Abstract
The spreading mechanisms of antibiotic resistance are related to many bacterial and environment factors. The overuse of antibiotics is leading to an unceasing emergence of new multidrug resistant strains. This problem also concerns uropathogenic Escherichia coli strains, which is the most common pathogen causing urinary tract infections. The aim of this study was the genetic analysis of antibiotic resistance in comparison to the phenotypic background of E. coli strains. The characterized collection of E. coli strains isolated 10 years ago from the urine samples of patients with urinary tract infections was used for antimicrobial susceptibility testing (the disc diffusion method) and analysis of antibiotic resistance genes (PCR reaction, sequencing). Additionally, the presence of ESBL strains was analyzed. Fourteen genes were associated with resistance to beta-lactams, aminoglycosides, sulfonamides and quinolones. The genetic analysis revealed that blaTEM-1 and sul2 were present in almost all of the studied strains. Other drug-resistance genes were very rare or non-existent. Otherwise, the phenotypic resistance to fluoroquinolones was well correlated with the genotypic background of the studied bacteria. The presence of particular genes and specific mutations indicate a high bacterial potential to multidrug resistance. On the other hand, it needs to be emphasized that the standard disk diffusion test for the routine antimicrobial susceptibility analysis is still the best way to estimate the current situation of bacterial drug-resistance.
Keywords: UPEC, Antibiotic resistance, Beta-lactamases, Quinolones
Introduction
Molecular analyses are increasingly being introduced into routine diagnostics. The identification of the genetic determinants of pathogenicity and antibiotic resistance is very important for prevention against the widespread of hazardous bacteria, especially Multiple Drug Resistant (MDR) strains [1]. In the case of urinary tract infections, where uropathogenic Escherichia coli strains (UPEC) are the main causative agent [2], the most attention is paid to beta-lactam antibiotics [3]. Among resistance genes that are often located on plasmids are those coding for multiple types of β-lactamases (bla genes) [4, 5]. In a significant part, it refers to extended-spectrum β-lactamases (ESBLs) that are one of the main problems in the epidemiology of infections caused by organisms from the family Enterobacteriaceae. ESBLs usually confer resistance to all penicillins, cephalosporins (except for cephamycins), and monobactams, being inhibited by β-lactam inhibitors [6, 7], and they are the predominant source of enterobacterial resistance to 3rd- and 4th-generation cephalosporins [8, 9]. Among ESBLs observed in E. coli, the most commonly identified are enzymes from the family CTX-M (mostly CTX-M-1 lineage), followed by SHVs, and recently the less and less frequent TEMs [10–13]. Another group of acquired β-lactamases responsible for resistance to newer generation β-lactams are cephalosporinases of the AmpC type [14]. These are derivatives of enzymes specific for organisms like Enterobacter cloacae, Citrobacter freundii or Morganella morganii [15]. In general they confer a similar resistance profile to ESBLs, except for resistance to cephamycins but not 4th-generation cephalosporins, and resistance to β-lactam-inhibitor combinations, mainly those with clavulanic acid [8]. Among the several families of the acquired AmpC-type enzymes identified so far, the group deriving from C. freundii is the largest, including CMY-2 which is the most common enzyme of this kind [16, 17].
Similarly, the E. coli resistance to other antibiotics like sulfonamides and aminoglycosides applied during UTI treatment is often associated with the presence of specific plasmids. Resistance to sulfonamides is determined by three genes (sul1, sul2 and sul3) [18]. Gene sul1 has usually been identified on large conjugative plasmids, opposite sul2 has mainly been detected on small non-conjugative plasmids. Recently, sul2 has been observed also on a large conjugative plasmids related to the streptomycin resistance [18–20]. sul3 is the least known and also the least frequently detected plasmid gene in E. coli [19]. At present, the knowledge about various sul genes and their carriers is poor and diverse depending on reservoirs (e.g. animals and human) [21]. Similarly, aminoglycosides resistance is also connected with few genes carried by plasmids. The aadB, aac(3)-II and aac(3)-IV genes are related to the gentamycin, tobramycin, neomycin resistance and other aminoglycosides. They belong to the most frequent genes detected in Escherichia coli strains and other Gram-negative bacteria [22].
The frequent antibiotic resistance of UPEC strains is also associated with fluoroquinolones. Primarily, they induce the mutation in DNA gyrase (gyrA) and topoisomerase IV (parC) genes. In the literature, the mutations of genes controlling fluoroquinolones accumulation are also describing [23]. Additionally, resistance to fluoroquinolones can also be facilitated by plasmids producing the Qnr protein (QnrA, QnrB, QnrS), which protects the antibiotic targets from quinolone treatment. Qnr plasmids induce resistance on low level, but it was observed also that their presence strongly enhance the quinolone resistance determined by other mechanisms [24, 25].
The uninterrupted increasing of resistance and the emergence of MDR strains are still monitored among UPEC [26]. Therefore, there is a need for periodic screening of common bacterial pathogens such as UPEC to control their antibiotic susceptibility profiles in different communities [27, 28]. It seems to be important to also monitor the distribution of genes associated with antibiotic resistance. This knowledge can allow us to prevent the spreading of strains with a high risk of MDR expression. In reference to this, the aim of the study was to investigate the prevalence of the genes encoding the resistance to the most popular antibiotics class (beta-lactams, aminoglycosides, sulfonamides and quinolones) used during therapy of UTI against E. coli [24, 26, 29].
Materials and methods
Bacterial strains
A previously characterized collection of 127 clinical Escherichia coli strains [30, 31] isolated from the urine of patients in different hospital wards in Lodz (Poland) in the years 2005–2007 was used. Additionally, reference E. coli strains producing different beta-lactamases (No. 3272/96–TEM-1, No. 3290/96–CTX-M-3, No. 3274/96–SHV-5, No. 394/06–CMY-2, No. 348/04–OXA-1) from the National Medicines Institute (Poland) and E. coli ATCC 25922 (Argenta) were used as a control during the antimicrobial disc diffusion test.
Susceptibility testing and phenotypic ESBL detection
Antimicrobial susceptibility testing was performed using the disk-diffusion method on Mueller–Hinton agar, using commercial disks (Oxoid, Wesel, Germany). The isolates were tested against 16 antimicrobials: amoxicillin (AMX, 25 µg), amoxicillin/clavulanate (AMC, 30 µg), piperacillin (PIP, 30 µg), cefoxitin (FOX, 30 µg), cefotaxime (CTX, 5 µg), ceftazidime (CAZ, 10 µg), imipenem (IMP, 10 µg), amikacin (AMK, 30 µg), tobramycin (TOB, 10 µg), gentamicin (GEN, 10 µg), netilmicin (NET, 10 µg), norfloxacin (NOR, 10 µg), ciprofloxacin (CIP, 5 µg), ofloxacin (OFX, 5 µg), trimethoprim (TMP, 5 µg), trimethoprim-sulfamethoxazole (STX, 25 µg). The results of susceptibility testing were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [32]. E. coli ATCC 25922 was used as a quality control strain. Resistance to newer generation cephalosporins was also confirmed on chromID® ESBL plates (bioMérieux). Additionally, all isolates were tested for ESBL production by the double-disk synergy test (DDST) with disks containing cefotaxime, ceftazidime, and amoxicillin with clavulanate [33, 34].
