Significance
Interpretation of genome-wide investigations of host–pathogen interactions are often obscured by analyses of mixed populations of infected and uninfected cells. Thus, we developed a system whereby we simultaneously characterize and compare genome-wide transcriptional and epigenetic changes in pure populations of virally infected and neighboring uninfected cells to identify viral-regulated host responses. Using patient-derived unmodified Zika viruses (ZIKV) infecting primary human macrophages, we reveal that ZIKV suppresses host transcription by multiple mechanisms. ZIKV infection causes both targeted suppression of type I interferon responses and general suppression by reducing RNA polymerase II protein levels and DNA occupancy. Simultaneous evaluation of transcriptomic and epigenetic features of infected and uninfected cells provides a powerful method for identifying coincident evolution of dominant proviral or antiviral mechanisms.
Keywords: Zika virus, macrophage, immune evasion, genomics, transcription
Abstract
Genome-wide investigations of host–pathogen interactions are often limited by analyses of mixed populations of infected and uninfected cells, which lower sensitivity and accuracy. To overcome these obstacles and identify key mechanisms by which Zika virus (ZIKV) manipulates host responses, we developed a system that enables simultaneous characterization of genome-wide transcriptional and epigenetic changes in ZIKV-infected and neighboring uninfected primary human macrophages. We demonstrate that transcriptional responses in ZIKV-infected macrophages differed radically from those in uninfected neighbors and that studying the cell population as a whole produces misleading results. Notably, the uninfected population of macrophages exhibits the most rapid and extensive changes in gene expression, related to type I IFN signaling. In contrast, infected macrophages exhibit a delayed and attenuated transcriptional response distinguished by preferential expression of IFNB1 at late time points. Biochemical and genomic studies of infected macrophages indicate that ZIKV infection causes both a targeted defect in the type I IFN response due to degradation of STAT2 and reduces RNA polymerase II protein levels and DNA occupancy, particularly at genes required for macrophage identity. Simultaneous evaluation of transcriptomic and epigenetic features of infected and uninfected macrophages thereby reveals the coincident evolution of dominant proviral or antiviral mechanisms, respectively, that determine the outcome of ZIKV exposure.
Viruses survive and cause disease by avoiding and suppressing innate and adaptive immune responses. Flaviviruses are small, enveloped, single-stranded positive-sense RNA viruses (1). They replicate using a viral RNA-dependent RNA polymerase that creates viral messenger RNA and new viral genomes using a negative-sense RNA intermediate (2). More than 40 species of flaviviruses cause human disease, some associated with extensive global morbidity and mortality. Most flaviviruses cause acute infections and therefore must inhibit detection and eradication by the innate immune system. Flaviviruses have evolved multiple mechanisms to inhibit innate immune responses. Improved understanding of how these important pathogens subvert innate immune responses could lead to novel strategies for the development of antiviral agents.
Zika virus (ZIKV) is a member of the mosquito-borne group of flaviviruses that are primarily transmitted by Aedes mosquitoes. ZIKV can also be also transmitted vertically from mother to fetus and between sexual partners (1, 3–7). Based on the presence of Aedes mosquitoes in nearly all tropical and subtropical areas of the world, it is estimated that ∼3.6 billion people worldwide are at risk for ZIKV infection (8, 9). Since 2007, ZIKV has caused outbreaks worldwide with documented transmission in at least 84 countries (10). Although the precise mechanisms are under investigation, there is strong evidence that ZIKV causes life-threatening fetal brain abnormalities and Guillain–Barré syndrome (11–15). ZIKV, like other human flaviviruses, naturally infects innate immune cells, including monocytes and macrophages (16). The ability to infect these cells is thought to increase viral dissemination and contribute to pathogenesis. However, how ZIKV infects macrophages and circumvents innate immune activation is not well understood.
Due to the central importance of IFNs in antiviral responses, most pathogenic viruses antagonize IFN production and/or IFN-dependent response pathways directly (17). However, many viruses also employ general host shut-off mechanisms that interfere with RNA transcription, RNA processing, and/or translation to inhibit host gene expression broadly (18, 19). This can both increase cellular resources available for the production of viral products and suppress host antiviral responses. To shut off host transcription, some viruses inhibit RNA polymerase II (RNApol2), a large protein complex responsible for catalyzing the synthesis of mRNAs, most snRNAs, and microRNAs (20–25). Although flaviviruses, including ZIKV, are known to inactivate the IFN pathway at multiple levels, none has been shown to inhibit transcription globally (26).
Genome-wide investigations of ZIKV–macrophage interactions have great potential to identify mechanisms by which ZIKV modulates macrophage response in an unbiased manner. A major limitation to understanding how ZIKV infects macrophages, or other cell types, is that the percentages of infected vs. uninfected bystander cells in cultures vary widely. Thus, population-level analyses are difficult to interpret because they include cells that are not productively infected but could potentially respond to signals from infected cells. Direct characterization of uninfected cells in comparison with infected cells is therefore required to disentangle proviral and antiviral responses and provide insights to viral infections in vivo, in which only a small fraction of cells is infected. Potential options to overcome this limitation include the use of reporter viruses, highly susceptible cell lines, or single-cell sequencing technologies. Reporter viruses help isolate infected cells, but the insertion of reporter proteins into flaviviruses, which are relatively small, may alter their virulence and prohibit studies comparing wild-type patient-derived viruses. Studying susceptible cell lines increases the percentage of infected cells within a population, but these cell lines often support robust viral replication because they lack important antiviral signaling responses. Single-cell profiling technologies are poised to circumvent many of these limitations; however, many methods, including ChIP sequencing (ChIP-seq), cannot yet be reliably applied at the single-cell level. Additionally, single-cell RNA-sequencing (RNA-seq) technologies that rely on oligo(dT) priming cannot distinguish flavivirus-infected from uninfected cells, as these viruses do not make polyadenylated RNA.
To overcome these obstacles, we developed an infection model that enables the application of unbiased genome-wide transcriptional and epigenetic analyses to identify how unmodified clinical isolates of ZIKV modulate host responses in pure populations of ZIKV-infected primary human macrophages, major cellular hosts of ZIKV, and other closely related flaviviruses. We observed strikingly divergent transcriptional and epigenetic responses between ZIKV-infected and uninfected bystander macrophages. Many of these differences would not be apparent if the population of cells were studied as a whole. ZIKV infection is associated with minimal gene activation and a nearly complete loss of type I IFN signaling primarily resulting from ZIKV-induced degradation of STAT2. In contrast, noninfected cells exhibit robust activation of the type I IFN pathway. Additionally, ChIP-seq of RNApol2 demonstrates that DNA occupancy is reduced during ZIKV infection, leading to general suppression of transcription and disproportionate loss of RNApol2 at core macrophage identity genes. This method of comparing genome-wide transcriptomic and epigenomic responses in neighboring infected and uninfected cells exposed to the same environment allows deconvolution of the specific molecular mechanisms by which ZIKV modulates macrophage responses.
Results
ZIKV Infection of Primary Human Macrophages.
To study ZIKV–macrophage interactions, we first developed an in vitro ZIKV infection model that utilizes unmodified viruses to infect primary human blood monocyte-derived macrophages (HMDMs). Infection of HMDMs with ZIKV FSS13025 (ZIKV FSS), an Asian-lineage ZIKV human isolate from Cambodia (2010), at a multiplicity of infection (MOI) of 1 (MOI1) resulted in only 1.5% infectivity on average at 24 h postinfection (PI) as measured by intracellular flow cytometry staining of the viral envelope (E) protein using anti-flavivirus E protein (clone 4G2) (SI Appendix, Fig. S1A). Antibodies from individuals previously infected with Dengue virus (DENV) can increase the infectivity of ZIKV by antibody-dependent enhancement (ADE) (27–29). When HMDMs were infected at the same MOI1 but in the presence of human DENV-immune serum (ADE1), 40% of macrophages were infected on average (SI Appendix, Fig. S1A). Even if the MOI was increased to greater than 20 (high-MOI), ADE1 infection still resulted in more infected macrophages than high-MOI conditions (SI Appendix, Fig. S1A). ZIKV SD001 is an Asian-lineage virus isolated in San Diego in 2016 during an acute ZIKV infection in a traveler returning from Venezuela. The frequency of infected HMDMs during infection with ZIKV FSS and SD001 was not significantly different using MOI1 or ADE1 conditions (SI Appendix, Fig. S1B). We next compared the production of new infectious virus using MOI1, ADE1, or high-MOI conditions by measuring the yield of secreted virus in the culture supernatant by focus-forming unit (FFU) assay. HMDMs infected by ADE1 produced 166-fold more infectious ZIKV than infection by MOI1 and 60-fold more infectious ZIKV than infection by high-MOI (SI Appendix, Fig. S1C). Compared with ZIKV SD001, ZIKV FSS produced slightly higher infectious titers at 24 h PI under ADE1 conditions (SI Appendix, Fig. S1D). To determine ZIKV growth kinetics in HMDMs, we first measured the production of viral RNA under ADE1 conditions. We detected intracellular ZIKV RNA starting at 8 h PI, with a rapid increase in viral RNA levels occurring at 12–24 h PI (SI Appendix, Fig. S1E). Intracellular staining of infected HMDMs using the anti-flavivirus E protein antibody 4G2 first detected ZIKV antigen-positive macrophages at 12 h PI (SI Appendix, Fig. S1F). Between 12 and 24 h PI both the percentage of ZIKV+ macrophages and the intensity of 4G2 staining increase (SI Appendix, Fig. S1F). Collectively, these results demonstrate robust ZIKV replication in HMDMs under ADE1 conditions.
