Abstract
Isolated hepatocytes and liver S9 fractions have been used to collect in vitro biotransformation data for fish as a means of improving modeled estimates of chemical bioaccumulation. To date, however, there have been few direct comparisons of these two methods. In the present study, cryopreserved trout hepatocytes were used to measure in vitro intrinsic clearance rates for 6 polycyclic aromatic hydrocarbons (PAHs). These rates were extrapolated to estimates of in vivo intrinsic clearance and used as inputs to a well-stirred liver model to predict hepatic clearance. Predicted rates of hepatic clearance were then evaluated by comparison to measured rates determined previously using isolated perfused livers. Hepatic clearance rates predicted using hepatocytes were in good agreement with measured values (< 2.1 fold difference for 5 of 6 compounds) under two competing binding assumptions. These findings, which may be attributed in part to high rates of PAH metabolism, are similar to those obtained previously using data from liver S9 fractions. For one compound (benzo[a]pyrene), the in vivo intrinsic clearance rate calculated using S9 data was 10-fold higher than that determined using hepatocytes, possibly due to a diffusion limitation on cellular uptake. Generally, however, there was good agreement between calculated in vivo intrinsic clearance rates obtained using either in vitro test system. These results suggest that both systems can be used to improve bioaccumulation assessments for fish, particularly when vitro rates of activity are relatively high, although additional work is needed to determine if the chemical domain of applicability for each system differs.
Keywords: Hepatocytes, Liver S9 fractions, Perfused liver, Intrinsic clearance, Hepatic clearance, Bioaccumulation
INTRODUCTION
Regulatory authorities in developed countries around the world require bioaccumulation assessments for hydrophobic organic compounds produced or imported in large quantities [1,2]. Because these compounds tend to partition out of water and into the lipid phase of an organism, such assessments are often focused on chemical accumulation in fish. Standardized aqueous and dietary testing protocols (e.g., OECD TG 305 [3]) may be used to directly measure this accumulation, which is typically expressed in terms of a steady-state bioconcentration factor (BCF; for water-only exposures) or dietary biomagnification factor (BMF). However, these approaches are costly, time-consuming, and use large numbers of animals. It is of interest, therefore, to develop predictive in silico and in vitro methods that can be used to perform screening-level bioaccumulation assessments [4].
One of the largest sources of uncertainty in bioaccumulation modeling efforts with fish relates to the role of chemical biotransformation [5]. Biotransformation may substantially reduce the extent to which a chemical accumulates in fish, but the rate of this activity cannot be easily predicted from a compound’s physicochemical characteristics (e.g., its octanol/water partition coefficient, KOW). One approach to this problem is to measure the rate of biotransformation using an in vitro assay and then extrapolate this information to the intact animal as a means of refining modeled bioaccumulation predictions. Several investigators have demonstrated the potential of this approach [6–12]. However, questions remain concerning the extrapolation procedure itself, and whether a particular in vitro system is best suited for this application.
The procedures used to extrapolate in vitro rates of biotransformation to the intact animal are based in part on methods developed by the pharmaceutical industry for preclinical screening of drug candidates [4, 7, 13]. Briefly, metabolic activity is characterized under non-saturating conditions using a substrate depletion approach. The resulting depletion rate constant is used to calculate an in vitro intrinsic clearance rate, which is then extrapolated to an estimate of hepatic clearance (CLH) using one or more extrapolation factors and a physiological model of the liver. Finally, the calculated CLH is extrapolated to a first-order whole-body metabolism rate constant (kMET), which is used as an input to well-known mass-balance models for chemical accumulation.
Most research in this area has been performed using isolated hepatocytes and liver S9 fractions from rainbow trout [6, 9, 11,12 14, 15]. This primary focus on a single species reflects the availability of established methods for collecting this biological material from trout, as well as the existence of physiological and anatomical information required to develop necessary extrapolation factors. The advantages conferred by each of these in vitro systems in terms of cost, ease of use, enzyme coverage, and potential working lifetime have been discussed elsewhere [16,17]. To our knowledge, however, there has only been one study which directly compared the ability of both systems to metabolize a set of chemical substrates [9]. In this earlier effort, in vitro rates of metabolism were determined for 5 compounds using trout liver microsomes and S9 fractions. These rates were then extrapolated to estimates of CLH and compared to CLH values determined previously [6] using data from freshly isolated cells. Although CLH rates determined using microsomes and S9 fractions showed excellent agreement, these values were 3.0- to 10.6-fold lower than those obtained using hepatocytes. Further, it was suggested that BCFs predicted using hepatocyte data were in better agreement with measured values than BCFs generated using S9 or microsomal data.
A recognized barrier to making these types of comparisons is the need to use well-matched biological material (hepatocytes, S9 fractions, etc.), ideally obtained from the same species, strain, and age of fish, which have been held under similar conditions. Additional challenges derive from uncertainties in the extrapolation factors required to predict CLH from in vitro intrinsic clearance (e.g., liver hepatocellularity, the S9 content of liver tissue, and hepatic blood flow), as well as potential effects of chemical binding on metabolic activity [4,7,13]. Finally, if measured levels of chemical accumulation (e.g., BCFs) are used to evaluate the accuracy of in vitro-in vivo extrapolations, uncertainties exist that relate to allometric scaling of biotransformation, the compound’s apparent volume of distribution (required to extrapolate CLH to kMET), and the unknown influence of extrahepatic metabolism [18].
To provide a more straightforward basis for evaluating in vitro-based metabolism predictions, Nichols et al. [19] measured the hepatic clearance of 6 polycyclic aromatic hydrocarbons (PAHs) by isolated perfused trout livers. Within the same study, liver S9 fractions from the same lot of fish were used to perform in vitro substrate depletion assays for the same chemicals at matched test concentrations. The in vitro data were then extrapolated to the intact liver using a well-stirred liver model. To evaluate the influence of chemical binding on clearance predictions, unbound chemical concentrations in vitro and in liver perfusates were measured using a solid phase microextraction (SPME) technique. The results showed that in vitro data derived from liver S9 fractions were, in general, predictive of measured clearance by perfused livers (< 3-fold prediction error for 5 of 6 compounds).
In the present study, we sought to extend these observations by performing in vitro substrate depletion assays for the same 6 PAHs using cryopreserved trout hepatocytes. Previous work has shown that cryopreserved trout hepatocytes retain > 70% of their initial (fresh) biotransformation capability for an extended period of time [15, 20]. This important finding means that one cell lot can be used to perform a substantial number of experiments. As in the study by Nichols et al. [19], binding effects on in vitro activity were accounted for by measuring unbound chemical concentrations in the test system. The primary goals of this effort were to directly compare in vivo intrinsic clearance values estimated using cryopreserved trout hepatocytes and liver S9 fractions, and to evaluate the accuracy of hepatocyte-based CLH predictions by comparison to measured levels of clearance by isolated perfused livers.
MATERIALS AND METHODS
Chemicals
Dulbecco’s modified Eagle’s medium (low glucose; DMEM), Hanks’ balanced salt solution with and without calcium chloride/magnesium sulfate (CaHBSS and HBSS, respectively), and Leibovitz-15 (L-15) medium were purchased from Life Technologies (Carlsbad, CA). All other chemicals, including fetal bovine serum (FBS), fraction V bovine serum albumin (BSA), collagenase IV, dimethyl sulfoxide (DMSO), trypan blue, naphthalene (NAPH), fluorene (FLU), anthracene (ANTH), phenanthrene (PHEN), pyrene (PYR), and benzo[a]pyrene (BAP) were obtained from Sigma-Aldrich (St. Louis, Missouri). All test chemicals were reagent grade (≥ 97% purity).
