Abstract
The human transcription factor TFIIH is a large complex composed of 10 subunits that form an intricate network of protein–protein interactions critical for regulating its transcriptional and DNA repair activities. The trichothiodystrophy group A protein (TTD-A or p8) is the smallest TFIIH subunit, shuttling between a free and a TFIIH-bound state. Its dimerization properties allow it to shift from a homodimeric state, in the absence of a functional partner, to a heterodimeric structure, enabling dynamic binding to TFIIH. Recruitment of p8 at TFIIH stabilizes the overall architecture of the complex, whereas p8's absence reduces its cellular steady-state concentration and consequently decreases basal transcription, highlighting that p8 dimerization may be an attractive target for down-regulating transcription in cancer cells. Here, using a combination of molecular dynamics simulations to study p8 conformational stability and a >3000-member library of chemical fragments, we identified small-molecule compounds that bind to the dimerization interface of p8 and provoke its destabilization, as assessed by biophysical studies. Using quantitative imaging of TFIIH in living mouse cells, we found that these molecules reduce the intracellular concentration of TFIIH and its transcriptional activity to levels similar to that observed in individuals with trichothiodystrophy owing to mutated TTD-A. Our results provide a proof of concept of fragment-based drug discovery, demonstrating the utility of small molecules for targeting p8 dimerization to modulate the transcriptional machinery, an approach that may help inform further development in anticancer therapies.
Keywords: protein-protein interaction, nuclear magnetic resonance (NMR), drug screening, DNA transcription, cancer, GTF2H5, quantitative imaging, STD-NMR, TFIIH, transcriptional regulation
Introduction
The general transcription factor TFIIH is a multisubunit complex essential for both transcription by RNA polymerase II and nucleotide excision repair. The seven subunits of the TFIIH core complex (p32, p44, p52, p62, XPD, XPB, and p8) plus the three subunits in the cyclin-activating kinase subcomplex (MAT1, Cdk7, and cyclin H) are assembled to constitute the molecular architecture of TFIIH (1). A deregulation in the network of protein–protein interactions within the complex can seriously alter structural integrity and cellular activities of this factor (1–3). In particular, p8 (also named trichothiodystrophy group A (TTD-A)4), the smallest and most recently discovered subunit of TFIIH, participates in the regulation of its steady-state level and of its DNA repair activity (4) by stabilizing TFIIH upon DNA damage (5).
A specific feature of the p8 subunit (Tfb5 in yeast) is that it is found in two distinct kinetic pools: (i) a free fraction shuttling between the cytoplasm and the nucleus and (ii) a TFIIH-bound fraction, which is exclusively nuclear (6). In line with this, p8 displays self-association properties that allow the protein to form a homodimer in solution in the absence of functional partners (7) while it shifts from homo- to heterodimer for associating with the p52 (Tfb2 in yeast) C-terminal domain in the TFIIH core complex (8). This transition enables recruitment of p8 into TFIIH, which governs the maintenance of TFIIH architecture and stability (9) and regulates its repair activity at UV-induced DNA damages (10, 11). In particular, the interaction between p8 and p52 up-regulates the ATPase activity of the XPB helicase (7). Recent cryo-EM studies revealed that p8 would act as a structural bridge between p52 and the helicase XPB (2).
Mutations in the p8 (TTD-A) gene cause the human syndrome trichothiodystrophy (TTD), one of the three photosensitive TFIIH-related syndromes that lead to severe premature aging. In cells isolated from the TTD-A patients (carrying mutations in p8) and the TTD-A knockout mouse model (complete deletion of the p8 gene), both transcription and DNA repair activities are severely affected, which may be explained by dramatically reduced steady-state levels of TFIIH (10, 12, 13). Remarkably, TTD-A patients, despite a high photosensitivity and a defect in DNA repair, do not develop skin cancers, unlike patients suffering from other nucleotide excision repair–deficient diseases, such as xeroderma pigmentosum. Interestingly, DNA repair function in TTD-A cells is not completely abolished but just slowed down (3, 14). Thus, the combination between a decreased DNA repair capacity and a reduced transcriptional activity, as observed in TTD-A patients, may explain the absence of sun-induced skin cancers.
In this work, we considered the dimerization interface of p8 as a “druggable” target for which, to our knowledge, no inhibitor has been described so far. With the aim to design molecules that destabilize the homodimer structure of p8 and alter its recruitment into TFIIH, we applied a fragment-screening strategy combined with quantitative imaging in living cells. Two small compounds were retained, accounting for significant destabilization of the p8 dimerization interface with a noticeable effect on concentration and transcription activity of TFIIH. Altogether, our approach validates the potential of targeting, with small molecules, transcription factor dimerization motifs (15) and offers p8 as an alternative pharmacological target that could be considered for an effective modulation of transcription.
Results
Selection of fragments potentially targeting dimerization interface
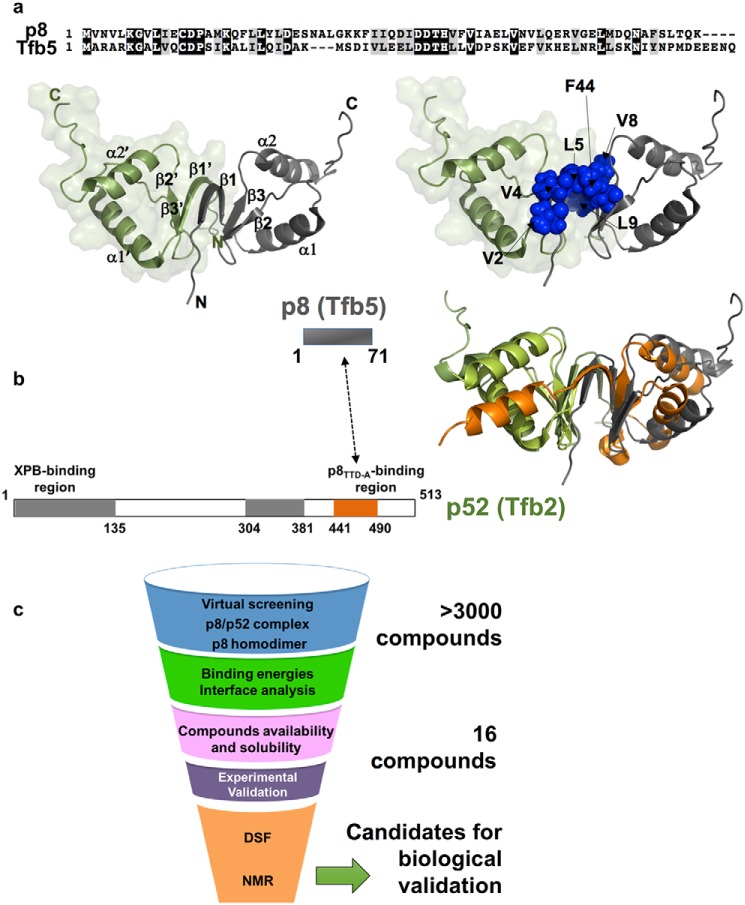
Before screening and in order to investigate the conformational stability of the human p8 subunit (Tfb5 in yeast) in the absence and presence of its functional partner, we performed molecular dynamics (MD) simulations by considering the solution structure of human p8 (7) and the crystal structure of the yeast complex formed between p8 (Tfb5) and the C terminus of p52 (Tfb2C) (10). As previously underlined, the two structures display a common fold due to conserved topology between Tfb5 and Tfb2C (Fig. 1) (8, 10). To provide a detailed view of the intermolecular persistent contacts, we followed the pairwise interactions between the two chains of the homodimer of p8 (below 3.5 Å for distances between atoms of each molecule) during the simulation, as described previously (16), and found that contacts involving residues Val2, Val4, Leu5, Val8, and Leu9 at the dimerization interface of p8 (and residues Ile452–Gln11, Tyr454–Leu9, Gly456–Gly7, Leu458–Arg5, and Ser460–Arg3 in the heterodimer Tfb5–Tfb2C) are present in more than 90% of the MD trajectory frames (Fig. 1 and Fig. S1). Phe44 was previously reported as critical in dimer stabilization (7). In almost 100% of the MD trajectory frames, close contacts were observed between ϵ aromatic protons of Phe44 from each chain, illustrating the importance of this amino acid on dimer stability.
Figure 1.
Structure of the TFIIH p8 subunit and protein–protein interface. a (top), sequence alignment of human p8 and yeast Tfb5. Left, solution structure of the human p8 homodimer (PDB code 2JNJ). The two monomers are depicted as green and gray ribbon, and their secondary structure elements are indicated. Right, the same view highlighting as blue spheres the residues located at the interface between the two monomers. b, schematic organization of p8 (residues 1–71, Tfb5 in yeast) and p52 (residues 1–513, Tfb2 in yeast) proteins. Gray boxes in p52 indicate the two regions that participate in the binding with the XPB helicase, whereas the orange box indicates the region of interaction with p8. Right, structure of p8 with the same color code as in a was superimposed onto the crystal structure of the Tfb5–Tfb2C complex (Tfb5 in green and Tfb2 in orange). Both structures could be superimposed for 67 atoms with a root mean square deviation value of 5.8 Å. c, flow chart showing the different steps of the screening process used in our work.
Both structures (p8 and Tfb5–Tfb2C) were used as targets for virtual screening of more than 3000 fragments provided by the Zenobia Therapeutics fragment library, the Enamine Golden fragment library, and our in-house molecules (Fig. 1c). Each docking provided a list of top 10 ligands with respect to the scoring function of AutoDock Vina. To select, among the top-ranked compounds, those that mainly occupy sites at the dimerization motif of either p8 or Tfb5–Tfb2C, a cut-off distance of 6–10 Å between ligands and dimerization interface was applied.