Divergent Transcriptional Responses in ZIKV-Infected and Bystander Cells.
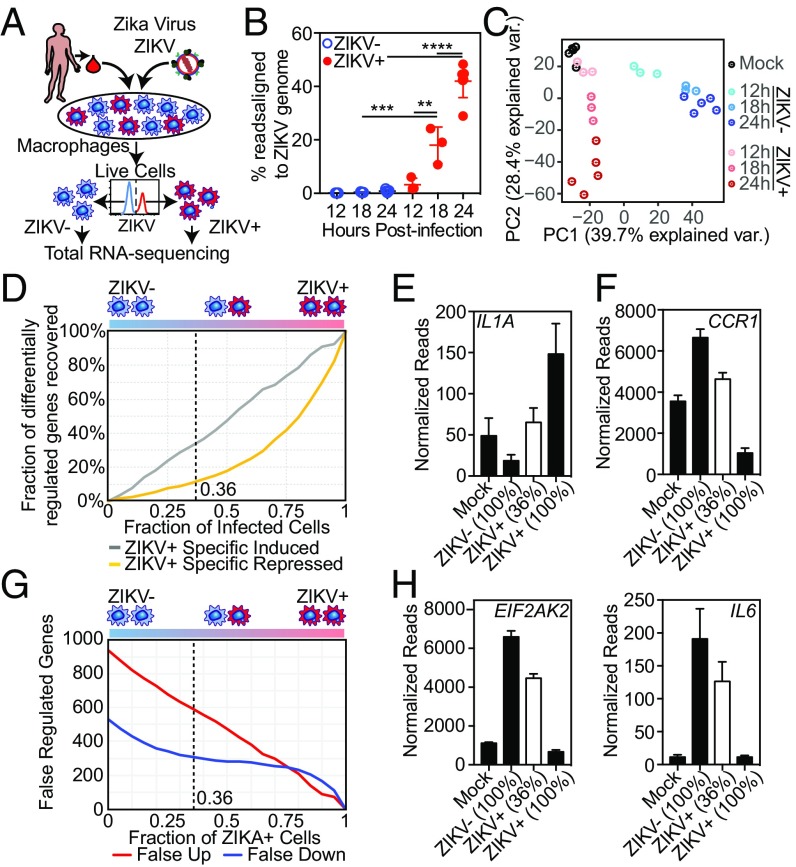
To identify how ZIKV modulates macrophage responses, we assessed genome-wide transcriptional responses in pure populations of HMDMs. HMDMs were infected with ZIKV using ADE1 conditions and were formaldehyde-fixed at different time points PI. HMDMs were then stained intracellularly for the flavivirus E protein using 4G2 antibody and were separated by FACS into infected ZIKV antigen-positive (ZIKV+) and uninfected ZIKV antigen-negative (ZIKV−) populations. After reverse cross-linking and protease digestion steps, high-quality total RNA was isolated and analyzed using stranded rRNA-depleted total RNA-seq (Fig. 1A). Our approach successfully segregated ZIKV+ from ZIKV− cells, as significant numbers of reads aligning to the ZIKV genome were found only in the ZIKV+ populations (Fig. 1B). As expected, the percentage of reads aligning to the ZIKV genome in ZIKV+ cells increased with time PI. By 24 h PI, more than 40% of all mapped RNA-seq reads in ZIKV+ HMDMs aligned to the ZIKV genome although the ZIKV genome is only ∼0.01% the size of all human nonoverlapping exons (Fig. 1B).
Fig. 1.
ZIKV modulates macrophage transcription during infection. (A) Diagram depicting the infection model. HMDMs are infected with ZIKV and stained for ZIKV group antigen followed by FACS isolation of live, productively infected ZIKV+ and bystander (ZIKV−) macrophages. (B) Percent of RNA-seq reads aligning to the ZIKV genome in FACS-isolated ZIKV+ and ZIKV− macrophages at the indicated time points PI. Percent was calculated as the total reads aligning to ZIKV alone vs. human + ZIKV genomes. Each data point (mean ± SEM) represents results from HMDMs derived from different human donors. All categories were compared using ANOVA with correction for multiple comparisons. (C) PCA biplot of the first two principal component dimensions comparing RNA-seq of FACS-isolated ZIKV+ or ZIKV− HMDMs from three (12 h and 18 h) or five (mock-infected and 24 h) individual donors. (D) Calculated loss of sensitivity in detecting ZIKV-regulated gene expression. Gene-expression levels were calculated using data from ZIKV+, ZIKV−, and mock-infected macrophage RNA-seq experiments 24 h PI performed with macrophages from five different donors. (E and F) Individual gene expression calculated by RNA-seq in pure populations of mock-infected, ZIKV+, and ZIKV− macrophages (24 h PI). Mean (± SEM) IL1A (E) and CCR1 (F) expressions of pure populations (black bars) were calculated based on RNA-seq from five different donors. Mean (± SEM) expression of a 36% mixed population (white bar) was calculated computationally based on mixing 36% ZIKV+ macrophages with 64% ZIKV− macrophages from each of those five independent RNA-seq experiments. (G) Calculated number of genes expected to be falsely attributed to ZIKV-dependent regulation based on percent infectivity. Calculations were performed as in D. (H) Mean (± SEM) EIF2AK2 and IL6 expression of pure populations (black bars) and a 36% mixed population (white bar) calculated as described in E and F.
We first compared the host transcriptomes of mock-infected (medium alone), ZIKV+, and ZIKV− HMDMs using principal component analysis (PCA). Even though each of the replicates was performed with cells from different individuals, PCA grouped replicates based on infection status and time point PI (Fig. 1C). This close association among replicates suggests that interindividual differences have less influence on the transcriptional response of HMDMs than infection status and time point PI. Strikingly, the first principal component (PC1) divided the data into two groups, separating ZIKV− macrophages from ZIKV+ and mock-infected HMDMs. This suggested that the transcriptional responses of ZIKV+ macrophages were more similar to mock-infected cells than to ZIKV− bystander cells (Fig. 1C). In fact, when considering only PC1 and PC2, which captured 68% of the overall transcriptomic differences between the samples, at 12 h PI, the transcriptome of ZIKV+ HMDMs largely overlapped with that of mock-infected macrophages. Only at later time points did the transcriptomes segregate along PC2 based on time PI. This suggests that ZIKV infection of macrophages, especially at early time points PI, does not elicit a strong transcriptional signature.
Transcriptome studies of in vitro infections have typically analyzed mixed populations of infected cells and uninfected bystanders. To determine how a traditional mixed-population transcriptome analysis would differ from our approach of isolating and analyzing pure populations of ZIKV+ and ZIKV− cells, we used our transcriptional profiles of pure ZIKV+ and ZIKV− HMDM populations derived from five different individuals to simulate the transcriptional profiles of hypothetical ZIKV-infected bulk populations over a range of 100–0% infected cells (Fig. 1D). We used these simulated profiles to calculate the recall rate of significantly changed genes identified in pure ZIKV+ cells. In our analysis, as the fraction of infected cells decreased from 100%, the ability to identify genes specifically repressed, and to a lesser degree induced, by ZIKV was lost rapidly (Fig. 1D). Using our highly efficient macrophage infection system, the average percent of infection at 24 h PI was 36%. Our model estimated that we would identify only 10% of the repressed and 32% of the induced genes in a 36% ZIKV+ HMDM population (Fig. 1D). Examples of genes that were induced by ZIKV that would have not been identified at 36% infectivity included IL1A, EIF2AK3/PERK, ASAP2, and KLF4 (Fig. 1E and SI Appendix, Fig. S1G). Similarly, we would have missed ZIKV-repressed genes such as CCR1, TNFRS11A, and CCL2 (Fig. 1F and SI Appendix, Fig. S1H). In addition to decreased sensitivity in identifying genes regulated by ZIKV, analysis of mixed populations can falsely identify genes as induced or suppressed by ZIKV. At 36% infectivity, we estimated that analysis of a mixed population would misidentify ∼600 genes as induced and 300 genes as repressed by ZIKV (Fig. 1G). For example, multiple inflammatory genes, including EIF2AK2/PKR, IL6, and IFITM2 were strongly induced only in ZIKV− bystander cells. Mixed population analysis would suggest that these genes were strongly up-regulated by ZIKV when in fact these genes were not induced or in many cases were even suppressed in cells productively infected with ZIKV (Fig. 1H and SI Appendix, Fig. S1I).
