Significance
Efficient synthesis and folding of proteins, avoiding misfolded states, are central to cell function. As folding may be initiated in parallel with translation, key experimental challenges are to map changes that occur in folding free energy landscapes as translation proceeds and to understand how these landscapes might be modulated by the ribosome and auxiliary factors. Here, we study the length-dependent folding of a domain from a tandem repeat protein and solve the structure of a stable folding intermediate. Although destabilized by the ribosome at equilibrium, modeling of the nonequilibrium folding pathway nevertheless indicates a significant role for proline isomerization during translation. We develop a simple model to explore the impact of cotranslational folding kinetics on misfolding hazards.
Keywords: cotranslational folding, protein misfolding, nuclear magnetic resonance, molecular dynamics simulations, tandem repeat protein
Abstract
Cotranslational folding (CTF) is a fundamental molecular process that ensures efficient protein biosynthesis and minimizes the formation of misfolded states. However, the complexity of this process makes it extremely challenging to obtain structural characterizations of CTF pathways. Here, we correlate observations of translationally arrested nascent chains with those of a systematic C-terminal truncation strategy. We create a detailed description of chain length-dependent free energy landscapes associated with folding of the FLN5 filamin domain, in isolation and on the ribosome, and thus, quantify a substantial destabilization of the native structure on the ribosome. We identify and characterize two folding intermediates formed in isolation, including a partially folded intermediate associated with the isomerization of a conserved cis proline residue. The slow folding associated with this process raises the prospect that neighboring unfolded domains might accumulate and misfold during biosynthesis. We develop a simple model to quantify the risk of misfolding in this situation and show that catalysis of folding by peptidyl-prolyl isomerases is sufficient to eliminate this hazard.
The misfolding and aggregation of proteins are implicated in a wide range of human disorders (1). It is, therefore, essential for cells to have mechanisms to ensure quality control in protein synthesis and maintain protein homeostasis (2). Increasing evidence indicates that cotranslational folding (CTF) can increase the efficiency of folding compared with the folding of full-length (FL) proteins in vitro (3), in some cases modulating the folding process itself (4, 5). In this work, we focus on the folding of multidomain proteins, which account for 70% of eukaryotic proteins, many of which are tandem repeat proteins (TRPs; i.e., are composed of multiple copies of similar domains) (6). Such sequences are at high risk of forming nonnative interdomain contacts during the folding process, potentially leading to misfolded states (7, 8), and therefore, a strong selective pressure exists to minimize the homology between adjacent domains (9).
A major obstacle to the folding of multidomain proteins is the exponential growth in the volume of conformational space that must be explored as the polypeptide chain length increases. CTF is widely believed to play a major role in the solution of this problem, as the sequential folding of emerging domains restricts the volume of conformational space, thus promoting rapid folding of the entire protein to its native state (while also minimizing the risk of interdomain misfolding). Therefore, in the context of the free energy landscape theory of protein folding, to study CTF, one should consider not a single free energy landscape but a series of nested landscapes of increasing dimensionality and polypeptide chain lengths (10). It is our goal here to map out these “cotranslational landscapes” to improve our understanding of the mechanisms by which the ribosome may contribute to efficient folding.
We have developed NMR spectroscopy as a tool for the observation of CTF in translationally arrested ribosome–nascent chain complexes (RNCs) (11, 12), in particular to investigate the emergence and folding of a pair of IgG-like domains, FLN5–6, from the TRP filamin (FLN) (Fig. 1 A and B) (5). Folding of FLN5 to the native state was observed, and a chemical shift-restrained ensemble structure was determined at a linker length (comprising the subsequent FLN6 domain) well beyond the point at which the entire domain was solvent exposed as probed by PEG mass-tagging assays.
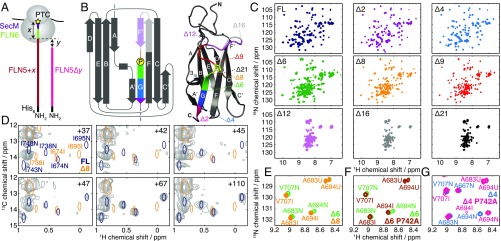
Fig. 1.
(A) Schematic and nomenclature of FLN5 RNC (FLN5 + ) and truncation (FLN5) constructs; and represent the FLN6 + secM linker length (measured from the PTC), and the extent of truncation, respectively. (B) Topology and crystal structure of FLN5 (1qfh) colored and labeled to indicate the truncated constructs used in this study. The cis proline P742 is highlighted in yellow (stick representation). (C) ,-selective optimized flip-angle short transient heteronuclear multiple quantum coherence (SOFAST-HMQC) spectra of FLN5 truncation variants (298 K, 600 MHz). (D) Comparison of ,-HMQC spectra of FL and 8 with spectra of [-Ile,]-labeled FLN5 + RNCs (5) (gray; 298 K, 700 MHz). (E) Comparison of ,-HSQC spectra of FLN56 and FLN58, showing a magnified view of representative residues (283 K, 700 MHz). FLN56 resonance assignments are not shown where overlapping with FLN58 assignments. (F) Comparison of ,-HSQC spectra of FLN56 and FLN56(P742A) (283 K, 700 MHz). (G) Comparison of ,-HSQC spectra of FLN54 and FLN54(P742A) (283 K, 700 MHz).
