Abstract
Muscleblind-like (MBNL) proteins are conserved RNA-binding factors involved in alternative splicing (AS) regulation during development. While AS is controlled by distribution of MBNL paralogs and isoforms, the affinity of these proteins for specific RNA-binding regions and their location within transcripts, it is currently unclear how RNA structure impacts MBNL-mediated AS regulation. Here, we defined the RNA structural determinants affecting MBNL-dependent AS activity using both cellular and biochemical assays. While enhanced inclusion of MBNL-regulated alternative exons is controlled by the arrangement and number of MBNL binding sites within unstructured RNA, when these sites are embedded in a RNA hairpin MBNL binds preferentially to one side of stem region. Surprisingly, binding of MBNL proteins to RNA targets did not entirely correlate with AS efficiency. Moreover, comparison of MBNL proteins revealed structure-dependent competitive behavior between the paralogs. Our results showed that the structure of targeted RNAs is a prevalent component of the mechanism of alternative splicing regulation by MBNLs.
INTRODUCTION
Alternative splicing (AS) is a complex process in pre-mRNA maturation, enabling the production of multiple protein isoforms distinguished by distinct functions from a single transcript. As much as 95% of pre-mRNAs in mammalian cells undergo AS governed by a unique set of RNA-binding proteins (RBPs) and their corresponding RNA binding sites (1). The pre-mRNA itself has a prominent impact on the activity of RBP on several levels, including RNA sequence specificity, distribution of RBP-binding motifs within target RNAs, the structural organization of RNA motifs and their position with respect to other RBP-binding motifs as well as the sequence composition of the local RBP binding environment (2–4). Along with ubiquitous splicing regulatory proteins, tissue-specific splicing factors with various RNA binding domains and RNA substrate preferences play a crucial role in developmental processes. One of these trans-acting factors is the muscleblind-like (MBNL) protein family composed of three paralogs, namely MBNL1, MBNL2 and MBNL3. Despite a primary function of MBNL proteins in the AS of hundreds of pre-mRNAs (5–7), MBNL proteins also regulate subcellular localization (7,8), mRNA stability (7,9,10) and alternative polyadenylation (11).
MBNL paralogs differ considerably in terms of function and expression level during embryonic and postnatal development (12–18). While MBNL1 is the major MBNL family member in adult skeletal muscles and governs myoblast differentiation, MBNL3 is mostly present at early stages of development as well as in the adult placenta, spleen and lung (15,19). MBNL1 is partially compensated by upregulation of MBNL2 but not by MBNL3 (7,20). In contrast, MBNL3 suppresses or delays muscle differentiation when it is constitutively expressed or knocked out in myoblasts, respectively (13,21). MBNL3 also plays a crucial role in a process of muscle regeneration (13,21,22). Mbnl1 knockout mice show retention of fetal splice isoforms in adult organs (23), which recapitulates the phenotype characteristic of myotonic dystrophy type 1 (DM1). DM1 is a multi-systemic disease associated with nuclear sequestration of MBNL proteins in nuclear RNA foci by CUG expansion RNAs transcribed from microsatellite CTG expansions in the 3′ untranslated region (3′UTR) of the dystrophia myotonica protein kinase gene (DMPK) (24,25). Another form of DM, DM2, is caused by structurally related CCTG expansion within intron 1 of the cellular nucleic-acid binding protein (CNBP) gene (26), and MBNL sequestration by CCUG expansion RNAs also occurs in the DM2 nuclei.
MBNL splicing factor activity generates an AS profile dependent on the pre-mRNA binding site location. For example, alternative cassette exon (CE) skipping is promoted by MBNL binding to the alternative exon or the upstream intron, whereas inclusion is favored by MBNL binding to the CE 3′-end or to the downstream intron (7,16). Current evidence indicates the strength of alternative exon exclusion and inclusion, which defines the expression profile of adult-specific mRNA isoforms during postnatal development (27–30), depends on the binding affinity, activity and relative concentration of MBNL paralogs (31,32). The degree to which the structural organization of MBNL-binding regulatory elements within RNA targets contributes to splicing regulation is currently unclear.
MBNL paralogs contain four zinc fingers (ZF1-4) (of the type CX7CX4–6CX3H) serving as RNA-binding domains which specifically recognize an RNA motif consisting of a GpC dinucleotide flanked by pyrimidines, YGCY (Y – U or C) (33). Moreover, a GpC flanking adenosine instead of a pyrimidine is an additional motif for MBNL3 binding (A/YGCY/A) (22,30). However, high affinity binding requires at least 9–10 nucleotides outside the YGCY motif with which ZFs also interact (34). The ZFs are arranged as two tandems not functionally equivalent for all splicing events (35,36) and which differ in specificity of YGCY recognition (37). These ZFs are separated by an ∼80 residue linker, which could facilitate binding to separated RNA motifs in cis or in trans (38,39).
Considering the structural organization of MBNL-binding RNA motifs, MBNL1 does not interact with fully double-stranded RNAs or CUG hairpins stabilized with pseudouridine (40–42). Recent whole transcriptome-based analysis highlighted the necessity of single stranded pyrimidines for MBNL binding, whereas two GpC steps may be either single or double stranded (28). The presence of several pyrimidines over purines in a neighborhood of YGCY motif might be relevant to MBNL binding preference due to their impact on destabilizing RNA structure (33,38). MBNL1 binding to a sequence within the RNA helical region may shift the RNA structure equilibrium towards partial unwinding (34,43). Although an increasing body of evidence indicates an essential role of RNA structure comprising YGCY motifs in MBNL1-binding, the RNA structural determinants have not yet been defined.
We previously showed that the three MBNL paralogs bind to similar RNA motifs with slightly different affinity and control the AS of the same alternative exons with different strength (30). The differences between MBNL paralogs in recognition of the canonical, near-optimal binding motifs and the structural organization of RNA targets may underlie the importance of expressing MBNL paralogs. Here, we examine the RNA structural features preferred by MBNL proteins and their impact on modulation of AS.
Our results demonstrate that MBNL-mediated alternative CE inclusion is modulated by the structural organization of RNA regulatory elements to a greater extent than by the number of MBNL-binding motifs. Strikingly, CE inclusion is promoted by UGCU motifs within unstructured regions or when these motifs are located on one side of an RNA hairpin structure, whereas their distribution on the opposite side of the hairpin weakens splicing activity. Additionally, the distance between these motifs, regardless of the RNA secondary structure, is less significant for MBNL-RNA complex formation compared to splicing regulation. We show that the splicing activity of all MBNL paralogs is fine-tuned by the same RNA structural features. Intriguingly, MBNL3 acts as a competitor of MBNL1 for specific RNA substrates under certain conditions and a similar relationship is observed between different splicing isoforms of MBNL1. Overall, our data suggest that MBNL-dependent splicing changes underlying both physiological and pathological development are fine-tuned by RNA substrate-dependent, tissue-specific and the relative expression levels of MBNL paralogs and their isoforms.
MATERIALS AND METHODS
Preparation of radiolabeled RNA fragments
The templates for transcription reactions for RNA fragments derived from pre-mRNAs (Mbnl1_1, Mbnl1_2, Atp2A1_1, Atp2A1_2, Pphln1_1, Pphln1_2, Calm3, Calm3 sequence mutants, neg_Ctrl, Mbnl2, Mbnl2 sequence mutants) were obtained in two rounds of PCR. First, longer products were amplified from mouse or human genomic DNA using specific _F/_R primer sets at certain Ta. Obtained PCR products constituted a template for the second round of PCR with specific _TF/_TR primer sets having a promoter for polymerase T7 and conducted at Ta = 55°C. The temples for short RNA fragments were either purchased (Future Synthesis) or prepared in a primer extension reaction using two ss DNA sequences (_TF and _TR) complementary to each other at 20 continuous nucleotides at the 3′ or 5′-end, respectively, or through the hybridization of one strand of DNA sequence with a short ss T7 promoter sequence. Prior to the 5 min denaturation step at 90°C, the DNA sequences were gel purified and ethanol precipitated. The templates for longer RNA sequences were prepared by PCR using one strand of DNA sequence and specific primers _TF/TR. The transcription reaction and 5′-end radiolabeling were performed as previously described (44). The sequences of all primers and Ta are listed in Supplementary Table S1.
Biochemical and in silico analysis of RNA structure
The analysis was conducted as previously described (45) with concentrations of S1, T1 and T2 endonucleases indicated in Supplementary Figures S1a, S3a, S8a and S9a. The analyzed RNA sequences were also run through the following secondary structure prediction software programs: RNAfold (46), Mfold (47), RNAstructure (48), using biochemically defined constrains or default settings. Each program predicted the same structures as the most energetically favorable.
Hybrid minigene preparation
Hybrid minigenes based on the Atp2a1 minigene were prepared as previously described (44). The sequences of interest which played a role of MBNL-responsive regulatory elements were either derived from particular fragments of genes or were artificially designed and amplified in PCR using set of primers (_cF/_cR) or in primer extension reaction using two ss DNA templates (_cF/_cR) complementary to each other at ∼20-nt-long continuous nucleotides at the 3′- or 5′-end, respectively and caring restriction sites for NotI and SalI enzymes. The sequences of primers and ss templates are listed in Supplementary Table S1.
Recombinant GST and (His)6-tagged MBNL1, MBNL2 and MBNL3
GST fusions with MBNL1, MBNL2 and MBNL3 proteins encoded by exons 1–4 of the MBNL1, MBNL2 and MBNL3 genes with a (His)6-tag at the C-terminus were prepared and purified as previously described (30,49,50). The protein concentration was measured using both the Bradford assay and Sypro Ruby staining using 10% SDS-PAGE.
Quantification of rMBNL–RNA interaction
A biochemical assay based on double-membrane filtration analysis was performed as previously described (44). The Kd of the rMBNL–RNA complexes was calculated based on the signal of free RNA and MBNL-RNA complexes in Graph-Pad using one site specific binding curve equation, Y = Bmax*X/(Kd + X).
Cell culture and transfection
The growth conditions of the human HeLa and Human Skeletal Myoblast (HSkM) cell lines were previously described (44), whereas those for Mouse Embryonic Fibroblasts (MEFs) and monkey Cos7 were the same as used for HeLa cells. Prior to transfection, the cells were plated in 12-well plates and transfected at 50–60% confluence with Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. Genes were knocked down with siRNAs against MBNL1 (51) or MBNL2 (52) (Future Synthesis and RiboTask™, respectively), or MBNL3 (22) with the ON-TARGET plus SMARTpool (Dharmacon), or siCtrl (44) (Future Synthesis) at 50 nM concentration each. Co-transfection was conducted with 200 ng of minigene and 500 ng of eGFP-MBNL, mCherry or eGFP expressing vectors (or as indicated in the figures). eGFP-MBNL constructs were previously described (30,45). Competition assay was conducted with 200 ng of minigene, 250 ng of MBNL1 (different isoforms) and increasing concentrations of MBNL3 or MBNL1 (different isoforms) as indicated in the figures, and substituted with eGFP. HSkMs or other cells were harvested 24 or 72 h after transfection with siRNA, respectively, or 48 h for the remaining experiments.