In addition to improved discovery and accuracy, a major advantage of our infection model is the ability to analyze transcriptional responses during infection with unmodified clinical isolates. Using our method, we compared the response of HMDMs during ADE1 infection with two ZIKV isolates derived from human infections, ZIKV FSS and ZIKV SD001. These strains infected the same percentage of HMDMs (SI Appendix, Fig. S1B), with FSS infection producing slightly higher infectious titers at 24 h PI (SI Appendix, Fig. S1D). The transcriptomes of the uninfected bystander ZIKV− cells from the FSS and SD001 infections were almost identical, with no genes called as significantly different between these strains (SI Appendix, Fig. S1 J and K). In contrast, the transcriptomes of ZIKV+ macrophages infected with either FSS or SD001 exhibited significant differences, pointing to biological differences between the virus strains (SI Appendix, Fig. S1L). In ZIKV+ cells, 88 genes were expressed at higher levels during SD001 infection than during FSS infection, while 22 genes were expressed at significantly higher levels during FSS infection than during SD001 infection (SI Appendix, Fig. S1K). Of these 110 genes, 71 genes were also significantly up- or down-regulated compared with mock-infected cells (SI Appendix, Fig. S1M). The set of genes down-regulated in ZIKV+ cells during FSS infection compared with SD001 infection and mock treatment was enriched for multiple genes involved in RNA processing and splicing, including the RNA helicases DDX5, DDX17, and DDX42 and the splicing factors SRRM2 and SUGP2 (SI Appendix, Fig. S1N). As shown by our simulation above, transcriptional analyses of mixed populations of ZIKV+ and ZIKV− cells have especially poor sensitivity in detecting down-regulated genes. We estimate that a mixed cell transcriptional analysis with 36% ZIKV+ cells would not identify 36 of the 46 genes specifically down-regulated during FSS infection, including DDX5, DDX17, DDX42, and SRRM2.
ZIKV Infection Elicits a Limited Transcriptional Response.
To evaluate ZIKV–macrophage interactions over time, we compared the transcriptional responses in ZIKV+ and ZIKV− cells at 12, 18, and 24 h PI to mock-infected HMDMs. ZIKV− cells up-regulated a larger number of genes than ZIKV+ cells at 12 h PI (420 genes in ZIKV− cells vs. 100 genes in ZIKV+ cells), 18 h PI (708 in ZIKV− cells vs. 244 in ZIKV+ cells), and 24 h PI (867 in ZIKV− vs. 657 in ZIKV+) (SI Appendix, Fig. S2A). While some inflammatory genes were induced in ZIKV+ cells (examples include CXCL10, RSAD2, IFIT1, IFIT2, CXCL11, IFITM1, IFI44L, CCL8, and TNFSF10/TRAIL), the degree of induction was greater in ZIKV− bystander cells (SI Appendix, Fig. S2 B–D). However, as time PI increased, more uniquely up-regulated genes were identified in ZIKV+ cells (SI Appendix, Fig. S2 C and D). For example, at 18 h PI, ZIKV+ cells up-regulated AP1S3 and ASAP2, involved in Golgi trafficking, and TNFRSF10D, a decoy TNF receptor that protects against TNFSF10/TRAIL-mediated apoptosis, more than ZIKV− cells (SI Appendix, Fig. S2C). After 24 h of infection, ZIKV+ cells up-regulated the gene encoding IFNβ, IFNB1, and the transcription factor KLF4, involved in cell-cycle control, more than their ZIKV− neighbors (SI Appendix, Fig. S2D).
ZIKV Suppresses Inflammatory Gene Activation in Macrophages.
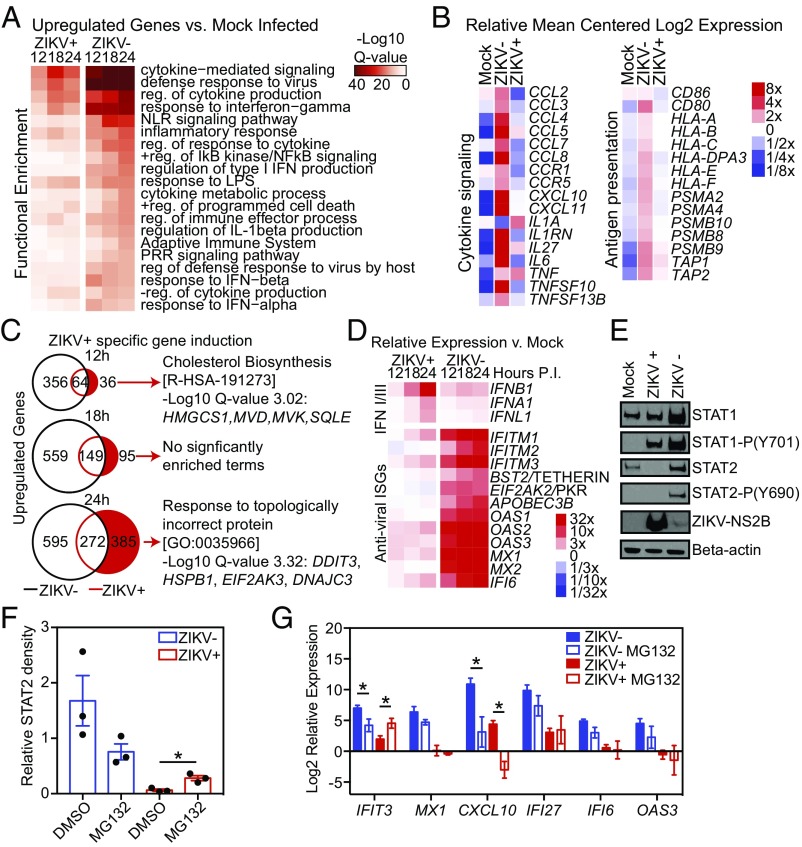
To identify signaling pathways affected by ZIKV infection of macrophages, we performed gene ontology and pathway analysis on ZIKV− and ZIKV+ cells and compared them with mock-infected HMDMs. At all time points ZIKV+ HMDMs showed a delayed and diminished induction of genes from multiple inflammation and immune response-related categories including IFN signaling, cytokine signaling, antigen presentation, and pattern recognition receptor (PRR) response pathways (Fig. 2 A and B). To identify how ZIKV manipulates the transcriptional responses in HMDMs, we identified the subset of genes uniquely induced in ZIKV+ cells at 12, 18, and 24 h PI. The number of uniquely induced genes at 12 h PI in ZIKV+ cells was limited to only 36 genes. However, these genes were significantly enriched for genes involved in cholesterol biosynthesis, such as HMGCS1, MVD, MVK, and SQLE (Fig. 2C). At 24 h PI, genes specifically induced in ZIKV+ macrophages were enriched for those involved in the unfolded protein response, including the endoplasmic reticulum (ER) stress response kinase EIF2AK3/PERK and the transcription factor DDIT3/CHOP (Fig. 2C).
Fig. 2.
Degradation of STAT2 by ZIKV impairs ISG activation. (A) Heat maps of the top enriched functional annotations of genes significantly up-regulated (fold change >2, FDR < 0.01) in ZIKV+ and ZIKV− macrophages compared with mock-infected macrophages at the indicated time points. (B) Heat map of the relative expression of selected genes involved in cytokine signaling and antigen presentation in mock-infected, ZIKV+ and ZIKV− macrophages 24 h PI. Data are the average of five experiments. (C) Venn diagrams showing the numbers of unique and shared up-regulated genes in ZIKV+ and ZIKV− macrophages compared with mock-infected macrophages at 12, 18, and 24 h PI. The top significantly enriched functional category for genes uniquely induced in ZIKV+ cells (red) at each time point is shown with examples of genes from that category. (D) Heat map depicting relative transcription of type I and III IFN genes and ISG in ZIKV− and ZIKV+ macrophages vs. control (Mock) over time. Data are the average of three experiments (12 h and 18 h) or five experiments (24 h). (E) Western blot of STAT1, phosphorylated-STAT1, STAT2, phosphorylated STAT2, ZIKV NS2B, and β-actin levels in equivalent numbers of mock-infected, ZIKV−, and ZIKV+ cells at 24 h PI. (F) Relative quantitation of Western blot STAT2 levels in ZIKV− and ZIKV+ cells treated with MG132 or vehicle control (Mock). STAT2 density is relative to β-actin. Data shown (mean ± SEM) are from three infections performed in HMDMs derived from three different individuals at 24 h PI. Data were analyzed by ANOVA with MG132 treatment groups compared with vehicle with correction for multiple comparisons. (G) Relative gene expression in ZIKV− and ZIKV+ cells treated with MG132 compared with DMSO. Data (mean ± SEM) show the relative gene expression by qRT-PCR in at least three independent experiments performed with FACS HMDMs derived from different donors at 24 h PI. Data for each gene were analyzed by ANOVA with MG132-treatment groups compared with vehicle with correction for multiple comparisons. Asterisks indicate statistically significant differences (*P < 0.05).
In addition to suppressed inflammatory gene induction, ZIKV+ macrophages also demonstrated increased down-regulation of genes involved in multiple immune pathways, including leukocyte activation/migration and cytokine signaling/secretion (SI Appendix, Fig. S2E). The top functional enrichment category for genes specifically down-regulated in ZIKV+ cells at all time points was histone deacetylases (HDACs) deacetylate histones (SI Appendix, Fig. S2F). The second most enriched pathway in genes uniquely down-regulated in ZIKV+ cells at 18 h PI was signaling by interleukins. Down-regulated genes in this category include multiple dual-specific protein phosphatases involved in regulating MAPK signaling as well as receptors and chemokines involved in immune activation. In ZIKV+ cells, there was specific down-regulation of many inflammatory response genes at 24 h PI, including the PRR signaling molecules CD14, TLR4, and LY96, cytokines such as CCL2 and CXCL8, and receptors such as C3AR1, C5AR1, and CCR1 (SI Appendix, Fig. S2F).
