Abstract
The protein phosphatase Sit4 has been shown to be required for lipogenesis and resistance against the acetyl-CoA carboxylase inhibitor soraphen A. Since Sit4 is also required for biosynthesis of Elongator dependent tRNA modifications such as 5-methoxycarbonylmethyluridine (mcm5U), we investigated the relevance of tRNA modifications in lipogenesis and soraphen A response. While sit4 and Elongator (elp3) mutants copy defects in mcm5U formation and stress sensitivity, they do not share soraphen A sensitivity and low lipid droplet (LD) phenotypes. In contrast to sit4, we found elp3 mutants to display partial soraphen A resistance and a high LD phenotype. Screening a collection of tRNA modification mutants additionally identified the tRNA pseudo-uridine synthase gene DEG1 to be required for soraphen A sensitivity. Since deg1 and elp3 share high LD and soraphen A resistance phenotypes, these are likely caused by translational defects. In support of this notion, we observe overexpression of tRNAGlnUUG suppresses lipolysis defects of deg1 mutants. Hence, the sit4 mutation results in a composite defect including tRNA modification deficiency and loss of Snf1 kinase dephosphorylation, which induce opposite effects on LD regulation. Importantly, however, the Snf1 kinase regulatory defects of the phosphatase mutant dominate over effects on LD regulation imposed by loss of the tRNA modification alone.
Keywords: soraphen A, Sit4, tRNA modification, Elongator complex
1. Introduction
A key step in lipogenesis is catalyzed by acetyl-CoA carboxylase, which in budding yeast Saccharomyces cerevisiae is encoded by the essential ACC1 gene [1]. Acc1 catalyzes the conversion of acetyl-CoA to malonyl-CoA and its activity is inhibited via phosphorylation by adenosine monophosphate (AMP)-activated protein kinase (AMPK) Snf1 [2,3]. Snf1 kinase activity is regulated by glucose availability and the nutrient sensing pathway (target of rapamycin, TORC1) [2,4,5]. The latter is thought to involve the Sit4-Sap190 protein phosphatase, which antagonizes Snf1 kinase activation. Consistent with this, absence of either Sit4 or Sap190 results in a constitutively phosphorylated and hyperactive Snf1 kinase which results in downregulation of Acc1 activity and a low lipid droplet phenotype, while a SNF1 gene deletion causes an opposite outcome [2]. The low lipid droplet phenotype of sit4 mutants was detected using boron-dipyrromethene (Bodipy) fluorescence measurements of stationary phase cultures [2]. In further support of this mutual relationship, cells lacking Snf1 activity are less sensitive towards soraphen A than wild-type yeast, and both sit4 and sap190 mutants display strong sensitivity towards the Acc1 inhibitor drug [2].
In addition to its role in regulating lipogenesis, the protein phosphatase complexes Sit4-Sap190 and/or Sit4-Sap185 are also required to maintain Elongator activity [6,7,8,9]. In yeast, the primary function of the Elongator complex, which is composed of subunits Elp1–Elp6 [10,11], lies with formation of the tRNA anticodon modifications 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 5-methoxycarbonylmethyluridine (mcm5U) and 5-carbamoylmethyluridine (ncm5U) [12,13]. Mcm5s2U synthesis additionally requires the presence of a functional sulfur transfer system consisting of Nfs1, Tum1, Urm1, Uba4, Ncs2 and Ncs6 [9]. Absence of Elongator, either alone or in combination with the sulfur transfer system, induces pleiotropic phenotypes including sensitivity against the target of rapamycin (TOR) inhibiting drugs rapamycin and caffeine, cell cycle delays, signaling defects and morphological abnormalities [12,14,15,16,17,18,19]. Most of these composite traits can be rescued by elevating the cellular abundance of tRNAs (i.e., tRNAGlnUUG, tRNAGluUUC and tRNALysUUU) naturally carrying the anticodon modification mcm5s2U [12,15,17]. Suppression of phenotypes by higher-than-normal levels of tRNAs that critically depend on the anticodon modifications is thought to restore near-normal translation by compensating for ribosomal A-site binding defects [20,21]. While sit4 and elp1–elp6 mutants display identical tRNA modification defects [13,22], it remained unknown whether the tRNA modification defect of sit4 mutants is related to lipid specific phenotypes. Here we compared mutants defective in both, Snf1 regulation and Elongator function (Sit4) with those defective in Elongator alone in relation to general growth and lipid metabolism phenotypes such as soraphen A sensitivity and lipid droplet (LD) content. We provide evidence that loss of the tRNA modification alone in Elongator mutants exerts opposite effects on lipid metabolism as compared to the defect in sit4. Further, we demonstrate that not only mcm5s2U defects but also deficiency in pseudo-uridine formation at tRNA position 38/39 in a deg1 mutant [23] are associated with a high lipid droplet phenotype that results from delayed lipolysis and confer partial resistance towards Acc1 inhibition by soraphen A.
