Abstract
Summary
Comparative metabolomics comes of age through commercial vendors offering metabolomics for translational researchers outside the mass spectrometry field. The MetaboDiff packages aims to provide a low-level entry to differential metabolomic analysis with R by starting off with the table of metabolite measurements. As a key functionality, MetaboDiffs offers the exploration of sample traits in a data-derived metabolic correlation network.
Availability and implementation
The MetaboDiff R package is platform-independent, available at http://github.com/andreasmock/MetaboDiff/ and released under the MIT licence. The package documentation comprises a step-by-step markdown tutorial.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
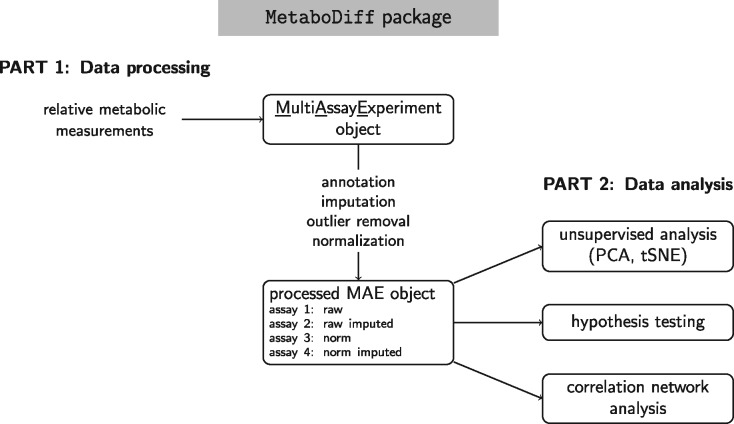
The comparative study of the metabolism promises more direct insights about health and disease phenotypes than obtained by genomic analyses (Johnson et al., 2016). However, metabolomic profiling requires ultra-high performance liquid chromatography/tandem mass spectrometry and gas chromatography/mass spectrometry. The acquisition and maintenance of this analytic setup is cost- and labor-intensive and requires expert-level knowledge in both the experimental and bioinformatical analysis. As a consequence, an increasing number of translational researchers are using the service of commercial vendors or core facilities for metabolomic profiling. The common result format researchers obtain from these vendors is a table of relative metabolic measurements that were identified through the process of peak picking and peak annotation. As much as the complexity and cost of running a metabolomics facility created the need for commercial vendors, there is a need for user-friendly computational solutions for R-using experimental biologist and bioinformaticians outside the mass spectrometry field. Comprehensive computational workflows for metabolomic analysis in R exists with the recent publication of ‘metaX’ leading the way including an extensive review of metabolomics tools since 2006 (Wen et al., 2017). However, we felt that available tools are still requiring too much expert-level knowledge about the details of processing metabolomic data. To this end, we developed ‘MetaboDiff’, an open source R package for differential metabolomic analysis (Fig. 1). The defining features what we believe makes MetaboDiff more user-friendly than previous tools are (i) the start of the analytic workflow from relative metabolic measurements, (ii) the storage of all metabolomic data within a single object, (iii) the usage of current gold standards for data imputation and normalization without the need to extensively study and compare methodologies and (iv) a step-by-step markdown tutorial.
Fig. 1.
Overview of data representation and analytic workflow of ’MetaboDiff’ package. Input is the table of relative metabolic measurements. The data and all its associated metadata are stored within a ’MultiAssayExperiment’ object. After processing, the object contains the four slots raw, raw imputed, norm and norm imputed. MAE, MultiAssayExperiment; PCA, Principal Component Analysis and tSNE, t-Distributed Stochastic Neighbor Embedding
2 Features and methods
2.1 Data processing
Within MetaboDiff, metabolic measurements and all related data are stored within a so called ‘MultiAssayExperiment’ object enabling the coordinated representation of multiple experiments and integrated sub-setting across experiments (Ramos et al., 2017; Supplementary Material). All common metabolic identifiers in the dataset (HMDB, KEGG and ChEBI) are used to query the Small Molecular Pathway Database (SMPDB 2.0; Jewison et al., 2014). In contrast to other high-throughput technologies, missing values are common in quantitative metabolomic datasets. K-nearest neighbor imputation is employed to minimize effects on the normality and variance of the data (Armitage et al., 2015). Combined hierarchical and k-means clustering can be used to determine outliers with the option to exclude individual samples or a cluster of samples from further analysis. Lastly, variance stabilizing normalization is used to ensure that the variance remains nearly constant over the measurement spectrum (Huber et al., 2002).
2.2 Data analysis
The data analysis section of ‘MetaboDiff’ starts by exploring the metabolome-wide difference between samples in an unsupervised fashion (see Table 1 for corresponding functions). Here, principal component analysis (PCA) and t-distributed stochastic neighbor embedding (tSNE) are at hand. Differential analysis (two or more groups) for individual metabolites is performed using Student’s t-Tests or ANOVA and corrected for multiple testing. The result of the comparative analysis can be visualized by a volcano plot. As a key functionality, ‘MetaboDiff’ offers the identification and exploration of metabolic correlation modules by the ‘weighted gene co-expression network analysis’ (WGCNA; Langfelder and Horvath, 2008) methodology. WGCNA is not limited by the need to define a priori metabolite sets for evaluation, factors in the topology of interactions and offers the possibility to relate modules to sample traits (Supplementary Material).
Table 1.
Biological questions that can be answered by MetaboDiff
| Question | Function |
|---|---|
| Missing measurements in dataset? | na_heatmap |
| Outliers in dataset? | outlier_heatmap |
| Metabolome-wide changes between samples? | pca_plot, tsne_plot |
| Differential metabolite abundance between groups? | diff_test |
| Differential sub-pathways between groups? | MS_plot |
| How do metabolites relate to each other in sub-pathway? | MOI_plot |
3 Usage scenario and benchmarking
The usability of ‘MetaboDiff’ is showcased in a case study of three datasets from a study by Priolo et al. (2014) and presented in the Supplementary Results. Here, a special emphasis is placed on the application and interpretation of the metabolic correlation network methodology.
4 Discussion
We present ‘MetaboDiff’, an R package for low-entry level differential metabolomic analysis. The functionality of the MultiAssayExperiment class opens up the possibility to incorporate other high-throughput data (e.g. expression data) from the same patient set (Ramos et al., 2017). ‘MetaboDiff’ will be continously updated as new evidence about metabolomic analysis arises.
Funding
This work was supported by the Anni Hofmann Stiftung (CHM).
Conflict of Interest: none declared.
Supplementary Material
References
- Armitage E.G. et al. (2015) Missing value imputation strategies for metabolomics data. Electrophoresis, 36, 3050–3060. [DOI] [PubMed] [Google Scholar]
- Huber W. et al. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics, 18, S96–104. [DOI] [PubMed] [Google Scholar]
- Jewison T. et al. (2014) SMPDB 2.0: big improvements to the small molecule pathway database. Nucleic Acids Res., 42(Database issue), D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.H. et al. (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol., 17, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo C. et al. (2014) AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer Res., 74, 7198–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M. et al. (2017) Software for the integration of multi-omics experiments in bioconductor. Cancer Res., 77, e39–e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B. et al. (2017) metaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics, 18, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.