In this work, using a C-terminal truncation system (13–15), we have conducted a systematic exploration of the thermodynamics and kinetics of folding in isolation to create a detailed description of the free energy landscape for CTF, from which the remarkably strong destabilizing effect of the ribosome on the native structure may be quantified. We also identify and determine the structure of an isolated intermediate associated with the isomerization of a conserved native-state cis proline, and we use this model to explore and eliminate the possible role of steric occlusion in perturbing the folding process on the ribosome. Lastly, we apply these data to a simple model to analyze the effect of folding kinetics and proline isomerization on the risk of misfolding in this multidomain protein.
Results
Variants of the FL FLN5 domain were designed to mimic CTF by truncating between 2 and 21 residues from the C terminus, denoted 2 to 21 (Fig. 1 A and B). Native PAGE showed 12 and 16 to have expanded conformations relative to FL, while the migration of 6 to 9 was approximately midway between these states (SI Appendix, Fig. S1A). A transition from -sheet to random coil structure was also observed at these lengths by far-UV CD spectroscopy (SI Appendix, Fig. S1 B and C).
To investigate the length dependence of folding further, 2D NMR correlation spectra of truncation variants were acquired (Fig. 1C and SI Appendix, Fig. S1D). The large chemical shift dispersion observed in FL 2 and 4 spectra indicates the presence of stable tertiary structure, while the narrow dispersion observed for 12, 16, and 21 is characteristic of a disordered state. At intermediate lengths (6, 8, and 9), multiple sets of cross-peaks were observed, indicating that both structured and unstructured states are populated in equilibrium. Two states, designated “I” (intermediate) and “U” (unfolded) were populated by 8 and 9, while an additional set of folded resonances [“N” (native like)] was observed in the 6 spectrum (Fig. 1E and SI Appendix, Fig. S2).
We hypothesized that the two sets of folded 6 resonances arise from cis/trans isomerization of the native-state cis proline P742 (Fig. 1B). To test this conjecture, we examined the 6 P742A variant, in which P742 was mutated to favor the nonnative trans conformation. The additional resonances observed in the 6 spectrum were absent in spectra of 6 P742A (Fig. 1F), and the remaining resonances corresponded closely with those observed for 9, in which P742 is absent altogether. We, therefore, conclude that there is a length-dependent transition from an intermediate I, in which P742, if present, is in a trans conformation, to a (near)-native state N, in which P742 is in a cis conformation. Consistent with this conclusion, in the 4 P742 A variant (Fig. 1G), the N state was destabilized, and both I and U resonances could additionally be observed with the same chemical shifts as observed in WT 6.
Structural Characterization of Folding Pathway.
Having identified a series of states populated within isolated FLN5 fragments, we sought to analyze their structures to understand and connect their folding behavior in isolation to that occurring cotranslationally within RNCs. Progressive changes in amide chemical shifts were observed from the FL native state to the 6 I state clustered around the A and F strands adjacent to the truncated G strand (SI Appendix, Fig. S3). The magnitude of chemical shift perturbations increased with the extent of truncation up to the 6 I state, but no large chemical shift differences were observed in shorter lengths, indicating that the intermediate structure does not change significantly beyond this point.
We then used chemical shift-restrained replica-averaged metadynamics simulations (16) to determine an ensemble structure of the 6 intermediate (together, as a control, with the FL native state) (Fig. 2 A and B and SI Appendix, Fig. S4). The quality of the ensembles was comparable with those previously determined by chemical shift methods (17) assessed by back-calculated chemical shift deviations, residual dipolar coupling Q factors (SI Appendix, Table S1), and a backbone rmsd of 1.8 Å between the FL ensemble and a crystal structure (18) (Fig. 2A). We found that the FL ensemble is generally well ordered, with C root-mean-square fluctuation (RMSF) values below 0.5 Å, except in some loop regions (Fig. 2A). In contrast, in the 6 intermediate, a native-like core is retained, comprising the A–F strands, but the truncated G strand is unfolded, with increased disorder also found in the adjacent A′ and F strands (Fig. 2B). No structure could be determined for the 6 N state due to broadening of resonances around the site of the truncation. relaxation dispersion measurements (19) of adjacent residues indicate that this is due to conformational exchange on a millisecond timescale (SI Appendix, Fig. S5), which we suggest may reflect transient association of the truncated G strand with the F strand that becomes stabilized as the chain length increases.
