Abstract
Fungi expressing P450nor, an unconventional nitric oxide (NO) reducing cytochrome P450, are considered significant contributors to environmental nitrous oxide (N2O) emissions. Despite extensive efforts, fungal contributions to N2O emissions remain uncertain. For example, the majority of N2O emitted from antibiotic-amended soil microcosms is attributed to fungal activity, yet axenic fungal cultures do not couple N-oxyanion respiration to growth and these fungi produce only minor quantities of N2O. To assist in reconciling these conflicting observations and produce a benchmark genomic analysis of fungal denitrifiers, genes underlying denitrification were examined in >700 fungal genomes. Of 167 p450nor—containing genomes identified, 0, 30, and 48 also harbored the denitrification genes narG, napA, or nirK, respectively. Compared with napA and nirK, p450nor was twice as abundant and exhibited 2–5-fold more gene duplications, losses, and transfers, indicating a disconnect between p450nor presence and denitrification potential. Furthermore, cooccurrence of p450nor with genes encoding NO-detoxifying flavohemoglobins (Spearman r = 0.87, p = 1.6e−10) confounds hypotheses regarding P450nor’s primary role in NO detoxification. Instead, ancestral state reconstruction united P450nor with actinobacterial cytochrome P450s (CYP105) involved in secondary metabolism (SM) and 19 (11%) p450nor-containing genomic regions were predicted to be SM clusters. Another 40 (24%) genomes harbored genes nearby p450nor predicted to encode hallmark SM functions, providing additional contextual evidence linking p450nor to SM. These findings underscore the potential physiological implications of widespread p450nor gene transfer, support the undiscovered affiliation of p450nor with fungal SM, and challenge the hypothesis of p450nor’s primary role in denitrification.
Keywords: P450nor, fungi, nitrous oxide, secondary metabolism, nitrogen cycle, horizontal gene transfer
Introduction
Since the early 20th century, increased human reliance on fixed nitrogen (N) for agricultural practices has contributed to a 20% increase in atmospheric nitrous oxide (N2O), a potent greenhouse gas with ozone destruction potential (Galloway et al. 2008; Canfield et al. 2010). N2O is primarily formed by denitrifying members of the Bacteria (Zumft 1997), a prevailing view that has been challenged by experiments reporting that abundant soil- and sediment-inhabiting fungi contribute up to 89% of the total N2O emitted from these systems (Cathrine and Raghukumar 2009; Mothapo et al. 2015; Wankel et al. 2017). Notably, fungi cannot convert N2O to inert N2 like many denitrifying bacteria (Shoun et al. 2012), suggesting their contributions to greenhouse effects and ozone destruction could be significant. Fungi are considered to be important sources of N2O emissions from agroecosystems (Mothapo et al. 2013; Chen et al. 2014), which are predicted to contribute up to two-thirds of the total N2O emissions by 2030 (Hu et al. 2015). Studies of model fungi show that N2O formation is due to P450nor, a heme-containing cytochrome P450, that catalyzes the two electron reduction of nitric oxide (NO) to N2O (Nakahara et al. 1993; Shiro et al. 1995; Shimizu et al. 2002). N2O formation by P450nor is thought to occur exclusively in fungi and the p450nor gene has been exploited as a distinctive biomarker in molecular assays to study fungal denitrifier diversity and abundance in the environment (Higgins et al. 2016; Li et al. 2016; Novinscak et al. 2016).
Despite these observations, the fungal contributions to N2O emissions remain uncertain. For example, fungi do not satisfy criteria set forth to classify microorganisms as respiratory denitrifiers (Mahne and Tiedje 1995). N2O-producing fungi in pure culture do not exhibit a balance between the inorganic N inputs and quantities of N2O formed (Bleakley and Tiedje 1982; Shoun 1992; Tsuruta et al. 1998) and possess three to six orders of magnitude lower rates of N2O production compared with denitrifying bacterial isolates under optimal conditions (Mothapo et al. 2015). Moreover, fungi fail to generate anoxic growth yields proportional to the quantity of inorganic N reduced in pure culture (Shoun and Tanimoto 1991; Zhou et al. 2001; Cathrine and Raghukumar 2009; Stief et al. 2014), and no significant relationship was detected between fungal denitrification activity and fungal biomass in anoxic soil incubations (Herold et al. 2012). Above all, partitioning techniques (antibiotic inhibition and isotope site preference) used to estimate fungal and bacterial contributions to N2O emissions are biased and often lack corroborating evidence in conjunction with their application, suggesting fungal contributions to N2O emissions are substantially inflated (Rousk et al. 2009; Ladan and Jacinthe 2016; Phillips et al. 2016; Wankel et al. 2017).
The capacity for N2O-production conferred by p450nor in fungi is a uniquely eukaryotic trait, yet previous investigations have hypothesized an actinobacterial origin for p450nor based on sequence comparisons (Kizawa et al. 1991; Moktali et al. 2012; Shoun et al. 2012; Chen et al. 2014). Of note, Actinobacteria are not considered canonical denitrifying bacteria, and only a few reports of their denitrification capacity exist (Shoun et al. 1998; Kumon et al. 2002; Shapleigh 2013). Most members of the Actinobacteria possess a truncated denitrification pathway or lack a canonical nitric oxide reductase gene (nor) (with the exception of Corynebacterium and Propionibacterium) (Kumon et al. 2002; Shapleigh 2013). Hence, members of the Fungi and Actinobacteria share an incomplete denitrification pathway with a potentially limited capacity to perform denitrification. Consistent with the horizontal gene transfer (HGT) hypothesis are sequence similarities between fungal P450nor and actinobacterial P450s of the CYP105 family, many of which have been investigated for their contributions to secondary metabolism (SM) (Shoun et al. 2012; Moody and Loveridge 2014). Despite these observations, the prevailing hypothesis regarding p450nor’s evolution and function was its acquisition from the Actinobacteria and subsequent evolution to fill a novel role in denitrification, specifically the reduction of NO to N2O (Kizawa et al. 1991; Shoun et al. 2012). The hypothesis that p450nor was acquired from one or more members of the Actinobacteria and retained an ancestral function in SM surprisingly remains unexplored.
Therefore, the overarching questions this study addresses are 1) what is the breadth of denitrification genes across fungal genomes and what are their evolutionary relationships, and 2) can phylogenomic analyses reconcile the conflict in fungal contributions to N2O formation observed in laboratory and environmental settings? Our comparative genomic and phylogenetic analyses identified a disconnect between p450nor and denitrification gene presence and supported a role for P450nor in SM rather than denitrification. Importantly, these results provide an explanation for the minor, nonrespiratory capacity of fungi to form N2O, and suggests N2O is a byproduct of active SM. These findings transform our understanding of the ecological significance and environmental consequences of p450nor presence/absence in fungal genomes.
Materials and Methods
Data Sets
Draft and complete fungal, algal, and bacterial genomes were accessed from the National Center for Biotechnology Information and the Joint Genome Institute on March 16, 2016 and downloaded from their respective database utilities. A list of fungal, algal, and bacterial genomes and their taxonomic and database affiliations can be found at https://doi.org/10.6084/m9.figshare.c.3845692.v4 (figshare Data Set S1, see Data Sharing).
Gene Marker Identification
To identify gene markers within fungal genomes suitable for phylogenetic analysis, a database of 1,438 amino acid sequences of fungal single copy orthologs from the BUSCO tool v1.1b (Simão et al. 2015) were provided as queries to the genblastG search tool v1.0.138 (She et al. 2011). The genblastG tool performs amino acid alignment of protein queries against a six frame translated nucleotide subject sequence (genome) to find significant alignments and uses heuristic analysis to piece the appropriate gene models back together from high-scoring segment pairs identified using BLAST (Altschul et al. 1990). Of the BUSCO gene models queried, 238 were used for phylogenetic tree reconstruction and were annotated using PfamScan against the Pfam A database and blastp against the uniprot database with default settings (Mistry et al. 2007; Finn et al. 2010, 2015; Li et al. 2015) (figshare Data Set S2, see Data Sharing). The genblastG tool was also used to detect gene sequences involved in denitrification (NapA, NarG, NirK/NirS, NorB, P450nor, and NosZ) from curated bacterial and fungal proteins in the FunGene repository (Fish et al. 2013) or proteins involved in NO detoxification (flavohemoglobins) identified in the literature (Wisecaver et al. 2016). Denitrification gene models used in downstream phylogenetic analysis were manually curated against full length fungal reference sequences to ensure that accurate gene models were predicted for each organism in which the gene was detected. After identification of these genes in fungal genomes, alignment of the fungal NapA, NirK, NarG, and P450nor amino acid sequences with blastp against the plant, archaea, bacteria, protozoa, and fungi RefSeq protein databases (Pruitt et al. 2007) was performed to identify similar sequences in each taxonomic group. Protein sequences demonstrating significant alignment (≥60% query coverage and ≥35% amino acid identity) to fungal proteins were used in subsequent phylogenetic reconstructions.
