Abstract
The Artemis nuclease and tyrosyl-DNA phosphodiesterase (TDP1) are each capable of resolving protruding 3′-phosphoglycolate (PG) termini of DNA double-strand breaks (DSBs). Consequently, both a knockout of Artemis and a knockout/knockdown of TDP1 rendered cells sensitive to the radiomimetic agent neocarzinostatin (NCS), which induces 3′-PG-terminated DSBs. Unexpectedly, however, a knockdown or knockout of TDP1 in Artemis-null cells did not confer any greater sensitivity than either deficiency alone, indicating a strict epistasis between TDP1 and Artemis. Moreover, a deficiency in Artemis, but not TDP1, resulted in a fraction of unrepaired DSBs, which were assessed as 53BP1 foci. Conversely, a deficiency in TDP1, but not Artemis, resulted in a dramatic increase in dicentric chromosomes following NCS treatment. An inhibitor of DNA-dependent protein kinase, a key regulator of the classical nonhomologous end joining (C-NHEJ) pathway sensitized cells to NCS, but eliminated the sensitizing effects of both TDP1 and Artemis deficiencies. These results suggest that TDP1 and Artemis perform different functions in the repair of terminally blocked DSBs by the C-NHEJ pathway, and that whereas an Artemis deficiency prevents end joining of some DSBs, a TDP1 deficiency tends to promote DSB mis-joining.
INTRODUCTION
Topoisomerase I (Top1) relaxes supercoiled DNA by inducing single-strand breaks (SSBs) in DNA by transiently linking itself covalently to the 3′ end of the DNA via its active site tyrosine (Y723) (1). However, certain DNA damaging agents, including Top1 poisons or intercalating agents, can lead to trapping of the 3′-tyrosyl-DNA covalent complexes, which prevents subsequent re-ligation and thereby undermines genomic integrity (1). Tyrosyl-DNA phosphodiesterase 1 (TDP1) resolves the 3′-tyrosyl-DNA covalent linkage and leaves a 3′-phosphate that can be subsequently processed by polynucleotide kinase/phosphatase (PNKP) to produce a hydroxyl terminus appropriate for ligation (2,3). In humans, a homozygous mutation in TDP1 (TDP1H493R) leads to an accumulation of residual unrepaired Top1-DNA lesions and is the molecular basis of the neurodegenerative disorder, spinocerebellar ataxia with axonal neuropathy (SCAN1) (4,5).
In addition to 3′-pTyr, TDP1 is biochemically competent in the processing of other 3′ end-blocking groups including 3′-phosphoglycolate (3′-PG) moieties formed in response to free radical-mediated DNA breaks (6–11). In extracts, processing of 3′-PG on DSB overhangs is completely dependent on TDP1 (8). Strangely, however, SCAN1 cells show neither hypersensitivity nor any deficit in the repair of DSBs induced by ionizing radiation (IR), expected to produce heterogeneous breaks including 3′-PG-ended DSBs (12,13). This argues for the presence of other enzymes functioning in parallel to TDP1 for the processing of 3′-PG DSBs.
Several candidate enzymes including apurinic/apyrimidinic endonuclease 1 (APE1) and the Artemis nuclease have been implicated in 3′-PG removal. However, although APE1 can process 3′-PG on blunt or recessed DSB ends, overhanging 3′-PGs are completely refractory to removal by APE1 (14). The Artemis nuclease is associated with the C-NHEJ pathway and is critical for hairpin opening during V(D)J recombination (15). In contrast to APE1, Artemis can effectively remove 3′-PG on overhanging DSB ends by endonucleolytic trimming in a DNA-dependent protein kinase catalytic subunit (DNA-PKcs)-, Ku- and ATP-dependent manner (16). Moreover, Artemis-deficient cells show increased sensitivity to IR as well as to neocarzinostatin (NCS) and bleomycin, radiomimetic agents that produce 3′-PG DSBs, and this sensitivity can be rescued by expressing wild-type, but not endonuclease-deficient (D165N) Artemis (16,17). Thus, Artemis, via its endonuclease function, is a likely candidate enzyme functioning in parallel to TDP1 for the repair of 3′-PG on DSB overhangs.
To investigate whether TDP1 and Artemis are alternative 3′-PG processing enzymes, clonogenic survival and DSB repair assays were performed in HCT116 cells doubly deficient in Artemis and TDP1, following treatment with radiation or radiomimetic drugs. These results demonstrated that TDP1 and Artemis function in the same pathway for the repair of 3′-PG-ended DSBs. In addition, TDP1 depletion did not lead to a DSB rejoining defect but caused DSB mis-joining partially via NHEJ. Artemis and TDP1 were epistatic with DNA-PK but not with ATM or PARP1. Taken together, these results strongly indicate a surprising epistatic interplay between Artemis and TDP1 for the repair of 3′-PG-ended DSBs and provide evidence for the involvement of TDP1 in DSB repair via C-NHEJ.
MATERIALS AND METHODS
Cell lines and reagents
HCT116 TDP1−/− cells, constructed in the laboratory of Dr. Yves Pommier, NIH, have been described (18). All cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (Atlanta Biologicals) and antibiotics (GIBCO) at 37°C in 5% CO2 atmosphere. Human Embryonic Kidney (HEK) 293 cells were obtained from American Type Culture Collection (ATCC). NU-7441 (aka KU-57788), KU-60019 and AZD-2287 were obtained from Selleckchem. Neocarzinostatin (NCS) was from Sigma or Nippon-Kayaku and Calicheamicin (CAL) was a gift of Wyeth Pharmaceuticals (now Pfizer).
HCT116 Artemis−/− cells
HCT116 Artemis−/− cells were constructed in the laboratory of Eric Hendrickson. rAAV-mediated gene targeting was performed using the rAAV-Artemis-Exon2-Neo virus to remove exon 2 of the Artemis gene. Positive correctly targeted first round clones (Artemisflox:NEO/+) were identified, the floxed neomycin selection cassette was removed using Cre recombinase followed by subjecting the Artemis−/+ sub-clone to a second round of targeting to obtain putatively Artemis-null clones. However, upon further examination it was determined that exon two of Artemis still remained. Consequently, a third round of gene targeting was then performed and two of the clones obtained, 18.1 and a Cre-treated sub-clone of 15.1 Cre1, were used for the subsequent characterizations described in this study. For details, see Supplementary materials.
Generation of Artemis and TDP1 doubly-deficient cells
TDP1 was knocked out in HCT116 Artemis−/− cells using CRISPR editing technique as described (18). Briefly, Artemis−/− cells were transfected with CRISPR editing reagents along with a vector harboring a cloned sequence targeting TDP1 exon 5 (GTTTAACTACTGCTTTGACGTGG) and a puromycin resistance gene flanked by homology arms upstream and downstream of the target site. Transfected cells were selected in 0.8 μg/ml puromycin for 4 days. Single cell clones were subsequently screened for TDP1 activity to obtain clones without any detectable TDP1 activity.
Knockdown of TDP1 and isolation of single-cell clones
shTDP1 lentiviral constructs (19) were transfected into HEK293T cells along with packaging plasmid psPAX2 and envelope plasmid pMD2.G using calcium chloride method. The viral supernatant was used to infect HCT116 WT and Art−/− target cells in the presence of 4 μg/ml polybrene. Puromycin-selected cells from each genotype were expanded under selection and cryogenic stocks were stored. Genotyping was performed by PCR amplifying the puromycin resistance gene. Single cell clones were harvested using dilution cloning, expanded under selection and were analyzed for knockdown efficiency using a TDP1 activity assay.
TDP1 knockouts in HEK293 and HEK293T cells
HEK293 and 293T cells were grown to 70% confluence and the medium was replaced with 1.5 ml OptiMem (GIBCO). A DNA mixture consisting of 12 μg hCAS9 (20), 2.4 μg pMaxGFP and 9.6 μg pUC19 targeting vector expressing a gRNA targeting bp 24880–24899 in exon 7 of the TDP1 genomic sequence (GCAAAGTTGGATATTGCGTT) from a U6 promoter (Supplementary Figure S1A) was prepared in 50 μl OptiMem, combined with a transfectant solution consisting of 60 μl Lipofectamine 2000 (ThermoFisher) and 50 μl OptiMem, incubated for 40 min at 22°C and then added to the medium in the dish. Cells were bathed in the transfection mixture for 4 h at 37°C on a rocker. The mixture was then replaced with 10 ml complete medium and the cells incubated for 16 h. Cells were harvested and single GFP+ cells were sorted into individual wells and expanded. DNA was isolated as above and clones with deletions/insertions at the target site in both alleles were identified by amplifying a 141-bp fragment encompassing the target using one Cy5-labeled (Cy5-AAATGACAATGCTTGAGGG) and one unlabeled primer (CCAGTAGATATGGATATTAGTGAG), and analyzing the products on a denaturing sequencing gel, with detection on a Typhoon imager. Extracts were prepared and screened for phosphodiesterase activity as above and a clone with no detectable activity (<0.1% of parental) was selected. RT-PCR (Supplementary Figure S1B) confirmed the presence of deletions in TDP1 mRNA for both the HEK293 TDP1−/− knockout (∼30 bases) and the HCT116 Artemis−/− • TDP1−/− double knockout (∼50 bases). However, attempts to generate a TDP1−/− derivative of patient-derived Artemis-deficient CJ179 fibroblasts using this method were unsuccessful.
