Summary
The approximately thirty core subunits of kinetochores assemble on centromeric chromatin containing the histone H3 variant CENP-A and connect chromosomes with spindle microtubules. The chromatin proximal 16-subunit CCAN (constitutive centromere associated network) creates a mechanically stable bridge between CENP-A and the kinetochore’s microtubule-binding machinery, the 10-subunit KMN assembly. Here, we reconstituted a stoichiometric 11-subunit human CCAN core that forms when the CENP-OPQUR complex binds to a joint interface on the CENP-HIKM and CENP-LN complexes. The resulting CCAN particle is globular and connects KMN and CENP-A in a 26-subunit recombinant particle. The disordered, basic N-terminal tail of CENP-Q binds microtubules and promotes accurate chromosome alignment, cooperating with KMN in microtubule binding. The N-terminal basic tail of the NDC80 complex, the microtubule-binding subunit of KMN, can functionally replace the CENP-Q tail. Our work dissects the connectivity and architecture of CCAN and reveals unexpected functional similarities between CENP-OPQUR and the NDC80 complex.
Keywords: kinetochore, centromere, CENP-U, CENP-P, CENP-O, CENP-R, constitutive centromere associated network, CCAN, microtubule, KMN network
Graphical Abstract
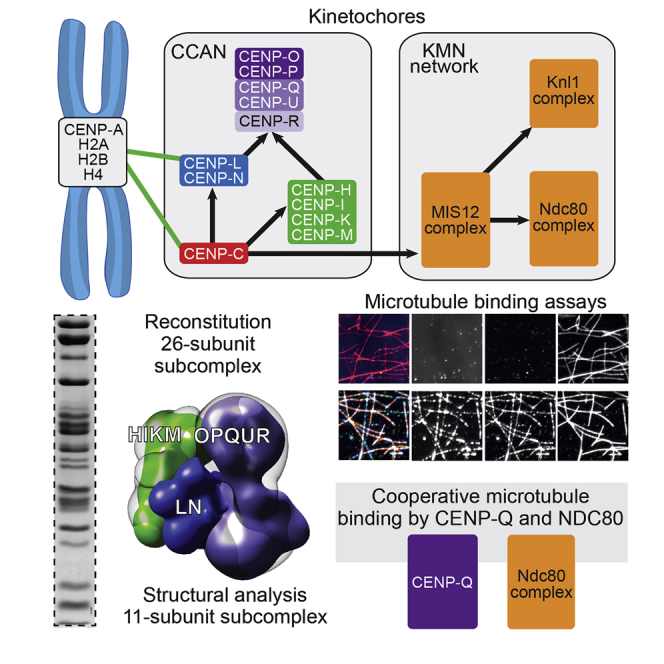
Highlights
-
•
The kinetochore CENP-OPQUR complex is reconstituted and functionally dissected
-
•
A kinetochore particle with 26 subunits and defined stoichiometry is reconstituted
-
•
EM structure of an 11-subunit inner kinetochore complex reveals globular shape
-
•
CENP-Q and the Ndc80 complex bind microtubules cooperatively
Kinetochores mediate chromosome attachment to the mitotic spindle. In a biochemical tour-de-force, Pesenti et al. reconstituted a 26-subunit kinetochore particle and characterized its structural organization. The CENP-Q subunit was shown to increase the microtubule-binding affinity of kinetochores, revealing that the kinetochore-spindle interaction is more complex than hitherto believed.
Introduction
Accurate chromosome segregation in mitosis and meiosis is of paramount importance for cellular and organismal viability. The ultimate goal of chromosome segregation is to endow the two daughter cells with a full complement of chromosomes, preventing the considerable burdens associated with whole-chromosome aneuploidy (Santaguida and Amon, 2015). Chromosome segregation requires the establishment of a structure named the mitotic spindle, which consists of microtubules, microtubule-associated proteins, and motors that harness the energy of ATP hydrolysis to organize microtubules in dense antiparallel arrays and to focus them at the spindle poles (Heald and Khodjakov, 2015).
Crucial for the chromosome segregation process is the attachment of chromosomes to the mitotic spindle. This takes place at kinetochores, large protein complexes built on a stretch of specialized chromatin named the centromere (Pesenti et al., 2016). A conserved feature of centromeric chromatin is the presence of a histone H3 variant named centromeric protein A (CENP-A, or CenH3), which interacts with histones H4, H2A, and H2B in a specialized nucleosome particle. Original work with anti-centromere antibodies, and more recent proteomic analyses of CENP-A and its binding partners, identified 16 vertebrate proteins now collectively identified as the constitutive-centromere-associated network (CCAN, Figure 1A), reflecting association of these proteins with kinetochores during the entire cell cycle (Earnshaw and Rothfield, 1985, Foltz et al., 2006, Hori et al., 2008a, Izuta et al., 2006, Obuse et al., 2004, Okada et al., 2006, Saitoh et al., 1992). These CENP-A proximal or “inner kinetochore” subunits include CENP-C, CENP-H, CENP-I, CENP-K, CENP-L, CENP-M, CENP-N, CENP-O, CENP-P, CENP-Q, CENP-R, CENP-S, CENP-T, CENP-U, CENP-W, and CENP-X. Most of these proteins are conserved in eukaryotes, including the yeast Saccharomyces cerevisiae, where they are generally identified as the Ctf19 complex (McAinsh and Meraldi, 2011, Perpelescu and Fukagawa, 2011, Westermann and Schleiffer, 2013, Westhorpe and Straight, 2013).
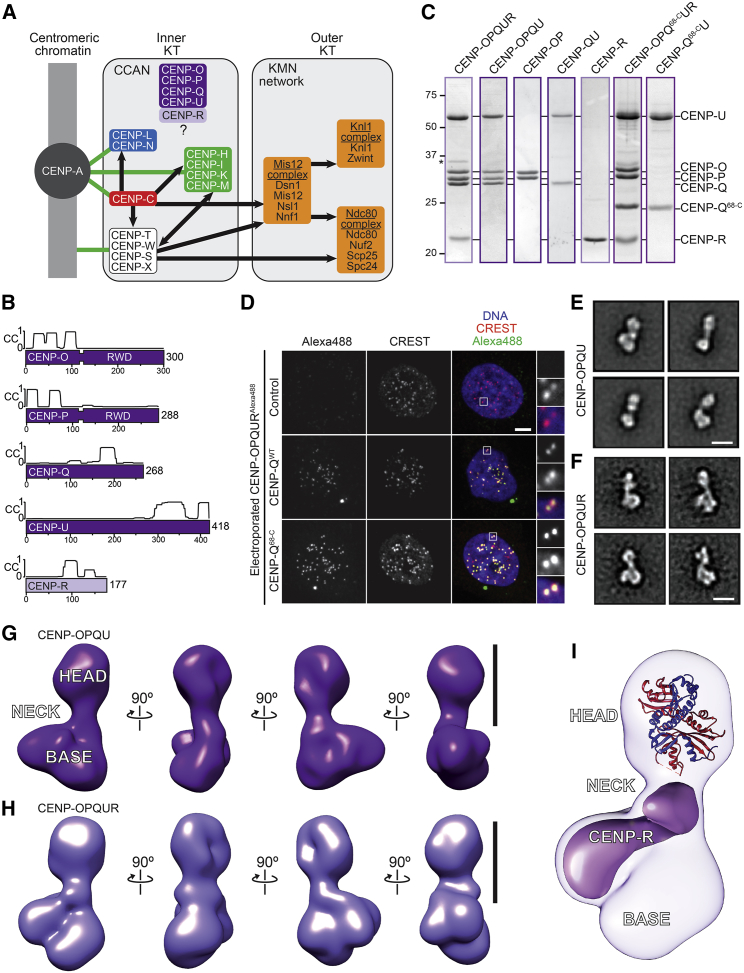
Figure 1.
Reconstitution and Characterization of the Human CENP-O Complex
(A) Layered organization of the human kinetochore with schematic depiction of subcomplexes. Green lines indicate direct connection with centromeric DNA or chromatin. Black lines indicate recruitment dependencies. CENP-C and the CENP-TWSX complex (not studied here) may create independent connections between centromeres and outer kinetochore.
(B) Schematic representation of the components of the CENP-O complex. Coiled-coil (CC) predictions calculated with program COILS (Lupas et al., 1991) are shown for subunits with partial or complete CC content. The C-terminal halves of CENP-O and CENP-P contain tandem RWD domains (Schmitzberger and Harrison, 2012).
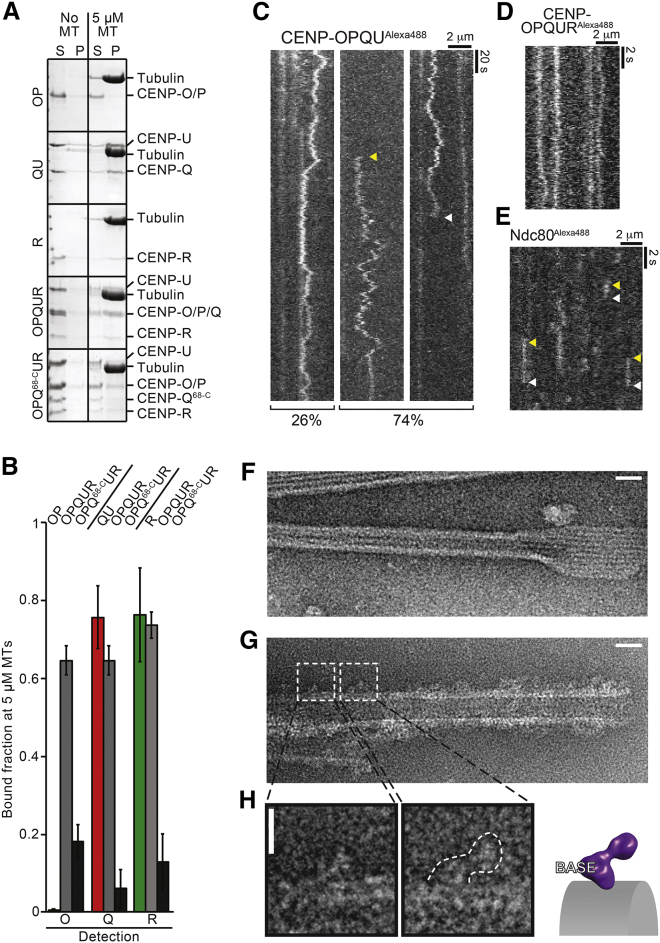
(C) Coomassie-stained SDS-PAGE gel of recombinant wild-type or mutant CENP-OPQUR complexes and subcomplexes used in this study. The asterisk indicates an impurity.
(D) Representative images show the interphase localization of recombinant CENP-OPQUR labeled with Alexa488 (green) after electroporation into HeLa cells. CREST immunostaining identifies kinetochores. Scale bar, 5 μm.
(E) Representative class averages of negatively stained CENP-OPQU.
(F) Representative class averages of negatively stained CENP-OPQUR. Scale bar in (E) and (F), 10 nm.
(G) 3D reconstruction from negatively stained particles of the CENP-OPQU complex at ∼22 Å resolution. Scale bar, 10 nm.
(H) 3D reconstruction of the CENP-OPQUR complex also at ∼22 Å resolution. Scale bar, 10 nm.
(I) A model of the Ctf19:Mcm21 complex (PDB ID 3ZXU), respectively homologous to CENP-P and CENP-O, was manually fitted in the head domain. The difference map between CENP-OPQUR and CENP-OPQU, corresponding to the density of CENP-R, is shown in purple. See also Figure S1.
The CCAN subunits are organized in distinct subcomplexes, including the CENP-T:CENP-W complex (herewith CENP-TW), proposed to form a nucleosome-like particle with CENP-S:CENP-X (CENP-SX) (Hori et al., 2008a, Nishino et al., 2012); the CENP-L:CENP-N (CENP-LN) complex (Carroll et al., 2009, Carroll et al., 2010); the CENP-H:CENP-I:CENP-K:CENP-M (CENP-HIKM) complex (Basilico et al., 2014, Klare et al., 2015, Okada et al., 2009, Pekgöz Altunkaya et al., 2016); and the CENP-O:CENP-P:CENP-Q:CENP-U:CENP-R (CENP-OPQUR) complex (Hori et al., 2008b) (Figure 1A).
CENP-C, which based on sequence prediction methods is largely intrinsically disordered, may function as a spatial organizer of the CCAN, binding to the CENP-HIKM and CENP-LN complexes (Hinshaw and Harrison, 2013, Klare et al., 2015, McKinley et al., 2015, Nagpal et al., 2015, Pentakota et al., 2017, Weir et al., 2016). The resulting 7-subunit assembly (CENP-CHIKMLN) establishes multiple contacts with the CENP-A nucleosome through CENP-A-selective binding regions in CENP-C and CENP-N (Cao et al., 2018, Carroll et al., 2009, Carroll et al., 2010, Chittori et al., 2018, Falk et al., 2016, Guo et al., 2017, Kato et al., 2013, Pentakota et al., 2017, Tian et al., 2018, Weir et al., 2016).
Acting as bridge for the microtubule binding “outer kinetochore”, CCAN contributes also to the mechanical stability of kinetochores (Suzuki et al., 2014). The CENP-TW complex, which binds directly to CENP-CHIKMLN and requires it for kinetochore targeting, contributes to the recruitment of the KMN (KNL1, MIS12, NDC80) assembly (Huis In 't Veld et al., 2016, Kim and Yu, 2015, Malvezzi et al., 2013, Nishino et al., 2013, Pekgöz Altunkaya et al., 2016, Rago et al., 2015, Schleiffer et al., 2012). Within the KMN, the 4-subunit NDC80 complex (NDC80C) is considered the major microtubule receptor of the kinetochore (Cheeseman, 2014, Pesenti et al., 2016). The CENP-TW pathway coexists with, and depends on, a second axis of outer kinetochore assembly centered on CENP-C (for an extended discussion, see Huis In 't Veld et al., 2016 and Pesenti et al., 2016). Besides interacting with CCAN, CENP-C also binds directly to the KMN assembly (Gascoigne et al., 2011, Przewloka et al., 2011, Screpanti et al., 2011) (Figure 1A).
CENP-CHIKMLN is also required for recruitment of a 5-subunit complex incorporating the CENP-O, CENP-P, CENP-Q, CENP-U, and CENP-R subunits (CENP-OPQUR, whose subunits are schematically shown in Figure 1B) (Eskat et al., 2012, Foltz et al., 2006, Hori et al., 2008b, McKinley et al., 2015, Minoshima et al., 2005, Okada et al., 2006, Samejima et al., 2015). CENP-OPQUR is related to the COMA complex of S. cerevisiae (De Wulf et al., 2003, Hori et al., 2008b, Hyland et al., 1999, Ortiz et al., 1999, Schmitzberger et al., 2017, Westermann et al., 2003). Its precise role at kinetochores remains poorly characterized, but it consists at least in part in the recruitment of other kinetochore residents, including the microtubule plus-end directed motor CENP-E and Polo-like kinase 1 (Plk1), the latter through phosphorylation of CENP-U (Bancroft et al., 2015, Hori et al., 2008b, Kang et al., 2006). Microtubule-binding activities have also been independently attributed to CENP-Q or CENP-U (Amaro et al., 2010, Hua et al., 2011).
In an effort to reconstitute kinetochores in vitro, we recently reported the assembly, entirely from recombinant material, of a 21-subunit assembly containing the CENP-A nucleosome, the CENP-CHIKMLN complex, and the 10-subunit KMN network (Weir et al., 2016). Biochemical reconstitution is crucial for unraveling the organization of kinetochores, as it facilitates the identification of stable modules of subunits, and for the characterization of their physical interactions, stoichiometries, and regulation. Furthermore, biochemical reconstitution can provide material for detailed structural analyses and for in vitro measurements of kinetochore function (e.g. force generation) under controlled conditions. Thus, our ultimate goal is to be able to reconstitute kinetochore particles that encompass the majority, or all, of constitutive subunits.
Here, we report the reconstitution of most of the CCAN complex, its structural characterization, its interactions within the human kinetochore, and its contributions to microtubule binding. We find that CCAN forms a globular particle, the topology of which we describe in detail. We also significantly extend our understanding of the mechanism of microtubule binding by the CCAN and its relationship to the previously characterized microtubule-binding site in the KMN network. Our studies provide strong mechanistic and structural insights into a fundamental and conserved component of the chromosome segregation machinery.
Results
Reconstitution and Structural Analysis of CENP-OPQUR
To investigate the requirements for stability of CENP-OPQUR subunits, we turned to in vitro reconstitution with recombinant components. CENP-O, -P, -Q, and -U were unstable when expressed individually in bacteria or insect cells and could not be recovered in soluble form (unpublished data). Co-expression in insect cells yielded two stable subcomplexes, CENP-OP and CENP-QU, which were purified to homogeneity and appeared monodisperse by size-exclusion chromatography (SEC; Figures 1C, S1A, and S1B). To generate the CENP-OPQU complex, we mixed stoichiometric amounts of CENP-OP and CENP-QU (Figure S1C) or co-infected insect cells (see STAR Methods; Figure S1D). Sedimentation velocity analytical ultracentrifugation (AUC) demonstrated that CENP-OP, CENP-QU, and CENP-OPQU contained single copies of each subunit (Table S1; Figure S1E).
