Tuberculosis (TB) is a significant global health threat, with one third of the world’s population infected with its causative agent, Mycobacterium tuberculosis (Mtb). The emergence of multi-drug resistant (MDR) Mtb resistant to the frontline anti-tubercular drugs, rifampicin and isoniazid, forces treatment with toxic second-line drugs. Currently ~4% of new and ~21% of previously treated TB cases are either rifampicin drug resistant or MDR Mtb infections1. The specific molecular host-pathogen interactions mediating the rapid world-wide spread of MDR Mtb strains remain poorly understood. W-Beijing Mtb strains are highly prevalent throughout the world and associated with increased drug resistance2. In the early 1990s, closely related MDR W-Beijing Mtb strains (strain W) were identified in large institutional outbreaks in New York City and caused high mortality rates3. Production of interleukin beta (IL-1β by macrophages coincides with the shift towards aerobic glycolysis, a metabolic process that mediates protection against drug susceptible Mtb4. Here, using a collection of MDR W-Mtb strains, we demonstrate that overexpression of Mtb cell wall lipids, phthiocerol dimycocerosates (PDIMs) bypasses the IL-1 receptor type I (IL-1R1) signaling pathway, instead driving the induction of interferon beta (IFN-β) to reprogram macrophage metabolism. Importantly, Mtb carrying a drug resistance conferring single nucleotide polymorphism (SNP) in rpoB (H445Y)5 can modulate host macrophage metabolic reprogramming. These findings transform our mechanistic understanding of how emerging MDR Mtb strains may acquire drug resistance SNPs altering Mtb surface lipid expression and modulating host macrophage metabolic reprogramming.
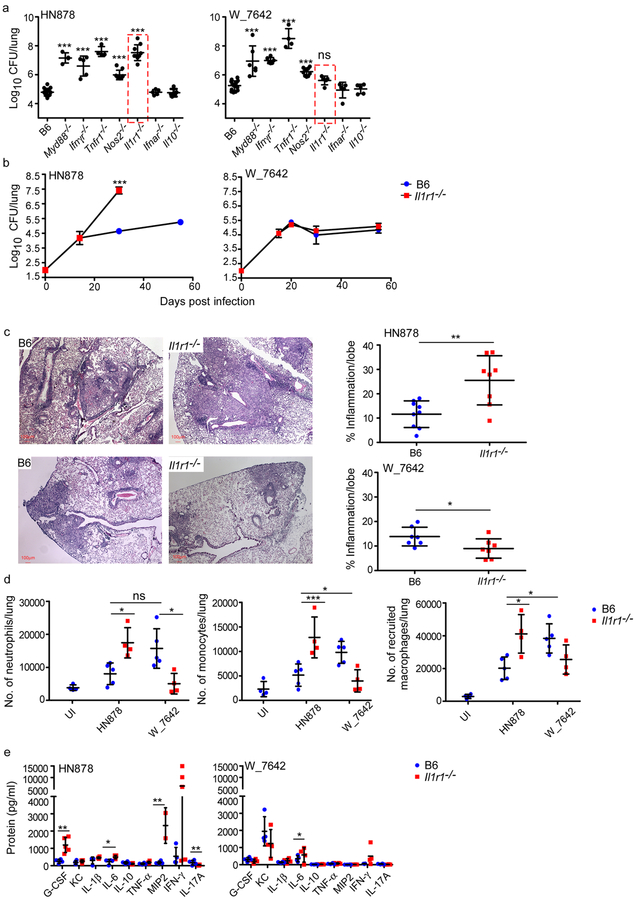
The interferon gamma (IFN-γ)6, tumor necrosis factor alpha (TNF-α)7, inducible nitric oxide synthase (iNOS)8, IL-1R19, and myeloid differentiation primary response gene 88 (Myd88)10 pathways are critical for host immunity to Mtb infection. We determined if these immune pathways are important for protection against both drug susceptible and drug resistant Mtb infection in vivo. As expected, Myd88−/−, Ifnγr−/−, Tnfr1−/−, Nos2−/−, and Il1r1−/− mice show increased lung burden upon infection with a drug susceptible W-Beijing Mtb strain, HN87811 (Fig. 1a,b). Mice deficient in IFN α/β receptor (Ifnar−/−)12 and IL-10 (Il10−/−)13 had similar lung burden when compared to C57BL/6J (B6) HN878-infected mice (Fig. 1a). Myd88−/−, Ifnγr−/−, Tnfr1−/−, and Nos2−/− mice also showed increased lung Mtb burden upon infection with an MDR strain, W_7642 (Fig. 1a). In contrast, Il1r1−/− mice, similar to Ifnar−/− and Il10−/− mice, controlled Mtb W_7642 infection (Fig. 1a,b). Increased susceptibility in HN878-infected Il1r1−/− mice resulted in exacerbated pulmonary inflammation (Fig. 1c-upper panel) and increased inflammatory myeloid cell accumulation (Fig. 1d, Supplementary Fig. 1a). Similar increased Mtb burden in Il1r1−/− mice was observed upon infection with a W-Beijing Mtb pyrazinamide resistant strain, HN563 (Supplementary Fig. 2). HN878 and W_7642 infection in B6 mice resulted in comparable pulmonary inflammation (Fig. 1c) but showed differences in recruitment of myeloid cell populations (Fig. 1d). In contrast, W_7642-infected Il1r1−/− mice did not exhibit exacerbated inflammation (Fig. 1c-lower panel), increased accumulation of inflammatory myeloid cells (Fig. 1d, Supplementary Fig. 1a), or altered accumulation of activated IFN-γ-producing CD4+ T cells (Supplementary Fig. 1b) when compared with W_7642-infected B6 mice. Only a small increase in lung IL-6 protein levels in Il1r1−/− W_7642-infected mice was observed when compared with B6 W_7642-infected mice (Fig. 1e). Thus, while several key protective immune pathways function in both drug susceptible and MDR Mtb infection, IL-1R1 signaling is critical for protection against drug susceptible Mtb infection but dispensable for immunity against an MDR Mtb strain, W_7642.
Figure 1. IL-1R1 is dispensable for protective immunity against MDR Mtb strain, W_7642.
B6 (n=10) and gene deficient mice (Myd88−/− HN878 n=3, W_7642 n=6; Ifngr−/− HN878 n=5, W_7642 n=6; Tnfr1−/− n=4; Nos2−/− n=5; Il1r1−/− HN878 n=9, W_7642 n=5; Ifnar−/− n=5; Il10−/− n=5) were aerosol infected with 100 CFU Mtb HN878 or W_7642. Lung bacterial burden was determined on 30 dpi (a) or at defined time points (b, B6 n=5, Il1r1−/− n=9 except W_7642 D15, D20 Il1r1−/− n=5). HN878-infected Il1r1−/− mice were sacrificed on 30 dpi due to severe TB disease (b). On 30 dpi, formalin-fixed, paraffin embedded (FFPE) lung sections from B6 and Il1r1−/− mice were stained with H&E and inflammatory area was measured. Micrographs are representative images - 5x magnification (c, HN878 B6 n=9, HN878 Il1r1−/− n=8, W_7642 n=7). Total number of lung neutrophils, monocytes, and recruited macrophages were determined on 30 dpi using flow cytometry (d, B6 n=5, Il1r1−/− n=4, UI B6 n=4). Cytokine and chemokine protein levels in lung homogenates were measured in B6 and Il1r1−/− mice at 30 dpi (e, n=5). UI-uninfected. (a) 1-way ANOVA with Tukey’s post-test, (b,d,e) 2-way ANOVA with Bonferroni post-test, (c) two tailed Student’s t-test. The data points represent the mean (±SD) of values. *p≤0.05, **p≤0.01, ***p≤0.001, ns-not significant (p>0.05).