Supplementary Table S1 summarizes the cell lines used and their derivations.
TDP1 activity assay
Cells (2 × 106) from each derivative cell line were collected using trypsinization and centrifuged at 1200 RPM for 5 min at room temperature (RT). The cell pellet was washed once in 1× PBS and treated with lysis buffer (10 mM HEPES at pH 7.8, 60 mM KCl, 1 mM EDTA, 0.5% NP-40) in the presence of 2 mM serine protease inhibitor phenylmethanesulfonyl fluoride (PMSF), 1 mM NaVO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin and 1 μg/ml pepstatin, vortexed until the pellet was disrupted, incubated on ice for 10 min and centrifuged at 13 000 RPM for 5 min at 4°C. The supernatant (‘cell extract’) was collected and serially diluted in dilution buffer (50 mM Tris at pH 8.0, 5 nM DTT, 100 mM NaCl, 5 mM EDTA, 10% glycerol, 500 μg/ml BSA). Serial dilutions of 1 μl of the extract were incubated with 100 attomoles of an 18-base 5′-Cy5 labeled 3′-phosphotyrosyl oligonucleotide with sequence TCCGTTGAAGCCTGCTTT (18Y) (Midland Certified Reagents Midland, TX, USA) in 1× reaction buffer (60 mM KOAc, 10 mM Mg(OAc)2, 50 mM triethanolamine-HOAc pH 7.5, 2 mM ATP, 1 mM DTT) in total 5 μl reaction volume and incubated at 37°C for 1 h, denatured at 95°C for 5 min and separated on 20% denaturing polyacrylamide sequencing gels by electrophoresis for around 4 h at 42 V/cm. Gels were then imaged on a Typhoon 9410 Variable Mode Imager (GE Healthcare) in Fluorescence Acquisition mode with a Cy5 Emission filter using a Red (633 nM) laser at PMT of 800 V and analyzed on ImageQuant 5.1 software.
Western blot analyses
For detection of TDP1, two million cells were fractionated using a nuclear/cytosol fractionation kit (BioVision #K266-25). Protein concentration estimation was performed using a BCA assay (Pierce). Laemmli buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.004% bromphenol blue, 0.125 M Tris–HCl pH 6.8) was added to the cytosolic and nuclear lysates and heated for 10 min at 95°C. 25 μg of the nuclear and 50 μg of the cytoplasmic lysates were separated on a 10% polyacrylamide gel at 120 V and transferred onto nitrocellulose membrane in transfer buffer at a constant current of 350 mA for 2 h. Membranes were blocked using 1% casein in PBS, probed with a mouse anti-TDP1 primary antibody (Abnova) at a 1:1000 dilution overnight at 4°C, then washed thrice with 1× TBST for 10 min each and incubated in peroxidase-conjugated goat anti-mouse secondary antibody at 1:2500 for 1 h at 22°C. Membranes were then washed thrice with 1× TBST for 10 min each and developed with ECL Super Signaling substrate. For the detection of Artemis, 100 μg of whole cell extract from each cell line tested was separated on a 4–20% Criterion gel (BioRad) and transferred to an Immobilon (Millipore) membrane at 100 V for 1 h. The resulting blot was incubated with anti-Artemis antibody at 1:200 overnight at 4°C before being exposed to a goat-anti- rabbit (IRDye800CW) secondary antibody at 1:15000. Imaging was performed using a Li-Cor Odyssey imaging system.
Clonogenic survival assays
Cells were seeded at densities ranging from 300 to 10 000 in 6 cm dishes and incubated for 12 h to allow attachment. Cells were then treated either with NCS (stock concentration 37 μM diluted to 2 μM in 20 mM sodium citrate buffer, pH 4.0) at concentrations ranging from 0.25 to 2 nM for 6 h or they were treated with CAL (stock concentration 20 μM diluted to 1 μM in 50% ethanol, further diluted in PBS to obtain a final working concentration of 1.2 nM) at concentrations ranging from 0.3 to 2.4 pM for 24 h. Following treatment, cells were incubated in fresh medium for 9–12 days to form colonies. Colonies were fixed with 100% methanol for 10 min (for HEK293 and HEK293T cells, colonies were fixed with formaldehyde solution, Sigma - #25249), stained with 0.5% crystal violet in 20% methanol for 10 min, washed under tap water, air dried and counted manually. Plating efficiency (PE) was calculated as the number of colonies formed/number of cells seeded × 100% for each dose. Surviving fraction (SF) was calculated as PE of treated/PE of control × 100%. Dose enhancement factor (DEF) was calculated as IC90 of control/IC90 of the mutant cell line. For experiments using IR, cells were irradiated using a MDS Nordion Gammacell 40 research irradiator (ON, Canada), with a 137Cs source. For experiments with KU-60019, NU-7441 and AZD-2287, the respective inhibitor was added 1 h prior to NCS treatment and left in the medium during and 24 h after NCS treatment.
Immunofluorescence
Twenty-five thousand cells were seeded in four-well chamber slides (Nunc Lab Tek) and incubated overnight. Cells were then serum-starved by incubating in 0.5% FBS/RPMI for 72 h. Cells were then treated with 4 nM NCS for 1 h and fixed at different time points using ice-cold 4% paraformaldehyde (PFA) in 1× PBS for 10 min. Cells were permeabilized in 0.5% Triton X-100/PBS for 10 min and blocked in 1× PBS 1% Casein blocker (Bio-Rad, 1610783) for 1 h at 22°C. Primary antibodies (mouse monoclonal anti-53BP1 at 1:1000 (BD Pharmingen) and mouse anti-TDP1 (Abnova) at 1:100 were added and incubated overnight at 4°C. Slides were washed four times with PBS for 15 min each and incubated with secondary goat anti-mouse CFL594 antibody at 1:1000 (sc-362277) for 2 h at 22°C. Slides were washed 4 times with PBS for 15 min each and post-fixed using ice-cold 4% PFA for 10 min. Nuclei were counterstained with Vectashield mounting medium containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, H-1200). Confocal images were obtained with the Zeiss LSM700 Confocal Laser Scanning Microscope equipped with a 63×, 1.4 NA oil immersion objective, located in the Virginia Commonwealth University Microscopy Core Facility using a 405 nm laser (DAPI) and a 555 nm laser (CFL594).
Cell cycle analysis by flow cytometry
Cells were harvested and fixed using ice-cold 70% ethanol for minimum 1 h. Cells were centrifuged at 2000 RPM for 5 min and washed with PBS. The pellet was dissolved in propidium iodide (PI) solution (3.8 mM sodium citrate, 0.05 mg/ml PI, 0.1% Triton X-100) in the presence of 10 μg/ml RNase A (Sigma) and stored in dark for 30 min. Cell cycle analysis was performed using Becton Dickinson (San Jose, CA, USA) FACS Canto II flow cytometer and ten thousand events were recorded for each sample.
Centromere-fluorescence in situ hybridization
Cells were treated with 1 mM caffeine to abrogate the G2/M block, just before treating with 2 nM NCS for 6 h. 1 μg/ml colchicine was added to the medium 2 h before harvesting. Metaphase spreads were prepared using standard procedures (8). For centromere labelling, slides were denatured in 70% formamide/2× SSC at 72°C for 2 min and then hybridized to 20 μl of 200 nM Cy3-labeled PNA CENP-B probe (PNA Bio – F3002) in hybridization buffer (20 mM Tris, pH 7.4, 60% formamide, 0.1 μg/ml salmon sperm DNA) for 2 h at 37°C in dark. Chromosomes were counterstained with DAPI and metaphases were imaged using Zeiss LSM700 Confocal Laser Scanning Microscope as mentioned above. For experiments using NU-7441, the inhibitor was added 1 h prior to NCS treatment.
