Abstract
The human oral mucosa hosts a diverse microbiome and is exposed to potentially toxic biomaterials from dental restoratives. Mucosal health is partly determined by cell and tissue responses to challenges such as dental materials and pathogenic bacteria. An in vitro model to rapidly determine potential layer-specific responses would lead to a better understanding of mucosal homeostasis and pathology. Therefore, this study aimed to develop a co-cultured microfluidic mucosal model on-a-chip to rapidly assess mucosal remodeling and the responses of epithelial and subepithelial layers to challenges typically found in the oral environment. A gingival fibroblast-laden collagen hydrogel was assembled in the central channel of a three-channel microfluidic chamber with interconnecting pores, followed by a keratinocyte layer attached to the collagen exposed in the pores. This configuration produced apical and subepithelial side channels capable of sustaining flow. Keratinocyte, fibroblast, and collagen densities were optimized to create a co-culture tissue-like construct stable over one week. Cells were stained and imaged with epifluorescence microscopy to confirm layer characteristics. As proof-of-concept, the mucosal construct was exposed separately to a dental monomer, 2-hydroxylethyl methacrylate (HEMA), and the oral bacteria Streptococcus mutans. Exposure to HEMA lowered mucosal cell viability, while exposure to the bacteria lowered trans-epithelial electrical resistance. These findings suggest that the oral mucosa-on-a-chip is useful for studying oral mucosal interactions with bacteria and biomaterials with a histology-like view of the tissue layers.
I. INTRODUCTION
The oral mucosa is a layered tissue with an active microbiome. Pathologies such as periodontitis, which affect 30%–50% of the population in the USA,1 stimulate interest in studying inter-kingdom and host-pathogen interactions in the oral mucosa. While over 600 taxa of bacteria colonize human oral mucosal surfaces,2,3 periodontitis involves a number of identified virulent bacterial strains that elicit an inflammatory and immune response.4–7 The complexity of the oral microbiome and its relationship to pathology warrants more study as new hypotheses of pathogenesis are considered, such as polymicrobial synergy and dysbiosis.4 Precise temporal and spatially-resolved assessments of human-bacterial cell-cell interactions, signaling events, and immune/inflammatory responses are helpful in determining the etiology of periodontal disease.8
Current biomaterial cytotoxicity testing standards provide guidelines to biologically evaluate medical/dental materials and devices. The goal of in vitro cytotoxicity testing is to efficiently and accurately determine biocompatibility of the tested biomaterial in the native tissue. Current standards to evaluate the biological effects of dental materials and devices are published by the International Organization for Standardization,9 the American National Standards Institute/American Dental Association,10 and the American Society for Testing Materials.11 Existing standards commonly use a single cell type that has been cultured on a solid substrate to subconfluency (∼80% confluency). A co-culture with conformation similar to the native mucosa is expected to have more physiological behaviors and responses than 2D monocultures12 and prove amenable to cytotoxicity test standardization.
Native oral mucosa consists of three primary layers, from apical to basal: stratified squamous epithelium, basement membrane, and subepithelial lamina propria containing fibroblasts.13,14 Cells in the stratified epithelium originate in the basal layer and differentiate toward the apical side, expressing a range of cytokeratins and connecting via desmosomes.15,16 The basement membrane is thin, consisting of type IV collagen, laminins, and other proteoglycans. The lamina propria is 0.3–0.4 mm thick13 and contains 200 × 106 fibroblasts/cm.3,14 Several key features of native oral mucosa that determine structure and function are possession of keratinocyte and fibroblast layers, expression of layer-appropriate protein markers, and possession of epithelial barrier function. Oral epithelium varies in thickness, cellularity, and keratin content, depending upon location within the oral cavity.17
Several tissue-engineered oral mucosa models have been developed to examine inter-kingdom interactions and biomaterial biocompatibility. These models include normal and transformed keratinocytes and fibroblasts from the gingiva, mucosa, and palate placed in natural extracellular matrix (ECM) hydrogels of (un)cross-linked, (un)contracted type I collagen and/or glycosaminoglycan,18–20 synthetic nylon mesh,21 Alloderm,22 decellularized mucosal tissue,23 and acellular dermal matrix.24 While some of these efforts were aimed at producing a mucosal substitute for implantation, others were used to test biocompatibility of metals used in dentistry, mercury chloride, and surfactants.16 In vitro models of mucosal pathology containing Porphyromonas gingivalis and Candida albicans demonstrated invasion of layers representing basement membrane and lamina propria, with concomitant release of pro-inflammatory cytokines by the mucosal cells.25 In these thick, 3D mucosal models, often only a fraction of the cells can be analyzed at a few timepoints using conventional methods such as histology or epifluorescence microscopy of the construct surface. In contrast, epithelial barrier function can be assessed repeatedly by transepithelial electrical resistance (TEER).26–30
Previous work establishing colonic mucosal models in microfluidic devices, or gut-on-a-chip,31–33 demonstrate the potential for development of other microfluidic mucosal models. The initial gut on-a-chip model, featuring a human intestinal epithelial cell line, featured fluid shear and cyclic strain to mimic the gut mechanical microenvironment and reported villus differentiation and epithelial barrier function maintained over 7 days.33 Epithelial barrier function was enhanced by the addition of the commensal bacteria Lactobacillus rhamnosus in flow culture with cyclic stretch. Later, enteroinvasive Escherichia coli and peripheral blood mononuclear cells were added to examine immune-microbiome interactions in a context resembling aspects of gut inflammation.31 These studies established guidelines for demonstrating functional microfluidic mucosal models: epithelial barrier function, maintenance of culture for multiple days, morphological and biomarker indicators of cell layer function, and appropriate tissue-level responses to bacteria and toxins. Subsequently, microfluidic models of airways,34–36 gastrointestinal,37 and nasal mucosa38,39 have been developed. Current microfluidic mucosal models are either monolayered or multi-layered, organized vertically. The vertical configuration is less efficient at monitoring layer-layer interactions, due to the need to change the vertical focus of the microscope, and poorer resolution of microscopy in the axial than lateral direction.
A horizontally-organized microfluidic platform with luminal, epithelial, and sub-epithelial compartments would address hypotheses involving inter-kingdom interactions of bacteria and oral mucosa as well as to test the biocompatibility of dental materials. Moreover, the interactions of each compartment would be easy to assess in a single field-of-view of an inverted microscope. In this study, an oral mucosa-on-a-chip with a lateral configuration was developed, consisting of undifferentiated keratinocytes and gingival fibroblasts. Further, this model’s ability to assess dental materials’ cytotoxicity and to study human-microbiome interactions was evaluated. This was achieved by (1) optimizing co-culture assembly for layer geometry and stability over 7 days; (2) determining the presence of epithelial and subepithelial layer morphology and biomarkers; and (3) determining culture response to dental leachables, and the oral bacteria Streptococcus mutans. Model geometry allowed facile repeated imaging during the culture period to track cell layer development and responses to challenge. Further characterization was performed with TEER measurements, as well as cell staining to assess viability and morphology, with epifluorescence and phase contrast microscopy. The results of these studies are significant in that they establish an oral mucosa-on-a-chip for determining the effects of dental materials and bacteria on mucosal cell health and tissue remodeling.
