Abstract
Intrinsic and/or acquired resistance represents one of the great challenges in targeted cancer therapy. A deeper understanding of the molecular biology of cancer has resulted in more efficient strategies, where one or multiple drugs are adopted in novel therapies to tackle resistance. This beneficial effect of using combination treatments has also been observed in colorectal cancer patients harboring the BRAF(V600E) mutation, whereby dual inhibition of BRAF(V600E) and EGFR increases antitumor activity. Notwithstanding this success, it is not clear whether this combination treatment is the only or most effective treatment to block intrinsic resistance to BRAF inhibitors. Here, we investigate molecular responses upon single and multi-target treatments, over time, using BRAF(V600E) mutant colorectal cancer cells as a model system. Through integration of transcriptomic, proteomic and phosphoproteomics data we obtain a comprehensive overview, revealing both known and novel responses. We primarily observe widespread up-regulation of receptor tyrosine kinases and metabolic pathways upon BRAF inhibition. These findings point to mechanisms by which the drug-treated cells switch energy sources and enter a quiescent-like state as a defensive response, while additionally compensating for the MAPK pathway inhibition.
Keywords: Colorectal cancer, Drug resistance, Systems biology, Signaling Circuits, Transcription, BRAF(V600E), EGFR, integrative analysis, multi-omics, PTPN11
Despite the development of novel drugs for personalized medicine, both intrinsic and acquired resistance remain major limitations of targeted anticancer therapies (1, 2). Most of these drugs target components of the mitogen-activated protein kinase (MAPK)1 signaling pathway, which contains oncogenes such as KRAS, BRAF and the epidermal growth factor receptor (EGFR) (3, 4). The use of monotherapy to inhibit these oncogenes has often been found to be ineffective due to reactivation of signaling pathways. For instance, upregulation of upstream components such as receptor tyrosine kinases (RTKs) in KRAS mutant lung and colorectal cancer (CRC) or of downstream components such as KRAS wild-type in CRC have been revealed to be responsible for intrinsic drug resistance (5, 6).
To overcome intrinsic and/or acquired resistance, combined drug treatments are frequently replacing single-agent targeted therapies (7–9). An elegant example of bypassing intrinsic resistance using a multi-target approach has been demonstrated in BRAF(V600E) mutant CRC (10). Whereas BRAF inhibitor (BRAFi) monotherapy is highly effective in BRAF(V600E) mutant melanoma, response rates in BRAF(V600E) mutant CRC are poor (11, 12). Multiple independent studies on CRC found a crucial role of EGFR as a key driver of resistance to BRAFi monotherapy (13–15). In congruence with the role of EGFR in conferring resistance to BRAFi, the suppression of tyrosine phosphatase nonreceptor type 11 (PTPN11)—which is required to transduce signals from EGFR and other RTKs to the downstream MAPK pathway—also sensitizes BRAF(V600E) CRC cells to BRAF inhibition (16). Consequently, the identification of EGFR as a mediator of intrinsic resistance to BRAFi in CRC has led to initiation of several clinical trials which combine inhibition of both EGFR and BRAF (BRAFi+EGFRi), or of other MAPK pathway members (10, 17).
Although the BRAFi+EGFRi combination treatment is more effective than BRAFi monotherapy in CRC (18), it remains unclear whether EGFR is the only or most potent synthetic lethal co-target of BRAF(V600E) in CRC. Addressing this issue requires an understanding of the cellular response to drug treatment across different molecular levels. Such multilayer approaches could elucidate different branches of the signaling network and track how perturbations propagate to gene and protein expression in driving resistance. Several studies have already highlighted the widespread responses to drug treatment in cancer using multi-omics approaches and adequate data integration (19–21). Advances in next-generation sequencing and proteomics approaches (22, 23) in combination with enhanced data integration solutions have paved the way for such important investigations (24, 25). Notably, the integrated use of transcriptomics and (phospho)proteomics has recently demonstrated its power in describing physiopathological processes through phenotype and proteotype analysis (19, 26–28).
In this study we analyze and integrate proteomics, phosphoproteomics, and transcriptomics data to track molecular responses over time upon perturbation with BRAFi, EGFRi, or their combination in CRC cell lines. We aim to study whether there are other post-translational or transcriptional mechanisms—in addition to EGFR and the MAPK pathway—that are activated upon treatment, to identify novel targets that may overcome innate resistance and prevent acquired resistance to BRAF inhibition.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
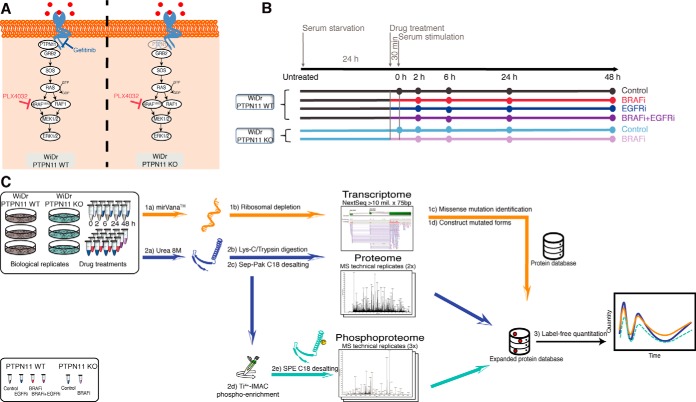
Colorectal tumor cell lines WiDr and WiDr PTPN11 KO were used as model to study resistance upon drug treatment. Both WiDr and WiDr PTPN11 KO cells were plated in 15-cm dishes for (phospho)proteomics and 10-cm dishes for transcriptomics. All cells were cultured in RPMI supplemented with 10% fetal calf serum (FCS) 1% l-Glutamine and 1% Penicillin/Streptomycin. Cells were grown to around 70–80% confluence and starved for 24 h in serum-free media, after which all plates were supplemented with serum-free media containing either no drugs (WiDr Control, WiDr PTPN11 KO control) or 3 μm PLX4032 (BRAFi, BRAFi in PTPN11 KO) or 3 μm gefitinib (EGFRi) or the combination of 3 μm PLX4032 and 3 μm gefitinib (BRAFi+EGFRi). Following 30 min incubation, both Controls (n = 24) and treated cells (n = 48) were stimulated by 10% FCS and then collected at four-time points: 2, 6, 24, and 48 h. Controls at T = 0 for both WiDr and WiDr PTPN11 KO were instead immediately harvested without any stimulation (n = 6). To increase the statistical power, the whole experiment was executed in three biological replicates for both (phospho)proteomics (n = 78) and transcriptomics (n = 78). At least two and three technical replicates were processed for proteomics (168 RAW spectra) and phosphoproteomics (243 RAW) respectively. No technical replicates were processed for the transcriptomics experiment. The reported driver mutations (BRAF(V600E), PIK3CA P449T, TP53 R273H) (29) were verified on 24 pooled samples and the ploidy of the chromosomes checked against previous characterizations of WiDr/HT-29 (30, 31) (supplemental Fig. S14). Single nucleotide variants were called by GATK v3.4–46 (32), and copy number was determined using CONTROL-FREEC v10.4 (33).
Cell Lysis
After each treatment, both WiDr and WiDr PTPN11 KO cells were harvested by washing twice with cold PBS and then resuspended in ice cold lysis buffer. For (phospho)proteomics analysis, protein extraction was obtained by adding 2 ml of buffer containing 8 m urea, 50 mm ammonium bicarbonate (pH 8.0), 1 mm sodium orthovanadate, complete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN), and phosSTOP phosphatase inhibitor mixture (Roche). Finally, cells were snap frozen in 15 ml Falcon Centrifuge Tubes and stored at −80 °C until use. For transcriptomics analysis, total RNA was extracted by adding 600 μl Lysis/Binding Buffer from the mirVana miRNA Isolation Kit by Ambion (Cat. AM1560), and then collecting cells by scraping. Next, cell lysate was transferred into a 1.5 ml tube and stored at −80 °C until time of total RNA isolation which was performed following the mirVana total RNA workflow.
Proteomics Analyses
Lysed cells were defrosted, and each tube was supplemented with 300 μl of fresh lysis buffer. Cells were further lysed by 10 rapid passages through 23G needle and by sonication on ice. Cell debris were removed by centrifugation at 20,000 × g for 30 min at 4 °C and cleared supernatants were stored at −80 °C. The total protein concentration was measured using Bradford assay (Bio-Rad, Hercules, CA).
Next, samples were split into 200 μg aliquots for quality control analysis via western blots and 2 mg aliquots for tryptic digestion. Proteins were reduced with 8 mm DTT (Sigma-Aldrich, Germany) for 1 h at room temperature, alkylated with 16 mm IAA (Sigma-Aldrich) for 30 min at room temperature in the dark and reduced again with 8 mm DTT at room temperature to prevent overalkylation. Later, proteins were first digested by Lys-C (Wako Chemicals, Virginia) (enzyme/substrate ratio 1:65) at 37 °C for 4 h. Subsequently, urea was diluted to 2 m with 50 mm ammonium bicarbonate and trypsin (Sigma-Aldrich) was added (enzyme/substrate ratio 1:50). The digestion was executed at 37 °C overnight and then quenched with 5% formic acid. Peptides were desalted using Sep-Pak C18 cartridges (Waters), dried and stored at −80 °C.
Phosphopeptide Enrichment
Ti4+-IMAC material was prepared as previously described (34). Briefly, the affinity material was loaded onto GELoader tips (Eppendorf, California) using a C8 plug. The columns were pre-equilibrated two times with 50 μl of Ti4+-IMAC loading buffer (80% ACN, 6% trifluoroacetic acid (TFA)). Next, samples were resuspended in loading buffer and 200 μg were loaded into each microcolumn. Columns were sequentially washed with 50 μl wash buffer A (50% ACN, 0.5% TFA, 200 mm NaCl) and 50 μl wash buffer B (50% ACN, 0.1% TFA). Bound peptides were first eluted by 30 μl of 10% ammonia into 30 μl of 10% FA. Finally, all remaining peptides were eluted with 2 μl of 80% ACN, 2% FA. The collected eluate was further acidified by adding 3 μl of 100% FA, and subsequently dried in vacuo and stored at −80 °C. The procedure was repeated in three technical replicates for each biological replicate. Later, phosphopeptides were further desalted using SPE C18 cartridge homemade. Stationary phase C18 beads were dissolved in 500 μl isopropanol and loaded onto GELoader tips using a C18 plug as previously described. The columns were washed with 50 μl wash buffer C (80% ACN, 0.1% TFA) and then conditioned with 0.1% TFA. Next, samples were resuspended in 30 μl of 10% TFA and loaded into columns which were further washed with 30 μl of 10% TFA. Finally, phosphopeptides were eluted with 30 μl of 80% ACN, 1% FA, dried and stored at −80 °C.