To perform experimental validation, fragment solubility in aqueous solution was first assessed by recording 1H NMR spectra of ligands using 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) as NMR standard at a concentration of 100 μm. Signal intensity for each ligand was compared with that of DSS, providing an estimation of ligand concentration in the protein buffer (1% DMSO-d6; see “Experimental procedures”).
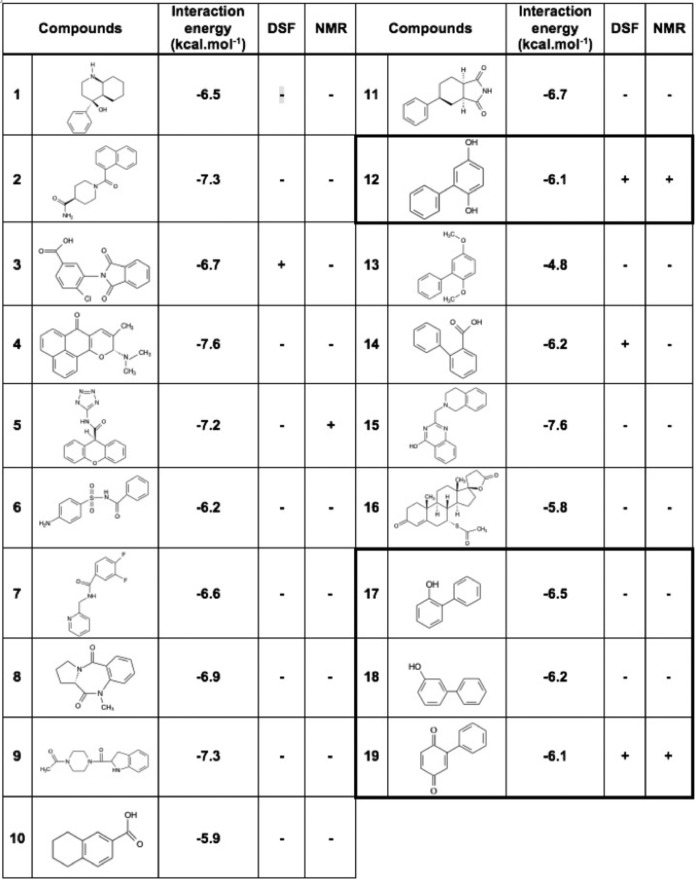
The ligands displaying poor solubility (<0.1 mm) were excluded from the validation assays. At this stage, 16 compound candidates (1–16) were retained for the experimental validation phase (Fig. 1c and Table 1).
Table 1.
Chemical structures of the compounds selected for the experimental validation
Chemical structure and results of analysis of DSF or NMR assays are given for each fragment (+/−). Energy values (in kcal·mol−1) corresponding to the score provided by the scoring function of AutoDock Vina are given. Bold boxes indicate compounds 17–19 that derive from compound 12 in a second round of screening.
Two fragments turned out to destabilize the dimerization interface of p8
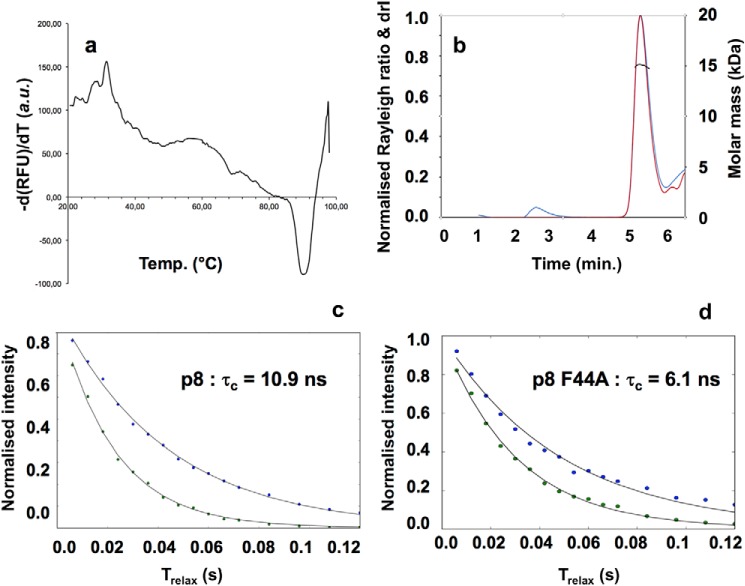
Taking advantage of the protein–protein interface preserved in the homodimer and heterodimer states, we sought to consider the p8 homodimer for experimental validation and biophysical assays. Fresh samples of recombinant protein were submitted to fluorescence-based thermal shift assays, also known as differential scanning fluorimetry (DSF), to examine the thermal stability of p8 in different buffers and varying salt concentrations. Heat denaturation of protein generally involves a change in its structure (thermal unfolding) and exposition of its hydrophobic residues. The DSF method takes advantage of this property by using an environmentally sensitive fluorescent dye, which exhibits high fluorescence when bound to the exposed hydrophobic residues upon protein thermal unfolding. Regardless of the tested conditions, p8 appeared highly stable, displaying elevated temperatures of unfolding (Tm) above 70 °C. In all tested conditions, the protein displayed a single melting transition, indicating a clear cooperative unfolding mechanism (Fig. 2a). SEC-MALS experiments gave a calculated average molar mass of 15,530 ± 120 g·mol−1 (Fig. 2b). Moreover, NMR 15N,1H TRACT experiments (17) were used to determine an overall rotational correlation time (τc), leading to a value of 10.9 ns (Fig. 2c), compatible with a 16-kDa protein at 25 °C. Therefore, p8 behaves like a dimer in solution, consistent with previous data (7) and particularly stable. The mutant F44A gave a much lower value of τc (6.1 ns), incompatible with dimeric species, probably reflecting the presence of monomer in solution (Fig. 2d). A unique melting transition centered at 55 °C was observed (compared with ∼88 °C for the WT in the same buffer), suggesting a lower thermal stability for this species with respect to the WT p8 protein (Fig. S2a).
Figure 2.
p8 is a dimer in solution. a, fluorescence signal (a.u., arbitrary units) as a function of temperature giving unfolding temperature of p8 in 50 mm Tris-HCl, 150 mm NaCl, 0.25 mm TCEP at pH 7.5, indicating high thermal stability (Tm ∼88 °C). The presence of 1 and 5% DMSO was found to leave this plot unaffected. b, elution profile obtained by SEC-MALS. The traces of the light scattering (blue), differential refractive index (red), and MALS-derived molar masses (black) are shown. The experimental SEC-MALS experiments gave a mean molar mass of 15,530 ± 120 g/mol−1, confirming the presence of homodimer species in solution (the calculated molecular mass of p8 is 8053.4 Da). c, NMR determination of the global rotational correlation times, at T = 298 K, for p8 and p8Phe44A (TRACT experiments (17)). The normalized intensity decay of the two 15N doublet coherences is plotted as a function of the relaxation delay. Fitting the data to mono-exponential functions returned the relaxation rate constants for α and β spin states. The difference between the two relaxation rates gave a global rotational correlation time τc of 10.9 ns (R2α = 23.3 Hz; R2β = 45.6 Hz) for p8, consistent with a dimer in solution. The τc value was significantly reduced in the case of p8Phe44A (6.1 ns; R2α = 20.3 Hz; R2β = 32.8 Hz), which is consistent with the expected value (5.7 ns) for a monomeric protein at this temperature (27).
