Abstract
Nucleic acid aptamers, especially RNA, exhibit valuable advantages compared to protein therapeutics in terms of size, affinity and specificity. However, the synthesis of libraries of large random RNAs is still difficult and expensive. The engineering of polymerases able to directly generate these libraries has the potential to replace the chemical synthesis approach. Here, we start with a DNA polymerase that already displays a significant template-free nucleotidyltransferase activity, human DNA polymerase theta, and we mutate it based on the knowledge of its three-dimensional structure as well as previous mutational studies on members of the same polA family. One mutant exhibited a high tolerance towards ribonucleotides (NTPs) and displayed an efficient ribonucleotidyltransferase activity that resulted in the assembly of long RNA polymers. HPLC analysis and RNA sequencing of the products were used to quantify the incorporation of the four NTPs as a function of initial NTP concentrations and established the randomness of each generated nucleic acid sequence. The same mutant revealed a propensity to accept other modified nucleotides and to extend them in long fragments. Hence, this mutant can deliver random natural and modified RNA polymers libraries ready to use for SELEX, with custom lengths and balanced or unbalanced ratios.
INTRODUCTION
In the field of therapeutic biotechnologies, nucleic acids have proved to be a useful tool for the regulation of gene expression, medical diagnostics, biological route modulation, molecular recognition strategies or drug design. Key strategies have been developed and have proved their efficiency for several years such as antisense nucleic acids (1,2), ribozymes and riboswitches (3,4) and aptamers (5,6). However, some innovative breakthrough in the generation of aptamer-based molecules remains needed to compete with medicinal chemistry and/or biological antibodies.
Nucleic acid aptamers consist of single-stranded DNA (ssDNA) or RNA that present defined 3D structures due to their propensity to form complementary base pairs, Hoogsteen base pairs, base triples or G-quartets. Their ability to fold into various secondary and tertiary structures (7) opens the possibility to design molecules that are capable of specific molecular recognition of their cognate targets. RNA aptamers are prone to generate more complex 3D structures than DNA aptamers and usually display a higher binding affinity and specificity (6). Van der Waals forces, hydrophobic and electrostatic interactions, triple base-pairs and base stacking all combine to generate folded structures and shielded active sites that determine their binding affinity and specificity. For all of these purposes, aptamers are listed among the most important classes of drug molecules and their development is facilitated by Systematic Evolution of Ligands by EXponential enrichment (SELEX) strategies (8,9). This efficient method of producing high-affinity aptamers relies on the generation of a combinatorial library of oligonucleotides. These libraries must contain a huge pool of random-sequence oligonucleotides to maximize the chances to select good candidates. As an alternative to the chemical synthesis of random oligonucleotides at the first step of aptamer selection, the engineering of DNA polymerases designed to synthesize nucleic acids (RNA and DNA) in a random fashion may provide a faster way to generate the initial libraries.
In vivo, DNA polymerases are responsible for the DNA replication and the maintenance of the genome and therefore their role is critical for the propagation of the genetic information. All DNA polymerases found in eukaryotes, prokaryotes, archaea and viruses have been classified into subfamilies according to their structure and primary sequence similarity (10–12). By construction, replicative DNA polymerases have evolved to copy the template DNA with high fidelity and consequently their native activity is of limited use for applications in modern synthetic biology, which seeks to build novel and versatile nucleic acid polymers. Indeed, DNA polymerases have an active site that is configured to incorporate the four canonical deoxyribonucleotides and to exclude ‘altered’ or modified nucleotides during cellular metabolism. However, the design of mutants based on the known three-dimensional structure of a DNA polymerase, if available, has the potential to lead to engineered enzyme(s) with the desired property in a relatively straightforward way.
DNA polymerases are divided into several families, based on sequence alignments (11,12). Classically, polA, polB, polY and reverse transcriptases (RT) all share the same folding type, while polC and polX share a different one. Nevertheless, they all have the same kind of catalytic site, based on the two-metal ion mechanism (13). To our knowledge, only members of the polA and polX families display a significant nucleotidyltransferase activity.
The first family of DNA polymerases that we will consider here is polA. Representative members of family A with a known crystal structure are Escherichia coli DNA pol (13) and Thermus aquaticus DNA pol (14) in prokaryotes, and phage T7 (15) DNA pol in viruses; in eukaryotes, several members of the polA family have been solved: in human, Pol θ takes part in the repair of DNA double-strand-breaks (DSB) in the alternative (Ku-independent) Non Homologous End Joining (alt-NHEJ) process (16,17), Pol ν in the repair of DNA crosslinks occurring during homologous recombination (18) and Pol γ is involved in mitochondrial DNA replication (19). Very recently, human Pol θ has been described to display a robust terminal transferase activity that is apparent when it switches between three different mechanisms (20,21). Indeed, human Pol θ is able to perform non-templated DNA extension, as well as instructed replication that is templated in cis or in trans of the DSB. The same study revealed that the non-templated transferase activity is enhanced in the presence of Mn2+ ions and can be combined with templated extension on the 3′-end of a nucleic acid primer. Furthermore, it was shown that human Pol θ could incorporate NTPs, leading to the synthesis of polymers of RNAs, although with low yields.
The other family of DNA polymerases which has some significant nucleotidyltransferase activity is the pol X family, especially those involved in classical (Ku-dependent) NHEJ in eukaryotes (Pol λ, Pol μ, and TdT) (22–24). This activity is usually enhanced in the presence of divalent transition metal ions such as Mn2+. Among them TdT (Terminal deoxynucleotidyl Transferase) is known to catalyze efficient, non-templated, random nucleotide addition at the V(D)J junctions to increase the diversity of the adaptive immune system repertoire (24). It was also recently shown that TdT can, in some conditions, have an in trans templated activity on a DNA synapsis (25,26). On top of this property, previous studies revealed that TdT indiscriminately incorporates ribonucleotides (NTPs) and deoxyribonucleotides (dNTPs) (27) but then fails to extend the primer beyond 4–5 ribonucleotides when the substrate sensed by the enzyme is no longer a DNA but an RNA. This observation is compatible with the known crystal structure of murine TdT which shows that the steric barrier close to the 2′ position of the incoming dATP sugar in pol β and λ, a tyrosine residue, is replaced by a glycine in pol μ and Tdt (28). However, engineering TdT to make it accept an RNA primer seems quite a challenge, given that the conformation of the primer is a B-DNA structure in the tertiary complex, contrary to what would be favored for an RNA primer (A-DNA). In other words, there seems to be no ‘gate’ to the ribonucleotides in the catalytic site of TdT but rather one or several ‘checkpoints’ in guiding the primer single-stranded oligonucleotide.
Taken together, the availability of crystal structures of Pol θ and TdT as well as their known predisposition to perform random nucleotides incorporation in a template-free manner suggest the possibility to engineer them so as to make them tolerate both NTPs and dNTPs, as well as other modified nucleotides, opening the way to create novel random nucleic acid synthesizing machines.
Given previous studies on the engineering of the gate of ribonucleotides in members of the polA family (29,30), it looks like it should be possible to engineer pol θ steric gate. Also pol θ was shown to be a very tolerant polymerase across lesions (31). Here, we propose to improve the original nucleotidyltransferase property of human DNA Pol θ by rational design with the aim of widening as much as possible the diversity of the nucleic acid analogs that it can incorporate and consequently sampling in a more extensive way the stability, the affinity and the specificity of RNA aptamers.