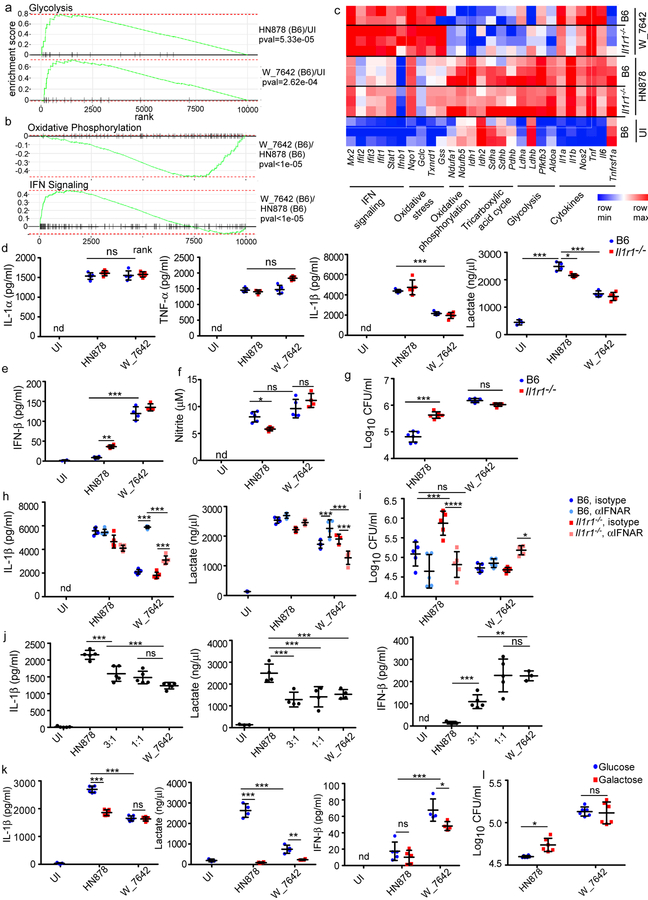
Mtb infection of macrophages induces Toll-like receptor 2 (TLR2) stimulation, activation of protein kinase B (PKB/Akt)/mammalian target of rapamycin (mTOR), and a shift towards aerobic glycolysis to mediate Mtb control4,14. Accordingly, macrophages infected with HN878 or W_7642 induced transcriptional pathways associated with a shift to aerobic glycolysis, including key enzymes such as lactate dehydrogenases (Ldha, Ldhb), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3) and aldolase A (Aldoa) (Fig. 2a,c). Of interest, W_7642 infection shut down transcriptional networks associated with macrophage oxidative phosphorylation, including downregulation of NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 (Ndufa1) and NADH:ubiquinone oxidoreductase subunit B5 (Ndufb5), key components of the macrophage oxidative phosphorylation complex (Fig. 2b,c). Importantly, W_7642 infection induced a distinct type I IFN transcriptional signature, including Ifnb1, and IFN-induced genes such as MX dynamin-like GTPase 2 (Mx2), interferon induced protein with tetratricopeptide repeats (Ifit) 1, 2 and 3, as well as signal transducer and activator of transcription 1 (Stat1) (Fig. 2b,c). Ifna transcripts were not detected by RNA sequencing. Increased lung IFN-β levels were found in W_7642-infected mice when compared to HN878-infected mice (Supplementary Fig. 3). While induction of IL-1α mRNA and protein was similar, IL-1β mRNA and protein levels were significantly lower in W_7642-infection when compared to HN878-infected macrophages (Fig. 2c,d) and coincided with increased IFN-β mRNA and protein levels in W_7642-infection, when compared to HN878-infected macrophages (Fig. 2c,e). These results were consistent at all time points measured and at varying multiplicity of infection (MOI, Supplementary Fig. 4a-c). The IL-1R1 pathway negatively regulated IFN-β in HN878 infection, but it did not regulate IFN-β production following W_7642 infection either in vitro or in vivo (Fig. 2e, Supplementary Fig. 3). Mtb-mediated induction of IL-1β occurred in a Tlr2-, apoptosis-associated speck-like protein containing a caspase recruitment domain (Asc)-, and NLR family pyrin domain containing 3 (Nlrp3)-dependent manner upon infection with either HN878 or W_7642 in myeloid cells (Supplementary Fig. 5a). W_7642 mediated induction of IFN-β was dependent on cyclic GMP/AMP synthase (cGAS)15, and minimally on TLR2 (Supplementary Fig. 5b,c). In contrast, mRNA and protein levels of TNF-α were comparable in HN878- and W_7642-infected macrophages (Fig. 2c,d). When macrophages use aerobic glycolysis to generate energy, lactate is secreted as a by-product4,16. W_7642 infection induced significantly lower accumulation of lactate than HN878 infection in macrophages (Fig. 2d). Nitrite levels used as a measure of macrophage activation were not significantly different between HN878- and W_7642-infected macrophages (Fig. 2f). In HN878 infection, lactate accumulation, macrophage activation and Mtb control was partly dependent on IL-1R1 signaling pathway while IL-1R1 signaling was dispensable for activation of macrophages and Mtb control in W_7642-infected macrophages (Fig. 2d-g). Blocking IFNAR did not impact IL-1β production in B6 HN878-infected macrophages, but it resulted in increased IL-1β and lactate production in B6 W_7642-infected macrophages (Fig. 2h). IFNAR blockade during W_7642 infection significantly reduced IL-1β and lactate production in Il1r1−/− macrophages compared to B6 macrophages (Fig. 2h). While IFNAR blockade in HN878-infected Il1r1−/− macrophages decreased Mtb CFU, IFNAR blockade in W_7642-infected Il1r1−/− macrophages increased Mtb CFU (Fig. 2i). Co-infection of W_7642 and HN878 strains in macrophages (even at 3 HN878:1 W_7642 ratio) decreased IL-1β and lactate production and increased IFN-β levels compared to HN878 infection alone (Fig. 2j). TNF-α levels remained unchanged in single and co-infected macrophages (Supplementary Fig. 6). Thus, the presence of W_7642 can limit HN878-induced IL-1β and lactate accumulation in macrophages. Furthermore, macrophages treated with heat killed (hk) W_7642, or with hkHN878 and hkW_7642 together, also decreased IL-1β and lactate and increased IFN-β levels, when compared to hkHN878 treatment alone (Supplementary Fig. 7a). Thus, while Mtb replication is not required, an MDR Mtb cellular component limited IL-1β and aerobic glycolysis, while inducing IFN-β in macrophages. Using extracellular acidification rate (ECAR) as an indicator of glycolysis17, hkHN878 treatment induced significant ECAR in macrophages where this response was partly IL-1R1 dependent (Supplementary Fig. 7b). However, hkW_7642 treatment, or co-treatment with hkHN878 and hkW_7642 induced significantly lower ECAR (Supplementary Fig. 7b). Additionally, HN878 infection induced lower IL-1β levels and negligible levels of lactate in glucose-deprived galactose-containing medium in comparison to infection in glucose-containing medium (Fig. 2k). In contrast, culturing W_7642 infected macrophages in galactose-containing medium minimally impacted IL-1β or intracellular Mtb CFU (Fig. 2k,l). Low IFN-β levels were induced in HN878-infected macrophages grown either in glucose- or galactose-containing media, when compared to significantly higher IFN-β production in W_7642-infected macrophages grown in glucose-containing media (Fig. 2k). Thus, HN878 infection induced aerobic glycolysis to activate macrophage to mediate Mtb control, partly through the IL-1R1 pathway. In contrast, W_7642 is a poor inducer of IL-1β, instead inducing a potent IFN-β response and driving a less effective shift to aerobic glycolysis.
Figure 2. MDR Mtb W_7642 infection induces the type I IFN pathway and distinctive host macrophage metabolism.
