Abstract
Replication of mitochondrial DNA is strictly regulated during differentiation and development allowing each cell type to acquire its required mtDNA copy number to meet its specific needs for energy. Undifferentiated cells establish the mtDNA set point, which provides low numbers of mtDNA copy but sufficient template for replication once cells commit to specific lineages. However, cancer cells, such as those from the human glioblastoma multiforme cell line, HSR-GBM1, cannot complete differentiation as they fail to enforce the mtDNA set point and are trapped in a ‘pseudo-differentiated’ state. Global DNA methylation is likely to be a major contributing factor, as DNA demethylation treatments promote differentiation of HSR-GBM1 cells. To determine the relationship between DNA methylation and mtDNA copy number in cancer cells, we applied whole genome MeDIP-Seq and RNA-Seq to HSR-GBM1 cells and following their treatment with the DNA demethylation agents 5-azacytidine and vitamin C. We identified key methylated regions modulated by the DNA demethylation agents that also induced synchronous changes to mtDNA copy number and nuclear gene expression. Our findings highlight the control exerted by DNA methylation on the expression of key genes, the regulation of mtDNA copy number and establishment of the mtDNA set point, which collectively contribute to tumorigenesis.
INTRODUCTION
The human mitochondrial genome (mitochondrial DNA, mtDNA) is a circular, double stranded molecule that is ∼16.6 kb in size (1). It is essential for the production of energy as it encodes 13 subunits of the electron transfer chain (ETC), which generates the vast majority of cellular energy through the biochemical process of oxidative phosphorylation (OXPHOS). mtDNA also encodes 22 transfer RNAs (tRNAs) and 2 ribosomal RNAs (rRNAs). The major non-coding region, the D-loop, is the site of interaction for the nuclear-encoded mtDNA transcription and replication factors that bind to the mitochondrial genome to regulate mitochondrial genomic processes (1). For example, this region contains the transcription start sites for the heavy and light strands of the genome, namely the heavy and light strand promoters (HSP1/2 and LSP), and the site for the initiation of heavy strand replication (OH).
Cells possess multiple copies of mtDNA with mtDNA copy number being cell-type specific. This is achieved during early development when mtDNA replication is strictly regulated in order that a cell acquires the appropriate numbers of mtDNA copy to meet its specific functions when fully differentiated (2,3). They are empowered to do this as they had previously established the mtDNA set point, which is defined as the number of mtDNA copies (∼200) that a naïve cell possesses before it initiates the process of differentiation. These copies are then used by undifferentiated cells as the template for mtDNA replication as they differentiate into distinct mature cell types (2–6). Indeed, during differentiation, mtDNA copy number increases in a synchronous manner as cells mature from a naïve state to a fully differentiated state, which is modulated by the expression of genes that are the master regulators of differentiation. They then express genes associated with terminal differentiation and possess their required numbers of mtDNA copy (4–6). However, a number of cancer cell types are unable to expand their mtDNA copy number as they mainly rely on aerobic glycolysis for energy production, which allows for higher rates of cellular proliferation and prevents differentiation from taking place (4). Indeed, cancer cells appear to be trapped in a ‘pseudo-differentiated’ state whereby they are unable to complete differentiation and cannot increase mtDNA copy number (7,8). As a result, they fail to maintain or reinforce the mtDNA set point.
mtDNA replication is dependent on transcription having first taken place and is driven by a group of nuclear-encoded mitochondrial-specific transcription and replication factors (9,10). There are several key mitochondrial-specific and direct-binding transcription factors including mitochondrial RNA polymerase (POLRMT) (11), mitochondrial transcription factor A (TFAM) (12,13) and mitochondrial transcription factors B1 (TFB1M) and B2 (TFB2M) (14). The initiation of replication requires a short transcript that is specifically transcribed for mtDNA replication (15) and is used by the direct-binding mitochondrial specific DNA Polymerase Gamma (POLG) to drive mtDNA replication (16,17). In the human, this is a heterotrimer enzyme composed of a catalytic subunit (POLGA) and two accessory subunits (POLGB) (18). POLGA (encoded by the gene POLG) acts as the DNA polymerase (19) whereas POLGB (encoded by the gene POLG2) recognizes the initial RNA primer and enhances the enzymatic activity of POLG (9,18,20). This process is supported by Twinkle (TWNK), the DNA helicase that separates the two strands allowing for replication to take place (21,22), and the mitochondrial single-stranded DNA binding protein (SSBP1), which acts to stabilize the transient status of single-stranded DNA. Furthermore, the double-stranded circular structure of the mitochondrial genome ensures the maintenance of DNA topology during mtDNA replication by allowing the two strands of the circular genome to unwind or rotate around each other. The mitochondrial topoisomerase, which is encoded by the TOP1MT gene, can facilitate this by transiently breaking one strand of mtDNA and religating the strand after having passed through the other strand (23).
A group of indirect factors also play vital roles in regulating mtDNA transcription and replication even though they do not directly bind to mtDNA as is the case for the direct-binding factors. The best-characterized regulators are the nuclear respiratory factors (NRF1 and NRF2), the peroxisome proliferator-activated receptor γ coactivator-1 family (PGC1α), the Sirtuin family, especially Sirtuin 1 and Sirtuin 3, the tumor suppressor p53 and signal transducer and activator of transcription 3 (STAT3). NRFs interact with PGC1α cofactors to regulate the expression of TFB1M and TFB2M by binding to their promoter regions (24). In addition, Sirtuin1 and 3, NAD+-dependent deacetylases encoded by the SIRT1/3 genes, are co-factors with PGC1α that co-localise with TFAM inside mitochondria (25,26). Given that thyroid hormone T3 is essential for mitochondrial biogenesis (27), it has been shown that T3 binding to T3 receptors affects mtDNA expression by either binding to mtDNA directly or interfering with the expression of the transcription factors in the nucleus, such as NRF1 and PCG1α (28–31).
We have previously shown that mtDNA copy number is associated with DNA methylation at exon 2 of POLG (4,32). It appears that tumor cells are extensively DNA methylated, which normally occurs at CpG dinucleotides, and converts the cytosine into 5-methylcytosine (5mC) (33,34). The glioblastoma multiforme HSR-GBM1 cell line is a high-grade malignant glioblastoma cell line (35). It is DNA hypermethylated, which is associated with oncomutations of the isocitrate dehydrogenase (IDH1/2) in the citric acid cycle (33,36–39). The hypermethylated genome, which leads to and supports its tumorigenic gene profile (37–41), can be reversed through the use of DNA demethylation agents such as 5-azacytidine (5Aza) and vitamin C (VitC) (42,43). 5Aza is a chemical analogue of cytosine that inhibits DNA methylation at low concentrations, and has been applied in cancer therapies (43,44). It blocks the DNA methyltransferase, DNMT1, which causes global DNA demethylation (45). VitC can also induce DNA demethylation by enhancing the activity of the TET1 enzyme and, thus, improves the conversion of 5mC to the DNA demethylated state of 5-hydroxy-methylcytosine (5hmC) (42).
The demethylation of HSR-GBM1 cells results in significant increases in mtDNA copy number, as they are able to modulate mtDNA copy number in synchrony with changes in chromosomal gene expression patterns during differentiation (4,6). Indeed, during development, the DNA methylation status of the genome undergoes significant changes to firstly erase the parental profiles by ten-eleven translocation methylcytosine dioxygenases, the TET family, and reestablish their own profiles through the DNA methyltransferase (DNMT) family of genes during differentiation (46,47). This highlights the cooperation that exists between the two genomes and the importance of the collective regulation of DNA methylation and mtDNA copy number in tumorigenesis and differentiation.
To further investigate the impact of DNA methylation on regulating the mtDNA-specific transcription and replication factors, we have used methylated DNA immunoprecipitation (MeDIP) in combination with whole-genome sequencing (MeDIP-Seq), whole genome RNA-sequencing (RNA-Seq), and a custom designed high-throughput Fluidigm real-time PCR array on the well characterised HSR-GBM1 tumor cell line and compared its DNA methylation profiles to its demethylated state when induced by 5Aza and VitC. Our results show that a number of tumor-specific genes and several key mtDNA transcription and replication factors are modulated by global DNA demethylation.
MATERIALS AND METHODS
Cell culture
HSR-GBM1 cells were cultured as neurospheres on ultra-low attachment plates (Sigma-Aldrich, MO, USA) in Dulbecco's Modified Eagle Medium /Nutrient Mixture (DMEM/F-12) Media (Thermo Fisher Scientific, MA, USA) containing 2% StemPro neural supplement (Thermo Fisher Scientific), 20 ng/ml basic fibroblast growth factor (bFGF; Merck Millipore, MO, USA) and 20 ng/ml epidermal growth factor (EGF; Merck Millipore) at 37°C, 5% CO2 and 95% humidity.
DNA demethylation assay
To induce DNA demethylation, HSR-GBM1 cells were cultured in the presence of 0.5 μM of 5Aza (Sigma-Aldrich) for 48 h or 100 μg/ml of VitC (l-ascorbic acid, Sigma-Aldrich) for 72 h. Culture media was changed every 24 h. Cells were harvested after the treatments for downstream experiments or stored as cell pellets at −80°C.
DNA and RNA extraction
Total genomic DNA and RNA were extracted from cultured cell pellets using the ISOLATE II Genomic DNA Kit (Bioline, London, UK) and RNeasy Mini Kit (Qiagen, CA, USA), respectively, according to the manufacturer's protocols with minor modifications. The DNA samples were treated with 3 μl of RNase solution (Qiagen) and Proteinase K Solution (20 μg/μl; Qiagen) at 65°C for 10 min and the RNA samples were treated with DNase I (3 kunitz units/ μl; Qiagen) for 20 min.
Determination of mtDNA copy number per cell
mtDNA copy number per cell was determined using an established protocol, as previously described (4). Quantitative real-time PCR (qPCR) was performed on purified total DNA targeting β-globin and mtDNA using a Rotor-Gene 3000 (Corbett Research, Cambridge, UK). mtDNA copy number was calculated using the formula: mtDNA copy number per cell = 2 × NmtDNA/Nβ-globin, where NmtDNA and Nβ-globin, determined by the formula: N = (qPCR product concentration × 6.023 × 1014)/(qPCR product size in bp × 660), as described in (32). The concentration of the qPCR product is determined using standard curves. Primer sequences and reaction conditions are listed in Supplementary Table S1. Biological triplicates (n = 3) were analysed.
Immunoprecipitation of methylated DNA (MeDIP)
Five microgram of genomic DNA was sheared into 200–1000 bp fragments in a precooled and degassed Covaris Adaptive Focused Acoustics (AFA™) S220 system. The dsDNA was denatured by incubation at 95°C for 10 min and then underwent MeDIP, as described in (48). 1.5 μg of anti-5mC antibody (Active Motif) were added to 3 μg of DNA fragments and 20 μl per sample of Dynabeads® Protein G (Thermo Fisher Scientific) in 500 μl of 1× IP buffer (100 mM sodium phosphate, pH 7.0; 1.4 M NaCl; 0.5% Triton X-100) at 4°C for 16 h under rotation. The beads were washed with 1 ml of 1× IP buffer three times and were resuspended in 250 μl proteinase K digestion buffer (50 mM Tris–HCl, pH 8.0; 10 M EDTA, pH 8.0; 1.0% SDS) with 10 μl of proteinase K (20 mg/ml; Bioline) at 50°C for 3 h on a thermo-shaker. The supernatant was then collected into a new tube and placed on a magnetic particle concentrator (Thermo Fisher Scientific). DNA was purified from the elutant using the QIAquick PCR Purification Kit (Qiagen). Purified DNA was eluted in 50 μl of autoclaved Milli-Q H2O.