PCR detection of resistance genes
Bacterial DNA was purified with the Gen Elute™ Bacterial Genomic DNA kit (Sigma Aldrich, Germany). The identification of the genes was carried out by PCR using previously described primers. The specific PCR parameters for all primers used in the study and their references have been shown in Table 1. In this study blaCTX-M-1-, blaTEM-, blaSHV-, blaOXA-1-, and blaCMY-2-like β-lactamase-encoding genes and also aac(3)-II, sul1, sul2, and sul3 were detected. The identification of the quinolones resistance was performed by PCR reaction for the qnrA, qnrB, qnrS genes detection and DNA sequencing of the PCR products of the gyrA and parC genes. The individual adjusted conditions of DNA amplification were carried out for each gene. After PCR amplification, products were visualized under the gel documentation system. The nucleic acid sequences of PCR products (Macrogen Europe) were compared to the original gene sequences accessed in the GenBank of the National Center for Biotechnology Information (NCBI) database. Nucleotide and amino-acid sequences were analyzed by searching the GenBank database of the NCBI with the Basic Local Alignment Search Tool (BLAST network service).
Table 1.
Oligonucleotides used in the study
| Primer | Sequence (5′ → 3′) | Locus | Ta [°C] | PCR [bp] | Ref. |
|---|---|---|---|---|---|
| TEM-A TEM-B |
ATAAAATTCTTGAAGAC TTACCAATGCTTAATCA |
Flank of blaTEM-like genes | 42 | 1181 | [12] |
| P1C P2D |
TTAATTCGTCTCTTCCAGA CAGCGCTTTTGCCGTCTAAG |
Flank of blaCTX-M1-like genes | 55 | 1042 | [11] |
| SHV-A SHV-B |
ACTGAATGAGGCGCTTCC ATCCCGCAGATAAATCACC |
Flank of blaSHV-like genes | 55 | 329 | [12] |
| OXA-1/F OXA-1/R |
ATGAAAAACACAATACATATCAAC TTTCCTGTAAGTGCGGACAC |
Internal fragment of blaOXA-1-related genes | 48 | 755 | [35] |
| CF-1 CF-2 |
ATGATGAAAAAATCGATATG TTATTGCAGTTTTTCAAGAATG |
Flank of blaCMY-like genes | 45 | 1146 | [15] |
| aac(3)-IIF aac(3)-II |
TGAAACGCTGACGGAGCCTC GTCGAACAGGTAGCACTGAG |
aac(3)-II | 55 | 369 | [36] |
| sul1-F sul1-R |
TGGTGACGGTGTTCGGCATTC GCGAAGGTTTCCGAGAAGGTG |
sulI | 56 | 790 | [37] |
| SUL2F SUL2R |
CGGCATCGTCAACATAACCT TGTGCGGATGAAGTCAGCTC |
sulII | 55 | 721 | [38] |
| SUL3F SUL3R |
CAACGGAAGTGGGCGTTGTGGA GCTGCACCAATTCGCTGAACG |
sulIII | 57 | 244 | [38] |
| gyrA-P1 gyrA-P3 |
TGT CCG AGA TGG CCT GAA GC TGC CGT CAT AGT TAT CAA CGA |
QRDR gyrA | 58 | 374 | [37] |
| parC-3 parC-4 |
CCG TGC GTT GCC GTT TAT TG AAGTGCCGTCGAAGTTTGGCA |
QRDR parC | 58 | 368 | [37] |
| qnrA-1 qnrA-2 |
ATTTCTCACGCCAGGATTTG GATCGGCAAAGGTTAGGTCA |
qnrA | Gradient | 516 | [39, 40] |
| qnrB-1 qnrB-2 |
GATCGTGAAAGCCAGAAAGG ACGATGCCTGGTAGTTGTCC |
qnrB | 469 | ||
| qnrS-1 qnrS-2 |
ACGACATTCGTCAACTGCAA TAAATTGGCACCCTGTAGGC |
qnrS | 417 |
Ta annealing temperature of PCR, Ref references, bp base pair
Results
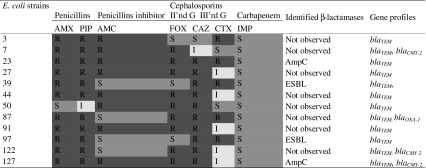
The collection of uropathogenic Escherichia coli strains was re-characterized based on antimicrobial susceptibility testing in order to verify the resistance profiles published previously [31]. Additionally, all isolates were tested for ESBL production. The whole collection of E. coli strains was analyzed based on the genetic conditioning of antibiotic resistance. The 5 genes encoding beta-lactamases (blaTEM, blaCTX-M-1, blaSHV, blaOXA-1, blaCMY) were detected at first in the reference group of E. coli ESBL strains and next, their presence was defined in the entire collection of clinical E. coli strains. Additionally, the presence of aac(3)-II determines resistance to aminoglycosides and three genes (sul1, sul2, sul3) encoding resistance to sulfonamides were studied.. Further, the strains resistant to quinolones [55] and the control representative group of strains [33]—intermediate sensitive and sensitive to quinolones—were analyzed based on the mutations in parC and gyrA genes and based on the presence of qnr genes.
β-Lactamase contents of E. coli isolates
The strains were selected based on the most beta-lactams resistant (Table 2). They were resistant to antibiotics from at least three different class of beta-lactams or/and resistant to III’rd generation of cephalosporins. Based on the phenotypic analysis only two strains from whole collection (1.5%) were identified as ESBL producers and two other strains (1.5%) were producers of AmpC. All the strains from Table 2 were blaTEM positive and it was also the most commonly identified gene present in 116 (91%) isolates (data not shown). It is worth adding that all blaTEM negative strains were sensitive to most beta-lactam antibiotics used, primarily amoxicillin. The next most frequently occurring gene was blaCMY-2 (19.5% of studied strains), but only one strain of them was producer of AmpC beta-lactamase (Table 2). The rest of them (blaCMY-2 positive) were mostly resistant to at least one beta-lactam antibiotic, but they were not detected as ESBL strains. Only one strain (E. coli No 87) possessed blaOXA, but despite its resistance to most of the analyzed beta-lactam antibiotics (except amoxicillin/clavulanate and imipenem), it was not detected as ESBL-positive. Also, one other ESBL-negative strain possessed blaSHV (E. coli No 108) and it was resistant to one beta-lactam antibiotic (ceftazidime). blaCTX-M-1 gene was not detected in any strains.
