Persistent HPV infection is responsible for most cases of cervical cancer. The transition from precancerous to cancerous stages of HPV infection is marked by a significant reduction in virus production. Most global gene expression studies of HPV infection have focused on the cancerous stages. Therefore, little is known about global gene expression changes at precancerous stages. For the first time, we measured global gene expression changes at the precancerous stages of HPV16 infection in human cervical tissue producing high levels of virus. We identified a group of genes that are typically overexpressed in cancerous stages to be significantly downregulated at the precancerous stage. Moreover, we identified significantly modulated genes that have not yet been studied in the context of HPV infection. Studying the role of these genes in HPV infection will help us understand what drives the transition from precancerous to cancerous stages and may lead to the development of new therapeutic targets.
KEYWORDS: HPV16, cervical cancer, epithelium, keratinocytes, microarrays, papillomavirus, transcriptome
ABSTRACT
Human papillomavirus (HPV) infection is the world's most common sexually transmitted infection and is responsible for most cases of cervical cancer. Previous studies of global gene expression changes induced by HPV infection have focused on the cancerous stages of infection, and therefore, not much is known about global gene expression changes at early preneoplastic stages of infection. We show for the first time the global gene expression changes during early-stage HPV16 infection in cervical tissue using 3-dimensional organotypic raft cultures, which produce high levels of progeny virions. cDNA microarray analysis showed that a total of 594 genes were upregulated and 651 genes were downregulated at least 1.5-fold with HPV16 infection. Gene ontology analysis showed that biological processes including cell cycle progression and DNA metabolism were upregulated, while skin development, immune response, and cell death were downregulated with HPV16 infection in cervical keratinocytes. Individual genes were selected for validation at the transcriptional and translational levels, including UBC, which was central to the protein association network of immune response genes, and top downregulated genes RPTN, SERPINB4, KRT23, and KLK8. In particular, KLK8 and SERPINB4 were shown to be upregulated in cancer, which contrasts with the gene regulation during the productive replication stage. Organotypic raft cultures, which allow full progression of the HPV life cycle, allowed us to identify novel gene modulations and potential therapeutic targets of early-stage HPV infection in cervical tissue. Additionally, our results suggest that early-stage productive infection and cancerous stages of infection are distinct disease states expressing different host transcriptomes.
IMPORTANCE Persistent HPV infection is responsible for most cases of cervical cancer. The transition from precancerous to cancerous stages of HPV infection is marked by a significant reduction in virus production. Most global gene expression studies of HPV infection have focused on the cancerous stages. Therefore, little is known about global gene expression changes at precancerous stages. For the first time, we measured global gene expression changes at the precancerous stages of HPV16 infection in human cervical tissue producing high levels of virus. We identified a group of genes that are typically overexpressed in cancerous stages to be significantly downregulated at the precancerous stage. Moreover, we identified significantly modulated genes that have not yet been studied in the context of HPV infection. Studying the role of these genes in HPV infection will help us understand what drives the transition from precancerous to cancerous stages and may lead to the development of new therapeutic targets.
INTRODUCTION
Human papillomavirus (HPV) infection is the world's most common sexually transmitted infection, with approximately 291 million women worldwide being infected with the virus at any given point in time (1). While low-risk HPV types cause benign warts in the anogenital area, persistent infection with high-risk HPV types can give rise to various cancers of epithelial origin. In particular, HPV is responsible for most cases of cervical cancer, which is the third most common cancer in women worldwide and the most common cancer in women in developing countries (2). For this reason, the mechanism of HPV infection and HPV-mediated oncogenesis has been extensively studied. Early viral proteins E6 and E7 have been identified to be oncoproteins that play a critical role in tumorigenesis and tumor maintenance by inhibiting the tumor suppressor genes TP53 and RB1, respectively (3–10).
HPV initially infects dividing cells of the basal layer of the epidermis via microabrasions. Most infections are naturally resolved, but in some cases, the virus establishes a persistent infection, which may subsequently progress to precancerous lesions that are histologically graded as cervical intraepithelial neoplasia (CIN) grades I to III. When left untreated, these neoplastic lesions may ultimately develop into carcinoma in situ and invasive cancer. In persistently infected cervical tissue with normal or low-grade dysplasia, the HPV genome is maintained episomally and infectious viral particles are produced. In contrast, progression to severe dysplasia and invasive cancer is marked by integration of the viral genome into the host genome, which typically results in disruption of the E2 gene, the subsequent upregulation of oncogenes E6 and E7, and abrogation of virion production (11, 12).
In the past, many studies have looked into changes in the whole-genome expression profiles of precancerous and cancerous lesions in order to better understand the progression of persistent HPV infection to cervical cancer. However, most of these studies focused on neoplastic lesions and cancerous lesions (13–21) because it is difficult to acquire clinical samples at earlier stages of infection when patients are largely asymptomatic. Therefore, there is a gap in knowledge of global gene expression in the earlier preneoplastic stages of the disease, when HPV establishes productive infection in the host. In two recent studies, human keratinocytes persistently infected with HPV16 were used to measure global gene expression changes (22, 23), but they used keratinocytes derived from foreskin, which may not be appropriate for modeling infection in cervical tissue since HPV may have tissue-specific effects (24). Furthermore, these studies used monolayer cell cultures that do not produce virions by disallowing the virus to progress through its differentiation-dependent replication life cycle (25, 26). In other studies, overexpression tools were used to examine the effect of specific HPV oncoproteins on global gene expression (27, 28). Since these studies examined the effect of only individual viral proteins, they did not account for the full picture of HPV infection in the natural environment.
In this study, we used oligonucleotide microarrays to measure global gene expression changes in early-passage HPV16-infected human cervical keratinocytes (16HCK). Organotypic raft cultures were used in order to allow the virus to go through its full life cycle. A total of 594 and 651 genes were at least 1.5-fold upregulated and downregulated, respectively. Gene ontology analysis of upregulated genes identified biological processes that were significantly represented, including the cell cycle process and DNA metabolism. In contrast, biological processes that were significantly represented by downregulated genes included epidermis development, extracellular matrix disassembly, and regulation of NF-κB signaling.
(This article was submitted to an online preprint archive [29].)
RESULTS
Microarray analysis of HPV16 infection in cervical tissue.
Global gene expression changes in cervical tissue infected with HPV16 at a preneoplastic state were measured by conducting cDNA microarray analysis (GEO accession no. GSE109039) on 10-day organotypic raft tissue from early-passage human cervical keratinocytes (HCK) persistently infected with HPV16 (16HCK). In order to create 16HCK cell lines, human cervical tissue from biopsy specimens were acquired, processed, and cultured with keratinocyte-selective medium to grow primary HCKs. The HCKs were then electroporated with the HPV16 genome to establish persistent infection and immortalization. We used organotypic raft cultures, as previously described, which allow us to observe the full HPV life cycle in 3-dimensional (3D) tissue at a precancerous stage of infection when HPV particles are maximally produced. The organotypic raft tissue was harvested for microarray analysis at 10 days of culture, when particle production is most active (30), and 20 days of culture, when particle maturation is at its maximum (31), to measure the viral titer showing that virus particles are produced at high levels (Table 1). By infecting each cell line with the same virus and subjecting the raft cultures to the same condition, we were able to minimize variables and observe the specific effect of HPV infection on cervical tissue.
TABLE 1.
Viral titers of organotypic rafts
| Raft identification | No. of viral particles/raft |
|---|---|
| 16HCK-1, replicate 1 | 9.38 × 107 |
| 16HCK-1, replicate 2 | 1.18 × 107 |
| 16HCK-2, replicate 1 | 5.25 × 108 |
| 16HCK-2, replicate 2 | 6.5 × 109 |
| 16HCK-3, replicate 1 | 5.81 × 107 |
| 16HCK-3, replicate 2 | 1.6 × 109 |
The experiment was conducted in six individually grown raft tissue specimens representing three donor 16HCK cell lines, and raft tissue from uninfected HCKs served as a control. Out of the 34,575 genes that were analyzed with the cDNA microarray, a total of 1,245 genes were modified at least 1.5-fold (P < 0.05) and 533 genes were modified at least 2-fold (P < 0.05) with HPV16 infection compared to their expression in the uninfected control (see Table S1 in the supplemental material). The average of the pairwise Pearson correlation of the primary HCK samples was 0.885, and that of the 16HCK samples was 0.941. Of those genes that were significantly modulated at least 1.5-fold, 594 genes were upregulated and 651 genes were downregulated. Tables 2 and 3 show the 50 most upregulated and the 50 most downregulated genes in the microarray analysis, respectively. Among the top 50 upregulated genes were those associated with the cell cycle (CDKN2A, CDC7, NASP, MDC1, NFIX, FOXQ1). In contrast, the 50 most downregulated genes span from those involved in differentiation (RPTN, LCE1D, LCE3C, LCE1E, S100A7), extracellular matrix (ECM) modulation (KLK6, KLK8, KLK10, KLK13, MMP9, and MMP10), and immune regulation (LCN2, SPNS2, FAM3D, IL1RN, PSG4, IL1F7) to the antimicrobial response (RNASE7, PRSS3, PRSS2). This suggests that HPV infection drives the cell cycle and disrupts epidermal differentiation and ECM homeostasis while evading the immune and antimicrobial responses by downregulating them.
TABLE 2.
Top 50 upregulated genes with productive HPV16 infection
| Gene | Description or functiona | Fold change in expression |
|---|---|---|
| MT1G | Metallothioneins have a high content of cysteine residues that bind various heavy metals; these proteins are transcriptionally regulated by both heavy metals and glucocorticoids | 12.032 |
| MT1H | Mineral absorption/metabolism | 7.679 |
| TGFBR3 | Binds to TGF-β; could be involved in capturing and retaining TGF-β for presentation to the signaling receptors | 7.295 |
| KLHL35 | Kelch-like family member; Kelch-repeat β-propellers are generally involved in protein-protein interactions | 5.814 |
| DLK2 | Calcium ion binding and protein dimerization activity | 5.057 |
| FBLN1 | Cell adhesion/migration/ECM architecture organization | 5.056 |
| ASS1 | Arginine biosynthetic pathway | 4.446 |
| GPER | G protein-coupled estrogen receptor; cAMP signaling, calcium mobilization | 4.446 |
| TDRD9 | Probable ATP binding RNA helicase | 4.393 |
| TWIST1 | Transcription factor, role in cell lineage determination and differentiation | 4.208 |
| FANCE | Member of the Fanconi anemia complementation group E | 4.183 |
| JAM3 | Cell-to-cell adhesion in epithelial and endothelial cells | 4.169 |
| CDKN2A | Cell cycle regulation | 4.122 |
| TSPAN4 | A cell surface protein that mediates cell development, activation, growth, and motility | 3.733 |
| LTBP4 | Involved in assembly, targeting, and activation of TGFB1 | 3.718 |
| ERAP2 | Aminopeptidase involved in peptide trimming, MHC class I presentation | 3.551 |
| LOC341230 | Similar to argininosuccinate synthetase | 3.454 |
| MXRA5 | Matrix remodeling-associated proteins | 3.281 |
| RPS23 | Component of the 40S ribosomal subunit | 3.23 |
| C14orf132 | Putative uncharacterized protein | 3.165 |
| CRIP2 | Putative transcription factor with two LIM zinc binding domains | 3.148 |
| ANGPTL2 | Member of the vascular endothelial growth factor family | 3.061 |
| GOLPH4 | Involved in endosome-to-Golgi apparatus protein trafficking | 3.058 |
| DPYSL2 | Cytoskeletal remodeling, endocytosis | 3.013 |
| CDC7 | Checkpoint control kinase critical for G1/S transition | 2.981 |
| FAM134B | ER-anchored autophagy receptor | 2.947 |
| NASP | Histone binding protein with a role in cell division, cell cycle progression, and proliferation | 2.885 |
| HEG1 | Calcium ion binding receptor component; may act through stabilization of endothelial cell junctions | 2.866 |
| ZCWPW1 | Zinc finger domain-containing protein | 2.8 |
| MTE | Metallothioneins have a high content of cysteine residues that bind various heavy metals | 2.786 |
| MDC1 | Required for checkpoint-mediated cell cycle arrest in response to DNA damage | 2.783 |
| CDH13 | Calcium-dependent cell adhesion proteins | 2.775 |
| OLFML2A | Extracellular matrix binding protein | 2.762 |
| LOC392871 | Undetermined gene ortholog | 2.728 |
| CYBRD1 | Ferric chelate reductase | 2.726 |
| RASIP1 | Ras-interacting protein required for the formation of vascular structures | 2.721 |
| LOC729137 | Undetermined gene ortholog | 2.7 |
| GPNMB | Type 1 transmembrane glycoprotein | 2.693 |
| MOBKL2B | May regulate the activity of kinases | 2.672 |
| LOC388494 | Undetermined gene ortholog | 2.645 |
| C10orf54 | Putative uncharacterized protein | 2.644 |
| E2F2 | E2F family of transcription factors | 2.572 |
| LFNG | Transferase enzyme | 2.571 |
| LOC100133866 | Undetermined gene ortholog | 2.568 |
| NFIX | DNA binding protein, capable of activating transcription and replication | 2.56 |
| P4HTM | Prolyl hydroxylases | 2.553 |
| RELL1 | Receptor expressed in lymphoid tissue-like 1 | 2.551 |
| FOXQ1 | FOX genes are involved in gene development, cell proliferation, and tissue-specific gene expression | 2.53 |
| TJAP1 | Tight junction-associated protein | 2.527 |
| HOXA11AS | Noncoding RNA gene | 2.525 |
| FN3KRP | Phosphorylates psicosamines and ribulosamines | 2.514 |
TGF-β, transforming growth factor β; MHC, major histocompatibility complex; ER, endoplasmic reticulum.
