Abstract
Mismatch repair (MMR) systems based on MutS eliminate mismatches originating from replication errors. Despite extensive conservation of mutS homologues throughout the three domains of life, Actinobacteria and some archaea do not have genes homologous to mutS. Here, we report that EndoMS/NucS of Corynebacterium glutamicum is the mismatch-specific endonuclease that functions cooperatively with a sliding clamp. EndoMS/NucS function in MMR was fully dependent on physical interaction between EndoMS/NucS and sliding clamp. A combination of endoMS/nucS gene disruption and a mutation in dnaE, which reduced the fidelity of DNA polymerase, increased the mutation rate synergistically and confirmed the participation of EndoMS in replication error correction. EndoMS specifically cleaved G/T, G/G and T/T mismatches in vitro, and such substrate specificity was consistent with the mutation spectrum observed in genome-wide analyses. The observed substrate specificity of EndoMS, together with the effects of endoMS gene disruption, led us to speculate that the MMR system, regardless of the types of proteins in the system, evolved to address asymmetrically occurring replication errors in which G/T mismatches occur much more frequently than C/A mismatches.
INTRODUCTION
All living organisms are equipped with a DNA replication and a DNA repair system for the maintenance of genomic information. Although the DNA replication system has high fidelity, it sometimes misincorporates incorrect nucleobases. The DNA mismatch repair (MMR) system is known to correct such replication errors (1). Dysfunction in the MMR system increases the spontaneous mutation rate and can lead to cancer in humans or drug resistance in pathogenic microorganisms. In the MMR system, cells utilize MutS to detect mismatches, and the binding of MutS to a mismatched nucleotide initiates the correction pathway by activating MutL protein, which works as an activator of MutH methyl-directed endonuclease (in Escherichia coli and closely related species) or as a nickase by itself (in other organisms) (2). One of the hallmarks of the MutS-MutL-dependent MMR system compared to other DNA repair systems is the breadth of target mutations. MutS has been reported to bind to a broad range of mismatched nucleotides (3,4) and to correct G/T, C/A, C/T, A/A, T/T and G/G mismatches with similar efficiencies (5). Such situations enable organisms to repair all types of mutations (6,7); however, the breadth of targets makes it difficult to determine the origin of mutations. For example, because G/T and C/A mismatches have possibility to lead to the same mutation depending on the strand in which the replication error occurs, the ability of MutS-based system to repair both types of mismatches prevents us from determining which of the two errors occurred in the absence of the system.
Although mutS gene is known to be widely conserved throughout the three domains of life, two bacterial groups, belonging to the phylum Actinobacteria and to the phylum mollicutes, and some Archaea has no gene homologous to mutS (8). Interestingly, despite the absence of mutS homolog, mutation rate of organisms belonging to phylum Actinobacteria is comparable to that of E. coli with intact MMR system (9). Recently, it has been revealed that NucS of Pyrococcus abyssi (an organism belonging to the archaeal phylum Euryarchaeota), which had been reported to be a flapped DNA endonuclease (10), has mismatch-dependent nuclease activity, and the name EndoMS (Endonuclease Mismatch-Specific) has been proposed for this enzyme (11). Because endoMS/nucS is preferentially conserved in organisms lacking mutS, the authors suggested that EndoMS/NucS works in non-canonical MMR systems. In addition, the disruption of endoMS/nucS gene in Mycobacterium smegmatis and Streptomyces coelicolor, which are both Actinobacteria, was recently reported to result in an increased spontaneous mutation rate, although biochemical analysis of the bacterial EndoMS/NucS protein failed to show enzymatic activity (12). Thus, although EndoMS/NucS has been suggested to function in a non-canonical MMR pathway, the link between the phenotype of the endoMS/nucS gene disruptant and the biochemical properties of the EndoMS/NucS protein is not clear. Finally, whether the EndoMS/NucS-dependent pathway corrects replication errors similar to the MutS-dependent canonical system has not been analyzed.
In the present study, we found that disruption of endoMS/nucS increased the spontaneous mutation rate in Corynebacterium glutamicum. Our biochemical and genetic analyses demonstrated that C. glutamicum EndoMS/NucS is a mismatch-specific endonuclease that works cooperatively with the sliding clamp of the replisome to correct replication errors. Although our data clearly indicated that the EndoMS-pathway is a counterpart of the MutS-dependent MMR system in Actinobacteria, unlike MutS-dependent systems, the EndoMS-pathway is highly specific to G/T, G/G and T/T mismatches. These findings led us to hypothesize about the nature of replication in vivo and about the biological significance of MMR systems.
MATERIALS AND METHODS
In silico screening
A protein database that included all proteins expected from 785 whole genome sequences of bacterial and archaeal type strains was constructed on the CLC Genomics workbench (CLC Bio). Among the 785 type strains, 105 strains belong to the phylum Actinobacteria. The type strains analyzed in this study are listed in Supplementary Table S1. M. tuberculosis H37Rv proteins were categorized according to Clusters of Orthologous Groups of proteins (COGs) using in silico molecular cloning (In Silico Biology, Inc.) as described by the manufacture's protocol. Using the M. tuberculosis nucleotide sequence of genes categorized to COG ‘L’ group as a query, a BLASTX search was performed on the CLC Genomics workbench against the protein database with BLOSUM45 matrix (E-value < 0.001, gap existence and extension penalties of 16 and 1, respectively). The number of strains with BLAST hits was counted for each query and used to find genes conserved among Actinobacteria. The threshold for genes predominant in Actinobacteria were those for which BLAST hits were found in >70% of actinobacterial species (>74 species) and in <35% of other species (<238 species).
Strains, plasmids, and growth conditions
Strains and plasmids used in this study are listed in Supplementary Table S2. Oligonucleotide primers used in this study are listed in Supplementary Table S3. Escherichia coli were grown at 37°C in Luria-Bertani medium while C. glutamicum was grown aerobically at 30°C in brain heart infusion (BHI; BD) medium with shaking at 200 rpm. When appropriate, kanamycin was added to the media at a concentration of 12.5 μg/ml (C. glutamicum) or 50 μg/ml (E. coli) and ampicillin at 100 μg/ml (E. coli). In-frame deletion mutants of C. glutamicum and codon exchanges in chromosomal genes were constructed by using a two-step homologous recombination protocol as described previously (13) using suicide vector.
Construction of plasmids
To construct pSBK3 the sacB gene preceded by tac promoter (14) was amplified from chromosomal DNA of Bacillus subtilis 168 by PCR with primers SalI-SpeI-Ptac-TSP-F, Ptac-sacR_F(BglII) and sacB-Cterm_R(SpeI). The whole region of pHSG298 (TaKaRa) was amplified by PCR with primers pHSG_ColE1_ori_F1(BglII-SpeI) and pHSG_KmR_F1(XhoI). Ptac-sacB fragment digested by SalI (TaKaRa) and BglII (TaKaRa) was ligated with PCR product of pHSG298 digested with BglII and XhoI (TaKaRa), yielding pSBK3. For construction of pColdIV-Cg-nucS derivatives, the endoMSCg gene and its derivatives were amplified from chromosomal DNA of C. glutamicum by PCR, and the PCR product was cloned into the NdeI–EcoRI site of expression vector pColdIV (Takara). The dnaNCg gene was amplified with primers NCgl0002_Nterm_NdeI_F and NCgl0002_Cterm_EcoRI-NotI_R, and the PCR product was cloned into NdeI/NotI site of pQE2, yielding pQE2-Cg-dnaN. The endoMSCg gene with upstream 400 bp was amplified with primer pair NCgl1167_F2(SacI)/NCgl1168(nucS)_R2(EcoRI-XhoI) and cloned into pYTKA1, yielding pYTKA1-PnucS-Cg-nucS (WT). The promoter-SD (Shine-Dargano sequence) region of NCgl1526 (gapA) was amplified with primer pair PgapA-PstI-F/PgapA_R and used for construction of pYTKA1-PgapA variants. The endoMSCg, endoMSMt and endMSTk genes were amplified with primer pairs PgapA-NCgl1168_F2/NCgl1168(nucS)_R2(EcoRI-XhoI), PgapA-Rv1321_F2/Mtb_Rv1321_R1(EcoRI) and PgapA-TK1898_F2/TK1898_R1(SalI), respectively. Genomic DNA of M. tuberculosis ATCC35801 was used as PCR template to amplify nucSMt. Genomic DNA of T. kodakarensis JCM12380T was purchased from RIKEN BRC. Appropriate two fragments were fused by overlap extension PCR (15) to obtain PgapA-Cg-nucS, PgapA-Mtb-nucS and PgapA-Tk-endoMS fragments and each fragment was cloned into pYTKA1. The NCgl2049 (dnaE) gene with 400 bp upstream region was amplified with primer pair NCgl2050_R1(SalI-SpeI)/NCgl2049_R1(SalI-SpeI) and cloned into pYTKA1. Site-directed mutagenesis was performed by inverse PCR using primers listed in Supplementary Table S3.
Overexpression and purification of His-tagged proteins
The endoMSCg gene and C-terminal region of the dnaNCg gene (S98-G394) were amplified from the chromosomal DNA of C. glutamicum ATCC13032 by PCR with primer pairs NCgl1168(nucS)_F2(NdeI)/NCgl1168(nucS)_R2(EcoRI-XhoI) and NCgl0002_F1(SphI)/NCgl0002_Cterm_EcoRI-NotI_R, respectively. The PCR product of endoMSCg was cloned into the NdeI-EcoRI site of expression vector pColdI (Takara), yielding pColdI-Cg-NucS. The PCR product of the C-terminal region of the dnaNCg gene was cloned into the BamHI-NotI site of the expression vector pColdI (Takara), yielding pColdI-Cg-dnaN-Cterm. His-tagged proteins were overexpressed in E. coli BL21-CodonPlus (DE3)-RIPL (Agilent Technologies) by cold shock and purified by affinity chromatography as described by the manufacture's protocol. The concentration of the purified protein was determined using a Bio-Rad protein assay (Bio-Rad Laboratories) with bovine serum albumin (BSA) as a standard.