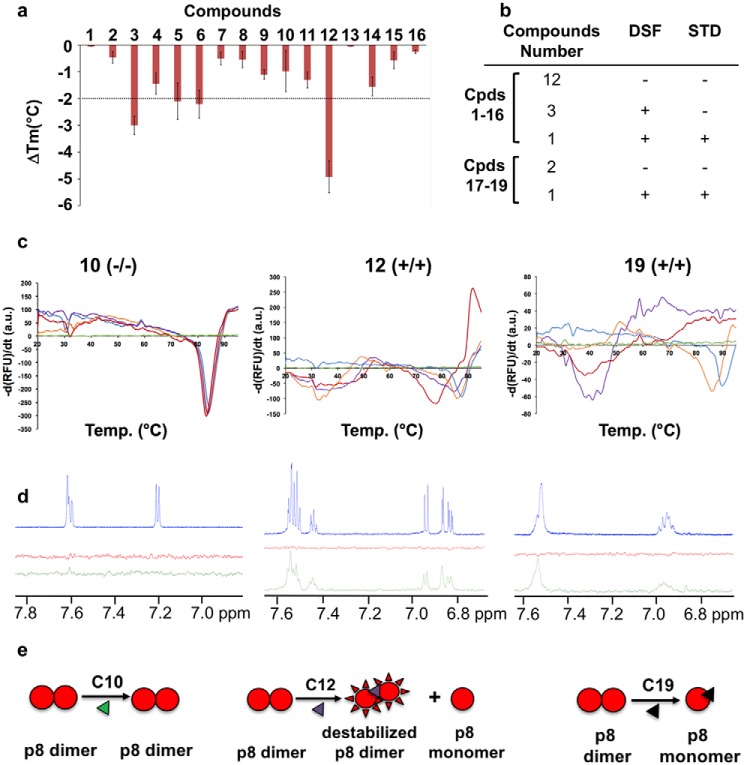
By using DSF, a binding can be inferred when the melting temperature of the protein changes in the presence of a ligand relative to that of the protein in the free state (18). If a ligand stabilizes (or destabilizes) the three-dimensional fold, a positive (or a negative) change in the inflection point of the transition (increase or decrease of the Tm) will be observed. Because ligands were diluted 100-fold to assay buffer from 100 mm DMSO stock solutions, the effect of increasing DMSO concentrations on protein thermal stability was first investigated. Only marginal shifts were observed for DMSO concentrations below 5% (v/v). Whereas the majority of the tested compounds did not alter the value at which p8 unfolds, four fragments (compounds 3, 5, 6, and 12) decreased the Tm by more than 2 °C compared with the DMSO control, suggesting an interaction with p8 (Fig. 3, a–c). Because compounds were initially selected for their capacity to occupy preferential sites on the dimerization interface of p8, the decrease in Tm values suggests that these four fragments would destabilize the homodimeric state (19). To complete the validation stage, we next employed the saturation transfer difference NMR (STD-NMR) approach (20). Upon saturation of the protein methyl resonances, the saturation propagates to the other protein protons by spin diffusion. Ligands that bind to the protein are then saturated by intermolecular saturation transfer and fast ligand exchange, giving rise to proton signals in a difference spectrum between the unsaturated and saturated spectra. To ensure that the STD signals are protein-dependent, a control experiment was performed for each ligand in the absence of protein (red spectrum, Fig. 3d). Saturation transfer efficiency was estimated from observation of resonance intensity enhancements in NMR aromatic regions compared with control experiments without protein. The majority of the evaluated candidates did not provide any significant intensity enhancement except for compound 12 (Table 1). Special care was also given to compounds 3, 5, and 6, which induced noticeable protein thermal shift in DSF assays (Fig. 3a). However, none of them resulted in significant saturation transfer efficiency compared with control experiments without protein, reinforcing the importance of combining two biophysical techniques for avoiding selection of false positives (Fig. 3b). Spironolactone (compound 16) was previously reported to target the XPB subunit of TFIIH (21). Our studies showed that this compound did not induce protein thermal shift or STD intensity enhancement, revealing no binding with p8 (Table 1).
Figure 3.
Evaluation of potential binders using thermal shift assays and NMR. a, histogram illustrating the ligand-induced changes in p8 thermal stability. The ΔTm values indicate the resulting shifts compared with the Tm signal of the free protein (around 90 °C; see Fig. 2a). DSF experiments were performed with a final protein concentration of 70 μm and final ligand concentration of 0.5 mm, respectively. The error bars are based at least on triplicate experiments. b, table summarizing the results obtained by DSF and STD for the 19 compounds tested (compound structures are given in Table 1). c, DSF melting curves of p8 dimer upon the addition of compounds 10 (left), 12 (middle), and 19 (right). Increasing concentrations of compounds (0.1 (orange), 0.25 (purple), and 0.5 mm (red)) were added to protein samples (70–90 μm) in buffer consisting of 50 mm Tris-HCl, 150 mm NaCl, and 0.25 mm TCEP at pH 7.5. Green and blue lines indicate DSF curves obtained for the compounds (0.5 mm) and p8 alone, respectively. d, 1H NMR spectra of aromatic regions of the compounds (blue) and STD spectra recorded for the compounds in the absence of protein (red) and in the presence of 10 μm of protein (green). The compounds were present at 1 mm concentration in phosphate buffer. The broader lines observed for compound 19 may arise from the formation of a covalent adduct with p8 (see Figs. S3 and S4). e, schematic drawing illustrating the effect of the three compounds 10, 12, and 19 on p8 dissociation; compound 10 has no effect, compound 12 leads to partial dissociation of the p8 homodimer, and compound 19 promotes complete dissociation leading to monomeric species in solution. At high protein/ligand ratios (>10) and upon long incubation times, compound 19 can form a covalent adduct with p8 that also destabilizes p8 (see Figs. S3 and S4).
On the other hand, compound 12 led to noticeable intensity enhancement in STD experiments (Fig. 3d). This fragment significantly decreased the Tm value of p8 (more than 5 °C), thus displaying an important negative effect on protein stability (Fig. 3a). Furthermore, an additional melting transition between 30 and 40 °C was observed, probably corresponding to the melting temperature of monomeric species. Taken together, these results revealed that compound 12 binds to p8 and targets the homodimer interface, which may result in equilibrium between monomeric and dimeric species in solution (Fig. 3c).
To investigate whether chemical derivatives of compound 12 could lead to the same or better behaviors, we selected three more ligands, namely 2-phenylphenol (compound 17), 3-phenylphenol (compound 18), and 2-phenyl-1,4-benzoquinone (compound 19), and evaluated their capacity to bind p8 using the same biophysical approaches (Table 1). Whereas compounds 17 and 18 did not induce any change in the Tm value of the protein, compound 19 strongly decreased the temperature at which p8 unfolds (Fig. 3, a and c). More interestingly, the addition of the compound at a concentration above 250 μm resulted in a unique and broad signal centered at 40 °C (Fig. 3c, purple line). Therefore, compound 19 corresponding to the fully oxidized form of compound 12 dramatically impacts protein stability and might lead to a complete dissociation of homodimeric species (Fig. 3e). Thus, the predominant species in solution would be monomers, displaying a much lower melting temperature than dimeric species.
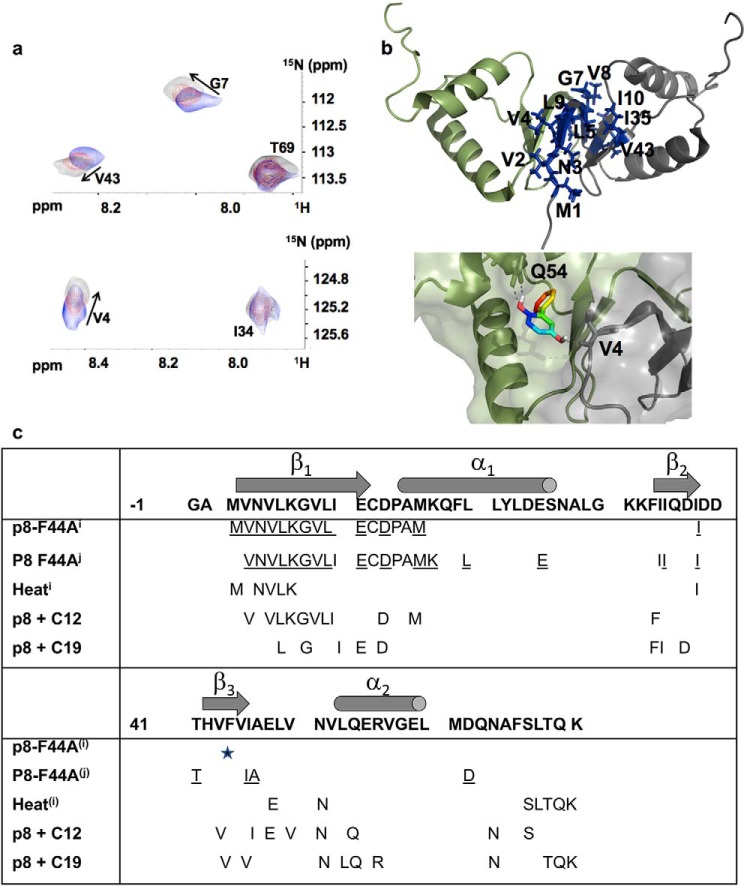
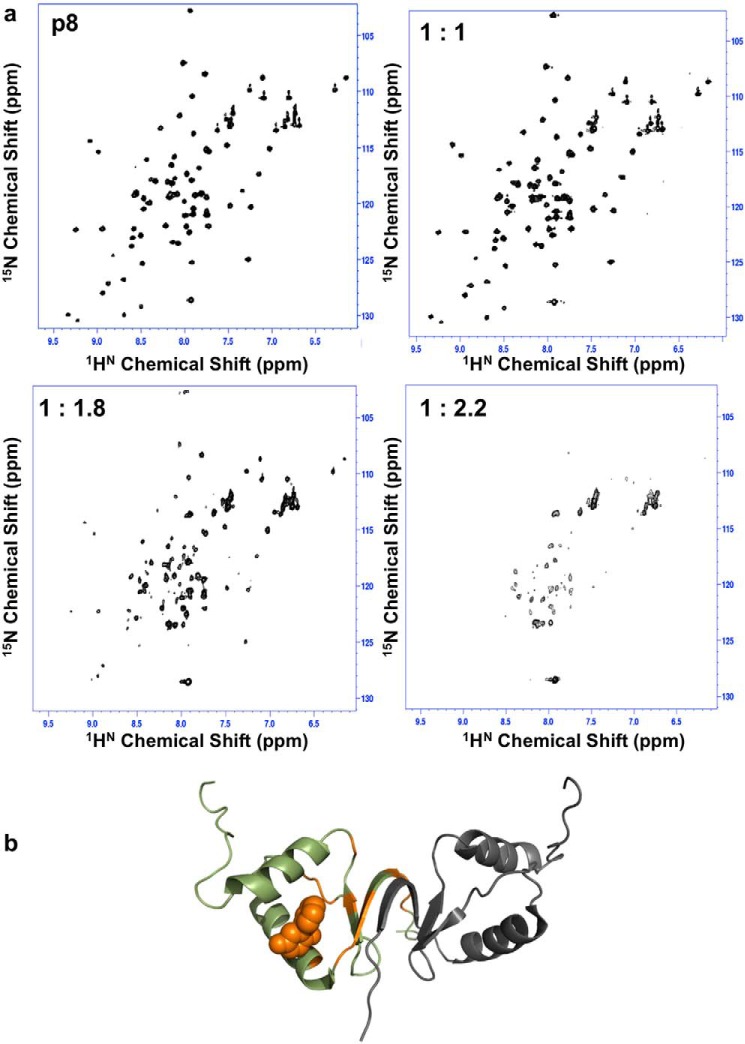
