Abstract
Despite their central importance in mammalian development, the mechanisms that regulate the DNA methylation machinery and thereby the generation of genomic methylation patterns are still poorly understood. Here, we identify the 5mC-binding protein MeCP2 as a direct and strong interactor of DNA methyltransferase 3 (DNMT3) proteins. We mapped the interaction interface to the transcriptional repression domain of MeCP2 and the ADD domain of DNMT3A and find that binding of MeCP2 strongly inhibits the activity of DNMT3A in vitro. This effect was reinforced by cellular studies where a global reduction of DNA methylation levels was observed after overexpression of MeCP2 in human cells. By engineering conformationally locked DNMT3A variants as novel tools to study the allosteric regulation of this enzyme, we show that MeCP2 stabilizes the closed, autoinhibitory conformation of DNMT3A. Interestingly, the interaction with MeCP2 and its resulting inhibition were relieved by the binding of K4 unmodified histone H3 N-terminal tail to the DNMT3A–ADD domain. Taken together, our data indicate that the localization and activity of DNMT3A are under the combined control of MeCP2 and H3 tail modifications where, depending on the modification status of the H3 tail at the binding sites, MeCP2 can act as either a repressor or activator of DNA methylation.
INTRODUCTION
The correct establishment and maintenance of DNA methylation patterns that are set during mammalian embryogenesis by the de novo DNA methyltransferases, DNMT3A and DNMT3B, depend on the accurate targeting and regulation of DNA methyltransferases (DNMTs) (1–3). Mammalian DNMTs comprise two parts, a large multi-domain N-terminal part and a C-terminal catalytic domain (CD) (4,5). The N-terminal parts of DNMTs are important for guiding the nuclear and sub-nuclear localization of the enzymes. They function as an interaction platform with other proteins, DNA and chromatin, thereby regulating the catalytic activity (1,6). Two defined sub-domains are present in the N-terminal part of DNMT3A and DNMT3B (Figure 1A): a PWWP domain, which recognizes H3K36me3-modified H3 tails (7–9), and an ADD domain, which binds the N-terminus of histone H3 if K4 is unmodified (10–12). Structural and biochemical work has shown that the ADD domain of DNMT3A forms contacts with the CD at two distinct interfaces (Supplementary Figure S1) (13), an autoinhibitory site, where ADD binding hinders DNA-binding and thereby inhibits the activity of the CD, and an allosteric site, where binding does not lead to inhibition. Different ADD residues are contacting the CD, depending on the conformation adopted by the enzyme. For instance, Y526 contacts the CD in the allosteric conformation, while D531 contacts the CD in the autoinhibitory conformation. The conversion of DNMT3A from the autoinhibitory to the open conformation was shown to be stimulated by binding of the unmodified H3 peptide to the ADD domain. Through its binding, H3 induces a large movement of the ADD domain, which leads to the activation of DNMT3A (12–14). Several studies have recently shown that the activity of DNMT3 enzymes is regulated by the interaction with the H3 tail not only in vitro, but also in cells (8,15–18).
Figure 1.
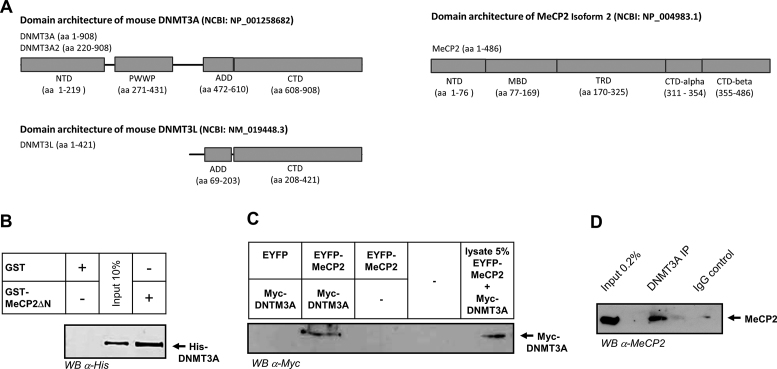
Structures of DNMT3 proteins and MeCP2 and interaction of MeCP2 with DNMT3A. (A) Domain architecture of mouse DNMT3A, DNMT3L and MeCP2 annotated with the domain boundaries used in this work. (B) Western blot detection of His-DNMT3A after its pull-down with the GST-tagged N-terminal truncated MeCP2 (MeCP2ΔN, residues 104–486). (C) Western blot detection of Myc-tagged DNMT3A after its pull-down by EYFP-tagged MeCP2 following transient co-expression of both proteins in human HEK293 cells. As a negative control, the pull-down was performed after co-expression of DNMT3A with EYFP. See also Supplementary Figure S10. (D) Western blot detection of endogenous MeCP2 after its pull-down by DNMT3A using mouse brain protein extracts. An immunoprecipitation using an IgG isotype control was included as negative control. NTD, N-terminal domain.
The biological role of DNA methylation is mediated by proteins, which specifically bind to DNA carrying methylated cytosine (19). One important reader of DNA methylation is the MeCP2 protein (Figure 1A) (20–23). It is the founding member of a group of proteins containing so called methyl-binding domains (MBDs), which bind to DNA in a methylation specific manner (24). MeCP2 recognizes methylated DNA with a preference for CpG dense islands (22,25), and its binding strength is strongly influenced by the flanking sequence of the methylated CpGs (26,27). Recently, binding of methylcytosine in non-CpG contexts was also reported (28,29). In addition to the MBD, MeCP2 contains a transcriptional repression domain (TRD), which serves as a protein recruitment platform and shows a weak methylation-independent DNA binding (23,30). MeCP2 is known to interact with various transcriptional repressors and co-repressors, including Histone Deacetylases, DNMT1, and the ATRX and Sin3A proteins (31–34). Functionally, MeCP2 is involved in numerous cellular processes, like methylation-induced gene repression (in particular of long genes), control of repetitive elements, chromatin compaction and looping and splice site regulation (23,29,30,35–38). In addition to its role in gene silencing, gene expression studies performed in specific brain sub-regions found altered expression levels of hundreds of genes after loss of MeCP2, most of which were upregulated by MeCP2 (39–41). These findings indicate that MeCP2 can function as a gene activator or repressor depending on the genomic context. Nevertheless, in spite of its high abundance in adult neurons, important role in chromatin organization and strong clinical relevance, the mechanistic details behind these opposing roles of MeCP2 are not fully understood (42).
Both DNMT3A and MeCP2 are highly expressed in neurons (35,43) (EBI expression atlas http://www.ebi.ac.uk/gxa/) and they have important functions in the brain. DNMT3A has been implicated in neuromuscular control, synaptic plasticity, learning and memory (44–46). MeCP2 functions as a structural protein and forms a specific type of chromatin, which is depleted of histone H1 (23). It plays an essential role in brain plasticity (35) and inactivating mutations of the X-linked MECP2 gene were shown to cause Rett syndrome, a severe neurodevelopmental disease associated with developmental disorders and autism-like symptoms in females (30,34).
Since functional crosstalks between several readers and writers of epigenetic modifications have been previously reported (47) and DNMT3A and MeCP2 were both known to be targeted to pericentromeric heterochromatin (48), we investigated their potential interaction in this work. We observed a strong binding of MeCP2 to DNMT3A in vitro, in cells, and in the mouse brain and mapped the interaction interface to the TRD domain of MeCP2 and the ADD domain of DNMT3A. We found that binding of MeCP2 strongly inhibits the activity of DNMT3A by stabilizing its autoinhibitory conformation. The inhibition of DNMT3A was relieved by binding of an H3 tail peptide unmodified at K4 to the ADD domain suggesting that MeCP2 controls DNMT3A localization and activity depending on the local chromatin context.
MATERIALS AND METHODS
Generation of DNMT3A2 mutants and MeCP2 domain constructs
The murine DNMT3A and MeCP2 proteins and protein domains were prepared as indicated in Figure 1A. The MeCP2 domains and mutant proteins were cloned in pGEX-6P2 vector using BamHI and XhoI cloning sites. All constructs were verified by DNA sequencing. Site-directed mutagenesis were carried out by rolling circle polymerase chain reaction (PCR) using a primer carrying point mutation (49). The presence of the mutations was confirmed by restriction marker analysis and by DNA sequencing.