Splicing and expression analyses of precursor and mature mRNA
Total RNA isolation, cDNA synthesis and RT-PCR were conducted as previously described (44). The primer sets and Ta are listed in Supplementary Table S2.
eGFP fluorescence detection
HeLa cells were lysed with RIPA buffer [150 mM NaCl, 50 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% NP-40, 0.5% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] supplemented with SIGMAFAST Protease Inhibitor Cocktail (Sigma). Lysates were incubated on ice for 30 min and vortexed every 10 min prior to centrifugation at 18 000 × g at 4°C for 10 min. Samples without heating were separated on 10% SDS polyacrylamide gels, and eGFP fluoresce was quantified on the Amersham Typhoon RGB laser scanner.
Immunoblotting
HeLa cells were lysed with RIPA buffer supplemented with SIGMAFAST Protease Inhibitor Cocktail (Sigma). Lysates were sonicated at 4°C and centrifuged at 18 000 × g at 4°C for 10 min. Samples were heated to 95°C for 5 min, separated on 10% SDS polyacrylamide gels and transferred to nitrocellulose (Protran BA 85, Whatman) using a wet transfer apparatus (1 h, 100 V, 4°C). Membranes were blocked for 1 h with 5% skim milk in PBST buffer (phosphate-buffered saline (PBS), 0.1% Tween-20) and incubated with a primary antibody against eGFP (1:1000, Santa Cruz cat. no. sc-8334) or human GAPDH (1:1000, Santa Cruz cat. no. sc-47724). Anti-rabbit (1:20 000, Sigma cat. no. A9169) and anti-mouse (1:2000, Millipore cat. no. 12-349) secondary antibodies were conjugated with horseradish peroxidase and detected using the Pierce ECL Plus Western Blotting Substrate detection kit (Thermo Scientific).
Statistic
Group data obtained in biochemical assays and in cellula are expressed as the mean ± standard deviation (SD) or as specified in the figures. Statistical significance of RT-PCR and the double membrane-filtration assay was determined by a two-tailed Student's t-test using Microsoft Excel (ns—non-significant; *P < 0.05; **P < 0.01 and ***P < 0.001). Statistical analysis was calculated using 2–4 biological replicates (n).
RESULTS
Different structures of the same RNA binding site affect MBNL1 binding and splicing activity
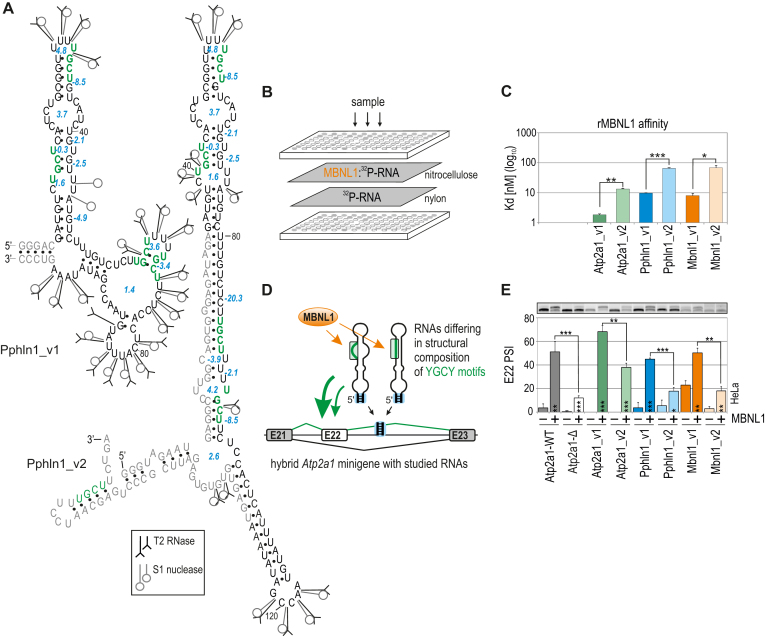
The significance of the structural organization of RNA regulatory elements for MBNL1-binding affinity has been noted (53,54), although the specific RNA structural determinants promoting or abrogating MBNL-RNA interactions remain unclear. To gain further insight, we first asked whether alterations of RNA secondary structure encompassing MBNL-binding motifs modify the MBNL1 affinity and its ability to regulate AS. For this purpose, we selected three MBNL-sensitive pre-mRNA regulatory elements from Mbnl1 exon1 (E1) (45), Atp2a1 intron 22 (I22) and Pphln1 intron 6 (I6) (44). Each of these AS substrates contains a few conserved MBNL-recognized 5′-YGCY-3′ motifs (33,55). Next, we transcribed in vitro 100 to 190-nt-long fragments of selected transcripts each in two variants containing the same MBNL-binding motifs but differing in the length of natural 5′ and/or 3′ ends (e.g. Pphln1_v1 and Pphln1_v2 having shorter and longer YGCY-flanking sequences, respectively). Then, for each transcript the RNA secondary structure was determined experimentally with the use of enzymatic probes (RNase T1 and T2 and nuclease S1) recognizing and cleaving single-stranded regions of 5′-end labelled RNAs (56). The experimental data was combined with the results of the RNA structure predictions using software programs (46–48). The secondary structures of RNA variants differed substantially, especially within the regions containing YGCY motifs (Figure 1A and Supplementary Figure S1). Next, we determined the association of recombinant MBNL1 (rMBNL1) to studied RNAs using a biochemical assay based on a double membrane filtration method and observed significant difference in dissociation constant (Kd) values between variants of the same transcript, which suggests the high sensitivity of MBNL1 to RNA secondary structural changes (Figure 1B, C, Supplementary Figure S1 and S2). The lowest Kd value, and the highest affinity, was uniformly characteristic of RNAs containing YGCY motifs within thermodynamically less stable structures.
Figure 1.
Different structures of the same RNA binding site affect MBNL1 binding and splicing activity. (A) Proposed secondary structures of variants of the Pphln1 transcript fragment with previously defined functional MBNL1-binding motifs marked in green and in bold (44). The optimal thermodynamic stability of YGCY-containing RNA regions is expressed in Gibbs free energy (ΔG) in kcal/mol for the reaction at 37°C using RNAfold software (46) and is marked in blue. T2 and S1 nucleases-specific cleavage sites and cleavage intensities are specified for each probe according to symbols shown in the legend. Unique nucleotides, not shared between RNA variants, are marked in grey. See Figure S1 for RNA structures of other transcript variants. (B) A simplified scheme of a biochemical assay based on double membrane filtration. The radioactivity on the upper nitrocellulose film (representing 32P-RNA bound to rMBNL1) and that on a lower nylon film (representing free 32P-RNA) was quantified for multiple protein concentrations and MBNL1 affinity (Kd value) for each transcript was calculated (see Figure S2 for more details). (C) The quantification of the biochemical assay showing different rMBNL1 binding affinities to two structural variants of three natural transcript fragments. The results are mean Kd values; n = 2 for each protein concentration (in the range of 0–250 nM of rMBNL1). (D) A model of a hybrid Atp2a1 minigene with the Atp2a1 gene fragment between exons 21 and 23 and with a natural MBNL-binding site within intron 22 (I22) in pre-mRNA (Atp2a1-WT) substituted with different YGCY motif-containing RNA fragments. These RNA fragments serve as artificial regulatory elements to test MBNL activity depending on RNA structural organization and YGCY arrangement. A stable helical region formed by Atp2a1 I22, restriction sites used for cloning and a 5-bp-long artificial sequence located at the base of an insert allowing the preservation of its experimentally and in silico-defined RNA secondary structure are marked with a blue box (see also Figure S2b). Alternative and constitutive exons are marked with white and grey boxes, respectively; E, exon. (E) Different efficiencies of alternative E22 inclusion into the mRNA of hybrid Atp2a1 minigenes depending on the studied RNA regulatory elements upon eGFP-control or MBNL1 overexpression (OE) in HeLa cells; n = 3.
We next sought to determine whether observed differences in structure-influenced affinities underlie MBNL-driven splicing changes in cellula. Thus, we substituted a natural MBNL-sensitive regulatory element within I22 of Atp2a1 minigene (44) with six of the above-described RNA fragments (hybrid Atp2a1 minigenes) (Figure 1D). The design of the minigenes preserves experimentally determined RNA secondary structures in cells by a slightly extended, natural and thermodynamically stable region within Atp2a1 I22 (Supplementary Figure S2b). Following co-transfection of HeLa cells with hybrid minigenes and an eGFP-control or MBNL1 expression vector, we measured the splicing activity of the endogenous MBNL pool or overexpressed MBNL1 (MBNL1 OE) as E22 percent spliced-in (PSI). A higher E22 inclusion level was elicited by RNA variants exhibiting a lower Kd and having less stable RNA secondary structures encompassing MBNL-binding motifs (Figure 1C, E and Supplementary Figure S1).
Although the Mbnl1 E1, Pphln1 E7 and Atp2a1 E22 RNAs contain multiple YGCY motifs, from 4 for Pphln1 to 11 for Mbnl1, the number of motifs was not the only determinant of MBNL1 binding affinity or splicing activity (Figure 1C, E, Supplementary Figures S1 and S2). Thus, we next addressed the question of the relationship between YGCY motif number and secondary structure context.