TABLE 3.
Top 50 downregulated genes with productive HPV16 infection
| Gene | Description or function | Fold change in expression |
|---|---|---|
| RPTN | Involved in cornified cell envelope formation; multifunctional epidermal matrix protein; reversibly binds calcium | −57.487 |
| SERPINB4 | May act as a protease inhibitor to modulate the host immune response against tumor cells | −48.016 |
| TCN1 | Binds vitamin B12 and protects it from the acidic environment of the stomach | −34.735 |
| CST6 | Active cysteine protease inhibitors; loss of function is associated with progression of a primary tumor to a metastatic phenotype | −27.758 |
| KLK8 | Serine proteases with diverse physiological functions | −25.804 |
| CEACAM6 | GPIa-anchored glycoprotein; plays a role in cell adhesion | −24.469 |
| KLK6 | Serine proteases regulated by steroids | −22.088 |
| KLK13 | Expression regulated by steroid hormones and an important marker for breast cancer | −21.464 |
| RNASE7 | Pancreatic ribosomal protein; broad-spectrum antimicrobial activity against bacteria and fungi | −20.029 |
| PRSS3 | Digestive protease specialized for the degradation of trypsin inhibitors; in the ileum, may be involved in processing of defensin, including DEFA5 | −19.968 |
| LCE1D | Precursors of the cornified layers of the stratum corneum | −17.868 |
| MMP9 | Zinc-dependent endopeptidases and major proteases involved in the degradation of the ECM | −17.142 |
| FLJ22662 | Weak phospholipase activity; may act as an amidase or peptidase | −14.394 |
| TMEM45B | Transmembrane protein 45B | −13.469 |
| DMKN | Expressed in differentiated layers of the skin; upregulated in inflammatory disease | −13.186 |
| C6orf15 | Uncharacterized protein-coding gene | −12.641 |
| TMEM45A | Paralog of TMEM45B | −12.629 |
| PNLIPRP3 | Triglyceride lipase activity | −11.232 |
| KRT23 | Intermediate filament protein | −11.074 |
| ANXA9 | Calcium-dependent phospholipid binding protein; binds ECM proteins | −10.728 |
| LCE3C | Precursors of the cornified layers of the stratum corneum | −10.044 |
| S100A7 | Calcium binding proteins involved in cell cycle progression and cellular differentiation | −9.88 |
| EPS8L1 | Substrate for the epidermal growth factor receptor | −9.659 |
| RORA | Member of NR1 family of nuclear hormone receptors | −9.136 |
| LCN2 | Iron-trafficking protein involved in apoptosis, innate immunity, and renal development | −9.134 |
| LCE1E | Precursors of the cornified layers of the stratum corneum | −9.067 |
| SPNS2 | Sphingolipid transporter with a critical function in cardiovascular, immunological, and neuronal development | −8.787 |
| FAM3D | Related to cytokine activity | −8.734 |
| KLK7 | Member of the kallikrein family of serine proteases | −8.61 |
| HMOX1 | Heme oxygenase, an essential enzyme in heme catabolism | −8.548 |
| RNF39 | Role in early phase of synaptic plasticity | −8.5 |
| SH3BGRL2 | Protein disulfide oxidoreductase activity | −8.478 |
| C1orf68 | Uncharacterized protein-coding gene | −8.417 |
| POF1B | Key role in organization of the epithelial monolayers by regulating the actin cytoskeleton | −8.233 |
| ATP6V1C2 | Subunit of the peripheral V1 complex of the vacuolar ATPase | −8.232 |
| MMP10 | Zinc-dependent endopeptidases and major proteases involved in the degradation of the ECM | −7.953 |
| LOC730833 | Undetermined gene ortholog | −7.935 |
| CAPNS2 | Calcium-regulated thiol protease; role in tissue remodeling and signal transduction | −7.64 |
| KLK10 | Member of the kallikrein family of serine proteases | −7.553 |
| IL1RN | Inhibits the activity of interleukin-1 | −7.499 |
| ABCG1 | Intracellular lipid transport | −7.408 |
| THEM5 | Role in mitochondrial fatty acid metabolism | −7.287 |
| CYP2J2 | Role in arachidonic acid metabolism | −7.259 |
| PRSS2 | Serine protease involved in defensin processing | −7.239 |
| LOC645869 | Undetermined gene ortholog | −6.803 |
| PSG4 | May play a role in regulation of innate immune system | −6.559 |
| FAM63A | Role in protein turnover, deubiquitinase activity | −6.52 |
| MAP2 | Stabilizing/stiffening of microtubules | −6.474 |
| SLC5A1 | Na+/glucose cotransporter | −6.468 |
| IL1F7 | Interleukin-1-related gene | −6.392 |
| EPB41L3 | Tumor suppressor that inhibits cell proliferation and promotes apoptosis | −6.356 |
GPI, glycosylphosphatidylinositol.
While most previous global gene expression studies have focused on neoplastic and cancerous stages of HPV infection (13–21), two previous studies have modeled preneoplastic, early-stage HPV infection, similar to our study (22, 23). However, most of the modulated genes in the two studies did not overlap our results. One of these studies reported that 135 genes were modulated with HPV infection, but only 38 (28.1%) of these were modulated in the same direction and 4 of them were modulated in the opposite direction in our microarray analysis (22). In the other study, a total of 966 genes were reported to be modulated with HPV infection, but only 15 (1.6%) of these were modulated in the same direction in our microarray analysis (23). The two major differences between our study and the two previous studies are that we used cervical keratinocytes instead of foreskin keratinocytes and that we used organotypic raft cultures instead of monolayer cultures, which do not allow HPV to complete its life cycle. The use of keratinocytes derived from cervical tissue and organotypic raft cultures that allow the virus to complete its life cycle in 3D tissue enables us to capture the whole picture of an early-stage HPV infection in the cervix.
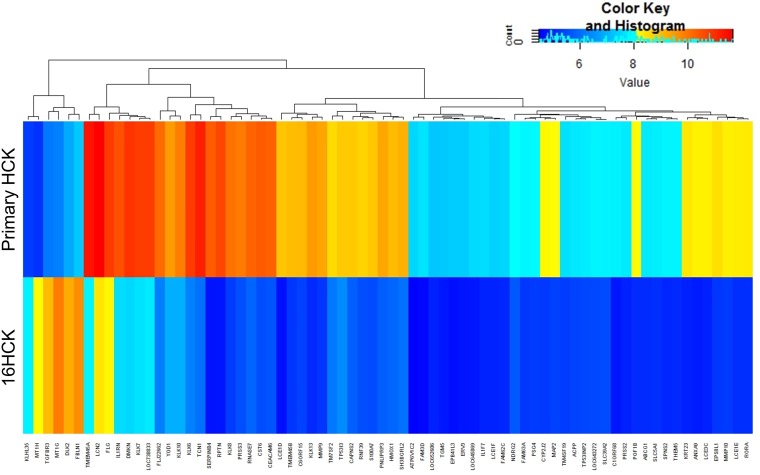
While the fold change in expression of the 50 most upregulated genes ranged from 12.1 to 2.5, that of the 50 most downregulated genes ranged from 57.5 to 6, indicating that a greater degree of gene modulation occurred in the downregulated genes than in the upregulated genes. Similarly, when the cutoff for inclusion was increased to at least a 5-fold modulation, only 6 genes were upregulated; in contrast, 72 genes were downregulated, as shown in Fig. 1. This suggests that HPV16 disrupts the host's physiology mostly by dampening many of the normal processes that may interfere with the virus's survival and replication.
FIG 1.
Heat map of genes whose expression was significantly modified. Six raft experiments, each with 16HCK and primary HCK cell lines representing three donors each, were set up. The tissues were harvested at 10 days of culture for RNA extraction and microarray analysis. The heat map shows genes whose expression was significantly modulated (P < 0.05) at least 5-fold with HPV16 infection.
Gene ontology analysis.
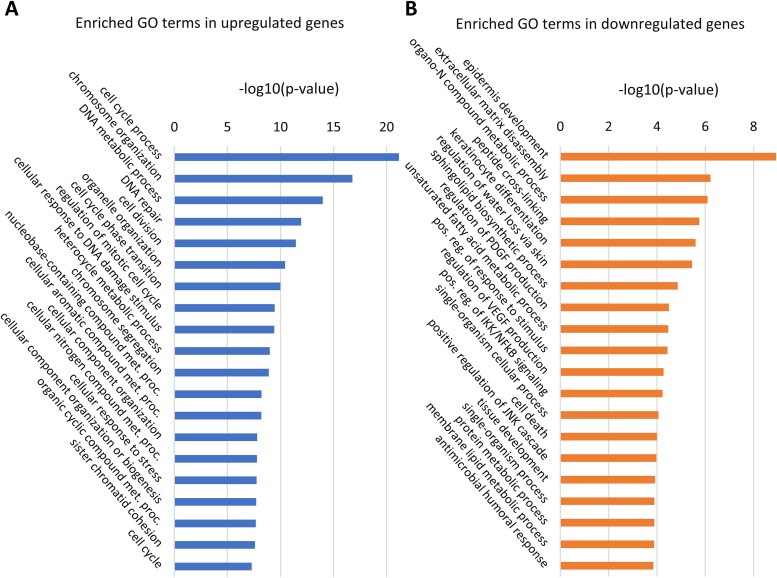
In order to identify the biological pathways that are significantly affected with HPV16 infection, we conducted gene ontology (GO) analysis using the online tool GOrilla (Tables S2 and S3) (32). The GO analysis result was then summarized using the REVIGO web server to combine similar GO terms for simplified visualization (33). A total of 144 GO terms that were significantly represented in upregulated genes were summarized to 82 GO terms using REVIGO (Table S4), and 77 GO terms that were significantly represented in downregulated genes were summarized to 52 GO terms using REVIGO (Table S5).
Among the genes that were upregulated with HPV16 infection, many of the represented GO terms are associated with cell cycle progression (the cell cycle process, cell division, cell cycle phase transition, regulation of mitotic cell cycle, and the cell cycle) and DNA metabolism (DNA metabolic process, DNA repair, cellular response to DNA damage stimulus), as shown in Fig. 2A. This suggests that persistent infection with HPV16 drives cellular proliferation in cervical tissue, as expected, due to the presence of viral oncogenes E6/E7, a finding consistent with the results of previous gene expression studies (16, 19, 27, 34). It is known that HPV oncoproteins E6 and E7 inhibit tumor suppressor genes TP53 and RB1, respectively, and that this is the main mechanism through which the virus promotes proliferation and tumorigenesis. “Translesion synthesis,” “neuron projection regeneration,” and “retina morphogenesis in camera-type eye” were among the upregulated DNA metabolism GO terms which have not yet been reported by previous global gene expression studies of HPV16 infection (Table S2). Translesion synthesis is a cellular DNA damage tolerance process of recovering from stalled replication forks by allowing DNA replication to bypass certain lesions (35, 36). Eight genes of the translesion synthesis GO category were upregulated in our analysis, including POLE2, UBA7, MAD2L2, and RPA2, which have not yet been reported in previous global gene expression studies of HPV infection. So far, not much is known about the exact role of translesion synthesis in HPV infection. One previous study speculated that HPV oncoprotein E6 may have inhibitory effects on translesion synthesis (37), while another study reported that p80, a cellular cofactor of HPV31 replication, may downregulate translesion synthesis (35). Our study shows for the first time that translesion synthesis is upregulated in a productive raft culture model of HPV16, suggesting that this process may facilitate viral replication and production.
