Abstract
Peripheral spondyloarthritis (SpA) is a common extra-intestinal manifestation in patients with active inflammatory bowel disease (IBD) characterized by inflammatory enthesitis, dactylitis, or synovitis of non-axial joints. However, a mechanistic understanding of the link between intestinal inflammation and SpA has yet to emerge. Here, we evaluated and functionally characterized the fecal microbiome of IBD patients with or without peripheral SpA. Coupling the sorting of IgA-coated microbiota with 16S rRNA-based analysis (IgA-seq) revealed a selective enrichment in IgA-coated E. coli in patients with Crohn’s disease-associated SpA (CD-SpA) compared to CD alone. E. coli isolates from CD-SpA-derived IgA-coated bacteria were similar in genotype and phenotype to an Adherent-invasive E. coli (AIEC) pathotype. In comparison to non-AIEC E. coli, colonization of germ-free mice with CD-SpA E. coli isolates induced Th17 mucosal immunity, which required the virulence-associated metabolic enzyme propanediol dehydratase (pduC). Modeling the increase in mucosal and systemic Th17 immunity we observed in CD-SpA patients, colonization of IL-10 deficient or K/BxN mice with CD-SpA-derived E. coli lead to more severe colitis or inflammatory arthritis, respectively. Collectively, these data reveal the power of IgA-seq to identify immune-reactive resident pathosymbionts that link mucosal and systemic Th17-dependent inflammation and offer microbial and immunophenotype stratification of CD-SpA that may guide medical and biologic therapy.
One Sentence Summary:
IgA-reactive E. coli in Crohn’s disease-associated spondyloarthritis links mucosal immunity and systemic inflammation.
Introduction
Inflammatory bowel disease (IBD) reflects a spectrum of intestinal disorders, principally Crohn’s disease (CD) and ulcerative colitis (UC), characterized by a dysregulated immune response to environmental and microbial antigens in genetically susceptible individuals (1). Although the characteristics of intestinal inflammation define the IBD phenotype, extra-intestinal manifestations (EIMs) of IBD frequently reveal distinct features underlying the immune disorder. IBD-associated spondyloarthritis (SpA), which includes both axial spinal inflammation and peripheral joint manifestations of synovitis, dactylitis, and enthesitis, is one of the most common EIMs of IBD (2). Peripheral manifestations alone carry a prevalence of 20% in CD and 10% in UC, predominantly affecting joints of the lower limbs (3). Clinical evidence linking intestinal inflammation with SpA (3-5) implicates the intestine as the source of aberrant systemic joint inflammation (6, 7). The independent identification of genetic variants in the IL23R locus in both IBD (8) and SpA (9) reinforces the biologic underpinnings of this association by highlighting a potential shared role for IL-23-dependent inflammation. Collectively, these data suggest that stratification of IBD using clinical EIM phenotypes may offer a strategy to define markers of disease that will improve diagnostic and therapeutic approaches to IBD.
While a mechanistic understanding of the link between intestinal inflammation and SpA has yet to emerge, notable alterations in the intestinal microbiome of IBD patients compared to non-IBD controls—including a consistent reduction in taxa-diversity and expansion of Enterobacteriaceae in new onset Crohn’s disease (CD) (10)—have implicated a role for the microbiome as the antigenic stimulus driving systemic inflammation. Seminal work supporting a role for the microbiome in systemic arthritis was performed using HLA-B27 transgenic rats that develop both spontaneous intestinal inflammation and arthritis (11). Animals reared under germ-free conditions failed to develop inflammatory disease, whereas the re-introduction of normal luminal bacteria (including Bacteroides spp.) was sufficient to trigger inflammation (12). Consistent with a role for intestinal microbiota in these animal models, early studies in ankylosing spondylitis (AS) patients observed an increased frequency of Klebsiella pneumoniae in fecal samples (13) coupled with cellular and humoral immunity to K. pneumoniae antigens (14). More recent studies using current technology for microbial sequencing have defined unique microbial communities in inflammatory arthritis, including distinct microbiomes in ankylosing spondylitis (15) and rheumatoid arthritis (RA) (16, 17). Recent characterization of the microbiome in psoriatic arthritis revealed decreased bacterial diversity similar to that seen in active IBD (18), but the microbiome in IBD-associated SpA has not been well characterized.
Compositional analysis of the microbiome, however, does not necessarily reflect the impact of the microbiome on host immunity. To address this limitation, a recently developed technique which couples the sorting of IgA-coated microbiota with 16S rRNA sequencing (called IgA-seq) focuses analysis on microbiota identified by the immune system (19). These analyses highlight the dominant IgA response to keystone species, such as segmented filamentous bacterium (SFB) in mice, that make up a small fraction of the total microbiome, yet exert a profound effect on IL-23/Th17 dependent mucosal and systemic immunity (20, 21). Recent studies in both mouse models (22) and humans (19, 23) revealed the ability of IgA-coated microbiota to serve as pathosymbionts that can exacerbate disease phenotype. Indeed, fecal samples from IBD patients have increased levels of IgA-bound microbes (24). Moreover, systemic sero-reactivity to microbial-derived antigens in patients with CD (25) suggest active immune recognition of microbial dysbiosis; however, the functional immune relevance of the IBD microbiome to disease phenotype is still emerging.
While a causative microbe has not been identified in IBD, clinical cohorts consistently report the expansion of Enterobacteriaceae in active CD (10, 26-28). These Enterobacteriaceae include a unique pathotype of E. coli designated as Adherent-invasive E. coli (AIEC) (29). Although these AIEC strains lack virulence factors associated with diarrheagenic E.coli, they are able to adhere to and invade cultured intestinal epithelial cells and replicate in macrophages (30, 31). Genetically, AIEC resemble extra-intestinal pathogenic E.coli (Expec) and harbor genes (including pduC and lpfA) that have been linked to invasion and intracellular persistence (32, 33).
Given the overlapping genetic variation in the IL23R locus in both IBD (8) and SpA (9) cohorts, IL-23 responsive lymphocytes may underlie a potential shared immunopathogenesis. A large component of these IL-23 responsive lymphocytes are CD4+ Th17 cells that express the transcription factor RORγt, produce the cytokine IL-17, and play a critical role in promoting homeostasis at the mucosal barrier at the steady (34); however, in genetically-susceptible animal models, microbial activation of Th17 cells by the mouse commensal segmented filamentous bacterium (SFB) (21, 35) is sufficient to drive pathogenic systemic inflammation (20, 36).
In this study, we investigated the hypothesis that functional analysis of the endogenous IgA-bound microbial repertoire in IBD-SpA will both stratify disease phenotype and provide insight into the immunopathogenesis. Compositional and functional analysis of the gut microbiome identified a significant expansion of IgA-coated E. coli, genotypically and functionally characterized as Adherent-invasive E. coli (AIEC). Given our findings of systemic Th17 activation in patients with CD-SpA reported here, gnotobiotic mice as well as mouse models of systemic Th17-dependent inflammatory arthritis were used to evaluate the role for CD-SpA-derived E. coli in linking mucosal and systemic immunity.
Results
Clinical and microbial parameters define CD-associated peripheral SpA
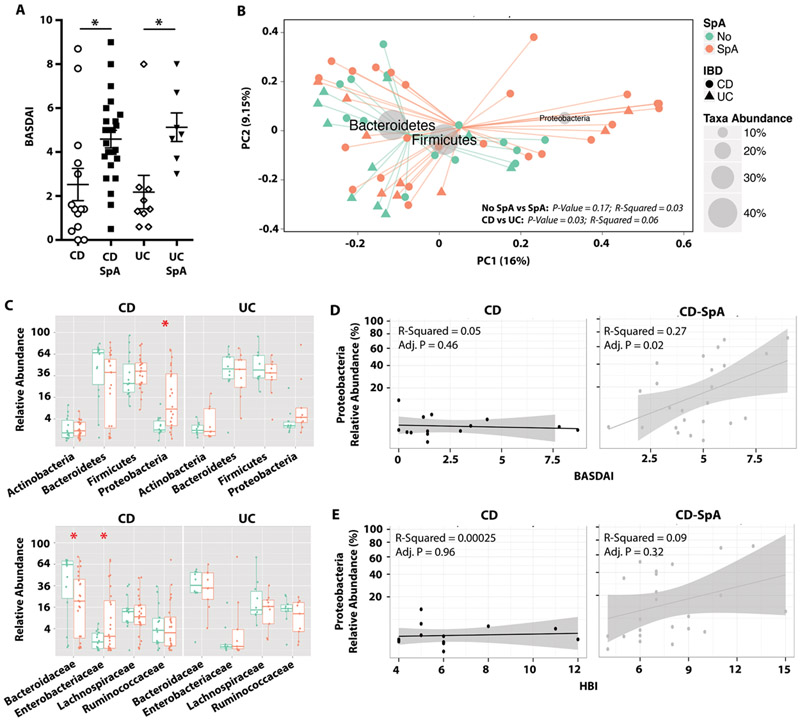
To test the hypothesis that clinical and microbial biomarkers correlate with IBD-SpA, we established a prospective cohort of 59 IBD patients with or without peripheral SpA (defined by the Assessment of Spondyloarthritis international Society (ASAS) criteria). To control for microbiome alterations associated with disease activity, antibiotic exposure, and intestinal surgery, only patients with active disease (defined by Harvey-Bradshaw Index > 4 or modified UC-DAI > 2), off antibiotics for at least 8 weeks, and with an intact ileocecal valve were included (Table S1). Only CD patients with ileal or ileocolonic disease (L1 or L3 disease by Montreal classification, Table S2) were included to target a clinical phenotype previously associated with changes in the microbiome (27). Furthermore, all patients were HLA-B27 negative to avoid potential overlap with axial spondyloarthritis. Groups were matched for age, gender, body-mass index (BMI), and intestinal disease activity indexes (Table S1). To evaluate the clinical utility of the Bath Ankylosing Spondylitis disease activity index (BASDAI)—a 6-question, patient-reported, clinically validated tool for assessing disease activity in AS (37)— in identifying joint inflammation in IBD-associated peripheral SpA, BASDAI scores were recorded for all patients at time of sample collection. BASDAI differentiated SpA from non-SpA for both CD and UC (Fig. 1A, Table S1). Using a cutoff of >4 to indicate disease, BASDAI provided 83% specificity compared to ASAS defined diagnosis (38). In contrast, no significant differences were observed between the cohorts in conventional serologic markers of disease activity including ESR, CRP, vitamin D level, and hemoglobin.