MBNL1 binding and splicing activity is regulated by both the number of UGCU motifs and RNA structural context
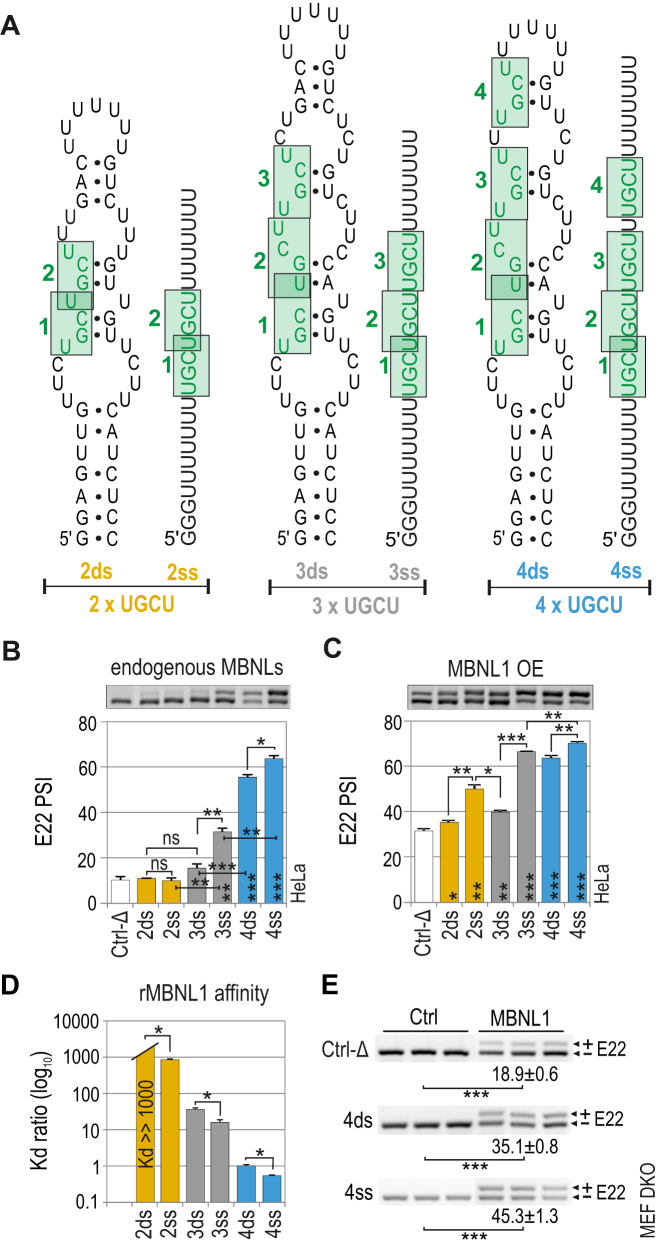
We designed several short artificial RNA sequences containing two to four 5′-UGCU-3′ motifs that either folded into semi-stable double-stranded (ds) RNA hairpin structures or remained single-stranded (ss) RNAs (Figure 2A) and subsequently cloned these sequences into the Atp2a1-Δ minigene followed by splicing analysis in cells. RT-PCR analysis showed that in the presence of endogenous MBNL proteins, the efficiency of E22 inclusion increased with increasing UGCU number when this motif was in either dsRNA or ssRNA regions (Figure 2B). While 3xUGCU and 4xUGCU, but not 2xYGCY, ssRNA motifs showed significantly higher activity. MBNL1 OE also induced E22 inclusion for all hybrid minigenes showing a saturated effect of E22 for the strongest regulatory elements (e.g. 4ss) (Figure 2C). We confirmed the linkage between observed splicing variations and MBNL1 activity following MBNL1 OE in Mbnl1; Mbnl2 double knockout (DKO) mouse embryonic fibroblasts (MEFs) (Figure 2E). MBNL1 binding affinities were in agreement with the in cellula splicing results for endogenous pool of MBNL proteins (Figure 2D).
Figure 2.
MBNL1 binding and splicing activity is regulated by both the number of UGCU motifs and structural context. (A) RNA secondary structures of semi-stable (ds; double-stranded) and unstructured (ss; single-stranded) artificial regulatory elements which are base paired at the 5′ and 3′ ends and internally (only ds RNA) to force the formation of one thermodynamically optimal secondary structure confirmed by us in several RNA structure prediction software programs (46–48). MBNL-binding motifs are numbered from 5′-end and marked in green. The RNA names correspond to the number of UGCU motifs (2, 3 or 4) and the structure type (ds or ss). The splicing response of pre-mRNAs of hybrid minigenes with ds or ss RNAs incorporated in I22 upon (B) endogenous level of MBNLs or (C) MBNL1 overexpression (OE) in HeLa cells. Vertical asterisks denote the statistical significance of results in comparison to a control experiment (Ctrl-Δ; transfection with Atp1a1-Δ minigene). Note a positive effect of the increasing UGCU motif number on alternative splicing - the PSI of E22 for 2ss < 3ss < 4ss; MBNL1 dose-dependency of weaker regulatory elements - the PSI of E22 for 2ds < 2ss > 3ds; and the structural organization of an RNA regulatory element surpassing the number of UGCU motifs - PSI of E22 for 4ds = 3ss; 2ss > 3ds; n = 2. (D) Quantification of the biochemical assay showing relative rMBNL1 binding affinity for RNA fragments normalized to Kd for the 4ds RNA molecule; n = 2 for each protein concentration (in the range of 0–200 nM of rMBNL1). (E) RT-PCR showing the splicing response of hybrid Atp2a1 minigenes representing ss and ds groups of RNAs with or without MBNL1 OE in Mbnl1; Mbnl2 DKO MEFs. The asterisks denote the statistical significance of results compared to cells treated with control eGFP construct (Ctrl). Ctrl-Δ, transfection with Atp1a1-Δ minigene; n = 3.
Our results demonstrated that the number and structural arrangement of RNA motifs recognized by MBNL proteins both impact MBNL-mediated splicing activity. Increasing the number of binding motifs, if analyzed in the same structural context, correlates with stronger affinity and splicing activity of MBNL1. Two UGCU motifs within a pyrimidine-rich ssRNA serve as a functional splicing regulatory element that responds solely to a high MBNL1 concentration.
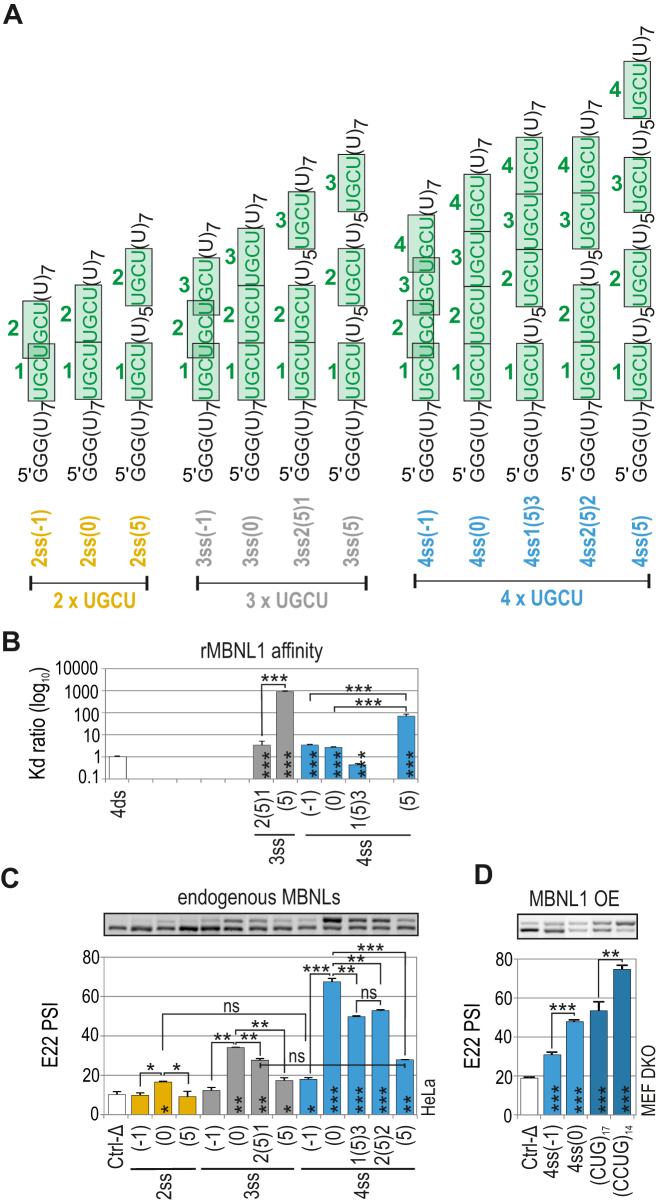
Distance between UGCU motifs modulates MBNL-mediated splicing regulation but not binding affinity
Next, we investigated the consequences of UGCU motif separation within unstructured RNAs on MBNL1 splicing activity. We designed three sets of RNA regulatory elements containing two, three or four UGCU motifs organized as either overlapping [(–1); …UGCUGCU…], sequential [(0); …UGCUUGCU…] or separated by five U residues [(5); …UGCUUUUUUUGCU…] (Figure 3A). We also experimentally confirmed the single-strand structure of these RNAs in solution (Supplementary Figure S3). Binding assays indicated that rMBNL1 slightly prefers to bind to contiguous UGCU motifs compared to interspaced motifs [e.g. 4ss(0) versus 4ss(5)] (Figure 3B). Subsequently, hybrid minigenes were prepared as previously described to investigate the regulatory properties of these RNAs in cells with endogenous MBNL levels. Interestingly, and in contrast to the results of the binding assays, all RNAs with overlapping UGCU motifs had no or reduced splicing activity [e.g. 4ss(–1)] (Figure 3C). On the other hand, RNAs with sequential UGCU motifs showed the highest splicing activity within sets of RNAs containing the same number of UGCU motifs [e.g. 4ss(0)] (Figure 3C). UGCU motif separation [e.g. 4ss2(5)2, 4ss1(5)3, 4ss(5)] always led to reduction of E22 inclusion compared to sequential UGCU motifs (Figure 3C).
Figure 3.
Distance between UGCU motifs modulates MBNL-mediated splicing regulation but not binding affinity. (A) The structures of ss RNA regulatory elements with distinctly interspaced UGCU motifs marked in green. The RNA names correspond to the distance between consecutive UGCU motifs, which is marked in brackets surrounded by the number of adjacent motifs. (B) As in Figure 2D but for a group of ss RNAs with interspaced UGCU motifs; n = 2–4 for each protein concentration (in the range of 0–400 nM of rMBNL1). (C) As in Figure 2C but for a group of ss RNAs with interspaced UGCU motifs; n = 2. Note no or very weak regulatory properties of RNAs with overlapping motifs - 2ss(–1), 3ss(–1) and 4ss(–1). RNAs with sequential motifs - 4ss(0), 3ss(0) and 2ss(0), exceeded all other tested regulatory elements within the UGCU number sets. UGCU separation had an adverse effect on E22 inclusion - the PSI of E22 for 4ss(0) > 4ss3(5)1 = 4ss2(5)2 > 4ss(5). Note the redundant impact of UGCU number - PSI of E22 for 3ss2(5)1 = 4ss(5). Vertical asterisks denote the statistical significance of results in comparison to a control experiment (Ctrl-Δ; transfection with Atp1a1-Δ minigene). (D) The splicing response of hybrid Atp2a1 minigenes with regulatory elements distinguished by either overlapping or sequential arrangement of distinct number of UGCU motifs. The graph shows RT-PCR results for DKO MEFs co-transfected with hybrid minigenes and the MBNL1 expression vector. Vertical asterisks denote the statistical significance of results in comparison to Ctrl-Δ construct; n = 3.