Animals
Rainbow trout (Oncorhynchus mykiss; Erwin strain) weighing approximately 50 g were obtained from the USGS Upper Midwest Environmental Sciences Center in La Crosse, WI, and grown to the desired size (482 ± 109 g) for these studies. The fish were fed a commercial trout chow (Classic Trout; Skretting USA) and maintained on a 16:8-h light:dark cycle at 11 ± 1 °C. Water used for fish holding was obtained directly from Lake Superior (single pass, sand filtered and UV treated) and had the following chemical characteristics: alkalinity, 41–44 mg/L as CaCO3; pH, 7.6–7.8; total ammonia, < 1 mg/L; dissolved O2, 85 to 100% of saturation. All animals were fasted for 24 h prior to use.
Isolation and cryopreservation of primary trout hepatocytes
Primary trout hepatocytes were obtained as described previously [17], with minor modifications. Livers were perfused with approximately 100 mL of HBSS containing 4.2 mM NaHCO3 and 2.3 mM EDTA, then 100 mL of CaHBSS containing 4.2 mM NaHCO3 and 0.25 mg/mL collagenase IV, and finally 25 mL of DMEM containing 4.2 mM NaHCO3 and 1% BSA. The cells were mechanically dissociated from the tissue, filtered through a 100 μm nylon filter (BD Falcon) and centrifuged at 52 g for 5 min (4 °C). Purification was achieved using a Percoll density gradient (DMEM containing 1% BSA, 4.2 mM NaHCO3 and 42% Percoll). The cells were then washed twice with CaHBSS, resuspended in L-15 medium and counted in the presence of 0.08% trypan blue. Cell viability was greater than 95% for all cell isolations. All buffers were pre-chilled to 11°C and adjusted to pH 7.8.
Primary cells were pelleted at 52 g for 5 min (4 °C) and suspended in a volume of icecold cryopreservation medium (DMEM containing 20% FBS and 0.25% BSA, pH 7.8) calculated to yield 10 × 106 cells/mL. The cells were centrifuged at 52 g for 5 min (4 °C), the supernatant was removed, and cells were resuspended in cryopreservation medium at a volume equal to one-third the previous volume. Cryopreservation medium containing 12% (v/v) of DMSO was slowly added to the cell suspension up to 50% of the previous volume. After 5 min on ice, cryopreservation medium containing 16% (v/v) of DMSO was slowly added up to the original volume of the cell suspension. The final suspension of approximately 10 × 106 cells/ml and 10% DMSO was kept on ice for an additional 15 min. The cells were suspended by gentle swirling, and aliquoted as 1.5 mL into pre-chilled 1.8 mL cryogenic vials. The vials were then placed in the vapor phase of liquid nitrogen for freezing and storage.
Thawing of cryopreserved trout hepatocytes
Cell suspensions in cryogenic vials were thawed in a room temperature water bath until only a small ice crystal remained. The contents of two vials were added to 42 mL of roomtemperature recovery medium (DMEM containing 10% FBS and 0.25% BSA, pH 7.8). One milliliter of recovery medium was then used to rinse each vial, and this volume was added to the bulk recovery medium. Cells used for characterization assays (EROD, UGT, GST) were pelleted at 52 g for 5 min (11 °C), the supernatant was aspirated, and cells were resuspended with 100 mM phosphate buffer (pH 7.8 at 11°C). Cells prepared for PAH depletion studies were washed twice with ice-cold L-15 after the initial pelleting step and resuspended in 1.5 mL of L-15. Subsamples of this suspension were stained with 0.08 % trypan blue and counted to determine cell concentration and viability. The suspension was diluted with additional ice-cold L-15 to obtain approximately 2 ×106 cells/mL. A final cell count was then performed to determine the cell concentration specific to each substrate depletion experiment.
Characterization assays
Cryopreserved cells from each lot were characterized for their ability to deethylate ethoxyresorufin to resorufin (EROD activity), which is a surrogate measurement for CYP1A activity. Additional assays were performed using 1-chloro-2,4-dinitrobenzene (CDNB) and pnitrophenol (p-NP) as substrates for glutathione-S-transferase (GST) and uridine diphosphate (UDP)-glucuronosyltransferase (UGT), respectively. All characterization assays were performed on cell lysates. The methods used to conduct these assays are described elsewhere [19, 20].
Determination of in vitro and in vivo intrinsic clearance (CLIN VITRO,INT,HEP and CLIN VIVO,INT,HEP)
In vitro intrinsic clearance rates (CLIN VITRO,INT,HEP; mL/h/106 cells) were determined for NAPH, FLU, ANTH, PHEN, PYR, and BAP using a substrate depletion approach. Live and heat-denatured (control) hepatocyte suspensions in L-15 medium (1.6 mL; 2×106 cells/mL) were incubated in a shaking water bath maintained at 11 °C. Samples were pre-incubated for 15 min prior to dosing with a test compound dissolved in acetone. PAH and solvent concentrations were designed to match those employed in previous studies with trout liver S9 fractions and isolated perfused livers (Supplemental Data, Table S1) [19]. The concentration of parent chemical within the reaction system at each time point was determined by removing an aliquot of the suspension (200 μL) and terminating the reaction in 600 μL of cold acetonitrile. Six aliquots were removed from each reaction vial over a time period of up to 4 h. Preliminary studies were conducted to determine a suitable time course for each chemical. Depletion experiments were performed as singlet assays with 9 lots of cells isolated from individual fish (n = 9), except for NAPH experiments, which were performed in duplicate or triplicate for each lot.
All studies were performed under conditions that reksulted in log-linear depletion kinetics. A first-order elimination rate constant (k; equal to −2.3 × slope) with units of inverse time was determined from log10-transformed concentration-time data. The CLIN VITRO,INT,HEP was calculated by dividing the elimination rate constant by the measured cell concentration [4, 7, 13]. Measured CLIN VITRO, INT,HEP values then were multiplied by the hepatocellularity value determined previously for sexually immature trout (510×106 hepatocytes/ g liver [20]) to obtain in vivo intrinsic clearance estimates, expressed per gram of liver tissue (CLIN VIVO,INT,HEP; mL/h/g liver), for direct comparison to extrapolated CLIN VIVO, INT,S9 values.
Assessment of chemical binding in the in vitro system
The free (unbound) fraction of each PAH in the hepatocyte suspension was determined by solid phase microextraction using glass fibers (Poly Micro Industries; 1–1.5 cm in length, 430 μm diameter) coated with polydimethylsiloxane (PDMS; 35 μm thickness) [21, 22]. Bulk suspensions of hepatocytes (2 for each PAH) were deactivated by the addition of 0.5 mM 1-aminobenzotriazole (1-ABT) and 10 μM ketoconazole. Deactivation conditions were initially developed using fluorene and confirmed for each test chemical (Supplemental Data, Figure S1). Deactivated suspensions (60 mL; 2 × 106 cells/mL) were dosed with a PAH at the concentration used in substrate depletion experiments and pre-incubated at 11 °C for 1 h using a Stuart roller mixer (Bibby Scientific Ltd.). Three subsamples (18 mL) of each suspension were transferred to 20 mL scintillation vials containing one PDMS fiber each. These subsamples were incubated at 11 °C on the roller mixer until steady state was achieved. Time to steady state was determined in preliminary studies as the time required to achieve a constant fiber/suspension concentration ratio.