FIG 2.
Enriched GO terms with the lowest P values. The online tool GOrilla was used to perform the initial GO analysis of our microarray data, and the REVIGO web server was used to summarize the enriched GO gene sets from the upregulated (A) and downregulated (B) lists of genes. The entire list of GO terms can be found in Tables S4 and S5 in the supplemental material. met. proc., metabolic process; PDGF, platelet-derived growth factor; VEGF, vascular endothelial growth factor; pos. reg., positive regulation.
Among the genes that were downregulated with HPV16 infection, the GO terms that were significantly represented are associated with skin development (epidermis development, keratinocyte differentiation), immune response (positive regulation of IκB kinase [IKK]/NF-κB signaling, antimicrobial humoral response), and cell death (cell death, positive regulation of Jun N-terminal protein kinase [JNK] cascade), as shown in Fig. 2B. The downregulation of genes associated with skin development has also been observed in previous studies of HPV-positive tumors (13, 17, 38). Similarly, the downregulation of genes under GO terms associated with the immune response and cell death has been shown in previous studies of HPV-positive tumors and E6 transgenic mice (15, 18, 22, 23, 28, 39). Overall, these results suggest that HPV16 perturbs normal epidermal development while having a hyperproliferative and immunosuppressive effect on cervical tissue. The GO terms for downregulated genes that have not yet been reported in previous global gene profiling studies of HPV infection included “positive regulation of cell migration” and “regulation of platelet-derived growth factor production” (Table S5). In contrast to our results, previous studies of global gene expression at cancerous stages of HPV infection have shown the upregulation of genes involved in cell migration, which is an indicator of invasive or metastatic cancer (15, 20, 40). Specifically, one study showed that MMP9, a well-established metastatic gene, is upregulated in cervical carcinoma cell lines and tissue samples, whereas our microarray analysis showed that this gene is downregulated 17.1-fold (40). Moreover, LCN2, which is overexpressed in various cancers and prevents the degradation of MMP9, was downregulated 9.1-fold in our microarray analysis (41–48). These opposing trends in modulation of cell migration genes highlight the fact that our study investigated early productive stages of HPV infection, whereas the other studies focused on the cancerous stages of infection, when viral production is significantly decreased. We speculate that cell migration genes interfere with efficient viral replication and assembly and, therefore, are suppressed during the productive stages of infection, whereas these genes are upregulated to facilitate tumor development and metastasis once the infection enters the cancerous stages.
Upregulation of the cell cycle and DNA metabolism.
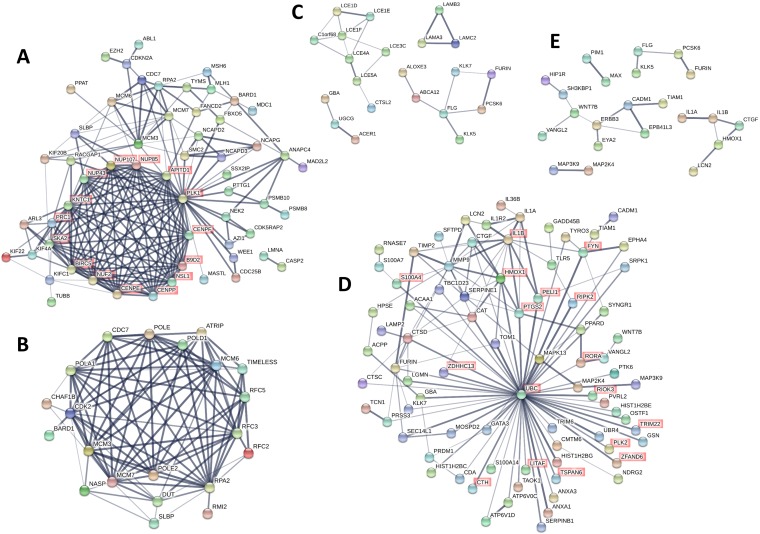
While GO analysis revealed biological processes that are significantly affected by persistent infection of HPV16, it does not provide information on interactions among the genes. Therefore, we further examined the interaction among individual proteins based on published data using the online tool STRING (49, 50). STRING protein-protein interaction network analysis allows us to identify signaling pathways, interactions among signaling pathways, and central proteins that have maximum interactions with other proteins within a category. A protein association network was created for each of the five aforementioned categories of biological processes that were broadly represented in the GO analysis: the cell cycle, DNA metabolism, skin development, immune response, and cell death.
From the upregulated genes, a protein association network was created with genes from 32 GO terms related to the cell cycle (Fig. 3A; Table S6). Genes that are not known to be associated with any other gene in this group were excluded from Fig. 3 for simplified visualization. The protein association analysis revealed 15 genes that are central to the network, as highlighted in red in Fig. 3A. These genes include nucleoporins (NUP43, NUP85, NUP107), centromere proteins (CENPE, CENPF, CENPP), and kinetochore-associated proteins (KNTC1, SKA2). This suggests that HPV infection drives the cell cycle and increases the expression of structural proteins involved in the cell cycle and cell division. Among these genes, NUP43, NUP85, NUP107, CENPP, and SKA2 have not yet been reported to be associated with HPV infection by previous gene expression studies.
FIG 3.
Protein association networks. The online tool STRING was used to create protein functional association networks of upregulated (A and B) and downregulated (C to E) genes with a >1.5-fold modulation of expression (P < 0.05). (A) Genes from GO categories related to cell cycle regulation were combined to create the network. (B) Genes from GO categories related to DNA metabolism were combined to create the network. (C) Genes from GO categories related to skin development and differentiation were combined to create the network. (D) Genes from GO categories related to inflammation and the immune response were combined to create the network. (E) Genes from GO categories related to apoptosis and cell death were combined to create the network. Thicker and darker lines represent greater confidence in the protein interaction on the basis of the supporting data. Genes that had no connection to any other gene within the network were excluded from the diagram.
Similarly, a protein association network was created with genes from 42 GO terms related to DNA metabolism (Fig. 3B; Table S7). The genes in this network included minichromosome maintenance (MCM) complex components (MCM3, MCM6, MCM7), replication factors (RFC2, RFC3, RFC5), and DNA polymerase subunits (POLA1, POLE, POLE2, POLD1). Among these genes, POLA1, POLE, and POLE2 have not yet been reported to be associated with HPV infection by previous gene expression studies. Previous studies have shown that overexpression of MCM genes is correlated with cervical carcinogenesis, and specifically, MCM7 has been shown to interact with the HPV18 E6 oncoprotein (51, 52). These results are consistent with the upregulation of cell cycle genes since cell division requires DNA replication and proteins associated with DNA metabolism. Additionally, our result suggests that the upregulation of MCM genes is initiated at early stages of HPV infection and sustained throughout carcinogenesis.
Downregulation of skin development, immune response, and cell death.
Within the downregulated gene sets, we combined genes from 7 GO terms in the protein association analysis for skin development, which gave 4 small networks (Fig. 3C; Table S8), including a network of late cornified envelope (LCE) genes (LCE1D, LCE1E, LCE1F, LCE3C, LCE4A, LCE5A) and a network of laminins (LAMA3, LAMB3, LAMC2). All eight genes in the network of LCEs were included in the GO term “epithelial cell differentiation,” while the three laminin genes were included in the GO term “epidermis development.” The LCE genes encode stratum corneum proteins of the epidermis and are all located in the same region of chromosome 1 (1q21.3), suggesting that epigenetic modifications might suppress the expression of this region as a whole. Our study shows for the first time the downregulation of a network of LCE genes in HPV infection. The two previous studies that analyzed global gene expression changes in early-stage HPV infection may not have observed changes in the expression of LCE genes since they used monolayer cultures, which do not allow the formation of the four strata of differentiating keratinocytes in the epidermis (22, 23). The three laminin genes LAMA3, LAMB3, and LAMC2 encode the three subunits that make up laminin 5, which plays an important role in wound healing, keratinocyte adhesion, motility, and proliferation (53, 54).
Genes from 16 GO terms were included in the protein association analysis for inflammation (Fig. 3D; Table S9). The gene for ubiquitin C (UBC), which is downregulated 2.48-fold with HPV16 infection, is at the center of this network and has the most associations with other genes related to inflammation. UBC is one of the two polyubiquitin genes that are involved in various cellular processes, including protein degradation, protein trafficking, cell cycle regulation, DNA repair, and apoptosis (55). Other ubiquitin-related genes that were significantly downregulated in our microarray analysis included those for proteins that are involved in ubiquitin conjugation (UBE2G1, UBE2F, UBR4, UBTD1), whereas two of the four ubiquitin-related genes that were significantly upregulated are involved in deubiquitination (USP1, USP13). This suggests that a decrease in ubiquitination may be important in the HPV16 life cycle and that the virus is trying to achieve this by decreasing ubiquitination and increasing deubiquitination. In previous studies, UBR4 has been shown to be a cellular target of the HPV16 E7 oncoprotein (56, 57), and we have shown that HPV16 upregulates the deubiquitinase UCHL1 in order to escape host immunity (58). Since ubiquitins are involved in many cellular processes, it is hard to identify which specific pathway is affected by the decrease in ubiquitination. It is possible that the virus prevents the degradation of proteins that are targeted by ubiquitin. Also, since our results suggest that HPV infection drives the cell cycle and downregulates cell death, it is possible that the downregulation of ubiquitination is involved in these processes. The network also included cytokines (IL1A, IL1B, IL36B), mitogen-activated protein kinases (MAPK13, MAP2K4, MAP3K9), proteases (CTSD, CTSC, KLK7, FURIN), serine protease inhibitors (SERPINB1, SERPINE1), and antimicrobial genes (S100A7, RNASE7, PRSS3, HIST1H2BC, HIST1H2BE, HIST1H2BG, LCN2). Among these genes, MAP3K9, CTSD, CTSC, SERPINE1, HIST1H2BC, HIST1H2BE, and HIST1H2BG have not yet been reported to be associated with HPV infection by previous gene expression studies. Of note, RNASE7 is a broad-spectrum antimicrobial protein that we have previously shown is downregulated by HPV infection (59). In terms of specific signaling pathways, the regulation of IκB kinase (IKK) and NF-κB signaling was significantly represented in the network (Fig. 3D, highlighted in red), which is consistent with our previous study that showed suppression of NF-κB activation by HPV16 (60). Most of these genes were shown to interact with UBC (Fig. 3D), and it is known that ubiquitination and proteolytic degradation of NF-κB inhibitor IκB can lead to NF-κB activation (61). This suggests that HPV16 evades the immune system by suppressing the NF-κB pathway and that this suppression may be mediated by downregulation of UBC.
Lastly, genes from 4 GO terms were included in the protein association analysis for cell death and gave 5 small networks (Fig. 3E; Table S10). The networks included genes from the JNK signaling pathway (TIAM1, IL1B, VANGL2, CTGF, WNT7B), suggesting that HPV16 may suppress cell death and promote transformation and tumorigenesis through this pathway. This is consistent with our previous study that showed the downregulation of cell death with HPV infection (62). Both the NF-κB and JNK pathways are downstream of tumor necrosis factor (TNF) signaling, and TNF ligands and receptors (TNFRSF19, LITAF, TNFSF9) were downregulated in the microarray. This suggests that HPV16 downregulates NF-κB and JNK pathways via TNF signaling downregulation. Among these genes, TIAM1, VANGL2, WNT7B, TNFRSF19, and LITAF have not yet been reported to be associated with HPV infection by previous gene expression studies.
Gene transcription changes correlate with changes in protein expression.