(Phospho)proteomics Mass Spectrometry
(Phospho)peptides were analyzed using an Agilent 1290 Infinity II LC system coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). The LC system was equipped with a 2 cm Aqua C18 (Phenomenex, Dr. Maisch GmbH, Germany) trapping column (packed in-house, i.d. 100 μm, resin 5 μm) and a 50 cm Poroshell 120 EC-C18 (Agilent Technologies, Netherlands) analytical column (packed in-house, i.d. 50 μm; resin 3 μm). (Phospho)peptides were first trapped at 5 μl/min in 100% solvent A (0.1% formic acid in water) for 10 min, and then eluted with solvent B (0.1% formic acid in ACN) at a flow rate of around 200 nL/min. Phoshoproteome analysis was performed in 120 min gradient as follows: 0–10 min 100% solvent A, 10–105 min 4% solvent B, 105–108 min 36% solvent B, 108–109 min 100% solvent B, 109–120 min 100% solvent A. For the proteome analysis instead a 180 min gradient was set as follows: 0–10 min 100% solvent A, 10–10.1 min 13% solvent B, 10.1–165 min 40% solvent B, 165–168 min 100% solvent B, 169–180 min 100% solvent A. The electrospray voltage was set to 1.7 kV using a coated SilicaTip P200P capillary (Thermo Scientific). The mass spectrometer was operated in data-dependent acquisition mode and was configured to perform a Fourier transform survey scan from 375 to 1600 m/z (resolution 35,000) followed by higher collision energy dissociation (HCD) fragmentation of the 10 most intense peaks (25% normalized collision energy at a target value of 50,000 ions, resolution 17,500). In total, 411 RAW spectra were collected: 168 for proteomics and 243 for phosphoproteomics.
(Phospho)proteomics Data Processing
A preliminary quality analysis was performed for each dataset by MaxQuant (version 1.5.1.2) selecting label-free quantification and using the integrated Andromeda search engine and Swiss-Prot Homo sapiens database (20,196 entries, released on 10_2014). A cut-off of R2 = 0.7 was applied on both proteomics and phosphoproteomics correlation matrix (data not shown) and, after this filtering step, 383 RAW phosphoproteomics and proteomics files were selected for the final analysis and analyzed by MaxQuant (version 1.5.2.8) with the integrated Andromeda search engine. Phosphoproteomics RAW data were classified as Group 0, whereas proteomics RAW data as Group 1. A mutant-modified Swiss-Prot database Homo sapiens (24,126 entries, released on 05_2015) was used for the database search: for the variant identification, all STAR aligned reads were merged into a single bam file, subsequent variant calling was performed using VarScan (35) (v2.3.8) with default settings; resulting variant positions were included in the extended database when covered by at least 100 reads with at least 20% of reads harboring the variant. Trypsin was specified as enzyme and up to two missed cleavages were allowed. Cysteine carbamidomethylation was set as a fixed modification, whereas methionine oxidation and protein N-term acetylation were set as variable modifications. Phosphorylation on serine, threonine and tyrosine was also selected as variable modification for the phosphoproteomics analysis. The mass tolerance was set to 4.5 ppm for precursor ions, and to 20 ppm (FTMS) for fragment ions. Fast Label free quantification (LFQ) was performed and 'match between runs' was enabled. Peptide and protein identification was set to 1% FDR, and the minimum score for modified peptides was set to 40. The quantified output (proteinGroups.txt, supplemental Table S7; phospho(STY)Sites.txt, supplemental Table S8) were processed using a custom in-house developed Python package (PaDuA). Potential contaminants and reverse peptides were removed. After filtering protein data for “only identified by site” and phospho-data for localization probability > 75% (Class I phosphosites), 6638 protein groups and 9055 phosphosites Class I were identified. Next, normalization was performed by subtracting the median of log2 transformed intensities from each column. For phospho-data, the 'expand side table' function was applied before normalization. Median of technical replicates was performed for each dataset and the resulting values were filtered to ensure each protein or phosphosite had valid measurements in at least one time point of any of the six cell culture conditions. For the final dataset, 5692 protein groups and 7141 phosphosites Class I were quantified. Enrichment analysis was calculated using modificationSpecificPeptides.txt table (supplemental Table S9). The final processed outputs were exported for subsequent analysis in R (https://bitbucket.org/evertbosdriesz/cgc-multi-omics).
Transcriptomics
Quality and quantity of isolated RNA was checked and measured with Agilent 2100 Bioanalyzer and RNA Nano 6000 chips (Agilent Technologies, Cat. 5067–1511). All samples had a RNA integrity number (RIN) value ≥ 9.5. After RNA purification, libraries were generated from 500 ng of Total RNA using the Truseq Stranded Total RNA kit with Ribo-Zero Human/Mouse/Rat set A and B by Illumina (Cat. RS-122–2201 and RS-122–2202, San Diego, CA). After the library preparation, libraries were checked with Bioanalyzer2100 DNA High Sensitivity chips (Agilent, Cat. 5067–4626) and with Qubit (Qubit® dsDNA HS Assay Kit, Cat. Q32854, Thermo Fisher Scientific). Libraries were equimolar pooled to 2 nm. Next, 0.8–1.4 pm of these pooled libraries were sequenced on the Illumina NextSeq, 2 × 75bp high output, and 1.0–1.4 pm of library pools was loaded. Mapping was performed using STAR_2.4.2a, read counting using ht-seq count and the v74 gencode definition for coding regions. Fragments were mapped against GRCh37 using STAR (36) (v 2.4.2a), reads within coding regions (ENSEMBL release 74) were counted using ht-seq count (37) (v0.6.1) and further normalized and analyzed using the DeSeq2 package (38) (v1.6.3). Libraries were sequenced on Illumina NextSeq to an average of 9.9 (±3.5) million reads per sample. From these RNA depleted RNA-seq libraries, 7.9 (±4.9) million reads mapped against the human reference genome (hg19), of which 36.6% (±8.6) correspond to mRNA regions. Full details and workflows are available online: https://github.com/UMCUGenetics/RNASeq (v.2.2.0 was used for this paper).
Western Blots
For both quality control and validation analysis, total cell extracts were quantified using the Pierce BCA Protein Assay (23227, Thermo Scientific) and the colorimetric reaction evaluated at 562 nm using the EnVision 2014 Microplate Reader (Perkin Elmer, Massachusetts). Equal amount of proteins was prepared for all samples adding 10× NuPage Sample Reducing Agent (NP0004, Thermo Scientific) and 4X NuPage LDS Sample Buffer (NP0007). Samples were subsequently incubated at 95 °C for 5 min to allow protein denaturation. Lysates were resolved by SDS-PAGE using NuPage 4–12% Bis-Tris precast gels and NuPAGE Gel Electrophoresis Systems (Thermo Scientific). The gels were run in 1× MOPS buffer (50 mm MOPS, 50 mm Tris base, 0.1% SDS, 1 mm EDTA) at a constant voltage of 165 V. Proteins were transferred on a methanol-activated PVDF membrane. Transfer was performed in 1X Transfer Buffer (25 mm Tris base, 122 mm glycine, 0.01% SDS, 10% methanol) using a Trans-Blot Cell apparatus (Bio-Rad) and applying a constant amperage of 70 mA. Blocking was performed by incubating the membranes in 5% BSA in TBS-T (0.1%) for 1 h. Primary antibodies were typically diluted 1:1000 in 5% BSA in TBS-T and incubated at 4 °C overnight while shaking. Membranes were washed 3 times during 10 min with TBS-T (0.1%). HRP-coniugated secondary antibodies (Bio-Rad) were diluted 1:10,000 in 5% BSA in TBS-T and incubated for 1 h at room temperature while shaking. Subsequently, membranes were washed additional 3 times during 10 min with TBS-T (0.1%). Final protein detection was performed using Clarity ECL Western blotting substrate (Bio-Rad) and blot imaging was performed using the Chemidoc Touch Imaging System (Bio-Rad).
Differential Expression Analysis
All differential expression analyses were performed using the limma R package (39). mRNA read counts were first transformed using voom (40). The transformed mRNA read counts and the log-normalized intensities of proteins and phosphosites were checked to be normally distributed. For the comparison of BRAFi+EGFRi to control samples, a linear model was fitted for each gene/protein/phosphosite with each condition (treatment and time point pair) as a separate variable. The contrast between the BRAFi+EGFRi and control samples at 48 h conditions was used for enrichment analysis. For the comparison of the PTPN11 KO cell line to the PTPN11 WT cell line, a linear model was fitted for each gene/protein using cell line, time point and treatment as variables, and subsequently contrasting the cell line coefficients. Because PTPN11 KO and EGFRi treatment are expected to have a similar biological effect, the treatment coefficient of PTPN11 KO controls and EGFRi-only treated samples were equated. Similarly, the treatment coefficient of PTPN11 KO cell line treated with BRAFi and BRAFi+EGFRi samples were equated.
Clustering
Hierarchical clustering was performed on the 1500 phosphosites, proteins, or mRNAs with the highest variance within each data set, calculated over all conditions. The mRNA expression data was first log-transformed and filtered for mRNAs that were differentially expressed compared with the T = 0 h Control at a false discovery rate (FDR) of 10−6. The pairwise distance between two phosphosites, proteins, or mRNAs i and j was calculated as (1-ρi,j)/2, where ρi,j is the Pearson correlation between the two. The obtained distances were used for hierarchical clustering using Ward's minimum variance method. For each data type, the resulting trees were cut into 8 groups, because consensus clustering (41) indicated that to be a reasonable number. Enrichment analysis of the clusters were done using Fisher's exact test, with all measured genes, proteins, or phosphosites as background. For the phosphoproteomic data, the enrichment for each kinase in each cluster was calculated based on its predicted substrates. Predicted kinase-substrate relations were obtained using KinomeXplorer (42). Similarly, for the mRNA expression data, enrichment of transcription factors in each cluster was calculated based on its known target genes. Transcription factor-target gene relations were obtained from the TransFac database (43, 44) (version 2_2016). Enrichment analysis of biological processes was done using the hallmarks gene sets from MSigDB (45). Multiple testing correction was performed using Benjamini and Hochberg's method.
Transcriptomics of Multiple BRAF(V600E) CRC Cell Lines
For this validation experiment, SNU-C5, VACO432 and KM-20 cell lines were plated in 10-cm dishes at a confluence of 2 × 106 cells per dish. All cell lines were cultured in RPMI 1640, supplemented with 10% FCS, 1% l-Glutamine and 1% Penicillin/Streptomycin. Cells were first incubated for 24 h, and subsequently starved for 24 h in serum-free media. Next, all plates were supplemented with serum-free media containing either no drugs (untreated) or 3 μm PLX4032 (BRAFi) or the combination of 3 μm PLX4032 and 3 μm gefitinib (BRAFi+EGFRi). Following 30 min incubation, both untreated (n = 3) and treated cells (n = 6) were stimulated with 10% FCS, and then collected after 48 h. Controls at T = 0 (n = 3) were instead collected immediately after starvation. Total RNA was extracted by adding 400 μl RLT buffer containing β-mercaptoethanol from the Qiagen RNaesy kit (Qiagen, Germany) and collected the cells by scraping. Cell lysates were stored in 2 ml tubes at −80 °C until time of total RNA isolation with the QIAsymphony machine (Qiagen), using the Qiasymphony RNA kit (Qiagen, Cat. 931636) and the miRNA CT400 workflow. For the differential expression analysis, a linear model using cell line (SNU-C5, VACO432, and KM-20) and treatment (T = 0 h, Control, BRAFi and BRAFi+EGFRi) was fitted. The contrast between BRAFi+EGFRi versus Control and BRAFi versus Control were used for the differential expression analysis. The contrast between T0 and all other treatments were used for the Rapid Feedback Inhibitor analysis.
Cell Proliferation Assays
All experiments were carried out culturing WiDr cells in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 1% Penicillin/streptomycin and 1% l-Glutamine, at 37 °C, 5% CO2.
