Abstract
We report Phoenics, a probabilistic global optimization algorithm identifying the set of conditions of an experimental or computational procedure which satisfies desired targets. Phoenics combines ideas from Bayesian optimization with concepts from Bayesian kernel density estimation. As such, Phoenics allows to tackle typical optimization problems in chemistry for which objective evaluations are limited, due to either budgeted resources or time-consuming evaluations of the conditions, including experimentation or enduring computations. Phoenics proposes new conditions based on all previous observations, avoiding, thus, redundant evaluations to locate the optimal conditions. It enables an efficient parallel search based on intuitive sampling strategies implicitly biasing toward exploration or exploitation of the search space. Our benchmarks indicate that Phoenics is less sensitive to the response surface than already established optimization algorithms. We showcase the applicability of Phoenics on the Oregonator, a complex case-study describing a nonlinear chemical reaction network. Despite the large search space, Phoenics quickly identifies the conditions which yield the desired target dynamic behavior. Overall, we recommend Phoenics for rapid optimization of unknown expensive-to-evaluate objective functions, such as experimentation or long-lasting computations.
Short abstract
Phoenics is a deep Bayesian optimizer satisfying defined goals in a broad range of chemical problems. It rapidly identifies optimal conditions by balancing exploration and exploitation strategies.
Introduction
Optimization problems are ubiquitous in a variety of disciplines ranging from science to engineering and can take various facets: finding the lowest energy state of a system, searching for the optimal set of conditions to improve experimental procedures, or identifying the best strategies to realize industrial processes. They also have a rich history in chemistry. For example, conditions for chemical reactions are optimized with systematic methods like design of experiments (DOE).1−3 More recently, optimization procedures assisted chemists in finding chemical derivatives of given molecules to best treat a given disease,4 finding candidates for organic photovoltaics,5 organic synthesis,6,7 predicting reaction paths,8−10 or automated experimentation.11−14 Often, these applications are subject to multiple local optima, and involve costly evaluations of proposed conditions in terms of required experimentation or extensive computations.
Optimization problems are typically formulated with an objective function, which for a given set of parameters returns a measure for the associated merit. This relates, for example, to measuring the yield of a chemical reaction conducted under specific experimental conditions. In the past, a variety of optimization algorithms have been developed. Gradient-based algorithms, such as gradient descent,15 conjugate gradient,16 or the more sophisticated BFGS,17 are efficient at finding local optima. However, they require numerous evaluations, i.e., conducted experiments or computations, and are thus not well-suited for optimization problems in chemistry where evaluations of the objectives are often costly.
Lately, the development of methods for finding the global optimum of nonconvex, expensive-to-evaluate objective functions has gained resurgence as an active field of research. Simplistic approaches consist in random searches, or systematic grid searches. While the advantages of random searches have been demonstrated in the context of hyperparameter optimization for machine learning models,18,19 systematic grid search approaches like DOE were successfully applied to real-life experimentation planning.1−3 More sophisticated methods include genetic algorithms20,21 based on evolution strategies,22,23 which are, for example, applied to the optimization of nanoalloy clusters,24 or to resolve electronic spectra of rotamers of organic compounds.25 Yet, such methods still require many evaluations of the objective function.26
Bayesian optimization approaches have emerged as a popular and efficient alternative during the past decade.27−33 The typical procedure of Bayesian optimization schemes consists of two major steps: First, an approximation (surrogate) to the merit landscape of the conditions is constructed. Second, a new set of conditions is proposed for the next evaluation based on this surrogate. As such, Bayesian optimization speculates about the experimental outcome using all previously conducted experiments, and verifies its speculations by requesting the evaluation of a new set of conditions. Several different models have been suggested for approximating the objective function landscape, ranging from random forests (RFs),33−35 over Gaussian processes (GPs),31,36 to Bayesian neural networks (BNNs).37,38 Likewise, a variety of methods for proposing new conditions from the surrogate exists.28,31,39−42
Bayesian optimization has been successfully employed for a variety of applications across chemistry. Examples include the property optimization of functional organic molecules,43 calibrating high-throughput virtual screening results for organic photovoltaics,44 or designing nanostructures for phonon transport.45 Nevertheless, we identify three challenges in the deployment of Bayesian optimization techniques:
-
(i)
One challenge is the domain specificity of the surrogate model: GPs typically perform well on continuous objective landscapes, while RFs are typically more suitable for discrete and quasidiscrete landscapes. Unknown experimental response landscapes are more amenable to methods which perform well on a large variety of possible landscapes.
-
(ii)
Another challenge is the parallelization capabilities: Traditionally, Bayesian optimization methods are sequential in nature, which prevents parallel evaluations of the objective function, for instance, by using multiple experimental platforms or computational resources. For parallel evaluation, however, a procedure enabling the generation of multiple informative parameter points is needed. Prior work on batched Bayesian optimization,46−50 and nonmyopic approaches,51 aimed to resolve this problem, but requires additional computation compared to sequential optimization.
-
(iii)
The third identified challenge is computational efficiency: Optimization strategies involving substantial additional computation are only applied efficiently if the time required to suggest a new set of conditions does not significantly exceed the execution time of the experiment. As such, the optimization procedure should be computationally efficient compared to the time required to evaluate the objectives of the considered problem, e.g., to run the experiment.
The Probabilistic Harvard Optimizer Exploring Non-Intuitive Complex Surfaces (Phoenics) algorithm introduced in this study tackles the aforementioned challenges by supplementing ideas from Bayesian optimization with concepts from Bayesian kernel density estimation. More technically, we use BNNs to estimate kernel distributions associated with a particular objective function value from observed parameter points. Consequently, our approach differs from the traditional use of BNNs in the Bayesian optimization context, where objective function values are predicted from BNNs directly. Employing the estimated kernel distributions, we can construct a simple functional form of the approximation to the objective function. As a consequence, the computational cost of Phoenics scales linearly with the dimensionality of the search space and the number of observations, without the cost of numerous full evaluations of the BNN. Phoenics is available for download on GitHub.52
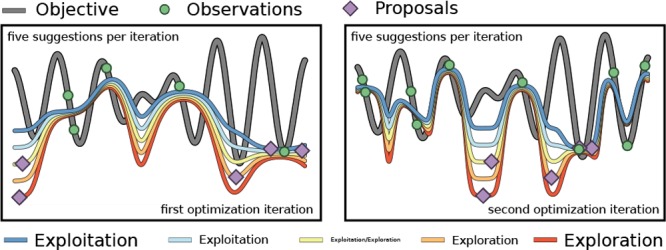
We propose an inexpensive acquisition function, which enables intuitive search strategies for efficient parallelization. This is achieved by simultaneously proposing multiple parameter points with different sampling policies at negligible additional cost. Those policies are biased toward exploration or exploitation of the search space tuned by an intuitive hyperparameter. A synergistic effect is observed when proposing batches of parameter points with different sampling policies. Our batching policy not only helps to accelerate the optimization process, but also reduces the total number of required function evaluations. It is therefore to be seen as an improvement over trivial parallelization.
In what follows, we start with a brief overview of related works. Then we detail the mathematical formulation of Phoenics. We further discuss performance results of Phoenics on analytic benchmark functions and compare to other Bayesian optimization methods. Before concluding we further highlight the applicability of our approach on the Oregonator, a model system for chemical reactions, on which we demonstrate the deployment of the proposed optimizer for practical problems in chemistry.
Background and Related Work
A plethora of global optimization algorithms has been developed to solve problems in different contexts, with different assumptions. The landscape of experimental responses could possibly be nonconvex, for instance, in scenarios where high reaction yields can be achieved with multiple different experimental conditions. Suitable optimization algorithms therefore need to be capable to overcome local optima to successfully find the global optimum.
The most straightforward approaches to global optimization include random searches, grid searches, and (fractional) factorial design strategies.1−3 While grid searches are embarrassingly parallel, they do not account for results obtained from recently executed experiments/computations. Computationally more involved methods, such as simulated annealing,53,54 particle swarm,55,56 or evolutionary strategies,22,23 are able to make informed decisions about the next conditions to evaluate by accounting for results from recent evaluations.
Recently, Bayesian optimization has gained increased attention as an alternative global optimization strategy as it was shown to reduce redundancy in the proposed conditions and, thus, locates global optima in fewer objective evaluations.57 Bayesian optimization is a gradient-free strategy for the global optimization of possibly noisy black-box functions, which we denote with f from hereon.27−30 It consists of two major steps: (i) construct a surrogate to f and (ii) propose new parameter points for querying f based on this probabilistic approximation.