Table 2.
The correlation between phenotypic resistance to beta-lactams (the strains were selected based on the resistance to antibiotics from at least 3 different class of beta-lactams or/and resistant to III’rd generation of cephalosporins) and ESBL-connected genes identified in the studied collection of E. coli strains
AMX amoxicillin, PIP piperacillin, AMC amoxicillin/clavulanate, FOX cefoxitin, CAZ ceftazidime, CTX cefotaxime, IMP imipenem, R resistance, S sensitive, I intermediate
Genes associated with aminoglycoside and sulfonamide resistance in E. coli strains
Further, the prevalence of genes connected with the resistance to aminoglycosides and sulfonamides was analyzed among the same clinical uropathogenic E. coli strains. The majority of studied bacteria (78% of strains) were aac(3)-II positive. These results did not correlate with resistance to aminoglycosides. Both group of strains resistant or sensitive have or have not carried aac(3)-II gene. We also analyzed the occurrences of sul genes. We wanted to see if there is any correlation between the phenotypic resistance to trimethoprim and/or cotrimoxazole and the occurrences of sul genes. The most popular gene in the studied collection of E. coli strains was sul2—96% of the studied strains were positive. Also, sul1 was popular in 86% of these strains, but only 33% of the studied strains possessed sul3. Sensitive strains were positive for at least one sul gene (sul1, sul2, sul3). These results also did not correlate with resistance to sulfonamides.
Genetic association with quinolone resistance in E. coli strains
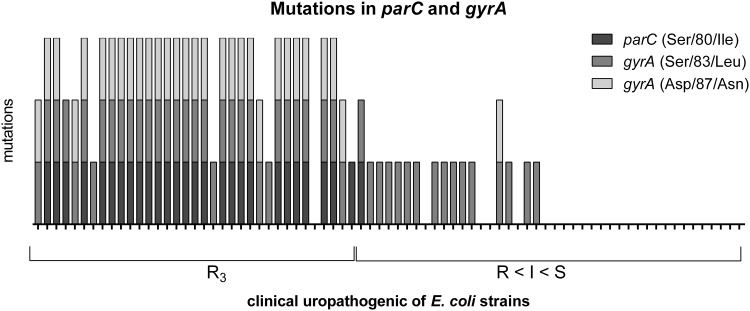
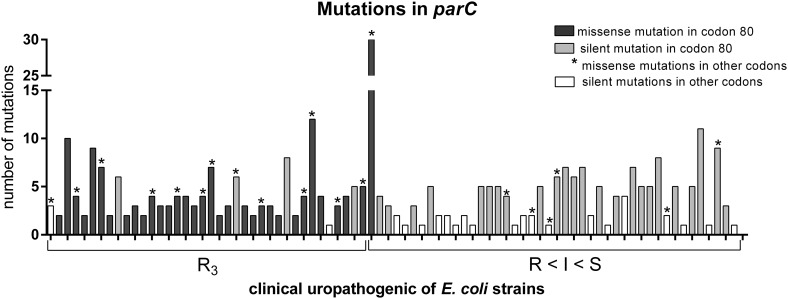
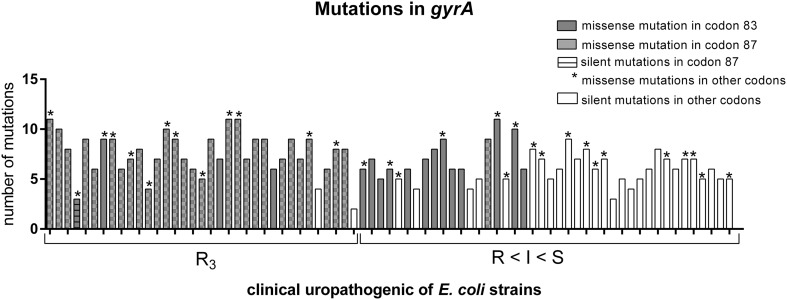
The mutations in parC and gyrA were also investigated among the studied E. coli strains. The point mutations correlated with phenotypic resistance to fluoroquinolones are well known as Ser/80/Ile in parC and also Ser/83/Leu and Asp/87/Asn in gyrA [37, 41, 42]. They were likewise recognized in this study (Fig. 3). The strains were divided into two groups: R3—strains resistant to ciprofloxacin, norfloxacin, ofloxacin, and strains with decreasing resistance to fluoroquinolones (R < I < S). The strains from the second group were arranged on the basis of changing susceptibility: from resistance to 2 or 1 quinolones (R), intermediate sensitivity (I) to sensitivity to all three quinolones (S). The most common were silent point mutations in different codons—transversion and transition in parC and mainly transition in gyrA. All strains from R3 group were positive for specific missense mutations mentioned above. Ser/80/Ile mutation in parC and then Ser/83/Leu and Asp/87/Asn mutations in gyrA were the most frequently identified. The parC gene had more frequent silent mutations in comparison to gyrA (Fig. 1). Additionally, silent mutations in codon 80 of parC were typical in the second group of strains (R < I < S). In contrast, the silent mutation in the hot spot of gyrA was observed only in one E. coli strain No 9 (Asp/87/Asp). On the other hand, generally there were more mutations in the gyrA gene compared to the parC gene (Fig. 2). Further, in case of gyrA gene—the occurrence of the Asp/87/Asn missense mutation always correlated with the occurrence of the Ser/83/Leu mutation. Ser/83/Leu mutation occurred alone in the case of strains from R < I < S group. Strains which were the most sensitive to fluoroquinolones possessed only missense and/or silent mutations in other codons.
Fig. 3.
Comparison of occurrence of specific mutations in the parC and gyrA genes of the studied uropathogenic E. coli strains
The diagram was made using GraphPad Prism6
Fig. 1.
Characteristics of identified mutations in the parC gene of the studied uropathogenic E. coli strains
The diagram was made using GraphPad Prism6
Fig. 2.
Characteristics of identified mutations in the gyrA gene of the studied uropathogenic E. coli strains
The diagram was made using GraphPad Prism6
Additionally, the strains were analyzed based on the presence of qnr genes and despite many attempt of optimizing PCR procedures, the results remained negative.