Fibers were removed from the suspensions, rinsed with water, blotted dry and extracted with 75% acetonitrile. The chemical mass was divided by the PDMS volume to give the concentration in the fiber ([PAH]fiber). The free concentration in each suspension was calculated under assumed equilibrium conditions as the concentration in the fiber divided by the chemical’s PDMS:water partitioning value (KPDMS; unitless), determined earlier by Nichols et al. [19].
| (1) |
The concentration of total PAH in the suspension was then determined, and the fraction of unbound chemical was calculated as
| (2) |
Measured clearance of PAHs by isolated perfused livers
Hepatic clearance rates for the 6 PAHs were determined previously using an isolated perfused trout liver preparation [19]. Livers from fish of mixed sex were isolated and perfused using procedures given by Nichols et al. [23]. Hepatic clearance (CLIPL; mL/h/g liver) was then calculated by measuring total concentrations of test chemicals in perfusate samples afferent (PAHAFF) and efferent (PAHEFF) to the tissue
| (3) |
| (4) |
where VPAH (μg/h/g liver) is the rate of substrate disappearance.
When the perfusion conditions were held constant, CLIPL changed little if at all in experiments lasting up to 8 h. This finding made it possible to change the experimental conditions as a means of exploring the concentration-dependence of the system as well as the effects of changes in chemical binding. The effect of chemical binding on clearance was explored by perfusing livers for 4 h with a solution containing 10 mg/mL BSA. The perfusate was then switched to one containing 1 mg/mL BSA, and the livers were perfused for an additional 4 h. Perfusate samples were analyzed to determine both free and total PAH concentrations. Free concentrations were determined using the same SPME sampling method described in the preceding section.
Extrapolation of CLIN VIVO,INT,HEP to CLIPL,PRED,HEP
In order to compare the results of the in vitro assays to measured rates of hepatic clearance, CLIN VIVO,INT,HEP values were used as inputs to a well-stirred liver model [24, 25]. This model accounts for potential rate limitations imposed by the rate of chemical delivery to the tissue, as well as the effects of chemical binding
| (5) |
Here the term QIPL (mL/h/g liver) refers to the measured perfusion rate of individual livers, and the binding term fU (unitless) is calculated as the ratio of unbound chemical fractions in the liver perfusate (fU,PERF; unitless) and hepatocyte cell suspension (fU,HEP; unitless). Implied by this approach is the assumption that biotransformation enzymes operate against the unbound chemical concentration in vitro and in situ. For this exercise, we used fU,PERF values given previously by Nichols et al. [19]. Finally, following the same authors, in vitro data were extrapolated a second time, assigning to the binding term a value of 1.0. This assignment does not imply an absence of chemical binding, but rather assumes that free fractions in vitro and in the liver perfusate are functionally the same. Noting further that PAH binding in the hepatocyte suspension is very similar to that in a solution containing 1.0 mg/mL S9 protein (see below), we adopted this binding assumption only for livers perfused with 1.0 mg/mL BSA (equivalent to 1.0 g/L).
Analysis of Test Chemicals
Samples (substrate depletion samples and SPME fiber extracts) were analyzed by a Beckman Model 126 HPLC system (Beckman Instruments) equipped with a Model 168 diode array detector and Waters Model 2475 multi-wavelength fluorescence detector (Waters Corp.), as described previously by Nichols et al. [19]. Chromatography was performed using a Hypersil Green PAH, 5 μm, 2.1 × 100 mm column (Thermo Scientific). The aqueous phase contained 90% deionized water and 10% acetonitrile while the organic phase consisted of 10% deionized water and 90% acetonitrile. Samples were typically eluted by isocratic flow (0.5 mL/min) using 75% organic phase. Minor variations from this method were made to accommodate chemicals with more extreme properties. The following excitation/emission wavelengths (nm) were used for detection: NAPH – 280/340; FLU – 280/340; ANTH – 250/390; PHEN – 250/390; PYR – 237/385; BAP – 255/420.
RESULTS
Characterization of cryopreserved hepatocytes
Hepatocytes were obtained from 5 male and 4 female trout. In terms of their age, size, and strain, these animals were well-matched to those used by Nichols et al. [19]. Each fish was evaluated to determine its gonadosomatic index (GSI; wet wt. of gonads/wet wt. of the animal x 100). The mean GSI for males was 0.04 ± 0.01, while that for females averaged 0.23 ± 0.14. These values are similar to those determined by Nichols et al. [19], and indicate that the fish were in very early stages of sexual development [26, 27].
Each lot of cryopreserved cells was characterized to determine EROD, GST, and UGT activity, as well as cell yield and viability. Measured activities for each lot are provided as Supplemental Data (Table S2). There were no significant gender-related differences in activity for any of the reaction pathways (Student’s t-test, p = 0.05). Calculated as overall means, EROD, GST, and UGT activities averaged 7.0 ± 2.8 pmol/min/mg protein, 363 ± 56 nmol/min/mg protein, and 330 ± 150 pmol/min/mg protein, respectively. The average cell yield after thawing (compared to the amount of freshly isolated cells initially frozen) was 30 ± 9%, and the average percentage of viable cells for 7 of the 9 lots was 80 ± 10%. Two cell lots (8 and 9) exhibited lower viability (~ 50%), although enzyme activities were in good agreement with those determined for other lots.
Determination of CLIN VITRO,INT,HEP and extrapolation to CLIN VIVO,INT,HEP
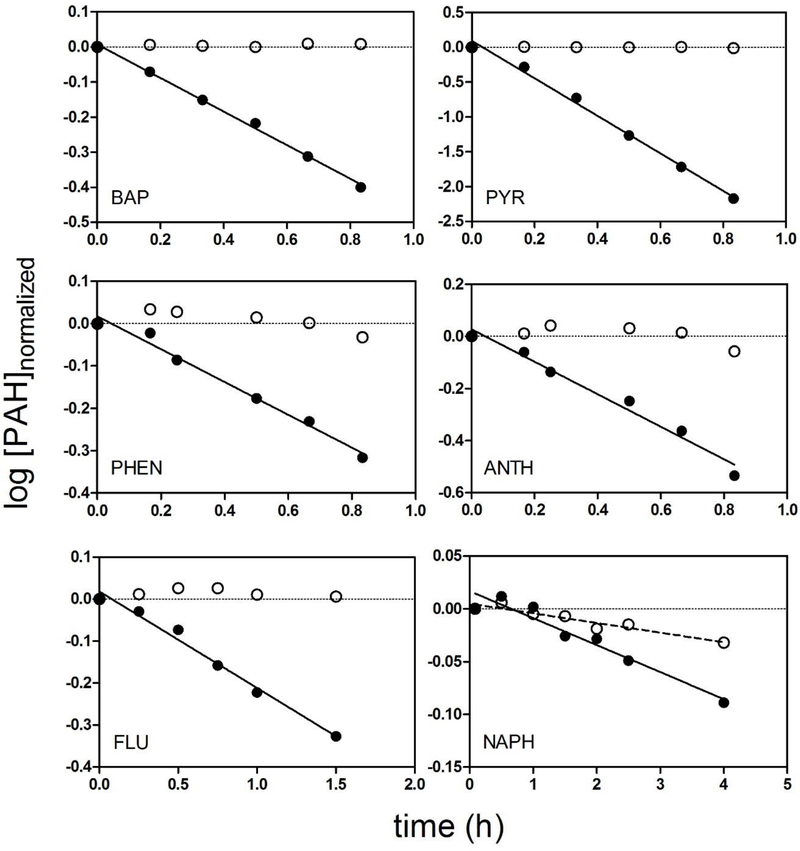
All substrate depletion experiments yielded measurable rates of CLIN VITRO, INT.HEP. Representative data for all 6 compounds are shown in Figure 1. In each case, the data shown correspond to the experiment which yielded the median rate of measured activity (from 9 independent experiments). In vitro intrinsic clearance values determined for each cell lot are given as Supplemental Data (Table S2). There were no significant differences in PAH clearance related to gender (Student’s t-test, p = 0.05). The NAPH depletion data were more variable than data obtained for the other PAHs and exhibited a loss of chemical from both live and control (heat-denatured) suspensions [19]. This loss from negative controls appeared to exhibit firstorder kinetics and was assumed to be due to an abiotic mechanism, presumably volatilization. The CLIN VITRO, INT,HEP for NAPH was therefore calculated from the difference between depletion rates in live cells and the heat-denatured suspensions from matched experiments. For the other PAHs, there was little or no loss of parent chemical from negative controls.
