Abstract
Members of the genus Campylobacter remain a leading cause of bacterial gastroenteritis worldwide. Infection is usually self-limiting but in severe cases may require antibiotic treatment. In a recent statement by the World Health Organization (WHO) Campylobacter was named as one of the 12 bacteria that pose the greatest threat to human health because they are resistant to antibiotics. In this mini review we describe recent trends in fluoroquinolone (FQ) (particularly ciprofloxacin) resistance in strains of members of the genus Campylobacter isolated from livestock and clinical samples from several countries. Using evidence from phenotyping surveys and putative resistance prediction from DNA sequence data, we discuss the acquisition and spread of FQ resistance and the role of horizontal gene transfer and describe trends in FQ-resistance in samples from livestock and clinical cases. This review emphasises that FQ resistance remains common among isolates of members of the genus Campylobacter from various sources.
Keywords: Campylobacter, fluoroquinolones, ciprofloxacin, resistance
Data Summary
All external data records cited in the text of this review are listed in the bibliography at the end of the document.
Impact Statement.
Fluoroquinolones (FQ), especially ciprofloxacin (CIP), are among the most common treatments for human and animal infection with Campylobacter – a common cause of gastroenteritis worldwide. Over time, this has resulted increased antimicrobial resistance (AMR), making these drugs much less effective. Here we review current literature to describe: (i) the mechanisms of FQ resistance in members of the genus Campylobacter; (ii) acquisition and spread; (iii) predicting resistance from whole-genome data; (iv) CIP resistance among clinical isolates; (v) CIP resistance in livestock; (vi) the potential for spread of resistance from animals to humans.
Introduction
Members of the genus Campylobacter are the leading cause of bacterial gastroenteritis in many countries [1, 2]. They are frequently isolated from the gut of warm-blooded animals (particularly poultry) [1, 2] and the common pathogenic species Campylobacter jejuni (approximately 90 %) and Campylobacter coli (approximately 10 %) [3, 4] cause infection in humans, most commonly after consumption of contaminated or under-cooked food, especially poultry. Campylobacteriosis is usually self-limiting and rarely requires antibiotic treatment, except in severe or prolonged cases [2, 5]. The most common drugs used to treat Campylobacter infections are macrolides (particularly erythromycin) for laboratory-confirmed cases. Members of the genus Campylobacter are generally susceptible to erythromycin and in Europe gentamicin (an aminoglycoside) is the antibiotic to which they have the second lowest antimicrobial resistance (AMR) rates, after gentamicin [6]. The fluoroquinolones (FQ) are broad-spectrum antimicrobials that are used to treat a multitude of infections including undiagnosed cases of diarrhoea, predominantly by using ciprofloxacin (CIP) [2, 5, 7–10].
Since the late 1980’s there has been an increasing trend in the proportion of FQ-resistant strains of members of the genus Campylobacter isolated from both clinical samples [11–13] and livestock, where FQs (particularly enrofloxacin) are frequently used to treat animals in intensive production [5, 7, 9, 12, 14]. In general it appears that resistance to FQs, and several other antimicrobials (AM’s), is more common in C. coli compared with C. jejuni (Table 1) [4, 6, 7, 15, 16] but the reason for this is not fully understood. However, FQ resistance remains common in both species [17]. The widespread acquisition of FQ resistance is the result of spontaneous independent mutations and is accelerated by the horizontal transfer of resistance-conferring DNA among strains of members of the genus Campylobacter [5, 18]. In response to the rising levels of FQ and other AMR, restrictions on the use of FQs in animal husbandry have been implemented in the EU (2003 and 2006) and the USA (2005) [13, 19, 20], and several specific government-led AMR surveillance and monitoring programs have been initiated [21]. However, comprehensive data assessing CIP resistance in members of the genus Campylobacter has only become available in recent years [6, 15] and FQs are already of limited use for treating infections with members of the genus Campylobacter in many countries [5]. The primary FQ in Campylobacter AMR testing schemes is CIP, probably due to its common use in the treatment of diarrhoea. In Europe, CIP is the only FQ listed, and is a mandatory AM to be tested under the harmonised methods scheme for the monitoring of AMR in isolates of members of the genus Campylobacter from humans [22]. For isolates of members of the genus Campylobacter obtained from the major food animals, nalidixic acid (NAL) (a quinolone) is also included on the mandatory list of AM’s to be tested for resistance [23]. Many other studies follow this, where other FQ’s are rarely included in the AMR testing of Campylobacter isolates. However, the resistance to quinolone and fluoroquinoline have a strong correlation [9]. For example, a study has revealed that 69.8 % of isolates of C. jejuni from poultry were resistant to CIP and 65.1 % were resistant to NAL [6]. In other studies, isolates of members of the genus Campylobacter had similar proportions that were resistant to CIP and NAL [16, 24, 25]. This is most likely to be due to a high level of resistance being conferred by just a single point mutation within the quinolone-resistance-determining region (QRDR) and it being the most common and recognised mechanism for FQ resistance in members of the genus Campylobacter [5, 7, 14]. If an isolate has developed a high level of resistance to a FQ drug, it is likely to have similar resistance levels to other FQ’s.
