Abstract
The role of water in the interaction of Hoechst 33258 with the minor groove binding site of the (AATT)2 sequence was investigated using calorimetric and equilibrium constant measurements. Using isothermal titration calorimetry measurements, the heat capacity change for the reaction is −256 ± 10 cal/(K mole Hoechst). Comparison with the heat capacity changes based on area models supports the expulsion of water from the interface of the Hoechst-DNA complex. To further consider the role of water, the osmotic stress method was used to determine if the Hoechst association with DNA was coupled with hydration changes. Using four osmolytes with varying molecular weights and chemical properties, the Hoechst affinity for DNA decreases with increasing osmolyte concentration. From the dependence of the equilibrium constant on the solution osmolality, 60 ± 13 waters are acquired in the complex relative to the reactants. It is proposed that the osmotic stress technique is measuring weakly bound waters that are not measured via the heat capacity changes.
Keywords: Hydration, Osmotic Stress, Isothermal Titration Calorimetry, DNA, Hoechst
Hydration is an important feature of DNA structure and function, and the challenge is to identify explicit roles for water molecules in the vast background of the aqueous solvent.1 Approximately 20 waters bind with each base pair in the primary hydration shell with varying strengths of interaction that are apparent from the infrared spectra.2 A second layer of water weakly associates with the primary layer, and it has properties that are similar to the bulk solvent. From a functional perspective, this bound water reorganizes when a ligand binds with DNA, and we are considering small molecules that bind in the minor groove of B-form DNA. These ligands have demonstrated great promise for the sequence specific binding needed to alter genetic expression,3,4,5 and this work describes the thermodynamic signatures of the hydration changes that accompany binding. Besides direct ligand-DNA contacts through electrostatic, van der Waals, and hydrogen bonding, the environment of the solution can influence complex formation. For example, ionic strength alters reactivity through electrostatic effects.6 Water can likewise participate in reactions involving DNA. One key idea is that water is expelled when sequence-specific bonds form. For both protein and small molecule complexes with DNA, largely dehydrated interfaces are revealed by X-ray diffraction and molecular dynamics studies.3,7,8 The hydration changes that accompany the loss of solvation are reflected in the change in the heat capacity of the reaction at constant pressure (∆Cp), and sequence-specific binding results in a decrease in ∆Cp.9,10,11 Besides the entropic contribution from solvent release, water can also mediate interactions between the ligand and DNA.12 In support, a number of protein and small molecule complexes with DNA possess strongly bound water molecules between interfacial functional groups.3,13,14,15 Recent attempts have clarified the contribution of this bound water to the thermodynamic properties of the reaction.16
Besides the strongly bound water apparent in X-ray diffraction, nuclear magnetic resonance, and molecular modeling studies, waters that are weakly associated with DNA should also be considered.17 A broader view of solvent exchange that encompasses weakly bound waters is accomplished through thermodynamic studies. For example, water associated with DNA has a different structure than water in the bulk solvent. Thus, volume and adiabatic compressibility measurements describe how ligand binding influences the number of bound waters and the strength of the solvent-DNA interactions. For example, when netropsin interacts with the minor groove of A/T rich oligonucleotides, the solvation changes reflect the different levels of hydration of the oligonucleotides and their complexes with DNA.18 Another approach for understanding hydration changes is to alter the properties of the solvent, as accomplished using osmotic and hydrostatic pressure.19 Osmotic pressure is applied using high concentrations of solutes that lower the water activity. When these solutes (osmolytes) are excluded from the vicinity of the DNA, osmotic pressure is exerted on water that is bound in the recessed sites of the DNA. Using this approach, nonspecific and specific complexes of proteins with DNA are shown to differ because the former retain their hydration shells while the latter cause significant solvent expulsion.19 This conclusion is supported by X-ray diffraction studies. A complementary technique is to apply hydrostatic pressure, to which a system responds by decreasing the volume and promoting hydration of the reaction components. An elegant demonstration of the complementary perspectives provided by hydrostatic and osmotic pressure studies is illustrated by studies of DNA cleavage at cognate and alternate (star) sites for restriction enzymes.19 Volumetric and pressure measurements often yield different numbers of exchanged waters, which may occur because different microscopic models are used to interpret the data. Recent theoretical efforts are resolving these discrepancies.20
The design of new sequence specific binding agents is motivated by the sequencing of the human genome. Thermodynamic and structural studies have demonstrated that a number of factors contribute to the affinity and selectivity of sequence specific binding of small molecules with DNA, and we investigate the role of water in the binding of Hoechst 33258 (hereafter referred to as Hoechst, Fig. 1) with DNA.21,22,23,24 Besides medicinal and diagnostic uses, Hoechst is an excellent model for understanding the factors that contribute to strong and sequence-specific binding with DNA.25,26 It has several features in common with other small molecules that preferentially bind in the minor groove: it is cationic, it is concave to closely match the shape of the minor groove, and it interacts with the minor groove through van der Waals and hydrogen bonding.27,28 Binding sites, three to five bases long, containing adenine (A) and thymine (T) are favored.29,30 In addition, the order of the A/T bases results in a large (≈200 fold) variation in the binding affinity, with 5’-TA-3’ steps decreasing the affinity.31 Calorimetric studies have provided significant insight into the forces that contribute to sequence specific binding of Hoechst.21 Of relevance to this work, Hoechst binds primarily as a monomer at low concentrations (nM), but it has multiple binding modes with DNA at higher concentrations (µM).32 In these studies, calorimetric and solvent perturbation studies are used to understand the role of hydration in the binding of Hoechst with the (AATT)2 minor groove binding site. Isothermal titration calorimetry measurements show that ∆Cp is negative for the reaction of Hoechst with (AATT)2, supporting the expulsion of water from the complex. In contrast, osmotic stress studies show that the amount of water associated with DNA increases when Hoechst binds with the DNA. We suggest that these techniques are probing waters with different strengths of interaction with DNA.
Figure 1:
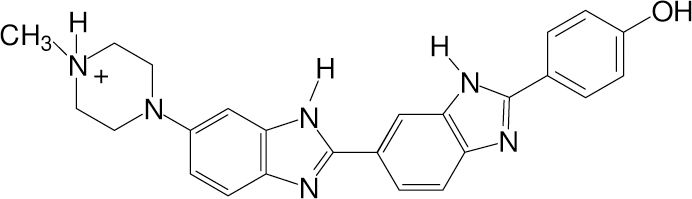
The structure of Hoechst 33258 (Hoechst) at neutral pH.
Experimental
The osmolytes, betaine (Fluka), acetamide (Baker Scientific), triethylene glycol (TCI-GR), and tetraethylene glycol (Aldrich), were used as received. The oligonucleotide 5′-CGCGCAATTGCGCG-3′ (Integrated DNA Technologies) was annealed by heating a 1 mM solution to 95 °C for 5 min and then slowly cooling to 10 °C over 60 hr. The melting profile indicated that predominantly one species formed with the expected melting temperature for the duplex form. The sterilized aqueous buffer contained 10 mM H2PO4−/HPO42− at pH = 7 and 50 mM NaCl.
Calorimetry studies were conducted using a Microcal VP-ITC (Northhampton, MA) controlled by Origin 7.0 software. Hoechst has a strong propensity to adsorb to glass surfaces,33 so high concentrations (500 µM) were used to minimize losses in the titrating syringe. The high concentration of Hoechst allowed the removal of 10–20 µL from the titrating syringe, and the concentration was checked immediately after loading the syringe and after the titration was complete. The loss was <10%, and the corrected concentration was used in the analysis. The Hoechst solution was titrated into a 5 µM solution of duplex oligonucleotide. All solutions were degassed prior to loading. The heat change associated with the titration was determined by integrating the power required to maintain the reference and sample cells at the same temperature. The heats of dilution of the concentrated Hoechst solution were empirically fit using a quadratic expression. Attempts to use a model based on a dimer dissociating to monomers were not successful, presumably because higher-order aggregates were present in the solution. The enthalpy change up to saturation of the minor groove binding site is constant, so the ∆H for minor groove binding was determined by averaging those 12–14 points.