B6 and Il1r1−/− macrophages were infected with HN878 or W_7642 (MOI1). RNA was extracted on 6 dpi and RNA sequencing was performed. The expression of genes in the glycolysis pathway in HN878- or W_7642-infected B6 macrophages over uninfected macrophages is shown (a, n=3, except W_7642 B6 n=2). The expression of genes in the oxidative phosphorylation and type I IFN pathways in W_7642- over HN878-infected B6 macrophages is shown (b, n=3, except W_7642 B6 n=2). The heat map of marker mRNAs from the annotated pathways are shown from individual samples within the different groups (c). Cytokine protein, lactate (d-e) and nitrite levels (f) were measured in supernatants, and intracellular Mtb CFU (g) was determined on 6 dpi (n=4–6). Infected macrophages were treated with IFNAR blocking antibody or isotype (25 μg/ml) at −1 and 3 dpi. IL-1β (n=5) and lactate levels (n=4) were measured in supernatants (h), and intracellular Mtb CFU (i, n=5) was determined on 6 dpi. B6 macrophages (n=5) were co-infected with HN878 and W_7642 (3 HN878:1 W_7642, 1 HN878:1 W_7642, total MOI1) for 6 dpi. IL-1β, lactate, and IFN-β levels were measured in supernatants (j). B6 macrophages were infected with HN878 or W_7642 while in glucose or galactose (25mM each)-containing media. IL-1β (n=6), lactate (n=4), and IFN-β levels (n=4) (k), and intracellular Mtb CFU (l, n=6) were determined 3 dpi. UN-untreated, UI-uninfected, nd-not detectable. (a-b) Gene set enrichment analysis was done using an FGSEA R package as in methods, (d-g,j,k) 1-way ANOVA with Tukey’s post-test, (h,i) 2-way ANOVA with Bonferroni post-test. The data points represent the mean (±SD) of values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, ns-not significant (p>0.05).
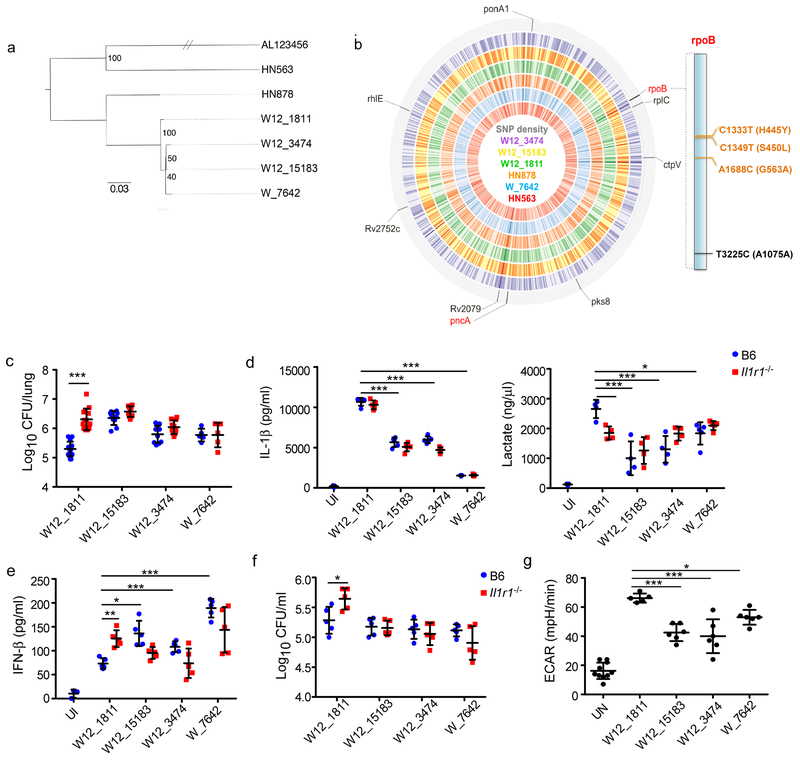
To delineate the specific molecular mechanism by which W_7642 activates macrophages, we used a collection of genetically conserved W-Mtb strains (Fig. 3a) that vary by a small number of SNPs by whole genome sequencing (WGS) (Fig. 3b, Supplementary Table 1). Of interest, W_7642 has two non-synonymous SNPs (H445Y and G563A) within the Mtb rpoB, which encodes the beta subunit of Mtb RNA polymerase5. A closely related MDR Mtb strain, W12_1811, encodes a mutation within rpoB at a distinct site (S450L). Infection of Il1r1−/− mice with W12_1811 resulted in increased lung Mtb CFU, pulmonary inflammation, and myeloid cell accumulation when compared to B6 mice infected with W12_1811 (Fig. 3c, Supplementary Fig. 8a,b). Additionally, W12_1811 infection in macrophages induced higher IL-1β and lactate production, and lower IFN-β levels when compared to W_7642-infected macrophages (Fig. 3d,e). Furthermore, Il1r1−/− macrophages were also less effective at controlling W12_1811 infection, when compared to B6 macrophages (Fig. 3f). Thus, similar to HN878, W12_1811 also uses the canonical IL-1R1 pathway to shift host metabolism towards aerobic glycolysis.
Figure 3. Identification of unique SNPs in rpoB in MDR Mtb strains that alter macrophage reprogramming.
WGS data was used to generate a SNP maximum-likelihood tree to determine the phylogenetic relationship between the Mtb strains. Reads were mapped onto the sequence of Mtb H37Rv (AL123456) (a). The WGS of the Mtb strains was used to generate a SNP density map. Loci where W12_1811 and W_7642 have different non-synonymous SNPs are indicated on the outer track and with labels (red labels indicate 2+ such loci per gene). On all tracks, darker shades indicate higher SNP rates. 1 tick = 100kb, total length = 4.4mbp, GC content = 65.6%. “Lolliplot” representation of rpoB (RVBD_0667) shows the positions of non-synonymous (orange) and synonymous (black) SNPs and the corresponding amino acid change (b). B6 and Il1r1−/− mice were aerosol infected with 100 CFU of Mtb strains and lung bacterial burden was determined on 30 dpi (W12_1811 n=15, W12_15183 and W12_3474 n=10, W_7642 n=5) (c). B6 and Il1r1−/− macrophages were infected with Mtb strains (MOI1) and IL-1β (d, n=5), lactate (d, n=4), and IFN-β (e, n=5), levels were measured in supernatants, and intracellular CFU (f, n=5) was determined by after 6 dpi. B6 macrophages were treated with hkMtb strains (20 μg/ml each) for 48 hours and ECAR was measured in treated cells (UN n=10, W12_1811 n=5, W12_15183 and W12_3474 n=6) (g). UN-untreated, UI-uninfected. (c-g) 1-way ANOVA with Tukey’s post-test. The data points represent the mean (±SD) of values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, ns-not significant (p>0.05).
W12_1811 represents a divergent point in the NYC MDR W-Mtb family where rifampicin resistance was gained3. We identified two other W12 strains, called W12_15183 and W12_3474, where similar to W_7642, W12_15183 contained both the rpoB-G563A and rpoB-H445Y SNPs, while W12_3474 only bore the rpoB-H445Y SNP. In vivo infection with W12_15183 and W12_3474 resulted in comparable Mtb lung CFU and pulmonary inflammation between B6 and Il1r1−/− infected mice (Fig. 3c, Supplementary Fig. 8a). Both strains also induced lower IL-1β and lactate levels, and higher IFN-β production when compared to W12_1811 (rpoB-S450L) infection (Fig. 3d,e). Also similar to W_7642 infection, in both W12_15183 and W12_3474 infections, Mtb control in macrophages was IL-1R1 independent (Fig. 3f). Further, while hkW12_1811 treatment induced significant ECAR in macrophages, both hkW12_15183 and hkW12_3474 treatment induced lower ECAR, and mirrored hkW_7642 ECAR induction (Fig. 3g). However, comparable Mtb growth kinetics in macrophages were observed (Supplementary Fig. 8c). From the 24 SNPs separating W12_1811 and W_7642, we eliminated SNPs where W_7642 had the same sequence as lab-adapted Mtb H37Rv or HN878 (Supplementary Table 1, highlighted blue-12 SNPs), as well as any SNPs that were unique to W_7642 alone (Supplementary Table 1, highlighted green-4 SNPs), or shared by only W_7642 and W12_15183, but not W12_3474 (Supplementary Table 1, highlighted red-6 SNPs). Thus we identified common SNPs in W_7642, W12_15183 and W12_3474 (Supplementary Table 1, highlighted yellow-2 SNPs), narrowing down our WGS results to two SNPs that may be functional in mediating macrophage metabolism reprograming: rpoB-H445Y and pykA-A369A, a pyruvate kinase A18. However, the pykA SNP is synonymous, thus implicating the rpoB-H445Y SNP as potentially causal in macrophage reprogramming.