Evolutionary and structural context for rational design of pol theta mutants with new properties
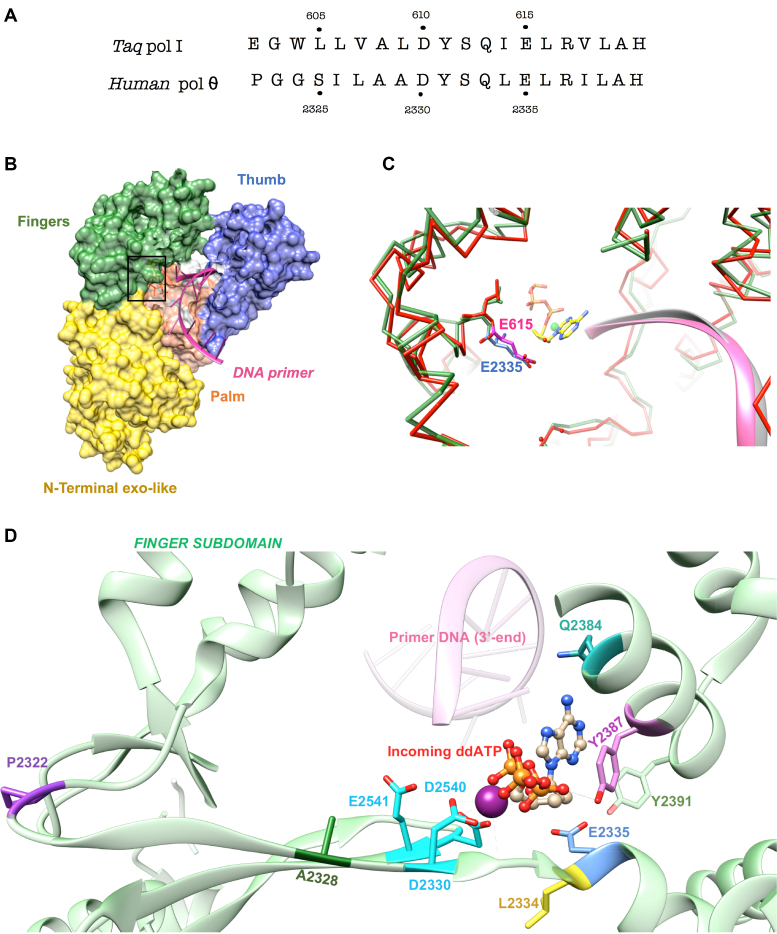
DNA polymerases contain an active site with highly conserved sequence motifs that are structurally superimposable within each family. In the polA family, several studies showed that most DNA polymerases share three highly conserved regions, called motifs A, B and C, that are located in the palm domain (10–12). Motif A contains a strictly conserved aspartate at the junction of a β-strand and α-helix, while motif C contains two carboxylate residues (Asp or Glu) at a beta-turn-beta structural motif (10). In the case of human pol θ the catalytic conserved aspartate (D2330) is located between β12 and α9 and is part of a strictly conserved motif A (DYSQLELR) in the different polA (Supplementary Figure S1). This catalytic aspartate corresponds to D610 in Thermus aquaticus Taq DNA polymerase I which interacts with the incoming dNTP through a Mg2+ ion and stabilizes the transition state that leads to new phosphodiester bond formation (32). This sequence motif (DYSQLELR) is exceptionally well conserved in A-family polymerases and within the Pol I family as described in the Supplementary Figure S1 for Human pol θ (PDB: 4X0P), Human pol ν (PDB: 4XVK), Human mitochondrial pol γ (PDB: 3IKM), Plasmodium falciparum DNA pol (PDB: 5DKT), Phage T7 DNA pol (PDB: 1T7P), Klenow fragment of E. coli DNA pol I (PDB: 1D8Y), Bacillus stearothermophilus DNA pol I (PDB: 1L3S) and T. aquaticus (Taq) DNA pol I (PDB: 1TAQ).
Previous studies in Taq DNA pol I showed that mutation of the catalytic aspartate of Motif A (D610) completely abolished the polymerase activity and was immutable (33) while libraries of mutants targeted on the other 13 residues (605 to 617) of motif A showed that some positions tolerated a wide spectrum of substitutions (A608, I614, R617). Otherwise, the remaining residues tolerated mainly conservative substitutions (Y611, E615). The residue S612 was highly mutable and accepted substitutions that are diverse in size and hydrophobicity while keeping WT-like activity. Alignment of human pol θ and Taq pol I sequences in Motif A (Figure 1A) illustrates the conservation of this Motif and displays the correspondence of the residues in Taq pol I and pol θ sequences (D610 corresponds to D2330, D615 to D2335). E615 forms a hydrogen bond with Y671, a residue located in helix O within the Motif B in the finger domain. From these early studies, we retain that the following positions are good candidates for site-directed mutations: L2334, E2335, A2328 within the motif A and residue Y2387 in motif B (corresponding to Y671 in Taq pol I).
Figure 1.
Structure of human pol θ polymerase domain (residues 1792–2590). (A) Sequence comparison of the motif A of T. aquaticus (Taq) pol I (1TAQ) and human pol θ (4X0P): the non-mutable residue D610 is colored in red while the residues that tolerate a wide spectrum of substitution are displayed in orange, green or blue according to their increasing tolerance of substitution. (B) Surface representation of human pol θ (PDB 4X0P) showing the general shape of a right hand with the three domains: the fingers domain in green, the palm domain in orange and the thumb domain in blue. The DNA primer is inserted between the palm and the fingers domains, with its 3′ end in the catalytic site. The N-terminal exonuclease domain is displayed in yellow. (C) Superposition of Taq pol I (1QSY) and human pol θ (4X0P), with both enzymes shown in stick models. A zoom-in stereo view of the fingers domain shows the residues E615 in pink (1QSY) compared to E2335 in blue (4X0P) in close proximity of the incoming nucleotide (ddATP, in yellow) facing the DNA primers (in pink for 4X0P and in grey for 1QSY). (D) Ribbon diagram of the ddATP-Ca2+ structure of pol θ (16) showing the environment of the catalytic site. The strictly conserved carboxylates (D2330, D2540 and E2541) coordinating the essential divalent metal cations, the mutable residues (P2322, A2328, L2334, E2335, Q2384, Y2387 and Y2391) are displayed as sticks of different colors. The incoming ddATP is showed in ball-and-stick model and the ssDNA primer (3′-end) is showed in light pink while the Ca2+ ion is shown as a purple sphere.
We note in passing that in polB family, the steric gate is located at the same place in Motif A as in polA (Y in the DxxLSYPSII motif) and the same is true for the polY family (30).
The crystal structure of the polymerase domain of pol θ (residues 1792–2590; PDB 4X0P) (16) reveals the same right hand-like topology seen in the homologs from bacteria and phages. The surface representation of pol θ (Figure 1B) illustrates the organization of the polymerase in three domains (fingers, palm and thumb) and a N-terminal exonuclease domain. The DNA strand slides into the catalytic site between the fingers and the palm domains where the motif A residues makes the junction between the two parts. When pol θ is superimposed with the large fragment of Taq DNA pol I (1QSY) (Figure 1c), both structures display the same overall conformation. The spatial organization of the catalytic site of human pol θ is shown in Figure 1d with the strictly three conserved carboxylates (D2330, D2540 and E2541) that chelate the essential catalytic metal divalent ions. In the immediate proximity of the catalytic triad the residue E2335 is facing the incoming nucleotide in close proximity to the C2′ of the ribose of the incoming nucleotide (ddATP), while its carboxylate group forms a hydrogen bond with the residues Y2391 (Motif B), Q2475 (Motif 6) and R2241 (TGR Motif). The residues E615 (1QSY) and E2335 (4X0P) are oriented in a similar manner in all polA structures around the incoming nucleotide and can interact with its sugar moiety and this confirms that E2335 should be one of the first positions to mutate. Nevertheless, both site-directed mutagenesis and early systematic mutational studies of Motif A noted that the best mutants, when successful in incorporating one or several ribonucleotides, were unable to add long stretches of RNA, namely to become a DNA-directed RNA polymerase (29).
To transform Taq pol I into a true RNA polymerase, several authors found that larger libraries and better ways to screen them were needed, such as short-patch compartmentalized self-replication (spCSR) selection (34). In this way, Taq pol I libraries were generated by mutating the residues 597–617 that include the nucleotide binding pocket and the steric gate residue E615. Among the different Taq pol I mutants, I614K and a multiple-site mutant called SFR3 (A597T, W604R, L605Q, I614T, E615G) acquired the ability to incorporate NTPs while no detectable primer extension by wt-Taq pol I was observed with NTPs. One of the best obtained mutants, called AA40, was able to incorporate up to six NTPs in the presence of Mg2+ and up to 14 NTPs when Mn2+ was used instead of Mg2+. This mutant involves four mutations located in (extended) Motif A: E602V, A608V, I614M and E615G.
In light of the close similarity of the Motif A region of Taq pol I and human pol θ (Figure 1A), all these results suggest to indeed focus on residue E2335 and the immediately close one L2334. We also considered the residues located in the proximal region of the nucleotide binding pocket (Y2387 and possibly Q2384, Y2391) and those found in mutant AA40 of Taq pol I described above (P2322, A2328).
MATERIALS AND METHODS
Materials and reagents
All chemicals and reagents were purchased from Sigma Aldrich (Saint-Quentin Fallavier, France) or Thermo Fisher Scientifics (Courtaboeuf, France) and were of the highest purity. The commercial enzymes T4 Polynucleotide kinase, T4 DNA ligase and T4 RNA ligase 1 were obtained from New England Biolabs (NEB). The enzymes used for the nucleosides digestion, Benzonase® nuclease, Phosphodiesterase I from Crotalus adamenteus venom and alkaline Phosphatase from bovine intestinal mucosa were purchased from Sigma Aldrich.
Nucleotides analogs
The following nucleotides were purchased from Jena Bioscience: 5-ethynyl-UTP, 2-aminopurine-ribonucleotide-5′-triphosphate, 2′-O-methyl-CTP, 2′-O-methyl-ATP, ara-CTP, ara-ATP, ϵ-ATP, 2′-fluoro-ATP, 2′-fluoro-CTP, 2′-fluoro-UTP and 2′-fluoro-GTP.
The following ones were purchased from Trilink Biotechnologies: 5-methyl-UTP, ATP, CTP, UTP, GTP.