Expression and purification of MeCP2 and DNMT3A proteins
The MeCP2 domains and mutant proteins were expressed in Escherichia coli BL-21 cells. Cells were cultivated in Lysogeny broth (LB) medium at 37°C while shaking until an OD (600 nm) of 0.6–0.7 was reached. Then, protein expression was induced by addition of 1 mM of isopropyl-β-d-thiogalactoside and the culture was incubated at 18°C shaking at 200 rpm overnight. The cells were harvested by centrifugation (15 min at 4600 rpm) and the pellet resuspended in sonication buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 500 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 10% glycerol) including protease inhibitor (Sigma). The cells were lysed by sonication and centrifuged at 18 000 rpm for 1 h to prepare a clear lysate, which was applied on a GST-sepharose column (GE Healthcare). After washing with sonication buffer, the protein was eluted with sonication buffer containing 50 mM reduced glutathione and dialyzed first against dialysis buffer I (20 mM HEPES (pH 7.5), 200 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) for 3 h, then against dialysis buffer II containing 60% glycerol overnight. The purified proteins were analyzed on 12% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel stained with colloidal Coomassie BB. The murine DNMT3A2 and DNMT3A–C proteins were expressed and purified as described previously (50–52). Since all DNMT3A structures were annotated with numbers for the human proteins, we use human numbering here. The residue numbers corresponding to human Q527, D528 and D531 in murine DNMT3A are Q523, D524 and D527, respectively. Examples of images of the purified proteins used in this work are shown in Supplementary Figures S2–S7.
GST pull-down experiments
For GST pull-down experiments, 20 μl of Glutathione-Sepharose 4B beads were washed with 200 μl of interaction buffer (25 mM Tris (pH 8.0), 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonident P-40 (NP-40), 200 μM Phenylmethanesulfonyl fluoride (PMSF)). The beads were incubated for 1 h at 4°C with 10–15 μg of the different GST-tagged proteins, washed three times with interaction buffer and incubated with His- or MBD-tagged proteins (15 μg) for 1 h at 4°C with shaking. Then, the beads were washed three times with wash buffer containing high salt (25 mM Tris (pH 8.0), 5 mM MgCl2, 300 mM KCl, 10% glycerol, 0.1% NP40, 200 μM PMSF). The interaction of DNMT3A–ADD and MeCP2 TRDs was also investigated using buffer containing up to 600 mM KCl. At last, the beads were resuspended in sodium dodecyl sulphate (SDS) gel loading buffer and incubated for 10 min at 95°C. After centrifugation of the beads at 14 000 rpm for 10 min, the supernatant was loaded on a 12% SDS-PAGE gel. Proteins were detected by western blotting or Coomassie BB staining as indicated. Some experiments were conducted in the presence of recombinant histone H3.1 (Cat. No. M2503S, New England Biolabs), as detailed in the ‘Results‘ section.
Co-immunoprecipitation assay
For co-immunoprecipitation of DNMT3A and MeCP2, pcDNA-DNMT3A (expressing myc tagged DNMT3A) and pEYFP-MeCP2 plasmids were co-transfected in HEK293 cells. The pEYFP plasmid was used as control. After 48 h, the cells were collected and the cell pellets stored at −80°C. The cells were lysed as recommended by the GFP trap protocol (ChromTek). Using GFP trap, YFP-tagged MeCP2 was pulled-down and the complex washed with buffer (10 mM Tris/Cl (pH 7.5), 0.5 mM EDTA, 0.5% NP-40, 200 mM NaCl). The MeCP2 and DNMT3A proteins were separated on a 12% SDS-PAGE and transferred to nitrocellulose membrane. To detect DNMT3A, the blot was probed with anti-myc antibody (Santa Cruz, 1:1000 dilution) for 1 h at room temperature.
For immunoprecipitation of endogenous DNMT3A and MeCP2, whole brains from 16-week-old C57Bl/N female mice were used. Following mechanical disruption, the tissue was lysed following a published protocol (53) with some modifications. Three brains were homogenized in NP-40 lysis buffer (10 mM HEPES (pH 7.9), 3 mM MgCl2, 10 mM KCl, 10 mM NaF, 1 mM Na3VO4, 0.5 mM DTT, 0.5% NP-40, 1× complete EDTA-free protease inhibitor cocktail (Roche)), by douncing 30× with a tight pestle, and pelleted at 1000 g. Lysates were next diluted 1:1 with Benzonase buffer (10 mM HEPES (pH 7.9), 3 mM MgCl2, 280 mM NaCl, 0.2 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 0.5 mM DTT, 0.5% NP-40) supplemented with 1× complete EDTA-free protease inhibitor cocktail (Roche) and sonicated with EpiShear (Active Motif) for 2 min 30 s (15 s ON, 30 s OFF cycles, 20% power, 3.2 mm microtip). The homogenate was then digested with 500 units of Benzonase (Novagen) for 2 h rotating at 4°C. Chromatin proteins were separated by centrifugation at 17 000 g for 20 min at 4°C. For each pull-down, 2.5 mg lysate were pre-cleared for 1 h at 4°C with 20 mg Protein A Sepharose CL-4B (GE Healthcare), followed by overnight incubation with 15 μg anti-DNMT3A antibody (sc-2070, Santa Cruz). For negative control, an equivalent amount of non-related rabbit IgG anti-myc (ab9106, Abcam) antibody was used. The antibody-bound proteins were immobilized to 100 mg Protein A Sepharose CL-4B, blocked in 10% bovine serum albumin (Roth) for 6 h rotating at 4°C. After five washes with immunoprecipitation buffer, the immune complexes were eluted from the beads by boiling in 100 μl Laemmli sample buffer. The samples were next analyzed by western blot as described above. For detection, anti-MeCP2 monoclonal primary antibody (#3456, Cell Signaling) was used, followed by incubation with HRP-linked anti-rabbit IgG light chain specific secondary antibody (211-032-171, Jackson ImmunoResearch). Western lighting Ultra (Perkin Elmer) was used as ECL HRP substrate.
Fluorescence microscopy
For sub-nuclear localization studies, NIH3T3 cells were seeded on glass slides and transfected with plasmids expressing CFP- and YFP-tagged DNMT3L, DNMT3A–ADD and MeCP2 using Fugene HD (Promega) according to the manufacturer’s instructions. After 48 h, the cells were fixed with 4% formaldehyde, mounted in Mowiol and Z stacks images were collected using a Zeiss LSM 710 confocal microscope. Fluorescence signals were collected in the YFP and CFP channels after confirming absence of crosstalk (Supplementary Figure S8).
Substrates used for DNA methylation
The following oligonucleotide substrates were used for DNA methylation assays: a biotinylated unmethylated 30 mer containing one CpG site (um30mer: TTG CAC TCT CCT CCC GGA AGT CCC AGC TTC / Bt-GAA GCT GGG ACT TCC GGG AGG AGA GTG CAA), the same sequence hemimethylated at the CpG site with the methylation in the lower DNA strand (hm30mer: TTG CAC TCT CCT CCC GGA AGT CCC AGC TTC / Bt-GAA GCT GGG ACT TCmC GGG AGG AGA GTG CAA), a biotinylated hemimethylated 30 mer with optimized flanks for methylation with DNMT3A (54) (hmF30mer: GAA GCT GGA CAG TAC GTC AAG AGA GTG CAA / Bt-TTG CAC TCT CTT GAmC GTA CTG TCC AGC TTC) and a non-CpG substrate (nonCpG: GAA GCT GGT CCA TTmC GAT GAT GGA GTG CAA / Bt-TTG CAC TCC ATC ATmC GAA TGG ACC AGC TTC). The oligonucleotides were annealed by heating to 86°C for 5 min and slowly cooling down to ambient temperature. In addition, a biotinylated 585-mer DNA substrate obtained by PCR was used that contains eight HpaII sites (CCGG) and 45 CpG sites (um585mer) (Supplementary Figure S9). The 585 mer was amplified from Lambda-phage DNA by PCR using the following primers: Bt-GAA GGA CAA CCT GAA GTC CAG GTTG and GTG TAT GAC CAC CAG AGC CTT TTGC and purified by PCR purification kits (Qiagen). To prepare partially methylated 585 mer (pm585mer), the DNA was methylated with M.HpaII (NEB) following the protocol of the provider and afterward purified by PCR purification kits. Successful pre-methylation at HpaII sites was confirmed by HpaII (NEB) restriction digestion (Supplementary Figure S9).