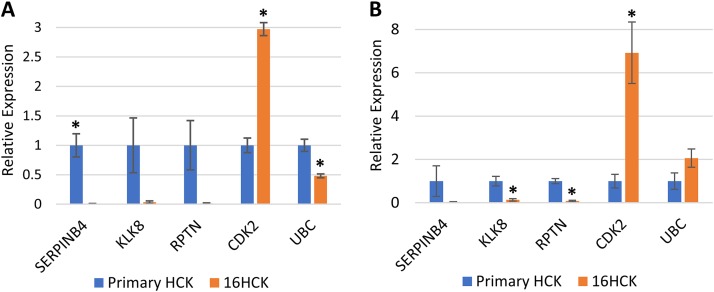
Of the numerous biological processes that were identified with GO analysis, we wanted to focus on processes and pathways that we felt were unique and most relevant to HPV infection and the life cycle. Therefore, four genes that are involved in processes involving epidermal development and differentiation (KLK8, RPTN, KRT23) and the immune response (SERPINB4) and that were modulated at least 10-fold were selected for validation. Additionally, UBC was selected for validation since it was shown to be central to the protein association network of inflammation (Fig. 3D), and cyclin-dependent kinase 2 (CDK2) was included for analysis as a marker of proliferation. In our microarray analysis, KLK8, RPTN, KRT23, SERPINB4, and UBC were downregulated 25.8-, 57.5-, 11.1-, 48-, and 2.48-fold, respectively, and CDK2 was upregulated 1.74-fold with HPV16 infection. KRT23 is a structural protein in epithelial cells, whereas KLK8 and RPTN are involved in the skin barrier proteolytic cascade and cornified cell envelope formation, respectively (63, 64). Recently, studies have reported that KRT23 may be involved in other cellular processes, including cell cycle regulation and apoptosis (65), which are key processes that were modulated by HPV infection in our study. KRT23 has not yet been reported by any other gene expression studies of HPV infection and, therefore, could be developed into a novel biomarker of productive HPV infection. In a recent gene expression profiling study, SERPINB4 was shown to be downregulated in early-stage HPV infection, consistent with the findings of our analysis (23). SERPINB4 is a serine protease inhibitor that is overexpressed in inflammatory skin diseases and various cancers, including squamous cell carcinomas, and may play a critical role in the immune response against HPV replication and virion production, as it has been shown that increased SERPINB4 expression can activate NF-κB (66–72). Similarly, KLK8 has been shown to be overexpressed in cervical cancer, ovarian cancer, and oral squamous cell carcinoma (73–76). So far, not much is known about these proteins in the context of preneoplastic HPV infections, and therefore, they could potentially become biomarkers or therapeutic targets of HPV infection at its early stages. In particular, overexpression of SERPINB4 and KLK8 in cancers prominently contrasts our microarray data, which include the two genes among the top downregulated genes. This contrast highlights the different microenvironments of the precancerous and cancerous states of HPV infection, and understanding the role of SERPINB4 and KLK8 may provide critical insight into the mechanism of HPV-induced carcinogenesis.
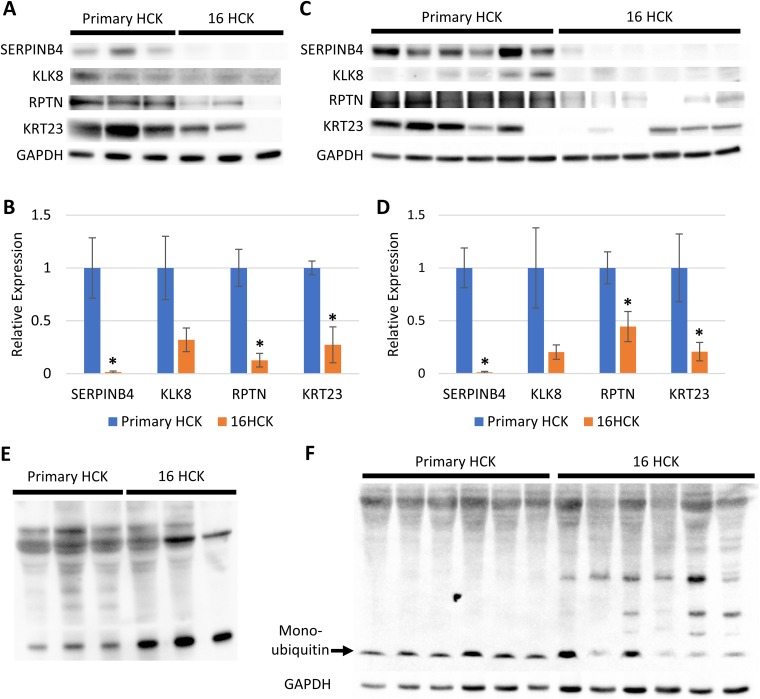
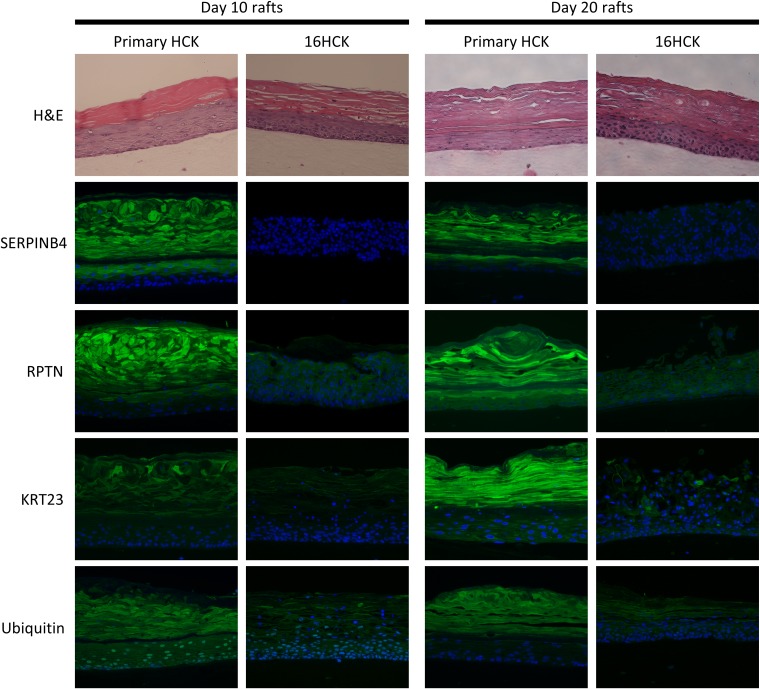
In order to increase the rigor of our data, all of the raft cultures for the validation experiments were set up with new cell lines, except for one set of 16HCK rafts. The downregulation of KLK8, RPTN, and SERPINB4 and the upregulation of CDK2 at the transcriptional level were observed with HPV16 infection at both 10 days and 20 days of culture, consistent with the results of microarray analysis (Fig. 4). Transcription of UBC was significantly decreased with HPV16 infection at 10 days of culture, as expected from our microarray analysis (Fig. 4A), but was slightly increased at 20 days of culture (Fig. 4B). We then further validated the downregulation of KLK8, RPTN, KRT23, and SERPINB4 at the translational level by Western blotting at 10 days (Fig. 5A and B) and 20 days (Fig. 5C and D) of culture. Although KLK8 expression was visibly downregulated with HPV16 infection by Western blotting, statistical significance was not reached on either 10 days or 20 days of culture. To measure UBC protein expression, we used an antiubiquitin antibody as a proxy for measuring UBC translation since UBC is simply a polyubiquitin protein that accounts for the majority of basal-level ubiquitin in cells (55, 77). Western blotting against ubiquitin showed the differential ubiquitination of various proteins with HPV16 infection (Fig. 5E and F) and the downregulation of monoubiquitin at 20 days of culture (Fig. 5F). Lastly, the downregulation of RPTN, KRT23, SERPINB4, and ubiquitin was validated with immunofluorescence staining (Fig. 6). In particular, RPTN, KRT23, and SERPINB4 were minimally expressed in the basal layers of both infected and uninfected controls, with no significant difference in expression between the two groups. In contrast, the three proteins were strongly expressed in the upper layers of uninfected tissues, and this expression was significantly decreased with HPV16 infection. The downregulation of the proteins may be attributed to the loss of the cornified layer in infected raft tissues.
FIG 4.
RT-PCR of genes of interest. RNA was extracted from day 10 (A) and day 20 (B) raft tissues, and RT-PCR was performed to measure the transcription levels of SERPINB4, KLK8, RPTN, CDK2, and UBC. For experiments with day 10 raft tissues (A), three raft experiments, each with 16HCK and primary HCK cell lines representing three donors each, were set up. For experiments with day 20 raft tissues (B), six raft experiments, each with 16HCK and primary HCK cell lines representing three and six donors, respectively, were set up. The transcription level of the TATA-binding protein (TBP) was used as a control to normalize each measurement (statistical significance, P < 0.05, two-tailed t test). The variance of relative expression ranged from 0.0001 to 0.65 for day 10 raft tissue and from 0.0012 to 2.96 for day 20 raft tissue.
FIG 5.
Western blot analysis of raft tissue. (A and C) Expression of the SERPINB4, KLK8, RPTN, and KRT23 protein was tested by Western blotting of primary HCK and 16HCK raft tissue harvested at day 10 (A) and day 20 (C). (B and D) Densitometry analysis was conducted on the Western blots at day 10 (B) and day 20 (D) (statistical significance, P < 0.05, two-tailed t test). The variance of relative expression ranged from 0.00037 to 0.27 for day 10 raft tissue and from 0.00029 to 0.86 for day 20 raft tissue. (E and F) Protein expression of ubiquitin was tested by Western blotting of primary HCK and 16HCK raft tissue harvested at 10 days (E) and 20 days (F) of culture. For experiments with day 10 raft tissues (A, B, E), three raft experiments, each with 16HCK and primary HCK cell lines representing three donors each, were set up. For experiments with day 20 raft tissues (C, D, F), six raft experiments, each with 16HCK and primary HCK cell lines representing three and six donors, respectively, were set up. GAPDH was used as a control. Images were acquired with a Bio-Rad ChemiDoc MP imaging system and Image Lab (v6.0.0) software.
FIG 6.
Immunofluorescence staining of raft tissue. The spatial protein expression of SERPINB4, RPTN, KRT23, and ubiquitin was observed by performing immunofluorescence staining of primary HCK and 16HCK raft tissue fixed at 10 days and 20 days of culture (green, target protein; blue, nuclear staining). For experiments with day 10 raft tissues, three raft experiments, each with 16HCK and primary HCK cell lines representing three donors each, were set up. For experiments with day 20 raft tissues, six raft experiments, each with 16HCK and primary HCK cell lines representing three and six donors, respectively, were set up. Magnifications, ×200. H&E, hematoxylin and eosin staining. Images were acquired with a Nikon Eclipse 80i microscope and NIS Elements (version 4.4) software.
DISCUSSION
In an attempt to understand HPV infection and its progression to cancer at a holistic level, many studies have investigated the global gene expression changes that occur with HPV infection. Most of these studies focus on neoplastic and cancerous lesions (13–21). Two previous studies used early-passage HPV16 cell lines, but only 28.1% and 1.6% of the genes reported to be modulated matched our results, and moreover, some of the genes were modulated in the opposite direction (22, 23). The discrepancies between our study and the two previous studies can be attributed to our use of an organotypic raft culture system and cervical cells instead of monolayer cell cultures and foreskin cells. Since the HPV life cycle spans all layers of the epidermis, monolayer cell cultures cannot produce progeny virus particles and, therefore, are limited models of HPV infection. Viral titers were measured on all 16HCK raft tissues to check for high levels of viral particle production (Table 1) and confirmed productive infection. Integration of the viral genome into the host genome and the subsequent reduction in viral particle production are hallmark events in the progression of precancerous to cancerous lesions, and thus, high levels of viral particle production indicate that the infection is in its earlier precancerous stages. Our study presents for the first time the global gene expression changes in cervical tissue with productive HPV infection.
Our microarray data showed that the majority of the modulated genes are downregulated (Fig. 1). Gene ontology analysis of the microarray data identified gene categories that were significantly represented, including cell cycle progression and DNA metabolism for the upregulated genes and skin development, immune response, and cell death for the downregulated genes (Fig. 2). The upregulation of cell cycle and DNA metabolism genes and the downregulation of cell death genes reflect the proliferative nature of persistent HPV16 infection. The downregulation of immune response and skin development genes can be understood in the context of the virus modulating the host environment to achieve efficient replication and virion production. The trends of modulation in these five gene categories are consistent with previously reported studies of global gene expression (13, 15–19, 22, 23, 27, 28, 34, 38, 39).