Protein purification
WT Pol θ (residues 1792–2590) was expressed from the pSUMO3 construct (obtained from S. Doublié & S. Wallace, Addgene plasmid #78462) in BL21 CodonPlus (DE3) RIPL cells (Agilent technologies). The expression was carried out by autoinduction in Terrific broth EZMix™ supplemented with α-lactose (2 g/l), d-glucose (0.5 g/l), glycerol (8 ml.l−1), 100 μg.ml−1 of ampicillin and 50 μg.ml−1 of chloramphenicol. Six liters of autoinducing medium were inoculated (starting OD600, 0.05) and the culture was grown for 60 h at 20°C, with saturated cultures reaching a final OD600 between 5 and 8. The following steps were performed at 4°C. Cells were harvested and resuspended at a ratio of 2.5–3 ml/g of cell pellet in lysis buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 10% glycerol, 1 mM TCEP, 5 mM imidazole, 1,5% (v/v) IGEPAL C6-30, 5 mM CaCl2, PIERCE™ EDTA-free protease inhibitor tablets and Benzonase® nuclease 500 U. Cell lysis was performed by using French press Cell-Disruptor at 20 000 psi. Following clarification by ultracentrifugation at 17 000 rpm for 1 h, two steps of column purification were performed. The supernatant was applied to a Ni-NTA resin through a HisTrap HP column (GE Healthcare Life sciences) which was equilibrated with buffer A (50 mM HEPES pH 7.4, 300 mM NaCl, 20 mM imidazole, 0.005% (v/v) IGEPAL C6-30, 1 mM TCEP, 10% (v/v) glycerol), and eluted with a gradient to 500 mM of imidazole with buffer B (50 mM HEPES pH 7.4, 300 mM NaCl, 500 mM imidazole, 0.005% (v/v) IGEPAL C6-30, 1 mM TCEP, 10% (v/v) glycerol). Fractions from Ni-NTA chromatography containing Pol θ were 2-fold diluted to decrease NaCl content using diluting buffer C (50 mM HEPES pH 7.4, 0.005% (v/v) IGEPAL C6–30, 1 mM TCEP, 10% (v/v) glycerol). Then, the diluted fraction was applied to Heparin affinity chromatography. The HiTrap Heparin column (GE Healthcare Life sciences) was equilibrated with buffer D (50 mM HEPES pH 7.4, 50 mM NaCl, 0.005% (v/v) IGEPAL C6-30, 1 mM TCEP, and 10% (v/v) glycerol) and the fraction was eluted with a gradient to 2 M of NaCl with buffer E (50 mM HEPES pH 7.4, 2 M NaCl, 0.005% (v/v) IGEPAL C6-30, 1 mM TCEP, and 10% (v/v) glycerol). The protein fraction was then concentrated and frozen rapidly in a liquid nitrogen bath prior to storage at –80°C.
Generation of mutants
Variant pol θ constructs were generated by site-directed mutagenesis by using the Quick-Change II XL kit (Agilent technologies) and were purified following the previously described protocol. The oligonucleotides used for the mutagenesis are listed in the supplementary Table S1.
Oligonucleotides
Oligonucleotides were purchased from Eurogentec with RP-HPLC purity and dissolved in Nuclease-free water. Concentrations were measured by UV absorbance using the absorption coefficient ϵ at 260 nm provided by Eurogentec.
ssDNA primer radiolabelling
Oligonucleotides were labelled as follows: 40 μM of ssDNA primer (14-mer) were incubated with [γ-32P] ATP (Perkin Elmer, 3000 Ci.mM−1) and T4 polynucleotide kinase (New England Biolabs) for 1 h at 37°C in a total volume of 25 μl. The reaction was stopped by heating the T4 polynucleotide kinase at 75°C for 10 min. 25 μM of label-free ssDNA primer was added to the mix and heated for 5 min up to 90°C, and slowly cooled to room temperature overnight.
Radioactive nucleotidyltransferase assay
5 μM of Pol θ was incubated with 50 nM of 5′ (33) P-labeled ssDNA in the presence of 5 mM of MnCl2 in a total volume of 10 μl of activity buffer (20 mM Tris pH 8, 150 mM NaCl, 10% glycerol, 0.01% IGEPAL C6-30, 0.1 mg.ml−1 BSA). The reaction was started by addition of 500 μM of canonical or modified NTPs and stopped after 0–60 min at 42°C by adding 10 mM EDTA and 98% formamide. The products of the reaction were resolved by gel electrophoresis on a 8% or 15% acrylamide gel and 8 M urea. The 0.4-mm wide gel was run for 3–4 h at 40 V/cm and scanned by Storm 860 Molecular Dynamics phosphorimager (GE Healthcare).
Non-radioactive nucleotidyltransferase assay
Different nucleotides ratios were tested in order to verify that each canonical ribonucleotide was equally incorporated by the polymerase. 5 μM of enzyme was incubated with 500 nM of non-labelled ssDNA primer and a ratio 1:1:1:1 of the four ribonucleotides (500 μM each) or with 500 μM of ATP, CTP and GTP, and 5 mM of UTP (1:1:1:10) or with 500 μM of ATP, CTP and GTP and 2.5 mM of UTP (1:1:1:5) or 500 μM of ATP and GTP and 2.5 mM of CTP and UTP (1:1:5:5). Additional mixtures were prepared with ATP/CTP and UTP/GTP (500 μM each). The reaction was performed in the same activity buffer in presence of 5 mM of MnCl2 in a total volume of 100 μl. Synthetic RNAs (5–80 μg) were first cleaned-up by using the RNA Clean & Concentrator™-5 kit (Zymo Research). The clean-up was carried out in two steps to allow the purification of small RNA fragments of size between 17 and 200 nucleotides (nt) and at the same time large RNAs (>200 nt). Purified synthetic RNAs were then used immediately for HPLC analysis, for RNA sequencing, for aptamer library construction or stored at –80°C.
Hydrolysis of synthetic RNA to nucleosides and HPLC analysis
Synthetic RNAs obtained after non-radioactive ssDNA primer extension were hydrolysed according to previous protocol (33) with slight modification. The purified RNA pool was treated with Benzonase®nuclease (20 U), Phosphatase alkaline (1 U), Phosphodiesterase I (0.05 U) in 50 μl of digestion buffer (50 mM Tris–HCl pH 8, 1 mM MgCl2, 0.1 mg.ml−1 BSA). The mix was incubated at 37°C for 3 h and the digestion was carried out overnight at room temperature to insure a total lysis. Ribonucleosides were then cleaned-up by using a 10 000 MWCO Vivaspin®-500 centrifugal concentrator (Sartorius) and centrifuging for 10 min at 4°C. The filtrates were transferred to a 100 μl-vial insert tube to be further analyzed by HPLC. Analytical HPLC were carried out on a reverse phase C18 column (Kromasil 100-5-C18, 150 × 4.6 mm, AIT France) using a linear gradient of 0–20% of acetonitrile in 10 mM TEAAc (triethylammonium acetate) buffer at a flow rate of 1 ml.min−1 over 15 min. A mixture of the four ribonucleosides (5 μl at 0.25 mM each in digestion buffer) was injected as standard. The linearity and reproducibility of the measurements were checked by injecting, separately, two more samples with 10 and 25 μl. The UV molar extinction coefficients of each nucleoside-5′-monophosphate (32) at 260 nm (15040, 12080, 7070 and 9660 l.mol−1.cm−1 for pA, pG, pC and pU respectively) were used to quantify the amount of bases injected in the HPLC system.
TruSeq RNA library preparation and sequencing
We used 100 ng of total synthetic RNA and constructed the sequencing libraries using the TruSeq Stranded mRNA LT Kit (Illumina, RS-122-2101, San Diego, CA, USA) as recommended by the manufacturer, except that the fragmentation step was omitted. All the reagents were added to the reaction but the incubation at 94°C was not performed. The directional libraries were controlled on Bioanalyzer DNA1000 Chips (Agilent Technologies, #5067-1504, Santa Clara, CA) and the concentration determined using the QuBit dsDNA HS kit (Q32854, Thermo Fisher Scientific). They were sequenced on an Illumina Hiseq 2500 sequencer using a HiSeq SR cluster kit v4 cBot HS (Illumina, # GD-401-4001) and a HiSeq SBS kit v4 50 cycles (Illumina, #FC-401-4002) in order to have around 50 millions single end reads of 65 bases per sample.
Different bases compositions were tested before RNA sequencing. A pool of the four nucleotides at a ratio of 1:1:1:1 (500 μM each, samples annotated as ‘N’) or with 500 μM of ATP, CTP and GTP, and 5 mM of UTP (ratio of 1:1:1:10, samples annotated as ‘10U’) or with 500 μM of ATP, CTP and GTP and 2.5 mM of UTP (ratio of 1:1:1:5, sample annotated as ‘5U’) or 500 μM of ATP and GTP and 2.5 mM of CTP and UTP (ratio of 1:1:5:5, sample annotated as ‘5U5C’).
Analysis of the sequencing reads
For each sample the quality of the reads was checked using FastQC v0.11.5 and reads containing TruSeq adapters were removed using cutadapt v1.14. Statistical analyses were performed using R v3.3.2, Shortread v1.32.0 and Biostrings v2.42.1.