CENP-R was stable when expressed in isolation, appeared monodisperse by SEC, and formed tetramers in AUC experiments (Figures S1E and S1F; Table S1). However, it did not interact with CENP-OP, CENP-QU, or CENP-OPQU in SEC experiments (Figures S1G and S1H). Conversely, CENP-R interacted with CENP-OPQU when co-expressed or when cell pellets where lysed together (co-lysis). The CENP-OPQUR complex, obtained by co-lysis, was monodisperse by SEC (Figure S1I), and AUC showed that it contained a single copy of each subunit (Figure S1E; Table S1). Thus, we suspect that CENP-R forms oligomers when it cannot interact in the CENP-OPQUR complex. This may not be an isolated case, because a previous study reported that CENP-Q, expressed and purified in isolation, forms octamers (Amaro et al., 2010). Our inability to obtain soluble CENP-Q in isolation prevented us from confirming this previous observation, but our results identify single copies of CENP-Q and CENP-R in the CENP-OPQUR complex. In co-expression experiments, we observed that CENP-R interacts predominantly with the CENP-QU subcomplex (Figure S1J).
Electroporated in interphase or mitotic HeLa cells, recombinant CENP-OPQUR covalently modified with Alexa488 fluorophore labeled kinetochores (marked by CREST auto-antibodies), indicating that the recombinant complex retains crucial properties of its endogenous counterpart (Figures 1D and S1K, representative of at least three independent experiments).
We studied the structural organization of the CENP-OPQU and CENP-OPQUR complexes by negative-stain electron microscopy (EM) (Figures 1E–1H, S2A, and S2B). Three-dimensional (3D) reconstructions showed that the structure of CENP-OPQU is bi-lobed, with a smaller head domain and a larger base domain. CENP-R did not grossly alter this organization, but an additional protuberance in the neck region and an enlargement in the base domain became evident (Figures 1G–1I).
Mechanism of Kinetochore Recruitment of the CENP-OPQUR Complex
Recently, we reconstituted a 7-subunit, CENP-A-associated CCAN subcomplex that includes CENP-C (and specifically its N-terminal region, residues 1–544: CENP-C1–544), the 4-subunit CENP-HIKM complex (containing a truncated form of CENP-I lacking its 56 N-terminal residues, and henceforth indicated as CENP-IΔ56), and the 2-subunit CENP-LN complex (Basilico et al., 2014, Klare et al., 2015, Weir et al., 2016). To assess the role of selected subunits of this CENP-A-associated complex (herewith referred to as CENP-CHIKMLN complex) in CENP-OPQUR recruitment, we created stable HeLa cell lines expressing, under an inducible promoter, each of the individual CENP-OPQUR subunits fused to green fluorescent protein (GFP), and we tested their kinetochore targeting in control cells and in cells depleted of CENP-H, CENP-L, or CENP-N by RNAi (Figures S2A–S2E). Each CENP-OPQUR subunit was lost from kinetochores under these conditions (shown in Figures 2A and 2B for CENP-Q and in Figures S2B–S2E for CENP-O, -P, -R, and -U). Thus, CENP-A proximal subunits of CCAN are required for recruitment of CENP-OPQUR subunits, in agreement with previous observations (Eskat et al., 2012, Foltz et al., 2006, Hori et al., 2008b, Minoshima et al., 2005, Okada et al., 2006). Furthermore, we suppressed the expression of individual CENP-O, -P, -Q, and -R subunits by RNAi, and we observed that reducing the level of any one of the four CENPs prevented localization of the other subunits (Figures S2F–S2H), indicating reciprocal support in kinetochore localization, as reported previously (Bancroft et al., 2015, Hori et al., 2008b, Kang et al., 2006).
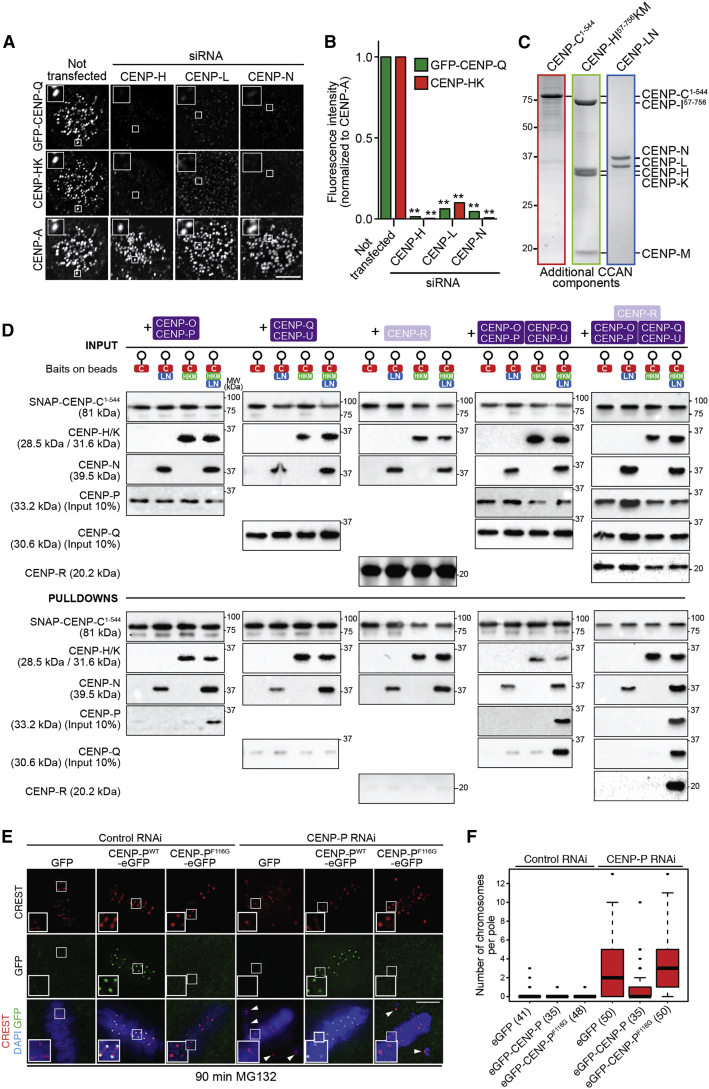
Figure 2.
Molecular Basis of Kinetochore Recruitment of CENP-OPQUR
(A) Depletion of CENP-H, CENP-L, or CENP-N prevented kinetochore localization of GFP-CENP-Q in HeLa FlpIn TRex cell lines stably expressing GFP-CENP-Q, as shown by representative images. CENP-HK complex is also lost from kinetochores during the aforementioned RNAi depletions. Scale bar, 5 μm.
(B) Quantification of the amount of GFP-CENP-Q (green bars) and CENP-HK (red bars) at kinetochores following CENP-H, CENP-L, or CENP-N depletion. ∗∗p ≤ 0.01. Graph shows representative results from one of three independent experiments. A minimum of 158 kinetochores was quantified.
(C) Coomassie-stained SDS-PAGE of recombinant CENP-C1–544, CENP-HIΔ56KM, and CENP-LN used in (D).
(D) Pull-down assays using SNAP-CENP-C1–544 bait. CENP-OP binds the solid phase only in the presence of CENP-HIΔ56KM and CENP-LN. Subsequently, CENP-QU and CENP-R can also be recruited. Shown are Western blots of the indicated species. The experiment shown is representative of three technical replicas.
(E) RNAi-resistant GFP-CENP-P localized to the kinetochore after depletion of endogenous CENP-P, while GFP-CENP-PF116G did not. CREST signal (red) was unaffected by CENP-P depletion or by impaired localization of GFP-CENP-PF118G. DAPI (DNA) is shown in blue. Arrowheads indicate misaligned chromosomes. MG132 (10 μM) was added to prevent mitotic exit. Scale bar, 5 μm.
(F) Quantification of the experiment in (E). The number of cells analyzed is in parentheses. Error bars represent standard deviations. See also Figure S2 and Figure S3.
We tried to reconstitute in vitro with purified proteins (Figure 2C and S3A–S3C) the recruitment hierarchy responsible for the dependencies observed in HeLa cells. CENP-C1–544 immobilized on solid phase was used as bait—in isolation or in presence of CENP-LN, CENP-HIΔ56KM, or both—to pull down CENP-OP, CENP-QU, or CENP-R (Figure 2D). Both CENP-LN and CENP-HIΔ56KM bound independently to CENP-C. When incubated together, an apparent increase in binding affinity, particularly evident for CENP-N, was observed. Neither CENP-OP nor CENP-QU or CENP-R bound to CENP-C1–544 beads in the presence of isolated CENP-LN or CENP-HIΔ56KM. CENP-OP, however, readily bound when CENP-LN and CENP-HIΔ56KM were added concomitantly to form the CENP-C1-544HIΔ56KMLN complex. Isolated CENP-QU or CENP-R, on the other hand, was unable to interact with the CENP-C1-544HIΔ56KMLN complex, and only did so in presence of CENP-OP or CENP-OPQU, respectively (Figure 2D). Thus, our data suggest that CENP-OP acts as a bridge between the CENP-C1–544HIΔ56KMLN complex and the other subunits of the CENP-OPQUR complex.
The previously determined crystal structure of the tandem RWD domains of KNL1 in complex with a linear peptide of Nsl1 from the MIS12 complex (MIS12C) (Petrovic et al., 2014) offers a possible model for the interaction of the CENP-OP RWD domains with CENP-C1-544HIΔ56KMLN. Tyr2125Knl1 lies at the interface with Nsl1, and its mutation impairs Nsl1 binding without destabilizing the Knl1 structure (Petrovic et al., 2014). Ctf19CENP-P and CENP-P also contain aromatic residues at the equivalent position (Phe138Ctf19 and Phe116CENP-P) (Figures S3D–S3F). Glycine point mutation of this residue (CENP-PF116G) did not apparently affect the stability of the CENP-OP dimer but largely abrogated its binding to the CENP-C1-544HIΔ56KMLN complex in vitro (Figure S3G). Furthermore, the mutant displayed reduced binding to CCAN components in immunoprecipitation (IP) assays from cell lysates (Figure S3H). Kinetochore localization of CENP-PF116G in HeLa cells was also impaired, regardless of whether the endogenous CENP-P protein had been depleted through RNAi (Figure 2E). Thus, the mechanism of kinetochore recruitment of CENP-OP may be structurally related to the mechanism of kinetochore recruitment of the RWD domains of KNL1 via an interaction with MIS12C.
The experiments in Figure 2E additionally demonstrated that depletion of CENP-P by RNAi, which destabilizes the entire CENP-OPQUR complex, interferes with chromosome congression, with several chromosomes occupying positions near the spindle poles and failing to complete alignment at the metaphase plate (white arrowheads in Figure 2E, quantification in Figure 2F). These effects of the CENP-P depletion were rescued by expression of wild type CENP-P, but not of CENP-PF116G mutant, confirming the specificity of the phenotype.
Organization of the CENP-HIΔ56KMLNOPQUR Complex
Two important conclusions so far are 1) that the CENP-C1-544HIΔ56KMLN complex recruits the CENP-OPQUR complex and 2) that this requires a direct interaction of CENP-OP with a composite interface created by CENP-HIΔ56KM and CENP-LN, as summarized in Figure 3A. Because solid-phase binding assays can suffer from absorption artifacts, we asked if we could reconstitute these interactions also in solution. When combined stoichiometrically, CENP-OPQUR, CENP-HIΔ56KM, and CENP-LN formed a single 11-subunit complex (CENP-HIΔ56KMLNOPQUR) with reduced retention volume in SEC (i.e., larger and/or more elongated) in comparison to the individual subcomplexes (Figure 3B). Omission of CENP-LN or CENP-HIΔ56KM prevented complex assembly (Figures S4A and S4B). By AUC, the CENP-HIΔ56KMLNOPQUR complex had an observed molecular mass of ∼404 kDa, in excellent agreement with the predicted molecular mass of ∼408 kD calculated on the assumption that each subunit is in single copy (Figures 3C and S5A; Table S1). This result matches results obtained with the solid-phase experiments in Figure 2D and demonstrates that CENP-OPQUR behaves as a coincidence detector for CENP-HIΔ56KM and CENP-LN complexes. Furthermore, these observations indicate that CENP-C1–544 is dispensable for the interaction, although we cannot exclude that it contributes to the stabilization of CENP-HIΔ56KMLNOPQUR. Importantly, CENP-C is required for kinetochore recruitment of CENP-HIKM and CENP-LN (Carroll et al., 2010, Gascoigne et al., 2011, Klare et al., 2015, Milks et al., 2009) and therefore it remains ultimately required for kinetochore targeting of CENP-OPQUR.
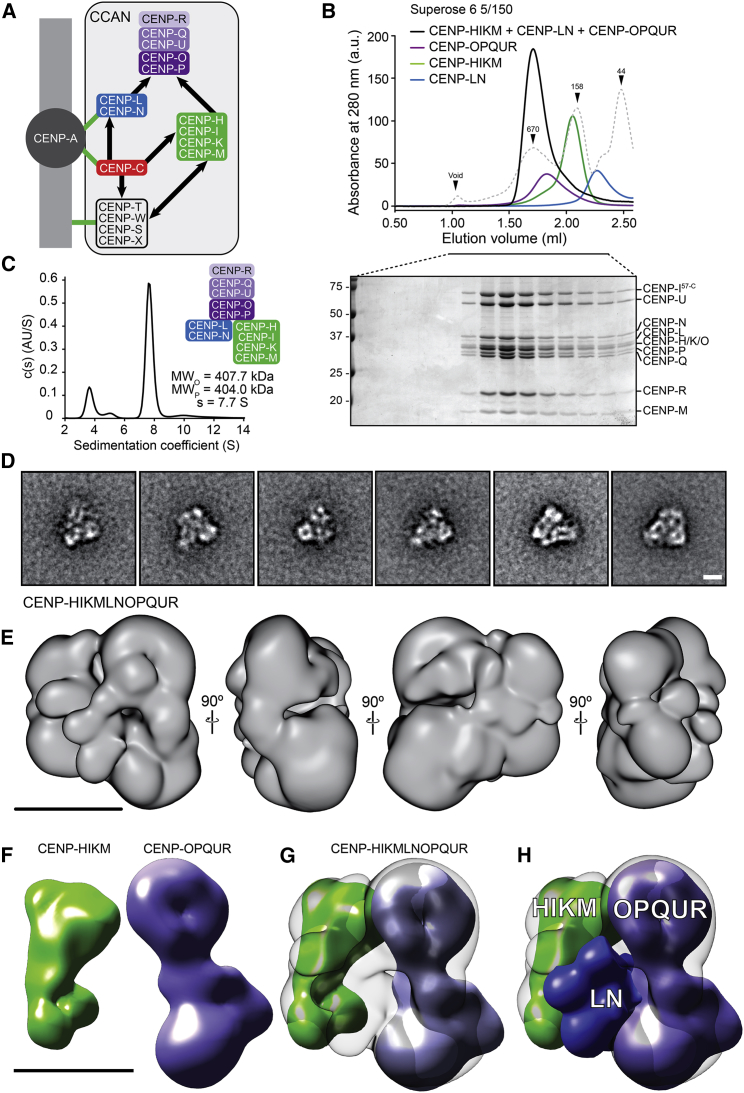
Figure 3.
A CENP-HIΔ56KMLNOPQUR Complex
(A) Model of CCAN assembly supported by our analysis. The presence of CENP-HIΔ56KM and CENP-LN at the centromere is necessary for recruitment of CENP-OPQUR complex.
(B) Elution profile and SDS-PAGE analysis of a stoichiometric mixture of CENP-OPQUR, CENP-LN, and CENP-HIΔ56KM (black). Elution profiles of individual complexes is also indicated; CENP-OPQUR (violet), CENP-HIΔ56KM (green), and CENP-LN (blue).
(C) Hydrodynamic analysis by sedimentation velocity AUC shows that CENP-HIΔ56KMLNOPQUR forms a compact, globular structure in which each subunit is represented once (see Table S1).
(D) Representative class averages of negatively stained CENP-HIΔ56KMLNOPQUR complex. Scale bar, 10 nm.
(E) 3D reconstruction from negatively stained particles at ∼22 Å resolution. Scale bar, 10 nm. See Figure S6G for additional class averages.
(F) 3D reconstructions of the CENP-HIΔ56KM complex (green; Basilico et al., 2014) and of the CENP-OPQUR complex (violet, see Figure 1H). Scale bar, 10 nm.
(G) Possible fitting of the 3D reconstructions of CENP-HIΔ56KM and CENP-OPQUR in the density of CENP-HIΔ56KMLNOPQUR.
(H) The unaccounted density was attributed to the CENP-LN complex. See also Figures S4, S5, and S6 and Table S1.
2D class averages of negatively stained CENP-HIΔ56KMLNOPQUR complex previously subjected to a mild crosslinking treatment with the GraFix procedure (Kastner et al., 2008; Figures S5B–S5D and S6G] are shown in Figure 3D. A 3D reconstruction of the complex at an approximate resolution of 23 Å showed that it has overall dimensions (in Å) of 180, 160, and 100 (Figures 3E and S5E), and is therefore largely globular. We previously described a negative-stain EM reconstruction of the CENP-HIΔ56KM complex (Basilico et al., 2014) (Figure 3F). We therefore fitted densities for the CENP-OPQUR and CENP-HIΔ56KM complex into the density of the CENP-HIΔ56KMLNOPQUR (Figure 3G, Figure S5E, and Video S1). The resulting model predicts that CENP-HIΔ56KM and CENP-OPQUR oppose each other, with a direct contact involving the head domain of CENP-OPQUR (containing CENP-OP). We attribute to CENP-LN the substantial residual unoccupied density sandwiched between CENP-HIΔ56KM and CENP-OPQUR (Figures 3H and S5F; Video S1). By predicting multiple contacts between CENP-LN and CENP-HIΔ56KM with CENP-OPQUR, this model is consistent with the binding data shown in Figures 2 and 3.