Several genes were selected for validation at the transcription and translational levels based on the degree of the fold change in expression, relevance to the HPV life cycle, and protein association network analysis. KLK8 and RPTN were selected for validation as they were among the top downregulated genes and are both involved in epithelium development. It is not surprising to see many genes involved in epithelium development to be affected by HPV infection since the virus infects, replicates, and assembles in the epithelium. In particular, HPV is not a lytic virus and is released from the epidermis via desquamation. Repetin was downregulated 57.5-fold with HPV16 infection in our microarray analysis, and this downregulation was validated by quantitative PCR (qPCR), Western blotting, and immunofluorescence (IF) staining. Repetin is a component of the epidermal differentiation complex and is involved in the formation of the cornified cell envelope (CE) (64). The CE is an insoluble matrix of covalently linked proteins formed beneath the plasma membrane of differentiating keratinocytes and plays an important role in the skin's function as a protective physical barrier against the external environment (78). In the context of HPV infection, the CE may hinder virion release because of its function as a physical barrier. Additionally, a previous study has shown that CEs of epithelial tissue infected with HPV11 are thinner and more fragile than those of healthy tissue (79). Therefore, it can be speculated that the downregulation of repetin by HPV may be a strategy to weaken the CE and increase the efficiency of virion release.
KLK8 was downregulated 25.8-fold in our microarray analysis, which was validated with qPCR and Western blotting. KLK8 was the most significantly downregulated gene of the seven kallikrein-related peptidase (KLK) genes that were downregulated with HPV16 infection; KLK3, KLK5, KLK6, KLK7, KLK10, and KLK13 were downregulated 3.6-, 4-, 22.1-, 8.6-, 7.5-, and 21.5-fold, respectively. A previous gene expression study also identified a cluster of KLK genes (KLK5, KLK6, KLK7, KLK10, KLK11) that are downregulated at early stages of HPV16 infection (22). KLKs are a family of 15 serine proteases that are clustered on chromosome 19q13.4, and one of their main functions is cleaving corneodesmosomal adhesion molecules in the cornified layer of the epidermis, which allows regulated desquamation of keratinocytes (63, 80–82). It is counterintuitive that HPV16 infection downregulates KLKs, since the virus is released via desquamation. We speculate that the rate at which normal epithelium desquamates is higher than the rate at which HPV virions mature in the cornified layer, and therefore, the virus may downregulate KLKs in order to impede desquamation and allow virions to adequately mature before being released to the surrounding environment. KLK8 also plays a role in activation of the antimicrobial peptide LL-37, and thus, HPV16 may downregulate the protein in order to prevent antimicrobial reaction (63, 83).
SERPINB4, also known as squamous cell carcinoma antigen 2 (SCCA2), is a member of the serpin family of serine protease inhibitors and was downregulated 48-fold in our microarray analysis. The downregulation in microarray analysis was validated with qPCR, Western blotting, and IF staining. SERPINB4, along with SERPINB3 (squamous cell carcinoma antigen 1 [SCCA1]), has been shown to be overexpressed in various types of cancers, including cervical, esophageal, lung, breast, and liver cancers (67, 69–71, 84–86). One of the mechanisms through which SERPINB4 contributes to tumor maintenance is the inhibition of granzyme M-induced cell death (87). Additionally, SERPINB4 overexpression is associated with inflammatory diseases, including psoriasis and atopic dermatitis (66, 68, 88–90). Remarkably, KLK8 shares the same expression pattern in these diseases: while our microarray analysis showed significant downregulation during productive HPV16 infection, the overexpression of KLK8 has also been associated with both squamous cell carcinomas and inflammatory skin diseases, such as psoriasis and atopic dermatitis (73–76, 91). We speculate that the virus downregulates SERPINB4 and KLK8 during productive infection as part of a broad effort to dampen the inflammatory response. Of particular note is that the two genes are overexpressed in various cancers, while in our study they are among the top downregulated genes during productive HPV infection. Additionally, we have identified nine other genes that were significantly downregulated in our microarray analysis but that have been shown to be overexpressed or contribute to disease progression in various types of cancers (Table 4). This highlights the fact that early productive stages of HPV infection present a microenvironment and disease state vastly different from those during the cancerous stages of infection, when virion production is significantly decreased. This suggests that KLK8 and SERPINB4 may interfere with the HPV life cycle or contribute to the immune surveillance against the virus and, therefore, are downregulated during productive infection. In contrast, the two proteins may be necessary for tumor maintenance and, therefore, are overexpressed during the cancerous stages of infection. However, since we did not measure the levels of expression of these proteins at cancerous stages, we cannot definitively compare the protein levels between precancerous and cancerous stages. A direct comparison would require creating organotypic raft cultures with cervical cancer cell lines and measuring SERPINB4 and KLK8 protein levels. In future studies, we aim to investigate the mechanism of KLK8 and SERPINB4 downregulation and how the two genes affect HPV entry, intracellular trafficking, replication, and assembly.
TABLE 4.
Differential gene expression between productive HPV16 infection and cancers
| Gene | Fold change in expression in productive HPV infection | Cancers in which gene expression increases and/or promotes disease progression [reference(s)]a |
|---|---|---|
| SERPINB4 | −48.0 | HNSCC (67), cervical (69, 71), esophageal (70), lung (99), liver (86), breast (85) |
| KLK5 | −3.2 | Bladder (100), ovarian (101), lung (102), breast (103), OSCC (75) |
| KLK6 | −22.1 | Bladder (100), ovarian (101), colorectal (104), HNSCC (105), pancreatic (106) |
| KLK8 | −25.8 | Bladder (100), ovarian (101), salivary gland (107), cervical (76), OSCC (75), colorectal (106) |
| KLK10 | −7.6 | Ovarian (101), OSCC (75), pancreatic (106), colorectal (106) |
| KLK13 | −21.5 | Lung (108), ovarian (109) |
| MMP9 | −17.1 | Cervical (40), breast (110), HNSCC (111), prostate (112) |
| MMP10 | −8.0 | OSCC (113), HNSCC (114), esophageal (115) |
| LCN2 | −9.1 | Breast (42, 43), esophageal (44), pancreatic (45), ovarian (46), colorectal (47), thyroid (48) |
| CTSD | −1.5 | Breast (116), ovarian (117) |
| CTSV | −2.8 | Breast (118), colorectal (118) |
OSCC, oral squamous cell carcinoma; HNSCC, head and neck squamous cell carcinoma.
UBC was the central protein in the network of proteins encoded by inflammatory genes that were significantly downregulated with persistent HPV16 infection (Fig. 3D). qPCR showed a statistically significant downregulation of UBC expression at 10 days of raft culture (Fig. 4A), while Western blotting of ubiquitin showed the downregulation of monoubiquitin at 20 days of culture and a differential pattern of protein ubiquitination (Fig. 5E and F). Additionally, IF staining showed the downregulation of the protein with persistent infection at both 10 days and 20 days of culture (Fig. 6). Since ubiquitin is involved in numerous biological processes, it is hard to conclude which specific pathways are affected by HPV16 infection. However, many proteins that are associated with UBC in the protein association network (Fig. 3D) are a part of the NF-κB pathway, and we speculate that UBC plays a central role in downregulating this pathway, especially since it has been shown that the ubiquitin-proteasome pathway plays a role in NF-κB activation. In future studies, we would like to investigate the role of UBC in relation to the top downregulated genes associated with inflammation, such as SERPINB4 and KLK8.
In conclusion, our study shows for the first time global gene expression changes with productive HPV16 infection in an organotypic raft culture model. With gene ontology analysis, broad gene categories that were significantly modulated with persistent HPV16 infection were identified, and these results were largely consistent with what previous studies have reported. However, we identified top downregulated genes that have not yet been extensively studied in the context of HPV infection and that have the potential to be developed as therapeutic targets or biomarkers. Moreover, the expression patterns of SERPINB4 and KLK8 highlighted that precancerous and cancerous stages of HPV infection are two distinct disease states. We attribute these new findings to our unique model of organotypic raft cultures at early-stage HPV16 infection, which allows the virus to go through its complete life cycle. Future studies investigating how the regulation of SERPINB4 and KLK8 changes throughout the different stages of infection may shed light on unidentified mechanisms of HPV persistence and tumorigenesis.
MATERIALS AND METHODS
Creating cervical cell lines and organotypic raft cultures.
Primary human cervical keratinocytes (HCK) were isolated from cervical biopsy specimens as previously described (92). The Human Subjects Protection Office of the Institutional Review Board (IRB) at the Penn State University College of Medicine screened our study design for exempt status according to the policies of this institution and the provisions of applicable federal regulations and, as submitted, found that it did not to require formal IRB review because no human participants were involved, as defined by the federal regulations.
HCK cell lines persistently infected with HPV16 (16HCK) were produced by electroporating primary HCK with HPV16 plasmid DNA as previously described (93, 94). The electroporated cells were cultured with mitomycin C-treated (Enzo Life Sciences) J2 3T3 feeder cells as previously described (92).
Organotypic raft cultures were grown as previously described (94, 95) at the first or second passage for primary HCK and the sixth to ninth passage for 16HCK. The raft tissues were harvested after 10 days for microarray analysis and 20 days for qPCR, Western blotting, and immunofluorescence staining. Viral gene expression has been shown to peak at between 10 and 12 days (30), while virion maturity reaches maximum stability at about 20 days (31).
For microarray analysis, three primary HCK cell lines and three 16HCK cell lines—each representing a different cervical tissue donor—were used to grow raft tissues. These experiments were repeated at separate time points to represent a total of six individually grown raft tissue specimens (n = 6) each for primary HCK and 16HCK.
For validation experiments (reverse transcription [RT]-qPCR, Western blotting, immunofluorescence staining), we used primary HCK and 16HCK cell lines from new donors in order to increase the rigor of our data. Only one 16HCK cell line that was used for the microarray analysis was also used for the validation experiments, but this raft tissue specimen was grown at a different time point in an experiment separate from that for tissue grown for the microarray analysis. For experiments with 10-day rafts, three primary HCK cell lines representing three donors (n = 3) and three 16HCK cell lines representing three different donors (n = 3) were used to grow raft tissues. For experiments with 20-day raft tissues, six raft experiments, each (n = 6) with 16HCK and primary HCK cell lines representing three and six donors, respectively, were set up.
Microarray analysis.
Raft tissue from primary HCK and 16HCK were harvested at 10 days, and RNA was extracted using an RNeasy fibrous tissue midikit (Qiagen). Microarray analysis was performed using an Illumina HumanHT-12 (v4) expression bead chip (Illumina, San Diego, CA), which targets over 31,000 annotated genes with more than 47,000 probes derived from the National Center for Biotechnology Information (NCBI) RefSeq Database, release 38 (7 November 2009), and other sources. RNA quality and concentration were assessed using an Agilent 2100 bioanalyzer with an RNA Nano LabChip (Agilent, Santa Clara, CA). cRNA was synthesized by TotalPrep amplification (Ambion, Austin, TX) from 500 ng of RNA according to the manufacturer's instructions. T7 oligo(dT)-primed reverse transcription was used to produce first-strand cDNA. The cDNA then underwent second-strand synthesis and RNA degradation by DNA polymerase and RNase H, followed by filtration clean-up. In vitro transcription (IVT) was employed to generate multiple copies of biotinylated cRNA. The labeled cRNA was purified using filtration and quantified by use of a NanoDrop spectrophotometer, and the volume was adjusted for a total of 750 ng/sample. Samples were fragmented and denatured before hybridization for 18 h at 58°C. Following hybridization, the bead chips were washed and fluorescently labeled. The bead chips were scanned with a BeadArray reader (Illumina, San Diego, CA).
The CLC Genomics Workbench (v4.8) package (Qiagen Bioinformatics) was used to determine the genes significantly differentially expressed between the HPV16-positive tissue and primary tissue. For each comparison, quantile normalization of six primary HCK samples (n = 6) and six 16HCK samples (n = 6) was performed, followed by a pairwise homogeneous t test, resulting in normalized fold changes and P values. Significantly differentially expressed genes were considered to be those with a P value for differential expression of <0.05 and an absolute fold change in expression of ≥1.5.
Gene ontology analysis and protein association network.
In order to categorize the significant gene expression changes into Gene Ontology (GO) groups, the GOrilla package was used (http://cbl-gorilla.cs.technion.ac.il/) (32). Two unranked lists of genes, the target (significantly modulated genes) and the background (all genes in the microarray), were used to identify significantly enriched GO terms. We focused on the subontology biological processes for our analysis. The REVIGO web server was used to further summarize the redundancy in the GO analysis (http://revigo.irb.hr/) (33). In our analysis, we used a similarity coefficient of 0.7 (medium-size list) to summarize the GO list.