MeDIP-Seq
To adapt MeDIP for next generation sequencing, minor changes were made to the MeDIP procedure described above. Two microgram of each genomic DNA sample were sheared into ∼500 bp in a precooled and degassed Covaris Adaptive Focused Acoustics (AFA™) S220 system. These fragments were then end repaired, A-tailed, ligated to Illumina adaptors (including the sample specific indexed) and purified without amplification to ensure conservation of methylated CpGs using the Illumina TruSeq DNA PCR-Free Kit, as described in the manufacturer's Low Sample (LS) protocol (Illumina Protocol part #15036187 Rev A Jan 2013). The dsDNA libraries then followed the procedure, as described above. After MeDIP, dsDNA libraries were generated and amplified for 10 cycles, as per the TruSeq ChIPSeq protocol with standard Illumina P7 and P5 primers (Illumina protocol part #15023092 Rev A August 2012). The resultant libraries were analysed using Qubit (Agilent) and Bioanalyzer (ThermoFisher Scientific) and quantitated by qPCR. The 12 pM library pool was prepared based upon qPCR and used for clustering by the cBot™ Hybridization system (Illumina, CA, USA). Cluster generation was performed according to the cBot™ user guide (part #15006165 Rev K October 2012). Clusters were in the optimal range (795 k/mm2; optimal 750–900 k/mm2). 50-bp single-read sequencing was then performed on the Illumina HiSeq1500 and HiSeq3000 sequencing platforms using the Illumina protocol (15035788 Rev D, April 2014; 15066493 v01, August 2015). The reads were converted to fastq format and adaptors were removed using the bcl2fastq software (Illumina). The number of reads per sample was evenly distributed. In total, >550 million reads passed the filter, and over 95% of the sequencing reads passed the Illumina sequencing quality score of Q30 that was deemed excellent quality for base-calling. Run quality parameters were deemed excellent with the PhiX spike-in having an error rate of < 0.2% (expected < 0.5%) and phasing/prephasing of < 0.08/0.1 (expected 0.2/< 0.4).
Bioinformatics analysis for MeDIP-Seq
Sequencing files in fastq format firstly underwent and passed the pre-alignment quality and remaining adaptor check using the FastQC software (v 0.11.5; downloaded from Babraham Bioinformatics public projects https://www.bioinformatics.babraham.ac.uk/projects/). The raw sequences were then mapped to the human reference genome GRCh38/hg38 (UCSC) using the Burrows-Wheeler Aligner (BWA) software (version 0.7.16a) (49). The commands of ‘aln’ and ‘samse’ of the BWA-backtrack algorithm were used with their default settings to undertake the mapping. The output files (*.sam) were then combined, filtered and converted to bam files (*.bam) by Samtools (version 1.4) using the commands of ‘view’, ‘sort’ and ‘merge’. Further filtering based on the mapping scores (MAPQ ≥ 30) was performed to remove ambiguous reads and only uniquely mapped reads were kept for further analysis. Mapping files (*.bam) were exported for downstream analysis. Mapping parameters are listed in Supplementary Table S2.
Analytical analysis was carried out using the MEDIPS package (version 1.24.0) on R programming software (version 3.3.2) (50,51). Command codes were based on the R script document of MEDIPS. MeDIP-Seq specific quality control analysis was performed on the mapping files for each sample, which included saturation analysis, CpG coverage analysis and CpG enrichment analysis. The parameters specified in this analysis were set to: ‘extend’ of 500, which means all short reads were extended to 500 nucleotides according to the reference genome; ‘shift’ to 0, which means there was no shift in the genome locations for the sequences; ‘window_size’ to 100, which indicates the analysis was performed every 100 bp of the genome; ‘uniq’ to 1, which indicates that the reads that mapped to exactly the same genomic positions were counted as one read. A coupling factor was set up based on the HSR-GBM1 samples (control group) for normalization. Biological triplicates were grouped into three cohorts: the GBM cohort, the GBM + VitC cohort and the GBM + 5Aza cohort. Differentially methylated windows of 100 bp were identified between groups using the ‘edgeR’ plug-in function. Among the six samples in each comparison, genomic windows that were covered by a minimal counts threshold of 100 reads were kept for further statistical analysis (minsrowsum = 100). Statistically significant results were selected using criteria for the adjusted P value ≤ 0.05. Continuous significant windows were merged as one differentially methylated region (DMR).
Comparisons between the GBM cohort and the GBM + VitC cohort and between the GBM cohort and the GBM + 5Aza cohort were performed. DMRs identified between the cohorts then underwent annotation based on their corresponding regions in the human genome (hg38) using the ChIPSeeker package (version 3.5) (52). The built-in function of region of interest (ROI) analysis in the MEDIPS package was specifically used to investigate the DNA methylation levels at the CpG islands (CGIs) found in gene bodies and promoter regions of the mtDNA replication factors. One-way ANOVA was used to statistically compare the mean relative methylation score of ROIs. Relative methylation score was developed specifically for MeDIP-Seq in order to normalize methylation scores for regions based on the concept of CpG coupling analysis (50,51). Relative methylation score is calculated using the formula of log 2 (mean MeDIP-Seq signal × 106/the corresponding estimated number of reads based on the coupling analysis × the total number of short reads) (50). It has been shown to correlate well with DNA methylation levels following bisulfite conversion methods, when applied to individual CpG sites within a region (50). In other words, relative methylation score offers a relatively high-throughput and accurate measurement of DNA methylation.
Purification of mtDNA from cells
To further verify whether DNA methylation exists within the mitochondrial genome, mtDNA was purified from cells to eliminate mtDNA pseudo-genes present in the nuclear genome from the analysis. Mitochondrial isolation was performed using a 5 ml Potter-Elvehjem tissue grinder set (Cat. No. 358034; Wheaton, USA). Freshly collected cells (∼10 million) were resuspended in 5 ml of solution A (20 mM HEPES–KOH, pH 7.6, 220 mM Mannitol, 70 mM sucrose, 1 mM EDTA and 2 mg/ml BSA freshly added) and placed on ice for 15 min to facilitate swelling. The cells were then homogenized at 4°C in a glass chamber with a drill-fitted pestle for 50 repetitions. The cell homogenate was centrifuged at 800g for 10 min to remove cell debris and nuclei. The supernatant was centrifuged at 10 000 g for 20 min to pellet the mitochondrial fraction. The pellet was then resuspended in 175 μl of solution B (20 mM HEPES–KOH, pH 7.6, 220 mM Mannitol, 70 mM sucrose, 10 mM MgCl2). 10 μl of DNase I (3 kunitz units/μl; Qiagen) in 20 μl of RDD buffer (Qiagen) were added to the mitochondrial suspension and incubated at 37°C for 30 min to further remove nuclear DNA. 1 ml of solution A without BSA was added to stop DNase activity and the suspension was centrifuged at 10 500 g for 20 min at 4°C to pellet the mitochondria. The pellet of mitochondria was resuspended in 200 μl of lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA and 1% SDS) with 1 μl of proteinase K (20 mg/ml, Bioline) and incubated at 50°C for 60 min. mtDNA was then purified using the phenol:chloroform method by adding 1 volume of phenol: chloroform: isoamyl alcohol (25:24:1, Sigma), vortexing for 1 min and centrifuging at 17 000g for 5 min. The upper layer was collected and underwent another round of chloroform extraction by adding 1 volume of chloroform: isoamyl alcohol (24:1, Sigma). The upper layer was collected and mtDNA was precipitated by adding 2.5 volumes of 100% ethanol, 0.1 volume of 3 M sodium acetate and 1 μl of glycogen (20 μg/μl), and incubated at −80°C for at least 2 h. Samples were centrifuged at 20 000g for 10 min at 4°C and the mtDNA pellets were washed with 500 μl of 70% ethanol. After further centrifugation at 12 000g for 5 min at 4°C, the mtDNA pellets were air-dried for 10 min and dissolved in 50 μl of autoclaved Milli-Q H2O.
Purified mtDNA samples firstly underwent the MeDIP-long-PCR protocol without sonication to demonstrate the presence of DNA methylation on mtDNA (described below in the section entitled ‘Whole-mtDNA MeDIP and amplification by long PCR’). Secondly, purified mtDNA samples and long PCR products underwent MeDIP (with sonication) followed by qPCR to determine the levels of DNA methylation at different regions of the mitochondrial genome (described below in the section entitled ‘Quantification of MeDIP products’).
Whole-mtDNA MeDIP and amplification by long PCR
Prior to performing MeDIP, a positive and a negative control were generated by long PCR to generate two overlapping long mtDNA fragments (∼8500 bp) spanning the whole mitochondrial genome. As PCR products are deemed to be unmethylated, the negative control (Neg-longPCR-5mC) comprised equal concentrations of two long mtDNA fragments generated by long PCR using primer set 1 (Supplementary Table S1). Each long PCR contained 1× High Fidelity PCR buffer, 100 mM MgSO4, 1 mM dNTPs (Bioline), 1 U of Platinum Taq High Fidelity (Invitrogen), 10 μM each forward and reverse primer (primer set 1; Supplementary Table S1) in a total volume of 50 μl. Reaction cycling profiles were 94°C for 2 min, 35 cycles of 94°C for 15 s, 63°C for 30 s and 68°C for 8 min 45 s. Similarly, the positive control (Pos-longPCR-5mC) comprised equal concentrations of the two long mtDNA fragments that had undergone DNA methylation treatment. DNA methylation treatment was carried out using the CpG methyltransferase M.SssI (New England Biolabs; USA), according to the manufacturer's instructions. Briefly, 1 μg of the combined mtDNA fragments was incubated with four units of the enzyme in the presence of 160 μM S-adenosylmethionine (SAM) at 37°C for 4 h followed by 65°C for 20 min.
The purified mtDNA sample (mtDNA-5mC), and the positive (Pos-longPCR-5mC) and negative (Neg-longPCR-5mC) control samples then underwent MeDIP, as described in the section of ‘Immunoprecipitation of methylated DNA (MeDIP)’, except that the sonication step was not included. This maintains the integrity of the mtDNA, which acts as the template for long PCR following MeDIP. Moreover, the purified mtDNA sample also underwent MeDIP without 5mC antibody (mtDNA-NAC) to account for non-specific pull-down. MeDIP products were purified using the phenol:chloroform method, and assessed by long PCR using primer set 2 (Supplementary Table S1) with the same reaction conditions, as described above. mtDNA sequences targeted by primer set 2 are located just downstream and just upstream of 5′ and 3′ ends of the sequences used for the negative and positive controls, respectively. A total DNA sample isolated from GBM cells (Total DNA) and a non-template control (H2O; NTC) were also amplified. PCR products were then run on a 0.8% agarose gel.
Quantification of MeDIP products
After performing MeDIP (with a sonication step), as described in the section ‘Immunoprecipitation of methylated DNA (MeDIP)’, on the purified mtDNA samples and long PCR products, the levels of DNA methylation for the regions of interest were quantified by qPCR on a Rotor-Gene 3000 machine under primer specific conditions (Supplementary Table S1). The levels of DNA methylation for the mitochondrial genome were determined by firstly normalizing the qPCR signals of the MeDIP products to their corresponding input samples (5mC/Input), and then weighting these values against the positive and negative controls that were used to represent 100% and 0% levels of DNA methylation, respectively. The levels of DNA methylation at exon 2 of POLG, and TOP1MT exon 8 and intron 10 from MeDIP-Seq were also validated by MeDIP-qPCR using total DNA. The results were determined by normalizing the qPCR signals of the MeDIP products to their corresponding input samples (5mC/Input).
Determination of copy number variation (CNV)
All samples were genotyped for CNV using the Illumina Human Global Screening Array Beadchip (Illumina), which covers ∼700K SNPs. Array data were processed using GenomeStudio 2.0 (Illumina), according to the user's manual. Data normalization, clustering and genotype calling were performed using the Genotype Module. The full data report containing log R ratios (LRR) and B allele frequencies (BAF) for each probe was then exported as input data to the Nexus 9.0 software (Biodiscovery Inc., CA, USA) for advanced analysis. The Nexus Copy Number Module was used to carry out the comparisons of DNA copy number between groups. Human genome build hg19 was chosen as the reference genome to assign genome locations. Statistical significance between groups in individual SNPs was determined by P value ≤ 0.05. Genomic regions were aligned to hg38 for overlapping analysis with DMRs.
