Abstract
Study Objective
Serotonergic adverse drug events (ADEs) are caused by enhanced intrasynaptic concentrations of 5‐hydroxytryptamine (5‐HT). No systematic process currently exists for evaluating cumulative 5‐HT and off‐target toxicity of serotonergic drugs. The primary study aim was to create a Serotonergic Expanded Bioactivity Matrix (SEBM) by using a molecular bioinformatics, polypharmacologic approach for assessment of the participation of individual 5‐HT drugs in serotonin syndrome (SS) reports.
Data Sources
Publicly available databases including the U.S. Food and Drug Administration (FDA) Adverse Event Reporting System (FAERS), ChEMBL, DrugBank, PubChem, and Kyoto Encyclopedia of Genes and Genomes (KEGG) were queried for computational and pharmacologic data.
Design
An in‐house bioinformatics TargetSearch program ( http://dxulab.org/software) was used to characterize 71 serotonergic drugs interacting at 13 serotonin receptor subtypes and serotonin reuptake transporter protein (SERT). In addition, off‐target interactions at norepinephrine transporter (NET), monoamine oxidase (MAO), and muscarinic receptors were included to define seven polypharmacological drug cohorts. Serotonin syndrome reports for each serotonergic drug were extracted from FAERS by using the Sternbach and Hunter criteria.
Measurements and Main Results
A proportional reporting adverse drug reaction (ADR) ratio (PRR) was calculated from each drug's total ADEs and SS case reports and aggregated by drug bioactivity cohorts. Triple‐receptor interactions had a disproportionately higher number of SS cases using both the Hunter criteria (mean PRR 1.72, 95% CI 1.05–2.39) and Sternbach (mean PRR 1.54, 95% CI 1.29–1.79). 5‐Hydroxytryptamine agonists were associated with a significantly lower proportion of SS cases using the Hunter and Sternbach criteria, respectively (mean PRR 0.49, 95% CI 0.17–0.81 and mean PRR 0.49, 95% CI 0.15–0.83). Drugs with disproportionately higher participation in SS vary considerably between the two diagnostic criteria.
Conclusion
The SEBM model suggests a possible polypharmacological role in SS. Although further research is needed, off‐target receptor activity may help explain differences in severity of toxicity and clinical presentation.
Keywords: adverse drug reactions, drug interaction, FDA, serotonin toxicity, serotonin syndrome, serotonin pharmacology
Serotonin, or 5‐hydroxytryptamine (5‐HT), is biochemically derived from tryptophan and found primarily in the gastrointestinal tract, platelets, and central nervous system (CNS). In the CNS, serotonin modulates attention, memory, behavior, cognition, and thermoregulation among other physiologic functions. In the peripheral nervous system, serotonin is produced primarily by intestinal enterochromaffin cells and is involved in regulating gastrointestinal motility, vasoconstriction, uterine contraction, and bronchoconstriction.1 Excessive CNS levels of serotonin produce a spectrum of adverse effects recognized clinically as serotonin syndrome (SS) that include cognitive, autonomic, and somatic effects. Symptoms may range from barely perceptible to fatal consequences.2, 3 At least seven serotonin receptor types and multiple subtypes have been identified. Although the stimulation of postsynaptic 5‐HT1A and 5‐HT2A receptors has been implicated in serotonin toxicity, recent evidence suggests that other receptors may also participate.4, 5
Numerous drugs and drug combinations have been reported to produce SS and may result from any combination of drugs that increases serotonergic neurotransmission. Although concurrently administered serotonergic drugs are believed to be the most common etiology, it may occur after the initiation of a single serotonergic drug or after a dosage increase in highly sensitive individuals. The combination of serotonergic drugs with monoamine oxidase inhibitors is especially dangerous, causing serious adverse outcomes, including death.6, 7, 8
In addition, clinical studies of serotonergic toxicity often mention potential off‐target effects, but no mechanism currently exists for evaluating their potential role. Therefore, a rational method for characterizing the potential off‐target interactions common to many serotonergic drugs may provide a useful foundation for predictive models of adverse drug event (ADE) risk, especially for concurrent serotonergic drug use. This may offer significant potential for reducing patient morbidity and mortality, in addition to decreasing the associated health care costs for their management.
The goals of the current investigation were to: (i) extract and/or calculate receptor interaction propensities of serotonergic drugs at 5‐HT and off‐target receptor sites using large bioactivity databases and molecular informatics techniques to create the Serotonin Expanded Bioactivity Matrix (SEBM), a publicly available repository; (ii) organize 71 United States Food and Drug Administration (FDA)‐approved drugs appearing in published lists of serotonergic agents into pharmacologically similar groups; and (iii) analyze the occurrence of SS cases represented by seven distinct bioactivity groups using the FDA Adverse Event Reporting System (FAERS) database.
Methods
Serotonergic Drugs and Polypharmacologic Activity
A list of 71 serotonergic drugs was compiled from the literature and used for the study.3, 8 The general product identifier (GPI‐8) code of each drug was obtained from findacode.com. The GPI‐8 encodes hierarchical information regarding drug group, class, subclass, and name. Using GPI‐8 allowed an efficient search of the GPI‐FAERS database (see FAERS section later) by identifying drugs of interest regardless of formulations, dosage forms, and strengths.
