMed] [Google Scholar]
- 10.Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcalpha receptor (FcalphaR) CD89. J Biol Chem. 1999;274:23508–23514. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- 11.Pleass RJ, Anderson CM, Dunlop JI, Woof JM. Probing the Fc alpha R binding site on IgA by mutagenesis of the IgA Fc region. Biochem Soc Trans. 1997;25:328S. doi: 10.1042/bst025328s. [DOI] [PubMed] [Google Scholar]
- 12.Morton HC, et al. Immunoglobulin-binding sites of human FcalphaRI (CD89) and bovine Fcgamma2R are located in their membrane-distal extracellular domains. J Exp Med. 1999;189:1715–1722. doi: 10.1084/jem.189.11.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes MM, et al. Analysis of IgA1 N-glycosylation and its contribution to FcalphaRI binding. Biochemistry. 2008;47:11285–11299. doi: 10.1021/bi801185b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herr AB, White CL, Milburn C, Wu C, Bjorkman PJ. Bivalent binding of IgA1 to FcalphaRI suggests a mechanism for cytokine activation of IgA phagocytosis. J Mol Biol. 2003;327:645–657. doi: 10.1016/s0022-2836(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 15.Peng M, et al. Ectodomain shedding of Fcalpha receptor is mediated by ADAM10 and ADAM17. Immunology. 2010;130:83–91. doi: 10.1111/j.1365-2567.2009.03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong MT, et al. Association of soluble CD89 levels with disease progression but not susceptibility in IgA nephropathy. Kidney Int. 2010;78:1281–1287. doi: 10.1038/ki.2010.314. [DOI] [PubMed] [Google Scholar]
- 17.Launay P, et al. Fcalpha receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger’s disease). Evidence for pathogenic soluble receptor-IgA complexes in patients and CD89 transgenic mice. J Exp Med. 2000;191:1999–2009. doi: 10.1084/jem.191.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres M, Fernandez-Fuentes N, Fiser A, Casadevall A. Exchanging murine and human immunoglobulin constant chains affects the kinetics and thermodynamics of antigen binding and chimeric antibody autoreactivity. PLoS One. 2007;2:e1310. doi: 10.1371/journal.pone.0001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton BJ, Beavil RL, Beavil AJ. Inhibition of IgE-receptor interactions. Br Med Bull. 2000;56:1004–1018. doi: 10.1258/0007142001903508. [DOI] [PubMed] [Google Scholar]
- 20.Chou KC. The biological functions of low-frequency vibrations (phonons). VI. A possible dynamic mechanism of allosteric transition in antibody molecules. Biopolymers. 1987;26:285–295. doi: 10.1002/bip.360260209. [DOI] [PubMed] [Google Scholar]
- 21.Hyeon C, Jennings PA, Adams JA, Onuchic JN. Ligand-induced global transitions in the catalytic domain of protein kinase A. Proc Natl Acad Sci USA. 2009;106:3023–3028. doi: 10.1073/pnas.0813266106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tehver R, Chen J, Thirumalai D. Allostery wiring diagrams in the transitions that drive the GroEL reaction cycle. J Mol Biol. 2009;387:390–406. doi: 10.1016/j.jmb.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 23.Zheng W, Brooks BR, Thirumalai D. Allosteric transitions in the chaperonin GroEL are captured by a dominant normal mode that is most robust to sequence variations. Biophys J. 2007;93:2289–2299. doi: 10.1529/biophysj.107.105270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng W, Thirumalai D. Coupling between normal modes drives protein conformational dynamics: Illustrations using allosteric transitions in myosin II. Biophys J. 2009;96:2128–2137. doi: 10.1016/j.bpj.2008.12.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nechushtai R, et al. Allostery in the ferredoxin protein motif does not involve a conformational switch. Proc Natl Acad Sci USA. 2011;108:2240–2245. doi: 10.1073/pnas.1019502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baxter EL, Jennings PA, Onuchic JN. Interdomain communication revealed in the diabetes drug target mitoNEET. Proc Natl Acad Sci USA. 2011;108:5266–5271. doi: 10.1073/pnas.1017604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capraro DT, Roy M, Onuchic JN, Gosavi S, Jennings PA. β-Bulge triggers route-switching on the functional landscape of interleukin-1β. Proc Natl Acad Sci USA. 2012;109:1490–1493. doi: 10.1073/pnas.1114430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 30.Wells JA. Binding in the growth hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells JA. Hormone mimicry. Science. 1996;273:449–450. doi: 10.1126/science.273.5274.449. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci USA. 2006;103:15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halperin I, Wolfson H, Nussinov R. Protein-protein interactions; coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure. 2004;12:1027–1038. doi: 10.1016/j.str.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 35.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 36.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 37.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 38.Hädge D, Ambrosius H. Evolution of low molecular weight immunoglobulins. V. Degree of antigenic relationship between the 7S immunoglobulins of mammals, birds, and lower vertebrates to the Turkey IgY. Dev Comp Immunol. 1986;10:377–385. doi: 10.1016/0145-305x(86)90027-3. [DOI] [PubMed] [Google Scholar]
- 39.Pürzel J, Schmitt R, Viertlboeck BC, Göbel TW. Chicken IgY binds its receptor at the CH3/CH4 interface similarly as the human IgA:Fc alpha RI interaction. J Immunol. 2009;183:4554–4559. doi: 10.4049/jimmunol.0901699. [DOI] [PubMed] [Google Scholar]
- 40.Viertlboeck BC, et al. The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci USA. 2007;104:11718–11723. doi: 10.1073/pnas.0702011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viertlboeck BC, et al. The chicken leukocyte receptor complex: A highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol. 2005;175:385–393. doi: 10.4049/jimmunol.175.1.385. [DOI] [PubMed] [Google Scholar]
- 42.Taylor AI, Fabiane SM, Sutton BJ, Calvert RA. The crystal structure of an avian IgY-Fc fragment reveals conservation with both mammalian IgG and IgE. Biochemistry. 2009;48:558–562. doi: 10.1021/bi8019993. [DOI] [PubMed] [Google Scholar]
- 43.Arnon TI, et al. The crystal structure of CHIR-AB1: A primordial avian classical Fc receptor. J Mol Biol. 2008;381:1012–1024. doi: 10.1016/j.jmb.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor AI, Sutton BJ, Calvert RA. Mutations in an avian IgY-Fc fragment reveal the locations of monocyte Fc receptor binding sites. Dev Comp Immunol. 2010;34:97–101. doi: 10.1016/j.dci.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 46.Renfrow MB, et al. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: Implications for IgA nephropathy. Anal Bioanal Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 47.Novak J, et al. Heterogeneity of O-glycosylation in the hinge region of human IgA1. Mol Immunol. 2000;37:1047–1056. doi: 10.1016/s0161-5890(01)00019-0. [DOI] [PubMed] [Google Scholar]
- 48.Mattu TS, et al. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J Biol Chem. 1998;273:2260–2272. doi: 10.1074/jbc.273.4.2260. [DOI] [PubMed] [Google Scholar]
- 49.Iwase H, et al. Estimation of the number of O-linked oligosaccharides per heavy chain of human serum IgA1 by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOFMS) analysis of the hinge glycopeptide. J Biochem. 1996;120:393–397. doi: 10.1093/oxfordjournals.jbchem.a021425. [DOI] [PubMed] [Google Scholar]
- 50.Field MC, Dwek RA, Edge CJ, Rademacher TW. O-linked oligosaccharides from human serum immunoglobulin A1. Biochem Soc Trans. 1989;17:1034–1035. doi: 10.1042/bst0171034. [DOI] [PubMed] [Google Scholar]
- 51.Field MC, Amatayakul-Chantler S, Rademacher TW, Rudd PM, Dwek RA. Structural analysis of the N-glycans from human immunoglobulin A1: Comparison of normal human serum immunoglobulin A1 with that isolated from patients with rheumatoid arthritis. Biochem J. 1994;299:261–275. doi: 10.1042/bj2990261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baenziger J, Kornfeld S. Structure of the carbohydrate units of IgA1 immunoglobulin. I. Composition, glycopeptide isolation, and structure of the asparagine-linked oligosaccharide units. J Biol Chem. 1974;249:7260–7269. [PubMed] [Google Scholar]
- 53.Novak J, Julian BA, Tomana M, Mestecky J. IgA glycosylation and IgA immune complexes in the pathogenesis of IgA nephropathy. Semin Nephrol. 2008;28:78–87. doi: 10.1016/j.semnephrol.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coppo R, Amore A. Aberrant glycosylation in IgA nephropathy (IgAN) Kidney Int. 2004;65:1544–1547. doi: 10.1111/j.1523-1755.2004.05407.x. [DOI] [PubMed] [Google Scholar]
