SHP2 (PTPN11) is a protein tyrosine phosphatase (PTP) containing two Src homology-2 domains (N-SH2, C-SH2), a PTP domain, and a carboxyl-terminal region.1 SHP2 is auto-inhibited by the N-SH2 domain and PTP domain interaction in the basal state. In cancer cells, SHP2 is activated by oncogenic protein tyrosine kinases (PTKs) or gain-of-function (GOF) mutations. GOF SHP2 mutations also link to Noonan Syndrome (NS).2
SHP2 plays important roles in tumor growth3, 4, in oncogenesis driven by PTK oncogenes such as FLT3-ITD in acute myeloid leukemia (AML)5, and in immune checkpoints mediated by PD-1 or BTLA.6 GOF SHP2 mutations are found in leukemias, particularly juvenile myelomonocytic leukemia (JMML), and in solid tumors.1
SHP2 has been targeted for cancer drug discovery.7, 8 However, it is challenging to develop potent, selective, and cell-permeable SHP2 inhibitors targeting the PTP catalytic site. Recently, a novel allosteric SHP2 inhibitor (SHP099) was identified.9, 10 SHP099 binds to a tunnel-like area comprised of all three domains in SHP2 (Fig. 1a), and stabilizes it in an inactive conformation. Several leukemia cell lines expressing PTK driver oncogenes, such as FLT3-ITD and PDGFRA, were the most sensitive to SHP099 (IC50 <10 μM).10 However, it was unclear whether SHP099 inhibits SHP2 mutants.
Figure 1.
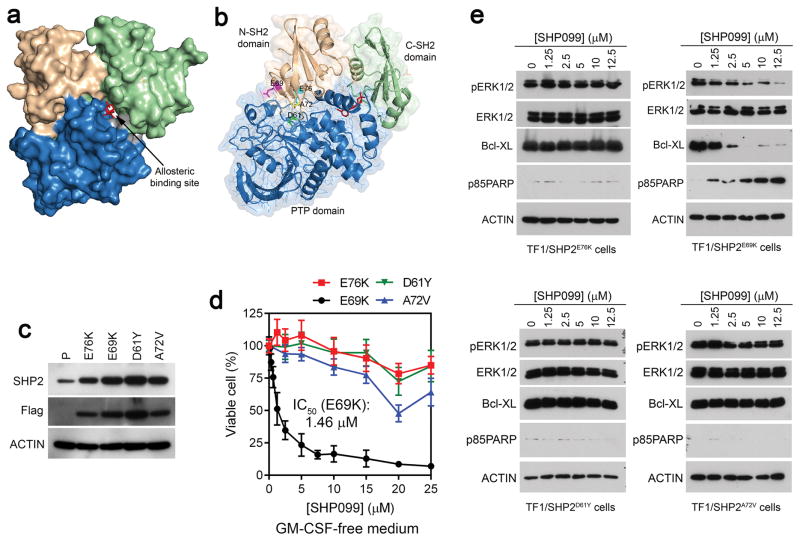
Effects of SHP099 in SHP2 mutants-dependent TF-1 cell lines. (a, b) Co-crystal structure of SHP2-SHP099 (PDB: 5EHR). N-SH2 domain (wheat), C-SH2 domain (green), PTP domain (blue), and residues D61 (green), E69 (magentas), A72 (yellow), and E76 (cyan) are colored. SHP099 is shown in red. (c) Analysis of SHP2 expression in the parental TF-1 cells and stable TF-1 cell lines expressing 4 SHP2 mutants by immunoblotting. The anti-SHP2 antibody reacts with both the endogenous wildtype and the exogenous SHP2 mutants. The anti-Flag antibody recognizes the exogenous Flag-tagged SHP2 mutants. (d) Growth inhibiting activity of SHP099 in the four SHP2 mutant cell lines. Cells were cultured in GM-CSF free medium with indicated concentrations of SHP099 and viable cells were measured on Day 5. (e) Immunoblot analysis of cell lysates after 2 days of SHP099 treatment.