II. MATERIALS AND METHODS
A. Fabrication of microfluidic channels
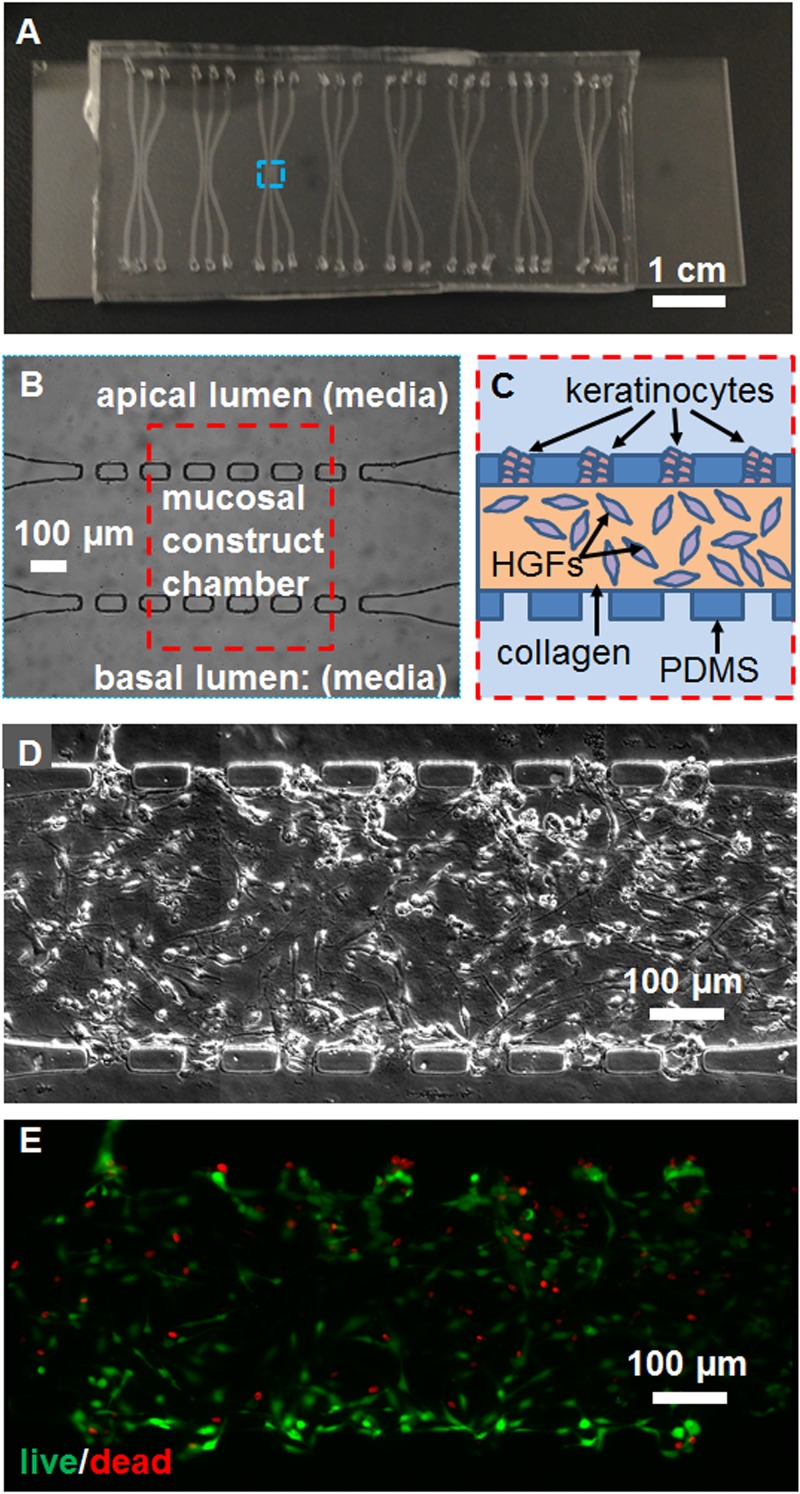
Each chip contained eight sets of microchannels [Fig. 1(a)], each set consisting of three channels separated by six PDMS posts and seven 50 μm × 50 μm square pores [Fig. 1(b)]. The SU-8 mold of microfluidic devices was custom-fabricated on a 4 in. silicon wafer using conventional photolithography with a spin coater (Laurell Technologies Co. WS-650-23NPP) and an exposure-masking system (Kloé UV-KUB 2). Polydimethylsiloxane (PDMS) elastomer Sylgard 184 (both from Dow Corning, Corp.) were mixed with curing agent at a ratio of 10:1 in a polystyrene 50 ml tube and stirred well. About 30 ml PDMS was used to create PDMS devices of around 3 mm thick. PDMS was added to the mold in a house-made aluminum foil container, degassed in a vacuum chamber (Zeny® Single-Stage) at −30 mm Hg pressure for 15 min, and was cured at 65 °C for 4 h. Then, the PDMS was peeled off the mold and placed on a clean surface, while fluid ports were punched at the ends of each channel using a 1 mm diameter dermal punch (Miltex 33-31AA). The PDMS slabs were bonded to glass microscope slides with oxygen plasma (200 mTorr, 10 psi gas source from an oxygen tank, 30 s, medium radiofrequency level) using a plasma cleaner (Harrick Plasma PDC-32G) and then baked at 150 °C overnight to make the channels hydrophobic. Each culture chamber consisted of a main channel and two parallel side channels [Fig. 1(a)], each 400 μm wide × 50 μm deep × 2.54 mm long. The side channels are connected to the middle channel in the central region by seven 50 μm × 50 μm pores spaced 100 μm apart [Fig. 1(b)].
FIG. 1.
Configuration of the microfluidic device and tissue construct. (a) A photograph of the chip containing eight 3-channel devices. (b) A phase contrast micrograph from the blue dashed box in (a), and (c) schematic of the construct culture chamber from the red dashed box in (b), showing channel and pore microstructure and cell placement. Mosaicked (d) phase contrast and (e) epifluorescence micrographs of the live-dead stained co-culture, showing the morphology of cells, viable cells in green, and dead cell nuclei in red. The scale bar is indicated.
B. Human cell line culture
Two immortalized human cell lines were cultured for the mucosa-on-a-chip. The Gie-No3B11 (keratinocyte) cell line40 and human gingival fibroblast (fibroblast)41,42 cell lines (Advanced Biological Materials, Inc.) were maintained in 25 cm2 polystyrene tissue culture flasks coated with type I collagen (Advanced Biological Materials, Inc.). Prigrow III and IV media (Advanced Biological Materials, Inc.) were used to culture fibroblasts and keratinocytes, respectively, supplemented with 10% fetal bovine serum, 500 U/ml penicillin, and 500 μg/ml streptomycin. Nutrient medium was replenished every other day. Cells were trypsinized, centrifuged (≤475×g, 3 min), resuspended in 1 ml of media, and counted before adding to the microfluidic devices. The rate of cell proliferation was monitored time from initial seeding to 80% confluency in culture on polystyrene. This time was ∼5 days for both cell types.
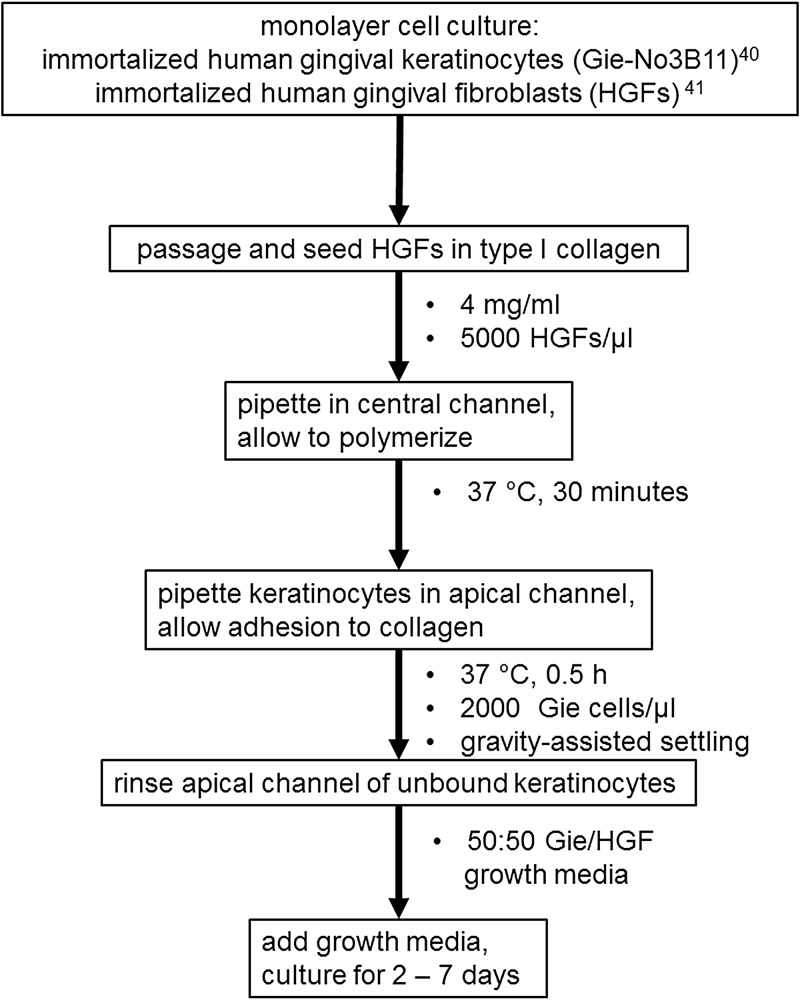
Human gingival keratinocyte and fibroblast cell lines introduced into a 3-channel microfluidic device formed a viable co-culture construct that was stable. The procedure for fabrication of the oral mucosa-on-a-chip in PDMS microchannels is presented in Fig. 2. Fibroblasts were embedded in collagen in the central channel, while keratinocytes were seeded in the pores by micropipette injection of a keratinocyte cell suspension in nutrient media into the luminal channel [Fig. 1(c)]. Deposition of keratinocytes in the pores on the side of the luminal channel occurred during 30 s of flow following the micropipette injection and was further enhanced by tilting the chip during that time period, using gravity to settle keratinocytes into pores. This transient flow following removal of the micropipette tip was caused by an equilibrating pressure gradient across the channel from the 3 μl injection into the 2 μl channel volume, a deliberate overfill to avoid air bubbles and ensure suspended keratinocyte circulation within the channel. The chips were then fed daily with media in static co-culture. Fresh media droplets of unequal volume (2 and 10 μl) were added on top of both ports connecting to the apical and basal channels, and fresh medium was introduced by surface tension-driven flow. Drying of media was avoided by using a humid incubator and enclosing the chips in the center of Petri dishes with sterile water droplets pipetted around the dish edges. Chips injected with collagen gels in the central channels, but no cells, and incubated for 1 week continuously in this manner, did not dry out.