WiDr cells were seeded in five 96-well plates at a density of 5000 cells/well. After 24 h of incubation (37 °C, 5% CO2), media was removed from all plates and replaced with media containing drugs. In three 96 wells plates, gefitinib, lapatinib or sapitinib were serially diluted to a final concentration range of 120 nm to 30 μm as a single treatment and in combination with PLX4032 at a fixed concentration of 3 μm (n = 4). In the fourth 96-well plate, PLX4032 was serially diluted to a final concentration range of 30 nm to 30 μm in combination with gefitinib at a fixed concentration of 3 μm and with either 1 mm DCA (n = 4) or 10 μm etomoxir (n = 4) respectively. In the last 96-well plate, PLX4032 was serially diluted to a final concentration range of 30 nm to 30 μm in combination with gefitinib at a fixed concentration of 3 μm (n = 8). After 96 h of treatment, 1 mm DCA was added to four replicates and 10 μm etomoxir was added to other four replicates. All plates contained a column with untreated cells as a reference sample and were treated for 7 days (37 °C, 5% CO2). Media containing the drugs was replaced after 72 h.
IC50 of single treatments was determined seeding WiDr cells in three 96-well plates at a density of 10,000 cells/well. After 24 h of incubation (37 °C, 5% CO2), media was removed from all plates and replaced with media containing either PLX4032 or etomoxir or DCA serially diluted in four replicates to a final concentration range 30 nm-30 μm, 200 μm-0.2 μm and 100 mm-0.1 mm, respectively. All plates contained a column with untreated cells as a reference sample and were treated until 72 h (37 °C, 5% CO2).
Cell growth inhibition was monitored in all assays using the IncuCyte™ automated microscope (Essen Bioscience, Ann Arbor, MI) and phase-contrast images were collected every 2 h using a 10x Nikon objective. Phase confluence percentage from each well at each time point was exported into GraphPad Prism 7.0 software. The area under the curve (AUC) was calculated for each concentration (n = 4), normalized in respect to untreated cells and fitted using a four-parameter logistic curve. Percentage of growth in single and combination treatments were visualized as dose response curves.
Drug Off Assays
All experiments were carried out culturing WiDr cells in RPMI 1640, supplemented with 10% FBS, 1% penicillin/streptomycin and 1% l-glutamine at 37 °C, 5% CO2.
For long-term assay, WiDr cells were seeded in one 6-well plate (200,000 cells/well) and grown to around 60% confluence. After 24 h starvation in serum-free media, cells were treated with PLX4032 and gefitinib both at a fixed concentration of 3 μm, in complete medium. Media was replaced two times per week and treatment was interrupted after 78 days. Pictures were acquired using an ECLIPSE Ti-e inverted microscope (Nikon, Tokyo, Japan) at a magnification of 10x. Cell confluence was measured using Fiji plugin in ImageJ software.
For short-term assay, WiDr cells were seeded in one 96-well plate at a density of 5000 cells/well. After 24 h of incubation (37 °C, 5% CO2), media was removed from all plates and replaced with media containing PLX4032 and gefitinib at fixed concentration of 3 μm. Media was replaced (once/twice) per week and treatment was interrupted after 5 days. Cell growth was monitored in the IncuCyte™ automated microscope and phase-contrast images were collected every 2 h using objective Nikon 10x.
Antibodies and Reagents
WiDr cells were purchased from American Type Culture Collection (ATCC) (13) and WiDr cells clone #B32 were used as knockout of PTPN11 (WiDr PTPN11KO) (16). Both RPMI 1640 medium (#12–167F), penicillin/streptomycin (#17–602E) and l-Glutamin (#17–605C) were purchased from Lonza, (Basel, Switzerland), whereas FBS (#16000044) was purchased from ThermoFisher.
PLX4032 (#S1267), gefitinib (#S1025) and Scientific sapitinib (#S2192) were purchased from Selleck Chemicals (Houston, TX), and lapatinib (#S1028) from MedKoo Bioscences Inc. (Chapel Hill, NC). Etomoxir (#11969) was purchased from Bio-Connect B.V. (TE Huissen, Netherlands), whereas dichloroacetic acid sodium salt (DCA) (#2156-56-1) from Sigma-Aldrich Chemie (Zwijndrecht, Netherlands).
Antibodies against HSP-90 (H-114), PTPN11 (SH-PTP2 C-18), ERK1 (C-16), ERK2 (C-14), and p-ERK1/2 (E4) were purchased from Santa Cruz. p-EGFR (Y1068, ab5644), p-SHP2 (Y542, ab62322) were purchased from Abcam. p-ERBB3 (Y1197, #4561), p-IGF1R (Y1135/1136, #3024), IGF1R (#3027), p90RSK (#8408), AKT 1/2 (#2920), p-AKT (S473, #4060) were purchased from Cell Signaling Technology. Anti-EGFR (#06–847), p-ERBB2 (Y1248, #06–229), ERBB2 (#OP15L), ERBB3 (#05–390) and p-p90RSK (T359/S363, #04–419) antibodies were from Millipore.
RESULTS
A Multi-omics Overview of BRAF Mutated Colorectal Cancer Cell Response to Targeted Drug Treatment
We selected the WiDr CRC cell line harboring the BRAF(V600E) mutation as a model system for our analyses (30, 31). To study the differences in signaling we treated the WiDr cells with either vemurafenib (BRAFi) or gefitinib (EGFRi), or with the combination treatment BRAFi+EGFRi. Additionally, we employed a WiDr PTPN11 knockout (KO) cell line treated with BRAFi (BRAFi in PTPN11 KO) to investigate if there are functional differences between PTPN11 KO and EGFRi when applied in combination with BRAFi (Fig. 1A). To reduce the signal background, cell growth was synchronized by serum starvation for 24 h (h), followed by 30 min (min) incubation with or without drugs before serum stimulation (Experimental Procedures). Unstimulated control and PTPN11 KO control cells were immediately harvested—indicated as T = 0 h throughout this study—whereas stimulated samples were collected in a time course at 2, 6, 24, and 48 h after treatment (Fig. 1B). We performed transcriptomic (RNA-seq) and (phospho)proteomic profiling at each of the time points as indicated in Fig. 1C. Our study was designed to capture the initial responses to the different drug treatments and the elicited RTK signaling, monitoring their effects on gene and protein expression, and the onset of feedback mechanisms. We therefore generated a customized protein sequence database using RNA-seq data to account for WiDr-specific non-synonymous variants (Experimental Procedures).
Fig. 1.
Study design. A, Biological model. Schematic representation of the MAPK signaling pathway whereby BRAF(V600E), the drugs and KO target sites are highlighted. B, Experimental design. WiDr and WiDr PTPN11 KO cells were cultured for transcriptome, proteome and phosphoproteome analysis. For each of the six treatments the time-course of events is indicated (Experimental Procedures). Color codes are utilized to differentiate the effect due to drug inhibition in PTPN11 WT with respect to the knock-out of PTPN11 KO by distinguishing BRAFi-treated samples (red and shades of purple) from not BRAFi-treated samples (gray and shades of blue). C, Workflow employed in this study. For the transcriptomics analysis, mRNA libraries were prepared and sequenced using Illumina NextSeq. Quantitation in proteomics and phosphoproteomics analysis was done by label-free quantitation. Phosphoproteomics was performed after Ti4+-IMAC phosphopeptide enrichment. For the integration of transcriptome, proteome and phosphoproteome approach, a customized protein database was developed to include identified missense mutations for peptide identification and quantification (Experimental Procedures).
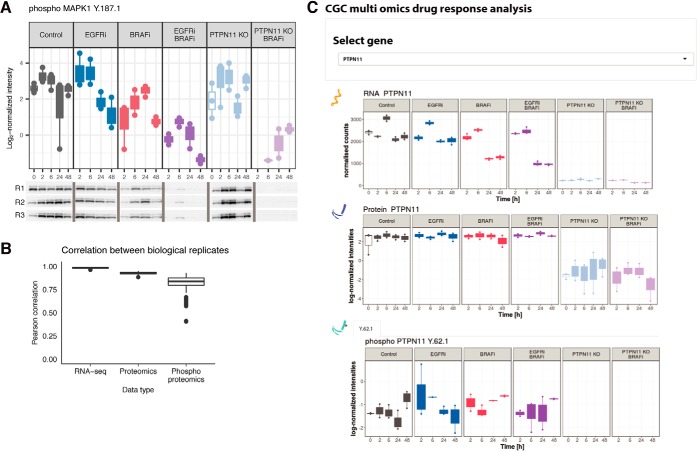
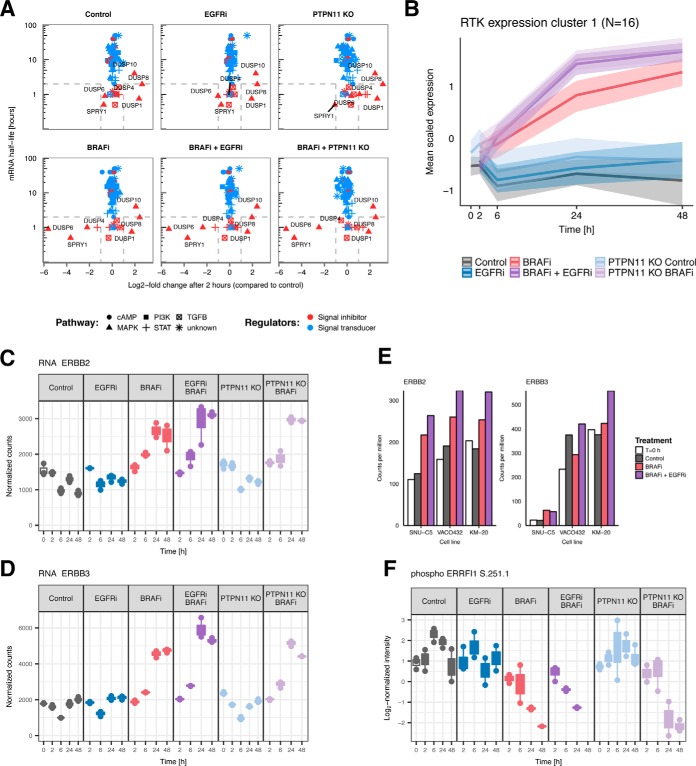
The phosphorylation profile of ERK (MAPK1) (Fig. 2A, top panel) was used as a positive control to verify the drug-induced regulation and overall quality of the label-free quantitative (phospho)proteomics approach. In concordance with previous studies (16), pERK is downregulated upon BRAFi, and this effect is enhanced by addition of EGFRi or in the PTPN11 KO cell line. Complementary Western blots (Fig. 2A, bottom panel) show excellent correlation with the label-free phosphoproteomics data. Further quality analysis demonstrates high correlation between respective biological replicates, with median Pearson correlation coefficients of 0.99, 0.93, and 0.83 for the transcriptomics, proteomics and phosphoproteomics data, respectively (Fig. 2B). As expected, the quantified proteomics data shows a slightly higher variability than the RNA-seq data (48), whereas phosphoproteomics data exhibits an even higher variability. The final (phospho) proteomics data set consisted of 5692 quantified protein groups and 7141 quantified Class I phosphosites (localization probability > 0.75), both of which were measured in at least one time point of any of the six applied conditions. The transcriptome dataset contained a final list of 21,446 genes (supplemental Fig. S1). We developed a Graphical User Interface (GUI) to facilitate rapid data comparison. The GUI enables selection of a specific gene to immediately visualize a comparison of its expression profile at transcriptomic and (phospho)proteomic levels (Fig. 2C).
Fig. 2.