- 55.Moore JS, et al. Reactivities of N-acetylgalactosamine-specific lectins with human IgA1 proteins. Mol Immunol. 2007;44:2598–2604. doi: 10.1016/j.molimm.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomes MM, et al. Recognition of galactose-deficient O-glycans in the hinge region of IgA1 by N-acetylgalactosamine-specific snail lectins: A comparative binding study. Biochemistry. 2010;49:5671–5682. doi: 10.1021/bi9019498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carayannopoulos L, Hexham JM, Capra JD. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Calpha2 and Calpha3 in human IgA1. J Exp Med. 1996;183:1579–1586. doi: 10.1084/jem.183.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langley R, et al. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J Immunol. 2005;174:2926–2933. doi: 10.4049/jimmunol.174.5.2926. [DOI] [PubMed] [Google Scholar]
- 59.Ramsland PA, et al. Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc Natl Acad Sci USA. 2007;104:15051–15056. doi: 10.1073/pnas.0706028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehm MK, Woof JM, Kerr MA, Perkins SJ. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: A study by X-ray and neutron solution scattering and homology modelling. J Mol Biol. 1999;286:1421–1447. doi: 10.1006/jmbi.1998.2556. [DOI] [PubMed] [Google Scholar]
- 61.Bonner A, Almogren A, Furtado PB, Kerr MA, Perkins SJ. Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity. Mucosal Immunol. 2009;2:74–84. doi: 10.1038/mi.2008.68. [DOI] [PubMed] [Google Scholar]
- 62.Bonner A, Furtado PB, Almogren A, Kerr MA, Perkins SJ. Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy. J Immunol. 2008;180:1008–1018. doi: 10.4049/jimmunol.180.2.1008. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki H, et al. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118:629–639. doi: 10.1172/JCI33189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monteiro RC. New insights in the pathogenesis of IgA nephropathy. Nefrologia. 2005;25:82–86. [PubMed] [Google Scholar]
- 65.Suzuki H, et al. Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest. 2009;119:1668–1677. doi: 10.1172/JCI38468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moura IC, et al. Identification of the transferrin receptor as a novel immunoglobulin (Ig)A1 receptor and its enhanced expression on mesangial cells in IgA nephropathy. J Exp Med. 2001;194:417–425. doi: 10.1084/jem.194.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moura IC, et al. Glycosylation and size of IgA1 are essential for interaction with mesangial transferrin receptor in IgA nephropathy. J Am Soc Nephrol. 2004;15:622–634. doi: 10.1097/01.asn.0000115401.07980.0c. [DOI] [PubMed] [Google Scholar]
- 68.Sagawa T, et al. Conformational changes in the antibody constant domains upon hapten-binding. Mol Immunol. 2005;42:9–18. doi: 10.1016/j.molimm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 69.Oda M, Kozono H, Morii H, Azuma T. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int Immunol. 2003;15:417–426. doi: 10.1093/intimm/dxg036. [DOI] [PubMed] [Google Scholar]
- 70.van Egmond M, et al. Enhancement of polymorphonuclear cell-mediated tumor cell killing on simultaneous engagement of FcgammaRI (CD64) and FcalphaRI (CD89) Cancer Res. 2001;61:4055–4060. [PubMed] [Google Scholar]
- 71.Stockmeyer B, et al. Triggering Fc alpha-receptor I (CD89) recruits neutrophils as effector cells for CD20-directed antibody therapy. J Immunol. 2000;165:5954–5961. doi: 10.4049/jimmunol.165.10.5954. [DOI] [PubMed] [Google Scholar]
- 72.Otten MA, et al. Immature neutrophils mediate tumor cell killing via IgA but not IgG Fc receptors. J Immunol. 2005;174:5472–5480. doi: 10.4049/jimmunol.174.9.5472. [DOI] [PubMed] [Google Scholar]
- 73.West AP, Jr, et al. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313:385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 74.Klimov DK, Thirumalai D. Native topology determines force-induced unfolding pathways in globular proteins. Proc Natl Acad Sci USA. 2000;97:7254–7259. doi: 10.1073/pnas.97.13.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolinski A, Godzik A, Skolnick J. A general method for the prediction of the three dimensional structure and folding pathway of globular proteins: Application to designed helical proteins. J Chem Phys. 1993;98:7420–7433. [Google Scholar]
- 76.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Veitshans T, Klimov D, Thirumalai D. Protein folding kinetics: Timescales, pathways and energy landscapes in terms of sequence-dependent properties. Fold Des. 1997;2:1–22. doi: 10.1016/S1359-0278(97)00002-3. [DOI] [PubMed] [Google Scholar]
- 78.Glykos NM. Software news and updates. Carma: A molecular dynamics analysis program. J Comput Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.