Fig. 1. Clinical and microbial markers segregate CD and CD-SpA.
A. Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) was calculated for each subject. ANOVA, **P<0.01. B. Subjects were clustered using PCoA with Bray-Curtis distances. Gray circles represent taxa abundance. P-value estimates calculated by Monte Carlo permutation test are indicated for CD vs. UC and No SpA vs. SpA. C. Relative abundances of top 4 phyla (top panel) and families (bottom panel) are shown for No SpA (green) and SpA (orange). Boxplots reflect 25 and 75% confidence interval and horizontal line reflects median. P-values < 0.05 are indicated with an asterisk, Mann-Whitney. D, E. Correlation between abundance of Proteobacteria is plotted against clinical joint disease activity score (BASDAI) (D) or Crohn’s disease activity (HBI) (E). Linear regression analysis and adjusted p-values are shown for both D and E. For all panels, CD N=15, CD-SpA N=25, UC N=10, UC-SpA N=9.
To determine if differences in the intestinal microbiota segregate individuals with IBD compared to IBD-SpA, we performed 16S rRNA sequencing of fecal DNA (all samples sequenced with >6500 reads/sample, Fig. S1A). While the alpha diversity metrics (including OTUs observed, Chao1, and Shannon entropy) revealed a contraction in diversity in CD compared to UC samples (Fig. S1B) similar to previous reports (27), no differences in diversity were noted between SpA and non-SpA groups (Fig. S1B). Subjects were clustered using principal coordinate analysis with Bray-Curtis distances (Fig. 1B). PERMANOVA analysis of beta-diversity showed significant differentiation between CD and UC cohorts (p=0.03), but no significant segregation between SpA and no SpA groups (p=0.17, Fig. 1B). The most discriminative axis (horizontal) largely reflected the abundance of Proteobacteria in these samples. Composite phyla and family-level relative abundances revealed an expansion of Proteobacteria and Enterobacteriaceae, respectively, in subjects with CD-SpA compared to CD (Fig. 1C). No differences in specific taxa were observed between UC and UC-SpA cohorts. Although analysis of genus-level relative abundances did not reveal any differences between CD and CD-SpA, linear regression analysis of BASDAI and relative bacterial abundance in patients with CD-SpA revealed a positive correlation with both Proteobacteria and Enterobacteriaceae abundance (Fig. 1D). Increase in Proteobacteria abundance has been reported to correlate with intestinal disease severity in new onset CD (10); however, in this cohort of patients with active L1/L3 CD, intestinal disease activity as measured by HBI was not sufficient to map this correlation by linear regression (Fig. 1E). Although we employed rigorous exclusion criteria to limit confounders, immunomodulator therapy varied in our patient cohort. Stratification by immunomodulator therapy (including thiopurines and biologics) did not account for differences in alpha-diversity, beta-diversity, or taxonomic abundances in either CD or UC (Fig. S2).
IgA-seq reveals immune-reactive E. coli in CD-SpA
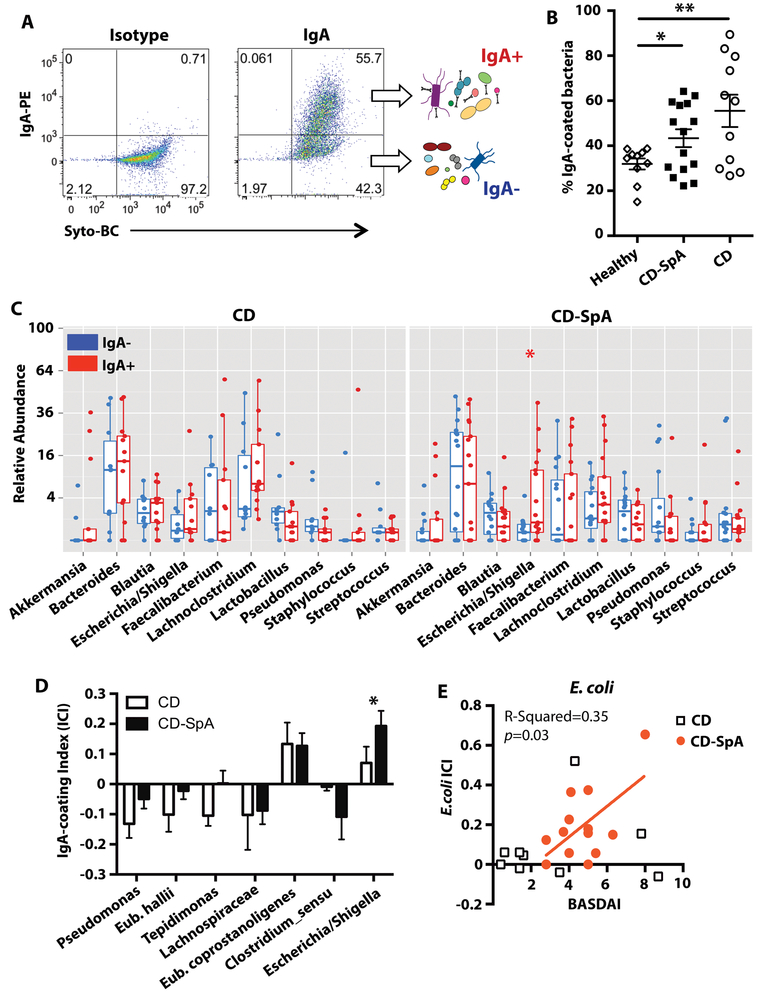
Although our compositional analysis revealed differential abundance of microbial communities, these analyses do not necessarily reflect the immunological impact of these microbiota. Morevoer, while no significant differences in genus or taxa level abundance between CD and CD-SpA cohorts were seen in our compositional analysis, we hypothesized that distinct immunologically reactive microbiota differentiate CD-SpA. To detect microbiota eliciting an immune response, we labeled and sorted IgA coated (IgA+) and non-coated (IgA-) bacteria from fecal homogenates of CD patients profiled above (IgA-Seq, Fig. 2A). Similar to previous reports (19) and consistent with a recent report suggesting that IgA responses primarily target commensal bacteria of the small intestine (39), the proportion of intestinal bacteria coated with IgA was increased in CD compared to healthy donors; however, no significant difference was seen in IgA-coating between CD-SpA and CD (Fig. 2B). Of the 40 CD samples used for 16S rRNA sequencing, 30 samples (14 CD and 16 CD-SpA) were available for IgA-seq. Rarefaction at sample depth of 1100 reads / sample left 23 samples (10 CD, 13 CD-SpA) for subsequent analysis (Fig. S3). Similar to compositional analysis, evaluation of the pre-sort population also revealed significantly increased relative abundance of Proteobacteria in CD-SpA compared to CD, but no significant differences at the genus level were observed (Fig. S4). We did not observe significant differences in alpha or beta diversity (PERMANOVA > 0.05) between IgA- and IgA+ microbial community (Fig. S5). Subject level variance dominated PCoA analysis of beta-diversity (PERMANOVA p < 0.001, Fig. S5B).
Fig. 2. IgA-coated microbiota in CD-SpA are enriched for E. coli.
A. The IgA-coated sort strategy is shown. Fecal homogenate was filtered, blocked and stained with cell permeable DNA dye Syto-BC and isotype or anti-IgA antibody. Samples were gated based on FSC/SSC. B. Percentage IgA-coated bacteria in fecal homogenate for individuals grouped by disease status. ANOVA, * P<0.05. CD N=11, CD-SpA N=15 C. Relative abundance of the top 10 most abundant taxa by genus are shown grouped by IgA negative (blue) or IgA positive (red) and faceted by CD or CD-associated SpA (CD-SpA). Boxplots reflect 25 and 75% confidence interval and horizontal line reflects median. P-values < 0.05 are indicated, Mann-Whitney D. IgA coating index (ICI) was calculated for each individual taxa as -(log(IgA+) – log(IgA-))/(log(IgA+) + log(IgA-)). Taxa with IgA index > 0.1 or <−0.01 are shown. CD (N=10) and CD-SpA (N=13). Asterisk indicates p-values < 0.05, Mann-Whitney. E. Correlation between ICI and clinical joint disease activity (BASDAI) is shown for both CD (square) and CD-SpA (orange circle). Linear regression analysis reveals significant correlation for CD-SpA (p=0.03). For C-E, CD N=10, CD-SpA N=13.