RESULTS
Efficient NTPs incorporation by unlocking the steric gate residue of pol θ
Human pol θ WT has been already described to have the ability to incorporate NTPs (20) in a template-free manner from the 3′-end of a ssDNA or ssRNA primer, albeit with a low yield. Therefore, human pol θ is a good starting point when compared to Terminal deoxynucleotidyltranferase (TdT), where the elongation of NTPs stops after 5–6 additions (27).
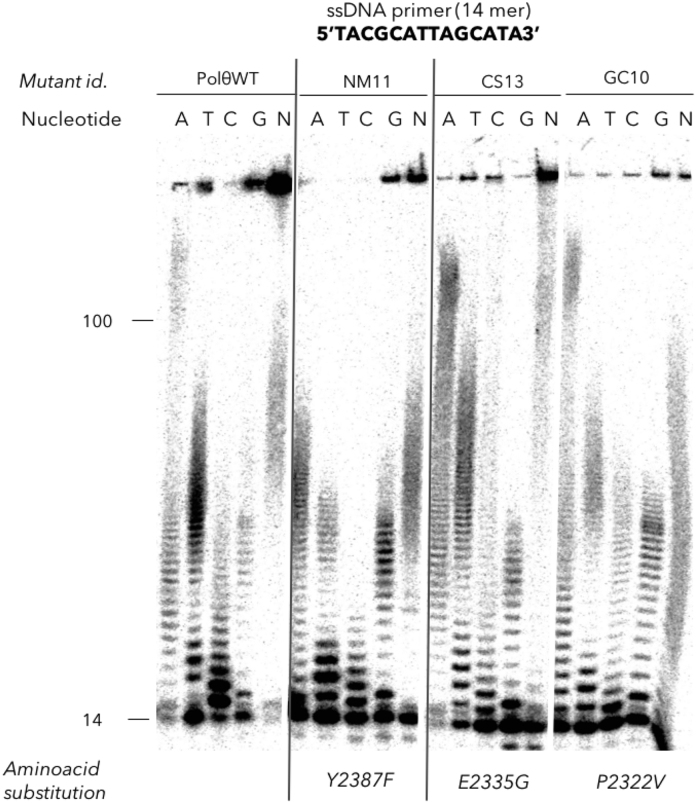
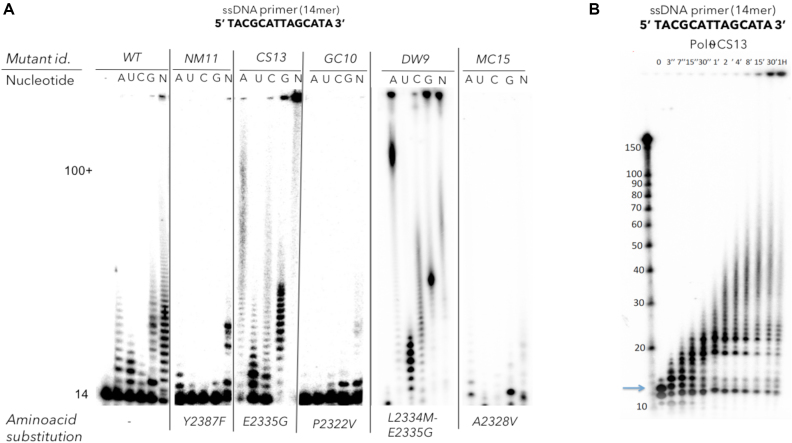
With the aim of selecting the best human pol θ mutant with an enhanced incorporation of NTPs, we evaluated the ability of several mutants to elongate a ssDNA primer through a nucleotidyltransferase assay. One or two-site directed mutagenesis were performed and led to generate the following mutants: NM11 (Y2387F), CS13 (E2335G), GC10 (P2322V), DW9 (L2334M-E2335G) and MC15 (A2328V). The nucleotidyltransferase activity was first tested with dNTPs (Figure 2) and all the mutants tested (NM11, CS13, GC10) retained the native activity as they incorporated the four natural dNTPs and generated fragments of length of up to 150 nt like the pol θ WT in the presence of 5 mM of Mn2+. When dNTPs were replaced by each NTP (ATP, UTP, CTP, GTP) the pol θ WT showed results similar to those previously described (20). ATP and GTP (purine bases) are well tolerated by pol θ WT and medium-length homopolymers of A and G (up to 50–70 nt) were obtained. UTP and CTP were incorporated but the elongation stopped after a few additions. The mutants NM11 and GC10 were unable to incorporate more than three or four NTPs. On the contrary, the mutant CS13 was particularly efficient as >50 of each ribonucleotide were readily incorporated, except for UTP that formed homopolymers whose length reached only up to 15 nt, still longer than the pol θ WT would add. Furthermore, when the four NTPs were mixed together the elongation was clearly enhanced compared to the wild-type enzyme, in that 100% of the primer were extended and the synthesized heteropolymers reached more than 150 nt in 30 min (Figure 3a). Mutants DW9 and MC15 were also tested for their incorporation rate of NTP: MC15 did not show better efficiency than the wild-type while DW9 showed an efficiency comparable to CS13 mutant. Overall, the best mutants are CS13 and DW9 which both carry the E2335G mutation, with a significantly improved substrate specificity towards ribonucleotides and an enhanced capability to promote RNA extension compared to the pol θ WT.
Figure 2.
Deoxynucleotidyltransferase activity of human pol θ WT and its respective mutants. Denaturing 15% acrylamide gel showing the activity of pol θ variants in the presence of Mn2+ cations and each of the four dNTPs (A, T, C, G) and the mix of all four dNTPs (N). A 14-mer ssDNA is used as a primer. The primer extension of three mutants (NM11 (Y2387F), CS13 (E2335G) and GC10 (P2322V)) are displayed in the same conditions as the pol θ WT. Reactions were stopped after 30 min by the addition of formamide blue.
Figure 3.
Ribonucleotidyltransferase activity of human pol θ WT and its respective mutants. (A) Denaturing 15% acrylamide gel showing the activity of pol θ variants in the presence of Mn2+ cations and each of the four NTPs (A, U, C, G at 0.5 mM each) and the mix of all four NTPs (N, at 0.5 mM each). A 14-mer ssDNA is used as a primer. The primer extension of five mutants: NM11(Y2387F), CS13 (E2335G), GC10 (P2322V), MC15 (A2328V) and DW9 (L2334M-E2335G) are also displayed in the same conditions as the pol θ WT. Reactions were stopped after 30 min. (B) Time-course of the elongation of a 14-mer ssDNA primer by CS13 mutant in the presence of a stoichiometric mix of the four NTPs (0.5 mM each) and separated in a 8% acrylamide gel. At each indicated time (0 s to 60 min), the reaction was stopped by the addition of formamide blue.
In the perspective of generating a library of random sequences of RNA with controlled fragment lengths the kinetics of primer extension was studied in the same conditions and allowed to monitor the length of the products as a function of the reaction time. In the case of 20–30 nt RNA fragments, 1 min-reaction is sufficient to complete the synthesis (Figure 3B).
Structural rationale of incorporation of ribonucleotides by CS13 mutant
CS13 mutant consists in the substitution of the charged glutamate residue E2335 by the small and flexible glycine amino-acid. The steric gate residue E2335 is located in close proximity to the sugar moiety of the incoming nucleotide. In the case of ddATP-pol θ structure (PDB 4X0P) the carboxylate group of the glutamate contributes indirectly, through its interaction with Y2387, to the positioning of the nucleotide (Supplementary Figure S2A). In the case of NTP, the presence of the hydroxyl group at the C2′ position creates both steric hindrance and electrostatic constraints between the sugar moiety and the pocket shaped by the residues E2335, Y2387 and Y2391 (Figure 1D and Supplementary Figure S2B). The mutation E2335G obviously enlarges the nucleotide binding pocket and is indeed compatible with an increased ability to incorporate a ribonucleotide into the 3′-end of the ssDNA primer (Supplementary Figure S2C). Similar observations were described for E. coli phage T7 DNA polymerase, where the ribose moiety of the incoming nucleotide is lodged between the aromatic ring of the residue Y526 and the aliphatic carbons of the E480 side chain, which itself is hydrogen-bonded to the hydroxyl of the strictly conserved Y530 (15). The spatial orientation of these residues forms a hydrophobic pocket at the C2′ position of the ribose that might exclude ribonucleotides from the active site, thus providing a structural basis for the strong discrimination against ribonucleotide incorporation by this DNA polymerase.
To understand how long RNA synthesis can occur in the CS13 mutant, we analyzed the primer strand conformation and residues in contact with it in pol θ, and compared it with the closed form of KlenTaq (PDB 3KTQ). Three out of four copies of the 4X0P pol θ structures superimposed very well, and also to KlenTaq, where the DNA is in a mixed A-B form, which would be compatible with binding an RNA strand. In addition, we found that two loops in close contact with the DNA in KlenTaq were either remodeled (586–589 in 3KTQ, 2254–2256 in pol θ) or longer and disordered (504–513 versus 2145–2175), possibly giving more conformational freedom to the DNA or RNA primer substrate in pol θ compared to KlenTaq, while still holding it with a very tight grip.