2. Materials and Methods
2.1. Strains
Mutants defective in individual subunits of the Elongator complex were obtained from the S. cerevisiae deletion collection (OpenBiosystems, Waltham, MA, USA) . Strains were transformed by the lithium acetate method [24]. The plasmids used were pRS425; pKQE (tRNAGlnUUG, tRNALysUUU and tRNAGluUUC genes cloned in pRS425); pRS423 and tQ (tRNAGlnUUG gene cloned in pRS423) [17,25,26].
2.2. Reagents
Soraphen A was a kind gift from Professor Rolf Müller (Helmoltz-Zentrum für Infektionsforschung, Saarbrücken, Germany) to M.M.L. and R.S. and was kept as a 0.1 mg/mL stock solution in 10% methanol. A crude preparation of zymocin was obtained from culture filtrates of the Kluyveromyces lactis killer strain and applied to yeast extract peptone dextrose (YPD) plates as described earlier [27,28]. All other reagents used were from Sigma-Aldrich (St. Louis, MO, USA).
2.3. Growth Conditions and Lipid Droplets Quantification
Yeast cells were grown to stationary phase for 48 h in liquid rich medium (YPD; 1% yeast extract, 2% peptone and 2% glucose) at 30 °C to achieve a high LD content. For LD dynamics, the stationary cells were seeded at low density (optical density (O.D) 600 nm = 0.25) in YPD. At the indicated times cells were fixed in 3.7% formaldehyde and the LD was measured by the liquid fluorescence recovery assay [2,29]. In this assay formaldehyde-fixed cells are added to a medium containing a quenched solution of the hydrophobic fluorescent probe Bodipy containing 5 µM Bodipy 493/503 (Invitrogen, Waltham, MA, USA) and 0.5 M of the fluorescence quencher potassium iodide (KI). After cell addition Bodipy 493/503 enters readily into the cells and fluorescence is measured with a Spectramax M5 fluorimeter (Molecular Devices, San Jose, CA, USA) at 495/510 nm. Fluorescence is normalized to O.D 600 nm and reported as lipid droplet index (LD index).
2.4. Drug Assays
S. cerevisiae strains were grown for 24 h to stationary phase in YPD or synthetic defined (SD) medium plus appropriate auxotrophic markers, and serial dilutions of the cultures were prepared in sterile-distilled water to 107, 106 and 105 cells/mL. Dilutions were spotted using a replica plater in solid medium in the presence of the drugs indicated. The microtiter based liquid growth assay was done as described [27].
3. Results
3.1. Analysis of Shared and Non-Shared Phenotypes of sit4 and elp3 Mutants
Mutants defective in individual subunits of the Elongator complex are known to exhibit pleiotropic phenotypes, which can routinely be suppressed by overexpression of the three tRNAs normally carrying the wobble uridine (U34) mcm5s2U modification. Since sit4 mutants also lack the ability to form modified nucleosides such as mcm5s2U, mcm5U and ncm5U [22], we first compared phenotypes of elp3 and sit4 mutants and tested their response to overexpression of the three tRNAs. We reasoned that phenotypes caused by mcm5s2U deficiency but not those resulting from additional impacts of the sit4 mutation on Snf1 regulation should respond to upregulated tRNA gene dosage.
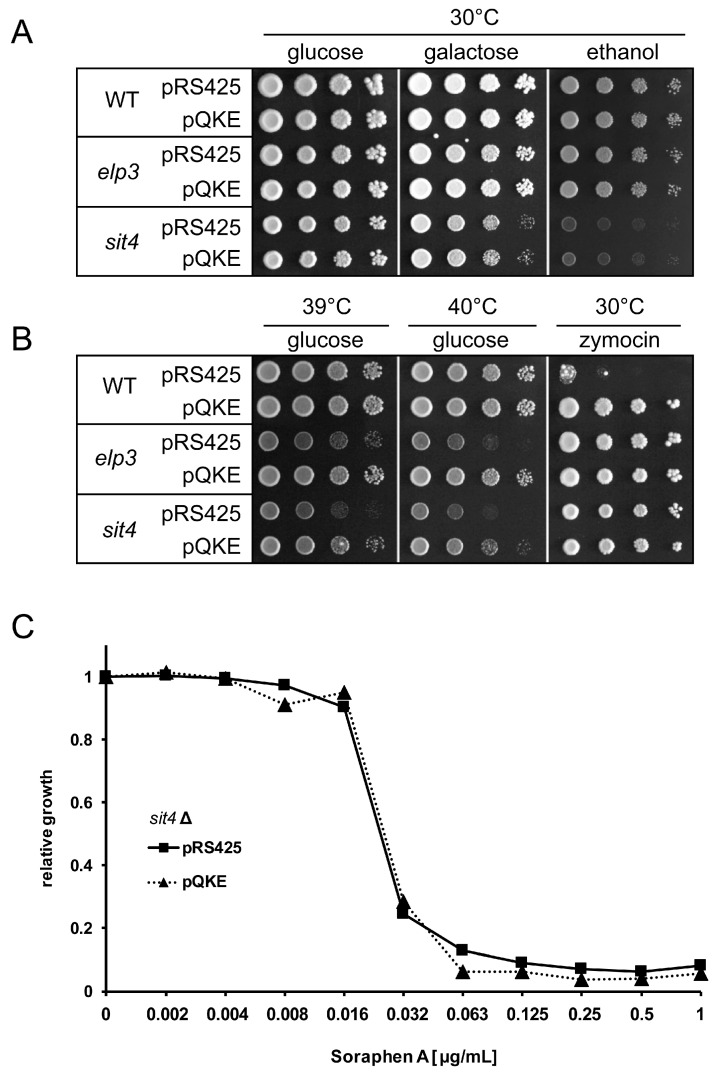
We transformed sit4 and elp3 mutant strains and the isogenic wild type BY4741 with either pRS425 or pQKE. The latter was previously shown to result in overexpression of tRNAGlnUUG, tRNALysUUU and tRNAGluUUC and to suppress most phenotypes of U34 modification defects, including temperature sensitivity [12,17]. In addition, overexpression of these three tRNAs or tRNAGluUUC alone results in resistance against the tRNA cleaving toxin zymocin [26,30]. As expected, the wild type carrying the pQKE construct displayed zymocin resistance, whereas the empty vector carrying strain was inhibited (Figure 1).