An in‐house bioinformatics Web service ( http://dxulab.org/software) was developed to mine the publicly available ChEMBL9 pharmacologic database for relevant drug–receptor interactions and functional activity. Each drug's molecular structure was retrieved from DrugBank10 and used as TargetSearch queries to search ChEMBL for known and off‐target interactions with 5‐HT receptors (1A‐F, 2A‐C, 3A, 4, 5A, 6, and 7), serotonin (SERT), and norepinephrine (NET) transporter proteins, monoamine oxidase (MAO) type A and B enzymes, and muscarinic receptors. The widely used three‐dimensional rapid overlay of chemical structures (ROCS) algorithm11 was used in the bioinformatics screening. A 10‐μmol/L activity cutoff was used to ensure a high‐level of confidence in identifying relationships within the human interactome. This computational approach efficiently accounts for the interaction of drugs at these receptors, and has been shown to capture drug off‐target interactions effectively12 and measure drug‐induced anticholinergic toxicity burden.13, 14
To further validate the computational method, ChEMBL, DrugBank, PubChem,15 and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases16 were searched for the confirmation of TargetSearch receptor interactions and provide functional drug information (e.g., agonism, inverse agonism, antagonism, and inhibition). Documented activity at each receptor subtype and computationally derived data were compared with estimate concordance between the two data sets.
Categorization of Serotonergic Drugs—SEBM Model)
Using their functional information and serotonergic activity, drugs were grouped into seven drug cohorts based on similar pharmacologic interactions. These were defined as:
Triple‐receptor drugs interact at SERT, NET, and muscarinic receptors.
Duo receptor drugs are limited to SERT and NET inhibition.
Mixed SERT are drugs whose primary target is SERT but in addition may have various agonist and antagonist interactions at 5‐HT subreceptors.
Most 5‐HT1 agonists are triptan antimigraine drugs and interact primarily at multiple 5‐HT1 receptors but have no identified interactions or other off‐target sites.
MAO inhibitors generally inhibit both A and B isoenzymes in the CNS, and some off‐target activity may occur but is poorly defined.
Second‐generation atypical antipsychotics are a diverse set of compounds interacting at multiple serotonergic receptor subtypes.
Miscellaneous drugs generally consist of several anticonvulsant and/or mood‐stabilizing drugs with ill‐defined mechanisms producing SS (see Discussion).
After eliminating drugs with fewer than five SS case reports, 56 drugs were identified using SDx and 52 by HDx criteria. Drugs were organized according to the seven defined drug cohorts, and the PRR was calculated for each individual drug based on the SS cases identified by both diagnostic criteria.
The SEBM model uses the Tanimoto coefficient to estimate the probability of a molecule fitting into a receptor complex by comparing a candidate drug with one that is known to interact at a specific receptor. It uses a continuous scale from 0 to 2.0 where a high propensity interaction is equal to 2.0 and < 2.0 represents lower probabilities of similarity. No data (ND) denotes an inability to identify any similar molecular entity (of 1.5 million molecular candidates) that possesses a known or inferred interaction at that specific receptor. This enhancement is evidenced by 355 receptor targets identified by the SEBM computational (TargetSearch) method versus known pharmacologic bioactivity data that revealed only 193 targets (Table S1).
SS Reports in FAERS
The FAERS public database for reporting adverse drug reactions (ADRs) is one of the largest repositories of ADR reports in the world, containing information voluntarily submitted by health care professionals, manufacturers, lawyers, and consumers in the United States and other countries.17 The FAERS database has been widely used in many postmarketing pharmacovigilance and drug safety studies.18, 19, 20, 21, 22, 23 An in‐house GPI‐enabled FAERS relational database (GPI‐FAERS, January 2004–June 2015) was used to detect and evaluate safety reports involving drug‐induced SS.
Two widely used serotonin toxicity criteria, Sternbach (SDx)24 and Hunter (HDx),6 were used to determine instances of drug‐induced SS in FAERS. The Sternbach criteria list 10 symptoms: mental status changes, agitation, myoclonus, hyperreflexia, diaphoresis, shivering, tremor, diarrhea, incoordination, and fever. At least 3 of the 10 symptoms are required to annotate an SS case. In contrast, HDx identifies SS by using an algorithm‐like decision tree25 that targets spontaneous clonus as a hallmark sign followed by inducible or ocular clonus or tremor in conjunction with additional symptoms of agitation, diaphoresis, hypertonia with pyrexia, or hyperreflexia. Symptoms listed in SDx and HDx were matched to the Medical Dictionary of Regulatory Activities (MedDRA)‐preferred terms by using the MedDRA online browser.26 The MedDRA terms used to identify SS signs and symptoms are provided in Table S1. The drug GPI‐8 codes and the MedDRA preferred terms matching SDx and HDx criteria were used in combination to query the GPI‐FAERS database. A visual inspection of 30 randomly selected case reports was performed to verify the accuracy of the computer algorithms used for both diagnostic criteria. All reviewed cases met the respective diagnostic criteria.