Table 1. Ciprofloxacin (CIP) resistance in members of the genus Campylobacter from clinical, poultry and livestock samples.
| Sample | Country | Area | Source | Sample type | Isolates year(s) | Species* | Resistance to CIP [% (n)] | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Clinical | Canada | Montreal | Human | 2002 | Cj | 41.6 | (440) | [17] | |
| 2013 | Cc | 50.0 | (38) | ||||||
| Canada | Ontario | Human | 2011 | Cj | 30.8 | (180) | [53] | ||
| 2013 | Cc | 41.0 | (39) | ||||||
| USA | Selected states | Human | 2011 | Cj | 25.8 | (5048) | [54] | ||
| 2015 | Cc | 36.3 | (576) | ||||||
| USA | USA | Human | 2011–12 | C. Spp. | 25.3 | (1962) | [55] | ||
| Peru | Lima | Human | Infant faeces | 2008–11 | Cj | 87.0 | (69) | [57] | |
| Cc | 91.3 | (46) | |||||||
| Europe | 13 countries | Human | 2014 | Cj | 60.2 | (11 585) | [6] | ||
| Cc | 68.9 | (1500) | |||||||
| Europe | 17 countries | Human | 2015 | Cj | 60.8 | (13 696) | [15] | ||
| 16 countries | Cc | 70.6 | (1754) | ||||||
| France | Human | 2014 | Cj | 55.6 | (1997) | [56] | |||
| 2015 | Cc | 64.4 | (419) | ||||||
| Poland | Bydgoszcz | Human | Child faeces | 2011–13 | Cj | 65.2 | (92) | [63] | |
| Cc | 71.4 | (7) | |||||||
| UK | Oxfordshire | Human | 2008 | C.spp. | 37.5 | (803) | [39, 58] | ||
| China | Beijing | Human | 1994/2010 | Cj | 86.7 | (203) | [25] | ||
| Broilers | Africa | Kenya | Chicken | Faeces/cloaca | Cj | 71.0 | (31) | [24] | |
| Cc | 75.0 | (4) | |||||||
| China | Multiple | Chicken | Caecae | 2008–09 | Cj | 99.4 | (971) | [46] | |
| Multiple | 2012–14 | Cc | 99.2 | (1021) | |||||
| China | Central | Chicken | Faeces/cloaca | 2012–16 | Cj | 100.0 | (166) | [61] | |
| CC | 100.0 | (40) | |||||||
| Europe | 25 countries | Chicken | Caecae | 2014 | Cj | 69.8 | (3317) | [6] | |
| 8 countries | Cc | 74.3 | (767) | ||||||
| Italy | Chicken | Cloaca/carcass | 2014 | Cj | 39.0 | (99) | [62] | ||
| 2015 | Cc | 69.7 | (41) | ||||||
| Poultry meat | China | Central | Chicken | Frozen/fresh | 2014 | Cj | 100.0 | (40) | [64] |
| 2015 | Cc | 100.0 | (12) | ||||||
| Europe | 3 countries | Chicken | Various | 2014 | Cj | 65.6 | (308) | [6] | |
| Cc | 85.8 | (134) | |||||||
| 2 countries | Turkey | Slaughter/retail | 2014 | Cj | 66.2 | (74) | [6] | ||
| Poland | Bydgoszcz | Poultry | Slaughter/retail | 2011 | Cj | 62.2 | (90) | [63] | |
| 2013 | Cc | 74.1 | (54) | ||||||
| UK | Multiple | Chicken | Retail | 2014 | Cj | 49.1* | (230) | [16] | |
| 2015 | Cc | 54.7* | (53) | ||||||
| Ruminants | Africa | Ghana | Cattle | Faeces/carcasses | 2013–14 | C. spp. | 42.6 | (54) | [60] |
| Sheep | Faeces/carcasses | 2013–14 | C. spp. | 32.8 | (64) | [60] | |||
| Goat | Faeces/carcasses | 2013–14 | C. spp. | 47.4 | (57) | [60] | |||
| USA | Michigan | Cattle | Faeces | 2012 | Cj | 16.3 | (22) | [40] | |
| USA | 5 States | Cattle | Faeces | 2012–13 | Cj | 35.4 | (320) | [33] | |
| Cc | 74.4 | (115) | |||||||
| Swine | China | Multiple | Pig | Faeces | 2008–14 | Cc | 97.0 | (970) | [39] |
| Africa | Ghana | Pig | Faeces/carcasses | 2013–14 | C. spp. | 30.3 | (66) | [55] | |
| Europe | 7 countries | Pig | Caecae | 2015 | Cc | 62.1 | (704) | [13] | |
*Cj (C. jejuni); Cc (C. coli); C. spp. (Unspecified species of the genus Campylobacter).
Here we describe the genetic basis of CIP resistance in members of the genus Campylobacter and the mechanisms of emergence and spread among strains and species. Drawing on recent publications that describe CIP-resistance in isolates from a number of different sources and countries, we assess if resistance in members of the genus Campylobacter is still rising and review the potential for using DNA sequence-based approaches to predict FQ-resistance in members of the genus Campylobacter.