ZIKV, like other human pathogenic flaviviruses, blocks IFN induction of antiviral IFN-stimulated genes (ISGs). However, the exact mechanisms by which and where in the IFN response pathway ZIKV blocks ISG activation are not fully understood. Moreover, the IFN response during flaviviral infections is highly context-dependent and varies in a virus-, cell type-, and host species-specific manner (30). Previous studies have demonstrated that ZIKV may block the induction and/or translation of IFNβ, thereby inhibiting the secretion of IFNs, and/or degrade STAT2 and inhibit the phosphorylation of both STAT1 and STAT2, which inhibits signaling downstream of the IFN α/β receptor and induction of ISGs (31–33). We therefore examined the expression of IFN and ISG genes in ZIKV+ and ZIKV− cells throughout ZIKV infection. ZIKV+ cells express type I and type III IFN genes equivalently to ZIKV− cells at 12 and 18 h PI and higher levels than ZIKV− cells at 24 h PI (Fig. 2D and SI Appendix, Fig. S2G). In contrast, ISG expression is markedly reduced in ZIKV+ cells compared with ZIKV− cells at 12, 18, and 24 h PI (Fig. 2D and SI Appendix, Fig. S2H). During ZIKV infection of human dendritic cells, IFNs are induced at the transcriptional level, but protein levels are not measurable in the supernatants, suggesting impaired IFN protein production (31). To determine if IFNβ is made and secreted following IFNB1 induction during ZIKV infection of macrophages, we measured the IFNβ protein levels in supernatants by ELISA. We consistently detected IFNβ in the supernatants of macrophages infected with ZIKV at 24 h PI (SI Appendix, Fig. S2I). Collectively, these data show that ZIKV broadly suppresses inflammatory gene expression in infected macrophages. Type I and type III IFN genes are induced in ZIKV+ macrophages, and IFNβ is produced, but ZIKV+ macrophages demonstrate markedly reduced ISG induction compared with ZIKV− cells. This suggests ZIKV blocks type I IFN signaling rather than type I IFN production in macrophages.
ZIKV Degrades STAT2 to Inhibit Type I IFN Signaling in Macrophages.
To explore ZIKV inhibition of type I IFN signaling in macrophages, we first asked at which time point PI ZIKV inhibits IFN signaling by treating ZIKV-infected HMDM cultures with exogenous type I IFN before infection or at multiple time points PI. Exogenous IFN inhibited ZIKV infection in the HMDM cultures when given at or before 4 h PI but not after 8 h PI (SI Appendix, Fig. S3A). We next measured the kinetics of type I IFN and ISG gene expression during ZIKV infection to determine how rapidly these genes are up-regulated. Compared with mock-infected cells, IFNB1 and ISGs, such as MX1, were consistently up-regulated only at 8–12 h PI in ZIKV-infected HMDM cultures (SI Appendix, Fig. S3 B and C). In contrast to IFNB1, which is induced similarly in ZIKV+ and ZIKV− cells at 12 h PI, there was no induction of MX1 in ZIKV+ cells (SI Appendix, Fig. S3 D and E). These findings demonstrate that ZIKV blocks type I IFN signaling in productively infected macrophages before IFNs can stimulate ISG production.
To identify the potential mechanisms by which ZIKV inhibits type I IFN signaling in macrophages, we examined levels of total and phosphorylated STAT1 and STAT2 proteins in ZIKV+, ZIKV−, and mock-infected control HMDMs. Western blots demonstrated decreased STAT2 levels in ZIKV+ cells compared with ZIKV− and mock-infected cells, consistent with STAT2 degradation (Fig. 2E and SI Appendix, Fig. S3F). We did not detect STAT2 phosphorylation in either ZIKV+ or mock cells. Levels of STAT1 were lower in ZIKV+ than in ZIKV− cells but were not statistically different from levels in mock-infected HMDMs (SI Appendix, Fig. S3G). Phosphorylated STAT1 was detectable in both ZIKV+ and ZIKV− cells but not in mock-infected cells (Fig. 2E and SI Appendix, Fig. S3F). Consistent with the data described above suggesting blockade of downstream IFN signaling early in infection, we observed (i) suppressed induction of STAT1, an ISG, and STAT1 phosphorylation by 12 h PI and (ii) up-regulation of STAT1 protein by 18 h PI in ZIKV− cells compared with ZIKV+ cells (SI Appendix, Fig. S3 H–J).
Proteasome Inhibition Rescues STAT2 Degradation but Minimally Restores ISG Induction.
Studies using cell lines have shown that ZIKV induces STAT2 degradation in a proteasome-dependent manner and that treatment with a proteasome inhibitor can restore STAT2 levels (32, 33). Given that STAT2 is degraded by ZIKV in human macrophages, we wanted to confirm that inhibiting proteasome function would block the degradation of STAT2 and additionally determine if this was sufficient to protect macrophages from ZIKV infection. We infected HMDMs with ZIKV and allowed the virus to establish infection for 12 h. We then added the proteasome inhibitor MG132 or DMSO control and isolated ZIKV+ and ZIKV− macrophages 12 h later (24 h PI) (SI Appendix, Fig. S3K). Addition of MG132, even at 12 h PI when ZIKV has inhibited IFN signaling, significantly, increased STAT2 protein levels (Fig. 2F and SI Appendix, Fig. S3L). However, while restoration of STAT2 levels by MG132 led to significantly increased IFIT3 gene expression in ZIKV+ cells, it did not increase MX1, CXCL10, IFI27, IFI6, and OAS3 expression (Fig. 2G).
ZIKV Degradation of STAT2 Suppresses the Genomic Activation Landscape of Infected Cells.
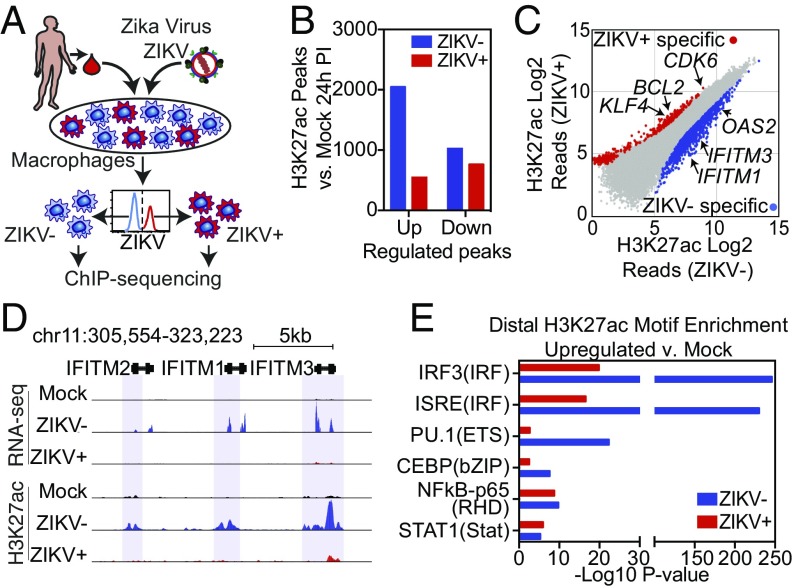
Signal-dependent transcription factors (SDTFs) play major roles in the regulation of gene expression by binding to distal enhancers (34). The activities of transcription factors at enhancer elements can be inferred by changes in H3K27ac, which correlates with transcriptional activity (35). Therefore, motif analysis of genomic regions exhibiting gain or loss of H3K27ac can be used to infer the activity states of the corresponding transcription factors in an unbiased manner. To implement this approach, we performed ChIP-seq for H3K27ac utilizing pure populations of ZIKV+ and ZIKV− cells 24 h PI as well as mock-infected controls from three independent infections using HMDMs derived from three different individuals (Fig. 3A).
Fig. 3.
ZIKV suppresses the activation of genomic regions containing ISRE/IRF motifs. (A) Diagram depicting the infection model for H3K27ac ChIP-seq. HMDMs are infected with ZIKV and stained for ZIKV group antigen followed by FACS isolation of productively infected ZIKV+ and bystander (ZIKV−) macrophages and then followed by H3K27ac ChIP-seq. (B) The number of regions with significantly increased or decreased H3K27ac (fold change >2 and FDR < 0.01) in ZIKV+ or ZIKV− cells compared with mock-infected cells. (C) Scatter plot of H3K27ac tag counts at genomic regions marked by significant H3K27ac in ZIKV+ vs. ZIKV− macrophages 24 h PI. Regions with significantly elevated levels of H3K27ac in ZIKV+ (red) and ZIKV− (blue) are colored. (D) UCSC browser visualization of H3K27ac near the IFITM gene locus in control, ZIKV+, and ZIKV− cells. The upper panel displays transcription as defined by RNA-seq. The lower panel displays H3K27ac abundance in control (Mock, black), ZIKV− (blue), or ZIKV+ (red) macrophages. Regions with significantly up-regulated H3K27ac in ZIKV− cells are marked with blue shading. (E) Comparative motif enrichment at promoter-distal active regulatory regions as defined by H3K27ac in ZIKV− (blue bars) vs. ZIKV+ (red bars) HMDMs.
