Abstract
This protocol describes reconstitution assays to study how the neurotransmitter release machinery triggers Ca2+-dependent synaptic vesicle fusion. The assays monitor fusion between proteoliposomes containing the synaptic vesicle SNARE synaptobrevin (with or without the Ca2+ sensor synaptotagmin-1) and proteoliposomes initially containing the plasma membrane SNAREs syntaxin-1 and SNAP-25. Lipid mixing (from fluorescence de-quenching of Marina Blue-labeled lipids) and content mixing (from development of FRET between Phycoerythrin-Biotin and Cy5-Streptavidin trapped in the two proteoliposome populations) are measured simultaneously to ensure that true, non-leaky membrane fusion is monitored, adapting a protocol previously developed to study yeast vacuolar fusion. In contrast to other protocols used to study the release machinery, these assays incorporate NSF and α-SNAP, which disassemble syntaxin-1 and SNAP-25 heterodimers. As a result, fusion requires Munc18–1, which binds to the released syntaxin-1, and Munc13–1, which orchestrates SNARE complex assembly together with Munc18–1. Total time required for one round is 4 days.
INTRODUCTION
The release of neurotransmitters by Ca2+-triggered synaptic vesicle exocytosis is a central event in communication between neurons. Elucidating the mechanism of neurotransmitter release is thus crucial to understand brain function, and in addition can yield key insights into intracellular membrane traffic in general because the core components of the release machinery have homologues in most intracellular membrane compartments and hence are believed to mediate a conserved basic mechanism of membrane fusion1. These core proteins include (Figure 1): i) the synaptic vesicle SNAP receptor (SNARE) synaptobrevin and the plasma membrane SNAREs syntaxin-1 and SNAP-25, which form a tight four-helix bundle called the SNARE complex that brings the two membranes together and is crucial for membrane fusion2–5; ii) N-ethylmaleimide sensitive factor (NSF) and soluble NSF attachments proteins (SNAPs), which disassemble the SNARE complex to recycle the SNAREs for another round of fusion2, 6, 7; iii) the Sec1-Munc18 (SM) protein Munc18–1, which orchestrates SNARE complex assembly together with Munc13s8, 9, likely providing a template to facilitate such assembly10; and iv) and Rab3s, small GTPases involved in vesicle docking through interactions with effectors that include RIMs11, 12. In addition, multiple specialized proteins confer the exquisite regulation of neurotransmitter release, including the aforementioned Munc13s13–16 and RIMs17, 18, which are large (ca. 200 kDa) multidomain proteins from presynaptic active zones19, the Ca2+ sensor synaptotagmin-120, CAPS21, 22 and complexins23, among others.
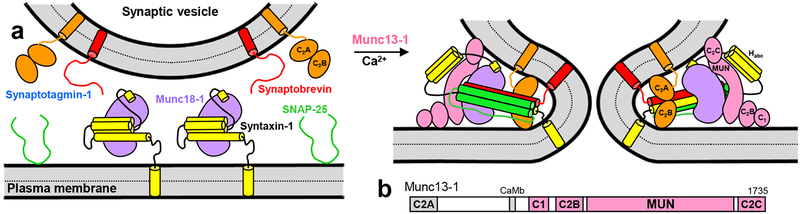
Figure 1.
Working model of neurotransmitter release. a. Model of how the neuronal SNAREs synaptobrevin (red), SNAP-25 (green) and syntaxin-1 (yellow) cooperate with Munc18–1 (purple), Munc13–1 (pink) and synaptotagmin-1 (orange) in triggering Ca2+-dependent membrane fusion. The model is based on many results described in the literature (see Introduction), particularly those from our reconstitution experiments9,29. The binary complex of Munc18–1 with syntaxin-1 folded into a closed conformation (left) is assumed to constitute the starting point for synaptic vesicle exocytosis. Formation of the SNARE complex (the four-helix bundle that brings the two membranes together) is orchestrated by Munc18–1 and Munc13–1 in an NSF-αSNAP-resistant manner. Munc13–1 (only the C1C2BMUNC2C region is shown) is proposed to bridge the two membranes through interactions of the C1-C2B region with the plasma membrane and the C2C domain with the vesicles29 (right), which together with the syntaxin-1-opening activity of the MUN domain8 and the templating activity of Munc18–110 facilitates SNARE complex assembly. Synaptotagmin-1 is proposed to accelerate fusion through interactions of its C2 domains with the membranes (right), but note that the orientation shown is arbitrary and is not consistent with structural studies performed with soluble fragments66,67, which have yielded very diverse binding modes. NSF and αSNAP are not shown, but note that they could be also part of the macromolecular complex that induces fusion, as other proteins that are not included in the model. b. Domain diagram of rat Munc13–1.
Because of the complexity of the neurotransmitter release machinery, reconstituting synaptic vesicle fusion in a meaningful fashion that recapitulates, at least partially, the molecular mechanisms that underlie release and its regulation in vivo constitutes a major challenge. Pioneering work toward this goal was initiated with the development of bulk assays that monitored lipid mixing between donor synaptobrevin-containing proteoliposomes and acceptor syntaxin-1-SNAP-25-containing proteoliposomes from de-quenching of the fluorescence of 7-nitrobenz-2-oxa-1,3-diazole (NBD)-labeled lipids on the donor liposomes due to loss of fluorescence resonance energy transfer (FRET) with rhodamine-labeled lipids24. The observation of lipid mixing in these experiments led to the proposal that the SNAREs constitute a minimal machinery for membrane fusion24, and other components of the release machinery are often considered in the literature as accessory proteins whose main role is to regulate SNARE activity. However, a wide variety of subsequent reconstitution studies incorporating various combinations of proteins and employing different types of methodologies25, some of which are discussed below, yielded considerably diverse results. For instance, proteoliposome fusion induced by a single neuronal SNARE complex has been reported26 whereas other experiments revealed that proteoliposomes can be docked by neuronal SNAREs for long periods of time without fusing27. Studies of other forms of intracellular membrane traffic such as yeast vacuolar fusion have also questioned whether SNAREs alone can yield physiological membrane fusion28. Clearly, including as many of the key components of the release machinery as possible in reconstitution experiments is critical to understand how, together, they mediate synaptic vesicle fusion and regulate release. At the same time, such an understanding will help to settle the debate as to whether physiological membrane fusion can be induced by SNAREs alone or requires in addition other proteins.
Development of the protocol
Our reconstitution studies9, 29, which led to the protocol described here, were pursued to complement structural and functional analyses of the release machinery and gain a detailed understanding of the mechanism of release. We were particularly interested in elucidating the functions of Munc18–1 and Munc13s because of their fundamental functional importance, as shown by the total abrogation of release observed in the absence of either Munc18–130 or Munc13s13–16. This essential nature of Munc18–1 and Munc13s in vivo contrasted with the proposal that the SNAREs alone constitute a minimal membrane fusion machinery based on the observation of lipid mixing between syntaxin-1-SNAP-25-proteoliposomes and synaptobrevin-proteoliposomes24. As mentioned above, widely diverse results have been obtained in subsequent studies with neuronal SNAREs alone31, but experiments including Ca2+ and a soluble fragment containing the two C2 domains of synaptotagmin-1 (C2AB fragment) consistently revealed a dramatic enhancement of SNARE-dependent lipid mixing32–34. Munc18–1 was later shown to moderately enhance SNARE-dependent lipid mixing35, but this finding did not explain why Munc18–1 and Munc13s are so critical in vivo, as the enhancement was much less pronounced than that induced by Ca2+-synaptotagmin-1 C2AB, and no Munc13 was included. An additional concern about these early studies is that most of them monitored only lipid mixing, but it is now well established that lipid mixing can occur without fusion, and hence that observation of content mixing without leakiness is crucial to demonstrate true membrane fusion27, 28, 36. From a methodological point of view, the introduction of assays that could monitor lipid mixing and/or content mixing between single vesicles and planar bilayers37–40, or between pairs of single vesicles27, 41, was also very important (see Comparison with other methods). For instance, this approach showed that, in the presence of Ca2+, synaptotagmin-1 and the SNAREs can indeed induce membrane fusion27, 42, 43. However, the question remained as to why fusion did not require Munc18–1 and Munc13s as it does in vivo.
Biochemical and structural studies showed that Munc18–1 binds tightly to syntaxin-1 folded into a closed conformation where an N-terminal regulatory domain of syntaxin-1 (the Habc domain) interacts intramolecularly with its SNARE motif, hindering SNARE complex formation44, 45 (Figure 1a). Subsequently, Munc18–1 was also shown to bind to the SNARE complex35, 46, leading to the notion that Munc18–1 inhibits release through its binary interaction with syntaxin-1 and plays an active role by binding to the SNARE complex. Munc13–1 was shown to mediate the transition from the Munc18–1-closed syntaxin-1 complex to the SNARE complex through a long helical domain called the MUN domain8, 47–49. This domain spans a major part of the conserved C-terminal region that is common in Munc13s, which in addition includes the C1 and C2B domains preceding the MUN domain and the C2C domain at the very C-terminus (Figure 1b). Overall, the available data suggested that Munc18–1 and Munc13–1 play key roles in coordinating the assembly of the SNARE complex and that the starting point of the pathway that leads to synaptic vesicle fusion is the binary Munc18–1-closed syntaxin-1 complex rather than the syntaxin-1-SNAP-25 heterodimer, in contrast to most models of neurotransmitter release. It might be argued that the Munc18–1-closed syntaxin-1 complex forms first and later SNAP-25 displaces Munc18–1 to form the syntaxin-1-SNAP-25 heterodimer, but competition assays monitored by NMR spectroscopy demonstrated that in fact Munc18–1 displaces SNAP-25 from syntaxin-1 in solution9. When starting with membrane-anchored syntaxin-1-SNAP-25 heterodimers, Munc18–1 also displaces SNAP-25 but full displacement requires the presence of NSF and α-SNAP9, presumably because NSF-αSNAP disassemble the heterodimers50 and Munc18–1 then binds to the released syntaxin-1.