Quantifying the amount of incorporation of the four ribonucleotides by CS13 mutant
The success of a SELEX method relies on the quality of the initial nucleic acids library. Therefore, the possibility to have access to a large collection of random-sequence RNA or DNA fragments is essential. The CS13 mutant demonstrated, as assessed by gel electrophoresis, a remarkable ability to elongate a DNA primer with ribonucleotides, but in these experiments the proportions of incorporation of the four NTPs are unknown. We used HPLC analysis in order to quantify the overall base composition of the newly synthesized RNAs. Prior to HPLC separation the RNAs were completely digested into ribonucleosides according to a published protocol (35).
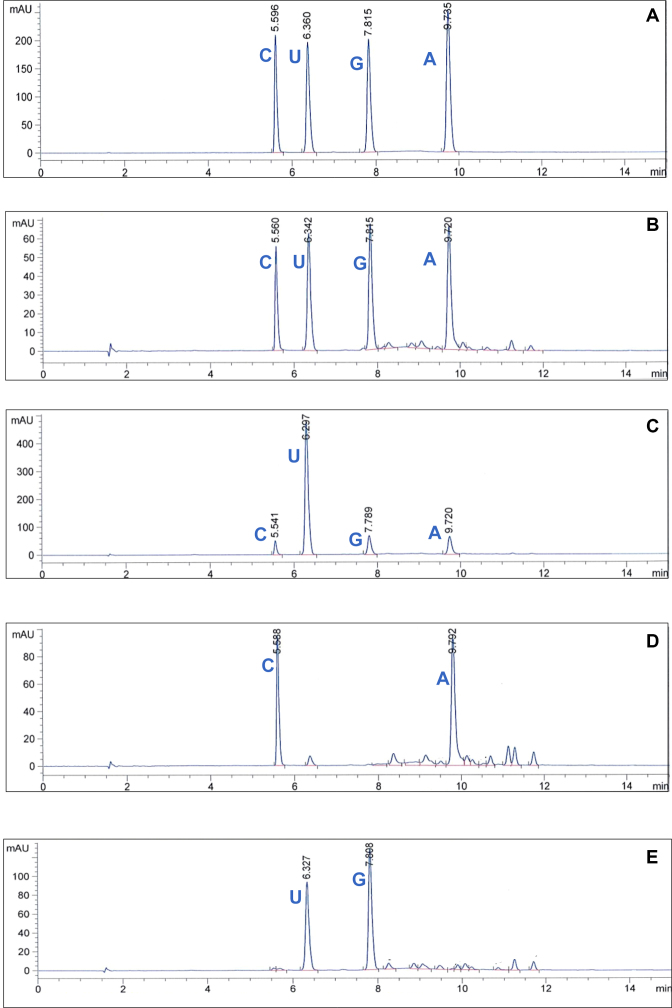
First a calibration of the method was performed: the chromatogram of the standards solutions of the four ribonucleosides at a final concentration of 0.25 mM in the digestion buffer indicated four peaks eluting at 5.60, 6.36, 7.82, 9.74 min respectively for cytidine, uridine, guanosine and adenosine (Figure 4A). The injected mix of standard nucleosides (in equal proportions) resulted in a global composition of 27.9%mol C, 22.5%mol G, 23.8%mol A and 25.8%mol U calculated from the UV extinction coefficient of each nucleoside-5′-monophosphates according to previous studies (35,36).
Figure 4.
HPLC separation and quantification of the ribonucleosides obtained after enzymatic hydrolysis of synthesized RNAs. (A) Chromatogram of the standard solutions of the four ribonucleosides (adenosine, guanosine, uridine and cytidine at concentrations of 0.25 mM each in the digestion buffer, the retention times and the base corresponding to each peak are displayed on top of each peak. (B) RNA hydrolysate obtained from RNA synthesis by CS13 and an equimolar mix of the four NTPs (C) RNA hydrolysate obtained from RNA synthesis by CS13 and a mix containing ATP/CTP/GTP/UTP at a molar ratio of 1:1:1:10. (D) RNA hydrolysate obtained from RNA synthesis by CS13 and an equimolar mix of ATP and CTP. (E) RNA hydrolysate obtained from RNA synthesis by CS13 and an equimolar mix of GTP and UTP. UV detection at 260 nm was used.
The denaturing gels of the products generated by the CS13 mutant (Figure 3A) indicated that in presence of an equal proportion of the four NTPs (1:1:1:1 at 0.5 mM each), long polymers of RNA were synthesized but they give no indication on the distribution of each nucleoside along the sequence. However, the incorporation of each of the four NTPs into homopolymers displayed different trends, especially UTP which seemed to be integrated less efficiently by the enzyme in the context of poly-U. To measure the proportion of nucleosides in long heteropolymers synthesized by CS13, the reaction products obtained by an initial mixture containing NTPs (1:1:1:1) were cleaned up and hydrolyzed, and the resulting ribonucleosides were analyzed by HPLC (Figure 4B). We found that the initial mix of 1:1:1:1 NTPs gave 24.8%mol C, 24.6%mol G, 22.1%mol A and 28.6%mol U in the generated polyribonucleotides, on average.
In addition, another synthesis condition was prepared that contained a tenfold molar excess of UTP (1:1:1:10) and analyzed in the same way (Figure 4). Again, four peaks were observed, corresponding to the retention time of the four nucleosides, but the calculated peak areas of each component revealed different distributions (Supplementary Table S2). We found that the initial mix of 1:1:1:10 NTPs gave 7.6%mol C, 9.0%mol G, 7.6%mol A and 75.9%mol U.
When the equimolar combination of only two NTPs (A/C; Figure 4D and U/G; Figure 4E) an approximately equal probability of incorporation of both substrates was obtained for C and A (55.8%mol C, 44.2%mol A) and for G and U (51.9%mol G, 48.1%mol U).
These results suggest the possibility to modify the initial substrates composition in order to modulate the incorporation of one or several specified nucleotide(s) in the final products. On average, it appears that CS13 mutant accepts the four natural NTPs with about the same efficiency.
Evaluation of the random character of the library of ssRNA generated by CS13 mutant
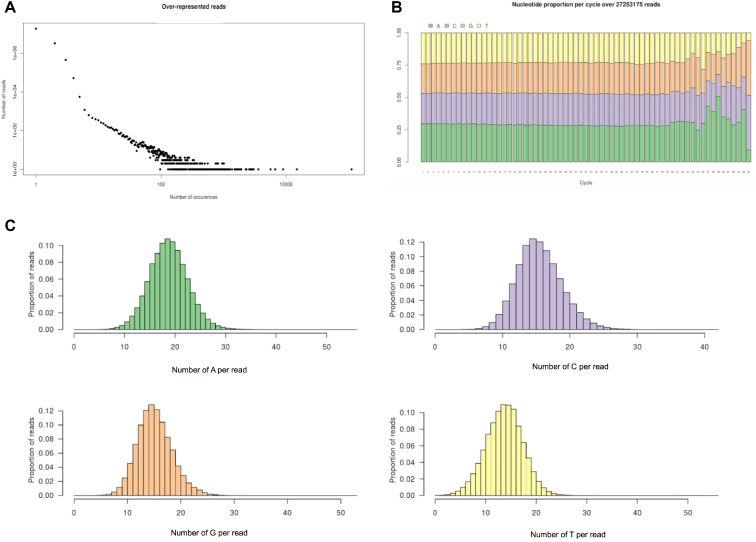
Next, the sequencing of RNA products was performed in order to reveal more details about the ribonucleotides incorporation by the mutant CS13. For the different conditions tested the RNA library displayed up to 31 millions of reads. For the whole library, the occurrence of the different sequences was estimated and was plotted as illustrated in Figure 5. Among 27 386 038 reads in the sample ‘N’, 18 626 379 reads had unique sequence (occurrence = 1), representing almost 68% of the total reads in the library (Figure 5A). The rest is dispersed between 2 (3 271 687 other reads with sequences repeated twice) and 111 670 occurrences (one sequence). To evaluate the distribution of the nucleotides along each sequence, we analyzed the proportion of the four nucleotides at each incorporation cycle (Figure 5B). The sequencing has been performed for the 65 first bases of each RNA fragment, and all the 65 positions of the 27 386 038 reads were taken in to account after cleaning the sequences from the DNA primer (14-mer) used for the elongation. The results indicate that when the four nucleotides were mixed at an equimolar ratio (sample ‘N’), a roughly equal incorporation is observed for the RNA synthesis by the CS13 mutant. Within the 65 cycles, the global proportion of added nucleotide remained constant up to position 50 with a value of 26.7%, 25.0%, 24.0% and 24.3% for A, C, G and U, respectively (Figure 5B). Additionally, the histograms representing the frequency of each nucleotide display an approximately Gaussian shape (Figure 5C).
Figure 5.