DNA methylation activity assay
The avidin–biotin microplate DNMT activity assay was used to monitor the activity of different DNMT3A variants in the methylation of biotinylated DNA substrates, basically as described (55,56). Each well of the microplate was coated with 1 μg of avidin dissolved in 100 μl of 100 mM NaHCO3 (pH 9.6) and incubated overnight at 4°C. Before starting the assay, the wells were washed five times with 200 μl of 1× PBST (140 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM K2HPO4, 0.05% v/v Tween 50, pH 7.2) containing 0.5 M NaCl. The reaction mixtures were prepared containing 2.5 μM DNMT3A2 or DNMT3A–C and 3 μM MeCP2 (or any of its domains) in methylation buffer (20 mM HEPES (pH 7.2), 1 mM EDTA, 50 mM KCl, 1.25 mg/ml bovine serum albumin). For the control reactions without MeCP2, the same volume of dialysis buffer II was added instead of the MeCP2. The reaction mixtures were incubated on ice for 20 min and the wells of the plate were filled with 5 μl of 0.5 M unlabeled AdoMet (Sigma) dispensed in 35 μl 1× PBST/0.5 M NaCl. Then, 1 μM 30-mer oligonucleotide DNA or 100 nM of 585-mer DNA and 0.76 μM [methyl-3H]-AdoMet (PerkinElmer Life Sciences) were added to the reaction mixture and the samples were incubated at 37°C. In order to follow the time course of the reaction, aliquots of 2 μl were removed from the reaction mixtures in duplicates at time points between 2 and 30 min and applied to one well of the microplate where the incorporation of labeled AdoMet was quenched by an excess of unlabeled AdoMet. This mixture was incubated while slightly shaking for 30 min to 1 h. The wells were washed five times with 200 μl of 1× PBST and 0.5 M NaCl. A buffer (100 μl; 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2) containing 0.7 μg unspecific nuclease from Serratia marcescens was added per well and the mixture was incubated for 30–60 min with slight shaking. At last, the released radioactivity was measured using liquid scintillation counting and the average count per minute of the duplicates was plotted against time. Linear regression was used to obtain the slopes of the initial linear parts of the time courses. The data are reported as averages and standard error of the mean (SEM) of at least two independent experiments.
DNA methylation analysis in HCT116 DNMT1 hyphomorph cells
To study the effect of MeCP2 on DNMT3A-mediated methylation in cells, we used HCT116 DNMT1 hyphomorphic cells (HCT116D1hypo, kindly provided by Prof. Bert Vogelstein, HHMI, USA), which have a reduced level of global DNA methylation (57,58). HCT116D1hypo cells were cultivated in McCoy’s 5A medium (Gibco catalog no.: 16 600) supplemented with 10% heat-inactivated calf serum, 2 mM l-glutamine (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a saturated humidity atmosphere containing 5% CO2. HCT116D1hypo cells were modified to express the ecotropic receptor and rtTA3 using retroviral transduction of pWPXLd-RIEP (pWPXLd-rtTA3-IRES-EcoR-PGK-Puro) followed by drug selection (0.8 ug/ml puromycin for 1 week, respectively) similarly as described (59). The resulting cell line was subsequently transduced with ecotropically packaged retroviruses containing the mecp2 gene fused to EYFP under control of a TRE3G promoter. Retroviral gene transfer was performed as previously described (60) using 10–20 μg plasmid DNA and 5 μg helper plasmid (pCMV-Gag-Pol, Cell Biolabs) for each calcium phosphate transfection. Retroviral packaging was performed using PlatiniumE cells (Cell Biolabs). Transduction efficiencies of retroviral constructs (TRE3G-MeCP2-EYFP-PGK-NEO and TRE3G-EYFP-PGK-NEO) (61) were measured 48 h post induction with 1 μg/ml doxycycline by flow cytometry (MACSQuant® VYB, Miltenyi Biotec GmbH, Germany). Transduced cell populations were selected 5 days post infection using 500 μg/ml G418 (Gibco Life technologies). After 14 days of induction, ∼1 million MeCP2 and the EYFP control expressing cells (as judged by being EYFP+) were sorted for each replicate by flow cytometry (FACSAria III, BD, USA). Genomic DNA was isolated using the DNA mini Kit (Qiagen) and followed by LC-ESI-MS/MS analysis of DNA methylation as described (62,63).
Quantitative RT-PCR
Quantitative RT-PCR assays were performed on a CFX96 Connect Real-Time detection system (Bio-Rad, Hercules, CA, USA) using SsoFast EvaGreen supermix (Bio-Rad, Hercules, CA, USA). For gene expression analysis, total RNA from 106 cells was isolated for each sample using RNeasy Mini Kit (Qiagen, Limburg, The Netherlands). Complementary DNA (cDNA) was prepared using MultiScribe™ Reverse Transcriptase (Applied Biosystems, Thermo Fisher, USA) with oligo d(T)18 primers (New England Biolabs, Ipswich, MA, USA) using 500 ng of RNA. After this, quantitative PCR (qPCR) was carried out using specific primers-sets for DNMT3A (CGA TTT CTC GAG TCC AAC CCT G, ACC GGG AAG GTT ACC CCA), DNMT3B (CAG TGA CAC GGG GCT TGA ATA TG, CTT TGA GGA CTA GGT AGC CTG TCG CG) and DNMT1 (GAG ACA CGA TGT CCG ACC TG, CCA ATG CAC TCA TGT CCT TAC AG), normalized to the housekeeping gene SDHA (TGG GAA CAA GAG GGC ATC TG, CCA CCA CTG CAT CAA ATT CAT). The relative DNMT expression was calculated using ΔCt (Expression = 2−ΔCt). Non-RT controls that did not undergo the cDNA synthesis step were included in all experiments. Error analysis was based on independent cDNA preparations.
RESULTS
MeCP2 interacts with DNMT3A
To test whether DNMT3A and MeCP2 interact, we first performed GST-pull-down assays using recombinant murine full-length (FL) proteins. MeCP2 could be obtained with good purity after generating a version lacking the N-terminal unstructured domain (MeCP2ΔN) (Supplementary Figure S2). As shown in Figure 1B, a robust pull-down of DNMT3A by GST-MeCP2ΔN could be detected. Encouraged by the strong and direct interaction detected in the in vitro assay, we next tested whether DNMT3A and MeCP2 can also interact in cells. For this, HEK293 cells were transiently co-transfected with Myc-tagged DNMT3A and EYFP-tagged MeCP2 (Supplementary Figure S10). To isolate immunocomplexes, the EYFP-tagged MeCP2 was immunoprecipitated and the pull-down material was tested for the presence of Myc-DNMT3A. As shown in Figure 1C, co-purified Myc-DNMT3A was detected after co-expression with EYFP-MeCP2, but not with the EYFP control. To exclude potential artifacts related to protein overexpression, we finally performed immunoprecipitation of endogenous DNMT3A from mouse brain, a tissue where both MeCP2 and DNMT3A are abundantly expressed. As shown in Figure 1D, MeCP2 could be specifically detected in the pulled-down material, but not in the IgG control. Together, these results demonstrate that in addition to their direct in vitro interaction DNMT3A and MeCP2 also associate after transient co-expression in mammalian cells as well as at endogenous levels in brain extracts.
MeCP2 interacts with the ADD of DNMT3 proteins
Having established that the interaction between DNMT3A and MeCP2 is not an artifact of the in vitro pull-down assay, but is also detected in biological relevant context, we were next interested to pinpoint the interaction interfaces between these two proteins. For this, we have undertaken a systematic domain mapping approach by performing in vitro pull-down experiments with serially truncated recombinant DNMT3A and MeCP2.