The overlapping and sequential arrangements of the UGCU motifs described above correspond to the arrangements of (CUG)n and (CCUG)n repeats occurring in DMPK and CNBP transcripts associated with DM1 and DM2 expansion mutations, respectively. Previously, we showed all MBNL paralogs have a high affinity, ∼10 nM, for RNA hairpin structures formed by a non-pathological number of repeats, (CUG)n and (CCUG)n (30). The question then arose as to whether the splicing regulatory properties of the repeats differ, as we observed for overlapping and sequential motif arrangements. We tested (CUG)17 and (CCUG)14, -containing regulatory elements within Atp2a1 hybrid minigenes in Mbnl1; Mbnl2 DKO MEFs to directly link the splicing response to MBNL1 activity. For MBNL1 OE, we detected higher increase in E22 splicing for Atp2a1-(CCUG)14 compared to Atp2a1-(CUG)17, which confirmed splicing enhancement for sequential motifs regardless of their number (Figure 3D and Supplementary Figure S4a).
Additionally, the impact of motif arrangement was greater than motif quantity on MBNL-mediated AS [e.g. 3ss(0) versus 4ss(5)] (Figure 3C). MBNL1 OE mostly enhanced the regulatory properties of weaker regulatory elements, indicating the susceptibility of these elements to a higher concentration of MBNL1 (Supplementary Figure S4b and c).
In summary, we have shown that the distance between motifs modulates MBNL1-driven splicing regulation, which is not a function of differences in affinity for targeted RNAs.
Effect of RNA structure context on MBNL splicing activity
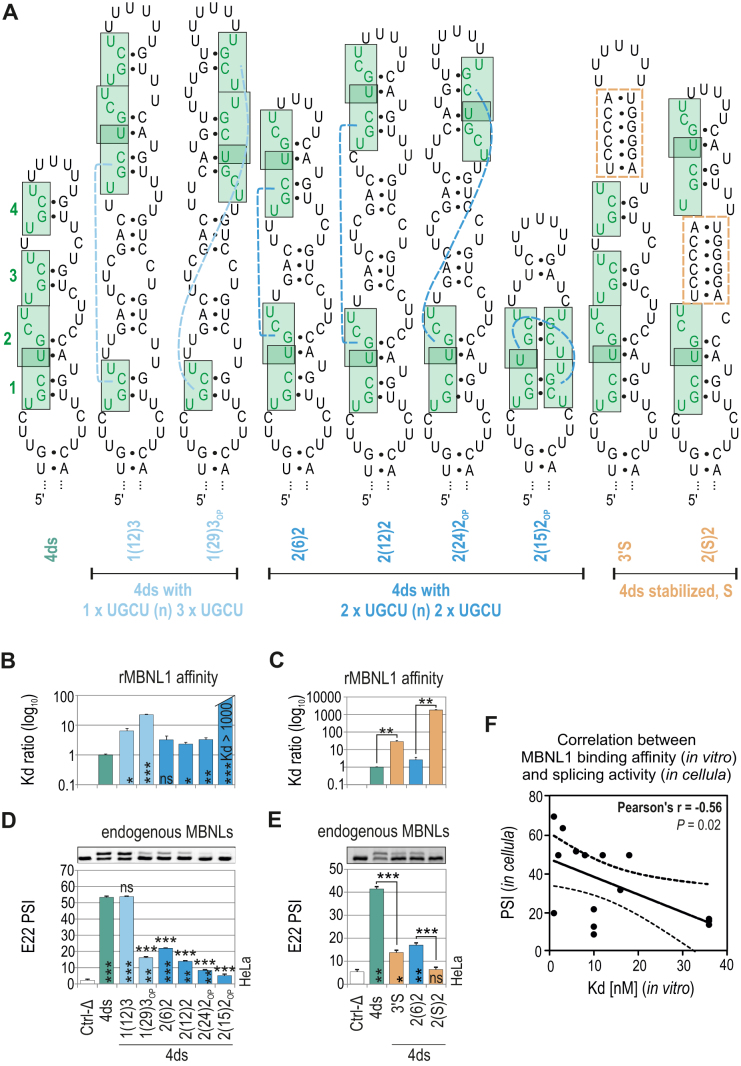
Another important question was the importance of YGCY motif arrangement within the semi-stable RNA structures that predominate at natural MBNL binding sites. We designed several regulatory elements of energetically optimal secondary structures comprising four UGCU motifs that were either: (i) contiguous or interspaced; (ii) located on one side or opposite sides of the RNA helix; (iii) interspaced with, or surrounded by, a stabilizing structure composed of six-base-pairs (Figure 4A).
Figure 4.
Effect of RNA structural context on MBNL splicing activity. (A) The RNA secondary structures of ds RNAs containing four UGCU motifs marked in green. Dashed blue lines depict a reciprocal position of UGCU motifs. Orange rectangles depict ds helical regions stabilizing the RNA secondary structure. The RNA names correspond to the distance between consecutive UGCU motifs which is marked in brackets surrounded by the number of adjacent motifs. (B and C) As in Figure 2D, but for a group of ds RNAs; n = 2–4 for each protein concentration (in the range of 0–200 nM of rMBNL1). (D and E) As in Figure 2C but for a group of ds RNAs; n = 2. Note the decreasing E22 inclusion upon separating tandems of motifs - the PSI of E22 for 4ds2(6)2 > 4ds2(12)2 > 4ds2(24)2. Vertical and horizontal asterisks denote the statistical significance of results in comparison to Ctrl-Δ and 4ds constructs, respectively. (F) The Pearson correlation coefficient of rMBNL1 binding affinity (Kd; nM) and splicing activity (PSI values) upon endogenous level of MBNLs for 20 artificial RNA structures analyzed in this study (based on the results shown in Figures 2–4). The RNAs represent two groups of structures (ss or ds) with the same sequence background which showed no effect on E22 inclusion in Mbnl1; Mbnl2 DKO cells (Figure 2E). Dashed lines represent the 95% confidence interval.
The rMBNL1 binding affinity for most of RNAs was only slightly reduced in comparison to a reference RNA with contiguous UGCU motifs [e.g., 4ds1(12)2] but strongly decreased for motifs located opposite to each other [e.g. 4ds2(15)2OP] and interspaced with, or surrounded by, six-base-pairs (e.g. 4ds3′S) (Figure 4B, C and Supplementary Figure S5a). Analogous regulatory elements with two or three UGCU motifs showed either no MBNL1–RNA complex formation or depicted the same rMBNL1 preferences to UGCU arrangement as it was shown for 4xUGCU-containing RNAs, respectively (Supplementary Figures S6 and S7).
The analysis of MBNL splicing activity for distinct arrangements of UGCU motifs under cellular conditions gave concordant results only for certain minigenes. MBNL splicing activity strongly decreased for UGCU motifs located on opposite sides of the RNA helix [e.g. 4ds1(29)3OP, 4ds2(15)2OP] (Figure 4D) and when separated or surrounded by stable helical regions [e.g. 4ds2(S)2] (Figure 4E). The same adverse effect was observed for overlapped or interspaced UGCU motifs, although the affinity of MBNL1 for these RNA examples remained high [e.g. 4ds2(6)2] (Figure 4B and D). Intrigued by these results, we next looked at designed RNA examples by correlating the defined values of MBNL1 affinity and the strength of E22 inclusion in cellula for the endogenous pool of MBNLs. As expected, the obtained correlation between these two parameters was moderate (r = –0.56), but many regulatory elements with low Kd were distinguished by a wide range of E22 inclusion (Figure 4F).
Interestingly, for all RNA regulatory elements, MBNL1 OE proportionally increased E22 inclusion, which confirmed that secondary structural features are sensitive to higher MBNL1 concentrations (Supplementary Figure S5b).
Overall, these results demonstrate that adjacent UGCU motifs located on one side of a hairpin structure are strong splicing regulatory elements while overlapping UGCU motifs impact MBNL splicing, but not binding activity. Furthermore, helix stabilization proximal to UGCU motifs markedly inhibits MBNL–RNA interactions. Since we defined the impact of artificial UGCU motifs within single-stranded and semi-stable RNA structures and the inter-motif distance, reciprocal location and RNA structure stability on MBNL splicing activity, we next examined these RNA features using natural RNA targets of MBNL proteins.
Natural RNA structural determinants that modulate MBNL activity
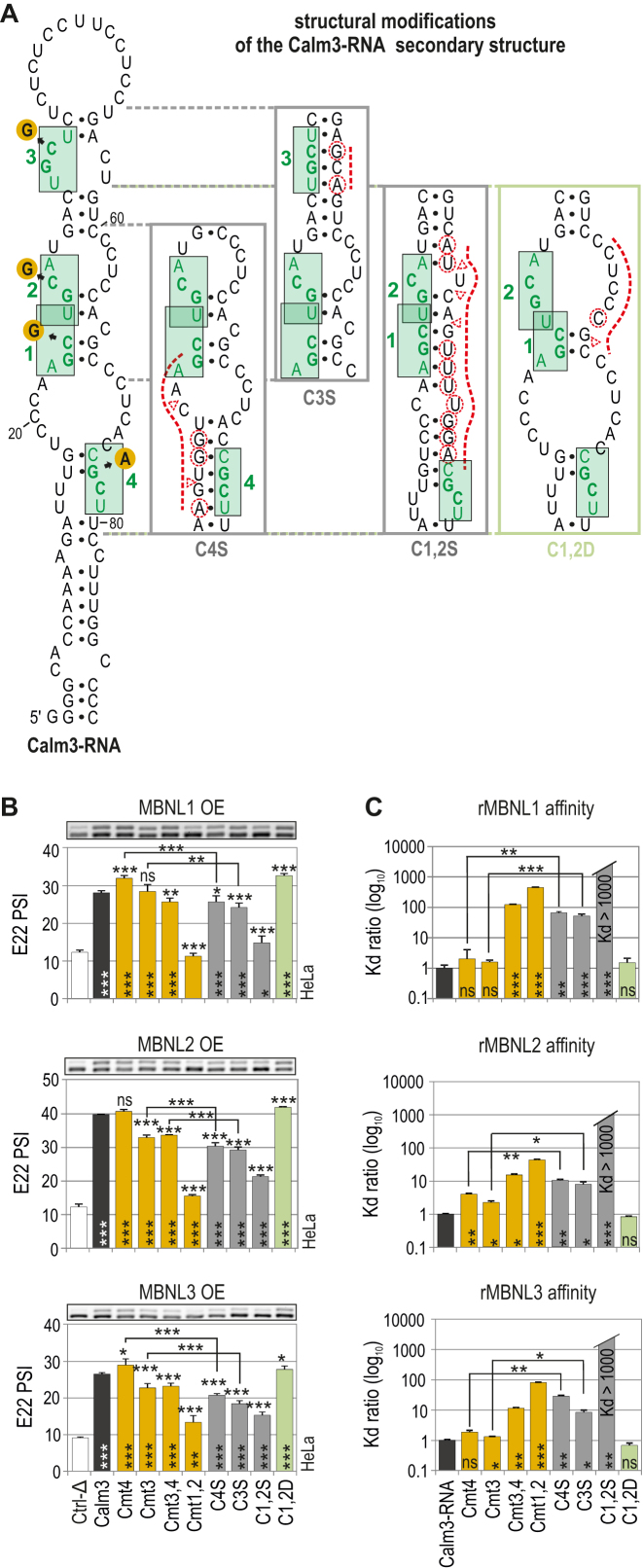
To study natural RNA structures that influence MBNL activity, we selected two RNA regions: (i) a subregion of the 3′UTR of the Calm3 transcript that undergoes MBNL-dependent selection of alternative polyadenylation sites; (ii) a subregion derived from Mbnl2 E5 that is downregulated by MBNL1 upon binding to intron 4 (Figure 5A, Supplementary Figures S8 and S9) (57). The secondary structure of an ∼100-nt-long sequence of Calm3 RNA was semi-stable (ΔG = −12.8 kcal/mol) with four internal loops and a long terminal loop (Figure 5A and Supplementary Figure S8). By combining a biochemical assay and mutagenesis, we previously defined two major (Cmt1,2) and two subsidiary (Cmt3,4) motifs in Calm3 RNA bound with high affinity by three rMBNL paralogs (30). We confirmed the functionality of these binding sites in AS regulation by incorporating WT and mutant variants of Calm3 in hybrid minigenes and co-expressing them along with MBNL1, MBNL2 and MBNL3. The percent of E22 inclusion decreased significantly upon mutations of the Cmt1 and 2 major motifs in contrast to mutations of the Cmt3 and 4 subsidiary motifs (Figure 5B). We noted certain determinants, including overlapping motifs and motifs located on opposite sides of the RNA helix, which most likely contribute to the reduced responsiveness of this RNA regulatory element. Moreover, it was not responsive to the endogenous MBNL level (Supplementary Figure S8c).