In the first step, the surrogate model is constructed
by conditioning f on a prior ϕprior(θ) over the functional form, which is described
by parameters θ. The parameters θ of the prior
distribution are refined based on observations of n pairs 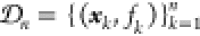 of parameter values xk, denoting, for instance,
experimental
conditions, and corresponding objective function values fk = f (xk), denoting the experimental
responses such as reaction yield. The functional prior ϕprior is updated based on observations
of parameter values xk, denoting, for instance,
experimental
conditions, and corresponding objective function values fk = f (xk), denoting the experimental
responses such as reaction yield. The functional prior ϕprior is updated based on observations 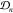 to yield
a more informative posterior ϕpost. With more and
more observations
to yield
a more informative posterior ϕpost. With more and
more observations 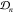 , the posterior ϕpost yields
a better approximation and eventually converges to the objective function
in the limit of infinitely many distinct observations, thus perfectly
reproducing the experimental response landscape.
, the posterior ϕpost yields
a better approximation and eventually converges to the objective function
in the limit of infinitely many distinct observations, thus perfectly
reproducing the experimental response landscape.
In the second step of the general Bayesian optimization procedure, this surrogate model is used to propose new conditions for future evaluations via an acquisition function. Bayesian optimization therefore relies on both an accurate approximation to the objective function and also the formulation of an efficient acquisition function.
Constructing the Objective Function Approximation
A
popular choice for modeling the functional prior ϕprior on the objective function are Gaussian processes (GPs),30,31,50,58 and random forests (RFs).33−35,59 GPs associate every point in the parameter domain with a normally
distributed random variable. These normal distributions are then constructed
via a similarity measure between observations given by a kernel function.
A GP therefore provides a flexible way of finding analytic approximations
to the objective function. Training a GP, however, is computationally
costly as it involves the inversion of a dense covariance matrix,
which scales cubically with the number of observations. Due to this
limitation, GPs are typically used in relatively low-dimensional problems
with an optimum that can be found in relatively few objective function
evaluations. RFs are a collection of regression trees, which, in contrast
to decision trees, have real numbers at their leaves. RFs have been
shown to perform particularly well for categorical input data and
classification tasks. RFs are therefore successfully applied to objective
functions with discrete or quasidiscrete codomain. The computational
cost of training a RF scales as 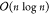 with the number of observations and linearly
with the dimensionality of the parameter space.
with the number of observations and linearly
with the dimensionality of the parameter space.
Recently, Bayesian neural networks (BNNs) have been employed for Bayesian optimization,37,38 retaining the flexibility of GPs at a computational scaling comparable to RFs. In contrast to traditional neural networks, weights and biases for neurons in BNNs are not single numbers but instead sampled from a distribution. BNNs are trained by updating the distributions from which weights and biases are sampled.
Acquisition Functions
The ideal acquisition function finds the adequate balance between exploration and exploitation. Exploration of the entire parameter space should be favored when no observations in vicinity to the global optimum have been made yet, and the acquisition function should only sample close to the global optimum once its general location has been determined.
One of the earliest and most widely applied acquisition functions is expected improvement and variants thereof.28,31,39 Expected improvement aims to measure the expected amount by which an observation of a point in parameter space improves over the current best value. Exploration and exploitation are implicitly balanced based on the posterior mean and the estimated uncertainty. More recently, alternative formulations of acquisition functions have been developed. The upper confidence bound method exploits confidence bounds for constructing an acquisition function which minimizes regret.42 Variants of this acquisition function have been designed specifically to be applied in higher-dimensional parameter spaces.49,50Predictive entropy estimates the negative differential entropy of the location of the global optimum given the observations,40,41 and has been shown to outperform expected improvement and upper confidence bound acquisitions.
Batched Bayesian Optimization
Many practical applications involve time-consuming evaluations of the objective function, but are amenable to parallelization. Examples include the execution of experiments on multiple experimental platforms or the distribution of computational models across multiple processors. Batched Bayesian optimization has been suggested with different assumptions and applicability scenarios. Marmin et. al proposed derivative-based expected improvement criterion for synchronous batch-sequential Bayesian optimization.48 Other methods include approaches for estimating the expected objective function value for future observations,31,50 or look ahead procedures.51 In addition, ensemble procedures have been proposed where new batches of samples are proposed from GPs trained on subsets of the available data.46,47 However, all proposed batch optimization methods require substantial additional computation compared to sequential evaluation strategies. A computationally less demanding batch optimization strategy has been proposed in the context of RF optimization.33−35 Due to the computational efficiency of RF models, batch optimization is realized by running several RF optimization instances in parallel, while sharing and contributing to the same observation data set.
Formulating Phoenics
In this section we present the mathematical formulation of Phoenics. We assume that evaluations of the objective are expensive, where the cost could be related to any budgeted resource such as required execution time, experimental synthesis of chemical compounds, computing resources, and others. Phoenics combines ideas from Bayesian optimization with concepts from Bayesian kernel density estimation (BKDE).60 The overall workflow of Phoenics is schematically represented in Figure 1 and follows the general principles of traditional Bayesian optimization. At each iteration, a surrogate model is constructed, from which new conditions are proposed. In the following we detail how the surrogate model is constructed (Figure 1a–c) and how the surrogate model can be biased toward particular sampling strategies, i.e., exploration or exploitation, using an intuitive sampling parameter (Figure 1d).
Figure 1.
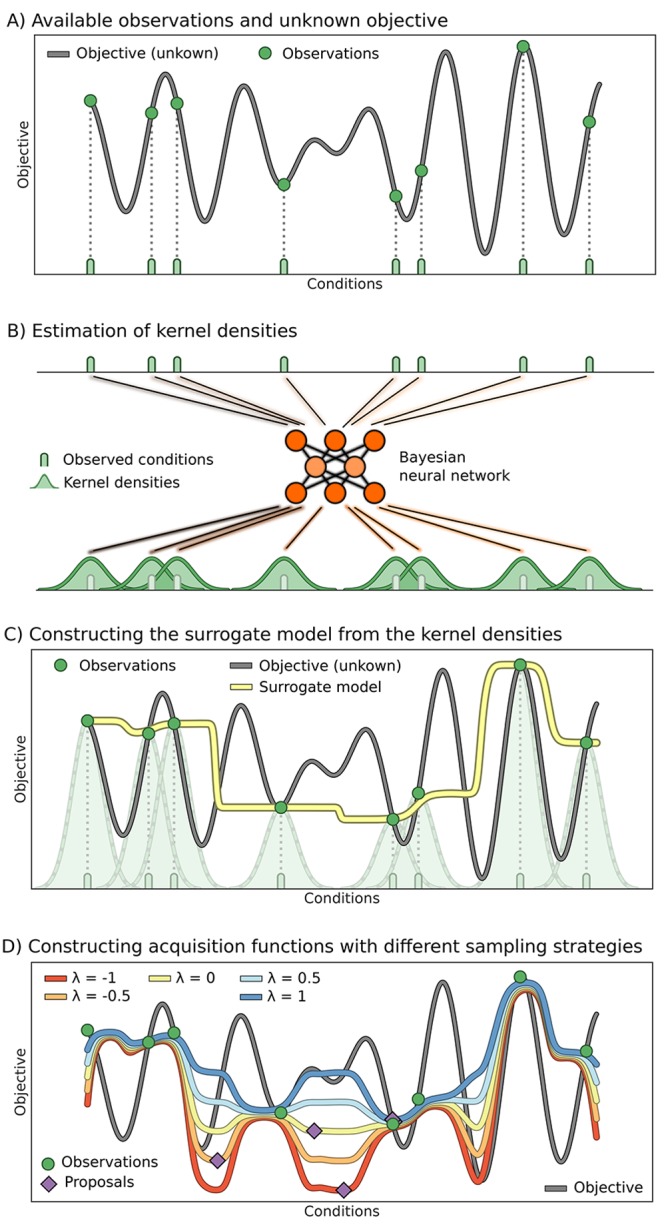
Illustration of the workflow of Phoenics. (A) Unknown, possibly high-dimensional, objective function of an experimental procedure or computation. The objective function has been evaluated at eight different conditions (green), which comprise the set of observations in this illustration. (B) The observed conditions are processed by a Bayesian neural network yielding a probabilistic model for estimating parameter kernel densities. Note that the probabilistic approach allows for a higher flexibility of our surrogate model compared to standard kernel density estimation. (C) The surrogate model is constructed by weighting the estimated parameter kernel densities with their associated observed objective values. (D) The surrogate can be globally reshaped using a single sampling parameter λ to favor exploration (red) or exploitation (blue) of the parameter space.
Approximating the Objective Function
We suggest to
use BNNs to estimate the parameter kernel density from the observed
parameter points in an autoencoder-like architecture. As such, the
BNN is used to nonlinearly estimate the density of the observed parameter
points x (see Figure 1b). A particular realization of the BNN represents
a map projecting parameter points into the parameter space, i.e, 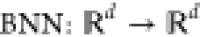 . Thereby, we can construct an estimate
to the parameter kernel density, which corresponds to a particular
observed objective function value. The use of a BNN for the construction
of the surrogate guarantees flexibility in the approximation as it
has already been reported that BNNs are versatile function approximators
at a favorable linear scaling with the number of observations.37,38 Details on the BNN architecture are reported in the Supporting Information (see Sec. S.3).