These results led us to perform reconstitution studies with proteoliposomes that contained Munc18–1-syntaxin-1 complexes as starting point9, instead of the syntaxin-1-SNAP-25 heterodimers used in previous reconstitution studies of the neurotransmitter release machinery. Using the standard lipid mixing assay that monitors NBD-fluorescence de-quenching and an assay that simultaneously monitors lipid mixing from fluorescence de-quenching of DiD-labeled lipids and content mixing from fluorescence de-quenching of sulforhodamine B trapped in synaptobrevin liposomes27, we showed that these liposomes fused efficiently with the Munc18–1-syntaxin-1-containing liposomes in a manner that required SNAP-25, the synaptotagmin-1 C2AB fragment and a fragment spanning the C1, C2B and MUN domains of Munc13–1 (C1C2BMUN)9. Moreover, we showed that in experiments started with syntaxin-1-SNAP-25-proteoliposomes, the efficient lipid mixing with synaptobrevin-proteoliposomes in the presence of Ca2+ and synaptotagming-1 C2AB was abolished by NSF-αSNAP because they disassemble the syntaxin-1-SNAP-25 heterodimers. In the presence of NSF-αSNAP, efficient lipid mixing required both Munc18–1 and the Munc13–1 C1C2BMUN fragment, indicating that Munc18–1 captures the syntaxin-1 released by NSF-αSNAP and, together with Munc13–1 C1C2BMUN, orchestrates SNARE complex assembly in an NSF-αSNAP-resistant manner9. These results suggested that our reconstitution system recapitulates basic steps of synaptic vesicle fusion with arguably the eight most central components of the release machinery, providing a natural explanation for the critical requirement of Munc18–1 and Munc13s for neurotransmitter release in vivo. These data were reminiscent of previous studies of yeast vacuolar fusion, which showed that the HOPS tethering complex mediates SNARE complex assembly in a manner that is resistant to the yeast homologues of NSF and αSNAP51.
In subsequent studies we have generally used reconstitutions that start with syntaxin-1-SNAP-25-proteoliposomes (instead of syntaxin-1-Munc18–1 proteoliposomes) and include NSF-αSNAP because Munc18–1 is sensitive to some detergents9, thus limiting the choices available to prepare proteoliposomes, and because NSF or particularly α-SNAP could potentially have a role in fusion [see 52]. For short, below we will refer to syntaxin-1-SNAP-25-proteoliposomes as T-liposomes and to synaptobrevin-proteoliposomes as V-liposomes. We have improved three aspects of our reconstitutions29. First, sulforhodamine B fluorescence de-quenching provides a powerful tool to monitor content mixing in single vesicle experiments where fusion can be distinguished from vesicle lysis27, but in bulk reconstitution assays such as those described here it is difficult to distinguish content mixing from vesicle leakiness. We attempted to perform control experiments where both liposome populations are loaded with sulforhodamine B to assess how much of the de-quenching arises from leakiness53, but we were unable to observe lipid mixing in these control experiments. We have not investigated in detail the reason for this result, but we speculate that it may arise from the low pH of the solutions of high sulforhodamine B concentrations trapped in the liposomes. We turned to another assay that was developed to study yeast vacuolar fusion54 and that simultaneously measures lipid mixing from de-quenching of the fluorescence of Marina Blue-labeled lipids in synaptobrevin liposomes and content mixing from the development of FRET between PhycoE-Biotin trapped in the T-liposomes and Cy5-Streptavidin trapped in the V-liposomes. The experiments are performed in the presence of external unlabeled streptavidin to ensure that the observed FRET arises only from content mixing, and controls without streptavidin reveal the extent of leakiness. This method (Figure 2) is the one described in this protocol and in our hands yields more consistent results than the sulforhodamine B approach. We found that in experiments including Munc18–1, Munc13–1 C1C2BMUN and NSF-αSNAP there was efficient Ca2+-dependent lipid mixing regardless of the addition of synaptotagmin-1 C2AB fragment, but efficient content mixing did require this fragment29.
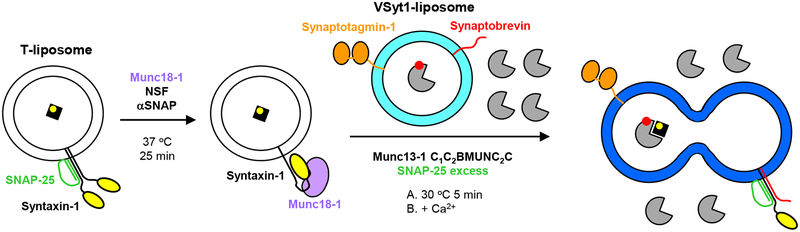
Figure 2.
Diagram summarizing the overall approach followed in the lipid and content mixing assays described here. The color coding is the same as in Figure 1a. PhycoE-Biotin trapped in the T-liposomes is shown as a black square with a yellow circle that represents the fluorescent probe. Cy5-Streptavidin trapped in the VSyt1-liposomes is shown in gray with a red circle that represents the fluorescent probe. Note that the T-liposomes are first incubated at 37 °C for 25 min with Munc18–1 and NSF-αSNAP to disassemble the syntaxin-1-SNAP-25 complexes and form the closed syntaxin-1-Munc18–1 complex. These liposomes are then mixed at 30 °C with the VSyt1-liposomes in the presence of other soluble components such as Munc13–1 C1C2BMUNC2C and excess SNAP-25 (step A), and Ca2+ is added after 5 min (step B). Unlabeled streptavidin (gray) is included outside the vesicles to ensure that the content mixing signal does not arise from leakiness.
A second improvement in our reconstitutions was the use of a Munc13–1 fragment that includes the C1, C2B, MUN and C2C domains (C1C2BMUNC2C). The presence of the C2C domain in this fragment leads to much more efficient lipid and content mixing than that observed with the Munc13–1 C1C2BMUN fragment, likely because C1C2BMUNC2C bridges the V- and T-liposomes more efficiently and specifically than C1C2BMUN29. These findings suggested that a key function of Munc13–1 is to bridge the synaptic vesicle and plasma membranes through interactions of the C2C domain with the vesicle and of the C1C2B region with the plasma membrane (Figure 1a), which together with the syntaxin-1 opening activity of the MUN domain facilitates SNARE complex formation. And a third improvement in our reconstitutions, with respect to our original experiments, was the incorporation of membrane-anchored synaptotagmin-1 into the V-liposomes. Assays performed with these liposomes reveal highly efficient Ca2+-dependent fusion with T-liposomes in the presence of NSF-αSNAP that strictly requires Munc18–1 and Munc13–1 C1C2BMUNC2C (Figure 3a,b). It should be noted that in experiments including C1C2BMUNC2C, Munc18–1 and NSF-αSNAP the presence of either membrane-anchored synaptotagmin-1 or soluble synaptotagmin-1 C2AB does not have a strong effect because fusion is already highly efficient in the time scale of these bulk assays29, but it seems likely that synaptotagmin-1 does accelerate fusion at faster time scales and its inclusion is critical for future experiments where additional components can be added.
Figure 3.
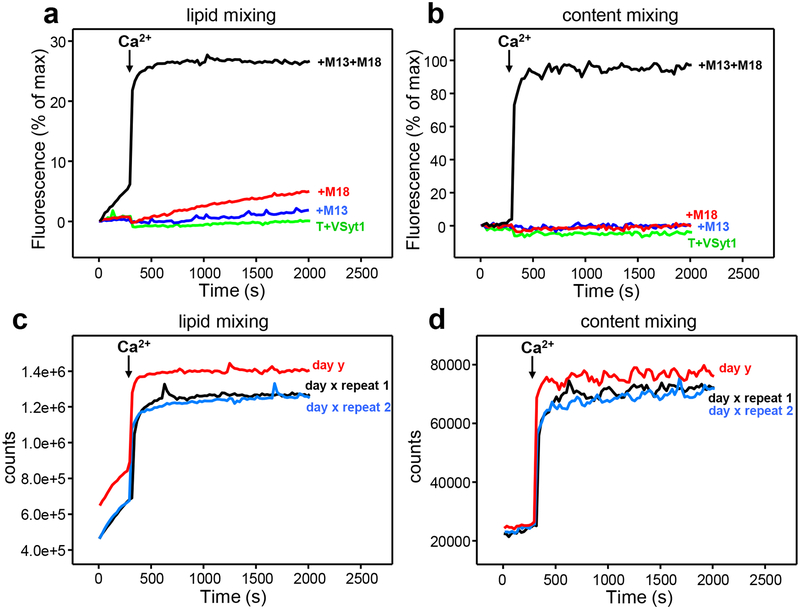
Typical results obtained in our reconstitution experiments. Lipid mixing (a) between VSyt1- and T-liposomes is monitored from the fluorescence de-quenching of Marina Blue-labeled lipids and content mixing (b) is monitored from the development of FRET between PhycoE-Biotin trapped in the T-liposomes and Cy5-Streptavidin trapped in the VSyt1-liposomes. The assays were performed in the presence of NSF-αSNAP without other additional proteins (T+VSyt1) or in the presence of Munc18–1 (M18) and/or Munc13–1 C1C2BMUNC2C (M13) as indicated. Experiments were started in the presence of 100 μM EGTA and 5 μM streptavidin, and Ca2+ (600 μM) was added after 300 s. The data were normalized as described in step 25. (c,d) Raw data (i.e. without normalization) obtained in analogous experiments performed with T- and VSyt1-liposomes in the presence of NSF-αSNAP, Munc18–1 and Munc13–1 C1C2BMUNC2C under the same conditions as those used in (a,b) to illustrate the typical variability observed in analogous experiments performed with the same liposome preparations in the same day (day × repeats 1 and 2) or in different days (compare data from day y with data from day x).