Discussion
The problem of bacterial antibiotic resistance belongs to the priorities of World Health Organization concerning threat to human health. The widespread use of antibiotics often without the antibiotic susceptibility testing is one of the reasons for the emergence of multidrug resistant pathogens, which seriously impedes therapeutic activities [43, 44]. This can also hinder other therapeutic successes as infectious complications appearing in patients undergoing chemotherapy for cancer or dialysis for renal failure. The effectiveness of secondary infections treatment is crucial also in surgery, especially organ transplantation [1, 45, 46]. Instead, urinary tract infections (UTIs) belong to the most common human infections in both hospital and community settings, where antibiotics are also usually applied [47]. Approximately every second woman and every twentieth man will suffer from UTI in their lifetime. Uropathogenic Escherichia coli (UPEC) is the primary agent causing uncomplicated and complicated UTIs [2, 47, 48]. Therapeutic difficulties result largely from the quick spread of multidrug resistance (MDR) among them. This problem concerns the large group of beta-lactam antibiotics but also other compounds, such as fluoroquinolones, aminoglycosides, etc., which are often used in UTI treatment [30]. The mechanisms of bacterial antibiotic resistance are often associated with the mobile properties of a genome, especially with horizontal gene transfer, the presence of plasmids or genomic mutations [6, 7, 49]. The genetic markers of bacterial antibiotic resistance are often described in the literature. The prevalence and differentiation of these genetic profiles vary depending on the countries, antibiotic policy, source and year of bacteria isolation.
In this study, we analyzed 14 genes related to resistance to antibiotics belonging to four different classes. We would like to check the relationships between genotypic and phenotypic resistance among uropathogenic Escherichia coli strains isolated from central Poland (Łódź). The obtained results represent the characteristic of the bacterial population from 10 years ago, which allowed us to look at the potential changes currently observed in a similar bacterial population. At present, the hot topic is ESBL strains. Among the studied bacterial collection we identified approx. 1.5% of ESBL-producing strains and 1.5% AmpC-producing strains, which was the standard level in those days (2005–2007). The situation in Poland seems to have been relatively stable for 10 years (at present approx. 2% ESBL of UPEC strains), but in many other countries the situation is much worse [3, 50–53]. However, the increase in bacterial antibiotic resistance is still observed. Considering the genetic background, these strains (ESBL- and AmpC-producing) carrying blaTEM gene and additionally blaCMY-2 was present only in one AmpC-producing E. coli strain, although blaCMY-2 belongs to a small family of plasmid-mediated AmpC-like enzymes [54]. The remaining blaCMY-2 positive strains did not correlate with the production of AmpC beta-lactamase. The mechanisms of resistance against beta-lactam antibiotics seem to be the most complicated and differential. This is also clearly visible in the case of genetic background analysis. Considering that the vast majority of the isolates were ESBL-negative despite the presence blaTEM in most strains (91%), it may be assumed that the blaTEM genes encoded broad-spectrum enzymes, most likely TEM-1. TEM-1 is the major determinant of E. coli resistance to amino-penicillins and the most common plasmid-encoded β-lactamase; it is estimated that this enzyme occurs in approximately 50% all of E. coli clinical isolates [12, 13, 55]. As it was mentioned above, the blaCMY-2 was the next most common gene in the studied bacterial collection (19.5%). The blaCMY-2 has been also often identified in E. coli, Klebsiella sp. and Salmonella spp. from different sources in the United States, Greece and Algeria [54, 56–58]. The prevalence of this gene did not correlate with the resistance to β-lactams in our study. In the studied E. coli collection the other genes were sought, too. Only one strain possessed blaOXA-1 and despite its resistance to all analyzed beta-lactam antibiotics (except amoxicillin/clavulanate and imipenem), it was not detected as ESBL-positive. A similar prevalence of blaOXA-1 was observed at present by other authors [59, 60]. Also, one other strain possessed blaSHV (E. coli No 108) and it was resistant to only one beta-lactam antibiotic (ceftazidime), which may suggest the presence of only broad-spectrum β-lactamase type SHV-1 or SHV-11 [61]. The SHV-2, SHV-5, SHV-7 or SHV-18 belong to the common ESBL variants that have been often observed in Poland [12, 13]. The blaCTX-M-1 gene was not detected in any strains, despite the first strain producing β-lactamase, CTX-M-3 was identified originally in Poland in 1996 [11], being the far predominant ESBL type in the country [11, 13]. Analyzing the occurrence of the blaCTX-M-1 gene, as reported in 2006, is not one of the most common genes in Poland like in the case of the genes encoding beta-lactamase from the CTX-M group: CTX-M-3, CTX-M-15, CTX-M-2 [62]. According to this, the lack of the blaCTX-M-1 gene in the collection of E. coli strains is not surprising. However, the distribution of the studied genes is very varied depending on the region, country, and the year of the strains isolation. The literature shows that CTX-M enzymes were identified in different locations in the second half of the twentieth century, among others, in Argentina, Israel, and Paraguay [62]. In Europe, blaCTX-M-1 genes were first identified in 1989 in Germany [62]. Results of antibiotic resistance from 2014 presented by Ojdana et al. [13] have shown that blaCTX-M-15 genes were identified in all of the 12 analyzed E. coli strains from a Polish patient (Białystok) and only two strains were positive for blaTEM-1 and blaSHV genes. Bailey et al. [55] presented antibiotic resistance among the collection of commensal E. coli strains. 35% of these strains were ampicillin-resistant and containing the blaTEM gene. In 2012, Korzeniewska et al. [63] published studies about the antibiotic resistance of E. coli strains from different sources (hospital and communal wastewater, river, air) and thereabouts 30% of studied strains were ESBL positive. In these strains genes blaCTX-M-1, blaCTX-M-3, blaCTX-M-5, blaCTX-M-15 were identified as the most common. Winokur et al. [14] analyzed the presence of the blaCMY-2 gene in E. coli strains isolated from people and animals in the USA. In the case of strains resistant to cephalosporins, blaCMY-2 was identified in 33% of human isolates and 94.8% of animal isolates [14]. It must be noted that the occurrence of antibiotic resistance genes usually differ between people and animals and they are determined by many other factors. It is very well known that bacteria can induce a lot of mechanisms against drugs, so the specific genes often do not correlate with the phenotypic antibiotic resistance, for example, structural changes at the site of the drug’s action or change of the action point for the antibiotic [43, 44]. Additionally, the efflux pumps system AcrAB-TolC, AcrEF-TolC, AcrABC-TolC is also often described in case of E. coli phenotypic resistance to beta-lactam antibiotic [64]. As we can see, the distribution of the analyzed genes is not correlated with the year in a global coverage, but in the Polish view we can see the rise of the number of antibiotic resistance genes in E. coli strains [10].