The changes in the surface areas were determined using GRASP 1.3.34 Hydrogen atoms were added to the pdb files 1d43 and 1d44 (Hoechst-(AATT)2 complexes) and 1bna ((AATT)2) using VegaZZ.35 The nonpolar atoms were carbon, hydrogen bound to carbon, and phosphorous, and the remaining atoms were defined as polar. A probe radius of 1.4 Å was used, and the atomic radii were obtained from the work of Cornell et al.36 The areas for Hoechst alone were obtained by removing it from the crystal structure data for 1d43 and 1d44.
Fluorescence measurements were acquired on a Jobin-Yvon Horiba Fluoromax-3 with an excitation wavelength of 350 nm and an emission wavelength of 465 nm. All spectroscopic measurements were conducted in polystyrene cuvettes.32 To measure the equilibrium constant for the reaction, the unbound and bound Hoechst concentrations were measured using their differences in the fluorescence. First, Hoechst was added to the buffer to give a 5 nM solution, thus providing the reference fluorescence of the unbound Hoechst. The reference fluorescence of the bound Hoechst was obtained after adding sufficient DNA. The observed fluorescence (F) at intermediate DNA concentrations is:
| Eq. 1 |
where [H]u is the unbound Hoechst during the DNA titration and Fu is the intrinsic fluorescence of the solely unbound Hoechst (for a 1 M concentration). [H]b and Fb are the corresponding parameters for the bound Hoechst. To calculate the amount of bound Hoechst, the equilibrium appropriate for the noncovalent binding of Hoechst in the minor groove was used:
| Eq. 2 |
Equilibrium analysis yields the equilibrium constant (K):
| Eq. 3 |
where [DNA] is the concentration of oligonucleotide during the titration and [H]t is the total concentration of Hoechst. After solving this equation for [H]b and using mass balance, least squares fitting using Eq. 1 determined K, Fb, and Fu from the dependence of the observed fluorescence on the concentration of the DNA.
Visible absorption spectra were acquired using a Cary 50 spectrometer (Varian Inc., Palo Alto, CA). Circular dichroism spectra were obtained from a Jasco J-710 spectropolarimeter (Jasco Inc, Easton, MD). Melting studies were conducted using a Cary 300 spectrometer (Varian Inc., Palo Alto, CA). For the continuous variation analysis, Hoechst was removed from the solution and replaced with an equivalent amount of DNA to maintain a constant total concentration of the two components.37 The fluorescence intensities were plotted as a function of the mole fraction of Hoechst to determine the complex stoichiometry.38 Osmolalities were determined using a Wecor 5520 (Wescor Inc., Logan, UT).
Results
Heat Capacity Studies
A number of studies have established that the binding of small molecules and proteins with DNA results in a negative change for ∆Cp, and a dominant contribution to ∆Cp comes from hydration changes.10,16,39 We have measured the variation of ∆H with temperature to determine the ∆Cp for the binding of Hoechst in the minor groove of the (5′-CGCGCAATTGCGCG-3′)2 sequence. Our flanking sequence differs from other studies, but as discussed later, we do not expect a large effect on the thermodynamic parameters. Thus, hereafter we will refer to this oligo as (AATT)2 to emphasize the binding site for the Hoechst. At the high concentrations used for the calorimetry studies, Hoechst tends to form self-aggregates and aggregates with DNA. To illustrate, at least three distinct reactions are apparent when a Hoechst solution is titrated into (AATT)2 up to a 22:1 molar ratio (Fig. 2). At low Hoechst stoichiometries, minor groove binding is favored, as indicated by the stoichiometry of 1.05 (± 0.05) Hoechst:oligonucleotide (Fig. 3). Furthermore, a continuous variation analysis showed that fluorescence is enhanced as expected for the minor groove complex, and the inflection point gives 1.15 ± 0.12 Hoechst/oligonucleotide (Fig. 4).40,41
Figure 2.
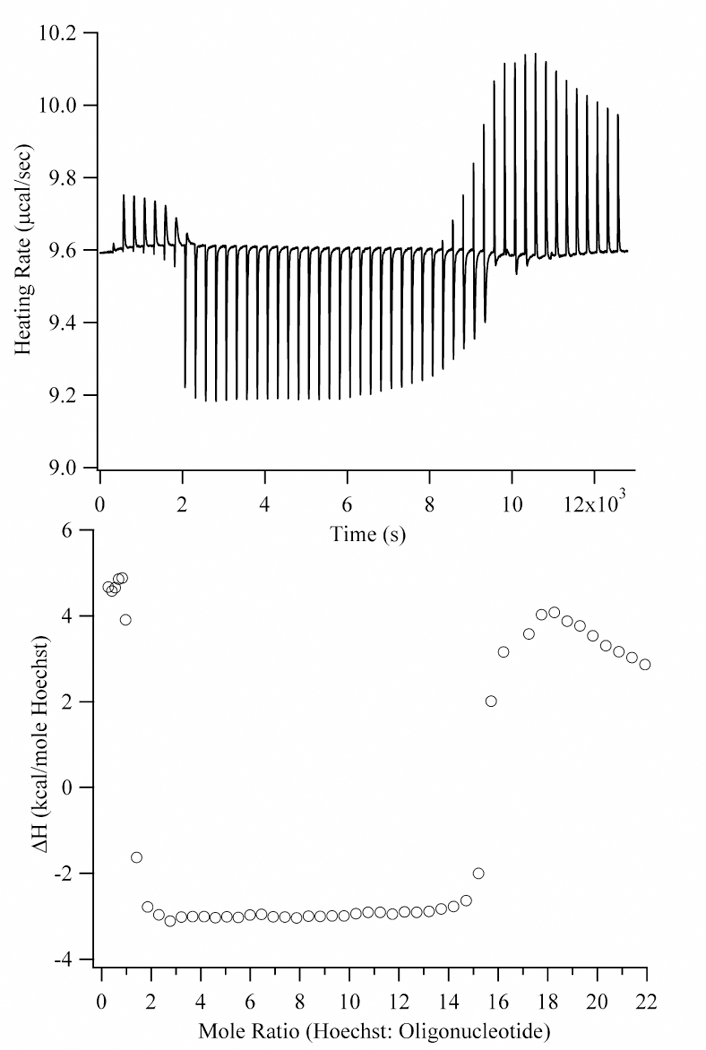
Calorimetric titration of 500 µM Hoechst into 5 µM AATT at 10 °C. The upper graph describes the heating rate as a function of Hoechst addition (2 µL additions for the first 7 injections and then 6.4 µL additions). The lower graph has the integrated heats from the upper graph plotted relative to the Hoechst:oligonucleotide ratio. The heat changes at low (0 to 1), intermediate (1 to 15), and high (>15) mole ratios are attributed to minor groove binding, Hoechst aggregation on the DNA, and Hoechst dilution, respectively. An expanded view at lower stoichiometries is shown in Fig. 3.
Figure 3.
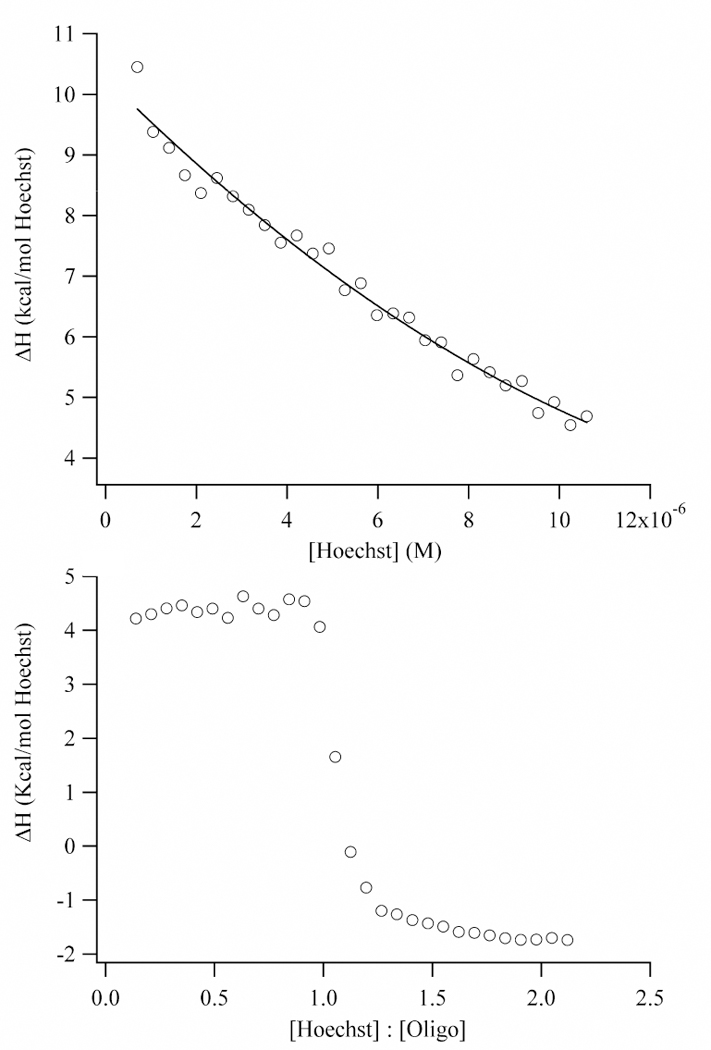
Integrated heats derived from the calorimetric titration of 500 µM Hoechst into buffer (top) and 5 µM AATT (bottom) at 10 °C. For both titrations, 1 µL of Hoechst was used. During the titration in the upper graph, Hoechst accumulates in the sample cell, which causes the enthalpy changes to decrease. The ∆H values were fit using a quadratic. In the lower graph, a one site model yielded a site size of 1.0 ± 0.1, which is consistent with minor groove binding.