RNA-Seq analysis
Total RNA was submitted to the Australian Genome Research Facility (AGRF; VIC, Australia) to perform RNA sequencing. An Agilent Bioanalysor Nanochip (Agilent) was used to determine the integrity of the RNA samples. cDNA libraries were constructed using the Illumina Truseq Standard mRNA Kit (Illumina), according to the manufacturer's instructions. The libraries were sequenced on the HiSeq 2500 platform (Illumina). Image analysis was performed in real time using the HiSeq Control Software (HCS) v2.2.68 and Real Time Analysis (RTA) program v1.18.66.3 running on the instrument's computer. RTA performed real-time base calling on the HiSeq instrument computer. The Illumina bcl2fastq 2.19.0.316 pipeline was then used to generate the sequence data at AGRF. The sequences were mapped to human genome hg38 by STAR aligner (2.5.3a) using default settings (53). The annotation was based on the comprehensive GENCODE gene annotation (v26). Only the uniquely mapped reads were used for downstream analysis. The reads were then summarized to counts for each gene by the ‘featureCounts’ function in the Rsubread package (1.26.0) using the inbuilt annotation hg38 (54). Identification of differentially expressed genes was performed using the edgeR package (3.16.5) employing the trimmed mean of M-values normalization method (TMM) application (55,56). Gene expression data were uploaded to Ingenuity Pathway Analysis software (IPA; Qiagen). Genes with false discovery rate (FDR) ≤0.05 and absolute fold change ≥ 2 were deemed to be significantly differentially expressed genes. These genes were used to perform pathway analysis for determining the most affected canonical pathways, biological functions and networks, and used for undertaking the DMR-overlapping analysis.
Gene expression analysis using the Fluidigm platform
cDNA was synthesized from 1 μg of total RNA using oligo (dT) primers and the Superscript III First-Strand synthesis system (Thermo Fisher Scientific), according to the manufacturer's instructions. Pre-amplification was performed to increase the number of copies of each gene to detectable levels, as detailed in Gene Expression Preamp with the Fluidigm Preamp MasterMix and Taqman Assays Quick Reference PN 100-5876 B1. All Taqman assays are listed in Supplementary Table S3. Taqman assays were pooled with C1 DNA suspension buffer to produce a final concentration for each assay of 180 nM. 1.25 μl of each cDNA sample as well as a non-template control underwent pre-amplification for 14 cycles with 3.75 μl of the pooled assays and Taqman PreAmp Master mix (Thermo Fisher Scientific), according to the manufacturer's instructions.
Following preamplification, reaction products were diluted 5-fold in C1 DNA suspension buffer. Assays and samples were combined in a 96.96 Dynamic array IFC plate, according to the Fluidigm® 96.96 Real-Time PCR Workflow Quick Reference PN 6800088. An Integrated Fluidic Circuit (IFC) controller HX was used to prime and load the plate. 5 μl of each preamplified sample were loaded in duplicate into each sample inlet and 5 μl of each Taqman assay (10×) were also loaded into each assay inlet of the plate. Gene expression was performed according to the Biomark GE 96.96 Standard v2 protocol. Data were exported as a spreadsheet (.csv) using the Fluidigm Real-Time PCR analysis software (v4.1.1). Differentially expressed genes were analysed using the HTqPCR package (version 1.28) (57). The package uses the ‘deltaCt’ method of normalization. To improve the analysis, three housekeeping genes were used for normalization. ActB, HPRT1 and OAZ1 were chosen as the housekeeping genes for the nuclear genes. They are widely used in glioblastoma studies (4,58). Rn18S, HPRT1 and OAZ1 were chosen as the housekeeping genes for the mitochondrial-related genes. Rn18S has higher levels of expression and is, therefore, more suitable for the normalization of the mitochondrial genes, which have much higher levels of expression than nuclear-encoded genes (58,59). The package uses the ‘limma’ method to perform statistical analysis, as stated in the user guide available from the website. Results were then exported and plotted using GraphPad Prism 7 (GraphPad Software, Inc., CA, USA).
Data deposition
MeDIP-Seq sequences (triplicates) for the HSR-GBM1 cells and HSR-GBM1 cells treated with 5Aza and VitC have been deposited in the Sequence Read Archive (SRA) under accession number SRP080899. CNV data and RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE98693.
RESULTS
Whole genome MeDIP-Seq, quality control and CNV analysis
To investigate the impact that DNA methylation has on the whole genome and how it affects mtDNA replication, we performed whole genome MeDIP-Seq on DNA samples from HSR-GBM1 cells (GBM) to identify sites that are DNA methylated, in order to then compare them with the DNA methylome of HSR-GBM1 cells cultured with either 5Aza (GBM + 5Aza) or VitC (GBM + VitC) to determine which sites undergo modulation. The HSR-GBM1 cell line was chosen as it is one of the best-characterized GBM cell lines and is indicative of an undifferentiated cancer cell line, which has the potential to be differentiated into neuronal- and astrocyte-like cells (6,35) once it has undergone DNA demethylation induced by 5Aza or VitC (4).
On average, >40.4 million uniquely mapped sequences per sample were obtained (Table 1). The mapped reads were cleaned to remove duplicates that mapped to exactly the same genomic position and were counted as one read to ensure that the repeat reads resulting from PCR amplification were not biasing the data. To check the reproducibility of the workflow, a Pearson correlation test was performed on the mapped reads within each cohort. On average, high correlation scores (0.94 for the GBM cohort, 0.95 for the GBM + VitC cohort and 0.95 for the GBM + 5Aza cohort) were achieved amongst the triplicates for each cohort (Table 1). The MEDIPS analysis package offers several quality control tests for MeDIP-Seq experiments. Saturation analysis gives the estimated saturation correlation score to assess the coverage of the genome. Overall, scores of 0.95 were achieved from the samples, which indicates sufficient coverage (Table 1). CpG coverage analysis specifically includes coverage levels of the 28 million CpG dinucleotides in the human genome. On average, >91% of total CpG dinucleotides in the human genome were covered with at least one read which indicates good enrichment of the targets for downstream analysis (Table 1) and the differences in coverage depth are shown in Supplementary Figure S1.
Table 1. MeDIP-Seq outputs and QC results following analysis using the MEDIPS package. Each number after a sample indicates a biological replicate.
| Sample | Reads without duplicates (millions) | Correlation between triplicates | Estimated Saturation Correlation | CpGs covered by the reads |
|---|---|---|---|---|
| GBM 1 | 41.8 | 0.95 | 91.7% | |
| GBM 2 | 37.3 | 0.94 | 0.95 | 91.51% |
| GBM 3 | 38.3 | 0.95 | 91.65% | |
| GBM+5Aza 1 | 37.6 | 0.95 | 91.23% | |
| GBM+5Aza 2 | 45.6 | 0.95 | 0.95 | 91.98% |
| GBM+5Aza 3 | 48.6 | 0.95 | 92.1% | |
| GBM+VitC 1 | 37.8 | 0.95 | 90.8% | |
| GBM+VitC 2 | 44.4 | 0.95 | 0.95 | 90.83% |
| GBM+VitC 3 | 32.5 | 0.94 | 89.56% |
Cancer cells frequently exhibit genomic instability. CNVs between cohorts can have a direct impact on MeDIP-Seq signals that are not reflective of true changes in DNA methylation and, therefore, result in false positive or negative results (60). To preclude the potential impact of CNVs on the MeDIP-Seq data, the three cohorts of cells were genotyped to screen for CNVs over the whole genome. By using an Illumina global screening array, 700 K SNPs were analyzed and statistical comparisons were performed using the Nexus 9.0 Copy Number module. Overall gains in copy number were observed in the GBM genome. In all, 247 regions were identified as copy number gains (Supplementary Table S4). However, no significant CNVs were identified between either the GBM and GBM + VitC or GBM + 5Aza cohorts (Supplementary Figure S2). In other words, the DNA demethylation treatments did not improve or intensify genomic instability among the cohorts of GBM cells, which means that the CNVs did not impact on the analysis by MeDIP-Seq.
Identification of differentially methylated regions (DMRs)
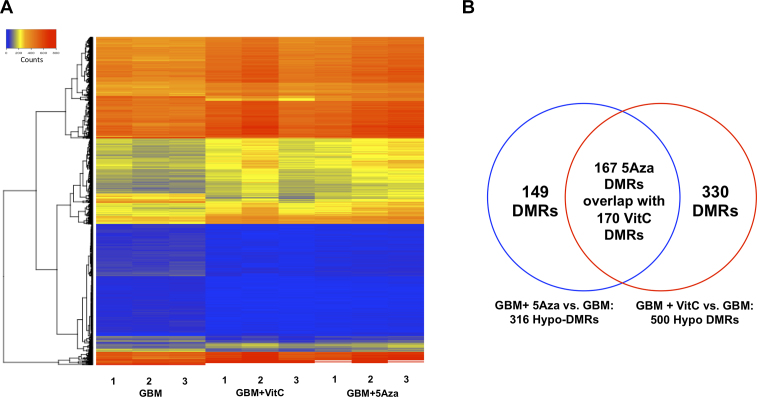
Using the MEDIPS package, 816 differentially methylated regions (DMRs) were identified between the GBM + 5Aza and the GBM cohorts and 1743 DMRs between the GBM + VitC and GBM cohorts (adjusted P value ≤ 0.05, minrowsum ≥ 100). The heatmap for the total number of DMRs indicated that the two cohorts that had undergone DNA demethylation treatment exhibited significant changes when compared with the GBM methylome (Figure 1A). Interestingly, both the GBM + 5Aza and the GBM + VitC cohorts exhibited hyper-DMRs and hypo-DMRs throughout the genome (Figure 1A). Indeed, 316 out of 816 DMRs (38.72%) were identified to be demethylated by 5Aza and 500 out of 1743 DMRs (28.68%) were demethylated by VitC. The occurrence of hypermethylation is expected, especially in the GBM + VitC cohort, as the rebound effect can result in the re-establishment of DNA methylation (4). We focused on the demethylated DMRs for further analysis, as they are indicative of the direct effects of the DNA demethylation treatments.
Figure 1.
Overview of the DMRs. (A) Heatmap of the DNA methylation profiles. Total identified DMRs were plotted using values in counts for the three biological replicates from the GBM cohort, the GBM + 5Aza cohort and the GBM + VitC cohort. The colour scheme from blue, yellow to red represents the level of DNA methylation from low to high. (B) Venn diagram of the DMRs identified in the two comparisons. The 316 DMRs identified in the comparison between GBM + 5Aza and GBM are shown in the blue circle; the 500 DMRs identified between GBM + VitC and GBM are shown in the red circle. 167 DMRs identified in the GBM + 5Aza cohort overlap or partially overlap with 170 DMRs identified in the GBM + VitC cohort.
Between the two cohorts of DMRs, 167 DMRs identified from the comparison between the GBM + 5Aza cohort and the GBM cohort were found to overlap with 170 DMRs identified from the comparison between GBM + VitC and GBM cohorts (the unequal distribution of DMRs is due to each of the treatments differentially affecting different regions resulting in some partial overlap) (Figure 1B). In all, 149 DMRs were uniquely identified from the comparison between the GBM + 5Aza and the GBM cohorts, whereas 330 DMRs were uniquely identified from the comparison between the GBM + VitC and the GBM cohorts (Figure 1B). The uniquely identified DMRs probably resulted from their different mechanisms of action for regulating DNA demethylation. As a result, the unique DMRs are likely to be the sites that were only affected by one of the agents. On the other hand, both of the agents could have demethylated the same DNA region but only one of them was able to significantly demethylate that region or maintain DNA demethylation.
As the function of DNA methylation varies dependent on the location of the methylated sites, the identified DMRs were then annotated according to their genome location using the ChIPSeeker package, which identified promoter regions (1000 bp upstream of transcription start site (TSS)), exons, introns, three UTRs and downstream regions (< 3 kb). These regions are collectively referred to as intragenic regions and are normally associated with transcriptional functions. However, the other DMRs (> 50%) are located at intergenic regions, which refer to non-coding genomic regions between genes (Supplementary Figure S3). Regardless of the number of DMRs, it appeared that VitC had a tendency to affect more intragenic regions when compared with 5Aza. This could arise from the enhanced activity of the TET1 enzyme modulated by VitC, as it is known to specifically target more intragenic regions (42), while DNMT1 is more likely to be inhibited by 5Aza (43), thus causing global DNA demethylation.
Furthermore, of the DMRs identified, only seven mapped with the copy number gain regions identified above, which further suggests the regions possessing CNVs were not affected by DNA demethylation and had only a very limited effect on the identification of DMRs (Supplementary Table S4).