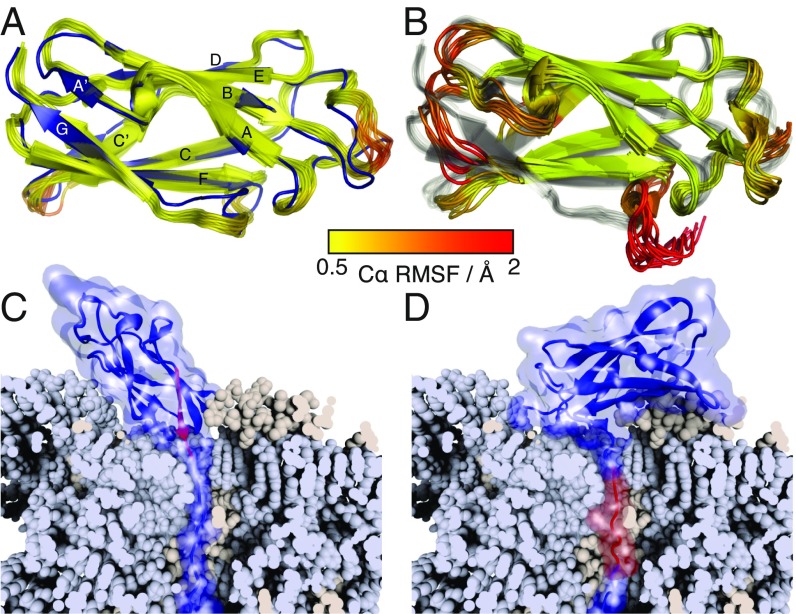
Fig. 2.
Structural analysis of FLN5 truncation variants. (A) Ensemble structure of FL FLN5 (yellow to red) aligned against the previously determined crystal structure (blue) (18). The FL ensemble is colored according to the C RMSF as indicated by the key. (B) Ensemble structure of the 6 trans intermediate (colored by the C RMSF) aligned against the FL ensemble (gray). (C and D) Modeling of the closest possible approach of (C) native and (D) intermediate FLN5 structures tethered to the ribosome (shown in a cutaway view to highlight the NC path through the exit tunnel). The G strand of FLN5 (disordered in the intermediate) is highlighted in red.
In these relaxation measurements, small dispersions ( ) were also observed in the unfolded state of 6 (SI Appendix, Fig. S6A). These data could be fitted to two-state exchange with a high-energy intermediate, populated to %, and designated “I*” (SI Appendix, Fig. S6B). Chemical shift differences were uniformly small ( ppm) for residues 646–670, but from residue 671 onward, they were correlated with those between 6 U and I states, indicating the formation of intermediate-like structure in this region. Thus, the I* intermediate comprises a native-like core formed from the B to F strands, while the A strand and the truncated G strand remain disordered (SI Appendix, Fig. S6C). The intermediate was also identified in 9, 12, and 16 variants, with a decreased population, %, in the shortest length, 16 (SI Appendix, Fig. S6 D–I).
In summary, therefore, the length-dependent folding pathway proceeds from the unfolded state via a high-energy intermediate with disordered A and G strands to a stable intermediate with a disordered G strand and P742, if present, in a trans conformation. Isomerization of P742 to cis, in either the intermediate or via the unfolded state, then allows association of the G strand to form the final native state.
If the structure of the intermediate is now considered in the context of CTF, the disordered nature of the C-terminal G strand suggests that this state should be accessible early in the translation process. To explore this, we ran simulations using structure-based models (20) with various linker lengths to study folding of the N and I states on the ribosome. Using this approach, we found that the native state requires a 20-aa linker (in an extended conformation) to fold on the ribosome, whereas only a 14-aa linker was needed for the intermediate (Fig. 2 C and D). By comparison with the linker conformation in an all-atom molecular dynamics (MD) simulation of the FLN5 + 110 RNC (5), we further estimated that these native and intermediate structures should be accessible with linker lengths (in a relaxed conformation) of ca. 26 and 18 aa, respectively (SI Appendix, Fig. S7).
The structured states of FLN5 RNCs can be observed with the greatest sensitivity using a specific isoleucine C methyl labeling strategy (Fig. 1D) (5). However, no resonances corresponding to the intermediate state were detected in RNC spectra of any linker length (although as it exists in isolation as a minor state in equilibrium with the unfolded state, it is not possible to exclude a small undetected population). Similarly, native resonances were not observed in these data (5) until a linker length of 42 aa (Fig. 1D), well beyond the 26 aa expected from our structural modeling. This indicates that there must be a specific inhibition of folding that arises from ribosomal tethering. To understand this further, we, therefore, sought to complement our structural description of the folding pathway during elongation with a detailed energetic description of the CTF landscape.
Thermodynamic Analysis.