Similar to the RNA-seq results that showed suppressed gene induction, ZIKV+ macrophages had far fewer significantly up-regulated H3K27ac peaks than ZIKV− cells (547 vs. 2,049 significant peaks) (Fig. 3B). In contrast to ZIKV− cells, ZIKV+ macrophages had more significantly down-regulated H3K27ac peaks than up-regulated peaks (Fig. 3B). Comparing H3K27ac in ZIKV+ and ZIKV− cells identified many H3K27ac regions at promoters and enhancers of genes that were specific to ZIKV− cells. These included ISGs, such as OAS2 and members of the IFITM family, as well as other inflammatory genes (Fig. 3 C and D). Cis-regulatory elements marked exclusively by H3K27ac in ZIKV+ cells were associated with genes involved in cell cycle, cell differentiation, delayed senescence, and apoptosis inhibition such as CDK6, BCL2, and KLF4 (Fig. 3C and SI Appendix, Fig. S4A). Genome-wide analysis of promoter-distal H3K27ac peaks demonstrated strikingly reduced enrichment of IFN regulatory factor (IRF)/IFN-stimulated response element (ISRE) motifs in ZIKV+ cells (Fig. 3E). In contrast, although enriched to a much lesser extent, NF-κB and STAT1 motifs were equally represented in promoter-distal H3K27ac peaks in ZIKV+ and ZIKV− macrophages. Motifs for PU.1 and C/EBP, two macrophage lineage-determining transcription factors (LDTFs), also showed increased representation in distal H3K27ac peaks in ZIKV− cells, likely reflecting their general requirement for binding of SDTFs (34). Collectively, these results provide evidence that the dominant transcriptional response of ZIKV− cells results from activation of type I IFN signaling and suggest that this response is impaired in ZIKV+ cells. STAT2 degradation in ZIKV+ cells leads to inhibition of genomic activation of ISRE/IRF-containing enhancers and promoters, thereby broadly suppressing inflammatory and ISG activation. These findings confirm and extend the literature by demonstrating that, in human macrophages, the type I IFN system is the dominant anti-ZIKV mechanism and that type I IFN signaling, rather than type I IFN production, is the major target of ZIKV antagonism.
ZIKV Infection Reduces RNApol2 Protein Levels and DNA Occupancy, Particularly at Genes Required for Macrophage Identity.
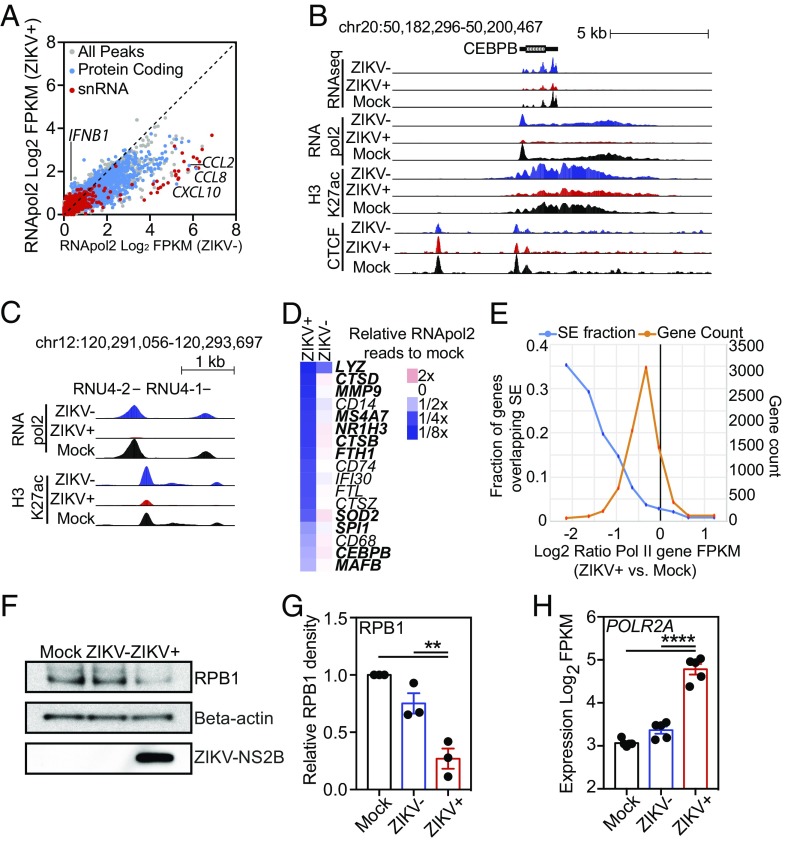
The RNA amounts recovered from ZIKV+ HMDMs were reproducibly lower than the amounts recovered from the corresponding ZIKV− cells [mean ZIKV+-to-ZIKV− ratio = 0.63, 95% CI (0.47–0.80)] (SI Appendix, Fig. S4B). Although the majority of cellular RNA is rRNA synthesized by RNApol1, we focused on RNApol2 because this polymerase produces the majority of protein-coding transcripts involved in antiviral responses. To determine the location and relative quantities of RNApol2 genome-wide, we performed ChIP-seq for RNApol2 in equivalent numbers of pure populations of ZIKV+ and ZIKV− cells 24 h PI as well as in mock-infected controls (Fig. 4A). In three independent infections, using HMDMs derived from three different individuals, we consistently detected less RNApol2 at many locations across the genome in ZIKV+ macrophages than in ZIKV− macrophages (Fig. 4A and SI Appendix, Fig. S4C). To ensure that the lower RNApol2 ChIP levels in ZIKV+ cells were not related to general problems performing ChIP-seq in ZIKV+ cells, we performed ChIP-seq for the CCCTC-binding factor (CTCF) transcription factor that is involved in regulating chromatin structure. In contrast to RNApol2, the levels of CTCF were roughly equivalent in both ZIKV+ and ZIVK− cells (SI Appendix, Fig. S4D). RNApol2 levels were increased at IFNB1 and decreased at MX1, MX2, CXLC10, CCL8, and CCL2 in ZIKV+ cells compared with ZIKV− cells, as expected (Fig. 4A and SI Appendix, Fig. S4E). However, RNApol2 was also significantly decreased at unexpected sites, such as CEBPB, and many snRNAs, such as RNU4-2 and RNU4-1 (Fig. 4 A–C). Additionally, ZIKV+ cells exhibited significantly lower levels of RNApol2 at many genes commonly associated with core macrophage functions such as the LDTFs SPI1, CEBPB, and MAFB, peptidases MMP9, CTSD, and CTSZ, lysozyme LYZ, ferritin light and heavy chain genes FTH1 and FTL, cell-surface receptors CD14 and CD68, and genes involved in antigen processing, i.e., IFI30 and CD74 (Fig. 4D and SI Appendix, Fig. S4F). Many of these genes are associated with superenhancers (SEs) in HMDMs. SEs are regions of disproportionately high densities of active chromatin regulatory marks and transcription factor binding close to genes that play essential roles in the identity and function of cell types (36). While the majority of genes have lower RNApol2 levels in ZIKV+ cells than in mock-infected macrophages, genes associated with SEs demonstrated a disproportionate reduction in RNApol2 signal (Fig. 4E).
Fig. 4.
ZIKV suppresses RNApol2 recruitment. (A) Scatter plot of log2 FPKM RNApol2 tag counts at genomic regions marked by significant RNApol2 in ZIKV+ vs. ZIKV− macrophages at 24 h PI. Color coding: gray, all genomic regions; blue, protein-coding regions; red, snRNA-coding regions. (B) UCSC browser visualization of RNA-seq (first panel), RNApol2 (second panel), H3K27ac (third panel), and CTCF (fourth panel) near the CEBPB gene locus in control (Mock, black), ZIKV− (blue), or ZIKV+ (red) macrophages. (C) UCSC browser visualization of RNApol2 (Upper) and H3K27ac (Lower) near two snRNA genes, RNU4-2 and RNU4-1, in control (Mock, black), ZIKV− (blue), or ZIKV+ (red) macrophages. (D) Heat map depicting relative RNApol2 levels at core macrophage genes in ZIKV− and ZIKV+ macrophages compared with control macrophages (mock). Genes associated with SEs are shown in bold type. Data are the average from three independent experiments. (E) Relationship between changes in RNApol2 and the presence of SEs. Shown is the log2 ratio of RNApol2 reads at individual genes in ZIKV+ cells compared with control cells (Mock). The orange trace shows the total number of genes associated with each ratio of RNApol2 change. The blue trace shows the fraction of genes overlapping SE as a function of their change in RNApol2. (F) Western blot of RPB1, β-actin, and ZIKV-NS2B levels extracted from FACS-isolated equivalent numbers of mock-infected, ZIKV−, and ZIKV+ cells at 24 h PI. (G) Relative quantitation of Western blot RPB1 levels. RPB1 density is relative to β-actin with control samples set to 1. Relative levels (mean ± SEM) of RPB1 in control (Mock), ZIKV−, and ZIKV+ cells are shown for three infections in different individuals at 24 h PI. (H) Log2-transformed FPKM RNA-seq counts for POLR2A in control (Mock), ZIKV−, and ZIKV+ cells 24 h PI. Data represent expression from RNA-seq performed in five different individuals. Data for G and H were analyzed by ANOVA with all-group comparison with correction for multiple comparisons. Asterisks indicate statistically significant differences (****P < 0.0001; **P < 0.01).
RNApol2 is a large protein complex made up of 12 subunits. The largest RNApol2 subunit, RPB1, contains the DNA-binding domain of RNApol2 and a C-terminal domain that is essential for regulating polymerase activity and associated processes. To determine if the decreased RNApol2 ChIP-seq levels in ZIKV+ cells were associated with decreased RNApol2 protein levels, we measured RPB1 levels in mock-infected, ZIKV−, and ZIKV+ cells. ZIKV+ cells had significantly lower levels of RPB1 protein than ZIKV− and mock-infected control macrophages (Fig. 4 F and G and SI Appendix, Fig. S4G). In contrast to RPB1 protein levels, transcription of the gene POLR2A that encodes RPB1 was significantly up-regulated in ZIKV+ cells compared with ZIKV− and mock-infected control macrophages (Fig. 4H). While this observation was similar to the discrepancy in STAT2 RNA and protein levels induced by ZIKV, RPB1 levels did not normalize after the addition of proteasome inhibitor, suggesting that ZIKV employs a different mechanism to lower host cell RPB1 levels. Collectively, these data demonstrate that productive ZIKV infection reduces RNApol2 DNA occupancy globally but that this effect is amplified at core macrophage genes, many of which are associated with SEs.