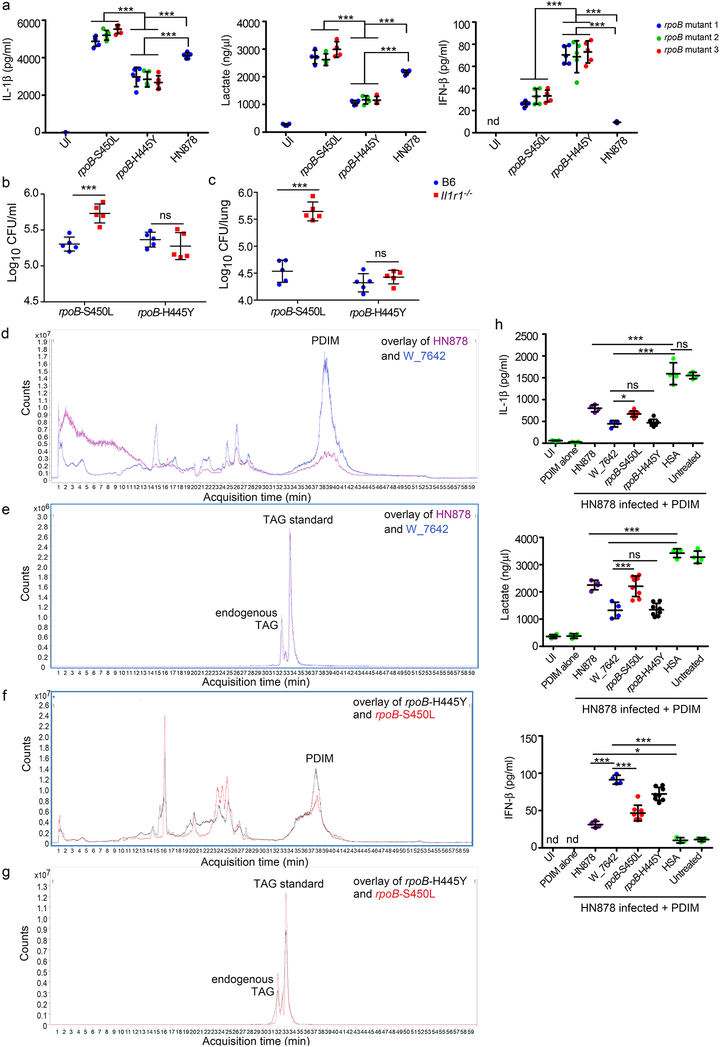
To validate a functional role for the rpoB-H445Y SNP in macrophage metabolic reprogramming, we therefore generated three HN878 clones independently under rifampicin selection carrying either the rpoB-H445Y SNP or the rpoB-S450L SNP, while exhibiting no changes in the pykA gene. Importantly, infection of macrophages with independent clones of HN878 rpoB-H445Y mutants, but not HN878 rpoB-S450L mutants, recapitulated effects of W_7642 infection, with lower production of IL-1β and lactate and increased induction of IFN-β, when compared to HN878 treatment and at varying MOIs (Fig. 3a, Supplementary Fig. 9a). Furthermore, infection with HN878 rpoB-S450L resulted in increased Mtb CFU in Il1r1−/− macrophages when compared with B6 macrophages at varying MOIs (Fig. 3b, Supplementary Fig. 9b). In contrast, there was no difference in the intracellular CFU of B6 and Il1r1−/− macrophages infected with HN878 rpoB-H445Y at varying MOIs (Fig. 3b, Supplementary Fig. 9b). Importantly, in vivo infection with HN878 rpoB-S450L resulted in increased pulmonary burden (Fig. 3c), recruited macrophage accumulation, and cytokine production in Il1r1−/− mice, compared with B6 infected mice (Supplementary Fig. 9c,e). In contrast, infection with HN878 rpoB-H445Y resulted in comparable pulmonary bacterial burden (Fig. 3c), and no increase in myeloid cellular recruitment or cytokine levels (Supplementary Fig. 9d,e) between B6 and Il1r1−/− mice. Additionally, HN878 rpoB-S450L but not HN878 rpoB-H445Y infection of macrophages induced robust IL-1β and lactate accumulation in glucose-containing medium but not very effectively in galactose-containing medium, suggesting that IL-1β induction is glucose dependent4 (Supplementary Fig. 9f). Similarly, low IFN-β levels were induced in HN878 rpoB-S450L-infected macrophages grown either in glucose- or galactose-containing media, and IFN-β production was significantly enhanced in HN878 rpoB-H445Y-infected macrophages grown in glucose-containing media (Supplementary Fig. 9f). These results demonstrate that the Mtb carrying the rpoB-H445Y SNP but not rpoB-S450L can modulate macrophage reprogramming to mediate Mtb control in the absence of IL-1R1 signaling.
SNPs in Mtb rpoB give rise to rifampicin resistance5 and are associated with broad transcriptomic changes, including the expression of secreted proteins and lipid biosynthetic intermediates19–21; the nature of lipid changes may depend on the location of SNPs within the rpoB gene21. Furthermore, upregulation of the PDIM biosynthetic operon19 and increased PDIM expression is reported in rpoB-H445Y resistant Mtb21. Thus, the rpoB-H445Y SNP in W_7642 may alter the composition of cell wall lipids, and impact host sensing of Mtb for reprogramming of macrophage metabolism. To address this, we identified PDIM changes in cell wall lipids between HN878 and the MDR Mtb strain W_7642. Cell wall lipids were purified from similar bacterial numbers, and an internal triacylglycerol (TAG) standard was used to obtain relative quantification of lipids between the different strains. W_7642 showed increased relative abundance of long-chain multimethyl-branched fatty acid PDIMs in cell wall lipid preparations (Fig. 3d,e, and Supplementary Fig. 10-structural characterization of PDIM subclasses, Supplementary Fig. 11a,b-PDIM spectra), when compared to the lower content of cell wall-associated PDIMs in HN878 (Fig. 3d,e and Supplementary Fig. 11a,b). Notably, as before21, when normalized to TAG, HN878 rpoB-H445Y Mtb recapitulated the presence of abundant long-chain multimethyl-branched fatty acid PDIMs in cell wall lipid preparations (Fig. 3f,g, Supplementary Fig. 11c,d), when compared to the PDIMs in HN878 rpoB-S450L (Fig. 3f,g, Supplementary Fig. 11c,d). Importantly, while the W12_1811 Mtb strain also expressed more short-chain fatty acid PDIMs (Supplementary Fig. 12a,b) in comparison to the long-chain fatty acid PDIMs present in cell wall lipid preparations from rpoB-H445Y containing MDR Mtb strains W12–15183 (Supplementary Fig. 12c), W12–3474 (Supplementary Fig. 12d), and W_7642 (Supplementary Fig. 12e). This coincided with increased expression of enzymes involved in PDIM synthesis such as phenolpthiocerol synthesis type-I polyketide synthase (ppsA, ppsB and ppsC) in W_7642 (Supplementary Fig. 11e). Both HN878 rpoB-H445Y and rpoB-S450L had increased mRNA expression of these enzymes (Supplementary Fig. 11f). Thus, different rpoB SNPs may upregulate PDIM biosynthetic pathways. To test the physiological effects of the PDIMs on macrophage metabolic responses, Mtb-infected macrophages were exposed to HN878-derived PDIM coated polystyrene beads and resulted in reduced IL-1β and lactate production, and increased IFN-β (Fig. 3h). Furthermore, while Mtb-infected macrophages exposed to W_7642-derived PDIM coated beads also suppressed IL-1β and lactate production and induced IFN-β, these changes were significantly pronounced and at lower PDIM coated bead doses when compared to effects of HN878-derived PDIM coated beads (Fig. 3h, Supplementary Fig. 11g). Similar to the W_7642-derived PDIM coated bead exposure, the PDIM-mediated metabolic rewiring also occurred more robustly and at lower doses when Mtb-infected macrophages were treated with HN878 rpoB-H445Y-derived PDIM coated beads, when compared with HN878 rpoB-S450L-derived PDIM coated beads (Fig. 3h, Supplementary Fig. 11g). PDIM-coated beads alone did not impact cytokine or metabolic changes in uninfected macrophages (Fig. 3h). Thus, while the abundance of PDIMs may directly induce IFN-β and limit IL-1β production and shift to aerobic glycolysis, PDIMs composition may likely also contribute towards reprogramming macrophage metabolism during Mtb infection and needs to be further tested.