A range of experimental techniques have been applied to create a detailed thermodynamic map of the length-dependent folding process. The stabilities of FL, 2, and 4 variants were measured by chemical denaturation (SI Appendix, Fig. S8), while in 6, 8 and 9, where multiple states were observed to be populated at equilibrium, NMR measurements of peak volumes in ,-heteronuclear single quantum correlation (HSQC) spectra were used to determine the relative populations of folded and unfolded states and hence, calculate free energies of folding directly (Fig. 3 and SI Appendix, Fig. S9). Where only the unfolded state could be observed (12 and 16), limits on the stability of the intermediate have been calculated based on a population below the ca. 1% limit of detection derived from signal-to-noise levels (dashed lines in Fig. 3). From these measurements, we found that the six C-terminal residues contribute almost the entire stability of the domain, kcal . The stability of the I state did not vary significantly between 6 and 9 ( kcal ), but in shorter lengths, the intermediate was further destabilized, with a population below the limit of detection.
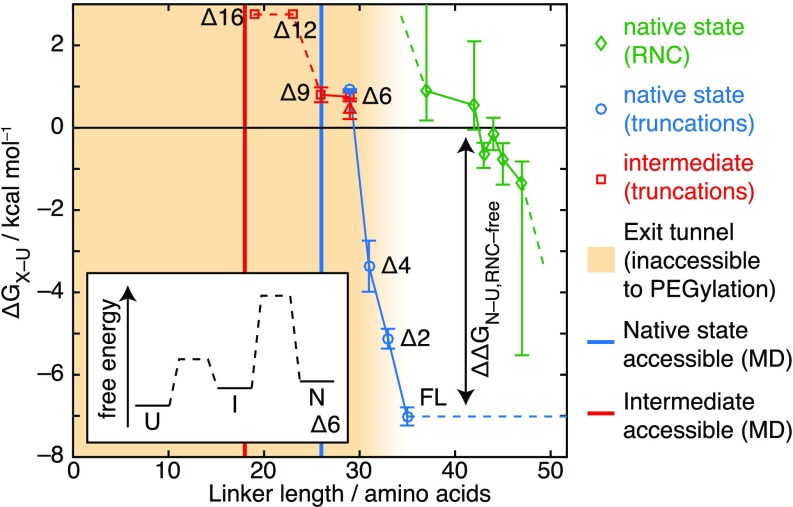
Fig. 3.
Thermodynamic characterization of the FLN5 CTF pathway. Free energies of folding to native (cis-P742) and intermediate (trans-P742) states at 298 K are shown in truncated variants and RNCs as indicated. As a specific example, the 6 folding pathway is illustrated in Inset. A red triangle indicates 6 intermediate stability as determined by magnetization transfer measurements (Fig. 4 A and B). Yellow shading indicates the extent of the ribosome exit tunnel determined by PEG accessibility (5) and used as the basis for alignment of the isolated and ribosome-associated folding pathways, while solid lines indicate the points at which native and intermediate states were found to be accessible in MD simulations (Fig. 2 C and D).
In a similar manner to the truncated variants, we then used decreases in unfolded-state peak volumes (5) to determine folding free energies in RNCs (Fig. 3), observing a folding midpoint at a 42- to 44-aa linker length, beyond which nascent chain (NC) stability rapidly increases. The alignment of chain lengths shown here is based on the accessibility to PEG observed for RNC residues 36 aa beyond the peptidyl-transferase center (PTC) (5), and therefore, we assigned the FL isolated state to this length. This is a conservative alignment compared with our structure-based modeling (Fig. 2 C and D), which predicts that linker lengths of ca. 18 and 26 aa are required for folding to the intermediate and native states, respectively (Fig. 3, solid lines). In either case, it is clear that the “delay” in NC folding after emergence from the exit tunnel must at the midpoint be associated with a substantial destabilization of the NC, with a comparable with the 7-kcal folding free energy of the isolated domain. In contrast, while no intermediate NC resonances have been detected (Fig. 1D), because the intermediate is only marginally stable in isolation, the extent to which it may be destabilized in RNCs is less clear. Nevertheless, its identification in isolation prompted us to consider more carefully the potential consequences of slow proline isomerization, whether occurring via the intermediate or directly from the unfolded state, on the CTF process.
Measurement of Folding Kinetics.
CTF is a nonequilibrium process, with folding occurring in parallel with elongation of the NC, and therefore, it is important to understand the rate of NC folding in addition to the thermodynamic characterization above. As direct measurements of NC folding kinetics are experimentally inaccessible, we have used the isolated 6 truncation as a proxy. We first used 2D -exchange measurements (21) to observe reversible folding between 6 U and I states (Fig. 4A), which was determined to occur on a timescale of 0.4 s at 298 K (Fig. 4B). We then carried out real-time NMR temperature jump measurements to measure the proline isomerization limited rate of refolding from a thermally unfolded state at 310 K (SI Appendix, Fig. S10A). At 298 K, trans-to-cis isomerization of P742 was observed with a time constant of 1.5 min (Fig. 4C), while at lower temperatures, an additional faster phase was resolved that we attribute to off-pathway cis-to-trans isomerization of seven native-state trans prolines in the unfolded state (SI Appendix, Fig. S10B). Consistent with this interpretation, trans-to-cis isomerization was not observed in measurements of 6 P742A refolding, while the refolding of 4 P742A at 283 K fitted to a single slow phase with a time constant of 17.3 min, indicating formation of a cis alanine (SI Appendix, Fig. S10 C and D). Activation energies of and kcal were determined for the fast and slow phases of 6 refolding (Fig. 4D), respectively, comparable with typical activation energies for proline isomerization (20–22 kcal ) (22).