Statistical Analysis
The proportional reporting ADR ratio (PRR) is a pharmacovigilance metric frequently used within adverse drug reports involving FAERS data.27 The PRR was calculated for each study drug having five or more SS case reports appearing in FAERS using both SDx and HDx criteria. For this study, PRR = a/(a+b)/c/(c+d), where “a” = all SS case reports of a specific serotonergic drug; “b” = all other ADE reports for that drug; “c” = all SS reports of all other serotonergic drugs; and “d” = all other ADE reports for serotonergic drugs. A 95% confidence interval (CI) was calculated for each PRR, and forest plots were constructed for each drug cohort. In addition, forest plots for the top 20 drugs associated with SS were calculated for both diagnostic criteria.
Results
Each of the 71 drugs (Table 1) were compiled into the SEBM format (Table 2) and assigned to a serotonergic cohort based on their relative ability to occupy specific receptor sites. For this preliminary investigation, the focus was on interactions at 5‐HT1A, 5‐HT2A‐C, SERT, NET, MAO enzymes, and muscarinic receptors. Computationally derived data had a 94.8% concordance with published pharmacologic data. Furthermore, 162 additional computationally derived receptor interactions were identified in which no pharmacologic data currently exist (i.e., novel off‐target interaction sites). Based on the high concordance rate with documented bioactivity, all computationally identified receptor interactions were incorporated and used to develop the SEBM.
Table 1.
Serotonergic Drug List Categorized by Similar Bioactive Sites of Action
| Drug Name | Category | Drug Name | Category |
|---|---|---|---|
| Amitriptylinea | Triple receptor | Selegiline | MAO inhibitor |
| Amoxapine | Triple receptor | Tranylcypromine | MAO inhibitor |
| Citaloprama | Triple receptor | Dihydroergotamine | 5‐HT1 agonists |
| Clomipraminea | Triple receptor | Eletriptan | 5‐HT1 agonists |
| Cyclobenzaprinea | Triple receptor | Frovatriptan | 5‐HT1 agonists |
| Cyproheptadine | Triple receptor | Pentazocine | 5‐HT1 agonists |
| Desipraminea | Triple receptor | Rizatriptan | 5‐HT1 agonists |
| Doxepina | Triple receptor | Sumatriptan | 5‐HT1 agonists |
| Fluoxetinea | Triple receptor | Zolmitriptan | 5‐HT1 agonists |
| Imipraminea | Triple receptor | Almotriptan | Mixed SERT |
| Mirtazapine | Triple receptor | Dextromethorphana | Mixed SERT |
| Nortriptyline | Triple receptor | Granisetron | Mixed SERT |
| Meperidinea | Triple receptor | Lorcaserin | Mixed SERT |
| Paroxetinea | Triple receptor | Naratriptan | Mixed SERT |
| Protriptyline | Triple receptor | Trazodone | Mixed SERT |
| Sertraline | Triple receptor | Vilazodone | Mixed SERT |
| Trimipramine | Triple receptor | Buspironea | Miscellaneous |
| Atomoxetinea | Duo receptor | Carbamazepine | Miscellaneous |
| Desvenlafaxinea | Duo receptor | Divalproex | Miscellaneous |
| Duloxetinea | Duo receptor | Lithium | Miscellaneous |
| Escitaloprama | Duo receptor | Ondansetrona | Miscellaneous |
| Fluvoxaminea | Duo receptor | Metoclopramide | Miscellaneous |
| Levomilnacipran | Duo receptor | Valproate | Miscellaneous |
| Maprotiline | Duo receptor | Valproic Acid | Miscellaneous |
| Methadonea | Duo receptor | Aripiprazolea | Atypical antipsychotic |
| Milnacipran | Duo receptor | Asenapine | Atypical antipsychotic |
| Nefazodone | Duo receptor | Brexpiprazole | Atypical antipsychotic |
| Tramadola | Duo receptor | Clozapinea | Atypical antipsychotic |
| Venlafaxinea | Duo receptor | Iloperidone | Atypical antipsychotic |
| Fentanyla | MAO inhibitor | Lurasidone | Atypical antipsychotic |
| Isocarboxazid | MAO inhibitor | Olanzapine | Atypical antipsychotic |
| Linezolid | MAO inhibitor | Paliperidone | Atypical antipsychotic |
| Methylene Blue | MAO inhibitor | Quetiapine | Atypical antipsychotic |
| Phenelzine | MAO inhibitor | Risperidonea | Atypical antipsychotic |
| Rasagiline | MAO inhibitor | Vortioxetine | Atypical antipsychotic |
| Ziprasidone | Atypical antipsychotic |
5‐HT1 = 5‐hydroxytryptamine receptor; MAO = monoamine oxidase; SERT = serotonin reuptake protein.
See Methods for description of pharmacologic drug categories.
Data taken from Flockhart P450 drug interaction table.42 Other listed drugs may also undergo hepatic metabolism but are less well documented.
Table 2.