FIG. 2.
Flow chart describing assembly of the microfluidic mucosal constructs. Parameter values such as cell seeding densities, collagen polymerization and construct incubation conditions, and nutrient media formulations are listed in bullet points beneath the box containing the relevant step.
C. HEMA exposure
The dental material monomer 2-hydroxyethyl methacrylate (HEMA, donated by Esstech, Inc.) was prepared as a sterile solution at 25 mM in 50:50 Prigrow III/IV by adding 0.01627 g of high purity liquid stock (molecular weight 130.14 g/mol) which was dissolved in 5 ml of the nutrient media and then sterile-filtered with a syringe by passing the solution through a 0.22 μm pore size filter unit. The media containing HEMA was immediately added to the apical side channel directly contacting the keratinocyte layer on the chip, with culture maintained for a subsequent 24 h.
D. Fluorescence staining and microscopy
The working area of each microfluidic device was imaged daily during the culture period with phase contrast microscopy and with epifluorescence microscopy at experimental endpoints (2–7 days). Overlapping micrographs were stitched together using MosaicJ, an ImageJ (Bethesda, MD) plugin developed in the Biomedical Imaging Group of École Polytechnique Fédérale de Lausanne (EPFL) (Lausanne, Switzerland).43 Microfluidic cultures were fixed in 4% paraformaldehyde for 1 h before staining for actin and nuclei using AlexaFluor 546-conjugated phalloidin and 4′,6-diamidino-2-phenylindole (DAPI), respectively (Life Technologies, Corp.). Epifluorescence microscopy was performed with an inverted microscope (Accu-Scope EXI-310, Ludesco, LLC) with a long working-distance, 20× plan objective, numerical aperture 0.4, and a fluorescence light source (LM-75 PhotoFluor, 89 North). The same stained specimens were also imaged with a confocal microscope (LSM 510, Zeiss) to capture z-stacks rendered as 3D images (supplementary material, Fig. S1, Movie) and image sequences (supplementary material, Fig. S2, Movie). Confocal microscopy was performed using a 20×, numerical aperture 0.7 objective, standard laser wavelengths for DAPI and AlexaFluor 546 excitation, and emission filters of 408–462 and 552–642 nm, acquired in two separate channels. Confocal images were 512 × 512 pixels, spanning an (800 × 800) μm2 field-of-view.
Living microfluidic co-cultures were stained using LIVE/DEAD® Viability/Cytotoxicity Kit (Life Technologies, Corp.) according to the manufacturer guidelines. Briefly, channels were rinsed with serum-free media before adding a mixture in phosphate-buffered saline (PBS) of 1:1500 = 2.67 μM calcein acetoxymethyl ester reagent and 1:250 = 8 μM ethidium homodimer-1. The incubation was 1 h at room temperature, in the dark, prior to epifluorescence imaging. Live and dead cells were counted by visual inspection of the epifluorescence images, spanning the entire working culture area of the microfluidic chip, in comparison to co-registered phase contrast images to delineate cell borders.
Epifluorescence microscopy of immunostained cells in the microfluidic chip was performed similar to the phalloidin-stained constructs (supplementary material, Fig. S3). The constructs were permeabilized for 30 min with 0.1% Triton-X in PBS before blocking with a solution of 1% bovine serum albumin in 0.1% Triton-X/PBS for 24 h. Primary antibodies against TE-7 (cbl271, EMD Millipore) and cytokeratin 19 (CK19, ab9221, Abcam) were diluted 1:25 and 1:50, respectively, and incubated in the channels for 1 h. While the anti-CK19 primary antibody was conjugated to a green fluorescent probe (AlexaFluor 488), the anti-TE-7 antibody was detected by further incubation with anti-mouse IgG secondary antibody (ab150116, Abcam, diluted 1:500 from stock) conjugated to a red fluorescent probe (AlexaFluor 594). Red and green channel images were obtained from stained constructs as well as constructs not exposed to the primary antibodies, at the same camera exposures for each channel.
E. Trans-epithelial electrical resistance
A custom-built impedance spectrometer was used to measure the TEER across the keratinocyte layer. The impedance spectrometer consists of two SR 830 lock-in amplifiers and a DS 340 function generator (both from Stanford Research Systems), which were controlled via general purpose interface bus ports with an automatic system and was able to measure the impedance of a signal at specific reference frequency and phase. Instrument output was calibrated across resistors in equivalent circuits ranging from 100 to 1000 kΩ, calculated from printed values and tolerances of individual resistors. The calibration equation for nominal vs. measured resistance was linear (R2 = 0.99) with a slope of 1.11 and an intercept of −24 kΩ and was used to adjust measured resistance values. For the TEER measurement, the instrument was set to operate at 50 Hz and 0.5 V. Two hollow metal catheter couplers (22 Ga, Instech Labs) were used as electrodes and were inserted through input ports connected to luminal and basal microchannels on opposite sides of the microfluidic chamber. The resistance of a similar microchannel with media but without cells and collagen was subtracted from the channel measurement. The resistance was multiplied by the surface area of interconnecting pores between the channels—seven 2500 μm2 pores—for TEER in units of Ω*cm2.
F. Bacterial culture
Streptococcus mutans (strain UA-159) were expanded from frozen glycerol stock by stab inoculation into 3 ml of brain-heart infusion (BHI) broth (Becton Dickinson) containing 1 mg/ml kanamycin. The S. mutans contain a kanamycin-resistance gene that reduces contamination during bacterial cell culture. Cells were cultured in 15 ml polypropylene tubes at 37 °C and in 5% CO2 for 24 h overnight, or until the optical density was between 0.2 and 0.3, assessed by a spectrophotometer (Implen P330 Nanophotometer). At that point, 2 μl of bacterial solution was injected into the apical (keratinocyte-containing) channel of the microfluidic device, and devices were cultured for another 24 h.
G. Statistical analysis
To assess differences in collagen contraction with fibroblast concentration or keratinocyte layer thickness, one-factor analysis of variance (ANOVA) was performed at each timepoint, and post hoc pairwise comparisons with Bonferroni corrections. Assumptions of normality and equal variance were evaluated using Kolmogorov-Smirnov and Levene’s tests, respectively. To compare TEER values pre- and post-bacterial incubation, paired Student’s t-tests were used to determine differences between groups. Significance was set at p < 0.05. Data were acquired from different culture chambers on n = 4–8 cultures per condition for each experiment. All statistical analysis was performed using Systat (version 13, Systat Software, Inc.).
III. RESULTS
A. Microfluidic co-culture of human oral mucosal cell lines
After 48 h, the fibroblasts achieved an elongated morphology in the collagen layer, while keratinocytes adhered to the collagen surface in the pores [Fig. 1(d)]. Cell viability after 48 h co-culture was 83 ± 6% (mean ± SD), assessed by epifluorescence microscopy of live-dead stained specimens, with some dead cells apparent in both layers [Fig. 1(e)].