Figure 4.
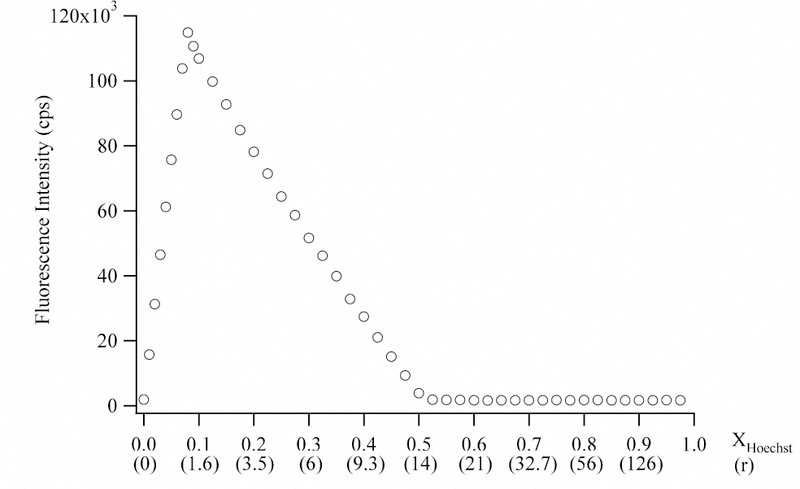
Continuous variation analysis using a total concentration of 70 µM Hoechst and base pairs (5 µM with respect to oligonucleotide) at 10 °C. The concentrations were also used in the titration shown in Fig. 2. The fluorescence intensities are plotted relative to the mole fraction with respect to Hoechst. The fluorescence emission was measured using an emission wavelength of 446 nm emission and excitation wavelength of 358 nm. In the parentheses on the x-axis, the r values (i.e. moles of Hoechst : moles oligonucleotide) are provided. The inflection point corresponding to minor groove binding occurs at 1.15 Hoechst:oligonucleotide (XHoechst = 0.076). The inflection point corresponding to aggregation occurs at 14 Hoechst : 1 oligonucleotide or 1 Hoechst : 1 base pair (XHoechst = 0.5).
After saturation of the minor groove binding site, an additional binding mode is accessed as demonstrated by the exothermic heat changes (Fig. 2). These binding sites are fully occupied at ≈15 Hoechst:DNA, which is similar to the number of base pairs in the 14 base pair oligonucleotide. (The slight discrepancy in the observed and expected stoichiometries may be due to the loss of Hoechst during the long time period for this experiment (see Experimental section)). One possible binding mode is intercalation, but the observed stoichiometry is inconsistent with the exclusion of neighboring intercalation sites.42,43 Fluorescence studies suggest that aggregation is occurring. H-aggregates occur when chromophores are cofacially stacked and are distinguished by their quenched fluorescence.44 In the continuous variation analysis using concentrations that are similar to those used in the calorimetry studies, the fluorescence decreases with increasing concentrations of Hoechst up to 14 Hoechst:oligonucleotide (Fig. 4). Such aggregation-induced fluorescence quenching is exhibited by a number of other aromatic chromophores.45,46 The inflection point at 14 Hoechst:oligonucleotide is similar to the stoichiometry observed in the calorimetry studies (Fig. 2) and in other studies of DNAHoechst aggregates.32
At higher concentrations, all the DNA binding sites are saturated, and the dissociation of Hoechst self-aggregates occurs as the concentrated Hoechst solution is diluted in the sample cell of the calorimeter. The continuous variation analysis exhibits a low fluorescence signal at these high concentrations, as expected for unbound Hoechst with its low fluorescence quantum yield (Fig. 4).40,41 Through additional experiments, similar behavior was observed when the concentrated Hoechst solution is titrated into buffer without DNA (Fig. 3). The progressively decreasing heat changes occur because accumulated Hoechst in the sample cell inhibits dissociation of the aggregates from the titrating solution. The tendency of Hoechst to aggregate in concentrated solutions is evident from the spectral shifts without isosbestic points that occur with increasing concentrations.32 In further studies, we conducted dilutions using a buffer containing 10% methanol. By lowering the dielectric constant of the buffer, self-aggregation is inhibited in the concentrated titration solution, thus lowering the heat of dilution.46 In a prior study, constant and exothermic heats of dilution were observed.21 The most significant difference that we can discern is the differing salt concentrations, and we are now investigating this effect on the heats of dilution.
Besides binding in the minor groove and with the phosphates, an additional reaction is observed at higher temperatures (Fig. 5). This complex forms after saturation of the minor groove site with a heat change that is relatively endothermic. In addition, the overall magnitude and width of the transition increase with temperature. It appears that this binding mode is not accessed at 10 °C because a stoichiometry of 1.05 ± 0.05 Hoechst/oligonucleotide was measured at this temperature. We note that the absolute magnitude of the transitions are similar (4–5 kcal) and are comparable to the heat change at 10 °C. Because the changes occur between 1 and 2 Hoechst:oligonucleotide, we consider that dimerization is occurring, a suggestion that is supported by other experiments. At µM - mM concentrations, spectroscopic studies are consistent with a dimeric binding mode of Hoechst with polynucleotides.32 Mass spectroscopic studies have identified 2:1 Hoechst complexes with oligonucleotides having the (AATT)2 binding site.47 Finally, derivatives of Hoechst have been shown to exhibit dimerization in the minor groove.48 We are now pursuing further studies with other sequences to clarify the nature of this binding mode.
Figure 5.
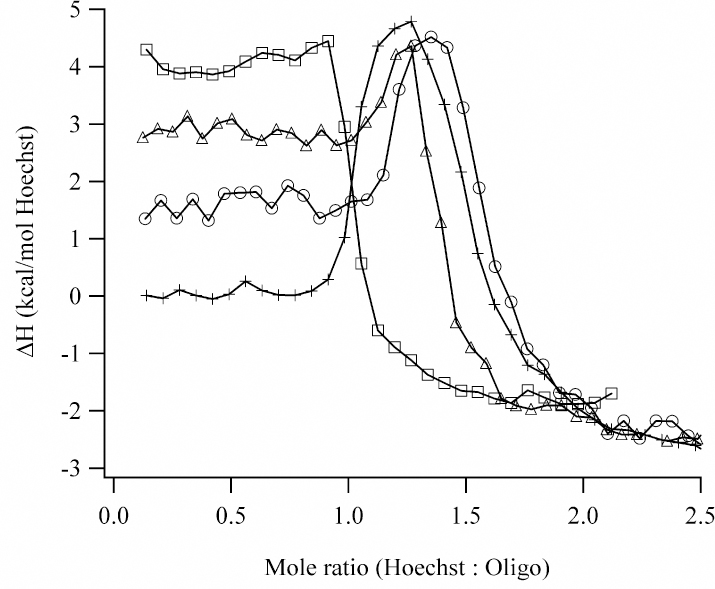
Integrated heats derived from the calorimetric titration of 500 µM Hoechst into 5 µM AATT at 10 °C (squares), 15 °C (triangles), 20 °C (circles), and 25 °C (crosses). The heat changes at low and high Hoechst:oligonucleotide stoichiometries correspond to minor groove and exterior binding, respectively. Between 15 °C and 25 °C, an additional binding mode is observed at intermediate stoichiometries. The lines between the data points are added for visual purposes.
