Abstract
Formation of longer root hairs under limiting phosphate (P) conditions can increase the inorganic P (Pi) uptake. Here, regulatory candidate genes for Pi deficiency-induced root hair growth were identified by comparison of massive analysis of cDNA ends (MACE) provided expression profiles of two Brassica carinata cultivars (cv.) differing in their root hair response to Pi deficiency: cv. Bale develops longer root hairs under Pi deficiency, but not cv. Bacho. A split-root experiment was conducted for the differentiation between locally and systemically regulated genes. Furthermore, plants were exposed to nitrogen and potassium deficiency to identify P-specific reacting genes. The latter were knocked out by CRISPR/Cas9 and the effect on the root hair length was determined. About 500 genes were differentially expressed under Pi deficiency in cv. Bale, while these genes did not respond to the low P supply in cv. Bacho. Thirty-three candidate genes with a potential regulatory role were selected and the transcriptional regulation of 30 genes was confirmed by quantitative PCR. Only five candidate genes seemed to be either exclusively regulated locally (two) or systemically (three), whereas 25 genes seemed to be involved in both local and systemic signaling pathways. Potassium deficiency affected neither the root hair length nor the expression of the 30 candidate genes. By contrast, both P and nitrogen deficiency increased the root hair length, and both affected the transcript levels in 26 cases. However, four genes reacted specifically to Pi starvation. These genes and, additionally, INORGANIC PHOSPHATE TRANSPORTER 1 (BcPHT1) were targeted by CRISPR/Cas9. However, even if the transcript levels of five of these genes were clearly decreased, FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 1 (BcFLA1) was the only gene whose downregulation reduced the root hair length in transgenic hairy roots under Pi-deficient conditions. To the best of our knowledge, this is the first study describing a fasciclin-like arabinogalactan protein with a predicted role in the Pi deficiency-induced root hair elongation.
Keywords: CRISPR/Cas9, BcFLA1, hairy root, K deficiency, local and systemic response, MACE, N deficiency, split-root
Introduction
Since phosphate (P) in the soil available to plants is often limited, plants rely on mechanisms increasing the inorganic P (Pi) uptake (Vance et al., 2003). One of these is the development of longer root hairs under Pi deficiency, which increases the depletion zone around the root and, thus, the acquisition capability (Jungk, 2001). Among the root hair development steps, the root hair elongation, during which the root hair grows by tip growth, is decisive for the final root hair size (Datta et al., 2011). The general root hair elongation mechanisms are, at least in Arabidopsis thaliana, relatively well known, but the regulating mechanisms underlying the P deficiency-induced root hair elongation are still not completely understood.
Recent studies in Arabidopsis revealed that transcription factors play an important role in the root hair elongation under Pi starvation. The bHLH transcription factor ROOT HAIR DEFECTIVE 6 induces the expression of another bHLH transcription factor, ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), which, in turn, regulates genes needed for cell growth (Yi et al., 2010; Vijayakumar et al., 2016). Interestingly, RSL4 synthesis under Pi deficiency and, therefore, its presence in the root hair was prolonged, so that the root hair growth duration and, accordingly, the final root hair size was enhanced (Datta et al., 2015). Moreover, some of the target genes of RSL4 can be upregulated by ETHYLENE INSENSITIVE 3, which is a key transcription factor in the ethylene signaling pathway that is upregulated by Pi deficiency, thus, also increasing the root hair length (Song et al., 2016). In addition, there is a functional paralog of RSL4, LOTUS JAPONICUS ROOTHAIRLESS1-LIKE 3 (LRL3), which was upregulated in response to Pi deficiency and shown to be necessary for a proper root hair elongation (Bruex et al., 2012; Salazar-Henao et al., 2016; Salazar-Henao and Schmidt, 2016). Furthermore, the homeodomain transcription factor ALFIN-LIKE 6 controls the transcription of several genes involved in root hair elongation under Pi deficiency (Chandrika et al., 2013).
In addition to transcription factors, protein kinases also seem to have key roles during root hair elongation under Pi deficiency, since many protein kinases which were regulated in response to Pi starvation were associated with root hair growth in a co-expression analysis (Lan et al., 2013). Phosphatidylinositol phosphate 5-kinases (PIP5Ks) were identified to promote root hair elongation in response to Pi starvation (Wada et al., 2015). The PIP5Ks produce a signaling peptide, phosphatidylinositol 4,5-bisphosphate, and the authors concluded that, at least at young seedling stages, the PIP5K genes are responsible for transferring the Pi deficiency signal to the root hair elongation pathway. However, the authors predicted signaling pathways independent of the PIP5K genes playing a role during prolonged Pi starvation.
Moreover, calcium signaling and plant RHO GTPases (ROPs) seem to be involved in the Pi deficiency-induced root hair elongation, since a calcium-binding EF-hand family protein (CALMODULIN-LIKE 25, CML25) and ROP GUANINE NUCLEOTIDE-EXCHANGE FACTOR 4 (ROPGEF4) seem to regulate the Pi deficiency-induced root hair elongation in Arabidopsis negatively (Lin et al., 2011). The ROPGEF4, in cooperation with the receptor-like kinase FERONIA (FER), plays a role in the activation of ROPs (Duan et al., 2010).
In spite of the identification of several players involved in the Pi deficiency-induced root hair elongation, especially the network of regulatory genes, which may act at the beginning of the signal transduction cascade leading to the longer root hairs under Pi deficiency, still exhibits major knowledge gaps. Therefore, the aim of the present study was to identify and functionally characterize new candidate genes for the regulation of the Pi deficiency-induced root hair elongation.
Preliminary studies revealed two cultivars (cv.) of Brassica carinata (Ethiopian mustard) with a difference in their Pi uptake efficiency (Eticha and Schenk, 2001). Cultivar Bale develops longer root hairs under Pi deficiency compared to sufficient Pi conditions, whereas cv. Bacho develops short root hairs under both conditions. We used massive analysis of cDNA ends (MACE) to compare expression profiles of the B. carinata cv.s under both sufficient and deficient Pi conditions. The MACE provides a high resolution, since one single cDNA fragment is analyzed per transcript molecule, consequently, medium and low level transcripts can also be detected (Kahl et al., 2012). We identified around 500 genes in two different annotations with a regulation that indicated a role in the root hair elongation under Pi deficiency. More than 30 of these were investigated further, because of a potential regulatory function, and the regulation of nearly all of them could be validated by quantitative PCR (qPCR). A split-root experiment showed that most of these genes may be involved in both local and systemic signaling pathways. Furthermore, a few genes were upregulated exclusively under Pi deficiency, although both Pi and nitrogen (N) deficiency increased root hair length in cv. Bale. One of these genes was FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 1 (BcFLA1), whose downregulation by CRISPR/Cas9 in a hairy root system led to a reduced root hair length compared to the wildtype. The role of BcFLA1 in Pi deficiency signaling is discussed.
Materials and Methods
Plant Material and Growth Conditions
MACE Experiment (1st Experiment)
Seeds of B. carinata cv. Bale and cv. Bacho were surface sterilized for 1 min in 1% sodium hypochlorite, put on 0.7% (w/v) phyto agar (pH 5.8; Duchefa Biochemie, Haarlem, Netherlands) and then stratified for 3 days in the dark at 4°C for the MACE experiment. Afterward, the seeds were germinated in vitro for five (Bale) and 6 days (Bacho), respectively, with a day/night interval of 16/8 h at 23°C. The plants were then transferred into a climate chamber (photoperiod, 16/8 h light/dark; temperature, 20/15°C day/night; relative humidity: 75%; light intensity: 220 μmol m-2 s-1) and cultivated in nutrient solutions containing (mM) 2.25 Ca(NO3)2 × 4 H2O, 2.5 K2SO4, 1 MgSO4 × 7 H2O, 0.25 KCl and exclusively in the control treatment 1 KH2PO4. Furthermore, the nutrient solutions contained the following micronutrient concentrations (μM): 25 H3BO3, 1.5 MnSO4, 1.5 ZnSO4, 0.5 CuSO4, 0.025 (NH4)6Mo7O24 and 35.8 Fe (FeIII-EDTA). The pH was adjusted to 5.3 and readjusted during the cultivation as needed. After 8 days, root tips with a length of 2 cm were cut for later MACE and qPCR analysis, frozen immediately in liquid nitrogen and then stored at -85°C. The root tips of 42 plants (two root tips per plant) were pooled for each treatment. Additionally, six replications, each consisting of seven plants (four root tips per plant), were harvested from the same plants. Before cutting the root tips, the roots were washed in diethylpyrocarbonate (DEPC) twice to minimize the contamination with RNAses. Furthermore, total shoots for dry matter (dm) and P determination were harvested and dried for 3 days at 60°C (four replications of four shoots each). Eight complete roots per treatment were incubated for 1 h in 70% (v/v) EtOH and then stored in tap water at 4°C for later root hair length measurement.
Independently Replicated (2nd) Experiment
The independently replicated experiment was identical to the MACE experiment, but 30 root tips were harvested per replication (six plants of five root tips each) for the expression analysis. Furthermore, six replications of three shoots each (for dm and P determination) and 12 total roots (for root hair length measurement) were harvested per treatment.
Split-Root Experiment
Brassica carinata cv. Bale plants were germinated as before and then hydroponically grown under the conditions above and in the above nutrient solution containing 0.05 mM KH2PO4 for 5 days for the split-root experiment. The main root was then cut and 1 day later, the plants were transferred into a split-root system consisting of nutrient solutions with (+ +) or without (--) 1 mM Pi on both sides and without Pi only on one side and sufficient (1 mM) Pi on the other side (+ -). After the appearance of P deficiency symptoms at the shoot and a distinct increase of the root hair length in the [--] plants, root tips (three replications consisting of ten plants each, of each plant two root tips were taken for expression analysis) and 30 total roots (+ + and --) and root halves (+ -), respectively (for root hair length measurement), were harvested and treated as before. Additionally, three replications of 21 shoots each and three replications of five total roots each (+ + and --) and five root halves each (+ -), respectively, were harvested for dm and P determination.