Figure 1.
Phenotypes of elp3 and sit4 mutants and their responses to overexpression of tRNAGlnUUG, tRNALysUUU and tRNAGluUUC. (A) BY4741 wild type (WT), elp3 or sit4 mutants were each transformed with empty plasmid pRS425 or pQKE (over-expression of tRNAGlnUUG, tRNALysUUU and tRNAGluUUC) and subjected to drop dilution spot assays involving yeast extract peptone (YP) based media with either glucose, galactose or ethanol as the carbon source. All plates were incubated at 30 °C for 3 days. (B) The same strains as in (A) were subjected to drop dilution spot assays on yeast extract peptone dextrose (YPD) medium (carbon source glucose) cultivated at 39 °C or 40 °C or on YPD medium supplemented with a crude zymocin preparation. (C) Microtiter based measurement of growth inhibition by soraphen A. Relative growth refers to optical density (O.D 600 nm) of microcultures with indicated concentrations of soraphen A as compared to soraphen A free cultures. Measurements were carried out in triplicate and presented as mean with standard deviation.
Since the mcm5U deficiency results in non-cleavage of tRNA by zymocin [26,31], elp3 and sit4 mutants displayed resistance against the toxin regardless of whether empty vector or pQKE was present, thereby confirming the shared modification defect of the two mutants. In addition, sit4 and elp3 mutants displayed comparable growth defects at elevated temperature (39 °C and 40 °C) which can be rescued by overexpression of the three tRNAs. In contrast to this, elp3 and sit4 mutants display divergent phenotypes on media with altered carbon sources, which may result from deregulation of Snf1 activity. While sit4 mutants show growth defects on galactose and ethanol media, the elp3 mutant did not (Figure 1A,B). Consistently, these sit4 specific phenotypes are not suppressed by tRNA overexpression, suggesting they are caused by loss of additional Sit4 functions not necessarily linked to the tRNA modification defects. We further investigated whether overexpression of the three tRNAs that rescued temperature sensitive growth of the sit4 mutant also suppressed the previously observed strong soraphen A sensitivity of this mutant [2]. Using a sensitive microtiter based liquid growth assay over a broad range of soraphen A concentrations, we found no difference in the soraphen A sensitivity between pRS425 and pQKE carrying sit4 strains (Figure 1C).
3.2. Role of 5-Methoxycarbonylmethyluridine Modification in Soraphen A Resistance and Lipid Droplet Dynamics
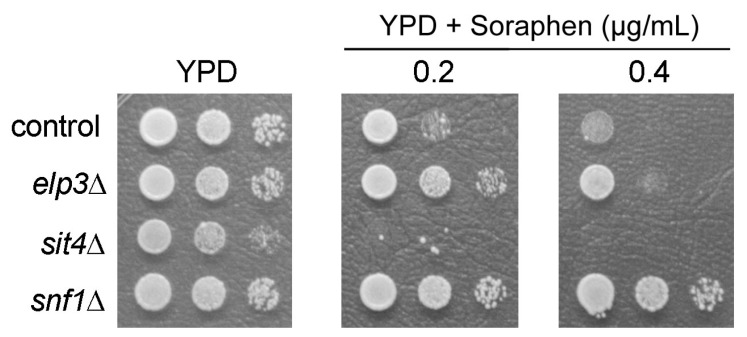
Since the tRNA overexpression results suggest that only part of the phenotypes of sit4 mutants are caused by the defect in tRNA modification we further investigated a possible role of the modification in regulation of soraphen A resistance and LD dynamics, which are known to be strongly affected in sit4 mutants [2]. We reasoned that a relevant contribution of the tRNA modification to these phenotypes should result in similar phenotypic changes in an elp3 mutant. However, when we analyzed the soraphen A response of elp3 mutants an opposite phenotype compared to that of sit4 was detected (Figure 2).
Figure 2.