RESULTS
TDP1 promotes cell survival following 3′-PG DSB formation
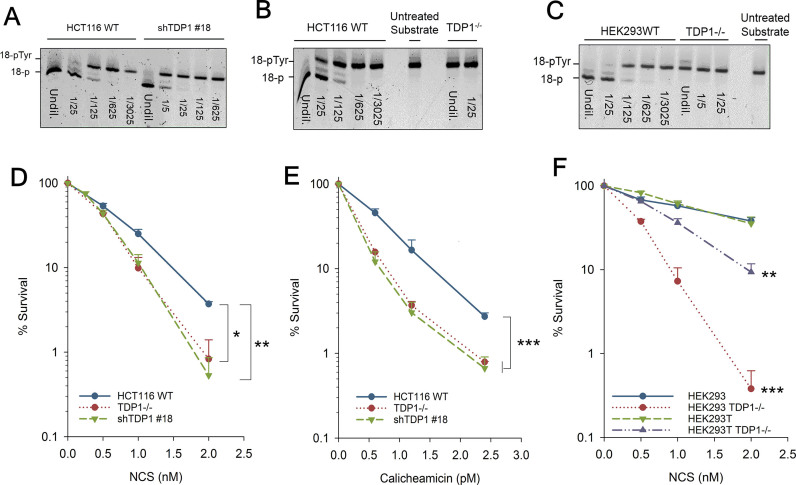
In vitro studies performed using whole cell extracts from TDP1-mutant SCAN1 cells showed that TDP1 is critical for processing the 3′-PG termini from DSB overhangs (7). To investigate the biological significance of this function of TDP1, shRNA-mediated knockdown of TDP1 was performed in HCT116 human colorectal adenocarcinoma cells. Stable single-cell clones were screened for maximum knockdown efficiency using a biochemical gel-based TDP1 activity assay, capable of detecting as little as 0.1% TDP1 activity. Due to the high specificity of TDP1 towards 3′-phosphotyrosyl oligonucleotides, TDP1 catalytic activity was measured in cell extracts by the extent of the conversion of a 5′-Cy5-labeled 18-base oligonucleotide bearing a 3′-phosphotyrosine (18Y) substrate to a 3′-phosphate (18P) product migrating with an increased electrophoretic mobility in a polyacrylamide gel (21). Clone #18 (shTDP1#18) showed maximum (94%) knockdown efficiency with just 6% residual TDP1 expression (Figure 1A). TDP1 assay performed in HCT116 TDP1−/− cells demonstrated a complete lack of conversion of 18Y to 18P (Figure 1B).
Figure 1.
TDP1 promotes cell survival following 3′-PG DSB formation. (A–C) The 3′-phosphotyrosyl oligonucleotide was incubated with serially-diluted whole cell extracts of HCT116 WT and shTDP1 (A) HCT116 WT and TDP1−/− (B) or HEK293 TDP1+/+ and TDP1−/− cells for 1 h. Processed (18-P) and unprocessed (18-pTyr) forms of the substrate are indicated. First lane for every sample is undiluted extract (Undil.). Substrate incubated in reaction buffer instead of cellular extract is represented as untreated substrate. (D, E, F) Clonogenic survival assays were performed on HCT116 (D, E) and HEK (293,293T) (F) cells treated with NCS (D, F) and Calicheamicin (E). Error bars represent SEM over at least three independent experiments for all except TDP1−/− HCT116 where n = 2. Data were analyzed using two-way ANOVA. *P< 0.05, **P< 0.005, ***P< 0.001.
Clonogenic survival assays were then performed in HCT116 WT and TDP1-knockdown cells using NCS and CAL, which are enediyne antitumor antibiotics that produce bi-stranded lesions, a substantial portion of which bear 3′-PG termini. TDP1-deficient cells showed significant hypersensitivity to both NCS (Figure 1D) and CAL (Figure 1E) with a DEF of 1.6× and 2× respectively, as compared to the parental cells.
HCT116 cells show reduced Mre11 expression (22). Mre11 functions in an alternative, TDP1-independent pathway for the repair of Top I-induced breaks (23). Thus, to investigate whether the hypersensitivity seen in TDP1-deficient HCT116 cells was specifically a function of TDP1 deficiency and not due to parallel TDP1-dependent and Mre11-dependent pathways being disrupted, TDP1 was knocked-out in HEK293 cells, which express normal levels of Mre11 (24). Initial attempts at CRISPR/Cas9-mediated knockout of TDP1 targeting the initiation codon resulted in several clones with homozygous deletions that nevertheless harbored a low (1–2% of parental) level of tyrosyl-DNA phosphodiesterase activity, suggesting a possible alternatively spliced or translated enzyme. Therefore, the TDP1 active site in exon 7 was targeted instead. Clones with deletions/insertions in both alleles were identified and whole-cell extracts were screened for phosphodiesterase activity. One TDP1−/− clone produced extracts that completely failed to hydrolyze the 18Y substrate to 18P product (<0.1% of WT activity, Figure 1C). Immunoblotting for TDP1 showed a clear absence of TDP1 protein in nuclear extracts of TDP1−/− compared to TDP1+/+ parental cells (Supplementary Figure S2A). In addition, immuno-labeling experiments demonstrated an absence of TDP1 expression in TDP1−/−, but not TDP1+/+, cells (Supplementary Figure S2B). Importantly, TDP1−/− derivatives of both HEK293 and HEK293T cells showed significant hypersensitivity to NCS compared to parental HEK293 and HEK293T TDP1+/+ cells (Figure 1F) as observed from clonogenic survival assays. In summary, these experiments demonstrated that TDP1 is not an essential gene, but that its absence results in significant cellular sensitivities consistent with observations from other laboratories (18,25).
TDP1 and Artemis are epistatic for the repair of 3′-PG DSBs via NHEJ
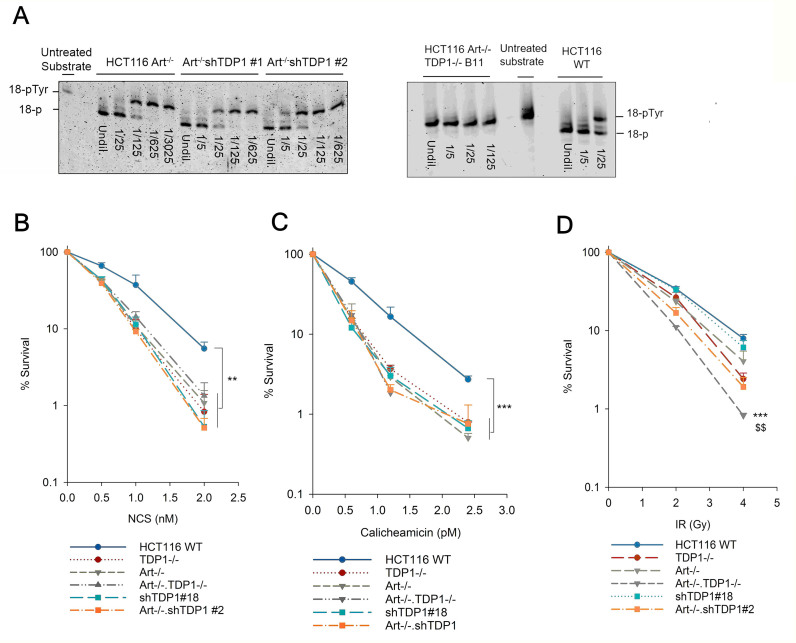
The Artemis nuclease functions in the NHEJ pathway of DSB repair and is biochemically competent in resolving 3′-PG termini by end trimming (16). Thus, to investigate whether Artemis and TDP1 are alternative end processing enzymes functioning in the same pathway for the resolution of 3′-PG termini, HCT116 Artemis-knockout cells were created (see Supplementary Materials and Methods; Supplementary Figure S3). These cells grew somewhat slowly (Supplementary Figure S4) and showed a mild increase in gene targeting efficiency (Supplementary Figure S5). More importantly, these cells showed the expected severe defect in V(D)J recombination coding junction formation but were only mildly affected for signal junction formation (Table 1, Supplementary Figure S6). Further, coding joint formation was rescued by targeted restoration of exon 2 to the knockout (Artemis−/KI cells, Table 1 and Supplementary Figure S6), confirming that the deficiency was due to loss of Artemis. To augment these cells, TDP1 was subsequently knocked out in Artemis−/− cells to generate Artemis−/−•TDP1−/− double knockout (DKO) mutants. Genotyping, performed by PCR amplification of exon 2 of the Artemis gene, showed that the Artemis−/− and Artemis−/−•TDP1−/− mutants had a larger amplicon than Artemis-proficient cells due to the insertion of the homology arms in between exons 1 and 3 (Supplementary Figure S7). A TDP1 assay demonstrated a complete lack of TDP1 activity in these cells (Figure 2A). In addition, TDP1 was also knocked down in Artemis−/− cells to create a double-mutant Artemis−/−•shTDP1 cell line. Single cell clones were isolated and a TDP1 activity assay performed on these clones identified clone #2 as displaying maximum knockdown efficiency with a loss of around 92% activity (Figure 2A and Supplementary Figure S8A). Consistent with a role for TDP1 in the repair of Top 1-mediated DNA lesions, Artemis−/−•shTDP1#2 double mutant cells were more sensitive to the Top 1 poison, camptothecin (CPT) than Artemis−/− cells (Supplementary Figure S8B).
Table 1.