Gene Prediction for Comparative Genomic Analyses
The ab initio gene predictor SNAP (Korf 2004) was used to predict gene models in fungal genomes where no such information was available (e.g., some draft genomes). In this case, one or several closely related fungal genomes containing gene models were selected based on phylogenetic affiliation to train SNAP for gene prediction. Although this methodology is limiting when closely related genomes are unavailable, gene models from close relatives were available for p450nor-containing genomes lacking gene predictions. All gene predictions performed with SNAP are provided in a FigShare repository (see Data Sharing below).
Analysis of SM Gene Clusters in Fungi
Genomic regions 50 kb up- and downstream of a p450nor gene in each genome were subjected to gene cluster prediction with the antiSMASH and CASSIS tools with default settings (Inglis et al. 2013; Wolf et al. 2016). Additionally, genes encoded ±10 genes up and downstream of p450nor were evaluated using PfamScan searches with default settings against the profile Hidden Markov Models (pHMMs) of curated SM genes identified by antiSMASH (figshare Data Set S3, see Data Sharing) (Inglis et al. 2013). Protein sequences with significant alignment to antiSMASH pHMMs were given an “automatic” SM function status and were colored blue. In order to supplement the automated SM annotation, additional functional annotation was performed by hmmscan searches with HMMER3 (Eddy 2009) against the eggNOG database (Powell et al. 2012). These functional annotations were manually flagged as related to SM if they possessed literature entries suggesting an involvement in SM or had functions related to methyl transfer, oxidation–reduction reactions, glycosyl transferases, fungal specific transcription factors, and other protein functions that may be important for SM outlined by antiSMASH (Inglis et al. 2013). All manual SM annotations were colored light blue to indicate potential involvement in SM. All other annotations were colored grey when no evidence connecting the function to SM could be identified.
Pairwise nucleotide alignments of p450nor-containing genomic regions were performed as previously described (Richter and Rosselló-Móra 2009). Briefly, the nucmer utility of MUMmer v3.0 (Kurtz et al. 2004) was used to align p450nor-containing genomic regions (∼100 kb) against whole genomes of fungi with and without p450nor. The average nucleotide identity (reported as ANIm) of the alignment was calculated from the resulting delta output file. The resulting data was plotted using Matplotlib (Hunter 2007) available for the python programming language (http://www.python.org).
Phylogenetic Analysis
Phylogenetic reconstruction of the fungal species tree was performed using concatenated amino acid sequences from 238 single copy orthologs found in ≥ 90% of all genomes (figshare Data Set S2, see Data Sharing). The genomes of Puccinia arachidis and Microbotryum lychnidis-dioicae strain p1A1 Lamole were excluded from further analysis due to an insufficient number of informative sites and inconsistent placement within the fungal tree. Alignment of amino acid sequences were performed individually on all 238 individual BUSCO gene models present within each organism using MAFFT v7.130b (Katoh et al. 2009) with linsi alignment tuning parameters (–maxiterate 1000 and –localpair settings used). Individual alignments were concatenated using in-house python scripts, resulting in a 65, 897 column alignment. Tree reconstruction was performed using FastTree2 (Price et al. 2010) with refined tree reconstruction settings for a slower, more exhaustive search of the tree space than default settings (-bionj -slow -gamma -spr 4 -lg -mlacc 2 and -slownni settings) (Wisecaver et al. 2016). For comparison to tree reconstruction using a concatenated alignment, individual trees from each BUSCO alignment were also constructed using FastTree2 with identical settings as above. The resultant alignments and trees were subjected to coalescent tree reconstruction using ASTRAL-II software (Mirarab and Warnow 2015). Overall, both phylogenies largely agreed except for branching patterns of some lineages (e.g., Zoopagomycota and Mucoromycota) and are available online in a Figshare repository (see Data Sharing below).
The predicted amino acid and intronless nucleotide sequences of fungal napA, nirK, and p450nor gene models were aligned using the MAFFT settings described above and manually refined in JalView and SeaView software (Galtier et al. 1996; Waterhouse et al. 2009). Maximum-likelihood (ML) and Bayesian phylogenetic tree reconstruction was performed on both nucleotide and amino acid alignments using RAxML and MrBayes, respectively (Stamatakis 2006; Ronquist et al. 2012). For details regarding phylogenetic tree parameters, please refer to Supplementary Material.
BayesTraits software was used to perform phylogenetically informed correlations between binary traits (i.e., the presence or absence of two denitrification markers) and ancestral state reconstruction (Pagel et al. 2004). Please refer to Supplementary Material for additional details on BayesTraits analyses.
Approximately unbiased (AU) tests were performed in the program CONSEL (Shimodaira and Hasegawa 2001) using default settings. The negative log likelihood values from the observed nucleotide phylogenies input into CONSEL were −140,261, −37,782, −111,531 for napA, nirK and p450nor, respectively. The observed negative log likelihood scores for amino acid phylogenies of NapA, NirK, and P450nor were −65,158, −14,197, and −44,171, respectively. Species-tree gene-tree reconciliation was performed using NOTUNG software v2.9 (Chen et al. 2000; Stolzer et al. 2012). Please see Supplementary Material for further details on NOTUNG parameters.
Statistical Analyses
All statistical analyses were carried out in R programming language (Team 2012) and significance of statistical tests were assessed using a P value cutoff ≤0.05.
Data Sharing
All Supplementary Material figures, tables, and data sets, as well as gene models, alignments, and trees reported in the manuscript are made available in a Figshare data collection prepared by S.A.H. at https://doi.org/10.6084/m9.figshare.c.3845692.v4, last accessed September 3, 2018.
Results
Infrequent Cooccurrence among Denitrification Genes in Fungi
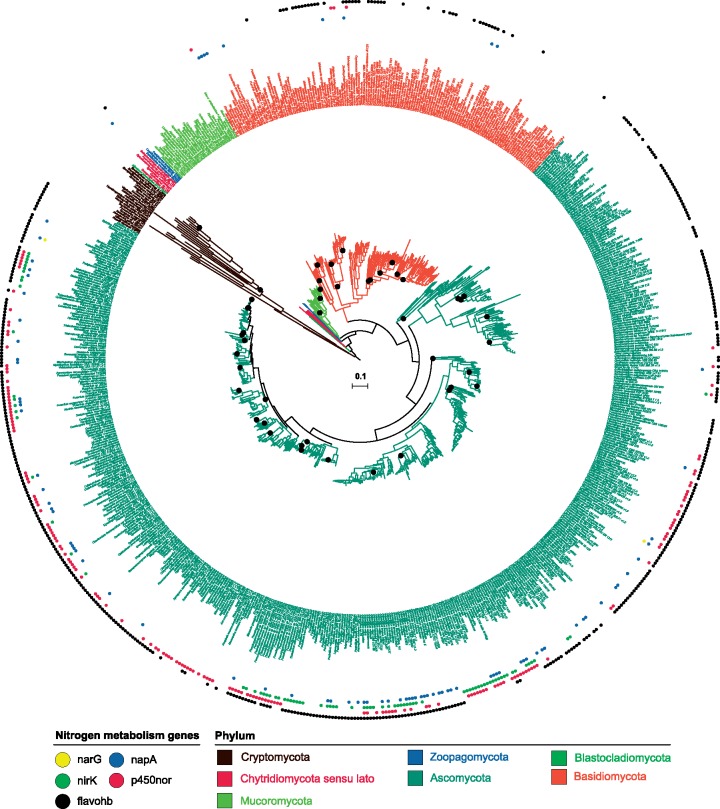
Bioinformatic analyses identified homologs of canonical bacterial and fungal denitrification genes (narG, napA, norB, nirK, nosZ, and p450nor) in 712 fungal genomes. Of the denitrification gene set investigated, only narG, napA, nirK, and p450nor were detected (fig. 1). Genes encoding the membrane bound respiratory nitrate reductase (narG) were detected in only three fungal genomes (0.42%) and were excluded from further analysis due to their low occurrence. The genes predicted to encode the periplasmic nitrate reductase (NapA) and the copper-containing nitrite reductase (NirK) were detected in 75 (10.5%) and 82 (11.5%) of the 712 fungal genomes analyzed, respectively (fig. 1 and supplementary table S1, Supplementary Material online). In contrast, P450nor gene sequences occurred at approximately twice the frequency in 167 (23%) of the fungal genomes analyzed, supporting the claim that P450nor-mediated N2O production may be widespread in fungi (fig. 1) (Maeda et al. 2015). A breakdown of genus- and family-level denitrification gene abundances in fungal genomes underscores the disparity in presence/absence of denitrification genes in fungi and is available in Supplementary Material (figshare Data Set S4, fig. S1).
Fig. 1.
—Maximum-Likelihood phylogeny of the kingdom Fungi inferred from a concatenated alignment of 238 single copy marker gene amino acid sequences (see Materials and Methods). Black circles marking branches indicate nodes with bootstrap percentages below 90%. Colored markers outside taxon names specify the presence or absence of each gene (narG, napA, nirK, p450nor, and flavohemoglobin) within a fungal genome. Flavohb, flavohemiglobin genes involved in NO detoxification. The scale bar (center of tree) represents amino acid substitutions per site.