Overexpression and purification of EndoMSCg and His-DnaNCg
EndoMSCg was overproduced in E. coli BL21 CodonPlus (DE3)-RIPL harboring pColdIV-Cg-nucS (WT). The cells were grown to OD600 = ∼0.8 at 37°C in LB medium containing 50 μg/ml carbenicillin, and further incubated with 0.4 mM IPTG at 16°C for 18 h. Cells were lysed in Lysis buffer (50 mM Tris–HCl, pH 8.0, 0.25 M NaCl, 10 mM spermidine, 2 mM dithiothreitol, 20 mM EDTA 0.1 mM PMSF, 0.2 mg/ml lysozyme). The cell lysate was treated with 0.25% polyethyleneimine in Lysis buffer, to remove nucleic acid. Then, the soluble fraction was precipitated by 0.30 g/ml ammonium-sulfate, followed by resuspension in column buffer (50 mM Tris–HCl, pH7.5, 1 mM EDTA, 20% glycerol, 4 mM dithiothreitol). The resuspended fraction was applied to a SOURCE 15Q 4.6/100 PE column (GE Healthcare), and EndoMSCg was eluted with linear gradient of 0.01–1.0 M NaCl in column buffer. EndoMSCg D144A and EndoMSCg -Cdel mutant proteins were purified by the same procedures as for EndoMSCg, using pColdIV-Cg-nucS (D144A) and pColdIV-Cg-nucS (Cdel), respectively.
His-DnaNCg was overproduced in E. coli BL21 CodonPlus (DE3)-RIPL harboring pQE2-Cg-dnaN. The cells were grown to OD600 = ∼0.8 at 37°C in LB medium containing 50 μg/ml carbenicillin, and further incubated with 1 mM IPTG at 37°C for 20 h. Cell lysis, polyethyleneimine treatment and ammonium-sulfate precipitation were performed by the same procedures as for EndoMSCg. The ammonium-sulfate precipitation was resuspended in Ni-column buffer (20 mM Tris–HCl, pH7.5, 500 mM NaCl, 2 mM 2-mercapt ethanol, 10% glycerol, 0.2 mM EDTA, 10 mM Imidazole). The resuspended fraction was applied to a HisTrap HP column (GE Healthcare), and His-DnaNCg was eluted with linear gradient of 10–500 mM Imidazole in Ni-column buffer. The protein concentrations were measured by the Bradford assay.
In vitro cleavage assay
Mismatch DNAs were prepared by annealing of oligonucleotides (Supplementary Table S3). The mismatch DNA (50 nM) was incubated at 37°C for 60 min in 10 μl of cleavage buffer (20 mM Tris–HCl, pH 8.0, 10 mM Mg[oAc]2, 4 mM dithiothreitol, 0.071 mg/ml BSA, and 100 mM [or indicated concentration] K[oAc]) in the presence of 1 μM EndoMSCg and/or 1 μM His-DnaNCg, unless otherwise indicated. The reaction was then stopped by the addition of an equal volume of 2 × Stop buffer (50 mM Tris–HCl, pH 8.0, 50 mM EDTA, 0.2% sodium dodecyl sulfate, 0.1 mg/ml proteinase K, 10% glycerol, 0.02% bromophenol blue) and further incubated at 37°C for 30 min. An aliquot (5 μl) of this mixture was analyzed by 12% native PAGE followed by SYBR Green I staining (Molecular Probes). For the denaturing PAGE analysis of 5′-FAM-labeled DNA, the reaction sample (1 μl) was added to 4 μl of denaturing buffer (100 mM Tris-borate, pH 8.3, 2 mM EDTA, 10 M urea), incubated 95°C for 5 min, and then analyzed by 8 M Urea 20% PAGE. Oligonucleotides used as 5′-FAM-labeled size markers are listed (Supplementary Table S5). The images were acquired with a Typhoon FLA 9500 (GE Healthcare).
Preparation of polyclonal antibodies
One hundred μg of purified His-EndoMSCg and His-DnaNCg-Cterm proteins was used as antigen to immunize rabbits or rats with Freund's adjuvant (Difco). After four times of booster, animals were sacrificed to prepare antiserum. The animal experiments were approved by the Ethical Committee for Animal Experiments at the Research Institute of the National Center for Global Health and Medicine.
Immunoprecipitation followed by immunoblot analysis
C. glutamicum ATCC13032 WT and nucS-Cdel strains were grown aerobically in 250 ml of BHI medium for 4h. Formaldehyde was added at final concentration of 1% (W/V) and incubated for 30 min at room temperature. Crosslinking reaction was quenched by adding of 4 ml of 1M Tris–HCl (pH7.5). Cells were collected by centrifugation (6000 × g, 10 min) and washed twice with 20 ml of cell wash buffer (50 mM Tris–HCl, H7.5, 50 mM NaCl). Cells were resuspended in HNTG buffer (20 mM HEPES buffer pH 7.5, containing 150 mM NaCl, 0.1% (w/v) Triton X-100 and 10% (w/v) glycerol) supplemented with cOmplete™ Protease Inhibitor Cocktail (Roche) and sonicated using handy-type sonicator (UR-20P, Tomy). Cell lysates were centrifuged (20 000 × g, 30 min) and supernatants were collected as cell free extracts (CFEs). CFEs were incubated with anti-EndoMSCg antibody raised in rats for at 4°C. After 1 h incubation, 50 μl of protein G-Sepharose 4FF (GE healthcare) was added for a further 1 h. The precipitates were washed five times with HNTG buffer and SDS-PAGE 4× sample buffer was added.
Cell-free extracts (Input) and immunoprecipitated fraction (IP) were analyzed on 12% polyacrylamide gels. After transfer to Immobilon-P polyvinylidene fluoride membranes (Millipore), EndoMSCg and DnaNCg were detected with anti-EndoMSCg and anti-DnaNCg antibodies raised in mice.
Bio-layer interferometry (BLI) analysis
The BLItz system with Ni-NTA (NTA) biosensor (Pall ForteBio) was used to monitor BLI response. Analysis steps were consists of 30 s initial baseline, 120 s DnaN-loading, 120 s baseline, 120 s EndoMS association and 120 s EndoMS dissociation followed by Ni-NTA biosensor regeneration steps (30 s 100 mM Glycin (pH 1.7), 10 sec H2O, 60 s 10 mM NiCl and 30 s cleavage buffer (20 mM Tris–HCl, pH 8.0, 10 mM Mg[oAc]2, 4 mM dithiothreitol, 0.071 mg/ml BSA and 100 mM K[oAc]). Cleavage buffer was used throughout analysis steps. In the DnaN loading step, 50 ng/μl purified His-DnaNCg was used.
Electrophoretic mobility shift assay (EMSA)
Double stranded DNA with or without G/T mismatch was incubated with indicated components at 37°C for 5 min in cleavage buffer. Then the samples were chilled on ice for 2 min and UV crosslink was conducted as described previously (3 × 105 μJ) (16). Resultant samples were subjected to native-PAGE in Tris-borate buffer (without EDTA).
Estimation of mutation rates by fluctuation tests
Mutation rates to rifampicin resistance was estimated using fluctuation tests as described (17). Mutant frequencies for each indicated strain were determined by starting 20 cultures (2 ml of BHI) from single colonies and growing them to saturation at 30°C. Appropriate dilutions of the cultures were plated on BHI plates containing 2 μg/ml rifampicin to determine the number of rifampicin resistant mutant and on BHI plates to determine the total cell count. The mutation rate was calculated using the Ma–Sandri–Sarkar maximum likelihood method (18) and confidence limits (CL) (17) were calculated as described using FALCOR web tool found at www.mitochondria.org/protocols/FALCOR.html (19).
Mutation accumulation
MA lines originated from single colonies isolated on BHI agar plates. Every day, a single colony isolated from each line was streaked onto a new BHI agar plate, with four lines per plate, and incubated at 30°C for 24 h. Culture was initiated with 40 lines of the wild-type strain, 20 lines of the ΔendoMS strain, 16 lines of the dnaE (D647G) strain, and 16 lines of the ΔendoMS -dnaE (D647G) strain. MA lines were passaged through 82 single-cell bottlenecks for the wild-type strain, 7 bottlenecks for the ΔendoMS strain, 7 bottlenecks for the dnaE (D647G) strain and 5 bottlenecks for the ΔendoMS -dnaE (D647G) strain. For all strains, the number of cells in a colony ranged from 2.2–3.8 × 107, with a log average of 3.0 × 107 cells, which indicated 24.8 generations.
Genomic DNA preparation and whole genome resequencing analysis
Single colonies from the endpoints of MA lines and from a parental stock of each strain were isolated and grown in BHI medium, and genomic DNA was extracted using a Qiagen DNeasy Blood & Tissue Kit. The genomes were subjected to MiSeq sequencing using Nextera XT library kits (Illumina, Inc., San Diego, CA, USA) according to the manufacturer's instructions. Approximately one million pair-end reads (301 base pairs [bp] × 2) were obtained from each genome. Sequence data obtained for the MA lines were archived in DDBJ under BioProject PRJDB6606. Data analyses were performed using CLC Genomics Workbench software (Qiagen, Hilden, Germany). The reads from each line were trimmed by screening for base quality (quality score limit = 0.05; reads that contained greater than two ambiguous nucleotides or that were less than 15 bp in length were removed). Trimmed reads were mapped to the C. glutamicum ATCC13032 genome (NC_003450) with the following parameters: no masking mode, match score = 3, mismatch cost = 3, linear gap cost, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity fraction = 0.99 and no global alignment. A basic variants detection analysis was performed with the following parameters: ploidy = 1, ignore positions with coverage above 100,000, restrict calling to target regions = not set, ignore broken pairs = no, ignore non-specific matches = reads, minimum coverage = 7, minimum count = 2, minimum frequency (%) = 90, base quality filter = yes, neighboring radius = 5, minimum central quality = 30, minimum neighboring quality = 30, read direction filter = no, relative read direction filter = yes, significance (%) = 1, read position filter = no and remove pyro-error variants = no. Single nucleotide polymorphisms (SNPs) detected in MA lines were compared with those in the parental stock, and SNPs detected only in MA lines were counted as mutations in the MA experiment. Because of the probable loss due to homologous recombination in some MA lines, mutations that occurred in the region from 1 958 664 to 1 994 456 were not counted. In the case of wild-type and ΔendoMS strains, mapping of all mutation sites was manually confirmed, and SNPs that detected in ambiguous mapping locations (probably because of the existence of highly homologous sequences) were ignored. In the case of strains dnaE (D647G) and ΔendoMS -dnaE (D647G), consensus bases across all MA lines were compared, and SNPs was counted as mutations if they were observed only in that line but not in the rest (6). To represent mutations as nucleotide changes in the leading strand, SNPs were categorized by the relative position to the ori (replication initiation site; position 1 or 3 309 401) and dif sites (replication termination site; position 1 551 515 (20,21)). When a mutation was located between positions 1 551 515 and 3 309 401 (the so-called left replicore), the mutation was represented as the mutational change of the bottom strand, which corresponded to the leading strand in the left replicore.