Discussion
In these studies, we identify the genome-wide signaling networks in primary human macrophages infected with ZIKV. We demonstrate that neighboring ZIKV-infected and uninfected cells have differing, often opposite, transcriptional responses that obscure analyses of mixed populations in which uninfected cells make up even a small percentage of the total population. Our transcriptomic and epigenomic profiles of pure populations of infected macrophages thus provide a more accurate map of the human macrophage signaling response during ZIKV infection. By comparing the transcriptomes and epigenomic features in neighboring ZIKV+ and ZIVK− macrophages that are exposed to identical environmental signals, the mechanisms employed by ZIKV to subvert macrophage immunity are revealed. Importantly, our findings demonstrate that studying a mixed culture of ZIKV-infected HMDMs would lead to the reasonable but erroneous conclusion that infection induces a type I IFN response when, in fact, this response is restricted to cells that do not become productively infected. The direct comparison of ZIKV+ and ZIKV− cells thus provides important insights into the balance of pro- and antiviral mechanisms and an unbiased assessment of the dominant regulatory pathways involved. This approach does not require viral modification and can be performed in difficult-to-infect primary human cells. Thus, it could be used to study genomic responses during infection with any virus and to directly compare host–viral interactions using clinical viral isolates in any cell type.
RNApol2 ChIP-seq in ZIKV+ and ZIKV− cells demonstrated that RNApol2 DNA occupancy is disrupted in cells productively infected with ZIKV. While RNApol2 levels are reduced at most genes, the loss is especially pronounced at many genes associated with SEs that play essential roles in macrophage identity and function. Loss of RNApol2 DNA occupancy in ZIKV+ cells is associated with decreases in protein levels of RPB1, the largest RNApol2 subunit that plays critical roles in both DNA binding and transcriptional elongation. Multiple pathogenic viruses, including poliovirus, some Old World alpha viruses, influenza virus, human herpesvirus 1, and Bunyamwera virus, inhibit RNApol2 during infection through either degradation or dephosphorylation of RPB1, preventing transcription initiation or elongation, respectively (20–24). Suppression of host transcription during viral infection would increase the cellular resources available for viral production and suppress host antiviral responses. The specific mechanism by which ZIKV infection leads to decreases in RPB1 and RNApol2 DNA occupancy and its effects on pathogenesis remain to be elucidated. Significantly smaller and more variable decreases in RNApol2 occupancy were also detected in ZIKV− bystander cells compared with mock-infected cells. Why this occurs in ZIKV− cells is unclear, but it may reflect a response to secreted host or viral environmental signals. Although the exact mechanisms of RNApol2 loss are still being identified, models of CNS infection and infection of glioblastoma cells have suggested that ZIKV causes loss of neural as well as cancer progenitor cells by blocking proliferation and inducing cell death (37, 38). Conceivably, loss of RNApol2 in these cell types could contribute to cell death in these tissues.
Our genome-wide studies of ZIKV infection in primary human macrophages demonstrate targeted suppression of ISRE/IRF-dependent signaling networks that can be explained by STAT2 degradation and are consistent with previous studies in cell lines demonstrating that ZIKV NS5 protein degrades STAT2 in proteasome-dependent manner (32, 33). In macrophages, proteasome inhibitor treatment given more than 4 h after ZIKV has disabled cellular IFN signaling partially restored induction of IFIT3 but not of other ISGs, such as MX1, CXCL10, IFI27, IFI6, and OAS3. While all six of these ISGs have at least one ISRE motif in their promoters [−300 to +100 bp from the transcription start site (TSS)], only CXCL10 and OAS3 have NFκB motifs in their promoters (SI Appendix, Fig. S4H). These and other antiviral ISGs that are not partially rescued by MG132 may also be regulated by NFκB at distal enhancer elements. Thus, although these results could reflect gene-specific requirements for NFκB, which is inhibited by proteosome blockade, they are also consistent with the inhibitory effects of ZIKV on RNApol2.
ZIKV was discovered in 1947 and has caused sporadic human infections for more than 50 y, but only during the recent outbreaks in French Polynesia and South America was ZIKV associated with congenital ZIKV syndrome and Guillain–Barré syndrome (13, 14, 39, 40). Although the specific mechanisms underlying this increased human pathogenicity are still under investigation, it has been suggested that evolution of the ZIKV genome may contribute to this increasing virulence (41, 42). In support of this hypothesis, experimental evidence has demonstrated strain-specific differences in viral pathogenesis and neurologic disease in a STAT2-deficient mouse model and single-amino acid substitutions that increase infectivity of Aedes aegypti mosquitos and human and mouse neural progenitor cells (43–45). Here, we performed genome-wide transcriptomic analysis comparing human macrophage responses to two patient-derived Asian ZIKV subtypes, SD001 and FSS. The specificity of our system is demonstrated by the fact that when bystander ZIKV− cells from SD001 and FSS infections are compared, we detect no significant differences in gene regulation. In contrast, when ZIKV+ cells infected with SD001 or FSS are compared, 110 genes are identified as being regulated in a strain-specific manner. The similar transcriptional response in bystander ZIKV− cells during SD001 and FSS infections suggests that the bystander ZIKV− cells principally respond to secreted environmental signals such as IFNs and cytokines and that these environmental signals are largely the same during FSS and SD001 infections. Indeed our transcriptional analysis shows that the majority of cytokines/chemokines are made in ZIKV− bystander cells. ZIKV+ cells also respond to environmental stimuli, but their response is modified by viral activation and repression of cellular response pathways. In the ZIKV+ populations, strain-specific differences can influence how the virus activates or represses these responses. In ZIKV+ cells, we identified differentially expressed genes with functional enrichment for RNA processing and RNA splicing and include multiple DEAD box RNA helicases (DDX5, DDX17, and DDX42) that are down-regulated in a strain-specific manner. Members of this protein family can act as PRRs that recognize viral RNA and initiate antiviral responses. Japanese encephalitis virus, another flavivirus, has been shown to prevent DDX42 from activating type I IFN signaling (46, 47). Additionally, RNA splicing is modulated by many human pathogenic viruses, and ZIKV was recently shown to cause alternative splicing events in infected neural progenitor cells (48). Given the accuracy of our method and its ability to study differences between unmodified patient-derived viruses in primary human cells, we anticipate that future studies comparing numerous ZIKV strains could identify key virulence mechanisms associated with viral genome evolution.
Although ZIKV largely suppresses inflammatory gene transcription activation, genes associated with specific pathways are selectively up-regulated in ZIKV+ cells. Genes involved in cholesterol biosynthesis (HMGCS1, MVD, MVK, and SQLE), ER/Golgi trafficking (AP1S3 and ASAP2), and cell survival (TNFRSF10D and KLF4) are all induced in ZIKV+ macrophages and, based on previous data, have the potential to increase ZIKV pathogenicity. Cholesterol synthesis plays critical roles in flavivirus innate immune evasion and replication (49). Flaviviruses bud into the ER lumen and require transport through the trans-Golgi to form mature infectious particles and viruses (50). Developing treatments that target these pathways could prove efficacious, as evidenced by the fact that treatment with the HMG-CoA reductase inhibitor lovastatin increased survival rates in a mouse model of DENV-2 infection (51).
In summary, our work demonstrates that ZIKV manipulates macrophage transcription at multiple levels. ZIVK specifically inhibits type I IFN signaling and suppresses global transcription by decreasing RNApol2 levels. Our method for identifying and comparing genome-wide transcriptional and epigenetic changes in neighboring infected and uninfected primary human macrophages using unmodified patient-derived viruses provides an approach to allow rapid deconvolution of complex host–pathogen interactions and directly compare the pathogenicity of clinical viral isolates.
Methods
HMDM Isolation and Differentiation.
Human blood for HMDM isolation was obtained from healthy volunteers and was deidentified under the La Jolla Institute Internal Review Board Protocol VD-057-0217. Donors were HIV, hepatitis B, and hepatitis C negative. Human peripheral blood was separated using Histopaque 1077 (catalog no. 10771; Sigma) spun at 400 × g for 30–60 min at 4 °C. The buffy coat was washed once, and then the red blood cells were lysed with molecular-grade water. Monocytes were then negatively selected from the buffy coat using the pan monocyte isolation kit (catalog no. 130-096-537; Miltenyi Biotech) as described by the manufacturer. Cells were seeded onto tissue-coated plates and differentiated for 7 d in complete macrophage medium [macrophage serum-free medium (catalog no. 12065; Gibco) supplemented with 1% penicillin/streptomycin, 1% Nutridoma-SP (catalog no. 11011375001; Roche), 1% fungizone (catalog no. 15290-018; Gibco), and 100 ng/mL human macrophage-colony stimulating factor (M-CSF) (catalog no. 300-25; PeproTech)] at 37 °C, 5% CO2. The medium was changed every 2–3 d.
ZIKV Infection and Treatments.