We synthesized SHP099 and tested it in the MV4-11 myeloid leukemia cells.10 SHP099 potently inhibited MV4-11 cell growth (IC50: 0.32 ± 0.11 μM; Supplementary Figure 1a), reduced pERK1/2 and Bcl-XL levels, and induced PARP cleavage (Supplementary Figure 1b). Thus, SHP099 synthesized by us had activity similar to that reported.10
SHP2E76K transformed the GM-CSF-dependent TF-1 myeloid cells into GM-CSF-independent cells, suggesting that SHP2 plays a major role in regulating the survival and proliferation of TF-1 cells.11 SHP099 inhibited TF-1 cell growth with an IC50 of 1.73 ± 0.52 μM (Supplementary Fig. 1c) and inhibited active ERK, reduced Bcl-XL, and induced PARP cleavage in these cells (Supplementary Figure 1b). Thus, TF-1 cells belong to a group of SHP099-sensitive cells that have SHP099 IC50 <10 μM.10 SHP099 had little effects in SHP2-knocked out TF-1 cell lines (Supplementary Figure 1b–d), demonstrating the specificity of SHP099.
Cancer-associated SHP2 mutations are located mostly in the N-SHP2-PTP domain interface (Supplementary Figure 2). Examination of the COSMIC database (http://cancer.sanger.ac.uk/cosmic) indicated that E76, A72, D61/G60, and E69 are the most frequently mutated amino acid residues in the N-SH2 domain (Supplementary Fig. 2). Therefore, we selected p.D61Y, p.E69K, p.A72V, and p.E76K (Supplementary Figure 2) to determine whether SHP099 is effective in inhibiting these common cancer-associated SHP2 mutations.
We previously established SHP2E76K-expressing TF-1 cells (TF1/SHP2E76K).11 SHP2E76K transformed the GM-CSF-dependent TF1 cells into GM-CSF-independence, a property resembling that of leukemic cells from patients with JMML.12 The SHP2E76K-induced cytokine-independent TF-1 cell survival is mediated by ERK1/2 and its downstream effector Bcl-XL.11 Here, we generated SHP2D61Y, SHP2E69K, and SHP2A72V mutations and established stable TF-1 cells expressing these SHP2 mutants (Figure 1c). Like TF1/SHP2E76K cells, TF1/SHP2D61Y, TF1/SHP2E69K, and TF1/SHP2A72V cells were able to survive and grow in RPMI-1640/10% FBS medium without supplementation with human GM-CSF.
These cells were then cultured in GM-CSF-free medium and treated with increasing concentrations of SHP099 (0–25 μM). Relative viable cells were measured after 5 days. SHP099 showed marginal effects on TF1/SHP2E76K, TF1/SHP2D61Y, and TF1/SHP2A72V cells (Figure 1d). Significantly, SHP099 potently inhibited TF1/SHP2E69K cell growth with an IC50 of 1.46 ± 0.46 μM (Figure 1d). Cell lysate analyses indicated that SHP099 had little effect on pERK1/2 or Bcl-XL levels in TF1/SHP2D61Y, TF1/SHP2A72V, and TF1/SHP2E76K cells. SHP099 also did not induce PARP cleavage in these cells. In contrast, SHP099 reduced pERK1/2 and Bcl-XL in TF1/SHP2E69K cells, with the corresponding induction of PARP cleavage (Figure 1e). These results showed that TF1/SHP2D61Y, TF1/SHP2A72V, and TF1/SHP2E76K cells were resistant to SHP099, whereas TF1/SHP2E69K cells underwent apoptosis in response to SHP099, suggesting that SHP2E69K is sensitive to SHP099.
