Abstract
Asthma is a chronic lung disease that has a high prevalence. The therapeutic intervention of this disease can be made more effective if genetic variability in patients’ response to medications is implemented. However, a clear picture of the genetic architecture of asthma intervention response remains elusive. We conducted a genome-wide association study (GWAS) to identify drug response-associated genes for asthma, in which 909,622 SNPs were genotyped for 120 randomized participants who inhaled multiple doses of glucocorticoids. By integrating pharmacodynamic properties of drug reactions, we implemented a mechanistic model to analyze the GWAS data, enhancing the scope of inference about the genetic architecture of asthma intervention. Our pharmacodynamic model observed associations of genome-wide significance between dose-dependent response to inhaled glucocorticoids (measured as %FEV1) and five loci (p = 5.315 × 10−7 to 3.924 × 10−9), many of which map to metabolic genes related to lung function and asthma risk. All significant SNPs detected indicate a recessive effect, at which the homozygotes for the mutant alleles drive variability in %FEV1. Significant associations were well replicated in three additional independent GWAS studies. Pooled together over these three trials, two SNPs, chr6 rs6924808 and chr11 rs1353649, display an increased significance level (p = 6.661 ×10−16 and 5.670×10−11). Our study reveals a general picture of pharmacogenomic control for asthma intervention. The results obtained help to tailor an optimal dose for individual patients to treat asthma based on their genetic makeup.
INTRODUCTION
Asthma is a chronic lung disease characterized by recurring periods of wheezing, chest tightness, shortness of breath, and coughing mediated through airway inflammation.1 Given its substantial societal cost,2 the discovery of any therapeutic intervention to treat this disease, especially severe asthma, has been a long-standing public health concern.3,4 Recent developments in molecular genetics provide an unprecedented opportunity to understand the genetic causes of asthma and identify targets that can be used to control the syndrome.4,5 More importantly, a systematic, large-scale survey of associations between common DNA sequence variants and disease has succeeded in identifying a set of specific genes that influence asthma.6–8 These asthma-associated genetic variants identified, distributed on various chromosomes, are found to affect this lung disease through altering key biochemical pathways that are related to lung function.9,10
Although there is no doubt that the identification of genes for disease risk facilitates the development of effective medications for its treatment,11 the efficient application of such medications will rely on our knowledge about pharmacogenomic effects on drug disposition, drug metabolism, and drug response, given the fact that inter-individual variation exists in the response to a particular drug.12,13 However, until recently, most pharmacogenomic studies have been carried out using candidate gene approaches. Specific genes that encode enzymes involved in drug metabolism as well as drug targets, typically receptors or enzymes, have been identified. A successful example of candidate gene studies is the identification of genes that control the effect of anticoagulant drugs, such as warfarin.14 Some pharmacogenomic studies have also identified genes responsible for adverse drug reactions,12 including those that encode metabolic enzymes and those that are related to the immune system and mitochondrial functions. In a recent review, Tse et al.15 described candidate genes and pathways detected thus far to control variability in response to three classes of asthma medications, β-adrenergic receptor agonists, inhaled corticosteroids and leukotriene modifiers.
Since 2007, genome-wide association studies (GWAS) have increasingly emerged as a powerful tool for pharmacogenomic studies. Significant associations through GWAS have been detected for the response to interferon-α,16–18 clopidogrel19 and warfarin,20–22 as well as for adverse drug reactions related to statin-induced myopathy23 and flucloxacillin-induced liver injury.24 All these studies may help shed light on the genetic control mechanisms of drug response and their clinical implications.
Unlike a case in mapping complex diseases, GWAS of drug response is often characterized by a small size of samples12 so that there may be insufficient power to detect small or moderate size effects. One approach to overcome this limitation is the use of family-based data25–29 and has recently been applied to asthma treatment response30. However, the vast majority of clinical trials of drug response do not involve DNA collection in non-trial family participants. Here, we present an alternative approach by incorporating the pharmacodynamic principle of drug response into a pharmacogenetic GWAS. The basic idea of such incorporation is to model drug effect-dose relationships through mathematical equations based on repeated measures of drug response at multiple dosages.30 By estimating and testing those mathematical parameters that define the effect-dose curves, one can determine how a specific gene affects drug effects at each dose or across a range of doses. Because of its statistical parsimony, i.e., the number of curve parameters is always less than the number of doses,31–34 the incorporation of mathematical equations can potentially increase the power of detecting significant associations, compared to traditional GWAS analysis based on a simple phenotype-genotype relationship. Although the theory of this incorporation has well been established in the previous studies,30–34 here we have for the first time reported a systematic implication of this theory for practical pharmacogenetic studies in asthma intervention.
The pharmacodynamic approach was applied to analyze a pharmacological GWAS trial derived from SNP Health Association Asthma Resource projects (SHARP),35–37 leading to the identification of five significant SNPs responsible for pulmonary response after asthma treatment. Associations between these SNPs and the same phenotype were well confirmed by analyzing three additional GWAS. In order to investigate how small sample sizes, characteristic of pharmacogenomics studies, impact on the estimation of genetic effects and the power of gene detection, we performed computer simulation by mimicking the data structure of SHARP. We found that the implementation and use of a pharmacodynamic model can overcome, to some extent, the limitation of small sample sizes in pharmacological GWAS.
PHARMACODYNAMIC MODELING OF GWAS
Statistical Design
Consider a clinical trial composed of n participants used for a pharmacological GWAS, in which each of the participants is genotyped for SNPs throughout the entire genome. These participants receive the administration of a drug under a multitude of doses, at each of which a pharmacological parameter that reflects drug effect is measured. Under this design, each participant (say i) has a series of dose-dependent pharmacological phenotypic data, expressed as yi = (yi(Cil), …, yi()), where (Ci1, …, ) are Mi doses of administration participant i receives. We allow different participants to possibly receive different number of doses in the clinical trial.