Nutrient Specificity Experiment
Seeds of B. carinata cv. Bale and Bacho were germinated between filter papers under the same climate chamber conditions as before for the nutrient specificity experiment. The composition of the nutrient solutions was the same as before for the control and Pi deficiency treatment, but Ca(NO3)2 × 4 H2O was substituted by CaCl2 × 2 H2O in the N deficiency treatment and K2SO4 and KCl were omitted and KH2PO4 was substituted by NaH2PO4 × 2 H2O in the potassium (K) deficiency treatment. After 8 days, root tips (six replications of six plants each of five root tips each for expression analysis), shoots (six replications of three shoots each for dm and P determination) and 12 complete roots (for root hair length measurement) were harvested and treated as before.
CRISPR Experiments
Plant cultivation and screening for transgenic roots was done as described in Kirchner et al. (2017) for the CRISPR experiments. As many 2-cm root tips as possible were harvested from each transgenic root for expression analysis. The remaining transgenic roots were used for the verification of the gene editing.
Determination of the Nutrient Concentrations
An amount of 50 mg dm was dry-ashed overnight at 480°C, dissolved in 1 mL 1:3 diluted HNO3, diluted with 9 mL H2O and filtrated (Rotilabo®-round filters, type 15A) for the extraction of total P and K.
The Pi was extracted, according to Grunwald et al. (2009), using 10 mg (shoot) and 5 mg (root) dm.
Both total P and Pi concentrations were photometrically determined, according to Gericke and Kurmies (1952).
Shoot P and K concentration in the nutrient specificity experiment were determined by inductively coupled plasma – mass spectrometry (ICP-MS) (Agilent 7500 cx, Agilent Technologies, Santa Clara, CA, United States). Shoot N concentration was determined by an elemental analyzer (Vario EL III, Elementar, Langenselbold, Germany).
Root Hair Length Measurement
Roots were stained in 0.05% (w/v) toluidine blue for 1–2 h. Subsequently, two 2-cm root tips were cut per plant, put into tap water and examined by the MZ10 F (Leica Mikrosysteme Vertrieb GmbH, Wetzlar, Germany). Root tips were pulled over 0.3% (w/v) agar–agar, until the root hairs were straight. Pictures were taken from the root sections with fully elongated root hairs and the root hair length was measured. For the nutrient specificity and CRISPR experiments pictures were taken using the ‘SMZ25’ (Nikon, Düsseldorf, Germany) and root hair length was measured using the corresponding software ‘NIS-Elements’ instead. Root tips of the transgenic first and second order lateral roots were cut for the CRISPR experiments.
RNA Isolation
Frozen pool samples were homogenized by mortar and pestle. The RNA was then isolated using the NucleoSpin miRNA kit (MACHEREY-NAGEL GmbH & Co., KG, Düren, Germany), following the manufacturer’s instructions (using the large RNA isolation procedure).
The frozen samples of the biological replications were homogenized in a swing mill at 28 s-1 for 2 min (MM 400, Retsch GmbH, Haan, Germany). The RNA was isolated using the GENEzolTM reagent (Geneaid Biotech Ltd., New Taipei City, Taiwan) (MACE, 2nd experiment and split-root experiment) and the peqGOLD TriFastTM reagent (VWR International GmbH, Darmstadt) (nutrient specificity and CRISPR experiments), respectively, according to the manufacturer’s instructions.
The RNA Quality was electrophoretically tested on a 1% (w/v) non-denaturating agarose gel containing 0.004% (v/v) Midori Green Advance (Nippon Genetics Europe, Dueren, Germany) and a runtime of 35 min at 6.9–7.6 V cm-1 in Tris-acetate-EDTA (TAE) buffer. The RNA quantity was determined using the NanoPhotometer® P-Class P 300 (Implen, Munich, Germany).
Massive Analysis of cDNA Ends (MACE)
One isolated RNA pool sample with a total amount of 6–12.7 μg and a concentration between 280 and 550 ng μL-1 for each treatment was used for MACE (GenXPro, Frankfurt am Main, Germany, according to Kahl et al., 2012).
The tags of the MACE were annotated to two related plant species. The first annotation process was done by GenXPro in which the tags were annotated against the genome and transcriptome of Brassica rapa (Ensembl, annotation release 20, Kersey et al., 2016), whereby non-annotatable tags were assembled de novo using TrinityRNASeq (version: trinityrnaseq_r20140717) and CAP3 (version date: 12/21/07). Consensus sequences were generated against the reference sequences (B. rapa genome and transcriptome and de novo assembly) using NovoAlign (V3.00.05). The second annotation process was performed with CLC Genomics Workbench (version 7.5.1; CLC Bio, Aarhus, Denmark). Firstly, the tags were trimmed in two steps using a 50 and subsequently a 25 bp poly-A adapter as template. Simultaneously, tags were trimmed to a quality limit of a maximum of 0.05 and to contain less than two ambiguous bases. Tags shorter than 50 bp were discarded. The tags were then annotated against Brassica napus (Brassica napus gene index (BnGI) release 5) using the RNAseq tool (settings: similarity fraction = 0.8, length fraction = 0.8, mismatch cost = 2, insertion cost = 3, deletion cost = 3) and consensus sequences were generated from the annotated tags.
The complete MACE dataset is accessible in the sequence and read archive under the accession number SRP124702.
A Gene Ontology (GO) enrichment analysis was performed using the data from the Brassica rapa annotation. Significantly enriched GO terms were determined by Fisher’s exact test (p < 0.01). Redundant GO terms were removed using REVIGO (Supek et al., 2011), whereby ‘medium’ (0.7) was selected for the resulting list and Arabidopsis thaliana was chosen as database.
Candidate Gene Selection
The list of annotated genes was filtered automatically using the criteria twofold regulation in Bale (I), at least three normalized tags in Bale (II), a regulation in Bacho between 0.7 and 1.5 (III), and the number of normalized tags in the Pi deficiency treatment had to be at least 1.5-fold higher (upregulated genes) or lower (downregulated genes) in Bale than in Bacho (IV) (Supplementary Table 1). Only genes with a potential regulatory function were chosen from the resulting list for the subsequent analyses. Additionally, BcPIP5K1 was selected from the MACE dataset, because Wada et al. (2015) uncovered a role in the Pi deficiency-induced root hair elongation for its counterpart in Arabidopsis thaliana, AtPIP5K3.
The candidate gene sequences obtained by MACE and 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) were submitted to the NCBI database under the accession numbers listed in Supplementary Table 2.
Expression Analysis
The cDNA synthesis and qPCR were performed, as described in Kirchner et al. (2017). Gene-specific primer pairs were designed on the consensus sequence of each candidate gene obtained by MACE in regions with high similarity between the four treatments (Supplementary Table 3). LOTUS JAPONICUS ROOTHAIRLESS1-LIKE 3-LIKE (BcLRL3L) primers were designed to bind both BcLRL3L1 and 2, whereby there was one mismatch in the reverse primer for BcLRL3L2 at position 16. Primers for INDUCED BY Pi STARVATION 2 (AtIPS2) (Shin et al., 2006) were designed on the corresponding Arabidopsis sequence (AT5G03545).
Primers with an efficiency between 85 and 115% were used for expression analysis if the discrepancy in the efficiency of the two cv.s was 10% at the most.
UBIQUITIN CONJUGATING ENZYME 9 (AtUBC9; AT4G27960) was used as an endogenous control in all the experiments, because of its invariant expression under nutrient stress conditions (Czechowski et al., 2005). ELGONGATION FACTOR 1 ALPHA 1 (BcEF-1-a1) was used as an additional control in the nutrient specificity experiment, whereupon the geometric mean of both endogenous controls was calculated and compared to the target genes.
The annealing temperature (Ta) was consistently 60°C in all qPCR runs.
Statistics were performed according to Steibel et al. (2009), using the R package ‘qpcrmix’.
Knockout of the Candidate Genes by CRISPR/Cas9
Construct Preparation
The construct preparation including the guide RNA (gRNA) design was carried out, as described in Kirchner et al. (2017). Briefly, the Bale gene sequence of selected candidate genes obtained by MACE was extended by 5′ RLM-RACE. A list of the gene-specific primers used for the RACE can be found in Supplementary Table 4. Based on the MACE and RACE sequences, primers were designed (Supplementary Table 5) for amplification and sequencing of the Bale gDNA sequences of the candidate genes. The cDNA and gDNA sequences were aligned to identify introns and exons within the genes. Introns and exons of INORGANIC PHOSPHATE TRANSPORTER 1 (BcPHT1) were identified based on the annotated gDNA sequence from B. rapa instead. Then, two gRNAs were designed in an exon near the 5′ end of each gene with a distance of (bp) 69 (BcFLA1), 34 (RAPID ALKALINIZATION FACTOR-LIKE 1, BcRALFL1), 56 (BcPHT1), 26 (ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, BcROPGEF1), 43 (BcLRL3L1) and 66 (BcLRL3L2), and cloned into pB-CRISPR+35S::GFP. The primers used additionally for the construct preparation for the different genes are shown in Supplementary Table 6.
Hairy Root Transformation
Constructs were introduced into B. carinata cv. Bale by application of the hairy root technique according to Kirchner et al. (2017). In short, constructs were introduced into Agrobacterium tumefaciens C58 (ARqua1). Then, the bacterial solution was injected into the hypocotyl of 3-day-old Bale plants, thus inducing hairy roots with an active CRISPR/Cas9 construct.
Verification of the Gene Editing
Gene editing of BcFLA1 was verified by the amplification of the potentially edited gene region (Ta = 60°C) and its separation by agarose gel electrophoresis (primers ‘TTCAAACGCAATGGCAACCA’ and ‘ACCCAAATCAAACGACGAGTTTC’). The PCR for it was performed with the KAPA Hifi HotStart PCR Kit (KAPA Biosystems, Wilmington, MA, United States) together with the GC buffer supplied and a primer concentration of 0.3 μM, according to the manufacturer’s instructions. Resulting products were purified from the agarose gel and sequenced. Additionally, indels were detected by polyacrylamide gel electrophoresis (PAGE) (primers ‘GTAAAACGACGGCCAGTTTCAAACGCAATGGCAACCA’ and ‘TTCCGACTCGTGTTGTGTTT’), as described in Kirchner et al. (2017). The primers were designed to bind two different alleles of BcFLA1 (BcFLA1a and BcFLA1b), whereby there was one mismatch for BcFLA1b in the reverse primer used for the validation by PAGE (base position 9).