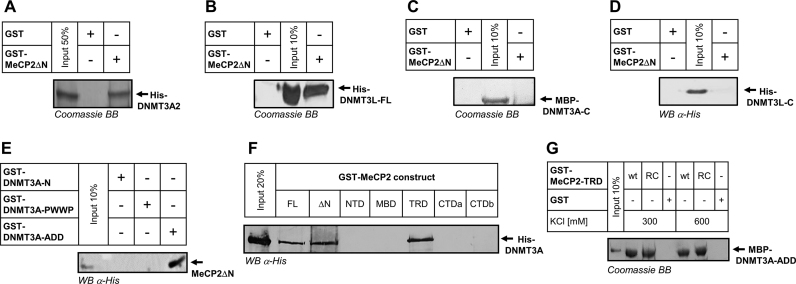
To define the interaction interface on DNMT3A, we first performed GST pull-downs using MeCP2ΔN-GST and DNMT3A2, a naturally occurring isoform of DNMT3A, which lacks 219 N-terminal amino acids reported to be involved in DNA binding (Figure 1A) (64,65). As documented in Figure 2A, a robust pull-down was detected with DNMT3A2 indicating that residues 1–219 of DNMT3A are not required for the interaction with MeCP2. To further define the interacting area, we next resorted to DNMT3L, an important regulatory member of the DNMT3 family, which shares the ADD domain with DNMT3A and DNMT3B, while containing a crippled CD and lacking the N-terminal part and the PWWP domains (Figure 1A). The direct interaction detected between DNMT3L and MeCP2ΔN (Figure 2B) suggested that the association with MeCP2 occurs through an interface that is shared between DNMT3L and DNMT3A. To test this hypothesis, we next performed pull-downs with the CDs of DNMT3A (Figure 2C) and DNMT3L (Figure 2D), respectively. Both of these did not show an interaction with MeCP2ΔN. By contrast, a robust interaction could be detected between the GST-tagged ADD domain of DNMT3A as bait and a GST-cleaved MeCP2ΔN as prey (Figure 2E). No signal was detectable for the NTD and the PWWP domains of DNMT3A in line with the results obtained for DNMT3L and DNMT3A2. Taken together, these results show that MeCP2 interacts with DNMT3 proteins via their ADD, a domain that has been already reported to serve as a protein–protein interaction platform and be essential for the regulation of DNMT3 proteins.
Figure 2.
The ADD domain of DNMT3 proteins interacts with the TRD of MeCP2. (A) Coomassie staining of the pull-down of His-DNMT3A2 by GST-MeCP2ΔN. (B) Coomassie staining of the pull-down of His-DNMT3L by GST-MeCP2ΔN. (C) Coomassie staining of the pull-down of MBP-DNMT3A–C by GST-MeCP2ΔN. A signal can be only detected in the input lane. See also Supplementary Figure S15. (D) Western blot detection of the pull-down of His-DNMT3L-C by GST-MeCP2ΔN. A signal is visible only in the input lane. (E) Western blot detection of the pull-down of MeCP2ΔN with different GST-tagged DNMT3A domains. For this experiment, GST-cleaved MeCP2ΔN was used. The endogenous His-tag in MeCP2-CTDb was used for detection. Out of the three tested DNMT3A domains, only the ADD domain displayed an interaction with MeCP2. (F) Western blot detection of the pull-down of DNMT3A by different GST-tagged MeCP2 domains documenting an interaction with the MeCP2 FL, MeCP2ΔN and MeCP2–TRD domain. (G) Coomassie detection of the pull-down of MBP–DNMT3A–ADD by wild-type GST-TRD domain (wt) or TRD containing the R306C Rett mutation (RC), under two different salt concentrations. See also Supplementary Figures S11–S14.
DNMT3A interacts with the TRD of MeCP2
Having successfully mapped the interaction interface on the DNMT3A side, we were next interested to dissect which part of MeCP2 is responsible for the binding to DNMT3A. To this end, we performed pull-downs with GST-tagged MeCP2 domains. As shown in Figure 2F, out of the five tested domains an interaction was detectable only for the TRD. Pull-downs using the isolated MeCP2–TRD and DNMT3A–ADD (Figure 2G) confirmed the direct and self-sufficient interaction between these domains. The interaction of MeCP2–TRD and DNMT3A–ADD was further confirmed by Alpha-assay (Supplementary Figure S11) and gel filtration (Supplementary Figure S12). Having identified the domains responsible for mediating the interaction between these two proteins, we next tested the stability and strength of the association between MeCP2–TRD and DNMT3A–ADD by performing pull-downs under increasingly high salt concentrations. Strikingly, we could retrieve comparable amounts of DNMT3A–ADD at both 300 and 600 mM KCl, indicating that its interaction with MeCP2 TRD is strong and not driven by electrostatic interactions (Figure 2G). A salt resistant and strong interaction of MeCP2–TRD was also observed with DNMT3A2 (Supplementary Figure S13). Pull-down experiments in the presence of the non-specific and highly active nuclease from S. marcescens demonstrated that the interaction of both proteins was not mediated by nucleic acids (Supplementary Figure S14). Since MeCP2 is mutated in the Rett syndrome, we also tested the effect of the R306C Rett mutation in the TRD domain of MeCP2 on the interaction with DNMT3A, but did not observe any change when compared with wild-type TRD (Figure 2G).
MeCP2 influences the sub-nuclear localization of DNMT3L and the DNMT3A–ADD domain
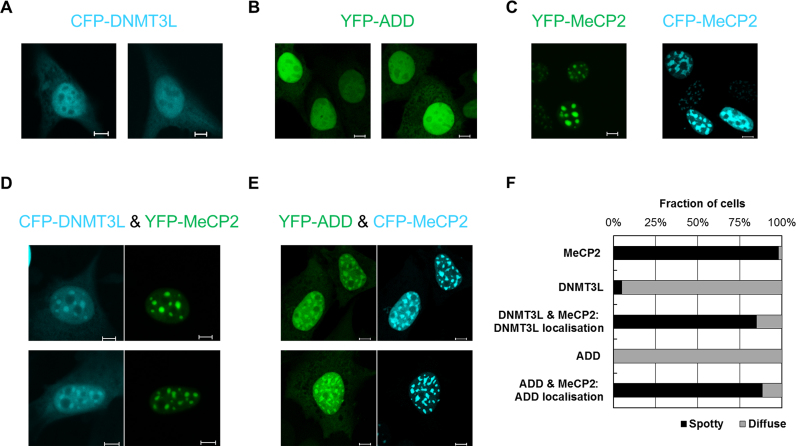
Having shown the direct interaction of MeCP2 with DNMT3 proteins, we next aimed to study the influence of this interaction on the cellular localization of these factors. MeCP2 is known to accumulate at pericentromeric heterochromatin (22,48,53), which clusters in characteristic DAPI-dense foci in mouse fibroblasts. Unlike MeCP2, the regulatory factor DNMT3L was shown to have an almost homogenous nuclear distribution (66). Expressing the fluorophore-tagged DNMT3L and MeCP2 in mouse fibroblasts confirmed the published localization patterns of both proteins (Figure 3A and C). Notably, co-expressing DNMT3L with MeCP2 led to a clear re-targeting of DNMT3L toward chromocenters (Figure 3D and F). This finding confirms the intracellular interaction of DNMT3L and MeCP2 and indicates that the expression of MeCP2 causes a re-distribution of the regulatory factor DNMT3L toward heterochromatin. Moreover, it shows that the MeCP2 interaction influences chromatin targeting of DNMT3L. However, the physiological role of the interaction of DNMT3L and MeCP2 is unclear, because DNMT3L does not appear to play a major role in the brain.
Figure 3.
Transient expression of MeCP2 alters the cellular localization of DNMT3L and DNMT3A–ADD in NIH3T3 cells. (A) Representative fluorescence microscopy images documenting the localization of CFP-DNMT3L upon its overexpression in mammalian cells. A predominantly homogenous nuclear distribution is observed. (B) Representative fluorescence microscopy images documenting the localization of YPF-ADD upon its overexpression in mammalian cells. A predominantly homogenous nuclear distribution is observed. (C) Representative fluorescence microscopy images documenting the chromocenter-enriched localization of YFP-MeCP2 (left) and CFP-MeCP2 (right) upon their overexpression in mammalian cells. (D) Representative fluorescence microscopy images showing that upon their co-expression DNMT3L is recruited to MeCP2 clusters. (E) Representative fluorescence microscopy images showing that upon their co-expression DNMT3A–ADD is recruited to MeCP2 clusters. The scale bars correspond to 10 μm. (F) Quantification of the fraction of cells showing spotty and diffuse localization patterns in the experiments shown in panels (A–E) (based on analysis of >20 individual cells in each case). See also Supplementary Figure S8.
Unlike DNMT3L, DNMT3A is strongly enriched at pericentromeric heterochromatin (52,66) (and references therein), where it co-localizes with MeCP2 (48). This natural co-localization precluded direct studies of the effect of MeCP2 on the localization of DNMT3A. To circumvent this caveat, we performed localization studies with the fluorescently tagged DNMT3A–ADD domain. This showed a diffuse nuclear localization when transfected alone in mouse fibroblasts (Figure 3B). Similar to DNMT3L, after its co-expression with CFP-tagged MeCP2, the ADD domain showed a preferential enrichment at heterochromatic foci, indicating a MeCP2-mediated targeting to heterochromatin (Figure 3E and F). These results confirm that the ADD domain interacts with MeCP2 in cells and demonstrate that through its binding MeCP2 recruits DNMT3L and DNMT3A–ADD to pericentromeric heterochromatin.