It is likely that drug response as a complex trait is controlled by many genes each with a different effect. The GWAS is motivated to identify all possible genes and estimate each gene’s effects on drug response. Assuming that there is such a gene with three genotypes and, also, considering the influence on drug response by other covariates, such as race, sex, life style, and age, the phenotypic value of participant i at dose m can be described by a regression model, expressed as,
| (1) |
where gj(Cim) is the genotypic value of participant i who carries SNP genotype j at dose Cim; zij is an indicator variable of participant i defined as 1 if this participant carries a genotype considered and 0 otherwise; uik is the value of the kth (k = 1, …, K) continuous covariate for participant i; αk is the effect of the kth continuous covariate; vls is the effect of the lth (l = 1, …, L) discrete covariate at its sth (s = 1, …, Sl) level; xils is the indicator variable that describes the sth level of the lth discrete covariate for participant i; and ei(Ciτ) is a random error.
We implement a maximum likelihood approach to estimate the parameters involved in model (1). The likelihood of all participants is constructed as
| (2) |
where nj is the number of participants with genotype j; and fj(yi) is assumed to follow a multivariate normal distribution with mean vector for genotype j as
| (3) |
and covariance matrix
| (4) |
We incorporated the pharmacodynamic model to estimate the genotypic values of a particular genotype j at different dosages30, as shown in equation (2). Emax equation38 that specifies drug effect E at a particular dose C is thought to be one of the most pharmacodynamics models, expressed as
| (5) |
where E0 is the baseline, Emax is the asymptotic (limiting) effect, EC50 is the drug concentration that results in 50% of the maximal effect, and H is the slope parameter that determines the slope of the concentration-response curve. The larger H, the steeper the linear phase of the log-concentration effect curve. The phenotypic longitudinal data were normalized to remove the baseline so that drug effect at different levels of dosage is defined by three parameters. By estimating these parameters for different SNP genotypes, we could draw the curves of drug response for each genotype and test how different genotypes vary in the form of curve.
We used the Emax equation (5) to model dose-dependent genotypic values for each SNP genotype j by pharmacological parameters (Emaxj, EC50j, Hj). In addition, considering the autocorrelation feature of the random error, we used the autoregressive regression model to estimate the across-dose covariance structure. Other approaches for modeling covariance structure are available in the literature,39–42 allowing a choice of the best fit model for a practical data set.
Hypothesis Tests
Whether a particular gene affects drug response can be tested by a log-likelihood ratio approach. This can be done on the basis of two alternative hypotheses:
| (6) |
Under each hypothesis, we calculate the likelihood and further calculate their ratio. This ratio is thought of being chi-square distributed with six degrees of freedom.
After a gene is confirmed to be significant among its three genotypes, we will further test how this gene acts to affect drug response. The action of a gene can be additive, dominant or recessive. Wu et al.30 provided a general procedure to test the mode of action under a pharmcodynamic model. A SNP is first analyzed by a genotypic model, i.e., testing differences among three genotypes based on hypothesis test (6), followed by the additive and dominant/recessive tests. We used model selection criteria, such as BIC, to choose an optimal model.
APPLICATIONS
Samples
The research with human participants has been approved by Penn State College of Medicine’s Review Board. Table 1 summarizes key population characteristics of several trials used for GWAS analysis. In the Dose of Inhaled Corticosteroids with Equisystemic Effects (DICE) trial35, 120 randomized participants (post-pubertal to 60 years of age) were recruited with mild-to-moderate asthma, defined as 12% change in forced expiratory volume in 1 second (FEV1) or ≤ 8 mg/ml methacholine provocative concentration causing a 20% drop in FEV1 (PC20) and baseline FEV1 65–90% of predicted. During enrollment, AM (prior to 9:30 AM) plasma cortisol concentration of ≥ 5 mcg/dl needed to be attained. After a 1-week run-in period, participants were randomized to one of six active inhaled corticosteroid (ICS) delivery systems (or the corresponding placebo). After each week of treatment, participants remained at the study center for an overnight stay and then received a medication supply with a doubled dose for the subsequent week. This process continues until there are a total of four dosages. At each of the dosages, participants were measured for the percent predicted value of the pre-bronchodilator forced expiratory volume in one second (%FEV1).
Table 1:
Population characteristics of the longitudinal DICE trial and other three independent trials.
| Characteristic | DICE | IMPACT | SOCS | SLIC | SOCS/SLIC |
|---|---|---|---|---|---|
| No. of subjects | 120 | 251 | 79 | 106 | 31 |
| Inhaled glucocorticoid | Budesonide | 1)Budesonide 2)Prednisone+ Budesonid+ Zafirlukast |
Triamcinolone |
1)Triamcinolone 2)Salmetrol+ Triamcinolone |
Triamcinolone |
| Age – yr | 30.6±8.1 | 34.2±10.8 | 30.4±10.9 | 35.9±12.4 | 32.7±11.4 |
| Sex – no. subject(%) | |||||
| Male | 51(56.7%) | 57(39.0%) | 30(41.1%) | 43(45.3%) | 11(52.4%) |
| Female | 39(43.3%) | 89(61.0%) | 43(58.9%) | 52(54.7%) | 10(47.6%) |
| Baseline FEV1-% of predicted | 79.0±7.4 | 88.8±13.3 | 85.6±14.0 | 67.6±10.9 | 73±15.9 |
| Change in FEV1-% | 6.5±10.3 | 1.3±7.4 | 7.3±11.8 | 4.5±9.8 | 2.6±16.6 |
The IMPACT trial35 includes 225 adult asthma participants (in age 18–65 years with mild, persistent asthma) with FEV1 ≥ 70% of predicted, displaying either ≥ 12% or > 200 ml improvement following albuterol inhalation or bronchial hyperreactivity (methacholine PC20 < 16 mg/ml). All participants were instructed to take open-label budesonide or prednisone as guided by the symptom-based action plan. The run-in and treatment phases both ended with a 14-day period of intense combined therapy. Salmeterol Off CorticoSteroids (SOCS)36 and Salmeterol ± Inhaled Corticosteroids (SLIC) trials37 include 79 and 106 asthma participants, respectively, which were conducted in tandem with a common 6-week run-in period on inhaled corticosteroid therapy (Table 1). At the end of the run-in period, the milder patients were allocated to SOCS (FEV1 > 80% predicted, PEF variability ≈ 20%) and the more moderate patients allocated to SLIC. The %FEV1 was measured for the three trials above.