In summary, aggregation is a significant feature of Hoechst and its interaction with DNA. These aggregated states are prominent at higher Hoechst:oligonucleotide stoichiometries. In this work, we focus on studies at lower concentrations of Hoechst where the enthalpy changes are due to the monomeric binding of Hoechst in the minor groove of the (AATT)2 sequence. Three separate measurements (in triplicate) were conducted to obtain the desired heat change (Eq. 7) with the appropriate dilution corrections:
| Eq. 4 |
| Eq. 5 |
| Eq. 6 |
| Eq. 7 |
Eq. 4 represents the titration of the concentrated Hoechst solution into the DNA solution. The reverse of Eq. 5 describes the titration of the concentrated Hoechst solution into the buffer without DNA. The reverse of Eq. 6 describes the addition of a small volume of buffer (a total of 3% of the volume in the sample cell) to the DNA solution in the sample cell. Eq. 7 incorporates the heats of dilution and describes the desired ∆H associated with binding in the minor groove.
The ∆H for the dilution of the concentrated Hoechst solution progressively decreases as the concentration of Hoechst increases in the sample cell (Fig. 3). Thus, the heat change associated with Eq. 5 is determined by assuming that each addition of Hoechst occurs into a solution without unbound Hoechst. Because of the high affinity (≈108 M−1) and the high concentrations of reactants (5 µM), the largest amount of dissociated complex is 2 – 3%, which occurs at saturation. In further support of near stoichiometric complex formation, the heat changes when Hoechst is added to DNA are constant up to saturation of the minor groove (Figs. 3 and 5). Data collected over the large range of Hoechst concentrations was extrapolated to more accurately estimate the heat change for the first injection, i.e. Hoechst dilution into pure buffer (Fig. 3). This approach avoids complications associated with the early injections.49
After accounting for the Hoechst and DNA dilutions (Eq. 4 and 5), the ∆H for minor groove binding at the different temperatures was calculated (Table 1). These exothermic heat changes are consistent with the −7.3 kcal/mole Hoechst at 25 °C determined for the reaction with (CCGGAATTCCGG)2 based on the variation of the equilibrium constants with temperatures.32 As expected for an exothermic reaction, we also observed that the affinity decreases with increasing temperature, with ∆G = −11.1 ± 0.1 kcal/mole Hoechst at 10 °C and ∆G = −10.2 ± 0.1 kcal/mole Hoechst at 25 °C, and these values are in good agreement with prior studies.31 However, the accuracy of enthalpy changes derived from van’t Hoff analysis is poor.21 For example, in the case of the binding of Hoechst with the (AATT)2 sequence, a large range from both negative to positive enthalpy changes are statistically significant from the limited data set used to generate the van’t Hoff plots. Previous studies measured endothermic heat changes for the reaction of Hoechst with A/T sequences.21,50 In Reference 21, the sequence (AAATTT)2 was used. In Reference 50, the heat after saturation of the minor groove binding site was used as the heat of dilution, but our studies indicate that this heat is due to the stacking of Hoechst on the exterior of DNA. As with other minor groove binders, molecular interactions account for a fraction of the free energy change for the reaction.21,22,51 Thus, enthalpic contributions that arise from hydrogen and van der Waals bonding must be considered in the context of other significant contributions to the overall free energy change, such as hydrophobic effects and the loss of translational and rotational degrees of freedom.
Table 1:
Enthalpy changes measured for the reaction of Hoechst with (AATT)2.i
| Temperature (K) |
Overall heat (Eq. 4) (kcal/mol) |
Hoechst dilution (Eq. 5) (kcal/mol) |
DNA dilution (Eq. 6) (kcal/mol) |
Net Heat (Eq. 7) (kcal/mole) |
|---|---|---|---|---|
| 283 | 4.3 ± 0.1 | 10.0 ± 0.1 | −0.26 ± 0.05 | −5.5 ± 0.1 |
| 289 | 2.8 ± 0.1 | 10.3 ± 0.3 | −0.17 ± 0.06 | −7.3 ± 0.1 |
| 293 | 1.0 ± 0.1 | 9.9 ± 0.2 | −0.08 ± 0.06 | −8.4 ± 0.1 |
| 298 | 0.06 ± 0.04 | 9.6 ± 0.1 | 0.16 ± 0.06 | −9.5 ± 0.1 |
Standard deviations are based on three separate measurements.
From the dependence of the enthalpy change with temperature, a ∆Cp = −256 ± 10 cal/(K mole Hoechst) was derived for the association reaction of Hoechst with (AATT)2 (Fig. 6). ∆Cp is assumed to be constant, as neither Hoechst nor DNA undergo major conformational changes over this temperature range.27,52 Despite the exothermic heat changes measured in this study, the ∆Cp value is similar to related studies.21,53 This agreement is expected if the enthalpy values differ by a heat of dilution that does not vary with temperature (Table 1). Importantly, the negative ∆Cp is consistent with the loss of solvent accessible surface area and the accompanying loss of water associated with the nonpolar and polar groups of the reactants. An empirical model derived from the intercalation of small molecules with DNA is appropriate for predicting the heat capacity changes for this reaction: ∆Cp = (0.382 ± 0.026)∆Anp - (0.121 ± 0.077)∆Ap, where ∆Anp and ∆Ap are the changes in the nonpolar and polar surface areas.54 These changes were determined using the X-ray crystal structures for the two orientations of Hoechst in the minor groove.28 For the piperazine pointing in the 3′ direction (1d44), ∆Anp = −791 Å2 and ∆Ap= −151 Å2, thus giving ∆Cp = −284 ± 24 cal/K mole Hoechst. Similar values were obtained for the piperazine pointing in the 5′ direction (1d43), with ∆Anp = −799 Å2 and ∆Ap= −137.5 Å2 resulting in ∆Cp = −288 ± 23 cal/K mole Hoechst. The deviations from the experimentally measured ∆Cp are consistent with previous studies.54
Figure 6.
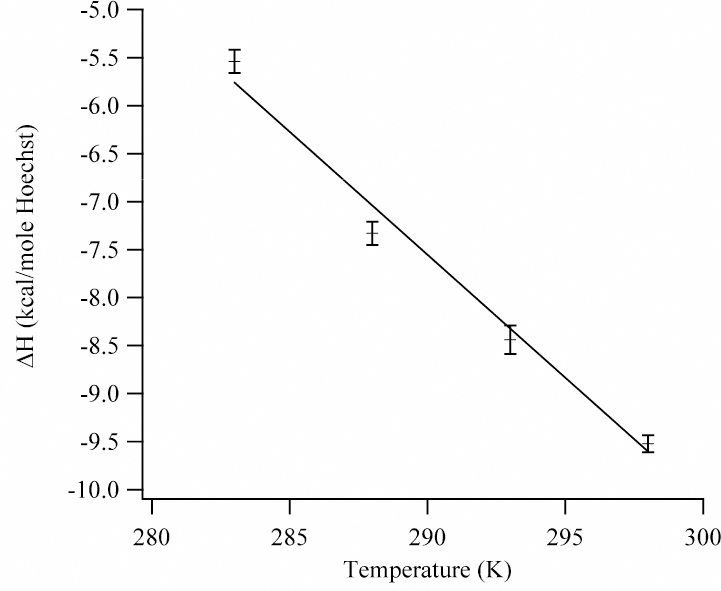
Temperature dependence of the enthalpy change for the binding of Hoechst in the minor groove of the (AATT)2 sequence. From the slope of the linear least squares fit, the heat capacity for the reaction is −256.3 ± 9.5 cal/(K mole Hoechst).