The video shows the first 3D reconstruction of the CENP-HIΔ56KMLNOPUR and focuses on how the separately reconstructed subcomplexes HIΔ56KM and OPQUR are oriented towards each other. With the potential caveat that the CENP-HIΔ56KMLNOPUR sample was stabilized by cross-linking with the GraFix method before processing for EM, which might cause it to undergo physical rearrangements, a substantial volume remains unoccupied that might account, at least partially, for the CENP-LN subcomplex.
Reconstitution of a 26-Subunit Kinetochore Particle Containing CENP-OPQUR
CCAN acts as a bridge between the CENP-A nucleosome in centromeric chromatin and the KMN assembly in the microtubule-binding portion of the kinetochore. We have recently reconstituted this bridge with 21 recombinant subunits covering the CENP-A nucleosome, the CENP-C1-544HIΔ56KMLN complex, and the KMN assembly (Weir et al., 2016) (referred to as rKT21, for recombinant kinetochore with 21 subunits). Our new evidence that CENP-OPQUR binds directly to the CENP-C1–544HIΔ56KMLN complex prompted us to ask if this interaction permitted inclusion of CENP-OPQUR in rKT21. CENP-OPQUR did not interact directly with the CENP-A nucleosome core particle (CENP-ANCP) but co-eluted with it when combined with CENP-C1-544HIΔ56KMLN, indicating that the latter mediates the interaction of CENP-OPQUR with CENP-ANCP (Figures 4A and 4B).
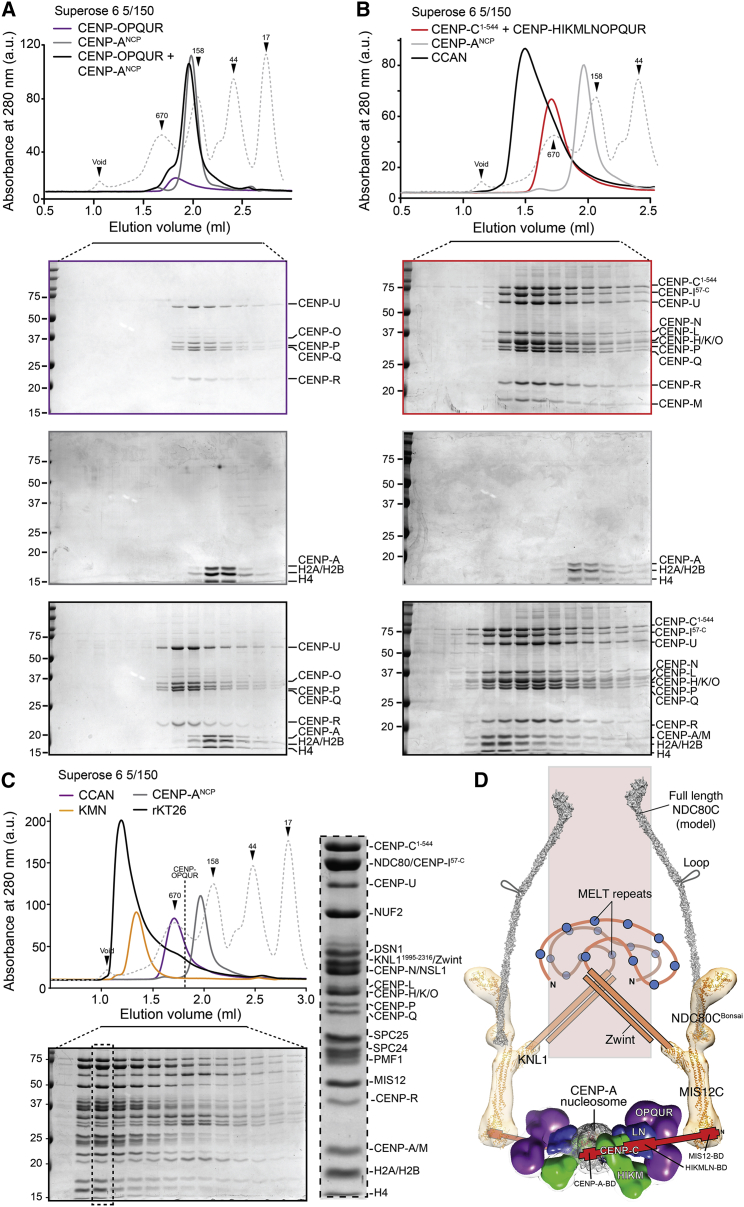
Figure 4.
Reconstitution of a 26-Subunit Kinetochore (rKT26)
(A) Elution profile from analytical SEC, and subsequent SDS-PAGE analysis, of a mixture of CENP-OPQUR (5 μM) complex and CENP-ANCPs (2.5 μM).
(B) Elution profile from analytical SEC, and subsequent SDS-PAGE analysis, of a stoichiometric mixture of CENP-C1–544, CENP-HIΔ56KMLN-OPQUR (each at 5 μM), and CENP-ANCPs (2.5 μM).
(C) Elution profile from analytical SEC, and subsequent SDS-PAGE analysis, of a mixture of CENP-C1-544 (red trace), the CCAN core (CENP-HIΔ56KMLN-OPQUR, violet trace), the KNM network (orange trace) (each at 5 μM), and the CENP-A nucleosomes (grey trace) (2.5 μM) resulting in the formation of a 26-subunit complex (black trace) that links the centromeric DNA to microtubules. Enlargement of the dotted lane of the SDS-PAGE gel demonstrates that the front of the peak contains all the indicated subunits.
(D) Structural and topological organization of the 26-subunit kinetochore (rKT26) based on current and previous work (Weir et al., 2016). The drawing is approximately in scale. A crystal structure of the CENP-A nucleosome has been reported previously (Tachiwana et al., 2011). Previously, we determined crystal structures of NDC80CBonsai (an engineered version of the NDC80 complex that retains microtubule-binding and kinetochore localization activities; Ciferri et al., 2008), of the kinetochore-targeting C-terminal domain of KNL1 (KNL1C) (Petrovic et al., 2014), and of the MIS12C (Petrovic et al., 2016), and we used negative-stain EM to obtain a first view in three dimensions of their complex (Petrovic et al., 2014). Shown in orange are experimental molecular models fitted into the EM density. The long axis of the NDC80CBonsai, MIS12C, KNL1 complex is approximately 35 nm, but the length of the actual complex is approximately 90 nm (Huis In 't Veld et al., 2016) due to the extensive coiled-coils of NDC80C that have been trimmed from NDC80CBonsai. This paper adds a view of CCAN to this scheme. CENP-C connects the CENP-A nucleosome, which it binds via a specific binding domain (CENP-BD), to the outer kinetochore, which it binds via a MIS12 binding domain. See also Figure S5.
Further inclusion of the KMN network resulted in the assembly of a particle with a retention volume smaller than any of those of the individual components (Figure 4C; individual complexes used for these experiments are shown in Figures S5G–S5J). The peak fractions of this species contained all 26 expected subunits (4 histones of the CENP-ANCP, 12 CCAN subunits, and 10 KMN subunits, named rKT26) (Figure 4C). The CENP-OPQUR complex appeared substoichiometric in this peak, and part of it eluted in a shoulder peak corresponding to the expected elution volume for CENP-OPQUR. This observation suggests either that CENP-OPQUR binds into this larger complex with reduced binding affinity, thus undergoing partial dissociation, or that its effective stoichiometry is lower. We have previously determined that two copies of CENP-CHIΔ56KMLN bind a single CENP-A nucleosome core particle (CENP-ANCP) (Weir et al., 2016), and it is possible that a single copy of CENP-OPQUR, rather than two, binds the CENP-C1–544HIΔ56KMLN:CENP-ANCP complex. However, given that CENP-C1–544HIΔ56KMLN and CENP-OPQUR form a stoichiometric complex (Figure 3B), the hypothesis that CENP-OPQUR undergoes partial dissociation from the larger complex seems more plausible. We speculate that this effect reflects a requirement for post-translational modifications that increase the binding affinity of CENP-OPQUR for rKT21 and that are still missing in our reconstitution.
Human CENP-OPQUR did not bind to the NDC80C or to the entire KMN (Figures S4C and S4D), nor did it bind to complexes of MIS12C or MIS12C:NDC80C with CENP-C1–544 (Figures S4E and S4F), a crucial link between the inner and outer kinetochore (Dimitrova et al., 2016, Gascoigne et al., 2011, Petrovic et al., 2016, Przewloka et al., 2011, Screpanti et al., 2011, Weir et al., 2016). Thus, CENP-OPQUR does not contribute to the stabilization of the connection between the inner and the outer kinetochore, a function that has instead been described for its ortholog in S. cerevisiae (the COMA complex, comprising the Ctf19CENP-P, Okp1CENP-Q, Mcm21CENP-O, and Ame1CENP-U subunits and lacking a clear CENP-R ortholog) (De Wulf et al., 2003, Dimitrova et al., 2016, Hornung et al., 2014, Hyland et al., 1999, Ortiz et al., 1999, Pekgöz Altunkaya et al., 2016, Schmitzberger and Harrison, 2012, Westermann et al., 2003).
The CENP-OPQUR Complex Binds Microtubules
Previous studies with isolated recombinant CENP-Q or CENP-U identified microtubule-binding activities in both subunits (Amaro et al., 2010, Hua et al., 2011). Because CENP-Q and CENP-U form a stable complex where CENP-Q is present in single copy, instead of the oligomers observed in isolation (Amaro et al., 2010), we wanted to revisit these results with reconstituted CENP-OPQUR subcomplexes. Only the CENP-QU and CENP-OPQUR complexes, but not CENP-OP, pelleted with Taxol-stabilized microtubules in co-sedimentation assays (Figures 5A, 5B, S6C, and S6D). CENP-OP, however, pelleted with microtubules when combined with CENP-QU (Figures S6C and S6D), suggesting that CENP-QU contains a microtubule-binding activity. In isolation, CENP-R also interacted with microtubules, but its incorporation in the CENP-OPQUR complex did not increase the apparent binding affinity of the CENP-OPQUR complex for microtubules, casting doubts on the significance of the interaction seen with isolated CENP-R oligomers (Figures 5B, S6C, and S6D; and unpublished data).
Figure 5.
Structural and Functional Analysis of the CENP-OPQUR Complex
(A) Representative SDS-PAGE of microtubule co-sedimentation assays with Taxol-stabilized microtubules and the indicated proteins. CENP-OP did not bind microtubules unless it was combined with CENP-QU and CENP-R. CENP-R bound microtubules autonomously. CENP-OPQ68-CUR does not sediment with microtubules.
(B) Quantification of experiments in (A). Error bars are standard deviations calculated from three technical replicas.
(C) Kymographs of CENP-OPQU particles labelled with Alexa488 imaged on Taxol-stabilized microtubules by TIRF microscopy (n = 279). Left side shows example of particles (26% of events) that remain bound for the full duration of the video; right sides (74% of events) show examples of particles landing (yellow arrow; n = 18) or unbinding (white arrow; n = 169).
(D) Kymographs of CENP-OPQUR particles labeled with Alexa488 at higher temporal resolution (100 ms/fr).
(E) Kymographs of Alexa488-labeled NDC80 particles labeled with Alexa488 showing binding and unbinding of the Alexa488-labeled NDC800 (100 mn/fr).
(F) Representative electron micrographs of negative-stained Taxol-stabilized microtubules. Scale, 100 nm.
(G) As in (F), with added CENP-OPQUR. Scale, 100 nm.
(H) The outline of the complex recognizable on the microtubule surface suggests that the microtubule-binding moiety is in the base domain. Scale, 10 nm. For additional examples, see Figure S6F.
Total internal reflection fluorescence (TIRF) microscopy was used to visualize single Alexa488-labeled CENP-OPQU particles at 1 nM concentration, allowing us to show that there is diffusive binding of the complex to the microtubule lattice (Figures 5C–5E). Of the diffusing particles, 26% remained bound to the lattice for the duration of the video (200 s), while 74% were observed to unbind or bind the microtubules during this time (mean time associated = 131 s). In contrast, the NDC80C, at 1 nM concentration, showed rapid binding and unbinding events (mean time associated = 1.3 s; SEM = 0.17 s; Figure 5E). Together, these data suggest that the CENP-OPQU complex is capable of mediating long-duration attachment to microtubules. In view of our recent observation that multimerization leads to a dramatic increase of the microtubule residency time of the NDC80C (Volkov et al., 2018), we cannot exclude that CENP-OPQU may form small oligomeric particles on microtubules under the condition of our assay, even if the AUC analysis with the isolated complex indicated absence of oligomerization. Further work will have to address this possibility.
At micromolar concentrations, CENP-OPQU had a very strong bundling effect on microtubules (Figure S6E). By negative-stain EM, microtubules incubated with CENP-OPQU appeared “rough” in comparison to naked microtubules (Figures 5F and 5G). In several cases, it was possible to visualize individual CENP-OPQU complexes docked on microtubules (Figure 5H and S6F). The interaction with the microtubule lattice appeared to involve the base of the CENP-OPQU complex, with the head pointing away. Because CENP-QU is responsible for microtubule binding, as shown above, we speculate that CENP-OP and CENP-QU reside in the head and base domains, respectively. Structural characterization of the complex of the S. cerevisiae homologs of CENP-O and CENP-P, the Ctf19CENP-P:Mcm21CENP-O complex, demonstrated that these subunits are structural paralogs, each containing tandem RWD (RING finger, WD repeat, DEAD-like helicases) domains (Schmitzberger and Harrison, 2012, Schmitzberger et al., 2017). In agreement with our model, the crystal structure of the Ctf19CENP-P:Mcm21CENP-O complex from S. cerevisiae fitted snugly into the density of the head domain (Figure 1I). This tentative structural model will require corroboration through high-resolution structural analyses in the future.
The CENP-Q Disordered and Basic N-Terminal Tail Promotes Microtubule Binding
In agreement with the sedimentation experiments, fluorescently labeled CENP-OP (with Alexa488 through maleimide thiol chemistry) did not bind microtubules in a flow cell (Figure 6A). Alexa647-labeled CENP-QU, on the other hand, decorated microtubules, and when combined with CENP-OP allowed it to decorate microtubules, indicating that the CENP-QU subcomplex binds microtubules and carries CENP-OP along (Figure 6A).
Figure 6.
CENP-OPQUR and NDC80 Complexes Bind Microtubules Cooperatively
In (A)–(C) and (E), Taxol-stabilized, rhodamine-labeled microtubules were tethered to glass coverslips and incubated in the presence of fluorescent recombinant proteins. The scale bar represents 1 μm.
(A) Alexa488-labeled CENP-OP (green channel) was unable to bind microtubules (red channel) in isolation and bound microtubules only in presence of Alexa647-labeled CENP-QU subcomplex (blue channel).
(B) NDC80-GFP complex (green channel) and Alexa-647-labeled CENP-OPQU (blue channel) interact with an overlapping surface to microtubules, as shown by reciprocal concentration-dependent competition.
(C) CENP-QU made deficient in microtubule binding by FAM labeling is translocated to microtubules through the interaction of CENP-OP with rKT21. Microtubules were incubated in the presence of CENP-QU N-terminally labeled with Fluorescein (green) and/or rKT21 in which CENP-AMN was fused to BFP (blue) and CENP-LN was labeled with Alexa-647 (purple).
(D) PrDos (Ishida and Kinoshita, 2007) disorder prediction of the CENP-Q (black) and NDC80/HEC1 (red) N-terminal tails. Dotted line indicates the disorder threshold; false positive rate 5%.
(E) CENP-OPQUR and rKT21 bind microtubules cooperatively. Microtubules (red channel) were incubated with the indicated concentrations of Alexa-647-labeled CENP-OPQUR (purple), rKT21 containing Alexa-488-labeled KMN (green), or combinations thereof. At the bottom, the same experiment carried out in presence of CENP-OPQ68-CUR shows that microtubule binding by the N-terminal region of CENP-Q is required for augmentation of microtubule binding affinity. Experiments in (A)–(C) and (E) are representative of at least 3 repeats. See also Figure S4 and Figure S7.
At least at high concentrations, NDC80C binds cooperatively to the microtubule lattice, interacting along protofilaments at the alternating αβ and βα tubulin interfaces with 4-nm spacing (Alushin et al., 2010, Ciferri et al., 2008). At a concentration of 100 nM, fluorescent recombinant NDC80CGFP strongly decorated the microtubule lattice in a flow cell (Figure 6B). Addition of CENP-OPQU labeled with Alexa647 at 400 nM caused an almost complete displacement of NDC80C (at 100 nM) from microtubules, with concomitant microtubule binding of fluorescent CENP-OPQU. Conversely, CENP-OPQU, at 100 nM, decorated microtubules, but was displaced upon addition of 400 nM NDC80C (Figure 6B). Thus, the footprints of NDC80C and of CENP-OPQU overlap on the microtubule lattice at least in part, implying that the mode of microtubule binding of these complexes is intrinsically competitive. Furthermore, based on the relative effects of competition in these single-point assays, the binding affinities of the two complexes for microtubules appear to be in a similar range. It has been proposed that CENP-Q and NDC80C interact directly and that this promotes cooperative microtubule binding (Hua et al., 2011), but we could not recapitulate this interaction in vitro, as already discussed (Figure S4C).