Identification of differentially expressed genes and the association with the DMRs
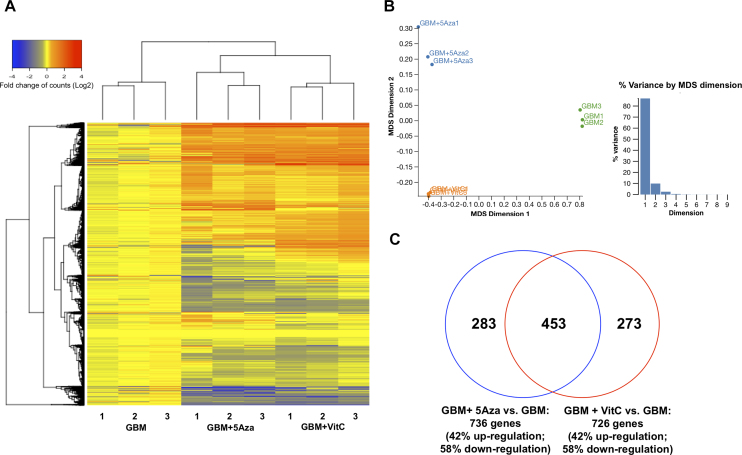
To investigate the changes to the overall gene expression profiles induced by DNA demethylation, total RNA samples from the three cohorts were sequenced and the total differentially expressed gene (FDR ≤ 0.05, absolute logFC ≥ 1) profiles are shown in the heatmap (fold change of counts in log2 scale) for each of the triplicates from the treated cohorts (Figure 2A). The multidimensional scaling (MDS) plot analysis shows that the three cohorts are distinct from each other (Figure 2B). The first dimension separated the untreated GBM and the DNA demethylated GBM cohorts, which indicates the primary factor that distinguishes the cohorts is their DNA methylation status. The differences between the GBM + 5Aza cohort and GBM + VitC cohort were also indicated by distance in the second dimension, which suggests that these two treatments caused different responses in gene expression despite the effects of DNA demethylation. Using edgeR for further analysis, 726 differentially expressed genes were identified from the GBM + VitC cohort and 736 differentially expressed genes were identified from the GBM + 5Aza cohort (FDR ≤ 0.05 and absolute logFC ≥ 1; Figure 2C). A total of 453 genes were commonly modulated by both treatments (Figure 2C). However, 273 genes were uniquely identified in the GBM + VitC gene list, whereas 283 genes were uniquely modulated by 5Aza (Figure 2C). Interestingly, both comparisons determined that 42% of genes were up-regulated and 58% were down-regulated (Figure 2C), which highlights the dual effects that DNA methylation has on transcription.
Figure 2.
Overview of differentially expressed genes. (A) Heatmap of the RNA-Seq profiles. Total identified differentially expressed genes (FDR ≤ 0.05, absolute logFC ≥ 1) for the GBM + 5Aza cohort and the GBM + VitC cohort were plotted based on the fold changes (log2) of read counts for the mean value of the GBM cohort. The color scheme from blue, yellow to red represents the level of expression from low to high. (B) MDS plot of the RNA profiles of the three cohorts. The MDS plot shows the similarity of the cohorts for each group indicated by the distance on the first two dimensions. Dimension 1 accounts for 83% of the total variance, and Dimension 2 accounts for 12% of the total variance. The GBM cohort was plotted in green, the GBM + VitC cohort was plotted in orange and the GBM + 5Aza cohort was plotted in blue. (C) Venn diagram of the differentially expressed genes identified in the two comparisons. 736 genes identified to be differentially expressed in the comparison between GBM + 5Aza and GBM are shown in the blue circle; 726 genes identified to be differentially expressed between GBM + VitC and GBM are shown in the red circle. 453 genes overlapped between these two comparisons. A 42% up-regulation and a 58% down-regulation were identified in both comparisons.
Differentially expressed genes were then used for pathway analysis using IPA software. The top canonical pathways, top diseases and bio-functions affected by each of the treatments are listed in Table 2. In the top canonical pathways, hepatic fibrosis and acute phase response signaling pathways were identified in both comparisons, which suggests the genes in these pathways are greatly sensitive to DNA demethylation. On the other hand, VitC modulated cellular pathways involved in cellular organization and cell cycle, including mitotic roles of the polo-like kinase, and G2/DNA damage checkpoints in cell cycle, whereas 5Aza modulated pathways associated with tumorigenesis and metabolism including colorectal cancer metastasis signaling, Wnt/-catenin signaling and MIF regulation of innate immunity. The most affected disease was cancer resulting from both treatments with 565 molecules being affected by VitC and 572 molecules affected by 5Aza. Interestingly, VitC tended to modulate cell growth by affecting DNA replication, recombination, and repair in the cell cycle and triggering cell death, whereas 5Aza tended to inhibit cellular proliferation. Indeed, the increases in cell number during tissue culture were slightly lower in the treated cohorts than the GBM cohort. From the same initial number of cells, the GBM cohort increased 2.9 (± 0.1) fold, whereas GBM+VitC gained 2.5 (± 0.14) fold and GBM + 5Aza gained 2.55 (± 0.1) fold (P > 0.05). The networks analyses described the gene ontology (GO) enrichment for the differentially expressed genes (Supplementary Figures S4–S7). The top network enriched in the VitC-induced differentially expressed genes was cell cycle, cellular assembly and organization, and DNA replication, recombination and repair (Supplementary Figure S4). Interestingly, the glioblastoma disease pathway was also found to be modulated by VitC with a range of associated genes up-regulated (Supplementary Figure S5). On the other hand, 5Aza more effectively influenced the: (i) cell morphology, cellular function and maintenance, and carbohydrate metabolism (Supplementary Figure S6); and (ii) cancer, neurological disease, and organismal injury and abnormalities (Supplementary Figure S7) networks. Even though there was similarity between the 5Aza- and VitC-induced differentially expressed genes and DMRs, the differences in the enrichment of cellular functions and networks indicated the extent of DNA demethylation. This is likely due to differences at different targets as a result of the different mechanisms associated with DNA demethylation resulting from the administration of VitC or 5Aza. This, in turn, leads to differences in transcription and, therefore, distinct responses from cellular pathways.
Table 2. Pathways affected by differentially expressed genes (FDR ≤ 0.05, absolute fold change ≥ 2).
| Name | P-value | Overlap/no. of molecules |
|---|---|---|
| Top canonical pathways affected by VitC | ||
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 5.61E–06 | 9.8% 18/183 |
| Mitotic Roles of Polo-Like Kinase | 1.05E–04 | 13.6% 9/66 |
| Acute Phase Response Signaling | 3.73E–04 | 8.3% 14/169 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 4.59E–04 | 14.3% 7/49 |
| Salvage Pathways of Pyrimidine Ribonucleotides | 1.60E–03 | 9.5% 9/95 |
| Diseases and disorders affected by VitC | ||
| Cancer | 2.57E–04–3.31E–16 | 565 |
| Organismal Injury and Abnormalities | 2.62E–04–3.31E–16 | 582 |
| Reproductive System Disease | 2.24E–04–3.53E–16 | 333 |
| Gastrointestinal Disease | 2.62E–04–4.33E–13 | 499 |
| Dermatological Diseases and Conditions | 1.01E–04–9.62E–11 | 380 |
| Molecular and cellular functions affected by VitC | ||
| Cellular Assembly and Organization | 2.68E–04–2.88E–14 | 122 |
| DNA Replication, Recombination, and Repair | 2.68E–04–2.88E–14 | 41 |
| Cell Cycle | 2.14E–04–5.22E–13 | 104 |
| Cell Death and Survival | 2.62E–04–2.84E–12 | 223 |
| Cellular Movement | 2.32E–04–3.75E–12 | 172 |
| Top canonical pathways affected by 5Aza | ||
| Hepatic Fibrosis / Hepatic Stellate Cell Activation | 7.52E–15 | 16.4% 30/183 |
| Colorectal Cancer Metastasis Signaling | 8.45E–06 | 8.5% 21/247 |
| Acute Phase Response Signaling | 2.68E–05 | 9.5% 16/169 |
| Wnt/-catenin Signaling | 9.92E–05 | 8.9% 15/169 |
| MIF Regulation of Innate Immunity | 1.39E–04 | 17.1% 7/41 |
| Diseases and disorders affected by 5Aza | ||
| Cancer | 2.38E–05–2.97E–22 | 572 |
| Organismal Injury and Abnormalities | 2.38E–05–2.97E–22 | 585 |
| Dermatological Diseases and Conditions | 4.48E–08–2.42E–19 | 399 |
| Gastrointestinal Disease | 1.18E–05–2.84E–18 | 510 |
| Metabolic Disease | 1.68E–05–1.05E–12 | 139 |
| Molecular and cellular functions affected by 5Aza | ||
| Cellular Movement | 2.39E–05–5.26E–18 | 173 |
| Cellular Development | 2.30E–05–2.14E–10 | 219 |
| Cellular Growth and Proliferation | 2.30E–05–2.14E-10 | 206 |
| Cell Morphology | 1.94E-05–5.13E-10 | 176 |
| Lipid Metabolism | 2.18E–05–6.79E–10 | 88 |
Validation of transcriptional changes to the DMR-overlapping differentially expressed genes
To specifically investigate the association between the intragenic DMRs and the differentially expressed genes, we mapped the intragenic DMRs with the differentially expressed genes (FDR ≤ 0.05, absolute logFC ≥ 1). In all, 51 genes overlapped with VitC DMRs; and 27 genes overlapped with 5Aza DMRs. There were 18 genes common to both lists (Table 3). However, due to the limited number of genes, no cellular pathways were significantly enriched (P < 0.001).
Table 3. Differentially expressed genes that were identified to overlap with DMRs. Differentially expressed genes that were mapped with DMRs in each comparison are listed: common DMR-overlapping differentially expressed genes that were identified in both comparisons and uniquely identified DMR-overlapping differentially expressed genes in each comparison.
| Comparison | DMR-overlapping differentially expressed genes | |
|---|---|---|
| GBM+5Aza vs. GBM | Unique | SLCO4A1; PRICKLE2; MEIS3; MAN1C1; HSD17B14; GAS6; FNDC10; FBXO17; COL22A1; |
| Common | TSNARE1; TLE3; SPPL2B; SMARCD3; SEC61G; RAB7B; PRDM16; PLXNA4; PLEC; NACAD; MEST; LINC01224; FZR1; FCGBP; ELN; CLU; CCDC71L; BCL3; | |
| GBM+VitC vs. GBM | Unique | ADGRB1; AGAP3; AGPAT4; ASL; ATP6V0E2-AS1; C2CD4C; C7orf57; CLIP2; COL4A2; CYP4F11; FAM20C; FAM222A; FAM49A; GTF2IRD1; HSPG2; KIF17; KIF21B; LRRC4B; LTBP4; LYPLA1; MASP2; MYCNUT; MYT1; NKAIN4; PALM3; PDGFA; PRKAG2; PTPRS; SHC2; SNX10; ST3GAL1; TTYH1; WNK2; |
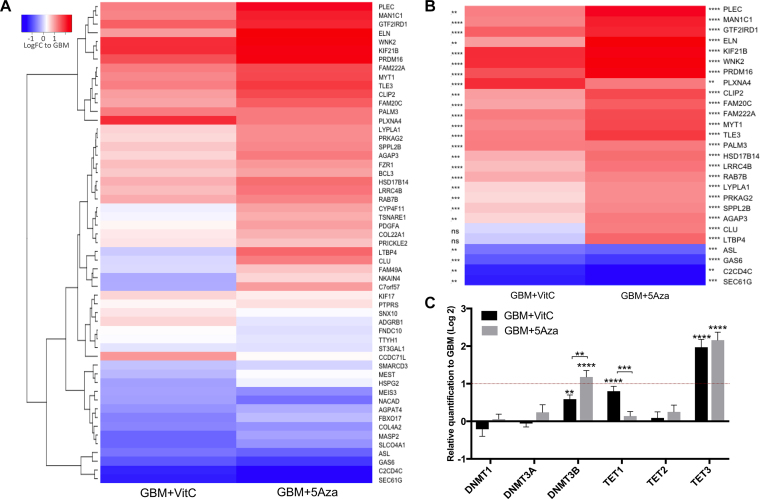
The 60 DMR-overlapping differentially expressed genes were further analysed using the Fluidigm platform to validate their changes in gene expression. Fold changes (log2) for each gene relative to the GBM cohort were used to generate the heatmap (Figure 3A; detailed results are shown in Supplementary Table S5). Genes with their levels of expression modulated by at least one of the treatments over absolute 2-fold are separately plotted in Figure 3B. The statistical significances are indicated on the sides of the plot. Overall, up-regulation in expression of these genes was observed in both treated cohorts with 5Aza triggering more significant changes than VitC (Figure 3B). MYT1, MAN1C1, PLEC, TLE3, PALM3, FAM222A, GTF2IRD1, PRDM16, WNK2, KIF21B and PLXNA4 were up-regulated by both treatments by over 2-fold, whilst SEC61G, C2CD4C and GAS6 were down-regulated by both treatments by over 2-fold, which indicates their expression levels are sensitively modulated by DNA demethylation. Interestingly, SEC61G encodes a subunit of the Sec61 complex as the central component of the protein translocation apparatus of the endoplasmic reticulum membrane. This gene is located upstream of another hallmark of GBM, EGFR, and is highly overexpressed in GBM to support tumour cell survival (61). WNK2 encodes the tumor suppressor WNK lysine deficient protein kinase 2, which is regulated by DNA methylation at its promoter region and loss of expression has been found in GBM as a result of promoter hypermethylation (62). PRDM16, PR domain containing 16, was found to be overexpressed when its promoter is hypomethylated and contributes to tumorigenesis by disrupting mitochondrial function in astrocytoma (63). Furthermore, 13 genes (ASL, LTBP4, CLU, PRKAG2, AGAP3, LYPLA1, SPPL2B, LRRC4B, HSD17B14, RAB7B, FAM20C, CLIP2 and ELN) were only significantly modulated by 5Aza to over 2-fold. They are likely to be more sensitive to the DNA demethylation mediated by the inhibition of DNMT1 activity resulting from the 5Aza treatment (Figure 3B).