In four trials, DICE, IMPACT, SOCS, and SLIC, subjects were genotyped for 909,622 SNPs throughout the entire genome. Genotyping was performed on Affymetrix 6.0 arrays. SNP genotypes were obtained after stringent quality-control filters.
Results
Gene detection:
We used the pharmacodynamic model to identify genes for drug response (%FEV1) to inhaled corticosteroids for asthma treatment by jointing estimating the effect due to covariates, age, BMI, race, gender and drug type. Dose levels of drugs were normalized to a range from 0 to 1. We implemented four genetic models to study the patterns of pharmacological inheritance: (1) genotypic model detecting the overall effect due to differences among three SNP genotypes, (2) additive model detecting the effect due to the substitution of the wild-type allele (common) by the mutant (minor), (3) dominant model in which the mutant allele is dominant over the wild-type allele, and (4) recessive model in which the expression of the mutant is masked by the wild-type allele. We excluded those SNPs from our GWAS analysis with minor allele frequency < 0.3; this threshold is larger than usual, aimed to assure sufficient samples for each genotype group given our modest sample size. To the end, a total of 266,944 SNPs were involved in the analysis.
The four genetic models were each used to scan SNPs throughout the entire genome for the DICE trial. After the Bonferroni correction, five significant SNPs were detected by the genotypic (Supplementary Fig. 1A) and recessive model (Supplementary Fig. 1D). The additive and dominant models did not identify significant associations (Supplementary Fig. 1B and 1C). Two SNPs on chromosomes 8 and 11 were detected by both genotypic and recessive models, but according to BIC values calculated, both SNPs conform to the recessive model better than the genotypic model. The following loci produce associations of genome-wide significance with physiological response to glucocorticoid therapy for asthma (Table 2); rs6924808 on chromosome 6 with wild-type allele C and mutant T (p = 5.315 × 10−7), rs10481450 on chromosome 8 with wild-type allele A and mutant T (p = 2.614 × 10−8), rs1353649 on chromosome 11 with wild-type allele G and mutant A (p = 3.924 × 10−9), rs12438740 on chromosome 15 with wild-type allele C and mutant T (p = 4.499 × 10−8), and rs2230155 on chromosome 15 with wild-type allele C and mutant T (p = 1.798 × 10−7). To evaluate the influence of population stratification, we calculated the ratio of the median of the log-likelihood ratios among all SNPs analyzed over the critical value of the chi-square distribution at the 0.05 significance level. If this ratio is near, or slightly less than, 1.0, this indicates that the effect due to population stratification is ignorable43. The ratios calculated are 0.82 – 0.88 for the genotypic, additive and dominant models used, suggesting that our results are not largely affected by population structure.
Table 2:
Power to correctly identify the pattern of pharmacological inheritance by the pharmacodynamic model. The data were simulated by mimicking the DICE data structure and the estimates obtained by BIC from 1000 simulation replicates.
| Sample Size | True Model | Estimated Model |
|||
|---|---|---|---|---|---|
| Full | Recessive | Dominant | Allelic | ||
| 100 | Full | 887 | 106 | 7 | 0 |
| Recessive | 1 | 993 | 1 | 5 | |
| Dominant | 0 | 0 | 954 | 46 | |
| Additive | 4 | 63 | 69 | 864 | |
| 200 | Full | 991 | 9 | 0 | 0 |
| Recessive | 14 | 986 | 0 | 0 | |
| Dominant | 5 | 0 | 983 | 12 | |
| Additive | 9 | 10 | 18 | 963 | |
| 400 | Full | 1000 | 0 | 0 | 0 |
| Recessive | 16 | 984 | 0 | 0 | |
| Dominant | 8 | 0 | 990 | 2 | |
| Additive | 2 | 1 | 1 | 996 | |
Pharmacodynamic pattern of genetic effects:
The pharmacodynamic model allows the estimates of genotype-specific curve parameters, (Emax, EC50, H), that define drug response (see Supplementary Table 1 for the maximum likelihood estimates of the parameters and the standard deviations of the estimates). Each of these parameters differs strikingly between two groups of genotypes, the homozygote for the mutant allele, and a mix of the homozygote for the wild-type allele and the heterozygote for the two different alleles, at significant SNPs. The overall influence of these parameters on variability in glucocorticoid response curve can be seen from response curves drawn for each genotype group (Fig. 1). It can be observed that individual SNPs fit raw longitudinal data reasonably well and also different SNPs affect drug response in different ways.