In this work, the (5′-CAATTG-3′)2 sequence was used, but the binding thermodynamics are not expected to be significantly different when compared with the (5’-GAATTC-3’)2 sequence. Specifically, Hoechst favors the (AATT)2 binding site, and flanking sequences should have a small effect.55 In support, the affinities measured in this work are comparable to previous studies.31 As shown above, the calculated ∆Cp values are insensitive to the orientation of the Hoechst. In addition, the X-ray crystal structures show equal population of the binding site by the two Hoechst orientations as the temperature is increased.28 Finally, the association kinetics support the occupation of the binding site by both orientations of Hoechst.31
Osmotic Stress Studies
Another approach to assess hydration changes is to alter the properties of the solution by using osmolytes. These solutes are used in high concentrations to reduce the activity of water.56 They are excluded from the vicinity of DNA because of their size and chemical properties. Thus, waters sequestered in the binding sites of DNA experience increasing osmotic pressure as the concentration of the osmolytes in the bulk solution increases. Equilibria that are coupled with hydration changes are influenced by the osmolyte concentration, as described by:57
| Eq. 8 |
where K is the measured equilibrium constant (for the association reaction of Hoechst with DNA in these studies), Os is the osmolality (moles solute/kg solvent) of the solution, and ∆Nw is the number of exchanged waters. Eq. 8 assumes that no partitioning of the osmolytes occurs in the vicinity of the DNA. Osmolytes have no net charge to eliminate electrostatic interactions, but even weak interactions can be significant at the high concentrations needed for osmotic stress. One approach for measuring the partition coefficients is through vapor pressure osmometry studies, but few studies have considered DNA.58 In one study, betaine is shown to be excluded from the vicinity of DNA, which makes it appropriate for osmotic stress studies.59
In addition to vapor pressure osmometry studies, a range of osmolytes with varying chemical properties and sizes is compared to determine if a common osmotic stress effect is being measured. For example, betaine and acetamide have opposite effects on the dielectric constant of the solution, yet their effect on the number of exchanged waters is similar (vide infra).60 Glycols are commonly used osmolytes, but they can interact with the hydroxyl groups of DNA, as suggested from duplex stability studies.61 So, two ethylene glycols with different sizes were used to examine this possible interference on the reaction of Hoechst with DNA.
Osmolytes can influence the properties of DNA and the Hoechst-DNA complex. For example, the conversion from B-form to A-form DNA is favored under dehydrating conditions.62 Using circular dichroism, the oligonucleotide conformation was shown to be unaltered at the highest concentrations of osmolyte (Fig. S1). Besides conformation, osmolytes could influence the stability of the duplex relative to the single-stranded state. At the highest osmolyte concentration, the melting temperature for the duplex decreases by 10 °C relative to the absence of osmolyte (Tm = 55 °C), but the duplex is stable at 25 °C, which was the temperature used in these studies (Fig. S2). Another factor is that the addition of osmolyte reduces the ionic strength of the buffer, which can enhance the affinity of the cationic Hoechst for DNA. However, the opposite effect was observed (Fig. 7). When a concentrated buffer solution was added to compensate for this dilution, no significant difference in the equilibrium constant was observed. Finally, continuous variation analysis showed that Hoechst still binds as a monomer in the minor groove in the osmolyte solutions (Fig. S3).
Figure 7.
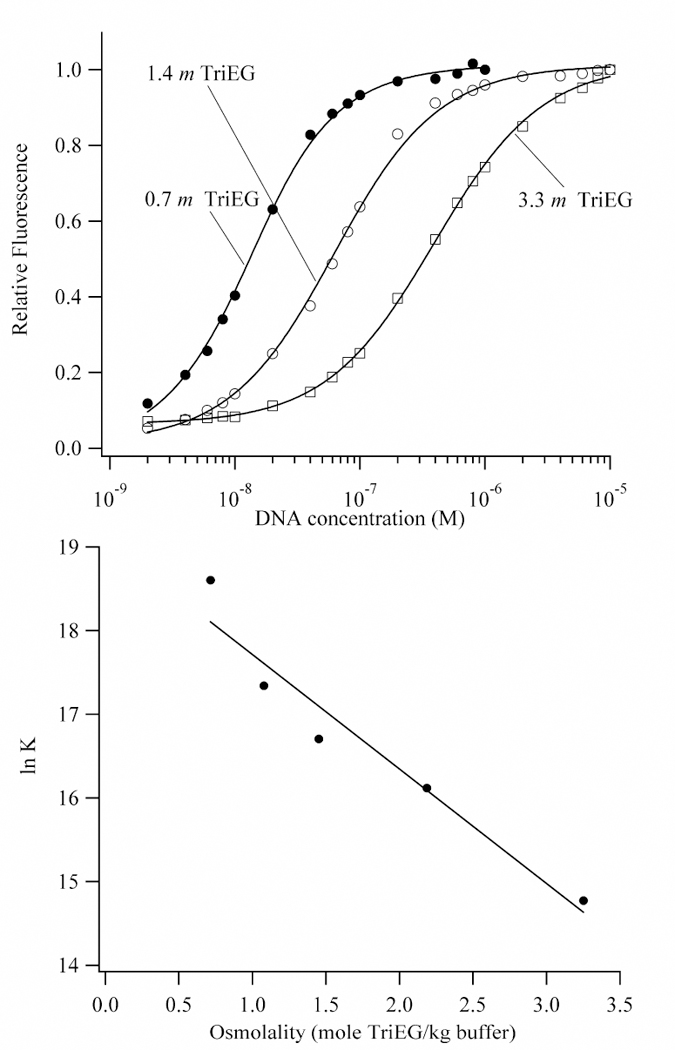
(Top) Binding isotherms for the titration of DNA into Hoechst in buffers with 0.7 m (closed circles), 1.4 m (open circles), and 3.3 m (open squares) triethylene glycol. The affinities of Hoechst for the AATT binding site decreases with increasing concentrations of triethylene glycol, which indicates that water is acquired in the complex. (Bottom) Plot of the natural logarithm of the observed equilibrium constant for the association of Hoechst with DNA as a function of the concentration of triethylene glycol. A linear least-squares fit using Eq. 8 (Kp = 0) gives 78 ± 11 waters that are acquired by the complex. The extrapolation to a 0.12 m for the buffer gives 1.6 (± 0.6) x 108 M−1, which is consistent with the direct measurement of the affinity in buffer alone (2.5 (± 0.4) x 108 M−1) at 25 °C.
As the concentration of osmolyte increases, the affinity of Hoechst for the (AATT)2 binding site decreases (Fig. 7), and this observation is consistent with the acquisition of water by the complex relative to the DNA. Using Eq. 8 with Kp = 0, the number of exchanged waters are 78 ± 11 for triethylene glycol, 51 ± 6 for acetamide, 51 ± 3 for betaine, and 67 ± 8 for tetraethylene glycol (Fig. 7 and S4). Extrapolation of the affinities to the osmolality of the buffer alone gave reasonable agreement with the directly measured affinity. This result indicates that the limiting behavior in osmolyte solutions describes the conditions in purely aqueous solution. A fit that includes the data from the measurements in all four osmolytes gives the acquisition of 60 ± 13 waters. Because osmotic stress studies measure the volume change, the density of 1 g/mL for water was used to calculate the number of exchanged waters.
The higher ∆Nw’s for the glycols could be due to their larger size: triethylene and tetraethylene glycol have diameters ≈10 Å and ≈12 Å, respectively, whereas vs. acetamide and betaine have diameters ≈4 Å and ≈5 Å, respectively. It is reasonable to expect that all these osmolytes would exert osmotic pressure on waters associated with the minor groove of the (AATT)2 sequence with a width of ≈ 4 Å.63 Because larger glycol osmolytes should be further removed from the DNA, DNA-bound water closer to the interface with the bulk solvent will be more effectively probed. This effect has been observed for protein-DNA interactions,19 and we are now investigating other ligands and DNA sequences. Vapor pressure osmometry experiments would determine if preferential interactions with DNA are occurring. In addition, we are conducting studies with alkylated glycols.61 In summary, these results with a chemically diverse range of osmolytes support a common osmotic stress effect on the binding of Hoechst with the (AATT)2 binding site. Relative to the DNA, the Hoechst-DNA complex is more hydrated with approximately 60 waters.
Discussion
The primary motivation for these studies is to understand the role of water in the sequence specific binding of Hoechst in the minor groove of the (AATT)2 sequence. Water is a prominent feature of this DNA sequence, as illustrated by the spine of hydration that emanates from the minor groove.64,65 Thus, it is expected that binding in the minor groove should disrupt the solvation of the DNA, and the calorimetry studies support this conclusion. A diverse range of reactions exhibit a linear relationship between the ∆Cp and the changes in the solvent exposure of the hydrophobic and hydrophilic groups.9,54 The binding of ligands with DNA is usually associated with a negative heat capacity change because of the loss of waters associated with the hydrophobic groups that have a larger heat capacity than bulk water.10 For the reaction of Hoechst with (AATT)2, the ∆Cp = −256 ± 10 cal/(K mole Hoechst) is consistent with this expulsion of water from DNA-Hoechst interface. This interpretation is supported by theoretical estimates of ∆Cp based on the loss of solvent exposed groups that accompanies the association reaction.