Taking into consideration the resistance to other antibiotics, aminoglycosides and sulfonamides also play a significant role during UTI treatment. The aac(3)-II is described as the most correlated gene with aminoglycosides resistance, which was not confirmed in our results. Both group of strains, resistant or sensitive, have or have not carried the aac(3)-II gene. Therefore, it should be stated that aac(3)-II cannot play the role of a marker for resistance to aminoglycosides or at least for one of them. A similar high prevalence of that gene was observed in other studied collection of E. coli isolated from Europe [36, 65, 66]. We also analyzed the occurrences of sul genes which are responsible for resistance to sulfonamides by changed activity of DHPS (dihydropteroate synthase). This enzyme shows affinity to PABA and when it is encoded by sul it remains insensible to sulfonamides. Trimethoprim, which we used in our study, binds to dihydrofolate reductase and inhibits the reduction of dihydrofolic acid (DHF) further upstream in the same pathway [67]. We wanted to check if there is any correlation between the phenotypic resistance to trimethoprim and the occurrences of sul genes—there was none. Furthermore, the presence of the studied sul genes also correlated with resistance to cotrimoxazole. These genes were very common for the studied E. coli strains, even in the case of strains sensitive to trimethoprim or cotrimoxazole, which can only mean that sulfonamides may enhance the expression of resistance to these antibiotics. The results presented by other authors show a similar distribution of these genes [68, 69]. Completely different results were presented by Mazurek et al. [70] where the presence of the studied genes was significantly lower. However, those E. coli strains were isolated from animals in Poland which could probably be an important reason for the differences [70]. The rare occurrence of sul3 may prove that the synthesis of modified DHPS due to the presence of sul3 gene has recently appeared in resistant bacteria strains [71]. This confirms the high incidence in this group of questionable and negative results obtained by PCR. Sulfonamides therapy is used in UTI as a combination of sulfamethoxazole with trimethoprim. This was the dominant therapy for UTI between 1995 and 1996. At present, due to the increasing prevalence of resistance to trimethoprim-sulfamethoxazole among E. coli strains, it should not be the first method of choice in the treatment of E. coli infection [47].
The most obvious results were obtained by analyzing the point mutations in parC and gyrA genes. Ser/80/Ile in parC and also Ser/83/Leu and Asp/87/Asn in gyrA give a strong correlation with phenotypic resistance to quinolones [37, 41, 42]. Our results confirm these findings (Fig. 1); however, there can be some other possible mechanisms involved in quinolones resistance, like the permeability effect, efflux pumps, and the decreased availability of quinolones at the target site can also be involved [9, 72, 73]. Looking more closely at the results we observed a specific tendency. The group of strains with reduced susceptibility to quinolones (R < I < S) carried a lot of atypical mutations (Figs. 2, 3). The silent mutations in the hot spot were characteristic for parC, and missense mutations in other codons were characteristic for gyrA. It can play a predictive role of imminent phenotypic resistance to fluoroquinolones and makes it suitable for epidemiological studies [74]. Silent mutation have also been identified by other authors [73, 75–77]. It could be interesting to observe the genetic background during the process of acquiring resistance to fluoroquinolones. This can be related to the accumulation of mutations because of the low specificity of fluoroquinolones action. Also, the antibiotics, especially fluoroquinolones can induce the response of SOS systems, which can be responsible for DNA changes in bacteria genome [78, 79]. There is some evidence that silence mutations might cause a phenotypic effect, they can especially have an influence on the regulation of transcription [80–82]; possibly they can also change the affinity of the hot spot to fluoroquinolones. Nevertheless, we can conclude that the hot spot of parC is more specific but less sensitive to fluoroquinolones (more silent mutations), whereas gyrA conversely—a lot of missense mutations give the phenotypic effect but not in the hot spot of gyrA. So, in this case, two hot spots were evolved. We have not identified more other specific mutations, but in the literature other mutations correlated with phenotypic resistance to quinolones were identified. These mutations were described generally in gyrA and parC genes, for example as new and rare mutations in gyrA—Ser/83/stop, Asp/82/Asn, Gly/81/Asp, Asp/82/Gly, Ser/431/Pro in resistant E. coli strains [83].
Additionally, none of the strains possessed the qnr genes. This similar low prevalence is still relevant in most cases [84–87]. Mammeri et al. in 2005 [72] published an analysis of the qnr gene in a collection of 297 of nalidixic-acid resistant E. coli strains. In this collection, only 1 strain with the qnr gene was identified. The low level of identification of the qnr gene can be due to the weak expression of the Qnr determinant [72]. Some authors present dissimilar results [88–90]. It probably depends on the local distribution of Qnr plasmid.
To conclude, the genetic background is not sufficient for identifying bacterial antibiotic resistance. This kind of analysis can play a role for predicting resistance and it may mark the high or low risk of the emergence of resistance. However, quinolones resistance is very strongly dependent on the specific mutations. As we can see, the distribution of the analyzed genes is very differentiated and shows a high adaptive potential of bacteria to a toxic (antibiotic) environment.
Acknowledgements
We would like to thank Marek Gniadkowski for participating in this study and the critical reading of the manuscript. This study was funded by the National Science Centre research Grant No. NCN 2011/01/D/NZ7/00107.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For this type of study formal consent is not required. This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study formal consent is not required.