Genomic DNA Isolation
The genomic DNA (gDNA) was isolated using a modified method from Oppermann (2004). All centrifugation steps were performed at 15000 U min-1 (4°C). Frozen samples were homogenized similarly to the RNA isolation and resuspended in 1 mL buffer, containing 0.2 M Tris-HCl (pH 9), 0.4 M LiCl, 25 mM EDTA and 1 % (w/v) SDS. After the addition of 50 μL RNase A (10 mg mL-1), samples were incubated for 10 min at 60°C, followed by a centrifugation step for 10 min. An amount of 1 mL of the supernatant was mixed for 10 s with 900 mL phenol/chlorophorm/isoamyl alcohol mix (25:24:1) (Roth, Karlsruhe, Germany). Samples were cooled on ice for 5–10 min and centrifuged for 10 min. An amount of 900 μL of the supernatant were mixed for 10 s with 1 mL phenol mix (Roth, Karlsruhe, Germany) and centrifuged for 5 min. Then, 900 μL of the supernatant was put into 1 mL cold isopropanol and the resulting precipitate was pelleted by a centrifugation step of 10 min. The pellet was washed in 1 mL 80% (v/v) EtOH by a centrifugation of 20 min. After drying, the pellet was resuspended in 50 μL TE buffer (pH 9), containing 10 mM Tris and 1 mM EDTA. The gDNA quality and quantity was tested, as described for RNA.
The gDNA sequences were submitted to the NCBI database under the accession numbers listed in Supplementary Table 2.
Results
Characterization of the Plant Material Used for MACE Regarding the P Deficiency and Root Hair Response
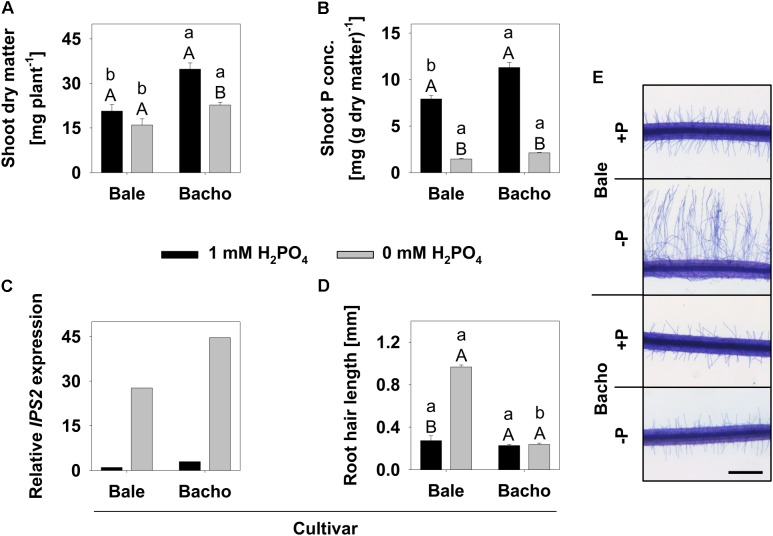
Shoot dm, shoot P concentration and expression of a marker gene for P deficiency, AtIPS2 (Shin et al., 2006), were determined to characterize the P deficiency in the 0 mM H2PO4 (-P) treatment. The -P treatment in Bacho resulted in a decrease of the shoot dm yield compared to the control (Figure 1A). In tendency, this effect also occurred in Bale. However, shoot P concentration in both cv.s was about 2 mg (g dm)-1 in the -P treatment and, thus, only one-fifth compared to the shoot P concentration in the 1 mM H2PO4 (+P) treatment (Figure 1B). Furthermore, the AtIPS2 expression was strongly enhanced during Pi starvation (Figure 1C). Bale exhibited a clear increase in the root hair length in the -P treatment compared to the +P treatment, whereas Bacho developed short root hairs under both P conditions (Figures 1D,E).
FIGURE 1.
Shoot dry matter (dm) (A), shoot P concentration (B), AtIPS2 expression (C), root hair length (D) and representative root hairs (E, scale = 0.5 mm) of B. carinata cultivar (cv.) Bale and cv. Bacho as affected by P supply in the 1st experiment (MACE experiment). Small letters denote significant differences at P < 0.05 between cv.s at the same P-level; capital letters between P treatments of the respective cv. (Tukey test). Columns represent means and bars indicate SE; n = 4 (A,B) and 8 (D) biological replications, whereas 3 technical replications of pooled samples were realized for (C).
Analysis of the MACE Data
Between 16 and 26 million tags were produced during the MACE (Supplementary Figure 1A). Around four million more tags were obtained in the 0 mM P treatment of both cv.s and six million more tags were produced in both treatments of Bacho compared to Bale. Around 70–80% of the total number of tags could be annotated to B. rapa (Supplementary Figure 1B), whereas only about 50–60% of the tags could be annotated to B. napus (Supplementary Figure 1C). About 10% more tags could be annotated in cv. Bacho compared to Bale in both annotations (Supplementary Figures 1B,C).
Bale and Bacho strongly differed in the significantly enriched GO terms (Supplementary Figure 2). Most of the GO terms significantly enriched in Bale were not significant in Bacho. Especially GO terms related to the root hair development were significantly enriched only in Bale. These categories were root (system) development, cell differentiation, cell wall (organization), Golgi apparatus and cell projection. In the GO categories cell projection, lysosome and negative regulation of cyclin-dependent protein serine/threonine kinase activity the percentage of significantly regulated genes was highest. Mostly more genes were downregulated than upregulated within the GO categories. In Bacho, downregulation was particularly pronounced.
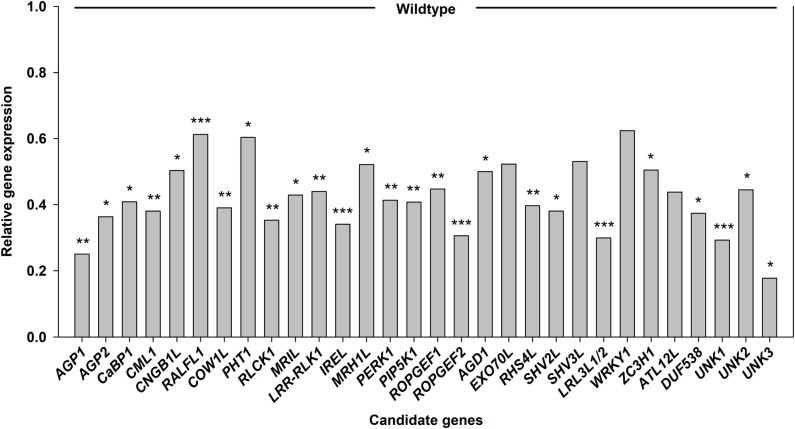
Criteria for the selection of candidate genes with a possible participation in the Pi deficiency-induced root hair elongation were, based on a representative number of tags: (i) significant up- or downregulation under Pi-deficient conditions compared to sufficient conditions in Bale, and (ii) no regulation in Bacho. (iii) Furthermore, the expression level in Bale should be clearly higher (upregulated genes) or lower (downregulated genes) compared to Bacho in the -P treatment. Filtering the dataset according to these criteria resulted in about 200–300 remaining genes in each filter process and annotation (Supplementary Table 1), from which 33 candidate genes with a potential regulatory role were selected encoding for arabinogalactan proteins, calcium-related proteins, proteins involved in P transport, protein kinases, GTPase-related proteins, transcription factors, zinc finger proteins, a protein phosphatase, proteins with unknown function and other proteins with a known root hair affecting role (Supplementary Table 7). The protein phosphatase was the only selected downregulated gene in Bale under Pi deficiency. Based on the results of the split-root and nutrient specificity experiment, five candidate genes were selected for the functional analysis by knockout and will be further described. Information on all genes is given in Supplementary Tables 7–11.
The five for the knockout selected candidate genes could be assigned to many of the significantly enriched GO terms, namely root (system) development and cell differentiation (BcLRL3L), cell wall organization (BcROPGEF1), ion transport and transporter activity (BcPHT1), extracellular region (BcRALFL1), plasma membrane (BcFLA1, BcPHT1 and BcROPGEF1), Golgi apparatus (BcFLA1) and cell wall (BcFLA1 and BcROPGEF1, Supplementary Figure 2). According to the MACE, most of the candidate genes selected for knockout exhibited a two- to three-times higher expression under deficient P conditions in Bale compared to sufficient P conditions, whereas BcPHT1 was even 15-fold upregulated (Figure 2). The expression of these genes in Bacho was nearly the same under both P conditions.
FIGURE 2.