B. Optimization of oral mucosa-on-a-chip
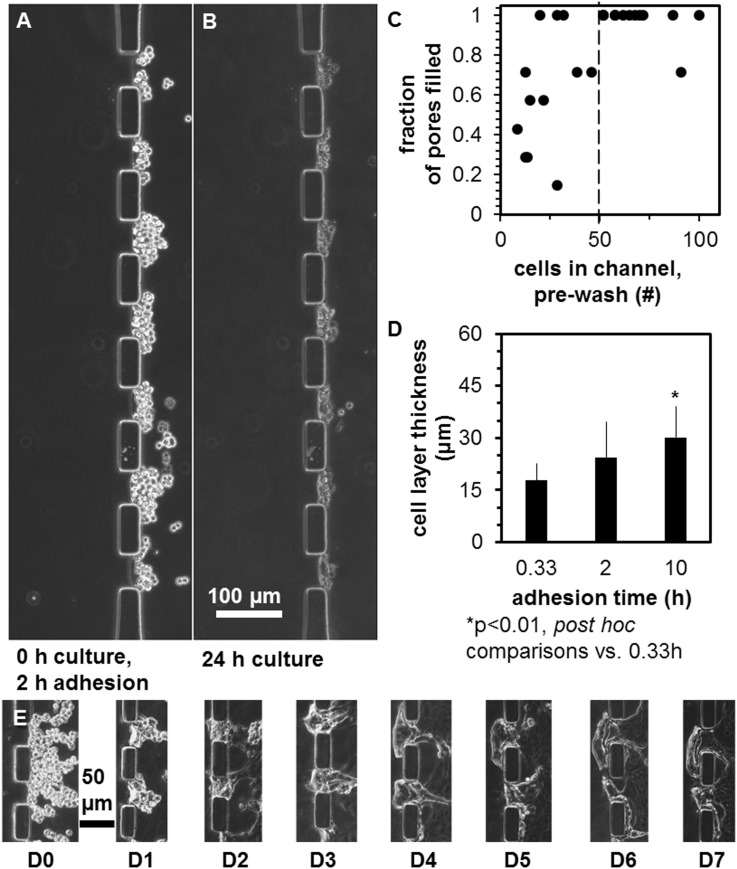
The epithelial layer was optimized by varying the method of gingival keratinocyte loading to maximize the percent of pores containing keratinocytes. Upon micropipette injection, keratinocytes flowed past the pores, while some were stopped by the PDMS posts and gradually settled into the pores during the ∼30 s of transient flow [Fig. 3(a)] and then stably attached over 24 h static culture [Fig. 3(b)]. A keratinocyte loading density of 2000 cells/μl led to 9–100 keratinocytes settling in the channel adjacent to the pores, with most often 7 of 7 pores filled when >50 cells were present [Fig. 3(c)]. After this initial experiment, gravity assistance by tipping the chips until flow stopped was found to increase the number of keratinocytes that settled into pores, leading to 97 ± 9% pore fill (mean ± SD), with initially 10.3 ± 5 keratinocytes/pore (mean ± SD). Pore loading efficiency did not depend upon incubation times between 20 min and 10 h (data not shown), although the keratinocyte layer was ∼69% thicker after 10 h than 20 min incubation [Fig. 3(d), ANOVA, post hoc comparison, p < 0.05]. Keratinocytes loaded into the side channel remained adherent to the collagen layer over 7 days [Fig. 3(e)].
FIG. 3.
The effect of keratinocyte loading parameters on layer features. Keratinocytes (a) immediately after seeding in the apical channel, incubation for 2 h to allow adhesion, and rinsing of the channel, and (b) after 24 h. (c) Dependence of pore fill with keratinocytes versus number of cells counted in the apical channel pre-wash. (d) Dependence of keratinocyte layer thickness on adhesion time. (e) Phase contrast micrographs of the same two keratinocyte-filled pores over 7 days monoculture in a microfluidic device. Scale bars are indicated.
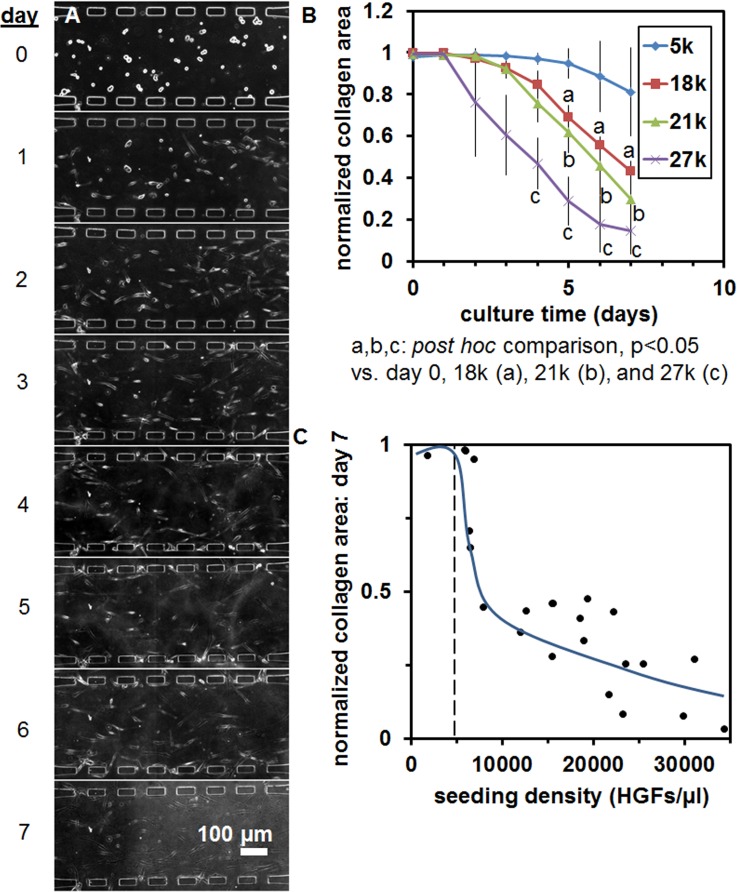
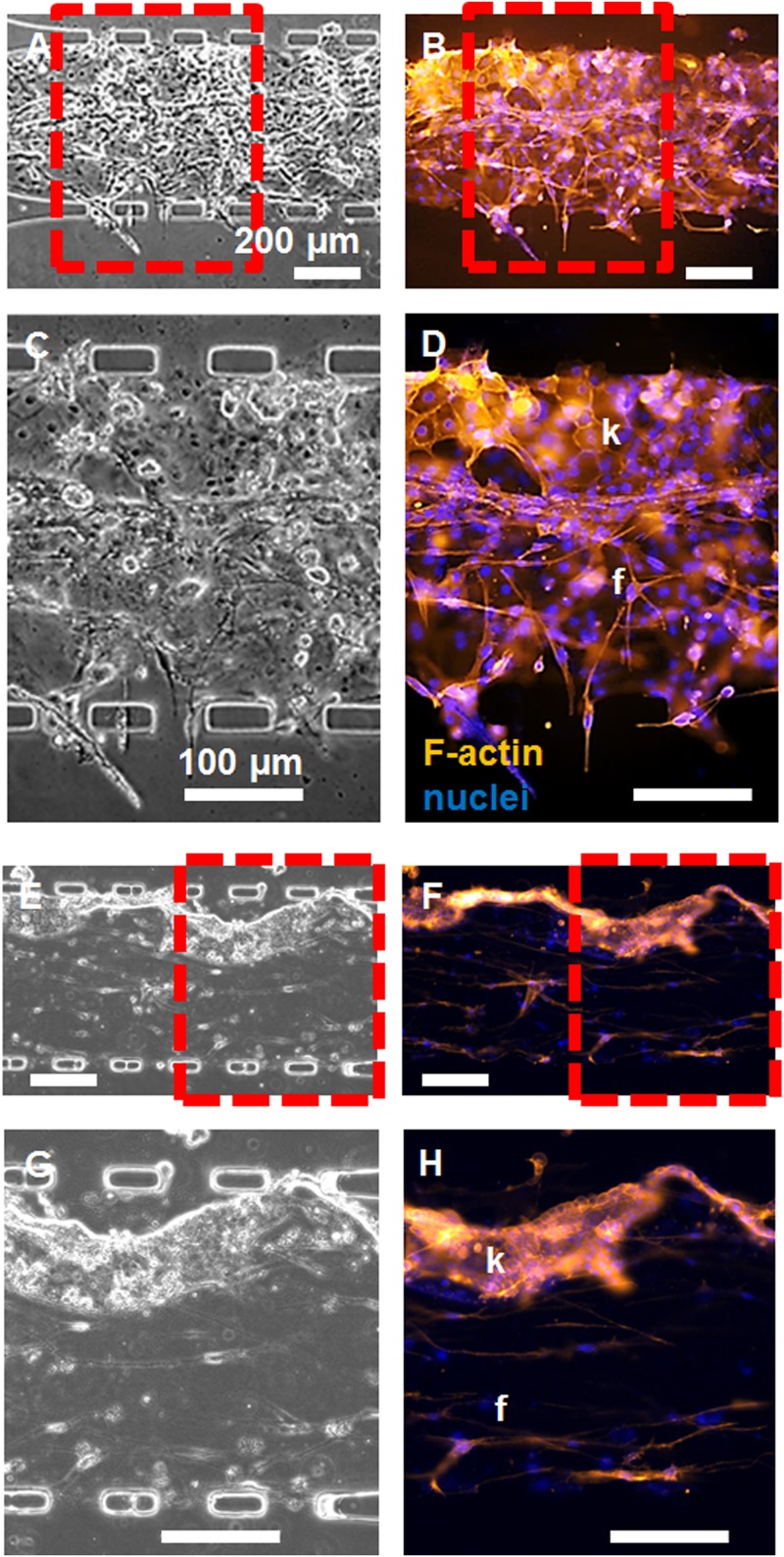
Mucosal construct stability was optimized over 7 days by varying fibroblast seeding density and initial collagen concentration in the hydrogel to avoid gel contraction. Fibroblasts at a nominal concentration of 5000 cells/μl, seeded in collagen hydrogels at 4 mg/ml in microfluidic monoculture, contracted the gel minimally over 7 days [Figs. 4(a) and 4(b)], compared to higher fibroblast concentrations [Fig. 4(b)]. Local fibroblast density in the critical culture region of the central pore adjacent to interconnecting pores varied, and contracted collagen area was found to drop sharply for local concentrations of fibroblasts >5000–6000 cells/μl [Fig. 4(c)]. Collagen hydrogel content of 2 mg/ml developed a thicker keratinocyte layer with more displacement of the collagen gel in the central channel [Figs. 5(a)–5(d)] than constructs with 4 mg/ml initial collagen concentration [Figs. 5(e)–5(h)].
FIG. 4.
The effect of fibroblast loading parameters on layer features. (a) Fibroblasts seeded in 4 mg/ml collagen gels in the microfluidic device, tracked over 7 days of monoculture. (b) Collagen gel contraction within the central channel versus culture time for fibroblast seeding densities of 5, 18, 21, and 27 × 103/μl. (c) Collagen gel area within the central channel after 7 days of monoculture versus local fibroblast seeding density determined from counting of cells within devices on day 0. The scale bar is indicated.