Soraphen A phenotype of elp3 mutants. Saccharomyces cerevisiae strains BY4741 (control) and its isogenic elp3, sit4 and snf1 mutants were grown to stationary phase and seeded at a concentration of 107, 106 or 105 cells/mL (from left to right) onto YPD medium plates containing the indicated soraphen A doses. Growth was recorded after three days. A representative result of three independent experiments is shown.
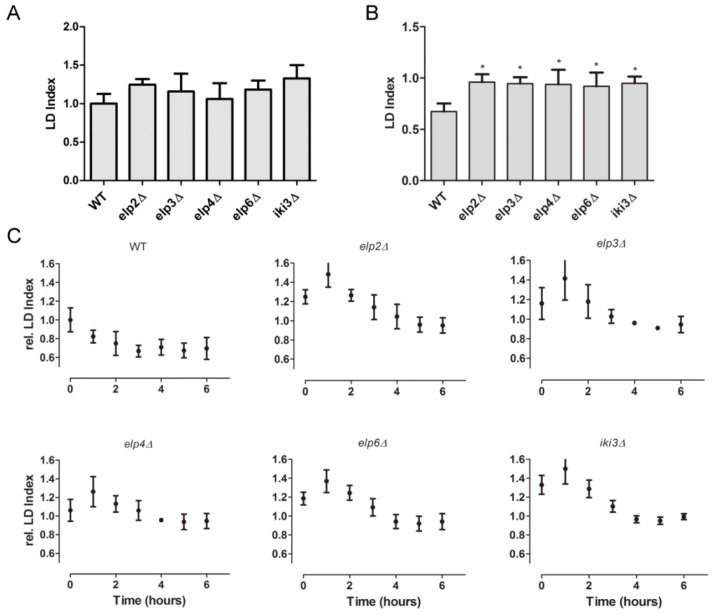
This result and the absence of rescue effects by pQKE on the soraphen A trait of sit4 (Figure 1C) suggest that this phenotype results from defects unrelated to the U34 modifications. However, the resistance of the elp3 mutant also suggested a possible role of the modification in regulation of the soraphen A target Acc1 and potentially, in lipid biogenesis or dynamics in general. Soraphen A resistance of the elp3 mutant was comparable to the snf1 mutant (Figure 2), lacking negative regulation of Acc1 activity by phosphorylation [2], potentially indicating a relevant effect of the Elongator dependent tRNA modification on lipid metabolism. To further investigate whether Elongator dependent tRNA modification may influence dynamic changes in LD content, we analyzed LD levels in mutants lacking different Elongator subunits in comparison to the wild type. We scored lipolysis after re-inoculation of stationary phase cells, which maintain a high lipid droplet level, into fresh media over a time range of 6 h as previously described [32]. While wild type cells displayed a significant decrease in LD levels already in the first 2 h, all the analyzed Elongator mutants showed a significant delay in lipolysis (Figure 3).
Figure 3.
Lipid droplet (LD) content and dynamic change in Elongator mutants as compared to the wild type (WT). Elongator mutant strains were pre-grown to stationary phase (48 h) to attain a high LD content and then seeded at low density in YPD medium at 30 °C (A) Aliquots were withdrawn at stationary phase (48 h) or (B) after 5h of growth. (C) Lipid droplet dynamics of Elongator mutants was studied during the exponential phase (lipolytic phase) of growth. Stationary cells were seeded at 0.25 O.D 600 nm in YPD. Aliquots were withdrawn at the times indicated and fixed in formaldehyde 3.7%. Afterwards the LD index was measured with the liquid fluorescent recovery (LFR) assay using Bodipy 493/503 as a probe [2] and normalized to WT. The mean ± (standard deviation) S.D. of three independent experiments is shown (* p < 0.05).
It was shown previously that loss of the Snf1 kinase also results in higher than normal LD levels and soraphen A resistance, whereas reg1, sap190 and sit4 mutants exhibit lowered LD levels and soraphen A sensitivity [2]. Thus, in several mutants (snf1, reg1, sap190, sit4), changes in soraphen A resistance go hand-in-hand with reciprocal changes in LD content.
3.3. Screening Other tRNA Modification Mutants for Soraphen A Resistance
Since our data revealed an unexpected role of the Elongator dependent tRNA modifications in regulating LD dynamics and soraphen A resistance, we investigated whether other tRNA modification mutants might display abnormal LD levels, which were identified in a previous screening for changes in soraphen A resistance [33]. We assembled a set of mutants defective in most of the known tRNA modifications (excluding essential ones) and classified their soraphen A response. Based on this screening, we identified deg1 and tum1 as soraphen A resistant (Table 1). While Tum1 is (along with the Elongator complex) involved in synthesis of the mcm5s2U modification [22,34], Deg1 represents a well characterized pseudo-uridine synthase mediating the isomerization of uridines to pseudo-uridine (Ψ) in tRNA at positions 38 and 39 [23]. As reference for soraphen A sensitivity, we included sit4 and sap185 in this table.
Table 1.