Figure 5.
Natural RNA structural determinants that modulate MBNL activity. (A) A secondary structure of Calm3-RNA containing MBNL1-binding motifs (HGCH – H stands for U, C or A) marked in green and numbered. Point mutations within the motifs are marked with orange circles. The structural modifications of particular Calm3-RNA regions are presented in boxes. Substitutions and insertions or deletions are marked with red dashed circles or triangles, respectively. (B) RT-PCR results showing the splicing response of pre-mRNAs of hybrid minigenes with incorporated different Calm3 regulatory elements carrying HGCH mutations or structural modifications upon MBNL1, MBNL2 and MBNL3 OE in HeLa cells. The MBNL paralogs are identical with respect to splicing isoforms (lacking residues encoded by alternative exons 5 and 7); n = 2. Vertical and horizontal asterisks denote the statistical significance of results in comparison to Ctrl-Δ and Calm3 WT constructs, respectively. Ctrl-Δ, Atp2a1-Δ minigene. (C) Quantification of the biochemical assay showing binding affinity of recombinant rMBNL1, rMBNL2 and rMBNL3 to intact Calm3-RNA, HGCH motif and RNA secondary structure mutants normalized to Calm3-RNA WT; n = 2–4 for each protein concentration (in the range of 0–200 nM of rMBNL1). Vertical asterisks denote the statistical significance of results in comparison to intact Calm3-RNA.
To assess the impact of the structural organization of the Calm3 regulatory element on MBNL splicing activity, we introduced point mutations solely into a hairpin side located opposite to MBNL-binding motifs preserving the primary sequence of the motifs and adjacent nucleotides (Figure 5A). Conceivably, the conversion of a semi-stable hairpin to a more single-stranded structure slightly increased the E22 inclusion rate for all MBNL paralogs (C1,2D; Figure 5B) exhibiting minor sensitivity to MBNL endogenous levels (Supplementary Figure S8c). In contrast, stabilizing the RNA secondary structure of subsidiary motifs (C4S, C3S) slightly reduced E22 inclusion, overcoming the effect of their point mutations (Cmt4, Cmt3), which was consistent with rMBNL affinity (Figure 5B, C, Supplementary Figure S8c and d). These results suggested that MBNL access to the major RNA motifs was constrained by the increased structural stability of flanking sequences. Base-pairing of the major motifs induced the strongest inhibitory effect (C1,2S). We also selected an ∼100-nt-long sequence of Mbnl2 I4 containing four YGCY motifs and confirmed, given certain structural alterations, the negative and positive impact of structural parameters on MBNL1 splicing activity (Supplementary Figure S9).
Thus, the binding and splicing activity of MBNL1, MBNL2 and MBNL3 are strongly modulated by the same RNA structural features in the same sequence contexts. We previously showed that depending on the AS event, the strength of splicing activities of MBNL paralogs and their AS isoforms may differ (30). Hence, we decided to investigate the reciprocal interactions of paralogs in the context of AS regulation and organization of RNA regulatory elements.
Competition between MBNL proteins is driven by RNA regulatory element organization
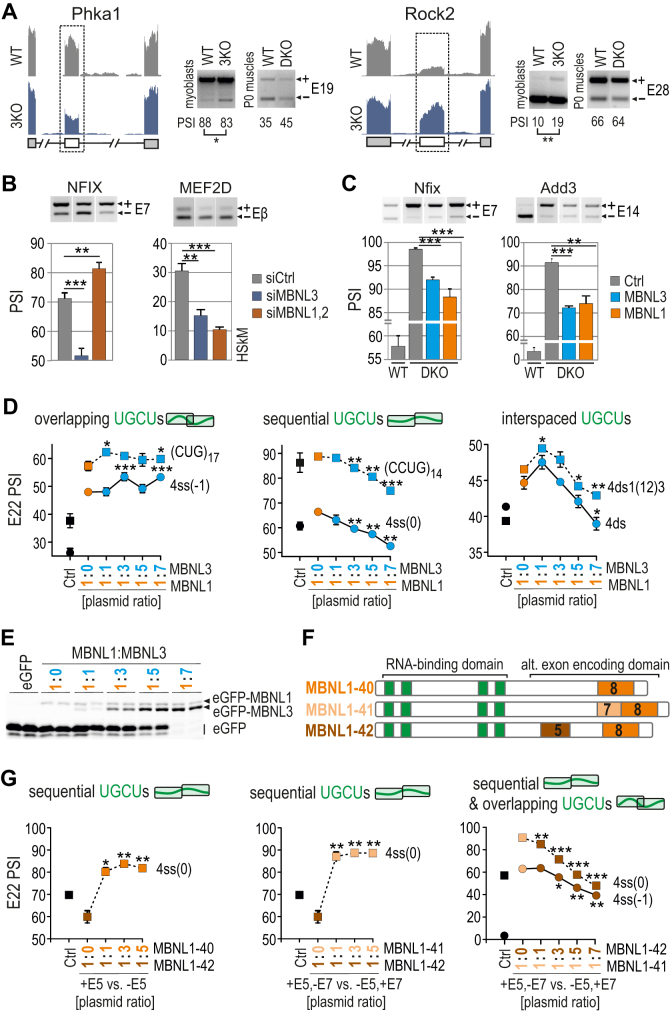
Due to distinct properties of MBNL1 and MBNL3, including a different developmentally regulated pattern of expression and specific function (22,23), we selected these two paralogs to investigate their reciprocal impact on AS regulation. We previously showed that Mbnl3 knockout in myoblasts (3KO) results in an increase in adult-like splicing profiles in hundreds of AS events compared to Mbnl1; Mbnl2 DKO muscles, as observed by whole transcriptome analysis (58). In this study, we used RT-PCR to verify several new AS events changed upon MBNL3 loss in 3KO myoblasts in favor of MBNL1 and MBNL2-mediated direction of AS (Figure 6A and Supplementary Figure S10a). To confirm the MBNL3-dependence of these events, we compared the splicing profile of several MBNL-sensitive transcripts in human myoblasts treated with either siRNAs targeting both full-length and short MBNL3 isoforms (3KD cells) or siRNAs against MBNL1 and MBNL2 (DKD cells). The results showed the same direction of splicing changes upon MBNL3 or MBNL1 and MBNL2 silencing, but mostly revealed a larger percentage of adult-like AS isoforms in 3KD cells, in contrast to DKD cells, in which expectedly, the reversion to a developmentally immature splicing profile was observed (Figure 6B and Supplementary Figure S10b). These results were in agreement with observations in 3KO mouse myoblasts (Figure 6A) and were recapitulated in HeLa cells (Figure S10b). In the light of these observations we suppose that in the absence of MBNL3 in cells, the activity of MBNL1 and MBNL2 is enhanced either due to loss of MBNL3 splicing antagonism or to occupation of released binding sites by more active MBNL1 and MBNL2 paralogs.
Figure 6.
Competition between MBNL proteins is driven by RNA regulatory element organization. (A) RNA-seq read coverage across two MBNL-dependent alternative exons indicating their adult-like splicing profiles in Mbnl3 knockout (3KO) myoblasts compared to wild-type cells and Mbnl1; Mbnl2 DKO quadriceps muscle (postnatal, day 0); alternative and constitutive exons are marked with white or gray boxes, respectively; n = 2. Note that the effect of 3KO is opposite to the effect of DKO. (B) RT-PCR results of splicing changes in two MBNL1-dependent alternative exons which direction was opposite upon MBNL3 silencing (3KD) compared to MBNL1; MBNL2 double knockdown (DKD) with respect to cells treated with control siRNA in human skeletal myoblasts (HSkM); n = 3. (C) RT-PCR results of splicing changes in MBNL1-dependent alternative exons which direction was similar upon MBNL3 and MBNL1 OE in Mbnl1; Mbnl2 DKO MEFs; Ctrl, the results from eGFP construct transfected cells (marked in grey); n = 3. (D) A competition assay between MBNL1 and MBNL3 paralogs. RT-PCR results showing the splicing response of pre-mRNAs with distinctly arranged UGCU motifs in intronic regulatory elements in constant background of MBNL1 OE (marked in orange) and with increasing concentration of MBNL3 (marked in blue) in HeLa cells. MBNL1 and MBNL3 paralogs are identical with respect to splicing isoforms (lacking residues encoded by alternative exons 5 and 7); Ctrl, the results from eGFP construct transfected cells (marked in black). The asterisks denote the statistical significance of results in comparison to cells with MBNL1 OE only (marked in orange); n = 2. Each graph represents the results obtained for two constructs belonging to the category of RNA regulatory elements with overlapping, sequential or interspaced UGCU motifs. (E) Image of polyacrylamide gels presenting the expression level of eGFP-MBNL1 and eGFP-MBNL3 fusion proteins and eGFP alone. After electrophoresis gels were scanned for eGFP fluorescence (protein samples were not heat-denatured before gel loading). (F) A schematic representation of MBNL1 splicing isoforms differing in the presence of residues encoded by alternative exons 5 and 7 (marked in brown and beige, respectively) but sharing the same RNA binding domain composed of four ZFs (marked in green). (G) As in Figure 6D but for MBNL1 isoform pairs. An equal amount of applied expression vectors resulted in 2:1 ratio of expressed proteins for MBNL1–42 versus MBNL1–40/41 (30). MBNL1–40, MBNL1–41 and MBNL1–42 are marked in orange, beige and brown, respectively. The asterisks denote the statistical significance of results in comparison to cells with overexpression of only one isoform; n = 2.