Mechanisms of resistance
In Campylobacter, FQs work by inhibiting a large enzyme, DNA gyrase, that is involved in DNA replication and transcription [5, 14, 26, 27]. It is now commonly recognised that the most frequent mechanism of CIP resistance in members of the genus Campylobacter is a single point mutation C257T in the gyrA gene, within the QRDR. This results in an amino acid substitution in the Gyrase A subunit at position 86, from threonine to isoleucine [5, 14, 28–31] and has been reported in all CIP-resistant strains of C. jejuni isolated from clinical samples in an example study [32]. Other mutations within the gryA gene have been associated with increased resistance to ciprofloxacin but at lower concentrations and frequency [5, 14, 32, 33]. The gyrA mutation works synergistically with the most common Campylobacter drug efflux pump CmeABC, where, when expression is elevated, the emergence of FQ-resistant strains is increased [5, 14]. However, in the absence of the gyrA gene mutation, over-expression of the CmeABC efflux pump does not generate ciprofloxacin resistance [5, 28, 32, 34, 35]. Other factors enhance the level of resistance further, such as the 16 bp inverted repeat (IR) in the cmeR–cmeABC intergenic region. When this mutation occurs in conjunction with the C257T-gyrA mutation, the proportion of resistant isolates increases and a higher mean ciprofloxacin minimum inhibitory concentration (MIC) is achieved. For example, in culture at a CIP concentration of 64–512 µg ml−1, 30 % resistance was observed in C257T-gyrA mutants compared with 97 % among IR-C257T mutants [28]. In addition a variant of the cmeABC gene that enhances CIP resistance (RE-cmeABC) has recently been identified and is increasing in prevalence [34].
Variation in other genes can also indirectly influence the level of CIP resistance. For example, variations in the mutant frequency decline gene (mfd) may have a role, since the inactivation of this gene has been shown to reduce mutations 100-fold [5, 36]. Unlike the more prolonged and often stepwise selection process for macrolide resistance [5], CIP resistance can accumulate rapidly in the population through mutation in different strains of members of the genus Campylobacter and selection pressure enriching for isolates that have a resistance mutations [14, 18, 27, 37].
Acquisition and spread of resistance
Spontaneous mutation is a major mechanism for acquisition of FQ-resistance. In an environment where resistance confers a selective advantage, clonal reproduction among resistant lineages will lead to local expansion. Consistent with this, there is evidence that CIP resistance phenotypes are more common among certain lineages or clonal complexes (CC) [38]. This lineage association has been observed among isolates from UK retail poultry [38] and clinical samples [39], and several studies have associated ciprofloxacin resistance with certain lineages of members of the genus Campylobacter, including CC-21, CC-206, CC-353 and CC-354 [40–45]. However, in contrasting studies in China there was no association between CC and CIP-resistance phenotypes, indicating that the acquisition of resistance had occurred in numerous distantly related strains of members of the genus Campylobacter [28, 46]. This indicates widespread dispersed resistance rather than clonal expansion of resistant strains. One explanation for this is the spread of resistance between strains by horizontal gene transfer (HGT) [5, 18].
In members of the genus Campylobacter, FQ-resistance is encoded on the chromosome and may therefore be expected to be less transmissible between lineages than AMR that are encoded on highly mobile plasmids. However, in highly recombining bacteria such as C. jejuni and C. coli, there is frequent natural transformation [5, 14] that may facilitate the spread of resistance genes. It is difficult to separate the role of mutation and HGT in conferring quinolone resistant phenotypes from DNA sequence data alone, but the distribution of resistance among relatively distant lineages (clonal complexes, CC’s) is consistent with widespread acquisition by mutation and recombination. The most likely evolutionary scenario is that CIP-resistance originates from independent point mutations in the gyrA gene or horizontal acquisition of resistance-encoding sequence(s). Mutants may proliferate locally but since the gyrA mutation does not incur a strong fitness cost on the recipient genotype, it persists in the absence of selective pressure [14]. When compared with C. jejuni, C. coli tends to have a greater proportion of isolates that are resistant to CIP, along with other AM’s [4, 6, 7, 15, 16]. There are several potential reasons for the higher FQ resistance in C. coli. First, specific mutations or natural transformations may occur at higher frequencies. For example, strains belonging to the C. coli 828 clonal complex show evidence of extremely high levels of interspecies recombination, with around 10 % of the genome introgressed from C. jejuni [47, 48]. Second, it is also possible that uncharacterized genes or adaptations may be present in C. coli, such as those associated with gentamicin resistance [49]. Third, the majority of C. coli isolated from clinical and agricultural sources belong to a single clonal complex (the ST-828 complex). Resistance mechanisms associated with this expansion will be shared by a large proportion of isolated strains. A more detailed understanding of the development and maintenance of AMR in strains and species of the genus Campylobacter will dependent upon analysis of exposure, transmission, strain mutation/recombination frequency and the fitness cost of adaptations to different AMs in the absence of selective pressure [7].
Predicting resistance from whole-genome data
Advances in whole-genome sequencing (WGS) technology and analysis have greatly improved understanding of the genetic basis of phenotypic variation. Large numbers of genomes of members of the genus Campylobacter are now routinely sequenced, and this has considerable potential for improving understanding of the evolution of FQ resistance [5]. A recent study correlated multiple in vitro antimicrobial resistance phenotypes with WGS genotype data [50]. Predictions based upon genomic variation were >99 % accurate, indicating that WGS may be a powerful tool for AMR surveillance programs. Other sequence-based approaches have also accurately detected the single point mutation in the gyrA gene in phenotypically confirmed CIP-resistant members of the genus Campylobacter [24] and the European Committee for Antimicrobial Susceptibility Testing (EUCAST) is currently reviewing the use and accuracy of WGS as a predictor of AMR [51] for surveillance and monitoring programs.