Our analyses also revealed a low cooccurrence between p450nor and additional fungal denitrification pathway markers. Since p450nor is regarded as the sole trait encoding N2O production in fungi, the cooccurrence of multiple denitrification gene markers would be indicative of a capacity for sequential respiratory denitrification, whereas isolated occurrences could be indicative of alternative processes such as detoxification. The three-gene set narG/nirK/p450nor did not cooccur in any of the fungal genomes examined, whereas cooccurrence of the gene set napA/nirK/p450nor was observed in 18 (10.8%) of 167 p450nor-containing fungal genomes. Sets of at least two cooccurring denitrification traits (i.e., narG/p450nor, napA/p450nor, and nirK/p450nor) were found in 0, 18 and 29% of fungal genomes, respectively. Of the napA-containing fungal genomes, 25 (33%) also contained a nirK gene, whereas 30% of the nirK-containing fungal genomes also harbored a napA gene. Evolutionary correlation was strongly supported for the gene sets napA/nirK, napA/p450nor, and nirK/p450nor, with average log Bayes Factor values of 31.9 ± 0.60, 12.2 ± 0.11, and 31.3 ± 0.04, respectively. Hence, the genes napA, nirK, and p450nor occur in related fungal taxa, but cooccurrences were infrequent within the individual fungal genomes analyzed.
Evolutionary Forces Acting upon Denitrification Traits within Fungi
To identify evolutionary forces shaping the observed distribution of denitrification traits within fungi, comparisons between gene and species trees were assessed with phylogenetic tests and parsimony-informed models to quantify evolutionary events. Visual inspection of p450nor gene and species trees indicated potential widespread HGT of p450nor within fungi, examples of which included HGT of p450nor from the phylum Ascomycota to members of the Basidiomycota and within and among classes of ascomycetes (supplementary fig. S2, Supplementary Material online). Furthermore, the monophyly of five fungal classes containing p450nor (Dothideomycetes, Eurotiomycetes, Leotiomycetes, Sordariomycetes, and Tremellomycetes) were not supported by approximately unbiased (AU) tests (P ≤ 0.05, supplementary table S2, Supplementary Material online), indicative of dynamic evolution of p450nor in most fungal lineages. Although cophylogeny plots are suggestive of HGT, additional analysis using NOTUNG software was performed to model potential gene duplication (GD), gene transfer (GT), and gene loss (GL) events (Stolzer et al. 2012). Of the napA, nirK, and p450nor genes analyzed, the p450nor phylogenies had the greatest number of predicted GT events, ranging from 4 to 15 GT events despite applying stringent GT costs within NOTUNG software (supplementary table S3, Supplementary Material online). At GT costs below 9, no temporally consistent optimal solutions were reached, suggesting that GD and GL alone are insufficient to describe the evolutionary dynamics of p450nor in fungi. Using the same stringent GT costs, the predicted number of GT events detected for napA and nirK were much lower, and ranged from 1 to 3 and 0 to 1 GT events for each gene, respectively (supplementary table S3, Supplementary Material online). The reduced number of GT events detected in napA and nirK phylogenies were also apparent from cophylogeny plots of each gene (supplementary figs. S3 and S4, Supplementary Material online) compared with cophylogenetic plots for p450nor (supplementary fig. S2, Supplementary Material online). Although GT events detected for napA were lower than p450nor at high GT costs, GT may still represent a significant evolutionary force contributing to the observed napA distribution in extant fungal lineages (supplementary table S3, Supplementary Material online). For example, AU tests rejected the monophyly of three Ascomycota (Dothideomycetes, Leotiomycetes, and Sordariomycetes) and one Basidiomycota (Pucciniomycetes) lineage within the napA phylogeny (supplementary table S2, Supplementary Material online, P ≤ 0.05). Specific instances of predicted HGT events for each gene are outlined in Supplementary Material (supplementary table S4 and fig. S5, Supplementary Material online).
Fungal P450nor Evolved from Actinobacterial P450s Involved in SM
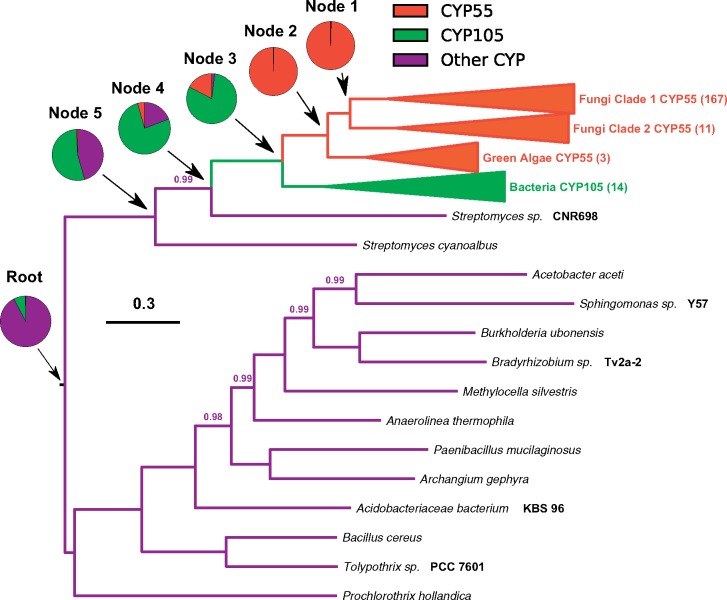
Previous investigations have hypothesized an actinobacterial origin for p450nor based on amino acid sequence alignments (Moktali et al. 2012; Shoun et al. 2012; Chen et al. 2014), but rigorous phylogenetic tests of p450nor’s origins were lacking to support this hypothesis. Alignment of fungal P450nor amino acid sequences to the NCBI RefSeq protein database identified 230 bacterial sequences with significant sequence alignment (≥65% query coverage and ≥35% amino acid identity) to P450nor. Of note, p450nor homologs were also detected within the genomes of three freshwater inhabiting green algae, Chlorella variabilis, Chlamydomonas reinhardtii, and Monoraphidium neglectum, expanding the known distribution of p450nor to photosynthetic eukaryotic microbes. Additional p450nor homologs were not detected in archaea, plant, protist, or other lineages housed within the RefSeq database. Of the bacterial cytochrome P450 (hereafter P450) sequences identified, approximately 6% (n = 13) were proteobacterial in origin, whereas the remaining sequences belonged to members of the bacterial phylum Actinobacteria (supplementary fig. S6, Supplementary Material online). Ancestral character state reconstruction of select P450 families on a subset of these sequences supported the monophyly of p450nor and bacterial P450 gene sequences of the P450 family CYP105 (fig. 2) (Nelson 2009). The same relationships were preserved when phylogenetic reconstruction was performed using the complete set of 408 P450 amino acid sequences (supplementary fig. S7, Supplementary Material online). Importantly, NO-utilizing P450 sequences from the CYP107 family belonging to members of the Streptomyces formed a larger monophyletic clade containing P450nor and other CYP105 sequences (supplementary fig. S7, Supplementary Material online). The CYP107 family includes txtE genes encoding nitrating enzymes that use NO as a substrate for the production of secondary metabolites and have no known role in respiratory denitrification or detoxification (Barry et al. 2012; Dodani et al. 2014). Thus, P450nor and TxtE are related (Barry et al. 2012; Dodani et al. 2014), yet TxtE is involved in SM and is the only other P450 observed to directly utilize NO as a substrate.
Fig. 2.
—Midpoint-rooted Bayesian phylogeny of select families of cytochrome P450 amino acid sequences from fungi, algae, and bacteria. Ancestral state reconstruction was performed using CYP55 (orange), CYP105 (green), and other CYPs (purple) to uncover the shared ancestry of algal and fungal N2O-producing cytochrome P450s with their most recent common bacterial ancestor. The scale bar indicates substitutions per site and posterior probability values <1 are displayed above branches of the Bayesian MCMC analysis. Numbers in parentheses next to collapsed clades indicate the number of sequences in the clade. Values in pie charts are average probabilities of each character state across one representative Bayesian MCMC analysis.