Figure 3.
The validation of gene expression using the Fluidigm array. (A) Heatmap of the gene expression levels of the DMR-overlapping differentially expressed genes. For each gene, fold changes to the mean value of the respective gene in the GBM cohort (log2) were plotted. (B) Heatmap of the gene expression levels of the DMR-overlapping differentially expressed genes that had more than 2-fold changes after either of the treatments. Statistical significances are indicated at the sides of the plot relevant to each gene: the left side is for the GBM+VitC cohort and the right side is for the GBM + 5Aza cohort. **, ***, **** indicate P values < 0.01, 0.001, 0.0001, respectively; ns = not significant. (C) Differential expression of the regulators of DNA methylation. Bars represent the mean of the relative quantification levels in log2 scale normalized to the GBM cohort. Detailed statistical results of the DMR-overlapping differentially expressed genes are shown in Supplementary Table S5. Bars in black represent GBM + VitC; bars in grey represent GBM + 5Aza. Error bars show SEM. The level of significance for each gene is labelled above each bar, **, ***, **** indicate P values < 0.01, 0.001, 0.0001, respectively. The red hash line at 1 indicates 2-fold change of up-regulation.
As there were different patterns of DNA methylation and gene expression induced by 5Aza and VitC, we investigated the levels of gene expression for the regulators of DNA methylation including the DNA methylation enzymes, namely methyltransferases DNMT1/3A/3B/3L, and the DNA demethylation enzymes, namely TET1/2/3. It is known that 5Aza inhibits DNA methylation through the DNMT1 enzyme, whereas VitC acts as the co-factor of the TET1 enzyme to enhance DNA demethylation (42,43). TET-mediated DNA demethylation converts 5mC to 5hmC, which is important for the regulation of the tumour- and differentiation-related genes. The DNMT3 family is known to mediate de novo DNA methylation and is maintained by DNMT1 during DNA replication. The expression of DNMT3L, the co-factor of DNMT3A, was not detected in each of the three cohorts. TET3 was significantly up-regulated following treatment with VitC by >2-fold compared with the GBM cohort (Figure 3C), whereas 5Aza enhanced the expression of DNMT3B and TET3 by >2-fold (Figure 3C). When we compared the two DNA demethylation agents, treatment with 5Aza resulted in significantly higher levels of gene expression for DNMT3B than VitC (P < 0.01; Figure 3C). VitC triggered significantly higher levels of expression for TET1 than 5Aza (P < 0.001; Figure 3C). The up-regulation of DNMT3B is likely to be a response to global DNA demethylation in order to compensate for the reduced levels of DNA methylation induced by the DNA demethylation agents. Indeed, this is a typical feature, which is similar to the rebound effect when DNA demethylation agents are withdrawn from cancer cells and they return to their hypermethylated state (4,64). It is also interesting to note that the levels of expression for TET1 were up-regulated by its co-factor VitC, and not by 5Aza. The differences shown in the methylation factors further indicated that these two treatments could mediate different DNA demethylation machineries as different targets in the genome were modulated.
The changes in DNA methylation to the genomic regions of the mtDNA transcription and replication factors
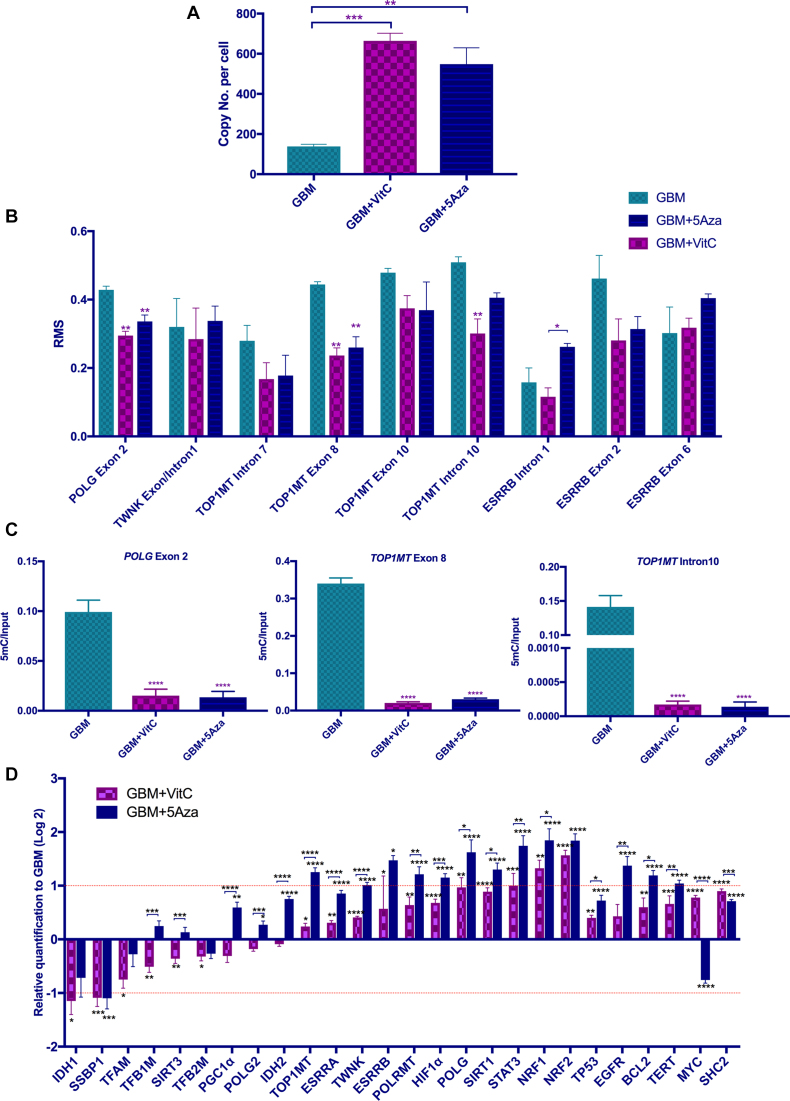
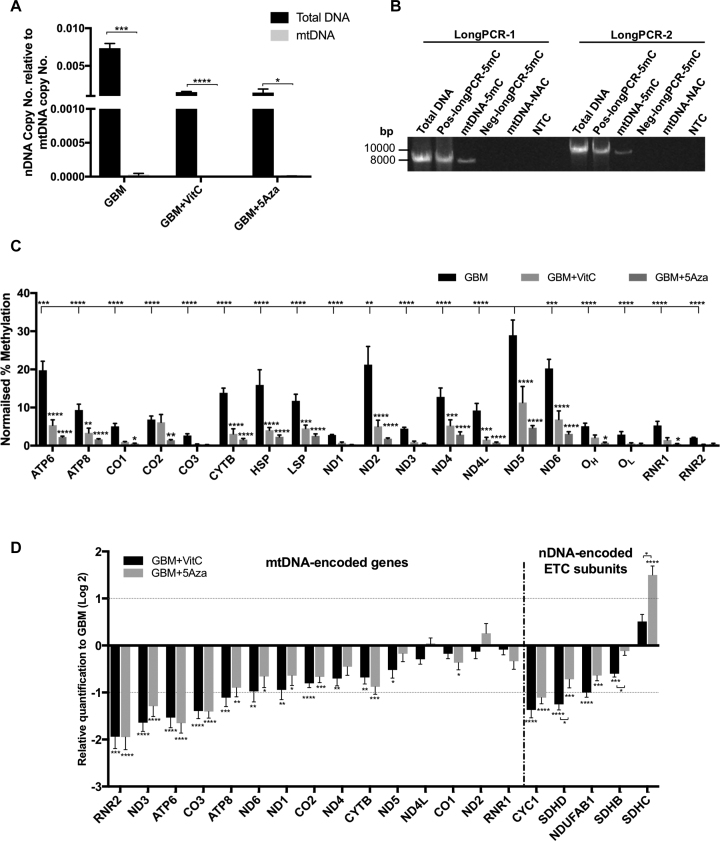
We specifically focused on investigating the changes in DNA methylation to the genomic regions of the mtDNA transcription and replication factors. Indeed, the rationale for undertaking this analysis is supported by the >3-fold increase in mtDNA copy number induced by the DNA demethylation treatments, which strongly indicates that mtDNA copy number is epigenetically regulated (Figure 4A) (4,32). Increased mtDNA copy number has also previously been shown to be tightly associated with the key regulator of mtDNA replication, POLG and more specifically associated with the DNA methylation status of a CpG island (CGI) in its second exon (4). Indeed, CGIs are genomic regions with repeated CpGs that usually exhibit low levels of DNA methylation. However, they are more tightly associated with regulatory roles in gene transcription than CpG dinucleotides, which are present throughout the whole genome (65).
Figure 4.
Analysis of mtDNA transcription and replication factor related regions. (A) mtDNA copy number for the three cohorts. mtDNA copy number per cell for the GBM, GBM + VitC and GBM + 5Aza cohorts (n = 3). (B) DNA methylation levels expressed as relative methylation score (RMS) over the CGIs located within the gene bodies of the mtDNA transcription and replication factors are shown (n = 3). The relative methylation score was determined using the MEDIPS package based on the MeDIP-Seq data. (C) Validation for DNA methylation using MeDIP-qPCR at exon 2 of POLG, and exon 8 and intron 10 of TOP1MT (n = 3). DNA methylation levels were determined by normalizing 5mC to the input (5mC/Input). (D) Validation for gene expression of the mtDNA transcription and replication factors (n = 6). Statistical analysis was performed using One-way ANOVA. Bars represent the mean ± SEM. * to **** indicate P values of < 0.05, 0.01, 0.001, 0.0001. The GBM cohort is shown by the light blue bars; the GBM + VitC cohort is shown by the purple bars, the GBM + 5Aza cohort is represented by the dark blue bars.
To further investigate changes in DNA methylation to the other mtDNA transcription and replication factors, the relative methylation score of CGIs found in the gene bodies and their promoter regions were analyzed using the MEDIPS package based on the MeDIP-Seq data. A total of 20 mtDNA transcription and replication factors were examined and had a CGI in the promoter regions of at least one transcript variant, which indicates their expression is highly likely to be affected by DNA methylation. Overall, reduced levels of DNA methylation were observed in the treated cohorts (Supplementary Figure S8). VitC significantly reduced the levels of DNA methylation in three promoter CGIs, TFB1M, TWNK and SSBP1 (Supplementary Figure S8).