V(D)J Recombination in HCT116 Artemis knockout cellsa
| PGG49 (SJ substrate) | PGG51 (CJ substrate) | |||||
|---|---|---|---|---|---|---|
| Cell Lines | DAC/DA | Signal joints (%) | Relative efficiency (%) | DAC/DA | Coding joints (%) | Relative efficiency (%) |
| HCT116 WT | 860/16700 | 0.51 | 100 | 274/77500 | 0.35 | 100 |
| 2018/449000 | 0.45 | 1263/447000 | 0.28 | |||
| 141/19000 | 0.74 | 273/38000 | 0.72 | |||
| 18.1 Artemis P/– | 604/231750 | 0.26 | 37 | 7/172500 | 0.0041 | 3.5 |
| 828/419750 | 0.2 | 21/540750 | 0.0039 | |||
| 32/18500 | 0.17 | 5/12500 | 0.04 | |||
| 15.1 Artemis –/– | 125/43500 | 0.29 | 53 | 8/170750 | 0.0047 | 0.6 |
| 309/159750 | 0.19 | 10/361000 | 0.0028 | |||
| 27/6500 | 0.42 | 0/6250 | 0 | |||
| 12–20 Artemis –/KI | 792/223500 | 0.35 | 65 | 34/26750 | 0.13 | 41 |
| 1150/295500 | 0.39 | 364/236500 | 0.15 | |||
| 35/9500 | 0.37 | 48/17250 | 0.28 | |||
aThe results of three separate experiments for parental HCT116 as well as for Artemis−/− (#18.1 and #15.1) are shown. DAC: number of ampicillin–chloramphenicol-resistant E. coli transformants after DpnI treatment; DA: number of ampicillin-resistant transformants after DpnI treatment. Transient V(D)J recombination assays were performed in the presence of RAG1 and RAG2 expression vectors (see Supplementary Figure S6 legend for details). Artemis P/–: Artemis knockout with a Puro gene in one allele, Artemis –/KI: Artemis knockout complemented by reintroduction of exon 2 (see Supplementary Figure S3B and C).
Figure 2.
TDP1 and Artemis are epistatic for the repair of radiomimetic double-strand breaks. (A) The 3′-phosphotyrosyl oligonucleotide was incubated with serially-diluted whole cell extracts of Art–/– cells with TDP1 knockdown (left panel) or TDP1 knockout (right panel) for 1 hour. Processed (18-P) and unprocessed (18-pTyr) forms of the substrate are indicated. (B–D) Clonogenic survival assays were performed on isogenic HCT116 WT, TDP1-deficient, Artemis-deficient, and TDP1/Artemis double-deficient cells treated with NCS (B), Calicheamicin (C) and ionizing radiation (D). Error bars represent SEM. n = 2 for TDP1–/– and Art–/–•TDP1–/– cells whereas n = 3 for WT, shTDP1, Art–/–, Art–/–•shTDP1. For (B), n = 5 for WT, shTDP1, Art–/–, Art–/–•shTDP1. Data were analyzed using two-way ANOVA. **P< 0.005, ***P< 0.001. In (D), *** indicates Art–/–•TDP1–/– comparison with WT, $$ indicates Art–/–•TDP1–/– comparison with Art–/– and TDP1–/– single mutants implying that the double mutants are more sensitive than the single mutants and WT (P< 0.005).
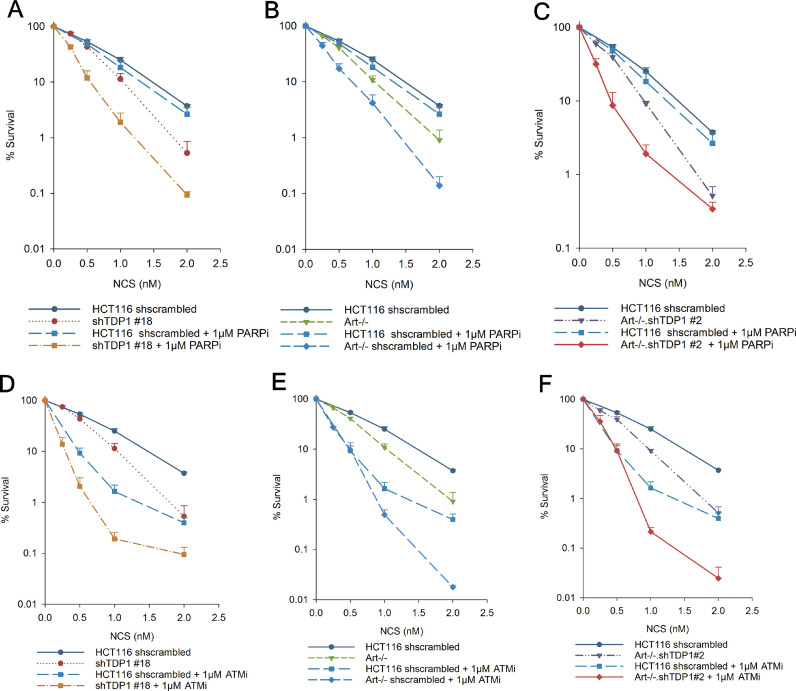
Clonogenic survival assays performed using NCS, CAL and IR showed that Artemis−/− cells were, as expected, hypersensitive to NCS and CAL (17) with a DEF of 1.5 (NCS) and 2 (CAL) compared to the parental Artemis+/+ cells. Surprisingly, however, an additional TDP1 deficiency in Artemis−/− cells (Artemis−/−•TDP1−/− or Artemis−/−•shTDP1#2) did not enhance the hypersensitivity to NCS and to CAL, indicating that TDP1 and Artemis are epistatic and function in the same pathway for the repair of NCS/CAL-induced DSBs (Figure 2B and C, respectively). In all cases, shTDP1 knockdown was as effective as TDP1 knockout in conferring NCS sensitivity. Thus, shTDP1 cells (which were used for most of the experiments described below, having been generated much earlier in the study) fully express TDP1 deficiency in terms of repair of NCS-induced damage. In contrast, the double mutants were more sensitive to IR than the single mutants (Figure 2D) suggesting that TDP1 and Artemis function in parallel for the repair of IR-induced DSBs.
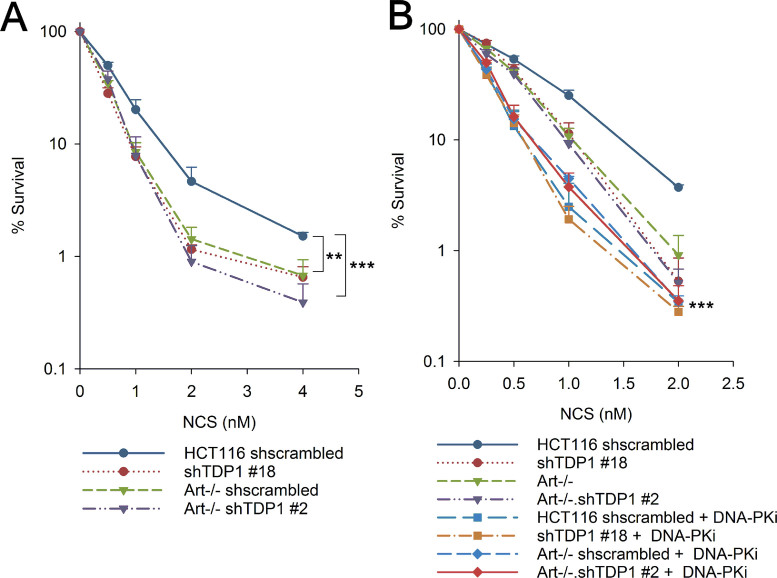
NHEJ is the pathway of choice for DSB repair in G1 phase (26). To investigate whether TDP1 and Artemis are also epistatic in the G1 phase for the repair of 3′-PG DSBs via NHEJ, clonogenic survival assays were performed using NCS on cells synchronized in G1 by mitotic shake-off. Approximately 80% cells were synchronized in G1 phase as determined by cell cycle analysis using PI staining (Supplementary Figure S9). Similar to the observation in exponentially growing cells, the hypersensitivity to NCS observed in G1-synchronized Artemis−/−•shTDP1#2 double-mutants was similar to the Artemis−/− and shTDP1#18 single-mutants (Figure 3A), suggesting that Artemis and TDP1 are epistatic for the repair of NCS-induced 3′-PG DSBs in G1 phase.
Figure 3.
Epistatic interplay between Artemis and TDP1 in G1-phase via canonical NHEJ. (A, B) Clonogenic survival assays were performed using NCS on G1-synchronized cells (A) or exponential cells in the presence of 1 μM DNA-PK inhibitor, NU-7441 (DNA-PKi) (B). Inhibitor was added 1 h prior to NCS treatment and left in the medium during and 24 h after NCS treatment. shTDP1, Art–/– and Art–/–•shTDP1 curves are the same as in Figure 2B. Error bars represent SEM for n = 4. Data were analyzed using two-way ANOVA, ***P< 0.001, **P< 0.005. For (B), *** represents significant statistical difference between all DNA-PK inhibitor treated cells versus all cells without DNA-PK inhibitor treatment.
DNA-PK is critical for the repair of DSBs via NHEJ. In order to investigate whether Artemis and TDP1 repair 3′-PG DSBs in G1 phase via NHEJ, clonogenic survival assays were performed using NCS in the presence of a DNA-PK inhibitor, NU-7441. WT cells showed an increased hypersensitivity to NCS upon DNA-PK inhibition whereas additional depletion/knockout of TDP1 (shTDP1#18), deficiency of Artemis (Artemis−/−) or both combined (Artemis−/−•shTDP1#2, Artemis−/−•TDP1−/−) did not further enhance this sensitivity (Figure 3B and Supplementary Figure S10). This result strongly suggests that Artemis and TDP1 are epistatic with DNA-PK and contribute to the repair of 3′-PG DSBs via the NHEJ pathway.