In order to identify protein-protein associations among the upregulated and downregulated genes, the online tool STRING (https://string-db.org/) was used (50). Genes from similar GO categories were pooled to form protein association networks of “cell cycle” and “DNA metabolism” for the upregulated genes, and “skin development,” “immune response,” and “cell death” for the downregulated genes.
Viral titers (DNA encapsidation assay).
The viral titers of each raft experiment were measured with the qPCR-based DNA encapsidation assay described previously (31, 96).
RT-qPCR.
RT-qPCR was used in order to measure the levels of transcription of SERPINB4, KLK8, RPTN, CDK2, and UBC. For SERPINB4, forward primer 5′-ATTTCCTGATGGGACTATTGGCAATG-3′, reverse primer 5′-CAGCAGCACAATCATGCTTAGA-3′, and probe 5′-/FAM/ACGACACTG/ZEN/GTTCTTGTGAACGCA/3IABkFQ/-3′ (where FAM represents 6-carboxyfluorescein and 3IABkFQ represents 3′ Iowa Black FQ) were used. For KLK8, forward primer 5′-TGGGTCCGAATCAGTAGGT-3′, reverse primer 5′-GCAGGAACATCCACGTCTT-3′, and probe 5′-/FAM/CCCTGGATT/ZEN/CTGGAAGACCTCACC/3IABkFQ/-3′ were used. For RPTN, forward primer 5′-CCACAAATATGCCAAAGGGAATG-3′, reverse primer 5′-GTCATTTGGTCTCTGGAGGATG-3′, and probe 5′-/FAM/ACTGCTCTT/ZEN/GGCTGAGTTTGGAGA/3IABkFQ/-3′ were used. For CDK2, forward primer 5′-GCCTGATTACAAGCCAAGTTTC-3′, reverse primer 5′-CGCTTGTTAGGGTCGTAGTG-3′, and probe 5′-/FAM/AGATGGACG/ZEN/GAGCTTGTTATCGCA/3IABkFQ/-3′ were used. For UBC, forward primer 5′-GGATTTGGGTCGCAGTTCTT-3′, reverse primer 5′-TGGATCTTTGCCTTGACATTCT-3′, and probe 5′-/FAM/AGGTTGAGC/ZEN/CCAGTGACACCATC/3IABkFQ/-3′ were used. TATA-binding protein (TBP) was used as a control, for which forward primer 5′-CACGGCACTGATTTTCAGTTCT-3′, reverse primer 5′-TTCTTGCTGCCAGTCTGGACT-3′, and probe 5′-HEX-TGTGCACAGGAGCCAAGAGTGAAGA-BHQ-1-3′ (where HEX represents 6-carboxy-2,4,4,5,7,7-hexachlorofluorescein and BHQ-1 represents black hole quencher 1) were used. All primers and probes were synthesized by Integrated DNA Technologies, and a QuantiTect Probe RT-PCR kit (Qiagen) was used for the PCRs. All RT-PCRs were performed using a C1000 thermal cycler (Bio-Rad). The thermal cycler was programmed for 30 min at 50°C, then 15 min at 95°C, and then 42 cycles of 15 s at 94°C and 1 min at 54.5°C.
Western blotting.
Raft tissues were harvested at 20 days and used to prepare total protein extracts as previously described (97). Total protein concentrations were measured using the Peterson protein assay as previously described (98). The total protein extracts were applied to a sodium dodecyl sulfate-polyacrylamide gel (8 to 10%) and transferred to a nitrocellulose membrane and then incubated overnight at 4°C with antibodies against SERPINB4 (1:2,000 dilution; catalog number LS-C172681; Life Span Biosciences), KLK8 (1:2,000 dilution; catalog number H00011202-M01; Abnova), RPTN (1:2,000 dilution; catalog number LS-B17, Life Span Biosciences), KRT23 (1:2,000 dilution; catalog number ab117590; Abcam), and ubiquitin (1:2,000 dilution; catalog number 3933S; Cell Signaling). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (1:1,000 dilution; catalog number sc-47724; Santa Cruz) was used as a control. The membranes were then washed and incubated with horseradish peroxidase-linked secondary antibody (catalog number NA931VS/NA934VS; GE Healthcare) and developed using the Amersham ECL Prime Western blotting detection reagent (GE Healthcare). Densitometry analysis was conducted by normalizing the protein expression levels to the GAPDH expression level.
Immunohistochemistry and immunofluorescence staining.
Raft cultures were fixed in 10% buffered formalin and embedded in paraffin, and 4-μm cross sections were prepared. A section from each sample was stained with hematoxylin and eosin as previously described (26).
For immunofluorescence staining, the slides were submerged in xylene for deparaffinization and then were rehydrated. Antigen retrieval was achieved by submerging the slides in Tris-EDTA buffer (pH 9) in a 90°C water bath for 10 min. The slides were then rinsed with Tris-buffered saline (TBS)–Tween and blocked with the Background Sniper blocking reagent (Biocare Medical). The slides were then stained with the primary antibody overnight at 4°C. Each sample was stained with antibodies against SERPINB4 (1:2,000 dilution; catalog number LS-C172681; Life Span Biosciences), RPTN (1:1,000 dilution; catalog number LS-B17; Life Span Biosciences), KRT23 (1:500 dilution; catalog number ab117590; Abcam), and ubiquitin (1:750 dilution; catalog number 3933S; Cell Signaling). The slides were then rinsed with TBS-Tween 3 times and stained with secondary antibody (Alexa Fluor 488; Life Technologies) diluted 1:200 for 1 h at room temperature. Next, the slides were stained with Hoechst nuclear stain (1:5,000 dilution) for 15 min and rinsed with TBS-Tween twice. All antibodies were diluted in Da Vinci Green diluent (Biocare Medical). The experiment was conducted with three primary HCK and three 16HCK samples in duplicate. A Nikon Eclipse 80i microscope and NIS Elements (v4.4) software were used to acquire images.
Statistical analysis.
The microarray data shown in Fig. 1 are quantile-normalized means for six primary HCK and six 16HCK samples determined using the CLC Genomics Workbench (v4.8) software package. Statistical significance was determined with the homogeneous t test, using the CLC Genomics Workbench (v4.8) software package.
In order to establish statistical significance in qPCR data and Western blotting densitometry analysis, a t test with a P value cutoff of <0.05 was used.
Accession number(s).
The GEO accession number for our microarray data is GSE109039.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lynn Budgeon for doing the raft tissue embedding, slicing of sections, and staining.
Work in C.M.'s group was supported by National Institutes of Health grants R01CA225268 and R01DE018305-03S1 (NIDCR-ARRA Supplement).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01261-18.
REFERENCES
- 1.de Sanjosé S, Díaz M, Castellsagué X, Clifford G, Bruni L. 2007. Worldwide prevalence and genotype distribution of cervical HPV in women with normal cytology. Lancet Infect Dis 7:453–459. doi: 10.1016/S1473-3099(07)70158-5. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 4.Werness BA, Levine AJ, Howley PM. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 5.Dyson N, Howley PM, Münger K, Harlow E. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 6.Boyer SN, Wazer DE, Band V. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res 56:4620–4624. [PubMed] [Google Scholar]
- 7.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol 75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu W, Putral L, Hengst K, Minto K, Saunders NA, Leggatt G, McMillan NAJ. 2006. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther 13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- 9.Hall AHS, Alexander KA. 2003. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol 77:6066–6069. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lea JS, Sunaga N, Sato M, Kalahasti G, Miller DS, Minna JD, Muller CY. 2007. Silencing of HPV 18 oncoproteins with RNA interference causes growth inhibition of cervical cancer cells. Reprod Sci 14:20–28. [DOI] [PubMed] [Google Scholar]
- 11.Cripe TP, Haugen TH, Turk JP, Tabatabai F, Schmid PG, Dürst M, Gissmann L, Roman A, Turek LP, Turek LP. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J 6:3745–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sang B-C, Barbosa MS. 1992. Increased E6/E7 transcription in HPV 18-immortalized human keratinocytes results from inactivation of E2 and additional cellular events. Virology 189:448–455. doi: 10.1016/0042-6822(92)90568-A. [DOI] [PubMed] [Google Scholar]
- 13.Wong YF, Cheung TH, Tsao GS, Lo KW, Yim SF, Wang VW, Heung MM, Chan SC, Chan LK, Ho TW, Wong KW, Li C, Guo Y, Chung TK, Smith DI. 2006. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int J Cancer 118:2461–2469. doi: 10.1002/ijc.21660. [DOI] [PubMed] [Google Scholar]
- 14.Sopov I, Sorensen T, Magbagbeolu M, Jansen L, Beer K, Kuhne-Heid R, Kirchmayr R, Schneider A, Dürst M. 2004. Detection of cancer-related gene expression profiles in severe cervical neoplasia. Int J Cancer 112:33–43. doi: 10.1002/ijc.20351. [DOI] [PubMed] [Google Scholar]
- 15.Gius D, Funk MC, Chuang EY, Feng S, Huettner PC, Nguyen L, Bradbury CM, Mishra M, Gao S, Buttin BM, Cohn DE, Powell MA, Horowitz NS, Whitcomb BP, Rader J. 2007. Profiling microdissected epithelium and stroma to model genomic signatures for cervical carcinogenesis accommodating for covariates. Cancer Res 67:7113–7123. doi: 10.1158/0008-5472.CAN-07-0260. [DOI] [PubMed] [Google Scholar]
- 16.Rosty C, Sheffer M, Tsafrir D, Stransky N, Tsafrir I, Peter M, de Crémoux P, de La Rochefordière A, Salmon R, Dorval T, Thiery JP, Couturier J, Radvanyi F, Domany E, Sastre-Garau X. 2005. Identification of a proliferation gene cluster associated with HPV E6/E7 expression level and viral DNA load in invasive cervical carcinoma. Oncogene 24:7094–7104. doi: 10.1038/sj.onc.1208854. [DOI] [PubMed] [Google Scholar]
- 17.Pyeon D, Newton MA, Lambert PF, Den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P. 2007. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res 67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, Gissmann L, Dürst M, Schneider A, Pothuri B, Mansukhani M, Basso K, Chaganti RSK, Murty VV. 2007. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Miller C, Mosher R, Zhao X, Deeds J, Morrissey M, Bryant B, Yang D, Meyer R, Cronin F, Gostout BS, Smith-McCune K, Schlegel R. 2003. Identification of cervical cancer markers by cDNA and tissue microarrays. Cancer Res 63:1927–1935. [PubMed] [Google Scholar]
- 20.Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, Fearon ER, Cho KR. 2007. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res 67:10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 21.Wong YF, Selvanayagam ZE, Wei N, Porter J, Vittal R, Hu R, Lin Y, Liao J, Shih JW, Cheung TH, Lo KWK, Yim SF, Yip SK, Ngong DT, Siu N, Chan KY, Chan CS, Kong T, Kutlina E, McKinnon RD, Denhardt DT, Chin K-V, Chung TKH. 2003. Expression genomics of cervical cancer: molecular classification and prediction of radiotherapy response by DNA microarray. Clin Cancer Res 9:5486–5492. [PubMed] [Google Scholar]
- 22.Wan F, Miao X, Quraishi I, Kennedy V, Creek KE, Pirisi L. 2008. Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int J Cancer 123:32–40. doi: 10.1002/ijc.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klymenko T, Gu Q, Herbert I, Stevenson A, Iliev V, Watkins G, Pollock C, Bhatia R, Cuschieri K, Herzyk P, Gatherer D, Graham SV. 2017. RNASeq analysis of differentiated keratinocytes reveals a massive response to late events during human papillomavirus type 16 infection, including loss of epithelial barrier function. J Virol 91:e01001-17. doi: 10.1128/JVI.01001-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arany I, Evans T, Tyring SK. 1998. Tissue specific HPV expression and downregulation of local immune responses in condylomas from HIV seropositive individuals. Sex Transm Infect 74:349–353. doi: 10.1136/sti.74.5.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedell MA, Hudson JB, Golub TR, Turyk ME, Hosken M, Wilbanks GD, Laimins LA. 1991. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol 65:2254–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyers C, Frattini MG, Hudson JB, Laimins LA. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 27.Garner-Hamrick PA, Fostel JM, Chien W-M, Banerjee NS, Chow LT, Broker TR, Fisher C. 2004. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J Virol 78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffy CL, Phillips SL, Klingelhutz AJ. 2003. Microarray analysis identifies differentiation-associated genes regulated by human papillomavirus type 16 E6. Virology 314:196–205. doi: 10.1016/S0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 29.Kang SD, Chatterjee S, Alam S, Salzberg AC, Milici J, van der Burg SH, Meyers C. 2018. The effect of productive HPV16 infection on global gene expression of cervical epithelium. bioRxiv doi: 10.1101/295402. [DOI] [PMC free article] [PubMed]