Applications
There are two basic aspects in these reconstitution experiments. One is the method of simultaneously monitoring lipid mixing and content mixing, while the other is the specific proteins used in the reconstitutions. As explained above, the method to simultaneously measure lipid mixing from Marina Blue-fluorescence de-quenching and content mixing from the development of FRET between PhycoE-Biotin and Cy5-Streptavidin was initially developed to study yeast vacuolar fusion54, and was adapted by us to analyze synaptic vesicle fusion. The method should be readily adaptable to investigate other types of intracellular membrane fusion by using the corresponding proteins.
With regard to studies of synaptic vesicle fusion, our reconstitution experiments provide a framework to further understand how fusion is executed and regulated. Thus, although much has been learned about the release machinery, fundamental questions remain about the mechanisms of release and its regulation55. This situation arises in part because, although the structures of some sub-complexes between the components of the release machinery have been solved, the overall architecture of the macromolecular complex formed between two membranes to induce fusion is still unknown. Our reconstitutions will facilitate structural studies to characterize such macromolecular complex. Moreover, the reconstitutions provide a basis to include additional components of the release machinery such as Rab3s, RIMs, complexins or CAPS. Note also that, for the moment our reconstitution assays have been used in bulk solution, but the basic principles can also be adapted for other types of fusion assays such as those monitoring fusion between single vesicles and planar bilayers or between pairs of single vesicles.
Experimental design
The protocol described below uses syntaxin-1, SNAP-25, synaptobrevin, Munc18–1, NSF, αSNAP, a Munc13–1 fragment (normally C1C2BMUNC2C) and the soluble synaptotagmin-1 C2AB fragment or a synaptotagmin-1 fragment that spans its transmembrane and cytoplasmic regions (referred to as Syt1 for simplicity). Expression and purification of these proteins has been described earlier9, 29, 44, 56, 57. The SNAREs are reconstituted into proteoliposomes using the standard method of mixing proteins, lipids and detergents followed by detergent removal through dialysis. Syntaxin-1 and SNAP-25 are reconstituted into T-liposomes that contain trapped PhycoE-biotin and have a lipid composition that resembles that of the plasma membrane. They normally include PIP2 and DAG, agents that activate Munc13–1 and in the case of PIP2 other proteins such as synaptotagmin-11, but these lipids can be omitted to study their effect on fusion. Synaptobrevin is reconstituted alone (V-liposomes) or together with Syt1 (referred to as VSyt1-liposomes), using a lipid composition that resembles that of synaptic vesicles and includes a FRET pair formed by Marina Blue-PE and NBD-PE. The V- or Vsyt1-liposomes contain trapped Cy5-streptavidin. The emission fluorescence of Marina Blue-PE at 465 nm (excitation at 370 nm) is measured to monitor lipid mixing, which causes dilution of the fluorescent lipids and hence Marina-Blue fluorescence de-quenching. The emission fluorescence of Cy5-streptavidin at 670 nm is measured with excitation of PhycoE-biotin at 565 nm to monitor content mixing, which leads to formation of the tight Cy5-streptavidin-PhycoE-Biotin complex and hence to increased Cy5-streptavidin emission due to the resulting FRET. The reactions are normally performed in the presence of 5 μM external unlabeled streptavidin to ensure that the FRET does not arise from leakiness. Control reactions performed in the absence of unlabeled streptavidin reveal the extent of leakiness (in our most efficient fusion experiments, about 10–20% of leakiness was observed in the beginning of the reaction, which likely arises from populations of small and/or unstable vesicles29). Experiments with V-liposomes allow the option of studying the effects of adding the soluble synaptotagmin-1 C2AB fragment, which contains the most central features of the full-length protein, most notably its Ca2+-binding sites29. Experiments with VSyt1-liposomes in principle provide a more physiologically relevant reconstitution than those with V-liposomes because synaptotagmin-1 is membrane-anchored as it occurs in vivo. In such experiments, it is important to keep the percentage of PS in the vesicles low, as it is in synaptic vesicles58 (we use 6.8%), to prevent inhibition of fusion by binding of synaptotagmin-1 in cis to its own VSyt1-liposome membrane59.
Fusion reactions are normally performed at 30 °C but, before mixing the two proteoliposome populations, the T-liposomes are incubated with Munc18–1, NSF and αSNAP at 37 °C to disassemble syntaxin-1-SNAP-25 heterodimers and enable formation of the syntaxin-1-Munc18–1 complex. In the most complete experiments that include T-liposomes, VSyt1-liposomes, Munc18–1, Munc13–1 C1C2BMUNC2C, NSF and αSNAP, we generally observe some lipid mixing but practically no content mixing in the absence of Ca2+, and Ca2+ addition induces fast lipid and content mixing (e.g. Figure 3a,b). These results suggest that a ‘primed state’ that is ready for fusion but does not fuse is generated before Ca2+ addition, likely resembling the primed state of synaptic vesicles29. Control experiments that omit Munc18–1, Munc13–1 C1C2BMUNC2C or both normally yield no content mixing even in the presence of Ca2+ (Figure 3a,b). The results obtained are highly reproducible. Figures 3c,d show raw data (i.e. not normalized) that illustrate the typical variability that we observe in repeated experiments performed the same day with the same liposome preparations, and in different days with different liposome preparations. Differences in the signals observed in different days arise in part from the natural variability expected in fluorescence spectroscopy (e.g. from slight differences in slit widths) and in part from the differences among the liposome preparations (e.g. different amounts of fluorescent lipids or of trapped fluorescence probes), but the overall profiles are similar and normally reveal fast completion of fusion upon Ca2+ addition. Occasionally we observe some slow content mixing before Ca2+ addition that is abolished by increasing the concentration of NSF-αSNAP, which we attribute to variable activity of different NSF preparations. Improving the method of purification of NSF60 may reduce this variability. In rare instances we observe content mixing in controls lacking Munc18–1 or Munc13–1 C1C2BMUNC2C, which may arise also from NSF inactivity and/or the existence of liposomes populations that have unusually high tendency to fuse. We speculate that such high fusogenicity may depend on the process of detergent removal by dialysis during proteoliposome preparation, which may lead to small liposomes or liposome populations with high protein densities61.
Different combinations of the soluble proteins can be used to examine how they affect membrane fusion. For instance, T-liposomes normally do not fuse with V-liposomes under our conditions, but inclusion of Munc13–1 C1C2BMUNC2C induces efficient Ca2+-independent fusion while inclusion of synaptotagmin-1 C2AB leads to Ca2+-dependent fusion; in both cases, addition of NSF-αSNAP abolishes such fusion29. T-liposomes do fuse with VSyt1-liposomes in a Ca2+-independent manner, which is also abolished by NSF-αSNAP. Another common control to test whether fusion under any of these conditions is SNARE dependent involves the addition of a large excess of soluble synaptobrevin cytoplasmic region, which is expected to abrogate fusion by forming cis-SNARE complexes with syntaxin-1-SNAP-25 on the T-liposomes.
The protocol described here has been successfully applied by several investigators, including undergraduate and graduate students, and should be readily accessible to laboratories with expertise in protein and lipid biochemistry. The overall method should be adaptable to study other forms of intracellular membrane fusion using the appropriate proteins, but it is key to have some understanding of the biochemical behavior of the proteins to design the experiments adequately.