Our limited knowledge of the immune parameters that mediate protection or drive disease progression during MDR Mtb infections is a significant hurdle to the current efforts to prevent world-wide emergence of MDR Mtb. We show here that Mtb carrying the widely prevalent rpoB-H445Y SNP5, can alter macrophage metabolism through the induction of IFN-β and bypass the requirement for IL-1R1 pathway signaling for protective immunity. While we did not carry out WGS on the independent rpoB-H445Y mutants, it is unlikely that the same type of secondary mutations would occur in each independent clone studied. Drug resistant Mtb strains with the rpoB-H445Y and rpoB-S450L SNPs are both associated with overexpression of PDIMs21. However, treatment with long-chain fatty acid PDIMs from rpoB-H445Y more stringently inhibited glycolysis and induced IFN-β in macrophages when compared to short-chain PDIMs from rpoB-S450L, suggesting that both abundance of PDIMs and composition of PDIMs may impact macrophage metabolic rewiring. The S450L SNP did not exhibit major structural changes in the rifampicin binding site, while the H445Y SNP mediated structural changes in the binding site of Mtb RNA polymerase, thus preventing any binding of rifampicin22. It is possible that these differential structural changes in RNA polymerase may regulate the differential expression of Mtb cell wall lipids including composition of PDIMs, as well other lipids21, and needs to be fully explored in future studies. PDIM presence has been associated with decreased phagosomal acidification, phagosomal permeabilization23, and also increased Mtb escape from the intracellular vacuole into the cytosol24, where it may mediate increased sensing by the cytosolic DNA sensor, cGAS and induction of IFN-β. A protective role for IL-1R1 signaling in Mtb infection is well known9, likely through the induction of the lipid mediator prostaglandin E225 and involvement in aerobic glycolysis within macrophages4. Type I IFN transcriptional signature is associated with pulmonary TB disease26, and considered detrimental to immunity against drug susceptible Mtb25. However, our results demonstrate that the IL-1R1 pathway is preferentially activated in drug susceptible Mtb infection, while MDR Mtb strains preferentially induce IFN-β that limits IL-1β induction, driving less effective aerobic glycolysis. Accordingly, even in pulmonary infection in mice, while both MDR and drug susceptible Mtb strains seemingly infect and induce TB disease, these infections recruit different inflammatory myeloid cells. Thus, the host immune metabolism induced in response to infection with drug resistant Mtb may be substantially different from responses induced upon infection with drug susceptible Mtb in hosts and need to more thoroughly studied. The implications of our findings are wide, as drug resistance in bacteria such as Staphlyococcus27, Klebsiella28 and Enterococci29 can induce cell surface lipid changes. Additionally, rifampicin resistance can occur in E. coli5, Streptococcus30, and Staphylococcus5, thus potentially impacting downstream host-pathogen interactions. Thus, our study emphasizes that fully understanding the mechanisms of pathogenesis and host immunity of drug resistant Mtb is critical for successful efforts to design new therapeutic targets and vaccines to prevent the spread of emerging MDR, as well as extensively and extremely drug resistant Mtb spread.
Methods
Mice
C57BL/6 (B6), Myd88−/−, Ifnar−/−, Nos2−/−, Il10−/− and Il1r1−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). The following mice were generously provided: Tlr2−/− mice (Dr. Laura Schuettpelz, Washington University in St. Louis), Tnfr1−/− mice (Dr. John H. Russell, Washington University in St. Louis), Ifngr−/− and cGas−/− mice (Dr. Herbert W. Virgin IV, Washington University in St. Louis) and bones from Asc−/− and Nlrp3−/− mice (Dr. Uma Nagarajan, University of North Carolina, Chapel Hill). Mice were used between the ages of 6 to 8 weeks, and both males and females were used. Sample sizes were chosen following empirical statistical power analysis based on previous studies31. Histological analysis following mouse experiments were subject to blinded analysis. All mice were maintained and used in accordance with approved Washington University in St. Louis IACUC guidelines.
Experimental infections
Mtb strains W_7642, W12_1811, W12_15183, and W12_3474 were from the Tuberculosis Center at the Public Health Research Institute, Newark, NJ. By whole genome sequencing (WGS), all MDR W-Mtb strains are resistant to isoniazid, rifampicin, ethambutol, streptomycin, pyrazinamide, and kanamycin, with the exception of W12_3474 which lacks resistance to pyrazinamide and W12_1811 which lacks resistance to ethambutol2. HN878 and HN563 were obtained from BEI resources under National Institutes of Health contract AI-75320. All Mtb strains were grown in Proskauer Beck (PB) medium with 0.05% Tween 80 and frozen at −80°C while in mid-log phase. Colony forming units (CFU) of bacterial stocks were calculated by plating serial dilutions on 7H11 agar plates. For Mtb aerosol infections, mice were infected with approximately 100 CFU of bacteria using a Glas-Col airborne infection system as previously described31. Pulmonary bacterial burden was determined at given time points through plating serial dilutions of lung homogenates on 7H11 agar plates.
Rifampicin susceptibility determination
Independent rifampicin resistant Mtb HN878 clones (biological replicates) were selected from rifampicin (2 μg/ml) containing 7H11 agar plates32. The sequences of rpoB and pykA in HN878 clones were confirmed by Sanger sequencing (Genewiz). Mtb stocks of 3 independent colonies for each SNP (rpoB-H445Y and –S450L) were grown, stocked, and the CFU determined as described above for further experimentation.
To confirm drug resistance, HN878, HN878 rpoB-H445Y and rpoB-S450L were grown on Middlebrook 7H10 agar plates at 35°C in a 10% CO2 atmosphere. Colonies not older than 2 weeks were transferred into a sterile tube containing 5.0 ml of water with 10 to 20 sterile glass beads. The suspension was vortexed for 1–2 mins, allowed to stand for 15 mins, then transferred to another tube and allowed to stand for 10 mins. The supernatant was transferred into a sterile tube and the turbidity was adjusted to 0.5 McFarland standard with water. A 1:5 dilution of this suspension in water was used. A volume of 0.1 ml of each final drug solution and 0.8 ml of oleic albumin dextrose catalase supplement were aseptically added into each MGIT containing 7.0 ml of broth followed by 0.5 ml of the final inoculum suspension. Lyophilized drugs (BACTEC streptomycin, isoniazid, rifampicin, ethambutol (S.I.R.E.) and pyrazinamide drug kit; BD Biosciences) were dissolved according to the manufacturer’s instructions. The final drug concentrations used were 0.1 μg/ml for isoniazid, 1.0 μg/ml for rifampin, 5 μg/ml for ethambutol, 1.0 μg/ml for streptomycin and 100 μg/ml for pyrazinamide. Mtb ATCC 27294 was used as control. Tubes were placed in the BACTEC™ MGIT™ 960 instrument, that automatically interprets the results as susceptible or resistant. HN878 and Mtb 27294 were susceptible to all drugs tested, while HN878 rpoB-H445Y, rpoB-S450L was only resistant to rifampin.
In vitro cell culture of myeloid cells
Bone marrow cells from the femur and tibia of B6 and gene deficient mice were extracted, and 1×107 cells were plated in 10 ml of complete Dulbecco’s modified eagle’s medium (cDMEM) supplemented with 20 ng/ml mouse recombinant (rm) granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech)31. Relevant conditions were tested for mycoplasma contamination using PCR33. Cells were then cultured at 37°C in 5% CO2. On day 3, 10 ml of cDMEM containing 20 ng/ml rmGM-CSF was added. On day 7, adherent cells were collected as macrophages and non-adherent cells were collected as dendritic cells (DCs).