High data quality enables reliable overview of gene-level response across omics datasets. A, Label-free phosphoproteomics and phosphoWB provide similar regulation patterns. The LC-MS/MS quality control was assessed by quantification of ERK phosphorylation (top panel) in all three biological replicates (R1, R2 and R3), and subsequent validation was done via western blots by using pERK1/2 (bottom panel) which confirmed down-regulation of pERK1 on Y187 as measured by mass spectrometry. B, Quality analysis of reproducibility at each omics level. Replicate consistency was assessed by inter-replicate Pearson correlation. In the phosphoproteomic analysis, selection of technical replicates involved discarding the poorest replicates before quantification. Median of technical replicates was performed for both phospho- and proteomics dataset, and the resulting processed data were further filtered to only include data that was quantified in at least one time point of any condition. Analysis indicates good median correlation (R > 0.8) among the three biological replicates for all three omics datasets. C, Cross-omics Graphical User Interface output. The GUI enables exploration of the multi-omics data for specific genes under all the tested conditions. Users can select a measured gene of interest from a dropdown menu, and visualize complete data at the transcript, protein, and phosphosite level, over the time-course and over all experimental conditions. An example output for the gene PTPN11 is shown.
Finally, we tested for potential off-target effects by looking at the substrates of kinases that had a dissociation constant for gefitinib or vemurafenib below the concentrations we used (ZAK, RIPK2, MAP2K5, PTK6, FECH, MAP4K5, RIPK3, GAP) (49). For all these kinases, we either did not observe any substrates, or their substrates did not show any coordinated dephosphorylation in response to the inhibitors (supplemental Fig. S2), providing no evidence of off-target effects.
EGFR Inhibition Amplifies Response to BRAFi in CRC Cells
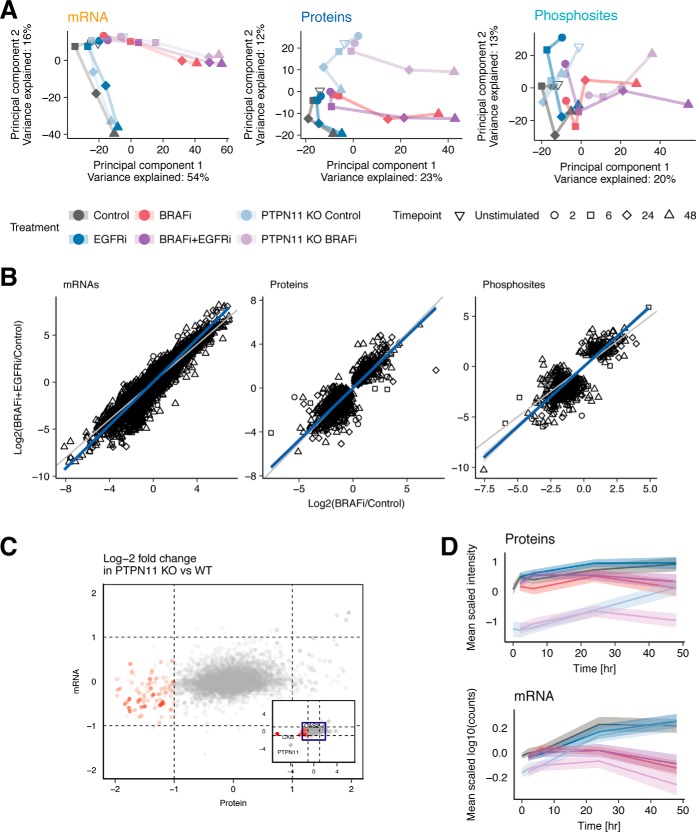
To extract an overview of the effect of the different treatments, we performed Principal Component Analysis (PCA) on each omics data type (Fig. 3A, supplemental Table S1). The trend along the first two principal components is similar for all data types. Principal Component 1 (PC1) represents the variation over time in the BRAFi treated samples (BRAFi, BRAFi+EGFRi, and BRAFi in PTPN11 KO). This variation is greater in BRAFi+EGFRi and BRAFi in PTPN11 KO samples compared with the samples treated with BRAFi only. Notably, the onset of the variation in the direction of PC1 occurs earlier in the transcriptomic data (after 6 h) than in the proteomics data (after 24 h), reflecting the delay from transcription to translation. Principal Component 2 (PC2) reflects the variation in the measurements over time in non-BRAFi treated samples (Control, EGFRi and PTPN11 KO control). In the proteomics data, PC2 also clearly separates the PTPN11 KO and WT samples.
Fig. 3.
Comparisons between drug treatments and omics data types. A, Principal Component Analysis (PCA) of transcriptomics, proteomics and phosphoproteomics data elucidates similar global trends in the data. B, Correlation plots of log2-fold-changes in BRAFi and BRAFi+EGFRi compared with Control, for each data set. The gene/protein/phosphosite-time point pairs selected are significantly differentially expressed for at least one of the two treatments. C, Scatterplot of log2-fold-changes of protein (x axis) and mRNA (y axis) levels in PTPN11 KO compared with PTPN11 WT cells reveals set of 77 genes downregulated at protein level but with no expression difference at RNA level. D, Average expression profiles of the 77 proteins (red dots in B) that are downregulated at the protein but not at the mRNA level. The expression of each mRNA/protein is mean-centered. The solid lines indicate mean expression of the 77 mRNA/proteins and the shaded area indicates the 95% confidence interval of the mean.
To further investigate the additional effect conferred by EGFRi in addition to BRAFi, we plotted log2(BRAFi+EGFRi/Control) against log2(BRAFi/Control) values for each gene/protein/phosphosite-time point pair that is significantly differentially expressed compared with untreated controls in at least one of the two treatments. The responses to BRAFi and BRAFi+EGFRi are highly correlated, as evidenced by Pearson correlation coefficients of 0.95, 0.86 and 0.82 for the transcriptomic, proteomic, and phosphoproteomic data, respectively (Fig. 3B). Interestingly, for both transcriptomic and phosphoproteomic data, the log2-fold-changes of the BRAFi+EGFRi treatment are larger, on average, than those of the BRAFi alone. This is clear from the linear regression line (blue) whose slope is steeper than the line with slope of 1 (gray line). This effect is statistically highly significant, because the 95% confidence intervals of these slopes are 1.14–1.15 and 1.14–1.25 for the mRNAs and phosphosites, respectively. This observation was further confirmed by analyzing mRNA expression data in three additional BRAF(V600E) CRC cell lines (SNU-C5, VACO432, KM-20) (supplemental Fig. S3A, supplemental Table S2). Furthermore, a similar trend is observed when correlating BRAFi in PTPN11 KO and BRAFi only treated samples in WiDr cell line (supplemental Fig. S3B), with the exception of a few proteins whose expression decreases in the PTPN11 KO cells but not upon EGFRi. Taken together, our data demonstrates that addition of EGFRi or the knockout of PTPN11 enhances the effect of BRAFi but does not produce qualitatively different molecular responses.
PTPN11 Knockout Induces Post-transcriptional Downregulation
Although the effect of PTPN11 KO and EGFRi mostly appears to be similar in our study, both the PCA (Fig. 3A) and the correlation of BRAFi in PTPN11 KO and BRAFi only (supplemental Fig. S3B) highlighted an effect of the PTPN11 KO on the proteomics data that was not observed in the transcriptomic data. To further investigate how PTPN11 KO affects proteins and mRNAs differently, we performed differential protein and mRNA expression analyses using a linear model to compare PTPN11 KO and wild type (WT) EGFRi treated cells overall, where the PTPN11 status (KO and WT+EGFRi), the drug treatment (BRAFi or not) and the time-points (0, 2, 6, 24 and 48 h) were used as variables (supplemental Table S3).
At the protein level, we identified 77 proteins that have (1) significant differential protein expression (FDR < 0.05) between PTPN11 KO and PTPN11 WT+EGFRi, (2) a large negative log2-fold-changes of < −1 at the protein level, and (3) a small absolute log2-fold-changes of < 1 at the mRNA level (Fig. 3C). The most disparate difference is observed in creatine kinase brain-type (CKB), with a log2-fold-change of −6.3 in PTPN11 KO cell line compared with PTPN11 WT+EGFRi and a remarkable minimal difference on average in mRNA expression (log2-fold-change = −0.65) (supplemental Fig. S4A).
These 77 proteins also demonstrate a consistent time-course profile in which expression at both the protein level and the mRNA level is downregulated in BRAFi treated conditions at T > 24 h (Fig. 3D, supplemental Fig. S4B, supplemental Fig. S4C). This observation suggests that expression of these 77 proteins still responds to mRNA expression level dynamics, despite an overall downregulation in an mRNA-expression independent manner upon PTPN11 KO.
We next investigated whether these 77 proteins are functionally related by performing an enrichment analysis using the MSigDB Hallmarks (45). Our analysis reveals a strong enrichment of the interferon alpha (IFN-α) and gamma (IFN-γ) response gene-sets (enrichment > 15-fold, p < 10−12, supplemental Table S3) which are known to suppress cell viability through the JAK/STAT pathway (50). Interestingly, PTPN11 negatively regulates the IFN-induced JAK/STAT pathway by dephosphorylating STAT1 on both residue Y701 and S727 (51). In line with this observation, STAT1 is also significantly downregulated at the protein level in the PTPN11 KO cells (FDR = 10−8) in our dataset and shows a time-course profile like the aforementioned 77 genes (supplemental Fig. S4D).
System-wide Propagation of Drug Perturbation
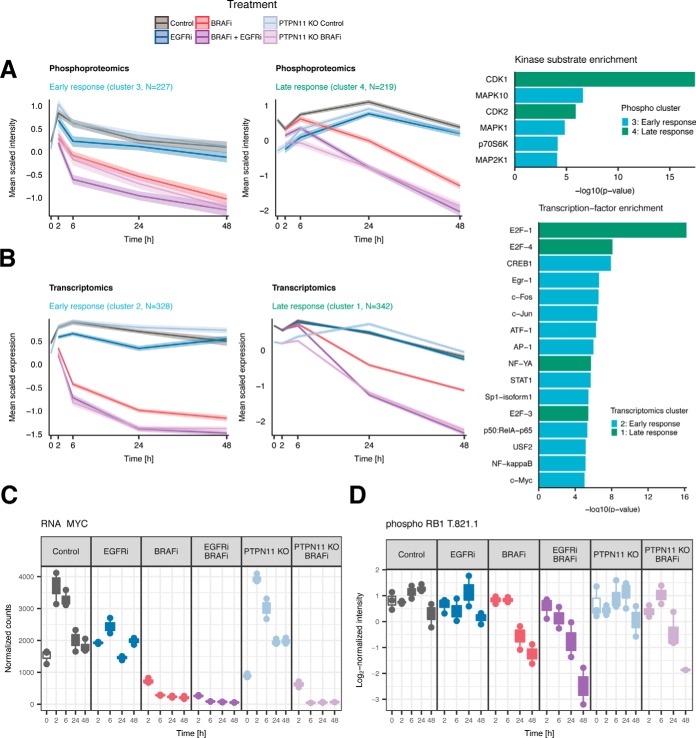
A key goal of our study was to provide a system-wide understanding of the propagation of cellular responses from signaling (phosphoproteomics) to gene transcription (transcriptomics) and then translation and protein expression (proteomics) in response to BRAF and/or EGFR inhibition. We therefore performed correlation-based hierarchical clustering on the 1500 phosphosites, mRNAs and proteins exhibiting the highest variance within each dataset and divided each omics dataset in eight clusters (supplemental Fig. S5). We observed that in the transcript-, phosphosite- and protein clusters, all BRAFi treated samples (BRAFi, BRAFi+EGFRi, and BRAFi in PTPN11 KO) exhibit similar clustering profiles, often distinct from non-BRAFi treated samples.