Analysis of the differential relative abundance of taxa at the phylum, family (Fig. S6), and genus level (Fig. 2C) from the IgA- and IgA+ microbial communities, however, revealed significant enrichment of Escherichia/Shigella in the IgA+ fraction of patients with CD-SpA. To identify bacteria enriched in either the IgA+ or IgA- fraction in CD, CD-SpA, or both, we used an average IgA coating index (ICI) (23) of > 0.1 or < −0.1, respectively (Fig. 2D). The ICI analysis revealed enrichment of E. coprostanoligenes in the IgA+ fraction from both CD and CD-SpA; however, similar to the analysis of the relative abundance data, only Escherichia / Shigella ICI score revealed differential enrichment in the IgA+ fraction of CD-SpA. Pseudomonas (CD-predominant), E. hallii (CD-predominant), Lachnospiraceae UCG008 (CD and CD-SpA), and C. sensu stricto (CD-SpA-predominant) were enriched in the IgA- fraction. Escherichia / Shigella ICI linearly correlated with joint disease activity measured by BASDAI for patients with CD-SpA, but did not correlate for patients with CD alone (Fig. 2E).
CD-SpA E. coli isolates are AIEC and initiate epithelial immunity
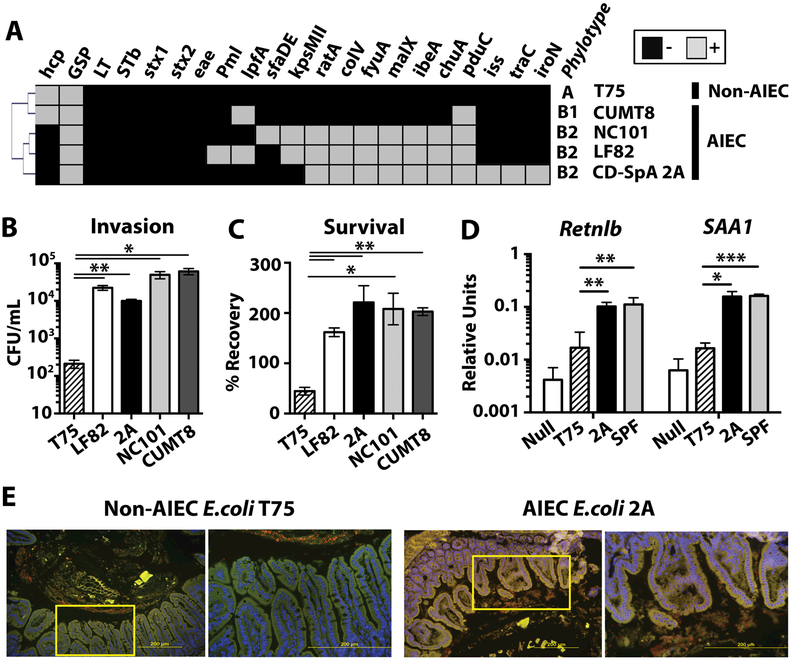
To determine the pathotype of the IgA-coated E. coli in CD-SpA, we generated patient-derived isolates from 3 CD-SpA donors with the highest Escherichia ICI score. RAPD PCR identified 15 unique isolates from these 3 CD-SpA-derived libraries (Table S3). PCR for phylogroup and key microbial virulence-associated genes revealed hierarchical clustering of all of these isolates with phylogroup B2 mouse (NC101) and CD-derived (LF82) AIEC isolates compared to a CD-derived non-AIEC E. coli strain (T75) (30) (Fig. 3A, Table S3). E. coli clones were also derived from the IgA-coated fraction from 3 CD patients with the highest ICI score. Virulence-associated genes were less frequent in CD-derived isolates including pduC, which was present in 11/15 CD-SpA-derived isolates, but only 4/15 CD-derived isolates (Table S3). Functional analysis of all CD-SpA-derived isolates revealed a high level of adherence and invasion of Caco-2 monolayers as well as persistence following macrophage invasion in vitro, consistent with their characterization as AIEC (Fig. 3B, C and Fig. S7). The complete genome of the B2 phylotype, CD-SpA derived E. coli isolate 2A, which is used in subsequent experiments, was sequenced using PacBio and assembled de novo into 3 contigs. The complete circular genome was 4,991,385 bp in length and most closely aligned to prototypical AIEC LF82 (Fig. S8).
Fig 3. CD-SpA-derived Adherent-invasive E. coli (AIEC) initiates epithelial immunity.
A. The presence (gray) or absence (black) of AIEC-associated virulence factors genes was determined by PCR for E. coli isolates T75 (non-AIEC), CUMT8 and NC101 (mouse-derived AIEC), LF82 (human-derived AIEC), and CD-SpA isolate 2A. Hierarchical clustering was performed using average linkage clustering and Pearson correlation distance metric. B. Invasion of Caco-2 cells by E. coli isolates was assessed by gentamicin protection assay. The mean colony forming units (cfu) / ml is presented. C. Survival in J774 macrophages by E. coli isolates was assessed by gentamicin protection assay. The percentage of input cfu recovered following gentamicin protection in J774 macrophages is shown. SEM is shown for each group. p-values *<0.05, **<0.01 are indicated, T-test. All samples were run in triplicate with N=3–5 biological replicates for each group. D. qPCR analysis of Retnlb and SAA1 expression by ileal epithelial cells 5 days after colonization with SPF microbiota, CD-SpA E. coli 2A, non-AIEC T75 or media control. Bar graph represent the geometric mean for 4–8 mice / group from 2 independent experiments. Error bars represent SEM. p-values *<0.05, **<0.01 are indicated, ANOVA. E. FISH of ileal mucosa 5 days after mono-colonization with E. coli isolate T75 or 2A. Nuclei are stained with DAPI (blue) and an E.coli are stained with a 16S rRNA probe (red). Tissue autofluorescence is in green. Yellow box indicates inset displayed in the right panel.
To evaluate the function of a representative CD-SpA-derived E. coli isolate 2A in vivo, we colonized 6–8 weeks old germ-free C57BL/6 mice with E. coli 2A and assessed colonization by qPCR 10 days later (Fig. S9A). Similar to previous studies, AIEC did not induce overt intestinal disease in gnotobiotic C57BL/6 mice despite robust colonization (Fig. S9B); however, real-time PCR of the terminal ileum epithelium revealed induction of resistin-like beta (Retnlb) and serum amyloid A1 (SAA1) (Fig. 3D) consistent with adhesion to the epithelial surface (21). To directly assess attachment to the epithelium in vivo, we performed fluorescence in situ hybridization (FISH) of the terminal ileum 5 days after colonization (Fig. 3E). Although CD-derived non-AIEC E. coli control T75 did not penetrate the mucus layer, CD-SpA E. coli 2A was broadly adherent to the epithelial mucosa.
CD-SpA derived AIEC promotes mucosal Th17 immunity
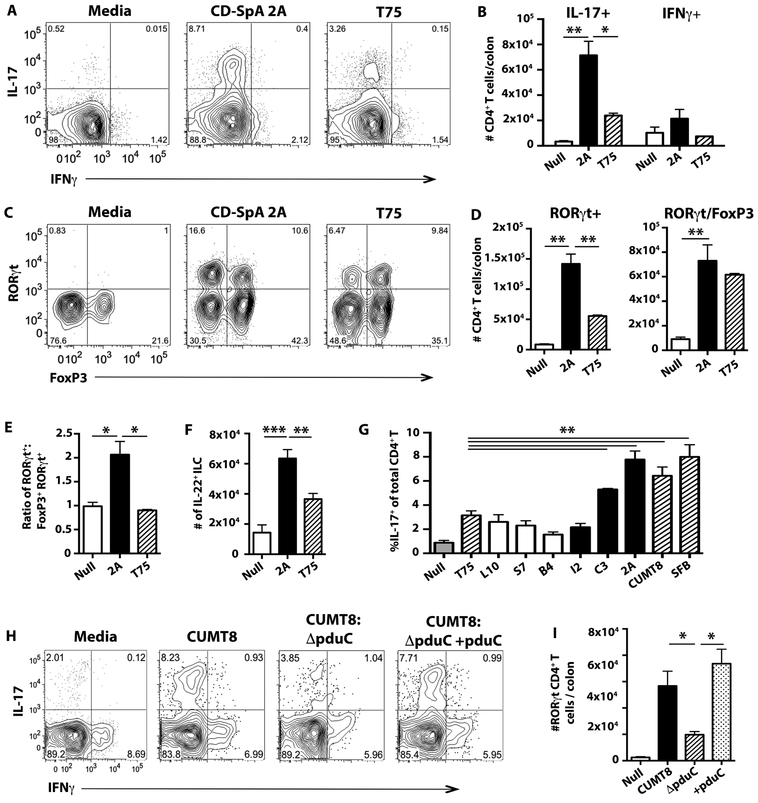
To evaluate the ability of CD-SpA-derived AIEC to promote Th17 mucosal immunity, we colonized 6–8 week old germ-free C57BL/6 mice with E. coli 2A. At 10 days post-colonization, the production of IL-17 was higher in colonic CD4+ T cells from E. coli 2A colonized mice compared to a non-AIEC control T75 (Fig. 4A, B), although both isolates robustly and equivalently colonized the intestine (Fig. S9A). No significant change in IFNγ production was observed. Consistent with the increase in IL-17+ CD4+ T cells, RORγt+ CD4+ T cells were increased in both the small intestinal (Fig. S10A, B) and colonic lamina propria (Fig. 4C) of mice colonized with E. coli 2A. Similar to other intestinal symbionts (40), both CD-SpA E. coli 2A and non-AIEC E. coli T75 were sufficient to induce RORγt/Foxp3+ CD4+ T cells (Fig. 4D); however, E. coli 2A induced significantly more Foxp3- RORγt CD4+ T cells (Fig. 4E). E. coli 2A induced Th17 polarization in the small intestinal lamina propria to a similar level seen following mono-colonization with SFB (Fig. S10A, B). Finally, to evaluate the ability of isolate 2A to induce Th17 immunity under non-gnotobiotic conditions, SFB-negative C57BL/6 weanlings were challenged with isolate 2A or T75. Similar to the mono-colonization experiments, colonization with isolate 2A was sufficient to induce RORγt CD4+ T cells compared to animals colonized with T75 (Fig. S10C). Thus, even in the presence of other commensal microbiota, CD-SpA-derived AIEC isolates induced robust Th17 polarization.