Soraphen A responses of tRNA modification and phosphatase mutants. Data were obtained from a soraphen A screening of a deletion library [33].
| Gene | Nomenclature | Description | Sensitivity Score | Response |
|---|---|---|---|---|
| DEG1 | YFL001W | depressed growth rate | +6 | RESISTANT |
| TUM1 | YOR251C | thiouridine modification | +2 | RESISTANT |
| SIT4 | YDL047W | suppressor initiation of transcription | −5 | SENSITIVE |
| SAP185 | YKR028W | Sit4 associated protein | −4 | SENSITIVE |
3.4. Changes in Lipid Droplet Dynamics Upon Loss of Ψ38/39
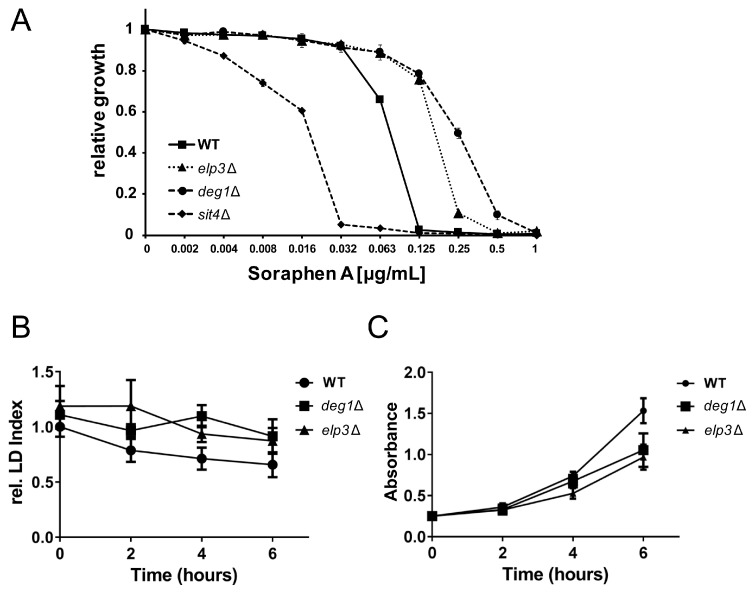
We confirmed the soraphen A resistance of the deg1 mutant by using the liquid growth inhibition assay, which revealed a soraphen A resistance level slightly exceeding the one of elp3 mutants (Figure 4A). Since a positive correlation between soraphen A resistance and LD content was observed in other mutants [2], we analyzed the LD dynamics in the deg1 mutant in comparison to wild type and elp3 cells. As shown in Figure 4B, elp3 and deg1 mutant cells indeed displayed a comparable defect in lipolysis upon re-inoculation from stationary phase to fresh media. Consistent with a slow growth phenotype of both mutants, a delay in resuming growth after refeeding in comparison to the wild type was observed by measuring the optical density concomitantly with LD quantification (Figure 4C).
Figure 4.
Soraphen A resistance and lipolysis defect in deg1 mutants. (A) Microtiter based measurement of growth inhibition of indicated strains by soraphen A. Relative growth refers to optical density (O.D 600 nm) of microcultures with indicated concentrations of soraphen A as compared to soraphen A free cultures. Measurements were carried out in triplicate and shown as mean plus standard deviation. (B) Lipid droplet dynamics and (C) growth of BY4741 (WT) and its isogenic elp3 and deg1 mutants was determined during exponential phase of growth (6 h). The mean ± S.D. of three independent experiments is shown.
3.5. Rescue of Lipolysis Defect of deg1 Mutants by tRNA Overexpression
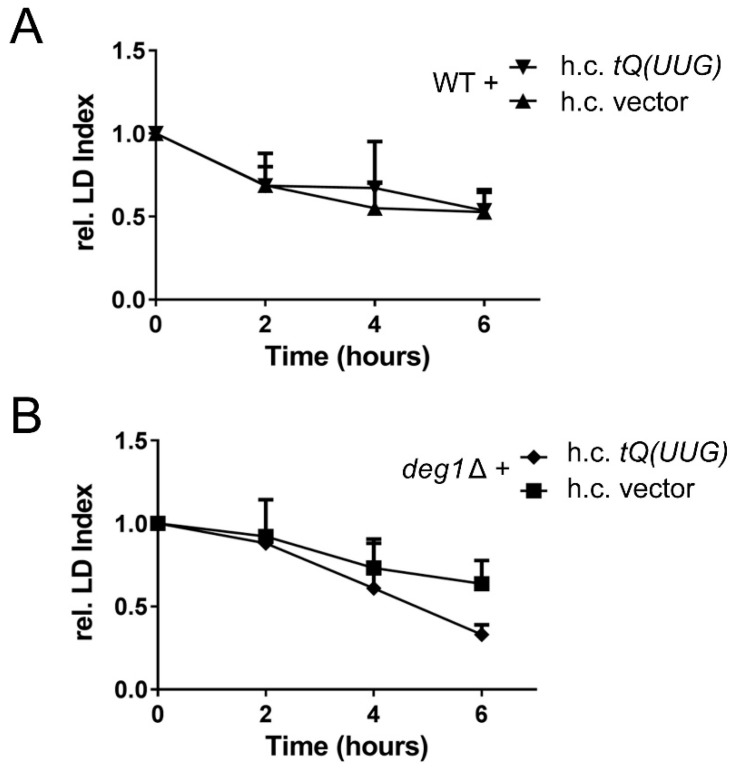
The identification of two different tRNA modifiers (Elongator and Deg1), which impact on soraphen A resistance and LD levels in a similar manner, suggests that translational defects are responsible for the observed LD and soraphen A phenotypes. Translational defects of different tRNA modification mutants can be rescued by overexpression of those tRNAs that become functionally affected in the absence of the modifications [12,18]. For Deg1, previous work identified a single tRNA (tRNAGlnUUG) to be functionally impaired in the absence of the modification Ψ38/39 [25,35]. Hence, we analyzed whether lipolysis defects in deg1 can be ameliorated upon overexpression of this tRNA. As shown in Figure 5, this is indeed the case. No significant difference in LD dynamics was observed in stationary phase wild type cells carrying empty vector or the tRNAGlnUUG overexpression construct upon refeeding (Figure 5A). In both, reduction of LD levels was observed. In contrast, deg1 mutants carrying the tRNA overexpression construct display an increased reduction of LD levels at 4 h and 6 h after refeeding as compared to the empty vector control (Figure 5B). These results support the interpretation that tRNA modification defects influence LD dynamics by translational defects, since the lipolysis defect of deg1 mutants can be ameliorated by overexpression of tRNAGlnUUG.