RESULTS
In silico screening of genes predominantly conserved in Actinobacteria
M. tuberculosis is the most important pathogen in Actinobacteria. Therefore, to identify alternative genes that compensate for the lack of a canonical mismatch repair system in Actinobacteria, BLAST searches against a protein database with whole genome sequences of 785 type strains of bacteria and archaea (Supplementary Table S1) were performed using the nucleotide sequence of M. tuberculosis H37Rv genes categorized as Clusters of Orthologous Groups (COGs) ‘L’ (replication, DNA repair and recombination) as queries, except for genes encoding mobile elements. From 142 queries, 20 candidate genes were identified that matched the defined criteria (see Methods and Supplementary Table S4).
Identification of genes working for the maintenance of low levels of spontaneous mutation
Because of the fast growth and simplicity of the method to construct a markerless gene deletion mutant, C. glutamicum was used as a model organism. Among 20 candidate genes, homologues of three genes (Rv1267c, Rv3114 and Rv3908) were not found in C. glutamicum. The C. glutamicum homologues of Rv2412 (NCgl2261) and Rv2738c (NCgl1806) were annotated as ribosomal protein and phage integrase, respectively, and were not analyzed further. We attempted to obtain a C. glutamicum in-frame deletion mutant strains with the remaining 15 genes, and deletion mutants of 12 genes, except for those of NCgl0004, NCgl2264 and NCgl2982, were obtained (Supplementary Table S4). An examination of the frequency of spontaneous rifampicin resistance mutations indicated that deletion of NCgl1168 (endoMS/nucS), corresponding to Rv1321 (endoMS/nucS) in M. tuberculosis (endoMS/nucSMt), increased the frequency of rifampicin resistance mutations (Supplementary Figure S1).
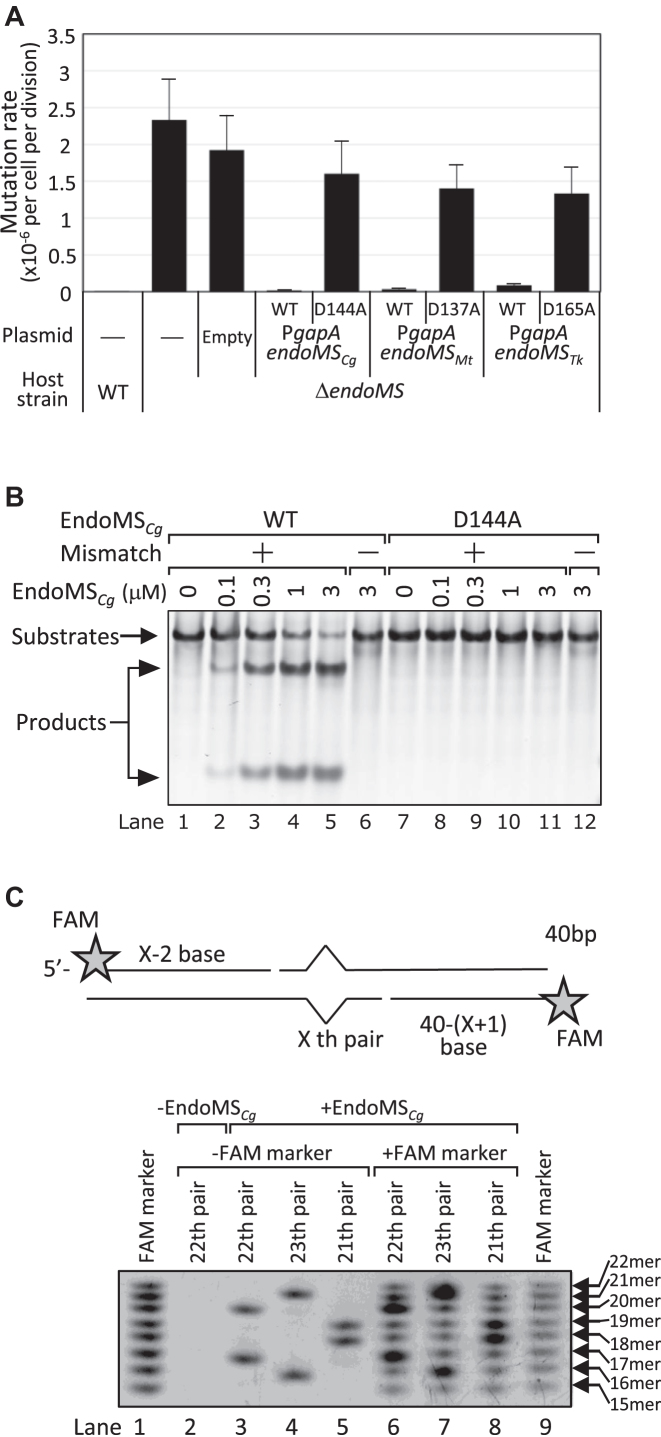
The rate of rifampicin resistance mutations estimated from the fluctuation assay was 8.2 × 10−9 per cell per division in wild-type C. glutamicum. Thus, deletion of NCgl1168 (endoMS/nucSCg) showed a mutator phenotype, in which the rate of spontaneous mutation increased ∼280-fold. Complementation with a plasmid containing endoMS/nucSCg completely restored the mutator phenotype, indicating the essentiality of endoMS/nucSCg in maintaining the low spontaneous mutation rate in C. glutamicum (Figure. 1A).
Figure 1.
Complementation of ΔendoMSCg by various EndoMS/NucS overexpression plasmids and in vitro DNA cleavage assay by EndoMS/NucSCg. (A) Rates of spontaneous rifampicin resistance mutations determined by fluctuation assay. Rates of mutations conferring rifampicin resistance to C. glutamicum ATCC13032 (WT) and its ΔendoMSCgderivatives harboring pYTKA1 plasmid with the indicated gene. Error bars show 95% confidence intervals (n = 20). (B, C) In vitro cleavage assay of mismatch-containing DNA by EndoMSCg. (B) DNA substrates with (+) or without (−) G/T mismatch (100-bp, 50 nM) were incubated with the indicated concentrations of wild-type EndoMSCg protein (lane 1–6) or D144A mutant (lane 7–12). (C) The cleavage site was determined using a G/T mismatch substrate (40-bp, 50 nM) with a FAM fluorescent label at the 5′ ends. Schematic drawing of substrates and cleavage products (upper panel). Results of denaturing PAGE of in vitro cleavage products (lower panel). FAM-labeled DNA substrates containing G/T mismatch at indicated position were subjected to in vitro cleavage assay and the products were separated by denaturing PAGE. To determine the cleavage product size, 5′-FAM-labeled oligonucleotides (15–22 mer) were mixed with cleavage products and analyzed (lane 6–8).
C. glutamicum EndoMS is mismatch-specific endonuclease and its nuclease activity is essential to maintaining a lower spontaneous mutation rate
EndoMS/NucS homologues have a RecB-like nuclease domain (10–12). Thermococcus kodakarensis EndoMS (EndoMSTk) has been reported to have mismatch-dependent double-stranded DNA (dsDNA) cleavage activity, and a mutational analysis showed that point mutations in the RecB-nuclease motif resulted in the loss of activity (11). In contrast, purified Mycobacterium smegmatis EndoMS/NucS (EndoMSMs) has been reported not to have the ability to cleave mismatch-containing dsDNA (12). To examine whether EndoMS/NucSCg has nuclease activity, we performed an in vitro cleavage assay using purified EndoMS/NucSCg protein (Supplementary Figure S2). An analysis of EndoMS/NucSCg cleavage products by native PAGE revealed that EndoMS/NucSCg digested dsDNA in a mismatch-dependent manner (Figure 1B, lanes 2–6). Although we found that the EndoMS/NucSCg-dependent cleavage of G/T mismatch was more efficient under low salt condition containing more than 0.3 mM Mn2+ ion, we used 100 mM potassium acetate with 10 mM Mg2+ condition to examine the enzymatic activity under physiologically possible condition (Supplementary Figure S3). As observed for EndoMSTk, our in vitro cleavage assay revealed that mutations in the RecB-nuclease motif (D144A) completely diminished the ability to cleave G/T mismatch-containing dsDNA (Figure 1B, lanes 8–11). These results clearly demonstrated that cleavage of mismatch containing dsDNA was dependent on intact EndoMS/NucSCg enzyme but not on any other contaminated nuclease in purified EndoMS/NucSCg fraction. Therefore, we use the name EndoMSCg to refer to this enzyme hereafter. An examination of overexpression of a D144A mutant of EndoMSCg in ΔendoMS cells showed that the D144A mutant protein did not restore the mutator phenotype (Figure 1A), although the expression level of the D144A mutant was comparable to that of the wild-type protein (Supplementary Figure S4), indicating the essentiality of nuclease activity to lower mutation rate. The number of EndoMSCg molecules in a wild-type cell was estimated to be 14 molecules per one cell, given that 1 bacterial cell contains 0.15 pg protein. Although this value was about one order lower than that of T. kodakarensis (11), our observation that the mutation rate of cells overexpressing EndoMSCg (Supplementary Figure S4) was not different from that of WT cells (Figure 1A, 15 versus 8.3 × 10−9 per cell per division, respectively, P = 0.17 (n = 20), Mann-Whitney test) suggests that the expression level of EndoMSCg in wild-type C. glutamicum cells is sufficient. To determine the cleavage sites, 5′-FAM-labeled dsDNA was subjected to an in vitro cleavage assay followed by denaturing PAGE (Figure 1C). The results showed that EndoMSCg cleaved the 5′ side of the mismatched base in both strands to produce a cohesive end with a three-nucleotide long 5′-protrusion.