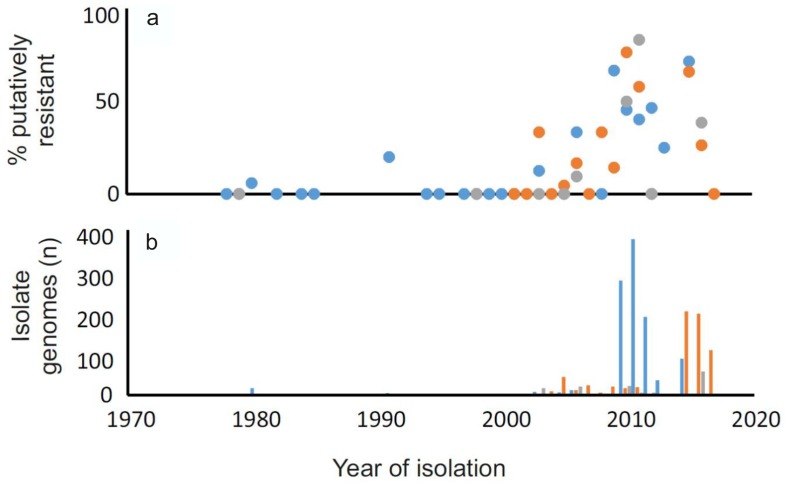
To demonstrate the utility of studying putative CIP resistance using genome data, we analysed assembled draft genomes archived on the Sheppardlab BIGSdb [52] (Table S1, available in the online version of this article). Briefly, gyrA allelic variants were identified among isolate genomes from human (clinical), chicken (faeces and meat) and ruminant (cattle and sheep faeces and meat) samples. Gene homology was defined using blast, with those found to have >70 % nucleotide identity in >50 % of the continuous sequence length, considered to be homologous. While the number of homologues identified would vary depending on the identity threshold, we used these as they are the default settings within the database [52]. Putative resistance genes and the proportion containing threonine and isoleucine at amino acid position 86 were compared after alignment using the mega program, version 7.0. This allowed comparison of putative CIP resistance in the genomes of 1844 isolates of members of the genus Campylobacter (Fig. 1). This demonstrates the utility of WGS for making predictions about resistance phenotypes on the basis of sequence variation in genes for which the putative function is known. There is some evidence for increasing resistance over time among strains from humans, chickens and ruminants but this is not a structured study so further analysis would be needed.
Fig. 1.
Proportion of genomes of isolates of members of the genus Campylobacter containing putative ciprofloxacin resistance. Isolate genomes that contained a complete gyrA gene sequence from 1038 human (Blue), 670 chicken (Orange) and 136 ruminant (Grey) samples collected from 1978 to 2017 were compared. (a) The percentage of isolates containing isoleucine at amino acid position 86 was determined and (b) the total number of samples was recorded.
Ciprofloxacin resistance among clinical isolates
Estimates of the proportion of CIP-resistant isolates of members of the genus Campylobacter from clinical samples vary considerably between published studies [6, 15, 17, 25, 39, 53–57] (Table 1). An increasing trend in the proportion of clinical CIP-resistant isolates of members of the genus Campylobacter has been reported in 2015 for 5 out of 17 (for C. jejuni) and 2 out of 13 (for C. coli) European member states [15]. In England, Scotland and Wales, CIP-resistant members of the genus Campylobacter increased from 7 % of clinical isolates in 1995 to 38 % in 2008, with a similar trend observed among C. jejuni in Northern Ireland (from 9 % in 1996 to 32 % in 2007) [58, 59]. While direct contextualization with isolates from the USA is not possible because of differences in data collection, the proportion of CIP-resistant clinical isolates was seen to increase from 22 % (2004–2010) to 25 % (2011–2012) [55] with a similar level of resistance observed in C. jejuni from human infections reported in Canada (2002–2014) [17].
Reports of reductions in the proportion of CIP-resistant members of the genus Campylobacter from human infections are comparatively rare. None of the EU countries with AMR data covering the previous three years (2013–2015) reported declining trends of CIP resistance in isolates of members of the genus Campylobacter from humans [15]. Furthermore, the members of the EU and the USA are thought to be among the countries with a lower resistance burden, and global trends indicate a higher proportion of CIP resistance in other countries. For example, among clinical samples from China a sustained increase has led to a rise of CIP resistance in C. jejuni from 78 % (1994–2002) to 90 % (2003–2010) [25]. Consistent with this, foreign travel is a major risk factor for infection with FQ-resistant Campylobacter. Studies in Canada, the USA and the UK revealed that individuals that had travelled abroad were more likely to have CIP-resistant strains [17, 39, 55]. In the USA 62.4 % of infections with members of the genus Campylobacteracquired abroad were CIP-resistant compared with 14.4 % of domestically acquired infections and CIP-resistant strains were five times more likely to have been acquired abroad than domestically in the UK [39, 55].