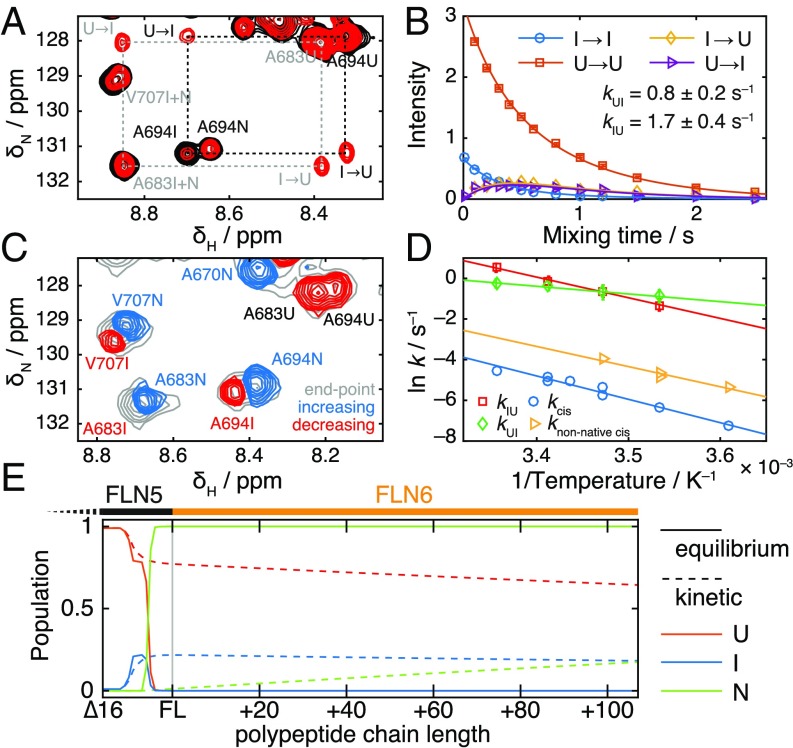
Fig. 4.
Analysis of folding kinetics. (A) 2D slices from magnetization transfer experiment (21) at 298 K, with exchange times of 10 ms (black) and 500 ms (red). Dashed rectangles highlight exchange cross-peaks between unfolded and intermediate states. Gray coloring indicates peaks excluded from additional analysis due to overlap of native and intermediate resonances. (B) Analysis and fitting of A694 cross-peak intensities from the magnetization transfer experiment shown in A. Data from multiple resonances were fitted globally to determine the indicated exchange rates. (C) Component spectrum determined from the analysis of real-time NMR data for the refolding of 6 after a temperature jump from 310 to 298 K, with a fitted time constant of 1.5 min. Peaks that increase or decrease over the reaction time course are shown in blue and red, respectively. The spectrum at equilibrium is shown in gray. (D) Arrhenius plot showing the temperature dependence of 6 folding kinetics. and are exchange rates determined by magnetization transfer measurements; and are proline isomerization rates determined by real-time NMR methods. (E) Populations of native, unfolded, and intermediate states based on characterization of isolated truncations plotted against polypeptide chain length at equilibrium (i.e., infinitely slow/stalled translation; solid lines) and under nonequilibrium conditions with a translation rate of 5 amino acids (dashed lines).
Lastly, we investigated the effect of the peptidyl-prolyl isomerase (PPIase) cyclophilin A. On addition of this enzyme, folding of 6 was no longer observed in real-time NMR experiments, but instead, new exchange cross-peaks were observed between and states in -exchange measurements (SI Appendix, Fig. S11). We, therefore, conclude that CypA can catalyze proline isomerization and folding to a timescale comparable with the nitrogen (ca. 1 s) and within the dead time of the real-time experiment. Exchange was not observed between and states, indicating that unfolding may be necessary for P742 to be sterically accessible for catalysis.
Reconstructing the Nonequilibrium CTF Pathway.
In this work, we have analyzed the thermodynamics and kinetics of FLN5 folding as a function of polypeptide chain length and identified states that are structurally compatible with tethering to the ribosome during translation (Figs. 2–4). This description can be combined with a known translation rate (ca. 5 amino acids in eukaryotes) to construct a Markov model of the CTF pathway (SI Appendix, Fig. S12), which we have integrated numerically to obtain quantitative predictions about the fundamentally nonequilibrium CTF process (23). From this model, we observe significant differences between populations calculated at thermodynamic equilibrium and those predicted under nonequilibrium conditions (Fig. 4E). In particular, as translation is more rapid than transcis isomerization (i.e., folding to the native state), only 17% of FLN5 NCs are predicted (based on our analysis of isolated truncations) to be fully folded by the point at which the subsequent FLN6 domain has been translated. Such an accumulation of adjacent unfolded domains seems likely to present a misfolding hazard (7, 8, 24), and this is explored further in the next section.