Fig. 4. CD-SpA AIEC promotes mucosal Th17 immunity.
A-F. Germ-free C57BL/6 mice were colonized with 2 × 109 CFU of CD-SpA-derived E. coli isolate 2A or non-adherent E. coli T75 and analyzed after 10 days. Data presented are 1 of 5 total experiments with at least 3 mice per experimental group (A-F). Error bars represent SEM. p-values indicated, ANOVA. A. Live, CD4+ T cells were identified by flow cytometry using a lymphocyte FSC gate and CD4+, TCRβ expression. Intracellular cytokine staining was used to evaluate IL-17 and IFNγ 4 hour stimulation with PMA/ionomycin with Brefeldin-A. B. Total number of IL-17 and IFNγ producing CD4+ T cells per colon for each experimental group. Bar graphs represent geometric mean for 3 mice per group C. Flow cytometry of live, CD4+ T cells was used to evaluate RORγt and Foxp3 expression in colonic lamina propria. D. Total number of RORγt or RORγt/Foxp3 CD4+ T cells per colon is shown in media gavage (null), CD-SpA E. coli 2A, or T75 colonized mice. Bar graph represents geometric mean of 3 mice / group. E. Ratio of RORγt+ cells: Foxp3+RORγt+ CD4+ T cells in the colonic lamina propria of mice colonized with the indicated microbes. Bar graphs represent geometric mean of ratio. F. Total number of live, lineage negative (CD19, CD3, CD11b, TCRγδ), CD127+ innate cells producing IL-22 in the colonic lamina propria. G. Non-colonized (Null, gray); non-AIEC E. coli T75 (hatched); CD-derived isolates L10, S7, B4 (white); CD-SpA derived isolates I2, C3, 2A (black); and mouse SFB or AIEC CUMT8 (hatched) were used to colonize germ-free C57BL/6 mice and analyzed after 10 days. Flow cytometry of live, CD4+ T cells was used to evaluate IL-17 expression in colonic lamina propria cells following 4 hour stimulation with PMA/ionomycin with Brefeldin-A. Bar graphs represent geometric mean of at least 3 mice / group. Error bars represent SEM. p-values indicated, ANOVA. H, I. Germ-free C57BL/6 mice were colonized with 2 × 109 CFU of mouse-derived AIEC isolate CUMT8, CUMT8:ΔpduC, or the complemented strain CUMT8:ΔpduC+pduC and analyzed after 10 days. Flow cytometry of live, CD4+ T cells was used to evaluate IL-17 and IFNγ expression in colonic lamina propria cells following 4 hour stimulation with PMA/ionomycin with Brefeldin-A (I). Total number of RORγt+ CD4+ T cells per colon for each experimental group was evaluated (I). Bar graphs represent geometric mean of at least 3 mice / group from 1 of 3 total experiments. Error bars represent SEM. p-values indicated, ANOVA.
In addition to Th17 cells, group 3 innate lymphoid cells (ILCs) also respond to IL-23 and produce IL-22 (41). To investigate the effect of E. coli isolate 2A on ILCs, colonic, lineage negative, CD127+ innate lymphocytes were also profiled by intracellular cytokine staining following PMA/ionomycin stimulation. E. coli 2A induced significant IL-22 production by ILCs, but no significant change in IFNγ production was observed (Fig. 4F, S11A, B). No differences were observed in the dendritic cell populations, CD8+ T cells, or T follicular helper cells in the Peyer’s patches following colonization with CD-SpA isolate 2A or T75 (Fig. S12).
To evaluate the ability of additional CD-SpA and CD derived E. coli isolates to induce Th17 polarization, CD-SpA derived isolates I2, C3, and 2A as well as CD-derived isolates L10, S7, and B4 (Table S3) were used to colonize germ-free C57BL/6 mice. At 10 days post-colonization, both isolate 2A (CD-SpA) and C3 (CD-SpA) induced significantly more Th17 polarization compared to control non-AIEC E. coli T75 (Fig. 4G). No significant induction of Th17 was observed following colonization with CD-derived isolates or CD-SpA isolate I2. Notably, isolate 2A and C3 both contain AIEC virulence-associated genes including iroN, ratA, and pduC (Table S3) not present in isolate I2, suggesting that genetic heterogeneity impacts the effect on mucosal immunity.
Recent reports suggest that mucosal adherence is a critical feature in microbial induction of Th17 cells (42, 43). Genetic deletion of the Intimin gene (eae), essential for epithelial attachment of Citrobacter rodentium and Enterohemorrhagic E. coli O157, resulted in decreased induction of Th17 (42). Although eae is not present in AIEC, pduC, which encodes the large subunit of propanediol dehydratase, enables AIEC to forage within the mucus layer using fucose-derived propanediol as an alternate carbon source and provides a competitive metabolic advantage for epithelial adherence (33, 44). To evaluate the role for pduC in AIEC induction of intestinal Th17 cells, we generated pduC deficient (ΔpduC) AIEC using the lambda red recombinase system (33) in a mouse-derived AIEC CUMT8 which only contains virulence-associated factors lpf and pduC. Similar to isolates 2A and C3, colonization with CUMT8 induced robust Th17 polarization (Fig. 4H, I). At 10 days following colonization, all strains efficiently and equivalently colonized the intestine (Fig. S13); however, deletion of pduC resulted in a significant decrease in colonic Th17 induction, which was restored with genetic complementation (ΔpduC + pduC) (Fig. 4H, I). Although the presence of pduC alone is not sufficient to afford Th17 cell induction in CD-derived isolates lacking additional virulence factors (Fig. 4G), these results identify pduC as a critical component of AIEC’s capacity to induce Th17 barrier immunity.
IL-10 and IL-23 regulate the effects of CD-SpA AIEC in DSS-induced colitis
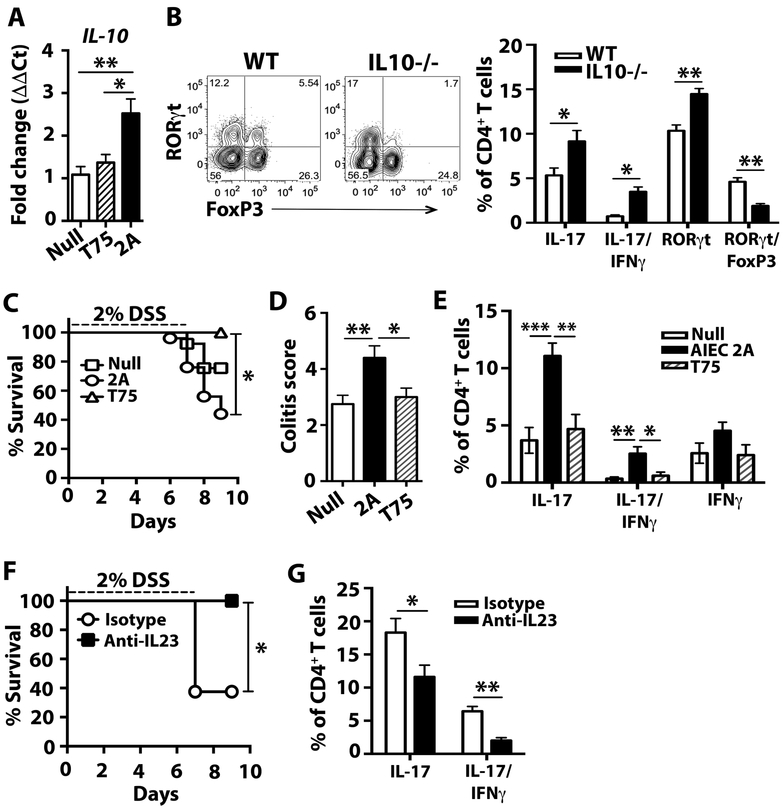
While colonization of WT germ-free mice did not induce intestinal histopathological abnormalities (Fig. S9B), we sought to evaluate the effect of colonization with CD-SpA E. coli 2A during colitis. Recent data suggests that lymphoid tissue resident commensal bacteria, which promote both Th17 and ILC3 immunity, protect host tissue from dextran sodium sulfate (DSS) via IL-10 (45). To test the protective ability of CD-SpA-derived AIEC, we induced colitis with DSS in 6–8 week old gnotobiotic C57BL/6 mice mono-colonized with CD-SpA E. coli 2A or control media. Whereas germ-free mice rapidly and uniformly succumbed to DSS-induced colitis by day 9, E. coli 2A as well as T75 colonization were sufficient to rescue these mice (Fig. S14). qPCR analysis of Il10 expression in lamina propria mononuclear cells from mono-colonized mice revealed a significant increase in gnotobiotic mice colonized with isolate 2A compared to T75 (Fig. 5A). To evaluate the role for this IL-10 induction in regulating the balance of Th17 immunity, we colonized germ-free IL-10-deficient mice with E. coli 2A or control media. In the absence of IL-10, we found a significant reduction in the RORγt/Foxp3+ CD4+ T cells and a more robust induction of Th17 cells and IL-17/IFNγ producing CD4+ T cells (Fig. 5B). Although no spontaneous colitis or histologic effect was seen at the basal state (consistent with the resistance of IL-10-deficient mice on a C57BL/6 background to develop spontaneous colitis (46)), exposure of IL10-deficient mice to DSS following colonization with E. coli 2A resulted in reduced survival compared to those mice gavaged with media or non-AIEC T75 (Fig. 5C). DSS-treated IL-10-deficient mice colonized with E. coli 2A developed more severe colonic inflammation compared to non-colonized controls or mice colonized with non-AIEC E. coli isolate T75 (Fig. 5D). Similar to basal state analysis, colonization with E. coli 2A prior to DSS-induced colitis resulted in increased production of both IL-17 and IL-17/IFNγ+ CD4+ T cells (Fig. 5E). These findings support the notion that CD-SpA E. coli 2A, like other AIEC, is a pathosymbiont that can opportunistically drive intestinal inflammation in a genetically susceptible host.