When M. tuberculosis EndoMS/NucS (EndoMSMt) or EndoMSTk was overexpressed in ΔendoMS of C. glutamicum, the mutator phenotype was partially restored, whereas the RecB-nuclease motif mutant (D137A of EndoMSMt or D165A of EndoMSTk) did not decrease mutation rate, confirming the importance of the RecB-nuclease motif maintaining the low mutation rate (Figure 1A).
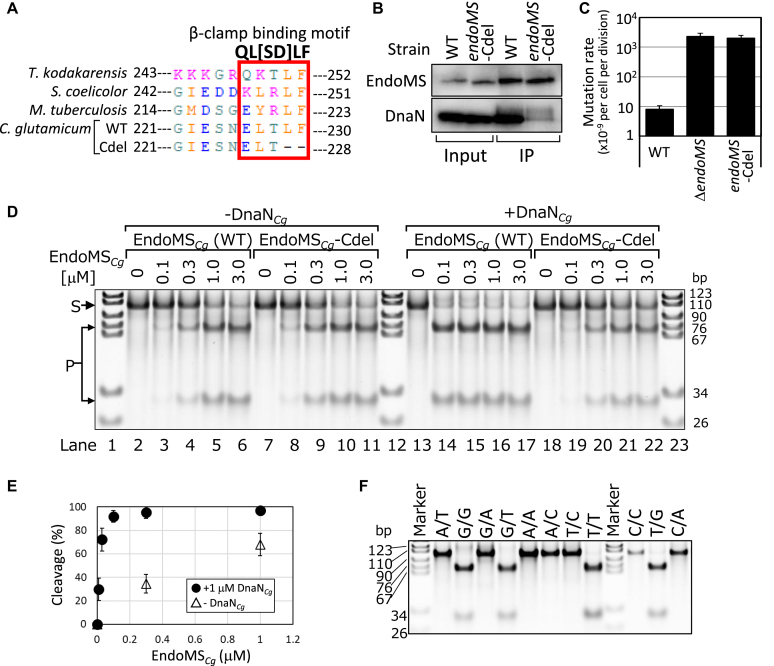
Interaction of EndoMS with the sliding clamp (beta-clamp) enhances EndoMS activity and is essential to maintaining a lower mutation rate
The existence of a beta-clamp (DnaN) binding motif in the C-terminus of EndoMS (Figure 2A) prompted us to examine whether EndoMS interacts with DnaN. Although an immunoblot analysis following immunoprecipitation of either EndoMSCg or DnaNCg under normal conditions did not result in the detection of their counterparts in the immunoprecipitated (IP) fraction (data not shown), crosslinking treatment of cells using formaldehyde enabled us to detect DnaNCg in the EndoMSCg IP fraction (Figure 2B, lower panel). As expected, when the endoMS-Cdel strain was subjected to immunoprecipitation assay, co-recovery of the DnaNCg protein with EndoMS-Cdel was severely reduced. Direct interaction between EndoMSCg and DnaNCg was confirmed by Bio-layer interferometry (BLI) analysis using purified proteins (Supplementary Figure S5). Loading of EndoMSCg wild-type and D144A mutant proteins after DnaNCg preloading showed BLI signal at association step (Supplementary Figure S5A and B). In contrast, as expected, such binding signal was not observed when DnaN pre-loading was not conducted or EndoMSCg Cdel mutant was used (Supplementary Figure S5C), although baseline shift caused by unknown reason was observed between association and dissociation steps. Estimated KD values of EndoMSCg wild-type and D144A mutant proteins were 185 nM and 182 nM, respectively, under our assay condition.
Figure 2.
The effects of EndoMS-sliding clamp interactions in vitro and in vivo. (A) Alignment of the C-terminal region of EndoMS/NucS proteins. (B) Immunoprecipitation analysis of EndoMS. Cell-free extracts (Input) of the indicated strains were subjected to immunoprecipitation (IP) with anti-EndoMSCg antibody followed by immunoprobing with anti-EndoMSCg (upper panel) and anti-DnaNCg (lower panel) antibodies. (C) The effect of deleting the sliding clamp binding motif in EndoMSCg on the mutation rate. The rate of spontaneous rifampicin resistance mutations was determined by fluctuation assay. Error bars show 95% confidence intervals (n = 20). (D) Enhancement of EndoMSCg activity with the addition of DnaNCg. An in vitro cleavage assay using intact EndoMSCg (WT) (lane 2–6 and 13–17) and the EndoMSCg-Cdel mutant (lane 7–11 and 18–22) was carried out in the absence (lane 2–11) or presence (lane 13–22) of 1 μM DnaNCg. The dsDNA with (+) or without (−) G/T mismatch flanking to A residues (100-bp, 50 nM) were used as substrates. The EndoMSCg protein concentration in the reaction mixture is indicated. Substrates and cleavage products were indicated by ‘S’ and ‘P’ on the left of panel, respectively. The percentage of cleaved substrates was indicated on the bottom of the panel. (E) Titration curve of relative cleavage activity and concentration of EndoMSCg. Relative cleavage activity was plotted against the concentration of EndoMSCg based on the data shown in Supplementary Figure S7A. Experiments were repeated three times and average with standard deviation was shown. (F) DNA substrates containing the indicated mismatch flanking to A residues (100-bp, 50 nM) were incubated with 3 μM EndoMSCg and 1 μM DnaNCg.
We next examined whether disruption of DnaN-EndoMS interactions affected the mutation rate in vivo. Surprisingly, the mutation rate of the endoMS-Cdel strain was almost the same as that of the endoMS gene disruptant (Figure 2C). We also confirmed that overexpression of EndoMSCg-Cdel mutant in ΔendoMSCg cells using strong gapA promoter could not restore the mutation rate at all (Supplementary Figures S4 and S6A). This result indicates that DnaN-EndoMS interaction itself is essential for EndoMS to maintain a lower spontaneous mutation rate. An in vitro cleavage assay using the EndoMSCg-Cdel mutant protein showed that EndoMSCg-Cdel cleaved mismatched dsDNA as efficiently as intact EndoMSCg did (Figure 2D, −DnaNCg). We next examined the effect of the DnaNCg protein on activity of the nuclease. As expected, the addition of DnaNCg considerably enhanced the cleavage efficiency of intact EndoMSCg but not that of EndoMSCg-Cdel (Figure 2D, +DnaNCg). Increasing concentration of DnaNCg in the reaction mixture increased the amount of cleavage product under both low salt (30 mM potassium acetate) and high salt (150 mM potassium acetate) conditions (Supplementary Figure S6B). In the presence of excess DnaNCg (2.7 μM DnaNCg against 50 nM DNA substrates), inhibition of the EndoMSCg activity was observed. When the substrate DNA concentration was reduced to 5 nM, the concentration of DnaNCg required for the inhibition decreased to 0.3 μM, suggesting that DNA-free DnaNCg titrate out of EndoMSCg from DNA substrates (Supplementary Figure S6C, D). Or, too much DnaNCg loaded on DNA may mask the mismatch site. Although DnaN loads onto DNA by the clamp loader complex (22), our electrophoretic mobility shift assay partially detected the assembly of DnaNCg onto the linear DNA substrate even in the absence of the clamp loader complex (Supplementary Figure S6E). In consistent with these idea, cleavage efficiency of EndoMSCg-Cdel mutant was slightly but reproducibly inhibited by addition of DnaNCg, probably due to the masking effect of DnaNCgloaded on to the substrate DNA (Figure 2D, lanes 7–11 and lanes 18–22). A quantitative analysis of the specific activity of EndoMSCg in the presence or absence of DnaNCg revealed that DnaNCg increased EndoMSCg activity about 30-fold (Figure 2E and Supplementary Figure S7A). We then tried to reveal the effects of DnaNCg-EndoMSCg interaction on the affinity of EndoMSCgto the DNA by performing electrophoretic mobility shift assay (EMSA). Although both wild-type (Supplementary Figure S7B) and D144A mutant (data not shown) of EndoMSCg were used, any upshifted bands were not detected. Instead, in the presence of DnaNCg, cleavage products were detected. Consequently, we could not clarify whether DnaNCg binding to EndoMSCg increase the affinity of EndoMSCg to DNA or not. However, it should be noted that these results were quite different from that of EndoMSTk in which band shift caused by EndoMSTk-DNA complex formation could be detected (11). These observations suggest that regardless of the presence or absence of DnaNCg, the dissociation of EndoMSCg from DNA is much faster than that of EndoMSTk.
When the preference for mismatch base-pair type was examined, EndoMSCg digested G–G, G–T and T–T mismatches flanking to A residues, but not others (Figure 2F). Even when nucleotides flanking to the mismatch were changed to G residues, the mismatch base-pair specificity was not changed (Supplementary Figure S7C), confirming that EndoMSCg is able to digest G–G, G–T and T–T mismatches independent of the flanking nucleotides.
The EndoMS-dependent pathway corrects replication errors
Since the canonical mismatch repair system works to repair replication errors, we attempted to determine whether the EndoMS-dependent mutation repair pathway was involved in the correction of replication errors. For this purpose, we examined the relationship between DNA polymerase fidelity and the effects of endoMS gene disruption. If EndoMS participates in the correction of replication errors, the combination of endoMS gene disruption and decreased DNA polymerase fidelity would have synergistic effects on the mutation rate (23). Conversely, if the EndoMS-dependent mutation repair pathway was independent of replication, the effects would be additive.