Selective Sortase modification of the CENP-Q N-terminus with a fluorescein amidite (FAM) peptide prevented CENP-QU binding to microtubules (Figure 6C). Conversely, fluorescently labeled rKT21 bound microtubules, as shown previously (Weir et al., 2016) (Figure 6C). However, rKT21 did not rescue microtubule binding of FAM-labeled CENP-QU (Figure 6C), in line with the inability of CENP-QU to interact with the C1–544HIΔ56KMLN complex in rKT21. Further addition of CENP-OP, on the other hand, promoted efficient translocation of FAM-labeled CENP-QU to microtubules, confirming the prediction, based on the experiments in Figure 2D, that CENP-OP promotes the interaction of CENP-QU with rKT21 required to generate rKT26.
We were interested in understanding why modification of the CENP-Q N-terminus interferes with microtubule binding. The sequence of the N-terminal region of CENP-Q is highly basic and predicted to be disordered due to its low complexity (Figure 6D). This is highly reminiscent of NDC80 (also known as HEC1), the microtubule-binding subunit of the NDC80C, where a similarly basic and disordered N-terminal region has been implicated in microtubule binding (Cheeseman et al., 2006, Ciferri et al., 2008, DeLuca et al., 2006) (Figure 6D). To test if the N-terminal region of CENP-Q contributes to microtubule binding, we generated a deletion mutant lacking 67 N-terminal residues (CENP-Q68-C) and co-expressed it with other subunits to generate CENP-Q68-CU and CENP-OPQ68-CUR complexes (Figure 1C). An Alexa488-labeled version of the latter labeled kinetochores robustly when electroporated in HeLa cells (Figures 1D and S1K). Importantly, however, CENP-OPQ68-CUR was largely unable to bind microtubules in the sedimentation and flow cell assays (Figures 5A, 5B, 6A, S6C, and S6D). Thus, the first 67 residues of CENP-Q are dispensable for kinetochore localization but necessary for microtubule binding.
Cooperative Microtubule Binding by rKT26
The competitive binding mode of CENP-QU and NDC80C shown in Figure 6B does not imply that their binding to microtubules within kinetochores is incompatible, as the number of binding sites on the microtubule lattice vastly exceeds the estimated number of NDC80C and CENP-QU binders within a microtubule-binding unit. Rather, it may be surmised that, if incorporated into the same particle, NDC80C and CENP-OPQUR may determine an increase in microtubule-binding affinity if they were concomitantly able to bind microtubules. To test this idea, we first confirmed that CENP-OPQUR or rKT21 (which contains NDC80C) bound to microtubules in isolation in a flow cell. Individually, both decorated microtubules tightly at 400 nM, but when their concentration was reduced to 25 nM, binding of CENP-OPQUR or rKT21 to microtubules appeared drastically reduced. When added together at 25 nM, however, both CENP-OPQUR and rKT21 bound strongly to microtubules (Figure 6E).
Thus, co-existence of CENP-OPQUR and NDC80C within the same complex strongly augments the microtubule-binding activity of rKT26. We reasoned that the augmented microtubule-binding activity of rKT26 ought to be abrogated in presence of CENP-OPQ68-CUR, which does not bind microtubules. In size-exclusion chromatography experiments, CENP-OPQ68-CUR readily bound the other CCAN subunits, indicating that the deletion of the N-terminal region of CENP-Q does not affect this interaction (Figure S7A); this is also in line with the ability of the electroporated mutant complex to reach kinetochores (Figure 1D). In agreement with the prediction, and contrarily with the observation with the wild-type CENP-OPQUR complex, no microtubule binding of rKT21 or of CENP-OPQ68-CUR was observed when these complexes were combined at 25 nM concentration (Figure 6E, bottom two rows). Thus, augmented microtubule binding of rKT26 requires the microtubule binding N-terminal region of CENP-Q.
The NDC80 N-Terminal Region Functionally Replaces CENP-Qs
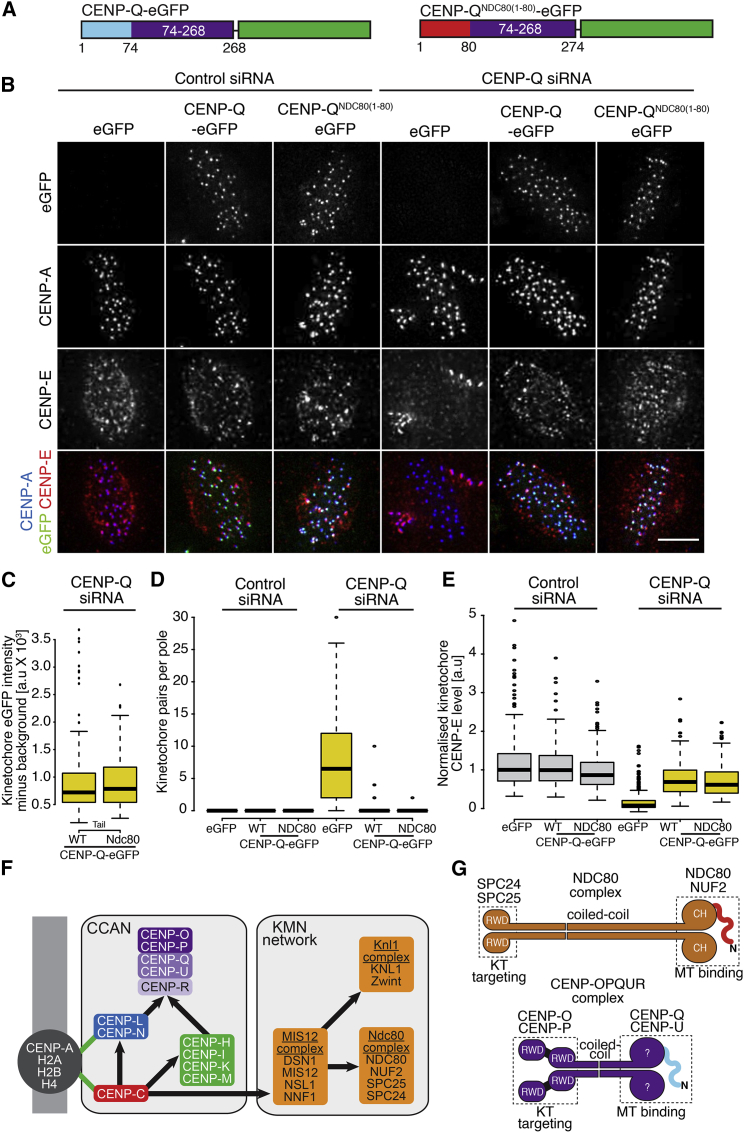
To further test the functional similarity of the CENP-Q and NDC80 N-terminal tails, we built a construct to express wild-type CENP-Q fused to a C-terminal eGFP (CENP-Q-eGFP) or an equivalent construct in which the N-terminal tail was replaced with that of NDC80 (CENP-QNDC80(1-80)-eGFP; Figure 7A). In cells depleted of endogenous CENP-Q, both constructs localized to kinetochores and to similar levels (Figures 7B and 7C). Importantly, depletion of CENP-Q led to a strong accumulation of chromosomes near spindle poles, indicative of congression errors (Figure 7D and Figures S7B and S7C). Expression of CENP-Q-eGFP or of CENP-QNDC80(1-80)-eGFP led to a near complete rescue of the congression phenotype, indicating that both constructs are functional. As a further control of the functionality of the CENP-Q construct, we observed that the strong reduction in CENP-E levels caused by CENP-Q depletion (originally reported by Bancroft et al., 2015) was also rescued in presence of both CENP-Q constructs (Figure 7E). Collectively, these and additional data in Figures 2E and 2F indicate that the CENP-OPQUR complex is required for chromosome alignment in HeLa cells and that the interaction with microtubules mediated by the N-terminal region of CENP-Q is important for this process. The disordered and basic N-terminal tail of NDC80, when grafted onto a CENP-Q mutant lacking its own N-terminal tail, can rescue a requirement for CENP-Q in chromosome alignment.
Figure 7.
The NDC80 N-Terminal Tail Functionally Replaces the CENP-Q Tail
(A) Schematic depicting the CENP-Q-eGFP and CENP-QNDC80(1-80)-eGFP constructs used for the tail-swap rescue experiments.
(B) Immunofluorescence images of the tail-swap rescue experiment. HeLa K cells were treated with control or CENP-Q siRNA and rescued with eGFP, CENP-Q-eGFP, or CENP-QNDC80(1-80)-eGFP transgenes. Cells were treated with MG132 for 90 min prior to fixation and stained with a CENP-E antibody (red) and CREST antisera (blue). Scale bar, 5 μm.
(C) Boxplot depicting the kinetochore eGFP intensity minus background in cells treated with CENP-Q siRNA and rescued with CENP-Q-eGFP or CENP-QNDC80(1-80)-eGFP.
(D) Boxplot showing the number of kinetochores per pole for each condition in the tail-swap rescue experiment.
(E) Boxplot depicting the kinetochore CENP-E intensity after background subtraction and normalisation to CREST for each condition in the tail-swap rescue experiment. In panels (C–E), the thick line represents the median, the box the 25th and 75th percentile, and the whiskers 1.5 times the interquartile range in each direction. Any data points beyond this are represented as single points (outliers).
(F) Schematic as in Figure 1A but after addition of interactions described in this study.
(G) Structural parallels between the NDC80 and CENP-OPQUR complexes. Both complexes have four subunits, with the RWD domain-containing subunits mediating kinetochore binding. In both complexes, basic and disordered N-terminal tails are involved in microtubule binding. A central CC shaft (much longer in NDC80) may separate functional moieties. There is no evidence that CENP-Q or CENP-U contain Calponin Homology (CH) domains also implicated in microtubule binding in the NDC80 complex. See also Figure S7.
Discussion
The comprehensive biochemical and structural analysis of the vertebrate CENP-OPQUR complex described here significantly extends previous studies (Hori et al., 2008b, Kang et al., 2011). After reconstituting rKT21 (Weir et al., 2016), we now report the reconstitution of rKT26, a 26-subunit “successor” that also incorporates the CENP-OPQUR complex. The 22 kinetochore subunits in this complex (the other four being histones) are all assembled on a single CENP-ANCP, with at least two subunits, CENP-C and CENP-N, being able to bind CENP-A directly and specifically (Cao et al., 2018, Carroll et al., 2009, Carroll et al., 2010, Chittori et al., 2018, Guo et al., 2017, Kato et al., 2013, Pentakota et al., 2017, Tian et al., 2018, Weir et al., 2016).
On the basis of the distributions of its binding sites for other kinetochore proteins, we have recently proposed that CENP-C, which is predicted to be largely unstructured, may act as a “blueprint” for the assembly of kinetochores (Klare et al., 2015). CENP-C binds MIS12C at its N terminus (Liu et al., 2016, Przewloka et al., 2011, Richter et al., 2016, Screpanti et al., 2011) and CENP-A via a motif in its central region (Kato et al., 2013). CENP-C also binds CCAN within a domain between the MIS12C and CENP-A binding sites (Hinshaw and Harrison, 2013, Klare et al., 2015, McKinley et al., 2015, Nagpal et al., 2015, Pentakota et al., 2017) (Figure 4D). CENP-C probably meanders on and through the structure of CCAN, creating multiple contacts that stabilize it, thus explaining why its presence is so crucial for kinetochore integrity (e.g., see Carroll et al., 2010, Milks et al., 2009), even if our studies clearly argue that CCAN is endowed with substantial structural stability even in the absence of CENP-C.
The 3D EM reconstruction of the CENP-HIΔ56KMLNOPQUR complex provides the first near-comprehensive structural analysis of the CCAN, albeit at low resolution, and extends our previous analysis of CENP-HIΔ56KM (Basilico et al., 2014). In spite of its low resolution, the reconstruction allows important conclusions on the organization of CCAN. Most notably, the CCAN subcomplex we have studied is very compact and globular, in contrast to the fibrous organization of the KMN in the outer kinetochore (Ciferri et al., 2008, Dimitrova et al., 2016, Petrovic et al., 2014, Petrovic et al., 2016, Valverde et al., 2016). Within CCAN, CENP-N binds directly to CENP-A nucleosomes in vitro, and structural information on this interaction has recently emerged (Chittori et al., 2018, Pentakota et al., 2017, Tian et al., 2018). Besides binding directly to the CENP-A nucleosome, CCAN may serve as a spacer to position KMN for a favorable interaction with the microtubule end. A nanometer-scale map of kinetochores concluded that the CCAN subunits are clustered within approximately 20 nm along the longitudinal (kinetochore-microtubule) axis of the kinetochore (Suzuki et al., 2014). Figure 4D presents a structural model of the kinetochore that incorporates available structural and functional information, including studies shown in Figures 1, 2, 3, and 4. The architecture shown in Figure 4D probably defines a conserved module of eukaryotic kinetochores present in single copy in the “point” kinetochores of S. cerevisiae (Pluta et al., 1995), which bind a single microtubule, or in multiple copies in the “regional” kinetochore of humans, which bind ∼25 microtubules (Musacchio and Desai, 2017).
Still missing from this reconstitution to completely represent the core subunits is CENP-TW (possibly operating in complex with CENP-SX; Nishino et al., 2012). Together with CENP-C, CENP-TW promotes outer kinetochore assembly by binding directly to KMN network components (Gascoigne et al., 2011, Hori et al., 2008a, Huis In 't Veld et al., 2016, Przewloka et al., 2011, Screpanti et al., 2011, Weir et al., 2016). The interaction of recombinant CENP-TW with the 26-subunit kinetochore complex we describe is weak (Basilico et al., 2014, Weir et al., 2016; and our unpublished observations; of note, this does not appear to be the case in S. cerevisiae, where Cnn1CENP-T interacts strongly with the ortholog of the CENP-HIKM complex: Pekgöz Altunkaya et al., 2016). Recent studies suggest that vertebrate CENP-TW docks on DNA that bridges neighboring nucleosomes (Takeuchi et al., 2014, Thakur and Henikoff, 2016), and therefore its incorporation into our recombinant particles may require the engineering of suitable high-affinity chromatin templates. CENP-TW is required for CENP-OPQUR recruitment to kinetochores (Gascoigne et al., 2011), but this likely reflects the established role of CENP-TW in stabilizing CENP-HIKMLN at the kinetochore (Basilico et al., 2014, Carroll et al., 2010, Foltz et al., 2006, Gascoigne et al., 2011, Hori et al., 2008a, Huis In 't Veld et al., 2016, Nishino et al., 2012, Okada et al., 2006, Pekgöz Altunkaya et al., 2016, Samejima et al., 2015, Wood et al., 2016), because we failed to observe a direct interaction of CENP-OPQUR with CENP-TW (M.E.P. and A.M., unpublished data).
An important conclusion is that CENP-OPQUR occupies an outermost position of the inner kinetochore, as its recruitment there requires concomitant binding to the centromere-proximal subunits CENP-LN and CENP-HIKM. Conversely, CENP-OPQUR is dispensable for recruitment of the proximal subunits (Eskat et al., 2012, Foltz et al., 2006, Hori et al., 2008b, Izuta et al., 2006, Kagawa et al., 2014, McClelland et al., 2007, McKinley et al., 2015, Minoshima et al., 2005, Okada et al., 2006, Samejima et al., 2015) (Figure 7F). This organization may differ significantly in S. cerevisiae, where the COMA complex appears to occupy an upstream position in the recruitment order of kinetochore subunits (Hinshaw et al., 2017, Pekgöz Altunkaya et al., 2016, Schmitzberger et al., 2017). Future studies will have to clarify what molecular adaptations underlie these differences. CENP-OP and CENP-QU form stable subcomplexes, which explains why CENP-O and CENP-P on the one hand, and CENP-Q and CENP-U on the other, are interdependent for physical stability in chicken DT40 cells (Hori et al., 2008b). In our work, CENP-OP emerged as the main factor promoting kinetochore targeting of CENP-OPQUR, even if CENP-Q and CENP-U have been shown to further stabilize it (Bancroft et al., 2015, Hori et al., 2008b, Kang et al., 2006). The interaction of Ame1CENP-U with the orthologs of CENP-HIKM and CENP-LN in S. cerevisiae was shown to require Ctf19CENP-P and Mcm21CENP-O (Pekgöz Altunkaya et al., 2016). A motif located near the C-terminus of Okp1CENP-Q mediates a physical interaction with the RWD domains of the Ctf19CENP-P:Mcm21CENP-O dimer (Schmitzberger et al., 2017). The Okp1 motif, however, does not appear to be conserved outside closely related yeast species, leading us to speculate that the interaction of the human CENP-OP and CENP-QU subcomplexes studied here uses different determinants.