Statistical analysis of random RNAs synthesized by CS13 mutant. (A) Occurrences of the reads illustrated by a log–log scatter plot chart. Condition ‘N’ represent RNA synthesis by CS13 and the mix of four nucleotides at a ratio of 1:1:1:1 (500 μM each). (B) Nucleotide proportion per incorporation cycle illustrated by a stacked bars chart for the same sample ‘N’. (C) Frequency of each number of nucleotides of type A, C, G or U per read, represented by a histogram.
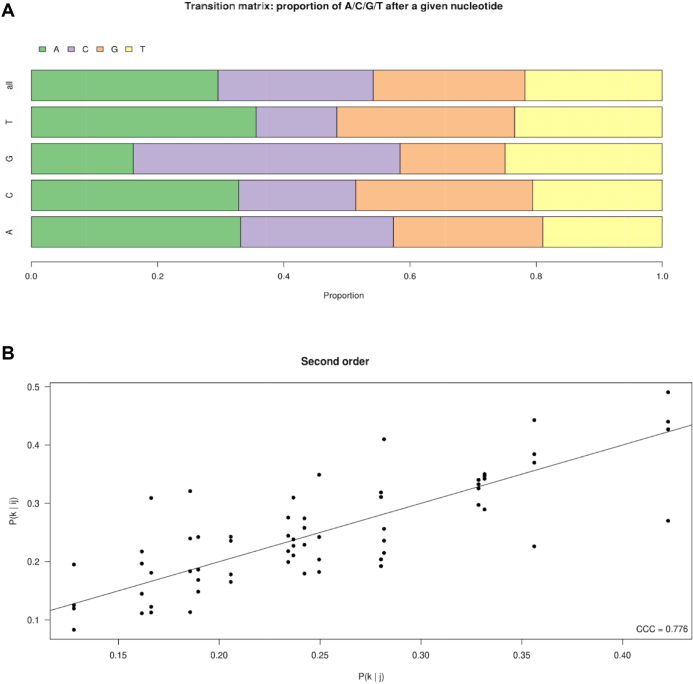
Furthermore, it is possible to estimate from the data the probability P(j | i) of adding one (A/C/G/U) nucleotide (j) in position N after a given nucleotide (i) in position N – 1. This quantity can then be used to test whether or not the nature of the added nucleotide in position N depends on the nature of the nucleotide present at the position N – 1. At first sight, there are indeed differences among the 16 possible dinucleotides, as, for instance, GC is the most probable occurrence when an equimolar ratio of ribonucleotides is present, while, to the opposite, it is less probable to form UC and GG dinucleotides in the same conditions (Figure 6a). We compare P(j | i) to P(j) by calculating their concordance correlation coefficient and find 0.252, 0.229 and 0.280 in three different replicates, meaning that the information on nucleotide frequencies is not enough to explain dinucleotide frequencies. These numbers change very little if one restricts the analysis to unique sequences, i.e. single reads (0.239, 0.226 and 0.278).
Figure 6.
Further statistical analysis of random RNAs synthesized by CS13 mutant. (A) Nucleotide transition matrix illustrating the proportion of A/C/G/U added after a given nucleotide, shown as a horizontal stacked bars chart for the condition ‘N’, which is the mix of four nucleotides at a ratio of 1:1:1:1 (500 μM each). (B) Scatter plot showing the correlation between the 64 trinucleotide frequencies P(k | j, i) as a function of each 16 P(k | j) frequency.
We next used information on tri-nucleotides P(k | j, i), the probability of observing nucleotide k at position N, given that nucleotide j is observed at position N – 1 and nucleotide i at position N – 2 and compare it to P(k | j). The concordance correlation coefficient between these two values is 0.776 (Figure 6B), 0.766 or 0.779 in three different replicates, indicating that most (but not all) of the information contained in trinucleotide (i, j, k) conditional frequencies is already contained in the knowledge of the dinucleotide (j, k). These numbers change very little if one restricts the analysis to unique sequences only (0.776, 0.769 and 0.781).
Modified nucleotides incorporation by the CS13 mutant
One of the main drawbacks of aptamers is their relative instability and their sensitivity to hydrolysis in biological fluids. The solution to overcome this point is to produce nuclease-resistant RNA molecules by different approaches (5,37,38). Modifications can be grafted on the nucleotide sugar moiety, the phosphodiester covalent link or on the base.
The incorporation of modified ribonucleotides has been also investigated for the CS13 mutant in comparison with pol θ WT. The following analogs were tested: 2′-fluoro-dUTP, 2′-fluoro-dATP, 2′-fluoro-dCTP, 2′-fluoro-dGTP, 2′-fluoro-dTTP, 2′-amino-dATP, 2′-amino-dCTP, 2′-amino-dGTP, 2′-amino-dTTP, 2′-O-methyl-dATP, 2′-O-methyl-dCTP, 2′-O-methyl-dGTP, 2′-O-methyl-dTTP, 2′-N3 -dATP, 2′-N3 -dCTP, 2′-N3 -dGTP, 2′-N3 -dTTP, ara-ATP, ara-CTP, ϵ-ATP and 2-aminopurine rTP.
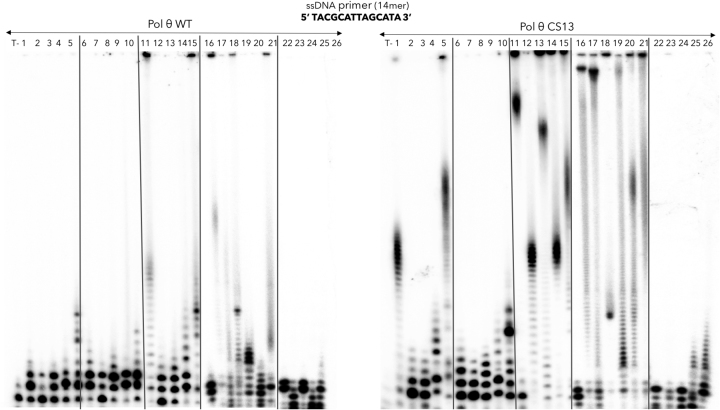
It has been described that the incorporation rate of 2′-fluoro-modified ribonucleotides by T7 RNA polymerase was ten-fold lower than that of the natural substrates (37). The templated incorporation of 2′-fluoro modified nucleotides by DNA polymerases were already assessed in several studies and found to be low (38), so that the incorporation of these modified nucleotides by enzymatic synthesis remains relatively limited compared to chemical synthesis. Here, we found that all the 2′-fluoro (Figure 7, lanes 16–21) were incorporated by CS13 mutant in the same experimental conditions as the primer extension by natural NTPs, with a much greater efficiency than pol θ WT. This modification is suitable for ribozyme development and aptamers selection as the 2′-fluoro-modified oligonucleotides have better ribonucleases resistance (39–41).
Figure 7.
Incorporation of modified nucleotides by pol θ WT in comparison with the pol θ CS13. The following nucleotide analogs were tested for the elongation of a ssDNA primer by pol θ CS13 compared to pol θ (1) 2′-amino-dATP, (2) 2′-amino-dUTP, (3) 2′-amino-dCTP, (4) 2′-amino-dGTP, (5) mix of 2′-amino-dATP/dUTP/dCTP/dGTP, (6) 2′-O-methyl-dATP, (7) 2′-O-methyl-dUTP, (8) 2′-O-methyl-dCTP, (9) 2′-O-methyl-dGTP, (10) mix of 2′-O-methyl-dATP/dUTP/dCTP/dGTP, (11) 2′-azido-2′-dATP, (12) 2′-azido-2′-dUTP, (13) 2′-azido-2′-dCTP, (14) 2′-azido-2′-dGTP, (15) mix of 2′-azido-2′-dATP/dUTP/dCTP/dGTP, (16) 2′-fluoro-dATP, (17) 2′-fluoro-dUTP, (18) 2′-fluoro-dCTP, (19) 2′-fluoro-dGTP, (20) 2′-fluoro-dTTP, (21) mix of 2′-fluoro-dATP/dUTP/dCTP/dGTP, (22) Ara-ATP (vidarabine triphosphate), (23) Ara-CTP (cytarabine triphosphate), (24) mix of Ara-ATP and Ara-CTP, (25) ϵ-ATP, (26) 2-aminopurine riboside triphosphate. The reactions were performed in the same conditions as the natural NTPs in presence of Mn2+.
The CS13 mutant failed to elongate the ssDNA primer in presence of 2′-O-methyl modified nucleotides further than just a few nucleotides (Figure 7, lanes 6–10). 2′-O-methyl-modified RNA display better stability against hydrolysis by ribonucleases as well as increased Tm values. This attribute may be useful to preserve the 3D conformation and to widen the diversity of a functional aptamers (42). Thus, the acceptance of 2′-O-methyl modified nucleotides need to be improved with CS13 mutant, perhaps by introducing a second mutation at residue 2334 with a non-hydrophobic residue (43).
The incorporation of 2′-amino dATP and 2′-amino dGTP (Figure 7, lane 1–5) appeared to be quite efficient with the CS13 mutant, more than 2′-amino dTTP and 2′-amino dCTP. For 2′-N3 dNTP, the incorporation was very good with the mutant CS13, almost as good as the 2′-F derivatives, allowing the possibility to perform click chemistry experiments with compounds containing an alkyne group (lanes 11–15).