- 30.Ozbun MA, Meyers C. 1997. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol 71:5161–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway MJ, Alam S, Ryndock EJ, Cruz L, Christensen ND, Roden RBS, Meyers C. 2009. Tissue-spanning redox gradient-dependent assembly of native human papillomavirus type 16 virions. J Virol 83:10515–10526. doi: 10.1128/JVI.00731-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J Virol 75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehoux M, Fradet-Turcotte A, Lussier-Price M, Omichinski JG, Archambault J. 2012. Inhibition of human papillomavirus DNA replication by an E1-derived p80/UAF1-binding peptide. J Virol 86:3486–3500. doi: 10.1128/JVI.07003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang TT, Nijman SMB, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8:341–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 37.Day T, Vaziri C. 2009. HPV E6 oncoprotein prevents recovery of stalled replication forks independently of p53 degradation. Cell Cycle 8:2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buitrago-Pérez A, Garaulet G, Vázquez-Carballo A, Paramio JM, García-Escudero R. 2009. Molecular signature of HPV-induced carcinogenesis: pRb, p53 and gene expression profiling. Curr Genomics 10:26–34. doi: 10.2174/138920209787581235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendoza-Villanueva D, Diaz-Chavez J, Uribe-Figueroa L, Rangel-Escareão C, Hidalgo-Miranda A, March-Mifsut S, Jimenez-Sanchez G, Lambert P, Gariglio P. 2008. Gene expression profile of cervical and skin tissues from human papillomavirus type 16 E6 transgenic mice. BMC Cancer 8:347. doi: 10.1186/1471-2407-8-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez-Ortiz G, Pina-Sanchez P, Vazquez K, Duenas A, Taja L, Mendoza P, Garcia JA, Salcedo M. 2005. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer 5:68. doi: 10.1186/1471-2407-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L, Borregaard N, Kjeldsen L, Moses MA. 2001. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J Biol Chem 276:37258–37265. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Bielenberg DR, Rodig SJ, Doiron R, Clifton MC, Kung AL, Strong RK, Zurakowski D, Moses MA. 2009. Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci U S A 106:3913–3918. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer M, Eickhoff JC, Gould MN, Mundhenke C, Maass N, Friedl A. 2008. Neutrophil gelatinase-associated lipocalin (NGAL) is a predictor of poor prognosis in human primary breast cancer. Breast Cancer Res Treat 108:389–397. doi: 10.1007/s10549-007-9619-3. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H, Xu L, Xiao D, Xie J, Zeng H, Wang Z, Zhang X, Niu Y, Shen Z, Shen J, Wu X, Li E. 2007. Upregulation of neutrophil gelatinase-associated lipocalin in oesophageal squamous cell carcinoma: significant correlation with cell differentiation and tumour invasion. J Clin Pathol 60:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedl A, Stoesz SP, Buckley P, Gould MN. 1999. Neutrophil gelatinase-associated lipocalin in normal and neoplastic human tissues. Cell type-specific pattern of expression. Histochem J 31:433–441. [DOI] [PubMed] [Google Scholar]
- 46.Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Pecorelli S. 2004. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer 112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- 47.Hu L, Hittelman W, Lu T, Ji P, Arlinghaus R, Shmulevich I, Hamilton SR, Zhang W. 2009. NGAL decreases E-cadherin-mediated cell-cell adhesion and increases cell motility and invasion through Rac1 in colon carcinoma cells. Lab Invest 89:531–548. doi: 10.1038/labinvest.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iannetti A, Pacifico F, Acquaviva R, Lavorgna A, Crescenzi E, Vascotto C, Tell G, Salzano AM, Scaloni A, Vuttariello E, Chiappetta G, Formisano S, Leonardi A. 2008. The neutrophil gelatinase-associated lipocalin (NGAL), a NF-kappaB-regulated gene, is a survival factor for thyroid neoplastic cells. Proc Natl Acad Sci U S A 105:14058–14063. doi: 10.1073/pnas.0710846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franceschini A, Lin J, Von Mering C, Jensen LJ. 2016. SVD-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics 32:1085–1087. doi: 10.1093/bioinformatics/btv696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, Von Mering C. 2015. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kühne C, Banks L. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem 273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- 52.Das M, Prasad SB, Yadav SS, Govardhan HB, Pandey LK, Singh S, Pradhan S, Narayan G. 2013. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS One 8:e69607. doi: 10.1371/journal.pone.0069607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider H, Mule C, Pacho F. 2007. Biological function of laminin-5 and pathogenic impact of its deficiency. Eur J Cell Biol 86:701–717. doi: 10.1016/j.ejcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Amano S, Akutsu N, Ogura Y, Nishiyama T. 2004. Increase of laminin 5 synthesis in human keratinocytes by acute wound fluid, inflammatory cytokines and growth factors, and lysophospholipids. Br J Dermatol 151:961–970. doi: 10.1111/j.1365-2133.2004.06175.x. [DOI] [PubMed] [Google Scholar]
- 55.Radici L, Bianchi M, Crinelli R, Magnani M. 2013. Ubiquitin C gene: structure, function, and transcriptional regulation. Adv Biosci Biotechnol 4:1057–1062. doi: 10.4236/abb.2013.412141. [DOI] [Google Scholar]
- 56.Huh K-W, Demasi J, Ogawa H, Nakatani Y, Howley PM, Mü K. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci U S A 102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White EA, Sowa ME, Tan MJA, Jeudy S, Hayes SD, Santha S, Münger K, Harper JW, Howley PM. 2012. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A 109:E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karim R, Tummers B, Meyers C, Biryukov JL, Alam S, Backendorf C, Jha V, Offringa R, van Ommen GJB, Melief CJM, Guardavaccaro D, Boer JM, van der Burg SH. 2013. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathog 9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karim R, Meyers C, Backendorf C, Ludigs K, Offringa R, van Ommen GJB, Melief CJM, van der Burg SH, Boer JM. 2011. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS One 6:e17848. doi: 10.1371/journal.pone.0017848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tummers B, Goedemans R, Pelascini LPL, Jordanova ES, van Esch EMG, Meyers C, Melief CJM, Boer JM, van der Burg SH. 2015. The interferon-related developmental regulator 1 is used by human papillomavirus to suppress NFκB activation. Nat Commun 6:6537. doi: 10.1038/ncomms7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Magnani M, Crinelli R, Bianchi M, Antonelli A. 2000. The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-kB (NF-kB). Curr Drug Targets 1:387–399. doi: 10.2174/1389450003349056. [DOI] [PubMed] [Google Scholar]
- 62.Ma W, Tummers B, van Esch EMG, Goedemans R, Melief CJM, Meyers C, Boer JM, van der Burg SH. 2016. Human papillomavirus downregulates the expression of IFITM1 and RIPK3 to escape from IFNγ- and TNFα-mediated antiproliferative effects and necroptosis. Front Immunol 7:496. doi: 10.3389/fimmu.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eissa A, Amodeo V, Smith CR, Diamandis EP. 2011. Kallikrein-related peptidase-8 (KLK8) is an active serine protease in human epidermis and sweat and is involved in a skin barrier proteolytic cascade. J Biol Chem 286:687–706. doi: 10.1074/jbc.M110.125310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huber M, Siegenthaler G, Mirancea N, Marenholz I, Nizetic D, Breitkreutz D, Mischke D, Hohl D. 2005. Isolation and characterization of human repetin, a member of the fused gene family of the epidermal differentiation complex. J Invest Dermatol 124:998–1007. doi: 10.1111/j.0022-202X.2005.23675.x. [DOI] [PubMed] [Google Scholar]
- 65.Liffers S-T, Maghnouj A, Munding JB, Jackstadt R, Herbrand U, Schulenborg T, Marcus K, Klein-Scory S, Schmiegel W, Schwarte-Waldhoff I, Meyer HE, Stühler K, Hahn SA. 2011. Keratin 23, a novel DPC4/Smad4 target gene which binds 14-3-3ε. BMC Cancer 11:137. doi: 10.1186/1471-2407-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mee JB, Cork MJ, Di Giovine FS, Duff GW, Groves RW. 2006. Interleukin-1: a key inflammatory mediator in psoriasis? Cytokine 33:72–78. doi: 10.1016/j.cyto.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 67.Ahmed ST, Darnell JE. 2009. Serpin B3/B4, activated by STAT3, promote survival of squamous carcinoma cells. Biochem Biophys Res Commun 378:821–825. doi: 10.1016/j.bbrc.2008.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iversen OJ, Lysvand H, Slupphaug G. 2017. Pso p27, a SERPINB3/B4-derived protein, is most likely a common autoantigen in chronic inflammatory diseases. Clin Immunol 174:10–17. doi: 10.1016/j.clim.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 69.Duk JM, de Bruijn HW, Groenier KH, Hollema H, ten Hoor KA, Krans M, Aalders JG. 1990. Cancer of the uterine cervix: sensitivity and specificity of serum squamous cell carcinoma antigen determinations. Gynecol Oncol 39:186–194. doi: 10.1016/0090-8258(90)90430-S. [DOI] [PubMed] [Google Scholar]
- 70.Shimada H, Nabeya Y, Okazumi S, Matsubara H, Shiratori T, Gunji Y, Kobayashi S, Hayashi H, Ochiai T. 2003. Prediction of survival with squamous cell carcinoma antigen in patients with resectable esophageal squamous cell carcinoma. Surgery 133:486–494. doi: 10.1067/msy.2003.139. [DOI] [PubMed] [Google Scholar]
- 71.Takeshima N, Hirai Y, Katase K, Yano K, Yamauchi K, Hasumi K. 1998. The value of squamous cell carcinoma antigen as a predictor of nodal metastasis in cervical cancer. Gynecol Oncol 68:263–266. doi: 10.1006/gyno.1998.4939. [DOI] [PubMed] [Google Scholar]
- 72.Catanzaro JM, Sheshadri N, Pan JA, Sun Y, Shi C, Li J, Powers RS, Crawford HC, Zong WX. 2014. Oncogenic Ras induces inflammatory cytokine production by upregulating the squamous cell carcinoma antigens SerpinB3/B4. Nat Commun 5:3729. doi: 10.1038/ncomms4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kishi T, Grass L, Soosaipillai A, Scorilas A, Harbeck N, Schmalfeldt B, Dorn J, Mysliwiec M, Schmitt M, Diamandis EP. 2003. Human kallikrein 8, a novel biomarker for ovarian carcinoma. Cancer Res 63:2771–2774. [PubMed] [Google Scholar]
- 74.Borgoño CA, Kishi T, Scorilas A, Harbeck N, Dorn J, Schmalfeldt B, Schmitt M, Diamandis EP. 2006. Human kallikrein 8 protein is a favorable prognostic marker in ovarian cancer. Clin Cancer Res 12:1487–1493. doi: 10.1158/1078-0432.CCR-05-2106. [DOI] [PubMed] [Google Scholar]