Flow cytometry
Lung cell suspensions were prepared as described before31. Briefly, after perfusion with heparin in PBS, lungs were minced, digested with DNAse/collagenase, lysed for red blood cells, and pressed through a 0.7 μm filter to generate a single cell suspension. Cells were stained with appropriate fluorochrome-labeled specific antibodies or isotype control antibodies. Intracellular cytokine staining was performed using the BD Cytofix/Cytoperm kit (BD Biosciences). Mouse antibodies used include anti-CD11b (clone M1/70; Tonbo Biosciences), anti-CD11c (clone HL3; BD Biosciences), anti-Gr-1 (clone RB6–8C5, eBioscience), anti-CD3 (clone 500A2; BD Biosciences), anti-CD4 (clone RM4–5; BD Biosciences), anti-CD44 (clone IM7; eBioscience), and anti-IFN-γ (XMG1.2; BD Biosciences). Cells were processed with the Becton Dickinson (BD) Fortessa flow cytometer using FACS Diva software, or the BD FACSJazz flow cytometer using FACS Sortware software (BD). Flow cytometry experiments were analyzed using FlowJo (Tree Star Inc). As before34, neutrophils were defined as CD11b+CD11c-Gr-1hi cells, monocytes were defined as CD11b+CD11c-Gr-1med cells, and recruited macrophages were defined as CD11b+CD11c-Gr-1low cells. Total numbers of cells within each gate were back calculated based on cell counts/individual lung sample.
In vitro Mtb infection
Macrophages or DCs were infected with Mtb (MOI1 or 5) in antibiotic-free cDMEM. After varying days post infection (dpi), supernatants were collected for analysis of proteins or metabolites, and RNA was extracted for downstream sequencing. Infected macrophages were washed rigorously with sterile PBS to remove non-phagocytosed Mtb, then lysed with 0.05% sterile sodium dodecyl sulfate (SDS) for 5 minutes, then plated in serial dilutions on 7H11 agar plates to estimate intracellular CFU. In some cases, macrophages were treated with IFNAR blocking antibody (clone MAR1–5A3 BioXcell) at 25 μg/ml, on both −1 and 3 dpi. In some experiments, macrophages were cultured in glucose-deprived cDMEM (Thermofisher) supplemented with D-glucose or D-galactose (Sigma, 25mM) for 24 hours prior to infection with Mtb (MOI1) as before4. Cells were maintained in D-glucose or D-galactose supplemented media for the duration of infection (72 hours).
Generation of hkMtb
hkMtb was generated by incubating Mtb cultures at 80°C for 30 minutes. The protein content of each hkMtb stock was determined by bicinchoninic acid (BCA) assay using the Pierce BCA Protein Assay Kit (Thermo Scientific), following manufacturer’s instructions. Macrophages were treated with hkMtb for 48 hours (20 μg/ml) and culture supernatants were used for analysis of proteins and metabolites.
Determination of proteins and metabolites
Cytokine and chemokine production in the lung homogenate of Mtb-infected B6 and Il1r1−/− mice were analyzed using Milliplex Multiplex Assays (Millipore), according to manufacturer’s protocol. IL-1α and TNF-α were measured using Duoset kits (R&D Systems), IL-1β was measured using a BD OptEIA IL-1β ELISA Set (BD Biosciences), IFN-β was measured using a Legend Max™ Mouse IFN-β ELISA Kit (BioLegend), lactate accumulation was measured using a Lactate Assay Kit (Sigma-Aldrich) and nitrite production was measured using the Griess Reagent System (Promega). All commercial kits followed manufacturer’s instructions.
Histology
Lung lobes were perfused with 10% neutral buffered formalin and embedded in paraffin (WUSM Elvie L. Taylor Histology Core Facility). Lung sections were stained with hematoxylin and eosin (H&E) and inflammatory features were evaluated by light microscopy. Inflammatory lesions were outlined with the automated tool of the Zeiss Axioplan 2 microscope (Carl Zeiss) and percentage of inflammation was calculated by dividing the inflammatory area by the total area of individual lung lobes.
DNA isolation and sequencing
DNA was extracted from Mtb cultures for sequencing35. Mtb cultures were incubated for 30 minutes at 80°C, then treated with 10% SDS and proteinase K for 1 hour at 60°C. Proteins were precipitated with 5M NaCl and 10% cetyl trimethylammonium bromide (CTAB) for 15 minutes at 60°C. DNA was purified through addition of chloroform:isoamyl alcohol (24:1) and precipitated with isopropanol at −20°C for 1 hour. DNA pellet was washed in 80% ethanol, and dissolved in nuclease-free water. The Nextera DNA Library Preparation Kit was used for genome library preparation, and WGS was performed using the Illumina NextSeq platform (Illumina) for Mtb strains, respectively. The resultant raw FASTQ data were trimmed using Sickle (https://github.com/ucdavis-bioinformatics/sickle) and reads alignment was performed by Burrows–Wheeler Aligner36 using Mtb H37Rv (GenBank: AL123456) as the reference. Duplicate marking was done by Picard (http://broadinstitute.github.io/picard), and local realignment was performed using Genome Analysis Tool Kit37. SNPs and insertion-deletions (InDels) were called using Samtools38 and VariScan39, followed by annotation using snpEff40. Potential variants were excluded if the mapping quality or the base quality score was below 20 or the minimum alternate fraction was below 0.75. SNPs located at the mobile genetic elements, PE, PPE and PE-PGRS gene regions that might cause incorrect read alignment were also excluded. SNP maximum-likelihood phylogenetic tree was produced by RAxML 8.2.4. using the GTRGAMMA model and 100 bootstrap replicates41. Relevant SNPs (rpoB, pykA) were reconfirmed by Sanger sequencing.
RNA isolation and quantitative real-time PCR (qRT-PCR)
RNA was extracted using the Qiagen RNeasy Mini kit (Qiagen) and DNase I treated (Qiagen). cDNA was generated using ABI reverse transcription reagents (ABI, ThermoFisher) and RT-PCR was run on a Viia7 Real-Time PCR system (Life Technologies, Thermo Fisher). The log10 fold induction of mRNA in W12_1811, W_7642, HN878 rpoB-H445Y, and HN878 rpoB-S450L was calculated over expression levels in HN878, determined using the ΔΔCt calculation recommended by the manufacturer, using esxA mRNA expression as baseline. The primer sequences for ppsA, ppsB, ppsC19 and esxA42 have been previously published.
RNA sequencing, differential gene expression analysis and enrichment
For cDNA synthesis, we used a custom oligo-dT primer with a barcode and adaptor-linker sequence (CCTACACGACGCTCTTCCGATCT-xrefXX-T15). After first-strand synthesis, samples were pooled together based on Actb qPCR values and RNA-DNA hybrids were degraded with consecutive acid-alkali treatment. Subsequently, a second sequencing linker (AGATCGGAAGAGCACACGTCTG) was ligated with T4 ligase (NEB) followed by clean up with solid phase reverse immobilization (SPRI)-beads (Agencourt AMPure XP, BeckmanCoulter). The mixture was enriched by PCR for 12 cycles and purified with SPRI-beads (Agencourt AMPure XP, BeckmanCoulter) to yield final strand-specific RNA sequencing libraries. Libraries were sequenced on the HiSeq 2500 platform (Illumina) using 50 bp x 25 bp paired-end sequencing. Second read (read-mate) was used for sample demultiplexing. Reads were aligned to the GRCm38.p2 assembly of the mouse genome using STAR aligner43. Aligned reads were quantified using quant3p script (https://github.com/ctlab/quant3p). GENCODE genome annotation was used and DESeq244 was used for differential gene expression analysis. Pre-ranked gene set enrichment analysis was done using fgsea R package45. Genes were ranked according to Wald-statistics from DESeq2 analysis, only top 10000 genes ordered by mean expression were considered. MSigDB C2 gene set collection was used.
Real-time Extracellular Flux Assay
7.5×104 cells per sample were stimulated with hkMtb strains (20 μg/ml) for 2 days. Real-time extracellular acidification rate (ECAR) was measured using XF-96 Extracellular Flux Analyzer (Seahorse Bioscience) as described before46. Three consecutive measurements were obtained under basal conditions.