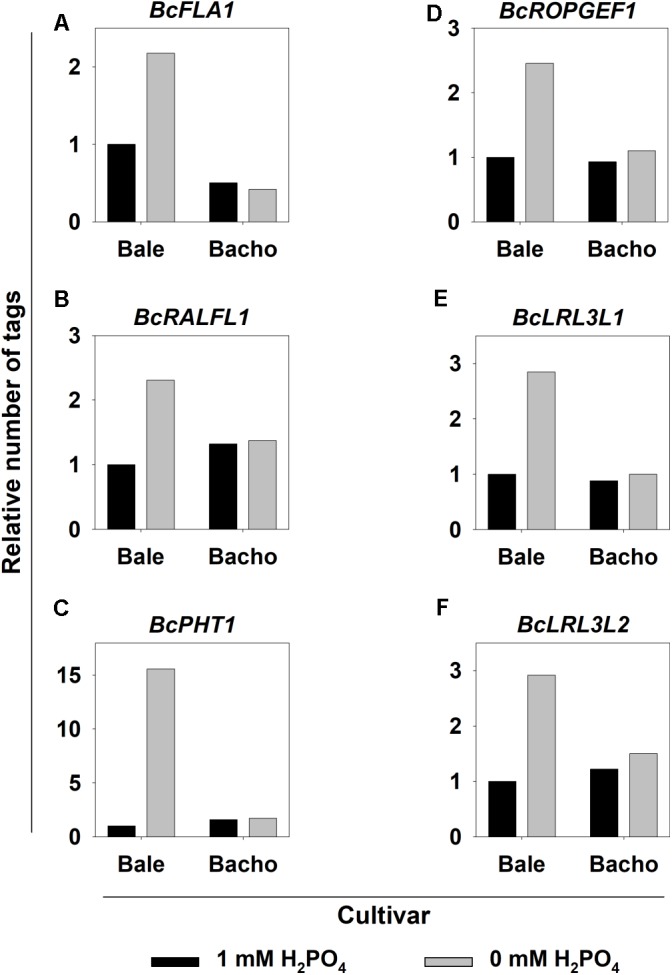
Relative number of normalized tags assigned to the selected candidate genes for increased root hair length under Pi deficiency (A–F) in B. carinata cv. Bale and cv. Bacho affected by P supply determined by MACE.
Validation of the Candidate Gene Regulation
In the next step, relative gene expression was tested by qPCR in three retained biological replicates originating from the same plant material utilized for the MACE (1st experiment) and in samples obtained from an independently conducted 2nd experiment with an equivalent design to the MACE experiment. For the replicated experiment, P deficiency was confirmed by decreased shoot dm and shoot P concentration (Supplementary Figures 3A,B), and root hair length was enhanced in the -P treatment in Bale similarly to the MACE experiment (Supplementary Figure 3C).
The up- or downregulated gene expression in Bale under P-deficient conditions was confirmed for 22 and 29 candidate genes in the samples from the MACE (1st) experiment and the 2nd experiment, respectively (Supplementary Tables 8, 9). Overall, the P deficiency effect on the gene regulation in Bale was stronger in the 2nd experiment. Every gene exhibited the expected regulation in Bale in at least one of both experiments. However, PHOSPHATE TRAFFIC FACILITATOR 1 (BcPHF1), DOMAIN OF UNKNOWN FUNCTION 620 (BcDUF620) and PROTEIN PHOSPHATASE 1 (BcPP1) were regulated in Bacho similar to Bale in both experiments, so that they were excluded from the following experiments.
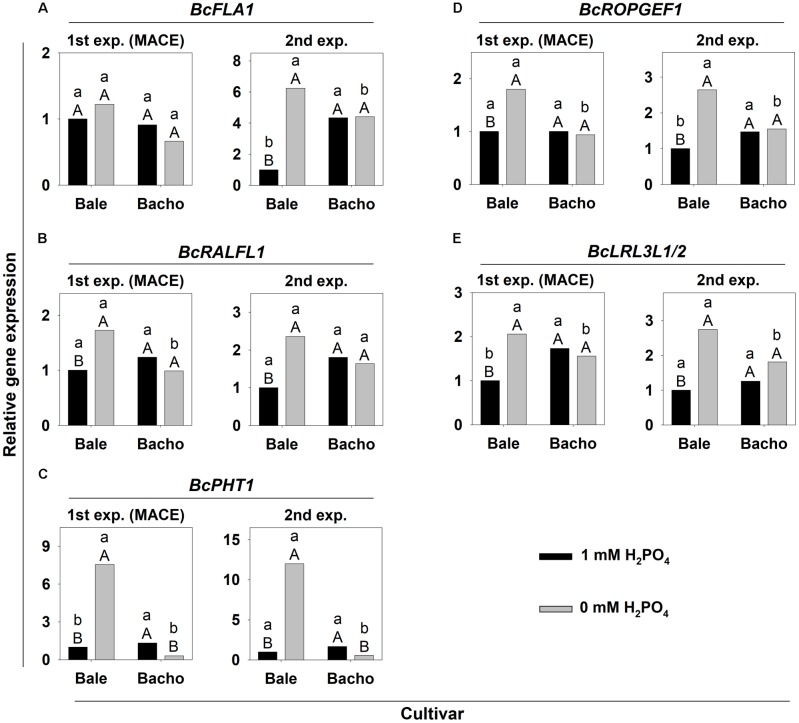
The upregulation of the five genes which were selected for knockout was confirmed in Bale under Pi deficiency (Figure 3). However, the upregulation of BcFLA1 was significant only in the 2nd experiment (Figure 3A). As predicted by MACE, BcPHT1 exhibited the strongest upregulation and, interestingly, a contrary regulation was observed in Bacho (Figure 3C).
FIGURE 3.
Relative expression of selected candidate genes for increased root hair length under Pi deficiency (A–E) in B. carinata cv. Bale and cv. Bacho affected by P supply determined by qPCR in samples obtained from the MACE (1st) experiment (exp.) and the independently conducted 2nd exp. AtUBC9 was used as an endogenous control. Significance was calculated according to Steibel et al. (2009). Small letters denote significant differences at P < 0.05 between cv.s at the same P-level; capital letters between P treatments of the respective cv. Columns represent means; n = 3.
Split-Root Experiment
Bale seedlings were grown in a split-root system in nutrient solutions with (+ +) or without (--) Pi on both sides and with Pi only on one side and without Pi on the other side (+ -) to test whether the candidate genes are involved more in local or systemic signaling pathways.
After 8 days in the split-root system, shoot growth of the [+ +] and [+ -] treatment was similar (Supplementary Figures 4A,B), but was clearly less in the [--] plants, which had small dark green leaves (Supplementary Figure 4C), typical P deficiency symptoms. Accordingly, the shoot dm yield of the [--] plants was about 50% lower than the other treatments (Figure 4A). However, the root dm yield was not significantly affected by any of the treatments (Figure 4B).
FIGURE 4.
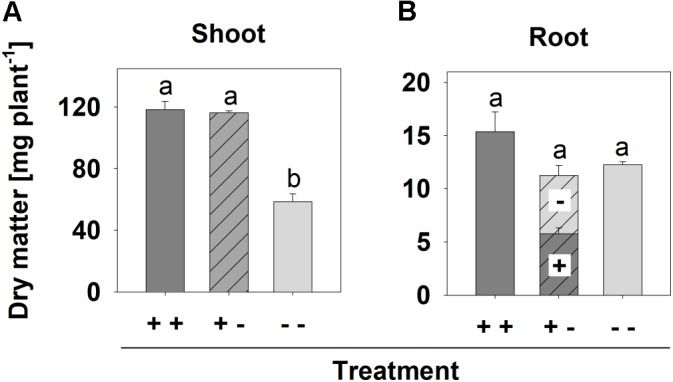
Shoot (A) and root (B) dm of B. carinata cv. Bale affected by P supply in a split-root system. Different letters denote significant differences at P < 0.05 (Tukey test). Columns represent means and bars indicate SE; n = 3.
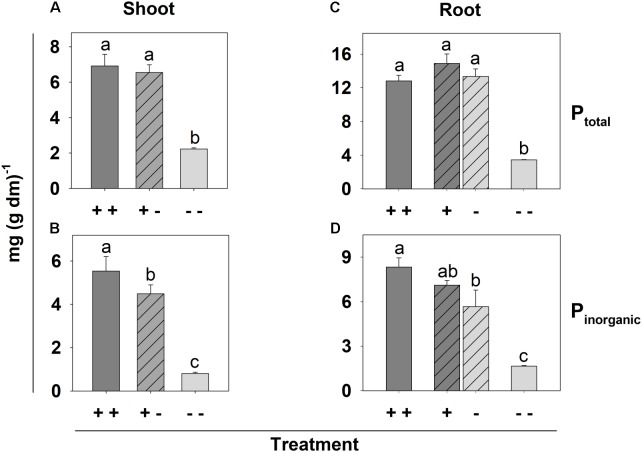
The similar shoot dm yield of the [+ -] and [+ +] plants indicated that the overall P status was sufficient in the [+ -] plants, which was in line with a high Ptotal concentration in shoot and root dm in both treatments (Figures 5A,B). By contrast, the Ptotal concentration in the shoot and root dm of the [--] plants was reduced to about 1/3. The Pinorganic concentration was reduced steepest in shoots and roots of the [--] plants (Figures 5C,D). However, the shoots of the [+ -] plants and the related root halves grown on the P deficient side ([-] roots) also exhibited a decrease of the Pi concentration compared to the [+ +] plants.
FIGURE 5.
Ptotal concentration in shoot (A) and root (B) as well as Pinorganic concentration in shoot (C) and root (D) of B. carinata cv. Bale affected by P supply in a split-root system. Different letters denote significant differences at P < 0.05 (Tukey test). Columns represent means and bars indicate SE; n = 3.
Root hair length of the [--] plants was enhanced by a factor of 2.5 compared to the [+ +] plants (Figure 6), whereas root hair length in the [+ -] treatment was increased by 30 % in the -P side, but not affected in the +P side compared to the [+ +] treatment.
FIGURE 6.
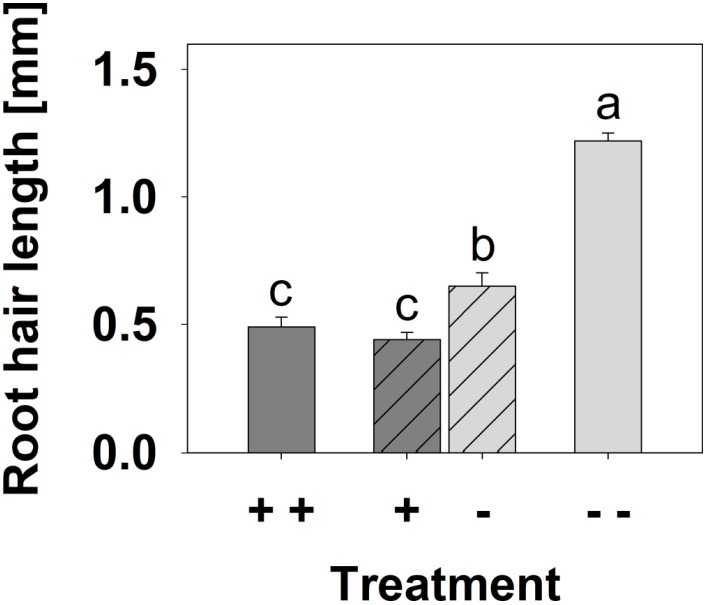
Root hair length of B. carinata cv. Bale affected by P supply in a split-root system. Different letters denote significant differences at P < 0.05 (Tukey test). Columns represent means and bars indicate SE; n = 30.