Absence of TDP1 does not confer a defect in DSB rejoining but causes NHEJ-dependent DSB mis-joining
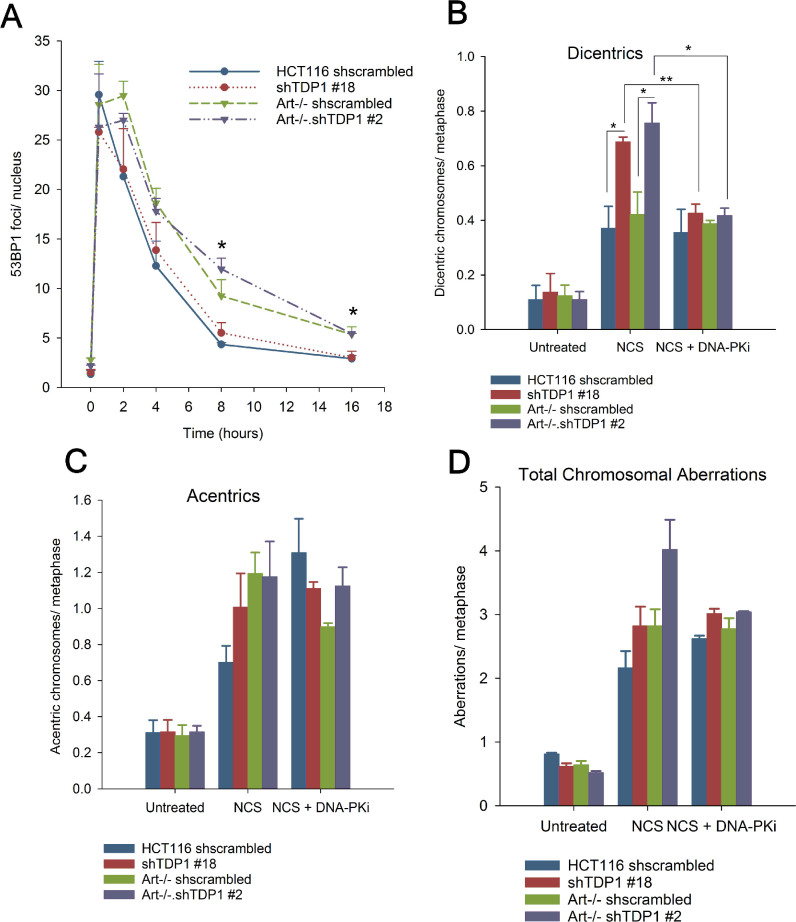
To directly assess whether the hypersensitivity shown by the mutants to NCS and CAL was due to a deficiency in repairing 3′-PG-ended DSBs, 53BP1 foci were quantified as representative markers of DSBs in these cells following NCS treatment. This assay was performed on serum-deprived G0/G1-phase cells to specifically analyze DSB repair in the context of NHEJ and to avoid including spontaneous focus formation at stalled replication forks. As expected (17), Artemis−/− cells showed an increased persistence and a delayed disappearance of 53BP1 foci, with a significant fraction of foci persisting even at 8 and 16 hr after NCS treatment compared to Artemis-proficient cells (Figure 4A and Supplementary Figure S11), suggesting a defect in DSB rejoining in the absence of Artemis. Surprisingly, depletion of TDP1 had no effect on the kinetics of 53BP1 resolution in either WT or Artemis−/− cells, indicating that TDP1 depletion does not confer a defect in DSB rejoining.
Figure 4.
Depletion of TDP1 does not cause a DSB rejoining defect but leads to DSB mis-joining. (A) 53BP1 foci were scored in serum-starved cells treated with 4 nM NCS for 1 h. (B–D) Cells were treated with 2 nM NCS for 6 h, and arrested in colchicine for 2 h. Metaphase spreads were hybridized with a centromeric probe. A total of 40–45 metaphases from 3 independent experiments were imaged, and dicentric chromosomes (B), acentric fragments (C) and total aberrations per metaphase were scored. Total aberrations include acentrics, dicentrics, breaks, gaps, radials and unstructured chromosomal regions. 1 μM NU-7441 was added 1 h prior to NCS treatment and left in the medium during the treatment. Error bars represent SEM. n = 2 for (B) with DNA-PKi, n = 3 for all other experiments. Data were analyzed using Students unpaired t-test. For (A), * indicates comparison between Artemis-proficient (HCT116 WT and shTDP1) and Artemis-deficient (Art–/– and Art–/–•shTDP1) cells.*P < 0.05, **P < 0.005
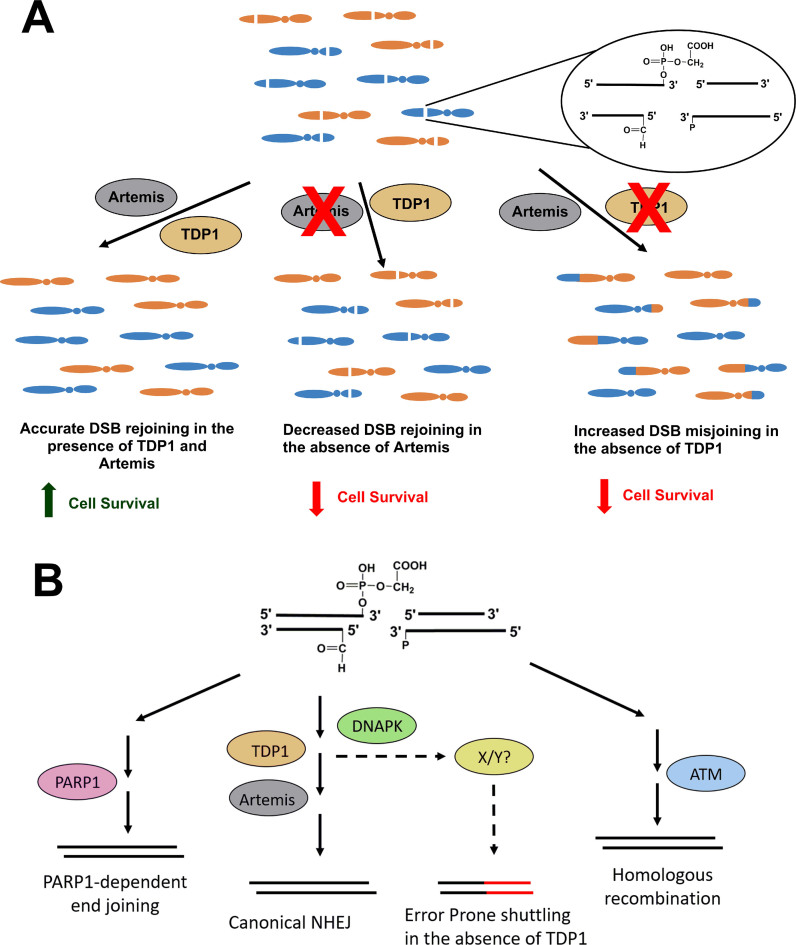
Together, these data presented a conundrum as it appeared as if 3′-PG-terminated DSBs were cytotoxic in the absence of TDP1 despite being rejoined with kinetics similar to cells proficient in TDP1. One possibility is that, rather than preventing end joining, TDP1 deficiency promotes mis-joining of DSB ends to ends of other DSBs in the cell. Thus, the toxicity of 3′-PG-ended DSBs in TDP1-depleted cells (shTDP1#18 and Artemis−/−•shTDP1#2) could be because of inaccurate and mis-joined DSBs due to repair being shunted to a more error-prone pathway. To test this hypothesis, centromere-fluorescence in situ hybridization (C-FISH) was performed on cells treated with 2 nM NCS. As with CAL-treated SCAN1 cells, shTDP1#18 single and Artemis−/−•shTDP1#2 double mutants showed a significant increase in the levels of NCS-induced dicentric chromosomes compared to WT and Artemis−/− cells, as measured on metaphase spreads with Cy3-labelled centromeres (Figure 4B and Supplementary Figure S12). Furthermore, all mutants showed an increase in the number of acentric chromosomal fragments (Figure 4C), as would be expected since both unjoined fragments and fragments mis-joined to each other would be scored as acentrics. In addition, the Artemis−/−•shTDP1#2 showed a statistically higher number of total aberrations than either WT or either single mutant alone (Figure 4D). Similarly, cytogenetic assays performed in HEK293 WT and TDP1−/– cells showed an increase in dicentric chromosomes, acentric fragments and total aberrations (Supplementary Figure S13). Thus, unlike for survival, where Artemis and TDP1 appear epistatic, for the formation of chromosomal aberrations the genes are additive and suggest that there are disparate repair processes at work (Figure 6).