Having identified compounds 12 and 19 as decreasing the stability of the p8 homodimer interface, we next used 2D 1H-15N NMR HSQC experiments and chemical shift perturbation (CSP) to characterize their binding site in solution (Fig. 4). Compound 12 was added to the 15N-labeled p8 sample at molar ratios ranging from 0 to 5. Upon the addition of the ligand, several residues (Met1–Leu5, Gly7–Ile10, Ile35, and Val43) exhibited either chemical shift changes or peak broadening (Fig. 4). These residues map the β-sheet region, which displayed a surface area of ∼250 Å2, at the interface between the two monomers of p8. In parallel, we performed docking to predict binding poses of this compound in the structure of p8 homodimer. A cluster of significant top-ranking poses was found at the interface between the β-sheet and helices α1–2, where the compound was predicted to give polar contacts with Val4 of one monomer and Gln54 of the other monomer (Fig. 4b). To determine the constant of dissociation, STD experiments were carried out using increasing concentrations of compound 12 (250 μm to 4 mm), leading to a rather modest constant of dissociation in the submillimolar range (Kd = 172 ± 65 μm) (Fig. S7). In the same manner, a titration experiment was performed with compound 19 by recording a series of 2D 1H-15N NMR HSQC experiments of p8 with increasing concentrations of ligand. As the concentration of compound 19 increased, severe line broadening was observed until the limit of detection for several residues (Figs. 4 and 5). Furthermore, beyond a ligand concentration of 300 μm (twice the protein concentration), severe partial protein unfolding occurred (Fig. 5a). This result was not observed with any of the other tested compounds. Attempts to determine the rotational correlation time of the protein in the presence of compounds 12 and 19 gave τc values of 12.5 and 19 ns, respectively (Fig. S2). The high τc value suggests that compound 19 could promote protein aggregation. This precluded the estimation of a Kd for this compound using STD titration, although DSF titration results are in favor of higher efficacy compared with compound 12 (Fig. 3). Compound 10, which did not induce protein thermal shift or STD enhancement (Fig. 3), was chosen as a negative control for subsequent studies. As compounds 12 and 19 are predicted to fall into the category of pan-assay interference compounds (22, 23), which may react with proteins, top-down nano-LC mass spectrometry (LC-MSMS) experiments were performed to check this possibility and to localize the potential covalent attachment to the p8 protein (Figs. S3–S5). At protein and ligand concentrations used for DSF assays, no adduct formation was observed with compounds 12 and 19 even after 12 h of incubation (protein/ligand ratios of 1:1 and 1:2; Fig. S4a). Similarly, NMR spectra of the ligands recorded during chemical shift perturbation assays were unchanged, showing that the ligand remained stable during these experiments. However, at high protein/ligand ratios (1:10 and 1:100), a specific covalent adduct was observed in the presence of compound 19 (C19), due to a nucleophilic addition of the Cys14 thiol side chain onto the C19 double bond (Figs. S3 and S4) (24). The kinetics of adduct formation was slower for C12 than for C19, and the molecular mass increase (184 Da) was the same in both cases, indicating that the reacting species is C19 in both cases (C12 is known to be slowly air-oxidized into C19), as shown in Fig. S4b. In line with this, we could notice changes in the spectra recorded for compound 19 in the absence and in the presence of p8, reflecting chemical modification of this ligand after incubation with p8 (Fig. S8).
Figure 4.
Compound 12 binds to the homodimer interface. a, selected view of the 1H-15N HSQC spectrum recorded for the homodimer of p8, illustrating chemical shift changes for residues Val4, Gly7, and Val43 at the interface of the dimer upon the addition of compound 12. b, cartoon representation of the homodimer structure of p8, indicating the residues exhibiting chemical shift perturbation (in blue). The two monomers are shown as gray and green ribbon representations. A representative docking pose of compound 12 indicates hydrogen bonding with Val4 from one subunit and Gln54 from the other subunit of the p8 homodimer. c, table summarizing the residues of p8 whose resonances are perturbed either in the Phe44 mutant (underlined residues; p8-F44A(i) (7) and p8-F44A(j) (our work)) or by heat(i) (7) or upon the addition of compounds 12 and 19 to the WT protein. Secondary structures are indicated above the p8 sequence.
Figure 5.
Partial unfolding of p8 in the presence of compound 19. a, 1H-15N HSQC spectra recorded for p8 in the presence of increasing concentrations of compound 19, revealing gradual alteration of the quality of NMR spectra until partial protein denaturation for protein/ligand ratios exceeding 1:1.8. b, cartoon representation of the homodimer structure of p8 mapping positions of residues exhibiting severe line-broadening until the limit of NMR detection (orange) in the presence of compound 19, shown as an orange sphere (predicted binding pose obtained from docking).
In cellulo TFIIH stability
Before testing the impact of the candidate molecules on TFIIH concentration and to estimate the timing of the biological experiments, we measured the stability of TFIIH in living cells. For this purpose, we used cells isolated from two mouse models. One of these models endogenously expressed a fluorescently tagged version of TFIIH. Specifically, the XPB subunit of TFIIH was fused with the fluorescent marker YFP (XPB-YFP+f/+f) (25), allowing a direct measure of the amount of XPB, and because XPB is only present within the TFIIH complex (26), this fluorescence is also a measure of the TFIIH steady-state level. Hereafter, cells isolated from this mouse model will be designated as XPB-YFP cells. This mouse model was crossed with a TTD-A knockout mouse model (13), creating a new mouse model that expresses a fluorescent TFIIH mutant complex (fluorescent XPB subunit and absent TTD-A: XPB-YFP.TTDA−/−). Cells isolated from this mouse model will be designated as XPB-YFPTTDA−/−). Like the TTDA−/− knockout mouse model, the XPB-YFP.TTDA−/− mouse model was embryonically lethal before day 13.5, and the only cells that could be isolated from it were embryonic stem (ES) cells at day 3.5 and mouse embryonic fibroblasts at day 10.5.
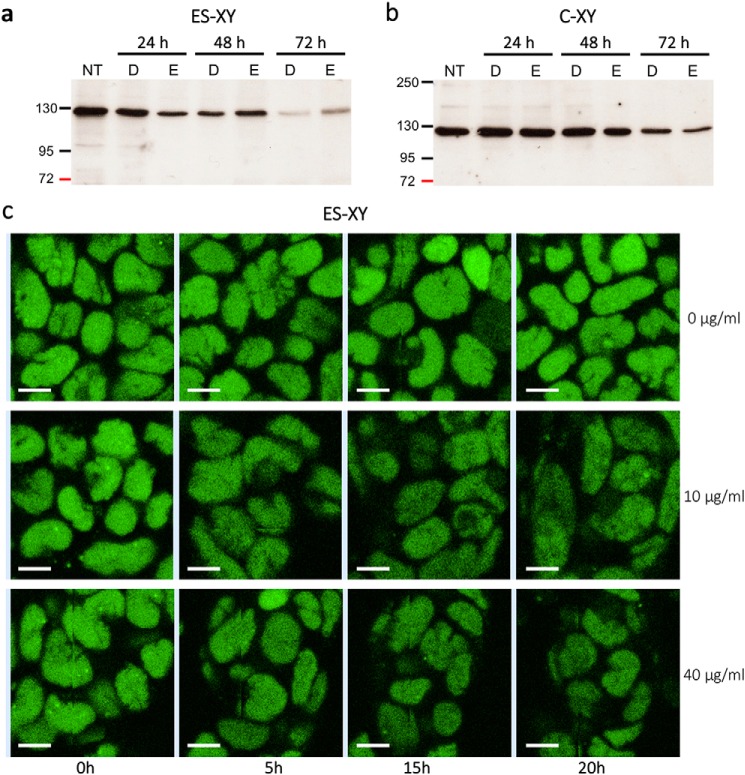
To measure the stability of TFIIH, we treated with cycloheximide (an inhibitor of protein synthesis) XPB-YFP ES cells and XPB-YFP chondrocytes and visualized the reduction of the XPB protein either by Western blotting (Fig. 6, a and b) or by direct imaging of the fluorescent XPB signal (Fig. 6c).
Figure 6.
Stability of TFIIH in living cells. a and b, Western blotting of the XPB subunit of TFIIH in ES cells (a) and chondrocytes (b) from the XPB-YFP mouse model (ES-XY and C-XY, respectively) after 24, 48, and 72 h of treatment with the protein synthesis inhibitor cycloheximide (133 μg/ml; NT, untreated; D, DMSO; E, ethanol). c, fluorescent nuclei of ES-XY cells at the 0-, 5-, 15-, and 20-h time points after treatment with different concentrations of cycloheximide (Scale bar: 10 μm.).
Our results showed that TFIIH degradation can take up to 72 h in ES cells and longer in chondrocytes, and that reduction of TFIIH steady-state levels can be easily visualized in living ES cells by confocal microscopy (Fig. 6).