To investigate the biological function of all the clusters, we performed enrichment analysis based on transcription factor-target gene (transcriptomic clusters) and kinase-substrate (phosphoproteomics cluster) relationships, and using the hallmarks gene-sets from MSigDB (all data types) (45) (supplemental Table S4): Two clusters, corresponding to an early treatment response within 2–6 h and late treatment response after 24–48 h at both the gene expression (transcriptomic) and signaling (phosphoproteomic) level are downregulated upon BRAFi (either alone or in combination with EGFRi or in PTPN11 KO) (Fig. 4A and 4B). In contrast, four clusters exhibit a distinct upregulation of proteins, phosphosites or transcripts in all BRAFi treatments, starting after 2 h and increasing throughout the time-course.
Fig. 4.
Clustering reveals a MAPK mediated early and CDK mediated late response to BRAF inhibition. A, Selected phosphoproteomics clusters show an early and late response of phosphosites in which phosphorylation decreases upon BRAF inhibition within 2–6 h (left panel) and after 24–48 h (middle panel). Enrichment analysis (right panel) indicates that the early response cluster is enriched for substrates of kinases located downstream of BRAF in the MAPK pathway, whereas the late response cluster is enriched for substrates of cyclin-dependent kinases. B, Selected transcriptomics clusters show a similar early (left panel) and late (middle panel) response. Enrichment analysis (right panel) reveals that the early response cluster is enriched for target genes of transcription factors that are downstream of the MAPK pathway, and the late response cluster is enriched for targets of the E2F-transcription factor. C, Deactivation of MYC is a consequence of MAPK pathway deactivation and corroborates the early-response TF-target gene enrichment analysis. D, Downregulation of RB1 at phosphosite T821 inhibits E2F1 activity and induces cell cycle arrest at late timepoints in BRAFi treated cells.
MAPK and PI3K-AKT Signaling Are Involved in Early Response
The early response phosphoproteomics cluster is enriched for substrates of kinases belonging to the MAPK and PI3K-AKT pathways (Fig. 4A, right panel). An immediate decrease in phosphorylation of the canonical MAPK-pathway members MAP2K1 (MEK1) and MAPK1 (ERK2) substrates occurs in all BRAFi treated samples, which is consistent with the immediate dephosphorylation of Y187 on MAPK1 and Y204 on MAPK3 (ERK1) (Fig. 2A and supplemental S6A). The early response cluster also includes substrates of p70S6K (RPS6KB1), a kinase downstream of AKT. Its reduced phosphorylation on S427 (supplemental Fig. S6B) is consistent with the dephosphorylation of AKT after 2–6 h upon BRAFi, and more strongly so in combination with EGFRi. Although we did not detect the relevant phosphosites of AKT1 and AKT2 in our dataset, the deactivation of AKT signaling is confirmed by Western blotting at later time points in the combination treatment (supplemental Fig. S7).
Consistent with the early deactivation of MAPK and AKT signaling observed in the phosphoproteomics cluster, the transcriptomics early response cluster is enriched for targets of transcription factors downstream of the MAPK and AKT pathways (Fig. 4B, right panel), such as CREB1, FOS, MYC, AP-1, and JUN. This is further corroborated by down-regulation of most of these transcription factors upon BRAFi (Fig. 4C and supplemental Fig. S6C). The early response cluster is absent in the proteomics data (supplemental Fig. S5), possibly reflecting a lag between transcription and translation.
BRAF(V600E) Inhibition Affects Cell Cycle and Cell Proliferation at Later Time Points
The late response cluster reflects the effect of drug treatment on cell cycle and proliferation. The phosphoproteomics late response cluster is highly enriched for substrates of the cell cycle regulators CDK1 and CDK2 (Fig. 4A, right panel). This finding is corroborated by decreased CDK1 (supplemental Fig. S6D) and CDK2 (supplemental Fig. S6E) protein expression. Similarly, the transcriptomic late response cluster is enriched for target genes of the cell cycle regulators E2F1, E2F4, and E2F3 (Fig. 4B, right panel). The connection between late response phospho-signaling and gene expression is mediated by down-regulation of the phospho-residue T821 on RB1 (Fig. 4D), a substrate of CDK2, whereas total RB1 protein expression remains relatively constant (supplemental Fig. S6F). This down-regulation induces binding of RB1 to E2F1 thereby inhibiting E2F1 activity (52) and promoting cell cycle arrest (53) in all BRAFi conditions at the late timepoints. Further enrichment analysis of the MSigDB hallmarks gene-sets revealed enrichment of proliferation-related gene sets including E2F targets, genes involved in the G2M checkpoint, and mitotic spindle genes (supplemental Table S4).
We also observe a late response cluster in the proteomics data (cluster 4), which is strongly enriched for targets of MYC and E2F (supplemental Table S4). MYC and E2F targets are also enriched in the transcriptome early response cluster, again indicative of the delay between transcription and translation.
Inhibition of BRAF(V600E) Induces Distinct Metabolic Response
In addition to the early and late response clusters, which show down-regulation upon BRAFi, we also observe a set of transcripts, proteins and phosphosite clusters that are up-regulated in all BRAFi treated samples compared with Control and EGFRi-only treated samples. These clusters include phosphoproteomics cluster 5, and transcriptomics clusters 5 and 7 (supplemental Fig. S5) and exhibit the strongest regulation at 48 h. Cluster 5 of the phosphoproteomics data was enriched for substrates of pyruvate dehydrogenase kinases (PDKs), which are known to regulate glucose and mitochondrial metabolism by phosphorylating pyruvate dehydrogenase E1 component subunit alpha (PDHA1) on serine residues (54, 55). Both clusters 5 and 7 of transcriptomics did not show any significant statistical enrichment.
Thus, to further interrogate which processes are up-regulated in BRAFi+EGFRi or BRAFi treated samples, we performed differential expression and subsequent enrichment analysis of transcripts and proteins that were significantly up-regulated at 48 h with respect to Control (log2-fold-changes > 0, FDR < 0.05), using the MSigDB Hallmarks, KEGG, and Reactome genesets. Our proteomics data demonstrates up-regulation of metabolic processes including the peroxisome, the TCA cycle and the fatty acid metabolism upon BRAFi and BRAFi+EGFRi, (supplemental Table S5, supplemental Table S6, respectively). Activation of the TCA cycle is corroborated by significant up-regulation of TCA cycle enzymes IDH1, IDH2 and SUCLG2, whereas the observed enrichment of the mitochondrial proteins involved in the β-oxidation of long chain fatty acids as CPT2, HADHA and HADHB suggests fatty acids may be used as energy source under the drug treatment inducing stress conditions (56) (supplemental Fig. S8 and supplemental Fig. S9A–S9D). These pathways are not significantly enriched in the transcriptomic differential expression analysis (supplemental Table S5 and S6), and the up-regulation of the RNA levels is much less pronounced or inconsistent in most cases (supplemental Fig. S9A–S9D). Nonetheless, the average mRNA log2-fold-changes of the genes in most of these genesets is significantly greater than 0 (supplemental Table S5 and S6), and similar outcomes were obtained from the analysis on transcriptome data of three additional BRAF(V600E) CRC cell lines (SNU-C5, VACO432, KM-20) (supplemental Table S2, supplemental Fig. S9E). Taken together, our omics data suggest that upon BRAF(V600E) inhibition, CRC cells activate a broad set of metabolic processes, mainly on the proteomic level, which may increase mitochondrial oxidative activity through fatty acid synthesis and uptake, suggesting a new possible mechanism as defensive response.
Feedback Responses Aim to Compensate for MAPK Pathway Inhibition
Next, we set out to find more specific feedback responses to drug inhibition and serum stimulation by focusing on known regulators of the major signaling pathways (MAPK, PI3K, cAMP, TGFβ, JAK/STAT). To achieve this, we obtained a data set compiled by Legewie et al., who collected mRNA half-lives and expression profiles in response to pathway stimulation of known signal transducers and inhibitors of the major signaling pathways (57). From plotting the expression changes against the half-lives, Legewie et al. observed that all regulators that changed expression upon pathway stimulation were short-lived signal inhibitors. They named these Rapid Feedback Inhibitors (RFIs), which constitute a fast, homeostatic response mechanism to pathway perturbation. To investigate if the responding signaling regulators in our data are also RFIs, we similarly plotted the log2-fold-changes in mRNA expression after 2 h—compared with Control T = 0 h in each condition—against the mRNA half-lives obtained by Legewie et al. (Fig. 5A). We observe four interesting features in this plot. Firstly, all responding regulators (DUSP1, DUSP4, DUSP6, DUSP8, DUSP10, and SPRY1) are indeed RFIs (i.e. short lived negative regulators). Secondly, only genes regulating MAPK pathway signaling show response, suggesting that no other signaling pathways are affected by either growth factor stimulation or BRAFi. Thirdly, all samples show strong up-regulation of DUSP1, DUSP8, and DUSP10; presumably in response to the serum stimulation at T = 0 h. Finally, all BRAFi treated samples exhibit strong downregulation of DUSP4, DUSP6, and SPRY1, demonstrating that RFIs can also be downregulated in response to pathway inhibition, to counteract pathway inhibition. Similar results are observed at the 48-h time point (supplemental Fig. S10A), and further corroborated in three other BRAF(V600E) CRC cell lines (SNU-C5, VACO432, KM-20) (supplemental Fig. S10B)—with the downregulation of RASGRF1 upon BRAFi in both WiDr and the validation cell lines as notable addition—indicating that the absence of in long-lived mRNAs is not due to their low degradation rates.
Fig. 5.
Feedback mechanisms aimed at restoring MAPK signaling activity. A, Log2-fold-changes of mRNA expression plotted against mRNA half-life of regulators of major signaling pathways at 2 h. Only short-lived negative regulators of MAPK signaling respond to growth factor stimulation or BRAF inhibition. In all conditions, DUSP1, DUSP8 and DUSP10 are up-regulated in response to serum stimulation at T = 0 h. DUSP4, DUSP6 and SPRY1 are downregulated only in BRAFi treated samples, in response to BRAF inhibition. B, Scaled mRNA expression levels of a cluster of RTKs. Of the 35 RTKs, 18 are up-regulated (INSRR, ROS1, STYK1, ERBB3, FGFR2, PTK7, EPHA4, TEK, EPHB2, ERBB2, LMTK3, MERTK, INSR, EPHA10, NTRK1, DDR1). C, mRNA expression of ERBB2. D, mRNA expression of ERBB3. E, mRNA expressions of ERBB2 and ERBB3 in SNU-C5, VACO432, and KM-20 BRAF(V600E) CRC cell lines. F, Phosphorylation expression of the negative regulator ERRFI1 is downregulated at residue S251 in BRAFi samples.