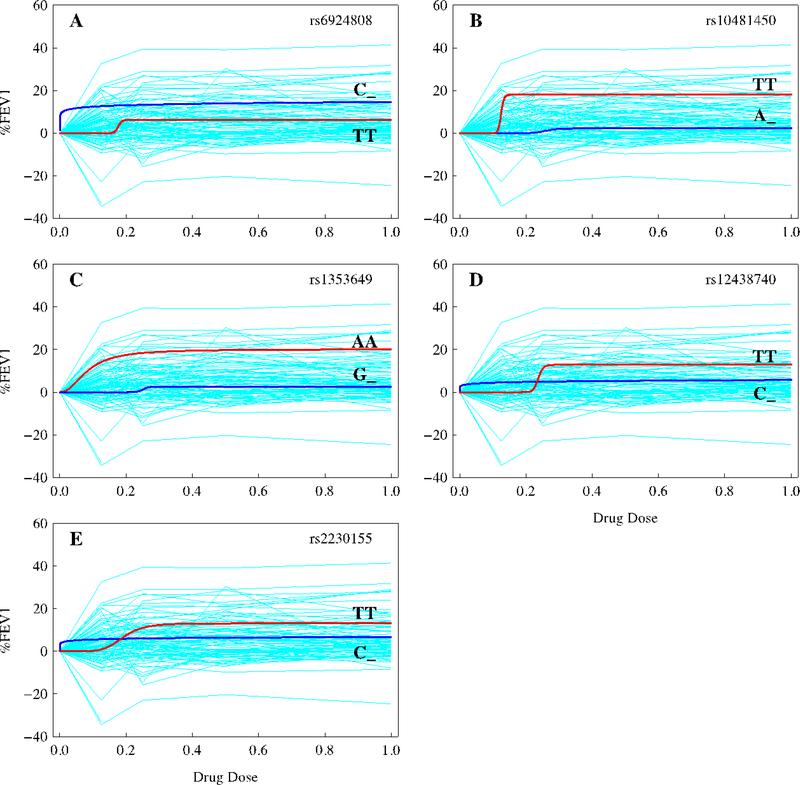
Figure 1.
Changes in pulmonary response to varying doses of inhaled corticosteroids as defined as prebronchodilator %FEV1 for two different groups of genotypes (i.e., the mutant homozygote, MM, and a mix of the homozygote for the wild-type allele and the heterozygote for the two different alleles, W_) at five significant SNPs, chr6 rs6924808 (A), chr8 rs10481450 (B), chr11 rs1353649 (C), chr15 rs12438740 (D), and chr15 rs2230155 (E), for the DICE trial detected by the recessive model. Blue thin lines in background are response curves of individual participants to varying dosages. The original data were normalized by removing the baselines and plotted against relative scales of corticosteroid dosages.
At all significant SNPs, the wild-type allele is dominant over the mutant for their actions in affecting glucocorticoid response, as revealed by the recessive model (Table 2). For chr6 rs6924808, the homozygote (CC) for the wild-type allele C and the heterozygote (CT) for the wild-type allele and mutant T are more responsive to changing doses of glucocorticoids than the homozygote (TT) for the mutant (Fig. 1A). At chr8 rs10481450 and chr11 rs1353649, the homozygotes for the mutant display remarkably greater sensitivity to drug dose than the genotypes containing the wild-type alleles (Fig. 1B and 1C). Chr15 rs12438740 and chr15 rs2230155 are located closely together on the same chromosome, with a high linkage disequilibrium (r = 0.99), exhibit a similar dynamic pattern of genetic effect (Fig. 1D and 1E); the homozygote for the mutant do not respond until a particular dose level is reached, whereas the genotypes containing the wild-type allele appear to be resistant to increasing dose. Figure 1 shows that some SNPs capture a wide variation like rs10481450 and rs1353649, whereas the others explain a narrow variation like rs6924808, rs12438740, and rs2230155. Some SNPs are sensitive to a small change in low doses of drug, such as rs6924808, and some display variation after a certain level of dose is reached, such as rs10481450 and rs12438740. The common feature of all the SNPs is that different groups of genotypes start to diverge at a lower level of dose and stabilize their variation during a wide range of dose.
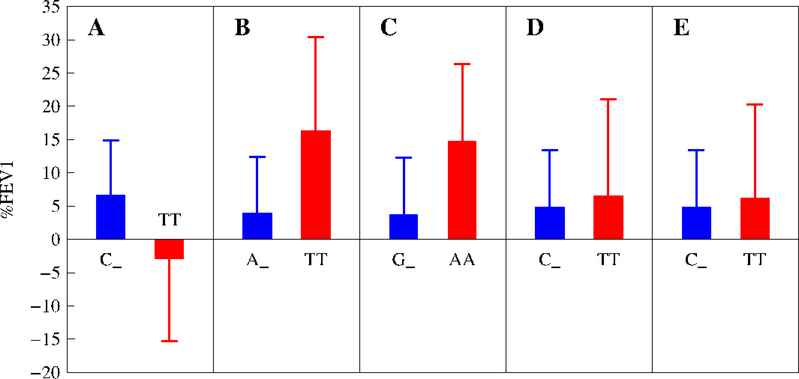
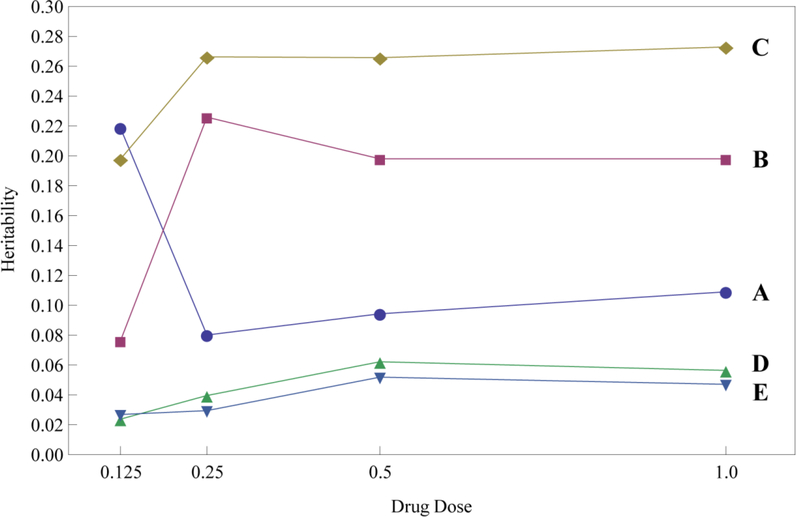
In sum, the mutant allele produces a pronounced increase in lung function after glucocorticoid treatment as compared with the wild-type allele for all SNPs, except for chr6 rs6924808. Overall, subjects who are homozygous for the mutant allele are 30–300% larger for %FEV1 values at an intermediate dose of glucocorticoids than those who are homozygous for the wile-type allele and heterozygous for the two alleles (Fig. 2; Supplementary Table 2). The differences between these two groups of genotypes are 30–245% of the mean of all treated subjects. There are striking differences in the heritability of %FEV1 response to glucocorticoid therapy explained by individual SNPs (Fig. 3). Chr11 rs1353649 accounts for 19–26% of the phenotypic variation, whereas these values are 8–20% for chr8 rs10481450 and chr6 rs6924808 and 2–5% for chr15 rs12438740 and chr15 rs2230155. All SNPs display a dynamic change of heritability over dose.