FIG. 5.
Epifluorescence microscopy of constructs cultured for two days, revealing epithelial and sub-epithelial layers. Constructs with (a)–(d) 2 mg/ml and (e)–(h) 4 mg/ml initial collagen concentrations in the central channel were imaged with [(a), (c), (e), and (g)] phase contrast and [(b), (d), (f), and (h)] epifluorescence microscopy after fixation and staining with phalloidin (red) and DAPI (blue) to visualize the cell organization within layers. Zoomed-in regions in (c), (d), (g), and (h) were taken from the red-dashed region of interest in (a), (b), (e), and (f), respectively. The keratinocyte (k) and fibroblast (f) layers are indicated. Scale bars are indicated.
C. Keratinocyte layer organization is affected by collagen concentration
Cell organization and subcellular features resembled the oral mucosa. Keratinocyte and fibroblast layers contained F-actin [Fig. 5]. In the keratinocyte layer, F-actin was distributed cortically in polygonal keratinocytes forming a layer up to four cells thick [Figs. 5(d) and 5(h), marked “k”]. In the fibroblast layer, elongated cells were aligned parallel and subadjacent to the keratinocyte layer [Figs. 5(d) and 5(h), marked “f”], forming a distinct interface. Keratinocyte layer thickness, keratinocyte invasion of the collagen gel, and fibroblast proliferation were higher with a 2 mg/ml initial concentration of collagen in the gel [Figs. 5(a)–5(d)], versus 4 mg/ml [Figs. 5(e)–5(h)]. Further, the keratinocytes on 4 mg/ml collagen appeared smaller and less spread laterally. Confocal z-stacks of phalloidin fluorescence signal from the construct reveal the vertical profile of the co-cultures in the 50 μm tall channels (supplemental material, Figs. S1, Movie, and S2, Movie).
D. Oral mucosa-on-a-chip exposure to HEMA and S. mutans
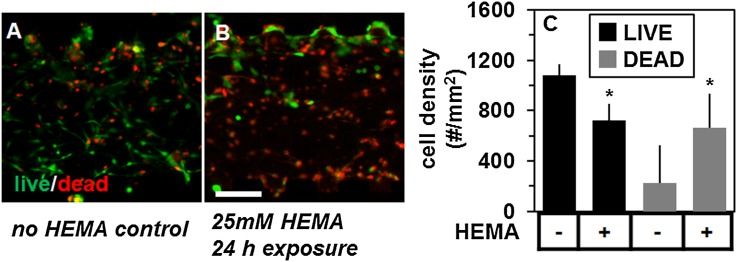
Cell viability in the oral mucosa construct was lower following 24 h exposure to 25 mM HEMA (53 ± 21%) than controls unexposed to HEMA (83 ± 6%), determined by epifluorescence microscopy of LIVE-DEAD™ stained cultures [Figs. 6(a) and 6(b), mean ± SD, Students t-test, p < 0.05). Specifically, the number of cells stained with calcein acetoxymethyl ester (AM) (“live”) was 33% lower, 229 ± 86 vs. 340 ± 42 and with ethidium homodimer (“dead”) was 300% higher, 70 ± 28 vs. 209 ± 96, in HEMA-exposed versus unexposed microfluidic co-cultures [Fig. 6(c), p < 0.05 for both, Student’s t-test, n = 4–6 constructs per condition, counted in a culture area of 0.315 mm2 on the chip]. Cell death occurred in both fibroblast and keratinocyte layers [Fig. 6(b)].
FIG. 6.
The effect of HEMA on layer viability of the mucosa-on-a-chip. Merged epifluorescence micrographs of calcein AM (live, green) and ethidium homodimer (dead, red) signal (a) before and (b) after 24 h exposure to 25 mM soluble HEMA from the apical (top) channel. (c) The density of live and dead cells counted within each condition. The (*) symbol indicates group differences by Student’s t-test, at p < 0.05. The scale bar is indicated.