The gene-body CGIs of the mtDNA transcription and replication factors were also analysed in the same way (Figure 4B). Firstly, the significant reduction in DNA methylation at exon 2 of POLG induced by both treatments was compatible with our previous findings (4). We also investigated the four CGIs that overlapped within intron 7, exon 8, exon 10 and intron 10 of TOP1MT, which would strongly indicate that the expression of TOP1MT is probably modulated by DNA methylation, as is the case for POLG (4,6). Both treatments caused a significant decrease in DNA methylation at exon 8 of TOP1MT, while the CGI at intron 10 was only significantly demethylated by VitC. CGIs at the boundary of exon 1 and intron 1 of TWNK and intron 1, exon 2/6 of ESRRB did not undergo significant DNA demethylation. Using MeDIP-qPCR, we confirmed that demethylation had taken place at exon 2 of POLG and exon 8 of TOP1MT through both 5Aza and VitC (Figure 4C). Interestingly, our validation also showed that the CGI at intron 10 of TOP1MT was also demethylated by both agents (Figure 4C).
These data show that the DNA methylation status at the promoter-located CGIs and the gene-body CGIs are modulated by the DNA demethylation treatments, which probably affect the gene expression profiles of these transcription and replication factors and their ability to regulate mtDNA copy number. To validate this, we determined the levels of expression of each of the transcription and replication factors. Firstly, we confirmed our previous finding that the expression of POLG was significantly up-regulated to ∼ 2-fold in both demethylated cohorts (Figure 4D). Along with POLG, the vast majority of the transcription and replication factors showed increased levels of expression after the DNA demethylation treatments. NRF1, NRF2 and STAT3 more than doubled their levels of expression after VitC treatment (Figure 4D) and even more so after 5Aza treatment. ESRRB, TWNK, TOP1MT, POLRMT, HIF1α and SIRT1 were significantly increased by <2-fold by VitC (Figure 4D). However, 5Aza increased expression for each of these genes to >2-fold (Figure 4D). SSBP1 was down-regulated in both cohorts, which indicates DNA demethylation treatments are likely to suppress its expression (Figure 4D). IDH1, involved in the TCA cycle and associated with TET-mediated DNA demethylation, was down-regulated by VitC by over 2-fold (Figure 4D). Interestingly, TFB1M, SIRT3, PGC1α and IDH2 had opposing results for the two treatments. Their levels of expression were decreased after VitC treatment but slightly up-regulated by 5Aza treatment (Figure 4D). From the comparisons between the two treated cohorts, 5Aza had significantly more effect on increasing the levels of gene expression, especially on the direct-binding transcription and replication factors, which included TFB1M, POLG2, TOP1MT, TWNK, POLRMT and POLG. The indirect transcription and replication factors, including SIRT3, PGC1α, IDH2, ESRRA, HIF1α, SIRT1, STAT3 and NRF1, were also more greatly affected by 5Aza than VitC (Figure 4D). Furthermore, we analysed other tumor markers of GBM including EGFR, BCL2, TP53, TERT, SHC2 and MYC that interact closely with the mtDNA transcription and replication factors (66–69). Similarly, overall up-regulation was observed after the treatments, but only 5Aza significantly up-regulated EGFR, BCL2 and TERT to ∼2-fold or above. However, 5Aza was found to repress MYC by over 1.5-fold (Figure 4D).
The relationship between mtDNA methylation and the expression of mtDNA-encoded genes
In the human mitochondrial genome (hg38), there are 435 CpG dinucleotides (70). Even though DNA methylation of mtDNA remains controversial, there is an increasing number of findings indicating that mtDNA methylation takes place, especially in the main non-coding region, the D-loop (70,71). Since the nuclear genome harbours mtDNA-pseudo genes, we performed MeDIP on purified populations of mtDNA to investigate the presence of mtDNA methylation.
Firstly, to determine the degree of nuclear DNA elimination from the purified mtDNA samples, we normalized the levels of nDNA (β-globin) to levels of mtDNA. The relative levels of nDNA in the purified mtDNA samples were significantly lower than in the total DNA samples isolated from cells that had not undergone mitochondrial isolation (Figure 5A). Indeed, significant reductions of nDNA (to nearly zero) in all three cohorts were achieved by the purification process. To determine whether mtDNA is DNA methylated, we tested purified mtDNA and long PCR fragments generated from the purified mtDNA, that had been treated with and without a CpG methylation specific enzyme. In this respect, we generated two overlapping fragments of mtDNA (∼8500 bp) by long PCR using primer set 1 (Supplementary Table S1) and treated these fragments with the CpG methylation enzyme, M.SssI to generate a positive control (Pos-longPCR-5mC) and without the enzyme to generate a negative control (Neg-longPCR-5mC), as PCR products alone do not maintain methylation. As can be seen from Figure 5B, the purified mtDNA sample (mtDNA-5mC) had weaker signals than the positive control (Pos-longPCR-5mC) and stronger signals than the negative control (Neg-longPCR-5mC). Additionally, no bands were observed for the non-antibody control for MeDIP (mtDNA-NAC), and the non-template control (NTC) indicating that the mitochondrial genome can undergo DNA methylation.
Figure 5.
Levels of DNA demethylation in the mitochondrial genome and expression levels of the mtDNA encoded genes. (A) Detection of nDNA contamination after mtDNA purification. Copy number for nDNA (β-globin) relative to the copy number for mtDNA was plotted. Multiple t-tests were performed between the purified mtDNA samples (gray bars) and corresponding total DNA samples (black bars) isolated from GBM cells. Bars represent the mean ± SEM (n = 3). (B) Agarose gel showing long-PCR products from purified mtDNA samples to demonstrate the presence of mtDNA methylation. Long PCR using two pairs of primers (primer set 2) spanning across the mitochondrial genome was performed on: (i) total DNA isolated from non-treated GBM cells (Total DNA); (ii) the positive control - MeDIP performed on the long PCR products previously generated with primer set 1 and treated with the M.SssI enzyme (Pos-longPCR-5mC); (iii) purified whole mtDNA having undergone MeDIP - (mtDNA-5mC); (iv) the negative control - MeDIP performed on the long PCR products previously generated with primer set 1 (Neg-longPCR-5mC); (v) the non-antibody control for MeDIP – MeDIP was performed on the purified mtDNA without using the 5mC antibody (mtDNA-NAC); and (vi) a non-template control (H2O) for PCR (NTC). Fragment sizes are indicated. (C) Normalized levels of DNA methylation within regions of the mitochondrial genome determined on purified mtDNA samples that had undergone MeDIP with sonication. The normalized levels of DNA methylation were determined by normalizing the MeDIP results (5mC/Input) against the positive and negative controls that were used to represent 100% and 0% of DNA methylation, respectively. Statistical significance was determined between the 5Aza and VitC treated cohorts and the GBM cohort and between ND5 and all the other genes by two-way ANOVA (n = 3). (D) Significant differential expression of the mitochondrial and chromosomal genes encoding the ETC subunits identified in the treated and non-treated cohorts using the Fluidigm array. Bars represent the mean of the relative quantification levels normalized to the GBM cohort (n = 6). *, **, ***, **** indicate P values of < 0.05, 0.01, 0.001, 0.0001, respectively.
Since the purified mtDNA sample had a weaker signal than the positive control, we performed the MeDIP assay (with sonication) on purified mtDNA samples and the Pos-longPCR-5mC and the Neg-longPCR-5mC controls to quantify the levels of DNA methylation at different regions of the mitochondrial genome. The levels of DNA methylation were determined by firstly normalizing the qPCR signals of the MeDIP products to their corresponding input samples (5mC/Input), and then weighting against the positive and negative controls that were used to represent 100% and 0% levels of DNA methylation, respectively (Figure 5C). We assessed the non-coding regions (OH, OL, HSP and LSP), 2 rRNAs and all 13 ETC subunits using region-specific primers (Figure 5C; Supplementary Table S1). In HSR-GBM1 cells, methylated mtDNA molecules represent ≤29% of the total population, and levels of DNA methylation in different regions varied. When comparing all regions with ND5, which was identified as having the highest level of DNA methylation, all the other regions were significantly lower (Figure 5C). Additionally, HSP and LSP were higher than the replication start sites OH and OL. DNA methylation levels at the two rRNAs were relatively low. Amongst the ETC subunits, ND5, ND6, ND2, ATP6 and CYTB had relatively higher levels of DNA methylation than the other ETC-encoding genes. Interestingly, following treatment with the DNA demethylation agents, 5Aza and VitC, there were significantly reduced levels of DNA methylation at most of the regions except for CO3, ND1, ND3, OL and RNR2 (Figure 5C). In contrast to the results observed for the nuclear genome, 5Aza induced more extensive DNA demethylation to the mitochondrial genome than VitC with CO1, CO2, OH and RNR1 being only demethylated by 5Aza.
As HSP and LSP are the transcription start sites of the heavy and light strands, we therefore investigated the levels of gene expression for the 13 mtDNA-encoded genes and 2 rRNAs (Figure 5D). In contrast to the overall up-regulation observed for nuclear gene expression, overall down-regulation was observed for the mitochondrial-related genes following induction of DNA demethylation. Through both agents, RNR2, ND3, ATP6 and CO3 were significantly down-regulated by ∼2-fold or more. 5Aza also triggered significant down-regulation in ATP8. However, there was no overall correlation between gene-specific DNA methylation and gene expression.
We also extended the scope to the chromosomal genes encoding the ETC subunits as they are also indicative of mitochondrial function and cellular metabolism. Through both agents, CYC1, encoding a subunit of complex III, had decreased levels of expression induced by both treatments, whereas SDHC, encoding a subunit of complex II, had increased levels of expression by >2-fold after 5Aza treatment (Figure 5D). Apart from this, the difference between these two agents reached significant levels in SDHB/C/D (encoding subunits of complex II) (Figure 5D). 5Aza promoted the expression of the tumor suppressors SDHC more than VitC, whereas VitC repressed the expression of SDHB/D.
DISCUSSION
The replication of mtDNA is strictly regulated during differentiation and tumorigenesis to ensure that cellular energy through OXPHOS is generated in a cell type-specific manner (5,6,32,72). Pluripotent and cancer cells have lower mtDNA copy number and, thus, primarily rely on glycolysis to promote their high rates of proliferation (4). HSR-GBM1 cells are stem-like cancer cells with restricted potential to differentiate and exhibit a hypermethylated genome (35). We have previously shown that the levels of DNA methylation at exon 2 of POLG are negatively correlated with mtDNA copy number in HSR-GBM1 cells to restrict their potential to complete differentiation (4). However, in the presence of DNA demethylation agents, such as 5Aza and VitC, global demethylation, including demethylation at exon 2 of POLG, promotes synchrony between mtDNA replication and chromosomal gene expression (4). Consequently, these cells are able to undergo differentiation in a manner similar to adult stem cells. Likewise, the partial and near complete depletion of mtDNA in cancer cells alters the levels of DNA methylation at exon 2 of POLG and the tumorigenic capacity of these cells (5).
Here, we have shown that the global DMRs and differentially expressed genes induced by DNA demethylation through 5Aza and VitC overlap greatly with associated cancer pathways, such as cell death, cell proliferation and glioblastoma, by altering the expression levels of tumorigenic-specific genes. Both up- and down-regulation were observed in the RNA-Seq datasets due to DNA demethylation, which probably not only resulted from the direct impact on the targeted genes but also from sequential responses. To this extent, we have shown that tumour markers that overlapped with DMRs included PRDM16 and WNK2, which were up-regulated by 5Aza and VitC, whereas the oncogene of GBM, SEC61G, was significantly down-regulated as a result of both treatments. SEC61G is located upstream of EGFR, which is a biomarker of GBM (61). Indeed, PRDM16, WNK2 and SEC61G were identified as DMR-overlapping differentially expressed genes that aligned with the pathway analysis to show the DNA demethylation treatments primarily target cell death, cell growth and cell differentiation in GBM.
In terms of the mtDNA-specific transcription and replication factors, the results firstly confirmed our previous finding that, by using DNA demethylation agents, DNA methylation levels decreased across the key regulatory region, exon 2, of POLG and, as a result, the expression of POLG was significantly increased. In addition, we have identified differentially methylated CGIs in the gene body of TOP1MT, especially at exon 8 and intron 10 by both treatments. The existence of several differentially methylated CGIs in TOP1MT strongly indicates the transcriptional control by DNA methylation on this gene. Indeed, TOP1MT was found to be significantly up-regulated by 5Aza to over 2-fold. However, TET-mediated DNA demethylation induced by VitC also increased its gene expression but not to significant levels.