To examine how RTKs upstream of the major signaling pathways responded to drug treatment, we performed hierarchical clustering of the mRNA expression of all RTKs based on Pearson correlation. This analysis revealed strong up-regulation of a subset of RTKs upon BRAF inhibition from the mid-timepoints (T = 6–24 h) onward. Of the 35 RTKs expressed in WiDr cells, 16 were up-regulated (Fig. 5B), including ERBB2 and ERBB3 (Fig. 5C and 5D). Selected targets were further quantified by performing western blots on the same WiDr and WiDr PTPN11 KO lysates used for omics analysis, confirming up-regulation of total ERBB3 upon BRAFi whereas the results for ERBB2 were less consistent (supplemental Fig. S7). To establish if transcriptional RTK up-regulation is a general response mechanism, we quantified their mRNA expressions in three additional BRAF(V600E) CRC cell lines. Up-regulation of several RTKs upon BRAFi+EGFRi was confirmed in all three cell lines (SNU-C5, VACO432 and KM-20) including ERBB2 which was consistently upregulated in all three lines, whereas ERBB3 was upregulated in KM-20 (Fig. 5E). Interestingly, in addition to the ERBB2/ERBB3 up-regulation, the regulator ERBB receptor feedback inhibitor 1 (ERRFI1), which interferes with ERBB family member homo- and hetero dimer formation (58), is downregulated in all BRAFi treated samples (Fig. 5F). Altogether, our data suggests the existence of an additional mechanism through which WiDr CRC cells might try to activate ERBB signaling to compensate for MAPK pathway inhibition.
ERBB Inhibitors Provide Limited Benefit in BRAF(V600E) CRC Treatment
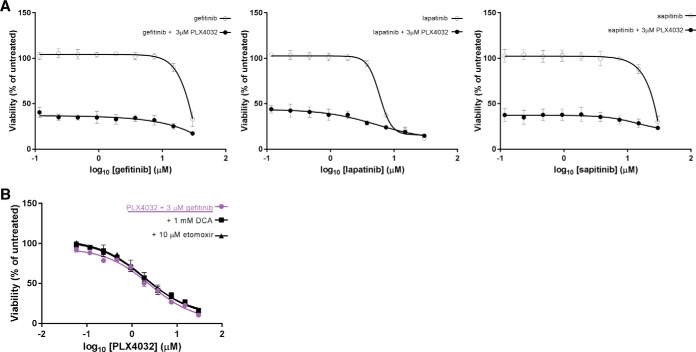
Next, we sought to determine if the observed up-regulation of ERBB2 and ERBB3 upon BRAFi+EGFRi treatment could be further exploited. We first studied whether inhibition of ERBB2 and ERBB3 in combination with BRAFi and EGFRi may lead to complete cell death by using gefitinib, lapatinib and sapitinib as known tyrosine-kinase inhibitors of EGFR, EGFR/ERBB2 and EGFR/ERBB2/ERBB3, respectively. We then explored if there was an optimal drug concentration, which would be synergistic with the maximum tolerated dose of PLX4032 (BRAFi) (Experimental Procedures). As expected (13), WiDr CRC cells are resistant to monotherapy of either gefitinib, lapatinib or sapitinib, with decreasing viability only at very high, i.e. cytotoxic, concentrations (Fig. 6A). All three viability curves depict a 60% decrease in cell viability upon addition of 3 μm PLX4032, with limited benefit from the combination with EGFRi. We did not observe a significant difference in growth inhibition across the different double treatments, suggesting that additional inhibition of ERBB2 and ERBB3 does not provide further synergy with BRAFi.
Fig. 6.
Assessment of WiDr CRC cell growth by combination treatments of BRAFi and ERBB or metabolic inhibitors. A, Comparison of mono- and double therapy on WiDr CRC cells growth. All three graphs show inhibition of either EGFR, EGFR/ERBB2 or EGFR/ERBB2/ERBB3 is ineffective as a monotherapy. Moreover, concomitant inhibition of ERBB2 and ERBB3 does not provide further benefit to the synergistic effect of BRAF(V600E) and EGFR inhibitors. B, WiDr cell confluence is measured comparing double and triple treatments. The addition of etomoxir or DCA as third metabolic inhibitors after 96 h does not show additional benefit to the BRAFi+EGFRi treatment.
Inhibiting Metabolic Enzymes Provides Limited Benefit in BRAF(V600E) CRC Treatment
We further evaluated combination treatments to target MAPK pathway together with the TCA cycle or with the fatty acid β-oxidation. For this purpose, we used two readily available metabolic drugs: etomoxir, a CPT1 inhibitor, and dichloroacetate (DCA), a pan-inhibitor of PDKs, to determine if inhibition of CPT1 or PDKs enhances sensitivity to therapy with PLX4032 and gefitinib (EGFRi). CPT1 is a site for intracellular regulation of lipid metabolism, transporting long-chain fatty acids into mitochondria for β-oxidation together with CPT2 (59, 60). CPT2 is significantly up-regulated in our experiments, nevertheless, to the best of our knowledge, a direct inhibitor of CPT2 has not been previously reported. PDKs are responsible for deactivation of PDHA1 through phosphorylation of serine residues in PDHA1 (54, 55) (supplemental Fig. S11). We hypothesized that inhibition of PDKs can increase the mitochondrial oxidative state and consequently the amount of reactive oxidative species (ROS) in the cytoplasm causing apoptosis due to high toxicity. We therefore evaluated the combination treatment of each metabolic inhibitor (etomoxir and DCA) with BRAFi and EGFRi on WiDr cell viability. After assessing IC50 concentrations of both metabolic drugs (supplemental Fig. S12A), we selected the clinical doses of 10 μm etomoxir (61–63) or 1 mm DCA (64, 65) and determined two dose-response curves in the presence of 3 μm gefitinib and increasing concentrations of PLX4032. The first curve was obtained by adding the metabolic drug simultaneously to the BRAFi and EGFRi (T = 0 h) (supplemental Fig. S12B), and the second by adding it 96 h after BRAFi and EGFRi (T = 96 h) (Fig. 6B), when metabolism is expected to be significantly up-regulated according to our data. In both cases, we do not observe any significant differences in the viability curves of the triple treatments compared with the double treatments. These findings suggest that inhibition of CPT1 or PDKs does not increase sensitivity to therapy with PLX4032 and gefitinib.
DISCUSSION
In this study we performed an integrated, quantitative, multi-omics analysis to obtain a system-wide molecular characterization of signaling perturbation over time in WiDr CRC and WiDr PTPN11 KO cell lines, after drug inhibition targeting either BRAF(V600E) and/or of EGFR. Our data reveal that all samples treated with BRAFi show similar response, with a more pronounced but qualitatively similar effect when BRAFi is combined with EGFRi or PTPN11 KO. This indicates that the main signaling responses depend on the inhibition of BRAF(V600E), and that additional inhibition of EGFR further amplifies the effect. Additionally, EGFRi-only treated cells exhibit similar responses to PTPN11 KO samples, confirming that suppression of this secondary signaling pathway confers sensitivity to BRAFi in CRC (16).
By comparing proteomics and transcriptomics data, we identified a set of genes that are exclusively downregulated at the protein level upon PTPN11 KO. These proteins are negative regulators of the interferon pathway (66), involved in controlling immune response. Down-regulation of negative regulators may support the immune response elicited by PTPN11 in vivo (67). This finding might be relevant for the development of therapeutic approaches that aim to inhibit PTPN11 activity. More generally, we have observed how mRNA expression levels do not always reflect protein dynamics and vice versa, underscoring the different but complementary information these data types can capture.
The integrated omics analysis enabled us to track the system-wide drug response upon treatment, from signaling - inactivation of kinases downstream of the MAPK pathway - through transcription - inhibition of genes downstream in the MAPK pathway. Shutting down MAPK signaling results in down-regulation of CDK signaling, inducing cell cycle arrest at a later stage. Besides the inactivation of the MAPK pathway, all three datasets show an increase of oxidative metabolic processes, with significant up-regulation of enzymes involved in lipid metabolism and the TCA cycle. We further observed that treatment with BRAFi induces up-regulation of RTKs, including ERBB2 and ERBB3, which was found to be more pronounced when co-treated with EGFRi or in PTPN11 KO cells.
We found evidence of metabolic rewiring in the proteomics data. One possible explanation of this finding is that these cells survive by utilizing different metabolic regimes, pointing at potential future avenues on how to target these cells. Although in our work combining inhibition of the MAPK pathway and specific metabolic processes did not result in any significant difference in cell viability within a 48 h time scale, this combination might still be relevant to therapy as demonstrated in two recent melanoma studies whereby resistant cells were found to be dependent on mitochondrial respiration (68, 69). Therefore, we further investigated the response of BRAF(V600E) mutant cells upon experimental BRAFi+EGFRi withdrawal. Similar approaches indeed has shown to induce massive cell death in melanoma and lung cancer cells (70). We observed that a subset of cells survived the combination treatment and could resume proliferation after stopping drug treatment (supplemental Fig. S13). However, the precise protective role of metabolic adaptation in the ability of cells to tolerate drug treatment remains elusive, and further studies are required. Although there is some evidence for transcriptional upregulation of these metabolic processes in our study, this finding would not have been evident from a transcriptomics-only analysis, highlighting the importance of studying the complete omics landscape.
Apart from the up-regulation of metabolic processes, all the adaptive responses we observed appear to be homeostatic responses aimed at compensating the MAPK pathway inhibition, but do not succeed in doing so within the 48 h time range of our study. Despite transcriptional up-regulation of ERBB family members, we do not observe additional benefit of inhibiting ERBB2 and/or ERBB3 in combination with EGFRi and BRAFi, suggesting that these homeostatic responses are not functional under the tested conditions. Activation of HER-family members has been recently reported in response to BRAFi in multiple CRC BRAF(V600E) cell lines (71), and increased expression of ERBB2 and ERBB3 is already known to confer acquired resistance (5, 72–74). In this regard, we hypothesize cells attempt to upregulate RTKs to overcome the inhibition of the MAPK pathway. Nevertheless, further studies are required to establish the more general implications of these findings, preferably by investigating different cell lines and several medium conditions that mimic physiological environments more closely. Importantly, we do not find any evidence of parallel signaling pathways being activated in response to drug treatment.
Taken together, we hypothesize that reactivation of the MAPK pathway is necessary for BRAF(V600E) mutation CRC cells to acquire resistance to BRAF inhibition. This view is supported by observations both in vivo and in the clinic whereby resistance to MAPK pathway inhibitors is typically mediated by mutations or amplification in the MAPK pathway in CRC patients (17, 75, 76).
The integrative multi omics approach employed here provides a time based in-depth view of the signaling mechanisms involved in drug response. Our findings highlight the importance of measuring these different levels simultaneously as exemplified by the RTKs regulation and PTPN11 specific signals. We expect our contribution to enable and accelerate future research into these mechanisms by making the data resource available.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (46) partner repository with the dataset identifier PXD007740. Sequence data has been deposited at the European Genome-phenome Archive (EGA) (47) which is hosted by the EBI and the CRG, under accession number EGAS00001002654. Annotated spectra can be inspected in MS-Viewer using the key azz8y3rc3q. Source code and data for all statistical analyses are available at https://bitbucket.org/evertbosdriesz/cgc-multi-omics. The Graphical User Interface (GUI) to visualize gene/protein/phosphosite expressions is available at https://cancergenomics.shinyapps.io/CGC_MultiOmics/. The FAIR data portal is available at https://cgc.fairdtls.surf-hosted.nl/demonstrator/.
Supplementary Material
Acknowledgments
We thank the PRIDE Team for assistance.
Footnotes
* This work was conducted under the framework of the Gravity Program CGC.nl, funded by The Netherlands Organisation for Scientific Research (NWO). AR, MA, AJRH were further supported by the Roadmap Initiative Proteins@Work (project number 184.032.201), funded by The Netherlands Organisation for Scientific Research (NWO).
 This article contains supplemental Figures and Tables. The authors declare that they have no conflict of interest.
This article contains supplemental Figures and Tables. The authors declare that they have no conflict of interest.