References
- 1.Anderson DI, Hughes D (2010) Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 10.1038/nrmicro2319 [DOI] [PubMed]
- 2.Nielubowicz GR, Mobley HLT. Host–pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 3.Stefaniuk E, Suchocka U, Bosacka K, Hryniewicz W. Etiology and antibiotic susceptibility of bacterial pathogens responsible for community-acquired urinary tract infections in Poland. Eur J Clin Microbiol Infect Dis. 2016;35:1363–1369. doi: 10.1007/s10096-016-2673-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 5.Chah KF, Agbo IC, Eze DC, Somalo S, Estepa V, Torres C. Antimicrobial resistance, integrons and plasmid replicon typing in multiresistant clinical Escherichia coli strains from Enugu State, Nigeria. J Basic Microbiol. 2010;50:S18–S24. doi: 10.1002/jobm.200900325. [DOI] [PubMed] [Google Scholar]
- 6.Li Y. Prevalence of plasmid-mediated AmpC β-lactamases in a Chinese University Hospital from 2003 to 2005: first report of CMY-2-type AmpC β-lactamase resistance in China. J Clin Microbiol. 2008;46(4):1317–1321. doi: 10.1128/JCM.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poole K. Resistance to β-lactam antibiotics. Cell Mol Life Sci. 2004;61:2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Empel L, Baraniak A, Literacka E, Mrówka A, Fiett J, Sadowy E, Hryniewicz W, Gniadkowski M, the Beta-PL Study Group Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother. 2008;52(7):2449–2454. doi: 10.1128/AAC.00043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charfi K, Grami R, Ben Jeddou A, Messaoudi A, Mani Y, Bouallegue O, Boujaafar N, Aouni M, Mammeri H, Mansour W. Extended-spectrum β-lactamases and plasmid-mediated quinolone resistance in enterobacterial clinical isolates from neonates in Tunisia. Microb Pathog. 2017;110:184–188. doi: 10.1016/j.micpath.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Mokracka J, Oszyńska A, Kaznowski A. Increased frequency of integrons and β-lactamase-coding genes among extraintestinal Escherichia coli isolated with a 7-year interval. Antonie Van Leeuwenhoek. 2013;103:163–174. doi: 10.1007/s10482-012-9797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gniadkowski M, Schneider I, Pałucha A, Jungwirth R, Mikiewicz B, Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob Agents Chemother. 1998;42(4):827–832. doi: 10.1128/aac.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gniadkowski M, Schneider I, Jungwirth R, Hryniewicz W, Bauernfeind A. Ceftazidime-resistant Enterobacteriaceae isolates from three polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1998;42(3):514–529. doi: 10.1128/aac.42.3.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojdana D, Sacha P, Wieczorek P, Czaban S, Michalska A, Jaworowska J, Jurczak A, Poniatowski B, Tryniszewska E. The occurrence of blaCTX-M, blaSHV, and blaTEM genes in extended-spectrum β-lactamase-positive strains of Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis in Poland. Int J Antibiot. 2014 [Google Scholar]
- 14.Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agent Chemother. 2001;45(10):2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Literacka E, Empel J, Baraniak A, Sadowy E, Hryniewicz W, Gniadkowski M. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15 in a Proteus mirabilis clone widespread in Poland. Antimicrob Agents Chemother. 2004;48(11):4136–4143. doi: 10.1128/AAC.48.11.4136-4143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaczmarczyk M, Wang J, Leonard N, Fanning S. Complete nucleotide sequence of a conjugative IncF plasmid from an Escherichia coli isolate of equine origin containing blaCMY-2 within a novel genetic context. FEMES Microbiol Lett. 2014;352(1):123–127. doi: 10.1111/1574-6968.12364. [DOI] [PubMed] [Google Scholar]
- 17.Seiffert SN, Tinguely R, Lupo A, Neuwirth C, Perreten X, Endimiani A. High prevalence of extended-spectrum-cephalosporin-resistant Enterobacteriaceae in poultry meat in Switzerland: emergence of CMY-2- and VEB-6-possessing Proteus mirabilis. Antimicrob Agents Chemother. 2013 doi: 10.1128/AAC.01773-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estrada A, Wright DL, Anderson AC. Antibacterial antifolates: from development through resistance to the next generation. Cold Spring Harb Perspect Med. 2016 doi: 10.1101/cshperspect.a028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bean DC, Livermore DM, Papa I, Lucinda M, Hall C. Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J Antimicrob Chemother. 2005 doi: 10.1093/jac/dki332. [DOI] [PubMed] [Google Scholar]
- 20.Frye JG, Jackson CR. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U. S. food animals. Front Microbiol. 2013;4:135. doi: 10.3389/fmicb.2013.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Dalsgaard A, Hammerum AM, Porsbo LJ, Jensen BL. Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet Scand. 2010;52:47. doi: 10.1186/1751-0147-52-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellaaj A, Bollet C, Ben-Mahrez K. Characterization of the 3-N-aminoglycoside acetyltransferase gene aac(3)-IIa of a clinical isolate of Escherichia coli. Ann Microbiol. 2003;53:211–217. [Google Scholar]
- 23.Sanchez-Cespedes J, Sáez-López E, Frimodt-Mřller N, Vila J, Soto S. Effects of a mutation in the gyrA gene on the virulence of uropathogenic Escherichia coli. Antimicrob Agents Chemother. 2015;59(8):4662–4668. doi: 10.1128/AAC.00665-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49(7):3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavaco LM, Aarestrup FM. Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J Clin Microbiol. 2009;47:2751–2758. doi: 10.1128/JCM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haguan EC, Mobley HLT. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect Immun. 2007;75(8):3941–3949. doi: 10.1128/IAI.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umolu PI, Ohenhen ER, Okwu IG, Ogiehor IS. Multiple antibiotics resistant index and plasmid of Escherichia coli in beef in Ekpoma. J Am Sci. 2006;2(3):22–28. [Google Scholar]
- 28.Soleimani N, Derakhshan S, Memariani M. Plasmid profile analysis of aminoglycoside-resistant Escherichia coli isolated from urinary tract infections. Int J Enteric Pathog. 2016;4(2):33806. [Google Scholar]
- 29.Fair JR, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adamus-Bialek W, Wojtasik A, Majchrzak M, Sosnowski M, Parniewski P. (CGG)4-based PCR as a novel tool for discrimination of uropathogenic Escherichia coli strains: comparison with enterobacterial repetitive intergenic consensus-PCR. J Clin Microbiol. 2009;47(12):3937–3944. doi: 10.1128/JCM.01036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamus-Białek W, Lechowicz Ł, Kubiak-Szeligowska AB, Wawszczak M, Kamińska E, Chrapek M. A new look at the drug-resistance investigation of uropathogenic E. coli strains. Mol Biol Rep. 2017;44(1):191–202. doi: 10.1007/s11033-017-4099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2017) Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf. Accessed 10 Mar 2017