In another experiment, we compared sensitivities of SHP099 in methylcellulose cultures11 of TF1/SHP2E76K and TF1/SHP2E69K cells. SHP099 had little effect on cytokine-independent TF1/SHP2E76K colonies and did not affect their GM-CSF hypersensitivity. In comparison, cytokine-independent colonies were eliminated and the GM-CSF hypersensitivity was lost when methylcellulose cultures of TF1/SHP2E69K were performed in the presence of SHP099 (Supplementary Figure 4).
We next compared inhibition of the wildtype and mutant SHP2 by SHP099 in vitro using recombinant proteins. It was reported that SHP099 displayed the highest potency on slightly activated SHP2 by exposure to low concentrations of an activating peptide analog 2P-IRS-1.10 We reasoned that oncogenic SHP2 mutants are partly activated. Therefore, we performed the in vitro inhibition experiments both in the absence of an activating phosphopeptide and in the presence of 1 μM PY627PY659 bisphosphopeptide derived from GAB1.13
SHP099 gave an IC50 of 0.69 ± 0.25 μM on the wildtype SHP2 in the absence of the activating phosphopeptide. This was lowered to 0.13 ± 0.04 μM in the presence of 1 μM PY627PY659 (Table 1). SHP099 displayed low micromolar IC50s on SHP2G61Y, SHP2A72V, and SHP2E76K in either the absence or the presence of PY627PY659 (Table 1). Interestingly, SHP099 showed a sub-micromolar IC50 of 0.42 ± 0.09 μM on SHP2E69K. In the presence of 1 μM PY627PY659, the IC50 of SHP099 against SHP2E69K increased to 1.1 ± 0.15 μM. However, this was 2–5 fold more potent than inhibition of SHP2G61Y, SHP2A72V, or SHP2E76K (Table 1). These data showed that, among the four common SHP2 N-SH2 domain mutations, SHP2E69K was selectively sensitive to SHP099 inhibition.
Table 1.
Inhibitory activities of SHP099 on the wildtype and mutant SHP2 in vitro
| Phosphatase | IC50 (μM)
|
|
|---|---|---|
| Without activating phosphopeptide | With activating phosphopeptide | |
|
| ||
| SHP2 | 0.690 ± 0.247 (n=3) | 0.128 ± 0.043 (n=3) |
| SHP2D61Y | 1.241 ± 0.342 (n=6) | 2.087 ± 0.345 (n=2) |
| SHP2E69K | 0.416 ± 0.093 (n=3) | 1.097 ± 0.148 (n=4) |
| SHP2A72V | 1.968 ± 0.483 (n=6) | 5.451 ± 1.639 (n=3) |
| SHP2E76K | 2.896 ± 0.810 (n=4) | 5.983 ± 0.437 (n=2) |
Inspection of the SHP2 crystal structure reveals a difference between the E69 residue and the D61/A72/E76 residues (Fig. 1b, Supplementary Figure 3), The E69 residue is located on the edge of the N-SH2 domain-PTP domain interface distal to the SHP099 binding site. The sidechain carboxylate of E69 is oriented toward the surface and interacts with the sidechain of K280 of the PTP domain, which also protrudes to the outer surface. In contrast, the D61, A72, and E76 residues are located in the middle of the N-SH2 domain-PTP domain interface and are closer to the SHP099 binding site. The p.E69K mutation, because of its localization on the distal edge of the N-SH2 domain-PTP domain interface, may cause less disturbance to the SHP099 binding tunnel. This may explain why SHP2E69K is sensitive to SHP099, while the other three mutants are not.