Fig. 5. CD-SpA E. coli 2A exacerbates DSS-induced colitis in IL10-deficient mice.
A. qPCR analysis of Il10 expression in lamina propria mononuclear cells from mono-colonized mice 5 days after colonization with CD-SpA E. coli 2A, T75, or no colonization control. Bar graphs represent geometric mean for 5–6 mice / group from 2 independent experiments. Error bars represent the SEM. P-values < 0.05 is indicated, T-test. B. Germ-free WT and IL-10−/− mice were colonized with 2 × 109 CFU of AIEC isolate 2A. Two weeks after colonization, flow cytometry of live, CD4+ T cells was used to evaluate RORγt and Foxp3 expression as well as IL-17 and IFNγ expression in colonic lamina propria cells following 4 hour stimulation with PMA/ionomycin with Brefeldin-A. Mean percentages are shown for each experimental group and error bars reflect SEM. Data are a composite of 2 independent experiments with 4–6 animals / group. C-E. Germ-free IL-10−/− mice were colonized with 2 × 109 CFU of CD-SpA-derived AIEC isolate 2A, non-AIEC T75 or media control. Ten days after colonization, mice were exposed to 2% DSS ad libitum for 7 days. C. Percent survival following DSS administration is shown. 2A N = 15, T75 N = 8, Media N = 13,. Log rank test, P<0.05. D. Colonic histopathology score on day 10 following DSS exposure. Mean and SEM are shown. p-values are indicated, ANOVA. E. Flow cytometry was used to evaluate RORγt and Foxp3 expression and IL-17 and IFNγ cytokine production in live, CD4+ T cells from the colonic lamina propria. F,G. IL10−/− mice mono-associated with AIEC isolate 2A were treated with anti-IL23 or isotype control on day 1 and 4 following DSS exposure. F. Percent survival following DSS administration is shown. Isotype N = 8, anti-IL-23 N = 8. Log rank test, P<0.05. G. Flow cytometry was used to evaluate RORγt and Foxp3 expression and IL-17 and IFNγ cytokine production in live, CD4+ T cells from the colonic lamina propria. Mean percentages are shown for each experimental group (N=5) and error bars reflect SEM. Data are from one of three independent experiments. P-values are indicated, ANOVA.
Although DSS-induced colitis is considered a non-T cell dependent colitis, IL-23 contributes to disease severity in a T-cell sufficient animal (47). To assess the contribution of IL-23 dependent colitis in our IL-10-deficient model, mice were treated with anti-IL-23 blocking antibody following induction of colitis with DSS. IL-23 blockade rescued mice from the reduced survival (Fig. 5F) and blockade resulted in a reduction in both IL-17 and IL-17/IFNγ+ CD4+ T cells (Fig. 5G). Collectively, in the genetically susceptible IL10-deficient background, CD-SpA-derived AIEC promotes IL-23-dependent inflammatory colitis.
Intestinal colonization with CD-SpA derived AIEC induces systemic Th17 immunity and promotes joint inflammation
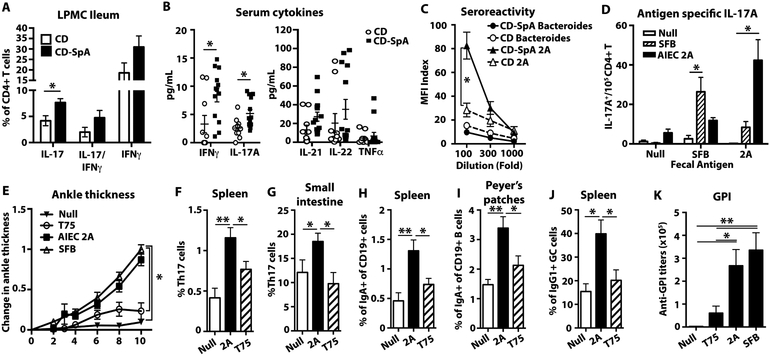
An increase in Th17 cells has been shown in CD patients with active ileal disease (48, 49). To evaluate mucosal Th17 cells in patients with CD-SpA, we phenotyped CD4+ T cells in ileal biopsies from patients with CD (N=8) or CD-SpA (N=9) undergoing endoscopic evaluation with active ileitis. Analysis of cytokine expression revealed a significant increase in IL-17 production from patients with CD-SpA compared to CD alone (Fig. 6A). No differences were seen in the expression of Foxp3 or IFNγ levels. To evaluate systemic immunity in our patient cohort, we measured inflammatory serum cytokines levels. Targeted multiplex analysis of inflammatory cytokines revealed a significant increase in IL-17A and IFNγ, but no difference in TNFα (Fig. 6B). These mucosal and systemic data reveal a Th17 immune phenotype in our CD-SpA cohort.
Fig. 6. CD-SpA E. coli 2A colonization promotes arthritis in K/BxN mice and models the systemic Th17 immunophenotype seen in CD-SpA patients.
A. CD4+ T cells from ileal biopsies of CD patients with (N=8) and without SpA (N=11) were analyzed for IL-17 or IFNγ production following PMA/ionomycin stimulation for 4 hours in the presence of Brefeldin A. Bar graph represents geometric mean. Error bars represent SEM. * p<0.05 are indicated, T-test. B. Serum cytokines was quantified by bead array (Biolegend). *P<0.05 are indicated, two-tailed t-test with Bonferroni correction. CD N=10, CD-SpA N=13. C. E. coli 2A (triangle) or B. vulgatus (circle) specific IgG titers determined by bacterial surface staining with serum from CD patients with (black, N=10) or without (white, N=10) SpA. MFI Index (MFI IgG/ MFI isotype) is shown for indicated serum dilution. Error bars represent SEM. P-values are indicated, ANOVA. D. CD4+ T cells were purified from the spleen of gnotobiotic mice colonized with or without AIEC 2A or SFB and re-stimulated with the indicated fecal antigen. Antigen specific IL-17 production was measured by ELISPOT. Data are a composite of 2 independent experiments with 3–6 mice / group performed in triplicate. Bars indicate geometric mean and error bars reflect SEM. P-values <0.05 are indicated, ANOVA. E-K. Antibiotic pre-treated SFB-free K/BxN mice were gavaged with media control (N=5), AIEC 2A (N=13), T75 (N=11), or SFB (N=9) and analyzed on day 10. E. Ankle thickening was measured daily following gavage. P-value <0.05 is indicated, ANOVA. F,G. Percentage of live, CD4+ T cells that produce IL-17 (Th17 cells) in both the spleen (F) and small intestinal lamina propria (G). H,I. Percentage of IgA+ cells within the total, live CD19+ B cell gate of the spleen (H) or Peyer’s patches (I). J. Percentage of IgG1+ cells within the total, live CD19+ germinal center (GC) B cells (Fas+ PNA+) in the spleen. K. Anti-GPI titer at day 10 following gavage. Bars indicate geometric mean and error bars reflect SEM. P-values are indicated, ANOVA. Data from E-K are a composite of three independent experiments.
To evaluate the systemic recognition of luminal AIEC in patients with CD-SpA, we determined IgG sero-reactivity to E. coli antigens. As previously described (50), the binding of total serum IgG from patients with or without SpA to E. coli was evaluated by flow cytometry. Patients with CD-SpA showed increased, titratable IgG sero-reactivity to E. coli antigens, but no increased reactivity against Bacteroides, in comparison to matched CD controls (Fig. 6C). This specific systemic sero-reactivity to E. coli antigens supports the potential role for AIEC in breaking the compartmentalization of the intestinal immune response to luminal microbes and promoting systemic Th17 immune phenotype in CD-SpA.
Given the systemic Th17 activation observed in CD-SpA, we sought to evaluate the ability of CD-SpA isolate 2A to support systemic Th17 immunity in vivo. To test if colonization with E. coli 2A induces systemic immunity, we evaluated the induction of antigen specific splenic CD4+ T cell response by ELISPOT following mono-colonization in C57BL/6 mice. Similar to SFB, colonization with E. coli 2A primed a systemic, antigen-specific Th17 response (Fig. 6D). To evaluate if colonization with E. coli 2A was sufficient to induce inflammatory arthritis, we employed a mouse model of arthritis (K/BxN) (20). Previous reports have shown the dependence of this model on Th17 cell induction and demonstrated the sufficiency of Th17-inducing microbiota to support disease. To test the sufficiency of E. coli 2A in promoting inflammatory arthritis, SPF K/BxN mice were colonized with E. coli 2A and ankle thickness was measured after colonization. Within 6 days, significant differences in ankle thickness were apparent and increased up to 10 days following colonization (Fig. 6E). Similar to colonization in gnotobiotic mice, we noted significant Th17 cell expansion in both the spleen and small intestinal lamina propria compared to controls (Fig. 6F, G). Colonization with CD-SpA E. coli 2A led to an increase in IgA+ B cells in both the spleen and Peyer’s patches (Fig. 6H, I), IgG1+ germinal center B cells in the spleen (Fig. 6J), and anti-GPI titers (Fig. 6K) compared to T75 control. Collectively, these data support the ability of IgA-coated AIEC that are enriched in patients with CD-SpA to support systemic Th17 inflammatory disease.