Figure 6.
Model of the epistasis between Artemis and TDP1. (A) NCS treatment leads to the production of double-strand breaks in the DNA having 3′-phosphoglycolate ends. In the presence of functional Artemis and TDP1, there is accurate DSB rejoining promoting cell survival. In the absence of Artemis, a fraction of DSBs remain unrejoined causing a survival defect. In the absence of TDP1, DSB ends are misjoined causing translocations and decrease in survival. Thus, although the two proteins function in the same pathway, loss of these proteins confers disparate phenotypes. (B) In the absence of TDP1, the breaks are shuttled to a more-error prone pathway regulated by the kinase activity of DNA-PK leading to mis-joining.
In order to examine the contribution of NHEJ towards the mis-joining of DSB ends in the absence of TDP1, C-FISH was performed on cells treated with NCS in the presence of NU-7441 (DNA-PKi). Treatment with NU-7441 prevents the inward translocation of DNA-PK from DSB ends, thereby limiting access to the nucleases and phosphodiesterases required for the processing of modified ends. DNA-PK inhibition had no effect on DSB mis-joining in WT and Artemis−/− cells but led to a decrease in DSB mis-joining in TDP1-depleted shTDP1#18 and Artemis−/−•shTDP1#2 cells down to the level of TDP1-proficient WT and Artemis−/− cells, as all four cell lines showed similar levels of dicentric chromosomes per metaphase (Figure 4B). Overall, the DNA-PK inhibitor data suggest that while there is a component of mis-joining in both WT and TDP1-deficient cells that is independent of C-NHEJ, the additional mis-joining in TDP1-deficient cells is C-NHEJ-dependent.
TDP1 and Artemis are not epistatic with PARP1 or ATM for the repair of 3′-PG-ended DSBs
TDP1 and PARP1 are epistatic for the repair of CPT-induced TopI-DNA lesions and TDP1 is poly(ADP)-ribosylated by PARP1 in this pathway (27). However, a recent study demonstrated a lack of apparent epistasis between TDP1 and PARP1 for the repair of sapacitabine, a chain-terminating nucleoside analog (CTNA) (18). Additionally, PARP1 is a key component in the alternative non-homologous end joining (a-EJ) pathway for the repair of DNA DSBs (28). Thus, to investigate the functional interaction between TDP1, Artemis and PARP1 for the repair of NCS-induced 3′-PG terminated DSBs, clonogenic survival assays were performed using the PARP inhibitor, AZD-2281 (Olaparib, PARPi), which strongly suppressed poly(ADP)-ribosylation in these cells (Supplementary Figure S14). Unlike WT cells, all the mutants showed an increase in sensitivity to NCS upon PARP inhibition (Figure 5A–C) suggesting that TDP1 and Artemis are not epistatic with PARP1 but are involved in a pathway parallel to the PARP1-dependent repair of NCS-induced 3′-PG terminated DSBs. These data are also consistent with the observation that Artemis-null cells show no deficit in microhomology-mediated DNA DSB repair (Supplementary Figure S15), a hallmark of a-EJ activity.
Figure 5.
ATM and PARP function in parallel to Artemis and TDP1 for the repair of NCS-induced double-strand breaks. Clonogenic survival assays were performed in cells treated with NCS in the presence of 1 μM PARP inhibitor, Olaparib (PARPi) (A–C) or 1 μM ATM inhibitor, KU60019 (ATMi) (D–F). shTDP1, Art–/– and Art–/–•shTDP1 curves are the same as in Figure 2B. Inhibitors were added 1 h prior to NCS treatment and left in the medium during and 24 hr after NCS treatment. Error bars represent SEM. n = 3 for experiments with ATMi and n = 4 for experiments with PARPi.
Finally, there is an epistatic interplay between Artemis and ATM for the repair of IR-induced breaks (29). Thus, to investigate the role of ATM in the epistasis between Artemis and TDP1, clonogenic survival assays were performed using NCS in the presence of an ATM inhibitor KU-60019 (ATMi). WT cells showed an increased sensitivity to NCS upon ATM inhibition suggesting an important role for ATM in the repair of 3′-PG DSBs (Figure 5D–F). Additional TDP1-depletion in these cells enhanced the sensitivity even further suggesting that ATM and TDP1 are involved in parallel pathways for the repair of NCS-induced DSBs (Figure 5D). Interestingly, compared to ATM-inhibited WT cells, inhibition of ATM in Artemis−/− single and Artemis–/−•shTDP1#2 double mutants led to an increase in sensitivity only at high doses of NCS (Figure 5E and F). In toto, these data indicated that TDP1 and Artemis perform both complementary and parallel functions in human DNA DSB repair.
DISCUSSION
IR and radiomimetic natural compounds like NCS, CAL and bleomycin induce terminally-occluded DNA DSBs by free-radical mechanisms (30). Although many 3′-blocked termini are unstable and spontaneously break down to 3′-phosphates, 3′-PGs formed by fragmentation of the deoxyribose by oxidation of the C-4′ position are stable and persistent (31). Gap-filling DNA polymerases and DNA ligases require 3′-OH DNA ends to efficiently add nucleotides and perform end ligation respectively, and therefore the resolution of a 3′-PG to a 3′-OH is an essential step in the repair of these DSBs. It is therefore not surprising that mammalian cells have evolved several enzymes—including TDP1, APE1 and Artemis—for such resolution (13).
In the current study, NCS and CAL were used to investigate the role of TDP1 in the repair of 3′-PG DSBs. In contrast to radiation-induced DSBs which are heterogeneous and bear 3′-PG only on 10% of the total sugar oxidation products (32), about one in every four DSBs induced by CAL and NCS bear a 3′-PG, making these antitumor antibiotics more informative than IR in studying the repair of 3′-PG (33). Supporting established evidence of TDP1’s critical role in 3′-PG removal (6–8), TDP1-deficient cells were hypersensitive to both NCS and CAL. Elimination of Artemis, a DSB end-trimming nuclease that can also resolve 3′-PGs, likewise sensitized cells to NCS. Surprisingly, however, the toxicity of 3′-PG DSBs observed in cells simultaneously deficient in Artemis and TDP1 compared to cells with individual deficiencies in these genes was similar, indicating that these two proteins function in the same DSB repair pathway. Furthermore, as DSBs in G1-phase are repaired almost exclusively by NHEJ (34), the epistasis between Artemis and TDP1 in cells synchronized in the G1-phase suggests an involvement of these two proteins in NHEJ. The fact that Artemis-null cells are completely proficient for a-EJ (Supplementary Figure S15), indicates that C-NHEJ is the pathway where Artemis and TDP1 cooperate. This conclusion is supported by the observation that the increased toxicity of 3′-PG DSBs upon DNA-PK inhibition was not further enhanced by an additional deficiency in TDP1, Artemis or both. Thus, when C-NHEJ is suppressed, TDP1 and Artemis no longer contribute to survival after NCS treatment. Taken together, these data strongly support a functional involvement of Artemis and TDP1 in C-NHEJ for the repair of NCS-induced 3′-PG DSBs.
The unexpected epistasis between Artemis and TDP1 in survival of NCS treatment suggests that these two proteins perform enzymatically distinct roles in the repair of NCS-induced DSBs. Considering the well-documented role of TDP1 in clean 3′-PG removal (8), one possibility is that, upon NCS/CAL-induced DSB formation, the overhanging 3′-PG on one end of the DSB is a substrate for TDP1 whereas Artemis is required for removing the 5′-aldehyde formed on the other strand. Since the other end bears a 3′-phosphate which is the canonical substrate of PNKP, a critical end processing enzyme that interacts with the X-ray cross-complementing 4 (XRCC4)•LigaseIV complex in the context of C-NHEJ (35), it is unlikely that either TDP1 or Artemis processes the 3′-phosphate ends. Alternatively, it can be surmised that if Artemis is important for 3′-PG end trimming via C-NHEJ, TDP1 might be playing a structural role in this pathway. Consistent with this possibility, TDP1 physically interacts with XLF and Ku70/80, key proteins in the C-NHEJ pathway and stimulates binding of XLF and Ku70/80 to DNA (36). In this manner, a TDP1 deficiency would abrogate C-NHEJ-mediated DSB repair and the simultaneous absence of Artemis would have no additional effect. On the other hand, Artemis has been proposed to play a role in DSB repair pathway choice, directing breaks into C-NHEJ when appropriate (37). Loss of this function could lead to suboptimal repair regardless of whether 3′-PG termini are resolved. In the future, it will be interesting to explore these possibilities by tracking the processing of 3′-PG ends using ligation-mediated PCR (38) or complementing our mutant cell lines with either an endonuclease-deficient Artemis or a phosphodiesterase-deficient TDP1, respectively.