Figure 5.
Suppression of lipolysis defects in deg1 mutants by overexpression of tRNAGlnUUG. Wild type (WT) (A) and deg1∆ (B) cells were transformed with empty vector (h.c vector) or tRNAGlnUUG multicopy plasmid (h.c.tQ(UUG)). Strains were seeded at low density and the lipid droplet dynamics were assayed during exponential phase (6 h) in YPD medium. The mean ± S.D. of three independent experiments is shown.
4. Discussion
Since deficiency of the protein phosphatase Sit4 induces multiple cellular effects, including the absence of mcm5s2U, mcm5U and ncm5U from tRNAs [22] which in itself already induces pleiotropic phenotypes [12,14,17], we aimed to dissect Sit4 functions relevant for regulating lipid metabolism and associated phenotypes. Our results demonstrate that Snf1 regulatory defects of sit4 mutants [2] dominate over lipid specific effects induced by loss of tRNA modification alone. This conclusion is based on the comparison of soraphen A resistance and dynamic changes in lipid content in Elongator mutants lacking n/mcm5/s2U with previously described effects of the sit4 mutation [2]. sit4 mutants lacking n/mcm5/s2U modification in tandem with Snf1 overactivation display low LD levels and soraphen A sensitivity whereas single loss of the same tRNA modification in elp3 mutants causes opposite effects (Figure 1 and Figure 3). Therefore, the former phenotypes appear to exclusively result from Snf1 overactivation in sit4 without participation of the concomitant tRNA modification defect. In support of this notion, we also show that the soraphen A sensitivity of a sit4 mutant cannot be rescued by tRNA overexpression, unlike other n/mcm5/s2U phenotypes (Figure 1) [12,17]. The same approach, however, also revealed a previously unknown soraphen A resistance and high lipid droplet phenotypes in Elongator mutants. The latter phenotype was specifically apparent when stationary phase cells were refed with fresh medium, which routinely induces fast mobilization of lipids from storage LDs [2,5]. Interestingly, a lipolysis defect was not only detected for mutants resulting in n/mcm5s2U deficiency but was also present in a deg1 mutant lacking an entirely different tRNA modification (Ψ38/39). However, recent work identified mcm5s2U and Ψ38 to be particularly relevant for functioning of tRNAGlnUUG [18,25,35]. Since the lipolysis defect of deg1 mutants was partially rescued by overexpression of tRNAGlnUUG, a specific translational defect of a Gln rich protein could be involved in the observed effects. Upon refeeding, triacylglycerol from lipid droplets is mobilized using Tgl3, Tgl4 and Tgl5 lipases [36]. Hence, a translation dependent defect in lipolysis at the level of refeeding could involve impaired expression of either of these proteins. However, none of these proteins are particularly Gln-rich (Tgl3 3.42% Gln, Tgl4 3.29% Gln, Tgl5, 4.53% Gln). Since the deg1 mutation was shown to impair function of tRNAGlnUUG rather than tRNAGlnCUG [18,25,35], we also compared the relative usage of the two alternative Gln codons (CAA/CAG) in the TGL3-5 genes. Relative CAA codon usage in TGL3 (68.2%) and TGL5 (67.6%) is close to the genome average (68.5%) and slightly increased in TGL4 (73.3%). Hence, TGL4 might be more sensitive to tRNAGlnUUG defects induced by deg1 mutation compared to the other two genes. Alternatively, the delay of elp3 and deg1 mutants to resume exponential growth upon refeeding (Figure 4) may indicate a more global translational defect that causes a reduced ability to remodel-/reactivate gene expression and hence result in a delay of storage lipid consumption. Such delayed entry into exponential phase may contribute to an apparent lipolysis defect but is unlikely the reason for increased resistance against Acc1 inhibition observed in both elp3 and deg1 mutants (Figure 4A). In addition to defects in translation, loss of tRNA modification may also result in widespread transcriptional changes in a concerted cellular response against translational pausing and subsequently induced protein homeostasis defects [20,37,38,39]. Whether or not this includes transcriptional induction of genes relevant for lipogenesis and/or repression of genes required for lipolysis, remains to be determined. In general, a delay of storage lipid content reduction upon refeeding might also result from upregulated de novo lipid biosynthesis. If this would imply upregulation of Acc1 activity in tRNA modification mutants, soraphen A resistance could be expected. Of note, both elp3 and deg1 deletions were found to induce widespread metabolic changes [40,41], which may also include complex alterations ultimately leading to decreased lipolysis or increased lipogenesis as cells enter from stationary to new exponential growth phase. In addition, loss of the Tum1 enzyme required for thiolation of tRNA was recently shown to affect lipid metabolism [42], providing additional support for a general functional link between tRNA modification and lipid homeostasis.