Sequences of the bacterial CYP105 family of P450s include diverse actinobacterial genera such as Streptomyces (n = 159), Amycolatopsis (n = 12), Saccharothrix (n = 5), Streptacidiphilus (n = 4), Frankia (n = 4), Kutzneria (n = 4), Nocardia (n = 3), and members from 17 additional actinobacterial genera (n = 39). The proteobacterial sequences were affiliated with members of the genera Burkholderia (n = 5), Paracoccus (n = 3), Bradyrhizobium (n = 3), Pseudomonas (n = 1), and Halomonas (n = 1). Bacterial P450 gene and species tree comparisons of 60% identity clustered P450 amino acid sequences (n = 57) and cognate 16S rRNA genes (n = 55) supported HGT of one or more actinobacterial P450 genes to members of the Alpha-, Beta-, and Gammaproteobacteria (supplementary fig. S6, Supplementary Material online). Furthermore, ancestral character state reconstruction overwhelmingly supported Actinobacteria as the root state (root probability = 0.99 ± 0.06) of the bacterial CYP105 family P450 phylogeny. When forcing the root state of the P450 phylogeny to be Proteobacteria (simple model) and comparing to the complex model where the root is allowed to vary, the simpler model with a proteobacterial root was not supported (average log Bayes Factor = 0.03 ± 0.18). Therefore, p450nor likely evolved from one or more CYP105 family P450 genes found in members of the Actinobacteria. This finding underscores p450nor’s distinct origin compared with the fungal denitrification traits napA and nirK, which exhibit a proteobacterial ancestry consistent with the majority of bacterial denitrifiers (supplementary fig. S8, Supplementary Material online).
Widespread Cooccurrence of p450nor and NO-Detoxifying Flavohemoglobins
Poor conversion of inorganic N-oxides to N2O by fungal isolates supports the hypothesis that P450nor is involved in NO detoxification (Morozkina and Kurakov 2007; Shoun et al. 2012). Fungi also possess NO-detoxifying flavohemoglobins responsible for detoxification of NO to under oxic conditions or NO to N2O under anoxic conditions (Poole and Hughes 2000; Morozkina and Kurakov 2007; Wisecaver et al. 2016). Flavohemoglobins were detected in 450 (63%) fungal genomes investigated and were widespread within ascomycete and basidiomycete fungi. Within p450nor-containing genomes, 125 (75%) also possessed a flavohemoglobin gene (fig. 1 and supplementary table S1, Supplementary Material online). Furthermore, the number of genomes in fungal families containing p450nor and NO-detoxifying flavohemoglobin genes were significantly correlated (Spearman r = 0.87, p = 1.6e−10), suggesting a competitive or compensatory functioning of P450nor with flavohemoglobins in fungi where they coexist.
Evidence of a Role for p450nor in Secondary Metabolism
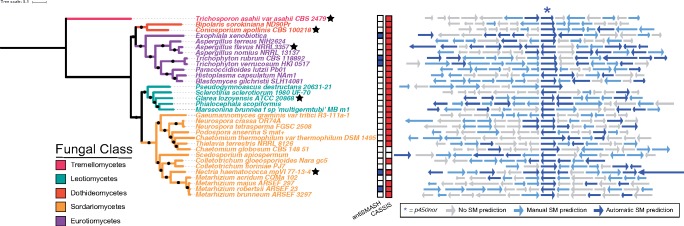
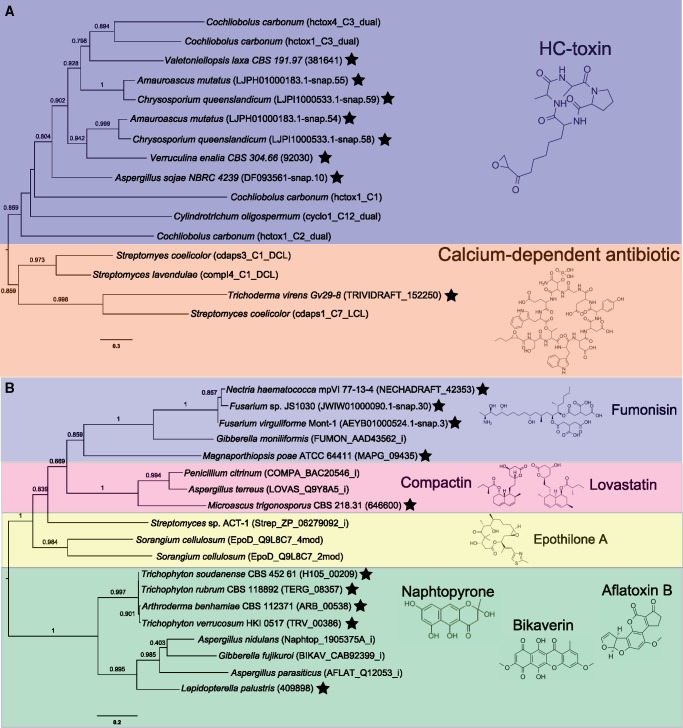
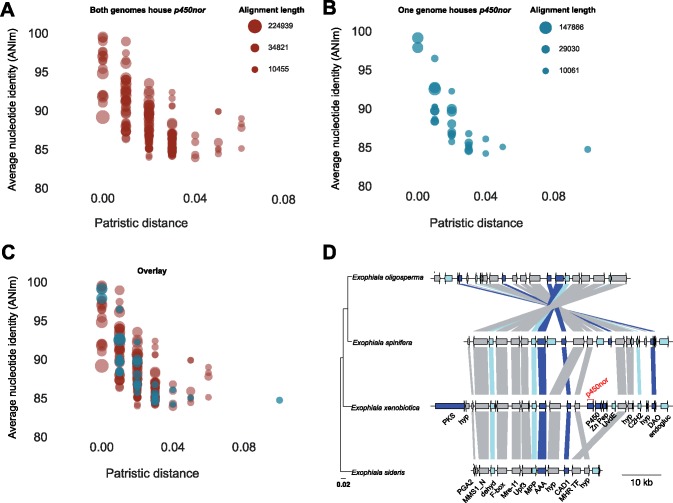
p450nor is actinobacterial in origin, yet Actinobacteria are not considered canonical denitrifiers and evidence for their role in denitrification was lacking when p450nor was initially identified (Kaspar 1982; Kizawa et al. 1991). Subsequent investigations did not posit a role for p450nor in SM despite the affiliation of p450nor and CYP105 P450s with documented roles in SM (O'Keefe and Harder 1991; Moody and Loveridge 2014). To assess genomic evidence linking p450nor to SM, we queried genes encoded within genomic regions approximately 50 kb on either side of p450nor for functions related to SM. The biosynthetic gene cluster (BGC) prediction tool antiSMASH detected putative BGCs containing p450nor in 19 (11%) of the 167 p450nor-containing genomes analyzed (figshare Data Set S3). The number of open reading frames in a predicted SM cluster ranged from 34 to 97, spanning 21,086 to 55,473 nucleotides in length. Inspection of protein-coding genes surrounding p450nor using curated antiSMASH pHMMs resulted in the identification of hallmark SM features (e.g., polyketide synthases (PKS), nonribosomal peptide synthases (NRPS), terpene cyclases, dimethylallyl tryptophan synthases) in an additional 40 (24%) of the 167 p450nor-containing genomes analyzed (figshare Data Set S3, see Materials and Methods for details). The distribution of automatic and manually curated protein-coding genes surrounding a subset of 32 p450nor-containing fungi suggests that p450nor-containing BGCs are structurally and functionally diverse (fig. 3). An additional BGC prediction tool, CASSIS, which detects BGCs based on shared transcription factor binding sites upstream and downstream of a user specified anchor gene (p450nor) (Wolf et al. 2016), predicted as many as 105 (63%) p450nor-containing gene regions to be BGCs (figshare Data Set S5). Furthermore, CASSIS analysis corroborated 74% of the 19 BGCs predicted by antiSMASH (figshare Data Set S5). A detailed accounting of antiSMASH and CASSIS predictions, gene annotations, and gene organization surrounding p450nor in all 167 p450nor-containing genomes is available in the Supplementary Material (supplementary fig. S9 and figshare Data Set S5). A diversity of secondary metabolite biosynthesis pathways were predicted to be encoded by p450nor-containing BGCs, including nonribosomal peptides (n = 7), polyketides (n = 5), terpenes (n = 2), hybrid terpene-polyketide-indoles (n = 2), indoles (n = 1), or currently unclassifiable compounds (n = 2). Phylogenetic reconstruction of C-type and ketosynthase domains encoded by NRPS and PKS genes surrounding p450nor enabled the prediction of potential secondary metabolites encoded by fungal p450nor-containing BGCs (fig. 4). Of the 94 fungal genera harboring p450nor, 21 (22%) genera contained species with and without a copy of p450nor (supplementary table S5, Supplementary Material online). Nucleotide alignments of p450nor-containing genomic regions (81.3 ± 27.8 kb in length) against other fungal genomes revealed a disproportionately high nucleotide identity and alignment length between genomes with and without p450nor from the same genus (fig. 5A–C). For example, genomic regions surrounding p450nor in Exophiala xenobiotica are highly conserved in other Exophiala species without p450nor (fig. 5D), and additional examples of large, high identity regions between closely related fungal genomes with and without p450nor are abundant (fig. 5A–C and figshare Data Set S6).
Fig. 3.
—SM gene cluster predictions for a subset of 32 (167 total) p450nor-containing fungi. The boxes to the right of the rooted Maximum-Likelihood phylogeny indicate whether the p450nor-containing genomic region was predicted by antiSMASH (blue squares) or CASSIS (red squares) to be an SM gene cluster. White squares indicate no prediction. Colored arrows indicate protein-coding genes surrounding p450nor that were automatically predicted (dark blue arrow), manually predicted (light blue arrow) or not predicted (grey arrow) to be involved in SM (see Materials and Methods for details). The black stars next to species names are individuals chosen for in depth presentation of the genes surrounding p450nor (supplementary fig. S9, Supplementary Material online).