. Thereby, we can construct an estimate
to the parameter kernel density, which corresponds to a particular
observed objective function value. The use of a BNN for the construction
of the surrogate guarantees flexibility in the approximation as it
has already been reported that BNNs are versatile function approximators
at a favorable linear scaling with the number of observations.37,38 Details on the BNN architecture are reported in the Supporting Information (see Sec. S.3).
The implementation of Phoenics supports the construction and training of the surrogate model using either the PyMC3 library,61,62 or the Edward library.63 In both cases, the model parameters θ of the BNN are trained via variational inference. We start the sampling procedure with 500 samples of burn-in followed by another 1000 iterations retaining every 10th sample. This protocol was fixed for all tasks.
We can construct
an approximation to the kernel density from the
distributions of BNN parameters learned from the sampling procedure.
In particular, for observed conditions 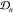 we compute
the kernel densities, which
are then used to approximate the objective function. The kernel density pk(x) generated from a single observed parameter point xk (see Figure 1b) can therefore be written
in closed form in eq 1, where ⟨·⟩ denotes the average over all sampled
BNN architectures, and xpred denotes the parameter points sampled from the BNN
we compute
the kernel densities, which
are then used to approximate the objective function. The kernel density pk(x) generated from a single observed parameter point xk (see Figure 1b) can therefore be written
in closed form in eq 1, where ⟨·⟩ denotes the average over all sampled
BNN architectures, and xpred denotes the parameter points sampled from the BNN
| 1 |
We formulate the approximation to the objective function as an ensemble average of the observed objective function values fk taken over the set of computed kernel densities pk(x) (see eq 2).64,65 In this ensemble average, each of the constructed distributions pk(x) is rescaled by the value of the objective function fk observed for the parameter point xk (Figure 1c). Note that α converges to the true objective f in the limit of infinitely many distinct function evaluations, as the kernel densities pk become more peaked due to the increasing precision in the Gaussian prior. Details are provided in the Supporting Information (see Sec. S.5).
| 2 |
This approximation to the objective function allows for inexpensive evaluations for any given parameter point x as repetitive, full BNN evaluations are avoided.
Acquisition Function
In the resulting approximation α we effectively model the expectation value of f for a given parameter point x based on prior observations. However, the parameter space could contain low-density regions, for which the objective function approximation α(x) is inaccurate (see Figure 1c).
With these
considerations, we propose an acquisition function based on the kernel
densities pk(x) for observations 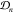 detailed
in eq 3. The acquisition
function differs from the
approximation to the objective function (see eq 2) by an additional term puniform(x) in the numerator
and the denominator, which denotes the uniform distribution on the
domain (see Figure 1d). In the numerator, puniform(x) is scaled by a factor λ, referred
to as the sampling parameter from hereon. Note that this modification
to α does not affect its convergence behavior (see the Supporting Information, Sec. S.5)
detailed
in eq 3. The acquisition
function differs from the
approximation to the objective function (see eq 2) by an additional term puniform(x) in the numerator
and the denominator, which denotes the uniform distribution on the
domain (see Figure 1d). In the numerator, puniform(x) is scaled by a factor λ, referred
to as the sampling parameter from hereon. Note that this modification
to α does not affect its convergence behavior (see the Supporting Information, Sec. S.5)
| 3 |
The introduced parameter λ effectively compares the cumulative height of each rescaled density estimate pk(x) to the uniform distribution. While the pk(x) are constructed from the knowledge we acquired from previous experiments, puniform is used as a reference to indicate the lack of knowledge in parameter space regions where little or no information is available yet. The sampling parameter therefore balances between acquired knowledge and the lack of knowledge, which effectively tunes the exploitative and explorative behavior of the algorithm.
Figure 1d illustrates the behavior of Phoenics on a one-dimensional objective function with different λ values. In this example, the acquisition function is constructed from eight observations indicated in green. Note that the acquisition function approximates the value of the objective function at observed parameter points. Acquisition functions which were constructed from a more positive λ show low values only in the vicinity of the observation with the lowest objective function value. In contrast, acquisition functions which were constructed from a more negative λ show low values far away from any observation. The choice for the value of the exploration parameter λ can therefore be directly related to explorative or exploitative behavior when proposing new conditions based on the global minimum of the surrogate. With a large positive value of λ, Phoenics favors exploitation, while a large negative value favors exploration. When λ = 0, the acquisition function shows no preference for a particular sampling strategy. Details on the global optimization of the surrogate are provided in the Supporting Information (see Sec. S.6).
From Figure 1d we see that distinct points in parameter space are proposed based on particular values of the exploration parameter λ. The best choice of λ for a given objective function is a priori unknown. However, with the possibility to rapidly construct several acquisition functions with biases toward exploration or exploitation, we can propose multiple parameter points in batches based on different sampling strategies. The newly proposed parameter points are then evaluated on the black-box optimization function in possibly parallel evaluation runs.
Results and Discussion
In this section we report the performance of Phoenics and compare it to four frequently used optimization algorithms: particle swarm optimization (PSO),55,56 covariance matrix adaptation evolution strategy (CMA-ES),22,23 as well as Bayesian optimization based on GPs and based on RFs.
PSO is implemented in the “pyswarms” python module.66 An implementation of CMA is also available in the “cma” module.67 Default settings as provided by the modules have been used for both optimization algorithms. The “spearmint” software package performs Bayesian optimization using GPs and the predictive entropy acquisition function.31,36 Batch optimization is implemented in spearmint via estimating the expected objective value for future evaluations. The SMAC software employs RF models and allows for batch optimization by running multiple RF instances sharing the same set of samples.33−35
Chemical systems can have complex, qualitatively different response surfaces. As chemical reactions are time-consuming to evaluate, we therefore assess the performance of each of these three algorithms on a set of 15 benchmark functions covering a large range of qualitatively diverse response surfaces for problems in chemistry. The employed functions are well-established benchmarks and include continuous and convex, nonconvex, or discrete functions with possibly multiple global minima. A complete list of the employed objective functions as well as their global minima is provided in the Supporting Information (see Table S.1).
For reliable performance estimates we executed 20 independent optimization runs initialized with different random seeds, unless noted otherwise. During each run we record the lowest achieved objective function value after each iteration. We compare the averaged lowest achieved objective function values by relating to results from simple random searches. Each random search was run for 104 objective function evaluations, and results were averaged over 50 independent runs initialized with different random seeds. The average lowest achieved objective function values of the random search runs are summarized in the Supporting Information (see Table S.2).
Our benchmark calculations indicate that Bayesian-based optimization algorithms outperform PSO and CMA-ES. As a matter of fact, after 200 function evaluations, PSO and CMA-ES yield significantly higher deviations from the global optimum than the three studied Bayesian optimization algorithms. Even when increasing the number of function evaluations by an order of magnitude, from 200 to 2000, PSO (and CMA-ES) fails to achieve lower deviations than the ones obtained with Phoenics after 200 evaluations for 12 (and 13) out of 15 objective functions. We therefore restrict our further analyses and comparisons to only Phoenics, GP optimization, and RF optimization. The benchmark results conducted with PSO and CMA-ES are detailed in the Supporting Information (see Figure S.6)
Analytic Benchmarks
Phoenics was set up with three different values for the sampling parameter, λ ∈ {−1, 0, 1}, to assess the effectiveness of a particular parameter choice. In Figure 2 we report the number of objective function evaluations required by each of the optimization algorithms to reach an objective function value lower than the average lowest value found in random searches. Optimization traces for these runs on all 15 objective functions are reported in the Supporting Information (see Figure S.3).
Figure 2.
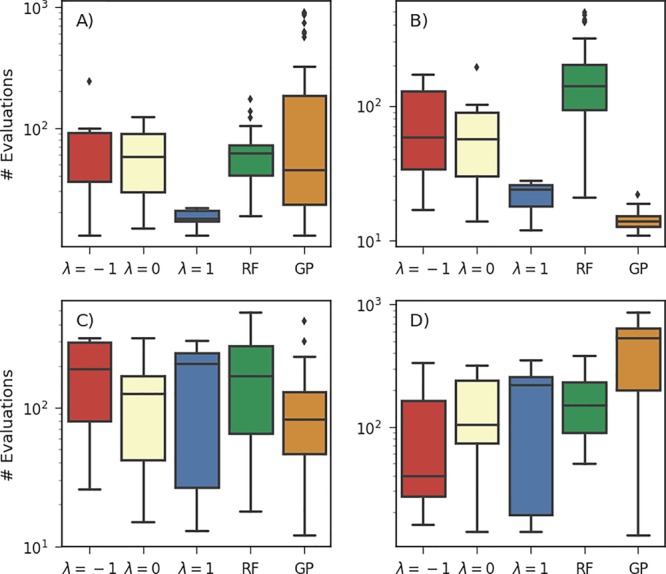
Number of objective function evaluations required to reach objective function values lower than the average lowest achieved values of random searches with 104 evaluations for Phoenics (λ ∈ {−1, 0, 1}), RFs, and GPs. Results are reported for the Ackley (A), Dejong (B), Schwefel (C), and dAckley (D) objective functions. Details on the benchmark functions are provided in the Supporting Information, Sec. S.1.