Selected Serotonin Drugs Aggregated by Receptor Interactions
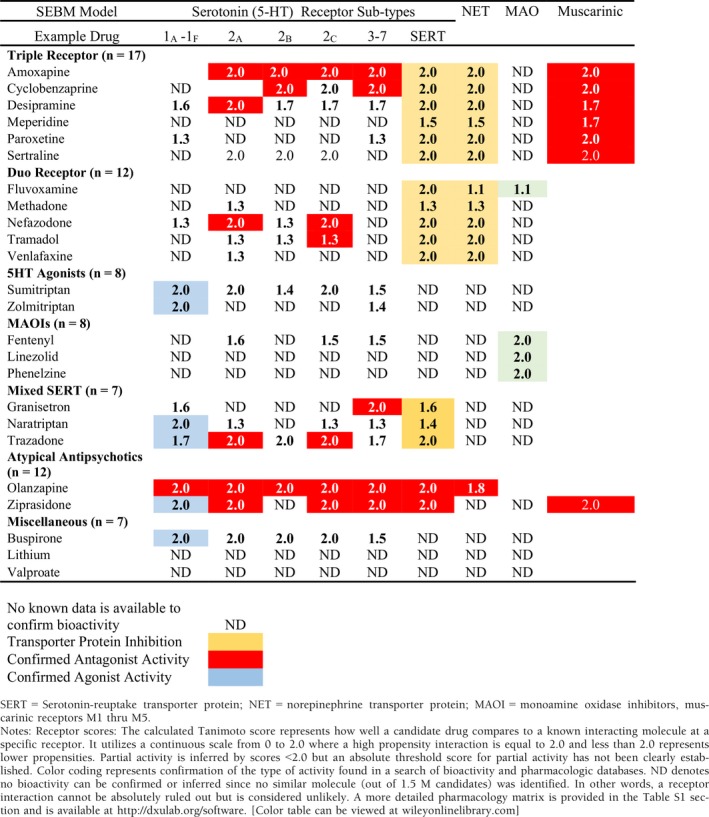
The Sternbach criteria identified 4164 unique SS reports that consisted of 2231 reports involving a single drug and 1933 (46%) reports of multiple drugs (range 2–12 drugs). Similarly, HDx criteria identified 3482 unique reports (45%, range 2–11 drugs). Due to size limitations, a truncated version incorporating several representative drugs is presented in Table 2. A more comprehensive compilation including 18 receptors may be found in Table S1 or at http://dxulab.org/software.
A mean PRR and 95% CI were calculated for each of the drug bioactivity cohorts using both SDx and HDx criteria and presented graphically in Figure 1. The lowest calculated PRR occurred with triptan 5‐HT agonists although only three drugs had more than five cases, making measurement less reliable. Nevertheless, the low prevalence of reports tends to support the clinical observation that severe toxicity is less likely with these drugs.28
Figure 1.
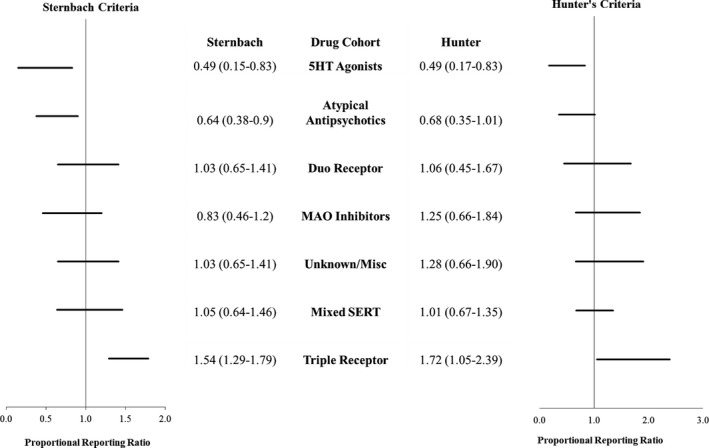
Proportional reporting ADE ratio (PRR) for 7 serotonergic drug cohorts based on the serotonin expanded bioactivity matrix (SEBM) model. Drug cohort mean PRR (95% confidence interval). See Table 1 for drugs included in each cohort. ADE = adverse drug event; 5‐HT = 5‐hydroxytryptamine; MAO = monoamine oxidase inhibitor; PRR = proportional reporting ratio; SERT = serotonin reuptake transporter protein.
Alternatively, drugs with triple‐receptor activity were associated with a significantly higher proportion of cases for both diagnostic criteria (SDx mean PRR = 1.54, 95% CI 1.29–1.79; HDx mean PRR = 1.72, 95% CI 1.05–2.39).
Aggregating all drugs with SERT inhibition regardless of other off‐target interactions, 36 of 52 drugs were represented in 5160 SS reports meeting HDx criteria versus only 2417 for non‐SERT drugs (n = 16). However, PRRs were surprisingly similar given the significance to which SERT inhibition is generally considered a key element in SS cases (i.e., SERT drugs HDx mean PRR = 1.25, 95% CI 0.92–1.57 versus non‐SERT drugs PRR = 1.10, 95% CI 0.62–1.58).
The top 20 serotonergic agents with the highest disproportionate ratio (PRR) are presented in Figure 2. Paroxetine was associated with the most cases (1019 by SDx but only 446 by HDx criteria) and had the highest SDx PRR (2.54, 95% CI 2.47–2.60). Conversely, citalopram was associated with most SS cases identified by HDx criteria (736 vs 601 using SDx criteria) and amoxapine had the highest HDx PRR (4.58, 95% CI 4.02–5.14).
Figure 2.