Assessing the Risk of Cotranslational Misfolding in TRPs.
Filamins are TRPs comprising up to 24 IgG domains, with a mean sequence identity between adjacent domains of % (SD) and 9% of pairs above the 40% threshold associated with misfolding in other IgG domains (SI Appendix, Table S2) (9). Given that P742 is conserved in a cis conformation in every known structure (SI Appendix, Table S2), the cotranslational accumulation of unfolded domains predicted above may indicate a more widespread risk of interdomain misfolding in filamins. More generally, a systematic analysis of Trp– structures [RepeatsDB (25)] shows that the abundance of cis prolines is comparable with that within the wider proteome [CATH S35 sets (26)] (SI Appendix, Fig. S14). Thus, proline isomerization limited CTF, and the potential accumulation of unfolded domains is likely to be an issue of relevance beyond filamins alone.
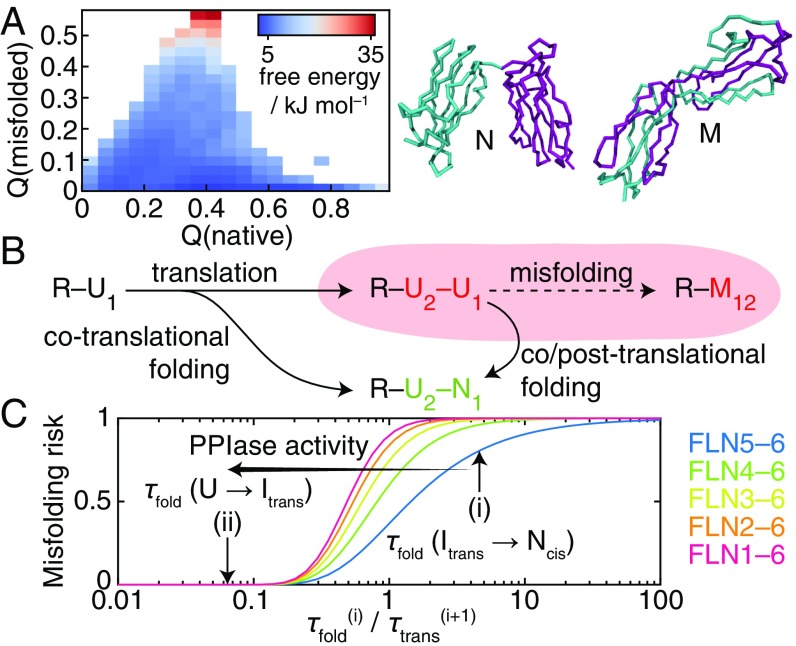
To explore the misfolding of filamin domains in more detail, we performed metadynamics simulations using structure-based models (7) to determine free energy landscapes for the folding and misfolding of pairs of filamin and titin IgG domains (Fig. 5A and SI Appendix, Fig. S13). From these calculations, we found that the population of domain-swapped misfolded states formed between filamin domains was 2.4% (Fig. 5A), comparable with that of the titin I27 dimer (0.7%) (SI Appendix, Fig. S13) for which misfolded states have been observed experimentally (7, 8).
Fig. 5.
Analysis of filamin misfolding during protein synthesis. (A) Free energy landscape calculated for an FLN4–5 dimer (cyan to purple) by coarse-grained MD simulations plotted against the fraction of native and misfolded contacts. Representative structures of native and misfolded states are shown as labeled. (B) A model for assessing the risk of cotranslational misfolding in which the rate of folding of a ribosome-bound unfolded NC (R–) to its native state () relative to the rate at which the following domain () is translated determines the likelihood of populating adjacent unfolded domains (R–), which may be at risk for forming a misfolded state (). (C) Estimated risk of misfolding in filamin as a function of the number of tandem repeat domains and the relative rates of folding and translation. Arrows indicate predicted risks where folding is (i) limited by proline isomerization to the native state or (ii) through the action of PPIases, limited by folding to the intermediate.
To investigate this further, we developed a simple model to quantify the risk of misfolding during biosynthesis (Fig. 5B). By analyzing the timescale of CTF, , relative to translation of the following domain, , we found (SI Appendix, SI Materials and Methods) that, for a protein with tandem repeat domains, the misfolding risk, , given by the probability of populating adjacent unfolded domains during biosynthesis is . This risk is plotted in Fig. 5C for increasing lengths of the filamin tandem repeat region (given a translation rate of 5 amino acids ). In isolation, when folding is limited by proline isomerization (Fig. 5C, i), the misfolding risk is high, particularly when the full multidomain protein is considered. However, if as shown this step is accelerated by PPIases (SI Appendix, Fig. S11), folding times will instead become limited by the rate of folding from the unfolded state (Fig. 5C, ii). Under this condition, we find that the risk of misfolding can be essentially eliminated.