Ciprofloxacin resistance in livestock
Consistent with the high levels of FQ resistance in clinical samples, numerous studies have reported CIP-resistant members of the genus Campylobacter in livestock, including ruminants [33, 40, 60], swine [15, 46, 60], poultry [6, 24, 46, 61, 62] and poultry retail meat [6, 16, 63, 64] (Table 1). In cattle from the USA, 16 % of C. jejuni isolated from gut and faecal samples (2014) were resistant to CIP [40] and in isolates from faeces and carcasses of cattle from Africa (2013–2014), the proportion of CIP-resistant species of the genus Campylobacter was 43 % [60]. Resistance was even more common among C. coli from fattening pigs in Europe (2015) with 62 % of isolates displaying CIP resistance [15].
As in clinical samples over a similar period, there is also evidence for an increasing trend in CIP-resistant members of the genus Campylobacter from beef and dairy cattle in the USA (2013–2014) [4] and broiler chickens from various European countries (2008–2014) [6]. In Austria and Spain the proportion of CIP-resistant C. jejuni from broilers remained stable over this period, as did resistance in C. coli isolated in France, although they remained at a relatively high prevalence. The only European country in which a reduction in the number of CIP-resistant C. jejuni and C. coli isolated from broilers was observed over this period (2008–2014) was the Netherlands [6].
Potential for spread of resistance from animals to humans
The transmission of FQ-resistant bacteria from agricultural animals to humans is difficult to prove and a recent global report on surveillance of antimicrobial resistance emphasised the need to collect more data on the effects of AMR in foodborne bacteria on animal and human health [21, 65]. Currently, there is little direct evidence of the transmission of FQ-resistant bacteria within livestock and to humans via food. The occurrence of resistant members of the genus Campylobacter on retail poultry meat is a major concern since this is a principal source of isolates infecting humans [3, 66]. Data covering the last three years reveals an overall increase in CIP-resistance among both C. jejuni and C. coli isolates from chicken meat [6]. In the USA, CIP-resistance among isolates from chicken breast meat increased from 15 to 17 % (C. jejuni) and from 10 to 26 % (C. coli) from 2002 to 2007 [67], and a separate study recorded a similar rise among C. jejuni isolates between 2013 and 2014 [4]. In UK studies, the proportion of CIP-resistant C. jejuni and C. coli isolated from retail chicken showed a similar increase between 2007–2008 and 2014–2015 with the proportion of resistant C. jejuni and C. coli isolates increasing from 21 to 49 % and from 35 to 53 %, respectively [16, 68]. Interestingly, samples from frozen chicken were more likely to contain ciprofloxacin-resistant isolates [68]
It has been noted that the proportion of CIP-resistant members of the genus Campylobacter in poultry meat is often strikingly similar to the proportion observed in human clinical cases [6, 16, 58, 69]. In China, broilers and chicken meat showed the highest Campylobacter CIP resistance at 99 to 100 %, which corresponds to the extremely high levels recorded in human isolates at 90 % between 2003 and 2010 [25, 61, 64]. Likewise, in Poland 66 % of human clinical isolates of members of the genus Campylobacter were CIP-resistant, which was similar to the 67 % sampled from chicken meat (Table 1) [63]. In the case where the direct consumption of FQ-resistant bacteria leads to human infection, it is compelling to conclude that the use of antimicrobials to treat animals potentially erodes the efficacy of important human drug treatments. The antimicrobial resistance in C. jejuni and C. coli of both animal and human origin, has a strong correlation with the amount of antimicrobial use in the animal production system (mg per kg of meat produced). Those with stricter guidelines tend to have lower proportions of resistant isolates (e.g. Nordic countries) [7, 70]. However, using WGS to identify the gyrA mutations in isolates of members of the genus Campylobacter from poultry failed to cluster isolates according to the country of origin or the current use of FQ in livestock production [45]. In addition, data from the USA Centre for Disease Control National Antimicrobial Resistance Monitoring System database [54] indicates that the 2005 USA ban on the use of FQ’s in livestock has had little effect on the increasing FQ-resistance among C. jejuni isolates from clinical samples.
The fate of genomic variation associated with resistance is at least partially determined by fitness. Substitutions that impose little or no fitness cost on the cell have a higher probability of persisting in the absence of antibiotic treatment [71]. It is known that FQs impose a fitness cost in members of the genus Campylobacter but this can vary depending on strain and study conditions, with resistance potentially persisting for some time in the absence of FQs [72], potentially associated with compensatory mutations that alleviate the costs of resistance. This means that livestock may not just act as transient vehicles of FQ resistance but are dissemination points where members of the genus Campylobacter resistant to FQ’s can persist and circulate within the animal population for several years following the decreased use of AM’s [7]. However, production systems that use AMs at a higher rate present an increased risk of resistant isolates spreading, at least on a local scale. Monitoring of AMR is seriously lacking in many countries [21] where this, along with different practices and regulations, is associated with wide variation in AMR between different countries [7]. Because of trade networks, global implementation of monitoring and restriction of the use of AM’s in livestock would be beneficial in the fight against AMR, with more detailed information on AM usage at local, regional and national scales to more accurately assess the potential risk of transfer to humans [7, 21, 70].