Figure 1.
Representative log10-transformed substrate depletion profiles for benzo[a]pyrene (BAP), pyrene (PYR), and phenanthrene (PHEN), anthracene (ANTH), fluorene (FLU), and naphthalene (NAPH). All data are from experiments which yielded the median rate of activity determined for each compound. To facilitate comparisons, concentration data from active (closed circles) and heat-denatured (open circles) cell suspensions were normalized to the starting concentration.
Measured CLIN VITRO, INT,HEP values and extrapolated CLIN VIVO,INT,HEP estimates for each PAH, averaged across all 9 fish, are provided in Table 1. For comparison, this table also includes extrapolated in vivo intrinsic clearance values (CLIN VIVO,INT,S9) determined previously using trout liver S9 fractions [19]. The units of estimated in vivo clearance are identical (i.e., mL/h/g liver), permitting direct comparisons between the two in vitro test systems. With the exception of BAP, extrapolated in vivo intrinsic clearance values obtained using either system were very similar (≤ 2.6 fold difference). For BAP, the clearance rate calculated from S9 data was nearly 10 times higher than that obtained using hepatocytes. However, it did not appear that either system consistently generated higher or lower in vivo intrinsic clearance values.
Table 1.
Intrinsic clearance of 6 polycyclic aromatic hydrocarbons by cryopreserved trout hepatocytes and trout liver S9 fractions
| Chemical | log Kowa | Substrate conc. (μΜ) | CLin vitro, int,hep (mL/h/106 cells) | CLin vivo,int,hep (mL/h/g liver) | CLin vivo, int,s9 (mL/h/g liver)b |
|---|---|---|---|---|---|
| Naphthalene | 3.30 | 2.7 ± 0.2 | 0.018 ± 0.009 | 9.4 ± 4.6 | 22.4 ± 2.5 |
| Fluorene | 4.18 | 0.93 ± 0.18 | 0.30 ± 0.11 | 151 ± 55 | 58.1 ± 2.3 |
| Anthracene | 4.45 | 0.14 ± 0.01 | 0.32 ± 0.10 | 162 ± 51 | 80.5 ± 5.8 |
| Phenanthrene | 4.47 | 1.5 ± 0.2 | 0.21 ± 0.04 | 109 ± 20 | 94.0 ± 2.7 |
| Pyrene | 4.88 | 0.25 ± 0.02 | 3.1 ± 1.0 | 1580± 500 | 1610 ± 160 |
| Benzo[a]pyrene | 6.13 | 0.16 ± 0.01 | 0.54 ± 0.10 | 273 ± 52 | 2660 ± 250 |
Kow = log10 octanol-water partition coefficient; CLin vitro, int,hep = in vitro intrinsic clearance measured using hepatocytes; CLin vivo,int,hep = in vivo intrinsic clearance calculated using data from hepatocytes; CLin vivo, int,s9 = in vivo intrinsic clearance calculated using data from liver S9 fractions; all data reported as mean ± SD
log Kow values are based on MLogP estimates (with preference given to measured values) in Bio-Loom for Windows
Data provided in Nichols et al. [19]
Chemical binding
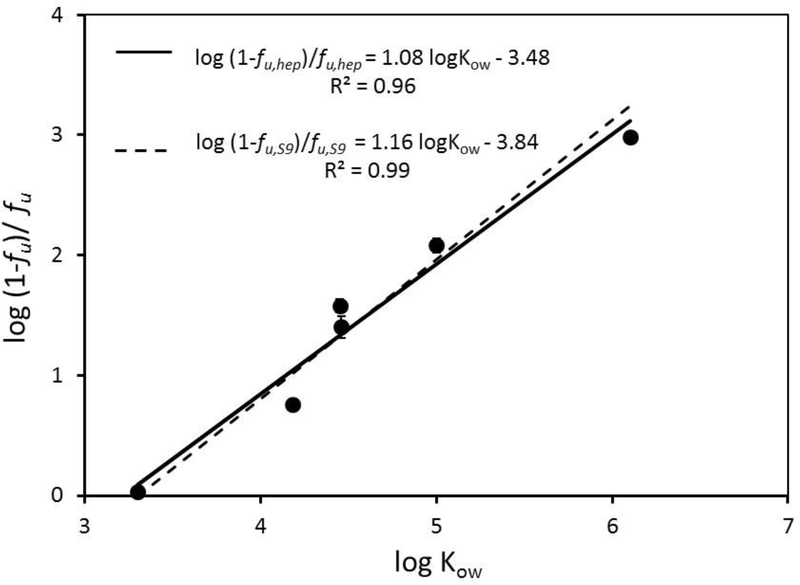
Unbound chemical fractions in the hepatocyte suspension (fU,HEP) decreased with increasing chemical log KOW. When plotted as log KOW verses the log of the bound-to-free ratio (i.e., 1-fU,HEP)/fU,HEP), these data yielded the linear relationship: log (1-fu, hep)/fu,hep = 1.08 log KOW – 3.48 (Figure 2, solid line). A similar relationship was reported previously [18] for PAH binding in the S9 reaction system: log (1-fU,S9)/fU,S9 = 1.16 log KOW – 3.84 (Figure 2, dashed line). Thus, PAH binding in a system containing 2×106 trout hepatocytes/mL is nearly identical to that in a system containing 1 mg/mL trout liver S9 protein. The protein content of a solution containing 2×106 trout hepatocytes/mL is less than half that of the S9 system (~0.3–0.4 mg/mL; Peterson’s modification of the Lowry method; Sigma technical bulletin TP0300); however, the cellular system contains more phospholipid (as a component of cell membranes).
Figure 2.
Binding of 6 polycyclic aromatic hydrocarbons (PAHs) in cryopreserved trout hepatocyte suspensions. The log of the bound-to-free ratio [(1-fu, hep)/ fu, hep] was linearly regressed against chemical log KOW. The best fit line that describes this relationship is shown as a solid line. The dashed line represents a relationship given previously [19] which describes PAH binding in trout S9 fractions. Each point represents the mean ± SE (n = 2 independent suspensions, each of which was evaluated in triplicate).