On day 7 of culture, macrophages were infected with Zika virus clinical isolates SD001 or FSS13025 at the indicated MOI with or without ADE [MOI = 1 + 0.6% (vol/vol) DENV human immune serum]. The virus was incubated with HMDMs for 2 h at 37 °C, 5% CO2 with rocking every 10–15 min. The supernatant was removed, and cells were washed three times with 1× PBS, before the addition of fresh warm complete macrophage medium. Both RNA-seq and flow cytometry confirmed that no infectious DENV was present in the human immune serum. When indicated, HMDMs were treated with 1,000 U of universal type I IFN (PBL Assay Science), 10 μM MG132, or carrier controls at the indicated time points.
Quantification of Infected HMDMs by Flow Cytometry.
HMDMs were lifted by gentle cell scraping (catalog no. 83.1830; Sarstedt) into PBS. Cells were washed once with FACS buffer (1× PBS, 3% FBS, and 2 mM EDTA). Cells were fixed and permeabilized with BD Cytofix/Cytoperm (BD Biosciences) and washed with BD Perm/Wash Buffer per the manufacturer’s instructions. Samples were blocked with Human Fc block (BioLegend) in BD Perm/Wash Buffer for 10 min and were incubated with 4G2-AF647–conjugated antibody for 30 min. Samples were washed twice with BD Perm/Wash Buffer followed by resuspension in FACS buffer and were analyzed by flow cytometry on a BD LSRII flow cytometer.
Quantitation of Virus Production by FFU.
Baby hamster kidney cells (BHKs) were seeded at 1–2E5 cells per well in a 24-well plate overnight. Supernatants harvested from infection experiments were serial diluted, added to BHKs, and incubated for 2 h with rocking. Supernatant was aspirated, and 1 mL of prewarmed carboxymethyl cellulose (CMC) medium (MEM-alpha containing 1% CMC, 1% Hepes, and 2% FBS) was added. At 2–3 d PI, 1 mL of 4% formalin was added directly to the CMC medium and was incubated for 30 min at room temperature. Cells were washed three times with 1× PBS, permeabilized with 1% Triton X-100, washed, blocked with 10% FBS, and stained with 4G2 primary antibody (1 μg/mL) supplemented with 1% FBS. Wells were washed with 1× PBS and stained with HRP-conjugated secondary antibody supplemented with 1% BSA at room temperature. Cells were washed three times with 1× PBS, and 0.2 mL of TrueBlue (SeraCare) was added and incubated for 20 min at room temperature or until foci became apparent. Plates were then washed and dried, and FFU were counted.
Viral RNA Quantitation by qRT-PCR.
Total RNA was isolated using the RNeasy Mini Kit (74106; Qiagen) as described by the manufacturer. A one-step qPCR was performed using ZIKV primers 853F: 5′-TTGGTCATGATACTGCTGATTGC-3′ and 911R: 5′-CCTTCCACAAAGTCCCTATTGC-3′ and 18S rRNA primers R: 5′-GCTGGAATTACCGCGGCT-3′ and F: 5′-CGGCTACCACATCCAAGGAA-3 using a Bio-Rad qRT-PCR Thermocycler. Viral RNA concentration was determined based on an internal standard curve composed of five 100-fold serial dilutions of in vitro-transcribed RNA from ZIKV strain FSS13025 and normalized to 18S rRNA.
STAT1 Flow Cytometry Analysis.
Macrophages were lifted by gentle cell scraping (catalog no. 83.1830; Sarstedt) into PBS, washed once with PBS, and incubated with Zombie live/dead stain (BioLegend) at 4 °C. After washing, cells were fixed in 4% formaldehyde, washed, and permeabilized in 90% ice-cold methanol. Samples were washed and blocked with Human Fc Block for 10 min at 4 °C followed by 30-min incubation at 4 °C with directly conjugated 4G2-AF647, AF488-Phospho-Stat1 (Tyr701) (58D6), PE-Stat1 (D1K9Y), or isotype controls (Cell Signaling). Samples were centrifuged, washed twice with PBS + 0.5% BSA, and analyzed by flow cytometry on a BD LSRI flow cytometer.
Separation of Infected and Uninfected Cells by FACS for RNA-Seq.
Macrophages were washed once with 1× PBS, gently scraped off in 1× PBS, pelleted at 4 °C, and stained with Zombie Violet viability dye (BioLegend) in the dark. Cells were washed once with FACS buffer (1× PBS, 3% FBS, and 2 mM EDTA) and were fixed and permeabilized with 4% paraformaldehyde (15710-S; Electron Microscopy Sciences) and 0.1% saponin (47036; Sigma-Aldrich) in molecular-grade PBS supplemented with 1:100 RNasin Plus RNase Inhibitor (N2615; Promega) for 30 min at 4 °C. Cells were washed in wash buffer (1× PBS containing 0.2% BSA, 0.1% saponin, and 1:100 RNasin Plus RNase inhibitor) and were blocked for 10 min with human Fc Block (1:500) (BioLegend) in staining buffer (1× PBS containing 1% BSA, 0.1% saponin, and RNasin Plus RNase inhibitor). HMDMs were then stained with 4G2 antibody (BioXcell) conjugated to AF647 (catalog no. A20186; Thermo Scientific) for 30 min at 4 °C, washed twice, resuspended at 5–10E6 cells/mL in sort buffer (PBS containing 0.5% BSA and 1:25 RNasin Plus RNase inhibitor) and sorted into ZIKV+ and ZIKV− cells on a FACSAria cell sorter (BD Biosciences) at the La Jolla Institute for Allergy and Immunology. Gates were set with reference to negative controls.
RNA Isolation and Library Preparation.
After sorting, cells were pelleted at 4 °C, the supernatant was discarded, and total RNA was isolated using the RecoverAll Total Nucleic Acid Isolation Kit (AM1975; Ambion), starting at the protease digestion step. All steps were performed per the manufacturer’s recommendations with the following modification. Cells were incubated in digestion buffer for 3 h at 50 °C supplemented with RNasin Plus RNase inhibitor. RNA was treated with in-column DNase per the manufacturer’s instructions and was eluted; RNA quality was determined by BioAnalyzer using the Eukaryote Total RNA Pico Chip. Samples with RNA integrity number (RIN) values greater than 8.0 were used for library preparation. RNA libraries were generated using the TruSeq stranded total RNA-seq kit (Illumina) per the manufacturer’s instructions and were single-end sequenced for 51 cycles on an Illumina Hi-Seq 2000 or NextSeq 500 system per the manufacturer’s instructions.
ChIP.
For ChIP-seq experiments, HMDMs were cross-linked with 1% formaldehyde for 15 min in the presence of 1 mM sodium butyrate and were quenched with 0.125 M glycine. Preparation of HMDMs for FACS was performed as described above for RNA-seq except that 1× cOmplete protease inhibitors (Roche) and 1 mM sodium butyrate (Sigma) were included in all buffers instead of RNasin Plus RNase inhibitor. Following FACS, cells were washed, pelleted, and snap-frozen. ChIP for histone modification H3K27ac was performed as previously described (52). Sequencing libraries were prepared from recovered DNA (ChIP) using the NEBNext Ultra II DNA library prep kit (New England Biolabs) using NEXTflex DNA Barcodes (Bioo Scientific). ChIP-seq libraries were single-end sequenced for 51 cycles on an Illumina HiSeq 4000 or NextSeq 500 system according to the manufacturer’s instruction.
Western Blot.
HMDMs were processed as described in the ChIP-seq protocol above except that cOmplete protease inhibitors (Roche) and PhosSTOP phosphatase inhibitors (Sigma) were included in all buffers. Following FACS, equal numbers of cells were aliquoted for each condition and were lysed using RIPA buffer. Samples were then boiled in Laemmli Sample Buffer (Bio-Rad) for 5–10 min. Protein lysates were separated by 10% SDS/PAGE, electrophoretically transferred to a nitrocellulose membrane, and immunoblotted at 4 °C overnight with antibodies (1:1,000) against STAT1, STAT1-P, β-actin (Cell Signaling Technology), and NS2B (GTX133308; GeneTex). Membranes were then incubated with an HRP-conjugated second antibody (1:10,000) for 1 h at room temperature followed by detection by enhanced chemiluminescence (Bio-Rad).
ELISA.
Supernatants from mock- or ZIKV-infected HMDMs were removed, aliquoted, snap-frozen, and stored at −80 °C. Aliquots were treated as single-use and did not undergo freeze–thaw cycles. Samples were diluted in sample buffer, and IFN-β ELISA was performed per the manufacturer’s instructions using the VeriKine high-sensitivity human IFN-β serum ELISA kit (PBL Assay Science).
qPCR.
Total RNA was isolated from unsorted cells using the Quick-RNA isolation kit (Zymo), with in-column DNase digestion per the manufacturer’s instructions. RNA from FACS-isolated cells was prepared as described above for the RNA-seq experiments. RNA was converted to cDNA by reverse transcription using the SuperScript III First-strand synthesis kit (Invitrogen). qPCR (SYBR GreenER SuperMix kit; Bio-Rad) analysis was performed on an Applied Biosystems 7300 Real-time PCR system (Invitrogen) using the following primers: RPLP0 forward 5′-GTGTTCGACAATGGCAGCAT-3′; RPLP0 reverse 5′-GACACCCTCCAGGGAGCGA-3′; IFNB1 forward 5′-AGTAGGCGACACTGTTCGTG-3′; IFNB1 reverse 5′-GCCTCCCATTCAATTGCCAC-3′; IFIT3 forward 5′-TCAGAAGTCTAGTCACTTGGGG-3′; IFIT3 reverse 5′-ACACCTTCGCCCTTTCATTTC-3′; MX1 forward 5′-GTGGCTGAGAACAACCTGTG-3′; MX1 reverse 5′-GGCATCTGGTCACGATCCC-3′.