9-β-d-Arabinofuranosyladenine (ara-ATP), an antiviral drug against Herpes simplex virus (44) and 1β-arabinofuranosylcytosine (Ara-CTP), an analogue of pyrimidine used in cancer chemotherapy (45), were also tested but the incorporation appeared to be very weak (Figure 7, lanes 22–24). Finally, two fluorescent analogues of adenine nucleotides were tested: etheno-adenine (Figure 7, lane 25) and 2-aminopurine (lane 26). Both were incorporated by CS13 mutant but formed only short polymers.
DISCUSSION AND PERSPECTIVES
To our knowledge, no RNA or DNA polymerase has been described to operate as a template-free ribonucleotidyltransferase and to synthesize long random RNAs. Since human DNA polymerase θ exhibits a versatile range of activities, including the capability to elongate nucleic acid primers without template requirement and to incorporate modified nucleotides (20,46) we took up the challenge of developing it into an RNA synthesizing machine. Among other members of the pol A family, the replicative human mitochondrial DNA polymerase γ has been described to incorporate NTPs both in vivo and in vitro, to maintain DNA replication when the dNTPs pool is poor (47) but this was in the presence of a DNA template.
On the basis of available structural information, we obtained two pol θ mutants, CS13 and DW9, that demonstrated a reliable ability to perform RNA random synthesis and to incorporate several other modified nucleotides. The randomness of the synthesized sequences and the Markov Model behind it have been assessed quantitatively and this new enzymatic activity should be useful for therapeutics applications. In that way, the continuous quest for ultra-effective, selective and non-toxic nucleic acids-based drugs might be pursued using optimized aptamer design strategies. Indeed, our work offers a viable biological alternative to the generation of RNA random sequences by chemical synthesis and library design. By establishing an efficient enzymatic SELEX procedure, the assay costs would be reduced at the same time as the duration of the selection cycle. Furthermore, human pol θ does not require toxic compounds (such as cacodylate buffer, cobalt divalent ions, which are needed for TdT) to operate RNA or RNA analogs synthesis, thereby conferring safer alternatives to library construction and biotechnological applications.
To make this work complete, it will be necessary to develop an efficient method of amplification of a set of selected sequences from the aptamer library, i.e. the pool of synthesized RNAs. As a matter of fact, SELEX procedure comprises a step where the aptamer candidates need to be amplified before the next selection step. In our case, the newly synthesized RNAs do not have a fixed 3′ region, which is necessary for their amplification by PCR. Our next goal is to enzymatically add a fixed sequence at the 3′ end of each RNA fragment (the 5′ end contains the constant known primer sequence). Thus, the resulting pool of RNAs would be suitable for self-amplification, possibly by using the same enzyme, human pol θ CS13. By doing this, it would be possible to execute the entire SELEX procedure in an all-in-one system. In some preliminary tests, we performed a ligation of a fixed fragment to each synthesized RNA by exploiting T4 RNA ligase I activity. The preliminary results are encouraging and indicate that the ligation of these fixed oligonucleotides indeed occurs with our protocol. It is now necessary to optimize the experimental conditions to ensure 100% of ligation.
We also showed that human pol θ CS13 mutant accepted modified nucleotides with different efficiencies. Recent studies already described that human pol θ was able to incorporate nucleotides analogs better than the DNA polymerases TdT, Pol η or Pol κ, which are also error-prone and show some ability to incorporate nucleotide analogs. For instance, base analogs conjugated on position 5 of the pyrimidine moiety by fluorescent compounds (20) (Cy3-dUTP, Texas Red-5-dCTP, Biotin-16AA-dUTP) or large benzo-expanded nucleotides (45) were found to be incorporated by pol θ. Here we show that ϵ-ATP and 2-aminopurine rTP that are intrinsically fluorescent, can be incorporated to some extend by CS13 mutant of human pol θ. It would be interesting to determine whether if it would be possible to control the addition of these analogs at the end of RNA fragments for specific applications such as nucleic acid labelling for cell imaging, click-chemistry or, improved target binding.
It should be mentioned here that Romesberg and coll. (48) managed to develop an engineered DNA polymerase capable of amplifying directly RNA and modified RNA such as 2′ fluoro RNA. Also, these authors developed a third type of base pair that could be tried in the near future to be incorporated by CS13 mutant (49). Moreover, DeStefano and colleagues (50) developed a SELEX procedure that was suitable to select FANA aptamer to HIV-1 reverse transcriptase (RT). Their work led to the first FANA aptamers to HIV-1 Reverse Transcriptase (RT) made by direct selection using all FANA nucleotides. The results demonstrated that XNA may be an excellent alternative for aptamer generation. Another example is illustrated by the work of S.A Benner (51) and colleagues who generated aptamer libraries from a six-letter genetic alphabet, that contain the standard nucleobases and two added nucleobases (2-amino-8H-imidazo(1,2-α)(1,3,5)triazine-4-one and 6-amino-5-nitopyridine-2-one). From this artificially expanded genetic information system (AEGIS) 8 aptamers were recovered after several rounds of selection on proteins expressed on the surface of an engineered liver cell-line.
The ability of Human pol θ CS13 mutant to generate random polyribonucleotides opens the way to a greater diversification of aptamer libraries. Not only larger pool of RNAs can be produced, but also more diverse secondary structures would be formed with modified nucleotides inserted along each RNA fragment. In this way, the use of pol θ CS13 could improve SELEX procedures and lead to the discovery of more efficient and more specific therapeutic compounds. In the immediate future, we will seek to make the proof-of-principle of the use of pol θ CS13 in a SELEX procedure by selecting a known target, such as the thrombin, a classical test-case (52,53).
DATA AVAILABILITY
T-Coffee is a freeware open source multiple sequence alignment package. It is available in the link below: http://tcoffee.crg.cat/apps/tcoffee/index.html
ESPript 3.0 is developed and maintained by Patrice GOUET and Xavier ROBERT in the ‘Retroviruses and Structural Biochemistry’ research team of the ‘Molecular Microbiology and Structural Biochemistry’ laboratory (UMR5086 CNRS / Lyon University). http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi
FastQC 0.11.5 is a software to control the quality of the sequencing reads and is available at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Cutadapt 1.14 is a tool to clean the sequencing reads and is available at https://github.com/marcelm/cutadapt
R 3.3.2 is a statistical software and is available at https://www.r-project.org/
Biostrings 2.42.1 is a R/Bioconductor package to handle biological sequences and is available at https://bioconductor.org/packages/3.4/bioc/html/Biostrings.html
ShortRead 1.32.0 is a R/Bioconductor package to manipulate fastq reads and is available at https://bioconductor.org/packages/3.4/bioc/html/ShortRead.html
Promals3D (54) is multi-alignment tool that uses three-dimensional information and is available at http://prodata.swmed.edu/promals3d/promals3d.php
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jerome Loc’h for help with 3D Figures (Figure 1 and Supplementary Figure S2) and Dariusz Czernecki for Supplementary Figure S1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Action Incitative Concertée from Institut Pasteur (to M.D. and S.P.). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. All the authors declare no competing interest. A provisional patent application no. 62/560693, about DNA POLYMERASE THETA MUTANTS, THE METHODS OF PRODUCING THESE MUTANTS, AND THEIR USES, has been filed and deposited to the UPSTO on 20 September 2017.