Fig. 4.
—Maximum-Likelihood phylogenetic trees of nonribosomal peptide synthase and polyketide synthase domains encoded within p450nor-containing genomic regions. Each phylogeny displays relationships of C-type condensation (C-type) (A) or ketosynthase (KS) domains (B) detected in nonribosomal peptide and polyketide synthase amino acid sequences, respectively, encoded within p450nor-containing genomic regions. A black star next to taxa indicates C-type or KS domains identified in fungal genomes nearby p450nor. The NCBI or JGI accession numbers are shown in parentheses next to taxa with black stars. Taxa without black stars are reference amino acid sequences of C-type and KS domains curated by the NAPDOS database, and their NAPDOS accession numbers are indicated in parentheses. Proteins from species without accession numbers were predicted ab initio using SNAP (Korf 2004) (see Materials and Methods for details). Chemical structures and names of secondary metabolites produced by NAPDOS reference sequences are indicated and highlighted distinct colors for clarity. Scale bars indicate amino acid substitutions per site. Values along branches indicate bootstrap support for the adjacent node.
Fig. 5.
—Within genera alignments of p450nor-containing genomic regions (N = 136) from species with and without p450nor. Alignment of two species with p450nor-containing genomic regions (red circles) (A) or between two species where only one member possesses p450nor (teal circles) (B). The overlay of the two plots in (C) indicates conservation of genomic architecture regardless of p450nor presence or absence. The size of the circles is proportional to the square root of the aligned length of the genomic regions. The gene synteny plot (D) highlights conservation of genomic architecture using a p450nor-containing genomic region from Exophiala xenobiotia aligned to three additional closely-related Exophiala species that do not possess p450nor. The arrows represent gene models within the gene region displayed. Both arrows and connecting lines above and below arrows are colored according to figure 3 (see Materials and Methods for details). Lines connecting arrows between species indicate the genes are homologous. The scale bars (left to right) in (D) indicate substitutions per site and genome size in kilobases, respectively. The location of p450nor in E. xenobiotica is indicated in red font. The labels in black font describes the putative functions of proteins encoded by each gene. PKS, polyketide synthase; hyp, hypothetical protein; PGA2, protein trafficking protein; MMS1_N, MMS1-like protein; dehyd, dehydrogenase; F-box, F-box domain containing protein; Mre-11, double strand break repair protein Mre-11; Upf3, nonsense mediated mRNA decay protein 3; MPP, metallophosphatase; AAA, ATPase, CAD1, cinnamyl alcohol dehydrogenase; MHR TF, middle homology region transcription factor; P450, cytochrome P450 reductase; Zn Pep, zinc peptidase superfamily protein; UvdE, UV-endonuclease; C2H2, zinc finger C2H2 type; DAO, D-amino acid oxidase; endogluc, endoglucanase.
Discussion
Hypotheses Regarding the Biological Role of p450nor
The leading hypotheses regarding the biological role of fungal p450nor are respiratory denitrification (Shoun et al. 2012), hybrid respiration of N-oxides and O2 (Takaya et al. 2003), NO detoxification (Morozkina and Kurakov 2007), and based on the work presented here, secondary metabolism. The respiratory denitrification hypothesis is misleading since evidence is lacking to classify fungi as respiratory denitrifiers (Shoun 1992; Mahne and Tiedje 1995; Tsuruta et al. 1998; Guengerich and Munro 2013; Mothapo et al. 2015). Furthermore, unaccounted for methodological biases inherent to partitioning techniques, especially antibiotic inhibition, raises substantial concerns over the validity of fungal N2O production in situ (Rousk et al. 2009; Mothapo et al. 2015; Ladan and Jacinthe 2016; Phillips et al. 2016). For example, site preference measurements of the intramolecular distribution of 15N within the linear N2O molecule (i.e., N2O isotopocules) of cultured microorganisms have been increasingly applied to partition microbial sources of N2O in situ (Toyoda et al. 2017; Wankel et al. 2017). Although promising, the limitations of N2O isotopocule measurements used in isolation are becoming apparent (Decock and Six 2013; Yang et al. 2014; Phillips et al. 2016). Of primary concern is the significant overlap in, and difficulty discretizing, site preference measurements of distinct processes or diverse microbial assemblages (Baggs 2008; Butterbach-Bahl et al. 2013; Yang et al. 2014). In addition to isotopocule measurements, antibiotic inhibition has been frequently used to assess the contributions of fungi and bacteria to N2O formation (Mothapo et al. 2015). For example, streptomycin, a widely applied bactericide used to inhibit bacterial N2O emissions, only inhibits a fraction of the active bacteria present in soil (Rousk et al. 2009; Ladan and Jacinthe 2016). The ineffectiveness of antibiotics to partition microbial respiration has been previously demonstrated (Rousk et al. 2009), yet antibiotics continue to be used to support the prevalence of fungal respiratory denitrification in situ. Therefore, the respiratory denitrification hypothesis is predicated on biased approaches often used in isolation that are unable to correctly assess fungal contributions to denitrification.
Another hypothesis for the retention or acquisition of denitrification traits in fungi is their role in hybrid respiration and survival under hypoxic conditions. Hybrid respiration involves the flow of electrons to two terminal electron acceptors (e.g., O2 and ) simultaneously when O2 is limiting (Takaya et al. 2003). Presumably, electron flow via terminal N-oxide reductases conserves additional energy under O2-limiting conditions (Chen and Strous 2013; Simon and Klotz 2013). Hybrid respiration has been investigated in Fusarium oxysporum strain MT-811 (Takaya et al. 2003), but the effect of denitrification trait presence/absence on fungal survival under hypoxic/anoxic conditions has yet to be determined. The only proteins encoded in the fungal genomes examined and linked to proton motive force generation were NarG and NirK (Takaya et al. 2003; Chen and Strous 2013; Simon and Klotz 2013), the former of which was only detected in three genomes, suggesting a limited role for in fungal hybrid respiration. NirK contributes indirectly to proton translocation via the activity of the cytochrome bc1 complex (Takaya et al. 2003), but investigations examining the contributions of NirK to proton motive force generation in fungi exposed to hypoxic conditions are lacking to support this hypothesis. The gene encoding the catalytic subunit of the periplasmic nitrate reductase, NapA, was detected at much greater frequency than NarG (10.5% of genomes examined), but NapA is not involved in proton translocation and energy conservation (Simon and Klotz 2013). Finally, P450nor is a soluble cytochrome P450 with no known role in energy conservation in fungi (Shoun et al. 2012). Hence, it remains to be determined whether denitrification trait acquisition/retention is driven by episodic O2 deficiency experienced by fungi in the environment.
Another alternative function suggested for P450nor is NO detoxification, which was initially postulated in experiments using the fungus Fusarium oxysporum strain 11n1 (Kurakov et al. 2000). This hypothesis was supported by low growth yields and a poor mass balance between the N-oxyanion inputs and N2O formed by the fungus (Shoun 1992; Tsuruta et al. 1998; Rohe et al. 2014). Although plausible, the NO detoxification hypothesis is confounded by extensive cooccurrence between p450nor and genes encoding canonical NO-detoxifying flavohemoglobins, which also produce N2O under anoxic conditions (Zumft 2005; Wisecaver et al. 2016) (fig. 1 and supplementary table S1, Supplementary Material online). The binding affinity of NO by P450nor is reported between 0.1 and 0.6 mM (Nakahara et al. 1993; Shiro et al. 1995) and is orders of magnitude higher than the 0.1 to 0.25 µM NO binding affinity reported for flavohemoglobins (Gardner et al. 2000), suggesting flavohemoglobins would outcompete P450nor for NO binding at physiological NO concentrations. Thus, the higher affinity of flavohemoglobins for NO and their greater distribution in fungi (supplementary table S1, Supplementary Material online) would suggest a limited role for P450nor in NO detoxification.
The SM hypothesis has traction considering that P450nor is derived from CYP105 P450s (fig. 2), all of which share a functional role in SM (O'Keefe and Harder 1991; Yasutake et al. 2007; Moody and Loveridge 2014). Thus, the adaptation of P450nor to a novel niche in NO reduction and denitrification is unlikely. A more parsimonious hypothesis is that P450nor has maintained a role in SM as observed for related actinobacterial enzymes. The monophyly of P450nor with the SM enzyme TxtE, the only other NO-utilizing P450, provides additional a priori support for P450nor’s role in SM (supplementary fig. S7, Supplementary Material online). P450nor’s role in SM is further corroborated by SM prediction tools where a sizeable proportion (35%) of gene regions surrounding p450nor contained genes predicted to encode hallmark SM functions, and as many as 105 (63%) p450nor-containing genomic regions were automatically predicted to be involved in SM (fig. 3). Although phylogenomic evidence supports a role for P450nor in the biosynthesis of secondary metabolites, direct physiological evidence should be a target for future research efforts. For example, transcriptomic approaches may provide corroborating evidence of P450nor’s role in SM, but experimental conditions used in investigations of p450nor-containing fungi are not optimized for p450nor gene expression (i.e., growth with low O2 and N-oxyanions present) (Delmas et al. 2012; Ehrlich and Mack 2014; Pullan et al. 2014). Further, transcriptomic analysis of fungal SM clusters requires knowledge of the specific growth conditions for their induction under laboratory conditions (Inglis et al. 2013). Emerging technologies enabling the expression of full length BGCs and metabolite identification should enable robust experimentation to test the SM hypothesis (Clevenger et al. 2017).