We find that GP optimization, as implemented in spearmint, generally quickly finds the global minimum if the objective function is strictly convex. In contrast, RF optimization, as implemented in SMAC, quickly finds the global minimum of objective functions with a discrete codomain. The performance of Phoenics varies with different values of the sampling parameter λ. When favoring exploitation over exploration, i.e., λ > 0, the algorithm performs better if the objective function features narrow and well-defined funnels (e.g., Ackley in Figure 2a or Schwefel in Figure 2c). With this choice for the sampling parameter, the algorithm is slightly biased toward exploring the local region around the current optimum. This behavior, however, is unfavorable in other cases, for instance, when the objective function has a discrete codomain (e.g., dAckley in Figure 2d). Since parameter points in the vicinity to the current optimum likely yield the same value if the objective function is discrete or quasidiscrete, Phoenics performs better on such objective functions when favoring exploration over exploitation, i.e., λ < 0.
Developing a Collective Sampling Strategy
The dependence of the performance of Phoenics on the sampling parameter λ could be eliminated by marginalizing over this parameter. Marginalization over the sampling parameter would effectively average out the advantageous effects of a bias toward exploitation for some objective functions and toward exploration for other objective functions.
The shape of the objective function is a priori unknown, so suitable choices of the sampling parameter cannot be determined beforehand. However, since the sampling parameter can be directly related to the explorative and exploitative behavior of the algorithm we follow an approach to take full advantage of the sampling policy. We suggest to propose parameter points based on multiple different sampling parameter values. Note that values of λ are chosen beforehand and are kept fixed throughout the optimization procedure.
Given a set of observations 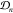 the construction of several objective
surrogates
with different values of λ is computationally cheap. This allows
us to suggest multiple parameter points at each optimization iteration,
which are proposed from more explorative and more exploitative parameter
values, at almost no additional cost. With the observations on the
simple benchmarks we would expect a synergistic effect of this batch
optimization over sequential optimization with a single sampling parameter
value. As parameter points can be proposed with both a bias toward
exploration and a bias toward exploitation, we expect the number of
required objective function evaluations to decrease. In addition,
suggesting a batch of parameter points in one optimization step allows
for the parallel evaluation of all proposed points, which accelerates
the optimization process.
the construction of several objective
surrogates
with different values of λ is computationally cheap. This allows
us to suggest multiple parameter points at each optimization iteration,
which are proposed from more explorative and more exploitative parameter
values, at almost no additional cost. With the observations on the
simple benchmarks we would expect a synergistic effect of this batch
optimization over sequential optimization with a single sampling parameter
value. As parameter points can be proposed with both a bias toward
exploration and a bias toward exploitation, we expect the number of
required objective function evaluations to decrease. In addition,
suggesting a batch of parameter points in one optimization step allows
for the parallel evaluation of all proposed points, which accelerates
the optimization process.
The behavior of the three studied optimization algorithms under parallel optimization on the Ackley objective function is highlighted in Figure 3. Full results on the entire benchmark set are reported in the Supporting Information (see Sec. S.8). Figure 3 depicts the minimum achieved objective function values for different runs with a different number of parallel evaluations of the objective function averaged over 20 independent runs. The minimum achieved objective function values are presented per number of objective function evaluations (left panel) and per batch evaluation (right panel).
Figure 3.
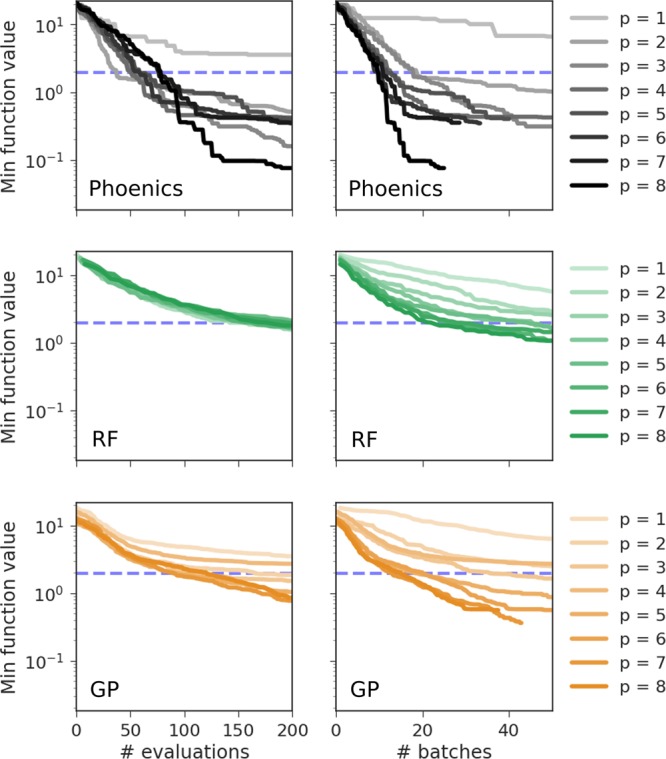
Average minimum objective function values for the Ackley function achieved in 20 independent runs of the three optimization algorithms studied in this work: our optimizer (Phoenics), spearmint (GP), and SMAC (RF). For each run a different number of proposed samples p was evaluated in parallel. Minimum achieved objective function values are reported with respect to the total number of objective function evaluations and the number of evaluated batches. The dashed blue lines denote the minimum achieved error after 104 of random search for reference.
We find that both spearmint and SMAC achieve low objective function values in fewer batches with an increasing number of points p proposed in each batch. While increasing the number of samples proposed per batch initially significantly improves the performance with respect to the number of proposed batches, this advantageous effect quickly levels off until there is no significant improvement beyond six samples per batch. However, when comparing the minimum achieved objective function values with respect to the total number of objective function evaluations, we did not observe any significant difference between runs with a different number of samples proposed per batch.
In contrast, Phoenics shows a different behavior. Our algorithm not only reaches lower objective function values in a fewer number of batches when proposing more samples per batch, but also shows a better performance when considering the total number of function evaluations. This synergistic effect demonstrates that Phoenics indeed benefits from proposing points with multiple sampling strategies in cases in which proposed samples are evaluated sequentially.
The performance improvement of Phoenics when proposing parameter points in batches at each optimization iteration is demonstrated on all 15 considered objective functions in the Supporting Information (see Sec. S.8). We ran our optimizer with four different sampling strategies using sampling parameter values evenly spaced across the [−1, 1] interval. All four proposed parameter points are then evaluated before we started another optimization iteration. For this particular batching protocol, we find that Phoenics outperforms RF-based optimization on all benchmark functions and GP-based optimization on 12 out of 15 benchmark functions. If and only if the objective function is convex, GP optimization finds lower objective function values. In addition, we observe the aforementioned synergistic effect of batch optimization for 12 out of 15 benchmark functions. Despite reducing the number of optimization iterations by a factor of 4, the achieved objective function values were found to be lower than values achieved in sequential optimizations with all three considered fixed sample parameter value.
We suggest that this improved performance of the algorithm is due to the trade-off between exploration and exploitation. The exploration samples systematically sample the parameter space and ensure that the algorithm does not get stuck in local minima, while the exploitation samples explore the local environment of the current global minimum. This sampling behavior is illustrated in Figure 4 for the Michalewicz function. The optimization runs on the Michalewicz function were all started from the same two random samples illustrated in black for all three investigated optimization algorithms. Bayesian optimization based on GPs as implemented in spearmint (lower panels) tends to sample many parameter points close to the boundaries of the domain space in this particular example. RF optimization as implemented in SMAC (central panels), however, shows a higher tendency of exploring the parameter space.
Figure 4.