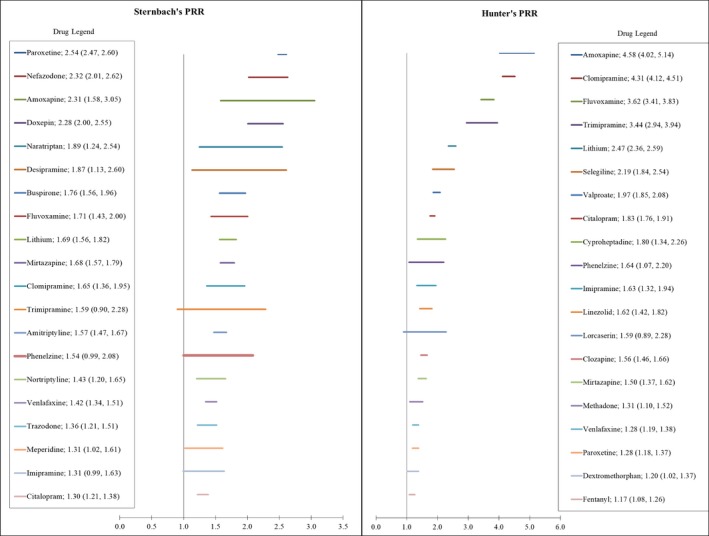
Top 20 drugs associated with serotonin syndrome identified by the Sternbach and Hunter criteria. Mean proportional reporting ratio (PRR) and 95% confidence interval.
Discussion
Serotonin Pharmacology Overview
The pharmacology of serotonin receptors and resulting physiologic responses is exceedingly complex and involves the orchestration, often simultaneously, of both stimulation and blockade at different 5‐HT receptor subtypes. Recent research has led to a deeper understanding and radical departure from the traditional agonist/antagonist pharmacology classifications. Terms such as receptor bias, pluridimensional efficacy, or functional selectivity are used to describe a variety of different drug responses depending on their affinities for differing receptor conformational states. Thus, one can no longer necessarily assume that two different agonists acting at the same receptor will elicit the same response.29 This is an emerging field of pharmacology research, and much remains unknown or is controversial.
Current serotonergic receptor pharmacology, especially that related to antidepressant activity, postulates that in the presence of SERT inhibition, 5‐HT is increased throughout serotonergic synapses, and antagonism at the 5‐HT2A receptor shunts elevated intrasynaptic 5‐HT levels toward the colocalized 5‐HT1A postsynaptic receptor. Thus, 5‐HT2A antagonism is generally considered an important receptor contributing to the antidepressant effects of SERT inhibition.30, 31 Although direct agonist effects at postsynaptic 5‐HT1A,1B,2C,4,6 receptors, antagonism at presynaptic 5‐HT1A/1B, as well as others, 32 may participate in clinical response. Their role in potential toxicity remains poorly defined, and therefore, additional methodologic approaches are needed to elucidate a better understanding.
Well‐established computational techniques were implemented in TargetSearch to categorize drug–receptor interactions as either strong or weak probability of pharmacologic response (Table 2). Although experimental drug‐receptor binding affinities are used in some studies as a basis to quantify relative pharmacodynamic activity, this approach has several shortcomings including (i) binding data collected from different sources may not have the same experimental consistency; (ii) it cannot identify off‐target polypharmacology; and (iii) binding experiments are time consuming and costly. Therefore, binding affinity data are limited to a small set of drugs or specific receptors. In contrast, the TargetSearch bioinformatics approach leverages large bioactivity databases and advanced molecular fingerprinting algorithms to detect and evaluate known interactions and unknown off‐target polypharmacology in an efficient and systematic fashion.
Further study is needed to explore the correlation between propensity threshold and clinical phenotype. Where TargetSearch did not include drug pharmacologic actions (i.e., agonist, antagonist), the TargetSearch receptor interaction data were supplemented with pharmacologic functions annotated in publicly available bioactivity databases such as PubChem, DrugBank, ChEMBL, and KEGG.
SS Diagnostic Criteria
Recent reviews of SS have provided a comprehensive discussion of its clinical manifestations and diagnosis.33, 34, 35 Although a potentially life‐threatening condition, severe cases of SS are generally easily recognized and involve a constellation of symptoms that include some combination of autonomic dysfunction, mental status changes, and/or neuromuscular hypertonicity. Much of our current understanding of severe SS comes from the Hunter Area Toxicology Service (HATS) in Australia. In 2006, their prospective toxicology database included over 2200 selective serotonin‐receptor inhibitor (SSRI) overdose cases. The HATS analysis has helped to establish several clinical caveats including specific SS diagnostic criteria, a dose–response relationship that is associated with increasing intrasynaptic 5‐HT levels in the CNS, and that coadministration of MAO inhibitors with SSRIs tends to produce the most serious cases.2
In addition to HDx6, two other diagnostic schemes24, 36 have been used to aid clinical diagnosis. Recently, one group challenged the superiority of HDx because it was derived solely from SSRI overdoses and called for more focus on potential etiologies.33 To that end, the SEBM uses a well‐established computational and bioinformatics approach that provides clinicians with additional information for consideration in clinical decision‐making.