Modulation of CTF by a Molecular Chaperone.
The interaction of molecular chaperones with NCs has previously been identified to be a key mechanism for suppressing long-range misfolding interactions (27) and may provide an additional defense against the misfolding hazard discussed above. Here, we have, therefore, investigated interactions of both and a disordered FLN5 RNC with the ribosome-associated holdase trigger factor (TF). Due to its marginal stability (Fig. 3), the intensities of , , and resonances in spectra are highly sensitive to intermolecular interactions. However, no changes in these intensities were observed in the presence of TF (SI Appendix, Fig. S15A). Likewise, only a small and uniform reduction in , resonance intensities was observed in an FLN5 + 21 RNC on binding of TF (SI Appendix, Fig. S15 B–E), comparable with that previously observed for an intrinsically disordered and weakly interacting -synuclein RNC (12). The observed absence of interactions for this folding-competent domain—in contrast to those between FLN5 and the ribosome surface (5)—indicates that only in some cases can individual chaperones provide protection against misfolding hazards.
Discussion
During protein synthesis on the ribosome, free energy landscapes for NC folding evolve as the chain length is increased by translation (10). Understanding CTF, therefore, requires characterizing a series of free energy landscapes rather than the single surface associated with traditional folding studies. Here, we have begun this process using observations of translationally arrested RNCs under equilibrium conditions complemented by a systematic C-terminal truncation strategy (13–15).
The truncation model developed in this work has two important roles. First, it provides an essential baseline against which the effects of ribosome interactions can be discerned. It is increasingly evident that the ribosome surface can be an active participant in CTF processes (4, 5), and this strategy has highlighted the large free energy changes ( kcal ) associated with the delay of FLN5 folding (Fig. 3). This is a substantially greater perturbation than observed using pulse proteolysis measurements in other RNC systems (28), and a key challenge for future work will be to develop a quantitative and structural understanding of these effects.
Second, our truncation model enabled us to apply high-resolution methods not currently feasible directly on the ribosome to analyze structure and folding kinetics. In doing so, we have identified a marginally stable intermediate associated with the isomerization of a native-state cis proline, and we have shown that this isolated structure can be used as a starting configuration for structural modeling of nascent chains (Fig. 2). From these results, we concluded that there are no steric barriers to formation of the intermediate early in the translation process, although its final stability on tethering to the ribosome remains an open question. Nevertheless, the slow trans-to-cis isomerization identified in isolation must still occur at some point during filamin biosynthesis, and a consideration of this led to the development of a simple model to assess the cotranslational misfolding hazard (Fig. 5).
It is still unclear whether ribosome–NC interactions, which necessarily act only over a short range after emergence from the exit tunnel, in anyway assist the folding of filamin. However, other situations can be envisaged where such interactions are more beneficial (e.g., sequestering aggregation or misfolding-prone sequences before recognition and engagement by chaperones or entry into secretion pathways). Likewise, while we have shown here that the ribosome surface and TF have at least partly different recognition motifs and therefore, client pools, it is possible that other molecular chaperones might effectively suppress long-range interactions and contribute to alleviating the misfolding hazard, even in the absence of PPIase activity.
It is interesting to compare the intermediate identified here with the aggregation-prone intermediate formed during -microglobulin folding (29). Both domains have IgG folds, and both intermediates are associated with isomerization of native cis proline residues; however, a key difference is that FLN5 is part of a multidomain protein, and therefore, the primary hazard is the intramolecular misfolding of adjacent unfolded states during translation rather than posttranslational intermolecular aggregation. We have found that cis prolines are as abundant within TRPs as within the broader proteome (SI Appendix, Fig. S14), and therefore, this hazard is clearly relevant to more than filamin proteins alone. A simple model (Fig. 5 B and C) has indicated that PPIase activity is necessary but also sufficient to eliminate this risk in filamins (as indeed could be other forms of chaperone intervention), and we expect that this model could be applicable to other cases and forms of cotranslational misfolding. Overall, our findings highlight that thermodynamically favorable CTF is not by itself sufficient to avoid misfolding hazards: kinetic requirements must also be satisfied.