Conclusions
The mechanisms by which FQ-resistance is acquired in Campylobacter (mutation and HGT) are well established, and routine surveillance of resistance phenotypes in clinical samples describe a continued and sustained increase in many countries. This trend is mirrored in isolates from livestock, especially chickens, which are a major source of human disease. It is difficult to quantify the extent to which the use of antimicrobials in agriculture reduces the efficacy of drugs such as fluoroquinolones in treating human infections. This is exacerbated by the slow rate at which FQ-resistance in Campylobacter is purged in the absence of AMs, meaning that, once acquired, resistance can be maintained in populations despite restrictions on the use of FQ’s in animal production. WGS offers a potential tool for improving understanding of the emergence and maintenance of AMR among members of the genus Campylobacter from multiple host species. On the basis of AMR phenotype predictions from large population genomic datasets it will be possible to more accurately characterize source and sink populations, environmental reservoirs and specific microevolutionary events associated with acquisition, maintenance and spread of resistance.
Data bibliography
Supporting external data for the prediction of putative CIP resistance using genome data is included as a supplementary table (Table S1) listing the isolates and a supplementary data file of gyrA sequences from all isolates.
Funding information
S. K. S. was supported by Medical Research Council (MRC) grants MR/L015080/1 and G0801929, Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I02464X/1 and the Wellcome Trust.
Acknowledgements
Thanks for input from members of the Sheppard Laboratory.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CIP, ciprofloxacin; FQ, Fluoroquinolones; WGS, whole-genome sequencing.
One supplementary table is available with the online version of this article.
Supplementary Data
References
- 1.Humphrey T, O'Brien S, Madsen M. Campylobacters as zoonotic pathogens: a food production perspective. Int J Food Microbiol. 2007;117:237–257. doi: 10.1016/j.ijfoodmicro.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Blaser MJ, Engberg J. Clinical aspects of Campylobacter jejuni and Campylobacter coli infections. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington, DC: ASM Press; 2008. pp. 99–121. (editors) [Google Scholar]
- 3.Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, et al. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol. 2009;134:96–103. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NARMS Integrated Report The National Antimicrobial Resistance Monitoring System: Enteric Bacteria. 2014 www.fda.gov/downloads/animalveterinary/safetyhealth/antimicrobialresistance/nationalantimicrobialresistancemonitoringsystem/ucm528861.pdf [accessed December 2017]
- 5.Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int. 2013;2013:1–12. doi: 10.1155/2013/340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J. 2016;14:1–207. doi: 10.2903/j.efsa.2018.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi M, Olkkola S, Roasto M, Kivistö R, Hänninen M, et al. Chapter 4: Antimicrobial resistance and Campylobacter jejuni and C. coli. In: Chen C, Yan X, Jackson CR, editors. Antimicrobial Resistance and Food Safety: Methods and Techniques. Amsterdam, Netherlands: Elsevier/Academic Press; 2015. pp. 55–75. (editors) [Google Scholar]
- 8.Guerrant RL, van Gilder T, Steiner TS, Thielman NM, Slutsker L, et al. Infectious Diseases Society of America: practice guidelines for the management of infectious diarrhea. Clin Infect Dis. 2001;32:331–351. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 9.Gupta A, Nelson JM, Barrett TJ, Tauxe RV, Rossiter SP, et al. Antimicrobial resistance among Campylobacter strains, United States, 1997–2001. Emerg Infect Dis. 2004;10:1102–1109. doi: 10.3201/eid1006.030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge B, Wang F, Sjölund-Karlsson M, McDermott PF. Antimicrobial resistance in campylobacter: susceptibility testing methods and resistance trends. J Microbiol Methods. 2013;95:57–67. doi: 10.1016/j.mimet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Allos BM. Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 12.Endtz HP, Ruijs GJ, van Klingeren B, Jansen WH, van der Reyden T, et al. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J Antimicrob Chemother. 1991;27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- 13.Nelson JM, Chiller TM, Powers JH, Angulo FJ. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin Infect Dis. 2007;44:977–980. doi: 10.1086/512369. [DOI] [PubMed] [Google Scholar]
- 14.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, et al. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 2009;4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control) The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017;15:1–212. doi: 10.2903/j.efsa.2017.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PHE (Public Health England) Antimicrobial resistance report for FSA Project FS241044. Forming part of the project: A Microbiological survey of Campylobacter contamination in fresh whole UK produced chilled chickens at retail sale (2014–15) 2016:1–50. www.food.gov.uk/sites/default/files/media/document/fs241044finreport.pdf [accessed December 2017]
- 17.Gaudreau C, Boucher F, Gilbert H, Bekal S. Antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli isolates obtained in Montreal, Quebec, Canada, from 2002 to 2013. J Clin Microbiol. 2014;52:2644–2646. doi: 10.1128/JCM.00362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey TJ, Jørgensen F, Frost JA, Wadda H, Domingue G, et al. Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob Agents Chemother. 2005;49:690–698. doi: 10.1128/AAC.49.2.690-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regulation (EC) No 1831/2003 of the European parliament and of the council of 22 September 2003 on additives for use in animal nutrition. http://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32003R1831&from=EN [accessed December 2017]