We further investigated the impact of DNA demethylation on the other mtDNA transcription and replication factors. Overall, up-regulation of expression profiles was observed, which is likely to be the major contributor to the up-regulation of mtDNA copy number. The transcription and replication factors including NRF1/2 andSTAT3 were up-regulated by 5Aza and VitC to over 2-fold, whilst 5Aza had greater effect in promoting 10 of the transcription and replication factors to over 2-fold including the direct-binding mtDNA transcription and replication factors TWNK, TOP1MT, POLRMT and POLG. Interestingly, SSBP1 was down-regulated by both treatments. However, it is the gene that had the highest level of methylation for the promoter CGIs amongst the mtDNA transcription and replication factors. Therefore, there is likely to be other regulatory machinery offsetting the up-regulatory role of demethylation at the promoter region of SSBP1 and causing the down-regulation of SSBP1. Furthermore, TFB1M, SIRT3, PGC1α and SIRT3 were down-regulated by VitC but slightly up-regulated by 5Aza. Although the DNA demethylation levels that occurred over their promoter regions were similar following the two treatments, the opposite patterns presented for gene expression likely resulting again from other patterns of regulation, such as to the upstream regulators of TFB1M, which include the NRFs and PGC1α (24), rather than from DNA demethylation to TFB1M itself.
Additionally, IDH is tightly involved in mitochondrial function through the TCA cycle and considered to contribute to the hypermethylation of many types of cancers including GBM (33,37–39). In the TCA cycle, mutant IDH can alter the metabolic pathway to generate (R)-2-hydroxyglutarate, known as an oncometabolite, instead of α-ketoglutarate (33,37,40). This metabolic change disrupts the normal function of the TET enzymes, and results in the hypermethylated genomes of cancers (33,37,40). Our results showed that there was an impact of DNA demethylation on the expression levels of IDH1 and IDH2. IDH2 could be significantly up-regulated by 5Aza, while IDH1 was down-regulated only by VitC. IDH1 encodes the cytosolic IDH isoform, whereas IDH2 encodes the mitochondrial isoform (39). Moreover, our finding also suggests that up-regulation of the TET family after DNA demethylation treatments compensates for or modulates DNA methylation, which was likely contributed by the up-regulation of IDH2 in the mitochondria. Thus, our finding suggests that the induced DNA demethylation could interfere with the TCA cycle in the mitochondria, and the differences in changes to gene expression between IDH1 and IDH2 might indicate a response to the treatments according to their different sites of action.
Apart from the DMR-overlapping differentially expressed genes, we also analysed other tumor markers including BCL2, TP53, EGFR, SHC2, TERT and MYC that interact tightly with the mtDNA transcription and replication factors in GBM (66–69,73) (Figure 4D). Up-regulation occurred in nearly all of these factors after both treatments, amongst which EGFR, BCL2 and TERT were up-regulated by 5Aza by over 2-fold while 5Aza repressed the expression of MYC (>1.5 fold). As an oncogene and an important transcription factor for cell proliferation, the different consequences of the up- or down- regulation of MYC by VitC and 5Aza could lead to various patterns of downstream metabolic reactions in cancer cells (64). PGC1α, repressed by MYC in cancer cells, plays vital roles in regulating mitochondrial function and metabolic features of cancer stem cells (66,74). Our findings show that the levels of gene expression for PGC1α were up-regulated by 5Aza, but not by VitC. Moreover, MYC is also reported to have a direct role in regulating mitochondrial biogenesis by binding to the promoter regions of several key mtDNA transcription and replication factors including POLG and POLG2 (75). POLG was significantly up-regulated after both DNA demethylation treatments. These changes could result from DNA demethylation through their direct effect on their own specific gene expression, for example the CGI at exon 2 of POLG, or indirectly via their upstream regulators, for example MYC.
Even though it is thought that mtDNA is not as extensively methylated as the nuclear genome, there are methylated sites in mtDNA that have previously been reported with unknown functions associated with mtDNA transcription (59,76). Our results confirmed the existence of DNA methylation in mtDNA by using a MeDIP assay. In general, the level of mtDNA methylation in GBM cells was <29% relative to the fully methylated positive control (Figure 5C). Even though mtDNA methylation was not present at a high level as shown by us and others (59,76), DNA demethylation agents were able to take effect in the mitochondrial genome. DNMT and TET enzymes have been found in mitochondria (59,77), which provide explanations as to why VitC, the activator of TET, and 5Aza, the inhibitor of DNMT, were able to significantly reduce the levels of mtDNA methylation. The control region of mtDNA underwent significant demethylation by 5Aza and VitC. The control region contains the start sites of transcription for both strands (HSP and LSP) and the origin of replication for the heavy strand (OH), where the mtDNA transcription and replication factors bind to initiate transcription and replication of mtDNA (1). Significant DNA demethylation was observed at HSP and LSP induced by both agents. 5Aza also significantly demethylated the OH site. The methylation status of this region could potentially affect the accessibility of the transcription and replication factors, similar to events observed in the nuclear genome, where, for example, the methylation status of exon 2 of Polg is associated with the binding of RNA polymerase II (32). Other than the control region, induced DNA demethylation at most of the mitochondrial genes was observed in both treated cohorts except for CO3, ND1, ND3, OL and RNR2.
In contrast to the negative correlation observed between DNA demethylation and up-regulation in nuclear gene expression, the patterns of mitochondrial gene expression showed overall down-regulation after DNA demethylation. Indeed, the down-regulation of mtDNA expression induced by the DNA demethylation agents is intriguing given that there are increased numbers of mtDNA copy induced by the DNA demethylation agents that would provide more template for mtDNA transcription. Consequently, in the presence of more templates, DNA demethylation repressed the transcription of the primary polycistronic transcripts of mtDNA. RNR2, ND3, ATP6, CO3 and ATP8 were significantly down-regulated to around 2-fold or more by both treatments. In synchrony with this, the transcription factors TFAM and TFB1/2M were also down-regulated or remained at similar levels of expression compared to the up-regulation of the other replication factors. Therefore, one or more factors could mediate the overall down-regulation of mtDNA expression.
Apart from the overall down-regulation, the change in expression for each gene varies, which led us to determine whether there were gene-specific correlations between the levels of DNA demethylation and transcriptional changes. However, we did not observe any gene-specific correlations. Gene-specific changes to mitochondrial genes that are associated with DNA methylation have been reported in p53-knockout mouse embryonic fibroblasts (59) and diabetic retinopathy (78). However, no validated mechanism has been fully described. The gene-specific expression did not result from the direct effect of DNA methylation, as it probably could only affect transcription at the level of the polycistronic RNA, likely from the potential indirect effects of DNA methylation on the post-transcriptional process. The tRNA punctuation model has been widely proposed to be the primary machinery of post-transcriptional processing of the polycistronic transcripts in human mitochondria (79,80). CO3, ATP6 and ATP8 are located sequentially on the mitochondrial genome without a recurring tRNA in between, which likely explains why the changes to gene expression induced by DNA demethylation remained similar over the three genes after processing. Indeed, whether mtDNA methylation adds an epigenetic layer in regulating mtDNA transcription has not been fully understood.
Whilst it has been reported that induced CpG methylation by the mitochondrial targeted M.SssI enzyme decreased mtDNA copy number in human colon carcinoma HCT116 cells (76,81), we have observed increases in mtDNA copy number resulting from DNA demethylation induced by 5Aza and VitC but decreases in mtDNA transcription. We propose that mtDNA demethylation ensures that the mitochondrial genome is accessible for replication through fewer CpG sites being methylated. DNA demethylation induced by these agents also results in DNA demethylation of key mtDNA replication factors, such as POLG, and increases in their expression to further promote mtDNA replication. Even though mtDNA transcription was down-regulated, the RNA-DNA hybrid primer would appear to be available to initiate replication, as recently shown (76,81). Whether transcription is by-passed or specifically down regulated remains to be determined. However, DNA demethylation induced the two genomes to promote mtDNA replication in synchrony with altered patterns of gene expression of the nuclear genome, which would be indicative of a cell going through differentiation unlike mature cells that would likely have less replication and prioritize transcription to promote energy production (6,32). Indeed, it is worth pointing out that there are short waves of mtDNA replication which do not reflect the need for ATP-derived by OXPHOS, as, for example, when mouse embryonic stem cells differentiate into mature cells types (2). As with DNA demethylated GBM cells, these events could represent a checking mechanism to ensure synchrony between the nuclear and mitochondrial genomes in order that cell differentiation can proceed. Consequently, during differentiation, it appears that mtDNA replication can take place independently of mtDNA transcription.
The differences observed for DNA demethylation and gene expression induced by 5Aza and VitC provide insights into their different modes of action in inducing DNA demethylation. 5Aza is an analogue of cytosine that competitively binds to DNMT1 so that DNMT1 could not bind to cytosines to modify methylation (43), whereas VitC acts as the cofactor of TET1 to enhance its activity (42). The unique DMRs and different proportions of overlapping regions of the genome provide evidence of their different modes of actions. Based on the use of the same methylome provided by the HSR-GBM1 cells, VitC significantly demethylated more genomic regions, especially intragenic regions. However, there were DMRs uniquely regulated by both agents. There is limited knowledge as to whether these agents have specific targets or preference over certain genomic regions. We have shown that the induced DNA demethylation caused by these two treatments differentially regulated gene expression. Even though 5Aza induced fewer DMRs compared with VitC, it affected gene expression more effectively. Indeed, VitC has been shown to have a faster rebound effect than 5Aza after its withdrawal, which allows cells to return to their original states (4). It is likely that, as HSR-GBM1 cells have fast rates of cell division (4), the inhibition of DNMT1 by 5Aza prevented the DNA methylation patterns in the newly formed cells from being reestablished, and allowed the transcription factors to bind to the genome to promote gene expression. However, the TET enzymes, enhanced by VitC, were interacting extensively with the genome to carry out DNA demethylation but the occupancy of the TET enzymes on the genome might result in less accessibility of the transcription factors to promote gene expression. Additionally, we also found several interesting differences in the effects induced by 5Aza and VitC: (i) the changes to the expression of IDH1 and IDH2 (discussed above); (ii) the changes to the expression of the key regulator of mitochondrial function and tumorigenesis, MYC, were contrasting where VitC up-regulated MYC and 5Aza down-regulated MYC; and (iii) 5Aza induced greater DNA demethylation in mtDNA than VitC, but the changes to mtDNA expression were more significantly modulated by VitC, which was opposite to what was found in the nucleus.
In all, we have shown that global demethylation of the GBM genome induces changes to several key tumor markers, such as SEC61G, PRDM16 and WNK2, and that several key pathways including glioblastoma, cell death, cell growth and cell differentiation are also affected as a result. We further show that increases in mtDNA copy number coincided with increases in the expression of many of the mtDNA replication factors, two of which appear to be modulated by DNA demethylation, namely POLG and TOP1MT, that were identified from our mtDNA replication factor-specific screen. Furthermore, the expression of the tumour markers EGFR, BCL2, TERT and MYC, which interact with the mtDNA transcription and replication factors, were also modulated by the DNA demethylation agents suggesting an association between the key regulators of cellular fate and mtDNA replication. These interactions likely empower HSR-GBM1 cells to undergo differentiation as we have previously shown after culture with both 5Aza and VitC (4) and to break free from their pseudo-differentiated state where they are trapped between two states, an undifferentiated and a fully differentiated state (7,8). It would, thus, appear that synchrony between nuclear gene expression and the regulation of mtDNA copy number, and thus resetting of the mtDNA set point, are important to cellular differentiation and that hypermethylation in HSR-GBM1 cells impinges on this process.