Predicting P450nor’s Role in Secondary Metabolism
A variety of metabolites containing nitro functional groups have been detected in fungal genera known to harbor denitrifying representatives (Chen et al. 2016), yet mechanistic explanations for nitration reactions in fungi remain elusive. The addition of a nitro functional group to a metabolite represents a potential mechanism for enhancing its toxicity or functional specificity (Kovacic and Somanathan 2014). The hypothesis of a role for P450nor in nitration, or possibly nitrosylation, of fungal metabolites is attractive given P450nor’s affiliation with the nitrating enzyme TxtE (Barry et al. 2012) (supplementary fig. S7, Supplementary Material online). The inclusion of p450nor within BGCs may be adaptive in fungal lineages in which this gene was acquired due to the augmenting effects nitro or nitroso groups impart on their substrates. Support for this hypothesis stems from the widespread distribution of p450nor within secondary metabolite producing members of the Ascomycota (Keller 2015; Macheleidt et al. 2016), and previous reports of HGT between members of Actinobacteria and fungi in enhancing fungal SM (Janssen et al. 2002). Furthermore, the high nucleotide identity shared between p450nor-containing genomic regions from closely related fungal species suggests p450nor gain or loss may have important consequences for the secondary metabolites potentially produced by the enzymatic machinery encoded in p450nor-containing BGCs (fig. 5D). The variability in p450nor gene presence/absence among closely related fungal species suggests p450nor is not essential for SM (supplementary table S5, Supplementary Material online). Indeed, genomic regions between closely related species with and without p450nor contain canonical SM elements (fig. 5D). Although p450nor may not be essential for SM generally, it may provide a selective advantage in specific contexts (e.g., plant pathogenesis) or environments (e.g., N-oxide rich [agricultural] soils). Specifically, the introduction of a nitro/nitroso group catalyzed by P450nor would result in a secondary metabolite with altered functionality with benefits to the host. Future research should explore the essential or ancillary role p450nor plays in fungal SM.
Additional unknowns related to P450nor’s role in SM is the identification of putative substrates and the sources of NO required to fuel the hypothesized nitration or nitrosylation reactions. To date, P450nor is solely reported to bind the electron donors NADH or NADPH and the electron acceptor NO (Shoun et al. 2012). However, N2O formation by P450nor is O2 dependent (Zhou et al. 2001; Mothapo et al. 2013), suggesting O2 may be an additional substrate as observed for TxtE (Barry et al. 2012). TxtE and NovI, both P450s affiliated with P450nor, bind to and transform L-tryptophan and L-tyrosine to produce the secondary metabolites thaxtomin A and novobiocin, respectively (Chen and Walsh 2001; Barry et al. 2012). It is conceivable that P450nor might also bind O2 and aromatic amino acids, but direct experimental evidence is required to confirm this hypothesis. A potential source of NO in fungi could result from nitrite reductase activity of the copper containing nitrite reductase, NirK. The NO synthase (TxtD) from Streptomyces turgidiscabies produces NO to fuel TxtE nitration of L-tryptophan (Barry et al. 2012), but txtD homologs were not detected in the fungal genomes examined. Although evidence of NO synthases in fungi exist, knowledge regarding their distribution is limited (Ninnemann and Maier 1996; Samalova et al. 2013). Given the functional redundancy between NO synthases and NirK, it is conceivable that one of NirK’s functions in fungi is to generate NO for use by P450nor in SM.
Causes and Consequences of p450nor Evolution in Fungi
A limited understanding of p450nor evolution represented an impediment to our knowledge of fungal N2O formation. For example, closely related fungi vary in their ability to produce N2O (Shoun 1992; Tsuruta et al. 1998; Maeda et al. 2015; Higgins et al. 2016), and the evolutionary forces (e.g., HGT, gene gain/loss, and incomplete lineage sorting) contributing to this observation were unexplored. For p450nor, many HGT events were observed between distantly related fungal lineages using gene and species tree comparisons (supplementary fig. S2, Supplementary Material online). Although HGT events are challenging to precisely quantify given the level of uncertainty in deeply branching nodes of the functional gene trees reported here, a signal of potentially double digit HGT events were observed using gene tree-species tree reconciliation (supplementary table S3, Supplementary Material online). Genetic elements encoding pogo family transposases (N = 9), retrotransposons (N = 4), and reverse transcriptases (N = 1) were in some cases detected adjacent to p450nor and may act as vehicles for dissemination of p450nor within fungi and between fungal chromosomes (figshare Data Set S3).
N2O production was previously coined a widespread trait in fungi (Maeda et al. 2015), yet genomic analysis suggests fortuitous N2O formation by fungi is largely restricted to members of the Ascomycota. N2O production has been reported for fungal isolates assigned to the recently revised phylum Mucoromycota (Mothapo et al. 2015; Spatafora et al. 2016), yet no evidence of genes underlying denitrification were detected in available genomes from members of this phylum (fig. 1). Denitrification markers were also absent from ascomycete yeast genomes (i.e., Candida, Yarrowia), though a number of N2O-producing ascomycete yeasts have been reported (Tsuruta et al. 1998). Even within the Basidiomycota, N2O formation is restricted to a few taxa within the Tremellomycetes and Agaricomycetes (Mothapo et al. 2015), and at least for members of the Tremellomycetes, was likely the result of HGT from one or more members of the Ascomycota (supplementary fig. S2, Supplementary Material online). The finding that genomes from fungi (e.g., ascomycete yeasts) previously observed to produce N2O did not possess denitrification traits was unexpected and suggests that experimental artifacts or other mechanisms, such as the NO-detoxifying activity of flavohemoglobins, may also contribute to N2O formation in fungi. In addition to fungi, species of green algae have been reported to produce small quantities of N2O, the production of which could, at least in part, be attributed to the presence of p450nor within this lineage (Weathers 1984; Guieysse et al. 2013; Plouviez et al. 2017). However, like fungi, respiratory denitrification by green algae is unlikely (Weathers 1984; Plouviez et al. 2017); thus, the SM hypothesis appears to be an attractive explanation for the presence of p450nor in green algae as well.
p450nor genes within fungi also have implications for fungal pathogenesis (Mothapo et al. 2015). At least for some bacteria (e.g., Neisseria, Brucella, and Mycobacterium), the presence of denitrification genes has been demonstrated to enhance virulence or detoxification of N-oxides produced by the host (Philippot 2005). Although the impact of denitrification gene acquisition on fungal pathogenesis is not well established, there is growing evidence for P450nor involvement in fungal virulence (Shoun et al. 2012; Mothapo et al. 2015). For example, p450nor gene expression is linked to Fusarium wilt in banana and cotton plants, yet mechanistic explanations of P450nor’s function during plant infection are lacking (McFadden et al. 2006; Sutherland et al. 2013). Notably, more than half of all p450nor-containing fungal species are known plant pathogens (Mothapo et al. 2015), and the involvement of p450nor in SM is consistent with and would support the plant pathogenic life history strategies of many p450nor-containing fungi.
The diversity of denitrifying microorganisms and the modularity of the pathway has led to the view of denitrification as a community function (Philippot 2002; Jones et al. 2011; Graf et al. 2014). Therefore, limited cooccurrence and correlated evolution between napA, nirK, and p450nor might suggest mutualistic interactions occur between fungal or bacterial species performing denitrification. However, gene cooccurrences and evolutionary correlations should be interpreted with caution as additional factors (e.g., shared ecological niche, selection pressures) related to fungal life history strategies may explain their distribution equally well. For example, N2O-producing fungi are frequently detected in, and cultivated from, highly disturbed, N-amended agricultural soils (Chen et al. 2014; Mothapo et al. 2015; Higgins et al. 2016) and detoxification or N-oxide utilization traits may merely cooccur more frequently due to selection imposed by environments with regular N inputs. Fungi also contain genes homologous to bacterial denitrifiers, but their presence does not guarantee a role in respiratory denitrification. For example, the presence of genes homologous to the bacterial NO reductase (norB) is not sufficient evidence for respiratory denitrification potential in pathogenic bacteria (Mahne and Tiedje 1995; Philippot 2005). The same is true of the abundant napA gene homologs detected in fungal genomes, which would suggest a robust capacity of fungi to perform dissimilatory nitrate reduction. Yet this is not the case, and many fungi only produce N2O when is present (Shoun 1992; Tsuruta et al. 1998; Mothapo et al. 2015). Hence, the role of the napA gene in fungal biology is intriguing since they exist in >10% of the 712 fungal genomes examined.