Here, we have also identified a novel microtubule-binding site in the basic N-terminal tail of CENP-Q. Many features of the CENP-Q N-terminal tail, most notably its highly basic isoelectric point and its tendency to structural disorder due to low sequence complexity, are highly reminiscent of the N-terminal tail of NDC80. The precise contribution of the latter to microtubule binding remains poorly understood from a mechanistic perspective, but there is ample evidence that it is required for biorientation and tight microtubule binding of NDC80 (Alushin et al., 2010, Alushin et al., 2012, Cheeseman et al., 2006, Ciferri et al., 2008, DeLuca et al., 2006, DeLuca et al., 2011, DeLuca et al., 2018, Zaytsev et al., 2014, Zaytsev et al., 2015). Two crucial differences between the CENP-Q and NDC80 tails are that 1) the NDC80 tail flanks a calponin homology (CH) domain that contributes to microtubule binding by NDC80 (Ciferri et al., 2008, Wei et al., 2007), and by structural modeling we found no conclusive evidence that CENP-Q (or CENP-U) contain calponin homology (CH) domains (unpublished data); and 2) the NDC80 tail is subject to regulation to phosphorylation by multiple kinases, including Aurora A and B, which may phosphorylate up to nine consensus sites in the tail, and Cdk1, for which there is at least one consensus site (Cheeseman et al., 2006, Ciferri et al., 2008, DeLuca et al., 2006, DeLuca et al., 2011, DeLuca et al., 2018, Zaytsev et al., 2014, Zaytsev et al., 2015). Inspection of the CENP-Q N-terminal tail reveals only one or two Aurora consensus sites and no Cdk1 site.
While future studies will have to address the functional implications of these differences, the ability of the NDC80 N-terminal tail to replace the CENP-Q tail and promote chromosome alignment is striking. Given that CENP-OPQUR occupies, with the CCAN, a position near the “bottom” of the kinetochore (Figure 4D), we surmise that its microtubule-binding activity may become especially important after establishment of end-on attachment. A previous study identified NDC80C as a passive force generator within vertebrate kinetochores and recognized the existence of an active force generator whose molecular identity remained unclear but whose position within kinetochores is compatible with that attributed to the CENP-OPQUR complex (Dumont et al., 2012). In TIRF microscopy experiments, we observed rare events in which individual CENP-OPQU complexes labeled with Alexa488 tracked depolymerizing microtubules (unpublished data), a property expected for an active force generator. The tools we describe here will enable a detailed study of this hypothesis.
The similarities between NDC80C and CENP-OPQU are not limited to the basic disordered N-terminal tails. Both complexes appear to “subdivide labor” by assigning kinetochore targeting and microtubule binding to different subcomplexes. In NDC80C, SPC24:SPC25 and NDC80:NUF2 subcomplexes mediate kinetochore recruitment and microtubule binding, respectively. In CENP-OPQU the same functions are attributed to CENP-OP and CENP-QU, respectively. CENP-O, CENP-P, SPC24, and SPC25 are structurally related proteins containing RWD domains (Ciferri et al., 2008, Petrovic et al., 2014, Schmitzberger and Harrison, 2012, Wei et al., 2006) (Figure 7G). On the other hand, the extent to which CENP-QU is structurally related to the NDC80:NUF2 dimer is unclear. CENP-Q and CENP-U are predicted to contain several α helices (Westermann and Schleiffer, 2013), the main secondary structure element of CH domains, and have predicted C-terminal coiled-coils (Figure 1B) like NDC80 and NUF2.
Although the phenotypic consequences of depleting CENP-OPQUR subunits vary in severity depending on the affected cell type (Kagawa et al., 2014, McKinley et al., 2015), our results are largely consistent with previous studies that identified severe chromosome alignment problems in cells depleted of CENP-OPQUR (Bancroft et al., 2015, Hori et al., 2008b, Hua et al., 2011, McAinsh et al., 2006, McClelland et al., 2007, Minoshima et al., 2005, Toso et al., 2009). We further document the chromosome alignment problems caused by CENP-P depletion in Figures S7D–S7F. The importance of CENP-OPQUR is further corroborated by a recent study that identified CENP-O and CENP-P in a group of gene products involved in chromosome alignment and whose depletion is compatible with cell viability but only in presence of a functional spindle assembly checkpoint (Raaijmakers et al., 2018). This is consistent with the idea that, in the absence of the CENP-OPQUR complex, achievement of bi-orientation is delayed and a call on the spindle checkpoint to delay mitotic exit is issued, without which cells undergo a catastrophic, lethal division. Our analysis indicates that, besides its contributions to the recruitment and regulation of Plk1 and CENP-E (Ahonen et al., 2005, Bancroft et al., 2015, Hori et al., 2008b, Kang et al., 2006, Kang et al., 2011, Nishino et al., 2006), CENP-OPQUR contributes to chromosome alignment through direct microtubule binding. In conclusion, our study fills an important gap in our understanding of human kinetochores and paves the way to full functional reconstitution of kinetochore function in the test tube.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal GFP | ABCAM | #AB6556; RRID: AB_305564 |
| Rabbit polyclonal anti-CENP-OP | Generated in house | N/A |
| Mouse monoclonal anti-CENP-P | ABCAM | #AB66058; RRID: AB_1523338 |
| Goat polyclonal anti-CENP-QU | Generated in house | N/A |
| Mouse monoclonal anti-CENP-Q | ABCAM | #AB57539; RRID: AB_940733 |
| Mouse monoclonal anti-CENP-R | ABCAM | #AB57098; RRID: AB_304751 |
| Rabbit polyclonal anti-CENP-HK | Generated in house | #SI0930 |
| Mouse polyclonal anti-CENP-A | Gene Tex | #GTX13939; RRID: AB_369391 |
| Mouse monoclonal anti-CENP-A | ABCAM | #AB13939; RRID: AB_300766 |
| Rabbit polyclonal anti-CENP-C(23-410) | Trazzi et al., 2009 | #SI410 |
| Rabbit polyclonal anti-CENP-N(1-212) | Generated in house | N/A |
| Rabbit polyclonal anti-CENP-E | Meraldi Lab; Meraldi et al., 2004 | N/A |
| Human anti-centromere (CREST) | Antibodies Inc. | Cat#15-234-0001; RRID: AB_2687472 |
| Bacterial and Virus Strains | ||
| E.coli: BL21(DE3)-RIL strain | Agilent Technologies | #230240 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| HRV 3C Prescision Protease | Generated in house | N/A |
| TEV Protease | Generated in house | N/A |
| Lambda phosphatase | Generated in house | N/A |
| Protease-inhibitor mix HP Plus | Serva | Cat#39107 |
| Sortase A delta 59 (S.aureus) | Hidde Ploegh Lab | Addgene:Cat#51139 |
| Alexa Fluor 488 C5 maleimide Protein labeling kit | ThermoFisher | #A10254 |
| Alexa Fluor 647 C2 maleimide Protein labeling kit | ThermoFisher | #A20347 |
| Alexa Fluor 405 Carboxylic acid, succinimidyl ester Protein labeling kit | ThermoFisher | #A30000 |
| NHS-Rhodamine labeling kit | Thermo Scientific | #46406 |
| FAM-LPETGG | Genscript | N/A |
| PhosSTOP phosphatase inhibitor | Roche | Cat#04906845001 |
| (+)-S-Trityl-L-cysteine (STLC) | Sigma Aldrich | #164739 |
| RO-3306 | Millipore | #217699 |
| MG-132 | Calbiochem | CAS 133407-82-6 |
| MG-132 | Sigma | SML1135 |
| Uranyl formate | SPI Supplies | CAS#16984-59-1 |
| Fetal bovine serum (FBS) | Clonetech | #631107 |
| Fetal bovine serum (FBS) | Sigma | Cat:f7524 Batch:111M3395 |
| Zeocin | Invitrogen | Cat#R25001 |
| L-glutamine | PAN Biotech | P04-80100 |
| Nocodazole | Sigma | Cat#M1404 |
| CENP-C(1-544) | Musacchio Lab; Screpanti et al., 2011 | N/A |
| CENP-HIKM | Musacchio Lab, Basilico et al., 2014 | N/A |
| CENP-LN | Musacchio Lab, Pentakota et al., 2017 | N/A |
| CENP-CHIKM | Musacchio Lab; Klare et al. 2015 | N/A |
| NDC80 complex | Musacchio Lab; Huis In 't Veld et al., 2016 | N/A |
| MIS12 complex | Musacchio Lab; Petrovic et al., 2014 | N/A |
| KNL1 complex | Musacchio Lab; Petrovic et al., 2014 | N/A |
| CENP-A NCP | Musacchio Lab; Weir et al., 2016 | N/A |
| Tubulin (in TIRF experiment) purified from pig brains | N/A | N/A |
| poly-L-lysine-poly-ethylene-glycol-biotin | SUSOS-AG | PLL (20) -g[3.5]- PEG(2)/PEGbi) |
| streptavidin | Sigma | S4762; CAS#: 9013-20-1 |
| GMP-CPP | Jena Biosciences | GpCpp, NU-405S CAS#: 14997-54-7 |
| Porcine tubulin (biotin labelled) | Cytoskeleton | Cat. # T333P-A |
| HiLyte 647 labeled porcine tubulin | Cytoskeleton | Cat. # TL670M-A |
| Paclitaxel (Taxol) | Sigma | T7402; CAS#: 33069-62-4 |
| Glucose oxidase | Sigma | G7141; CAS#:9001-37-1 |
| Catalase | Sigma | SRE0041 CAS#:9001-05-2 |
| DAPI | Sigma | D9542 |
| PenStrep | Gibco | 15-140 |
| Fugene6 | Promega | E2691 |
| Oligofectamine | Invitrogen | 12252011 |
| Critical Commercial Assays | ||
| QIAquick Kit | Qiagen | 28704 |
| Mini-prep kit | Qiagen | 27104 |
| Maxi-prep kit | Qiagen | 10023 |
| Experimental Models: Cell Lines | ||
| Trichoplusia ni:BTI-Tnao38 | Garry W Blissard Lab | N/A |
| S.frugiperda:Sf9 cells | ThermoFisher | Cat#12659017 |
| HeLa cells | IEO Milan | N/A |
| Human: Flp-IN T-Rex HeLa | S.S. Taylor, University of Manchester | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-O | This paper | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-P | This paper | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-P(F116G) | This paper | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-Q | This paper | N/A |
| Human: HeLa K cells | Meraldi Lab | N/A |
| Human: HeLa K-CENP-Q-eGFP | This paper | N/A |
| Human: HeLa K-CENP-Q(Ndc80(1-80))-eGFP | This paper | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-U | This paper | N/A |
| Human: Flp-In T-Rex HeLa-GFP-CENP-R | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| E.coli: BL21(DE3)-RIL strain | Agilent Technologies | #230240 |
| E.coli: BL21CodonPlus(DE3)-RIL strain | Agilent Technologies | #230280 |
| Oligonucleotides | ||
| CENP-O siRNA: 5’-UAGGAGACCAGACUCAUAU-3’ |
Dharmacon | |
| CENP-P SMARTpool: 5’-GUGCAAGAGAGAACAACUA-3’ 5’-AGUCAUUGUUUGGAGGAUA-3’ 5’-UAUCGUAAGCGCACGUUUA-3’ 5-CCUAAGUGCUAUAUCGAUC-3’ |
Dharmacon | |
| CENP-P siRNA 5’-GAACCCTGGTAGGACTGCTTGGAAT-3’ |
Invitrogen Stealth; Amaro et al., 2010 | |
| CENP-Q SMARTpool: 5’-GAGUUAAUGACUGGGAAUA-3’, 5’-AUGGAAAGGGCACGAGACA-3’ 5’-ACAAAGCACACUAACCUAA-3’ 5’-UGUCAGAGAAAUAAGGUUAG-3’ |
Dharmacon | |
| CENP-Q siRNA: 5‘-GGUCUGGCAUUACUACAGGAAGAAA-3‘ |
Invitrogen Stealth; Bancroft et al., 2015 | |
| CENP-R SMARTpool: 5’-GAAGUUGGAUGGUCUGUUA-3’ 5’-UGACAGCUAUGAAUUCCUU-3’ 5’-UAAGUAGUAUACAGGCUUU-3’ 5’-GAAUUCAUGAUGUUGCUAU-3’ |
Dharmacon | |
| CENP-H siRNA: 5’-CUAGUGUGCUCAUGGAUAA-3’ |
Dharmacon | |
| CENP-L siRNA: 5’-UUUAUCAGCCACAAGAUUA-3’ |
Dharmacon | |
| CENP-N SMARTpool: 5’-CUACCUACGUGGUGUUACUA-3’ 5’-GUUCAGCACUUGAUCCAUC-3’ 5’-AUACACCGCUUCUGGGUCA-3’ 5’-ACACAAAGCCAAACCAGUA-3’ |
Dharmacon | |
| Recombinant DNA | ||
| pGEX-2rbs | Musacchio Lab | N/A |
| pGEX-2rbs-CENP-R | This study | N/A |
| pGEX-2rbs-CENP-C(1-544) | Screpanti et al., 2011 | N/A |
| pGEX-2rbs-CENP-C(1-544)-SNAP | This study | N/A |
| MultiBac | Geneva Biotech | N/A |
| pFL-6His-CENP-P:CENP-O | This study | N/A |
| pFL-6His-CENP-P(F116G):CENP-O | This study | N/A |
| pFL-6His-CENP-Q:CENP-U | This study | N/A |
| pUCDM-CENP-R | This study | N/A |
| pLIB | Peters Lab. | Addgene #80610 |
| pBIG1A | Peters Lab. | Addgene #80611 |
| pLIB CENP-O | This study | N/A |
| pLIB CENP-P | This study | N/A |
| pLIB CENP-Q | This study | N/A |
| pLIB CENP-Q(68-C) | This study | N/A |
| pLIB CENP-U | This study | N/A |
| pLIB CENP-R | This study | N/A |
| pBIG1A with CENP-OPQUR (His-CENP-Q) | This study | N/A |
| pBIG1A with CENP-OPQ(68-C)UR (His-CENP-Q) | This study | N/A |
| pCDNA 5/FRT/TO | Invitrogen | Cat#V6520-20 |
| pCDNA 5/FRT/TO EGFP | Musacchio Lab; Krenn et al., 2012 | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-O | This study | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-P | This study | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-P(F116G) | This study | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-Q | This study | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-U | This study | N/A |
| pCDNA 5/FRT/TO EGFP-CENP-R | This study | N/A |
| peGFP-C1 | Clontech | #6085-1 |
| pCENP-Q-eGFP | McAinsh Lab; Bancroft et al., 2015 | N/A |
| pCENP-Q(NDC80(1-80))-eGFP | This study | N/A |
| Software and Algorithms | ||
| GraphPad Prism 6.0 | GraphPad software | http://www.graphpad.com |
| Imaris 7.3.4 32-bit | Bitplane | http://www.bitplane.com/imaris |
| ImageJ 1.46 r | NIH | http://imageJ.nih.gov/ij/ |
| SoftWorx | Applied Precision | NA |
| Image Lab | Bio-rad | https://www.bio-rad.com/de-de/product/image-lab-software?ID=KRE6P5E8Z |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera |
| SPHIRE suit | Moriya et al., 2017 | http://www.sphire.mpg.de |
| SEDFIT | Schuck, 2000 | http://www.analyticalultracentrifugation.com/default.htm |
| SEDNTERP | Laue et al., 1992 | http://bitcwiki.sr.unh.edu/index.php/Main_Page |
| GUSSI | Chad Brautigam | http://biophysics.swmed.edu/MBR/software.html |
| PrDOS | Ishida and Kinoshita, 2007 | http://prdos.hgc.jp/cgi-bin/top.cgi |
| COILS | Lupas et al., 1991 | https://embnet.vital-it.ch/software/COILS_form.html |
| Deposited Data | ||
| Mendeley data | This paper | https://doi.org/10.17632/yv552m8s98.1 |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by Andrea Musacchio (andrea.musacchio@mpi-dortmund.mpg.de).
Experimental Model and Subject Details
cDNAs used for expression of recombinant proteins were either of human origin, or generated synthetically based on human sequences. HeLa (female Cervix Adenocarcinoma) and USOS cells were grown in DMEM (PAN Biotech) supplemented with 10% FBS, penicillin and streptomycin and 2 mM L-glutamine. Cells were grown in a humidified atmosphere of 37 °C and 5% CO2.
Method Details
Plasmids
For expression and purification of recombinant proteins, synthetic codon optimized DNA (Genscript) encoding human CENP-O, CENP-P, CENP-Q, CENP-U and CENP-C were used. The gene encoding for CENP-R was PCR amplified from human cDNA. CENP-R was subcloned in pGEX-6P-2rbs, a modified pGEX-6P vector (GE Healthcare) as a C-terminal 3C precision cleavable tag fusion to the sequence encoding GST and in Multibac pUCDM vector with no tag. CENP-P and CENP-PF116G were subcloned in a MultiBac pFL-derived vector (Bieniossek et al., 2012) with an N-terminal TEV cleavable 6xHis tag, under the control of the polh promoter. A codon optimized human CENP-O was subcloned in the 2nd MCS of the same vector, under the control of the p10 promoter. Simultaneously, others pFL-based vectors were created with an N-terminal TEV cleavable 6xHis tag on CENP-Q, or CENP-Q68-C and CENP-U under the control of the polh and p10 promoters, respectively. CENP-C (residues 1 to 544) was PCR amplified using a forward primer carrying BglII and a reverse primer carrying BamH1-Stop-SalI and subsequently cloned in the first cassette of pGEX-6P-2rbs. The SNAP tag was amplified with primers carrying BamH1 (forward) and SalI (reverse) and was cloned into the pGEX-CENPC1-544 construct using the same sites resulting in C-terminal tagging. Site- directed mutagenesis, performed with QuickChange Mutagenesis kit (Agilent Technologies) was used to generate mutant versions of recombinant proteins. Constructs were sequence verified.