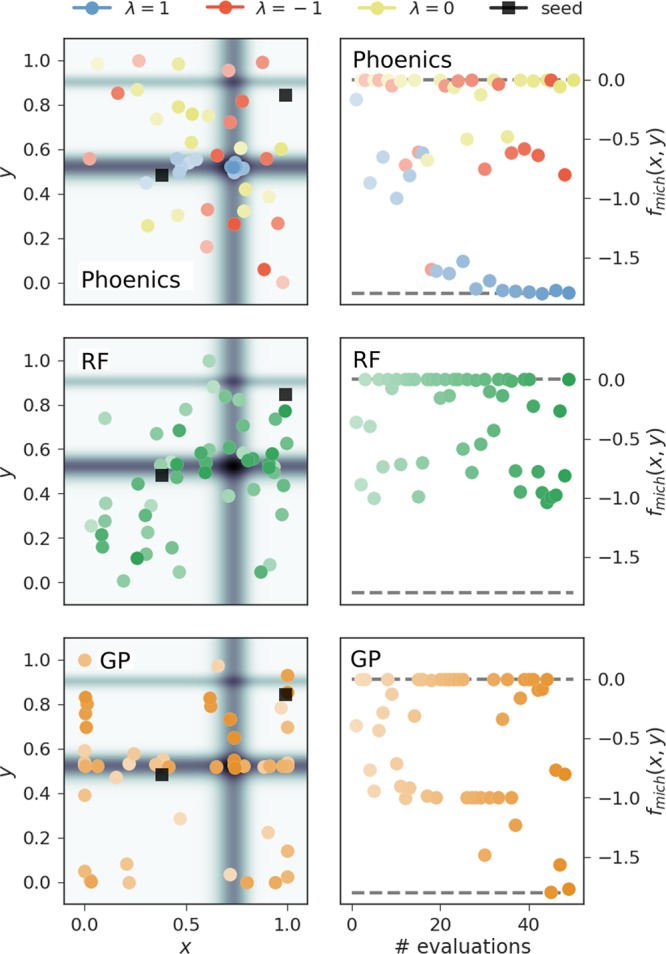
Progress of sample optimization runs of the three studied optimization algorithms on the two-dimensional Michalewicz function. Phoenics proposed a total of three samples per batch, which were then evaluated in parallel. Each sample was suggested based on a particular value of the exploration parameter λ ∈ {−1, 0, 1}. Left panels illustrate the parameter points proposed at each optimization iteration while right panels depict the achieved objective function values. Depicted points are more transparent at the beginning of the optimization and more opaque toward the end. Starting points for the optimization runs are drawn as black squares.
Phoenics (Figure 4, upper panels) starts exploring the space and quickly finds a local minimum in vicinity of one of the initial samples. After finding this local minimum, samples which are proposed based on a more exploitative (positive) value of the sampling parameter λ explore the local environment of this local minimum while samples proposed from more explorative (negative) values of λ explore the entire parameter space. As soon as the exploration points find a point in parameter space with a lower value of the objective function, the exploitation points jump to this new region in parameter space and locally explore the region around the current best to quickly converge to the global minimum.
Overall we have demonstrated that the value of the sampling parameter λ in the proposed acquisition function clearly influences the behavior of the optimization procedure toward a more explorative behavior for more negative values of this parameter and a more exploitative behavior for more positive parameter values. Batched optimization improves the performance of Phoenics even in terms of total objective function evaluations and reduces the number of required optimization iterations.
Increasing the Number of Dimensions
Practical chemistry problems are typically concerned with more than just two parameters. In fact, chemical reactions can be influenced by environmental conditions and experimental device settings, and computational studies frequently employ parameters to describe the system of interest. In this section we illustrate the performance of Phoenics in parameter spaces with dimensions k > 2.
We evaluate the performance of the three optimization algorithms on the same objective function subset, but now successively increase the dimensionality of the parameter space from 2 to 20. Based on the results on batch optimization we ran GP optimization and RF optimization with one point per batch and the optimization algorithm introduced in this study with four points per batch on each considered benchmark function. Exploration parameter values were chosen to be evenly spaced across the [−1, 1] interval.
For better comparisons we report the average deviation of the lowest encountered objective function value from the global minimum of each function taken over 20 independent optimization runs. Average deviations achieved by each of the optimization algorithms after 200 objective function evaluations are depicted in Figure 5.
Figure 5.
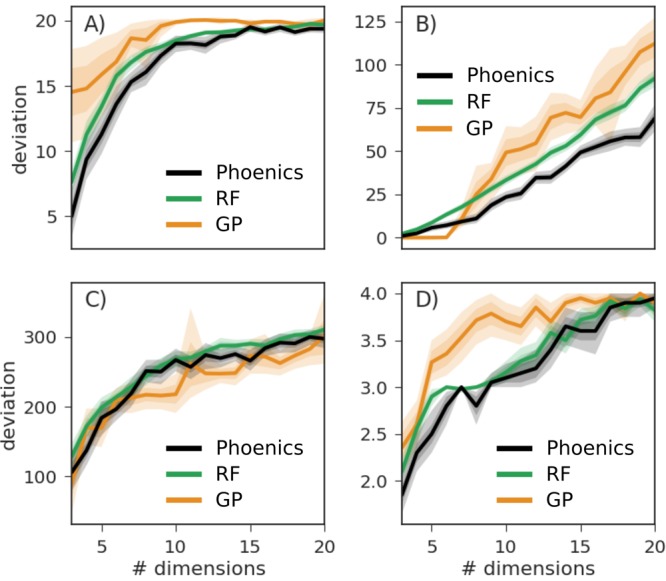
Average deviations taken over 20 independent runs between the lowest encountered objective function value and the global minimum achieved after 200 objective function evaluations for different parameter set dimensions. Results are reported for Ackley (A), Dejong (B), Schwefel (C), and dAckley (D). Uncertainty bands illustrate bootstrapped estimates of the deviation of the means with one and two standard deviations.
We observe that Phoenics maintains its rapid optimization properties for a variety of different objective functions even when increasing the number of dimensions. In the case of the Ackley function (Figure 5a) Phoenics appears to find and explore the major funnel close to the global optimum faster than the other two optimization algorithms regardless of the number of dimensions. The paraboloid (Figure 5b) is an easy case for the GP in low dimensions, but is optimized the fastest by Phoenics when considering parameter spaces with seven or more dimensions. No major differences are observed for the Schwefel function (Figure 5c). However, in the case of a discrete objective function (Figure 5d) Phoenics seems to have a slight advantage over the other two optimizers for lower dimensions and performs about as well as random forest optimization for higher dimensions.
Application to Chemistry
In this section, we demonstrate the applicability of Phoenics on the Oregonator, a model system of a chemical reaction described by a set of nonlinear coupled differential equations.68 In particular, we demonstrate how Phoenics can be employed to propose a set of conditions for an experimental procedure. The experimental procedure can then be executed with the proposed conditions, and the results of the procedure are reported back to Phoenics. With this feedback, Phoenics can make more informed decisions and, thus, provides more promising sets of experimental conditions, to eventually result in the discovery of the optimal set of conditions.
Most chemical reactions lead to a steady-state, i.e., a state in which the concentrations of involved compounds are constant in time. While chemical reactions described by linear differential equations always feature such a steady-state, more complicated dynamics phenomena can arise for reactions described by sets of nonlinear coupled differential equations. With the right choice of parameters, such differential equations may have a stable limit cycle, leading to periodic oscillations in the concentrations of involved compounds.69,70
One of the earliest discovered reactions featuring a stable limit cycle for a set of reaction conditions is the Belousov–Zhabotinsky reaction.71,72 This network of chemical reactions involves temporal oscillations of [CeIV] and [CeIII]. The entire reaction network can be written as a set of three subreactions listed in Scheme 1. For details on the mechanism we refer to a brief summary in the Supporting Information (see Sec. S.10) as well as to the literature.68,72−75
Scheme 1. Subreactions of the Belousov–Zhabotinsky Reaction78.
Models at different levels of complexity have been developed to describe the dynamic behavior of the Belousov–Zhabotinsky reaction.69,75−77 One of the simplest models of this reaction is the Oregonator.68 The Oregonator consists of a set of three coupled first-order nonlinear differential equations for three model compounds X, Y, and Z, which are shown in eqs 4–6. These equations involve five reaction constants ki, a stoichiometric factor f, determined by the prevalence of one subreaction over another subreaction, and the concentration of two additional chemical compounds A and B. A map of eqs 4–6 to Scheme 1 is outlined in ref (68).
| 4 |
| 5 |
| 6 |
The set of differential equations in the Oregonator can be reduced into a dimensionless form, such that the number of correlated parameters is reduced to a smaller set of independent parameters. This reduced version of the Oregonator, presented in the Supporting Information (see Sec. S.10), includes three dimensionless variables α, η, and ρ, which describe the concentration of chemical species, and four dimensionless reaction constants.
Phoenics is used to reverse engineer the set of reaction conditions consisting of three initial concentrations (α0, η0, and ρ0) and four reaction constants (q, s, w, and f) from the concentration traces computed in the original publication.68 Parameter values yielding the target concentration traces are reported in Table 1. The goal is 2-fold: (i) find a set of conditions for which the dynamical behavior qualitatively agrees with the behavior of the target, i.e., find chemical oscillations, and (ii) fine-tune these conditions such that we reproduce the dynamical behavior on a quantitative level. We aim to achieve these two goals while keeping the number of function evaluations, i.e., the number of experiments to run, to a minimum. Note that this constraint along with the dimensionality of the parameter space implies that grid searches or gradient-based algorithms are not suited for this problem.