In contrast to NCS and CAL, the repair of IR-induced breaks seems to require either TDP1 or Artemis as cells deficient in both are more radiosensitive than cells deficient in either individual protein. On one hand, it is known that the endonuclease function of Artemis is required in IR-induced DSB repair (17) and that Artemis is required for the repair of a subset of DSBs formed in heterochromatin (29). On the other hand, TDP1-mutant SCAN1 cells are defective in IR-induced SSB repair (39). Due to the heterogeneous nature of damage induced by radiation, TDP1 may thus be required for the repair of IR-induced SSBs while Artemis repairs the subset of DSBs in the heterochromatin. Accordingly, simultaneous absence of both proteins makes the cells more radiosensitive than their individual absence. Another possible basis for the positive epistasis is that PGs formed on short overhangs (<3 bases) might be repaired by TDP1 whereas longer overhangs—due to clustered damage—might require Artemis (16).
Another unanticipated finding was the apparent lack of a DSB repair defect in TDP1-depleted cells, despite their hypersensitivity to NCS. Assays of 53BP1 foci, a surrogate marker of unrepaired DSBs, indicated that these repair foci disappear with similar kinetics in WT and TDP1-depleted cells. Although this assay is standard for measuring the fraction of DSBs that remain unrejoined, it is limited by its inability to differentiate between DSB ends that are correctly rejoined, and DSB ends that are mis-joined to ends of other DSBs in the cell. Thus, it was hypothesized that TDP1-depleted cells might be hypersensitive to NCS due to inaccurate DSB joining and consequent formation of lethal chromosome aberrations. Indeed, C-FISH experiments demonstrated a significant increase in the number of dicentric chromosomes upon TDP1 depletion suggesting that, in these cells, many 3′-PG DSBs are mis-joined. Although C-NHEJ is generally regarded as being protective against the mis-joining mediated by the more error-prone a-EJ pathway, mis-joining that is mediated by C¬NHEJ and is suppressed by DNA-PK inhibitors has been reported previously in other contexts (34). Conversely, in agreement with a DSB rejoining defect observed in the 53BP1 assay, Artemis-deficient cells showed an increase in the level of acentric chromosomal fragments, representing unjoined DSBs, but not in dicentrics. The increased incidence of dicentrics in TDP1-depleted cells is reminiscent of increased dicentrics observed in TDP1-mutant SCAN1 cells following treatment with CAL (8). Our conclusion that TDP1 is involved predominately in mis-joining is consistent with a recent study that demonstrated that TDP1 is required for efficient C-NHEJ in human cells (25). In this study, a deficiency of TDP1 reduced the fidelity of end joining with an increase in insertions at repair junctions of I-SceI-induced DSBs, which could be completely restored by WT TDP1 but only partially by catalytically-inactive TDP1H263A (25). These insertions at DSB sites could reflect mis-joining of persistent DSB ends to small pieces of unrelated DNA. Even in yeast, in the absence of TDP1, restriction enzyme-induced DSBs are inaccurately repaired with an increase in C-NHEJ-dependent insertions possibly via the mutagenic polymerase, Pol IV (40).
In contrast to the recent findings that TDP1 knockout in HEK293 cells caused a decrease in end joining efficiency of I-SceI induced-DSBs as observed from the EJ5-GFP assay (25), results in the current study do not show a DSB rejoining defect in TDP1-depleted cells. A limitation of the EJ5-GFP assay is that end-joining efficiency cannot be strictly correlated with GFP-expression since only rejoining of two ends of the I-SceI-induced DSB to each other would result in GFP expression and any inaccurate mis-joining of these DSBs to other unrelated DSB sites would yield GFP-negative cells. Therefore, the decrease in NHEJ observed by Li et al. could be attributed to misjoining of I-SceI-induced DSBs to other unrelated DSB sites. Other possible explanations for this discrepancy include the difference in the nature and the number of breaks as well as the use of different cellular systems. DSBs induced by I-SceI contain unmodified, 3′-OH ends compared to those induced by NCS. In the absence of TDP1, the terminal nucleoside on the restriction enzyme-induced DSB ends would not be removed, possibly leading to insertions by mutagenic polymerases decreasing the overall end joining efficiency (25,40). Conversely, the 3′-modification in NCS-induced DSBs precludes the activity of these polymerases averting aberrant insertions and thus may prevent a DSB rejoining defect. Moreover, the use of different cellular systems impedes direct comparison of the results between the two studies.
In the context of C-NHEJ, inhibition of DNA-PK blocks its auto-phosphorylation and prevents its inward translocation from DSB ends and thereby restricts access of other end-processing enzymes to the DSB end. The finding that DNA-PK inhibition rescues mis-joining in TDP1-depleted cells (both shTDP1#18 and Artemis−/−•shTDP1#2) to levels seen in TDP1-proficient cells suggests that a TDP1 deficiency renders the C-NHEJ pathway more error-prone, rather than invoking a separate backup pathway. Although Artemis-mediated resection-dependent C-NHEJ can cause translocations (34), in the current study, dicentric chromosomes were still persistent in Artemis−/−•shTDP1#2 cells, refuting the role of Artemis in this error-prone mis-joining. The a-EJ-associated factor, C-terminal interacting protein (CtIP) is another possible candidate mediating error-prone joining in the absence of TDP1. Indeed, CtIP functions in parallel to TDP1 for the repair of Top1-induced SSBs and methyl methane sulfonate (MMS)-induced lesions (11). Moreover, CtIP is phosphorylated by Polo-like kinase 3 (Plk3), promotes IR-induced chromosomal translocations (41,42) and the depletion of CtIP decreases translocations in mouse cells (43). Taken together, our results clearly indicate that TDP1 is required for accurate joining of DSB ends and, in its absence, a more error-prone DNA-PK-dependent process inaccurately repairs NCS-induced DSBs.
Survival assays performed to investigate the relationship between TDP1 and Artemis with ATM strongly suggest that ATM is critical for the repair of 3′-PG DSBs as WT cells show enhanced sensitivity to NCS upon ATM inhibition. ATM and TDP1 appear to function in parallel pathways for the repair of 3′-PG DSBs since TDP1-depleted cells showed increased sensitivity to NCS upon ATM inhibition compared to ATM inhibition in WT cells (Figure 5D). More interestingly, compared to ATM inhibition in WT, ATM inhibition in Artemis-deficient cells (Artemis−/− single mutants and Artemis−/−•shTDP1 double mutants) did not sensitize them further to low doses of NCS (Figure 5E and F). One possible explanation for this discrepancy is that, at lower doses of NCS, in the absence of ATM, Artemis drives the repair of 3′-PG DSBs towards a TDP1-dependent repair pathway. In this way, a combined absence of ATM and TDP1 would make the cells more sensitive than the absence of the individual proteins as both TDP1-dependent and ATM-dependent repair of 3′-PG DSBs would be abolished. However, in the absence of Artemis, the drive towards TDP1- dependent repair would be lost, allowing 3′-PG DSB repair via a backup repair pathway. This backup process would then prevent Artemis deficiency upon ATM inhibition from being more deleterious than ATM inhibition alone at low doses of NCS. The fact that the initially superimposable survival curves diverge at higher NCS concentrations, combined with the slight downward concavity of the curves for Artemis−/− cells, is consistent with the proposed backup pathway being saturable, so that at higher levels of damage, repair again becomes Artemis-dependent.
In contrast to a previous report showing radiosensitization of HCT116 WT cells by a PARP inhibitor (44), PARP1 inhibition did not sensitize these cells to NCS (Figure 5A–C). This differential response could be attributed to the type of damage induced by the different agents. Since PARP1 is a key protein in base excision repair (BER), PARP1 inhibition upon IR, which forms DSBs to SSBs at a ratio of around 1:20 in addition to base damage, would cause accumulation of residual SSBs and damaged bases that upon replication would be converted to one-ended DSBs (33). Due to the higher proportion of DSBs formed by NCS (DSB/SSB ratio ∼1:5) and the absence of base damage, PARP1 inhibition is less likely to significantly sensitize the cells through interference with SSB repair. The inference that SSBs contribute little to NCS cytotoxicity, even in PARP1-inhibited cells, provides further support for the conclusion that the NCS sensitivity of shTDP1 knockdown cells reflects a role for TDP1 in DSB repair, not its well-documented role in SSB repair. However, the toxicity of 3′-PG DSBs in PARP1-inhibited cells was slightly enhanced by a deficiency of TDP1 (Figure 5A), Artemis (Figure 5B) and both (Figure 5C). This relationship between PARP1 and TDP1/Artemis is consistent with PARP1-mediated DSB repair (i.e. a-EJ) acting as a backup for NHEJ in the absence of TDP1 or Artemis.