The number of identified SS cases varies considerably depending on which diagnostic criteria are used. This is highlighted by 1140 additional cases identified by SDx, which supports the argument that it is less specific for serious SS.2, 4, 8, 37 Thus, SDx may detect cases where off‐target neurotransmitter interactions contribute to a broader toxicity presentation in which serotoninergic drugs are participatory. The striking overlap of autonomic and mental status symptoms (e.g., blood pressure instability, tachycardia, tachypnea, tremor, mydriasis, confusion, and agitation), which are also well‐known effects of norepinephrine and anticholinergic drugs, is intriguing. The highly variable clinical presentation of SS may result from a complex neurotransmitter interplay. This is supported by the observation that 67% of SS implicated drugs have identified polypharmacologic off‐target sites. Moreover, those drugs without off‐target activity, in all likelihood, had concurrently administered polypharmacologic agents in many cases.
Comparison of SEBM drug cohorts (Figure 1) shows both criteria identified a higher‐than‐expected association of SS cases in the triple‐receptor drug cohort lending support for a multiple receptor hypothesis. Thus, the additional contribution of antimuscarinic activity may result in more SS cases than is observed with drugs having SERT and NET inhibition alone. Conversely, a trend toward lower associations was observed for triptans and atypical antipsychotics. Radomski hypothesis36 of three toxicity states (i.e., mild, full‐blown SS, and toxic) provides an interesting perspective. The finding that approximately half of FAERS SS reports involve a single drug is surprising and different from the common perception of a multidrug etiology. Further research is needed to validate and better characterize whether off‐target receptor interactions play an important role in the presentation and/or severity of serotonin toxicity.
The important role of SERT inhibition as a major pharmacologic mechanism necessary for SS is evident from the SEBM bioactivity target data. At least 46 drugs have potential interactions at SERT, which serves to highlight the fact that many drugs, not just antidepressants (i.e., SSRIs, serotonin‐norepinephrine inhibitors), have potentially important interactions at SERT. The SEBM provides clinicians with an additional tool for identifying less well‐known SERT inhibitors.
Although the similar PRRs between SERT and non‐SERT cohorts were surprising, most of the non‐SERT cases came from MAO inhibitors, which are known to be more toxic in combination with serotonergic drugs.2 Drug combinations are an especially important consideration in SS cases. Because investigation of every potential serotoninergic drug combination is not feasible, further research using SEBM bioactivity data may provide some insight into drug combinations representing different bioactivity cohorts.
The top 20 drugs within each diagnostic category associated with a disproportionately higher number of SS cases (highest PRRs) are provided in Figure 2. Although interpretation for any specific drug is difficult due to the possible contribution of multiple serotonergic drugs, it does permit an overview of FDA‐approved drugs implicated in SS.
Previous reports have indicated that tricyclic and tetracyclic antidepressants (TCAs), with the exception of imipramine and clomipramine, have little serotonergic toxicity potential.2, 38 However, SDx criteria implicated a higher proportion of SS cases for nearly all TCAs (Figure 2). Conversely, HDx is represented by only four TCAs, suggesting that SDx may identify milder, nonspecific toxicity versus more severe toxicity using HDx.
Atypical Antipsychotic Drugs
Because several atypical antipsychotics are commonly listed as potential contributors to serotonin toxicity, the identification of target receptors is provided in Table S1. The recent characterization of atypical antipsychotic interactions at serotonin receptors in an inverse agonist manner, rather than as antagonists as previously classified, may have important clinical interpretations. Thus, rather than simply blocking 5‐HT actions, inverse agonism at constitutive 5‐HT receptors may result in an opposite action that may serve to further augment or alternatively mitigate 5‐HT toxicity. A growing body of experimental evidence suggests that 5‐HT1A agonism and 5‐HT2A antagonism may contribute to serotonin toxicity.39 Of interest, both chlorpromazine40 and olanzapine41 have been shown to antagonize serotonin toxicity. How this translates to clinical response is unclear and further complicated by the similar presentation between SS and neuromuscular malignant syndrome. Data extraction of FAERS reports using the megaterms presented in this report may not always differentiate between the two.
Other Mechanisms
Metabolism of at least 25 serotonergic drugs42 occurs primarily through cytochrome P450 enzymes (Table 1). Of the top 20 drugs (Figure 2), 50% have known pharmacokinetic interactions in which coadministration of cytochrome P‐450 inhibitors may elevate drug concentrations to toxic levels. These drugs collectively participated in >70% of all FAERS SS reports. Thus, pharmacokinetic drug interactions must be given consideration as an important contributing mechanism.
Limitations
The self‐reporting nature of FAERS case reports is highly subjective, and therefore, potential reporting bias may skew reports toward perceived offenders. In addition, computer queries used to identify SS cases may have missed atypical cases or incorrectly identified some cases (e.g., malignant neuroleptic syndrome).
The relative propensity of a drug to occupy a receptor does not necessarily convey activity, and the bioactivity databases used may contain incomplete and occasionally even contradictory information. Nevertheless, the SEBM model provides a reproducible and biologically consistent rationale for assessing serotonin toxicity. The computational techniques employed allow estimation of receptor interactions where no bioactivity data currently exist. The validation of these predictions will require further research, but the high internal concordance of predicted versus documented receptor interactions is encouraging.