Table 1. Reaction Parametersa of the Reduced Oregonator Model for the Belousov–Zhabotinsky Reaction.
| parameter | target value | range |
|---|---|---|
| s | 77.27 | 0 ... 100 |
| w | 0.1610 | 0 ... 1 |
| q | 8.375 × 10–6 | 10–8 ... 10–4 |
| f | 1 | 0 ... 5 |
| α0 | 2.0 × 107 | 104 ... 109 |
| η0 | 3.3 × 103 | 103 ... 105 |
| ρ0 | 4.1 × 104 | 103 ... 106 |
Target parameters induce the existence of a limit cycle, from which chemical oscillations emerge. For finding these target parameters via optimization we constrained the domain space to the reported ranges. All reported quantities are dimensionless.
Phoenics was run in parallel proposing four samples per batch with λ equally spaced on the [−1, 1] interval. We compare the performance to PSO in the pyswarms module, CMA-ES in the cma module, GP optimization in spearmint, and RF optimization in SMAC. Each of the five optimization algorithms was used in 50 independent optimization runs for 150 evaluations.
All optimization procedures were carried out on a constrained parameter space reported in Table 1. Note that different choices of parameter sets within this bounded domain can result in quantitatively and qualitatively different dynamical behavior. In particular, parameter choices close to the target result in oscillatory behavior, for which the reduced concentrations α, ρ, and η change periodically over time, while other parameter choices can break the limit cycle and create a stable fixed point instead.68,74,77
Concentration traces for a sampled set of reaction parameters were computed with a fourth-order Runge–Kutta integrator with adaptive time stepping. The integrator was run for a total of 107 integration steps covering 12 full concentration oscillations for the target parameter set. Sampled concentration traces are compared to the target concentration traces after a cubic spline interpolation. The distance (loss) between the sampled traces and the target traces is calculated as the euclidean distance between the points in time at which a concentration trace reaches a dimensionless concentration value of 100.
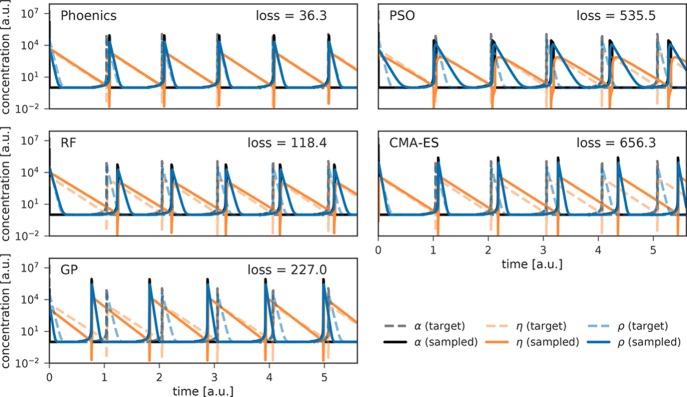
Average achieved losses for all three optimization algorithms are displayed in Figure 6. Loss values between 300 and 500 indicate that the periodicity of the predicted concentration traces resembles the periodicity of the target traces; i.e., the predicted traces qualitatively agree with the target. Quantitative agreement, i.e., matching traces, is only achieved for loss values lower than about 100. Examples for concentration traces yielding different losses are presented in the Supporting Information (see Figure S.7).
Figure 6.
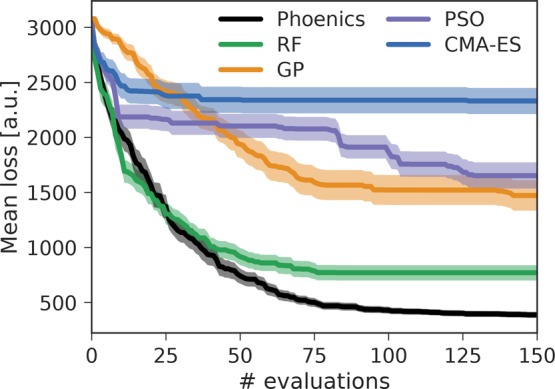
Average achieved losses for finding reaction parameters of the reduced Oregonator model achieved by the five optimization algorithms employed in this study. Correct periodicities of the concentration traces are achieved for losses lower than 500. Uncertainty bands illustrate bootstrapped deviations on the mean for one standard deviation.
Figure 7 shows concentration traces associated with the lowest loss achieved by each of the three optimization algorithms across all 50 independent runs. Phoenics is the only algorithm reproducing qualitatively and quantitatively target dynamic behavior within 150 optimization iterations. RF optimization only finds parameter sets which qualitatively agree with the target. GP optimization finds only in rare occasions concentration traces in qualitative agreement with the target. Both PSO and CMA-ES consistently yield high losses for the first 75 evaluations, after which PSO slightly improves but never reaches the degree of agreement achieved by Phoenics.
Figure 7.
Time traces of dimensionless concentrations of compounds in the Oregonator model. Target traces are depicted with solid, transparent lines while predicted traces are shown in dashed, opaque lines. Traces were simulated for a total of 12 dimensionless time units, but are only shown for the first six time units for clarity.
Conclusion and Outlook
We introduced Phoenics, an algorithm for global optimization in the context chemistry and experimentation. Phoenics is designed for scenarios where the merit of a set of conditions is evaluated via experimentation or expensive computations, which can possibly be parallelized. Our probabilistic optimizer combines Bayesian optimization with conceptual aspects of Bayesian kernel density estimation. As such, our algorithm is well-suited for applications where evaluations of the objective function are expensive with respect to budgeted resources such as time or money. Through an exhaustive benchmark study, we showed that Phoenics improves over optimization strategies based on particle swarms or evolutionary approaches, as well as on existing Bayesian global optimization methods and avoids redundant evaluations of the objective.
We formulate an inexpensive acquisition function balancing the explorative and exploitative behavior of the algorithm. This acquisition function enables intuitive sampling policies for an efficient parallel search of global minima. By leveraging synergistic effects from running multiple sampling policies in batches, the performance of the algorithm improves, and requires a reduced total number of objective function evaluations.
The applicability of Phoenics was highlighted on the Oregonator, a model system describing a complex chemical reaction network. Phoenics was able to determine the set of seven experimental conditions reproducing a target dynamic behavior in the concentrations of involved chemical species. High degrees of qualitative and quantitative agreement could be achieved with only 100 merit-evaluations of proposed conditions despite the rich solution space containing both steady-state systems and chemical oscillators.
We believe that Phoenics has the potential to be applied to a wide range of problems, from optimization of reaction conditions and material properties, over control of robotics systems, to circuit design for quantum computing.79,80 All in all, we recommend Phoenics for an efficient optimization of scalar, possibly nonconvex, black-box unknown objective functions.
Acknowledgments
We thank Dr. S. K. Saikin for fruitful discussions and helpful comments. F.H. was supported by the Herchel Smith Graduate Fellowship. L.M.R. and A.A.-G. were supported by the Tata Sons Limited—Alliance Agreement (A32391). C.K. and A.A.-G were supported by the National Science Foundation under Award CHE-1464862. All computations reported in this paper were completed on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00307.