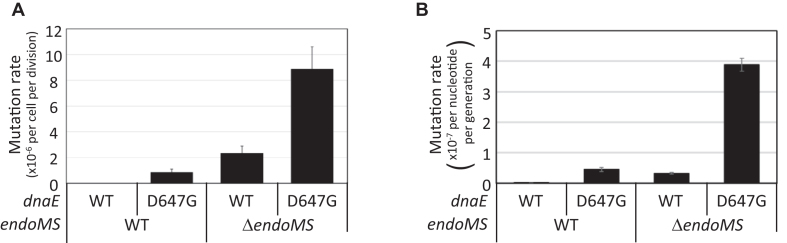
Overexpression of the dnaE E612K mutant in E. coli has been reported to increase mutation frequency (24) and confer hyper-processivity to the polymerase (25,26). In C. glutamicum dnaE, although the amino acid residue of the corresponding position was aspartic acid (D647) rather than glutamate, overexpression of the D647G mutant protein increased mutation frequency (Supplementary Figure S8). In the course of our examinations, we noticed that the plasmid containing the D647G mutant was instable. Therefore, we introduced the D647G mutation into the chromosomal dnaE locus. The mutation rate of the resulting strain, determined by fluctuation assay, was 0.85 × 10−6 per cell per division, which was ∼100-fold higher than the rate of the wild-type strain (Figure 3A). The mutation rate in the dnaE (D647G) and ΔendoMS double mutant strain was 8.9 × 10−6 per cell per division, which was ∼2.7-fold higher than the sum of the mutation rate in the dnaE (D647G) strain and the ΔendoMS strain (0.85 × 10−6 and 2.3 × 10−6 per cell per division, respectively), indicating that the effects of reducing replication fidelity and disrupting the endoMS gene on the mutation rate are synergistic (23).
Figure 3.
Synergistic effect of the D647G mutation in dnaE and endoMS disruption on the mutation rate. (A) The rate of spontaneous rifampicin resistance mutations was determined by fluctuation assay. Error bars show 95% confidence intervals (n = 20). (B) Rates of transition mutations determined by MA assay (Table 2). Overall transition mutation rates are shown. Average and standard errors for all lines are presented.
The EndoMS-dependent pathway is specific to transitions and some transversions
A mutation accumulation (MA) assay enabled us to detect neutral mutations, including base-pair substitutions (BPSs) and short insertions and deletions (indels) that occur throughout the genome (6,9,27–29). Therefore, the MA assay was performed in wild-type, dnaE (D647G), ΔendoMS, and ΔendoMS-dnaE (D647G) strains. The results of the MA assay are summarized in Supplementary Table S5 and Table 1, and the rates of each mutation are presented (Tables 2 and 3). A whole genome re-sequencing analysis of 37 lines of wild-type strains passaged for about 2000 generations resulted in the detection of 240 BPSs and 25 indels. The overall mutation rate was 1.1 × 10−9 per nucleotide per generation (Table 2). This value was almost comparable to that observed in M. smegmatis (9). The ratio of the number of BPSs that occurred in the coding versus non-coding DNA was 4.3 (195/45), which is similar to the expected value of 6.6 (chi-squared test χ2 = 2.6, P = 0.11). In addition, the ratio of nonsynonymous to synonymous nucleotide changes determined by the MA assay was 2.0 (131/64), which was not significantly different from that expected based on the codon usage in C. glutamicum ATCC13032 strain (3.1 [nonsynonymous/synonymous], chi-squared test χ2 = 3.4, P = 0.07) (30). These observations suggest that the effects of selective pressure were minimal in the MA assay. Two-thirds of the BPSs (168/240) observed in the analysis of the wild-type strain were transitions, indicating a bias towards transitions as observed in other species (6,9,31,32). The remaining one-third of BPSs (72/240) were transversion mutations, and, among them, 75% (54/72) were A:T ↔ G:C mutations (Table 1).
Table 1. Results of MA assay.
| Strain | WT | ΔendoMS | dnaE (D647G) | ΔendoMS-dnaE (D647G) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of lines | 37 | 20 | 16 | 16 | ||||||||||
| Generation per line | 2034 | 174 | 174 | 124 | ||||||||||
| Mutation types | Number of mutations* | % of each mutation | Number of mutations* | % of each mutation | Number of mutations* | % of each mutation | Number of mutations* | % of each mutation | ||||||
| Transition | A:T→G:C | A→G | 48 | 61 | 18.1 | 137 | 236 | 25.8 | 76 | 112 | 13.2 | 626 | 1400 | 22.7 |
| T→C | 13 | 4.9 | 99 | 18.7 | 36 | 6.3 | 774 | 28 | ||||||
| G:C→A:T | G→A | 40 | 107 | 15.1 | 105 | 290 | 19.8 | 61 | 195 | 10.6 | 643 | 1149 | 23.3 | |
| C→T | 67 | 25.3 | 185 | 34.9 | 134 | 23.3 | 506 | 18.3 | ||||||
| Transversion | A:T→C:G | A→C | 11 | 27 | 4.2 | 1 | 3 | 0.2 | 36 | 68 | 6.3 | 22 | 42 | 0.8 |
| T→G | 16 | 6 | 2 | 0.4 | 32 | 5.6 | 20 | 0.7 | ||||||
| G:C→T:A | G→T | 11 | 27 | 4.2 | 0 | 1 | 0 | 28 | 69 | 4.9 | 19 | 51 | 0.7 | |
| C→A | 16 | 6 | 1 | 0.2 | 41 | 7.1 | 32 | 1.2 | ||||||
| A:T→T:A | A→T | 2 | 7 | 0.8 | 0 | 0 | 0 | 10 | 15 | 1.7 | 9 | 19 | 0.3 | |
| T→A | 5 | 1.9 | 0 | 0 | 5 | 0.9 | 10 | 0.4 | ||||||
| G:C→C:G | G→C | 5 | 11 | 1.9 | 0 | 0 | 0 | 1 | 5 | 0.2 | 9 | 16 | 0.3 | |
| C→G | 6 | 2.3 | 0 | 0 | 4 | 0.7 | 7 | 0.3 | ||||||
| Indels | Deletion | 14 | 5.3 | 0 | 0 | 45 | 7.8 | 37 | 1.3 | |||||
| Insertion | 11 | 4.2 | 0 | 0 | 65 | 11.3 | 48 | 1.7 | ||||||
| Overall | 265 | 530 | 574 | 2762 | ||||||||||
*Mutations are represented as that occurred in leading strand.
Table 2. Mutation rates of BPSs.
| Mutation rate (× 10–9 per nucleotide per generation) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation type* | WT | ΔendoMS | dnaE(D647G) | ΔendoMS-dnaE (D647G) | ||||||
| Transition | A:T→G:C | A→G | 0.84 | 0.53 | 52 | 44 | 37 | 26 | 430 | 460 |
| T→C | 0.22 | 37 | 17 | 520 | ||||||
| G:C→A:T | G→A | 0.57 | 0.80 | 33 | 47 | 24 | 39 | 360 | 330 | |
| C→T | 1.04 | 62 | 56 | 310 | ||||||
| Transversion | A:T→C:G | A→C | 0.19 | 0.24 | 0.38 | 0.57 | 17 | 16 | 16 | 14 |
| T→G | 0.28 | 0.75 | 15 | 14 | ||||||
| G:C→T:A | G→T | 0.16 | 0.20 | - | 0.16 | 11 | 14 | 11 | 14 | |
| C→A | 0.25 | 0.20 | 17 | 21 | ||||||
| A:T→T:A | A→T | 0.04 | 0.06 | - | <0.19 | 4.7 | 3.5 | 6.0 | 6.3 | |
| T→A | 0.09 | - | 2.3 | 8.5 | ||||||
| G:C→C:G | G→C | 0.07 | 0.08 | - | <0.16 | 0.39 | 1.0 | 6.0 | 4.5 | |
| C→G | 0.09 | - | 1.7 | 4.1 | ||||||
| Indels | Deletion | 0.06 | <0.07 | 4.9 | 5.6 | |||||
| Insertion | 0.04 | <0.07 | 7.1 | 7.3 | ||||||
| Overall | 1.1 | 46 | 62 | 420 | ||||||
*Mutations are represented as that occurred in leading strand.
Table 3. Local sequence context of indels.
| Number of single nucleotide indels | Percentage | Mutation rate per nucleotide | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Length of identical nucleotide repeat (nt) | WT | dnaE (D647G) | ΔendoMS−dnaE (D647G) | WT | dnaE (D647G) | ΔendoMS−dnaE (D647G) | WT | dnaE (D647G) | ΔendoMS−dnaE (D647G) |
| 1 | 1 | 3 | 10 | 4.5 | 2.7 | 11.0 | 4.0 × 10–12 | 3.3 × 10–10 | 1.5 × 10–9 |
| 2 | 1 | 11 | 11 | 4.5 | 10.0 | 12.1 | 1.6 × 10–11 | 4.6 × 10–9 | 6.5 × 10–9 |
| 3 | 0 | 22 | 12 | 0 | 20.0 | 13.2 | 0 | 4.0 × 10–8 | 3.1 × 10–8 |
| 4 | 1 | 16 | 7 | 4.5 | 14.5 | 7.7 | 3.2 × 10–10 | 1.4 × 10–7 | 8.5 × 10–8 |
| 5 | 5 | 23 | 21 | 22.7 | 20.9 | 23.1 | 5.3 × 10–9 | 6.6 × 10–7 | 8.5 × 10–7 |
| 6 | 5 | 18 | 14 | 22.7 | 16.4 | 15.4 | 4.5 × 10–8 | 4.3 × 10–6 | 4.7 × 10–6 |
| 7 | 5 | 13 | 9 | 22.7 | 11.8 | 9.9 | 3.9 × 10–7 | 2.7 × 10–5 | 2.7 × 10–5 |
| 8 | 3 | 4 | 1 | 13.6 | 3.6 | 1.1 | 1.7 × 10–6 | 2.2 × 10–5 | 2.2 × 10–5 |
| 9 | 1 | 0 | 0 | 4.5 | 0 | 0 | 1.3 × 10–5 | 0 | 0 |
No indels were detected in MA assay of ΔendoMS and the column for ΔendoMS is not shown.