The upregulation of the candidate genes by Pi deficiency ([--] treatment) was confirmed (Supplementary Table 10 and Figure 7). Twenty-five of the 30 candidate genes in the [+ -] treatment were upregulated in the -P side. However, in most of the cases, this upregulation was not as high as in the [--] treatment. Three candidate genes, namely ARABINOGALACTAN PROTEIN 2 (BcAGP2), BcRALFL1 and SHAVEN3-LIKE (BcSHV3L), were not affected by the P supply in the split-root system (Supplementary Table 10 and Figure 7B). Conversely, a few genes, most of all CALCIUM-BINDING EF-HAND FAMILY PROTEIN 1 (BcCaBP1) and EXOCYST COMPLEX COMPONENT EXO70-LIKE (BcEXO70L), were nearly as strongly upregulated in the roots of the -P side as in the [--] treatment (Supplementary Table 10). BcPHT1 exhibited the strongest upregulation in the [--] treatment, whereas the upregulation in the split-root variant was only 2–4-fold in the [+] and [-] side, respectively (Figure 7C). The expression level of BcFLA1, BcROPGEF1 and BcLRL3L1/2 in the [-] side of the [+ -] treatment was higher than in the sufficiently supplied roots, but lower compared to the [--] treatment (Figures 7A,D,E).
FIGURE 7.
Relative expression of candidate genes for increased root hair length during Pi deficiency (A–E) in B. carinata cv. Bale affected by P supply in a split-root system. AtUBC9 was used as an endogenous control. Significance was calculated according to Steibel et al. (2009). Different letters denote significant differences at P < 0.05. Columns represent means; n = 3.
Investigation of the Nutrient Specificity of the Candidate Genes
Bale and Bacho were also grown in nutrient solutions lacking N and K, respectively, to check whether the successfully validated candidate genes are regulated specifically during root hair growth under Pi starvation.
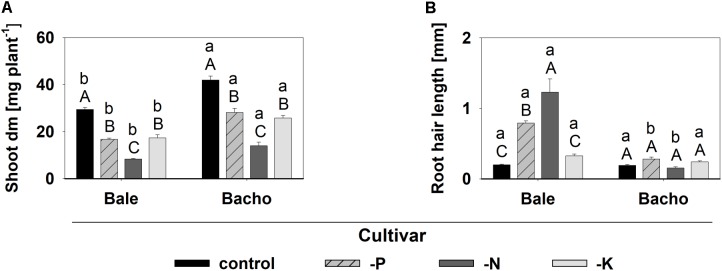
The lower shoot dm yield (Figure 8A) and nutrient concentration in the shoot dm (Supplementary Figure 5) in the deficiency variants compared to the control with sufficient nutrient supply indicated deficiency for all three nutrients in both Bale and Bacho. Root hair length was increased under P and N deficiency in Bale, but not in Bacho (Figure 8B and Supplementary Figure 6) as was expected from previous studies (Bremer, 2010). The N deficiency even led to a higher root hair length than P deficiency in Bale. The K deficiency did not affect the root hair length in either cv.
FIGURE 8.
Shoot dm (A) and root hair length (B) affected by nutrient deficiency. Significant differences at P < 0.05 are denoted by small letters between cv.s for the same nutrient treatment and capital letters between nutrient treatments for the same cv. (Tukey test); columns represent means and bars indicate SE; n = 6 (A) and n = 12 (B), respectively.
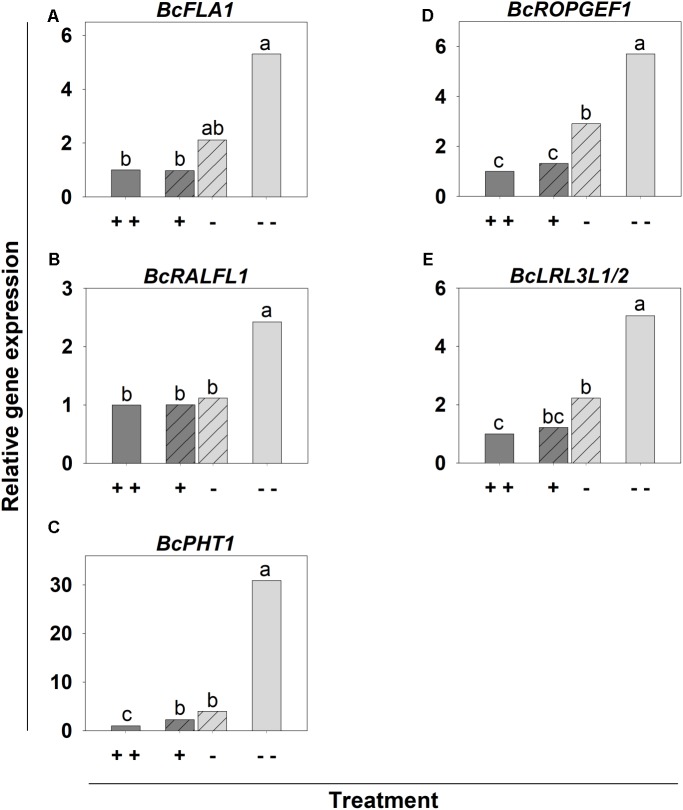
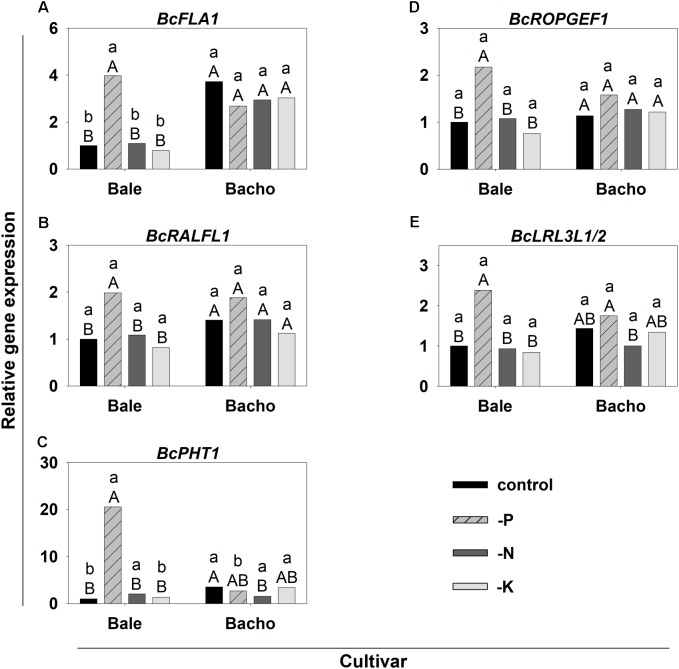
Most of the 30 candidate genes for Pi deficiency-induced root hair growth were upregulated in Bale by both Pi and N deficiency compared to sufficient conditions (Supplementary Table 11 and Figure 9). However, the increase of the expression in most of the cases was at least higher in tendency under Pi deficiency compared to N deficiency. Interestingly, four genes (BcFLA1, BcRALFL1, BcROPGEF1, and BcLRL3L1/2) were specifically upregulated by Pi deficiency (Figures 9A,B,D,E) and the upregulation of BcPHT1 under Pi deficiency was many times over that during N deficiency (Figure 9C). By contrast, K deficiency did not affect the expression of the candidate genes. No significant upregulation in Bacho was observed for any genes and nutrient deficiencies compared to sufficient conditions, except for an upregulation of the expression of CYCLIC NUCLEOTIDE-GATED CATION CHANNEL BETA-1-LIKE (CNGB1L) under N deficiency.
FIGURE 9.
Relative expression of candidate genes for increased root hair length during Pi deficiency (A–E) in B. carinata cv. Bale and cv. Bacho affected by nutrient deficiency determined by qPCR. AtUBC9 and BcEF-1-a1 were used as endogenous controls. Significance was calculated according to Steibel et al. (2009). Significant differences at P < 0.05 are denoted by small letters between cv.s for the same nutrient treatment and capital letters between nutrient treatments for the same cv. Columns represent means; n = 3.
Knockout of Candidate Genes by CRISPR/Cas9
According to the prior experiments, five candidate genes were selected for further functional investigation by knockout using CRISPR/Cas9. These included the four P-specific reacting genes BcFLA1, BcRALFL1, BcROPGEF1 and BcLRL3L1/2 (Figures 9A,B,D,E). Furthermore, BcPHT1 was selected because of its very strong upregulation under Pi deficiency in Bale, while it was downregulated in Bacho (Figure 3C).
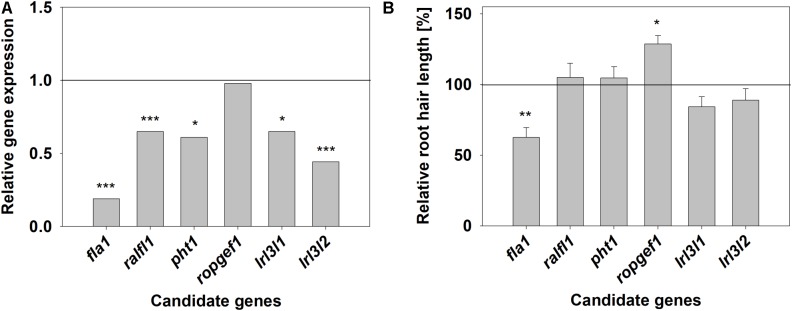
The gene expression of all candidate genes, except for BcROPGEF1, was reduced in the transgenic CRISPR roots compared to the control (Figure 10A). Expression of BcFLA1 was reduced to 20% and BcLRL3L2 expression to 45%. BcRALFL1, BcPHT1 and BcLRL3L1 expression was reduced to about 60%. The reduction in case of BcFLA1 (Supplementary Figure 7A), BcRALFL1 (Supplementary Figure 7B) and both BcLRL3L isoforms (Supplementary Figures 7E,F) was relatively homogenous in the different transgenic roots, whereas the expression of the other genes exhibited strong fluctuations (Supplementary Figures 7C,D).