Comparison with other methods and limitations
The most critical innovation of our approach compared to previous reconstitution studies of the release machinery was the incorporation of what we consider the eight most central components, i.e. the three neuronal SNAREs syntaxin-1, synaptobrevin and SNAP-25, plus Munc18–1, Munc13–1, synaptotagmin-1, NSF and αSNAP. Previous studies had included the SNAREs and one or two additional components but, as explained above, did not explain the vital importance of Munc18–1 and Munc13s for neurotransmitter release. Particularly important was the inclusion of Munc13–1, which had not been incorporated in previous reconstitution studies. Preparation of soluble, well-behaved fragments corresponding to the conserved C-terminal region of this large protein involved extensive work that started in 1998 and gradually allowed us to obtain increasingly longer fragments9, 29, 48, 62. Thus, large fragments such as C1C2BMUN and C1C2BMUNC2C can now be prepared readily. Another key aspect of our reconstitutions was the inclusion of NSF and αSNAP. One early report had shown that NSF-αSNAP inhibit the lipid mixing induced by the neuronal SNAREs alone because it disassembles syntaxin-1-SNAP-25 heterodimers50, but these factors were omitted from most reconstitution studies and their inclusion is crucial to render fusion strictly dependent on both Munc18–1 and Munc13–1 (e.g. Figure 3). Note also that inclusion of NSF-αSNAP is also required for the tight Ca2+ dependence of fusion in this system, as efficient Ca2+-independent fusion can be observed for instance between VSyt1-liposomes and T-liposomes without Munc18–1 and Munc13–1, or between V-liposomes and T-liposomes in the presence of Munc13–1 C1C2BMUNC2C29.
It is worth noting that the tight Ca2+ dependence of fusion in our system is not fully understood. It is plausible that Ca2+ binding to synaptotagmin-1 contributes to accelerate fusion at faster time scales than those associated with our bulk assays, but in these assays the Ca2+-dependence of fusion arises largely from Ca2+ binding to the Munc13–1 C2B domain29. Mutations in the Ca2+ binding sites of the Munc13–2 C2B domain suggested that Ca2+ binding to this domain is not critical for release evoked by a single action potential but is important for release during repetitive stimulation63 (note that Munc13–2 is very closely related to Munc13–1). These observations suggest that our reconstitutions may be recapitulating an activate state of the release machinery formed during repetitive stimulation. However, there is also some evidence that the Munc13–2 C2B domain acts as a Ca2+ sensor in release63, and it is plausible that the mutations in the Munc13–2 C2B domain Ca2+ binding sites did not influence release evoked by a single action potential because of partial functional redundancy with another protein (e.g. CAPS)29. Indeed, the fact that our reconstitutions reproduce many features of neurotransmitter release implies that they constitute a powerful tool to discover novel features of the mechanism of release that may have not been found in studies performed in neurons because of functional redundancy.
Another important feature of our reconstitutions is the simultaneous monitoring of lipid and content mixing using the approach pioneered by the lab of Bill Wickner to study yeast vacuolar fusion, which emphasized the critical importance of measuring content mixing to monitor real membrane fusion28, 54. This conclusion was also drawn from work with single vesicle assays27, 43. Compared with these assays, whether they monitor fusion between pairs of vesicles27, 41 or between single vesicles and planar bilayers37–40, our bulk solution experiments offer the advantage that they are easy to perform and in terms of instrumentation they only require a fluorimeter. Single vesicle assays are more technically demanding but they offer a much better time resolution and allow distinction of different steps such as docking, fusion and the potential formation of intermediates in the fusion pathway25, 41. In bulk experiments, dynamic light scattering can be used to monitor vesicle clustering that reflects docking, but it is much more informative to be able to dissect the kinetics of docking and the kinetics from docking to fusion at the single vesicle level. Note also that in bulk fusion assays signals reporting fluorescence de-quenching are highly non-linear, which can lead to misinterpretation of the fusion kinetics, and single rounds of fusion are not easy to distinguish from multiple rounds of fusion. In contrast, single-vesicle assays normally do not suffer from this limitation because fusion events are counted ‘digitally’. If instrumentation is available to perform both bulk assays and single vesicle experiments, we believe that it is most productive to start with the former to optimize conditions and have an idea of the activities of the different reagents, and then continue with the latter to gain more detailed information.
MATERIALS
REAGENTS
!CRITICAL All lipid reagents must be stored at −20C.
Marina Blue® 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine (MB-DHPE) (Invitrogen, cat. No. M12652)
1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt), 18:1 PS (DOPS) (Avanti Polar Lipids, Inc. cat. No. 840035C)
1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) (ammonium salt) 16:0 NBD-PE (Avanti Polar Lipids, cat. No. 810144C)
Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 16:0–18:1 PC (POPC) (Avanti Polar Lipids, Inc. cat. No. 850457C)
Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 16:0–18:1 PE (POPE) (Avanti Polar Lipids, Inc. cat. No. 850757C)
Cholesterol (Avanti Polar Lipids, cat. No. 70000P)
Palmitoyl-2-oleoyl-sn-glycerol, 16:0–18:1 DG (DAG) (Avanti Polar Lipids, Inc. cat. No. 800815C)
L-α-phosphatidylinositol-4,5-bisphosphate (Brain, Porcine) (ammonium salt), Brain PI(4,5)P2 (PIP2) (Avanti Polar Lipids, Inc. cat. 840046X)
Amberlite® XAD®−2 beads (Sigma-Aldrich, cat. No.10357)
ATP (Sigma-Aldrich, cat. No.A3377)
Calcium chloride dehydrate (Sigma-Aldrich, cat. No. C3881)
Chloroform (anhydrous) (Fisher Scientific, cat. No. C606–1)! CAUTION Chloroform is a carcinogen. All sample preparations involving chloroform should be performed under a chemical hood
Cy5-streptavidin (KPL, cat. No. 072-02-30-00)
EGTA [Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid] (Research Products International, cat. No. E57060–100.0)
Glycerol (Fisher BioReagents™ cat. No. BP229–4)
HEPES, Free Acid [N-(2-Hydroxyethyl) piperazine N’-(2-ethanesulfonic acid)] (Research Products International, cat. No. H75030–1000.0)! CRITICAL Once any buffer including HEPES is prepared, it should be stored at 4 °C and used within 1 month.
Histodenz (Sigma-Aldrich, cat. No. D-2158)
Liquid nitrogen (Airgas, cat. No. NINF160LT50) !CAUTION Liquid Nitrogen is extremely cold. Personal protective equipment required while handling.
Magnesium chloride (hexahydrate) (Fisher Scientific, cat. No. BP214–500)
Octylglucoside, ultra pure (Gold Biotechnology, cat. No. O-110–50)
Potassium chloride (Sigma-Aldrich, cat. No. P3911–500G)
Potassium hydroxide (pellets/certified ACS) (Fisher Scientific, cat. No. P250–1)
Protease Inhibitor Cocktail powder (Sigma-Aldrich, cat. No. P2714)
Proteins purified by the procedures described in the indicated references: full length rat syxtaxin-1A9,56; full length rat SNAP-25A (with its four cysteines mutated to serines)56; full-length rat Synaptobrevin56, rat synaptotagamin-1 C2AB fragment (residues 131–421)64 or rat synaptotagamin-1 (residues 57–421 with the following cysteine mutations: C74S, C75A, C77S, C79I, C82L (referred to as Synaptotagmin-1 for simplicity) (the expression vector was a kind gift from Thomas Söllner)29; full length Cricetulus griseus NSF V155M mutant9; full-length Bos taurus αSNAP9; full-length rat Munc18–144; and rat Munc13–1 fragments spanning the C1C2BMUN and C1C2BMUNC2C regions (residues 529–1531 and 529–1735, respectively, all with residues 1408–1452 from a flexible loop replaced by the sequence EF)9,29.
R-Phycoerythrin, Biotin-XX Conjugate (Thermo Fisher Scientific, cat. No. P811)
Streptavidin (Thermo Fisher Scientific, cat. No. 21125)
TCEP-HCl (GoldBioTechnology, Inc. cat. No. TCEP10) !CAUTION TCEP can cause severe skin burns and eye damage. Personal protective equipment required while handling.
EQUIPMENT
PTI QM-400 QuantaMaster Spectrofluorometer (Photon technology international) equipped with Four-Position Cuvette Holder (TURRET 400™) and water circulator (EHEIM ecco pro).
DynaPro instrument to measure dynamic light scattering (Wyatt Protein Solutions).
8mm Assembled Clear Autosampler Vial Kits (Thermo Fisher Scientific, cat. No. C4013–57)
Aluminum foil (Fisher Scientific, cat. No. 01-213-102)
Analytic Digital Scale (Denver Instruments Company, model: A-160)
Centrifuge tubes (Thinwall, Polypropylene, 4 ml, 11 × 60 mm, Beckman, item no. 328874)
Disposable Borosilicate Glass Tubes with Plain End (16mmX100mmm, Fisher Scientific, cat. No. 14-961-29)
Magnetic Stir plate (Corning, mfr. No. 6795–611)
Microcentrifuge tubes (1.5 ml, USA Scientific, cat. No. 1615–5500; 0.2 ml, Thermo Fisher Scientific, cat. No. AB0620)
Milili-Q water purification system (Barnstead)
Needle for syringe (20 G × 1 1/2 in., BD, cat. No. 305176)
Parafilm (Fisher Scientific, cat. No. 13-374-10)
Positive Displacement Digital Microdispensers (Drummond Scientific Company, cat. No. 3-000-575)
Replacement Bores for Microdispensers (Drummond Scientific Company, cat. No. 3-000-275-G)
PYREX™ Griffin Beakers (Thermo Fisher Scientific, cat. No. 02-540)
Special Glass Fluorometer Cuvettes (Starna Cells, Inc. cat No. 3-3.45-SOG-3)
Sterile disposable syringe (1ml, BD, cat. No. 309602)
Swinging-Bucket Rotor for ultracentrifuge (Beckman Coulter, SW 60 Ti)
Thermo Scientific™ Slide-A-Lyzer™ 10K MWCO Dialysis Cassettes (Thermo Fisher Scientific cat. No. 66383)
Ultracentrifuge (Optima L-90K, Beckman Coulter, Item No: 365670)
Ultrasonic Cleaner (Branson, Model 2510)
Vacuum Desiccator (BEL-ART, cat. No. F42025–0000)
Vacuum Pump or house vacuum line
Vortex mixer (Barnstead, Model 16715)
Water bath (Precision Scientific, Model 180 SERIES, cat. No. 51221065)
REAGENT SETUP
Marina-Blue DHPE stock
Prepare the stock in chloroform (1 mg/ml) in a dark vial (8mm Assembled Clear Autosampler Vial Kits). The solution can be stored at −20 °C for up to 6 months.