When replication fidelity was reduced by the D647G mutation in dnaE gene, rates of all types of mutations increased more than 50-folds, except for that of G:C ↔ C:G transversions (Table 2). Although the extent of the mutation rate increase was slightly larger in indels, a correlation analysis of mutation compositions between the wild-type and dnaE (D647G) strains showed good correlation (Supplementary Figure S9, r = 0.85). Especially with respect to BPSs, the correlation between the mutational composition of the two strains was high (Supplementary Figure S9, r = 0.96). These results indicate that the D647G mutation in dnaE successfully reduced replication fidelity to a similar extent in most BPS types. The observed ratio of BPSs in coding versus non-coding regions was 5.6 (395/71), which was not significantly different from the expected value of 6.6 (chi-squared test χ2 = 0.7, P = 0.41). However, we detected significant differences between observed and expected ratios of nonsynonymous to synonymous nucleotide changes (265/130 [nonsynonymous/synonymous], chi-squared test χ2 = 7, P = 0.008), implying that we could not exclude the possibility of some selection pressure during culture for ∼170 generations. This was probably due to the high mutation rate, which caused one mutation to accumulate per every six generations, suggesting that the dnaE (D647G) strain would be an appropriate model to reduce the time to accumulate natural mutations in our culture conditions.
Disruption of endoMS in the wild-type background increased the transition mutation rate more than 50-fold, but the effect on transversions and indels was not obvious (Table 2). As a result, almost all mutations detected were transitions (526/530). Although a 2- to 3-fold increase was observed for some transversions, the number of detected mutations was very small and statistical power was insufficient to obtain a precise mutation rate for such mutations (Table 1).
When endoMS was disrupted in the background of dnaE (D647G), the rate of transitions increased 5- to 31-fold. As a result, the overall transition mutation rate in the ΔendoMS-dnaE (D647G) strain was 3.8 × 10−7 per nucleotide per generation, which was ∼4.9-fold higher than the sum of the mutation rates in the ΔendoMS and dnaE (D647G) strains (4.6 × 10−8 and 3.3 × 10−8 per nucleotide per generation, respectively; Figure 3B). Thus, by focusing on the preferred target for the EndoMS-dependent pathway, the synergistic effects of replication fidelity reduction and endoMS gene disruption were more obvious. In contrast to transition mutations, the mutation rates of A:T ↔ C:G transversions and indels were not affected by ΔendoMS. However, although the numbers of mutations detected were small (Table 1), the rates of A ↔ T and G ↔ C transversions tended to increase slightly with the loss of endoMS (Table 2).
Effects of local sequence context on the occurrence of mutations
To verify the influence of flanking nucleotide sequences on the occurrence of BPSs, we focused on nucleotides immediately 5′ (upstream) or 3′ (downstream) to the BPS site on the leading strand (Supplementary Figure S10). When the nucleotides upstream of the BPS site were evaluated, 67% (161/240) and 69% (365/530) were G or C in the wild-type and ΔendoMS, respectively (Supplementary Figure S10A and S10B). Similarly, the base pairs immediately downstream of the BPS site were also biased toward GC pairs; 64% (154/240) and 71% (376/530) of all BPSs in the wild-type and ΔendoMS, respectively, were located immediately 5′ to a GC base pair. As a result, most BPSs (91% [218/240] in the wild-type and 91% [484/530] in ΔendoMS) immediately flanked the GC base-pair site (Supplementary Figure S10). Such a bias toward GC base pairing in neighboring nucleotides was also observed in the dnaE background (D647G); however, the extent of the bias was slightly decreased (Supplementary Figure S10). The fractions of BPSs that immediately flanked the GC base pair site were 84% (392/464) in dnaE (D647G) and 83% (2242/2677) in ΔendoMS-dnaE (D647G).
When 1-base pair indels were considered, 91% (20/22) were insertions or deletions in single mononucleotide repeat sites of more than four bases in the wild type and 68% (15/22) were located at five to seven base repeat sites (Table 3). These results were consistent with previous observations that indels are caused by slippage of the DNA polymerase at mononucleotide repeat sites (33,34). In the dnaE (D647G) strain, the occurrence of indels at shorter mononucleotide repeat sites increased. These results indicate that the D647G mutation in dnaE increased the frequency of slippage at short mononucleotide repeat sites. Interestingly, although the mutation rate of indels at mononucleotide repeat sites in the ΔendoMS-dnaE (D647G) strain was comparable to that in the dnaE (D647G) strain, indels at no repeat sites increased 4.5-fold with the endoMS disruption (Table 3). These observations are consistent with the results of the in vitro cleavage assay of EndoMSCg, in which an indel was cleaved in non-repeat sequences in the presence of DnaNCg, but not in repeat sequences (Supplementary Figure S11).
DISCUSSION
Very recently, disruption of the endoMS/nucS gene in M. smegmatis and S. coelicolor was reported to considerably increase the spontaneous mutation rate of these bacteria (12); however, a biochemical analysis of mycobacterial EndoMS/NucS failed to detect DNA cleavage activity. In contrast, here, we showed that purified C. glutamicum EndoMS/NucS was able to cleave dsDNA containing mismatches by itself (Figure 1B). A mutation in the RecB nuclease motif resulted in the loss of both nuclease activity and the ability to complement endoMS gene disruption in C. glutamicum (Figure 1A and B). We also showed that overexpression of M. tuberculosis endoMS/nucS complemented the loss of endoMSCg in C. glutamicum. However, mutation in the RecB nuclease motif completely diminished the complementation ability of endoMS/nucSMt (Figure 1A). Based on these observations, we suggest that mycobacterial EndoMS/NucS also has mismatch-specific nuclease activity and that some experimental differences may have impaired the ability to detect this activity in the previous study.
In addition to demonstrating EndoMS nuclease activity, we revealed that direct interaction of EndoMS with DnaN was essential to maintaining a lower mutation rate (Figure 2, Supplementary Figures S5 and S6). We also succeeded in demonstrating DnaN-dependent enhancement of cleavage efficiency of EndoMS. Then, how DnaN clamp enhance the EndoMS cleavage efficiency? One possibility is that DnaN interaction with EndoMS allosterically activates the EndoMS enzymatic activity. In the case of MutL of B. subtilis, it has been suggested that DnaN–MutL interaction, following MutL recruitment to the mismatch site by MutS–MutL interaction, coordinately causes large conformational change of MutL dimer, licensing the endonuclease activity of MutL (35). In the case of human flap endonuclease 1 (FEN1) which functions in both replication and repair, it has been reported that sliding clamp alters both Km and Vmax of FEN1 nuclease activity. The authors suggested that in addition to large decrease in Km by tethering the enzyme near the cleavage site, sliding clamp alters the conformation or orientation of FEN1, resulting in a small improvement of catalysis (36). Thus, there are various mechanisms in activation of nuclease by sliding clamp. Another possibility is that DnaN tethers EndoMS to the zone at where replication, and mismatches caused thereby, occurs and this tethering enhances the mismatch detection efficiency. In B. subtilis, it has been reported that elimination of DnaN–MutS interaction by deleting MutS C-terminal clamp binding region almost completely disrupted in vivo MMR and that the interaction is required for coupling of MutS with replisome prior to mismatch recognition (37,38). The authors also demonstrated that overexpression of this DnaN-interaction defective MutS mutant restored MMR, indicating MutS–DnaN interaction is required to increase accessibility of MutS to mismatch (37). Here, we demonstrated that overexpression of EndoMSCg-Cdel mutant in ΔendoMSCg could not restore the mutation rate at all (Supplementary Figure S6A), suggesting that facilitation of mismatch detection is not the only work of EndoMS-DnaN interaction. Therefore, DnaN interaction with EndoMS may cause some conformational change of EndoMS to facilitate the cleavage activity and the allosteric activation of EndoMS may be the essential work of DnaN-EndoMS interaction in EndoMS-type MMR system. More detailed biochemical, structural and imaging analyses will uncover the role of DnaN in EndoMS-type MMR system.
Together with results showing that disruption of endoMS and a mutation in DNA polymerase DnaE had synergistic effects on the mutation rate (Figure 3), we conclude that EndoMS functions in cooperation with DNA polymerase to correct replication errors. These observations clearly indicated that the EndoMS-dependent MMR system conserved in organisms lacking mutS and mutL genes is the counterpart of the MutS-MutL-dependent canonical MMR system. Since the first report of the absence of mutS and mutL genes in M. tuberculosis (39,40), as well as the demonstration of comparable mutation rates between M. tuberculosis and other organisms with canonical MMR systems (41), the gap between the absence of a canonical MMR system and the presence of comparable mutation rate in Actinobacteria and organisms equipped with canonical MMR system has been an intriguing area for inquiry. Here, we established the first evidence that EndoMS plays the central role in the repair of replication errors, the most important task of a canonical MMR system, addressed the gap that has existed for two decades.
Interestingly, archaeal endoMS (endoMSTk) was able to partially complement the loss of endoMS in C. glutamicum in C-terminal sliding clamp binding motif-dependent manner (Supplementary Figure S6A). These results suggest that EndoMSTk is able to interact with the bacterial sliding clamp as bacterial EndoMS does. We also observed that EndoMSCg cleaved both strands of DNA in miamatch-dependent manner as observed in archaeal EndoMS. These observations strongly suggest that the bacterial and archaeal EndoMS-dependent MMR pathways utilize the same repair system after mismatch DNA cleavage. Because archaeal EndoMS can cause mismatch-dependent double-stranded breaks in vitro, together with the observation that T. kodakarensis genome is polyploid (42), homologous recombination repair has been postulated to be the downstream repair pathway (11,43). Alternatively, interaction with sliding clamp may provide strand information for EndoMS and the interaction may inhibit the cleavage of template strand as observed in eukaryotic MMR system (44). Although the possibility that sliding clamp-dependent strand discrimination should be tested as performed previously (44), we favor the homologous recombination hypothesis, because of the advantage of using homologous recombination is that there is no need to discriminate the nascent strand. Although, polyploidy has advantages in utilizing homologous recombination as a pathway to repair double strand breaks, it is well known that non-polyploid bacteria such as E. coli utilize homologous recombination to repair double strand breaks, suggesting that polyploidy is not essential to repair double strand breaks by homologous recombination. It is also interesting that, in some species harboring canonical MMR systems, the mechanism by which these systems discriminates the nascent strand has not been uncovered to date (45). For example, reconstitution experiments of a Thermus thermophilus MMR system revealed that MutL of this species can cleave both strands of mismatched DNA and that this activity is not affected by the sliding clamp (46). From these observations, together with previous and present findings in non-canonical MMR systems, it seems reasonable to consider homologous recombination as the alternative pathway to complete MMR without strand discrimination.