However, the osmotic stress method shows that when Hoechst binds with DNA, 60 ± 13 waters are acquired by the DNA. This result is in accord with previous studies of small molecules interacting with DNA. The association of the minor groove binder netropsin with a natural source of DNA (chicken erythrocyte) is accompanied by an uptake of 50–60 waters.66 Intercalator complexes with DNA also exhibit enhanced hydration, and the amount of water acquired increases with the size of their pendant groups.67 Of relevance to these studies, the peptide side chain of actinomycin fills a significant amount of the minor groove adjacent to the intercalation site, and the complex has 30 more waters than the DNA alone. Recent volumetric and compressibility studies have measured the acquisition of 21–34 waters in the Hoechst-DNA complex, and these values are comparable to the 60 ± 13 measured in this work.50 In addition, the volume changes measured using hydrostatic pressure indicated the enhanced hydration of the Hoechst complex with poly(dA)-poly(dT).68
A possible explanation for the disparate results from the calorimetry and osmotic stress studies is that they are sensitive to different types of bound water. A variety of experimental and theoretical studies suggest that there are different degrees of hydration of DNA. For example, waters observed by X-ray crystallography and molecular modeling are the most strongly bound.69 Thermodynamic techniques are sensitive to a broader population of bound waters, as illustrated by volumetric and compressibility measurements that account for the strength and number of intermolecular bonds between DNA and water.18 However, different types of thermodynamic measurements distinguish different degrees of hydration. For example, hydrostatic and osmotic pressure studies usually yield hydration changes that are not equivalent because of how the volume of exchanged waters is treated.70 In calorimetry studies, waters that are closest to the DNA will have thermodynamic properties that are most different from the bulk solvent, as illustrated by the high heat capacity of the clathrate-like waters associated with hydrophobic groups.10,39 The localized nature of these heat capacity differences is supported by the linear relationship between the ∆Cp and the change in the solvent accessible surface area for reactions ranging from the dissolution of small molecules in water to the unfolding of proteins.9,54 The need to consider waters beyond the immediate hydration shell of DNA is demonstrated by both experimental and theoretical studies.18,71 For example, using the Kirkwood-Buff theory, no assumptions are made about the range of solvation, and better agreement between volumetric and osmotic stress studies has been obtained.20
The use of water to direct complexation reactions is a potentially powerful tool for sequence selective binding of small molecules with DNA, and prior studies offer important insight for these studies with Hoechst.72,73,74 For example, ligands whose shapes are not complementary to the DNA binding site can still have high affinities by using water to bridge the functional groups of the DNA and ligand.14,72 Because Hoechst has a tight fit in the minor groove binding site, the acquired waters are most likely outside the interface with DNA.27,28 For example, the propamidine complex in the (AATT)2 minor groove has a spine of hydration on the exterior of the complex. These waters are possibly associated with the hydrophobic groups of the propamidine, analogous to the water associated with the methyl groups of thymine bases.75,76 For Hoechst and Hoechst analogs bound with A/T rich sequences, water is associated with the polar terminal groups - phenol and piperazine.73,77,78,79 For example, when the phenol has an hydroxyl at the meta position (see Fig. 1 for the p-OH derivative), this OH associates with a network of approximately 30 waters that continues into the major groove.79 The higher affinity of meta-OH derivatives of Hoechst suggests that this hydration network provides an enthalpic anchor for the ligand.74 Femtosecond resolved fluorescence studies indicate the presence of weakly bound waters in the Hoechst-DNA complex, but the contribution to the fluorescence changes from the overall dynamics of the DNA is also being considered.80,81 It is possible that the acquired water is not in the vicinity of the complex. Hoechst causes small changes in the DNA conformation, and the resulting changes in the solvent exposure of neighboring nucleotides may contribute to the hydration changes.28,82
In summary, the negative heat capacity change for the association of Hoechst with the (AATT)2 binding site supports the expected desolvation of the interfacial functional groups in the complex. Osmotic stress studies show that complexation with Hoechst causes the DNA to become more hydrated. We consider that the waters measured via osmotic stress are weakly bound and thus do not make a large contribution to the thermodynamic properties. While it is reasonable to expect that water can participate in biochemical reactions, the rational use of water to direct biochemical reactions is challenging.83,84 Through a variety of techniques, including ITC and osmotic stress, the role of water can be clarified. Towards this goal, we are now studying other sequences and ligands to understand the nature of this bound water.85
Supplementary Material
ACKNOWLEDGMENT
We thank the National Institutes of Health and the National Science Foundation for financial support. We also thank C. Bruce, B. Chaires, D. Matthews, B. Nguyen, B. Wallace, and D. Wilson for their assistance and helpful discussions. The comments and suggestions of the reviewers are greatly appreciated.
Abbreviations
- ∆Cp
Heat capacity change for the reaction
- ∆Nw
Number of exchanged waters
- ∆Anp
Change in the nonpolar surface area
- ∆Ap
Change in the polar surface area
References:
- 1.Saenger W, Principles of nucleic acid structure Springer-Verlag: New York, 1988. [Google Scholar]
- 2.Bloomfield VA; Crothers DM; Ignacio Tinoco Jr., Nucleic Acids: Structures, Properties, and Functions University Science Books: Sausaltio, CA, 2000. [Google Scholar]
- 3.Neidle S, DNA minor-groove recognition by small molecules. Natural Product Reports 2001, 18, (3), 291–309. [DOI] [PubMed] [Google Scholar]
- 4.Dervan PB; Poulin-Kerstien AT; Fechter EJ; Edelson BS, Regulation of Gene Expression by Synthetic DNA-Binding Ligands. Topics in Current Chemistry 2005, 253, 1–31. [Google Scholar]
- 5.Reddy BS; Sharma SK; Lown JW, Recent developments in sequence selective minor groove DNA effectors. Current Medicinal Chemistry 2001, 8, (5), 475–508. [DOI] [PubMed] [Google Scholar]
- 6.Record MT Jr.; Anderson CF; Lohman TM, Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Quarterly Reviews of Biophysics 1978, 11, (2), 103–78. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg JM, Structure and function of restriction endonucleases. Current Opinion in Structural Biology 1991, 1, (1), 104–113. [DOI] [PubMed] [Google Scholar]
- 8.Shaikh SA; Ahmed SR; Jayaram B, A molecular thermodynamic view of DNA-drug interactions: a case study of 25 minor-groove binders. Archives of Biochemistry and Biophysics 2004, 429, (1), 81–99. [DOI] [PubMed] [Google Scholar]
- 9.Spolar RS; Record MT Jr., Coupling of local folding to site-specific binding of proteins to DNA. Science 1994, 263, (5148), 777–784. [DOI] [PubMed] [Google Scholar]
- 10.Prabhu NV; Sharp KA, Heat Capacity in Proteins. Annual Review of Physical Chemistry 2005, 56, (1), 521–548. [DOI] [PubMed] [Google Scholar]
- 11.Jayaram B; Jain T, The Role of Water in Protein-DNA Recognition. Annual Review of Biophysics and Biomolecular Structure 2004, 33, (1), 343–361. [DOI] [PubMed] [Google Scholar]
- 12.Ladbury JE, Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chemistry and Biology 1996, 3, (12), 973–80. [DOI] [PubMed] [Google Scholar]