Extrapolation of CLIN VIVO, INT to CLIPL using measured binding values
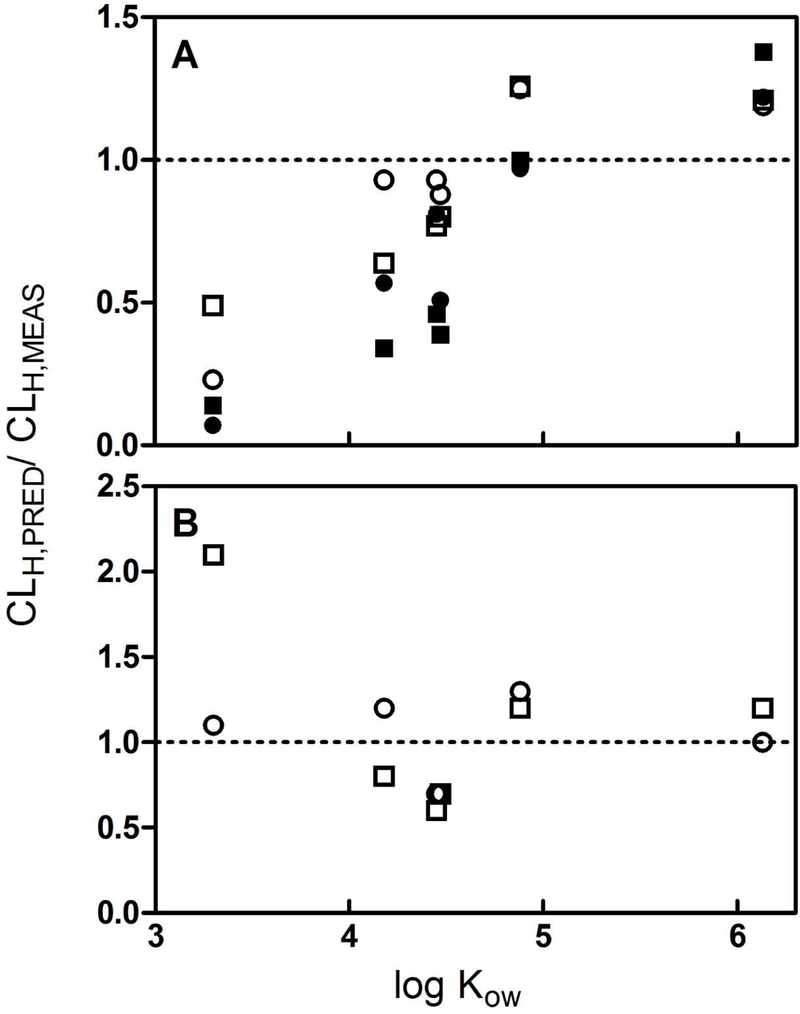
Calculated CLIN VIVO,INT,HEP values were used to predict the clearance of PAHs by isolated perfused livers (CLIPL,PRED,HEP) using the “full model” which accounts for chemical binding in both the hepatocyte suspension and perfusates delivering chemical to the tissue (Eq. 5). These predictions are given in Table 2, along with the measured rates of hepatic clearance (CLIPL,MEAS). To visualize the quality of these in vitro-based hepatic clearance predictions, we plotted the ratio CLIPL,PRED/CLIPL,MEAS as a function of chemical log KOW for each of the tested substances (circles, Figure 3A). An examination of these ratios suggests that, for all compounds except NAPH, predicted clearance rates were within a factor of 2.1 of measured clearance values. Marginally greater prediction accuracy was obtained for livers perfused with 1 mg/mL BSA. In addition, there appeared to be a trend toward overestimation of measured clearance values for higher log KOW compounds (PYR and BAP) and underestimation of clearance for the lower log KOW compounds. For NAPH, hepatic clearance rates predicted using hepatocyte data underestimated measured values by a factor of 4.6 to 15.8, depending on the perfusate BSA concentration.
Table 2.
Extrapolation of in vitro intrinsic clearance by isolated hepatocytes to hepatic clearance by isolated perfused livers under different assumptions with respect to chemical binding
| Chemical | BSA protein (g/L) | fu,MEAS= fu,PERF/ fu, HEP (unitless) | CLipl,pred,hep; fu = fu,MEAS (mL/h/g liver) | CLipl,pred,hep; fu = 1.0 (mL/h/g liver) | CLipl,meas (mL/h/g liver) |
|---|---|---|---|---|---|
| Naphthalene | 1 | 0.20 | 1.8 ± 0.8 | 8.5 ± 3.7 | 8.3 ± 0.1 |
| 10 | 0.02 | 0.17 ± 0.08 | 2.7 ± 0.2 | ||
| Fluorene | 1 | 0.51 | 38.8 ± 7.6 | 52.5 ± 7.2 | 41.8 ± 10.9 |
| 10 | 0.12 | 15.1 ± 4.6 | 26.2 ± 9.1 | ||
| Anthracene | 1 | 2.91 | 64.1 ± 3.4 | 50.3 ± 5.7 | 66.6 ± 13.3 |
| 10 | 0.58 | 41 ± 6.3 | 48 ± 7.6 | ||
| phenanthrene | 1 | 1.69 | 53.8 ± 5.7 | 44.1 ± 6.2 | 61.9 ± 15.2 |
| 10 | 0.28 | 21 ± 4.4 | 43.4 ± 11.7 | ||
| pyrene | 1 | 1.20 | 78.4 ± 1.4 | 82.1 ± 0.02 | 62.7 ± 2.9 |
| 10 | 0.25 | 67.0 ± 4.6 | 68.2 ± 3.3 | ||
| Benzo[a]pyrene | 1 | 20.31 | 69.2 ± 0.2 | 54.8 ± 3.1 | 58 ± 13 |
| 10 | 1.94 | 61.7 ± 1.7 | 50.6 ± 4.2 |
BSA = bovine serum albumin; fu,meas = free fraction in the liver perfusate (fu,PERF)/free fraction in the hepatocyte suspension (fu,HEp); CLipl,pred,hep = predicted hepatic clearance by perfused livers; CLipl,meas = measured hepatic clearance by perfused livers; all data reported as mean ± SD
Figure 3.
Ratio of predicted (CLIPL,PRED) to measured (CLIPL,MEAS) hepatic clearance for 6 polycyclic aromatic hydrocarbons, plotted as a function of chemical log KOW. Predicted values were obtained by extrapolating in vitro intrinsic clearance rates measured using isolated trout hepatocytes (open and filled circles) and liver S9 fractions (open and filled squares). Measured values were determined using isolated trout livers perfused with solutions containing 1 g/L (open symbols) or 10 g/L (filled symbols) bovine serum albumin. A. Predictions generated using a liver model which corrects for unbound chemical fractions in vitro and in liver perfusates (i.e., fU = fU,PERF/fU,HEP or S9). B. Predictions obtained assuming fU = 1.0.
Extrapolation of CLIN VITRO, INT,HEP to CLIPL,PRED,HEP, assuming fU = 1.0
When the binding term fU was set equal to 1.0, the apparent trend of increasing CLIPL,PRED/CLIPL, MEAS with increasing log KOW was eliminated. The predicted hepatic clearance rate (CLIPL,PRED,HEP) for BAP exhibited somewhat better agreement with the measured rate of clearance (CLIPL,MEAS), while the correspondence between predicted and measured values for FLU, ANTH, and PHEN was made slightly worse. In each case, however, these changes were relatively small (< 35%, relative to values calculated using the full model; Table 2, Figure 3B). In contrast, setting fU equal to 1.0 substantially increased the predicted hepatic clearance of NAPH and resulted in much better agreement with CLIPL,MEAS.