In conclusion, our results suggest a strong epistasis between Artemis and TDP1 for survival against the toxicity of 3′-PG terminated DSBs induced by NCS and CAL. Importantly, however, although Artemis and TDP1 are involved in the same pathway, they perform non-overlapping functions and thereby, mediate survival through different mechanisms. Artemis promotes survival by promoting DSB rejoining, whereas TDP1 promotes the accurate joining of the DSB ends.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health, USDDHS [CA40615 and CA166264 to K.A., K.V., L.F.P.; CA154461 and GM088351 to E.A.H., B.R.]; NCI-NIH intramural research program and Z01 BC006150 (to S.N.H.,Y.P.); a Dissertation Assistantship from VCU School of Medicine (to A.S.K.); VCU Department of Anatomy and Neurobiology Microscopy Facility supported, in part, by NIH-NINDS Center core grant [5P30NS047463]. Open access charge: (in part) CA154461, GM088351.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pommier Y., Sun Y., Huang S., Nitiss J.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davies D., Interthal H., Champoux J., Hol W.. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002; 10:237–248. [DOI] [PubMed] [Google Scholar]
- 3. Pouliot J.J., Yao K.C., Robertson C.A., Nash H.A.. Yeast gene for a tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999; 286:552–555. [DOI] [PubMed] [Google Scholar]
- 4. Takashima H., Boerkoel C., John J., Saifi G.M., Salih M., Armstrong D., Mao Y., Quiocho F., Roa B., Nakagawa M. et al. Mutation of TDP1, encoding a topoisomerase i–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 2002; 32:267–272. [DOI] [PubMed] [Google Scholar]
- 5. Kawale A.S., Povirk L.F.. Tyrosyl-DNA phosphodiesterases: Rescuing the genome from the risks of relaxation. Nucleic Acids Res. 2018; 46:520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Inamdar K.V., Pouliot J.J., Zhou T., Lees-Miller S.P., Rasouli-Nia A., Povirk L.F.. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 2002; 277:27162–27168. [DOI] [PubMed] [Google Scholar]
- 7. Hawkins A.J., Subler M.A., Akopiants K., Wiley J.L., Taylor S.M., Rice A.C., Windle J.J., Valerie K., Povirk L.F.. In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair (Amst.). 2009; 8:654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou T., Akopiants K., Mohapatra S., Lin P., Valerie K., Ramsden D.A., Lees-Miller S.P., Povirk L.F.. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair (Amst.). 2009; 8:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Interthal H., Chen H.J., Champoux J.J.. Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 2005; 280:36518–36528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang S.Y., Murai J., Dalla Rosa I., Dexheimer T.S., Naumova A., Gmeiner W.H., Pommier Y.. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013; 41:7793–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murai J., Huang S.N., Das B.B., Dexheimer T.S., Takeda S., Pommier Y.. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 2012; 287:12848–12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El-Khamisy S.F., Saifi G.M., Weinfeld M., Johansson F., Helleday T., Lupski J.R., Caldecott K.W.. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005; 434:108–113. [DOI] [PubMed] [Google Scholar]
- 13. Zhou T., Lee J.W., Tatavarthi H., Lupski J.R., Valerie K., Povirk L.F.. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1). Nucleic Acids Res. 2005; 33:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suh D., Wilson D.M., Povirk L.F.. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997; 25:2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma Y., Pannicke U., Schwarz K., Lieber M.R.. Hairpin opening and overhang processing by an artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002; 108:781–794. [DOI] [PubMed] [Google Scholar]
- 16. Povirk L.F., Zhou T., Zhou R., Cowan M.J., Yannone S.M.. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by artemis nuclease. J. Biol. Chem. 2007; 282:3547–3558. [DOI] [PubMed] [Google Scholar]
- 17. Mohapatra S., Kawahara M., Khan I.S., Yannone S.M., Povirk L.F.. Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient artemis. Nucleic Acids Res. 2011; 39:6500–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abo M.A., Sasanuma H., Liu X., Rajapakse V.N., Huang S., Kiselev E., Takeda S., Plunkett W., Pommier Y.. TDP1 is critical for the repair of DNA breaks induced by sapacitabine, a nucleoside also targeting ATM- and BRCA-deficient tumors. Mol. Cancer Ther. 2017; 16:2543–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Budanov A.V., Sablina A.A., Feinstein E., Koonin E.V., Chumakov P.M.. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004; 304:596–600. [DOI] [PubMed] [Google Scholar]
- 20. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao R., Das B.B., Chatterjee R., Abaan O.D., Agama K., Matuo R., Vinson C., Meltzer P.S., Pommier Y.. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair (Amst.). 2014; 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takemura H., Rao V.A., Sordet O., Furuta T., Miao Z.H., Meng L., Zhang H., Pommier Y.. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 2006; 281:30814–30823. [DOI] [PubMed] [Google Scholar]
- 23. Sacho E.J., Maizels N.. DNA repair factor MRE11/RAD50 cleaves 3′-phosphotyrosyl bonds and resects DNA to repair damage caused by topoisomerase 1 poisons. J. Biol. Chem. 2011; 286:44945–44951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Staples C.J., Barone G., Myers K.N., Ganesh A., Gibbs-Seymour I., Patil A.A., Beveridge R.D., Daye C., Beniston R., Maslen S. et al. MRNIP/C5orf45 interacts with the MRN complex and contributes to the DNA damage response. Cell. Rep. 2016; 16:2565–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Summerlin M., Nitiss K.C., Nitiss J.L., Hanakahi L.A.. TDP1 is required for efficient non-homologous end joining in human cells. DNA Repair (Amst.). 2017; 60:40–49. [DOI] [PubMed] [Google Scholar]
- 26. Lee S.E., Mitchell R.A., Cheng A., Hendrickson E.A.. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 1997; 17:1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Das B.B., Huang S.Y., Murai J., Rehman I., Ame J.C., Sengupta S., Das S.K., Majumdar P., Zhang H., Biard D. et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014; 42:4435–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mansour W.Y., Rhein Tim, Dahm-Daphi Jochen. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010; 38:6065–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riballo E., Kuhne M., Rief N., Doherty A., Smith G.C., Recio M.J., Reis C., Dahm K., Fricke A., Krempler A. et al. A pathway of double-strand break rejoining dependent upon ATM, artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004; 16:715–724. [DOI] [PubMed] [Google Scholar]
- 30. Menon V., Povirk L.F.. End-processing nucleases and phosphodiesterases: An elite supporting cast for the non-homologous end joining pathway of DNA double-strand break repair. DNA Repair (Amst.). 2016; 43:57–68. [DOI] [PubMed] [Google Scholar]
- 31. Povirk L.F. Processing of damaged DNA ends for double-strand break repair in mammalian cells. ISRN Mol. Biol. 2012; 2012: 10.5402/2012/345805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen B., Zhou X., Taghizadeh K., Chen J., Stubbe J., Dedon P.C.. GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3′-phosphoglycolate pathways of 4′ oxidation of 2-deoxyribose in DNA: Application to DNA damage produced by gamma radiation and bleomycin. Chem. Res. Toxicol. 2007; 20:1701–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Povirk L.F. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 1996; 355:71–89. [DOI] [PubMed] [Google Scholar]
- 34. Biehs R., Steinlage M., Barton O., Juhasz S., Kunzel J., Spies J., Shibata A., Jeggo P.A., Lobrich M.. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell. 2017; 65:671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aceytuno R.D., Piett C.G., Havali-Shahriari Z., Edwards R.A., Rey M., Ye R., Javed F., Fang S., Mani R., Weinfeld M. et al. Structural and functional characterization of the PNKP-XRCC4-LigIV DNA repair complex. Nucleic Acids Res. 2017; 45:6238–6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heo J., Li J., Summerlin M., Hays A., Katyal S., McKinnon P.J., Nitiss K.C., Nitiss J.L., Hanakahi L.A.. TDP1 promotes assembly of non-homologous end joining protein complexes on DNA. DNA Repair (Amst.). 2015; 30:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang J., Aroumougame A., Lobrich M., Li Y., Chen D., Chen J., Gong Z.. PTIP associates with artemis to dictate DNA repair pathway choice. Genes Dev. 2014; 28:2693–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Akopiants K., Mohapatra S., Menon V., Zhou T., Valerie K., Povirk L.F.. Tracking the processing of damaged DNA double-strand break ends by ligation-mediated PCR: Increased persistence of 3′-phosphoglycolate termini in SCAN1 cells. Nucleic Acids Res. 2014; 42:3125–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. El-Khamisy S.F., Hartsuiker E., Caldecott K.W.. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst.). 2007; 6:1485–1495. [DOI] [PubMed] [Google Scholar]
- 40. Bahmed K., Nitiss K.C., Nitiss J.L.. Yeast Tdp1 regulates the fidelity of nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barton O., Naumann S.C., Diemer-Biehs R., Kunzel J., Steinlage M., Conrad S., Makharashvili N., Wang J., Feng L., Lopez B.S. et al. Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J. Cell Biol. 2014; 206:877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Makharashvili N., Paull T.T.. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair (Amst.). 2015; 32:75–81. [DOI] [PubMed] [Google Scholar]
- 43. Helmink B.A., Tubbs A.T., Dorsett Y., Bednarski J.J., Walker L.M., Feng Z., Sharma G.G., McKinnon P.J., Zhang J., Bassing C.H. et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011; 469:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alotaibi M., Sharma K., Saleh T., Povirk L.F., Hendrickson E.A., Gewirtz D.A.. Radiosensitization by PARP inhibition in DNA repair proficient and deficient tumor cells: Proliferative recovery in senescent cells. Radiat. Res. 2016; 185:229–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.