- 75.Pettus JR, Johnson JJ, Shi Z, Davis JW, Koblinski J, Ghosh S, Liu Y, Ravosa MJ, Frazier S, Stack MS. 2009. Multiple kallikrein (KLK 5, 7, 8, and 10) expression in squamous cell carcinoma of the oral cavity. Histol Histopathol 24:197–207. doi: 10.14670/HH-24.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cané S, Bignotti E, Bellone S, Palmieri M, De Las Casas L, Roman JJ, Pecorelli S, Cannon MJ, O'Brien T, Santin AD. 2004. The novel serine protease tumor-associated differentially expressed gene-14 (KLK8/Neuropsin/Ovasin) is highly overexpressed in cervical cancer. Am J Obstet Gynecol 190:60–66. doi: 10.1016/j.ajog.2003.07.020. [DOI] [PubMed] [Google Scholar]
- 77.Ryu K-Y, Maehr R, Gilchrist CA, Long MA, Bouley DM, Mueller B, Ploegh HL, Kopito RR. 2007. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J 26:2693–2706. doi: 10.1038/sj.emboj.7601722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalinin A, Marekov LN, Steinert PM. 2001. Assembly of the epidermal cornified cell envelope. J Cell Sci 114:3069–3070. [DOI] [PubMed] [Google Scholar]
- 79.Brown DR, Bryan JT. 2000. Abnormalities of cornified cell envelopes isolated from human papillomavirus type 11-infected genital epithelium. Virology 271:65–70. doi: 10.1006/viro.2000.0317. [DOI] [PubMed] [Google Scholar]
- 80.Lundstrom A, Egelrud T. 1991. Stratum corneum chymotryptic enzyme: a proteinase which may be generally present in the stratum corneum and with a possible involvement in desquamation. Acta Derm Venereol 71:471–474. [PubMed] [Google Scholar]
- 81.Egelrud T, Lundström A. 1991. A chymotrypsin-like proteinase that may be involved in desquamation in plantar stratum corneum. Arch Dermatol Res 283:108–112. doi: 10.1007/BF00371618. [DOI] [PubMed] [Google Scholar]
- 82.Egelrud T, Hofer PA, Lundström A. 1988. Proteolytic degradation of desmosomes in plantar stratum corneum leads to cell dissociation in vitro. Acta Derm Venereol 68:93–97. [PubMed] [Google Scholar]
- 83.Yamasaki K. 2006. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J 20:2068–2080. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 84.Petty RD, Kerr KM, Murray GI, Nicolson MC, Rooney PH, Bissett D, Collie-Duguid ESR. 2006. Tumor transcriptome reveals the predictive and prognostic impact of lysosomal protease inhibitors in non-small-cell lung cancer. J Clin Oncol 24:1729–1744. doi: 10.1200/JCO.2005.03.3399. [DOI] [PubMed] [Google Scholar]
- 85.Catanzaro JM, Guerriero JL, Liu J, Ullman E, Sheshadri N, Chen JJ, Zong WX. 2011. Elevated expression of squamous cell carcinoma antigen (SCCA) is associated with human breast carcinoma. PLoS One 6:e19096. doi: 10.1371/journal.pone.0019096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pontisso P, Calabrese F, Benvegnù L, Lise M, Belluco C, Ruvoletto MG, Marino M, Valente M, Nitti D, Gatta A, Fassina G, Fassina G. 2004. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br J Cancer 90:833–837. doi: 10.1038/sj.bjc.6601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Koning PJA, Kummer JA, de Poot SAH, Quadir R, Broekhuizen R, McGettrick AF, Higgins WJ, Devreese B, Worrall DM, Bovenschen N. 2011. Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death. PLoS One 6:e22645. doi: 10.1371/journal.pone.0022645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun Y, Sheshadri N, Zong WX. 2017. SERPINB3 and B4: from biochemistry to biology. Semin Cell Dev Biol 62:170–177. doi: 10.1016/j.semcdb.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitsuishi K, Nakamura T, Sakata Y, Yuyama N, Arima K, Sugita Y, Suto H, Izuhara K, Ogawa H. 2005. The squamous cell carcinoma antigens as relevant biomarkers of atopic dermatitis. Clin Exp Allergy 35:1327–1333. doi: 10.1111/j.1365-2222.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- 90.Takeda A, Higuchi D, Takahashi T, Ogo M, Baciu P, Goetinck PF, Hibino T. 2002. Overexpression of serpin squamous cell carcinoma antigens in psoriatic skin. J Invest Dermatol 118:147–154. doi: 10.1046/j.0022-202x.2001.01610.x. [DOI] [PubMed] [Google Scholar]
- 91.Kuwae K, Matsumoto-Miyai K, Yoshida S, Sadayama T, Yoshikawa K, Hosokawa K, Shiosaka S. 2002. Epidermal expression of serine protease, neuropsin (KLK8) in normal and pathological skin samples. J Clin Pathol Mol Pathol 55:235–241. doi: 10.1136/mp.55.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McLaughlin-Drubin ME, Wilson S, Mullikin B, Suzich J, Meyers C. 2003. Human papillomavirus type 45 propagation, infection, and neutralization. Virology 312:1–7. doi: 10.1016/S0042-6822(03)00312-X. [DOI] [PubMed] [Google Scholar]
- 93.Meyers C, Mayer TJ, Ozbun MA. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol 71:7381–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McLaughlin-Drubin ME, Meyers C. 2005. Propagation of infectious, high-risk HPV in organotypic “raft” culture, p 171–186. In Human papillomaviruses. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 95.Meyers C. 1996. Organotypic (raft) epithelial tissue culture system for the differentiation-dependent replication of papillomavirus. Methods Cell Sci 18:201–210. doi: 10.1007/BF00132885. [DOI] [Google Scholar]
- 96.Biryukov J, Cruz L, Ryndock EJ, Meyers C. 2015. Native human papillomavirus production, quantification, and infectivity analysis, p 317–331. In Keppler D, Lin AW (ed), Cervical cancer: methods and protocols. Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 97.Meyers C, Alam S, Mane M, Hermonat PL. 2001. Altered biology of adeno-associated virus type 2 and human papillomavirus during dual infection of natural host tissue. Virology 287:30–39. doi: 10.1006/viro.2001.0968. [DOI] [PubMed] [Google Scholar]
- 98.Peterson GL. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 99.Calabrese F, Lunardi F, Balestro E, Marulli G, Perissinotto E, Loy M, Nannini N, Valente M, Saetta M, Agostini C, Rea F. 2012. Serpin B4 isoform overexpression is associated with aberrant epithelial proliferation and lung cancer in idiopathic pulmonary fibrosis. Pathology 44:192–198. doi: 10.1097/PAT.0b013e3283511b61. [DOI] [PubMed] [Google Scholar]
- 100.Shinoda Y, Kozaki K, Imoto I, Obara W, Tsuda H, Mizutani Y, Shuin T, Fujioka T, Miki T, Inazawa J. 2007. Association of KLK5 overexpression with invasiveness of urinary bladder carcinoma cells. Cancer Sci 98:1078–1086. doi: 10.1111/j.1349-7006.2007.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yousef GM, Polymeris ME, Yacoub GM, Scorilas A, Soosaipillai A, Popalis C, Fracchioli S, Katsaros D, Diamandis EP. 2003. Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res 63:2223–2227. [PubMed] [Google Scholar]
- 102.Planque C, de Monte M, Guyetant S, Rollin J, Desmazes C, Panel V, Lemarié E, Courty Y. 2005. KLK5 and KLK7, two members of the human tissue kallikrein family, are differentially expressed in lung cancer. Biochem Biophys Res Commun 329:1260–1266. doi: 10.1016/j.bbrc.2005.02.100. [DOI] [PubMed] [Google Scholar]
- 103.Yousef GM, Scorilas A, Kyriakopoulou LG, Rendl L, Diamandis M, Ponzone R, Biglia N, Giai M, Roagna R, Sismondi P, Diamandis EP. 2002. Human kallikrein gene 5 (KLK5) expression by quantitative PCR: an independent indicator of poor prognosis in breast cancer. Clin Chem 48:1241–1250. [PubMed] [Google Scholar]
- 104.Ogawa K, Utsunomiya T, Mimori K, Tanaka F, Inoue H, Nagahara H, Murayama S, Mori M. 2005. Clinical significance of human kallikrein gene 6 messenger RNA expression in colorectal cancer. Clin Cancer Res 11:2889–2893. doi: 10.1158/1078-0432.CCR-04-2281. [DOI] [PubMed] [Google Scholar]
- 105.Marimuthu A, Chavan S, Sathe G, Sahasrabuddhe NA, Srikanth SM, Renuse S, Ahmad S, Radhakrishnan A, Barbhuiya MA, Kumar RV, Harsha HC, Sidransky D, Califano J, Pandey A, Chatterjee A. 2013. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim Biophys Acta 1834:2308–2316. doi: 10.1016/j.bbapap.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 106.Yousef GM, Borgoño CA, Popalis C, Yacoub GM, Polymeris M-E, Soosaipillai A, Diamandis EP. 2004. In-silico analysis of kallikrein gene expression in pancreatic and colon cancers. Anticancer Res 24:43–51. [PubMed] [Google Scholar]
- 107.Darling MR, Tsai S, Jackson-Boeters L, Daley TD, Diamandis EP. 2008. Human kallikrein 8 expression in salivary gland tumors. Head Neck Pathol 2:169–174. doi: 10.1007/s12105-008-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Planque C, Li L, Zheng Y, Soosaipillai A, Reckamp K, Chia D, Diamandis EP, Goodglick L. 2008. A multiparametric serum kallikrein panel for diagnosis of non-small cell lung carcinoma. Clin Cancer Res 14:1355–1362. doi: 10.1158/1078-0432.CCR-07-4117. [DOI] [PubMed] [Google Scholar]
- 109.White NMA, Mathews M, Yousef GM, Prizada A, Fontaine D, Ghatage P, Popadiuk C, Dawson L, Doré JJE. 2009. Human kallikrein related peptidases 6 and 13 in combination with CA125 is a more sensitive test for ovarian cancer than CA125 alone. Cancer Biomarkers 5:279–287. doi: 10.3233/CBM-2009-0113. [DOI] [PubMed] [Google Scholar]
- 110.Wu Z-S, Wu Q, Yang J-H, Wang H-Q, Ding X-D, Yang F, Xu X-C. 2008. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int J Cancer 122:2050–2056. doi: 10.1002/ijc.23337. [DOI] [PubMed] [Google Scholar]
- 111.Riedel F, Götte K, Schwalb J, Hörmann K. 2000. Serum levels of matrix metalloproteinase-2 and -9 in patients with head and neck squamous cell carcinoma. Anticancer Res 20:3045–3049. [PubMed] [Google Scholar]
- 112.Morgia G, Falsaperla M, Malaponte G, Madonia M, Indelicato M, Travali S, Mazzarino MC. 2005. Matrix metalloproteinases as diagnostic (MMP-13) and prognostic (MMP-2, MMP-9) markers of prostate cancer. Urol Res 33:44–50. doi: 10.1007/s00240-004-0440-8. [DOI] [PubMed] [Google Scholar]
- 113.Yen C-Y, Chen C-H, Chang C-H, Tseng H-F, Liu S-Y, Chuang L-Y, Wen C-H, Chang H-W. 2009. Matrix metalloproteinases (MMP) 1 and MMP10 but not MMP12 are potential oral cancer markers. Biomarkers 14:244–249. doi: 10.1080/13547500902829375. [DOI] [PubMed] [Google Scholar]
- 114.Deraz EM, Kudo Y, Yoshida M, Obayashi M, Tsunematsu T, Tani H, Siriwardena SBSM, Kiekhaee MR, Qi G, Iizuka S, Ogawa I, Campisi G, Muzio L Lo, Abiko Y, Kikuchi A, Takata T. 2011. MMP-10/stromelysin-2 promotes invasion of head and neck cancer. PLoS One 6:e25438. doi: 10.1371/journal.pone.0025438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mathew R, Khanna R, Kumar R, Mathur M, Shukla NK, Ralhan R. 2002. Stromelysin-2 overexpression in human esophageal squamous cell carcinoma: potential clinical implications. Cancer Detect Prev 26:222–228. doi: 10.1016/S0361-090X(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 116.Rochefort H, Cavailles V, Augereau P, Capony F, Maudelonde T, Touitou I, Garcia M. 1989. Overexpression and hormonal regulation of pro-cathepsin D in mammary and endometrial cancer. J Steroid Biochem 34:177–182. doi: 10.1016/0022-4731(89)90080-0. [DOI] [PubMed] [Google Scholar]
- 117.Athanassiadou P, Sakellariou V, Petrakakou E, Athanassiades P, Zerva C, Liossi A, Michalas S. 1998. Cathepsin D immunoreactivity in ovarian cancer: correlation with prognostic factors. Pathol Oncol Res 4:103–107. doi: 10.1007/BF02904702. [DOI] [PubMed] [Google Scholar]
- 118.Santamaría I, Velasco G, Cazorla M, Fueyo A, Campo E, López-Otín C. 1998. Cathepsin L2, a novel human cysteine proteinase produced by breast and colorectal carcinomas. Cancer Res 58:1624–1630. [PubMed] [Google Scholar]
- 119.Reference deleted.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.