Lipid extraction and characterization of PDIMs
500 mg of Mtb (5×1011 bacteria47) was collected from cultures grown on solid agar 7H11 plates and boiled at 80°C for 30 minutes (min). In some experiments, we added 10 μg 20:0/20:0/20:0 triacylglycerol (TAG, Cayman Chemical) as internal standard to the Mtb cell pellets before lipid extraction. The added 20:0/20:0/20:0-TAG is a synthetic compound, m/z 992 as a [M + NH4]+ ion, that does not exist in nature. Endogenous Mtb TAG was represented by two well-separated peaks on the ion chromatograms, one of which was identified as13C2-18:1/16:0/26:0-TAG from 18:1/16:0/26:0-TAG. Therefore, the exogenously added TAG can be used as an internal standard. As before48, HN878 total lipids were sequentially extracted by chloroform:methanol (C:M, 2:1 and 1:2, v/v) and by chloroform:methanol:water (C:M:W, 10:10:3, v/v/v). Liquid chromatography/mass spectrometry (LC/MS) analysis was carried out using a Thermo Scientific TSQ Vantage mass spectrometer with Thermo Accela UPLC operated by Xcalibur software, or an Agilent 6550 A QTOF instrument with an Agilent 1290 HPLC, operated by Agilent Masshunter software. Separation of lipids was achieved by a Supelco 100 × 2.1 mm (2.7 μm particle size) Ascentis Express C-8 column at a flow rate of 300 μl/min. The mobile phase contained 5 mM ammonium formate (pH 5.0) both in solvent A, acetonitrile:water (60:40, v/v), and solvent B, isopropanol:acetonitrile (90:10, v/v). A gradient elution in the following manner was applied: 68% A, 0–1.5 min; 68–55% A, 1.5–4 min; 55–48% A, 4–5 min; 48–42% A, 5–8 min; 42–34% A, 8–11 min; 34–30% A, 11–14 min; 30–25% A, 14–18 min; 25–3% A, 18–23 min; 3–0% A, 25–30 min; 0% A, 30–35 min; 68% A, 35–40 min. The PDIM fraction was eluted at 25.7–29.5 min. The electrospray ionization (ESI) MS spectra of PDIMs were the signal average of the eluted peak. The resultant PDIM spectra were normalized to bacterial numbers, and relative abundance determined according to the 20:0/20:0/20:0-TAG internal standard, which gave rise to the [M+NH4]+ ions of m/z 992.98 and was eluted at 33–34 min. PDIM spectra and the internal TAG standards are shown in Supplementary Fig. 11. The normalized spectra were presented in Figure 3, where the ESI/MS spectra contain the homologous [M+NH4]+ ions of PDIM, ranging from m/z 1250 to 1550 (i.e., ions of m/z 1343, 1357, 1371, 1385, 1399, 1413, etc)49.
For structural characterization of PDIMs, shown in Supplementary Fig. 10, the lipid extract from Mtb W_7642 was dissolved in 1/1 chloroform/methanol with 0.5% NH4OH, and was infused into a Thermo Obitrap Velos mass spectrometer at a rate of 3ul/min. One in each methoxy (ion at m/z 1385) and keto (ion at m/z 1369) PDIM family was selected for structural characterization56.
PDIM isolation
HN878 total lipids were sequentially extracted by C:M (2:1 and 1:2, v/v) and by C:M:W (10:10:3, v/v/v). Extractions were combined, dried and kept at −20°C until use. Preparative TLCs (20 × 20 cm) were loaded with 7.5 mg of total lipid extract. Preparative thin-layer chromatography (TLC) to purify PDIMs were resolved by petroleum ether:acetone (96:4, v/v) as a solvent system as described50. PDIMs were identified, scraped from TLC plates, extracted from the silica using petroleum ether, and their purity verified by TLC, resolved as above, and visualized using 10% sulfuric acid in ethanol as described51. The preparation of 1 μm polysterine beads coated with human serum albumin (HSA, sham control) or PDIMs was performed as previously described for other Mtb antigens52. Briefly, 1.5×109 Polybead polystyrene beads (Polysciences Inc., Warrington, PA) were washed twice in 0.05 M carbonate-bicarbonate buffer (pH 9.6) and then incubated with 50 μg of purified PDIMs from various Mtb strains or buffer alone for 1 h at 37°C. Beads were then blocked with 5% HSA, washed repeatedly with 0.5% HSA, and finally adjusted to 4.0 × 108/ml in 0.5% HSA before being used in macrophage assays. Mtb-infected macrophages were treated with PDIM coated polystyrene beads at varying concentrations of beads:cells (25:1, 50:1, 100:1, or 200:1). HSA coated beads were used as a control. Cells were incubated with the beads overnight prior to infection with Mtb (MOI1) for 6 days.
Data availability
All relevant data are available from the authors. DNA sequencing data have been submitted under BioProject ID PRJNA353361. RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE115495).
Statistical analysis
Differences between the means of groups were analyzed using the two tailed Student’s t-test. Differences between the means of more than two groups were analyzed using 1-way ANOVA with Tukey’s post-test. For comparisons between two or more groups with two independent variables, 2-way ANOVA with Bonferroni post-test was used. All statistical analyses were done in GraphPad Prism 5. A p value <0.05 was considered significant. The data points across figures represent the mean (±SD) of values. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, ns-not significant (p>0.05). All experiments were replicated for reproducibility.
Supplementary Material
Figure 4. W-Beijing Mtb strains carrying the rpoB-H445Y SNP overexpress PDIM and bypass the IL-1R1 signaling pathway for protective immunity.
B6 and Il1r1−/− macrophages were infected with HN878 or three independently isolated HN878 rpoB-S450L or HN878 rpoB-H445Y mutants (MOI1) for 6 days (n=4). IL-1β, lactate, and IFN-β levels were measured in supernatants (a) and intracellular CFU was determined (b). B6 and Il1r1−/− mice (n=5) were aerosol infected with 100 CFU Mtb HN878 rpoB-S450L or HN878 rpoB-H445Y. Lung bacterial burden was determined on 30 dpi (c). The LC/MS reconstructed ion chromatogram (RIC, m/z 1330–1450) of PDIM (d) and the internal standard TAG (e) from HN878 and W_7642 are overlayed (the two earlier elution peaks of the TAG chromatogram are from endogenous Mtb 18:1/16:0/26-TAG (second isotope) and 18:0/16:0/26:0-TAG). The amounts of 20:0/20:0/20:0-TAG internal standard in the two samples are nearly equal. The LC/MS RIC (m/z 1330–1450) of PDIM (f) and the internal standard TAG (g) from rpoB-H445Y and rpoB-S450L are overlayed. The 20:0/20:0/20:0-TAG internal standard in the rpoB-S450L sample is about 1.5 times of the amount in rpoB-H445Y. Trace data is representative of at least two replicates. PDIM was isolated from each Mtb strain and coated onto polystyrene beads. B6 macrophages (n=4, except rpoB-H445Y and rpoB-S450L PDIM n=8) were treated with PDIM coated beads (200:1) from different Mtb strains or control HSA coated beads (200:1) alone or in combination with HN878 infection (MOI1). IL-1β, lactate and IFN-β protein levels were determined in 6 dpi supernatants (h). UN-untreated, UI-uninfected, nd-not detectable. (a-c) 2-way ANOVA with Bonferroni post-test, (h) 1-way ANOVA with Tukey’s post-test. The data points represent the mean (±SD) of values. *p≤0.05, ***p≤0.001, ns-not significant (p>0.05).
Acknowledgements
This work was supported by Washington University in St. Louis, NIH grant HL105427, AI123780 and AI111914 to S.A.K., NIH/NHLBI T32 HL007317–37 to N.H. A.S. was supported by the Ministry of Education and Science of Russian Federation (Project 2.3300.2017/4.6). J.R.-M. was supported by funds of the Department of Medicine, University of Rochester, and U19 AI91036. The protein identifications and LC-MS analyses were generated at the Washington University Proteomics Shared Resource (WU-PSR). The WU-PSR is supported by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the WU Mass Spectrometry Research Resource (NIGMS P41 GM103422, P60-DK-20579, P30-DK56341) and the Siteman Comprehensive Cancer Center (NCI P30 CA091842). The authors thank Drs. Laura Schuettpelz (Washington University in St. Louis), Uma Nagarajan (University of North Carolina, Chapel Hill), John H. Russell (Washington University in St. Louis) and Herbert W. Virgin IV (Washington University in St. Louis) for generously providing mice, Julia M. Scordo (Texas Biomed) and Racquel Domingo-Gonzalez (WashU) for technical help, and Sarah Squires and Lan Lu for animal breeding. Authors thank Drs. Thaddeus Stappenbeck and Jennifer Phillips (WashU) for critical reading of the manuscript.