5. Conclusions
The multifunctional protein phosphatase Sit4 is required for tRNA modification and modification independent regulatory processes in S. cerevisiae. Absence of Sit4 lowers cellular LD levels and results in increased sensitivity to acetyl-CoA carboxylase (Acc1) inhibition. It remained unknown whether these effects are due to loss of tRNA modification or due to modification independent functions of Sit4. We demonstrate in here that the effects on LD levels and susceptibility to Acc1 inhibition are caused by the tRNA modification independent effects of a SIT4 gene deletion. However, absence of two distinct tRNA modifications results in defective lipolysis causing LD accumulation and resistance against Acc1 inhibition, which represent opposite effects of sit4 deletion. Since the lipolysis defect in a tRNA modification mutant can be ameliorated upon overexpression of the tRNA affected by loss of the modification, translational inefficiency is assumed to induce these effects.
Acknowledgments
We greatly acknowledge generous gift of soraphen A from Rolf Müller (Helmoltz-Zentrum für Infektionsforschung, Saarbrücken, Germany).
Author Contributions
Conceptualization, B.L.B.-M., R.K., R.S. and M.M.-L.; Investigation, B.L.B.-M., L.V., V.D., M.M.-L. and R.K.; Writing-Original Draft Preparation, R.K., R.S. and M.M.-L.; Writing-Review & Editing, B.L.B.-M., L.V., V.D., R.S., R.K. and M.M.-L.; Funding Acquisition, R.S., R.K., B.L.B.-M. and M.M.-L.
Funding
A cooperation initiation grant between Deutsche Forschungsgemeinschaft (DFG) to RS (SCHA750/18 and SCHA750/19) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) to MML is greatly acknowledged. In addition, the work was supported by grants from the Fundação de Amparo a Pesquisa do Rio de Janeiro (FAPERJ-Cientistas do Nosso Estado) to MML, a postdoctoral CAPES-PNPD Institutional fellowship to BLB, joint DFG SPP1784 funding to RS (SCHA750/20-2) and RK (KL2937/1-2) and the research consortium PhosMOrg of University of Kassel.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hasslacher M., Ivessa A.S., Paltauf F., Kohlwein S.D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 2.Bozaquel-Morais B.L., Madeira J.B., Maya-Monteiro C.M., Masuda C.A., Montero-Lomeli M. A new fluorescence-based method identifies protein phosphatases regulating lipid droplet metabolism. PLoS ONE. 2010;5:e13692. doi: 10.1371/journal.pone.0013692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods A., Munday M.R., Scott J., Yang X., Carlson M., Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- 4.Conrad M., Schothorst J., Kankipati H.N., van Zeebroeck G., Rubio-Texeira M., Thevelein J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madeira J.B., Masuda C.A., Maya-Monteiro C.M., Matos G.S., Montero-Lomelí M., Bozaquel-Morais B.L. TORC1 inhibition induces lipid droplet replenishment in yeast. Mol. Cell. Biol. 2015;35:737–746. doi: 10.1128/MCB.01314-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jablonowski D., Fichtner L., Stark M.J.R., Schaffrath R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell. 2004;15:1459–1469. doi: 10.1091/mbc.e03-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jablonowski D., Täubert J.-E., Bär C., Stark M.J.R., Schaffrath R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryot. Cell. 2009;8:1637–1647. doi: 10.1128/EC.00205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehlgarten C., Jablonowski D., Breunig K.D., Stark M.J.R., Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- 9.Schaffrath R., Leidel S.A. Wobble uridine modifications–a reason to live, a reason to die?! RNA Biol. 2017;14:1209–1222. doi: 10.1080/15476286.2017.1295204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauden M.I., Kosinski J., Kolaj-Robin O., Desfosses A., Ori A., Faux C., Hoffmann N.A., Onuma O.F., Breunig K.D., Beck M., et al. Architecture of the yeast Elongator complex. EMBO Rep. 2017;18:264–279. doi: 10.15252/embr.201643353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setiaputra D.T., Cheng D.T., Lu S., Hansen J.M., Dalwadi U., Lam C.H., To J.L., Dong M.-Q., Yip C.K. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017;18:280–291. doi: 10.15252/embr.201642548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esberg A., Huang B., Johansson M.J.O., Byström A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Huang B., Johansson M.J.O., Byström A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frohloff F., Fichtner L., Jablonowski D., Breunig K.D., Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leidel S., Pedrioli P.G.A., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 16.Scheidt V., Jüdes A., Bär C., Klassen R., Schaffrath R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell. 2014;1:416–424. doi: 10.15698/mic2014.12.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klassen R., Grunewald P., Thüring K.L., Eichler C., Helm M., Schaffrath R. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS ONE. 2015;10:e0119261. doi: 10.1371/journal.pone.0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klassen R., Ciftci A., Funk J., Bruch A., Butter F., Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016;44:10946–10959. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinshteyn B., Gilbert W.V. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet. 2013;9:e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedialkova D.D., Leidel S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjan N., Rodnina M.V. Thio-Modification of tRNA at the Wobble Position as Regulator of the Kinetics of Decoding and Translocation on the Ribosome. J. Am. Chem. Soc. 2017;139:5857–5864. doi: 10.1021/jacs.7b00727. [DOI] [PubMed] [Google Scholar]
- 22.Huang B., Lu J., Byström A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecointe F., Simos G., Sauer A., Hurt E.C., Motorin Y., Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of Ψ38 and Ψ39 in tRNA anticodon loop. J. Biol. Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 24.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 25.Klassen R., Schaffrath R. Role of Pseudouridine Formation by Deg1 for Functionality of Two Glutamine Isoacceptor tRNAs. Biomolecules. 2017;7:8. doi: 10.3390/biom7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J., Huang B., Esberg A., Johansson M.J.O., Byström A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klassen R., Krampe S., Meinhardt F. Homologous recombination and the yKu70/80 complex exert opposite roles in resistance against the killer toxin from Pichia acaciae. DNA Repair. 2007;6:1864–1875. doi: 10.1016/j.dnarep.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Klassen R., Wemhoff S., Krause J., Meinhardt F. DNA repair defects sensitize cells to anticodon nuclease yeast killer toxins. Mol. Genet. Genom. MGG. 2011;285:185–195. doi: 10.1007/s00438-010-0597-5. [DOI] [PubMed] [Google Scholar]
- 29.Romero-Aguilar L., Montero-Lomeli M., Pardo J.P., Guerra-Sánchez G. Lipid Index Determination by Liquid Fluorescence Recovery in the Fungal Pathogen Ustilago Maydis. J. Vis. Exp. JoVE. 2018;134 doi: 10.3791/57279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler A.R., White J.H., Folawiyo Y., Edlin A., Gardiner D., Stark M.J. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 1994;14:6306–6316. doi: 10.1128/MCB.14.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator’s key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- 32.Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S.D. Obese yeast: Triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 33.Bozaquel-Morais B.L., Madeira J.B., Venâncio T.M., Pacheco-Rosa T., Masuda C.A., Montero-Lomeli M. A Chemogenomic Screen Reveals Novel Snf1p/AMPK Independent Regulators of Acetyl-CoA Carboxylase. PLoS ONE. 2017;12:e0169682. doi: 10.1371/journal.pone.0169682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jüdes A., Bruch A., Klassen R., Helm M., Schaffrath R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb. Cell. 2016;3:554–564. doi: 10.15698/mic2016.11.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han L., Kon Y., Phizicky E.M. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA. 2015;21:188–201. doi: 10.1261/rna.048173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajakumari S., Daum G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2010;285:15769–15776. doi: 10.1074/jbc.M109.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiaville P.C., Legendre R., Rojas-Benítez D., Baudin-Baillieu A., Hatin I., Chalancon G., Glavic A., Namy O., de Crécy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell. 2016;3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou H.-J., Donnard E., Gustafsson H.T., Garber M., Rando O.J. Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol. Cell. 2017;68:978–992.e4. doi: 10.1016/j.molcel.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokołowski M., Klassen R., Bruch A., Schaffrath R., Glatt S. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta. 2018;4:409–418. doi: 10.1016/j.bbagrm.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Karlsborn T., Mahmud A.K.M.F., Tükenmez H., Byström A.S. Loss of ncm5 and mcm5 wobble uridine side chains results in an altered metabolic profile. Metabol. Off. J. Metabol. Soc. 2016;12:177. doi: 10.1007/s11306-016-1120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mülleder M., Calvani E., Alam M.T., Wang R.K., Eckerstorfer F., Zelezniak A., Ralser M. Functional Metabolomics Describes the Yeast Biosynthetic Regulome. Cell. 2016;167:553–565.e12. doi: 10.1016/j.cell.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uršič K., Ogrizović M., Kordiš D., Natter K., Petrovič U. Tum1 is involved in the metabolism of sterol esters in Saccharomyces cerevisiae. BMC Microbiol. 2017;17:181. doi: 10.1186/s12866-017-1088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]