The interaction with MeCP2 inhibits the catalytic activity of DNMT3A
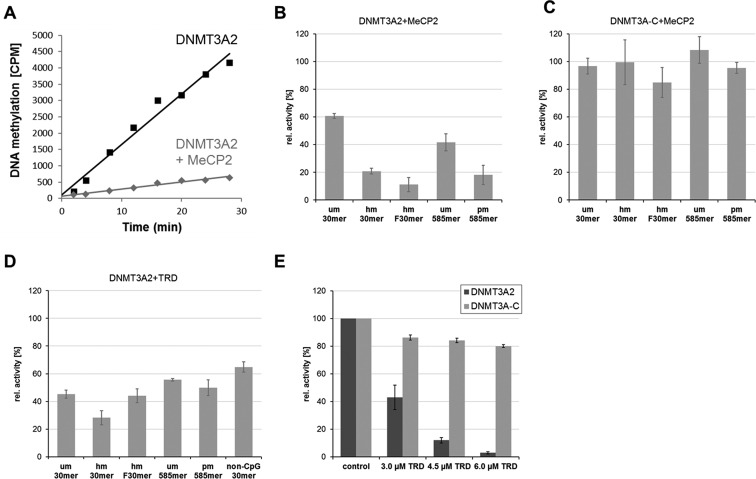
To elucidate the function of the interaction between MeCP2 and DNMT3A, we next measured the in vitro rates of DNA methylation by DNMT3A2 in the presence of MeCP2 or its TRD domain. The activity of DNMT3A has been shown to be modulated by the target site, where CpG is preferred over CpA (67–69), the flanking sequence of CpG sites (70,71) and the length of the DNA substrates (72). To study all these different properties, six different DNA substrates were used for our DNA methylation experiments (Figure 4): (i) an unmethylated 30-mer oligonucleotide (um30mer), (ii) the same substrate in hemimethylated form (hm30mer), (iii) a hemimethylated 30 mer with an optimized flank for DNMT3A (hmF30mer) (54), (iv) a 585-mer PCR fragment (um585mer), (v) the 585-mer PCR fragment pre-methylated at HpaII sites (pm585mer) and (vi) a 30-mer oligonucleotide non-CpG substrate (non-CpG 30 mer).
Figure 4.
MeCP2 (A–C) and MeCP2–TRD (D and E) inhibit the activity of DNMT3A2 on a broad range of substrates. In vitro DNA methylation kinetics was conducted with 2.5 μM DNMT3A2 (A and B) or DNMT3A–C (C) in the presence of 3 μM MeCP2 (A–C) or MeCP2–TRD (D). In each panel, identical control reactions without addition of MeCP2 or MeCP2–TRD were used to calculate relative activities. Different DNA substrates were used as indicated. (E) Inhibition of DNMT3A2 at increasing concentrations of MeCP2–TRD in kinetics using um30mer as substrate. Reactions with DNMT3A–C were performed in parallel since this domain does not interact with MeCP2–TRD. Control refers to reactions without added TRD. Panel (A) shows methylation kinetics of hmF30mer as an exemplary primary data set. Bars show averages and SEM based on 2–3 independent experiments. See also Supplementary Figures S6, S13 and S14.
Using 2.5 μM DNMT3A2 and 3 μM MeCP2, we consistently observed that the interaction of MeCP2 with DNMT3A2 resulted in ∼40–60% reduction in DNMT3A2 activity with the unmethylated substrates (Figure 4A and B). Similar results were obtained with a truncated form of DNMT3B corresponding to DNMT3A2 (Supplementary Figure S6). The activity of DNMT3A2 was further reduced by ∼80% with methylated substrates, which can be attributed to the better binding of MeCP2 to methylated DNA via its MBD. We speculated that binding of MeCP2 to pre-methylated DNA might target DNMT3A2. To test this hypothesis, a partially methylated 585-mer DNA substrate was prepared by methylation with HpaII, an enzyme that exclusively methylates CG sites found within CCGG motifs (Supplementary Figure S9). However, even on this substrate, we observed inhibition of the activity of DNMT3A2 by MeCP2 (Figure 4B, pm585mer). As a control, we used DNMT3A–C, which lacks the ADD domain and does not interact with the MeCP2–TRD (Supplementary Figure S15), and observed that MeCP2 did not inhibit its activity (Figure 4C).
Having mapped the interaction interface to the TRD of MeCP2, we next tested whether this isolated domain can also inhibit the activity of DNMT3A2 and observed 40–70% inhibition with the different substrates (Figure 4D). Non-CpG methylation has recently been detected in considerable amounts in human ES cells and neurons, and it was connected to DNMT3A activity (28,73–75). We, therefore, also investigated the influence of MeCP2–TRD on the non-CpG methylation activity of DNMT3A2 using a 30-mer oligonucleotide substrate that contains one already methylated CpG site, such that additional methylation could only occur at non-CpG sites. As with the other DNA substrates, we observed a similar inhibition of DNMT3A2 activity by TRD (Figure 4D, non-CpG 30 mer), indicating that CpG and non-CpG methylation are equally inhibited by the TRD interaction.
At last, we tested the activity of DNMT3A2 in the presence of increasing amounts of TRD and observed that the methyltransferase activity was strongly inhibited (>95%) using a 2.4-fold excess of TRD (6 μM with 2.5 μM DNMT3A2) (Figure 4E). These results were fitted by a binding constant (KD) of TRD to DNMT3A2 of 2.8 μM under catalytic conditions. As a control, the same experiments were conducted with DNMT3A–C, but only a very weak reduction of activity was observed (Figure 4E) indicating that the inhibition of DNMT3A2 by TRD is not caused by competition for the DNA substrate. This is an important control, since TRD was reported to weakly bind DNA (76,77). In summary, our results indicate that the interaction between the TRD domain of MeCP2 and the ADD domains of DNMT3A and DNMT3B results in a direct and very strong inhibition of the DNMT activity at both CpG and non-CpG sites.
MeCP2 overexpression reduces DNA methylation in HCT116 cells
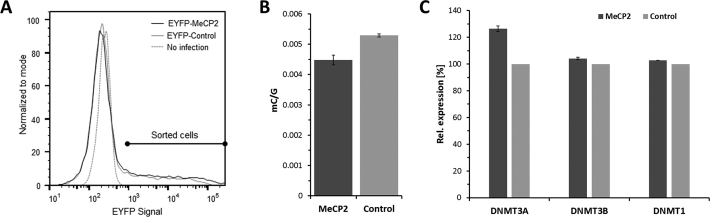
Based on the strong influence of MeCP2 on the activity of DNMT3 proteins, we were next interested to see if the inhibitory effects on DNMT3A activity observed in in vitro assays are also re-capitulated in a cellular context. For this we resorted to the HCT116 DNMT1 hypomorphic colon cancer cell line, which contains a truncated DNMT1 with reduced activity, but active copies of DNMT3A and DNMT3B (57,58). Because of the impaired maintenance DNA methylation activity, these cells have an ∼20% reduced amount of DNA methylation, which is more dependent on the activity of DNMT3A and DNMT3B. This makes the HCT116D1hypo cell line a suitable model system to study the effect of the inhibition of DNMT3A by MeCP2. We genomically integrated EYFP-fused MeCP2 or only the fluorophore as control by viral transduction and selected for stably expressing clones. Expression of MeCP2 or fluorophore control was induced for 14 days. Afterward, EYFP-MeCP2 or fluorophore expressing clones were enriched by fluorescence-activated cell sorting (Figure 5A). Genomic DNA was isolated and the global levels of 5-methylcytosine were quantified by liquid chromatography-mass spectrometry (LC-MS/MS). As shown in Figure 5B, we observed a 15% decrease in global DNA methylation. Expression levels of all DNMTs were determined by qPCR indicating a slight increase in DNMT3A expression, and no changes in DNMT1 and DNMT3B (Figure 5C), showing that the reduction in DNA methylation was not caused by reduced expression of any of the DNMTs. Since DNMT1 interaction with MeCP2 was reported not to cause a reduction in catalytic activity (32), this result indicates that overexpression of MeCP2 reduces the activity of DNMT3 enzymes in cells.