FIGURE 10.
Relative candidate gene expression (A) and relative root hair length of the transgenic hairy roots (B) after targeting the respective candidate genes for increased root hair length under Pi deficiency by CRISPR/Cas9. Horizontal lines represent the average wildtype expression and root hair length, respectively. Stars denote significant differences, determined according to Steibel et al. (2009) (A) and Tukey test (B) (significance codes: 0 ‘∗∗∗’ 0.001 ‘∗∗’ 0.01 ‘∗’ 0.05). Columns represent means for n = 4–8 (A) and 5–14 (B), respectively. Bars in (B) indicate SE.
The root hair length was reduced only in the bcfla1 roots (to about 60% compared to the control) (Figure 10B and Supplementary Figure 8). By contrast, the bcropgef1 roots exhibited about 30% longer root hairs.
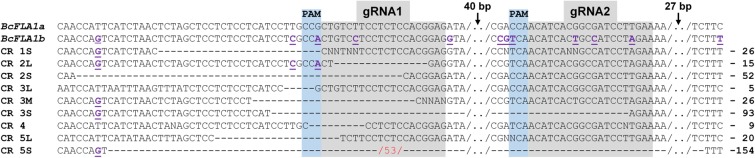
Since the strong downregulation of BcFLA1 was accompanied by a reduction in the root hair length, we took a closer look at the gene-editing events caused by CRISPR/Cas9. Sanger sequencing detected deletions in BcFLA1 in all replications ranging from 5 to 154 bp (Figure 11). Both alleles of BcFLA1 were affected. Up to three different deletions were observed per replication (transgenic root). However, only two deletions (CR 3S and CR 5S) spanned both gRNA regions. All other deletions occurred exclusively in the gRNA1 region. Furthermore, a 53° bp insertion occurred in CR 5S, the replication with the largest deletion of 154 bp. There was often an overlapping of at least two sequences in the sequencing chromatograms starting at gRNA1, which indicates that several amplicons with different gene-editing events were purified at once.
FIGURE 11.
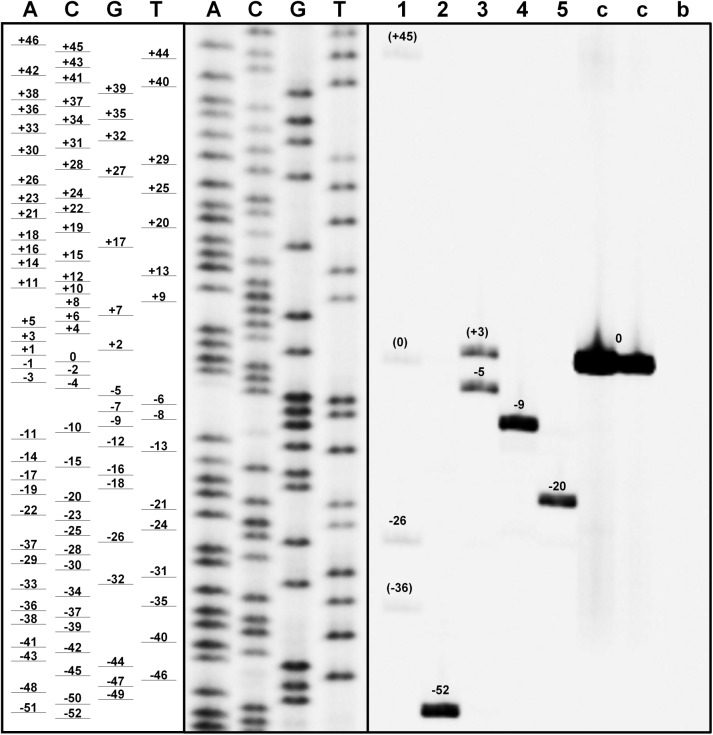
CRISPR/Cas9-induced deletions and insertions in BcFLA1. gRNA regions (gray), PAM sequence (blue), number of bp inserted (red) and number of bp deleted (on the right). Numbers on the left indicate the respective transgenic roots, whereby ‘L’ indicates a long fragment (small deletion), ‘M’ a medium fragment and ‘S’ a small fragment (large deletion). Nucleotides used for differentiation of BcFLA1b from BcFLA1a (violet, underlined). CR = CRISPR, /../ = break.
The fragment analysis by PAGE uncovered indels ranging from -52 to +45 bp (Figure 12). Three replications (2, 4, and 5) exhibited only one gene-editing event, whereas one insertion and one deletion occurred in replication 3. Replication 1 even exhibited two deletions and one insertion, whereby there was some of the wildtype sequence left. The intensity of the amplicon bands and therefore, the frequency of the gene-editing events was similar within the replications.
FIGURE 12.
Validation of the CRISPR/Cas9-induced gene-editing of BcFLA1 by fragment analysis by PAGE. A, C, G and T = Sanger sequencing reactions with the corresponding dideoxynucleotides; c, control; b, blank (no template control). Lane numbers indicate the respective transgenic roots. Numbers close to the bands indicate the bp deleted (–) and inserted (+) compared to the wildtype size (0). Brackets indicate indels with approximated size, which did not occur during sequencing.
The expression of the other candidate genes was analyzed in the bcfla1 roots to investigate where BcFLA1 may be working in the signal transduction cascade leading to the Pi deficiency-induced root hair elongation. Most of them exhibited a clear decrease in their expression (Figure 13), whereby the deepest reduction occurred for UNKNOWN 3 (BcUNK3) (to a level below 20%).
FIGURE 13.
Relative expression of candidate genes for Pi deficiency-induced root hair growth in bcfla1 roots grown under Pi-deficient conditions. AtUBC9 was used as an endogenous control. Significance was calculated according to Steibel et al. (2009) (significance codes: 0 ‘∗∗∗’ 0.001 ‘∗∗’ 0.01 ‘∗’ 0.05). Columns represent means; n = 3.
Discussion
Identification of Regulatory Candidate Genes for Pi Deficiency-Induced Root Hair Elongation
The aim of the present study was to identify candidate genes for the Pi deficiency-induced root hair elongation in B. carinata. Consequently, we used two different cv. of B. carinata differing in their root hair length under Pi starvation. cv. Bale develops long root hairs in response to Pi deficiency, whereas cv. Bacho develops short root hairs under both sufficient and deficient Pi conditions (Eticha and Schenk, 2001). Genome-wide expression profiles of Bale and Bacho under low (0 mM) and high (1 mM) Pi conditions were received by MACE. Therefore, RNA was extracted from 2-cm root tips, because this length ensures the receipt of root hairs with different developmental stages from the initiation to the termination of the root hair growth, so that the genes participating in the root hair elongation should also be expressed in the harvested plant material. The reduced shoot dm and shoot P concentration in both Bale and Bacho and the upregulation of the marker gene expression (Figures 1A–C) confirmed P starvation in the respective treatment of the MACE experiment. Additionally, longer root hairs were induced exclusively in Bale and not in Bacho under Pi deficiency (Figure 1D), as was expected from preliminary studies (Eticha and Schenk, 2001; Bremer, 2010), so that the plant material was defined as suitable for the subsequent MACE.
Since no database was available for B. carinata, the tags produced in the MACE were annotated to B. rapa and B. napus, which are, as B. carinata, a member of the Triangle of U (Li et al., 2017). The latter predicts a higher similarity between B. carinata and B. napus. However, more tags could be annotated to B. rapa than B. napus (Supplementary Figure 1). This may be due to the different annotation methods and the degree of completeness of the databases. Nevertheless, we used both annotations to have a greater basis for the candidate gene selection.
According to the root hair response, we selected genes exhibiting a clear down- or upregulation under Pi deficiency in Bale and no regulation in Bacho. Furthermore, only genes with a potential regulatory function were chosen, because we were interested in genes regulating the root hair elongation according to the Pi availability rather than genes involved in the general root hair elongation process. According to their function or structure, the 33 candidate genes were assigned to nine groups. Finally, five candidate genes were selected for functional analysis by knockout: (i) Since we were most interested in regulatory genes transmitting the signal of low P to the general root hair elongation pathway, we chose the four P-specific reacting candidate genes BcFLA1, BcRALFL1, BcROPGEF1 and BcLRL3L1/2 (Figures 9A,B,D,E). (ii) BcPHT1 was selected because of its very strong upregulation under Pi deficiency in Bale and the contrary regulation in Bacho (Figure 3C), which may indicate a role in P-sensing analogous to nitrate-sensing by nitrate transporter NRT1.1 (Krouk et al., 2010). A more detailed description of these genes can be found in Supplementary Text 1.
There was a good overall agreement between the regulation of the candidate genes observed in the MACE and in the qPCR (Supplementary Tables 7, 8). Even stronger upregulations were observed in the 2nd experiment, so that it actually correlated better to the MACE than the biological replications from the MACE (1st) experiment itself (Figure 3 and Supplementary Table 9). Furthermore, the regulation of the candidate genes was confirmed in the split-root and nutrient specificity experiments (Supplementary Tables 10, 11).
In addition to the candidate genes which were selected after filtering the MACE data, BcPIP5K1 was investigated, because its counterpart in A. thaliana (AtPIP5K3) was predicted to have a key role in increasing the root hair elongation response to Pi starvation, at least at young seedling stages. However, in this study, BcPIP5K1 was upregulated by Pi deficiency both in Bale and Bacho (Supplementary Table 8), indicating that it has another or an additional role in B. carinata or, at least, in Bacho.