Additional methods, data, and figures including contour plots, illustration of convergence behavior, lowest discovered objective function values, deviation values, and concentration traces (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Fisher R. A.The Design of Experiments; Oliver and Boyd: Edinburgh, London, 1937. [Google Scholar]
- Box G. E. P.; Hunter J. S.; Hunter W. G.. Statistics for Experimenters: Design, Innovation and Discovery; Wiley-Interscience: New York, 2005; Vol. 2. [Google Scholar]
- Anderson M. J.; Whitcomb P. J.. DOE Simplified: Pratical Tools for Effective Experimentation; CRC Press: New York, 2016. [Google Scholar]
- Negoescu D. M.; Frazier P. I.; Powell W. B. The Knowledge-Gradient Algorithm for Sequencing Experiments in Drug Discovery. INFORMS J. Comput. 2011, 23, 346–363. 10.1287/ijoc.1100.0417. [DOI] [Google Scholar]
- Lopez S. A.; Sanchez-Lengeling B.; de Goes Soares J.; Aspuru-Guzik A. Design Principles and Top Non-Fullerence Acceptor Candidates for Organic Photovoltaics. Joule 2017, 1, 857–870. 10.1016/j.joule.2017.10.006. [DOI] [Google Scholar]
- Walker B. E.; Bannock J. H.; Nightingale A. M.; deMello J. C. Tuning Reaction Products by Constrained Optimisation. React. Chem. Eng. 2017, 2, 785–798. 10.1039/C7RE00123A. [DOI] [Google Scholar]
- Jensen K. F.; Reizman B. J.; Newman S. G. Tools for Chemical Synthesis in Microsystems. Lab Chip 2014, 14, 3206–3212. 10.1039/C4LC00330F. [DOI] [PubMed] [Google Scholar]
- Wei J. N.; Duvenaud D.; Aspuru-Guzik A. Neural Networks for the Prediction of Organic Chemistry Reactions. ACS Cent. Sci. 2016, 2, 725–732. 10.1021/acscentsci.6b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley C. W.; Barzilay R.; Jaakkola T. S.; Green W. H.; Jensen K. F. Prediction of Organic Reaction Outcomes Using Machine Learning. ACS Cent. Sci. 2017, 3, 434–443. 10.1021/acscentsci.7b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segler M. H. S.; Waller M. P. Neural-Symbolic Machine Learning for Retrosynthesis and Reaction Prediction. Chem. - Eur. J. 2017, 23, 5966–5971. 10.1002/chem.201605499. [DOI] [PubMed] [Google Scholar]
- Nikolaev P.; Hooper D.; Perea-Lopez N.; Terrones M.; Maruyama B. Discovery of Wall-Selective Carbon Nanotube Growth Conditions via Automated Experimentation. ACS Nano 2014, 8, 10214–10222. 10.1021/nn503347a. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D. E.; Battilocchio C.; Ley S. V. A Novel Internet-Based Reaction Monitoring, Control and Autonomous Self-Optimization Platform for Chemical Synthesis. Org. Process Res. Dev. 2016, 20, 386–394. 10.1021/acs.oprd.5b00313. [DOI] [Google Scholar]
- Nikolaev P.; Hooper D.; Webber F.; Rao R.; Decker K.; Krein M.; Poleski J.; Barto R.; Maruyama B. Autonomy in Materials Research: A Case Study in Carbon Nanotube Growth. npj Comput. Mate. 2016, 2, 16031. 10.1038/npjcompumats.2016.31. [DOI] [Google Scholar]
- Duros V.; Grizou J.; Xuan W.; Hosni Z.; Long D. L.; Miras H.; Cronin L. Human vs Robots in the Discovery and Crystallization of Gigantic Polyoxometalates. Angew. Chem., Int. Ed. 2017, 56, 10815–10820. 10.1002/anie.201705721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder S.An Overview of Gradient Descent Optimization Algorithms. 2004, arXiv:1609.04747. arXiv.org e-Print archive. https://arxiv.org/abs/1609.04747.
- Hestenes M. R.; Stiefel E.. Methods of Conjugate Gradients for Solving Linear Systems; NBS: Washington, DC, 1952; Vol. 49. [Google Scholar]
- Fletcher R.Methods of Optimization; John Wiley & Sons: New York, 1987. [Google Scholar]
- Bergstra J. A.; Badenet R.; Bengio Y.; Kégl B. Algorithms for Hyper-Parameter Optimization. NIPS 2011, 24, 2546–2554. [Google Scholar]
- Bergstra J.; Bengio Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Back T.Evolutionary Algorithms in Theory and Practice: Evolution Strategies, Evolutionary Programming, Genetic Algorithms; Oxford University Press: New York, 1996. [Google Scholar]
- Simon D.Evolutionary Optimization Algorithms; John Wiley & Sons: New York, 2013. [Google Scholar]
- Hansen N.; Ostermeier A. Completely Derandomized Self-Adaptation in Evolution Strategies. Evol. Comput. 2001, 9, 159–195. 10.1162/106365601750190398. [DOI] [PubMed] [Google Scholar]
- Hansen N.; Müller S. D.; Koumoutsakos P. Reducing the Time Complexity of the Derandomized Evolution Strategy with Covariance Matrix Adaptation (CMA-ES). Evol. Comput. 2003, 11, 1–18. 10.1162/106365603321828970. [DOI] [PubMed] [Google Scholar]
- Shayeghi A.; Götz D.; Davis J. B. A.; Schäfer R.; Johnston R. L. Pool-BCGA: A Parallelised Generation-Free Genetic Algorithm for the Ab Initio Global Optimisation of Nanoalloy Clusters. Phys. Chem. Chem. Phys. 2015, 17, 2104–2112. 10.1039/C4CP04323E. [DOI] [PubMed] [Google Scholar]
- Schneider M.; Wilke M.; Hebestreit M. L.; Ruiz-Santoyo J. A.; Álvarez-Valtierra L.; John T. Y.; Meerts W. L.; Pratt D. W.; Schmitt M. Rotationally Resolved Electronic Spectroscopy of the Rotamers of 1, 3-Dimethoxybenzene. Phys. Chem. Chem. Phys. 2017, 19, 21364–21372. 10.1039/C7CP04401A. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Li X.; Zare R. N. Optimizing Chemical Reactions with Deep Reinforcement Learning. ACS Cent. Sci. 2017, 3, 1337–1344. 10.1021/acscentsci.7b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner H. J. A New Method of Locating the Maximum Point of an Arbitrary Multipeak Curve in the Presence of Noise. J. Basic Eng. 1964, 86, 97–106. 10.1115/1.3653121. [DOI] [Google Scholar]
- Močkus J.On Bayesian Methods for Seeking the Extremum, Optimization Techniques IFIP Technical Conference; 1975; pp 400–404.
- Močkus J. The Bayesian Approach to Global Optimization. Sys. Model. Optim. 1982, 38, 473–481. 10.1007/BFb0006170. [DOI] [Google Scholar]
- Osborne M. A.; Garnett R.; Roberts S. J.. Gaussian Processes for Global Optimization, Internat. Conf. Learn. Intell. Optim.; 2009; pp 1–15.
- Snoek J.; Larochelle H.; Adams R. P. Practical Bayesian Optimization of Machine Learning Algorithms. NIPS 2012, 4, 2951–2959. [Google Scholar]
- Srinivas N.; Krause A.; Kakade S. M.; Seeger M. W. Information-Theoretic Regret Bounds for Gaussian Process Optimization in the Bandit Setting. IEEE Trans. Inf. Theory 2012, 58, 3250–3265. 10.1109/TIT.2011.2182033. [DOI] [Google Scholar]
- Lindauer M.; Eggensperger K.; Feurer M.; Falkner S.; Biedenkapp A.; Hutter F.. SMAC v3: Algorithm Configuration in Python. https://github.com/automl/SMAC3 (2017).
- Hutter F.; Hoos H. H.; Leyton-Brown K. Sequential Model-Based Optimization for General Algorithm Configuration. Internat. Conf. Learn. Intel. Optim. 2011, 6683, 507–523. 10.1007/978-3-642-25566-3_40. [DOI] [Google Scholar]
- Hutter F.; Hoos H.; Leyton-Brown K. Parallel Algorithm Configuration. Learn. Intell. Optim. 2012, 7219, 55–70. 10.1007/978-3-642-34413-8_5. [DOI] [Google Scholar]
- Snoek J.; Swersky K.; Zemel R.; Adams R. P.. Input Warping for Bayesian Optimization of Non-Stationary Functions, Internat. Conf. Mach. Learn.; 2014; pp 1674–1682.
- Snoek J.; Rippel O.; Swersky K.; Kiros R.; Satish N.; Sundaram N.; Patwary M.; Prabhat; Adams R.. Scalable Bayesian Optimization using Deep Neural Networks, Internat. Conf. Mach. Learn.; 2015; pp 2171–2180.
- Springenberg J. R.; Klein A.; Falkner S.; Hutter F.. Bayesian Optimization with Robust Bayesian Neural Networks, NIPS; 2016; pp 4134–4142.
- Jones D. R.; Schonlau M.; Welch W. J. Efficient Global Optimization of Expensive Black-Box Functions. J. Glob. Opt. 1998, 13, 455–492. 10.1023/A:1008306431147. [DOI] [Google Scholar]
- Hernández-Lobato J. M.; Gelbart M.; Hoffman M.; Adams R. P.; Ghahramani Z.. Predictive Entropy Search for Efficient Global Optimization of Black-Box Functions, NIPS; 2014; pp 918–926.
- Hernández-Lobato J. M.; Gelbart M.; Hoffman M.; Adams R. P.; Ghahramani Z.. Predictive Entropy Search for Bayesian Optimziation with Unknown Constraints, Internat. Conf. Mach. Learn.; 2015; pp 1699–1707.
- Srivinas N.; Krause A.; Seeger M.; Kakade S. M.. Gaussian Process Optimization in the Bandit Setting: No Regret and Experimental Design, Proc. Int. Conf. Mach. Learn.; 2010; pp 1015–1022.