Figure 5.
MeCP2 reduces global DNA methylation in HCT116 cells containing a DNMT1 hypomorphic allele. (A) Flow cytometry analysis of EYFP-MeCP2 and EYFP expression in HCT116D1hypo cells. The signal range used for sorting is indicated. (B) Global DNA methylation levels after overexpression of EYFP-MeCP2 in human HCT116D1hypo cells. EYFP-transfected cells were used as control. (C) Expression levels of all DNMTs in the HCT116D1hypo cells expressing EYFP-MeCP2 or EYFP control. SDHA was used as reference gene. The error bars represent the SD based on two repeats.
The TRD domain inhibits DNMT3A2 activity by an allosteric mechanism
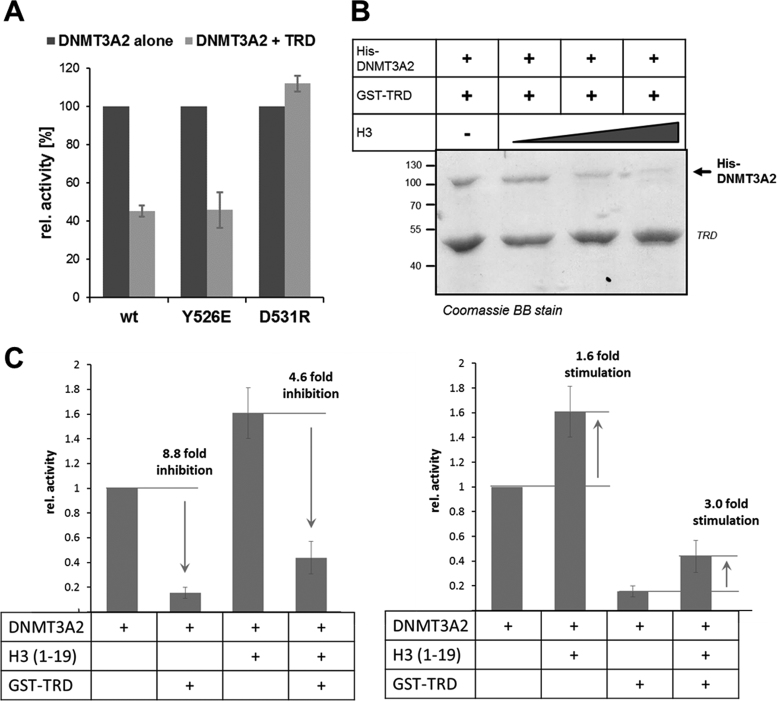
We next aimed to mechanistically analyze the striking inhibitory effect of MeCP2 on DNMT3A. As described above, structural studies showed that the ADD domain of DNMT3A can dock on the CD at two alternative sites: an allosteric and an autoinhibitory one (13). Binding of the H3 tail peptide to the ADD domain was shown to stabilize the allosteric conformation and thereby activate DNMT3A (13,14). To investigate the mechanism of the repression of DNMT3A by the TRD domain, we engineered DNMT3A2 variants containing mutations at Y526 or D531 in the ADD domain, two critical residues involved in the two binding sites at the CD (Supplementary Figure S1) in order to selectively disrupt or strongly destabilize one of the two DNMT3A conformations. Y526E was introduced to disrupt the allosteric and D531R to disrupt the autoinhibitory conformation. After confirming that both mutants still interact with MeCP2–TRD (Supplementary Figure S16), we investigated if these conformationally locked DNMT3A variants still respond to the presence of the TRD. As shown in Figure 6A, the inhibitory effect of MeCP2 was specifically lost in the D531R variant that can no longer adopt the autoinhibitory conformation. This finding suggests that MeCP2 reduces the activity of DNMT3A by an allosteric mechanism, in which TRD binding stabilizes the autoinhibitory conformation of the enzyme.
Figure 6.
Mechanism of the inhibition of DNMT3A by the MeCP2–TRD. (A) Relative in vitro activity of DNMT3A2 wild-type and its conformational variants in the absence (dark gray) or presence (light gray) of MeCP2–TRD. The inhibition by the TRD is lost in the DNMT3A2 D531R variant, which carries a mutation that disrupts the autoinhibitory conformation. Error bars indicate the SEM based on three independent experiments. (B) Coomassie staining of the pull-down of His-DNMT3A2 (0.25 μM) by GST-TRD in the presence of increasing concentrations of recombinant histone H3 (0, 0.26, 1.3 and 3.9 μM) indicating that the H3 binding to DNMT3A2 interferes with the TRD interaction. (C) DNA methylation activity of DNMT3A2 (1 μM) in the absence or presence of the H3 peptide (amino acid sequence 1–19, 5 μM) or TRD (1.2 μM). The two panels show the same data in different representation. Error bars indicate the SEM based on three independent experiments. See also Supplementary Figures S16 and S17.
Since through its binding to the ADD domain, unmodified histone H3 was reported to allosterically activate DNMT3A, we next investigated if TRD and histone H3 binding to the ADD domain influence each other. For this, we conducted pull-down experiments using GST-TRD and DNMT3A2 in the presence of increasing concentrations of recombinant histone H3. As shown in Figure 6B, addition of histone H3 abolished the ADD–TRD interaction, suggesting that H3 and TRD binding to the ADD domain is mutually exclusive. To investigate whether histone H3 can rescue the TRD-mediated inhibition, we next conducted DNA methylation experiments with DNMT3A2 and DNMT3A2 pre-incubated with the unmodified H3 (1–19) peptide in the absence and presence of GST-TRD (Figure 6C). In line with the previous experiments, we observed that the TRD-mediated inhibition is alleviated in the presence of H3 peptide (5 μM). Correspondingly, the activation of DNMT3A2–TRD complexes (which are predominantly in the autoinhibitory conformation) is stronger than the activation of free DNMT3A2 (which is in a mixed conformation state). Moreover, we observed that the inhibitory effect of the TRD domain on DNMT3A activity was completely lost at higher concentrations of the H3 peptide (25 μM, Supplementary Figure S17). This indicates that the binding of H3 to the ADD domain can disrupt the DNMT3A–TRD interaction and relieve the associated enzymatic inhibition.
DISCUSSION
During the past decade compelling experimental evidence has accumulated, indicating that DNA methylation patterns are highly dynamic and result from ongoing de novo methylation and demethylation events (3). This dynamic landscape plays particularly important roles in non-dividing cells, such as terminally differentiated neurons (78–80). In the absence of cell division and DNA replication, the DNA methylation profiles in these cells can only be controlled through a tight regulation of the targeting and activity of DNA methylating and demethylating enzymes. However, despite their importance, the details of this regulatory network have remained mysterious so far. In this work, we took a closer look at the DNMT3 methyltransferases, factors that play essential roles in mammalian development and disease (1,2,81), their targeting and allosteric regulation (6). We find that recombinant DNMT3A and DNMT3L proteins directly and strongly interact with the chromatin regulator MeCP2 in vitro. We confirmed this interaction under overexpression conditions in mammalian cells, as well as at endogenous expression levels in mouse brain lysates. By performing systematic domain mapping, we find that the interaction of MeCP2 and DNMT3 proteins is mediated by their TRD and the ADD domains, respectively. Based on the fact that MeCP2 is highly expressed in neurons and it has important functions in this cell type, the newly discovered DNMT3A–MeCP2 interaction likely plays an important role in controlling DNA methylation patterns in the brain.