Figure 2.
Changes of normalized %FEV1 mean (±SE) by the mutant homozygote (MM) over a mix of the homozygote for the wild-type allele and the heterozygote for the two different alleles (W_) at an intermediate dose of glucocorticoids for significant SNPs, chr6 rs6924808 (A), chr8 rs10481450 (B), chr11 rs1353649 (C), chr15 rs12438740 (D), and chr15 rs2230155 (E), for the DICE trial detected by the recessive model.
Figure 3.
Dynamic changes of the heritability for %FEV1 over relative scales of corticosteroid dosages explained by individual SNPs, chr6 rs6924808 (A), chr8 rs10481450 (B), chr11 rs1353649 (C), chr15 rs12438740 (D), and chr15 rs2230155 (E) for the DICE trial.
Cross validation:
We performed an additional analysis to cross-validate the results detected by randomly splitting the population into two equally-sized sub-groups. In Supplementary Table 3, we summarized the results about the estimates of parameters from 100 resampling replicates for each subgroup at a significant SNP rs10481450, in a comparison with those from the whole population. The parameter estimates of each subgroup are quite consistent, and they are consistent with those using the whole population. Although standard errors of the estimates for some parameters are large due to a small sample size, they are reasonably within the space of estimates.
Computer Simulation
The statistical properties of the pharmacodynamic model to analyze GWAS data were investigated through computer simulation. The simulation mimicked the DICE trial in terms of sample size, dose level, and demographical attributes of participants. The phenotypic data of drug response were simulated using parameters estimated for SNP rs10481450 detected from the DICE trial by assuming normally-distributed residuals. The data were simulated using the genotypic, additive, dominant and recessive models and then analyzed by each model.
The pharmacological model has good power to detect a correct pattern of genetic action (Table 2). When a correct model was used, genotype-specific pharmacodynamic parameters and covariate effects can be reasonably estimated. Table 3 gives the estimates of parameters and their sampling errors under different sample sizes when the genotypic model is assumed. In Supplementary Table 4, estimates of parameters by the dominant, recessive and additive models are given. The estimates of three pharmacological parameters, Emax, EC50 and H, each have a reasonably small sampling error for each genotype, even when the sample size is modest (100). The estimation precision of these parameters increases dramatically when sample size increases to 200 or 400.
Table 3:
Means of the estimates of parameters for the pharmacodynamic approach and their standard errors (in parentheses) from simulated data by mimicking the DICE data structure with different sample sizes based on 1000 simulation replicates (full genotypic model).
| Parameter | True Value | Sample Size |
||
|---|---|---|---|---|
| 100 | 200 | 400 | ||
| Emax1 | 9.934 | 10.511 (1.653) | 10.465 (1.417) | 10.332 (1.167) |
| H1 | 1.472 | 1.441 (0.589) | 1.435( 0.562) | 1.455 (0.517) |
| EC50(1) | 0.069 | 0.094 (0.089) | 0.082 (0.050) | 0.076 (0.024) |
| Emax2 | 17.637 | 17.317 (2.539) | 17.515 (2.228) | 17.668 (1.884) |
| H2 | 2.509 | 2.864 (0.796) | 2.723 (0.662) | 2.618 (0.477) |
| E50(2) | 0.551 | 0.544 (0.091) | 0.549 (0.068) | 0.552 (0.053) |
| Emax3 | 19.858 | 20.185 (2.422) | 20.060 (2.019) | 19.976 (1.567) |
| H3 | 2.2 | 2.580 (0.955) | 2.430 (0.779) | 2.340 (0.583) |
| EC50(3) | 0.13 | 0.144 (0.047) | 0.137 (0.028) | 0.134 (0.019) |
| α1 | −0.075 | −0.077 (0.086) | −0.074 (0.058) | −0.074 (0.040) |
| α2 | 0.113 | 0.116 (0.109) | 0.111 (0.072) | 0.112 (0.050) |
| v11 | 2.135 | 2.178 (1.100) | 2.150 (0.775) | 2.117 (0.527) |
| v12 | −1.065 | −1.074 (1.407) | −1.104 (0.966) | −1.104 (0.681) |
| v13 | −3.553 | −3.531 (1.427) | −3.532 (0.944) | −3.521 (0.668) |
| v14 | 3.037 | 2.978 (1.641) | 3.014 (1.124) | 3.039 (0.822) |
| v21 | 3.855 | 3.943 (1.666) | 3.925 (0.956) | 3.872 (0.627) |
| v22 | −6.059 | −6.031 (2.081) | −6.039(1.279) | −6.044 (0.874) |
| v23 | 4.904 | 5.001 (2.418) | 4.943 (1.417) | 4.902 (0.998) |
| v31 | −0.798 | −0.829 (0.699) | −0.814 (0.464) | −0.816 (0.322) |
| ρ | 0.754 | 0.738 (0.027) | 0.746 (0.020) | 0.750 (0.014) |
| σ2 | 71.401 | 66.294 (6.071) | 68.769 (4.415) | 69.995 (3.103) |
Note: α1 and α2 are the effects due to two continuous covariates; v1, v2 and v3 are the effects due to three discrete covariates each with a different level; and ρ and σ2 are the autoregressive parameters that model the covariance structure.