- 20.Castanon JI. History of the use of antibiotic as growth promoters in European poultry feeds. Poult Sci. 2007;86:2466–2471. doi: 10.3382/ps.2007-00249. [DOI] [PubMed] [Google Scholar]
- 21.WHO (World Health Organisation) Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland: World Health Organization; 2014. pp. 1–232.http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1 [accessed December 2017] [Google Scholar]
- 22.ECDC (European Centre for Disease Prevention and Control) EU Protocol for Harmonised Monitoring of Antimicrobial Resistance in Human Salmonella and Campylobacter Isolates. Technical Document, ECDC; 2016. pp. 1–14.https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/antimicrobial-resistance-Salmonella-Campylobacter-harmonised-monitoring.pdf [accessed May 2018] [Google Scholar]
- 23.EFSA (European Food Safety Authority) Manual for Reporting on Antimicrobial Resistance Within The framework of Directive 2003/99/EC and Decision 2013/652/EU for Information Deriving from The year 2015. EFSA Supporting Publication; 2016. pp. 1–35.https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/sp.efsa.2016.EN-990 EN990. [accessed May 2018] [Google Scholar]
- 24.Nguyen TN, Hotzel H, Njeru J, Mwituria J, El-Adawy H, et al. Antimicrobial resistance of Campylobacter isolates from small scale and backyard chicken in Kenya. Gut Pathog. 2016;8:39. doi: 10.1186/s13099-016-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, Zhang M, Yang W, Fang Y, Wang G, et al. A seventeen-year observation of the antimicrobial susceptibility of clinical Campylobacter jejuni and the molecular mechanisms of erythromycin-resistant isolates in Beijing, China. Int J Infect Dis. 2016;42:28–33. doi: 10.1016/j.ijid.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41:S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 27.Smith JL, Fratamico PM. Fluoroquinolone resistance in Campylobacter. J Food Prot. 2010;73:1141–1152. doi: 10.4315/0362-028X-73.6.1141. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Zhang M, Zhou J, Pang L, Wang G, et al. The molecular mechanisms of ciprofloxacin resistance in clinical Campylobacter jejuni and their genotyping characteristics in Beijing, China. Foodborne Pathog Dis. 2017;14:386–392. doi: 10.1089/fpd.2016.2223. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Huang WM, Taylor DE. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother. 1993;37:457–463. doi: 10.1128/AAC.37.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, et al. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006;8:1967–1971. doi: 10.1016/j.micinf.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 31.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001;7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan M, Sahin O, Lin J, Zhang Q. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother. 2006;58:1154–1159. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 33.Tang Y, Sahin O, Pavlovic N, Lejeune J, Carlson J, et al. Rising fluoroquinolone resistance in Campylobacter isolated from feedlot cattle in the United States. Sci Rep. 2017;7:494. doi: 10.1038/s41598-017-00584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao H, Shen Z, Wang Y, Deng F, Liu D, et al. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. MBio. 2016;7:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge B, McDermott PF, White DG, Meng J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005;49:3347–3354. doi: 10.1128/AAC.49.8.3347-3354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J, Sahin O, Barton YW, Zhang Q. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog. 2008;4:e1000083. doi: 10.1371/journal.ppat.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griggs DJ, Johnson MM, Frost JA, Humphrey T, Jørgensen F, et al. Incidence and mechanism of ciprofloxacin resistance in Campylobacter spp. isolated from commercial poultry flocks in the United Kingdom before, during, and after fluoroquinolone treatment. Antimicrob Agents Chemother. 2005;49:699–707. doi: 10.1128/AAC.49.2.699-707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wimalarathna HM, Richardson JF, Lawson AJ, Elson R, Meldrum R, et al. Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol. 2013;13:160. doi: 10.1186/1471-2180-13-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cody AJ, McCarthy NM, Wimalarathna HL, Colles FM, Clark L, et al. A longitudinal 6-year study of the molecular epidemiology of clinical Campylobacter isolates in Oxfordshire, United kingdom. J Clin Microbiol. 2012;50:3193–3201. doi: 10.1128/JCM.01086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cha W, Mosci RE, Wengert SL, Venegas Vargas C, Rust SR, et al. Comparing the genetic diversity and antimicrobial resistance profiles of Campylobacter jejuni recovered from cattle and humans. Front Microbiol. 2017;8:818. doi: 10.3389/fmicb.2017.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Habib I, Miller WG, Uyttendaele M, Houf K, de Zutter L. Clonal population structure and antimicrobial resistance of Campylobacter jejuni in chicken meat from Belgium. Appl Environ Microbiol. 2009;75:4264–4272. doi: 10.1128/AEM.00168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinana AD, Cardinale E, Tall F, Bahsoun I, Sire JM, et al. Genetic diversity and quinolone resistance in Campylobacter jejuni isolates from poultry in Senegal. Appl Environ Microbiol. 2006;72:3309–3313. doi: 10.1128/AEM.72.5.3309-3313.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kittl S, Kuhnert P, Hächler H, Korczak BM. Comparison of genotypes and antibiotic resistance of Campylobacter jejuni isolated from humans and slaughtered chickens in Switzerland. J Appl Microbiol. 2011;110:513–520. doi: 10.1111/j.1365-2672.2010.04906.x. [DOI] [PubMed] [Google Scholar]