DATA AVAILABILITY
MeDIP-Seq sequences in triplicates for HSR-GBM1 cells and the cells treated with 5Aza and VitC have been deposited in the Sequence Read Archive (SRA) under accession number SRP080899. CNV data and RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE98693.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Trevor Wilson and Dr Selva Kumari Ramasubramanian, The Medical Genomics Facility Monash Health Translation Precinct, for assistance in performing next generation sequencing and the Fluidigm high-throughput real-time PCR array, respectively. We are grateful to BioDiscovery, Inc. for the use of Nexus Copy Number software in our data analysis. We thank Dr Aidan Sudbury from the School of Mathematical Sciences, Monash University for assistance with statistical analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Hudson Institute of Medical Research Discretionary Funds; Victorian Government's Operational Infrastructure Support Program; Australian Postgraduate Award (to X.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. [DOI] [PubMed] [Google Scholar]
- 2. Facucho-Oliveira J.M., Alderson J., Splikings E.C., Egginton S., St. John J.C.. Mitochondria DNA replication during differentiation of murine embryonic stem cells. J. Cell Sci. 2007; 15:4025–4034. [DOI] [PubMed] [Google Scholar]
- 3. Facucho-Oliveira J.M., St John J.C.. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev. 2009; 5:140–158. [DOI] [PubMed] [Google Scholar]
- 4. Lee W., Johnson J., Gough D.J., Donoghue J., Cagnone G.L., Vaghjiani V., Brown K.A., Johns T.G., St John J.C.. Mitochondrial DNA copy number is regulated by DNA methylation and demethylation of POLGA in stem and cancer cells and their differentiated progeny. Cell Death Dis. 2015; 6:e1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee W.T., Cain J.E., Cuddihy A., Johnson J., Dickinson A., Yeung K.Y., Kumar B., Johns T.G., Watkins D.N., Spencer A. et al. Mitochondrial DNA plasticity is an essential inducer of tumorigenesis. Cell Death Discov. 2016; 2:16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dickinson A., Yeung K.Y., Donoghue J., Baker M.J., Kelly R.D., McKenzie M., Johns T.G., St John J.C.. The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death Differ. 2013; 20:1644–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee W.T., St John J.. The control of mitochondrial DNA replication during development and tumorigenesis. Ann. N. Y. Acad. Sci. 2015; 1350:95–106. [DOI] [PubMed] [Google Scholar]
- 8. Sun X., St John J.C.. The role of the mtDNA set point in differentiation, development and tumorigenesis. Biochem. J. 2016; 473:2955–2971. [DOI] [PubMed] [Google Scholar]
- 9. Xu B., Clayton D.A.. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. EMBO J. 1996; 15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 10. Xu B., Clayton D.A.. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol. Cell. Biol. 1995; 15:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tiranti V., Savoia A., Forti F., D’Apolito M.F., Centra M., Rocchi M., Zeviani M.. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 1997; 6:615–625. [DOI] [PubMed] [Google Scholar]
- 12. Fisher R.P., Clayton D.A.. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J. Biol. Chem. 1985; 260:11330–11338. [PubMed] [Google Scholar]
- 13. Fisher R.P., Clayton D.A.. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 1988; 8:3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M.. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002; 31:289–294. [DOI] [PubMed] [Google Scholar]
- 15. Hillen H.S., Parshin A.V., Agaronyan K., Morozov Y.I., Graber J.J., Chernev A., Schwinghammer K., Urlaub H., Anikin M., Cramer P. et al. Mechanism of transcription anti-termination in human mitochondria. Cell. 2017; 171:1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kasamatsu H., Grossman L.I., Robberson D.L., Watson R., Vinograd J.. The replication and structure of mitochondrial DNA in animal cells. Cold Spring Harb. Symp. Quant. Biol. 1974; 38:281–288. [DOI] [PubMed] [Google Scholar]
- 17. Clayton D.A. Replication of animal mitochondrial DNA. Cell. 1982; 28:693–705. [DOI] [PubMed] [Google Scholar]
- 18. Carrodeguas J.A., Theis K., Bogenhagen D.F., Kisker C.. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol. Cell. 2001; 7:43–54. [DOI] [PubMed] [Google Scholar]
- 19. Kaguni L.S., Olson M.W.. Mismatch-specific 3′—-5′ exonuclease associated with the mitochondrial DNA polymerase from Drosophila embryos. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:6469–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim S.E., Longley M.J., Copeland W.C.. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999; 274:38197–38203. [DOI] [PubMed] [Google Scholar]
- 21. Takamatsu C., Umeda S., Ohsato T., Ohno T., Abe Y., Fukuoh A., Shinagawa H., Hamasaki N., Kang D.. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002; 3:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korhonen J.A., Gaspari M., Falkenberg M.. TWINKLE Has 5′ → 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003; 278:48627–48632. [DOI] [PubMed] [Google Scholar]
- 23. Zhang H., Meng L.H., Pommier Y.. Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie. 2007; 89:474–481. [DOI] [PubMed] [Google Scholar]
- 24. Gleyzer N., Vercauteren K., Scarpulla R.C.. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005; 25:1354–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., Ciriolo M.R.. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010; 285:21590–21599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y.. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010; 5:e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weitzel J.M., Iwen K.A., Seitz H.J.. Regulation of mitochondrial biogenesis by thyroid hormone. Exp. Physiol. 2003; 88:121–128. [DOI] [PubMed] [Google Scholar]
- 28. Enriquez J.A., Fernandez-Silva P., Garrido-Perez N., Lopez-Perez M.J., Perez-Martos A., Montoya J.. Direct regulation of mitochondrial RNA synthesis by thyroid hormone. Mol. Cell. Biol. 1999; 19:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato I., Miyado M., Miwa Y., Sunohara M.. Expression of nuclear and mitochondrial thyroid hormone receptors in postnatal rat tongue muscle. Cells Tissues Organs. 2006; 183:195–205. [DOI] [PubMed] [Google Scholar]
- 30. Casas F., Rochard P., Rodier A., Cassar-Malek I., Marchal-Victorion S., Wiesner R.J., Cabello G., Wrutniak C.. A variant form of the nuclear triiodothyronine receptor c-ErbAalpha1 plays a direct role in regulation of mitochondrial RNA synthesis. Mol. Cell. Biol. 1999; 19:7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fernandez-Vizarra E., Enriquez J.A., Perez-Martos A., Montoya J., Fernandez-Silva P.. Mitochondrial gene expression is regulated at multiple levels and differentially in the heart and liver by thyroid hormones. Curr. Genet. 2008; 54:13–22. [DOI] [PubMed] [Google Scholar]
- 32. Kelly R.D., Mahmud A., McKenzie M., Trounce I.A., St John J.C.. Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A. Nucleic Acids Res. 2012; 40:10124–10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010; 18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W.M., Lu C., Ward P.S.. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012; 483:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A.. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004; 64:7011–7021. [DOI] [PubMed] [Google Scholar]
- 36. Hess K.R., Broglio K.R., Bondy M.L.. Adult glioma incidence trends in the United States, 1977–2000. Cancer. 2004; 101:2293–2299. [DOI] [PubMed] [Google Scholar]
- 37. Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W., Lu C., Ward P.S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012; 483:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009; 360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen A.L., Holmen S.L., Colman H.. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013; 13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haseeb A., Makki M.S., Haqqi T.M.. Modulation of ten-eleven translocation 1 (TET1), Isocitrate Dehydrogenase (IDH) expression, alpha-Ketoglutarate (alpha-KG), and DNA hydroxymethylation levels by interleukin-1beta in primary human chondrocytes. J. Biol. Chem. 2014; 289:6877–6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marie S.K., Shinjo S.M.. Metabolism and brain cancer. Clinics (Sao Paulo). 2011; 66(Suppl. 1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. et al. Vitamin[thinsp]C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013; 500:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones P.A., Taylor S.M.. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980; 20:85–93. [DOI] [PubMed] [Google Scholar]
- 44. Kaminskas E., Farrell A.T., Wang Y.C., Sridhara R., Pazdur R.. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005; 10:176–182. [DOI] [PubMed] [Google Scholar]
- 45. Kelly T.K., De Carvalho D.D., Jones P.A.. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010; 28:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.P., Hu B. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014; 15:447–459. [DOI] [PubMed] [Google Scholar]
- 47. Brunner A.L., Johnson D.S., Kim S.W., Valouev A., Reddy T.E., Neff N.F., Anton E., Medina C., Nguyen L., Chiao E. et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009; 19:1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D.. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005; 37:853–862. [DOI] [PubMed] [Google Scholar]
- 49. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chavez L., Jozefczuk J., Grimm C., Dietrich J., Timmermann B., Lehrach H., Herwig R., Adjaye J.. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010; 20:1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lienhard M., Grimm C., Morkel M., Herwig R., Chavez L.. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics. 2014; 30:284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 53. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liao Y., Smyth G.K., Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013; 41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McCarthy D.J., Chen Y., Smyth G.K.. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012; 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dvinge H., Bertone P.. HTqPCR: high-throughput analysis and visualization of quantitative real-time PCR data in R. Bioinformatics. 2009; 25:3325–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Valente V., Teixeira S.A., Neder L., Okamoto O.K., Oba-Shinjo S.M., Marie S.K., Scrideli C.A., Paco-Larson M.L., Carlotti C.G. Jr. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. Ann. Neurosci. 2014; 21:62–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shock L.S., Thakkar P.V., Peterson E.J., Moran R.G., Taylor S.M.. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3630–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robinson M.D., Stirzaker C., Statham A.L., Coolen M.W., Song J.Z., Nair S.S., Strbenac D., Speed T.P., Clark S.J.. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res. 2010; 20:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu Z., Zhou L., Killela P., Rasheed A.B., Di C., Poe W.E., McLendon R.E., Bigner D.D., Nicchitta C., Yan H.. Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res. 2009; 69:9105–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moniz S., Martinho O., Pinto F., Sousa B., Loureiro C., Oliveira M.J., Moita L.F., Honavar M., Pinheiro C., Pires M. et al. Loss of WNK2 expression by promoter gene methylation occurs in adult gliomas and triggers Rac1-mediated tumour cell invasiveness. Hum. Mol. Genet. 2013; 22:84–95. [DOI] [PubMed] [Google Scholar]
- 63. Lei Q., Liu X., Fu H., Sun Y., Wang L., Xu G., Wang W., Yu Z., Liu C., Li P. et al. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget. 2016; 7:5007–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deaton A.M., Bird A.. CpG islands and the regulation of transcription. Genes Dev. 2011; 25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morrish F., Hockenbery D.. MYC and mitochondrial biogenesis. Cold Spring Harb. Perspect. Med. 2014; 4:a014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sahin E., Colla S., Liesa M., Moslehi J., Muller F.L., Guo M., Cooper M., Kotton D., Fabian A.J., Walkey C. et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011; 470:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Autret A., Martin S.J.. Emerging role for members of the Bcl-2 family in mitochondrial morphogenesis. Mol. Cell. 2009; 36:355–363. [DOI] [PubMed] [Google Scholar]
- 69. Suen D.F., Norris K.L., Youle R.J.. Mitochondrial dynamics and apoptosis. Genes Dev. 2008; 22:1577–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu B., Du Q., Chen L., Fu G., Li S., Fu L., Zhang X., Ma C., Bin C.. CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 2016; 6:23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mposhi A., Van der Wijst M.G., Faber K.N., Rots M.G.. Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. (Landmark Ed). 2017; 22:1099–1113. [DOI] [PubMed] [Google Scholar]
- 72. Kelly R.D., Rodda A.E., Dickinson A., Mahmud A., Nefzger C.M., Lee W., Forsythe J.S., Polo J.M., Trounce I.A., McKenzie M. et al. Mitochondrial DNA haplotypes define gene expression patterns in pluripotent and differentiating embryonic stem cells. Stem Cells. 2013; 31:703–716. [DOI] [PubMed] [Google Scholar]
- 73. Taylor T.E., Furnari F.B., Cavenee W.K.. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr. Cancer Drug Targets. 2012; 12:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sancho P., Burgos-Ramos E., Tavera A., Kheir T., Jagust P., Schoenhals M., Barneda D., Sellers K., Campos-Olivas R., Graña O.. MYC/PGC-1 balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015; 22:590–605. [DOI] [PubMed] [Google Scholar]
- 75. Kim J., Lee J.H., Iyer V.R.. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008; 3:e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bellizzi D., D’Aquila P., Scafone T., Giordano M., Riso V., Riccio A., Passarino G.. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013; 20:537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dzitoyeva S., Chen H., Manev H.. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging. 2012; 33:2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mishra M., Kowluru R.A.. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2015; 56:5133–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. [DOI] [PubMed] [Google Scholar]
- 80. Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A. et al. The human mitochondrial transcriptome. Cell. 2011; 146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van der Wijst M.G., van Tilburg A.Y., Ruiters M.H., Rots M.G.. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 2017; 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MeDIP-Seq sequences in triplicates for HSR-GBM1 cells and the cells treated with 5Aza and VitC have been deposited in the Sequence Read Archive (SRA) under accession number SRP080899. CNV data and RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE98693.