Cholesterol stock
Prepare the stock in chloroform (10 mg/ml) in a dark vial (8mm Assembled Clear Autosampler Vial Kits). The solution can be stored at −20 °C for up to 1 month.
HEPES Buffer
The solution consists of 25 mM HEPES, pH 7.4 with KOH, 150 mM KCL, 10% (vol/vol) Glycerol, and 0.5 mM TCEP. Freshly prepared each time and filtered through a 0.2 μm filter.
Histodenz stock
Prepare 70% (wt/vol) histodenz in HEPES buffer but only with 2% (vol/vol) glycerol. It can be stored at 4°C for 6 months. Dilute with HEPES buffer to obtain 25% (wt/vol) histodenz solution when needed.
OG stock
Prepare 20% (wt/vol) Octylglucoside stock in water. It can be stored at −20 °C for 6 months.
Protease inhibitors stock
Prepare 200× protease inhibitor stock by dissolving one bottle of Protease Inhibitor Cocktail powder in 20 mL water. This mixture contains 10 mM AEBSF, 1.5 μM Aprotinin, 580 μM Bestatin, 70 μM E-64, 5 μM Leupeptin and 5 mM EDTA. It can be stored at −20 °C for 6 months.
TCEP stock
Prepare 1 M TCEP stock in water. It can be stored at −20 °C for 6 months.
EQUIPMENT SETUP
Samples were premixed in the cells of the PTI spectrofluorometer, which is controlled by the Felix GX software. The system has a Four-Position Cuvette Holder, which allows rapid position changes among four samples during data acquisition. The temperature can be changed rapidly and precisely with the water circulation system. All the data were collected at 30 °C. Multi Dye acquisition is set up to look at the de-quenching of Marine Blue (excitation at 370 nm, emission at 465 nm) and increase in Cy5 emission through the buildup of FRET between PhycoE and Cy5 (excitation at 565 nm, emission at 670 nm) simultaneously.
PROCEDURE
Prepare lipids. TIMING 1h plus vacuum time (~12h)
1| Take vials with lipid stock solutions out of −20 °C freezer and keep them in a dark place to equilibrate to room temperature (~20 °C).
2| While wearing the appropriate gloves, rinse the glass tubes (16mm × 10mm) with chloroform at least three times to clean them.
! CAUTION Chloroform is a carcinogen. The experiment should be performed in a chemical hood.
3| Combine the following lipid solutions in a cleaned glass tube to make: T-liposomes (containing syntaxin-1 and SNAP25) (PC/ PS/ PE/ PIP2/ DAG/ Cholesterol = 38: 18: 20: 2: 2: 20 mol%); V-liposomes (containing synaptobrevin) (PC/ PS/ PE/ Cholesterol/ NBD-PE/ MB-DHPE = 39: 19: 19: 20: 1.5: 1.5 mol%) or VSyt1-liposomes (containing synaptobrevin and synaptotagmin-1) (PC/ PS/ PE/ Cholesterol/ NBD-PE/ MB-DHPE = 40: 6.8: 30.2: 20: 1.5: 1.5 mol%), respectively. Use microdispensers with positive replacement glass capillary for transferring the lipid solutions. Mix lipids according to the following table (for 10 mM total lipid concentration).
| T-liposomes | V-liposomes | VSyt1-liposomes | ||||
|---|---|---|---|---|---|---|
| % | Volume (μl) | % | Volume (μl) | % | Volume (μl) | |
| POPC (25 mg/ml) | 38 | 48.5 | 39 | 49.8 | 40 | 51.1 |
| DOPS (10 mg/ml) | 18 | 61.2 | 19 | 64.6 | 6.8 | 23.1 |
| POPE (10 mg/ml) | 20 | 60.3 | 19 | 57.3 | 30.2 | 91.1 |
| Cholesterol (10 mg/ml) | 20 | 32.5 | 20 | 32.5 | 20 | 32.5 |
| PIP2 (1 mg/ml) | 2 | 92.2 | - | - | - | - |
| DAG (2 mg/ml) | 2 | 25.0 | - | - | - | - |
| NBD-PE (1 mg/ml) | - | - | 1.5 | 54.9 | 1.5 | 54.9 |
| MB-DHPE (1 mg/ml) | - | - | 1.5 | 59.5 | 1.5 | 59.5 |
! CRITICAL STEP VSy1-liposomes were prepared with a smaller percentage of PS (6.8%) to prevent inhibition of fusion due to interactions of Syt1 with the membrane where it is anchored59.
! CAUTION Chloroform is a carcinogen. The experiment should be performed in a chemical hood. The lipid composition can be changed according to the purpose of the experiment.
4| Blow-dry lipids carefully under a gentle stream of nitrogen. Tilt and rotate the tube continuously until a thin film is formed on the wall near the bottom of the tube.
! CRITICAL STEP If precipitation is observed in the lipid mixture because of PIP2 addition, dry the lipids in the water bath at 45°C. Adjust the nitrogen stream strength to prevent the lipid solution from spilling out of the glass tube. Make sure lipids dried on the wall near the bottom, otherwise it will be difficult to harvest all the lipids in the hydration step.
! CAUTION Chloroform is a carcinogen. The experiment should be performed in a chemical hood.
5| Wrap the tubes with foil with a small hole on the top for chloroform evaporation and keep the tubes in the vacuum desiccator overnight for further drying.
! CRITICAL STEP Protect the lipids from exposure to ambient light by aluminum foil in order to minimize the photobleaching of dyes.
6| Add 420 μl HEPES buffer containing 2% βOG to hydrate the lipid film. Vortex vigorously for complete suspension (about 2 min). Incubate the lipids at room temperature for 15 min and afterwards sonicate in a bath-sonicator for 5 min twice.
! CRITICAL STEP The amount of buffer can be adjusted depending on the desired lipid concentration. The critical micellar concentration of βOG is ca. 25 mM (ca. 0.7% w/v). Therefore, the detergent is added well above this concentration, but an even higher concentration may be required for higher lipid concentrations.
! PAUSE POINT The lipids can be snap-frozen in liquid nitrogen and stored at −80°C for at least one month.
Reconstitution. TIMING 2h plus dialysis time (14–16 h)
7| Follow the steps in A for reconstitution of T-liposomes containing PhycoE-Biotin dye at protein to lipid ratio Syx:SNAP25:Lipids = 1:5:800, B for V-liposomes containing Cy5-Strepavidin dye at protein to lipid ratio Syb:lipids = 1:500, and C for VSyt1-liposomes containing Cy5-Strepavidin dye at protein to lipid ratio Syt:Syb:Lipids = 1:2:1000.
! CAUTION The protein to lipid ratio can be changed according to the purpose of the experiment. The proteins reconstituted in this way are expected to be oriented both outside and inside of the vesicles.
(A) Reconstitution of T-liposomes
| T-liposomes (500 μl) | ||
|---|---|---|
| final concentration | volume (μl) | |
| Syntaxin-1 (36 μM) | 5 μM | 69.4 |
| βOG 1 (20 %) | ~1% | 3.3 |
| SNAP-25 (200 μM) | 25 μM | 62.5 |
| TCEP (1 mM) | 1 μM | 0.5 |
| Protease inhibitors (200×) | 1× | 2.5 |
| Incubate the above reagents at r.t. for 25 min | ||
| Lipids for T-liposomes (10 mM) | 4 mM | 200.0 |
| βOG 2 (20 %) | ~1% | 6.0 |
| PhycoE-Biotin (16.7 μM) | 4 μM | 119.8 |
| HEPES buffer with 1 % βOG | 36.0 | |
(i) In a 1.5 ml microcentrifuge tube, mix Syntaxin-1, βOG and SNAP-25 according to the table above. Incubate the mixture at room temperature for 25 min.
! CRITICAL STEP SNAP-25 with all the cysteine mutated to serine is used to avoid polymerization through disulfide bonds. Syntaxin-1 has a transmembrane domain. Extra βOG was added before adding SNAP-25 to keep always at least 1 % βOG in the solution. TCEP and protease inhibitors are added at this step.
(ii) To the Syntaxin-1-βOG-SNAP-25 mixture, add lipids, βOG, PhycoE-Biotin and buffer according to the table above. Wrap the tube with foil and incubate the mixture at room temperature for 25 min.