Footnotes
Author Information
The authors declare no competing financial interests.
References
- 1.Global tuberculosis report 2015: WHO/HTM/TB/2015.22. Geneva: World Health Organization; (2015). [Google Scholar]
- 2.Bifani PJ, Mathema B, Kurepina NE & Kreiswirth BN Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends in Microbiology 10, 45–52, doi:S0966842X01022776 [pii] (2002). [DOI] [PubMed] [Google Scholar]
- 3.Bifani PJ et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275, doi: 10.1001/jama.1996.03530300036037 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Gleeson LE et al. Cutting Edge: Mycobacterium tuberculosis Induces Aerobic Glycolysis in Human Alveolar Macrophages That Is Required for Control of Intracellular Bacillary Replication. J Immunol 196, 2444–2449, doi: 10.4049/jimmunol.1501612 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Goldstein BP Resistance to rifampicin: a review.J. Antibiot.(Tokyo) 67, 625–630, doi: 10.1038/ja.2014.107 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Cooper AM et al. Disseminated tuberculosis in interferon gamma gene-disrupted mice. Journal of Experimental Medicine 178, 2243–2247 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn JL et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2, 561–572 (1995). [DOI] [PubMed] [Google Scholar]
- 8.MacMicking JD et al. Indentification of NOS2 as a protective locus against tuberculosis. Proceedings of the National Academy of the Sciences U.S.A. 94, 5243–5248 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juffermans NP et al. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. The Journal of infectious diseases 182, 902–908, doi: 10.1086/315771 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Scanga CA et al. MyD88-deficient mice display a profound loss in resistance to Mycobacterium tuberculosis associated with partially impaired Th1 cytokine and nitric oxide synthase 2 expression. Infection and immunity 72, 2400–2404 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manca C et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proceedings of the National Academy of Sciences of the United States of America 98, 5752–5757, doi: 10.1073/pnas.091096998 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desvignes L, Wolf AJ & Ernst JD Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J Immunol 188, 6205–6215, doi: 10.4049/jimmunol.1200255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redford PS et al. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur J Immunol, doi: 10.1002/eji.201040433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lachmandas E et al. Rewiring cellular metabolism via the AKT/mTOR pathway contributes to host defence against Mycobacterium tuberculosis in human and murine cells. Eur. J. Immunol. 46, 2574–2586, doi: 10.1002/eji.201546259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson RO et al. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe 17, 811–819, doi: 10.1016/j.chom.2015.05.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L, Nazarova EV, Tan S, Liu Y & Russell DG Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med, doi: 10.1084/jem.20172020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill LA & Pearce EJ Immunometabolism governs dendritic cell and macrophage function. J Exp Med 213, 15–23, doi: 10.1084/jem.20151570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noy T et al. Central Role of Pyruvate Kinase in Carbon Co-catabolism of Mycobacterium tuberculosis. J Biol Chem 291, 7060–7069, doi: 10.1074/jbc.M115.707430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bisson GP et al. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. J. Bacteriol.194, 6441–6452, doi: 10.1128/JB.01013-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Knegt GJ et al. Rifampicin-induced transcriptome response in rifampicin-resistant Mycobacterium tuberculosis. Tuberculosis 93, 96–101, doi: 10.1016/j.tube.2012.10.013 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Lahiri N et al. Rifampin Resistance Mutations Are Associated with Broad Chemical Remodeling of Mycobacterium tuberculosis. J. Biol. Chem. 291, 14248–14256, doi: 10.1074/jbc.M116.716704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molodtsov V, Scharf NT, Stefan MA, Garcia GA & Murakami KS Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis. Mol Microbiol 103, 1034–1045, doi: 10.1111/mmi.13606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barczak AK et al. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 13, e1006363, doi: 10.1371/journal.ppat.1006363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quigley J et al. The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis. MBio 8, doi: 10.1128/mBio.00148-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer-Barber KD et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature 511, 99–103, doi: 10.1038/nature13489, http://www.nature.com/nature/journal/v511/n7507/abs/nature13489.html#supplementary-information (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry MP et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977, doi:nature09247 [pii], 10.1038/nature09247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewelt-Belka W et al. Untargeted Lipidomics Reveals Differences in the Lipid Pattern among Clinical Isolates of Staphylococcus aureus Resistant and Sensitive to Antibiotics. J Proteome Res 15, 914–922, doi: 10.1021/acs.jproteome.5b00915 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kidd TJ et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9, 430–447, doi: 10.15252/emmm.201607336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra NN et al. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PloS one 7, e43958, doi: 10.1371/journal.pone.0043958 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright M, Zawadski P, Pickerill P & Dowson CG Molecular evolution of rifampicin resistance in Streptococcus pneumoniae. Microb Drug Resist 4, 65–70, doi: 10.1089/mdr.1998.4.65 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Khader SA et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8, 369–377 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Ford CB et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet 45, 784–790, doi: 10.1038/ng.2656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young L, Sung J, Stacey G & Masters JR Detection of Mycoplasma in cell cultures. Nat. Protoc 5, 929, doi: 10.1038/nprot.2010.43 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Treerat P et al. Novel role for IL-22 in protection during chronic Mycobacterium tuberculosis HN878 infection. Mucosal Immunol, doi: 10.1038/mi.2017.15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Soolingen D, Hermans PW, de Haas PE, Soll DR & van Embden JD Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29, 2578–2586 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson ND et al. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Frontiers in Genetics 6, 235, doi: 10.3389/fgene.2015.00235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutter S, Vilella AJ & Rozas J Genome-wide DNA polymorphism analyses using VariScan. BMC Bioinformatics 7, 409, doi: 10.1186/1471-2105-7-409 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cingolani P et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92, doi: 10.4161/fly.19695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313, doi: 10.1093/bioinformatics/btu033 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogerson BJ et al. Expression levels of Mycobacterium tuberculosis antigen-encoding genes versus production levels of antigen-specific T cells during stationary level lung infection in mice. Immunology 118, 195–201, doi: 10.1111/j.1365-2567.2006.02355.x (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sergushichev A An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv doi: 10.1101/060012, http://biorxiv.org/content/early/2016/06/20/060012 (2016). [DOI] [Google Scholar]
- 46.Huang SC-C et al. Cell-intrinsic lysosomal lipolysis is essential for macrophage alternative activation. Nat. Immunol 15, 846–855, doi: 10.1038/ni.2956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarthy Travis R et al. Overexpression of Mycobacterium tuberculosis manB, a phosphomannomutase that increases phosphatidylinositol mannoside biosynthesis in Mycobacterium smegmatis and mycobacterial association with human macrophages. Mol. Microbiol 58, 774–790, doi: 10.1111/j.1365-2958.2005.04862.x (2005). [DOI] [PubMed] [Google Scholar]
- 48.Torrelles JB et al. Truncated structural variants of lipoarabinomannan in Mycobacterium leprae and an ethambutol-resistant strain of Mycobacterium tuberculosis. J Biol Chem 279, 41227–41239, doi: 10.1074/jbc.M405180200 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Flentie KN, Stallings CL, Turk J, Minnaard AJ & Hsu F-F Characterization of phthiocerol and phthiodiolone dimycocerosate esters of M. tuberculosis by multiple-stage linear ion-trap MS. Journal of Lipid Research 57, 142–155, doi: 10.1194/jlr.D063735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torrelles JB et al. Identification of Mycobacterium tuberculosis clinical isolates with altered phagocytosis by human macrophages due to a truncated lipoarabinomannan. J Biol Chem 283, 31417–31428, doi: 10.1074/jbc.M806350200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slayden RA & Barry CE 3rd. Analysis of the Lipids of Mycobacterium tuberculosis. Methods Mol Med 54, 229–245, doi: 10.1385/1-59259-147-7:229 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Torrelles JB, Azad AK & Schlesinger LS Fine discrimination in the recognition of individual species of phosphatidyl-myo-inositol mannosides from Mycobacterium tuberculosis by C-type lectin pattern recognition receptors. J Immunol 177, 1805–1816 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors. DNA sequencing data have been submitted under BioProject ID PRJNA353361. RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession number GSE115495).