Investigation of the Candidate Gene Characteristics Regarding Their Role in Signaling and Nutrient Specificity
Genes, whose regulation according to MACE was confirmed in at least one of the two independent experiments, were further investigated regarding their role in signaling and nutrient specificity.
Bale seedlings were grown in a split-root system containing sufficient (+ +) or deficient (--) Pi on both sides or sufficient Pi only on one and deficient Pi on the other side (+ -) to investigate whether the effect of Pi deficiency on root hair growth in B. carinata cv. Bale is mediated locally or systemically. Reduced shoot dm yield and low shoot Ptotal conc. indicated strong Pi deficiency in the [--] treatment (Figures 4A, 5A), accompanied by clear Pi deficiency symptoms on these plants (Supplementary Figure 4). However, shoot dm yield, Ptotal concentration and phenotype in the [+ -] treatment were similar to the [+ +] treatment, indicating that the Pi uptake on the +P side was enough for an overall sufficient P status of the plants. In addition to the Ptotal concentrations, we measured the Pinorganic concentrations in the shoots and roots, because Pi itself could serve as signaling molecule in transmitting the low Pi signal (Chiou and Lin, 2011). In contrast to Ptotal, Pinorganic also decreased in the shoots and in the -P side roots of the [+ -] treatment compared to the [+ +] treatment (Figures 5C,D). Pinorganic represents the storage form contained in the vacuoles (Bieleski, 1973). In this storage pool, a lower P supply in the [+ -] treatment compared to the [+ +] treatment was reflected. However, the root halves of the [+ -] treatment were not different, suggesting that the slightly enhanced root hair length (Figure 6) and increased gene expression in the -P side roots of this treatment were not caused by Pinorganic, but local Pi deficiency in the medium. This fits with the results of Thibaud et al. (2010), who concluded that local P deficiency responses are mediated by the external Pi. However, systemic signaling also seems to have an influence, because the root hair length of the roots from the -P side was not as strongly increased as in the [- -] treatment (Figure 6). The assumption that both local and systemic signaling play a role in the Pi deficiency-induced root hair elongation was supported by the expression level of most of the candidate genes in the root tips from the -P side of the split-root variant, which was between the level in the root tips grown in the sufficient P solutions and that of the [--] treatment (Supplementary Table 10). However, three candidate genes, among them BcRALFL1 (Figure 7B), did not exhibit any local P response in the split-root variant, so they may be involved exclusively in systemic signaling pathways. Alternatively, two genes (BcCaBP1 and BcEXO70L) exhibited a strong local P deficiency response, which was nearly as high as in the [--] treatment, thus, they may particularly play a role in local signaling pathways (Supplementary Table 10). Therefore, both local and systemic signaling may play a role in the Pi deficiency-induced root hair elongation in B. carinata.
Bale and Bacho were cultivated additionally under N- and K deficiency to uncover candidate genes which are specifically reacting in response to Pi deficiency. After validation of the nutrient deficiencies by a reduced shoot dm (Figure 8A) and nutrient concentration (Supplementary Figure 5), the expression of the candidate genes was determined (Supplementary Table 11). Interestingly, none of the candidate genes reacted in response to K deficiency, which was the only deficiency that did not lead to an enhanced root hair length (Figure 8B). This strongly indicates that the candidate genes are indeed directly or indirectly involved in the root hair elongation and not in general nutrient deficiency pathways. However, most of the candidate genes were upregulated by both P and N deficiency in Bale (Supplementary Table 11), which indicates that these genes are more involved in the general root hair elongation pathway. By contrast, the four candidate genes specifically upregulated under Pi deficiency: BcFLA1, BcRALFL1, BcROPGEF1 and BcLRL3L1/2 (Figures 9A,B,D,E) may play a role in the regulation of the root hair elongation pathway dependent on the Pi status. The FLAs have not been associated with Pi deficiency so far. However, it has been shown in several studies that they have the ability to counteract abiotic stress by altering gene expression (Zang et al., 2015).
Functional Characterization of the Candidate Genes by Knockout via CRISPR/Cas9
The most promising candidate genes were targeted by CRISPR/Cas9 to cause a knockout of the gene function and the effects on the root hair length under Pi deficiency were investigated. Out of the targeted genes, downregulation of BcFLA1 led to a significantly reduced root hair length (Figure 10), which is a strong evidence for a role of BcFLA1 in the Pi deficiency-induced root hair elongation in B. carinata.
Since the number of transcripts derived from a functional gene may be overestimated by qPCR, because of an ineffective nonsense-mediated mRNA decay in the case of an absent frameshift mutation, we further investigated the CRISPR/Cas9-induced gene-editing events in BcFLA1 by Sanger sequencing and fragment analysis by PAGE. In most of the cases, relatively large deletions occurred in both alleles of BcFLA1, indicating that the knockout of the gene function was successful (Figure 11). However, not all gene editing events could be separated by agarose gel electrophoresis. Therefore, a PAGE was performed to separate the amplicons of the potentially edited gene region with a higher resolution and to estimate their frequency. Indeed, if the loading of the gel was sufficiently high, some additional bands became visible on the polyacrylamide gel compared to Sanger sequencing (Figure 12). Hence, we were able to separate all amplicons and determine the indel sizes more precisely. Furthermore, the intensity of the amplicon bands allowed the estimation of the quantity of every indel. Even if the bands of the first replication were weak due to a low amount of DNA, four bands of similar intensity were observed from which one may represent the wildtype sequence. By contrast, the wildtype sequence in the other replications was completely absent. Some of the indels may not have induced a frameshift mutation, but it can be assumed that the gene function was knocked out due to the large size of most of the indels. This fits with the strong and relatively homogenous downregulation of the BcFLA1 gene expression measured by qPCR (Figure 10A and Supplementary Figure 7A), since a possible self-stimulation of the BcFLA1 expression may be impeded due to a defective BcFLA1 protein. Additionally, as the small indels around gRNA1 affected the N-terminal signal peptide of BcFLA1 (Supplementary Table 12), this may have caused a mislocalization of BcFLA1, so that it could not perform its normal function in the cell wall and/or the extracellular matrix.
The decreased transcript level of BcFLA1 was accompanied by a reduced root hair length of the bcfla1 roots (Figure 10B and Supplementary Figure 8A). However, even if the specificity of the CRISPR/Cas9 mechanism is relatively high in plants (Feng et al., 2014; Zhang et al., 2014; Jacobs et al., 2015), off-target effects cannot be excluded, especially if non-model plants without the full genome information are used, as in our study. Therefore, we developed a complementation variant with an additional overexpressed version of BcFLA1, which was resistant to the CRISPR/Cas9 mechanism in combination with the two gRNAs used in this study (for details, see Kirchner et al., 2017). The complementation of the BcFLA1 gene function led to a significant increase of the root hair length compared to the CRISPR variant, thus, it was confirmed that the shorter root hairs of the transgenic CRISPR roots arose from the downregulation of BcFLA1.
In the last step, we wanted to test whether the downregulation of BcFLA1 had an influence on the expression of the other candidate genes. Unexpectedly, nearly all candidate genes were strongly downregulated in the bcfla1 root tips (Figure 13). This may indicate that BcFLA1 indeed has a role at the beginning of the signal transduction cascade leading to a higher root hair length under Pi deficiency. However, this result can be falsified, because the number of root hairs may also be affected in bcfla1. A lower number of root hairs would automatically lead to an overall reduced expression of root hair-related genes in the sample. Therefore, the number of root hairs should be taken into account in further analyses to confirm this result.
Considering all results, we suggest a regulating role for BcFLA1 in the Pi deficiency-induced root hair elongation. The specific upregulation of BcFLA1 in response to Pi deficiency and the effect of its altered expression on the root hair length indicates that BcFLA1 acts in a P-specific pathway which may translocate the low Pi signal to the root hair elongation pathway. This P-specific pathway may merge with a pathway translocating environmental cues to a putative root hair-specific transcription factor which senses both developmental and environmental signals and controls downstream root hair genes, thus, leading to a balanced root hair elongation (Hwang et al., 2016).
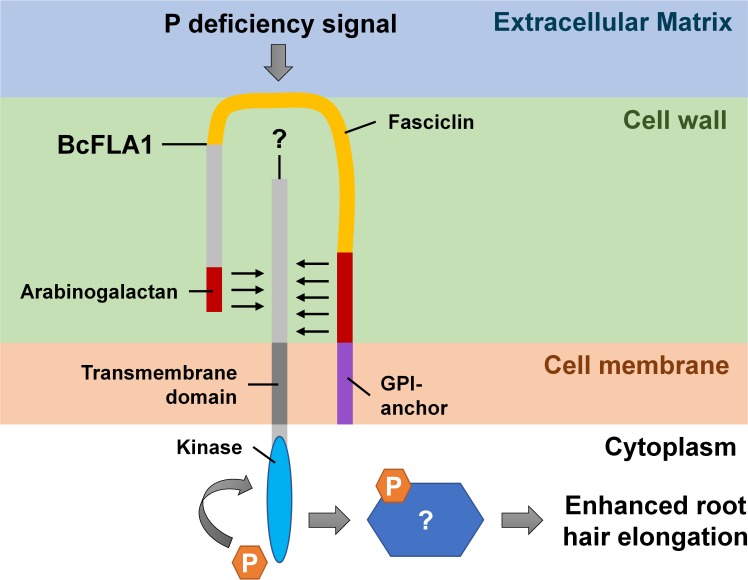
Another open question is the functional mechanism of the putative signal perception and transduction by BcFLA1. The FLAs can have a key function in transmitting environmental changes to the cell, as they connect the cell with neighboring cells or the extracellular matrix physically (Faik et al., 2006). Via their fasciclin (FAS) domains, they may mediate protein-protein interactions by facilitating the adhesion of plasma membrane or cell wall-associated ligands (Tan et al., 2012). A topology prediction of BcFLA1 revealed that the C-terminal end of BcFLA1 is embedded in the plasma membrane via a GPI-anchor and that the rest of the protein is outside the cell (Figure 14). Since the protein consists of two AG-rich regions surrounding the FAS domain (Supplementary Table 12), it could be speculated that the first AG-rich region of BcFLA1 passes through the cell wall, the FAS domain is located on the surface of the cell wall with contact to the extracellular matrix and the second AG-rich region is again located in the cell wall (Figure 14). Low Pi signal perception could, therefore, be mediated by the interaction of the FAS domain with signal molecules in the extracellular matrix. The signal may then be transmitted to a putative cell wall protein kinase, whose extracellular domain may be connected to the AG regions of BcFLA1 within the cell wall, which was proposed in a model regarding another FLA protein in Arabidopsis (Basu et al., 2016). This may, in turn, lead to autophosphorylation of the cytosolic kinase domain of the putative protein kinase, leading to downstream signaling events for the Pi deficiency-induced root hair elongation. Furthermore, the GPI anchor could be cleaved from BcFLA1 so that it is released from the plasma membrane, possibly leading to further signaling events. Interestingly, a role in elongation processes was documented for several FLAs, for example, the involvement of GhFLA1 in the initiation and elongation of cotton fibers (Huang et al., 2013) and AtFLA4, which maintains cell expansion during salt stress (Tan et al., 2012). Furthermore, another FLA was preferentially expressed in root hairs of Arabidopsis and was upregulated at the protein level in response to Pi starvation (Salazar-Henao and Schmidt, 2016), which may indicate the involvement of several FLAs in the Pi deficiency-induced root hair growth.