Discussion
The immunologic relevance of compositional changes in the microbiome to disease phenotype remains a major question in the study of human disease. Initial studies introducing new approaches to identify immunogenic components of the microbiome, including IgA-Seq (19) and Bug-FACS (23), identified intestinal pathosymbionts within a microbial community that promote disease under specific genetic or environmental circumstances. Our identification here of IgA-coated E. coli as a pathosymbiont in CD-SpA illustrates that these technologies can be applied more broadly to help stratify complex clinical phenotypes, particularly those in which antigenic stimuli from the intestine are thought to be a key component. While previous studies in HLA-B27 transgenic rats and mouse models revealed a role for luminal microbes in supporting systemic inflammatory arthritis, our results now identify AIEC as a pathosymbiont in CD-associated SpA.
Although our analysis identified the enriched IgA-coated E. coli in CD-SpA as an AIEC pathotype, AIEC are present in the microbiome of healthy controls as well as Crohn’s disease patients without spondyloarthritis. Our experimental models highlight two features of the host-pathogen interaction that must be considered to understand the specificity of pathogenetic mechanisms of AIEC in SpA, namely host susceptibility and strain variability.
First, although microbial activation of the IL-23 pathway plays a critical role in maintaining homeostasis and promoting mucosal healing, Th17 cell activation may act as a double-edged sword by promoting the inflammatory response in cases of environmental or host genetic susceptibility to inflammatory disease (47). Similar to SFB protection from Citrobacter rodentium-induced colitis (21) or lymphoid tissue resident commensal bacteria protection from DSS-induced colitis (45), CD-SpA-derived AIEC protect against acute injury and death from DSS-induced colitis in wild-type mice. Resident microbiota, including AIEC, induce colonic RORγt/Foxp3+ CD4+ T cells, which play an important role in restraining inflammatory colitis (40). Consistent with the protective effect illustrated by our data, a higher Enterobacteriaceae IgA-coating index in 6-month old infants correlated with better nutritional status (23). Thus, in situations of nutritional sufficiency or immune competence, the response to Enterobacteriaceae may have co-evolved to protect the host; however, persistent nutritional (23) or genetic susceptibility (modeled in IL10-deficient and K/BxN mice) evokes maladaptive responses, which, in turn, promote more severe inflammatory Th17 disease. Likewise, our data link the shared genetic susceptibility in the IL23R locus in both CD and SpA (9) with increased systemic E. coli sero-reactivity and Th17 inflammatory cytokines. With the recent approval of IL-23 blockade therapy for Crohn’s disease, the clinical phenotype of peripheral SpA may help stratify an immune phenotype in a susceptible host that may benefit from targeted anti-IL-23 therapy.
Second, unique strain-specific features of epithelial cell attachment are critical in dictating the immune outcome. Epithelial cell attachment is a critical feature of the mouse commensal SFB and is required to induce Th17 immunity (42). A recent report, which identified the ability of the human commensal Bifidobacter adolescentis to induce Th17 cells, similarly requires attachment to the intestinal epithelium despite triggering distinct epithelial transcriptional changes (51). In human fecal samples, PCR detection of virulence genes eae and aggR required for adherence of enteropathogenic and enteroaggregative E. coli, respectively, identified individuals with a higher Enterobacteriaceae IgA-coating index and impaired nutritional status (23). eae-dependent attachment to the epithelial cell barrier is a critical determinant of Th17 induction by Citrobacter rodentium and Enterohemorrhagic E. coli O157 (42). CD-SpA associated E.coli are distinct from Enteropathogenic E.coli and C. rodentium in that they lack type III secretion system effectors (including eae) and aggR associated with diarrheagenic Enterobacteriaceae; however, the majority of CD-SpA E. coli isolates expressed the AIEC virulence-associated gene pduC. The metabolic advantage provided by propanediol dehydratase promotes epithelial cell adhesion seen in both clinical and environmental AIEC isolates containing pduC (33, 44). This requirement of pduC for AIEC induction of Th17 cells in the lamina propria offers a potential targetable link of microbial metabolic capacity to Th17-dependent mucosal immunity.
Despite the role of pathosymbionts such as AIEC as keystone species, dynamic interactions with other commensals may modulate the immune pathology. In mouse models of gut barrier disruption, diverse microbial community members including Bacteroidetes were required for the pathogenic effect of Enterobacteriaceae isolated from nutritionally deficient individuals, while Akkermansia is protective (23). The outcome of these microbial interactions may depend on environmental context. While reduced levels of A. muciniphila in both IBD and metabolic disease (52, 53) support an anti-inflammatory role for this mucin-degrading microbe, recent data revealed a positive correlation of A. muciniphila IgA-coating index with arthritis severity in the HLA-B27 rat model (22). The interaction of AIEC with other intestinal pathosymbionts may offer a more complete understanding of its impact on mucosal immunity and inflammation.
Our study has important limitations. First, our immune phenotype and microbiome results reflect findings in an HLA-B27 negative peripheral IBD-SpA cohort. Despite increased IL-23 in intestinal tissue, previous reports of axial SpA did not show an increase in ileal IL-17 (54). Moreover, HLA-B27 has been reported to alter the intestinal microbiome (55) and further studies are needed to evaluate the microbiome in HLA-B27+ axial and peripheral IBD-SpA. Second, no compositional changes were noted between UC and UC-SpA patients suggesting that other environmental or genetic factors may account for peripheral SpA in UC. Third, strain variability even within AIEC isolates can be diverse. While our analysis primarily reflects colonization with CD-SpA isolate 2A and shows dependence on pduC, additional isolates and genetic elements may also contribute to systemic Th17 inflammatory disease. Finally, our study does not address the immune response within the inflamed joint. Although previous studies have highlighted a potential role for molecular mimicry in microbial induction of spondyloarthritis (56), indirect support of alternative IL-23 dependent immune effector cells (57) still need to be characterized in IBD-SpA.
These results highlight the functional implication of IgA-coated E.coli enriched in CD-SpA and identify a Th17 immune phenotype characteristic of this extra-intestinal manifestation. This mechanistic link between intestinal microbiota and systemic inflammation may underlie the clinical efficacy of sulfasalazine in peripheral joint symptoms (58). Anti-TNFα therapy improves axial symptoms in patients with active CD (59), but our data highlight the activation of the IL-23/IL-17 pathway in CD patients with peripheral symptoms. Although blockade of IL-17A yielded disappointing results in the overall treatment of CD, the lack of clinical response correlated with the absence of polymorphisms in genes associated with Th17 immunity (60). With the recent approval of anti-IL23 therapy for CD, our data now offer a method of clinical, microbial, and immune phenotyping of CD-SpA that can be used to guide precision medical and biologic therapy.
Materials and Methods
Study Design.
A prospective cohort of IBD patients was evaluated for spondyloarthritis (SpA) at the Jill Roberts Center for IBD under IRB-approved protocol (1103011578) at Weill-Cornell Medical College. The overall objective of this observational study was to determine differences in microbial composition in IBD patients with SpA compared to IBD patients without SpA. Based on preliminary and published data, 25 subjects / group were required to detect a 10% difference relative abundance of Proteobacteria. Subjects between the ages of 18 – 80 with biopsy-proven active ileal or ileo-colonic (Crohn’s disease (CD) (HBI >4) or ulcerative colitis (UC) (modified UC-DAI >2) with or without clinical evidence of peripheral SpA assessed by a rheumatologist (according to the Assessment of Spondyloarthritis international Society (ASAS) guidelines as dactylitis, enthesitis, or arthritis) (61) were recruited. All subjects were free of other rheumatic disease, HLA-B27 negative, off antibiotics (including sulfasalazine) for at least 8 weeks at enrollment, and had an intact colon and ileocecal valve. All patients completed the Bath Ankylosing Spondylitis (AS) disease activity index (BASDAI)—a clinically validated exam for disease activity in AS (37)—and a 44-point joint assessment. Fresh fecal samples and serum were obtained and stored at −80°C until further analyses. For patients undergoing endoscopic evaluation, cells isolated from mucosal biopsies were obtained and stored at −160°C until further analyses. Sequencing and immune cell analyses were performed blinded with respect to patient phenotype.
BugFACS.
As previously described (19), 100mg of fecal contents were homogenized in PBS on ice. Clarified supernatants were washed in PBS-1% BSA and incubated with blocking buffer (20% normal mouse serum in PBS) for 20 minutes. Samples were subsequently stained with mouse anti-human IgA (IS11–8E10) or isotype (Mouse IgG1) and sorted on a FACS Aria (BD Biosciences). Samples were co-stained for nucleic acids with SYTO BC (Invitrogen).
Animal Models
Mice.
Germ-free C57BL/6 wild-type (WT) and IL10−/− mice were bred and maintained at Weill Cornell Medical College. K/BxN mice were generated by crossing KRN TCR transgenic mice on the B6 background with NOD mice and maintained at the University of Arizona. All treatments were in accordance with WCMC and the University of Arizona Institutional Animal Care and Use Committee guidelines.
Colonization of Germ-free Mice.
For mono-colonization experiments, 6–8 week old germ-free mice were gavaged with 2 × 109 CFU log-phase bacteria grown under anaerobic conditions in PYG liquid media (Anaerobe Systems, CA). 16S rRNA qPCR in fecal and cecal content-derived DNA was used to confirm colonization. For K/BxN colonization experiments, three-week-old SPF K/BxN mice were treated with ampicillin (1g/L), neomycin (1g/L), and metronidazole (1g/L) for 10 days. After a one-day washout, the mice were then gavaged with media or 2 × 109 CFU of AIEC 2A daily for three days. Ankle thickness was measured daily after bacterial gavage and tissues were collected 10 days after bacterial challenge for flow cytometry analysis.