Compounds 12 and 19 reduce TFIIH steady-state and transcriptional activity
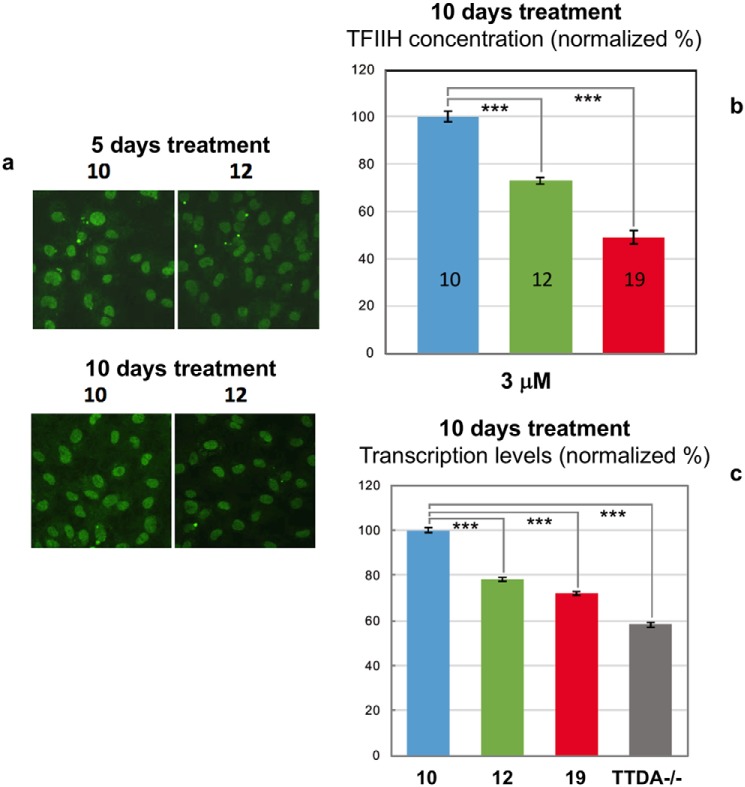
We first treated XPB-YFP ES cells and chondrocytes with compounds 10 and 12, and we observed that concentrations above 3 μm of compound 12 induced a cytotoxic effect compared with compound 10 (used as negative control). Because of this cytotoxicity, we decided to test concentrations below 3 μm, where cells were treated for 5–10 days to measure both TFIIH steady-state level and transcription. A clear reduction of the TFIIH steady state could be observed already after 5 days post-treatment with 3 μm compound 12 but not with compound 10 (Fig. 7a and Fig. S6). A longer incubation time was needed to observe a reduction with 1 and 2 μm concentrations of compounds (Fig. S6). We then tested compound 19 and measured that at a longer treatment time (10 days), and a strong reduction of the TFIIH steady-state level was observed (Fig. 7b). This TFIIH reduction level corresponded to a reduction in transcriptional activity measured by RNA synthesis 5-ethynyl uridine incorporation (Fig. 7c), showing that compound 19 has a stronger effect in destabilizing the TFIIH complex and in reducing transcriptional activity compared with compound 12. This reduction is not as high as the one observed in TTD-A mutant cells (isolated from human patients) and TTD-A knockout cells (isolated from knockout XPB-YFP.TTDA−/− mice). In the latter cells, p8 is completely absent, and consequent to this absence, the amount of TFIIH is about 30% lower than in WT cells.
Figure 7.
Reduction of TFIIH steady-state and transcriptional activity. a, typical examples of XPB-YFP–expressing cells, 5 and 10 days after continued treatment with compounds 10 and 12 at 1 μm concentration. b, results for the quantification of TFIIH levels via measurement of total XPB-YFP intensities in the nucleus of cells treated with 3 μm compounds 10, 12, and 19 after 10 days of treatment. c, results for the quantification of transcriptional activity in cells treated 10 days with compounds 10, 12, and 19 and in XPB-YFPTTDA−/− cells. Error bars, S.E. ***, p < 0.001, Student's t test (two tailed, assuming unequal variances).
Discussion
It is widely acknowledged that high levels of transcription are required to maintain a proliferative state in cancer cells; therefore, targeting transcription factors constitutes an interesting way to directly modulate transcription activity in cancer treatment (15), and this approach has led to some success stories (28–31). In this context, TFIIH has been established as an important target for cancer therapy (32), and the network of protein–protein interactions (PPIs) present in this transcription factor appears to be a promising class of drug targets.
In the last decade, the fragment-based drug discovery approach has emerged as a successful method for the design of low-affinity ligands that target PPIs, providing efficient starting points for the development of higher-affinity inhibitors (33–35). Although this approach to target PPIs is particularly challenging compared with more canonical and “druggable” protein targets (36–38), its success relies on chemically relevant low-molecular weight fragments together with sensitive and robust biophysical techniques for screening and detecting small-molecule drug candidates (39, 40). Here, we performed DSF and ligand-based NMR spectroscopy, which have proven to be efficient for the identification of weak binders, such as fragments (41–43), and we applied quantitative imaging of TFIIH in living cells to validate the promising candidates.
Our work provides evidence that the TTD-A/p8 subunit of TFIIH is a pharmacological target of high potential. Although in the cellular context, there is no evidence that it can dimerize (11), our work indicates that the integrity of the dimerization interface of p8 is critical for TFIIH transcription activities. We identified two compounds that target the dimerization interface of p8. One of them, compound 19, dramatically destabilizes the subunit and promotes partial protein unfolding and aggregation. As for compound 12, it is able to destabilize the homodimer, which probably leads to an equilibrium between monomeric species and dimers in solution. Interestingly, most of the chemical shift changes caused by binding of this compound to p8 involve residues located at the protein–protein interface that were previously shown to be affected when the conserved Phe44 was substituted into alanine, as found in TTD-A patients. Previous 15N experiments suggested that this mutation shifts the p8 population toward monomeric species in solution (7).
Our results indicate that p8 in its monomeric form is less thermostable in solution compared with dimeric species (an unfolding temperature of 40 °C indicates that at physiological temperature, about half of the protein is folded and half denatured). We previously reported that p8 is present in two distinct kinetic pools and navigates between a free form and a fraction bound to TFIIH (6), where it interacts with p52 and XPB subunits (5). The dimerization interface of p8 would serve as a scaffold to exchange partners during homo- to heterodimerization of p8 for its incorporation into TFIIH (7, 10, 11). Furthermore, we demonstrated the interaction between p8 and p52 in vivo using a tripartite split-GFP system (11), showing that the structural integrity of p8 is essential for its interaction with p52 and consequently for TFIIH activities. It is therefore conceivable that by thermally destabilizing p8 and by shifting the equilibrium toward monomeric species, less stable in solution, compounds 12 and 19 may shorten the lifetime of p8 in vivo and alter its recognition with p52 and consequently its incorporation into TFIIH. More drastically, compound 19 promotes important protein denaturation, probably resulting in low concentration of folded and active p8 protein. Considering the structural mimicry between p8 homodimer and p8/p52C heterodimer, the two promising compounds may also affect the stability of p8/p52C heterodimer.
Compounds 12 and 19 turned out to give a decrease of TFIIH concentration levels of 30–50% in vivo. Similar reduction in steady-state levels of TFIIH was observed in cultured cells from TTD-A patients (4, 12). Furthermore, our results indicated that after 10 days of treatment, transcriptional activity was reduced with a more pronounced effect when cells were treated with compound 19 (∼30%). This finding shows that these two compounds are able to promote significant reduction in transcriptional activity, as it is observed in the cells lacking p8 or in TTD-A cells (12, 13). Compound 12 (2-phenylhydroquinone) is a metabolite of ortho-phenylphenol by cytochrome P450 that was shown to induce cell cycle arrest and apoptosis (44, 45). The mechanism of its carcinogenic effect is unknown, and no link with TFIIH and transcription complex has been described previously for this compound.
Compound 19 was shown in vitro to bind specifically to p8 and to form a covalent adduct at long incubation time. Considering that the dissociation constant of the noncovalent complex is well above the in vivo concentrations, the activity in vivo may be due to the covalent adduct. This covalent adduct is formed specifically on Cys14 (it should be noted that none of the lysine residues reacted with compound 19, Figs. S3 and S4). This reaction was observed (both by NMR and MS) to destabilize p8 dimer and cause its precipitation in vitro. Both mechanisms, covalent and noncovalent binding, lead to the same effect, the reduction of functional TFIIH concentration. Furthermore, it is likely that compound 12 exerts its in vivo activity after oxidation into compound 19. In a way, compound 12 could be described as a “prodrug” capable of slowly delivering the oxidized product, compound 19, acting as killer of p8 in vivo.
Conclusion
In conclusion, using a strategy based on virtual, biophysical, and NMR screening, we have identified two compounds that bind to and may react with the dimerization motif of p8, provoking destabilization of its protein–protein interface and its precipitation. We have previously shown that this subunit of the basal transcription factor TFIIH plays a key role in the steady-state level of TFIIH in vivo. In the present work, using a knock-in mouse model (with endogenous expression of a fluorescent version of TFIIH), we validated the potential of these two fragments to modulate the steady-state level of TFIIH and the cellular basal transcription. These compounds have helped to reveal an original mechanism of action.
In a more general context where tumor cells require high levels of transcription for proliferation and survival, our work demonstrates the potential of small molecules in targeting protein–protein interactions that are critical for basal transcription machinery, opening new perspectives to design modulators of TFIIH-associated transcription. Moreover, our study offers another subunit of TFIIH besides Cdk7 and XPD to be considered as an alternative pharmacological target in cancer treatment.