1 The abbreviations used are:
- MAPK
- mitogen-activated protein kinase
- KO
- knockout
- RTK
- receptor tyrosine kinase
- CRC
- colorectal cancer
- FDR
- false discovery rate
- GUI
- graphical user interface
- PC
- Principal component
- RFI
- rapid feedback inhibitor
- IMAC
- immobilized metal ion affinity chromatography
- LC
- liquid chromatography
- HCD
- higher energy collision induced dissociation.
REFERENCES
- 1. Groenendijk F. H., and Bernards R. (2014) Drug resistance to targeted therapies: Déjà vu all over again. Mol. Oncol. 8, 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahronian L. G., and Corcoran R. B. (2017) Strategies for monitoring and combating resistance to combination kinase inhibitors for cancer therapy. Genome Med. 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dhillon A. S., Hagan S., Rath O., and Kolch W. (2007) MAP kinase signalling pathways in cancer. Oncogene 26, 3279–3290 [DOI] [PubMed] [Google Scholar]
- 4. Miyamoto Y., Suyama K., and Baba H. (2017) Recent advances in targeting the EGFR signaling pathway for the treatment of metastatic colorectal cancer. Int. J. Mol. Sci. 18, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun C., Hobor S., Bertotti A., Zecchin D., Huang S., Galimi F., Cottino F., Prahallad A., Grernrum W., Tzani A., Schlicker A., Wessels L. F. A., Smit E. F., Thunnissen E., Halonen P., Lieftink C., Beijersbergen R. L., Di Nicolantonio F., Bardelli A., Trusolino L., and Bernards R. (2014) Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep. 7, 86–93 [DOI] [PubMed] [Google Scholar]
- 6. Karapetis C. S., Khambata-Ford S., Jonker D. J., O'Callaghan C. J., Tu D., Tebbutt N. C., Simes R. J., Chalchal H., Shapiro J. D., Robitaille S., Price T. J., Shepherd L., Au H.-J., Langer C., Moore M. J., and Zalcberg J. R. (2008) K-ras Mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 359, 1757–1765 [DOI] [PubMed] [Google Scholar]
- 7. Komarova N. L., and Boland C. R. (2013) Calculated treatment. Nature 499, 291–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Webster R. M. (2016) Combination therapies in oncology. Nat. Rev. Drug Discov. 15, 81–82 [DOI] [PubMed] [Google Scholar]
- 9. Tong C. W. S., Wu W. K. K., Loong H. H. F., Cho W. C. S., and To K. K. W. (2017) Drug combination approach to overcome resistance to EGFR tyrosine kinase inhibitors in lung cancer. Cancer Lett. 405, 100–110 [DOI] [PubMed] [Google Scholar]
- 10. Sundar R., Hong D. S., Kopetz S., and Yap T. A. (2017) TargetingBRAF-mutant colorectal cancer: progress in combination strategies. Cancer Discov. 7, 558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapman P. B., Hauschild A., Robert C., Haanen J. B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S. J., Sosman J. A., Kirkwood J. M., Eggermont A. M. M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R. J., Flaherty K. T., and McArthur G. A. (2011) Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kopetz S., Desai J., Chan E., Hecht J. R., O'Dwyer P. J., Lee R. J., Nolop K. B., and Saltz L. (2010) PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J. Clin. Oncol. 28, 3534–3534 [Google Scholar]
- 13. Prahallad A., Sun C., Huang S., Di Nicolantonio F., Salazar R., Zecchin D., Beijersbergen R. L., Bardelli A., and Bernards R. (2012) Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 [DOI] [PubMed] [Google Scholar]
- 14. Corcoran R. B., Ebi H., Turke A. B., Coffee E. M., Nishino M., Cogdill A. P., Brown R. D., Della Pelle P., Dias-Santagata D., Hung K. E., Flaherty K. T., Piris A., Wargo J. A., Settleman J., Mino-Kenudson M., and Engelman J. A. (2012) EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2, 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klinger B., Sieber A., Fritsche-Guenther R., Witzel F., Berry L., Schumacher D., Yan Y., Durek P., Merchant M., Schäfer R., Sers C., and Blüthgen N. (2013) Network quantification of EGFR signaling unveils potential for targeted combination therapy. Mol. Syst. Biol. 9, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prahallad A., Heynen G. J. J. E., Germano G., Willems S. M., Evers B., Vecchione L., Gambino V., Lieftink C., Beijersbergen R. L., Di Nicolantonio F., Bardelli A., and Bernards R. (2015) PTPN11 is a central node in intrinsic and acquired resistance to targeted cancer drugs. Cell Rep. 12, 1978–1985 [DOI] [PubMed] [Google Scholar]
- 17. Corcoran R. B., André T., Atreya C. E., Schellens J. H. M., Yoshino T., Bendell J. C., Hollebecque A., McRee A. J., Siena S., Middleton G., Muro K., Gordon M. S., Tabernero J., Yaeger R., O'Dwyer P. J., Humblet Y., De Vos F., Jung A. S., Brase J. C., Jaeger S., Bettinger S., Mookerjee B., Rangwala F., and Van Cutsem E. (2018) Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 8, 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopetz S., McDonough S. L., Morris V. K., Lenz H.-J., Magliocco A. M., Atreya C. E., Diaz L. A., Allegra C. J., Wang S. E., Lieu C. H., Eckhardt S. G., Semrad T. J., Kaberle K., Guthrie K. A., and Hochster H. S. (2017) Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG 1406). J. Clin. Oncol. 35, 520–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyö D., Ellis M. J., Carr S. A., and CPTACN. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J.-Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K.-H., Tabb D. L., Fenyö D., Bafna V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I.-M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., Rodland K. D., Investigators CPTAC SA, Gillette M. A., Klauser K. R., Kuhn E., Mani D. R., Mertins P., Ketchum K. A., Thangudu R., Cai S., Oberti M., Paulovich A. G., Whiteaker J. R., Edwards N. J., McGarvey P. B., Madhavan S., Wang P., Chan D. W., Pandey A., Shih I.-M., Zhang H., Zhang Z., Zhu H., Cope L., Whiteley G. A., Skates S. J., White F. M., Levine D. A., Boja E. S., Kinsinger C. R., Hiltke T., Mesri M., Rivers R. C., Rodriguez H., Shaw K. M., Stein S. E., Fenyo D., Liu T., McDermott J. E., Payne S. H., Rodland K. D., Smith R. D., Rudnick P., Snyder M., Zhao Y., Chen X., Ransohoff D. F., Hoofnagle A. N., Liebler D. C., Sanders M. E., Shi Z., Slebos R. J. C., Tabb D. L., Zhang B., Zimmerman L. J., Wang Y., Davies S. R., Ding L., Ellis M. J. C., and Townsend R. R. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., and Zhang B. (2014) Integrating genomic, transcriptomic, and interactome data to improve peptide and protein identification in shotgun proteomics. J. Proteome Res. 13, 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Altelaar A. F. M., Munoz J., and Heck A. J. R. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 23. Mathivanan S., Ahmed M., Ahn N. G., Alexandre H., Amanchy R., Andrews P. C., Bader J. S., Balgley B. M., Bantscheff M., Bennett K. L., Björling E., Blagoev B., Bose R., Brahmachari S. K., Burlingame A. S., Bustelo X. R., Cagney G., Cantin G. T., Cardasis H. L., Celis J. E., Chaerkady R., Chu F., Cole P. A., Costello C. E., Cotter R. J., Crockett D., DeLany J. P., De Marzo A. M., DeSouza L. V., Deutsch E. W., Dransfield E., Drewes G., Droit A., Dunn M. J., Elenitoba-Johnson K., Ewing R. M., Eyk Van J., Faca V., Falkner J., Fang X., Fenselau C., Figeys D., Gagné P., Gelfi C., Gevaert K., Gimble J. M., Gnad F., Goel R., Gromov P., Hanash S. M., Hancock W. S., Harsha H., Hart G., Hays F., He F., Hebbar P., Helsens K., Hermeking H., Hide W., Hjernø K., Hochstrasser D. F., Hofmann O., Horn D. M., Hruban R. H., Ibarrola N., James P., Jensen O. N., Jensen P. H., Jung P., Kandasamy K., Kheterpal I., Kikuno R. F., Korf U., Körner R., Kuster B., Kwon M.-S., Lee H.-J., Lee Y.-J., Lefevre M., Lehvaslaiho M., Lescuyer P., Levander F., Lim M. S., Löbke C., Loo J. A., Mann M., Martens L., Martinez-Heredia J., McComb M., McRedmond J., Mehrle A., Menon R., Miller C. A., Mischak H., Mohan S. S., Mohmood R., Molina H., Moran M. F., Morgan J. D., Moritz R., Morzel M., Muddiman D. C., Nalli A., Navarro J. D., Neubert T. A., Ohara O., Oliva R., Omenn G. S., Oyama M., Paik Y.-K., Pennington K., Pepperkok R., Periaswamy B., Petricoin E. F., Poirier G. G., Prasad T. S. K., Purvine S. O., Rahiman B. A., Ramachandran P., Ramachandra Y. L., Rice R. H., Rick J., Ronnholm R. H., Salonen J., Sanchez J.-C., Sayd T., Seshi B., Shankari K., Sheng S. J., Shetty V., Shivakumar K., Simpson R. J., Sirdeshmukh R., Michael Siu K. W., Smith J. C., Smith R. D., States D. J., Sugano S., Sullivan M., Superti-Furga G., Takatalo M., Thongboonkerd V., Trinidad J. C., Uhlen M., Vandekerckhove J., Vasilescu J., Veenstra T. D., Vidal-Taboada J.-M., Vihinen M., Wait R., Wang X., Wiemann S., Wu B., Xu T., Yates J. R., Zhong J., Zhou M., Zhu Y., Zurbig P., and Pandey A. (2008) Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 26, 164–167 [DOI] [PubMed] [Google Scholar]
- 24. Nesvizhskii A. I. (2014) Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J. C., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J. C., Liebler D. C., Carr S. A., Gillette M. A., Klauser K. R., Kuhn E., Mani D. R., Mertins P., Ketchum K. A., Paulovich A. G., Whiteaker J. R., Edwards N. J., McGarvey P. B., Madhavan S., Wang P., Chan D., Pandey A., Shih I.-M., Zhang H., Zhang Z., Zhu H., Whiteley G. A., Skates S. J., White F. M., Levine D. A., Boja E. S., Kinsinger C. R., Hiltke T., Mesri M., Rivers R. C., Rodriguez H., Shaw K. M., Stein S. E., Fenyo D., Liu T., McDermott J. E., Payne S. H., Rodland K. D., Smith R. D., Rudnick P., Snyder M., Zhao Y., Chen X., Ransohoff D. F., Hoofnagle A. N., Liebler D. C., Sanders M. E., Shi Z., Slebos R. J. C., Tabb D. L., Zhang B., Zimmerman L. J., Wang Y., Davies S. R., Ding L., Ellis M. J. C., and Reid Townsend R. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Faulkner S., Dun M. D., and Hondermarck H. (2015) Proteogenomics: emergence and promise. Cell. Mol. Life Sci. 72, 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Low T. Y., van Heesch S., van den Toorn H., Giansanti P., Cristobal A., Toonen P., Schafer S., Hübner N., van Breukelen B., Mohammed S., Cuppen E., Heck A. J. R., and Guryev V. (2013) Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 5, 1469–1478 [DOI] [PubMed] [Google Scholar]
- 28. Cutillas P. R. (2015) Role of phosphoproteomics in the development of personalized cancer therapies. PROTEOMICS - Clin. Appl. 9, 383–395 [DOI] [PubMed] [Google Scholar]
- 29. Ahmed D., Eide P. W., Eilertsen I. A., Danielsen S. A., Eknæs M., Hektoen M., Lind G. E., and Lothe R. A. (2013) Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Noguchi P., Wallace R., Johnson J., Earley E. M., O'Brien S., Ferrone S., Pellegrino M. A., Milstien J., Needy C., Browne W., and Petricciani J. (1979) Characterization of the WIDR: a human colon carcinoma cell line. In Vitro 15, 401–408 [DOI] [PubMed] [Google Scholar]
- 31. Chen T. R., Drabkowski D., Hay R. J., Macy M., and Peterson W. (1987) WiDr is a derivative of another colon adenocarcinoma cell line, HT-29. Cancer Genet. Cytogenet. 27, 125–134 [DOI] [PubMed] [Google Scholar]