- 13.Schwabe JW, The role of water in protein-DNA interactions. Current Opinion in Structural Biology 1997, 7, (1), 126–134. [DOI] [PubMed] [Google Scholar]
- 14.Bailly C; Chessari G; Carrasco C; Joubert A; Mann J; Wilson WD; Neidle S, Sequence-specific minor groove binding by bis-benzimidazoles: water molecules in ligand recognition. Nucleic Acids Research 2003, 31, (5), 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams HEL; Searle MS, Structure, Dynamics and Hydration of the Nogalamycind(ATGCAT)2 Complex Determined by NMR and Molecular Dynamics Simulations in Solution,. Journal of Molecular Biology 1999, 290, (3), 699–716. [DOI] [PubMed] [Google Scholar]
- 16.Cooper A, Heat capacity effects in protein folding and ligand binding: a re-evaluation of the role of water in biomolecular thermodynamics. Biophysical Chemistry 2005, 115, (2–3), 89–97. [DOI] [PubMed] [Google Scholar]
- 17 (a).Colombo MF; Rau DC; Parsegian VA, Protein Solvation in Allosteric Regulation: A Water Effect on Hemoglobin. Science 1992, 256, (5057), 655–659. [DOI] [PubMed] [Google Scholar]
- (b).Rand RP, Raising Water to New Heights. Science 1992, 256, (5057), 618. [DOI] [PubMed] [Google Scholar]
- 18 (a).Chalikian TV; Breslauer KJ, Volumetric properties of nucleic acids. Biopolymers 1998, 48, (4), 264–280. [DOI] [PubMed] [Google Scholar]
- (b).Chalikian TV, Volumetric Properties of Proteins. Annual Review of Biophysics and Biomolecular Structure 2003, 32, (1), 207–235. [DOI] [PubMed] [Google Scholar]
- 19.Robinson CR; Sligar SG, Hydrostatic and osmotic pressure as tools to study macromolecular recognition. Methods in Enzymology 1995, 259, 395–427. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu S, Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, (5), 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haq I; Ladbury JE; Chowdhry BZ; Jenkins TC; Chaires JB, Specific Binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 Duplex: Calorimetric and Spectroscopic Studies. Journal of Molecular Biology 1997, 271, (2), 244–257. [DOI] [PubMed] [Google Scholar]
- 22.Haq I, Thermodynamics of drug-DNA interactions. Archives of Biochemistry and Biophysics 2002, 403, (1), 1–15. [DOI] [PubMed] [Google Scholar]
- 23.Lane AN; Jenkins TC, Thermodynamics of nucleic acids and their interactions with ligands. Quarterly Reviews of Biophysics 2000, 33, (3), 255–306. [DOI] [PubMed] [Google Scholar]
- 24.Ladbury JE; Williams MA, The extended interface: measuring non-local effects in biomolecular interactions. Current Opinion in Structural Biology 2004, 14, (5), 562–569. [DOI] [PubMed] [Google Scholar]
- 25.Rao KE; Lown JW, Molecular Recognition between Ligands and Nucleic Acids: DNA Binding Characteristics of Analogues of Hoechst 33258 Designed To Exhibit Altered Base and Sequence Recognition. Chemical Research in Toxicology 1991, 4, 661–669. [DOI] [PubMed] [Google Scholar]
- 26.Bostock-Smith C; Searle M, DNA minor groove recognition by bis-benzimidazole analogues of Hoechst 33258: insights into structure-DNA affinity relationships assessed by fluorescence titration measurements. Nucleic Acids Research 1999, 27, (7), 1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pjura PE; Grzeskowiak K; Dickerson RE, Binding of Hoechst 33258 to the minor groove of B-DNA. Journal of Molecular Biology 1987, 197, (2), 257–71. [DOI] [PubMed] [Google Scholar]
- 28.Quintana JR; Lipanov AA; Dickerson RE, Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G. Biochemistry 1991, 30, (42), 10294–306. [DOI] [PubMed] [Google Scholar]
- 29.Abu-Daya A; Fox K, Interaction of minor groove binding ligands with long AT tracts. Nucleic Acids Research 1997, 25, (24), 4962–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boger DL; Fink BE; Brunette SR; Tse WC; Hedrick MP, A Simple, High-Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. Journal of the American Chemical Society 2001, 123, (25), 5878–5891. [DOI] [PubMed] [Google Scholar]
- 31.Breusegem SY; Clegg RM; Loontiens FG, Base-sequence specificity of Hoechst 33258 and DAPI binding to five (A/T)4 DNA sites with kinetic evidence for more than one high-affinity Hoechst 33258-AATT complex. Journal of Molecular Biology 2002, 315, (5), 1049–1061. [DOI] [PubMed] [Google Scholar]
- 32.Loontiens FG; Regenfuss P; Zechel A; Dumortier L; Clegg RM, Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)] and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry 1990, 29, (38), 9029–9039. [DOI] [PubMed] [Google Scholar]
- 33.Breusegem SY; Loontiens FG; Regenfuss P; Clegg RM, Kinetics of binding of Hoechst dyes to DNA studied by stopped-flow fluorescence techniques. Methods in Enzymology(Drug-Nucleic Acid Interactions) 2001, 340, 212–233. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls A; Sharp KA; Honig B, Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 1991, 11, (4), 281–96. [DOI] [PubMed] [Google Scholar]
- 35.Pedretti A; Villa L; Vistoli G, VEGA - An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. Journal of Computer-Aided Molecular Design 2004, 18, 167–173. [DOI] [PubMed] [Google Scholar]
- 36.Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz J, Kenneth M ; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA, A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. Journal of the American Chemical Society 1995, 117, (19), 5179–5197. [Google Scholar]
- 37.Polak M; Hud NV, Complete disproportionation of duplex poly(dT)*poly(dA) into triplex poly(dT)*poly(dA)*poly(dT) and poly(dA) by coralyne. Nucleic Acids Research 2002, 30, (4), 983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CY, Determination of binding stoichiometry by the continuous variation method: the Job plot. Methods in Enzymology 1982, 87, 509–25. [DOI] [PubMed] [Google Scholar]
- 39.Sturtevant JM, Heat capacity and entropy changes in processes involving proteins. Proceedings of the National Academy of Sciences of the United States of America 1977, 74, (6), 2236–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latt SA; Wohlleb JC, Optical studies of the interaction of 33258 Hoechst with DNA, chromatin, and metaphase chromosomes. Chromosoma 1975, 52, (4), 297–316. [DOI] [PubMed] [Google Scholar]
- 41.Adhikary A; Buschmann V; Muller C; Sauer M, Ensemble and single-molecule fluorescence spectroscopic study of the binding modes of the bis-benzimidazole derivative Hoechst 33258 with DNA. Nucleic Acids Research 2003, 31, (8), 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berman HM; Young PR, The interaction of intercalating drugs with nucleic acids. Annual Review of Biophysics and Bioengineering 1981, 10, 87–114. [DOI] [PubMed] [Google Scholar]
- 43 (a).Bailly C; Colson P; Henichart JP; Houssier C, The different binding modes of Hoechst 33258 to DNA studied by electric linear dichroism. Nucleic Acids Research 1993, 21, (16), 3705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (b).Colson P; Houssier C; Bailly C, Use of electric linear dichroism and competition experiments with intercalating drugs to investigate the mode of binding of Hoechst 33258, berenil and DAPI to GC sequences. Journal of Biomolecular Structure & Dynamics 1995, 13, (2), 351–66. [DOI] [PubMed] [Google Scholar]
- 44.McRae EG; Kasha ML, The molecular exciton model. In Physical Processes in Radiation Biology, ed.; Augenstein L; Mason R; Rosenberg B, Academic Press: New York, 1964, pp. 23–42. [Google Scholar]
- 45.Seifert JL; Connor RE; Kushon SA; Wang M; Armitage BA, Spontaneous assembly of helical cyanine dye aggregates on DNA nanotemplates. Journal of the American Chemical Society 1999, 121, (13), 2987–2995. [Google Scholar]
- 46.Sovenyhazy KM; Bordelon JA; Petty JT, Spectroscopic studies of the multiple binding modes of a trimethine-bridged cyanine dye with DNA. Nucleic Acids Research 2003, 31, (10), 2561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosu F; Gabelica V; Houssier C; De Pauw E, Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Research 2002, 30, (16), e82–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satz AL; White CM; Beerman TA; Bruice TC, Double-stranded DNA binding characteristics and subcellular distribution of a minor groove binding diphenyl ether bisbenzimidazole. Biochemistry 2001, 40, (21), 6465–74. [DOI] [PubMed] [Google Scholar]