- 33.Soltani R, Khalili H, Shafiee F. Double-disk synergy test for detection of synergistic effect between antibiotics against nosocomial strains of staphylococcus aureus. J Res Pharm Pract. 2012;1(1):21–24. doi: 10.4103/2279-042X.99675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begum N, Shamsuzzaman SM. Emergence of CTX-M-15 producing E. coli O25b-ST131 clone in a tertiary care hospital of Bangladesh. Malays J Pathol. 2016;38(3):241–249. [PubMed] [Google Scholar]
- 35.Edelstein M, Pimkin M, Dmitrachenko T, Semenov T, Kozlova N, Gladin D, Baraniak A, Stratchounski L. Multiple outbreaks of nosocomial Salmonellosis in Russia and Belarus caused by a single clone of Salmonella enterica serovar Typhimurium producing an extended-spectrum β-lactamase. Antimicrob Agents Chemother. 2004;48:2808–2815. doi: 10.1128/AAC.48.8.2808-2815.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakobsen L, Sandvang D, Hansen LH, Bagger-Skjøt L, Westh H, Jørgensen C, Hansen DS, Pedersen BM, Monnet DL, Frimodt-Møller N, Sørensen SJ, Hammerum AM. Characterization, dissemination and persistence of gentamicin resistant Escherichia coli from Danish university hospital to the waste water environment. Environ Int. 2008;34:108–115. doi: 10.1016/j.envint.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 37.McDonald LC, Chen FJ, Lo HJ, Yin HC, Lu PL, Huang CH, Chen P, Lauderdale TL, Ho M. Emergence of reduced susceptibility and resistance to fluoroquinolones in Escherichia coli in Taiwan and contributions of distinct selective pressures. Antimicrob Agents Chemother. 2001;45:3084–3091. doi: 10.1128/AAC.45.11.3084-3091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin SW, Byun JV, Jung M, Shin MK, Yoo HS. Antimicrobial resistance, virulence genes and PFGE-profiling of Escherichia coli isolates from South Korean cattle farms. J Microbiol. 2014;52(9):785–793. doi: 10.1007/s12275-014-4166-1. [DOI] [PubMed] [Google Scholar]
- 39.Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. Qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50(8):2872–2874. doi: 10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pazhani GP, Chakraborty S, Fujihara K, Yamasaki S, Ghosh A, Nair GB, Ramamurthy T. QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad, India. Indian J Med Res. 2011;134:214–223. [PMC free article] [PubMed] [Google Scholar]
- 41.Maidhof H, Guerra B, Abbas S, Elsheikha HM, Whittam TS, Beutin L. A multiresistant clone of shiga toxin-producing Escherichia coli O118:[H16] is spread in cattle and humans over different european countries. Appl Environ Microb. 2002;68(12):5834–5842. doi: 10.1128/AEM.68.12.5834-5842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White DG, Piddock LJ, Maurer JJ, Zhao S, Ricci V, Thayer SG. Characterization of fluoroquinolone resistance among veterinary isolates of avian Escherichia coli. Antimicrob Agents Chemother. 2000;44:2897–2899. doi: 10.1128/AAC.44.10.2897-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6A):S3–S10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Martinez JL, Fajardo A, Garmendia L, Hernandez A, Linares JF, Martınez-Solano J, Sanchez MB. A global view of antibiotic resistance. FEMS Microbiol Rev. 2009;33:44–65. doi: 10.1111/j.1574-6976.2008.00142.x. [DOI] [PubMed] [Google Scholar]
- 45.Burke JP (2003) Infection control: a problem for patient safety. N Engl J Med. 10.1056/NEJMhpr020557 [DOI] [PubMed]
- 46.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 47.Kallen A, Welch HG, Sirovich BE. Current antibiotic therapy for isolated urinary tract infections in women. Arch Intern Med. 2006;166(6):635–639. doi: 10.1001/archinte.166.6.635. [DOI] [PubMed] [Google Scholar]
- 48.Brumbaugh AR, Mobley TLT. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines. 2012;11(6):663–676. doi: 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frost LS, Leplae R, Anne O, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 50.Wu YH, Chen PL, Hung YP, Ko WC. Risk factors and clinical impact of levofloxacin or cefazolin nonsusceptibility or ESBL production among uropathogens in adults with community onset urinary tract infections. J Microbiol Immunol Infect. 2014;47:197–203. doi: 10.1016/j.jmii.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Mashwal FA, El Safi SH, George SK, Adam AA, Jebakumar AZ. Incidence and molecular characterization of the extended spectrum beta lactamase-producing Escherichia coli isolated from urinary tract infections in Eastern Saudi Arabia. Saudi Med J. 2017;38(8):811–815. doi: 10.15537/smj.2017.8.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jena J, Sahoo RK, Debata NK, Subudhi E. Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum β-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. 3 Biotech. 2017;7(4):244. doi: 10.1007/s13205-017-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parajuli NP, Maharjan P, Parajuli H, Joshi G, Paudel D, Sayami S, Khanal PR. High rates of multidrug resistance among uropathogenic Escherichia coli in children and analyses of ESBL producers from Nepal. Antimicrob Resist Infect Control. 2017;11:6–9. doi: 10.1186/s13756-016-0168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40(1):221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey JK, Pinyon JL, Anantham S, Hall RM. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother. 2011;66:745–751. doi: 10.1093/jac/dkq529. [DOI] [PubMed] [Google Scholar]
- 56.Koeck JL, Arlet G, Philippon A, Basmaciogullari S, Thien HV, Buisson Y, Cavallo JD. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol Lett. 1997;152(2):255–260. doi: 10.1016/S0378-1097(97)00206-1. [DOI] [PubMed] [Google Scholar]
- 57.Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradford PA, Angulo FJ, Hinrichs SH. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med. 2000;342(17):1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 58.Winokur PL, Brueggemann A, DeSalvo DL, Hoffmann L, Apley MD, Uhlenhopp EK, Pfaller MA, Doern GV. Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob Agents Chemother. 2000;44(10):2777–2783. doi: 10.1128/AAC.44.10.2777-2783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao K, Chen Y, Wang M, Guo P, Yang Q, Ni Y, Yu Y, Hu B, Sun Z, Huang W, Wang Y, Wu A, Feng X, Luo Y, Hu Z, Chu Y, Chen S, Cao B, Su J, Gui B, Duan Q, Zhang S, Shao H, Kong H, Xu Y. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae causing intra-abdominal infections from 9 tertiary hospitals in China. Diagn Microbiol Infect Dis. 2017;87(1):45–48. doi: 10.1016/j.diagmicrobio.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Alizade H, Fallah F, Ghanbarpour R, Aflatoonian MR, Goudarzi H, Sharifi H. Phylogenetic groups, extended-spectrum β-lactamases and metallo-β-lactamase in Escherichia coli isolated from fecal samples of patients with diarrhea in Iran. Gastroenterol Hepatol Bed Bench. 2015;8(3):207–214. [PMC free article] [PubMed] [Google Scholar]