Comparison to predicted clearance values obtained by extrapolating data from liver S9 fractions
Figures 3A and3B (squares) also show predicted ratios (CLIPL,PRED/CLIPL,MEAS), obtained by extrapolating measured rates of activity in liver S9 fractions. These ratios, which were reported previously by Nichols et al. [19], follow the same general trends noted above with regard to log KOW and the amount of BSA in the perfusate solution. Setting fU = 1.0 again resulted in increased predicted clearance of NAPH, although in this case the prediction was 2.1 times greater than the measured value.
DISCUSSION
The purpose of this study was to further evaluate the utility of cryopreserved trout hepatocytes as a model system for predicting metabolism impacts on chemical accumulation in fish. Substrate depletion assays were performed using cells isolated from 9 individual fish, which provided an opportunity to assess variability among cell preparations, and by extension individual animals. Such an assessment is useful for addressing both methodological concerns (e.g., what are the principal sources of variability when using in vitro data to estimate in vivo rates of biotransformation) as well as questions inherent to the practice of bioaccumulation assessment (e.g., what is the expected variance in BCFs determined in whole-animal studies due to individual differences in rates of chemical biotransformation).
Coefficients of variation (% CV) for clearance rates determined using the 9 cell lots ranged from 18% (BAP) to 50% (NAPH). Excepting NAPH, which presented unique methodological challenges, the mean of these values was 27 ± 8%. Similar CVs were obtained for EROD (36%), UGT (22%), and GST (17%) activity. In a recent ring trial involving three laboratories, cryopreserved trout hepatocytes were used to measure the intrinsic clearance of 5 chemical substrates [11]. Each compound was evaluated using the same cell lot, and the analysis of each chemical was performed in one laboratory. Intra-laboratory CVs averaged 16% across the 3 labs and 5 test chemicals, attesting to the high degree of reproducibility that can be expected when the assay is performed by an experienced user. The inter-laboratory CVs determined for these 5 compounds ranged from 27% to 61%. Thus, variability associated with different users of the assay, while still relatively small, may be similar to or even larger than variability among individuals of a well-defined trout strain.
When in vitro intrinsic clearance values (CLIN VITRO,INT,HEP) determined in this study were extrapolated to in vivo intrinsic clearance rates (CLIN VIVO, INT,HEP), the values for all compounds except BAP were in close agreement with those determined previously using S9 fractions [19] and exhibited the same rank order of activity (NAPH< FLU ≈ ANTH ≈ PHEN < PYR). For BAP, the CLIN VIVO, INT,HEP determined using S9 fractions was approximately 10-fold higher than that obtained using isolated hepatocytes. Previously, Hallifax et al. [28] assembled in vitro intrinsic clearance data for a large number of drugs, obtained using human microsomes or isolated hepatocytes. These data were used to predict in vivo rates of intrinsic clearance which were then compared to apparent in vivo values, calculated based on observed rates of hepatic clearance. In general, in vivo intrinsic clearance rates predicted using in vitro data tended to under-predict apparent in vivo values. Averaged across all compounds, this bias was about 5- and 4-fold for predictions based on microsomal and hepatocyte data, respectively. For the microsomal data, the magnitude of this bias did not vary with the rate of activity. In contrast, the bias for predictions based on hepatocyte data increased for high clearance substrates. Similar observations were subsequently reported in a study that employed liver microsomes and suspended hepatocytes from the same donor livers [29]. Based on these findings, it was suggested that co-factor limitations and/or rate-limiting cell permeability may limit in vitro clearance of some highly metabolized substrates by isolated hepatocytes. In the present study, BAP was the largest and most hydrophobic of the compounds tested. It is also a relatively high clearance compound. We may speculate, therefore, that restricted diffusion of BAP across the hepatocyte membrane limits its rate of metabolism, which raises questions about the utility of different in vitro test systems for specific substrates.
In vivo intrinsic clearance rates determined in this study were used as inputs to a wellstirred liver model to predict clearance by isolated perfused livers (CLIPL) under two different binding assumptions. Predicted CLIPL values were then evaluated by comparison to measured rates determined previously for the same compounds [19]. With the exception of NAPH, predictions obtained using the full model were in good agreement with measured CLIPL values (< 1.25 fold difference for 1 g/L BSA and < 2.1 fold difference for 10g/L BSA; Figure 3A). Under the second binding scenario (fu = 1.0; Figure 3B), the predictions for NAPH and BAP improved, while predictions for the other chemicals were made slightly worse. Generally, however, both binding assumptions yielded similar results. Figures 3A and3B also show the correspondence between measured levels of CLIPL and clearance values predicted using in vitro data from S9 fractions [19]. The resulting patterns are quite similar to those obtained using data from hepatocytes, attesting to similarities between the two systems with respect to measured rates of activity and in vitro chemical binding.
The close agreement between observed and predicted CLIPL values for compounds other than NAPH can be attributed, in part, to the fact that CLIN VIVO,INT,HEP rates determined using both in vitro systems often approached the perfusate flow rate to isolated livers. When intrinsic clearance rates are high, the well-stirred liver model (Equation 5) predicts that CLIPL will approach the rate of chemical delivery to the tissue, resulting in near first-pass clearance. To illustrate this point, measured intrinsic clearance values were divided by liver perfusion rates and expressed as a percentage (Table 3). For PHEN, ANTH, PYR, and BAP, these percentages range from approximately 55% to nearly 90%. Under these circumstances, binding effects on clearance become relatively unimportant as long as the product term fU CLIN VIVO,INT approaches or exceeds QIPL. The existence of a flow rate limitation on QIPL also explains why hepatic clearance predictions for BAP obtained using data from trout hepatocytes were comparable to those obtained using S9 fractions, even though activity levels were 10-fold lower; in both cases, the rate of activity was high enough to create a flow-limited system.
Table 3.
Comparison of liver perfusion rates to measured rates of hepatic clearance by isolated perfused livers
| Chemical | Qipl (mL/h/g liver) | BSA protein (g/L) | CLipl,meas (mL/h/g liver) | CLipl,meas/Qipl (as %) |
|---|---|---|---|---|
| Naphthalene | 74.3 | 1 | 8.3 ± 0.1 | 11.2 |
| 10 | 2.7 ± 0.2 | 3.6 | ||
| Fluorene | 85.8 | 1 | 41.8 ± 10.9 | 48.7 |
| 10 | 26.2 ± 9.1 | 30.5 | ||
| Anthracene | 75.2 | 1 | 66.6 ± 13.3 | 88.5 |
| 10 | 48 ± 7.6 | 63.8 | ||
| Phenanthrene | 79.7 | 1 | 61.9 ± 15.2 | 77.7 |
| 10 | 43.4 ± 11.7 | 54.5 | ||
| Pyrene | 82.1 | 1 | 62.7 ± 2.9 | 76.4 |
| 10 | 68.2 ± 3.3 | 83.0 | ||
| Benzo[a]pyrene | 70.1 | 1 | 58 ± 13 | 82.8 |
| 10 | 50.6 ± 4.2 | 72.2 |
Qipl = average perfusion rate; BSA = bovine serum albumin; CLipl,meas = measured hepatic clearance by perfused livers; data reported as mean ± SD
A better test of binding effects on hepatic clearance would require working with compounds that are both highly bound and metabolized at lower rates. The results shown here do, however, illustrate a point made previously, which is that confidence in predicted impacts of biotransformation on chemical accumulation is related in part to the rate of activity [18], and is generally higher when metabolism rates are rapid. The fact that high rates of activity drive the system to a flow-limited condition also explains why variability in BCFs predicted using in vitro data from different sources may be substantially smaller than variability in the in vitro datasets themselves [11, 18].