In summary, fungi often produce little or no gaseous N from reduction of N-oxyanions and do not grow proportionally to the quantity of N-oxyanions consumed; thus, fungi cannot be classified as respiratory denitrifiers (Mahne and Tiedje 1995). Given the limited accounting of methodological bias in the study of N2O production by fungi (Rousk et al. 2009; Ladan and Jacinthe 2016; Phillips et al. 2016), alternative explanations for the biological function of p450nor in fungi are likely and raises concerns over the validity of these techniques in estimating fungal contributions to N2O emissions. NO detoxification by P450nor is plausible, but their cooccurrence with flavohemoglobins and weak binding affinity for NO suggests insufficient evidence exists to support the NO detoxification hypothesis. The added benefit of hybrid respiration using N-oxyanions in concert with O2 is also a possible explanation for the acquisition/retention of denitrification traits in fungi, but evidence of growth or cell maintenance conferred by these traits under hypoxic conditions is currently lacking. The analyses presented here support the affiliation of p450nor with nondenitrifying actinobacterial sequences involved in SM and provide multiple lines of evidence for their inclusion in candidate BGCs. Nevertheless, a premier goal of future research should be direct experimentation to support or refute a role for P450nor in SM. Recent developments in technologies geared at characterizing fungal SM products and identification of novel fungal BGCs should facilitate these experiments. Considering that many putatively denitrifying fungi are also plant disease causing secondary metabolite producers and agricultural pests, our results strongly endorse a biological role for p450nor in SM rather than denitrification.
Supplementary Material
Acknowledgments
We thank Gerald Bills, Gregory Bonito, Pedro Crous, Kathryn Bushley, Colleen Hansel, Patrik Inderbitzin, Gabor Kovacs, Bjorn Lindahl, Jon Magnuson, Francis Martin, Kerry O’Donnell, Nancy Nichols, Minou Nowrousian, and Joseph Spatafora for providing access to unpublished genome data produced by the U.S. Department of Energy Joint Genome Institute. The authors thank A. Frank for assistance with manuscript revisions. S.A.H. would like to acknowledge U.S. Department of Energy, Office of Science, Office of Workforce Development for Teachers and Scientists, Office of Science Graduate Student Research (SCGSR) program for support during preparation of the manuscript. The SCGSR program is administered by the Oak Ridge Institute for Science and Education for the DOE under contract number DE‐AC05‐06OR23100. This work was supported by the US Department of Energy, Office of Biological and Environmental Research, Genomic Science Program [DE-SC0006662]. Participation of C.W.S and F.E.L. was partially sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the U. S. Department of Energy.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Baggs EM. 2008. A review of stable isotope techniques for N2O source partitioning in soils: recent progress, remaining challenges and future considerations. Rapid Commun Mass Spectrom. 22(11):1664–1672. [DOI] [PubMed] [Google Scholar]
- Barry SM, et al. 2012. Cytochrome P450—catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat Chem Biol. 8(10):814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleakley BH, Tiedje JM.. 1982. Nitrous oxide production by organisms other than nitrifiers or denitrifiers. Appl Environ Microbiol. 44(6):1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterbach-Bahl K, Baggs EM, Dannenmann M, Kiese R, Zechmeister-Boltenstern S.. 2013. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos Trans Biol Sci USA. 368(1621):20130122–20130113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield DE, Glazer AN, Falkowski PG.. 2010. The evolution and future of Earth’s nitrogen cycle. Science 330(6001):192–196. [DOI] [PubMed] [Google Scholar]
- Cathrine SJ, Raghukumar C.. 2009. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycol Res. 113(1):100–109. [DOI] [PubMed] [Google Scholar]
- Chen H, Mothapo NV, Shi W.. 2014. The significant contribution of fungi to soil N2O production across diverse ecosystems. Appl Soil Ecol. 73:70–77. [Google Scholar]
- Chen H, Walsh CT.. 2001. Coumarin formation in novobiocin biosynthesis: β-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem Biol. 8(4):301–312. [DOI] [PubMed] [Google Scholar]
- Chen J, Strous M.. 2013. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim Biophys Acta 1827(2):136–144. [DOI] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M.. 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol. 7(3–4):429–447. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. 2016. Chrysamides A–C, three dimeric nitrophenyl trans-epoxyamides produced by the deep-sea-derived fungus Penicillium chrysogenum SCSIO41001. Org Lett. 18(15):3650–3653. [DOI] [PubMed] [Google Scholar]
- Chen W, et al. 2014. Fungal cytochrome P450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol Evol. 6(7):1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger KD, et al. 2017. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol. 13(8):895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decock C, Six J.. 2013. How reliable is the intramolecular distribution of 15N in N2O to source partition N2O emitted from soil? Soil Biol Biochem. 65:114–127. [Google Scholar]
- Delmas S, et al. 2012. Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet. 8(8):e1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodani SC, et al. 2014. Structural, functional, and spectroscopic characterization of the substrate scope of the novel nitrating cytochrome P450 TxtE. Chembiochem 15(15):2259–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23(1):205–211. [PubMed] [Google Scholar]
- Ehrlich CK, Mack MB.. 2014. Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins (Basel) 6(6):1916.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38(Database issue):D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J, et al. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol. 4:291.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JN, et al. 2008. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320(5878):889–892. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C.. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 12(6):543–548. [DOI] [PubMed] [Google Scholar]
- Gardner PR, et al. 2000. Nitric-oxide dioxygenase activity and function of flavohemoglobins: sensitivity to nitric oxide and carbon monoxide inhibition. J Biol Chem. 275(41):31581–31587. [DOI] [PubMed] [Google Scholar]
- Graf DRH, Jones CM, Hallin S.. 2014. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9(12):e114118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP, Munro AW.. 2013. Unusual cytochrome P450 enzymes and reactions. J Biol Chem. 288(24):17065–17073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guieysse B, Plouviez M, Coilhac M, Cazali L.. 2013. Nitrous oxide (N2O) production in axenic Chlorella vulgaris microalgae cultures: evidence, putative pathways, and potential environmental impacts. Biogeosciences 10(10):6737–6746. [Google Scholar]
- Herold MB, Baggs EM, Daniell TJ.. 2012. Fungal and bacterial denitrification are differently affected by long-term pH amendment and cultivation of arable soil. Soil Biol Biochem. 54:25–35. [Google Scholar]
- Higgins SA, et al. 2016. Detection and diversity of fungal nitric oxide reductase genes (p450nor) in agricultural soils. Appl Environ Microbiol. 82(10):2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H-W, Chen D, He J-Z.. 2015. Microbial regulation of terrestrial nitrous oxide formation: understanding the biological pathways for prediction of emission rates. FEMS Microbiol Rev. 39(5):729–749. [DOI] [PubMed] [Google Scholar]
- Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng. 9(3):90–95. [Google Scholar]
- Inglis D, et al. 2013. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 13(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M.. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 68(5):2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, et al. 2011. Phenotypic and genotypic heterogeneity among closely related soil-borne N2- and N2O-producing Bacillus isolates harboring the nosZ gene. FEMS Microbiol Ecol. 76(3):541–552. [DOI] [PubMed] [Google Scholar]
- Kaspar HF. 1982. Nitrite reduction to nitrous oxide by propionibacteria: detoxification mechanism. Arch Microbiol. 133(2):126–130. [Google Scholar]
- Katoh K, Asimenos G, Toh H.. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 537:39–64. [DOI] [PubMed] [Google Scholar]
- Keller NP. 2015. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol. 11(9):671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizawa H, et al. 1991. Nucleotide sequence of the unique nitrate/nitrite-inducible cytochrome P-450 cDNA from Fusarium oxysporum. J Biol Chem. 266:10632–10637. [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic P, Somanathan R.. 2014. Nitroaromatic compounds: environmental toxicity, carcinogenicity, mutagenicity, therapy and mechanism. J Appl Toxicol. 34(8):810–824. [DOI] [PubMed] [Google Scholar]
- Kumon Y, et al. 2002. Codenitrification and denitrification are dual metabolic pathways through which dinitrogen evolves from nitrate in Streptomyces antibioticus. J Bacteriol. 184(11):2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakov AV, Nosikov AN, Skrynnikova EV, L’vov NP.. 2000. Nitrate reductase and nitrous oxide production by Fusarium oxysporum 11dn1 under aerobic and anaerobic conditions. Curr Microbiol. 41(2):114–119. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladan S, Jacinthe P-A.. 2016. Evaluation of antibacterial and antifungal compounds for selective inhibition of denitrification in soils. Environ Sci Process Impacts. 18:1519–1529. [DOI] [PubMed] [Google Scholar]