Plasmids for stable cell lines were generated in pCDNA5/FRT/TO-EGFP-IRES, a modified version of the pCDNA5/FRT/TO vector (Invitrogen, Carlsbad, CA). The control plasmid for EGFP expression was created by PCR amplifying the EGFP sequence from pEGFP-C1 (Takara Bio Inc.) and cloning it into the pcDNA5/FRT/TO vector previously modified to carry an internal ribosomal entry site (IRES) sequence to create the pcDNA5/FRT/TO EGFP-IRES vector (Petrovic et al., 2010). All plasmids used in the study for mammalian expression were derived from the pCDNA5/FRT/TO-EGFP-IRES and used for genomic integration and expression of human CENP-OPQUR proteins. To create all EGFP tagged proteins, we amplified individual CENP-OPQUR full-length proteins by PCR from full-length human cDNA. RNAi-resistant CENP-O and CENP-P were amplified from codon-optimized cDNA synthesized by GeneArt (Life Technologies) and then subcloned into the pcDNA5/FRT/TO EGFP-IRES vector using the restriction sites BamHI and XhoI. Mutant construct, CENP-P F116G, was created by site-directed mutagenesis of CENP-O and CENP-P siRNA resistant constructs respectively. All clones were sequence verified.
Protein expression and purification
Escherichia coli BL21 (DE3) cells harboring vectors expressing CENP-R were grown in Terrific Broth at 37°C to an OD600 of 0.6 - 0.8, when 0.2 mM IPTG was added and the culture was grown at 18°C for ∼15 hours. Cell pellets were resuspended in lysis buffer (20 mM Tris/HCl pH 6.8, 300 mM NaCl, 10% glycerol and 5 mM 2-mercaptoethanol) supplemented with protease inhibitor cocktail (Serva), lysed by sonication and cleared by centrifugation at 108,000 g at 4°C for 1 hour. The cleared lysate was filtered (0.8 μm) and applied to Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) pre-equilibrated in lysis buffer, incubated at 4°C for 2 hours, washed with 50 volumes of lysis buffer and subjected to an overnight cleavage reaction with HRV 3C Prescision Protease (in house generated) to separate CENP-R from GST. The sample containing CENP-R was loaded on 5ml HiTrap Heparin HP column (GE Healthcare) pre-equilibrated in 20 mM Tris/HCl pH 6.8, 300 mM NaCl, 5% glycerol, 0.5 mM EDTA and 1 mM DTT. The sample was eluted with a linear gradient of 300 - 1000 mM NaCl in 15 bed column volumes. Fractions containing CENP-R were pooled, concentrated and loaded onto a Superdex 200 16/60 SEC column (GE Healthcare) pre-equilibrated in SEC buffer (20 mM Tris/HCl pH 6.8, 500 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP). Fractions containing CENP-R were concentrated, flash-frozen in liquid nitrogen and stored at -80°C.
Expression and purification of CENP-OPQUR (and CENP-OPQ68-CUR, CENP-OPQU, CENP-OP (and CENP-OPF116G), and CENP-QU (and CENP-Q68-CU) complexes was carried out in insect cells using a MultiBac system. Production of high-titer V2 virus was carried out separately for pFL-CENP-P-6xHis:CENP-O, pFL-CENP-Q-6xHis:CENP-U and pUCDM-CENP-R in Sf9 cells. Tnao38 insect cells (Hashimoto et al., 2012) were used for expression (96 hours, 27°C) after which the cells were centrifuged, washed once in PBS, and resuspended in lysis buffer. Cell pellets infected with CENP-OP (or its mutants) virus were resuspended in lysis buffer (20 mM Tris/HCl pH 8.0, 300 mM NaCl, 5 mM imidazole, 5% (v/v) glycerol and 5 mM 2-mercaptoethanol) supplemented with protease inhibitor cocktail, lysed and cleared. The cleared lysate was applied to 5 ml HisTALON Cartridges pre-packed with TALON Superflow Resin (Clontech) pre-equilibrated in lysis buffer, washed with 10 volumes of lysis buffer. Bound proteins were eluted with lysis buffer supplemented with 250 mM imidazole. Tag cleavage with TEV protease (in house production) was performed for 15 hours at 4°C and the fractions containing the CENP-OP complex were then diluted in 10 volumes of 20 mM Tris/HCl pH 8.0, 5% glycerol, 0.5 mM EDTA and 1 mM DTT. Resource Q anion exchange chromatography column (GE Healthcare) was pre-equilibrated in 20 mM Tris/HCl pH 8.0, 30 mM NaCl, 5% (v/v) glycerol, 0.5 mM EDTA and 1 mM DTT. The sample now adjusted to a salt concentration of 30 mM was loaded onto the Resource Q column and eluted with a linear gradient of 30 - 500 mM NaCl in 15 bed column volumes. Fractions containing CENP-OP complex were pooled and de-phosphorylated by Lambda-phosphatase (in house production) for ∼15 hours at 4C. The sample was after concentrated and loaded onto a Superdex 200 10/300 or 16/60 SEC column (GE Healthcare) pre-equilibrated in SEC buffer (20 mM Tris pH 8.0, 150 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP). Fractions containing CENP-OP complex were concentrated, flash-frozen in liquid nitrogen and stored at -80°C.
Cell pellets infected with CENP-QU, CENP-Q68-C, CENP-OPQU, CENP-OPQUR and CENP-OPQ68-CUR viruses were resuspended in lysis buffer (20 mM Tris/HCl pH 6.8, 300 mM NaCl, 5 mM imidazole, 5% glycerol and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail, lysed and cleared. The cleared lysate was applied to 5ml HisTALON column pre- equilibrated in lysis buffer, washed with 10 volumes of lysis buffer. Bound proteins were eluted with lysis buffer supplemented with 250 mM imidazole. Tag cleavage was performed with TEV protease. The sample was loaded on 5 ml HiTrap Heparin HP column (GE Healthcare) pre-equilibrated in 20 mM Tris/HCl pH 6.8, 300 mM NaCl, 5% glycerol, 0.5 mM EDTA and 1 mM DTT. The sample was eluted with a linear gradient of 300 mM - 1M NaCl in 15 bed column volumes. Fractions containing the complexes of interest were pooled and de-phosphorylated. Following which the sample was concentrated and loaded onto a Superdex 200 10/30 or 16/60 SEC column pre-equilibrated in SEC buffer (20 mM Tris pH 6.8, 500 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP). The relevant fractions were pooled, concentrated, flash-frozen in liquid nitrogen and stored at -80°C.
Escherichia coli BL21 (DE3) cells harboring vectors expressing CENP-C1-544-SNAP were grown in Terrific Broth at 37°C to an OD600 of 0.6 - 0.8, at which time 0.2 mM IPTG was added and the culture was grown at 18°C for ∼15 hours. Cell pellets were resuspended in lysis buffer (25 mM Na-Hepes pH 7.5, 300 mM NaCl, 1 mM DTT and 1 mM PMSF) supplemented with protease inhibitor cocktail (Serva), lysed by sonication and cleared by centrifugation at 108,000 g at 4°C for 1 hour. The cleared lysate was filtered (0.8 μm) and applied to Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) pre-equilibrated in lysis buffer, incubated at 4°C for 2 hours, washed with 50 volumes of lysis buffer and subjected to an overnight cleavage reaction with HRV 3C Prescision Protease to separate CENP-C1-544-SNAP from GST. The sample containing CENP-C1-544 was loaded onto a Resource S 6 ml column column (GE Healthcare) pre-equilibrated in 20 mM Tris/HCl pH 6.8, 300 mM NaCl, 5% glycerol, 0.5 mM EDTA and 1 mM DTT. The sample was eluted with a linear gradient of 300 - 2000 mM NaCl in 15 bed column volumes. Fractions containing CENP-C1-544 were pooled, concentrated and loaded onto a Superdex200 16/60 SEC column (GE Healthcare) pre-equilibrated in SEC buffer (20 mM Tris/HCl pH 6.8, 500 mM NaCl, 5% (v/v) glycerol and 1 mM TCEP). Fractions containing CENP-C were concentrated, flash-frozen in liquid nitrogen and stored at -80°C. Other proteins were purified with detailed protocols (Basilico et al., 2014, Klare et al., 2015, Petrovic et al., 2014, Petrovic et al., 2010, Tachiwana et al., 2015, Weir et al., 2016).
Protein fluorescence labeling
CENP-OP, CENP-QU, CENP-Q68-CU, CENP-OPQU, CENP-OPQUR, CENP-OPQ68-CUR, CENP-LN, and the KMN complex were labeled using different Alexa Fluor protein labeling kit (Thermo Fisher Scientific) according to the manufacturer instructions. Purified S. aureus Sortase (Guimaraes et al., 2013) was used to label CENP-QU with LPETGG peptides with a N-terminally conjugated fluorescein amidite (FAM) (Genscript). Labeling was performed for ∼14 hr at 4°C in the presence of 10 mM CaCl2 using molar ratios of Sortase, CENP-QU, and peptide of approximately 1:20:200. CENP-QUFAM was separated from Sortase and the unreacted peptides by size-exclusion chromatography.
Electroporation of CENP-OPQUR into mitotic and interphase cells
HeLa cells depleted for both CENP-P and CENP-Q were harvested, washed in PBS and electroporated with 3.5 μM of either recombinant Alexa-488 labeled CENP-OPQUR or CENP-OPQ68-CUR. As control we used Alexa-488. Following electroporation (Neon Transfection System, Thermo Fisher) and recovery, cells were either fixed for IF, or synchronized in G2 with 9 μM RO-3306 (Millipore) for 16 hours and then released from G2 in the presence of 5 μM STLC for 2 hours (Sigma-Aldrich). Following STLC wash-out, cells were grown for 150 minutes in media containing 5 μM MG132 (Calbiochem), fixed, prepared for immunofluorescence analysis and then scored for the presence of uncongressed choromosomes. Results are representing the average and standard deviation of two replicated experiments. In total, between 603 and 731 cells were scored for each condition. Experiments were imaged on a Deltavision Elite System (see below for description). Scale bar is 5 μm.
Analytical SEC analysis
Analytical size exclusion chromatography was carried out on a Superdex 200 5/150 or Superose 6 5/150 in a buffer containing 20 mM Na-HEPES pH 7.5, 300 mM NaCl, 5% glycerol and 1 mM TCEP on an ÄKTA micro system. All samples were eluted under isocratic conditions at 4°C in SEC buffer (20 mM Hepes pH 7.5, 300 mM NaCl, 5% Glycerol and 1 mM TCEP) at a flow rate of 0.2 ml/min. Elution of proteins was monitored at 280 nm. 100 μl fractions were collected and analysed by SDS-PAGE and Coomassie blue staining. To detect the formation of a complex, proteins were mixed at the concentrations of 5 μM (except CENP-ANCP, with a concentration of 2.5 μM) in 50 μl, incubated for at least 1 hr on ice and then subjected to SEC.
Co-infection and co-purification of CENP-R with CENP-OPQUR subunits
For each His-pull-down experiment, 50 ml of freshly diluted Tnao38 cells at a density of 106 cells/ml in serum-free medium (Sf-900 II SFM, Life Technologies) were co-infected with CENP-R virus and CENP-O/His-CENP-P or/and CENP-U/HisCENP-Q viruses using a virus:culture ratio of 1:30 for each virus at 27°C for 96 hours. Cell pellets were resuspended in lysis buffer (20 mM Tris pH 6.8, 500 mM NaCl, 5 mM imidazole, 10% (v/v) glycerol and 1 mM TCEP) supplemented with protease inhibitor cocktail (Serva), lysed by sonication and cleared by centrifugation at 108,000 g at 4°C for 30 min. The cleared lysate was applied to 1 ml HisTALON column (GE Healthcare) pre-equilibrated in lysis buffer, washed with 10 volumes of lysis buffer and eluted with lysis buffer supplemented with 250 mM imidazole. Samples of total lysate, supernatant, flow through and elution were analysed by SDS-PAGE, Coomassie blue staining and by western blotting.
Sample preparation for electron microscopy
The protein samples (CENP-OPQU and CENP-OPQUR) were separated on a Superdex 200 10/300 or Superose 6 10/300 SEC column (pre-equilibrated in 20 mM Na-HEPES pH 7.5, 300 mM NaCl and 1 mM TCEP). The fractions of interest were pooled and concentrated to 15 μM. Purified CENP-HIΔ56KM, CENP-LN and CENP-OPQUR complexes were incubated at 15 μM in 500 μl for 1 hour at 4C and separated on a Superose 6 10/300. The fractions containing the 11 proteins complex were pooled and concentrated to 15 μM. Samples were purified and stabilized via the GraFix method (Kastner et al., 2008): two 2 ml gradients ranging from 20 to 50% glycerol in 20 mM Na-HEPES pH 7.5, 300 mM NaCl and 1 mM TCEP were set up. In one of the gradients, the 50% solution contained 0.025% of glutaraldehyde. 30 μl of sample was applied to each gradient and centrifuged by ultracentrifugation at 150,000 g at 4°C for 16 hours. The samples from the cross-linked gradient and the non-cross-linked gradient were fractionated and analyzed by SDS-PAGE, Coomassie blue staining and immunoblotting. The fractions of interest were buffer exchanged to 20 mM HEPES pH 7.5, 300 mM NaCl, 2.5% (v/v) glycerol and 1mM TCEP with PD SpinTrap G-25 column (GE Healthcare) prior EM experiment. 4 μl of the samples was used non-diluted. To analyze microtubule bundling by CENP-OPQUR, 4 μl of the pellets fraction of the microtubule co-sedimentation assay (prepared as described below) was used non-diluted.
Preparation of negative stain specimens and electron microscopy
Negative stain specimens were prepared as described previously (Bröcker et al., 2012). 4 μl of the cross-linked sample were absorbed at 25 °C for 1 min onto freshly glow-discharged 400 mesh carbon-coated copper grids (G2400C, Plano GmbH, Wetzlar, Germany). Depending on particle density incubation time was elongated to 30 min in a humidity-controlled environment. Excess sample was blotted by touching a Whatman filter paper and washed with four droplets of SEC buffer and exposed to freshly prepared 0.75% uranyl formate solution (SPI Supplies/Structure Probe, West Chester, PA) for about 1 min. Excess negative stain solution was blotted and the specimen air-dried. Specimens were inspected with a JEM1400 microscope (Jeol, Tokio, Japan) equipped with a LaB6 cathode and operated at an acceleration voltage of 120 kV. Digital micrographs were recorded at a corrected magnification of 82,524x using a 4k x 4k CMOS camera F416 (TVIPS, Gauting, Germany). Single particles were manually selected, aligned and classified using the Iterative Stable Alignment and Clustering (ISAC) approach implemented in SPHIRE (www.sphire.mpg.de) (Moriya et al., 2017). The initial datasets contained 4745, 3260, 10515 single particles for the CENP-OPQU, -OPQUR, -HIΔ56KMLNOPQUR complexes respectively. The best ISAC class averages were used to calculate the 3D reconstruction using the VIPER approach (Penczek et al., 2014). The complete dataset of raw particles was used to refine the initial models. For CENP-OPQU and -OPQUR the iterative projection matching (sxali3d) implemented in SPHIRE was used until convergence was achieved. The CENP-HIΔ56KMLNOPQUR complex was refined using the Meridien implemented in SPHIRE (Moriya et al., 2017). The resolution of the final reconstructions was estimated by the Fourier shell correlation (FSC) criterion 0.5 to be 22-23 Å. UCSF Chimera (Pettersen et al., 2004) was used to visualize and analyze the EM data and to prepare EM figures. Fitting of the subcomplexes was done using the implemented “fit in map” function of Chimera. The 3D reconstruction of the CENP-HIΔ56KM complex (green) was published before (Basilico et al., 2014).