Because humans cannot be controlled, like plants or animals, it is unavoidable to include many covariates, such as different demographic factors, in human GWAS. These covariates would often confound the identification of significant genes. However, the deployment of a multiple regression model that incorporates covariate effects can filter some of these confounders. As shown in Table 4, the estimates of genotype-specific pharmacodynamics parameters are not affected by covariates. Furthermore, model (1) can provide an estimate of the effects of each covariates including continuous and discrete. Estimates of some of the covariate effects are not very precise under a small sample size (100), but this situation can improve dramatically when the sample size increases to 400.
Table 4.
Significant SNPs detected for inhaled corticosteroid drug response to asthma treatment from the DICE trials and confirmed in three additional trials. The chromosomal positions, alleles, minor allele (in parentheses) frequencies (MAF), and significance levels of these SNPs are given.
| Model | SNP | Chr | Position | Alleles | MAF |
p-value |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| DICE | IMPACT | SOCS | SLIC | Pooled | ||||||
| Recessive | rs6924808 | 6 | 98465296 | C/T | 0.477 (T) |
5.315 ×10−7 |
5.908 ×10−11 |
0.034 | 0.001 | 6.661 ×10−16* |
| Recessive | rs10481450 | 8 | 9835656 | A/T | 0.310 (T) |
2.614 ×10−8 |
0.072 | 0.057 | 1.154 ×10−6 |
0.023 |
| Recessive | rs1353649 | 11 | 20210175 | A/G | 0.320 (A) |
3.924 ×10−9 |
0.003 | 0.089 | 0.024 |
5.670 ×10−11* |
| Recessive | rs12438740 | 15 | 57303059 | C/T | 0.348 (T) |
4.499 ×10−8 |
0.004 | 0.0003 | 2.883 ×10−5 |
0.040 |
| Recessive | rs2230155 | 15 | 57297481 | C/T | 0.350 (T) |
1.798 ×10−7 |
0.043 | 0.211 | 0.056 | 0.132 |
These p-values were obtained from an optimal model, genotypic model.
Wu et al.30 showed that the pharmacodynamics model displays increased power of gene detection compared to traditional GWAS analysis based on a single static phenotype. To confirm their result, we used the simulated data above that mimics the DICE trial to estimate the empirical power of gene detection at each of four doses. This power was observed to be substantially lower (by 1.5 to 8.5 times) than the power of gene detection by the pharmacodynamics model that makes use of data at all doses at one time, consistent with Wu et al.’s finding.
Replication
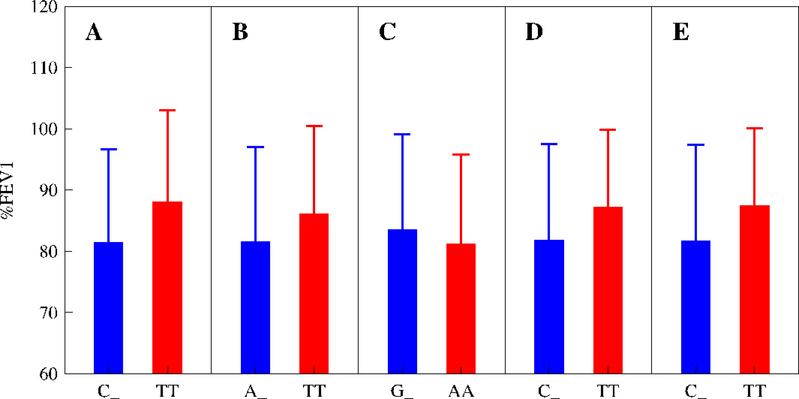
Here, we used three other trials to validate the results. These trials are the IMProving Asthma Control Trial (IMPACT),35 Salmeterol Off CorticoSteroids (SOCS) trial,36 and Salmeterol ± Inhaled Corticosteroids (SLIC) trials.37 Because of one single dose of corticosteroid used, we analyzed associations between %FEV1 values and five significant SNPs detected from DICE using a univariate model (Table 4). All the five SNPs were found to be significant in the three trials, except for chr15 rs2230155 being non-significant for SOCS and chr8 rs10481450 and chr11 rs1353649 being marginally significant for IMPACT and SOCS, respectively (Table 4). Pooled together over IMPACT, SOCS, and SLIC of a similar design, all SNPs, except for chr15 rs2230155, display significant associations with glucocorticoid response. Figure 4 compares the differences of %FEV1 between two groups of genotypes, the mutant homozygote and a mix of the homozygote for the wild-type allele and the heterozygote for the two different alleles, at each SNP for the pooled three trials. For chr6 rs6924808 and chr11 rs1353649, such differences have different directions between the pooled trials and DICE. When an optimal model, i.e., genotypic model, was used, these two SNPs produce a very high level of significance for associations (p = 6.661 ×10−16 and 5.670×10−11; Table 4).
Figure 4.