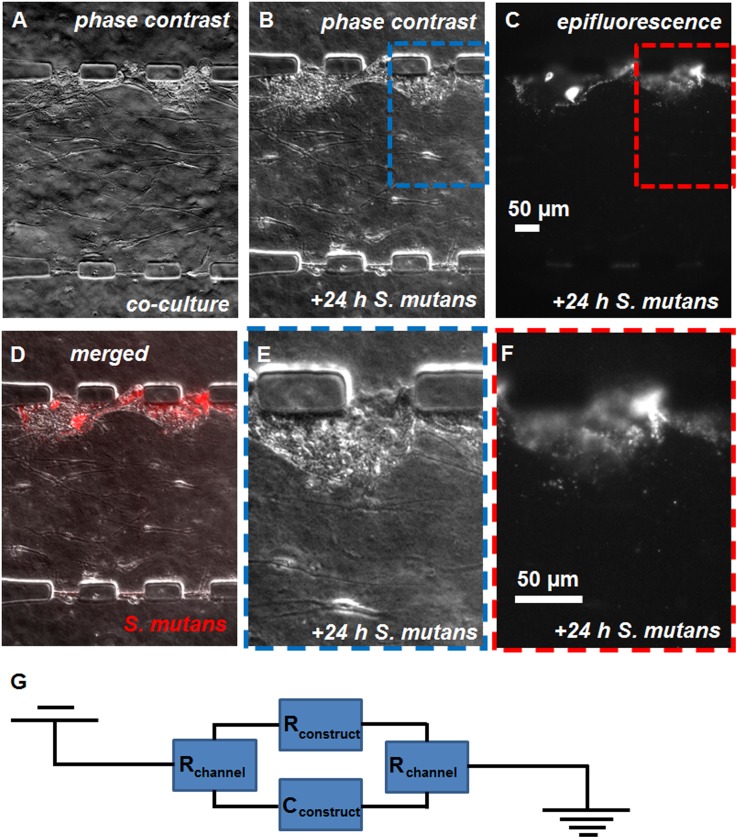
The oral bacteria S. mutans, introduced into the luminal channel, infiltrated the keratinocyte layer. Compared to before exposure [Fig. 7(a)], the mucosal cell layers appeared intact following 24 h of bacterial cell culture in the chip [Fig. 7(b)], although bacteria associated with the keratinocytes were visible as a mottled phase contrast texture of the epithelial layer. The bacterial fluorescence signal [Fig. 7(c)] co-localized primarily with the keratinocyte layer [Fig. 7(d)]. In zoomed-in views of the co-localized phase contrast [Fig. 7(e)] and fluorescence [Fig. 7(f)] micrographs, most bacterial fluorescence overlaps the keratinocyte layer, but some fluorescent bacteria are present in the collagen and fibroblast layer. Infiltration of the keratinocyte layer by S. mutans was associated with a 64% lower TEER (66 ± 49 to 24 ± 32 Ω cm2, p < 0.05, Student’s t-test), assessed across the tissue construct and modeled as the equivalent circuit in Fig. 7(g).
FIG. 7.
The exposure of the mucosa-on-a-chip to S. mutans. [(a) and (b)] Phase contrast and (c) epifluorescence micrographs of microfluidic constructs (a) before and [(b) and (c)] after 24 h culture of intravitally-stained (DiI) S. mutans in the apical channel adjacent to keratinocytes. (d) Overlay of phase contrast (gray scale) and epifluorescence micrographs (red signal, DiI fluorescence) from (b) and (c), respectively. (e) Zoomed-in region of (b). (f) Zoomed-in region of (c). The scale bars are indicated. (g) Equivalent circuit of device used to measure transepithelial resistance, connected across tissue construct from the luminal to basal channels, including components contributing to resistance (R) and capacitance (C).
IV. DISCUSSION
The fabrication of an oral mucosa-on-a-chip occurred through the creation of a submucosal layer of fibroblasts embedded in collagen in a central channel, followed by trans-channel seeding of keratinocytes into pores between PDMS posts. This configuration allowed convenient and precise tracking of keratinocytes and fibroblasts with conventional microscopy, with posts allowing for dual attachment of keratinocytes to apical and basal surfaces, as occurs in the junctional epithelium. Fabrication parameters of keratinocyte and fibroblast loading densities were optimized at 2000 and 5000 cells/μl, respectively. Collagen densities of 2 and 4 mg/ml were found to reduce gel contraction and keratinocyte invasion of the adjacent gel over one week of culture, although the keratinocyte layer was thicker adjacent to 2 mg/ml collagen. Cells maintained keratinocyte and fibroblast-like characteristics and formed two adjacent layers with distinct borders. Cell viability was lower following HEMA exposure, and TEER was lower following S. mutans culture from the luminal channel. These data establish a microfluidic culture on-a-chip approach to test hypotheses about the oral mucosa.
There are strengths and weaknesses to the microfluidic chamber design that influence potential experiments on and applications of the oral mucosa-on-a-chip. Three channels with interconnecting pores lead to the facile assembly of adjacent layers and create an apical-basal geometry that mimics the lumen of the oral cavity and a subepithelial space. This geometry will allow future studies of mucosal responses to exposure to dental biomaterials and oral bacteria from the keratinocyte side, and cytokines and immune cells from the subepithelial channel. A perpendicular configuration to the present model, as in the gut on-a-chip33 or a transwell assay,16 would increase layer surface area but layer-layer interactions would be less easily visualized by conventional microscopy. Keratinocyte layer assembly is also controlled by the number and surface area of pores connecting the middle and side channels, which is an important factor in limiting keratinocyte invasion of the collagen layer and restricting collagen gel contraction. In contrast to static wellplate culture, this microfluidic configuration allows flow in the luminal channels, which in the future can occur during culture, mimicking the flow of saliva or gingival crevicular fluid, which is known to slough keratinocytes and maintain epithelial layer morphology.44 Keratinocyte invasion of the collagen gel, and gel contraction might limit culture beyond seven days, the longest culture period tested. Future efforts will seek to introduce flow, construct a more rigid and/or denser collagen layer, and examine the expression of markers of differentiation and inflammation in response to normal culture conditions and pathological treatments. Finally, it is noted that the current mucosa on-a-chip was cultured in a normal oxygen atmosphere and colonized with oxygen-tolerant S. mutans, and thus best represents bacterial association with tissue at the gingival margins.45 To represent periodontitis in the gingival sulcus, the current model should be altered to allow culture in anaerobic conditions. One possible alteration requiring both future numerical modeling and experimentation in culture is the addition of a channel parallel to the luminal side of the construct for the flow of nitrogen gas, which would decrease the local oxygen concentration in the keratinocyte layer and luminal channel.
The microfluidic mucosa-on-a-chip recapitulated some, but not all features of the native oral epithelium. In the microfluidic device, polygonal keratinocytes grew into a layer 1–4 cells deep [Figs. 3 and 5], similar to the native oral epithelium.17 Further, the keratinocyte layers on 4 mg/ml collagen were notably thinner, indicating the potential for collagen concentration to control the thickness of the adherent epithelial layer. In 2D monoculture and in the microfluidic channel, the fibroblast cell line stained positively for fibroblast marker TE-7, and keratinocytes for cytokeratin 19 (CK19), indicating stable expression of these respective mesoderm-derived and epithelial markers on tissue culture plastic and in the microfluidic culture chamber (supplementary material, Fig. S3). In the future, the presence of a proliferative basal layer of keratinocytes along with a less proliferative, differentiated apical layer may be tested by staining with Ki-67.46 Apical shear flow is also known to affect keratinocyte layer morphology and cell turnover in the oral epithelium.47 In future experiments, the site-specific differentiation status of keratinocytes and formation of a basement membrane in the microfluidic co-culture will be tested in response to altered media formulations and shear flow. The TEER of 66 ± 49 Ω cm2 is less than reported values for the Gie keratinocyte cell line, which found are in the range of 110–160 Ω cm2 in wellplate systems.40,48 This lower TEER value may be caused by tight junction and/or basal cell-substrate attachment disruption near the PDMS posts and represents an area for improvement. For example, the PDMS surfaces could be coated with basement membrane-derived proteins such as Matrigel to improve keratinocyte adhesion.
The fibroblast-containing collagen gel layer of the microfluidic construct resembled a simplified in vivo lamina propria. While gingival fibroblasts were present, fibroblast density, initially at 5000 cells/μl, was 40 times lower than in vivo.14 Higher initial fibroblast densities caused collagen layer contraction, likely due to extracellular matrix remodeling associated with enzymatic degradation and/or actin-mediated contraction of the collagen network. Further work is needed to examine fibroblast and keratinocyte proliferation in response to initial collagen concentration of the hydrogel, co-culture, and flow of nutrient media. However, in static culture, the fibroblasts expressed F-actin, became elongated, and aligned parallel to the keratinocyte layer (Fig. 5), indicating some development of a spatial arrangement resembling that found in vivo. During periodontitis, the subepithelial layer is a site of inflammation, immune cell recruitment, tissue remodeling, and edema.1 This could be simulated in the future by adding human neutrophils or lymphocytes to the subepithelial channel.