Analytical ultracentrifugation (AUC)
Sedimentation velocity experiments were performed in an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Palo Alto, US-CA) with Epon charcoal-filled double-sector quartz cells and an An-60 Ti rotor (Beckman Coulter, Palo Alto, US-CA). Samples were centrifuged at 203,000xg at 20°C and 500 radial absorbance scans at either 280 nm and collected with a time interval of 1 min. Data was analysed using the SEDFIT software (Schuck, 2000) in terms of continuous distribution function of sedimentation coefficients (c(S)). The protein partial specific volume was estimated from the amino acid sequence using the program SEDNTERP. Data were plotted using the program GUSSI, which is freely available from http://biophysics.swmed.edu/MBR/software.html
Analysis of CENP-R and CENP-QU were carried out at 20°C in 20 mM Tris pH 6.8, 5% glycerol, 300 mM NaCl and 1 mM TCEP (leading to values of buffer density of 1.02542 g/ml and viscosity of 1.199 cP). Analysis of CENP-OP was carried out at 20°C in 20 mM Tris pH 8.0, 5% glycerol, 300 mM NaCl and 1 mM TCEP (leading to values of buffer density of 1.02542 g/ml and viscosity of 1.199 cP). Analysis for CENP-OPQU and CENP-HIΔ56KMLNOPQUR were performed in 20 mM Na-HEPES, 5% glycerol, 300 mM NaCl and 1 mM TCEP (leading to values of buffer density of 1.04039 g/ml and viscosity of 1.300 cP). Analysis of CENP-OPQUR was carried out at 20°C in 20 mM Tris pH 6.8, 5% glycerol, 500 mM NaCl and 1 mM TCEP (leading to values of buffer density of 1.03352 g/ml and viscosity of 1.217 cP). The calculate values of the partial specific volume [V(bar), inverse of density] at 20°C for CENP-R is 0.73707 ml/g, CENP-OP is 0.73725 ml/g, CENP-QU is 0.73376 ml/g, CENP-OPQU is 0.73537 ml/g, CENP-OPQUR is 0.73558 ml/g and for CENP-HIΔ56KMLNOPQUR is 0.73975 ml/g.
Microtubule co-sedimentation assays
Tubulin was purchased from Cytoskeleton, Inc. (Denver, CO) and was polymerized according to manufacturer’s instructions. Microtubules and proteins were mixed in a final volume of 20 μl in 80 mM Pipes, pH 6.8, 125 mM KCl, 2 mM MgCl2, 1 mM EGTA and 10 μM Taxol. 0 and 5 μM taxol-stabilized microtubules (tubulin dimer concentration), and 1 μM protein of interest (protein monomer concentration) were mixed in 20 μl reactions. Reaction mixtures were incubated for 10 min at room temperature, transferred onto 120 μl of cushion buffer (80 mM Pipes, pH 6.8, 125 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50% glycerol and 10 μM taxol) and ultracentrifuged at 350,000 g for 10 min at 25 °C. Supernatants and pellets were analyzed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Quantification was carried out as described previously (Ciferri et al., 2008). Briefly, gel densitometry was carried out with Image Lab (Biorad). Bound fractions were obtained by dividing values of the pellet fraction by the sum of pellet and supernatant. Normalized binding data was fitted using GraphPad Prism (GraphPad software, Inc.).
Microtubule binding assay on spinning disc confocal microscope
Cover slips and glass slides were cleaned by sonication in isopropanol and 1 M KOH or 1% Hellmanex and 70% ethanol, respectively. After functionalization of cover slips with 5% biotinylated poly-L-lysine- PEG for 30 min, flow cells were created with a volume of 10-15 μl. Flow cells were passivated with 1% pluronic F-127 for 1 h and coated with avidin for 30-45 min. After incubation with 100 nM microtubules (10% biotinylated, 10% Rhodamine labeled, Cytoskeleton, Inc., polymerized according to manufacturer’s instructions) for 10-20 min. Proteins of interest were added in 80 mM Pipes (pH 6.8), 125 mM KCl, 1 mM EGTA, 1 mM MgCl2 and 20 μM Taxol). Flow cells were sealed with wax and imaged with spinning disk confocal microscopy on a 3i Marianas system.
TIRF microscopy
The experiments were performed and analysed as described previously (Drechsler and McAinsh, 2016) with the following modifications: Each flow cell contained either 1 nM Alexa488-labeled CENP-OPQU, 1 nM Alexa488-labeled CENP-OPQUR, or 1 nM Alexa488-labeled NDC80 in 80 mM Pipes, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 20 mM glucose, 0.1 mg/ml k-casein, 0.54 mg/ml glucose oxidase and 0.27 mg/ml catalase. Taxol-stabilised microtubules were labeled 1:30 with biotin and Hilyte647 (Cytoskeleton) and bound to the glass surface of a flow chamber. The assay mix was then flowed into the chamber and sealed with VALAP. Time-lapse sequences for Alexa488-labeled CENP-OPQU (488 nm excitation, exposure 100 ms, 1fr/s) were recorded for a duration of 200 s at 35°C; longer videos were not possible because of bleaching. Time lapse for Alexa488-labeled NDC80 (including Alexa488-labeled CENP-OPQUR for direct comparison) were recorded for 40 s (488 nm excitation, exposure 60 ms, 100 ms/frame). All experiments were done on a CELLR/TIRF microscope (Olympus) equipped with an ImageEM emCCD camera (Hamamatsu photonics) and a 100x 1.49NA objective.
SNAP-CENP-C pull-down experiments
CENP-C1-544-SNAP was covalently labeled with biotinylated benzylguanin (“Snap-biotin” reagent, New England Biolabs) according to manufacturer’s protocols. In a typical assay, 20 μl of streptavidin (STV)-coated beads (Pierce Streptavidin UltraLink Resin, Thermo Scientific) were used, per sample, and washed two times with 300 μl bead buffer (10 mM Hepes pH 7.5, 300 mM NaCl, 5% glycerol, 2 mM TCEP and 0.05% Triton X-100). The beads were re- suspended in 25 μl solution containing the proteins of interest at 2 μM and the mixture was incubated for 10 min on ice. To remove unbound materials from the beads, they were washed two times with 200 μl bead buffer. Samples were boiled in SDS loading buffer and subsequently analyzed by western blot. The following antibodies were used: CENP-P (Mouse monoclonal ABCAM ab66058, 1:1000), CENP-Q (Mouse monoclonal ABCAM ab57539, 1:1000), CENP-R (Mouse monoclonal ABCAM ab57098, 1:1000), CENP-HK (rabbit polyclonal antibody SI0930 raised against the full-length human CENP-HK complex; 1:1000 (Klare et al., 2015)), CENP-N (rabbit polyclonal antibody SI0930 raised against CENP-N1-212 peptide; 1:1000), CENP-C (rabbit polyclonal antibody SI410 raised against CENP-C23-410 peptide; 1:1000; (Trazzi et al., 2009)). Secondary antibodies were anti-mouse, and anti-rabbit (Amersham, part of GE Healthcare) affinity purified with horseradish peroxidase conjugate (working dilution 1:10000).
Cell culture and transfection
U2OS cells, a gift from A. Bird (MPI-Dortmund, Germany), were grown in DMEM (PAN Biotech, Aidenbach, Germany) supplemented with 10% FBS (Clontech, part of Takara Bio group, Shiga, Japan), penicillin and streptomycin (GIBCO, Carlsbad, CA), and 2 mM L-glutamine (PAN Biotech).
FlpIn T-REx HeLa cells used to generate stable doxycycline-inducible cell lines were a gift from SS Taylor (University of Manchester, Manchester, England, UK). Flp-In T-REx host cell lines were maintained in DMEM (PAN Biotech, Aidenbach, Germany) with 10% tetracycline-free FBS (Clontech) supplemented with 50 μg/ml Zeocin (Invitrogen) and 2 mM L-glutamine (PAN Biotech). Flp-In T-REx HeLa expression cell lines were generated as previously described (Krenn et al., 2012). Gene expression was induced by addition of 0.2–0.5 μg/ml doxycycline (Sigma, St. Louis, MO) for 48 to 72 hr.
Proteins were depleted by siRNA, transfected into cells using HiPerFect transfection reagent (Qiagen) as per the manufactures instructions. Cells were treated with CENP-O siRNA (Dharmacon; 5’-UAGGAGACCAGACUCAUAU-3’) or CENP-P siRNA (Dharmacon SMARTpool; 5’-GUGCAAGAGAGAACAACUA-3’, 5’-AGUCAUUGUUUGGAGGAUA-3’, 5’-UAUCGUAAGCGCACGUUUA-3’, and 5-CCUAAGUGCUAUAUCGAUC-3’) for 48 h. While treatments of CENP-Q siRNA (Dharmacon SMARTpool; 5’-GAGUUAAUGACUGGGAAUA-3’, 5’-AUGGAAAGGGCACGAGACA-3’, 5’-ACAAAGCACACUAACCUAA-3’, and 5’-UGUCAGAGAAAUAAGGUUAG-3’), CENP-R (ITGB3BP, Dharmacon SMARTpool; 5’-GAAGUUGGAUGGUCUGUUA-3’, 5’-UGACAGCUAUGAAUUCCUU-3’, 5’-UAAGUAGUAUACAGGCUUU-3’, and 5’-GAAUUCAUGAUGUUGCUAU-3’), CENP-H (Dharmacon; 5’-CUAGUGUGCUCAUGGAUAA-3’), CENP-L (Dharmacon; 5’-UUUAUCAGCCACAAGAUUA-3’), or CENP-N (Dharmacon SMARTpool; 5’-CUACCUACGUGGUGUUACUA-3’, 5’-GUUCAGCACUUGAUCCAUC-3’, 5’-AUACACCGCUUCUGGGUCA-3’, and 5’-ACACAAAGCCAAACCAGUA-3’) were for 72 h.
Immunofluorescence
FlpIn T-REx HeLa cells were grown on coverslips precoated with poly-D-Lysine (Millipore, 15 μg/ml). For co-localization experiments, FlpIn T-REx HeLa cells expressing full length GFP tagged CENP-R were synchronized overnight, 32 h or 56 h after siRNA transfection, in RO-3306 (Calbiochem) then released into 3.3 μM nocodazole (Sigma–Aldrich) for 2-3 h before fixation. For monopolar spindle recovery experiments U2OS cells were treated overnight in 5 μM (+)-S-Trityl-L-Cysteine (STLC, Sigma-Aldrich), then released and fixed after 3 h. To determine how CENP-O F158G and CENP-P F116G localize in metaphase, cells were fixed following their respective 48 h siRNA treatment. To investigate CENP-OPQUR recruitment following CENP-H, -L, or –N siRNA treatment cells were synchronized using 330 nM nocodazole for 15 h, 56 h after siRNA transfection. Cells were fixed as described (De Antoni et al., 2012). Alternatively cells were fixed using 4% paraformaldehyde, and permeablised using 0.25% Triton X-100. Cells were stained for GFP (Goat polyclonal, 1:500 (Poser et al., 2008) or GFP-Boost, Chromoteck gba-488, 1:400), CENP-OP (Rabbit polyclonal, raised against full length CENP-OP complex, 1:500), CENP-QU (Goat, raised against full length CENP-QU complex, 1:500), CENP-HK (rabbit polyclonal antibody SI0930 raised against the full-length human CENP-HK complex; 1:400, CREST/anti-centromere antibodies (Antibodies, Inc., Davis, CA, 1:100), CENP-A (Rabbit, Ossolengo, 1:500 or Mouse, Gene Tex GTX13939, 1:500) diluted in 5% boiled donkey serum in PHEM (Pipes, Hepes, EGTA, and MgCl2) for 2 h (PFA fixation) or over night (PHEM fixation) (De Antoni et al., 2012).
Donkey anti-human and donkey anti-goat Alexa Fluor 647, donkey anti-mouse, donkey anti-rabbit, and donkey anti-goat rhodamine, donkey anti-mouse and donkey anti-goat Alexa Fluor 488, donkey anti-human, donkey anti-rabbit, and donkey anti-mouse Alexa Fluor 405 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), as well as donkey anti-mouse and chicken anti-rabbit Alexa Fluor 647, and donkey anti-rabbit Alexa Fluor 488 (Invitrogen) were used as secondary antibodies. DNA was stained with 0.5 μg/ml DAPI (Serva), and coverslips were mounted with Mowiol mounting media (Calbiochem). Cells were imaged using either a3i Marianas system or a Deltavision Elyte System. The spinning disk confocal device on the 3i Marianas system equipped with an Axio Observer Z1 microscope (Zeiss), a CSU-X1 confocal scanner unit (Yokogawa Electric Corporation, Tokyo, Japan), Plan-Apochromat 63× or 100×/1.4NA Oil Objectives (Zeiss), and Orca Flash 4.0 sCMOS Camera (Hamamatsu). Images were acquired as z-sections at 0.2 μm. Images were converted into maximal intensity projections, exported, and converted into 8-bit. Quantification of kinetochore signals was performed on unmodified 16-bit z-series images using Imaris 7.3.4 32-bit software (Bitplane, Zurich, Switzerland). After background subtraction, all signals were normalized to CREST or CENP-A. Measurements were exported in Excel (Microsoft) and graphed with GraphPad Prism 6.0 (GraphPad Software, San Diego California USA). The Deltavision Elite System (GE Healthcare, UK) is equipped with an IX-71 inverted microscope (Olympus, Japan), UPlanFLN 40x/1.3NA objective or a PLAPON 60x/1.42NA objective (Olympus) and a pco.edge sCMOS camera (PCO-TECH Inc., USA).
Live cell imaging
Cells were plated on an 8 well 15 μ-Slide (Ibidi, Martinsried, Germany). CENP-P was depleted as previously described for 48 h prior to imaging. For asynchronous cells, FlpIn T-REx HeLa cells were transferred into CO2 Independent Medium (Gibco) 16 h before timelapse. Two hours before imaging SiR-Hoechst DNA dye (Spirochrome) was added. Timelapse demonstrating recovery from nocodazole treatment utilized U2OS cells drugged overnight with 330 nM nocodazole in CO2 Independent Medium (Gibco). SiR-Hoechst DNA dye (Spirochrome) was added into the nocodazole containing media 2 h before imaging. Cells were released from nocodazole by washing three times with PBS and then placed in CO2 Independent Media (Gibco) containing SiR-Hoechst DNA dye (Spirochrome). Where indicated, 0.5 μM Reversine was added to cells after release. Cells were imaged every 2 min for 12 h in a heated chamber (37°C) with a Deltavision Elite System. Images were acquired as Z-sections (using the softWoRx software from Deltavision) and converted into maximal intensity projections TIFF files for illustrative purposes.
Immunoprecipitation and immunoblotting
To generate mitotic populations for immunoprecipitation experiments, cells were treated with 330 nM nocodazole for 16 hr. Mitotic cells were then harvested by mitotic shake off and lysed in lysis buffer (25 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl, 10% Glycerol, 0.2% NP-40, 1 mM NaF, Benzonase (Sigma), supplemented with protease inhibitor cocktail (Serva, Heidelberg, Germany) and PhosSTOP phosphatase inhibitors (Roche)). Extracts were precleared using protein A–Sepharose (CL-4B; GE Healthcare) for 1 hr at 4°C. Subsequently, extracts were incubated with GFP-Traps (ChromoTek, Martinsried, Germany; 2 μl/mg of extract) for 2 hr at 4°C. Immunoprecipitates were washed with wash buffer (25 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl, 10% Glycerol, supplemented with protease inhibitor cocktail [Serva, Heidelberg, Germany] and PhosSTOP phosphatase inhibitors [Roche]). To elute the proteins beads were incubated with 0.1 M glycine pH 2.0 for 10 min, 1 M Tris pH 9.2 was then added to neutralize eluates. Sample buffer was added, samples boiled and analyzed by SDS-PAGE and Western blotting using 14% tricine gels.
The following antibodies were used: anti-GFP (in house made rabbit polyclonal antibody; 1:1,000), CENP-OP (Rabbit polyclonal, raised against full length CENP-OP complex, 1:1,000), CENP-QU (Goat, raised against full length CENP-QU complex, 1:1,000), CENP-HK [rabbit polyclonal antibody SI0930 raised against the full-length human CENP-HK complex; 1:1.000 (Klare et al., 2015)], anti-Tubulin (mouse monoclonal; Sigma; 1:8000). Secondary antibodies were anti-mouse, anti-goat, and anti-rabbit (Amersham, part of GE Healthcare) affinity purified with horseradish peroxidase conjugate (working dilution 1:10000). After incubation with ECL Western blotting system (GE Healthcare), images were acquired using a BioRAD chemiDoc MP Imaging System (BioRAD). Images were acquired using Image Lab software Version 5.2 (BioRAD). Images were adjusted using the image lab software then exported in 8-bit tiff format for publication.
Tail-swap experiment
A cDNA encoding full-length, siRNA protected CENP-QNDC80(1-80) was ordered from GeneArt (Thermo Fisher) and subcloned into pMC273 using EcoRI and ScaI sites (replacing wild-type CENP-Q) to create CENP-QNDC80(1-80)-eGFP (pMC614). Correct insertion was confirmed by sequencing with the following primer 5’ ttgacgcaaatgggcggtag 3’. Production of wild-type CENP-Q-eGFP and protection against the CENP-Q siRNA oligonucleotide are reported in (Bancroft et al., 2015). HeLa K cells (MC009) were grown on 22 mm coverslips in DMEM until 40% confluency. Cells were then transfected with control 5’ GGACCUGGAGGUCUGCUGU 3’ (Sigma) or CENP-Q 5’ GGUCUGGCAUUACUACAGGAAGAAA 3’ (Invitrogen Stealth) siRNA using oligofectamine and incubated in 1.5 ml MEM for 24 h. The media was replaced with 1.5 ml DMEM and the cells transfected with 1 μg of eGFP-N1 (pMC005), CENP-Q-eGFP (pMC308) or CENP-QNDC80(1-80)-eGFP (pMC614) using Fugene6 at 1:3 according to the manufacturers guidelines. Cells were incubated for a further 48 h and treated with 1 μM MG132 for 90 min before fixation. Cells were fixed at room temperature for 10 min in 20 mM PIPES pH6.8, 10 mM EGTA, 1 mM MgCl2, 0.2% Triton X-100 and 4% formaldehyde. Cells were then were then washed three times with PBS before blocking with 3% BSA in PBS for 30 min. After blocking, the fixed cells were incubated for 1hr at room temperature in rabbit anti-CENP-E antibody (1/1500, Meraldi lab) and CREST antisera (1/250, Antibodies Incorporated). Cells were then washed three times in PBS and incubated for 1 h with AlexaFluor-conjugated highly cross-adsorbed secondary antibodies (Invitrogen) before mounting on coverslips in vectashield (VectorLabs). Three dimensional image stacks of mitotic cells were acquired in 0.2 μm steps using a 100X oil-immersion 1.4 NA objective lens on an Olympus DeltaVision Elite microscope (Applied Precision, LLC) equipped with a DAPI, fluorescein isothiocyanate (FITC), Rhodamine or Texas Red, CY5 filter set (Chroma), solid state light source and a CoolSNAP HQ camera (Roper Scientific). Image stacks were deconvolved using SoftWorx (Applied Precision, LLC). Fluorescence-intensity measurements were taken manually using SoftWorx and data visualized in R or Excel (Microsoft). Figures were prepared in illustrator (Adobe). CENP-Q and NDC80 tail disorder predictions were generated using PrDOS (Ishida and Kinoshita, 2007) with a 5% false positive rate.