REFERENCES
- 1. DeVos S.L., Miller T.M.. Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics. 2013; 10:486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crooke S.T. Antisense Drug Technology: Principles, Strategies, and Applications. 2008; Boca Raton: CRC Press, Taylor and Francis group [Google Scholar]
- 3. Breaker R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter N.G., Engelke D.R.. Ribozymes: catalytic RNAs that cut things, make things, and do odd and useful jobs. Biologist (London). 2002; 49:199–203. [PMC free article] [PubMed] [Google Scholar]
- 5. Diafa S., Hollenstein M.. Generation of aptamers with an expanded chemical repertoire. Molecules. 2015; 20:16643–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou J., Rossi J.. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017; 16:440. [DOI] [PubMed] [Google Scholar]
- 7. Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. 2009; 48:2672–2689. [DOI] [PubMed] [Google Scholar]
- 8. Lipi F., Chen S., Chakravarthy M., Rakesh S., Veedu R.N.. In vitro evolution of chemically-modified nucleic acid aptamers: pros and cons, and comprehensive selection strategies. RNA Biol. 2016; 13:1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoltenburg R., Reinemann C., Strehlitz B.. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007; 24:381–403. [DOI] [PubMed] [Google Scholar]
- 10. Delarue M., Poch O., Tordo N., Moras D., Argos P.. An attempt to unify the structure of polymerases. Protein Eng. Des. Sel. 1990; 3:461–467. [DOI] [PubMed] [Google Scholar]
- 11. Patel P.H., Loeb L.A.. Getting a grip on how DNA polymerases function. Nat. Struct. Biol. 2001; 8:656–659. [DOI] [PubMed] [Google Scholar]
- 12. Braithwaite D.K., Ito J.. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993; 21:787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999; 274:17395–17398. [DOI] [PubMed] [Google Scholar]
- 14. Eom S.H., Wang J., Steitz T.A.. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996; 382:278–281. [DOI] [PubMed] [Google Scholar]
- 15. Ellenberger T., Doublié S., Tabor S., Long A.M., Richardson C.C.. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2|[thinsp]||[angst]|resolution. Nature. 1998; 391:251–258. [DOI] [PubMed] [Google Scholar]
- 16. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublié S.. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wood R.D., Doublié S.. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair (Amst). 2016; 44:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee Y.-S., Gao Y., Yang W.. How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis. Nat. Struct. Mol. Biol. 2015; 22:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Longley M.J., Prasad R., Srivastava D.K., Wilson S.H., Copeland W.C.. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12244–12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kent T., Mateos-Gomez P.A., Sfeir A., Pomerantz R.T.. Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. Elife. 2016; 5:16203–16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Black S.J., Kashkina E., Kent T., Pomerantz R.T.. DNA polymerase θ: a unique multifunctional end-joining machine. Genes (Basel). 2016; 7:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andrade P., Martin M.J., Juarez R., Lopez de Saro F., Blanco L.. Limited terminal transferase in human DNA polymerase defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16203–16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramadan K., Maga G., Shevelev IV., Villani G., Blanco L., Hübscher U.. Human DNA polymerase λ possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003; 328:63–72. [DOI] [PubMed] [Google Scholar]
- 24. Fowler J.D., Suo Z.. Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chem. Rev. 2006; 106:2092–2110. [DOI] [PubMed] [Google Scholar]
- 25. Loc’h J., Rosario S., Delarue M.. Structural basis for a new templated activity by terminal deoxynucleotidyl Transferase: Implications for V(D)J Recombination. Structure. 2016; 24:1452–1463. [DOI] [PubMed] [Google Scholar]
- 26. Gouge J., Rosario S., Romain F., Poitevin F., Béguin P., Delarue M.. Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair. EMBO J. 2015; 34:1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boulé J.-B., Rougeon F., Papanicolaou C.. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001; 276:31388–31393. [DOI] [PubMed] [Google Scholar]
- 28. Delarue M., Boulé J.B., Lescar J., Expert-Bezançon N., Jourdan N., Sukumar N., Rougeon F., Papanicolaou C.. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 2002; 21:427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Astatke M., Ng K., Grindley N.D.F., Joyce C.M.. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown J.A., Suo Z.. Unlocking the sugar “Steric Gate” of DNA polymerases. Biochemistry. 2011; 50:1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hogg M., Seki M., Wood R.D., Doublié S., Wallace S.S.. Lesion bypass activity of DNA polymerase θ (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J. Mol. Biol. 2011; 405:642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Korolev S., Waksman G.. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998; 17:7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel P.H., Loeb L.A.. DNA polymerase active site is highly mutable: evolutionary consequences. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ong J.L., Loakes D., Jaroslawski S., Too K., Holliger P.. Directed evolution of DNA polymerase, RNA polymerase and reverse transcriptase activity in a single polypeptide. J. Mol. Biol. 2006; 361:537–550. [DOI] [PubMed] [Google Scholar]
- 35. Su D., Chan C.T., Gu C., Lim K.S., Chionh Y.H., McBee M.E., Russell B.S., Babu I.R., Begley T.J., Dedon P.C. et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 2014; 9:828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cavaluzzi M.J., Borer P.N.. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004; 32:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bunka D.H., Platonova O., Stockley P.G.. Development of aptamer therapeutics. Curr. Opin. Pharmacol. 2010; 10:557–562. [DOI] [PubMed] [Google Scholar]
- 38. Lauridsen L.H., Rothnagel J.A., Veedu R.N.. Enzymatic recognition of 2′-modified ribonucleoside 5′-triphosphates: towards the evolution of versatile aptamers. ChemBioChem. 2012; 13:19–25. [DOI] [PubMed] [Google Scholar]
- 39. Ono T., Scalf M., Smith L.M.. 2′-Fluoro modified nucleic acids: polymerase-directed synthesis, properties and stability to analysis by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 1997; 25:4581–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pieken W.A., Olsen D.B., Benseler F., Aurup H., Eckstein F.. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991; 253:314–317. [DOI] [PubMed] [Google Scholar]
- 41. Rhie A., Kirby L., Sayer N., Wellesley R., Disterer P., Sylvester I., Gill A., Hope J., James W., Tahiri-Alaoui A. et al. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to Disease-Associated conformations of Prion protein and inhibit conversion. J. Biol. Chem. 2003; 278:39697–39705. [DOI] [PubMed] [Google Scholar]
- 42. Dellafiore M.A., Montserrat J.M., Iribarren A.M.. Modified nucleoside triphosphates for In-vitro selection techniques. Front. Chem. 2016; 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schultz H.J., Gochi A.M., Chia H.E., Ogonowsky A.L., Chiang S., Filipovic N., Weiden A.G., Hadley E.E., Gabriel S.E., Leconte A.M. et al. Taq DNA polymerase mutants and 2′-modified sugar recognition. Biochemistry. 2015; 54:5999–6008. [DOI] [PubMed] [Google Scholar]
- 44. Whitley R.J., Alford C.A., Hirsch M.S., Schooley R.T., Luby J.P., Aoki F.Y., Hanley D., Nahmias A.J., Soong S.J.. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N. Engl. J. Med. 1986; 314:144–149. [DOI] [PubMed] [Google Scholar]
- 45. Tilly H., Castaigne S., Bordessoule D., Casassus P., Le Prisé P.Y., Tertian G., Desablens B., Henry-Amar M., Degos L.. Low-dose cytarabine versus intensive chemotherapy in the treatment of acute nonlymphocytic leukemia in the elderly. J. Clin. Oncol. 1990; 8:272–279. [DOI] [PubMed] [Google Scholar]
- 46. Kent T., Rusanov T.D., Hoang T.M., Velema W.A., Krueger A.T., Copeland W.C., Kool E.T., Pomerantz R.T.. DNA polymerase θ specializes in incorporating synthetic expanded-size (xDNA) nucleotides. Nucleic Acids Res. 2016; 44:9381–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berglund A.K., Navarrete C., Engqvist M.K., Hoberg E., Szilagyi Z., Taylor R.W., Gustafsson C.M., Falkenberg M., Clausen A.R.. Nucleotide pools dictate the identity and frequency of ribonucleotide incorporation in mitochondrial DNA. PLOS Genet. 2017; 13:e1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen T., Romesberg F.E.. Polymerase chain transcription: exponential synthesis of RNA and modified RNA. J. Am. Chem. Soc. 2017; 139:9949–9954. [DOI] [PubMed] [Google Scholar]
- 49. Malyshev D.A., Dhami K., Quach H.T., Lavergne T., Ordoukhanian P., Torkamani A., Romesberg F.E.. Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alves Ferreira-Bravo I., Cozens C., Holliger P., DeStefano J.J.. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015; 43:gkv1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang L., Yang Z., Le Trinh T., Teng I.T., Wang S., Bradley K.M., Hoshika S., Wu Q., Cansiz S., Rowold D.J. et al. Aptamers against cells overexpressing Glypican 3 from expanded genetic systems combined with cell engineering and laboratory evolution. Angew. Chemie Int. Ed. 2016; 55:12372–12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu X., Zhang D., Cao G., Yang G., Ding H., Liu G., Fan M., Shen B., Shao N.. RNA aptamers specific for bovine thrombin. J. Mol. Recognit. 2003; 16:23–27. [DOI] [PubMed] [Google Scholar]
- 53. Deng B., Lin Y., Wang C., Li F., Wang Z., Zhang H., Li X.F., Le X.C.. Aptamer binding assays for proteins: the thrombin example—a review. Anal. Chim. Acta. 2014; 837:1–15. [DOI] [PubMed] [Google Scholar]
- 54. Pei J., Kim B.-H., Grishin N.V.. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008; 36:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
T-Coffee is a freeware open source multiple sequence alignment package. It is available in the link below: http://tcoffee.crg.cat/apps/tcoffee/index.html
ESPript 3.0 is developed and maintained by Patrice GOUET and Xavier ROBERT in the ‘Retroviruses and Structural Biochemistry’ research team of the ‘Molecular Microbiology and Structural Biochemistry’ laboratory (UMR5086 CNRS / Lyon University). http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi
FastQC 0.11.5 is a software to control the quality of the sequencing reads and is available at https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Cutadapt 1.14 is a tool to clean the sequencing reads and is available at https://github.com/marcelm/cutadapt
R 3.3.2 is a statistical software and is available at https://www.r-project.org/
Biostrings 2.42.1 is a R/Bioconductor package to handle biological sequences and is available at https://bioconductor.org/packages/3.4/bioc/html/Biostrings.html
ShortRead 1.32.0 is a R/Bioconductor package to manipulate fastq reads and is available at https://bioconductor.org/packages/3.4/bioc/html/ShortRead.html
Promals3D (54) is multi-alignment tool that uses three-dimensional information and is available at http://prodata.swmed.edu/promals3d/promals3d.php