Materials and Methods
FLN5 truncations, mutational variants, and TF were cloned, expressed, and purified in Tico buffer as described previously (5, 12). RNCs were prepared, and their stability was monitored during acquisition as described previously (5, 11). CD spectra were acquired at protein concentrations of ca. 0.5 mg m using a Jasco J-810 spectropolarimeter. To determine protein stability, CD signals were measured at 234 nm for urea concentrations between 0 and 8.2 M and fitted to a two-state unfolding process. NMR data were acquired at 283 K on a 700-MHz Bruker Avance III spectrometer with TXI cryoprobe unless otherwise indicated. Spectra were assigned with standard triple-resonance methods, and magnetization transfer, real-time kinetics, and Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion data were acquired as described in SI Appendix, SI Materials and Methods. FL and 6 FLN5 structures were determined by chemical shift-restrained replica-averaged metadynamics (16) using Gromacs as described in SI Appendix, SI Materials and Methods. Simulations of folding on the ribosome and interdomain misfolding were conducted in Gromacs with structure-based models generated in SMOG 2 (20) and misfolding potentials derived following the approach of refs. 7 and 8 as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Flemming Hansen for helpful discussions, Dr. John Kirkpatrick and Dr. Tom Frenkiel for assistance with NMR experiments, and Dr. Leo James for a gift of CypA. We acknowledge the use of Legion@UCL and the University College London NMR Center, the Medical Research Council for access to the Biomedical NMR Center at the Francis Crick Institute, and the staff for their support. This work was supported by a Wellcome Trust Investigator Award (to J.C.). T.W. was supported by an European Molecular Biology Organization fellowship, and M.-E.K. and S.H.S.C. were supported by Biotechnology and Biological Sciences Research Council studentships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716252115/-/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 3.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrita LD, et al. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol. 2016;23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 7.Borgia MB, et al. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgia A, et al. Transient misfolding dominates multidomain protein folding. Nat Commun. 2015;6:8861. doi: 10.1038/ncomms9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 10.Clark PL. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Cassaignau AME, et al. A strategy for co-translational folding studies of ribosome-bound nascent chain complexes using NMR spectroscopy. Nat Protoc. 2016;11:1492–1507. doi: 10.1038/nprot.2016.101. [DOI] [PubMed] [Google Scholar]
- 12.Deckert A, et al. Structural characterization of the interaction of -synuclein nascent chains with the ribosomal surface and trigger factor. Proc Natl Acad Sci USA. 2016;113:5012–5017. doi: 10.1073/pnas.1519124113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shortle D, Meeker AK. Residual structure in large fragments of staphylococcal nuclease: Effects of amino acid substitutions. Biochemistry. 1989;28:936–944. doi: 10.1021/bi00429a003. [DOI] [PubMed] [Google Scholar]
- 14.De Prat Gay G, et al. Conformational pathway of the polypeptide chain of chymotrypsin inhibitor-2 growing from its N terminus in vitro. Parallels with the protein folding pathway. J Mol Biol. 1995;254:968–979. doi: 10.1006/jmbi.1995.0669. [DOI] [PubMed] [Google Scholar]
- 15.Neira JL, Fersht AR. Exploring the folding funnel of a polypeptide chain by biophysical studies on protein fragments. J Mol Biol. 1999;285:1309–1333. doi: 10.1006/jmbi.1998.2249. [DOI] [PubMed] [Google Scholar]
- 16.Camilloni C, Vendruscolo M. Statistical mechanics of the denatured state of a protein using replica-averaged metadynamics. J Am Chem Soc. 2014;136:8982–8991. doi: 10.1021/ja5027584. [DOI] [PubMed] [Google Scholar]
- 17.Rosato A, et al. Blind testing of routine, fully automated determination of protein structures from NMR data. Structure. 2012;20:227–236. doi: 10.1016/j.str.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy AJ, Fucini P, Noegel AA, Stewart M. Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod. Nat Struct Biol. 1999;6:836–841. doi: 10.1038/12296. [DOI] [PubMed] [Google Scholar]
- 19.Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J Phys Chem B. 2008;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- 20.Noel JK, et al. SMOG 2: A Versatile software package for generating structure-based models. PLoS Comput Biol. 2016;12:e1004794. doi: 10.1371/journal.pcbi.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 22.Balbach J, Schmid FX. Proline iomerization and its catalysis in protein folding. In: Pain RH, editor. Mechanisms of Protein Folding. Oxford Univ Press; Oxford: 2000. pp. 212–249. [Google Scholar]
- 23.O’Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W, Schafer NP, Wolynes PG. Frustration in the energy landscapes of multidomain protein misfolding. Proc Natl Acad Sci USA. 2013;110:1680–1685. doi: 10.1073/pnas.1222130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paladin L, et al. RepeatsDB 2.0: Improved annotation, classification, search and visualization of repeat protein structures. Nucleic Acids Res. 2017;45:D308–D312. doi: 10.1093/nar/gkw1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson NL, et al. Cath: An expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45:D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashaghi A, et al. Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature. 2013;500:98–101. doi: 10.1038/nature12293. [DOI] [PubMed] [Google Scholar]
- 28.Samelson AJ, Jensen MK, Soto RA, Cate JHD, Marqusee S. Quantitative determination of ribosome nascent chain stability. Proc Natl Acad Sci USA. 2016;113:13402–13407. doi: 10.1073/pnas.1610272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rennella E, et al. Real-time NMR characterization of structure and dynamics in a transiently populated protein folding intermediate. J Am Chem Soc. 2012;134:8066–8069. doi: 10.1021/ja302598j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.