Quantification and Statistical Analysis
Statistical analyses are described in the Figure legends and in the Method Details.
Data and Software Availability
The data have been uploaded on the Mendeley server at https://doi.org/10.17632/yv552m8s98.1.
Acknowledgments
We thank all members of the Musacchio laboratory for helpful discussions and comments. A.M. gratefully acknowledges funding by the Max Planck Society, the European Research Council (ERC) Advanced Investigator Grant RECEPIANCE (proposal number n° 669686), and the DFG’s Collaborative Research Centre (CRC) 1093. A.D.M. is supported by a Wellcome Trust Senior Investigator Award (grant number 106151/Z/14/Z) and a Royal Society Wolfson Research Merit Award (grant number WM150020). S.R. gratefully acknowledges the Max Planck Society and the European Research Council under the European Union’s Seventh Framework Programme (FP7/ 2007–2013; grant no. 615984).
Author Contributions
M.E.P. carried out biochemical purifications, in vitro binding and reconstitution assays, and microtubule-sedimentation assays. M.E.P. and D.P. carried out EM data preparations and collection, and D.P. performed EM data analysis and reconstructions. C.M.S. carried out kinetochore localization and alignment assays, RNAi depletions except in Figures 2E, 2F, and 7, and IP experiments in Figure S3H. A.C.F. and M.E.P. carried out microtubule binding analyses by fluorescence microscopy with the help of Y.-C.L., except TIRF assays in Figures 5C–5E. M.E. carried out TIRF assays. P.A. carried out localization and rescue experiments with CENP-P mutant and the CENP-Q tail-swap experiments. A.P. carried out ultracentrifugation analyses. S.M. carried out electroporation experiments and contributed to quantitative data analyses. J.R.W. and S.P. contributed essential reagents. M.E.P., D.P., C.M.S., S.M., M.E., and P.A. contributed to data visualization. S.R. supervised the EM work. A.D.M. supervised TIRF experiments and the cell-biological analysis of CENP-P and CENP-Q mutants. A.M. supervised and administered the research team, contributed to data visualization, and wrote the paper with contributions from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: August 30, 2018
Footnotes
Supplemental Information includes seven figures and one table and one video and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.07.038.
Contributor Information
Stefan Raunser, Email: stefan.raunser@mpi-dortmund.mpg.de.
Andrew D. McAinsh, Email: a.d.mcainsh@warwick.ac.uk.
Andrea Musacchio, Email: andrea.musacchio@mpi-dortmund.mpg.de.
Supplemental Information
References
- Ahonen L.J., Kallio M.J., Daum J.R., Bolton M., Manke I.A., Yaffe M.B., Stukenberg P.T., Gorbsky G.J. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr. Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G.M., Musinipally V., Matson D., Tooley J., Stukenberg P.T., Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat. Struct. Mol. Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro A.C., Samora C.P., Holtackers R., Wang E., Kingston I.J., Alonso M., Lampson M., McAinsh A.D., Meraldi P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft J., Auckland P., Samora C.P., McAinsh A.D. Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J. Cell Sci. 2015;128:171–184. doi: 10.1242/jcs.163659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 2014;3:e02978. doi: 10.7554/eLife.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C., Imasaki T., Takagi Y., Berger I. MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem. Sci. 2012;37:49–57. doi: 10.1016/j.tibs.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H.J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA. 2012;109:1991–1996. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Zhou K., Zhang Z., Luger K., Straight A.F. Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Mol. Biol. Cell. 2018;29:751–762. doi: 10.1091/mbc.E17-10-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Silva M.C., Godek K.M., Jansen L.E., Straight A.F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., Straight A.F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M. The kinetochore. Cold Spring Harb. Perspect. Biol. 2014;6:a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chittori S., Hong J., Saunders H., Feng H., Ghirlando R., Kelly A.E., Bai Y., Subramaniam S. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 2018;359:339–343. doi: 10.1126/science.aar2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A., Maffini S., Knapp S., Musacchio A., Santaguida S. A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. J. Cell Biol. 2012;199:269–284. doi: 10.1083/jcb.201205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A.D., Sorger P.K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., DeLuca J.G. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca K.F., Meppelink A., Broad A.J., Mick J.E., Peersen O.B., Pektas S., Lens S.M.A., DeLuca J.G. Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J. Cell Biol. 2018;217:163–177. doi: 10.1083/jcb.201707160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova Y.N., Jenni S., Valverde R., Khin Y., Harrison S.C. Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell. 2016;167:1014–1027 e1012. doi: 10.1016/j.cell.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H., McAinsh A.D. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc. Natl. Acad. Sci. USA. 2016;113:E1635–E1644. doi: 10.1073/pnas.1516370113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., Salmon E.D., Mitchison T.J. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Eskat A., Deng W., Hofmeister A., Rudolphi S., Emmerth S., Hellwig D., Ulbricht T., Döring V., Bancroft J.M., McAinsh A.D. Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex. PLoS ONE. 2012;7:e44717. doi: 10.1371/journal.pone.0044717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S.J., Lee J., Sekulic N., Sennett M.A., Lee T.H., Black B.E. CENP-C directs a structural transition of CENP-A nucleosomes mainly through sliding of DNA gyres. Nat. Struct. Mol. Biol. 2016;23:204–208. doi: 10.1038/nsmb.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., 3rd, Cleveland D.W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., Cheeseman I.M. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes C.P., Witte M.D., Theile C.S., Bozkurt G., Kundrat L., Blom A.E., Ploegh H.L. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 2013;8:1787–1799. doi: 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.Y., Allu P.K., Zandarashvili L., McKinley K.L., Sekulic N., Dawicki-McKenna J.M., Fachinetti D., Logsdon G.A., Jamiolkowski R.M., Cleveland D.W. Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun. 2017;8:15775. doi: 10.1038/ncomms15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Zhang S., Zhang S., Chen Y.R., Blissard G.W. Correction: BTI-Tnao38, a new cell line derived from Trichoplusia ni, is permissive for AcMNPV infection and produces high levels of recombinant proteins. BMC Biotechnol. 2012;12:12. doi: 10.1186/1472-6750-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Khodjakov A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J. Cell Biol. 2015;211:1103–1111. doi: 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., Harrison S.C. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 2013;5:29–36. doi: 10.1016/j.celrep.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S.M., Makrantoni V., Harrison S.C., Marston A.L. The Kinetochore Receptor for the Cohesin Loading Complex. Cell. 2017;171:72–84 e13. doi: 10.1016/j.cell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung P., Troc P., Malvezzi F., Maier M., Demianova Z., Zimniak T., Litos G., Lampert F., Schleiffer A., Brunner M. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 2014;206:509–524. doi: 10.1083/jcb.201403081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S., Wang Z., Jiang K., Huang Y., Ward T., Zhao L., Dou Z., Yao X. CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment. J. Biol. Chem. 2011;286:1627–1638. doi: 10.1074/jbc.M110.174946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis In 't Veld P.J., Jeganathan S., Petrovic A., Singh P., John J., Krenn V., Weissmann F., Bange T., Musacchio A. Molecular basis of outer kinetochore assembly on CENP-T. eLife. 2016;5:e21007. doi: 10.7554/eLife.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland K.M., Kingsbury J., Koshland D., Hieter P. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Kagawa N., Hori T., Hoki Y., Hosoya O., Tsutsui K., Saga Y., Sado T., Fukagawa T. The CENP-O complex requirement varies among different cell types. Chromosome Res. 2014;22:293–303. doi: 10.1007/s10577-014-9404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.H., Park J.E., Yu L.R., Soung N.K., Yun S.M., Bang J.K., Seong Y.S., Yu H., Garfield S., Veenstra T.D., Lee K.S. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Kang Y.H., Park C.H., Kim T.S., Soung N.K., Bang J.K., Kim B.Y., Park J.E., Lee K.S. Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores. J. Biol. Chem. 2011;286:19744–19757. doi: 10.1074/jbc.M111.224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B., Fischer N., Golas M.M., Sander B., Dube P., Boehringer D., Hartmuth K., Deckert J., Hauer F., Wolf E. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods. 2008;5:53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- Kato H., Jiang J., Zhou B.R., Rozendaal M., Feng H., Ghirlando R., Xiao T.S., Straight A.F., Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 2015;208:181–196. doi: 10.1083/jcb.201407074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K., Weir J.R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 2012;196:451–467. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue T.M., Shah B.D., Ridgeway T.M., Pelletier S.L. Analytical Ultracentrifugation in Biochemistry and Polymer Science. In: Harding S.E., Rowe A.J., Horton J., editors. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- Liu Y., Petrovic A., Rombaut P., Mosalaganti S., Keller J., Raunser S., Herzog F., Musacchio A. Insights from the reconstitution of the divergent outer kinetochore of Drosophila melanogaster. Open Biol. 2016;6:150236. doi: 10.1098/rsob.150236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 2013;32:409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A.D., Meraldi P. The CCAN complex: linking centromere specification to control of kinetochore-microtubule dynamics. Semin. Cell Dev. Biol. 2011;22:946–952. doi: 10.1016/j.semcdb.2011.09.016. [DOI] [PubMed] [Google Scholar]
- McAinsh A.D., Meraldi P., Draviam V.M., Toso A., Sorger P.K. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., Sekulic N., Guo L.Y., Tsinman T., Black B.E., Cheeseman I.M. The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface. Mol. Cell. 2015;60:886–898. doi: 10.1016/j.molcel.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Milks K.J., Moree B., Straight A.F. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y.C., Kawashima T., Kitamura T., Fukagawa T. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol. Cell. Biol. 2005;25:10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya T., Saur M., Stabrin M., Merino F., Voicu H., Huang Z., Penczek P.A., Raunser S., Gatsogiannis C. High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE. J. Vis. Exp. 2017;123 doi: 10.3791/55448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Desai A. A molecular view of kinetochore assembly and function. Biology (Basel) 2017;6:E5. doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal H., Hori T., Furukawa A., Sugase K., Osakabe A., Kurumizaka H., Fukagawa T. Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell. 2015;26:3768–3776. doi: 10.1091/mbc.E15-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino M., Kurasawa Y., Evans R., Lin S.H., Brinkley B.R., Yu-Lee L.Y. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr. Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., 3rd, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Okada M., Okawa K., Isobe T., Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol. Biol. Cell. 2009;20:3986–3995. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekgöz Altunkaya G., Malvezzi F., Demianova Z., Zimniak T., Litos G., Weissmann F., Mechtler K., Herzog F., Westermann S. CCAN Assembly Configures Composite Binding Interfaces to Promote Cross-Linking of Ndc80 Complexes at the Kinetochore. Curr. Biol. 2016;26:2370–2378. doi: 10.1016/j.cub.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Penczek P.A., Fang J., Li X., Cheng Y., Loerke J., Spahn C.M. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy. 2014;140:9–19. doi: 10.1016/j.ultramic.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentakota S., Zhou K., Smith C., Maffini S., Petrovic A., Morgan G.P., Weir J.R., Vetter I.R., Musacchio A., Luger K. Decoding the centromeric nucleosome through CENP-N. eLife. 2017;6:e33442. doi: 10.7554/eLife.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Pesenti M.E., Weir J.R., Musacchio A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 2016;37:152–163. doi: 10.1016/j.sbi.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Petrovic A., Keller J., Liu Y., Overlack K., John J., Dimitrova Y.N., Jenni S., van Gerwen S., Stege P., Wohlgemuth S. Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores. Cell. 2016;167:1028–1040 e1015. doi: 10.1016/j.cell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., Glover D.M. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Raaijmakers J.A., van Heesbeen R.G.H.P., Blomen V.A., Janssen L.M.E., van Diemen F., Brummelkamp T.R., Medema R.H. BUB1 Is Essential for the Viability of Human Cells in which the Spindle Assembly Checkpoint Is Compromised. Cell Rep. 2018;22:1424–1438. doi: 10.1016/j.celrep.2018.01.034. [DOI] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., Cheeseman I.M. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M.M., Poznanski J., Zdziarska A., Czarnocki-Cieciura M., Lipinszki Z., Dadlez M., Glover D.M., Przewloka M.R. Network of protein interactions within the Drosophila inner kinetochore. Open Biol. 2016;6:150238. doi: 10.1098/rsob.150238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., 3rd, Maurer M., Rothfield N.F., Earnshaw W.C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Samejima I., Spanos C., Alves Fde.L., Hori T., Perpelescu M., Zou J., Rappsilber J., Fukagawa T., Earnshaw W.C. Whole-proteome genetic analysis of dependencies in assembly of a vertebrate kinetochore. J. Cell Biol. 2015;211:1141–1156. doi: 10.1083/jcb.201508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- Schmitzberger F., Harrison S.C. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012;13:216–222. doi: 10.1038/embor.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzberger F., Richter M.M., Gordiyenko Y., Robinson C.V., Dadlez M., Westermann S. Molecular basis for inner kinetochore configuration through RWD domain-peptide interactions. EMBO J. 2017;36:3458–3482. doi: 10.15252/embj.201796636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Badger B.L., Wan X., DeLuca J.G., Salmon E.D. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev. Cell. 2014;30:717–730. doi: 10.1016/j.devcel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S.Y. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Tachiwana H., Müller S., Blümer J., Klare K., Musacchio A., Almouzni G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Nishino T., Mayanagi K., Horikoshi N., Osakabe A., Tachiwana H., Hori T., Kurumizaka H., Fukagawa T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res. 2014;42:1644–1655. doi: 10.1093/nar/gkt1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J., Henikoff S. CENPT bridges adjacent CENPA nucleosomes on young human α-satellite dimers. Genome Res Epub. ahead of print. 2016 doi: 10.1101/gr.204784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Li X., Liu Y., Wang C., Liu X., Bi G., Zhang X., Yao X., Zhou Z.H., Zang J. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res. 2018;28:374–378. doi: 10.1038/cr.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., McAinsh A.D. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., Della Valle G. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R., Ingram J., Harrison S.C. Conserved Tetramer Junction in the Kinetochore Ndc80 Complex. Cell Rep. 2016;17:1915–1922. doi: 10.1016/j.celrep.2016.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V.A., Huis In ’t Veld P.J., Dogterom M., Musacchio A. Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. eLife. 2018;7:e36764. doi: 10.7554/eLife.36764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.R., Schnell J.R., Larsen N.A., Sorger P.K., Chou J.J., Harrison S.C. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Wei R.R., Al-Bassam J., Harrison S.C. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Weir J.R., Faesen A.C., Klare K., Petrovic A., Basilico F., Fischböck J., Pentakota S., Keller J., Pesenti M.E., Pan D. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature. 2016;537:249–253. doi: 10.1038/nature19333. [DOI] [PubMed] [Google Scholar]
- Westermann S., Schleiffer A. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 2013;23:260–269. doi: 10.1016/j.tcb.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I.M., Anderson S., Yates J.R., 3rd, Drubin D.G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., Straight A.F. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood L., Booth D.G., Vargiu G., Ohta S., deLima Alves F., Samejima K., Fukagawa T., Rappsilber J., Earnshaw W.C. Auxin/AID versus conventional knockouts: distinguishing the roles of CENP-T/W in mitotic kinetochore assembly and stability. Open Biol. 2016;6:150230. doi: 10.1098/rsob.150230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev A.V., Sundin L.J., DeLuca K.F., Grishchuk E.L., DeLuca J.G. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev A.V., Mick J.E., Maslennikov E., Nikashin B., DeLuca J.G., Grishchuk E.L. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell. 2015;26:1829–1844. doi: 10.1091/mbc.E14-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video shows the first 3D reconstruction of the CENP-HIΔ56KMLNOPUR and focuses on how the separately reconstructed subcomplexes HIΔ56KM and OPQUR are oriented towards each other. With the potential caveat that the CENP-HIΔ56KMLNOPUR sample was stabilized by cross-linking with the GraFix method before processing for EM, which might cause it to undergo physical rearrangements, a substantial volume remains unoccupied that might account, at least partially, for the CENP-LN subcomplex.