! CRITICAL STEP Add detergent βOG before adding the content dye PhycoE-Biotin, to make sure the solution always contain at least 1 % βOG. Protecting the sample from exposure to light can reduce photobleaching. Never Freeze the PhycoE-Biotin to prevent precipitation.
(iii) Transfer the sample mixture to a pre-hydrated dialysis cassette using a Syringe with needle. Dialyze the sample against HEPES buffer containing detergent absorbing beads as indicated in the table below.
| HEPES buffer | XAD-2 beads | Temperature | Time | |
|---|---|---|---|---|
| 1 | 0.5 Liter | 1 g | 4 °C | 1 hour |
| 2 | 0.5 Liter | 1 g | 4 °C | 2 hour |
| 3 | 1 Liter | 2 g | 4 °C | ~12 hour |
!CAUTION Protect the sample from light to reduce photobleaching. Using different kinds of dialysis cassettes, different amounts of detergent absorbing beads, different dialysis temperature, and different volume of dialysis solution might change the dialysis speed and compromise the reconstitution.
(B) Reconstitution of V-liposomes
| V-liposomes (500 μl) | ||
|---|---|---|
| final concentration | volume (μl) | |
| Lipids for V-liposomes (10 mM) | 4 mM | 200.0 |
| Synaptobrevin (32 μM) | 8 μM | 125.0 |
| βOG (20 %) | ~1% | 6.4 |
| Cy5-Streptavidin (32 μM) | 8 μM | 125.0 |
| HEPES buffer with 1 % βOG | 40.6 | |
| TCEP (1 mM) | 1 μM | 0.5 |
| Protease inhibitors (200×) | 1× | 2.5 |
(i) In a 1.5 ml microcentrifuge tube, mix lipids, Synaptobrevin, βOG and Cy5-Streptavidin according to the table above. Incubate the mixture at room temperature for 25 min. TCEP and protease inhibitors are added at this step.
! CRITICAL STEP Add detergent βOG before adding the content dye Cy5-Streptavidin, to make sure that the solution always contains at least 1 % βOG. Protecting the sample from exposure to light can reduce photobleaching.
(ii) Transfer the sample mixture to a pre-hydrated dialysis cassette using a Syringe with needle. Dialyze the sample against HEPES buffer containing detergent absorbing beads as described for the T-liposomes.
(C) Reconstitution of VSyt1-liposomes
| VSyt1-liposomes (500 μl) | ||
|---|---|---|
| final concentration | volume (μl) | |
| Synaptotagmin-1 (18 μM) | 2.5 μM | 69.4 |
| βOG 1 (20 %) | ~1% | 1.0 |
| KCl 1 (3000 mM) | 600 mM | 20.5 |
| Synaptobrevin (32 μM) | 5 μM | 78.1 |
| TCEP (1 mM) | 1 μM | 0.5 |
| Protease inhibitors (200×) | 1× | 2.5 |
| Incubate the above reagents on ice for 15 min | ||
| βOG 2 (20 %) | ~1% | 9.5 |
| KCl 2 (3000 mM) | 600 mM | 65.6 |
| Lipids for VSyt1-liposomes (10 mM) | 2.5 mM | 125.0 |
| Cy5-Streptavidin (32 μM) | 8 μM | 125.0 |
| HEPES buffer with 1 % βOG | 2.9 | |
(i) In a 1.5 ml microcentrifuge tube, mix Synaptotagmin-1, Synaptobrevin, βOG and KCl according to the table above. Incubate the mixture on ice for 15 min.
! CRITICAL STEP The high salt concentration can prevent Synaptotagmin-1 aggregation. Add detergent βOG before adding KCl and Synaptobrevin, to make sure that the solution always contain at least 1 % βOG. TCEP and protease inhibitors are added at this step.
(ii) To the pre-incubation mixture, add βOG, KCl, lipids, Cy5-Streptavidin and buffer according to the table above. Wrap the tube with foil and incubate the sample on ice for 15 min.
! CRITICAL STEP Add detergent βOG first to keep 1 % βOG in the solution. The KCl concentration should be 600 mM to prevent Synaptotagmin-1 aggregation. Protecting the sample from exposure to light can reduce photobleaching.
(iii) Transfer the sample mixture to a preydrated dialysis cassette using a Syringe with needle. Dialyze the sample against HEPES buffer containing detergent absorbing beads as described for the T-liposomes.
?TROUBLESHOOTING
Purification of proteoliposomes. TIMING up to 3 hours
8| Harvest proteoliposome suspensions from dialysis cassette using a 1 ml syringe with needle (~0.5 ml). Transfer the proteoliposome solution into a 2 ml microcentrifuge tube. Add HEPES buffer to each sample to make it 1 ml.
! CRITICAL STEP The volume of the proteoliposome solution may change during dialysis. Measure the sample volume using a pipetman and add the corresponding volume of HEPES buffer to make it 1 ml.
9| Add 1 ml 70% Histodenz into the proteoliposome solution and mix.
10| Transfer the proteoliposome solution to 11×60 mm Beckman ultracentrifuge tubes.
11| Overlay the sample with 1.8 ml 25% Histodenz to the 3.8 ml mark.
! CRITICAL STEP Premark the ultracentrifuge tube at 3.8 ml. Add the Histodenz solution slowly and gently along the tube wall.
12| Overlay the sample with 600 μl HEPES buffer.
! CRITICAL STEP Add the HEPES buffer slowly and gently along the tube wall. The solution should just fill the tube but not overflow.
13| Carefully put the ultracentrifuge tube into the bucket. Match numbered caps to numbered buckets and assemble them.
! CRITICAL STEP Before putting the ultracentrifuge tube inside the bucket, check whether all the caps can be assembled with the buckets. Moreover, hook all the empty buckets on the rotor and see whether they can swing smoothly.
14| Balance all the buckets, empty or loaded, within 2 mg difference.
! CRITICAL STEP Loaded buckets must be arranged symmetrically in the rotor. Opposing tubes must be filled to the same level with liquid of the same density.
15| Hook buckets to SW60ti rotor and spin at 406,849g (55,000 rpm with Rmax = 120.3mm in our centrifuge) at 4 °C for 1.5 hours.
! CRITICAL STEP All buckets must be attached to the rotor, whether loaded or empty. Use slow stop mode.
16| Harvest the solution (~600 μl) at the top interface.
! CRITICAL STEP Protect the samples from light.
Characterization of proteoliposomes. TIMING up to 5 hours
17| Determine the concentration for liposome samples. For V- or Vsyt1-liposomes, the lipid concentration can be estimated from the NBD absorbance at 465nm comparing to a standard curve. For T-liposomes, the lipid concentration can be measured by the Stewart method65.
18| Characterize the size distribution of the reconstituted liposomes. We normally perform this analysis by dynamic light scattering56, and the average liposome radii that we observe is typically around 60–95 nm (Figure 4). It is advisable to in addition analyze the liposomes by cryo-electron microscopy56 if an appropriate electron microscope is available, as this technique yields a more faithful representation of the liposome size distribution.
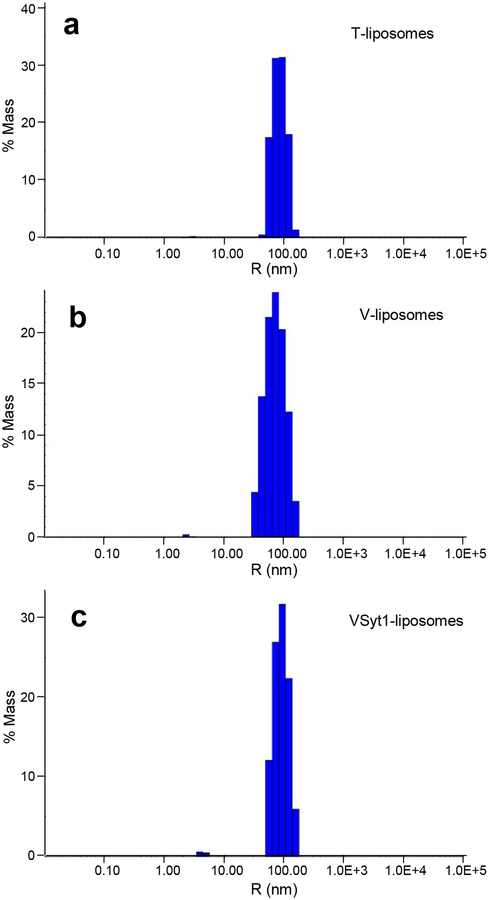
Figure 4.
Analysis of liposome size distribution by DLS. The diagrams show typical results obtained in analyses of particle size for T-liposomes (a), V-liposomes (b) and VSyt1-liposomes (c) by DLS. The average liposome radii calculated in these experiments were 85, 75 and 92 nm, respectively.