In the presence of S. mutans, the response of the microfluidic construct suggests a compromised keratinocyte layer. The close association of bacteria and keratinocytes, and reduced TEER is similar to the response of a colonic mucosa-on-a-chip to pathogenic E. coli49 and of in vitro gingival constructs to culture with Porphyromonas gingivalis.50 The differing culture media requirements of bacteria and human mucosal cells can make long-term inter-kingdom co-culture difficult. Further, static nutrient media represents a departure from the physiological environment of the oral mucosa. Two separate approaches might address these issues. First, multispecies bacterial culture on the mucosal chip may be achieved by compartmentalizing the mucosal and bacterial culture spaces on the chip, while still retaining soluble communication.51 Second, artificial saliva52 rather than nutrient media may be used to bathe the apical channel containing both keratinocytes and bacteria.
The microfluidic mucosal model represents an alternative to 3D organotypic models30,53 or well-plate cultures for cytotoxicity testing.42 Importantly, the viability of keratinocytes and fibroblasts can be assessed in a single microscope field of view following exposure to a soluble dental material at specified concentrations and durations [Fig. 6]. The 25 mM HEMA concentration used was intended to exceed the EC50 of HEMA,54 producing a severe cytotoxic response, to demonstrate feasibility, and is well above the levels of HEMA likely encountered by a patient’s tissues during material leaching.55 Current standards test materials in contact with monocultures of oral mucosal cell lines.9–11 A thorough comparison of microfluidic construct viability to that of current standards used for cytotoxicity testing is needed at physiologically relevant levels of dental material monomers to establish the oral mucosa-on-a-chip as an alternative technique. However, advantages of the chip construct are the capture of keratinocyte and fibroblast layer morphology and labeled signals in a single field-of-view, as well as the capability for control over fluid flow in the luminal channel.
Several future research directions depend on the oral mucosa-on-a-chip platform described here. The flow rate of media or a synthetic analog of saliva will be incorporated into the apical channel to determine the effects of flow rate on the keratinocyte layer morphology, permeability, and differentiation. The platform geometry is appropriate for image-based assessment of rapid or transient calcium signaling in keratinocytes and fibroblasts loaded with calcium-sensitive fluorescent dyes. Such experiments would aid understanding of the role of calcium signaling in mucosal response to bacteria, biomaterials, and inflammatory triggers. In a personalized medicine approach, the mucosa-on-a-chip may be adapted for use with primary cells and bacteria cultured from patients with periodontal disease. Accurate on-chip modeling of periodontitis would likely need to incorporate the patient’s immune cells, pathogenic anaerobic bacteria cultured from the patient’s mouth, and a modified local oxygen environment on the chip. Such models could be useful in choosing the most effective of several treatment options. The initial efforts, described here, to optimize and characterize an oral mucosa-on-a-chip, represent a significant step toward these clinically-relevant goals.
SUPPLEMENTARY MATERIAL
See supplementary material for two videos and one figure, with captions, presenting confocal and epifluorescence microscopy results from the stained tissue constructs. The videos are rendered from confocal microscopy data. The first video is a 3D rotating view of the mucosa on-a-chip, including overlaid blue and red fluorescence channels representing DAPI signal from nuclei and phalloidin-AlexaFluor 546 signal from F-actin respectively (Fig. S1, Movie). The second video is the same confocal image depth stack presented as serial video frames (Fig. S2, Movie). The final figure presents epifluorescence microscopy images from the immunostaining experiment for CK19 and TE-7 within the microfluidic constructs, as well as from control conditions (Fig. S3).
ACKNOWLEDGMENTS
This work was supported in part by the National Science Foundation CAREER (Award No. 1553330), the School of Engineering at the Catholic University of America, the American Dental Association (ADA), ADA Foundation, and the National Institute of Dental and Craniofacial Research (Grant No. R01-DE26122-02).
References
- 1.Newman M., Takei H., Klokkevold P., and Carranza F., Carranza’s Clinical Periodontology (Saunders, 2014). [Google Scholar]
- 2.Avila M., Ojcius D. M., and Yilmaz O., DNA Cell Biol. 28(8), 405–411 (2009). 10.1089/dna.2009.0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T., Yu W. H., Izard J., Baranova O. V., Lakshmanan A., and Dewhirst F. E., Database (Oxford) 2010, baq013 (2010). 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G. and Lamont R. J., Mol. Oral Microbiol. 27(6), 409–419 (2012). 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones B. D. and Falkow S., Annu. Rev. Immunol. 14, 533–561 (1996). 10.1146/annurev.immunol.14.1.533 [DOI] [PubMed] [Google Scholar]
- 6.Ding Y., Uitto V. J., Firth J., Salo T., Haapasalo M., Konttinen Y. T., and Sorsa T., Oral Dis. 1(4), 279–286 (1995). 10.1111/j.1601-0825.1995.tb00194.x [DOI] [PubMed] [Google Scholar]
- 7.Scott D. A. and Krauss J., Front Oral Biol. 15, 56–83 (2012). 10.1159/000329672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Socransky S. S. and Haffajee A. D., J. Periodontol. 63(4 Suppl), 322–331 (1992). 10.1902/jop.1992.63.4s.322 [DOI] [PubMed] [Google Scholar]
- 9.ISO 10993-5:2009, Biological evaluation of medical devices–Part 5: Tests for in vitro cytotoxicity, June, 2009, International Organization for Standardization, Geneva, Switzerland: https://www.iso.org/standard/36406.html. [Google Scholar]
- 10.ANSI/ADA Standard No. 41 for ADA41-2015, Evaluation of Biocompatibility of Medical Devices, December, 2015, American National Standards Institute/American Dental Association, Washington, D.C; http://ebusiness.ada.org/productcatalog/503/Restorative-and-Orthodontic-Materials/ANSIADA-Standard-No-41-for-Recommended-Standard-Practices-fo/ADA41-2005D. [Google Scholar]
- 11.ASTM F1027-86(2017), Standard Practice for Assessment of Tissue and Cell Compatibility of Orofacial Prosthetic Materials and Devices, 2017, ASTM International, West Conshohocken, PA. http://www.astm.org/cgi-bin/resolver.cgi?F1027.