Experimental procedures
Human p8 was expressed in Escherichia coli with a TEV-cleavable histidine tag in its N terminus. Cells were grown either in Luria-Bertani (LB) broth medium or in minimal (M9) medium containing 15NH4Cl. The His tag fusion protein was purified in the first step of affinity chromatography using a nickel-nitrilotriacetic acid column (HiTrap, Amersham Biosciences) followed by dialysis overnight against a buffer consisting of 50 mm Tris-HCl, 150 mm NaCl, 0.25 mm TCEP at pH 7.5 (gel filtration buffer). The protein was further purified on a Sephadex size-exclusion chromatography column (HiLoad 16/60 Superdex 75; Amersham Biosciences) equilibrated with the gel filtration buffer. Cleavage of the histidine tag was performed using TEV protease (AcTEV, Life Technologies, Inc.) for 3 h at room temperature. The cleaved sample was applied on a nickel-nitrilotriacetic acid column, and the protein of interest was collected in the flowthrough and concentrated in a final buffer using VivaspinTM ultrafiltration columns (GE Healthcare). The protein concentration was determined by measuring the absorbance at 280 nm using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and an ϵ value of 1490 m−1 cm−1.
MD simulations were carried out with the program Amber12 and using the all-atom ff03 force field in explicit solvent, represented by the TIP3P water model. A representative structure of the ensemble of NMR structures for the p8 homodimer (7) was used as the initial structure (PDB code 2JNJ) as well as the yeast Tfb5–Tfb2C (p8/p52C) crystal structure (PDB code 3DOM). The tleap module of AMBER12 software (46) was used to create a periodic box containing the complex, six Na+ ions, and 25,134 water molecules. The system minimization and the molecular dynamics simulations were performed with the parallel version of the PMEMD module of AMBER12, as described previously (16). A total of 20 ns of MD were run at 300 K at constant pressure of 1 bar with the atomic coordinates saved every 1 ps. All of the calculations required over 56 CPU hours on an Intel Xeon E5540 at 2.53 GHz.
The fragment libraries were docked against the representative NMR structure of human p8 and the crystallographic structure of yeast Tfb5–Tfb2C. The docking calculations were performed using Autodock Vina version 1.1.2 (47). The pdbqt coordinates of the proteins were built with the addition of explicit polar hydrogen and Gasteiger partial charges. The grid module of AutoDockTools was used to define the docking active site, by considering either the full structure or the residues located at the interface. The docking studies were performed on human p8, employing either a grid box with sizes of 74, 46, and 50 Å in the x, y, and z dimensions for the full protein or a smaller grid (28, 52, and 32 Å) restricted to the residues at the interface between the two monomers. The same procedure was used to perform docking against the p8–p52 complex, with a grid box of dimensions set to 54, 52, and 58 Å in x, y, and z dimensions, respectively. For the interface, a grid box was built with x, y, and z sizes of 13, 15, and 4 Å. The pdbqt files of the fragments were translated into three-dimensional coordinates using the Open Babel version 2.2.3 program (48). The PDB files of each ligand were prepared, following the procedure described previously (16). Several fragment libraries were filtered according to physical and chemical criteria such as molecular mass (<250 Da), clogP (<2.5), and number of hydrogen bond donors (<2) while removing non-lead-like compounds and selected. Following these criteria, 678 molecules of the Zenobia Therapeutics fragment library, 1500 compounds of the Enamine Golden fragment library, and 978 in-house molecules were used for virtual screening. For each calculation, 10 resulting docking poses were ranked according to the scoring function of Autodock Vina (47) and visualized with PyMOL version 0.99rc6 (49). Chemical structures were analyzed with MarvinSketch from ChemAxon (Marvin version 6.1.5) and drawn using Chemwindow.
DSF experiments were carried out using a CFX96 real-time PCR detection system (Bio-Rad), and the fluorescence of the Sypro®Orange dye (Invitrogen Life Technology) as a function of temperature was recorded using the HEX channel (excitation, 450–490 nm; emission, 560/580 nm). The samples were submitted to a temperature ramp from 20 to 95 °C with a heating rate of 0.3 °C/min and 0.5 °C increments. Data were analyzed using the CFX ManagerTM software (Bio-Rad). A total of four measurements were recorded for each condition. The Tm value was estimated from the transition midpoint of the fluorescence curve. The stability of p8 was monitored in different buffers distributed in 96-well plates (200 mm acetate (pH 5.5), 200 mm MES (pH 6.0 and pH 6.5), 200 mm phosphate (pH 6.5 and pH 7.0), 200 mm HEPES (pH 7.5 and pH 8.0), and 200 mm Tris (pH 8.0)). For each buffer, six salt conditions were tested (0 mm NaCl, 50 mm NaCl, 100 mm NaCl, 200 mm NaCl, 50 mm arginine/glutamic acid, and 50 mm Na2SO4). To perform the DSF screening, 10 μl of protein and 10 μl of stock buffer solution were mixed before the addition of 2 μl of Sypro®Orange (final dilution 1:1000).
To monitor the binding of compounds to p8, DSF was used where each well contained protein (70–90 μm in gel filtration buffer), ligand (from 4 to 1000 μm; 1% (v/v) final DMSO), and Sypro®Orange (1:1000 final dilution). The control experiments were performed with buffer instead of protein solution in the same conditions. For each protein/ligand ratio, duplicates were performed, and each titration was repeated at least three times.
The p8 (histidine tag–cleaved) protein sample buffered in 50 mm Tris, pH 7.4, 150 mm NaCl, and 0.25 mm TCEP was loaded on a Superdex 200 Increase 5/150 column (GE Healthcare) using an Agilent 1260 Infinity LC chromatographic system (Agilent Technology) coupled to a multiangle light scattering (MALS) detection system. The column was equilibrated with a 0.1-mm filtered 50 mm Tris buffer, pH 7.5, 150 mm NaCl, 0.5 mm TCEP buffer. Data were collected using a DAWN HELEOS 8+ (8-angle) and Optilab T-rEX refractive index detector (Wyatt Technology). 100 μl of protein sample at 140 μm were loaded on the column, and the separation was performed at a flow rate of 0.4 ml/min at 15 °C. Results were analyzed using ASTRA version 6.1 software (Wyatt Technology).
All NMR experiments were recorded at 298 K with 3-mm NMR tubes on a Bruker Avance 600-MHz spectrometer (Wissembourg, France) equipped with a proton-optimized triple resonance cryoprobe. Purity and solubility of selected compounds were checked using NMR. The ligands were first dissolved in deuterated DMSO (DMSO-d6) at 100 mm concentration to prepare stock solutions before dilution to 1 mm in the NMR buffer (unless reduced solubility required us to decrease ligand concentration). The final DMSO-d6 concentration in the NMR buffer (50 mm phosphate, 150 mm NaCl, pH 6.9) was 1%. To enable a quantitative determination of ligand concentration and solubility, one-dimensional NMR spectra were recorded in NMR buffer containing DSS as NMR reference at a concentration of 100 μm. Signal intensity for each ligand was compared with that of DSS.
The ligand-based STD experiments (20) were performed with 10 μm protein and a 1 mm concentration for ligand. On-resonance and off-resonance irradiation frequencies were set to −104 Hz (−0.17 ppm) and 18,000 Hz (30 ppm), respectively, using selective 50-ms Gaussian-shaped pulses at a radiofrequency field amplitude of 86 Hz with a 100-ms delay between each pulse. The total duration of the saturation time was 2 s. A watergate sequence was used to suppress the residual water signal (50).
To determine the dissociation constant, STD experiments were performed using a p8 concentration of 10 μm and increasing concentrations of compound 12 from 250 μm to 4 mm. The STD factor, defined as ASTD = (I0 − Isat)/I0 = ISTD/I0 (where I0 is the intensity of the signal in the reference experiment and Isat is the intensity of the same signal in the saturated spectrum), was determined for individual protons.
Protein-based 2D 1H-15N NMR HSQC experiments were recorded on a 0.4 mm 15N-labeled p8 sample with 256 points in indirect dimension and eight scans. CSP experiments were performed by recording a series of 2D 1H-15N HSQC experiments of 0.4 mm p8 and increasing concentrations of ligands (0.25, 0.75, 1.0, 2.0, 3.0, and 4.0 mm). To ensure that the NMR chemical shift changes were not due to the introduction of DMSO, a similar titration was performed by adding increasing concentrations of DMSO-d6, from 0 to 10% (v/v), to the solution of p8. Significant chemical shift changes were only observed for DMSO concentrations exceeding 5% (v/v), which is beyond the final concentration of DMSO present in the protein sample at the end of titration (<2%). Overall CSP experiments were calculated from the 15N and HN chemical shift changes according to the following formula,
| (Eq. 1) |
All data were acquired and processed using topspin version 3.5 software (Bruker Biospin). Overall rotational correlation times of the proteins were estimated using 15N,1H TRACT experiments that measure the difference between the two relaxation rates for α and β spin states (17).