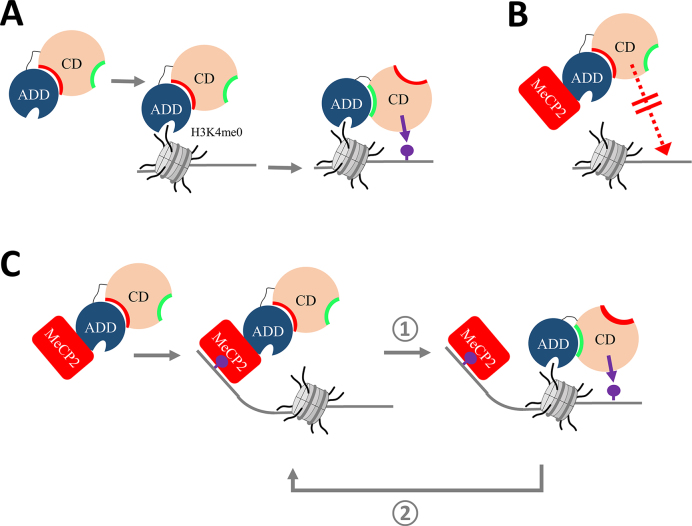
By employing in vitro methyltransferase assays using recombinant proteins and a variety of DNA substrates, we observed an almost complete, concentration-dependent inhibitory effect caused by MeCP2 binding to DNMT3A. Inhibition of DNMT3A was observed on both CpG and non-CpG substrates. Furthermore, DNMT3B activity was comparably reduced, proposing a conserved mode of action. To our knowledge, MeCP2 is the first interactor of DNMT3 proteins shown to have a direct inhibitory effect on the enzymatic activity of these proteins. As MeCP2 is an important reader of 5mC and 5hmC, this interaction might be required for mediating the crosstalk between 5mC/hmC sites and DNMT3 proteins and for preventing ectopic de novo methylation. By using engineered conformationally locked DNMT3A variants as a novel tool to investigate DNMT3A regulation, we show that the inhibition of DNMT3A by MeCP2 occurs by an allosteric mechanism, in which binding of MeCP2 stabilizes the autoinhibitory conformation of DNMT3A. Interestingly, binding of the unmodified H3 N-terminal tail peptide to the ADD domain of DNMT3A was shown to have the opposite effect, by precluding the autoinhibitory conformation and leading to the activation of DNMT3A (Figure 7A) (13,14). We mechanistically addressed this crosstalk and show that binding of H3 and TRD to DNMT3A are mutually exclusive and the MeCP2-mediated inhibition of DNMT3A2 can be overcome by addition of the unmodified H3 tail peptide.
Figure 7.
Model of the dual role of MeCP2 in the regulation and targeting of DNMT3A. (A) Binding of unmodified H3K4 to the DNMT3A–ADD domain triggers a conformational change where the ADD moves from the autoinhibitory (red) to the allosteric (green) interaction site and DNA methylation can take place (purple lollipop). Model based on (13). (B) Inhibitory role of MeCP2 on DNMT3A. Binding of MeCP2 (red) to DNMT3A–ADD inhibits the methyltransferase by stabilizing the autoinhibitory conformation, thereby preventing untargeted activity. (C) Role of MeCP2 in targeting of DNA methylation. At genomic sites with unmodified H3K4, H3 binding to the ADD disrupts the interaction between MeCP2 and DNMT3A leading to the activation of the enzyme and DNA methylation (step  ). MeCP2 can next bind to the methylated CpG sites and recruit additional DNMT3A, thereby initiating a positive feedback loop (step
). MeCP2 can next bind to the methylated CpG sites and recruit additional DNMT3A, thereby initiating a positive feedback loop (step  ).
).
In summary, our data unravel one part of the intricate regulatory network, which controls DNA methylation by suggesting a model in which DNMT3A is under the combined control of MeCP2 and the modification state of histone H3 tails. On the one hand, the interaction with MeCP2 globally inhibits DNMT3A activity after overexpression of MeCP2 in tissue culture (Figure 7B). This may act as a safeguard mechanism to protect the genome from aberrant DNA methylation. On the other hand, at specific target sites such as repetitive sequences, where histone H3 lacks activating marks, MeCP2 can function as a recruiter of DNMT3 enzymes (Figure 7C). As shown by our biochemical data, unmodified histone H3 can disrupt the MeCP2–DNMT3A interaction, subsequently leading to the relief of the allosteric inhibition. Therefore, the specific delivery of DNMT3A to such regions by MeCP2 as visualized in the cellular localization experiments can target DNA methylation. Afterward, the elevated DNA methylation may increase the methylcytosine-dependent MeCP2 recruitment to these loci, initiating a positive feedback loop, which can contribute to the stable maintenance of methylation at these sites. In neurons, this process may be further supported by the non-CpG (mainly CpA) methylation introduced by DNMT3A, which is bound by MeCP2 as well. The opposing effect of MeCP2 on DNMT3A as potential inhibitor and stimulator depending on the genomic context agrees well with the dual role of MeCP2 in gene control, either as gene repressor or activator (23,30,39,42).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Sylke Lutz and Roland Kontermann (Institute of Cell Biology and Immunology, University Stuttgart) for providing the animal brain materials and to Dieter Wolf (Department of Biochemistry, University Stuttgart) for providing the anti-rabbit light chain specific antibody. We also thank Ingo Amm and Nicole Berner (Department of Biochemistry, University Stuttgart) for technical advice on the endogenous co-immunoprecipitation protocol and Benjamin Hackner (Department of Chemistry and Pharmacy, Ludwig-Maximilians-Universität München) for help with the LC-MS measurements. We are very grateful to the Central Facility for Advanced Microscopy of the Stuttgart Research Center Systems Biology at the University of Stuttgart, for providing access to the laser scanning microscope.
Author contributions: A.J., A.R., C.L. and R.Z.J. devised the project and analyzed the data. C.L. and A.R. conducted the biochemical assays with contributions from I.H., M.D., A.B. and M.E. C.L. and A.R. performed the fluorescence microscopy experiments. C.L. and M.E. performed the cell culture and biochemical work for the data shown in Figure 5 with contribution from P.R. and J.B. S.S., E.P. and T.C. performed the LC-ESI-MS/MS. All authors contributed to data interpretation and discussion, read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [Je 252/10 to A.J.]; Carl Zeiss Foundation (to R.Z.J.). Funding for open access charge: University funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jurkowska R.Z., Jurkowski T.P., Jeltsch A.. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011; 12:206–222. [DOI] [PubMed] [Google Scholar]
- 2. Bergman Y., Cedar H.. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013; 20:274–281. [DOI] [PubMed] [Google Scholar]
- 3. Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. [DOI] [PubMed] [Google Scholar]
- 4. Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002; 3:274–293. [DOI] [PubMed] [Google Scholar]
- 5. Cheng X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995; 24:293–318. [DOI] [PubMed] [Google Scholar]
- 6. Jeltsch A., Jurkowska R.Z.. Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Res. 2016; 44:8556–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R.Z., Ragozin S., Jeltsch A.. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010; 285:26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schubeler D.. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015; 520:243–247. [DOI] [PubMed] [Google Scholar]
- 9. Rondelet G., Dal Maso T., Willems L., Wouters J.. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 2016; 194:357–367. [DOI] [PubMed] [Google Scholar]
- 10. Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007; 448:714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M.. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009; 10:1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W. et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010; 38:4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo X., Wang L., Li J., Ding Z., Xiao J., Yin X., He S., Shi P., Dong L., Li G. et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015; 517:640–644. [DOI] [PubMed] [Google Scholar]
- 14. Li B.Z., Huang Z., Cui Q.Y., Song X.H., Du L., Jeltsch A., Chen P., Li G., Li E., Xu G.L.. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011; 21:1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morselli M., Pastor W.A., Montanini B., Nee K., Ferrari R., Fu K., Bonora G., Rubbi L., Clark A.T., Ottonello S. et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife. 2015; 4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noh K.M., Wang H., Kim H.R., Wenderski W., Fang F., Li C.H., Dewell S., Hughes S.H., Melnick A.M., Patel D.J. et al. Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Mol. Cell. 2015; 59:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart K.R., Veselovska L., Kim J., Huang J., Saadeh H., Tomizawa S., Smallwood S.A., Chen T., Kelsey G.. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015; 29:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petell C.J., Alabdi L., He M., San Miguel P., Rose R., Gowher H.. An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 2016; 44:7605–7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long H.K., Blackledge N.P., Klose R.J.. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013; 41:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A.. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992; 69:905–914. [DOI] [PubMed] [Google Scholar]
- 21. Meehan R.R., Lewis J.D., Bird A.P.. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992; 20:5085–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nan X., Tate P., Li E., Bird A.. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 1996; 16:414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ausio J., de Paz A.M., Esteller M.. MeCP2: the long trip from a chromatin protein to neurological disorders. Trends Mol. Med. 2014; 20:487–498. [DOI] [PubMed] [Google Scholar]
- 24. Klose R.J., Bird A.P.. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006; 31:89–97. [DOI] [PubMed] [Google Scholar]
- 25. Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P.. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998; 19:187–191. [DOI] [PubMed] [Google Scholar]
- 26. Klose R.J., Sarraf S.A., Schmiedeberg L., McDermott S.M., Stancheva I., Bird A.P.. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005; 19:667–678. [DOI] [PubMed] [Google Scholar]