Changes of %FEV1 mean (±SE) by the mutant homozygote (MM) over a mix of the homozygote for the wild-type allele and the heterozygote for the two different alleles (W_) at an intermediate dose of glucocorticoids for significant SNPs, chr6 rs6924808 (A), chr8 rs10481450 (B), chr11 rs1353649 (C), chr15 rs12438740 (D), and chr15 rs2230155 (E), for pooled IMPACT, SOCS and SLIC trials detected by the recessive model. Note that %FEV1 was not normalized because these trials contain only one dose of glucocorticoids.
DISCUSSION
The pharmacodynamic model has successfully detected five loci of significant effects on response curves of corticosteroids for asthma by integrating the biochemical processes of drug response into GWAS. This integration has proven to be statistically more powerful for gene detection than traditional approaches based on a single dose30. The identification of significant SNPs by our model has been validated by resampling and simulation studies. Furthermore, these five SNPs demonstrate good replication in three independent clinical trial populations for the same phenotype. In general, the mutant alleles at most SNPs tend to increase pulmonary function of asthma participants by 30–300% after inhaled glucocorticoid treatment relative to the wild-type alleles, although the expression of the mutant may be masked by the wild-type allele. In another study, Tantisira et al.29 found that the mutant homozygote at chr7 rs37972 displays 120–330% decrease of lung function through glucocorticoid treatment compared with the wild-type homozygote. In both our and Tantisira et al.’s studies, the heritabilities of glucocorticoid response explained by individual SNPs are much larger than those detected for disease and physiological traits.8–11 This may be due to the fact that drug response is evolutionarily a “young” trait, which has not experienced yet a long history of natural selection as the other traits have.12
High heritability detection should benefit from the statistical merit of our pharmacodynamic model which was derived from parsimonious modeling of the mean-covariance structures for longitudinal data of drug response across a series of doses. For example, four parameters are needed to describe drug response of a genotype at four dose levels if traditional approaches are used, while the pharmacodynamic model only uses three parameters to do the same thing. Moreover, the pharmacodynamic model, such as the Emax model36 and differential equations, 44,45 contains biologically meaningful aspects of drug response in terms of body-drug interactions. Applied to GWAS of response to corticosteroids for asthma intervention, this model can not only facilitate the interpretation and elucidation of the pharmacogenomics architecture of this important clinical problem, but also increase the statistical power of significant association detection.
Except for SNP rs6924808 on chromosome 6, the other four detected are located in the vicinity of candidate genes associated with cellular functions. It appears that SNP rs10481450 on chromosome 8 is related to gene TNKS, a PARP member localized predominantly in the cytosol, that regulates cellular viability and NAD(+) metabolism46 and gene MSRA that has a function to repair oxidative damage to proteins to restore biological activity.47 SNP rs1353649 on chromosome 11 is nearby many candidate genes, such as DBX1,48 NAV2,49 HTATIP2,50 and PRMT3,51 some of which determine nicotine dependence. MYO1E is also a nicotine dependence-related gene52 in which two associated SNPs, rs12438740 and rs2230155, on chromosome 15 were identified.
A modest sample size used may overestimate genetic effects of SNPs. However, our pharmacodynamic model makes use of the longitudinal feature of phenotypic data measured repeatedly for the same subjects, which has proven to be powerful for increasing the precision of parameter estimation.30 Our finding here shows a promise to utilize the genetic results obtained to predict individual patients’ performance in asthma intervention. Recent studies showed that asthma may be affected by DNA methylation through regulating gene expression.53 It is straightforward to integrate methylation variants into the model to better reveal the genetic and epigenetic basis of asthma intervention. To the end, by incorporating our new model with genetic and epigenetic observations for asthma6,7 and associated alteration in lung function by asthma,9,10 we may better determine and design the optimal doses for individual patients13, maximizing drug efficacy for optimal pulmonary function response while minimizing drug toxicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kwangmi Ahn for her assistance in data management. This research was funded by grants U01 HL119178, NIH/NHLBI-1U10HL098115, UL1 TR000127, U10 HL-51810, U10 HL-51834, U10 HL-51831, U10 HL-51823, U10 HL-51845, U10 HL-51843, and U10 HL-56443. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Fanta CH. Asthma. N Engl J Med 2009; 360:1002–1014. [DOI] [PubMed] [Google Scholar]
- 2.Szefler SJ. Advancing asthma care: The glass is only half full!. J Allergy Immunol 2011;128:485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apter AJ. Advances in adult asthma diagnosis and treatment and HEDQ in 2010. J Allergy Clin Immunol 2011;127:116–22. [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health, National Heart, Lung, and Blood Institute, National Asthma Education and Prevention Program. Expert panel report 3: guidelines for the diagnosis and management of asthma. NIH Publication no. 07–4051. Available at: http://www.nhlbi.nih. gov/guidelines/asthma/index.htm. Accessed July 16, 2011.