Data Processing.
Preprocessing.
FASTQ files from sequencing experiments were mapped to the University of California, Santa Cruz (UCSC) genome build hg38 (for human) and access KU955593.1 (for the ZIKV genome). STAR with default parameters was used to map RNA-seq experiments. Bowtie2 with default parameters was used to map ChIP-seq experiments. HOMER was used to convert uniquely aligned reads into tag directories for further analysis.
RNA-seq.
RNA-seq reads aligned to a combined GRCh38/hg38 and ZIKV genome were used to calculate the percentage of reads aligned to the ZIKV genome: [(number of reads aligned to the ZIKV genome/number of reads aligned to hg38+ZIKV genomes) × 100 − the average number of reads aligning to the ZIKV genome in mock-infected cells]. RNA-seq reads aligned to the GRCh38/hg38 assembly were used to generate gene expression FPKM (fragments per kilobase of transcript per million mapped reads) values using HOMER (53). Genes with less than 0.5 FPKM in all conditions were defined as not expressed. To measure gene expression, HOMER’s analyzeRepeats.pl utility was used to quantify reads in transcript exons defined by GENCODE. Differentially expressed genes and regularized logarithm (rlog) normalization values for each gene were calculated using DESeq2 while accounting for individual donors in the design matrix. Functional enrichment calculations were performed using Metascape (54), and promoter DNA motif enrichment calculations were performed using HOMER (53). Enrichment values (logP) were clustered using Cluster 3.0 (55) and were visualized using Java TreeView (56). PCA was performed in R (57). In silico modeling of the detection of differentially expressed genes in mixed bystander/infected populations was performed by proportionally blending gene-based read counts from each ZIKV− (bystander) and ZIKV+ RNA-seq replicate experiment into a single mixed experiment for each donor and repeating the differential expression calculations using DESeq2. This was repeated for different fractions of ZIKV+ cells (every 5%) and compared with the lists of differentially expressed genes derived from the pure sorted populations.
ChIP-seq.
ChIP-seq peaks were called on the tags from pooled experiments in each condition with the input DNA as background using HOMER’s findPeaks command using the -style histone (H3K27ac) or -style factor (RNApol2/CTCF) options and were merged using the mergePeaks command. To get differentially bound peaks, tags were counted across the merged peak set for each replicate experiment and analyzed using DESeq2, accounting for donor-matched samples in the design matrix [twofold difference and false-discovery rate (FDR) <0.01]. Motif finding in promoter-distal H3K27ac regions (>3 kb from the TSS) was performed using the findMotifsGenome.pl command using a region size of 1,000 bp. SEs were called using HOMER on pooled H3K27ac samples for mock-infected, ZIKV−, and ZIKV+ conditions. Putative H3K27ac peaks within 12,500 bp were stitched together during SE calling, and peaks overlapping GENCODE-defined promoter regions were excluded during this step. RNApol2-defined expression levels were calculated by treating RNApol2 ChIP-seq data as unstranded RNA-seq and quantifying gene-body FPKM values using HOMER’s analyzeRepeats.pl script.
Statistical Analysis.
Statistical analysis was conducted using GraphPad Prism 7 (GraphPad Software) using the recommended multiple comparison test. Values of P < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Aravinda de Silva (University of North Carolina School of Medicine, Chapel Hill) for providing dengue-immune sera. This research was supported by National Institute of Diabetes and Digestive and Kidney Diseases/NIH Grants R01 DK091183 and PPG DK074868 (to C.K.G.), National Institute of Allergy and Infectious Diseases/NIH Grants R01AI116813, R21NS100477, and R21AI127988 and an Interactive Fund Grant from Kyowa Kirin Pharmaceutical Research (to S.S.), and NIH KL2 Grant 1KL2TR001444 of the Clinical and Translational Science Awards (CTSA) Program and a Career Award for Medical Scientists from the Burroughs Wellcome Fund (to A.F.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-sequencing and ChIP-sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE118305).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807690115/-/DCSupplemental.
References
- 1.Musso D, Gubler DJ. Zika virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 3.Davidson A, Slavinski S, Komoto K, Rakeman J, Weiss D. Suspected female-to-male sexual transmission of Zika virus–New York City, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:716–717. doi: 10.15585/mmwr.mm6528e2. [DOI] [PubMed] [Google Scholar]
- 4.Musso D, et al. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehnert MJ, et al. Screening of blood donations for Zika virus infection–Puerto Rico, April 3-June 11, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:627–628. doi: 10.15585/mmwr.mm6524e2. [DOI] [PubMed] [Google Scholar]
- 6.Calvet G, et al. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: A case study. Lancet Infect Dis. 2016;16:653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 7.Besnard M, Lastere S, Teissier A, Cao-Lormeau V, Musso D. Evidence of perinatal transmission of Zika virus, French Polynesia, December 2013 and February 2014. Euro Surveill. 2014;19:20751. [PubMed] [Google Scholar]
- 8.Gubler DJ. Dengue, urbanization and globalization: The unholy trinity of the 21(st) century. Trop Med Health. 2011;39(Suppl 4):3–11. doi: 10.2149/tmh.2011-S05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO 2017. Zika situation report. 10 March 2017 (WHO, Geneva)
- 11.Driggers RW, et al. Zika virus infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 12.Mlakar J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika virus and birth defects–Reviewing the evidence for causality. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 14.Cao-Lormeau VM, et al. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dos Santos T, et al. Zika virus and the Guillain-Barré syndrome–Case series from seven countries. N Engl J Med. 2016;375:1598–1601. doi: 10.1056/NEJMc1609015. [DOI] [PubMed] [Google Scholar]
- 16.Michlmayr D, Andrade P, Gonzalez K, Balmaseda A, Harris E. CD14+CD16+ monocytes are the main target of Zika virus infection in peripheral blood mononuclear cells in a paediatric study in Nicaragua. Nat Microbiol. 2017;2:1462–1470. doi: 10.1038/s41564-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Sastre A. Ten strategies of interferon evasion by viruses. Cell Host Microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwig A, Landick R, Berkhout B. The battle of RNA synthesis: Virus versus host. Viruses. 2017;9:E309. doi: 10.3390/v9100309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyles DS. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev. 2000;64:709–724. doi: 10.1128/mmbr.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhrymuk I, Kulemzin SV, Frolova EI. Evasion of the innate immune response: The Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol. 2012;86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford N, Fire A, Samuels M, Sharp PA, Baltimore D. Inhibition of transcription factor activity by poliovirus. Cell. 1981;27:555–561. doi: 10.1016/0092-8674(81)90397-4. [DOI] [PubMed] [Google Scholar]
- 22.Fraser KA, Rice SA. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J Virol. 2007;81:5091–5101. doi: 10.1128/JVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas D, et al. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J Biol Chem. 2004;279:31471–31477. doi: 10.1074/jbc.M400938200. [DOI] [PubMed] [Google Scholar]
- 24.Vreede FT, Chan AY, Sharps J, Fodor E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology. 2010;396:125–134. doi: 10.1016/j.virol.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miorin L, Maestre AM, Fernandez-Sesma A, García-Sastre A. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun. 2017;492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardina SV, et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356:175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priyamvada L, et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci USA. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dejnirattisai W, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ngono AE, Shresta S. Immune response to dengue and Zika. Annu Rev Immunol. 2018;36:279–308. doi: 10.1146/annurev-immunol-042617-053142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen JR, et al. Zika virus antagonizes type I interferon responses during infection of human dendritic cells. PLoS Pathog. 2017;13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant A, et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, et al. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep. 2016;17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Saucedo-Cuevas L, Shresta S, Gleeson JG. The neurobiology of Zika virus. Neuron. 2016;92:949–958. doi: 10.1016/j.neuron.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Z, et al. Zika virus has oncolytic activity against glioblastoma stem cells. J Exp Med. 2017;214:2843–2857. doi: 10.1084/jem.20171093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 40.Weaver SC, et al. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metsky HC, et al. Zika virus evolution and spread in the Americas. Nature. 2017;546:411–415. doi: 10.1038/nature22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver SC. Emergence of epidemic Zika virus transmission and congenital Zika syndrome: Are recently evolved traits to blame? MBio. 2017;8:e02063-16. doi: 10.1128/mBio.02063-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripathi S, et al. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog. 2017;13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature. 2017;545:482–486. doi: 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan L, et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017;358:933–936. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- 46.Lin CW, et al. Interferon antagonist function of Japanese encephalitis virus NS4A and its interaction with DEAD-box RNA helicase DDX42. Virus Res. 2008;137:49–55. doi: 10.1016/j.virusres.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B, et al. ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virol J. 2017;14:217. doi: 10.1186/s12985-017-0882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martín-Acebes MA, Vázquez-Calvo Á, Saiz JC. Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses. Prog Lipid Res. 2016;64:123–137. doi: 10.1016/j.plipres.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. Pathogenesis of flavivirus infections: Using and abusing the host cell. Cell Host Microbe. 2009;5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Gutierrez M, Correa-Londoño LA, Castellanos JE, Gallego-Gómez JC, Osorio JE. Lovastatin delays infection and increases survival rates in AG129 mice infected with dengue virus serotype 2. PLoS One. 2014;9:e87412. doi: 10.1371/journal.pone.0087412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gosselin D, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tripathi S, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 56.Saldanha AJ. Java Treeview–Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 57.The R Development Core Team 2013. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.