To test the possibility that MBNL3 antagonizes MBNL1 activity, we analyzed several MBNL-mediated AS events upon MBNL3 and MBNL1 OE in Mbnl1; Mbnl2 DKO fibroblasts and Cos7 cells. The direction of splicing changes was the same for all analyzed AS events (Figure 6C and Supplementary Figure S10c). These results rule out MBNL3 antagonism but support its competitive behavior in an RNA substrate dependent manner. In consistence with that, nuclear factor IX (NFIX) fetal exon 7 splicing decreased both when MBNL3 was silenced in myoblasts and upon MBNL3 OE in DKO fibroblasts (Figure 6B and C).
We next investigated whether MBNL3 occupancy impedes recruitment of MBNL1 to target RNAs, or vice versa, and whether the structural organization of RNA regulatory elements modulates competitive interactions between these paralogs. Because the primary structures of the examined endogenous pre-mRNAs vary in the number of MBNL binding motifs and nucleotide sequence composition, we utilized hybrid minigenes expressing RNAs with structurally distinct regulatory elements. We performed an in cellula competition assay, in which in the background of MBNL1 OE promoting E22 inclusion, we titrated different amounts of MBNL3 (Figure 6E). For pre-mRNAs with sequential and interspaced, but not for overlapping UGCU motifs, MBNL1-mediated splicing changes were strongly suppressed in an MBNL3 dose-dependent manner (Figure 6D and Supplementary Figure S11a). A control experiment did not show a negative effect of increasing levels of MBNL1 on splicing in of E22 (Supplementary Figure S11b). For some minigenes, MBNL3 suppressed endogenous MBNL-induced inclusion of E22 [e.g. 4ss(0); Figure 6D], which was recapitulated in other cell lines (Supplementary Figure S11c). To further demonstrate the competitive behavior of MBNL proteins, we overexpressed MBNL3 together with hybrid minigenes in Mbnl1; Mbnl2 DKD cells, and noted only the MBNL1-mediated direction of splicing changes (Supplementary Figure S11d).
We next asked whether the RNA structure-dependent competition between paralogs may be a resultant of their different binding affinity. We conducted biochemical studies with truncated (RNA binding domain only) and full-length recombinant MBNL3, although the results showed the reminiscent affinity of both rMBNL3 proteins to selected RNAs (Supplementary Figure S11e) consistently with the results for rMBNL1 (Figure 3).
Intrigued by the competitive behavior observed between MBNL paralogs, we tested if this phenomenon is an attribute of MBNL splicing isoforms as well. Hence, we examined the MBNL1 isoforms differing in the presence of MBNL1 exon 5, which encodes part of a multipartite nuclear localization signal (NLS) (39) and exon 7, encoding a sequence that is responsible for homotypic dimerization (59) (Figure 6F). To monitor differences in the splicing activity of MBNL1 isoforms, we applied a hybrid minigene-based cellular assay with overexpression of pairs of MBNL1 isoforms (Figure 6G and Supplementary Figure S11f). For pre-mRNAs with sequential UGCU motifs, isoform MBNL1–42 (containing exon 5) substantially suppressed E22 inclusion promoted by the endogenous MBNL pool, whereas titrations of MBNL1-40 or MBNL1-41 (which lack exon 5) were equally effective in eliminating this effect, leading to a large increase in E22 inclusion at a relatively low dose (Figure 6G, left and middle chart). Additionally, increasing levels of MBNL1–42 considerably mitigated the activity of MBNL1–41 for sequential and overlapping UGCU motifs (Figure 6G, right chart). A difference in the splicing activity of MBNL1–40 and MBNL1–41 varying in the presence of exon 7 was significant but exclusive to sequential UGCU motifs (Supplementary Figure S11f).
MBNL paralogs and their isoforms compete for the same RNA binding sites but the resultant splicing profile is a function of distinct paralog and isoform splicing activities and the variability of their C-terminal domains.
DISCUSSION
MBNL activity controlled by the number and distribution of YGCY RNA sequence motifs
A central question in RNA splicing regulation is the relative importance of primary sequence composition and structural organization of RNA regulatory elements within pre-mRNAs on splicing factor affinity and AS regulation. Here, we addressed that question for the MBNL family of developmental splicing regulators using hybrid minigenes with the same pre-mRNA backbone but differing in MBNL-binding UGCU regulatory cassettes to minimize interference by other cis- and trans-acting factors (44).
The results reported here show a positive correlation between the number of UGCU motifs (2–4) and the binding affinity and splicing activity of MBNL proteins in a sequence/structural context-dependent manner. At least 2xUGCU motifs are required for MBNL binding but the affinity is at least 10-times lower than for 4xUGCU motif-containing RNAs. Expectedly, the splicing of pre-mRNAs expressing 2xUGCU responds solely to high MBNL concentrations, in contrast to 4xUGCU motifs, which are sensitive to a low dose of MBNL. Our findings partially agree with previous results showing the sufficiency of just one such motif but the frequent requirement for multiple motifs (33,38) and the primary sequence composition dependency of MBNL1 binding (33,34). Generally, multiple copies of RNA-binding modules present in RBPs cooperate functionally and structurally (2); hence, the presence of several RNA motifs may allow binding of consecutive ZFs or ZF tandems and enhance interaction affinity as well as the specificity and stability of subsequent RBP-RNA complexes (34,60). However, in whole-transcriptome analyses, many MBNL-binding sites have been identified, but only a small percentage of them are functional and important for RNA processing pathways. For experimentally confirmed natural RNA targets, the number of YGCY motifs is often high and ranges from 9 for INSR I11 (61), 12 for MBNL1 E1 (45) and 15 for LDB3 E10 (44). Our studies show that YGCY arrangement and RNA structure have sometimes a greater impact on AS regulation than the number of YGCY motifs and that these three RNA hallmarks determine the functionality of potential RNA motifs. We observed, that MBNL1 affinity remains within the low-nanomolar Kd range and its splicing activity is high for contiguous 3x or 4xUGCU motifs as well as upon the separation of UGCU tandems within more and less stable structures (Figures 3 and 4) most likely due to a flexible linker which combines ZF tandems and enables their binding to separated RNA motifs (2,38,39). In contrast, overlapping UGCU motifs decrease MBNL1 splicing activity but not binding affinity, whereas separation of individual motifs affects both properties of MBNL1. Hence, we expect that further scattering of individual UGCU motifs will worsen MBNL1 function. ZF tandems of MBNL are not functionally equivalent but regulate splicing in an exclusive or mutually compensating manner (35). Most recent crystallographic studies report that the second ZF in each tandem specifically binds the YGCY motif, whereas the other ZF may interact with RNA less specifically (34). In contrast, another in-depth analysis describes ZF1-2 as a domain specifically recognizing the YGCY motif while ZF3-4 acts as a more general RNA binding domain (37). It could suggest that different YGCY motif arrangement may lead to the formation of MBNL-RNA complexes of distinct splicing capacities.
MBNL activity controlled by RNA secondary structures
Consistently with previous findings (28,34,38), we show that MBNL1 preferentially binds single-stranded RNAs, rather than more stable RNA structures with the same arrangement of YGCY motifs. Base-pairing of YGCY motifs (53) or their structural stabilization via substitution of uridines with pseudouridines (42,54) was shown to impair splicing. Complementing these previous works, we showed that greater stability of RNA structure in regions adjacent to UGCU motifs introduced via mutations or occurring through natural interaction of RNA strands, plays a predominant role in modulation of MBNL-mediated AS regulation (Figures 1 and 4). The analyses of RNA structural determinants revealed high preferences of MBNL to contiguous UGCU motifs located on one side of the RNA hairpin. In contrast, neither binding nor splicing activity of MBNL1 was observed when 3x or 4xUGCU motifs are distributed on opposite sides of a stem of the RNA hairpin. Hence, we could expect that the splicing profile of MBNL-regulated alternative exons may result from the composition and arrangement of different local RNA structures as well as binding of other RBPs (44,62–65), the splicing of precursor RNAs and epigenetic parameters (66), which modulate the affinity and activity of MBNL proteins.
MBNL’s splicing activity is not solely driven by the binding affinity
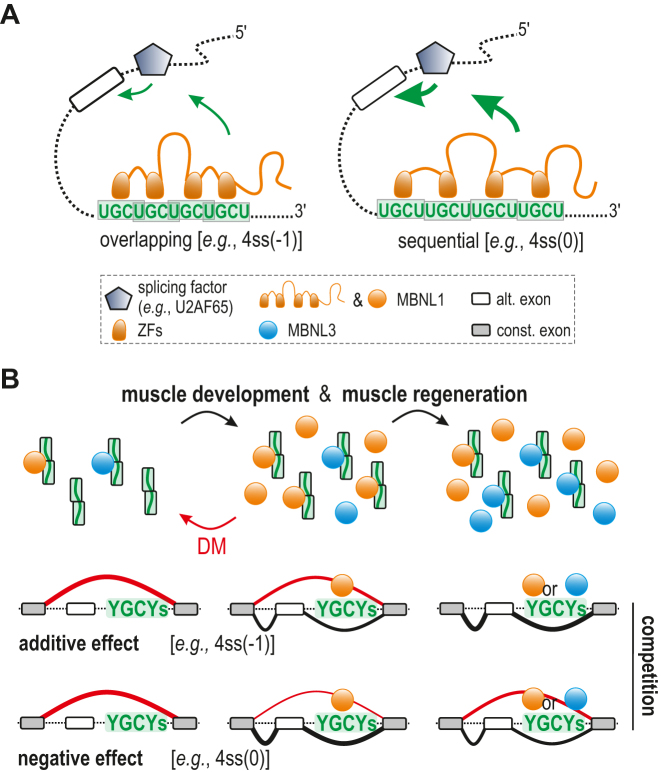
Our analysis of UGCU motif distribution in various RNA structural contexts revealed discrepancies between MBNL1 and MBNL3 binding affinity and splicing activity. The most prominent differences were observed for sequential arrangement of 4xUGCU motifs (5′…UGCUUGCU…), which in contrast to overlapping arrangement (5′…UGCUGCU…), highly promoted MBNL-dependent inclusion of alternative exons in different cellular models, although both served as preferable binding sites in biochemical assays in the present and previous studies (Figures 3–6 and Supplementary Figure S11) (28,30). Similar discrepancies have been described (35), but these reports did not correlate their observations with regulatory element organization and these prior analyses examined six different minigenes potentially introducing interfering variables. Moreover, we noticed that the same localization of distinct RNA regulatory elements in the same pre-mRNA backbone of the minigene induces various MBNL-determined maximal alternative exon inclusion rates, which differ between MBNL paralogs and their splicing isoforms. Our observations lead us to speculate that the process of MBNL binding to different UGCU motif arrangements and RNA structures alters the availability of protein regions previously reported as important in AS regulation. These MBNL regions include a linker sequence between ZF tandems (39,67), proline-rich elements (68), glutamine-rich regions downstream of ZF2 (35), alternative exon encoding sequences (39,67) and ZFs themselves (35) (Figure 7A). However, the observed lack of correlation between RNA binding in vitro and MBNL splicing activity in cellula could also result from different local RNA structural variations, including the formation of higher order RNA structures due to the impact of various cellular factors or biochemical conditions. The in vitro conditions described here are simplified and often limited to the presence of one major RNA structure. Moreover, certain structural/primary sequence arrangements and mutations may alter direct or indirect competition between MBNL1 and other RBPs. For example, polypyrimidine tract-binding protein 1 (PTB) and U2AF65 may potentially recognize U-rich regions present in our single-stranded RNA models (62,63). RNA-binding fox-1 homolog (RBFOX) could interact with UGCAUG motif within our Calm3-derived models (69), whereas binding of CUGBP elav-like family member 1 (CELF1) or serine/arginine-rich splicing factor 1 (SRSF1) may modulate MBNL1 binding to RNA (44,64). However, our control experiments using Mbnl1; Mbnl2 DKO cells do not indicate an essential role of other splicing factors in MBNL-mediated splicing (Figure 2 and Supplementary Figure S4).