Statistics.
Analysis and visualization of microbiome communities was conducted in R, utilizing the phyloseq package (62) to import sample data and calculate α- and β-diversity metrics. For microbiome analysis, significance of relative abundance was calculated using the non-parametric Mann-Whitney test. Principal coordinate plots employed the Monte Carlo permutation test to estimate p-values. All p-values were adjusted for multiple comparisons with the FDR algorithm. Correlation between two continuous variables was determined with linear regression models, where p-values indicate the probability that the slope of the regression line is zero. Significance of categorical variables such as gene expression, cell populations, or histology, were calculated by t-test for comparison of two groups and two-way ANOVA for comparison of more than two groups. The significance of survival curves was determined by log rank test.
Supplementary Material
Fig. S1. Alpha and beta diversity metrics of 16S rRNA compositional data.
Fig S2. Alpha and beta diversity metrics stratified by immunomodulator exposure.
Fig S3. 16S rRNA IgA-seq sample rarefaction.
Fig S4. Relative abundance by phylum, family and genus in the pre-sort fraction.
Fig S5. Alpha and beta diversity of 16S rRNA IgA-seq data.
Fig S6. Relative abundance by phylum and family in the sorted IgA+ and IgA- fractions.
Fig. S7. CD-SpA-derived, IgA-coated E. coli isolates are AIEC.
Fig. S8. Circular genome map of CD-SpA-derived E. coli isolate 2A.
Fig. S9. Colonization of germ-free C57BL/6 mice with CD-SpA E. coli 2A.
Fig. S10. CD-SpA E. coli 2A induces Th17 mucosal immunity.
Fig. S11. CD-SpA E. coli 2A induces ILC3 mucosal immunity.
Fig. S12. Characterization of dendritic cells, CD8+ T cells and Tfh.
Fig. S13. Assessment of colonization.
Fig. S14. CD-SpA AIEC 2A protects WT mice against DSS.
Table S1. Demographics, clinical parameters and disease activity indexes of study subjects.
Table S2. Montreal classification of CD cohort.
Table S3. AIEC marker genes in reference strains and clonal isolates from CD and CD-SpA patients.
Acknowledgments:
We thank David Artis, Dan Littman, and Steven Lipkin for helpful comments and discussion, Rielmer Pinedor for technical assistance with gnotobiotic husbandry, and Gregory Putzel for bioinformatics help.
Funding:
This work was supported by grants from the NIH (K08 DK099381 (R.S.L.), R56AI107117 (HJ.W.), and RO1AI107117 (HJ.W.)), Crohn’s and Colitis Foundation of America #323814 (B.D., K.W.S.), NSF Research Fellowship 1144247 (A.S.), the Southwest Clinic and Research Institute Fund (HJ.W.), the Center for Advanced Digestive Care / Roberts Institute for IBD Research (R.S.L.), the Charina Foundation (R.S.L.), and NY Crohn’s Disease Foundation (R.S.L.).
Footnotes
Competing interests: None.
Data and materials availability: The sequencing data used for this study have been deposited at NCBI BioProject ID PRJNA349809.
Supplementary Materials
Supplementary Materials and Methods
For all experiments where n < 20, please provide individual data points in tabular format. This can be accomplished with a separate Excel file, organized with tabs for each figure. (see Checklist)
If necessary, please superscript anything in the figures that will be superscripted in the final copy-edited text (e.g., IgA+, IL-10−/−, etc)
References and Notes:
- 1.DeFilippis EM, Longman R, Harbus M, Dannenberg K, Scherl EJ, Crohn’s Disease: Evolution, Epigenetics, and the Emerging Role of Microbiome-Targeted Therapies. Curr Gastroenterol Rep 18, 13 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Longman RS, E., in Medical Therapy of Ulcerative Colitis, Lichtenstein G, Ed. (Springer Science, New York, 2015). [Google Scholar]
- 3.Peluso R, Di Minno MND, Iervolino S, Manguso F, Tramontano G, Ambrosino P, Esposito C, Scalera A, Castiglione F, Scarpa R, Enteropathic Spondyloarthritis: From Diagnosis to Treatment. Clin Dev Immunol, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bargen JA, Complications and sequelae of chronic ulcerative colitis. Ann Intern Med 3, 335–352 (1929). [Google Scholar]
- 5.Mielants H, Veys EM, Devos M, Stefan CC, Goemaere S, Declercq L, Schatteman L, Elewaut D, The Evolution of Spondyloarthropathies in Relation to Gut Histology .1. Clinical Aspects. Journal of Rheumatology 22, 2266–2272 (1995). [PubMed] [Google Scholar]
- 6.Van Praet L, Jans L, Carron P, Jacques P, Glorieus E, Colman R, Cypers H, Mielants H, De Vos M, Cuvelier C, Van den Bosch F, Elewaut D, Degree of bone marrow oedema in sacroiliac joints of patients with axial spondyloarthritis is linked to gut inflammation and male sex: results from the GIANT cohort. Ann Rheum Dis 73, 1186–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Van Praet L, Van den Bosch FE, Jacques P, Carron P, Jans L, Colman R, Glorieus E, Peeters H, Mielants H, De Vos M, Cuvelier C, Elewaut D, Microscopic gut inflammation in axial spondyloarthritis: a multiparametric predictive model. Ann Rheum Dis 72, 414–417 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH, A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wellcome C Trust Case Control, C. Australo-Anglo-American Spondylitis, Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, Barrett JC, Davison D, Easton D, Evans DM, Leung HT, Marchini JL, Morris AP, Spencer CC, Tobin MD, Attwood AP, Boorman JP, Cant B, Everson U, Hussey JM, Jolley JD, Knight AS, Koch K, Meech E, Nutland S, Prowse CV, Stevens HE, Taylor NC, Walters GR, Walker NM, Watkins NA, Winzer T, Jones RW, McArdle WL, Ring SM, Strachan DP, Pembrey M, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskivina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Farmer A, Williamson R, McGuffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop TD, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Braund PS, Dixon RJ, Mangino M, Stevens S, Thompson JR, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Forbes A, Lewis CM, Onnie CM, Prescott NJ, Sanderson J, Matthew CG, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, Webster J, Brown MJ, Lathrop MG, Connell J, Dominiczak A, Marcano CA, Burke B, Dobson R, Gungadoo J, Lee KL, Munroe PB, Newhouse SJ, Onipinla A, Wallace C, Xue M, Caulfield M, Farrall M, Barton A, R. A. G. Biologics in, C. Genomics Study Syndicate Steering, Bruce IN, Donovan H, Eyre S, Gilbert PD, Hilder SL, Hinks AM, John SL, Potter C, Silman AJ, Symmons DP, Thomson W, Worthington J, Dunger DB, Widmer B, Frayling TM, Freathy RM, Lango H, Perry JR, Shields BM, Weedon MN, Hattersley AT, Hitman GA, Walker M, Elliott KS, Groves CJ, Lindgren CM, Rayner NW, Timpson NJ, Zeggini E, Newport M, Sirugo G, Lyons E, Vannberg F, Hill AV, Bradbury LA, Farrar C, Pointon JJ, Wordsworth P, Brown MA, Franklyn JA, Heward JM, Simmonds MJ, Gough SC, Seal S, C. Breast Cancer Susceptibility, Stratton MR, Rahman N, Ban M, Goris A, Sawcer SJ, Compston A, Conway D, Jallow M, Newport M, Sirugo G, Rockett KA, S. J. Bumpstead, Chaney A, Downes K, Ghori MJ, Gwilliam R, Hunt SE, Inouye M, Keniry A, King E, McGinnis R, Potter S, Ravindrarajah R, Whittaker P, Widden C, Withers D, Cardin NJ, Davison D, Ferreira T, Pereira-Gale J, Hallgrimsdo’ttir IB, Howie BN, Z. Su, Teo YY, Vukcevic D, Bentley D, Brown MA, Compston A, Farrall M, Hall AS, Hattersley AT, Hill AV, Parkes M, Pembrey M, Stratton MR, Mitchell SL, Newby PR, Brand OJ, Carr-Smith J, Pearce SH, McGinnis R, Keniry A, Deloukas P, Reveille JD, Zhou X, Sims AM, Dowling A, Taylor J, Doan T, Davis JC, Savage L, Ward MM, Learch TL, Weisman MH, Brown M, Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 39, 1329–1337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ, The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taurog JD, Hammer RE, Experimental spondyloarthropathy in HLA-B27 transgenic rats. Clinical rheumatology 15 Suppl 1, 22–27 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE Jr., Balish E, Taurog JD, Hammer RE, Wilson KH, Sartor RB, Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest 98, 945–953 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowling P, Ebringer R, Cawdell D, Ishii M, Ebringer A, C-reactive protein, ESR, and klebsiella in ankylosing spondylitis. Ann Rheum Dis 39, 45–49 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowling P, Ebringer R, Ebringer A, Association of inflammation with raised serum IgA in ankylosing spondylitis. Ann Rheum Dis 39, 545–549 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, Marshall M, Kenna TJ, Triolo G, Brown MA, Intestinal dysbiosis in ankylosing spondylitis. Arthritis & rheumatology, (2014). [DOI] [PubMed] [Google Scholar]
- 16.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR, Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2, e01202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J, The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, Manasson J, Pamer EG, Littman DR, Abramson SB, Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis & rheumatology 67, 128–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA, Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D, Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR, Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asquith M, Stauffer P, Davin S, Mitchell C, Lin MP, Rosenbaum JT, Perturbed mucosal immunity and dysbiosis accompany clinical disease in a rat model of spondyloarthritis. Arthritis & rheumatology, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, Ashorn P, Dewey KG, Houpt ER, Hsieh CS, Gordon JI, Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7, 276ra224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Waaij LA, Kroese FG, Visser A, Nelis GF, Westerveld BD, Jansen PL, Hunter JO, Immunoglobulin coating of faecal bacteria in inflammatory bowel disease. Eur J Gastroenterol Hepatol 16, 669–674 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Longman RS, E. , Biomarkers Stratify Disease Phenotype and Therapeutic Response in IBD. European Gastroenterology and Hepatology Reviews 7, 224–228 (2012). [Google Scholar]