Results obtained for NAPH using either in vitro test system are difficult to interpret. Hepatic clearance predictions obtained by setting fU = 1.0 were substantially higher than those obtained using the full model. In one case (hepatocyte data), this resulted in much better agreement between predicted and measured values, while in the other case (S9 data) the relative difference between predicted and measured values remained essentially the same (~ 2 fold error). When using either test system, low rates of activity combined with a significant non-enzymatic loss process made it difficult to accurately estimate clearance of NAPH. Volatilization of NAPH from isolated perfused livers (increasing the apparent CLIPL,MEAS) also may have contributed to under-prediction of CLIPL,MEAS by in vitro test systems (full model). These findings underscore the challenge of performing such studies with volatile substrates.
Interestingly, the results of the present study appear to contradict the findings of an earlier study by Han et al. [9], which compared in vitro intrinsic clearance rates measured using trout liver microsomes, S9 fractions, and isolated hepatocytes (Table 4 of the cited work). Clearances rates determined by these authors using S9 fractions were 4.5 to 10.5 times lower than those measured using hepatocytes. The only scaling factor which differs substantially between the present work and that given by Han et al. [9] relates to the S9 content of liver tissue. Here we used a value of 163 mg/g [19] while Han et al. [9] used a factor of 95 mg/g. If the scaling factor used it the present study is applied to data given by Han et al. [9], the agreement between CLH values predicted using S9 fractions and hepatocytes improves somewhat (1.8- to 6.3-fold difference across all test chemical); nevertheless, hepatocytes still yield higher rates of activity.
Most of the compounds evaluated by Han et al. [9] were metabolized at relatively low rates, requiring that S9 and microsomal incubations be performed for 4 h. We note also that substrate concentrations employed by these authors (2 or 5 μM) were, in general, higher than those tested in the present study. With the exception of NAPH, all of the incubations performed in the present study as well as those given by Nichols et al. [19] were conducted in 1.5 h or less. The working lifetime of different in vitro preparations may differ, and it is possible that one system is better suited than another for use with slowly metabolized substances. In this regard, we take special note of the work by Uchea et al. [30] which showed that liver spheroids, obtained by culturing trout hepatoctyes for 10 d, exhibit metabolic activity similar to that of freshly isolated cells. Liver spheroids, or a similar system, may be particularly useful for performing in vitro biotransformation assays over very long (hours to days) time periods.
Based on the results of the present study, it cannot be said that one in vitro system (hepatocytes or S9 fractions) or the other is clearly preferred for measuring intrinsic clearance rates in trout. Instead, these findings provide strong support for use of either test system to estimate hepatic clearance rates, and by extension whole-body metabolism rate constants, particularly when measured rates of in vitro activity are relatively high. This information can be used as an input to established computational models to perform screening-level bioaccumulation assessments for fish [4]. Alternatively, measured rates of in vitro activity could be used directly to focus limited whole-animal testing resources and inform the selection of more acceptable chemical alternatives.
Supplementary Material
Effect of aminobenzotriazole (ABT) and ketoconazole (Ket) on biotransformation activity of hepatocyte suspensions. Enzymatic inhibition using either 1 mM ABT (dots) or 0.5 uM Ket (up triangles) was incomplete, as evidenced by diminished but measurable clearance of fluorene (FLU). The use of 1 mM ABT + 0.5 uM Ket resulted in complete inhibition of fluorene metabolism up to 4 h (down triangles). Data for the active depletion of fluorene by the same lot of hepatocytes (no inhibitors) is also provided (squares). The depletion of benzo[a]pyrene (BAP) was similarly inhibited by the combination of ABT and Ket for up to 8 h (diamonds). BAP was the most rapidly depleted test chemical used in this study.
Measured substrate concentrations (μM; mean ± SD) across studies
Enzymatic characterization of cryopreserved hepatocytes obtained from individual fish, including measured in vitro intrinsic clearance (CLIN VITRO,INT,HEP) of 6 polycyclic aromatic hydrocarbons
Acknowledgments
This document has been subjected to review by the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
REFERENCES
- 1.Gobas FAPC, de Wolf W, Burkhard LP, Verbruggen E, Plotzke K 2009. Revisiting bioaccumulation criteria for POPs and PBT assessments. Integr Environ Assess Manage 5:624–637. [DOI] [PubMed] [Google Scholar]
- 2.van Wijk D, Chénier R, Henry T, Hernando MD, Schulte C. 2009. Integrated approach to PBT and POP prioritization and risk assessment. Integr Environ Assess Manage 5:697–711. [DOI] [PubMed] [Google Scholar]
- 3.OECD Guidelines for the Testing of Chemicals, Section 3: Degradation and Accumulation Test no. 305: Bioaccumulation in Fish: Aqueous and Dietary Exposure, last updated October 2, 2012. Organization for Economic Co-operation and Development, Paris, France. [Google Scholar]
- 4.Nichols JW, Bonnell M, Dimitrov SD, Escher B, Han X, Kramer NI. 2009. Bioaccumulation assessment using predictive approaches. Integr Environ Assess Manage 5:577–597. [DOI] [PubMed] [Google Scholar]
- 5.Nichols JW, Erhardt S, Dyer S, James M, Moore M, Plotzke K, Segner H, Schultz I, Thomas K, Vasiluk L, Weisbrod A. 2007. Workshop report: Use of in vitro absorption, distribution, metabolism and excretion (ADME) data in bioaccumulation assessments for fish. Human and Ecological Risk Assessment 13: 1164–1191. [Google Scholar]
- 6.Han X, Nabb DL, Mingoia RT, Yang C-H. 2007. Determination of xenobiotic intrinsic clearance in freshly isolated hepatocytes from rainbow trout (Oncorhynchus mykiss) and rat and its application in bioaccumulation assessment. Environ Sci Technol 41:3269–3276. [DOI] [PubMed] [Google Scholar]
- 7.Cowan-Ellsberry CE, Dyer SD, Erhardt S, Bernhard MJ, Roe AL, Dowty ME, Weisbrod AV. 2008. Approach for extrapolating in vitro metabolism data to refine bioconcentration factor estimates. Chemosphere 70:1804–1817. [DOI] [PubMed] [Google Scholar]
- 8.Dyer SD, Bernhard MJ, Cowan-Ellsberry C, Perdu-Durand E, Demmerle S, Cravedi J-P. 2008. In vitro biotransformation of surfactants in fish. Part I: Linear alkylbenze sulfonate (C12-LAS) and alcohol ethoxylate (C13EO8). Chemosphere 72:850–862. [DOI] [PubMed] [Google Scholar]