Figure 7.
RNA regulatory element organization influences the outcome of MBNL-determined AS profile. (A) Upon MBNL binding to distinctly arranged or structured RNA motifs, changes in availability or conformation of splice-activation elements of the protein may occur and lead to alterations in the strength of the MBNL interaction with other splicing factors including U2 auxiliary factor 65 kDa subunit (U2AF65), resulting in weakened or enhanced alternative CE inclusion (depicted by the size of green arrows). (B) A model in which MBNL1 and MBNL3 compete at a dose- and RNA substrate-dependent manner during muscle development and regeneration as well as in pathological conditions (DM) modulating the AS profile; more explanation in the text. To simplify, MBNL splicing isoforms and MBNL2 paralog were omitted in the scheme.
Competition between MBNLs
Since the presence of three MBNL paralogs was discovered (13,70), much attention has focused on determining the cellular functions and distinguishable features of these paralogs. In contrast to a previously described antagonistic effect of MBNL3 on MBNL1 activity (13,17,71), in the current study, we discovered a competitive interaction between MBNL paralogs and their isoforms that is determined by the organization of RNA regulatory elements and modulates AS profile (Figure 6). Initially, we observed that the loss of MBNL3 in myoblasts promotes the MBNL-mediated splicing profile (Figure 6). This splicing phenomenon was not evident either in adult heart or muscles of Mbnl3 KO mice, probably due to the relatively low contribution of MBNL3 to the total pool of MBNL in adult tissues (17). However, while MBNL3 predominates in cells lacking MBNL1 and MBNL2, the paralog shifts the AS profile towards MBNL-induced (Figure 6 and Supplementary Figure S11) what confirms a convergent splicing activity of MBNL paralogs and suggests their competitive interaction while present concomitantly in cells. The competition seems to be RNA structure-influenced, since MBNL paralogs and isoforms competed with endogenous and overexpressed pool of MBNL for binding to both sequential and overlapping UGCU motifs within regulatory elements triggering a negative effect on splicing for mostly sequential UGCU arrangement (Figure 6d). One possible explanation of this phenomenon is that the splice-activation domains of MBNL paralogs vary, including a linker between ZF tandems that is merely 50% identical between the paralogs (67) and the distinct arrangement of proline-rich regions considered as a unique platform for association with other splicing factors (71). Moreover, MBNL3 lacks an alanine-rich region potentially important for RNA binding (72). MBNL isoforms share the same RNA-binding domain but have distinct C-terminal domains encoded by alternative exons which contribute to the protein activity either directly or by modulating protein regions involved in AS regulation (39,67,68,72).
The RNA organization-based competition of RBPs has not been described yet. However, a reminiscent observation has been reported for proteins belonging to the nuclear factor I (NFI) protein family, which binds to a specific sequence of DNA. NFI comprises four variants regulating gene expression in an antagonistic and synergistic way depending on their activities and distribution in cells (73). Future studies focused on MBNL1- and MBNL3-associated splicing factors should shed more light on the competitive mode of MBNL function.
In the light of the collected evidence, we propose a model for the competitive mode of action of MBNL paralogs dependent on RNA organization (Figure 7b). Due to the low active pool of MBNL paralogs occurring in initial stages of development (23) or under pathological conditions (25), MBNL-sensitive splicing isoforms are not promoted. After MBNL1 and MBNL3 levels increase during muscle development, MBNL-dependent splicing favors transcripts distinguished by a sequential-like YGCY arrangement. Under increased MBNL3 production during muscle regeneration and the saturation of many binding regions, MBNL3 displaces other MBNLs in protein-RNA complexes and causes either an additive effect on splicing (e.g., for RNAs with the YGCY-overlapping arrangement) or a reverse direction of splicing changes (e.g., for RNAs with the YGCY-sequential arrangement).
The discovery of complementary MBNL paralogs that become competitors for certain types of RNAs and in particular environments provides deeper insight into the molecular changes underlain by the sequestration and functional insufficiency of MBNLs in DM. Revealing the RNA structural features enabling MBNLs to efficiently bind the substrates with a low impact on MBNL splicing activity have potential therapeutic implications. It may lead to the development of an MBNL-derived molecule with high affinity to toxic CUG and CCUG repeats in DM, but negligible cellular activity due to limitations of interactions with other trans-acting factors.
Supplementary Material
ACKNOWLEDGEMENTS
Authors Contributions: Conception and design of experiments: K.S., K.T.. Acquisition, analysis and interpretation of data: P.C. (western blots), Ł.J.S. (RT-PCR verification of splicing profile in WT and 3KO myoblasts), J.D.T. (RNA-seq data processing), K.T. (the rest of experiments). Drafting or revising the manuscript: K.S., K.T., M.S.S., Ł.J.S., J.D.T.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Foundation for Polish Science TEAM program co-financed by the European Union within the European Regional Development Fund [TEAM/2011-7/10 to K.S.]; Polish National Science Centre [2011/01/B/NZ1/01603, 2014/15/B/NZ2/02453 to K.S., 2017/24/C/NZ1/00112 to K.T.]; post-doctoral fellowship award from the Myotonic Dystrophy and Wyck Foundations (to Ł.J.S.). Funding for open access charge: Ministry of Science and Higher Education of the Republic of Poland, from the quality promoting subsidy, under the Leading National Research Centre (KNOW) program for the years 2014–2019.
Conflict of interest statement. M.S.S. is a member of the scientific advisory board of Locana Bio. The other authors declare no competing interests.
REFERENCES
- 1. Kornblihtt A.R., Schor I.E., Allo M., Dujardin G., Petrillo E., Munoz M.J.. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013; 14:153–165. [DOI] [PubMed] [Google Scholar]
- 2. Lunde B.M., Moore C., Varani G.. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007; 8:479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang X.L., Vukovic L., Koh H.R., Schulten K., Myong S.. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res. 2015; 43:7566–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lewis C.J.T., Pan T., Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017; 18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho T.H., Charlet-B N., Poulos M.G., Singh G., Swanson M.S., Cooper T.A.. Muscleblind proteins regulate alternative splicing. EMBO J. 2004; 23:3103–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fleming V.A., Geng C., Ladd A.N., Lou H.. Alternative splicing of the neurofibromatosis type 1 pre-mRNA is regulated by the muscleblind-like proteins and the CUG-BP and ELAV-like factors. BMC Mol. Biol. 2012; 13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang E.T., Cody N.A.L., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S.J., Schroth G.P., Housman D.E., Reddy S. et al. . Transcriptome-wide regulation of Pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012; 150:710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adereth Y., Dammai V., Kose N., Li R.Z., Hsu T.. RNA-dependent integrin alpha(3) protein localization regulated by the Muscleblind-like protein MLP1. Nat. Cell Biol. 2005; 7:1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Du H.Q., Cline M.S., Osborne R.J., Tuttle D.L., Clark T.A., Donohue J.P., Hall M.P., Shiue L., Swanson M.S., Thornton C.A. et al. . Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010; 17:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masuda A., Andersen H.S., Doktor T.K., Okamoto T., Ito M., Andresen B.S., Ohno K.. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Scientific Rep. 2012; 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Batra R., Manchanda M., Swanson M.S.. Global insights into alternative polyadenylation regulation. RNA Biol. 2015; 12:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D.. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002; 11:805–814. [DOI] [PubMed] [Google Scholar]
- 13. Squillace R.M., Chenault D.M., Wang E.H.. Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. Dev. Biol. 2002; 250:218–230. [DOI] [PubMed] [Google Scholar]
- 14. Kanadia R.N., Johnstone K.A., Mankodi A., Lungu C., Thornton C.A., Esson D., Timmers A.M., Hauswirth W.W., Swanson M.S.. A muscleblind knockout model for myotonic dystrophy. Science. 2003; 302:1978–1980. [DOI] [PubMed] [Google Scholar]
- 15. Holt I., Jacquemin V., Fardaei M., Sewry C.A., Butler-Browne G.S., Furling D., Brook J.D., Morris G.E.. Muscleblind-Like proteins similarities and differences in normal and myotonic dystrophy muscle. Am. J. Pathol. 2009; 174:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charizanis K., Lee K.Y., Batra R., Goodwin M., Zhang C.L., Yuan Y., Shiue L., Cline M., Scotti M.M., Xia G.B. et al. . Muscleblind-like 2-Mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012; 75:437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi J., Dixon D.M., Dansithong W., Abdallah W.F., Roos K.P., Jordan M.C., Trac B., Lee H.S., Comai L., Reddy S.. Muscleblind-like 3 deficit results in a spectrum of age-associated pathologies observed in myotonic dystrophy. Scientific Rep. 2016; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang B.W., Cai H.F., Wei X.F., Sun J.J., Lan X.Y., Lei C.Z., Lin F.P., Qi X.L., Plath M., Chen H.. miR-30-5p regulates muscle differentiation and alternative splicing of Muscle-Related genes by targeting MBNL. Int. J. Mol. Sci. 2016; 17:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee K.S., Squillace R.M., Wang E.H.. Expression pattern of muscleblind-like proteins differs in differentiating myoblasts. Biochem. Biophys. Res. Commun. 2007; 361:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee K.Y., Li M.Y., Manchanda M., Batra R., Charizanis K., Mohan A., Warren S.A., Chamberlain C.M., Finn D., Hong H. et al. . Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013; 5:1887–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee K.S., Smith K., Amieux P.S., Wang E.H.. MBNL3/CHCR prevents myogenic differentiation by inhibiting MyoD-dependent gene transcription. Differentiation. 2008; 76:299–309. [DOI] [PubMed] [Google Scholar]