- 27. Hansen J.C., Ghosh R.P., Woodcock C.L.. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010; 62:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014; 17:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., Hemberg M., Ebert D.H., Greenberg M.E.. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015; 522:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyst M.J., Bird A.. Rett syndrome: a complex disorder with simple roots. Nat. Rev. Genet. 2015; 16:261–275. [DOI] [PubMed] [Google Scholar]
- 31. Fuks F., Hurd P.J., Wolf D., Nan X., Bird A.P., Kouzarides T.. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003; 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- 32. Kimura H., Shiota K.. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003; 278:4806–4812. [DOI] [PubMed] [Google Scholar]
- 33. Nan X., Hou J., Maclean A., Nasir J., Lafuente M.J., Shu X., Kriaucionis S., Bird A.. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bienvenu T., Chelly J.. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat. Rev. Genet. 2006; 7:415–426. [DOI] [PubMed] [Google Scholar]
- 35. Guy J., Cheval H., Selfridge J., Bird A.. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011; 27:631–652. [DOI] [PubMed] [Google Scholar]
- 36. Hite K.C., Adams V.H., Hansen J.C.. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009; 87:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muotri A.R., Marchetto M.C., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H.. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010; 468:443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adkins N.L., Georgel P.T.. MeCP2: structure and function. Biochem. Cell Biol. 2011; 89:1–11. [DOI] [PubMed] [Google Scholar]
- 39. Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y.. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008; 320:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ben-Shachar S., Chahrour M., Thaller C., Shaw C.A., Zoghbi H.Y.. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum. Mol. Genet. 2009; 18:2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugino K., Hempel C.M., Okaty B.W., Arnson H.A., Kato S., Dani V.S., Nelson S.B.. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 2014; 34:12877–12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Della Ragione F., Vacca M., Fioriniello S., Pepe G., D’Esposito M.. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genomics. 2016; 15:420–431. [DOI] [PubMed] [Google Scholar]
- 43. Feng J., Chang H., Li E., Fan G.. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005; 79:734–746. [DOI] [PubMed] [Google Scholar]
- 44. Nguyen S., Meletis K., Fu D., Jhaveri S., Jaenisch R.. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 2007; 236:1663–1676. [DOI] [PubMed] [Google Scholar]
- 45. Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G.. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010; 13:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris M.J., Adachi M., Na E.S., Monteggia L.M.. Selective role for DNMT3a in learning and memory. Neurobiol. Learn. Mem. 2014; 115:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang T., Cooper S., Brockdorff N.. The interplay of histone modifications - writers that read. EMBO Rep. 2015; 16:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bachman K.E., Rountree M.R., Baylin S.B.. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001; 276:32282–32287. [DOI] [PubMed] [Google Scholar]
- 49. Jeltsch A., Lanio T.. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002; 182:85–94. [DOI] [PubMed] [Google Scholar]
- 50. Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X.. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007; 449:248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jurkowska R.Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A.. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008; 36:6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rajavelu A., Jurkowska R.Z., Fritz J., Jeltsch A.. Function and disruption of DNA methyltransferase 3a cooperative DNA binding and nucleoprotein filament formation. Nucleic Acids Res. 2012; 40:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ebert D.H., Gabel H.W., Robinson N.D., Kastan N.R., Hu L.S., Cohen S., Navarro A.J., Lyst M.J., Ekiert R., Bird A.P. et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013; 499:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jurkowska R.Z., Siddique A.N., Jurkowski T.P., Jeltsch A.. Approaches to enzyme and substrate design of the murine Dnmt3a DNA methyltransferase. ChemBioChem. 2011; 12:1589–1594. [DOI] [PubMed] [Google Scholar]
- 55. Roth M., Jeltsch A.. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 2000; 381:269–272. [DOI] [PubMed] [Google Scholar]
- 56. Jurkowska R.Z., Ceccaldi A., Zhang Y., Arimondo P.B., Jeltsch A.. DNA methyltransferase assays. Methods Mol. Biol. 2011; 791:157–177. [DOI] [PubMed] [Google Scholar]
- 57. Rhee I., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W. et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002; 416:552–556. [DOI] [PubMed] [Google Scholar]
- 58. Egger G., Jeong S., Escobar S.G., Cortez C.C., Li T.W., Saito Y., Yoo C.B., Jones P.A., Liang G.. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rathert P., Roth M., Neumann T., Muerdter F., Roe J.S., Muhar M., Deswal S., Cerny-Reiterer S., Peter B., Jude J. et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015; 525:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013; 5:1704–1713. [DOI] [PubMed] [Google Scholar]
- 61. Liu G.J., Cimmino L., Jude J.G., Hu Y., Witkowski M.T., McKenzie M.D., Kartal-Kaess M., Best S.A., Tuohey L., Liao Y. et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev. 2014; 28:1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schiesser S., Pfaffeneder T., Sadeghian K., Hackner B., Steigenberger B., Schroder A.S., Steinbacher J., Kashiwazaki G., Hofner G., Wanner K.T. et al. Deamination, oxidation, and C-C bond cleavage reactivity of 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine. J. Am. Chem. Soc. 2013; 135:14593–14599. [DOI] [PubMed] [Google Scholar]
- 63. Bashtrykov P., Rajavelu A., Hackner B., Ragozin S., Carell T., Jeltsch A.. Targeted mutagenesis results in an activation of DNA methyltransferase 1 and confirms an autoinhibitory role of its RFTS domain. ChemBioChem. 2014; 15:743–748. [DOI] [PubMed] [Google Scholar]
- 64. Chen T., Ueda Y., Xie S., Li E.. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002; 277:38746–38754. [DOI] [PubMed] [Google Scholar]
- 65. Suetake I., Mishima Y., Kimura H., Lee Y.H., Goto Y., Takeshima H., Ikegami T., Tajima S.. Characterization of DNA-binding activity in the N-terminal domain of the DNA methyltransferase Dnmt3a. Biochem. J. 2011; 437:141–148. [DOI] [PubMed] [Google Scholar]
- 66. Jurkowska R.Z., Rajavelu A., Anspach N., Urbanke C., Jankevicius G., Ragozin S., Nellen W., Jeltsch A.. Oligomerization and binding of the Dnmt3a DNA methyltransferase to parallel DNA molecules: heterochromatic localization and role of Dnmt3L. J. Biol. Chem. 2011; 286:24200–24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ramsahoye B.H., Biniszkiewicz D., Lyko F., Clark V., Bird A.P., Jaenisch R.. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gowher H., Jeltsch A.. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J. Mol. Biol. 2001; 309:1201–1208. [DOI] [PubMed] [Google Scholar]
- 69. Dodge J.E., Ramsahoye B.H., Wo Z.G., Okano M., Li E.. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002; 289:41–48. [DOI] [PubMed] [Google Scholar]
- 70. Lin I.G., Han L., Taghva A., O’Brien L.E., Hsieh C.L.. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 2002; 22:704–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Handa V., Jeltsch A.. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005; 348:1103–1112. [DOI] [PubMed] [Google Scholar]
- 72. Emperle M., Rajavelu A., Reinhardt R., Jurkowska R.Z., Jeltsch A.. Cooperative DNA binding and protein/DNA fiber formation increases the activity of the Dnmt3a DNA methyltransferase. J. Biol. Chem. 2014; 289:29602–29613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arand J., Spieler D., Karius T., Branco M.R., Meilinger D., Meissner A., Jenuwein T., Xu G., Leonhardt H., Wolf V. et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012; 8:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ghosh R.P., Nikitina T., Horowitz-Scherer R.A., Gierasch L.M., Uversky V.N., Hite K., Hansen J.C., Woodcock C.L.. Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry. 2010; 49:4395–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baker S.A., Chen L., Wilkins A.D., Yu P., Lichtarge O., Zoghbi H.Y.. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013; 152:984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heyward F.D., Sweatt J.D.. DNA Methylation in Memory Formation: Emerging Insights. Neuroscientist. 2015; 21:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weaver I.C. Integrating early life experience, gene expression, brain development, and emergent phenotypes: unraveling the thread of nature via nurture. Adv. Genet. 2014; 86:277–307. [DOI] [PubMed] [Google Scholar]
- 80. Shin J., Ming G.L., Song H.. DNA modifications in the mammalian brain. Philos. Trans. R. Soc. Lond. Series B, Biol. Sci. 2014; 369:20130512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yang L., Rau R., Goodell M.A.. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015; 15:152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.