19| Analyze the amount of PhycoE-Biotin or Cy5-Streptavidin molecules trapped in the different liposome preparations by measuring their emission fluorescence intensity at 579 nm or 676 nm, respectively, and comparing the measurement with a standard curve obtained by measuring the emission fluorescence intensity of solutions with known PhycoE-Biotin or Cy5-Streptavidin concentrations. The average numbers of PhycoE-Biotin or Cy5-Streptavidin molecules trapped per liposome are then calculated based on these measurements, the lipid concentrations measured in step 17 and the average liposome radii determined in step 18, assuming that all PhycoE-Biotin or Cy5-Streptavidin molecules are located in the volume inside the liposomes (which can be confirmed by verifying that no FRET develops quickly when the T- and V- or VSyt1 are mixed). Typically, the average numbers of trapped molecules that we observe are similar or somewhat lower to those expected according to the concentrations of PhycoE-Biotin or Cy5-Streptavidin used during reconstitution (4 μM and 8 μM, respectively) and the average radii measured by DLS. For example, the expected average numbers of trapped PhycoE-Biotin or Cy5-Streptavidin molecules in the preparations analyzed by DLS in Figure 4 were 6.2, 8.5 and 15.7, respectively, and those measured were 6.1, 8.1 and 6.4, respectively.
! PAUSE POINT The reconstituted proteoliposomes can be stored at 4 °C for at least 24 hours. They can be snap frozen as 10 μl drop beads in liquid nitrogen. They can be stored in liquid nitrogen or at −80°C but with some (ca. 10%) activity loss that may be due to content release during frezzing-de-freezing (fresh liposomes gave the best efficiency)
?TROUBLESHOOTING
FRET assay to test lipid mixing and content mixing TIMING up to 70 min per reaction.
20| Set up the PTI spectrofluorometer. In the FelixGX control software, choose the “QM400/Single Em” configure and “multi dye” mode. Set observation of two dyes at the same time. Excitation at 370 nm and emission at 465 nm for monitoring lipid mixing; excitation at 565 nm and emission at 670 nm to monitor content mixing. Turn on the temperature controller to the target temperature (30 °C).
! CRITICAL STEP The temperature can be adjusted according to the experiments required. Glass cuvettes that are transparent to these wavelengths should be used to monitor the fusion reactions.
21| In a 200 μl microcentrifuge tube, prepare the sample by mixing proteins, reagents and liposomes according to the experiments required. The following table shows one reaction (T+V+NSF+αSNAP+C2AB+M18+M13) as example. First, mix 250 μM T-liposomes with 2.5 mM Mg2+, 0.5 mM TCEP, 100 μM EGTA, 5 μM Streptavidin, 0.8 μM NSF, 2 μM αSNAP, 2 mM ATP, and 1 μM Munc18. Incubate the sample mixture at 37 °C for 25 min to allow disassembly of Syntaxin-1-SNAP-25 complexes by NSF-αSNAP and binding of Munc18–1 to the released Syntaxin-19 (Figure 2). Then mix the preincubation sample with 125 μM VSyt1-liposomes, 0.5 μM Munc13–1 C1C2BMUNC2C fragment and 1 μM SNAP-25.
| Final concentration | Volume to add (μl) | |
|---|---|---|
| HEPES buffer 1 | 36.4 | |
| TCEP (10 mM) | 0.5 mM | 10.0 |
| Mg2+ (50 mM) | 2.5 mM | 10.0 |
| T-liposomes (3.2 mM) | 250 μM | 15.6 |
| ATP (200 mM) | 2 mM | 2.0 |
| αSNAP (50 μM) | 2 μM | 8.0 |
| NSF (20 μM) | 0.8 μM | 8.0 |
| Munc18–1 (20 μM) | 1 μM | 10.0 |
| EGTA (10 mM) | 100 μM | 2.0 |
| Streptavidin (100 μM) | 5 μM | 10.0 |
| Incubate the components above at 37 °C for 25 min | ||
| HEPES buffer 2 | 77.5 | |
| V-Syt1 liposomes (2 mM) | 125 μM | 12.5 |
| Munc13–1 C1C2BMUNC2C (20 μM) | 0.5 μM | 5.0 |
| SNAP-25 (40 μM) | 1 μM | 5.0 |
| Transfer 190ul to cuvette for data acquisition. | ||
| CaCl2 (10 mM) at 5min | 0.5 mM | 10.0 |
| βOG (20 %) at the end | 1% | 10.0 |
! CRITICAL STEP 100 μM EGTA was added to chelate any trace Ca2+ in the beginning, and 5 μM streptavidin was used to make sure no content mixing signal arise from vesicle leakiness. The concentrations of purified liposomes and protein stocks are different for different batches. The table above described just an example of typical concentrations and volumes used. The volume of different components to add in the reaction need to be calculated according to the concentrations obtained in each case.
?TROUBLESHOOTING
22| Transfer 190 ul of the sample mixture to the fluorometer cuvette. Place the cuvette in the PTI spectrofluorometer with the temperature controlled at 30 °C. Start to acquire the data.
23| 0.6 mM Ca2+ is added at 300 s if needed.
! CRITICAL STEP We add 0.6 mM Ca2+ into the sample containing 0.1 mM EGTA to make the final concentration of Ca2+ about 0.5 mM, but the Ca2+ concentration can be adjusted as needed.
24| At the end of each reaction, 1% w/v β-OG is added to solubilize the liposomes, and measure the maximum lipid mixing signal. Control experiments without streptavidin are performed to estimate the maximum content mixing signal upon detergent addition.
! CRITICAL STEP Mix the detergent well enough with the sample. Keep collecting data for a few minutes after detergent addition to make sure that the maximum signal is obtained.
?TROUBLESHOOTING
Data Analysis TIMING up to 30 min for each data set.
25| For both lipid and content mixing, subtract the intensity of the initial point of the fluorescence curve (I0) from all the intensities of all the points of the curve (It). Normalize the lipid mixing curve to the fluorescence emission intensity obtained after detergent addition (Imax). For content mixing, the initial fluorescence is also subtracted from all the points of the curve, but for normalization the Imax-I0 value should ideally be derived from the control experiments performed without external streptavidin. However, there is sometimes variability in the maximum Cy5 fluorescence values observed upon detergent addition, perhaps because of binding of the dye to the detergent. As an alternative, the Imax-I0 value can also be derived from the maximum Cy5 fluorescence observed at the end of the most efficient fusion reactions (T+ VS+ NSF/αSNAP+ Munc18+ Munc13–1 C1C2BMUNC2C), which is similar to that observed upon detergent addition but more reproducible29.
There are multiple ways to analyze the data quantitatively. The fluorescence intensity values measured toward the end of the reaction inform on the overall extent of fusion but may not provide much information on the kinetics of the reaction if it is relatively fast. For our experiments29, we quantified the fluorescence intensity at 500 s (i.e. 200 s after addition of Ca2+). For each condition, we performed the experiments at least in triplicate and calculate average signal intensity as well as standard deviation. In repeated experiments performed under the same conditions but with different preparations of reconstituted proteoliposomes, there is some variability in terms of the absolute values of the extent of fusion as a function of time, but the relative results normally remain comparable.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table.
| Step | Problem | Possible Reason | Possible Solution |
|---|---|---|---|
| 7,17 | Low fusion efficiency after Ca2+ addition for the reaction of T+VSyt1+NSF/αSNAP+M18+M13 | a) Overestimated liposome concentration b) Not enough functional SNARE proteins reconstituted on the liposomes, either because of degradation or wrong concentration c) Low activity of one or more of the proteins and/or poor lipid quality. |
a) Measure the liposome concentration. b) Use SDS-PAGE to check both the proteoliposomes and the SNARE protein stocks used for reconstitution. If degradation is found in the protein stocks, then purify new proteins and add protease inhibitors during storage. If degradation happened during reconstitution, make sure protease inhibitors are included. If too little protein is incorporated on the liposomes, check whether the protein stock concentration is overestimated. Prepare fresh proteins and/or buy new lipids. |
| 21 | Too much content mixing before Ca2+ addition in the reaction of T+VSyt1+NSF/αSNAP+M18+M13 | Not enough EGTA | Check the concentration of EGTA. Make sure100 μM EGTA is included in the reaction. |
| 21 | Too much fusion for the control experiments T+VSyt1+NSF/αSNAP+M18, T+VSyt1+NSF/αSNAP+M13 and T+VSyt1+NSF/αSNAP |
NSF is not sufficiently active | NSF can become partially inactive after storage at −80 °C. Freshly purified NSF gives the best activity. |
| 24 | Signal keeps increasing slowly after adding detergent | The detergent did not mix well with the sample | Pipette the sample up and the down at least 10 times after detergent addition to make sure the detergent is well mixed with the sample. Keep collecting the data for a few minutes, until the signal intensity becomes stable. |
TIMING
Prepare lipids. TIMING 1h plus vacuum time (~12h)
Step 7, Reconstitution. TIMING 2h plus dialysis time (14–16 h)
Steps 8–16, Purification of proteoliposomes. TIMING up to 3 hours
Steps 17–19, Characterization of proteoliposomes. TIMING up to 5 hours
Steps 20–24, FRET assay to test lipid mixing and content mixing. TIMING up to 70 min per reaction.
Steps 25, Data Analysis. TIMING up to 30 min for each data set.