The allosteric SHP2 inhibitor SHP099 represents a breakthrough in the PTP inhibitor field.14 It had shown in vivo efficacy in inhibiting tumors expressing the wildtype SHP2,10 and may be used to block the PD-1 immune checkpoint in T cells.6 However, whether SHP099 is effective in inhibiting oncogenic SHP2 mutants is an important unanswered question. Our findings here identify SHP2E69K as a SHP099-sensitive mutation. Three of four most common oncogenic SHP2 mutants studied here are resistant to SHP099. These findings point to a need to further develop SHP099 analogs and identify other allosteric SHP2 inhibitors. For instance, an allosteric inhibitor (Compound-23) specific to the less common SHP2E76A was reported recently.15 Moreover, whether NS-associated SHP2 mutants are sensitive to SHP099 remained to be tested. Encouragingly, the most prevalent SHP2 mutant found in NS, SHP2N308D, is not located in the N-SHP2-PTP domain interface.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health (NIH) grant R01CA17456 (to J. Wu), and by the Chemical Biology Core facilities at the H. Lee Moffitt Cancer Center supported by NIH grant P30CA076292, and the shared resources at the University of Oklahoma Health Sciences Center supported by NIH grant P20GM103639.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 2008;27:179–192. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- 2.Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet. 2006;15(Spec No 2):R220–226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- 3.Ren Y, Chen Z, Chen L, Fang B, Win-Piazza H, Haura E, et al. Critical role of Shp2 in tumor growth involving regulation of c-Myc. Genes Cancer. 2010;1:994–1007. doi: 10.1177/1947601910395582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneeberger VE, Ren Y, Luetteke N, Huang Q, Chen L, Lawrence HR, et al. Inhibition of Shp2 suppresses mutant EGFR-induced lung tumors in transgenic mouse model of lung adenocarcinoma. Oncotarget. 2015;6:6191–6202. doi: 10.18632/oncotarget.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nabinger SC, Li XJ, Ramdas B, He Y, Zhang X, Zeng L, et al. The protein tyrosine phosphatase, Shp2, positively contributes to FLT3-ITD-induced hematopoietic progenitor hyperproliferation and malignant disease in vivo. Leukemia. 2013;27:398–408. doi: 10.1038/leu.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott LM, Lawrence HR, Sebti SM, Lawrence NJ, Wu J. Targeting protein tyrosine phosphatases for anticancer drug discovery. Curr Pharm Des. 2010;16:1843–1862. doi: 10.2174/138161210791209027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He RJ, Yu ZH, Zhang RY, Zhang ZY. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin. 2014;35:1227–1246. doi: 10.1038/aps.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Fortanet J, Chen CH, Chen YN, Chen Z, Deng Z, Firestone B, et al. Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. J Med Chem. 2016;59:7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 10.Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 11.Ren Y, Chen Z, Chen L, Woods NT, Reuther GW, Cheng JQ, et al. Shp2E76K mutant confers cytokine-independent survival of TF-1 myeloid cells by up-regulating Bcl-XL. J Biol Chem. 2007;282:36463–36473. doi: 10.1074/jbc.M705789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emanuel PD, Shannon KM, Castleberry RP. Juvenile myelomonocytic leukemia: molecular understanding and prospects for therapy. Mol Med Today. 1996;2:468–475. doi: 10.1016/1357-4310(96)10044-7. [DOI] [PubMed] [Google Scholar]
- 13.Cunnick JM, Mei L, Doupnik CA, Wu J. Phosphotyrosines 627 and 659 of Gab1 constitute a bisphosphoryl tyrosine-based activation motif (BTAM) conferring binding and activation of SHP2. J Biol Chem. 2001;276:24380–24387. doi: 10.1074/jbc.M010275200. [DOI] [PubMed] [Google Scholar]
- 14.Salamoun JM, Wipf P. Allosteric Modulation of Phosphatase Activity May Redefine Therapeutic Value. J Med Chem. 2016;59:7771–7772. doi: 10.1021/acs.jmedchem.6b01210. [DOI] [PubMed] [Google Scholar]
- 15.Xie J, Si X, Gu S, Wang M, Shen J, Li H, et al. Allosteric inhibitors of SHP2 with therapeutic potential for cancer treatment. J Med Chem. 2017 doi: 10.1021/acs.jmedchem.7b01520. epub: Nov 20, 2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.