- 61.Shahi SK, Singh VK, Kumar A. Detection of Escherichia coli and associated β-lactamases genes from diabetic foot ulcers by multiplex PCR and molecular modeling and docking of SHV-1, TEM-1, and OXA-1 β-lactamases with clindamycin and piperacillin-tazobactam. PLoS ONE. 2013;8(7):e68234. doi: 10.1371/journal.pone.0068234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Canton R, Coque TM. The CTX-M-β-lactamase pandemic. Curr Opt Microbiol. 2006;9:466–475. doi: 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 63.Korzeniewska E, Korzeniewska A, Harnisz M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol Environ Saf. 2013;91:96–102. doi: 10.1016/j.ecoenv.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 64.Sun J, Deng Z, Aixin Y. Bacterial multidrug efflu pumps: mechanisms physiology and pharmacological exploitions. BBRC. 2014;453:254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 65.Soleimani N, Aganj M, Ali L, Shokoohizadeh L, Sakinc T. Frequency distribution of genes encoding aminoglycoside modifying enzymes in uropathogenic E. coli isolated from Iranian hospital. BMC Res Notes. 2014;7:842. doi: 10.1186/1756-0500-7-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haldorsen BC, Simonsen GS, Sundsfjord A, Samuelsen O, Norwegian Study Group on Aminoglycoside Resistance Increased prevalence of aminoglycoside resistance in clinical isolates of Escherichia coli and Klebsiella spp. in Norway is associated with the acquisition of AAC(3)-II and AAC(6′)-Ib. Diagn Microbiol Infect Dis. 2014;78(1):66–69. doi: 10.1016/j.diagmicrobio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Caron F, Wehrle V, Etienne M. The comeback of trimethoprim in France. Med Mal Infect. 2017;47(4):253–260. doi: 10.1016/j.medmal.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Huovinen P, Sundstrom L, Swedberg G, Skold O. Trimethoprim and sulfonamide resistance. Antimicrob Agent Chemother. 1995;39(2):279–289. doi: 10.1128/AAC.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sköld O. Resistance to trimethoprim and sulfonamides. Vet Res. 2001;32:261–273. doi: 10.1051/vetres:2001123. [DOI] [PubMed] [Google Scholar]
- 70.Mazurek J, Pusz P, Bok E, Stosik M, Baldy-Chudzik K. The phenotypic and genotypic characteristics of antibiotic resistance in Escherichia coli populations isolated from farm animals with different exposure to antimicrobial agents. Pol J Microbiol. 2013;62(2):173–179. [PubMed] [Google Scholar]
- 71.Grape M, Sundström L, Kronvall G. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J Antimicrob Chemother. 2003;52:1022–1024. doi: 10.1093/jac/dkg473. [DOI] [PubMed] [Google Scholar]
- 72.Mammeri H, Loo M, Poirel L, Martinez-Martinez L, Nordmann P. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agent Chemother. 2005 doi: 10.1128/AAC.49.1.71-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hashem RA, Yassin AS, Zedan HH, Amin MA. Fluoroquinolone resistant mechanisms in methicillin-resistant Staphylococcus aureus clinical isolates in Cairo, Egypt. J Infect Dev Ctries. 2013;7(11):796–803. doi: 10.3855/jidc.3105. [DOI] [PubMed] [Google Scholar]
- 74.Supek F. The code of silence: widespread associations between synonymous codon biases and gene function. J Mol Evol. 2016;82(1):65–73. doi: 10.1007/s00239-015-9714-8. [DOI] [PubMed] [Google Scholar]
- 75.Yang W, Zhang M, Zhou J, Pang L, Wang G, Hou F. The molecular mechanisms of ciprofloxacin resistance in clinical Campylobacter jejuni and their genotyping characteristics in Beijing. China Foodborne Pathog Dis. 2017;4(7):386–392. doi: 10.1089/fpd.2016.2223. [DOI] [PubMed] [Google Scholar]
- 76.Thong KL, Ngoi ST, Chai LC, Teh CS. Quinolone resistance mechanisms among Salmonella enterica in Malaysia. Microb Drug Resist. 2016;22(4):259–272. doi: 10.1089/mdr.2015.0158. [DOI] [PubMed] [Google Scholar]
- 77.Yu X, Susa M, Knabbe C, Schmid RD, Bachmann TT. Development and validation of a diagnostic DNA microarray to detect quinolone-resistant Escherichia coli among clinical isolates. J Clin Microbiol. 2004;42(9):4083–4091. doi: 10.1128/JCM.42.9.4083-4091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baharoglu Z, Mazel D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother. 2011;55(5):2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2007;305(5690):1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 80.Yadegari H, Biswas A, Akhter MS, Driesen J, Ivaskevicius V, Marquardt N, Oldenburg J. Intron retention resulting from a silent mutation in the VWF gene that structurally influences the 5′ splice site. Blood. 2016;128(17):2144–2152. doi: 10.1182/blood-2016-02-699686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agashe D, Sane M, Phalnikar K, Diwan GD, Habibullah A, Mrtinez-Gomez NC, Sahasrabuddhe V, Polachek W, Wang J, Chubiz LM, Marx CJ. Large-effect beneficial synonymous mutations mediate rapid and parallel adaptation in a bacterium. Mol Biol Evol. 2016;33(6):1542–1553. doi: 10.1093/molbev/msw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hauber DJ, Grogan DW, DeBry RW. Mutations to less-preferred synonymous codons in a highly expressed gene of Escherichia coli: fitness and epistatic interactions. PLoS ONE. 2016;11(1):e0146375. doi: 10.1371/journal.pone.0146375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cesaro A, Bettoni R, Lascols C, Merens A, Soussy CJ, Cambau E. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J Antimicrob Chemother. 2008;61(5):1007–1015. doi: 10.1093/jac/dkn077. [DOI] [PubMed] [Google Scholar]
- 84.De Silva BCJ, Hossain S, Wimalasena SHMP, Pathirana HNKS, Wendt M, Heo GJ. Quinolone susceptibility and genetic characterization of Salmonella enterica subsp. enterica isolated from pet turtles. Lab Anim Res. 2017;33(2):49–56. doi: 10.5625/lar.2017.33.2.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang HY, Nam YS, Lee HJ. Prevalence of plasmid mediated quinolone resistance genes among ciprofloxacin nonsusceptible Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in Korea. Can J Infect Dis Med Microbiol. 2014;25(3):163–169. doi: 10.1155/2014/329541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jlili NEH, R´ejiba S, SmaouiH, et al. Trend of plasmid mediated quinolone resistance gene sat the Children’s Hospital in Tunisia. J Med Microbiol. 2014;63(2):195–202. doi: 10.1099/jmm.0.062216-0. [DOI] [PubMed] [Google Scholar]
- 87.Wang Q, Li B, Tsang AKL, Yi Y, Woo PCY, Liu CH. Genotypic analysis of Klebsiella pneumoniae isolates in a Beijing Hospital reveals high genetic diversity and clonal population structure of drug-resistant isolates. PLoS ONE. 2013;8(2):e57091. doi: 10.1371/journal.pone.0057091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Badawy MF, Tawakol WM, El-Far SW, Maghrabi IA, Al-Ghamdi SA, Mansy MS, Ashour MS, Shohayeb MM. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int J Microbiol. 2017 doi: 10.1155/2017/8050432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferreira S, Toleman M, Ramalheira E, DaSilva GJ, Walsh T, Mendo S. First description of Klebsiella pneumoniae clinical isolates carrying both qnrA and qnrB genes in Portugal. Int J Antimicrob Agents. 2010;35(6):584–586. doi: 10.1016/j.ijantimicag.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 90.Esmat MM, Mohamed HH (2016) Quinolone resistance among extended-spectrum β-lactamases producing Klebsiella pneumoniae in Sohag University Hospital, Upper Egypt. EJMM 25(1):69–76