Application of a mutation accumulation assay revealed the following points. First and most importantly, a synergistic effect was observed with the combination of ΔendoMS and the D647G mutation in dnaE, confirming that EndoMS is involved in the repair of replication errors (Figure 3B).
Second, the types of mutations that increased with the endoMS deletion were consistent with the substrate specificity of EndoMS. As shown in Figure 2F and Supplementary Figure S7C, EndoMS was able to cleave mismatches leading to transitions (G/T mismatch) and A ↔ T and G ↔ C transversions (T/T and G/G mismatches, respectively), but not those leading to A:T ↔ C:G transversions. In the case of indels, EndoMSCg activated by DnaNCg was able to cleave DNA substrates containing a 1-base indel in neighboring DNA in a nucleotide-dependent manner (Supplementary Figure S11). These results are consistent with the in vivo mutation spectrum (Tables 1–3). These observations strongly indicate that the biochemical properties of EndoMSCg observed in this study reflect the in vivo function of the system.
Third, an MA assay of the ΔendoMS strain of C. glutamicum revealed that most BPSs occurred at the site immediately flanking a GC base pair. These cases are very similar to those reported previously, in which E. coli, Pseudomonas fluorescens, and B. subtilis strains lacking canonical MMR system were analyzed (6,32,47). This consistency suggests that the basis for these mismatches is the same in each of these organisms. In addition, the fact that the mechanisms underlying proofreading differs between these organisms suggests that pre-proofreading mismatch generation mechanisms such as new nucleotide incorporation may be similar between these organisms. Unlike E. coli, in which the presence of an MMR system affects the occurrence of mutations in specific local sequence contexts (6), the presence of endoMS had no effects on the sequences neighboring mutations detected in C. glutamicum (Supplementary Figure S10A and B). These results suggest that, whereas a MutS-dependent canonical MMR system has a preference for a local sequence context and efficiently corrects mismatches to the sides of a GC base pair, a EndoMS-dependent system has no such preference for nucleotides immediately flanking the mismatch. These observations were consistent with our biochemical results that neighboring sequences did not affect the mismatch-type specificity of EndoMSCg (Figure 2F and Supplementary Figure S7C). These results are also consistent with the biochemical properties of T. kodakarensis EndoMS, whose efficiency in cleaving G/T mismatches is not affected by neighboring sequences (11,43).
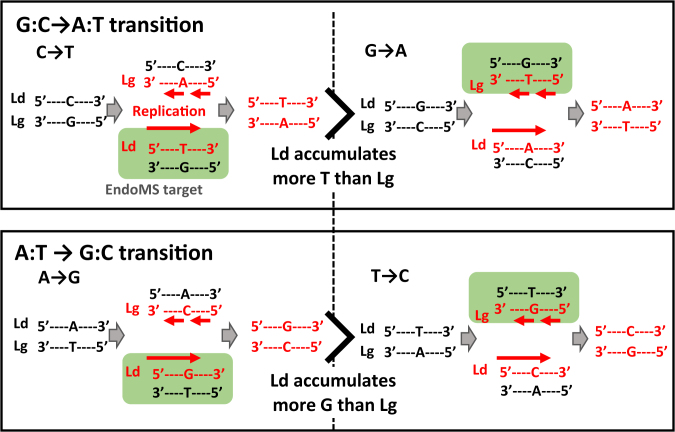
It is noteworthy that unlike canonical MMR system in which broad types of mismatches are targeted (3–5), the EndoMS enzyme showed high specificity for G/T mismatches; therefore, we were able to discriminate whether C/A or G/T mismatches were likely to form during replication (Figure 4). For example, when the origin of C→T transition in the leading strand is considered, there are two possibilities; C/A mismatch generated during lagging strand synthesis and G/T mismatch generated during leading strand synthesis. Because EndoMS is able to cleave only G/T mismatch, C→T transition mutation increased by disruption of endoMS gene reflects the frequency of G/T mismatch occurrence during leading strand synthesis (Figure 4, green box). Residual transition mutations in the presence of endoMS are caused by non-EndoMS target C/A and G/T mismatches that escaped the EndoMS-dependent correction pathway. If the EndoMS-dependent pathway functions perfectly and is able to correct all of the G/T mismatches, the rate of transition mutations in the wild-type is expected to correspond to the frequency of replication errors that generates C/A mismatches. Inversely, a lower efficiency of the EndoMS-dependent pathway means a lower frequency of C/A mismatches. With respect to the C/A mismatch elimination pathway, although the effect on the mutation rate is much smaller than that of the MutS-dependent MMR system, the MutY N-glycosylase/AP (apurinic/apyrimidinic) endonuclease eliminates C/A mismatches in the absence of a MutS-dependent MMR system in E. coli (48,49). In C. glutamicum, we evaluated the effect of the mutY gene on the mutation rate and revealed that the mutY gene disruption only had a minor effect (Supplementary Figure S12). Similarly, in M. smegmatis, the lack of a mutY gene only slightly increased the rate of AT → GC transitions, suggesting the successful correction of C/A mismatches by MutY, the extent of increase was much smaller than the disruption of the EndoMS-dependent pathway (12,50). Although we are unable to rule out the possibility that some other mechanism functioned to eliminate C/A mismatches, our data, together with previous reports, indicate that the dnaE-type DNA polymerase is prone to generating many more G/T mismatches than C/A mismatches. Thus, by analyzing a G/T mismatch-specific MMR system, we succeeded in determining fundamental properties of DNA replication in vivo. Application of this non-canonical and asymmetric MMR system in other species will be required to demonstrate the generality of our hypothesis that G/T mismatches are much more likely to occur in vivo. Elimination of frequent G/T mismatches might be the most important task for the MMR system, regardless of the types of proteins in the system. The higher binding specificity of MutS for G/T mismatches than for other mismatches (3) seems to be an important property that allows this system to address the high frequency of G/T mismatches. Indeed, it has been reported that G/T mismatches are the most stable of all mismatched bases (51). Also, a biochemical analysis of the E. coli PolII α subunit without the proofreading ϵ subunit showed that the misincorporations that generate G/T mismatches are about 3-fold more frequent than the misincorporations that generate C/A mismatches (52). In addition to such fundamental properties of the polymerase, some other factors such as local concentrations of nucleotide substrates might lead to higher frequencies of G/T mismatches (53–55).
Figure 4.
Schematic drawing of the asymmetric specificity of the EndoMS-dependent MMR system and asymmetric occurrence of replication errors. The origin of transition mutations was considered (G:C → A:T transitions; upper panel and A:T → G:C transitions; lower panel). The cases of C → T and A → G mutations in the leading strand are shown on the left side, and the cases of G → A and T → C mutations in the leading strand side are shown on the right side. Possible replication errors (mismatches) leading to each mutation during lagging (Lg)- and leading (Ld)-strand synthesis are shown in the upper and lower parts of each panel, respectively. The direction of DNA synthesis is indicated by a red arrow, and the nascent strand is indicated by red letters. Replication errors targeted by EndoMS-dependent pathway are indicated by a green box.
The results of the MA assay of the ΔendoMS strain, in which EndoMS-dependent post replication repair is absent, showed that the number of C → T transitions detected was significantly more than that of G → A transitions in the leading strand (Figure 4, upper panel and Table 1, 185 versus 105, chi-squared test χ2 = 11.2, P < 0.001). Because G → A mutations in the leading strand correspond to C → T mutations in the lagging strand, C → T mutations are more likely to occur in the leading strand than in the lagging strand in the ΔendoMS strain (Figure 4, upper panel). Together with the fact that EndoMS is able to cleave G/T mismatches but not C/A mismatches (Figure 4, green box) and that these mutations arise mainly because of the loss of the EndoMS-dependent pathway, the frequency of misincorporation of T at the template G site is expected to be higher in the leading strand than in the lagging strand. Similarly, although there was no statistical significance, A → G transitions seemed to be more likely to occur than T → C transitions in the leading strand (Figure 4, lower panel, and Table 1, 137 versus 99, chi-squared test χ2 = 3.1, P = 0.08), suggesting that the frequency of misincorporation of G at template T site may be higher in the leading strand than in the lagging strand. In the presence of intact endoMS, a similar preference for C → T and A → G mutations in the leading strand was observed, although the extent of this preference was slightly different from that in the ΔendoMS strain. In the wild-type, the preference for A → G to T → C mutation was statistically significant (48 versus 13, chi-squared test χ2 = 10.9, P < 0.001), but that for C → T to G → A was not (67 versus 40, chi-squared test χ2 = 3.5, P = 0.06). In the case of AT ↔ CG transversions, no bias was observed. Comparison of the mutation rates in all BPS types revealed that the mutational pattern would likely produce more Ts and Gs than As and Cs, respectively, in the leading strand (Table 2 and Figure 4). This bias is consistent with the composition of nucleotides in the C. glutamicum genome and might be contributes to the formation of GC skew. These results are similar to those of E. coli and S. cerevisiae, in which the patterns of neutral mutations occurring in synonymous sites and intergenic regions are consistent with the nucleotide compositions of the genomes (6,56). Interestingly, in contrast to previous reports, our results enabled us to speculate about the origin of this mutational skew. When the misincorporation frequency was increased with the D647G mutation of dnaE, although the preference for C → T and A → G mutations in the leading strand was observed as in the wild-type, disruption of endoMS completely abolished such a strand-specific preference in these mutations (Tables 1 and 2). These observations suggest that a strand-specific preference in mutations is based on the combination of biases in nucleotide misincorporation and in the mismatch repair by the EndoMS pathway. Together with these observations, future analyses such as experimental evolution of these strains or precise measurement of strand-specific mutation rates will lead to new insights into the relationship between GC skewing and the strand-specific mutational spectrum.