- Gómez-Bombarelli R.; Wei J. N.; Duvenaud D.; Hernández-Lobato J. M.; Sánchez-Lengeling B.; Sheberla D.; Aguilera-Iparraguirre J.; Hirzel T. D.; Adams R. P.; Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018, 4, 268–276. 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyzer-Knapp E. O.; Simm G. N.; Aspuru-Guzik A. A Bayesian Approach to Calibrating High-Throughput Virtual Screening Results and Application to Organic Photovoltaic Materials. Mater. Horiz. 2016, 3, 226–233. 10.1039/C5MH00282F. [DOI] [Google Scholar]
- Ju S.; Shiga T.; Feng L.; Hou Z.; Tsuda K.; Shiomi J. Designing Nanostructures for Phonon Transport via Bayesian Optimization. Phys. Rev. X 2017, 7, 021024. 10.1103/PhysRevX.7.021024. [DOI] [Google Scholar]
- Wang Z.; Li C.; Jegelka S.; Kohli P. Batched High-Dimensional Bayesian Optimization via Structural Kernel Learning. 2017, arXiv:1703.01973. arXiv.org e-Print archive. https://arxiv.org/abs/1703.01973.
- Wang Z.; Gehring C.; Kohli P.; Jegelka S.. Batched Large-Scale Bayesian Optimization in High-Dimensional Spaces, Internat. Conf. Artif. Intell. Stat.; 2018; pp 745–754.
- Marmin S.; Chevalier C.; Ginsbourger D. Efficient Batch-Sequential Bayesian Optimization with Moments of Truncated Gaussian Vectors. 2016, arXiv:1609.02700. arXiv.org e-Print archive. https://arxiv.org/abs/1609.02700.
- Contal E.; Buffoni D.; Robicquet A.; Vayatis N. Parallel Gaussian Process Optimization with Upper Confidence Bound and Pure Exploration. Eur. Conf. Mach. Learn. Know. Discovery Databases. 2013, 8188, 225–240. 10.1007/978-3-642-40988-2_15. [DOI] [Google Scholar]
- Desautels T.; Krause A.; Burdick J. W. Parallelizing Exploration-Exploitation Tradeoffs in Gaussian Process Bandit Optimization. J. Mach. Learn. Res. 2014, 15, 3873–3923. [Google Scholar]
- González J.; Osborne M.; Lawrence N.. GLASSES: Relieving the myopia of Bayesian optimization, Artif. Intell. Stat.; 2016; pp 790–799.
- Häse F.; Roch L. M.; Kreisbeck C.; Aspuru-Guzik A.. PHOENICS: A Universal Deep Bayesian Optimizer. https://github.com/aspuru-guzik-group/phoenics (2018). [DOI] [PMC free article] [PubMed]
- Kirkpatrick S.; Gelatt C. D.; Vecchi M. P. Optimization by Simulated Annealing. Science 1983, 220, 671–680. 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Černỳ V. Thermodynamical Approach to the Traveling Salesman Problem: An Efficient Simulation Algorithm. J. Optim. Theory Appl. 1985, 45, 41–51. 10.1007/BF00940812. [DOI] [Google Scholar]
- Eberhart R.; Kennedy J. A New Optimizer Using Particle Swarm Theory. Proc. Int. Symp. - Micro Mach. Hum. Sci. 1995, 39–43. 10.1109/MHS.1995.494215. [DOI] [Google Scholar]
- Shi Y.; Eberhart R. A Modified Particle Swarm Optimizer. IEEE World Cong. Comput. Intell. - Evol. Comput. Proc. 1998, 69–73. 10.1109/ICEC.1998.699146. [DOI] [Google Scholar]
- Jones D. R. A Taxonomy of Global Optimization Methods Based on Response Surfaces. J. Glob. Optim. 2001, 21, 345–383. 10.1023/A:1012771025575. [DOI] [Google Scholar]
- Martinez-Cantin R.; de Freitas N.; Brochu E.; Castellanor J.; Doucet A. A Bayesian Exploration-Exploitation Approach for Optimal Online Sensing and Planning with a Visually Guided Mobile Robot. Autonom. Robots 2009, 27, 29–103. 10.1007/s10514-009-9130-2. [DOI] [Google Scholar]
- Breiman L. Random Forests. Mach. Learn. 2001, 45, 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Escobar M. D.; West M. Bayesian Density Estimation and Inference Using Mixtures. J. Am. Stat. Assoc. 1995, 90, 577–588. 10.1080/01621459.1995.10476550. [DOI] [Google Scholar]
- Hoffman M. D.; Gelman A. The No-U-turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res. 2014, 15, 1593–1623. [Google Scholar]
- Salvatier J.; Wiecki T. V.; Fonnesbeck C. Probabilistic Programming in Python Using PyMC3. PeerJ. Comp. Sci. 2016, 2, e55. 10.7717/peerj-cs.55. [DOI] [Google Scholar]
- Tran D.; Kucukelbir A.; Dieng A. B.; Rudolph M.; Liang D.; Blei D. M.. Edward: A Library for Probabilistic Modeling, Inference, and Criticism. 2016, arXiv:1610.09787. arXiv.org e-Print archive. https://arxiv.org/abs/1610.09787.
- Nadaraya E. A. On Estimating Regression. Theory Probab. Its Appl. 1964, 9, 141–142. 10.1137/1109020. [DOI] [Google Scholar]
- Watson G. S. Smooth Regression Analysis. Sankhya A 1964, 26 (4), 359–372. [Google Scholar]
- Miranda L. J. V. PySwarms, a research-toolkit for Particle Swarm Optimization in Python. JOSS 2018, 3 (21), 433. 10.21105/joss.00433. [DOI] [Google Scholar]
- Hansen N.A Python implementation of CMA-ES. https://github.com/CMA-ES/pycma (2018).
- Field R. J.; Noyes R. M. Oscillations in Chemical Systems. IV. Limit cycle Behavior in a Model of a Real Chemical Reaction. J. Chem. Phys. 1974, 60, 1877–1884. 10.1063/1.1681288. [DOI] [Google Scholar]
- Van Kampen N. G.Stochastic Processes in Physics and Chemistry; Elsevier: Amsterdam, 1992; Vol. 1. [Google Scholar]
- Zhabotinsky A. M. A History of Chemical Oscillations and Waves. Chaos 1991, 1, 379–386. 10.1063/1.165848. [DOI] [PubMed] [Google Scholar]
- Degn H. Effect of Bromine Derivatives of Malonic Acid on the Oscillating Reaction of Malonic Acid, Cerium Ions and Bromate. Nature 1967, 213, 589–590. 10.1038/213589a0. [DOI] [Google Scholar]
- Zhabotinsky A. M.; Zaikin A. N.. Oscillatory Processes in Biological and Chemical Systems; Izdatelstro “Nauka” Publishers: Moscow, 1967. [Google Scholar]
- Field R. J.; Koros E.; Boyes R. M. Oscillations in Chemical Systems. II. Thorough Analysis of Temporal Oscillation in the Bromate-Cerium-Malonic Acid System. J. Am. Chem. Soc. 1972, 94, 8649–8664. 10.1021/ja00780a001. [DOI] [Google Scholar]
- Gyorgyi L.; Turányi R.; Field R. J. Mechanistic Details of the Oscillatory Belousov-Zhabotinski Reaction. J. Phys. Chem. 1990, 94, 7162–7170. 10.1021/j100381a039. [DOI] [Google Scholar]
- Field R. J. Das Experiment: Eine oszillierende Reaktion. Chem. Unserer Zeit 1973, 7, 171–176. 10.1002/ciuz.19730070603. [DOI] [Google Scholar]
- Bar-Eli K. The Minimal Bromate Oscillator Simplified. J. Phys. Chem. 1985, 89, 2855–2860. 10.1021/j100259a030. [DOI] [Google Scholar]
- Voorsluijs V.; Kevrekidis I. G.; De Decker Y. Nonlinear Behavior and Fluctuation-Induced Dynamics in the Photosensitive Belousov-Zhabotinsky Reaction. Phys. Chem. Chem. Phys. 2017, 19, 22528–22537. 10.1039/C7CP03260A. [DOI] [PubMed] [Google Scholar]
- Kuhnert L.Selbstorganisation Chemischer Strukturen: Arbeiten von Friedlieb Ferdinand Runge, Raphael Eduard Liesegang, Boris Pavlovich Belousov u. Anatol Markovich Zhabotinsky; Geest & Portig: Leipzig, 1987. [Google Scholar]
- Peruzzo A.; McClean J.; Shadbolt P.; Yung M. H.; Zhou X. Q.; Love P. J.; Aspuru-Guzik A.; O’brien J. L. A Variational Eigenvalue Solver on a Photonic Quantum Processor. Nat. Commun. 2014, 5, 4213. 10.1038/ncomms5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J.; Olson J. P.; Aspuru-Guzik A. Quantum Autoencoders for Efficient Compression of Quantum Data. Quant. Sci. Technol. 2017, 2, 045001. 10.1088/2058-9565/aa8072. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.