Nano-LC-MS and MSMS analyses of intact p8 were performed on a nanoRS UHPLC system (Dionex) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). After incubation of p8 with compounds 12 or 19, samples were diluted to 1 μm in 2% acetonitrile (ACN), 0.2 formic acid (FA), and 5 μl were loaded on a C4-precolumn (300-μm inner diameter × 5 mm, Thermo Fisher Scientific) at 20 μl/min in 5% ACN, 0.05% TFA. After a 5-min desalting, the precolumn was switched online with the analytical C4 nanocolumn (75-μm inner diameter × 15 cm, in-house packed with C4 Reprosil) equilibrated in 95% solvent A (5% ACN, 0.2% FA) and 5% solvent B (0.2% FA in ACN). Proteins were eluted using the following gradient of solvent B at a 300-nl/min flow rate: 5–40% during 0.5 min; 40–99% during 7.5 min, followed by 6 min at 99% and 2 min at 5%. The LTQ-Orbitrap Velos was operated either in single MS or in data-dependent acquisition mode with the XCalibur software. MS scans were acquired in the 500–2000 m/z range with the resolution set to a value of 60,000. For top-down experiments, survey scan MS were acquired in the same way, with a resolution at 60,000. Precursor ions were selected from an inclusion list established thanks to previous MS analyses and were fragmented by collision-induced dissociation or higher-energy collisional dissociation, and the resulting fragment ions were analyzed in the Orbitrap, at a resolution of 60,000. Isolation width was set at 5 m/z, and normalized collisional energy was set at a value of 35% for collision-induced dissociation fragmentation and 30% for higher-energy collisional dissociation. MS spectra were deconvoluted with the Xtract algorithm, and MSMS spectra were analyzed manually.
Mouse embryonic fibroblasts were isolated from the XPB-YFP+f/+f and XPB-YFP+f/+f TTDA−/− mouse models were isolated at 10.5 days post-coitus and cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F10 (Lonza) with 10% fetal calf serum and 1% penicillin-streptomycin (Gibco) at 37 °C, 3% O2, and 5% CO2. Cells were treated with compounds 10 (mock), 12, and 19 for 1–6 days with doses ranging from 1 to 3 μm (higher concentrations were found to be cytotoxic). A 10-day treatment at 3 μm was chosen as an optimum for effect size.
ES cells and chondrocytes were isolated as described previously (25). Cells were treated with protein synthesis inhibitor cycloheximide (133 μg/ml) for 24, 48, and 72 h. Proteins were separated on SDS-PAGE composed of bisacrylamide (37.5:1), blotted onto a polyvinylidene difluoride membrane (0.45 μm, Millipore), and analyzed using the following primary antibody: anti-HA 3F10 (rat IgG1, Roche Applied Science). A fixed number of cells (200,000) were loaded in each well. Western blotting was performed as described previously (51), and protein bands were visualized via ECL (enhanced chemoluminescence; Pierce ECL Western Blotting Substrate) using horseradish peroxidase–conjugated secondary antibodies.
Quantification of fluorescence levels followed a procedure similar to that described previously (52). In brief, cells expressing XPB-YFP were imaged using a Zeiss LSM 710 confocal microscope with a ×40, 1.3 numerical aperture oil immersion objective. All images were obtained at constant acquisition parameters so as to preserve relative fluorescent quantities within a given set of experiments. The resulting images were analyzed using the ImageJ program (53) as follows; after background subtraction, the total fluorescence signal present in the nucleus of individual cells was estimated and then averaged along with more than 90 cells over two or three independent experiments. The resulting data for the different conditions were then normalized to the compound 10 (mock)-treated case.
RNA detection was performed using the Click-iT RNA Alexa Fluor imaging kit (Invitrogen), according to the manufacturer's instructions. Briefly, cells were treated with the different drugs for 6 days and incubated for 16 h at 37 °C, 3% O2, and 5% CO2. Then cells were incubated for 2 h with 5-ethynyl uridine, fixed, and permeabilized. Cells were incubated for 30 min with the Click-iT reaction mixture containing Alexa Fluor Azide 594. After washing, the coverslips were mounted with Vectashield (Vector). Images of the cells were obtained with an LSM 710 Zeiss confocal microscope and constant acquisition parameters, and then the average fluorescence intensity per nucleus was estimated after background subtraction (ImageJ) and normalized to the mock-treated cells. For each sample, at least 100 nuclei were analyzed from three independent experiments.
Author contributions
V. Gervais, A. Milon, and G. G.-M. conceptualization; V. Gervais, I. M., A. Mourcet, K. T. M., J. M., and V. Guillet formal analysis; V. Gervais, P. R., O. B.-S., G. C., A. Milon, and G. G.-M. supervision; V. Gervais, I. M., P.-O. M., S. J., and G. G.-M. validation; V. Gervais investigation; V. Gervais, I. M., P.-O. M., A. Mourcet, K. T. M., and S. J. visualization; V. Gervais, I. M., P.-O. M., P. R., J. M., V. Guillet, and G. C. methodology; V. Gervais writing-original draft; V. Gervais writing-review and editing; O. B.-S. and G. G.-M. funding acquisition; A. Milon project administration.
Supplementary Material
Acknowledgments
We acknowledge the use of PICT-Genotoul-Ibisa facility, funded by Ibisa, CNRS, European structural funds, and the Région Midi-Pyrénées and the use of the imaging facility CIQLE (Centre d'imagerie quantitative Lyon-Est, Université Claude Bernard Lyon 1, Lyon, France). We are grateful to M. Trivedi and K. Rottier for input at the beginning of the project, O. Saurel for NMR support, and L. Mourey for insightful comments during the study. We acknowledge B. Brutscher for support with TRACT experiments, and we thank B. Kieffer and A. Poterszman for stimulating discussions and the generous gift of plasmids.
This work was funded in part by the RITC Foundation and in part by French Ministry of Research (Investissement d'Avenir Program, Proteomics French Infrastructure) Grant ANR-10-INBS-08 (to O. B.-S.), the FEDER (Fonds Européens de développement Régional), Toulouse Métropole, and the Région Midi-Pyrénées. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting information and Figs. S1–S8.
- TTD-A
- trichothiodystrophy group A
- MD
- molecular dynamics
- DSF
- differential scanning fluorimetry
- TRACT
- TROSY for rotational correlation times
- τc
- rotational correlation time
- SEC
- size-exclusion chromatography
- MALS
- multiangle light scattering
- STD
- saturation transfer difference
- ES cells
- embryonic stem cells
- PPI
- protein–protein interaction
- 2D
- two-dimensional
- CSP
- chemical shift perturbation
- HSQC
- heteronuclear single quantum coherence
- TEV
- tobacco etch virus
- TCEP
- tris(2-carboxyethyl)phosphine
- PDB
- Protein Data Bank
- ACN
- acetonitrile
- FA
- formic acid
- YFP
- yellow fluorescent protein.
References
- 1. Luo J., Cimermancic P., Viswanath S., Ebmeier C. C., Kim B., Dehecq M., Raman V., Greenberg C. H., Pellarin R., Sali A., Taatjes D. J., Hahn S., and Ranish J. (2015) Architecture of the human and yeast general transcription and DNA repair factor TFIIH. Mol. Cell 59, 794–806 10.1016/j.molcel.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greber B. J., Nguyen T. H. D., Fang J., Afonine P. V., Adams P. D., and Nogales E. (2017) The cryo-electron microscopy structure of human transcription factor IIH. Nature 549, 414–417 10.1038/nature23903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Theil A. F., Nonnekens J., Wijgers N., Vermeulen W., and Giglia-Mari G. (2011) Slowly progressing nucleotide excision repair in trichothiodystrophy group A patient fibroblasts. Mol. Cell Biol. 31, 3630–3638 10.1128/MCB.01462-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giglia-Mari G., Coin F., Ranish J. A., Hoogstraten D., Theil A., Wijgers N., Jaspers N. G. J., Raams A., Argentini M., van der Spek P. J., Botta E., Stefanini M., Egly J. M., Aebersold R., Hoeijmakers J. H. J., and Vermeulen W. (2004) A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36, 714–719 10.1038/ng1387 [DOI] [PubMed] [Google Scholar]
- 5. Coin F., Proietti De Santis L., Nardo T., Zlobinskaya O., Stefanini M., and Egly J. M. (2006) P8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell 21, 215–226 10.1016/j.molcel.2005.10.024 [DOI] [PubMed] [Google Scholar]
- 6. Giglia-Mari G., Miquel C., Theil A. F., Mari P. O., Hoogstraten D., Ng J. M. Y., Dinant C., Hoeijmakers J. H. J., and Vermeulen W. (2006) Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 4, e156 10.1371/journal.pbio.0040156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vitorino M., Coin F., Zlobinskaya O., Atkinson R. A., Moras D., Egly J. M., Poterszman A., and Kieffer B. (2007) Solution structure and self-association properties of the p8 TFIIH subunit responsible for trichothiodystrophy. J. Mol. Biol. 368, 473–480 10.1016/j.jmb.2007.02.020 [DOI] [PubMed] [Google Scholar]
- 8. Kainov D. E., Cura V., Vitorino M., Nierengarten H., Poussin P., Kieffer B., Cavarelli J., and Poterszman A. (2010) Structure determination of the minimal complex between Tfb5 and Tfb2, two subunits of the yeast transcription/DNA-repair factor TFIIH: a retrospective study. Acta Crystallogr. D Biol. Crystallogr. 66, 745–755 10.1107/S0907444910009844 [DOI] [PubMed] [Google Scholar]