DATA AVAILABILITY
Sequence data obtained for the MA lines were archived in DDBJ under BioProject PRJDB6606.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Yota Tsuge for kindly providing the pYTKA1 plasmid. We also thank Yu Sakurai for technical assistance and Dr. Masanori Ogawa for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant for International Health Research [29A1028] from the Ministry of Health, Labour and Welfare of Japan (to N.T.); JSPS KAKENHI [18K06190 to N.T.]; Council for Science, Technology and Innovation (CSTI) ImPACT Program (to M.S.). Funding for open access charge: JSPS KAKENHI [18K06190 to N.T.); Grant for International Health Research [29A1028] from the Ministry of Health, Labour and Welfare of Japan (to N.T.); Council for Science, Technology and Innovation (CSTI) ImPACT Program (to M.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Kunkel T.A., Erie D.A.. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lenhart J.S., Schroeder J.W., Walsh B.W., Simmons L.A.. DNA repair and genome maintenance in bacillus subtilis. Microbiol. Mol. Biol. Rev. 2012; 76:530–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Su S.S., Modrich P.. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:5057–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fishel R., Ewel A., Lescoe M.K.. Purified human MSH2 protein binds to DNA containing. Cancer Res. 1994; 54:5539–5542. [PubMed] [Google Scholar]
- 5. Su S.S., Lahue R.S., Au K.G., Modrich P.. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J. Biol. Chem. 1988; 263:6829–6835. [PubMed] [Google Scholar]
- 6. Lee H., Popodi E., Tang H., Foster P.L.. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2774–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Serero A., Jubin C., Loeillet S., Legoix-Né P., Nicolas A.G.. Mutational landscape of yeast mutator strains. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sachadyn P. Conservation and diversity of MutS proteins. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2010; 694:20–30. [DOI] [PubMed] [Google Scholar]
- 9. Kucukyildirim S., Long H., Sung W., Miller S.F., Doak T.G., Lynch M.. The rate and spectrum of spontaneous mutations in mycobacterium smegmatis, a bacterium naturally devoid of the postreplicative mismatch repair pathway. G3. 2016; 6:2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ren B., Kühn J., Meslet-Cladiere L., Briffotaux J., Norais C., Lavigne R., Flament D., Ladenstein R., Myllykallio H.. Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 2009; 28:2479–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishino S., Nishi Y., Oda S., Uemori T., Sagara T., Takatsu N., Yamagami T., Shirai T., Ishino Y.. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res. 2016; 44:2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castañeda-Garciá A., Prieto A.I., Rodríguez-Beltrán J., Alonso N., Cantillon D., Costas C., Pérez-Lago L., Zegeye E.D., Herranz M., Plociński P. et al. . A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 2017; 8:14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inui M., Kawaguchi H., Murakami S., Vertès A.A., Yukawa H.. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004; 8:243–254. [DOI] [PubMed] [Google Scholar]
- 14. de Boer H.A., Comstock L.J., Vasser M.. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higuchi R., Krummel B., Saiki R.K.. Volume 16 Number 15 1988 Nucleic Acids Research. Nucleic Acids Res. 1988; 16:7351–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malmqvist T., Spickett C., Gallo J.-M., Anthony K.. A UV cross-linking method combined with infrared imaging to analyse RNA–protein interactions. Biol. Methods Protoc. 2017; 2:bpx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foster P.L. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006; 409:195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarkar S., Ma W.T., Sandri G.H.. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992; 85:173–179. [DOI] [PubMed] [Google Scholar]
- 19. Hall B.M., Ma C.-X., Liang P., Singh K.K.. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009; 25:1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Higgins N.P. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites: What's the Dif. Mol. Microbiol. 2007; 64:1–4. [DOI] [PubMed] [Google Scholar]
- 21. Hendrickson H., Lawrence J.G.. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol. Microbiol. 2007; 64:42–56. [DOI] [PubMed] [Google Scholar]
- 22. Stukenberg P.T., Studwell-Vaughan P.S., O’Donnell M., O’Donnell M.. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 1991; 266:11328–11334. [PubMed] [Google Scholar]
- 23. Paschalis V., Le Chatelier E., Green M., Képès F., Soultanas P., Janniere L.. Interactions of the Bacillus subtilis DnaE polymerase with replisomal proteins modulate its activity and fidelity. Open Biol. 2017; 7:170146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maki H., Mo J.Y., Sekiguchi M.. A strong mutator effect caused by an amino acid change in the alpha subunit of DNA polymerase III of Escherichia coli. J. Biol. Chem. 1991; 266:5055–5061. [PubMed] [Google Scholar]
- 25. Sugaya Y., Ihara K., Masuda Y., Ohtsubo E., Maki H.. Hyper-processive and slower DNA chain elongation catalysed by DNA polymerase III holoenzyme purified from the dnaE173 mutator mutant of Escherichia coli. Genes Cells. 2002; 7:385–399. [DOI] [PubMed] [Google Scholar]
- 26. Yanagihara F., Yoshida S., Sugaya Y.. The dnaE173 mutator mutation confers on the α subunit of Escherichia coli DNA polymerase III a capacity for highly processive DNA synthesis and stable binding to primer / template DNA. Genes Genet Syst. 2007; 82:273–280. [DOI] [PubMed] [Google Scholar]
- 27. Lind P.A., Andersson D.I.. Whole-genome mutational biases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:17878–17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dettman J.R., Sztepanacz J.L., Kassen R.. The properties of spontaneous mutations in the opportunistic pathogen Pseudomonas aeruginosa. BMC Genomics. 2016; 17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heilbron K., Toll-Riera M., Kojadinovic M., MacLean R.C.. Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment. Genetics. 2014; 197:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishio Y., Nakamura Y., Kawarabayasi Y., Usuda Y., Kimura E., Sugimoto S., Matsui K., Yamagishi A., Kikuchi H., Ikeo K. et al. . Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 2003; 13:1572–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long H., Kucukyildirim S., Sung W., Williams E., Lee H., Ackerman M., Doak T.G., Tang H., Lynch M.. Background mutational features of the radiation-resistant bacterium deinococcus radiodurans. Mol. Biol. Evol. 2015; 32:2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sung W., Ackerman M.S., Gout J.F., Miller S.F., Williams E., Foster P.L., Lynch M.. Asymmetric context-dependent mutation patterns revealed through mutation-accumulation experiments. Mol. Biol. Evol. 2015; 32:1672–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujii S., Akiyama M., Aoki K., Sugaya Y., Higuchi K., Hiraoka M., Miki Y., Saitoh N., Yoshiyama K., Ihara K. et al. . DNA replication errors produced by the replicative apparatus of Escherichia coli. J. Mol. Biol. 1999; 289:835–850. [DOI] [PubMed] [Google Scholar]
- 34. Viguera E., Canceill D., Ehrlich S.D.. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001; 20:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pillon M.C., Lorenowicz J.J., Uckelmann M., Klocko A.D., Mitchell R.R., Chung Y.S., Modrich P., Walker G.C., Simmons L.A., Friedhoff P. et al. . Structure of the endonuclease domain of MutL: Unlicensed to cut. Mol. Cell. 2010; 39:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tom S., Henricksen L.A., Bambara R.A.. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease1. J. Biol. Chem. 2000; 275:10498–10505. [DOI] [PubMed] [Google Scholar]
- 37. Lenhart J.S., Sharma A., Hingorani M.M., Simmons L.A.. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol. Microbiol. 2013; 87:553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simmons L.A., Davies B.W., Grossman A.D., Walker G.C.. β clamp directs localization of mismatch repair in bacillus subtilis. Mol. Cell. 2008; 29:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E. et al. . Deciphering the biology of mycobacterium tuberculosis from the complete genome sequence. Nature. 1998; 393:537–544. [DOI] [PubMed] [Google Scholar]
- 40. Mizrahi V., Andersen S.J.. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence. Mol. Microbiol. 1998; 29:1331–1339. [DOI] [PubMed] [Google Scholar]
- 41. David H.L. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 1970; 20:810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spaans S.K., van der Oost J., Kengen S.W.M.. The chromosome copy number of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Extremophiles. 2015; 19:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakae S., Hijikata A., Tsuji T., Yonezawa K., Kouyama K.I., Mayanagi K., Ishino S., Ishino Y., Shirai T.. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure. 2016; 24:1960–1971. [DOI] [PubMed] [Google Scholar]
- 44. Kawasoe Y., Tsurimoto T., Nakagawa T., Masukata H., Takahashi T.S.. MutSα maintains the mismatch repair capability by inhibiting PCNA unloading. Elife. 2016; 5:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fukui K. DNA mismatch repair in eukaryotes and bacteria. J. Nucleic Acids. 2010; 2010:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shimada A., Kawasoe Y., Hata Y., Takahashi T.S., Masui R., Kuramitsu S., Fukui K.. MutS stimulates the endonuclease activity of MutL in an ATP-hydrolysis-dependent manner. FEBS J. 2013; 280:3467–3479. [DOI] [PubMed] [Google Scholar]
- 47. Long H., Sung W., Miller S.F., Ackerman M.S., Doak T.G., Lynch M.. Mutation rate, spectrum, topology, and context-dependency in the DNA mismatch repair-deficient Pseudomonas fluorescens ATCC948. Genome Biol. Evol. 2014; 7:262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bai H., Lu A.L.. Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS. J. Bacteriol. 2007; 189:902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsai-Wu J.J., Liu H.F., Lu A.L.. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kurthkoti K., Srinath T., Kumar P., Malshetty V.S., Sang P.B., Jain R., Manjunath R., Varshney U.. A distinct physiological role of MutY in mutation prevention in mycobacteria. Microbiology. 2010; 156:88–98. [DOI] [PubMed] [Google Scholar]
- 51. Ke S.H., Wartell R.M.. Influence of nearest neighbor sequence on the stability of base pair mismatches in long DNA: Determination by temperature-gradient gel electrophoresis. Nucleic Acids Res. 1993; 21:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sloane D.L., Goodman M.F., Echols H.. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988; 16:6465–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ganai R.A., Johansson E.. DNA Replication—a matter of fidelity. Mol. Cell. 2016; 62:745–755. [DOI] [PubMed] [Google Scholar]
- 54. Watt D.L., Buckland R.J., Lujan S.A., Kunkel T.A., Chabes A.. Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools. Nucleic Acids Res. 2015; 44:1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gawel D., Fijalkowska I.J., Jonczyk P., Schaaper R.M.. Effect of dNTP pool alterations on fidelity of leading and lagging strand DNA replication in E. coli. Mutat. Res. 2014; 759:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Agier N., Fischer G.. The mutational profile of the yeast genome is shaped by replication. Mol. Biol. Evol. 2012; 29:905–913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data obtained for the MA lines were archived in DDBJ under BioProject PRJDB6606.