- 22. Poulos M.G., Batra R., Li M.Y., Yuan Y., Zhang C.L., Darnell R.B., Swanson M.S.. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum. Mol. Genet. 2013; 22:3547–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han H., Irimia M., Ross P.J., Sung H.K., Alipanahi B., David L., Golipour A., Gabut M., Michael I.P., Nachman E.N. et al. . MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013; 498:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. et al. . Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992; 68:799–808. [DOI] [PubMed] [Google Scholar]
- 25. Miller J.W., Urbinati C.R., Teng-umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S.. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000; 19:4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ranum L.P.W., Rasmussen P.F., Benzow K.A., Koob M.D., Day J.W.. Genetic mapping of a second myotonic dystrophy locus. Nat. Genet. 1998; 19:196–198. [DOI] [PubMed] [Google Scholar]
- 27. Paul S., Dansithong W., Jog S.P., Holt I., Mittal S., Brook J.D., Morris G.E., Comai L., Reddy S.. Expanded CUG repeats dysregulate RNA splicing by altering the stoichiometry of the muscleblind 1 complex. J. Biol. Chem. 2011; 286:38427–38438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagner S.D., Struck A.J., Gupta R., Farnsworth D.R., Mahady A.E., Eichinger K., Thornton C.A., Wang E.T., Berglund J.A.. Dose-Dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 2016; 12:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sznajder L.J., Michalak M., Taylor K., Cywoniuk P., Kabza M., Wojtkowiak-Szlachcic A., Matloka M., Konieczny P., Sobczak K.. Mechanistic determinants of MBNL activity. Nucleic Acids Res. 2016; 44:10326–10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalsotra A., Xiao X.S., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A.. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Solana J., Irimia M., Ayoub S., Orejuela M.R., Zywitza V., Jens M., Tapial J., Ray D., Morris Q.D., Hughes T.R. et al. . Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. Elife. 2016; 5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goers E.S., Purcell J., Voelker R.B., Gates D.P., Berglund J.A.. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010; 38:2467–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park S., Phukan P.D., Zeeb M., Martinez-Yamout M.A., Dyson H.J., Wright P.E.. Structural basis for interaction of the tandem zinc finger domains of human muscleblind with cognate RNA from human cardiac troponin T. Biochemistry. 2017; 56:4154–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purcell J., Oddo J.C., Wang E.T., Berglund J.A.. Combinatorial mutagenesis of MBNL1 zinc fingers elucidates distinct classes of regulatory events. Mol. Cell. Biol. 2012; 32:4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He F., Dang W., Abe C., Tsuda K., Inoue M., Watanabe S., Kobayashi N., Kigawa T., Matsuda T., Yabuki T. et al. . Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci. 2009; 18:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hale M., Richardson J., Day R., McConnell O., Arboleda J., Wang E., Berglund J.. An engineered RNA binding protein with improved splicing regulation. Nucleic Acids Res. 2018; 46:3152–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cass D., Hotchko R., Barber P., Jones K., Gates D.P., Berglund J.A.. The four Zn fingers of MBNL1 provide a flexible platform for recognition of its RNA binding elements. BMC Mol. Biol. 2011; 12:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tran H., Gourrier N., Lemercier-Neuillet C., Dhaenens C.M., Vautrin A., Fernandez-Gomez F.J., Arandel L., Carpentier C., Obriot H., Eddarkaoui S. et al. . Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J. Biol. Chem. 2011; 286:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S.. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004; 13:495–507. [DOI] [PubMed] [Google Scholar]
- 41. Goers E.S., Voelker R.B., Gates D.P., Berglund J.A.. RNA binding specificity of Drosophila muscleblind. Biochemistry. 2008; 47:7284–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. deLorimier E., Coonrod L.A., Copperman J., Taber A., Reister E.E., Sharma K., Todd P.K., Guenza M.G., Berglund J.A.. Modifications to toxic CUG RNAs induce structural stability, rescue mis-splicing in a myotonic dystrophy cell model and reduce toxicity in a myotonic dystrophy zebrafish model. Nucleic Acids Res. 2014; 42:12768–12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fu Y., Ramisetty S.R., Hussain N., Baranger A.M.. MBNL1-RNA Recognition: Contributions of MBNL1 sequence and RNA conformation. Chembiochem. 2012; 13:112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cywoniuk P., Taylor K., Sznajder L.J., Sobczak K.. Hybrid splicing minigene and antisense oligonucleotides as efficient tools to determine functional protein/RNA interactions. Scientific Rep. 2017; 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Konieczny P., Stepniak-Konieczna E., Taylor K., Sznajder L.J., Sobczak K.. Autoregulation of MBNL1 function by exon 1 exclusion from MBNL1 transcript. Nucleic Acids Res. 2017; 45:1760–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gruber A.R., Lorenz R., Bernhart S.H., Neuboock R., Hofacker I.L.. The vienna RNA websuite. Nucleic Acids Res. 2008; 36:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen C.Z., Sobczak K., Hoskins J., Southall N., Marugan J.J., Zheng W., Thornton C.A., Austin C.P.. Two high-throughput screening assays for aberrant RNA-protein interactions in myotonic dystrophy type 1. Analyt. Bioanalyt. Chem. 2012; 402:1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wojciechowska M., Taylor K., Sobczak K., Napierala M., Krzyzosiak W.J.. Small molecule kinase inhibitors alleviate different molecular features of myotonic dystrophy type 1. RNA Biol. 2014; 11:742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Childs-Disney J.L., Stepniak-Konieczna E., Tran T., Yildirim I., Park H., Chen C.Z., Hoskins J., Southall N., Marugan J.J., Patnaik S. et al. . Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat. Commun. 2013; 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dansithong W., Paul S., Comai L., Reddy S.. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005; 280:5773–5780. [DOI] [PubMed] [Google Scholar]
- 53. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C.A., Burge C.B.. RNA Sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. deLorimier E., Hinman M.N., Copperman J., Datta K., Guenza M., Berglund J.A.. Pseudouridine modification inhibits Muscleblind-like 1 (MBNL1) Binding to CCUG repeats and minimally structured RNA through reduced RNA flexibility. J. Biol. Chem. 2017; 292:4350–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Warf M.B., Berglund J.A.. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007; 13:2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sobczak K., Michlewski G., de Mezer M., Krol J., Krzyzosiak W.J.. Trinucleotide repeat system for sequence specificity analysis of RNA structure probing reagents. Analyt. Biochem. 2010; 402:40–46. [DOI] [PubMed] [Google Scholar]
- 57. Batra R., Charizanis K., Manchanda M., Mohan A., Li M.Y., Finn D.J., Goodwin M., Zhang C.L., Sobczak K., Thornton C.A. et al. . Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-Mediated disease. Mol. Cell. 2014; 56:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thomas J.D., Sznajder L.J., Bardhi O., Aslam F.N., Anastasiadis Z.P., Scotti M.M., Nishino I., Nakamori M., Wang E.T., Swanson M.S.. Disrupted prenatal RNA processing and myogenesis in congenital myotonic dystrophy. Genes Dev. 2017; 31:1122–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan Y., Compton S.A., Sobczak K., Stenberg M.G., Thornton C.A., Griffith J.D., Swanson M.S.. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007; 35:5474–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Edge C., Gooding C., Smith C.W.J.. Dissecting domains necessary for activation and repression of splicing by muscleblind-like protein 1. BMC Mol. Biol. 2013; 14:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sen S., Talukdar I., Liu Y., Tam J., Reddy S., Webster N.J.G.. Muscleblind-like 1 (Mbnl1) Promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J. Biol. Chem. 2010; 285:25426–25437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Warf M.B., Diegel J.V., von Hippel P.H., Berglund J.A.. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9203–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Echeverria G.V., Cooper T.A.. Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron. Nucleic Acids Res. 2014; 42:1893–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang E.T., Ward A.J., Cherone J.M., Giudice J., Wang T.T., Treacy D.J., Lambert N.J., Freese P., Saxena T., Cooper T.A. et al. . Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015; 25:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sellier C., Cerro-Herreros E., Blatter M., Freyermuth F., Gaucherot A., Ruffenach F., Sarkar P., Puymirat J., Bjarne U., Day J.W. et al. . RbFOX1/MBNL1 competition for CCUG RNA repeats binding contributes to myotonic dystrophy type1/type 2 differences. Nat. Commun. 2018; 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Luco R.F., Allo M., Schor I.E., Kornblihtt A.R., Misteli T.. Epigenetics in Alternative Pre-mRNA Splicing. Cell. 2011; 144:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grammatikakis I., Goo Y.H., Echeverria G.V., Cooper T.A.. Identification of MBNL1 and MBNL3 domains required for splicing activation and repression. Nucleic Acids Res. 2011; 39:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Botta A., Malena A., Tibaldi E., Rocchi L., Loro E., Pena E., Cenci L., Ambrosi E., Bellocchi M.C., Pagano M.A. et al. . MBNL1(42) and MBNL1(43) gene isoforms, overexpressed in DM1-patient muscle, encode for nuclear proteins interacting with Src family kinases. Cell Death Dis. 2013; 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Conboy J.G. Developmental regulation of RNA processing by Rbfox proteins. Wiley Interdiscip. Rev.-RNA. 2017; 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fardaei M., Larkin K., Brook J.D., Hamshere M.G.. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001; 29:2766–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee K.S., Cao Y., Witwicka H.E., Tom S., Tapscott S.J., Wang E.H.. RNA-binding protein muscleblind-like 3 (MBNL3) disrupts myocyte enhancer factor 2 (Mef2) beta-exon splicing. J. Biol. Chem. 2010; 285:33779–33787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pascual M., Vicente M., Monferrer L., Artero R.. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006; 74:65–80. [DOI] [PubMed] [Google Scholar]
- 73. Perez-Casellas L.A., Wang X.Y., Howard K.D., Rehage M.W., Strong D.D., Linkhart T.A.. Nuclear Factor I transcription factors regulate IGF binding protein 5 gene transcription in human osteoblasts. Biochim. Biophys. Acta-Gene Regul. Mech. 2009; 1789:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.