- 26.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL, IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 9, 219–230 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, Tysk C, Jansson JK, Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis 15, 653–660 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR, Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104, 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolhion N, Darfeuille-Michaud A, Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis 13, 1277–1283 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW, Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. Isme J 1, 403–418 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang ZD, Dupont HL, Garneau P, Harel J, Rishniw M, Simpson KW, Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm Bowel Dis, (2012). [DOI] [PubMed] [Google Scholar]
- 32.Miquel S, Peyretaillade E, Claret L, de Vallee A, Dossat C, Vacherie B, Zineb E, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel JF, Medigue C, Mojica FJM, Peyret P, Bonnet R, Darfeuille-Michaud A, Complete Genome Sequence of Crohn’s Disease-Associated Adherent-Invasive E. coli Strain LF82. PLoS One 5, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, Stewart K, Scherl EJ, Araz Y, Bitar PP, Lefebure T, Chandler B, Schukken YH, Stanhope MJ, Simpson KW, Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis 20, 1919–1932 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Longman RS, Yang Y, Diehl GE, Kim SV, Littman DR, Microbiota: host interactions in mucosal homeostasis and systemic autoimmunity. Cold Spring Harb Symp Quant Biol 78, 193–201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N, The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK, Microbes and Health Sackler Colloquium: Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calin A, Nakache JP, Gueguen A, Zeidler H, Mielants H, Dougados M, Defining disease activity in ankylosing spondylitis: is a combination of variables (Bath Ankylosing Spondylitis Disease Activity Index) an appropriate instrument? Rheumatology 38, 878–882 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Rudwaleit M, van der Heijde D, Landewe R, Akkoc N, Brandt J, Chou CT, Dougados M, Huang F, Gu J, Kirazli Y, Van den Bosch F, Olivieri I, Roussou E, Scarpato S, Sorensen IJ, Valle-Onate R, Weber U, Wei J, Sieper J, The Assessment of SpondyloArthritis International Society classification criteria for peripheral spondyloarthritis and for spondyloarthritis in general. Ann Rheum Dis 70, 25–31 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, Antonopoulos DA, Bendelac A, Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C, MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnenberg GF, Artis D, Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med 21, 698–708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nunez G, Camp JG, Hattori M, Umesaki Y, Honda K, Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, Bonneau R, Littman DR, An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell 163, 381–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vignaroli C, Di Sante L, Magi G, Luna GM, Di Cesare A, Pasquaroli S, Facinelli B, Biavasco F, Adhesion of marine cryptic Escherichia isolates to human intestinal epithelial cells. Isme J 9, 508–515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, Wang K, Ziegler CG, Goc J, Shima T, Umesaki Y, Sartor RB, Sullivan KV, Lawley TD, Kunisawa J, Kiyono H, Sonnenberg GF, Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity 44, 634–646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eun CS, Mishima Y, Wohlgemuth S, Liu B, Bower M, Carroll IM, Sartor RB, Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10−/− mice. Infect Immun 82, 2239–2246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox JH, Kljavin NM, Ota N, Leonard J, Roose-Girma M, Diehl L, Ouyang W, Ghilardi N, Opposing consequences of IL-23 signaling mediated by innate and adaptive cells in chemically induced colitis in mice. Mucosal Immunol 5, 99–109 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Zorzi F, Monteleone I, Sarra M, Calabrese E, Marafini I, Cretella M, Sedda S, Biancone L, Pallone F, Monteleone G, Distinct profiles of effector cytokines mark the different phases of Crohn’s disease. PLoS One 8, e54562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Ueno A, Fort Gasia M, Luider J, Wang T, Hirota C, Jijon HB, Deane M, Tom M, Chan R, Barkema HW, Beck PL, Kaplan GG, Panaccione R, Qian J, Iacucci M, Gui X, Ghosh S, Profiles of Lamina Propria T Helper Cell Subsets Discriminate Between Ulcerative Colitis and Crohn’s Disease. Inflamm Bowel Dis 22, 1779–1792 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder TF, Hardt WD, Bercik P, Verdu EF, McCoy KD, Macpherson AJ, Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, Kasper DL, Benoist C, Mathis D, Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derrien M, Belzer C, de Vos WM, Akkermansia muciniphila and its role in regulating host functions. Microb Pathog, (2016). [DOI] [PubMed] [Google Scholar]
- 53.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song YM, Lee K, Franzosa EA, Morgan XC, Gevers D, Lander ES, Xavier RJ, Birren BW, Ko G, Huttenhower C, Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med 8, 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciccia F, Bombardieri M, Principato A, Giardina A, Tripodo C, Porcasi R, Peralta S, Franco V, Giardina E, Craxi A, Pitzalis C, Triolo G, Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis and rheumatism 60, 955–965 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, Davin S, Pan Y, Cambronne ED, Dorris M, Debelius JW, Lauber CL, Ackermann G, Baeza YV, Gill T, Knight R, Colbert RA, Taurog JD, Van Gelder RN, Rosenbaum JT, HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One 9, e105684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rashid T, Ebringer A, Autoimmunity in Rheumatic Diseases Is Induced by Microbial Infections via Crossreactivity or Molecular Mimicry. Autoimmune Dis 2012, 539282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A, Cannizzaro A, Sireci G, De Leo G, Alessandro R, Triolo G, Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis 74, 1739–1747 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Clegg DO, Reda DJ, Weisman MH, Blackburn WD, Cush JJ, Cannon GW, Mahowald ML, Schumacher HR Jr., Taylor T, Budiman-Mak E, Cohen MR, Vasey FB, Luggen ME, Mejias E, Silverman SL, Makkena R, Alepa FP, Buxbaum J, Haakenson CM, Ward RH, Manaster BJ, Anderson RJ, Ward JR, Henderson WG, Comparison of sulfasalazine and placebo in the treatment of ankylosing spondylitis. A Department of Veterans Affairs Cooperative Study. Arthritis and rheumatism 39, 2004–2012 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Generini S, Giacomelli R, Fedi R, Fulminis A, Pignone A, Frieri G, Del Rosso A, Viscido A, Galletti B, Fazzi M, Tonelli F, Matucci-Cerinic M, Infliximab in spondyloarthropathy associated with Crohn’s disease: an open study on the efficacy of inducing and maintaining remission of musculoskeletal and gut manifestations. Ann Rheum Dis 63, 1664–1669 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, Wehkamp J, Feagan BG, Yao MD, Karczewski M, Karczewski J, Pezous N, Bek S, Bruin G, Mellgard B, Berger C, Londei M, Bertolino AP, Tougas G, Travis SP, Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 61, 1693–1700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Braun J, van den Berg R, Baraliakos X, Boehm H, Burgos-Vargas R, Collantes-Estevez E, Dagfinrud H, Dijkmans B, Dougados M, Emery P, Geher P, Hammoudeh M, Inman RD, Jongkees M, Khan MA, Kiltz U, Kvien T, Leirisalo-Repo M, Maksymowych WP, Olivieri I, Pavelka K, Sieper J, Stanislawska-Biernat E, Wendling D, Ozgocmen S, van Drogen C, van Royen B, van der Heijde D, 2010 update of the ASAS/EULAR recommendations for the management of ankylosing spondylitis. Ann Rheum Dis 70, 896–904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMurdie PJ, Holmes S, phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Human Microbiome Project C, Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J, Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O, The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delcher AL, Bratke KA, Powers EC, Salzberg SL, Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grin I, Linke D, GCView: the genomic context viewer for protein homology searches. Nucleic Acids Res 39, W353–356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erben U, Loddenkemper C, Doerfel K, Spieckermann S, Haller D, Heimesaat MM, Zeitz M, Siegmund B, Kuhl AA, A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int J Clin Exp Pathol 7, 4557–4576 (2014). [PMC free article] [PubMed] [Google Scholar]
- 69.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR, Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 494, 116–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, Umesaki Y, Wu HJ, Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity 44, 875–888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Alpha and beta diversity metrics of 16S rRNA compositional data.
Fig S2. Alpha and beta diversity metrics stratified by immunomodulator exposure.
Fig S3. 16S rRNA IgA-seq sample rarefaction.
Fig S4. Relative abundance by phylum, family and genus in the pre-sort fraction.
Fig S5. Alpha and beta diversity of 16S rRNA IgA-seq data.
Fig S6. Relative abundance by phylum and family in the sorted IgA+ and IgA- fractions.
Fig. S7. CD-SpA-derived, IgA-coated E. coli isolates are AIEC.
Fig. S8. Circular genome map of CD-SpA-derived E. coli isolate 2A.
Fig. S9. Colonization of germ-free C57BL/6 mice with CD-SpA E. coli 2A.
Fig. S10. CD-SpA E. coli 2A induces Th17 mucosal immunity.
Fig. S11. CD-SpA E. coli 2A induces ILC3 mucosal immunity.
Fig. S12. Characterization of dendritic cells, CD8+ T cells and Tfh.
Fig. S13. Assessment of colonization.
Fig. S14. CD-SpA AIEC 2A protects WT mice against DSS.
Table S1. Demographics, clinical parameters and disease activity indexes of study subjects.
Table S2. Montreal classification of CD cohort.
Table S3. AIEC marker genes in reference strains and clonal isolates from CD and CD-SpA patients.