Anticipated Results
For experiments that include membrane-anchored synaptotagmin-1, i.e. using Vsyt1-liposomes, these liposomes are expected to fuse in a slow, Ca2+-independent manner with T-liposomes. Such fusion should be abolished in the presence of NSF-αSNAP. When NSF-αSNAP are included, highly efficient, Ca2+-dependent lipid and content mixing are observed in the presence of both Munc18–1 and Munc13–1 C1C2BMUNC2C, and no fusion is expected when either Munc18–1 or Munc13–1 C1C2BMUNC2C is omitted (Figure 3). In the ‘complete’ conditions that include Munc18–1, Munc13–1 C1C2BMUNC2C and NSF-αSNAP, there is lipid mixing but practically no content mixing before Ca2+ addition (Figure 3).
The high efficiency of membrane fusion in these complete experiments may prevent the observation of active roles for additional components of the release machinery that can be included in these assays. As mentioned above, such active roles may be observable using single-vesicle assays that can dissect docking from membrane fusion. Using the bulk solution approach described here, active roles may be uncovered if fusion is less efficient. For instance, synaptotagmin-1 C2AB has no clear effect in fusion assays performed with T- and V-liposomes in the presence of NSF-αSNAP, Munc18–1 and Munc13–1 C1C2BMUNC2C, but analogous experiments performed with the less active C1C2BMUN fragment revealed that synaptotagmin-1 did have a strong effect in promoting content mixing29. Since the use of the Munc13–1 C1C2BMUNC2C fragment and membrane-anchored synaptotagmin-1 are expected to be more physiologically relevant, it is advisable to keep these components as part of the system and lower the concentrations of soluble proteins and/or proteoliposomes, or decrease the protein-to-lipid ratios to decrease the activity in investigations of the active roles of other proteins. More discussion of the anticipated results can be found in our previous published paper29.
Acknowledgments
We thank Michael Zick and William Wickner for discussions on how to set up the simultaneous lipid and content mixing assays. This work was supported by grant I-1304 from the Welch Foundation (to JR) and by NIH Research Project Award R35 NS097333 (to JR), which continues work supported previously by NIH grants NS037200 and NS049044 (to JR).
Footnotes
Competing financial interests The authors declare that they have no competing financial interests.
REFERENCES
- 1.Rizo J & Sudhof TC The Membrane Fusion Enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices-Guilty as Charged? Annu. Rev. Cell Dev. Biol 28, 279–308 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, & Rothman JE A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Hanson PI, Roth R, Morisaki H, Jahn R, & Heuser JE Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Poirier MA et al. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol 5, 765–769 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Sutton RB, Fasshauer D, Jahn R, & Brunger AT Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Mayer A, Wickner W, & Haas A Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Banerjee A, Barry VA, DasGupta BR, & Martin TF N-Ethylmaleimide-sensitive factor acts at a prefusion ATP-dependent step in Ca2+-activated exocytosis. J. Biol. Chem 271, 20223–20226 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Ma C, Li W, Xu Y, & Rizo J Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol 18, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma C, Su L, Seven AB, Xu Y, & Rizo J Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker RW et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gracheva EO, Hadwiger G, Nonet ML, & Richmond JE Direct interactions between C. elegans RAB-3 and Rim provide a mechanism to target vesicles to the presynaptic density. Neurosci. Lett. 444, 137–142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y, Kaeser PS, Sudhof TC, & Schneggenburger R RIM determines Ca(2)+ channel density and vesicle docking at the presynaptic active zone. Neuron 69, 304–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Augustin I, Rosenmund C, Sudhof TC, & Brose N Munc13–1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400, 457–461 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Richmond JE, Davis WS, & Jorgensen EM UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravamudan B, Fergestad T, Davis WS, Rodesch CK, & Broadie K Drosophila UNC-13 is essential for synaptic transmission. Nat. Neurosci 2, 965–971 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Varoqueaux F et al. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc. Natl. Acad. Sci. U. S. A 99, 9037–9042 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koushika SP et al. A post-docking role for active zone protein Rim. Nat. Neurosci 4, 997–1005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoch S et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415, 321–326 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Sudhof TC The presynaptic active zone. Neuron 75, 11–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Chacon R et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature 410, 41–49 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Ann K, Kowalchyk JA, Loyet KM, & Martin TF Novel Ca2+-binding protein (CAPS) related to UNC-31 required for Ca2+-activated exocytosis. J. Biol. Chem 272, 19637–19640 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Jockusch WJ et al. CAPS-1 and CAPS-2 are essential synaptic vesicle priming proteins. Cell 131, 796–808 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Reim K et al. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell 104, 71–81 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Weber T et al. SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT, Cipriano DJ, & Diao J Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Crit Rev. Biochem. Mol. Biol 50, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Bogaart G et al. One SNARE complex is sufficient for membrane fusion. Nat. Struct. Mol. Biol 17, 358–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyoung M et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc. Natl. Acad. Sci. U. S. A(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zick M & Wickner WT A distinct tethering step is vital for vacuole membrane fusion. elife. 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X et al. Functional synergy between the Munc13 C-terminal C1 and C2 domains. elife. 5, e13696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhage M et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Rizo J & Xu J The Synaptic Vesicle Release Machinery. Annu. Rev. Biophys 44, 339–367 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Tucker WC, Weber T, & Chapman ER Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science 304, 435–438 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Xue M, Ma C, Craig TK, Rosenmund C, & Rizo J The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat. Struct. Mol. Biol 15, 1160–1168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chicka MC, Hui E, Liu H, & Chapman ER Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+. Nat. Struct. Mol. Biol 15, 827–835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J, Tareste DC, Paumet F, Rothman JE, & Melia TJ Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell 128, 183–195 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Chan YH, van LB, & Boxer SG Effects of linker sequences on vesicle fusion mediated by lipid-anchored DNA oligonucleotides. Proc. Natl. Acad. Sci. U. S. A 106, 979–984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fix M et al. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. U. S. A 101, 7311–7316 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Smith EA, Chapman ER, & Weisshaar JC Lipid mixing and content release in single-vesicle, SNARE-driven fusion assay with 1–5 ms resolution. Biophys. J 96, 4122–4131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowen ME, Weninger K, Brunger AT, & Chu S Single molecule observation of liposome-bilayer fusion thermally induced by soluble N-ethyl maleimide sensitive-factor attachment protein receptors (SNAREs). Biophys. J 87, 3569–3584 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domanska MK, Kiessling V, Stein A, Fasshauer D, & Tamm LK Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J. Biol. Chem 284, 32158–32166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon TY, Okumus B, Zhang F, Shin YK, & Ha T Multiple intermediates in SNARE-induced membrane fusion. Proc. Natl. Acad. Sci. U. S. A 103, 19731–19736 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HK et al. Dynamic Ca2+-dependent stimulation of vesicle fusion by membrane-anchored synaptotagmin 1. Science 328, 760–763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai Y et al. Fusion pore formation and expansion induced by Ca2+ and synaptotagmin 1. Proc. Natl. Acad. Sci. U. S. A 110, 1333–1338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulubova I et al. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 18, 4372–4382 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misura KM, Scheller RH, & Weis WI Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Dulubova I et al. Munc18–1 binds directly to the neuronal SNARE complex. Proc. Natl. Acad. Sci. U. S. A 104, 2697–2702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richmond JE, Weimer RM, & Jorgensen EM An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature 412, 338–341 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu J et al. A minimal domain responsible for Munc13 activity. Nat. Struct. Mol. Biol 12, 1017–1018 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Yang X et al. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat. Struct. Mol. Biol 22, 547–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber T et al. SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J. Cell Biol. 149, 1063–1072 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H, Jun Y, Thompson J, Yates J, & Wickner W HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29, 1948–1960 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zick M, Orr A, Schwartz ML, Merz AJ, & Wickner WT Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc. Natl. Acad. Sci. U. S. A 112, E2290–E2297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu H et al. Comparative studies of Munc18c and Munc18–1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. U. S. A 110, E3271–E3280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zucchi PC & Zick M Membrane fusion catalyzed by a Rab, SNAREs, and SNARE chaperones is accompanied by enhanced permeability to small molecules and by lysis. Mol. Biol. Cell 22, 4635–4646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudhof TC Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X et al. SNARE-Mediated Lipid Mixing Depends on the Physical State of the Vesicles. Biophys. J 90, 2062–2074 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arac D et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat. Struct. Mol. Biol 13, 209–217 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Takamori S et al. Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Stein A, Radhakrishnan A, Riedel D, Fasshauer D, & Jahn R Synaptotagmin activates membrane fusion through a Ca(2+)-dependent trans interaction with phospholipids. Nat. Struct. Mol. Biol 14, 904–911 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Zhao M et al. Mechanistic insights into the recycling machine of the SNARE complex. Nature 518, 61–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rigaud JL, Pitard B, & Levy D Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223–246 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Li W et al. The Crystal Structure of a Munc13 C-terminal Module Exhibits a Remarkable Similarity to Vesicle Tethering Factors. Structure. 19, 1443–1455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin OH et al. Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat. Struct. Mol. Biol 17, 280–288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu J, Brewer KD, Perez-Castillejos R, & Rizo J Subtle Interplay between Synaptotagmin and Complexin Binding to the SNARE Complex. J. Mol. Biol 425, 3461–3475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart JCM Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem 104, 10–14 (1980). [DOI] [PubMed] [Google Scholar]
- 66.Brewer KD et al. Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution. Nat. Struct. Mol. Biol 22, 555–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Q et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature 525, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]