- 9.Han X, Nabb DL, Yang C-H, Snajdr SI, Mingoia RT. 2009. Liver microsomes and S9 from rainbow trout (Oncorhynchus mykiss): Comparison of basal-level enzyme activities with rat and determination of xenobiotic intrinsic clearance in support of bioaccumulation assessment. Environ Toxicol Chem 28:481–488. [DOI] [PubMed] [Google Scholar]
- 10.Gomez CF, Constantine L, Huggett DB. 2010. The influence of gill and liver metabolism on the predicted bioconcentration of three pharmaceuticals in fish. Chemosphere 81:1189–1195. [DOI] [PubMed] [Google Scholar]
- 11.Fay KA, Mingoia RT, Goeritz I, Nabb DL, Hoffman AD, Ferrell BD, Peterson HM, Nichols JW, Segner H, Han X. 2104. Intra- and interlaboratory reliability of a cryopreserved trout hepatocyte assay for the prediction of chemical bioaccumulation potential. Environ Sci Technol 48:8170–8178. [DOI] [PubMed] [Google Scholar]
- 12.Laue H, Gfeller H, Jenner KJ, Nichols JW, Kern S, Natsch A. 2014. Predicting the bioconcentration of fragrance ingredients by rainbow trout using measured rates of in vitro intrinsic clearance. Environ Sci Technol 48:9486–9495. [DOI] [PubMed] [Google Scholar]
- 13.Nichols JW, Schultz IR, Fitzsimmons PN. 2006. In vitro-in vivo extrapolation of hepatic biotransformation data for fish. I. A review of methods, and strategies for incorporating intrinsic clearance estimates into chemical kinetic models. Aquat Toxicol 78:74–90. [DOI] [PubMed] [Google Scholar]
- 14.Han X, Mingoia RT, Nabb DL, Yang C-H, Snajdr SI, Hoke RA. 2008. Xenobiotic intrinsic clearance in freshly isolated hepatocytes from rainbow trout (Oncohynchus mykiss): determination of trout hepatocellularity, optimization of cell concentrations and comparison to serum and serum-free incubations. Aquat Toxicol 89:11–17. [DOI] [PubMed] [Google Scholar]
- 15.Mingoia RT, Glover KP, Nabb DL, Yang C-H, Snajdr SI, Han X. 2010. Cryopreserved hepatocytes from rainbow trout (Oncorhynchus mykiss): A validation study to support their application in bioaccumulation assessments. Environ Sci Technol 44:3052–3058. [DOI] [PubMed] [Google Scholar]
- 16.Johanning K, Hancock G, Escher B, Adekola A, Bernhard MJ, Cowan-Ellsberry C, Domoradzki J, Dyer S, Eickhoff C, Embry M, Erhardt S, Fitzsimmons P, Halder M, Hill J, Holden D, Johnson R, Rutishauser S, Segner H, Schultz I, Nichols J. 2012. Assessment of metabolic stability using the rainbow trout (Oncorhynchus mykiss) liver S9 fraction. Curr Prot Toxicol 14101–28. [DOI] [PubMed] [Google Scholar]
- 17.Fay KA, Nabb DL, Mingoia RT, Bischof I, Nichols JW, Segner H, Johannning K, Han X. 2015. Determination of metabolic stability using cryopreserved hepatocytes from rainbow trout (Oncorhynchus mykiss). Curr Prot Toxicol 4421–29. [DOI] [PubMed] [Google Scholar]
- 18.Nichols JW, Huggett DB, Arnot JA, Fitzsimmons PN, Cowan-Ellsberry CE. 2013. Toward improved models for predicting bioconcentration of well-metabolized compounds by rainbow trout using measured rates of in vitro intrinsic clearance. Environ Toxicol Chem 32:1611–1622. [DOI] [PubMed] [Google Scholar]
- 19.Nichols JW, Hoffman AD, ter Laak TL, Fitzsimmons PN. 2013. Hepatic clearance of 6 polycyclic aromatic hydrocarbons by isolated perfused trout livers: prediction from in vitro clearance by liver S9 fractions. Toxicol Sci 136:359–372. [DOI] [PubMed] [Google Scholar]
- 20.Fay KA, Fitzsimmons PN, Hoffman AD, Nichols JW. 2014. Optimizing the use of rainbow trout hepatocytes for bioaccumulation assessments with fish. Xenobiotica 44:345–351. [DOI] [PubMed] [Google Scholar]
- 21.Vaes WHJ, Urrestarazu Ramos E, Vehaar HJM, Seinen W, Hermens JLM. 1996. Measurement of the free concentration using solid-phase microextraction: binding to protein. Anal Chem 68:4463–4467. [Google Scholar]
- 22.Heringa MB, Hermens JLM. 2003. Measurement of free concentrations using negligible depletion solid-phase microextraction (nd-SPME). Trends Anal Chem 22:575–587. [DOI] [PubMed] [Google Scholar]
- 23.Nichols JW, Hoffman AD, Fitzsimmons PN. 2009. Optimization of an isolated perfused rainbow trout liver model: clearance studies with 7-ethoxycoumarin. Aquat Toxicol 95:182–194. [DOI] [PubMed] [Google Scholar]
- 24.Rowland M, Benet LZ, Graham GG. 1973. Clearance concepts in pharmacokinetics. J Pharmacokinet Biopharm 1:123–136. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson GR, Shand DG. 1975. Commentary: a physiological approach to hepatic drug clearance. Clin Pharmacol Ther 18:377–390. [DOI] [PubMed] [Google Scholar]
- 26.Gomez JM, Mourot b, Fostier A, Le Gac F. 1999. Growth hormone receptors in ovary and liver during gametogenesis in female rainbow trout (Oncorhynchus mykiss). J Reprod Fertil 115:275–285. [DOI] [PubMed] [Google Scholar]
- 27.Le Gac F, Thomas JL, Mourot B, Loir M. 2001. In vivo and in vitro effects of prochloraz and nonylphenol ethyoxylates on trout spermatogenesis. Aquat Toxicol 53:187–200. [DOI] [PubMed] [Google Scholar]
- 28.Hallifax D, Foster JA, Houston JB. 2010. Prediction of human metabolic clearance from in vitro systems: retrospective analysis and prospective view. Pharm Res 27:2150–2161. [DOI] [PubMed] [Google Scholar]
- 29.Foster JA, Houston JB, Hallifax D. 2011. Comparison of intrinsic clearances in human liver microsomes and suspended hepatocytes from the same donor livers: clearancedependent relationship and implications for prediction of in vivo clearance. Xenobiotica 41:124–136. [DOI] [PubMed] [Google Scholar]
- 30.Uchea C, Sarda S, Schulz-Utermoehl T, Owen S, Chipman KJ. 2013. In vitro models of xenobiotic metabolism in trout for use in environmental bioaccumulation studies. Xenobiotica 43:421–431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of aminobenzotriazole (ABT) and ketoconazole (Ket) on biotransformation activity of hepatocyte suspensions. Enzymatic inhibition using either 1 mM ABT (dots) or 0.5 uM Ket (up triangles) was incomplete, as evidenced by diminished but measurable clearance of fluorene (FLU). The use of 1 mM ABT + 0.5 uM Ket resulted in complete inhibition of fluorene metabolism up to 4 h (down triangles). Data for the active depletion of fluorene by the same lot of hepatocytes (no inhibitors) is also provided (squares). The depletion of benzo[a]pyrene (BAP) was similarly inhibited by the combination of ABT and Ket for up to 8 h (diamonds). BAP was the most rapidly depleted test chemical used in this study.
Measured substrate concentrations (μM; mean ± SD) across studies
Enzymatic characterization of cryopreserved hepatocytes obtained from individual fish, including measured in vitro intrinsic clearance (CLIN VITRO,INT,HEP) of 6 polycyclic aromatic hydrocarbons