- 9. Hashimoto S., and Egly J. M. (2009) Trichothiodystrophy view from the molecular basis of DNA repair/transcription factor TFIIH. Hum. Mol. Genet. 18, R224–R230 10.1093/hmg/ddp390 [DOI] [PubMed] [Google Scholar]
- 10. Kainov D. E., Vitorino M., Cavarelli J., Poterszman A., and Egly J. M. (2008) Structural basis for group A trichothiodystrophy. Nat. Struct. Mol. Biol. 15, 980–984 10.1038/nsmb.1478 [DOI] [PubMed] [Google Scholar]
- 11. Nonnekens J., Cabantous S., Slingerland J., Mari P. O., and Giglia-Mari G. (2013) In vivo interactions of TTDA mutant proteins within TFIIH. J. Cell Sci. 126, 3278–3283 10.1242/jcs.126839 [DOI] [PubMed] [Google Scholar]
- 12. Vermeulen W., Bergmann E., Auriol J., Rademakers S., Frit P., Appeldoorn E., Hoeijmakers J. H. J., and Egly J. M. (2000) Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet. 26, 307–313 10.1038/81603 [DOI] [PubMed] [Google Scholar]
- 13. Theil A. F., Nonnekens J., Steurer B., Mari P. O., de Wit J., Lemaitre C., Marteijn J. A., Raams A., Maas A., Vermeij M., Essers J., Hoeijmakers J. H., Giglia-Mari G., and Vermeulen W. (2013) Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality. PLoS Genet. 9, e1003431 10.1371/journal.pgen.1003431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nonnekens J., Perez-Fernandez J., Theil A. F., Gadal O., Bonnart C., and Giglia-Mari G. (2013) Mutations in TFIIH causing trichothiodystrophy are responsible for defects in ribosomal RNA production and processing. Hum. Mol. Genet. 22, 2881–2893 10.1093/hmg/ddt143 [DOI] [PubMed] [Google Scholar]
- 15. Mapp A. K., Pricer R., and Sturlis S. (2015) Targeting transcription is no longer a quixotic quest. Nat. Chem. Biol. 11, 891–894 10.1038/nchembio.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cala O., Remy M. H., Guillet V., Merdes A., Mourey L., Milon A., and Czaplicki G. (2013) Virtual and biophysical screening targeting the γ-tubulin complex: a new target for the inhibition of microtubule nucleation. PLoS One 8, e63908 10.1371/journal.pone.0063908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee D., Hilty C., Wider G., and Wüthrich K. (2006) Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 178, 72–76 10.1016/j.jmr.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 18. Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 10.1038/nprot.2007.321 [DOI] [PubMed] [Google Scholar]
- 19. Seetoh W. G., and Abell C. (2016) Disrupting the constitutive, homodimeric protein-protein interface in CK2β using a biophysical fragment-based approach. J. Am. Chem. Soc. 138, 14303–14311 10.1021/jacs.6b07440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mayer M., and Meyer B. (2001) Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 10.1021/ja0100120 [DOI] [PubMed] [Google Scholar]
- 21. Alekseev S., Ayadi M., Brino L., Egly J. M., Larsen A. K., and Coin F. (2014) A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem. Biol. 21, 398–407 10.1016/j.chembiol.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 22. Baell J. B., and Holloway G. A. (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- 23. Dahlin J. L., and Walters M. A. (2016) How to triage PAINS-full research. Assay Drug Dev. Technol. 14, 168–174 10.1089/adt.2015.674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W. W., Heinze J., and Haehnel W. (2005) Site-specific binding of quinones to proteins through thiol addition and addition-elimination reactions. J. Am. Chem. Soc. 127, 6140–6141 10.1021/ja050974x [DOI] [PubMed] [Google Scholar]
- 25. Giglia-Mari G., Theil A. F., Mari P. O., Mourgues S., Nonnekens J., Andrieux L. O., de Wit J., Miquel C., Wijgers N., Maas A., Fousteri M., Hoeijmakers J. H. J., and Vermeulen W. (2009) Differentiation driven changes in the dynamic organization of basal transcription initiation. PLoS Biol. 7, e1000220 10.1371/journal.pbio.1000220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoogstraten D., Nigg A. L., Heath H., Mullenders L. H. F., van Driel R., Hoeijmakers J. H. J., Vermeulen W., and Houtsmuller A. B. (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10, 1163–1174 10.1016/S1097-2765(02)00709-8 [DOI] [PubMed] [Google Scholar]
- 27. Daragan V. A., and Mayo K. H. (1997) Motional model analyses of protein and peptide dynamics using C-13 and N-15 NMR relaxation. Prog. Nucl. Magn. Reson. Spectrosc. 31, 63–105 10.1016/S0079-6565(97)00006-X [DOI] [Google Scholar]
- 28. Villicaña C., Cruz G., and Zurita M. (2014) The basal transcription machinery as a target for cancer therapy. Cancer Cell Int. 14, 18 10.1186/1475-2867-14-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Darnell J. E., Jr. (2002) Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2, 740–749 10.1038/nrc906 [DOI] [PubMed] [Google Scholar]
- 30. Johnston S. J., and Carroll J. S. (2015) Transcription factors and chromatin proteins as therapeutic targets in cancer. Biochim. Biophys. Acta 1855, 183–192 [DOI] [PubMed] [Google Scholar]
- 31. Hagenbuchner J., and Ausserlechner M. J. (2016) Targeting transcription factors by small compounds: current strategies and future implications. Biochem. Pharmacol. 107, 1–13 10.1016/j.bcp.2015.12.006 [DOI] [PubMed] [Google Scholar]
- 32. Zurita M., and Cruz-Becerra G. (2016) TFIIH: new discoveries regarding its mechanisms and impact on cancer treatment. J. Cancer 7, 2258–2265 10.7150/jca.16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray C. W., and Blundell T. L. (2010) Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 20, 497–507 10.1016/j.sbi.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 34. Hajduk P. J., and Greer J. (2007) A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 10.1038/nrd2220 [DOI] [PubMed] [Google Scholar]
- 35. Scott D. E., Coyne A. G., Hudson S. A., and Abell C. (2012) Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51, 4990–5003 10.1021/bi3005126 [DOI] [PubMed] [Google Scholar]
- 36. Mullard A. (2012) Protein-protein interaction inhibitors get into the groove. Nat. Rev. Drug Discov. 11, 173–175 10.1038/nrd3680 [DOI] [PubMed] [Google Scholar]
- 37. Scott D. E., Ehebauer M. T., Pukala T., Marsh M., Blundell T. L., Venkitaraman A. R., Abell C., and Hyvönen M. (2013) Using a fragment-based approach to target protein-protein interactions. Chembiochem 14, 332–342 10.1002/cbic.201200521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scott D. E., Bayly A. R., Abell C., and Skidmore J. (2016) Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550 10.1038/nrd.2016.29 [DOI] [PubMed] [Google Scholar]
- 39. Murray C. W., and Rees D. C. (2009) The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192 10.1038/nchem.217 [DOI] [PubMed] [Google Scholar]
- 40. Sheng C., Dong G., Miao Z., Zhang W., and Wang W. (2015) Correction: state-of-the-art strategies for targeting protein-protein interactions by small-molecule inhibitors. Chem. Soc. Rev. 44, 8375 10.1039/C5CS90090E [DOI] [PubMed] [Google Scholar]
- 41. Dalvit C. (2009) NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov. Today 14, 1051–1057 10.1016/j.drudis.2009.07.013 [DOI] [PubMed] [Google Scholar]
- 42. Silvestre H. L., Blundell T. L., Abell C., and Ciulli A. (2013) Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery. Proc. Natl. Acad. Sci. U.S.A. 110, 12984–12989 10.1073/pnas.1304045110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Campos-Olivas R. (2011) NMR screening and hit validation in fragment based drug discovery. Curr. Top. Med. Chem. 11, 43–67 10.2174/156802611793611887 [DOI] [PubMed] [Google Scholar]
- 44. Nakata Y., Nishi K., Nishimoto S., and Sugahara T. (2013) Phenylhydroquinone induces loss of thymocytes through cell cycle arrest and apoptosis elevation in p53-dependent pathway. J. Toxicol. Sci. 38, 325–335 10.2131/jts.38.325 [DOI] [PubMed] [Google Scholar]
- 45. Imai M., Matsuno R., Komura J., Ono T., and Yamamoto K. (2009) Induction of mitosis delay, apoptosis and aneuploidy in human cells by phenyl hydroquinone, an Ames test-negative carcinogen. Genes Genet. Syst. 84, 73–79 10.1266/ggs.84.73 [DOI] [PubMed] [Google Scholar]
- 46. Case D. A., Darden T. A., Cheatham T. E., Simmerling I. C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., et al. (2012) AMBER 12, University of California, San Francisco [Google Scholar]
- 47. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., and Hutchison G. R. (2011) Open Babel: an open chemical toolbox. J. Cheminform. 3, 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Delano W. (2002) The PyMOL Molecular Graphics System, version 0.99rc6, Schroedinger LLC, New York [Google Scholar]
- 50. Piotto M., Saudek V., and Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 10.1007/BF02192855 [DOI] [PubMed] [Google Scholar]
- 51. Rockx D. A., Mason R., van Hoffen A., Barton M. C., Citterio E., Bregman D. B., van Zeeland A. A., Vrieling H., and Mullenders L. H. (2000) UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 97, 10503–10508 10.1073/pnas.180169797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mourgues S., Gautier V., Lagarou A., Bordier C., Mourcet A., Slingerland J., Kaddoum L., Coin F., Vermeulen W., Gonzales de Peredo A., Monsarrat B., Mari P. O., and Giglia-Mari G. (2013) ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc. Natl. Acad. Sci. U.S.A. 110, 17927–17932 10.1073/pnas.1305009110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.