As a very early step in addressing serotonin toxicity, no attempt to address potential drug combinations contributing to SS was made. Thus, individual SS cases may be represented by multiple drugs and contribute to more than one drug category. High prescription volume drugs (e.g., paroxetine) may also overrepresent an individual category, or conversely, newer drugs may underrepresent the participation rate. Although these issues limit definitive conclusions, the analysis characterizes a comprehensive list of commonly implicated drugs and provides a bioactivity foundation from which further research may help elucidate a better understanding of the full spectrum of serotonin toxicity, (i.e., from early, mild symptoms to severe life‐threatening toxicity).
Conclusions
Although several different diagnostic criteria exist, the evaluation of serotonergic toxicity, especially as it relates to differentiating serious from milder degrees of toxicity, remains clinically challenging. Analysis of SS reports in FAERS suggests that many serotonergic drugs have important off‐target receptor interactions that may contribute to the highly variable clinical presentation of serotonin toxicity. Development of clinical tools for predicting toxicity risk resulting from complex multidrug, polypharmacologic regimens is needed. The SEBM model is a rational, systematic, and efficient approach for characterizing potential polypharmacologic activity and may provide a useful platform for investigating ADEs arising from the cumulative target and off‐target drug toxicity.
Supporting information
Table S1. Serotonin Expanded Bioactivity Matrix (SEBM)
Acknowledgments
The authors thank the NIGMS Mountain West Clinical and Translational Infrastructure Network (CTR‐IN), the Idaho INBRE program, the Idaho State University Office of Research, the Idaho State University College of Pharmacy, and the Idaho State University Career Path Intern Program for their research support.
Funding sources: This work is supported, in part, by the National Institutes of Health NIGMS Mountain West Clinical and Translational Infrastructure Network (CTR‐IN) Mini and Pilot Grants (1U54GM104944‐02/15‐746Q‐ISU‐MG5‐00, 5U54GM104944‐03/16‐746Q‐ISU‐PG44‐00) to D.X., a subaward from the Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (Grants P20GM103408 and P20GM109095) to D.X., the Idaho State University Faculty Seed Grant to D.X., the Idaho State University College of Pharmacy Graduate Student Assistantships to S.R., and the Idaho State University Career Path Interns (CPI) funding to G.C.B. None of the funders had any role in study design, data collection, data analysis, interpretation, or writing of the report.
Conflict of interest: The authors have declared no conflict of interests for this article.
Contributor Information
Vaughn L. Culbertson, Email: culbvaug@isu.edu.
Dong Xu, Email: xudong@isu.edu.
References
- 1. Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Annu Rev Med 2009;1:355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gillman PK. A review of serotonin toxicity data: implications for the mechanisms of antidepressant drug action. Biol Psychiatry 2006;11:1046–51. [DOI] [PubMed] [Google Scholar]
- 3. Boyer EW, Shannon M. The serotonin syndrome. N Engl J Med 2005;11:1112–20. [DOI] [PubMed] [Google Scholar]
- 4. Isbister GK, Buckley NA. The pathophysiology of serotonin toxicity in animals and humans: implications for diagnosis and treatment. Clin Neuropharmacol 2005;5:205–14. [DOI] [PubMed] [Google Scholar]
- 5. Nisijima K, Yoshino T, Yui K, Katoh S. Potent serotonin (5‐HT) 2A receptor antagonists completely prevent the development of hyperthermia in an animal model of the 5‐HT syndrome. Brain Res 2001;1:23–31. [DOI] [PubMed] [Google Scholar]
- 6. Dunkley E, Isbister G, Sibbritt D, Dawson A, Whyte I. The Hunter serotonin toxicity criteria: simple and accurate diagnostic decision rules for serotonin toxicity. QJM 2003;9:635–42. [DOI] [PubMed] [Google Scholar]
- 7. Isbister GK, Bowe SJ, Dawson A, Whyte IM. Relative toxicity of selective serotonin reuptake inhibitors (SSRIs) in overdose. J Toxicol Clin Toxicol 2004;3:277–85. [DOI] [PubMed] [Google Scholar]
- 8. Buckley NA, Dawson AH, Isbister GK. Serotonin syndrome. BMJ 2014;348:g1626. [DOI] [PubMed] [Google Scholar]
- 9. Bento AP, Gaulton A, Hersey A, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res 2014;D1:D1083–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2017;46:D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hawkins PC, Skillman AG, Nicholls A. Comparison of shape‐matching and docking as virtual screening tools. J Med Chem 2007;1:74–82. [DOI] [PubMed] [Google Scholar]
- 12. Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature 2009;7270:175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu D, Anderson HD, Tao A, et al. Assessing and predicting drug‐induced anticholinergic risks: an integrated computational approach. Ther Adv Drug Saf 2017;11:361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu D, Ham AG, Tivis RD, et al. MSBIS: a multi‐step biomedical informatics screening approach for identifying medications that mitigate the risks of metoclopramide‐induced tardive dyskinesia. EBioMedicine 2017;26:132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 2009;37(Suppl 2):W623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2016;D1:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiss‐Smith S, Deshpande G, Chung S, Gogolak V. The FDA drug safety surveillance program: adverse event reporting trends. Arch Intern Med 2011;6:591–3. [DOI] [PubMed] [Google Scholar]