- 49.Tellinghuisen J, Volume errors in isothermal titration calorimetry. Analytical Biochemistry 2004, 333, (2), 405–406. [DOI] [PubMed] [Google Scholar]
- 50.Han F; Taulier N; Chalikian TV, Association of the Minor Groove Binding Drug Hoechst 33258 with d(CGCGAATTCGCG)2: Volumetric, Calorimetric, and Spectroscopic Characterizations. Biochemistry 2005, 44, (28), 9785–94. [DOI] [PubMed] [Google Scholar]
- 51.Lane AN; Jenkins TC, Thermodynamics of nucleic acids and their interactions with ligands. Quarterly Reviews of Biophysics 2000, 33, (3), 255–306. [DOI] [PubMed] [Google Scholar]
- 52.Milev S; Gorfe AA; Karshikoff A; Clubb RT; Bosshard HR; Jelesarov I, Energetics of sequence-specific protein-DNA association: binding of integrase Tn916 to its target DNA. Biochemistry 2003, 42, (12), 3481–91. [DOI] [PubMed] [Google Scholar]
- 53.Haq I, Thermodynamics of drug-DNA interactions. Archives of Biochemistry and Biophysics 2002, 403, (1), 1–15. [DOI] [PubMed] [Google Scholar]
- 54.Ren JS; Jenkins TC; Chaires JB, Energetics of DNA intercalation reactions. Biochemistry 2000, 39, (29), 8439–8447. [DOI] [PubMed] [Google Scholar]
- 55.Teng M; Usman N; Frederick C; Wang A, The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Research 1988, 16, (6), 2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rand RP; Parsegian VA; Rau DC, Intracellular osmotic action. Cellular and Molecular Life Sciences 2000, 57, (7), 1018–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsegian VA; Rand RP; Rau DC, Macromolecules and water: probing with osmotic stress. Methods in Enzymology 1995, 259, 43–94. [DOI] [PubMed] [Google Scholar]
- 58.Courtenay ES; Capp MW; Anderson CF; Record MT Jr., Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry 2000, 39, (15), 4455–71. [DOI] [PubMed] [Google Scholar]
- 59.Hong J; Capp MW; Anderson CF; Saecker RM; Felitsky DJ; Anderson MW; Record M. Thomas Jr. , Preferential Interactions of Glycine Betaine and of Urea with DNA: Implications for DNA Hydration and for Effects of These Solutes on DNA Stability. Biochemistry 2004, 43, (46), 14744–14758. [DOI] [PubMed] [Google Scholar]
- 60.Cohn EJ; Edsall JT, Proteins, Amino Acids, and Peptides as Ions and Dipolar Ions Reinhold: New York, 1943; pp 140–154. [Google Scholar]
- 61.Nakano S; Karimata H; Ohmichi T; Kawakami J; Sugimoto N, The Effect of Molecular Crowding with Nucleotide Length and Cosolute Structure on DNA Duplex Stability. Journal of the American Chemical Society 2004, 126, (44), 14330–14331. [DOI] [PubMed] [Google Scholar]
- 62.Egli M; Tereshko V; Teplova M; Minasov G; Joachimiak A; Sanishvili R; Weeks CM; Miller R; Maier MA; An H; Dan Cook P; Manoharan M, X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers 1998, 48, (4), 234–52. [DOI] [PubMed] [Google Scholar]
- 63.Wood A; Nunn C; Czarny A; Boykin D; Neidle S, Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2. Nucleic Acids Research 1995, 23, (18), 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drew HR; Dickerson RE, Structure of a B-DNA dodecamer. III. Geometry of hydration. Journal of Molecular Biology 1981, 151, (3), 535–56. [DOI] [PubMed] [Google Scholar]
- 65.Shui X; McFail-Isom L; Hu GG; Williams LD, The B-DNA Dodecamer at High Resolution Reveals a Spine of Water on Sodium. Biochemistry 1998, 37, (23), 8341–8355. [DOI] [PubMed] [Google Scholar]
- 66.Sidorova NY; Rau DC, The osmotic sensitivity of netropsin analogue binding to DNA. Biopolymers 1995, 35, 377–384. [DOI] [PubMed] [Google Scholar]
- 67.Qu X; Chaires JB, Hydration changes for DNA intercalation reactions. Journal of the American Chemical Society 2001, 123, (1), 1–7. [DOI] [PubMed] [Google Scholar]
- 68.Tang GQ; Tanaka N; Kunugi S, Effects of pressure on the DNA minor groove binding of Hoechst 33258. Bulletin of the Chemical Society of Japan 1998, 71, (7), 1725–1730. [Google Scholar]
- 69.Williams HEL; Searle MS, Structure, Dynamics and Hydration of the Nogalamycind(ATGCAT)2 Complex Determined by NMR and Molecular Dynamics Simulations in Solution. Journal of Molecular Biology 1999, 290, (3), 699–716. [DOI] [PubMed] [Google Scholar]
- 70.Robinson CR; Sligar SG, Hydrostatic and osmotic pressure as tools to study macromolecular recognition. Methods in Enzymology (Energetics of Biological Macromolecules) 1995, 259, 395–427. [DOI] [PubMed] [Google Scholar]
- 71.Shimizu S, Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, (5), 1195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown D; Sanderson M; Skelly J; Jenkins T; Brown T; Garman E; Stuart D; Neidle S, Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding. The EMBO Journal 1990, 9, (4), 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailly C; Chessari G; Carrasco C; Joubert A; Mann J; Wilson WD; Neidle S, Sequence-specific minor groove binding by bis-benzimidazoles: water molecules in ligand recognition. Nucleic Acids Research 2003, 31, (5), 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Breusegem SY; Sadat-Ebrahimi SE; Douglas KT; Bichenkova EV; Clegg RM; Loontiens FG, Experimental Precedent for the Need To Involve the Primary Hydration Layer of DNA in Lead Drug Design. Journal of Medicinal Chemistry 2001, 44, (16), 2503–2506. [DOI] [PubMed] [Google Scholar]
- 75.Nunn CM; Jenkins TC; Neidle S, Crystal-Structure of d(CGCGAATTCGCG) Complexed with Propamidine, a Short-Chain Homolog of the Drug Pentamidine. Biochemistry 1993, 32, (50), 13838–13843. [DOI] [PubMed] [Google Scholar]
- 76.Schneider B; Cohen D; Berman HM, Hydration of DNA bases: analysis of crystallographic data. Biopolymers 1992, 32, (7), 725–50. [DOI] [PubMed] [Google Scholar]
- 77.Wood A; Nunn C; Czarny A; Boykin D; Neidle S, Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2. Nucleic Acids Research 1995, 23, (18), 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spink N; Brown DG; Skelly JV; Neidle S, Sequence-Dependent Effects in Drug-DNA Interaction - the Crystal-Structure of Hoechst-33258 Bound to the d(CGCAAATTTGCG)2 Duplex. Nucleic Acids Research 1994, 22, (9), 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clark G; Squire C; Gray E; Leupin W; Neidle S, Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2. Nucleic Acids Research 1996, 24, (24), 4882–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pal SK; Zhao L; Zewail AH, Water at DNA surfaces: Ultrafast dynamics in minor groove recognition. Proceedings of the National Academy of Sciences of the United States of America 2003, 100, (14), 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Andreatta D; Perez Lustres JL; Kovalenko SA; Ernsting NP; Murphy CJ; Coleman RS; Berg MA, Power-Law Solvation Dynamics in DNA over Six Decades in Time. Journal of the American Chemical Society 2005, 127, (20), 7270–7271. [DOI] [PubMed] [Google Scholar]
- 82.Parkinson JA; Barber J; Douglas KT; Rosamond J; Sharples D, Minor-groove recognition of the self-complementary duplex d(CGCGAATTCGCG)2 by Hoechst 33258: a high-field NMR study. Biochemistry 1990, 29, (44), 10181–90. [DOI] [PubMed] [Google Scholar]
- 83.Lam PY; Jadhav PK; Eyermann CJ; Hodge CN; Ru Y; Bacheler LT; Meek JL; Otto MJ; Rayner MM; Wong YN; et al. , Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science 1994, 263, (5145), 380–4. [DOI] [PubMed] [Google Scholar]
- 84.Nguyen B; Hamelberg D; Bailly C; Colson P; Stanek J; Brun R; Neidle S; Wilson WD, Characterization of a Novel DNA Minor-Groove Complex. Biophysical Journal 2004, 86, (2), 1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carrondo MA; Coll M; Aymami J; Wang AH; van der Marel GA; van Boom JH; Rich A, Binding of a Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry 1989, 28, (19), 7849–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