- 32. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M. A. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., and Barillot E. (2012) Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28, 423–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J. R., Zou H., and Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere–based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 35. Koboldt D. C., Chen K., Wylie T., Larson D. E., McLellan M. D., Mardis E. R., Weinstock G. M., Wilson R. K., and Ding L. (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anders S., Pyl P. T., and Huber W. (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., and Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilkerson M. D., and Hayes D. N. (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horn H., Schoof E. M., Kim J., Robin X., Miller M. L., Diella F., Palma A., Cesareni G., Jensen L. J., and Linding R. (2014) KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods 11, 603–604 [DOI] [PubMed] [Google Scholar]
- 43. Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., Voss N., Stegmaier P., Lewicki-Potapov B., Saxel H., Kel A. E., and Wingender E. (2006) TRANSFAC(R) and its module TRANSCompel(R): transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wingender E., Dietze P., Karas H., and Knüppel R. (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., and Tamayo P. (2015) The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lappalainen I., Almeida-King J., Kumanduri V., Senf A., Spalding J. D., ur Rehman -S., Saunders G., Kandasamy J., Caccamo M., Leinonen R., Vaughan B., Laurent T., Rowland F., Marin-Garcia P., Barker J., Jokinen P., Torres A. C., de Argila J. R., Llobet O. M., Medina I., Puy M. S., Alberich M., de la Torre S., Navarro A., Paschall J., and Flicek P. (2015) The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 47, 692–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haider S., and Pal R. (2013) Integrated analysis of transcriptomic and proteomic data. Curr. Genomics 14, 91–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.-A., Reinecke M., Ruprecht B., Petzoldt S., Meng C., Zecha J., Reiter K., Qiao H., Helm D., Koch H., Schoof M., Canevari G., Casale E., Depaolini S. R., Feuchtinger A., Wu Z., Schmidt T., Rueckert L., Becker W., Huenges J., Garz A.-K., Gohlke B.-O., Zolg D. P., Kayser G., Vooder T., Preissner R., Hahne H., Tõnisson N., Kramer K., Götze K., Bassermann F., Schlegl J., Ehrlich H.-C., Aiche S., Walch A., Greif P. A., Schneider S., Felder E. R., Ruland J., Médard G., Jeremias I., Spiekermann K., and Kuster B. (2017) The target landscape of clinical kinase drugs. Science 358, eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ivashkiv L. B., and Donlin L. T. (2014) Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Du Z., Shen Y., Yang W., Mecklenbrauker I., Neel B. G., and Ivashkiv L. B. (2005) Inhibition of IFN-alpha signaling by a PKC- and protein tyrosine phosphatase SHP-2-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 10267–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lentine B., Antonucci L., Hunce R., Edwards J., Marallano V., and Krucher N. A. (2012) Dephosphorylation of threonine-821 of the retinoblastoma tumor suppressor protein (Rb) is required for apoptosis induced by UV and Cdk inhibition. Cell Cycle 11, 3324–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Henley S. A., and Dick F. A. (2012) The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harris R. A., Bowker-Kinley M. M., Huang B., and Wu P. (2002) Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzyme Regul. 42, 249–259 [DOI] [PubMed] [Google Scholar]
- 55. Dupuy F., Tabariès S., Andrzejewski S., Dong Z., Blagih J., Annis M. G., Omeroglu A., Gao D., Leung S., Amir E., Clemons M., Aguilar-Mahecha A., Basik M., Vincent E. E., St-Pierre J., Jones R. G., and Siegel P. M. (2015) PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 22, 577–589 [DOI] [PubMed] [Google Scholar]
- 56. Röhrig F., and Schulze A. (2016) The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 [DOI] [PubMed] [Google Scholar]
- 57. Legewie S., Herzel H., Westerhoff H. V., and Blüthgen N. (2008) Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol. Syst. Biol. 4, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu N., Matsumoto M., Kitagawa K., Kotake Y., Suzuki S., Shirasawa S., Nakayama K. I., Nakanishi M., Niida H., and Kitagawa M. (2012) Chk1 phosphorylates the tumour suppressor Mig-6, regulating the activation of EGF signalling. EMBO J. 31, 2365–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lodhi I. J., and Semenkovich C. F. (2014) Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pucci S., Zonetti M. J., Fisco T., Polidoro C., Bocchinfuso G., Palleschi A., Novelli G., Spagnoli L. G., Mazzarelli P., Pucci S., Zonetti M. J., Fisco T., Polidoro C., Bocchinfuso G., Palleschi A., Novelli G., Spagnoli L. G., and Mazzarelli P. (2016) Carnitine palmitoyl transferase-1A (CPT1A): a new tumor specific target in human breast cancer. Oncotarget 7, 19982–19996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D. E., Schafer Z. T., Evans R. M., Suda T., Lee C.-H., and Pandolfi P. P. (2012) A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18, 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Holubarsch C. J. F., Rohrbach M., Karrasch M., Boehm E., Polonski L., Ponikowski P., and Rhein S. (2007) A double-blind randomized multicentre clinical trial to evaluate the efficacy and safety of two doses of etomoxir in comparison with placebo in patients with moderate congestive heart failure: the ERGO (etomoxir for the recovery of glucose oxidation) study. Clin. Sci. (Lond). 113, 205–212 [DOI] [PubMed] [Google Scholar]
- 63. Samudio I., Harmancey R., Fiegl M., Kantarjian H., Konopleva M., Korchin B., Kaluarachchi K., Bornmann W., Duvvuri S., Taegtmeyer H., and Andreeff M. (2010) Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Invest. 120, 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dunbar E. M., Coats B. S., Shroads A. L., Langaee T., Lew A., Forder J. R., Shuster J. J., Wagner D. A., and Stacpoole P. W. (2014) Phase 1 trial of dichloroacetate (DCA) in adults with recurrent malignant brain tumors. Invest. New Drugs 32, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fox A. W., Sullivan B. W., Buffini J. D., Neichin M. L., Nicora R., Hoehler F. K., O'Rourke R., and Stoltz R. R. (1996) Reduction of serum lactate by sodium dichloroacetate, and human pharmacokinetic-pharmacodynamic relationships. J. Pharmacol. Exp. Ther. 279, [PubMed] [Google Scholar]
- 66. Porritt R. A., and Hertzog P. J. (2015) Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 36, 150–160 [DOI] [PubMed] [Google Scholar]
- 67. Mainardi S., Mulero-Sánchez A., Prahallad A., Germano G., Bosma A., Krimpenfort P., Lieftink C., Steinberg J. D., de Wit N., Gonçalves-Ribeiro S., Nadal E., Bardelli A., Villanueva A., and Bernards R. (2018) SHP2 is required for growth of KRAS mutant Non Small Cell Lung Cancer in vivo. Nat. Med. 24, 961–996 [DOI] [PubMed] [Google Scholar]
- 68. Corazao-Rozas P., Guerreschi P., Jendoubi M., André F., Jonneaux A., Scalbert C., Garçon G., Mallet-Martino M., Balayssac S., Rocchi S., Savina A., Formstecher P., Mortier L., Kluza J., Marchetti P., Corazao-Rozas P., Guerreschi P., Jendoubi M., André F., Jonneaux A., Scalbert C., Garçon G., Mallet-Martino M., Balayssac S., Rocchi S., Savina A., Formstecher P., Mortier L., Kluza J., and Marchetti P. (2013) Mitochondrial oxidative stress is the achille's heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget 4, 1986–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cesi G., Walbrecq G., Zimmer A., Kreis S., and Haan C. (2017) ROS production induced by BRAF inhibitor treatment rewires metabolic processes affecting cell growth of melanoma cells. Mol. Cancer 16, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kong X., Kuilman T., Shahrabi A., Boshuizen J., Kemper K., Song J.-Y., Niessen H. W. M., Rozeman E. A., Geukes Foppen M. H., Blank C. U., and Peeper D. S. (2017) Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550, 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Herr R., Halbach S., Heizmann M., Busch H., Boerries M., and Brummer T. (2018) BRAF inhibition upregulates a variety of receptor tyrosine kinases and their downstream effector Gab2 in colorectal cancer cell lines. Oncogene 37, 1576. [DOI] [PubMed] [Google Scholar]
- 72. Montero-Conde C., Ruiz-Llorente S., Dominguez J. M., Knauf J. A., Viale A., Sherman E. J., Ryder M., Ghossein R. A., Rosen N., and Fagin J. A. (2013) Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 3, 520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jain A., Penuel E., Mink S., Schmidt J., Hodge A., Favero K., Tindell C., and Agus D. B. (2010) HER kinase axis receptor dimer partner switching occurs in response to EGFR tyrosine kinase inhibition despite failure to block cellular proliferation. Cancer Res. 70, 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Temraz S., Mukherji D., and Shamseddine A. (2016) Dual targeting of HER3 and EGFR in colorectal tumors might overcome anti-EGFR resistance. Crit. Rev. Oncol. Hematol. 101, 151–157 [DOI] [PubMed] [Google Scholar]
- 75. Ahronian L. G., Sennott E. M., Van Allen E. M., Wagle N., Kwak E. L., Faris J. E., Godfrey J. T., Nishimura K., Lynch K. D., Mermel C. H., Lockerman E. L., Kalsy A., Gurski J. M., Bahl S., Anderka K., Green L. M., Lennon N. J., Huynh T. G., Mino-Kenudson M., Getz G., Dias-Santagata D., Iafrate A. J., Engelman J. A., Garraway L. A., and Corcoran R. B. (2015) Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 5, 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hazar-Rethinam M., Kleyman M., Han G. C., Liu D., Ahronian L. G., Shahzade H. A., Chen L., Parikh A. R., Allen J. N., Clark J. W., Kwak E. L., Faris J. E., Murphy J. E., Hong T. S., Van Seventer E. E., Nadres B., Hong C. B., Gurski J. M., Jessop N. A., Dias-Santagata D., Iafrate A. J., Van Allen E. M., and Corcoran R. B. (2018) Convergent therapeutic strategies to overcome the heterogeneity of acquired resistance in BRAFV600E colorectal cancer. Cancer Discov. 8, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (46) partner repository with the dataset identifier PXD007740. Sequence data has been deposited at the European Genome-phenome Archive (EGA) (47) which is hosted by the EBI and the CRG, under accession number EGAS00001002654. Annotated spectra can be inspected in MS-Viewer using the key azz8y3rc3q. Source code and data for all statistical analyses are available at https://bitbucket.org/evertbosdriesz/cgc-multi-omics. The Graphical User Interface (GUI) to visualize gene/protein/phosphosite expressions is available at https://cancergenomics.shinyapps.io/CGC_MultiOmics/. The FAIR data portal is available at https://cgc.fairdtls.surf-hosted.nl/demonstrator/.