- Li W, et al. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43(W1):W580–W584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. 2016. A new fluorescence biosensor for nitric oxide detection based on cytochrome P450 55B1. Sensors Actuators B Chem. 230:405–410. [Google Scholar]
- Macheleidt J, et al. 2016. Regulation and role of fungal secondary metabolites. Annu Rev Genet. 50(1):371–392. [DOI] [PubMed] [Google Scholar]
- Maeda K, et al. 2015. N2O production, a widespread trait in fungi. Sci Rep. 5(1):9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahne I, Tiedje JM.. 1995. Criteria and methodology for identifying respiratory denitrifiers. Appl Environ Microbiol. 61(3):1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden HG, Wilson IW, Chapple RM, Dowd C.. 2006. Fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) genes expressed during infection of cotton (Gossypium hirsutum). Mol. Plant Pathol. 7(2):87–101. [DOI] [PubMed] [Google Scholar]
- Mirarab S, Warnow T.. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31(12):i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Bateman A, Finn RD.. 2007. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 8(1):298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moktali V, et al. 2012. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics 13:525.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody SC, Loveridge EJ.. 2014. CYP105—diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J Appl Microbiol. 117(6):1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozkina EV, Kurakov AV.. 2007. Dissimilatory nitrate reduction in fungi under conditions of hypoxia and anoxia: a review. Appl Biochem Microbiol. 43(5):544–549. doi: 10.1134/S0003683807050079. [PubMed] [Google Scholar]
- Mothapo N, et al. 2015. Phylogenetic, taxonomic and functional diversity of fungal denitrifiers and associated N2O production efficacy. Soil Biol Biochem. 83:160–175. [Google Scholar]
- Mothapo NV, Chen H, Cubeta MA, Shi W.. 2013. Nitrous oxide producing activity of diverse fungi from distinct agroecosystems. Soil Biol Biochem. 66:94–101. [Google Scholar]
- Nakahara K, Tanimoto T, Hatano K, Usuda K, Shoun H.. 1993. Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J Biol Chem. 268:8350–8355. http://www.jbc.org/content/268/11/8350.abstract. [PubMed] [Google Scholar]
- Nelson DR. 2009. The cytochrome P450 homepage. Hum Genomics 4(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninnemann H, Maier J.. 1996. Indications for the occurrence of nitric oxide synthases in fungi and plants and the involvement in photoconidiation of Neurospora crassa. Photochem Photobiol. 64(2):393–398. [DOI] [PubMed] [Google Scholar]
- Novinscak A, et al. 2016. Novel P450nor gene detection assay used to characterize the prevalence and diversity of soil fungal denitrifiers. Appl Environ Microbiol. 82(15):4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe DP, Harder PA.. 1991. Occurrence and biological function of cytochrome P450 monooxygenases in the actinomycetes. Mol Microbiol. 5(9):2099–2105. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D.. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 53(5):673–684. [DOI] [PubMed] [Google Scholar]
- Philippot L. 2005. Denitrification in pathogenic bacteria: for better or worst? Trends Microbiol. 13(5):191–192. [DOI] [PubMed] [Google Scholar]
- Philippot L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim Biophys Acta 1577(3):355–376. http://www.sciencedirect.com/science/article/pii/S0167478102004207. [DOI] [PubMed] [Google Scholar]
- Phillips RL, et al. 2016. Chemical formation of hybrid di-nitrogen calls fungal codenitrification into question. Sci Rep. 6(1):39077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouviez M, et al. 2017. The biosynthesis of nitrous oxide in the green alga Chlamydomonas reinhardtii. Plant J. 91:45–56. [DOI] [PubMed] [Google Scholar]
- Poole RK, Hughes MN.. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 36(4):775–783. [DOI] [PubMed] [Google Scholar]
- Powell S, et al. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40(D1):D284–D289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR.. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35(Database):D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullan ST, et al. 2014. RNA-sequencing reveals the complexities of the transcriptional response to lignocellulosic biofuel substrates in Aspergillus niger. Fungal Biol Biotechnol. 1(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 106(45):19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe L, et al. 2014. Dual isotope and isotopomer signatures of nitrous oxide from fungal denitrification—a pure culture study. Rapid Commun Mass Spectrom. 28(17):1893–1903. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Demoling LA, Bååth E.. 2009. Contrasting short-term antibiotic effects on respiration and bacterial growth compromises the validity of the selective respiratory inhibition technique to distinguish fungi and bacteria. Microb Ecol. 58(1):75–85. [DOI] [PubMed] [Google Scholar]
- Samalova M, et al. 2013. Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytol. 197(1):207–222. [DOI] [PubMed] [Google Scholar]
- Shapleigh JP. 2013. Denitrifying prokaryotes In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: prokaryotic physiology and biochemistry. Berlin, Heidelberg: Springer: p. 405–425. [Google Scholar]
- She R, et al. 2011. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics 27(15):2141–2143. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Park S-Y, Shiro Y, Adachi S.. 2002. X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution. Acta Crystallogr. D 58(1):81–89. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Shiro Y, et al. 1995. Spectroscopic and kinetic studies on reaction of cytochrome P450nor with nitric oxide: implication for its nitric oxide reduction mechanism. J. Biol. Chem 270(4):1617–1623. [DOI] [PubMed] [Google Scholar]
- Shoun H. 1992. Denitrification by fungi. FEMS Microbiol Lett. 94(3):277–281. [DOI] [PubMed] [Google Scholar]
- Shoun H. 1992. Fungal denitrification and cytochrome P-450. J Agric Chem Soc Jpn. 66:154–157. [Google Scholar]
- Shoun H, Fushinobu S, Jiang L, Kim S-W, Wakagi T.. 2012. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos Trans R Soc Lond B Biol Sci. 367(1593):1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoun H, Kano M, Baba I, Takaya N, Matsuo M.. 1998. Denitrification by actinomycetes and purification of dissimilatory nitrite reductase and azurin from Streptomyces thioluteus. J Bacteriol. 180(17):4413–4415. http://jb.asm.org/content/180/17/4413.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoun H, Tanimoto T.. 1991. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J Biol Chem. 266(17):11078–11082. [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Simon J, Klotz MG.. 2013. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta 1827(2):114–135. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, et al. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycology 108(5):1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stief P, et al. 2014. Dissimilatory nitrate reduction by Aspergillus terreus isolated from the seasonal oxygen minimum zone in the Arabian Sea. BMC Microbiol. 14:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzer M, et al. 2012. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28(18):i409–i415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland R, Viljoen A, Myburg AA, van den Berg N.. 2013. Pathogenicity associated genes in Fusarium oxysporum f. sp. cubense race 4. S. Afr J Sci. 109(5/6):1–10. [Google Scholar]
- Takaya N, et al. 2003. Hybrid respiration in the denitrifying mitochondria of Fusarium oxysporum. J Biochem. 133(4):461–465. [DOI] [PubMed] [Google Scholar]
- Team RDC. 2012. R: A language and environment for statistical computing R Foundation for Statistical Computing: Vienna, Austria. http://www.r-project.org/.
- Toyoda S, Yoshida N, Koba K.. 2017. Isotopocule analysis of biologically produced nitrous oxide in various environments. Mass Spectrom Rev. 36(2):135–160. [DOI] [PubMed] [Google Scholar]
- Tsuruta S, et al. 1998. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168(1):105–110. [DOI] [PubMed] [Google Scholar]
- Wankel SD, et al. 2017. Evidence for fungal and chemodenitrification based N2O flux from nitrogen impacted coastal sediments. 8:15595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weathers PJ. 1984. N2O evolution by green algae. Appl Environ Microbiol. 48(6):1251–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver JH, Alexander WG, King SB, Todd Hittinger C, Rokas A.. 2016. Dynamic evolution of nitric oxide detoxifying flavohemoglobins, a family of single-protein metabolic modules in Bacteria and Eukaryotes. Mol Biol Evol. 33:1979–1987. [DOI] [PubMed] [Google Scholar]
- Wolf T, Shelest V, Nath N, Shelest E.. 2016. CASSIS and SMIPS: promoter-based prediction of secondary metabolite gene clusters in eukaryotic genomes. Bioinformatics 32(8):1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gandhi H, Ostrom NE, Hegg EL.. 2014. Isotopic fractionation by a fungal P450 nitric oxide reductase during the production of N2O. Environ Sci Technol. 48(18):10707–10715. [DOI] [PubMed] [Google Scholar]
- Yasutake Y, et al. 2007. Crystal structure of cytochrome P450 MoxA from Nonomuraea recticatena (CYP105). Biochem Biophys Res Commun. 361(4):876–882. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Takaya N, Sakairi MAC, Shoun H.. 2001. Oxygen requirement for denitrification by the fungus Fusarium oxysporum. Arch Microbiol. 175(1):19–25. [DOI] [PubMed] [Google Scholar]
- Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 61(4):533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. 2005. Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme–copper oxidase type. J Inorg Biochem. 99(1):194–215. [DOI] [PubMed] [Google Scholar]
- 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43(D1):D204–D212. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.