- 18. Yue Z, Shi J, Jiang P, Sun H. Acute kidney injury during concomitant use of valacyclovir and loxoprofen: detecting drug–drug interactions in a spontaneous reporting system. Pharmacoepidemiol Drug Saf 2014;11:1154–9. [DOI] [PubMed] [Google Scholar]
- 19. Lorberbaum T, Sampson KJ, Woosley RL, Kass RS, Tatonetti NP. An integrative data science pipeline to identify novel drug interactions that prolong the QT interval. Drug Saf 2016;5:433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimura G, Kadoyama K, Brown JB, et al. Antipsychotics‐associated serious adverse events in children: an analysis of the FAERS database. Int J Med Sci 2015;2:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deepak P, Sifuentes H, Sherid M, Stobaugh D, Sadozai Y, Ehrenpreis ED. T‐cell non‐Hodgkin's lymphomas reported to the FDA AERS with tumor necrosis factor‐alpha (TNF‐α) inhibitors: results of the REFURBISH study. Am J Gastroenterol 2013;1:99–105. [DOI] [PubMed] [Google Scholar]
- 22. Piccinni C, Motola D, Marchesini G, Poluzzi E. Assessing the association of pioglitazone use and bladder cancer through drug adverse event reporting. Diabetes Care 2011;6:1369–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao S, Nishimura T, Chen Y, et al. Systems pharmacology of adverse event mitigation by drug combinations. Science Transl Med 2013;206:206ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sternbach H. The serotonin syndrome. Am J Psychiatry 1991;6:705–13. [DOI] [PubMed] [Google Scholar]
- 25. Isbister GK, Buckley NA, Whyte IM. Serotonin toxicity: a practical approach to diagnosis and treatment. Med J Aust 2007;6:361. [DOI] [PubMed] [Google Scholar]
- 26. MedDRA . World Health Organization. Available from https://tools.meddra.org/wbb. Accessed December 1, 2017.
- 27. Evans S, Waller PC, Davis S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol Drug Saf 2001;6:483–6. [DOI] [PubMed] [Google Scholar]
- 28. Orlova Y, Rizzoli P, Loder E. Association of coprescription of triptan antimigraine drugs and selective serotonin reuptake inhibitor or selective norepinephrine reuptake inhibitor antidepressants with serotonin syndrome. JAMA Neurol 2018;5:566–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kenakin T. Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther 2011;2:296–302. [DOI] [PubMed] [Google Scholar]
- 30. Celada P, Puig MV, Amargós‐Bosch M, Adell A, Artigas F. The therapeutic role of 5‐HT1A and 5‐HT2A receptors in depression. J Psychiatry Neurosci 2004;4:252. [PMC free article] [PubMed] [Google Scholar]
- 31. Artigas F. Serotonin receptors involved in antidepressant effects. Pharmacol Ther 2013;1:119–31. [DOI] [PubMed] [Google Scholar]
- 32. Carr GV, Lucki I. The role of serotonin receptor subtypes in treating depression: a review of animal studies. Psychopharmacology 2011;2–3:265–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Werneke U, Jamshidi F, Taylor DM, Ott M. Conundrums in neurology: diagnosing serotonin syndrome – a meta‐analysis of cases. BMC Neurol 2016;1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uddin MF, Alweis R, Shah SR, et al. Controversies in serotonin syndrome diagnosis and management: a review. J Clin Diagn Res 2017;11(9):OE05–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Volpi‐Abadie J, Kaye AM, Kaye AD. Serotonin syndrome. Ochsner J 2013;4:533–40. [PMC free article] [PubMed] [Google Scholar]
- 36. Radomski J, Dursun S, Reveley M, Kutcher S. An exploratory approach to the serotonin syndrome: an update of clinical phenomenology and revised diagnostic criteria. Med Hypotheses 2000;3:218–24. [DOI] [PubMed] [Google Scholar]
- 37. Evans RW, Tepper SJ, Shapiro RE, Sun‐Edelstein C, Tietjen GE. The FDA alert on serotonin syndrome with use of triptans combined with selective serotonin reuptake inhibitors or selective serotonin‐norepinephrine reuptake inhibitors: American Headache Society Position Paper. Headache 2010;6:1089–99. [DOI] [PubMed] [Google Scholar]
- 38. Gillman P. Tricyclic antidepressant pharmacology and therapeutic drug interactions updated. Br J Pharmacol 2007;6:737–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Racz R, Soldatos TG, Jackson D, Burkhart K. Association between serotonin syndrome and second‐generation antipsychotics via pharmacological target‐adverse event analysis. Clin Transl Sci 2018;11:322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Graham PM. Successful treatment of the toxic serotonin syndrome with chlorpromazine. Med J Aust 1997;3:166. [DOI] [PubMed] [Google Scholar]
- 41. Boddy R, Dowsett R, Jeganathan D. Sublingual olanzapine for the treatment of serotonin syndrome. Clin Toxicol 2006;4:439. [Google Scholar]
- 42. Indiana School of Medicine, Department of Medicine. http://medicineiupuiedu/clinpharm/ddis/main-table. Accessed January 30, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Serotonin Expanded Bioactivity Matrix (SEBM)