FIGURE 14.
Model of the potential position and working mechanism of BcFLA1.
Taken together, the results of this study in combination with the proposed functional abilities of fasciclin-like AGPs strongly indicate a role of BcFLA1 in the Pi deficiency-induced root hair elongation.
Author Contributions
TK and MS planned the experiments. TK, MN, KR, and TL conducted the experiments. MH provided the CRISPR vector. TK performed the data analysis, visualization, and writing the manuscript. MS and HK supervised and revised the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Thomas Debener, Marcus Linde, and Ina Menz, Department of Molecular Plant Breeding of the Institute of Plant Genetics, for their support with the PAGE.
Footnotes
Funding. This work was funded by the German Research Foundation (DFG, http://www.dfg.de) as part of the Research Training Group (GRK1798) “Signaling at the Plant-Soil Interface.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01372/full#supplementary-material
References
- Basu D., Tian L., Debrosse T., Poirier E., Emch K., Herock H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-Protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. 10.1371/journal.pone.0145092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski R. L. (1973). Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 24 225–252. [Google Scholar]
- Bremer M. (2010). Molecular Mechanisms of Root Hair Growth Induced by Pi Deficiency in Brassica carinata. Doctoral thesis, Gottfried Wilhelm Leibniz Universität Hannover, Hannover. [Google Scholar]
- Bruex A., Kainkaryam R. M., Wieckowski Y., Kang Y. H., Bernhardt C., Xia Y., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8:e1002446. 10.1371/journal.pgen.1002446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrika N. N. P., Sundaravelpandian K., Yu S. M., Schmidt W. (2013). ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol. 198 709–720. 10.1111/nph.12194 [DOI] [PubMed] [Google Scholar]
- Chiou T. J., Lin S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62 185–206. 10.1146/annurev-arplant-042110-103849 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Kim C. M., Pernas M., Pires N. D., Proust H., Tam T., et al. (2011). Root hairs: development, growth and evolution at the plant-soil interface. Plant Soil 346 1–14. 10.1007/s11104-011-0845-4 22730024 [DOI] [Google Scholar]
- Datta S., Prescott H., Dolan L. (2015). Intensity of a pulse of RSL4 transcription factor synthesis determines Arabidopsis root hair cell size. Nat. Plants 1:15138. 10.1038/nplants.2015.138 [DOI] [PubMed] [Google Scholar]
- Duan Q., Kita D., Li C., Cheung A. Y., Wu H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107 17821–17826. 10.1073/pnas.1005366107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eticha D., Schenk M. K. (2001). “Phosphorus efficiency of cabbage varieties,” in Plant Nutrition – Food Security and Sustainability of Agro-Ecosystems Through Basic and Applied Research, eds Horst W. J., Schenk M. K., Bürkert A., Claasen N., Flessa H., Frommer W. B., et al. (Netherlands: Kluwer Academic Publishers; ), 542–543. [Google Scholar]
- Faik A., Abouzouhair J., Sarhan F. (2006). Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analyses. Mol. Genet. Genomics 276 478–494. 10.1007/s00438-006-0159-z [DOI] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 4632–4637. 10.1073/pnas.1400822111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke S., Kurmies B. (1952). Die kolorimetrische phosphorsäurebestimmung mit ammonium-vanadat-molybdat und ihre anwendung in der pflanzenanalyse. J. Plant Nutr. Soil Sci. 59 235–247. [Google Scholar]
- Grunwald U., Guo W., Fischer K., Isayenkov S., Ludwig-Müller J., Hause B., et al. (2009). Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 229 1023–1034. 10.1007/s00425-008-0877-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. Q., Gong S. Y., Xu W. L., Li W., Li P., Zhang C. J., et al. (2013). A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol. 161 1278–1290. 10.1104/pp.112.203760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y., Lee H., Lee Y. S., Cho H. T. (2016). Cell wall-associated ROOT HAIR SPECIFIC 10, a proline-rich receptor-like kinase, is a negative modulator of Arabidopsis root hair growth. J. Exp. Bot. 67 2007–2022. 10.1093/jxb/erw031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T. B., LaFayette P. R., Schmitz R. J., Parrott W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. 10.1186/s12896-015-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungk A. (2001). Root hairs and the acquisition of plant nutrients from soil. J. Plant Nutr. Soil Sci. 164 121–129. [Google Scholar]
- Kahl G., Molina C., Rotter B., Jüngling R., Frank A., Krezdorn N., et al. (2012). Reduced representation sequencing of plant stress transcriptomes. J. Plant Biochem. Biotechnol. 21 119–127. 10.1007/s13562-012-0129-y [DOI] [Google Scholar]
- Kersey P. J., Allen J. E., Armean I., Boddu S., Bolt B. J., Carvalho-Silva D., et al. (2016). Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44 D574–D580. 10.1093/nar/gkv1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner T. W., Niehaus M., Debener T., Schenk M. K., Herde M. (2017). Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PLoS One 12:e0185429. 10.1371/journal.pone.0185429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Lacombe B., Bielach A., Perrine-Walker F., Malinska K., Mounier E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18 927–937. 10.1016/j.devcel.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Lan P., Li W., Schmidt W. (2013). Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genomics 14:210. 10.1186/1471-2164-14-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang S., Li F., Zhang S., Zhang H., Wang X., et al. (2017). A phylogenetic analysis of chloroplast genomes elucidates the relationships of the six economically important Brassica species comprising the Triangle of U. Front. Plant Sci. 8:111. 10.3389/fpls.2017.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. D., Liao Y. Y., Yang T. J., Pan C. Y., Buckhout T. J., Schmidt W. (2011). Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 155 1383–1402. 10.1104/pp.110.166520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann E. (2004). Funktionelle Diversifizierung in Auxin-Gesteuerten Wurzelfunktionen Bei Einer Tandem-Duplikation in Der Genfamilie Der Phospholipase A Von Arabidopsis Thaliana. Doctoral thesis, Gottfried Wilhelm Leibniz Universität Hannover, Hannover. [Google Scholar]
- Salazar-Henao J. E., Schmidt W. (2016). An inventory of nutrient-responsive genes in Arabidopsis root hairs. Front. Plant Sci. 7:237. 10.3389/fpls.2016.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Henao J. E., Velez-Bermudez I. C., Schmidt W. (2016). The regulation and plasticity of root hair patterning and morphogenesis. Development 143 1848–1858. 10.1242/dev.132845 [DOI] [PubMed] [Google Scholar]
- Shin H., Shin H. S., Chen R., Harrison M. J. (2006). Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 45 712–726. 10.1111/j.1365-313X.2005.02629.x [DOI] [PubMed] [Google Scholar]
- Song L., Yu H., Dong J., Che X., Jiao Y., Liu D. (2016). The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 12:e1006194. 10.1371/journal.pgen.1006194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steibel J. P., Poletto R., Coussens P. M., Rosa G. J. M. (2009). A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94 146–152. 10.1016/j.ygeno.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Showalter A. M., Egelund J., Hernandez-Sanchez A., Doblin M. S., Bacic A. (2012). Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3:140. 10.3389/fpls.2012.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud M. C., Arrighi J. F., Bayle V., Chiarenza S., Creff A., Bustos R., et al. (2010). Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64 775–789. 10.1111/j.1365-313X.2010.04375.x [DOI] [PubMed] [Google Scholar]
- Vance C. P., Uhde-Stone C., Allan D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157 423–447. 10.1046/j.1469-8137.2003.00695.x [DOI] [PubMed] [Google Scholar]
- Vijayakumar P., Datta S., Dolan L. (2016). ROOT HAIR DEFECTIVE SIX-LIKE4 (RSL4) promotes root hair elongation by transcriptionally regulating the expression of genes required for cell growth. New Phytol. 212 944–953. 10.1111/nph.14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y., Kusano H., Tsuge T., Aoyama T. (2015). Phosphatidylinositol phosphate 5-kinase genes respond to phosphate deficiency for root hair elongation in Arabidopsis thaliana. Plant J. 81 426–437. 10.1111/tpj.12741 [DOI] [PubMed] [Google Scholar]
- Yi K., Menand B., Bell E., Dolan L. (2010). A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat. Genet. 42 264–267. 10.1038/ng.529 [DOI] [PubMed] [Google Scholar]
- Zang L., Zheng T., Chu Y., Ding C., Zhang W., Huang Q., et al. (2015). Genome-wide analysis of the fasciclin-like arabinogalactan protein gene family reveals differential expression patterns, localization, and salt stress response in Populus. Front. Plant Sci. 6:1140. 10.3389/fpls.2015.01140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.