- 44.Kovač J, Čadež N, Stessl B, Stingl K, Gruntar I, et al. High genetic similarity of ciprofloxacin-resistant Campylobacter jejuni in central Europe. Front Microbiol. 2015;6:1169. doi: 10.3389/fmicb.2015.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Technical University of Denmark - National Food Institute . Comparative Genomics of Quinolone-Resistant and Susceptible Campylobacter Jejuni of Poultry Origin from Major Poultry Producing European Countries (GENCAMP) EFSA supporting publication; 2018. p. 35. EN-1398. [Google Scholar]
- 46.Wang Y, Dong Y, Deng F, Liu D, Yao H, et al. Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–14. J Antimicrob Chemother. 2016;71:666–669. doi: 10.1093/jac/dkv382. [DOI] [PubMed] [Google Scholar]
- 47.Sheppard SK, McCarthy ND, Falush D, Maiden MC. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- 48.Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, et al. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol. 2013;22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Mukherjee S, Hoffmann M, Kotewicz ML, Young S, et al. Whole-genome sequencing of gentamicin-resistant Campylobacter coli isolated from U.S. retail meats reveals novel plasmid-mediated aminoglycoside resistance genes. Antimicrob Agents Chemother. 2013;57:5398–5405. doi: 10.1128/AAC.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, et al. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol. 2015;82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect. 2017;23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 52.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riley A, Eshaghi A, Olsha R, Allen VG, Patel SN. Antibiotic susceptibility of clinical isolates of Campylobacter jejuni and Campylobacter coli in Ontario, Canada during 2011–2013. Diagn Microbiol Infect Dis. 2015;83:292–294. doi: 10.1016/j.diagmicrobio.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 54.NARMS Now: Human Data. www.cdc.gov/narmsnow/ [accessed December 2017]
- 55.Geissler AL, Bustos Carrillo F, Swanson K, Patrick ME, Fullerton KE, et al. Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004–2012. Clin Infect Dis. 2017;65:1624–1631. doi: 10.1093/cid/cix624. [DOI] [PubMed] [Google Scholar]
- 56.Sifré E, Salha BA, Ducournau A, Floch P, Chardon H, et al. EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J Microbiol Methods. 2015;119:206–213. doi: 10.1016/j.mimet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Lluque A, Riveros M, Prada A, Ochoa TJ, Ruiz J. Virulence and antimicrobial resistance in Campylobacter spp. from a Peruvian pediatric cohort. Scientifica. 2017;2017:1–8. doi: 10.1155/2017/7848926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cody AJ, Clarke L, Bowler IC, Dingle KE. Ciprofloxacin-resistant campylobacteriosis in the UK. Lancet. 2010;376:1987. doi: 10.1016/S0140-6736(10)62261-1. [DOI] [PubMed] [Google Scholar]
- 59.Moore JE, Millar BC, McMahon MA, McDowell DA, Rooney PJ. Antimicrobial resistance in campylobacter isolates from sporadic cases of acute human gastroenteritis in Northern Ireland. Ulster Med J. 2009;78:139. [PMC free article] [PubMed] [Google Scholar]
- 60.Karikari AB, Obiri-Danso K, Frimpong EH, Krogfelt KA. Antibiotic resistance of Campylobacter recovered from faeces and carcasses of healthy livestock. Biomed Res Int. 2017;2017:1–9. doi: 10.1155/2017/4091856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang T, Dong J, Cheng Y, Lu Q, Luo Q, et al. Genotypic diversity, antimicrobial resistance and biofilm-forming abilities of Campylobacter isolated from chicken in Central China. Gut Pathog. 2017;9:62. doi: 10.1186/s13099-017-0209-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pergola S, Franciosini MP, Comitini F, Ciani M, de Luca S, et al. Genetic diversity and antimicrobial resistance profiles of Campylobacter coli and Campylobacter jejuni isolated from broiler chicken in farms and at time of slaughter in central Italy. J Appl Microbiol. 2017;122:1348–1356. doi: 10.1111/jam.13419. [DOI] [PubMed] [Google Scholar]
- 63.Szczepanska B, Andrzejewska M, Spica D, Klawe JJ. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017;17:80. doi: 10.1186/s12866-017-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang T, Luo Q, Chen Y, Li T, Wen G, et al. Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in Central China. Gut Pathog. 2016;8:48. doi: 10.1186/s13099-016-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. MBio. 2016;7:e02227-15. doi: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, et al. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis. 2009;48:1072–1078. doi: 10.1086/597402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao S, Young SR, Tong E, Abbott JW, Womack N, et al. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl Environ Microbiol. 2010;76:7949–7956. doi: 10.1128/AEM.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.FSA (Food Standards Agency) FSA report for the UK survey of Campylobacter and Salmonella contamination of fresh chicken at retail sale. FSA Project B18025. 2009:1–97. http://webarchive.nationalarchives.gov.uk/20140403130456/http://www.foodbase.org.uk/admintools/reportdocuments/351-1-676_B18025.pdf [accessed December 2017] [Google Scholar]
- 69.Nichols GL, Richardson JF, Sheppard SK, Lane C, Sarran C. Campylobacter epidemiology: a descriptive study reviewing 1 million cases in England and Wales between 1989 and 2011. BMJ Open. 2012;2:e001179. doi: 10.1136/bmjopen-2012-001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Migura L, Hendriksen RS, Fraile L, Aarestrup FM. Antimicrobial resistance of zoonotic and commensal bacteria in Europe: the missing link between consumption and resistance in veterinary medicine. Vet Microbiol. 2014;170:1–9. doi: 10.1016/j.vetmic.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 71.Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeitouni S, Kempf I. Fitness cost of fluoroquinolone resistance in Campylobacter coli and Campylobacter jejuni. Microb Drug Resist. 2011;17:171–179. doi: 10.1089/mdr.2010.0139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.