- 5.Yoshihara S Early intervention for infantile and childhood asthma. Expert Rev Clin Immunol 2010;6:247–255. [DOI] [PubMed] [Google Scholar]
- 6.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med 2008;359:1985–1994. [DOI] [PubMed] [Google Scholar]
- 7.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–473. [DOI] [PubMed] [Google Scholar]
- 8.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010;363:1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 2010;42:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 2010;42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med 2010;363:166–176. [DOI] [PubMed] [Google Scholar]
- 12.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet 2010;11:241–246. [DOI] [PubMed] [Google Scholar]
- 13.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009;360:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daly AK. Pharmacogenomics of anticoagulants: steps toward personal dosage. Genome Med 2009;1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tse SM, Tantisira K, Weiss ST. The pharmacogenetics and pharmacogenomics of asthma therapy. Pharmacogenomics J 2011;1–10 doi: 10.1038/tpj.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009;461:399–401. [DOI] [PubMed] [Google Scholar]
- 17.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet 2009;41:1100–1104. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet 2009;41:1105–1109. [DOI] [PubMed] [Google Scholar]
- 19.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009;302:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper GM, Johnson JA, Langaee TY, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 2008;112:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichert M, Eijgelsheim M, Rivadeneira F, et al. A genome-wide association study of acenocoumarol maintenance dosage. Hum Mol Genet 2009;18:3758–3768. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association stud confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 2009;5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link E, Parish S, Armitage J, et al. SLCO1B1 variants and statin-induced myopathy – a genomewide study. N Engl J Med 2008;359:789–799. [DOI] [PubMed] [Google Scholar]
- 24.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA.B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet 2009;41:816–819. [DOI] [PubMed] [Google Scholar]
- 25.Van Steen K, McQueen MB, Herbert A, et al. Genomic screening and replication using the same data set in family based association testing. Nat Genet 2005; 37:683–91. [DOI] [PubMed] [Google Scholar]
- 26.Herbert A, Gerry NP, McQueen MB, et al. A common genetic variant is associated with adult and childhood obesity. Science 2006;312:279–83. [DOI] [PubMed] [Google Scholar]
- 27.Lasky-Su J, Lyon HN, Emilsson V, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet 2008;82:849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertram L, Lange C, Mullin K, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet 2008;83:623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J of Med 2011;365:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu RL, Tong CF, Mauger D, et al. A conceptual framework for pharmacodynamic genome-wide association studies in pharmacogenomics. Drug Discov Today 2011;16:804–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong Y, Wang Z, Liu T, et al. A statistical model for functional mapping of quantitative trait loci regulating drug response. Pharmacogenomics J 2004;4:315–321. [DOI] [PubMed] [Google Scholar]
- 32.Lin M, Aqvilonte C, Johnson JA, et al. Sequencing drug response with HapMap. Pharmacogenomics J 2005;5:149–156. [DOI] [PubMed] [Google Scholar]
- 33.Lin M, Hou W, Li HY, et al. Modeling sequence-sequence interactions for drug response. Bioinformatics 2007;23:1251–1257. [DOI] [PubMed] [Google Scholar]
- 34.Wu RL, Lin M. Statistical and Computational Pharmacogenomics. Chapman & Hall/CRC, London, 2008. [Google Scholar]
- 35.Martin RJ, Szefler SJ, Chinchilli VM, et al. Systemic effect comparisons of six inhaled corticosteroid preparations. Am J Resp Crit Care Med 2002;165:1377–1383. [DOI] [PubMed] [Google Scholar]
- 36.Boushey HA, Sorkness CA, King TS, et al. Regular vs. intermittent controller therapy for mild persistent asthma. New Engl J Med 2005;352:1519–1528. [DOI] [PubMed] [Google Scholar]
- 37.Lazarus SC, Boushey HA, Fahy JV, et al. Long-acting beta2-agonist monotherapy vs continued therapy with inhaled corticosteroids in patients with persistent asthma: a randomized controlled trial. JAMA 2001;285(20):2583–2593. [DOI] [PubMed] [Google Scholar]
- 38.Giraldo J Empirical models and Hill coefficients. Trend Pharmacolog Sci 2003;24:63–65. [DOI] [PubMed] [Google Scholar]
- 39.Diggle PJ, Heagerty P, Liang KY, et al. Analysis of Longitudinal Data. Oxford University Press, Oxford, 2002. [Google Scholar]
- 40.Zimmerman DL, Nunez-Anton V. Parametric modeling of growth curve data: an overview (with discussion). Test 2001;10:1–73. [Google Scholar]
- 41.Zhao W, Hou W, Littell RC, et al. Structured antedependence models for functional mapping of multivariate longitudinal quantitative traits. Stat Appl Mol Genet Biol 2005;4(1):Article 33. [DOI] [PubMed] [Google Scholar]
- 42.Yap JS, Li Y, Das K, et al. Functional mapping of reaction norms to multiple environmental signals through nonparametric covariance estimation. BMC Plant Biol 2011;11:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin B, Roeder K. Genomic control for association studies, Biometrics 1999;55: 997–1004. [DOI] [PubMed] [Google Scholar]
- 44.Mustavich LF, Miller P, Kidd KK, et al. Using a pharmacokinetic model to relate an individual’s susceptibility to alcohol dependence to genotypes. Hum Hered 2010;70:177–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn K, Luo JT, Berg A, et al. Functional mapping of drug response with pharmacodynamic-pharmcokinetic principles. Trend Pharmacol Sci 2010;31:306–311. [DOI] [PubMed] [Google Scholar]
- 46.Yeh TY, Sbodio JI, Nguyen MT, et al. Tankyrase-1 overexpression reduces genotoxin-induced cell death by inhibiting PARP1. Mol Cell Biochem 2005;276:183–192. [DOI] [PubMed] [Google Scholar]
- 47.Kuschel L, Hansel A, Schonherr R, et al. Molecular cloning and functional expression of a human peptide methionine sulfoxide reductase (hMsrA). FEBS Lett 1999;456:17–21. [DOI] [PubMed] [Google Scholar]
- 48.Lanuza GM, Gosgnach S, Pierani A, et al. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron 2004; 42:375–386. [DOI] [PubMed] [Google Scholar]
- 49.Maes T, Barcelo A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics 2002;80:21–30. [DOI] [PubMed] [Google Scholar]
- 50.Persson B, Kallberg Y, Bray JE, et al. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem Biol Interact 2009;178:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang J, Gary JD, Clarke S, et al. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem 1998;273:16935–16945. [DOI] [PubMed] [Google Scholar]
- 52.Hasson T, Skowron JF, Gilbert DJ, et al. Mapping of unconventional myosins in mouse and human. Genomics 1997;36:431–439. [DOI] [PubMed] [Google Scholar]
- 53.Rastogi D, Suzuki M, Greally JM. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci Rep 2013;3:2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.