- 12.Duval K., Grover H., Han L. H., Mou Y., Pegoraro A. F., Fredberg J., and Chen Z., Physiology (Bethesda) 32(4), 266–277 (2017). 10.1152/physiol.00036.2016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 13.Cho K. H., Yu S. K., Lee M. H., Lee D. S., and Kim H. J., Anat. Cell Biol. 46(3), 171–176 (2013). 10.5115/acb.2013.46.3.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroder H., The Periodontium. Handbook of Microscopic Anatomy (Springer-Verlag, New York, 1986). [Google Scholar]
- 15.Garzon I., Serrato D., Roda O., Del Carmen Sanchez-Quevedo M., Gonzales-Jaranay M., Moreu G., Nieto-Aguilar R., Alaminos M., and Campos A., Int. J. Artif. Organs 32(10), 711–719 (2009). 10.1177/039139880903201002 [DOI] [PubMed] [Google Scholar]
- 16.Moharamzadeh K., Brook I. M., Van Noort R., Scutt A. M., and Thornhill M. H., J. Dent. Res. 86(2), 115–124 (2007). 10.1177/154405910708600203 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M., Jpn. Dent. Sci. Rev. 54(2), 59–65 (2018). 10.1016/j.jdsr.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro F. A., Mizuno S., Huertas J. C., Glowacki J., and Orgill D. P., Wound Repair Regen. 9(6), 507–512 (2001). 10.1046/j.1524-475x.2001.00507.x [DOI] [PubMed] [Google Scholar]
- 19.Masuda I., Kokubyo Gakkai Zasshi 63(2), 334–353 (1996). 10.5357/koubyou.63.334 [DOI] [PubMed] [Google Scholar]
- 20.Moriyama T., Asahina I., Ishii M., Oda M., Ishii Y., and Enomoto S., Tissue Eng. 7(4), 415–427 (2001). 10.1089/10763270152436472 [DOI] [PubMed] [Google Scholar]
- 21.Schmalz G., Schweikl H., and Hiller K. A., Eur. J. Oral Sci. 108(5), 442–448 (2000). 10.1034/j.1600-0722.2000.108005442.x [DOI] [PubMed] [Google Scholar]
- 22.Izumi K., Takacs G., Terashi H., and Feinberg S. E., J. Oral Maxillofac. Surg. 57(5), 571–577 (1999). 10.1016/S0278-2391(99)90077-0 [DOI] [PubMed] [Google Scholar]
- 23.Hildebrand H. C., Hakkinen L., Wiebe C. B., and Larjava H. S., Histol. Histopathol. 17(1), 151–163 (2002). 10.14670/HH-17.151 [DOI] [PubMed] [Google Scholar]
- 24.Cho K. H., Ahn H. T., Park K. C., Chung J. H., Kim S. W., Sung M. W., Kim K. H., Chung P. H., Eun H. C., and Youn J. I., J. Dermatol. Sci. 22(2), 117–124 (2000). 10.1016/S0923-1811(99)00056-0 [DOI] [PubMed] [Google Scholar]
- 25.Dongari-Bagtzoglou A., Kashleva H., Dwivedi P., Diaz P., and Vasilakos J., PLoS One 4(11), e7967 (2009). 10.1371/journal.pone.0007967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moharamzadeh K., Franklin K. L., Brook I. M., and van Noort R., J. Periodontol. 80(5), 769–775 (2009). 10.1902/jop.2009.080610 [DOI] [PubMed] [Google Scholar]
- 27.Moharamzadeh K., Brook I. M., Van Noort R., Scutt A. M., Smith K. G., and Thornhill M. H., J. Mater. Sci. Mater. Med. 19(4), 1793–1801 (2008). 10.1007/s10856-007-3321-1 [DOI] [PubMed] [Google Scholar]
- 28.Anderson M. J., Parks P. J., and Peterson M. L., J. Microbiol. Methods 92(2), 201–208 (2013). 10.1016/j.mimet.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Ryck T., Grootaert C., Jaspaert L., Kerckhof F. M., Van Gele M., De Schrijver J., Van den Abbeele P., Swift S., Bracke M., Van de Wiele T., and Vanhoecke B., Appl. Microbiol. Biotechnol. 98(15), 6831–6846 (2014). 10.1007/s00253-014-5841-1 [DOI] [PubMed] [Google Scholar]
- 30.Dongari-Bagtzoglou A. and Kashleva H., Nat. Protoc. 1(4), 2012–2018 (2006). 10.1038/nprot.2006.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H. J., Li H., Collins J. J., and Ingber D. E., Proc. Natl. Acad. Sci. U.S.A. 113(1), E7–E15 (2016). 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H. J. and Ingber D. E., Integr. Biol. (Camb) 5(9), 1130–1140 (2013). 10.1039/c3ib40126j [DOI] [PubMed] [Google Scholar]
- 33.Kim H. J., Huh D., Hamilton G., and Ingber D. E., Lab Chip 12(12), 2165–2174 (2012). 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 34.Sellgren K. L., Butala E. J., Gilmour B. P., Randell S. H., and Grego S., Lab Chip 14(17), 3349–3358 (2014). 10.1039/C4LC00552J [DOI] [PubMed] [Google Scholar]
- 35.Skolimowski M., Weiss Nielsen M., Abeille F., Skafte-Pedersen P., Sabourin D., Fercher A., Papkovsky D., Molin S., Taboryski R., Sternberg C., Dufva M., Geschke O., and Emneus J., Biomicrofluidics 6(3), 34109 (2012). 10.1063/1.4742911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barr J. J., Auro R., Sam-Soon N., Kassegne S., Peters G., Bonilla N., Hatay M., Mourtada S., Bailey B., Youle M., Felts B., Baljon A., Nulton J., Salamon P., and Rohwer F., Proc. Natl. Acad. Sci. U.S.A. 112(44), 13675–13680 (2015). 10.1073/pnas.1508355112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah P., Fritz J. V., Glaab E., Desai M. S., Greenhalgh K., Frachet A., Niegowska M., Estes M., Jager C., Seguin-Devaux C., Zenhausern F., and Wilmes P., Nat. Commun. 7, 11535 (2016). 10.1038/ncomms11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W., Yan Y., Li C. W., Xia H. M., Chao S. S., Wang de Y., and Wang Z. P., Lab Chip 14(4), 677–680 (2014). 10.1039/C3LC51208H [DOI] [PubMed] [Google Scholar]
- 39.Na K., Lee M., Shin H. W., and Chung S., Lab Chip 17(9), 1578–1584 (2017). 10.1039/C6LC01564F [DOI] [PubMed] [Google Scholar]
- 40.Groger S., Michel J., and Meyle J., J. Periodontal. Res. 43(6), 604–614 (2008). 10.1111/j.1600-0765.2007.01019.x [DOI] [PubMed] [Google Scholar]
- 41.Vardar-Sengul S., Arora S., Baylas H., and Mercola D., J. Periodontol. 80(5), 833–849 (2009). 10.1902/jop.2009.080483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Illeperuma R. P., Park Y. J., Kim J. M., Bae J. Y., Che Z. M., Son H. K., Han M. R., Kim K. M., and Kim J., J. Mater. Sci. Mater. Med. 23(3), 753–762 (2012). 10.1007/s10856-011-4473-6 [DOI] [PubMed] [Google Scholar]
- 43.Thevenaz P. and Unser M., Microsc. Res. Tech. 70(2), 135–146 (2007). 10.1002/jemt.20393 [DOI] [PubMed] [Google Scholar]
- 44.Jones K. B. and Klein O. D., Int. J. Oral Sci. 5(3), 121–129 (2013). 10.1038/ijos.2013.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law V., Seow W. K., and Townsend G., Aust. Dent. J. 52(2), 93–100 (2007), quiz 159. 10.1111/j.1834-7819.2007.tb00471.x [DOI] [PubMed] [Google Scholar]
- 46.Birajdar S. S., Radhika M., Paremala K., Sudhakara M., Soumya M., and Gadivan M., J. Oral Maxillofac. Pathol. 18(2), 169–176 (2014). 10.4103/0973-029X.140729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dawes C., Arch. Oral Biol. 48(5), 329–336 (2003). 10.1016/S0003-9969(03)00014-1 [DOI] [PubMed] [Google Scholar]
- 48.Groeger S., Jarzina F., Windhorst A., and Meyle J., J. Periodontal. Res. 51(6), 748–757 (2016). 10.1111/jre.12351 [DOI] [PubMed] [Google Scholar]
- 49.Kim H. J., Lee J., Choi J. H., Bahinski A., and Ingber D. E., J. Vis. Exp. (114), e54344 (2016). 10.3791/54344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Groeger S., Doman E., Chakraborty T., and Meyle J., Eur. J. Oral Sci. 118(6), 582–589 (2010). 10.1111/j.1600-0722.2010.00782.x [DOI] [PubMed] [Google Scholar]
- 51.Pham P. L. H., Rooholghodos S. A., Choy J. S., and Luo X., Adv. Biosyst. 2(3), 1700180 (2018). 10.1002/adbi.201700180 [DOI] [Google Scholar]
- 52.Seo S. H., Han I., Lee H. S., Choi J. J., Choi E. H., Kim K. N., Park G., and Kim K. M., Sci. Rep. 7(1), 8395 (2017). 10.1038/s41598-017-08725-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buskermolen J. K., Reijnders C. M., Spiekstra S. W., Steinberg T., Kleverlaan C. J., Feilzer A. J., Bakker A. D., and Gibbs S., Tissue Eng. Part C Methods 22(8), 781–791 (2016). 10.1089/ten.tec.2016.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urcan E., Scherthan H., Styllou M., Haertel U., Hickel R., and Reichl F. X., Biomaterials 31(8), 2010–2014 (2010). 10.1016/j.biomaterials.2009.11.065 [DOI] [PubMed] [Google Scholar]
- 55.Urban V. M., Machado A. L., Vergani C. E., Giampaolo E. T., Pavarina A. C., de Almeida F. G., and Cass Q. B., Dent. Mater. 25(5), 662–671 (2009). 10.1016/j.dental.2008.10.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for two videos and one figure, with captions, presenting confocal and epifluorescence microscopy results from the stained tissue constructs. The videos are rendered from confocal microscopy data. The first video is a 3D rotating view of the mucosa on-a-chip, including overlaid blue and red fluorescence channels representing DAPI signal from nuclei and phalloidin-AlexaFluor 546 signal from F-actin respectively (Fig. S1, Movie). The second video is the same confocal image depth stack presented as serial video frames (Fig. S2, Movie). The final figure presents epifluorescence microscopy images from the immunostaining experiment for CK19 and TE-7 within the microfluidic constructs, as well as from control conditions (Fig. S3).