Abstract
Most analyses of randomised trials with incomplete outcomes make untestable assumptions and should therefore be subjected to sensitivity analyses. However, methods for sensitivity analyses are not widely used. We propose a mean score approach for exploring global sensitivity to departures from missing at random or other assumptions about incomplete outcome data in a randomised trial. We assume a single outcome analysed under a generalised linear model. One or more sensitivity parameters, specified by the user, measure the degree of departure from missing at random in a pattern mixture model. Advantages of our method are that its sensitivity parameters are relatively easy to interpret and so can be elicited from subject matter experts; it is fast and non-stochastic; and its point estimate, standard error and confidence interval agree perfectly with standard methods when particular values of the sensitivity parameters make those standard methods appropriate. We illustrate the method using data from a mental health trial.
Keywords: Intention-to-treat analysis, Longitudinal data analysis, Mean score, Missing data, Randomised trials, Sensitivity analysis
1. Introduction
Missing outcome data are a threat to the validity of randomised controlled trials, and they usually require untestable assumptions to be made in the analysis. One common assumption is that data are missing at random (MAR) (Little and Rubin, 2002). Other possible assumptions may be less implausible in particular clinical settings. For example, in smoking cessation trials, the outcome is binary, indicating whether an individual quit over a given period, and it is common to assume that missing values are failures — “missing=failure” (West et al., 2005); while in weight loss trials, missing data is sometimes assumed to be unchanged since baseline — “baseline observation carried forward” (Ware, 2003).
The US National Research Council (2010) suggested measures that should be taken to minimise the amount of missing outcome data in randomised trials, and described analysis strategies based on various assumptions about the missing data. This report recommended that “Sensitivity analyses should be part of the primary reporting of finding from clinical trials. Examining sensitivity to the assumptions about the missing data mechanism should be a mandatory component of reporting.” However, among “several important areas where progress is particularly needed”, the first was “methods for sensitivity analysis and principled decision making based on the results from sensitivity analyses”. Sensitivity analysis is also an essential part of an intention-to-treat analysis strategy, which includes all randomised individuals in the analysis strategy (White et al., 2011a, 2012): even if the main analysis is performed under MAR and hence draws no information from individuals with no outcome data, such individuals are included in sensitivity analysis and hence in the analysis strategy.
Sensitivity analysis is often done by performing two different analyses, such as an analysis assuming MAR and an analysis by last observation carried forward, and concluding that inference is robust if the results are similar (Wood et al., 2004). Better is a principled sensitivity analysis, where the data analyst typically formulates a model including unidentified ‘sensitivity parameter(s)’ that govern the degree of departure from the main assumption (e.g. from MAR), and explores how the estimate of interest varies as the sensitivity parameter(s) are varied (Rotnitzky et al., 1998; Kenward et al., 2001). We consider global sensitivity analyses where the sensitivity parameter(s) are varied over a range of numerical values that subject-matter experts consider plausible.
Likelihood-based analyses assuming MAR can usually ignore the missing data mechanism and simply analyse the observed data (Little and Rubin, 2002). Under a missing not at random (MNAR) assumption, however, it is usually necessary to model the data of interest jointly with the assumed missing data mechanism. The joint model can be specified as a pattern-mixture model, which explicitly describes the differences between profiles of patients who complete and drop out (Little, 1993, 1994), or as a selection model, which relates the chance of drop-out to the (possibly missing) response values either directly (Diggle and Kenward, 1994; Kenward, 1998) or indirectly through a random effect (Follmann and Wu, 1995; Roy, 2003). Rotnitzky et al. (1998) proposed a selection model for incomplete repeated measures data and showed how to estimate it by inverse probability weighting, given values of the sensitivity parameters. Scharfstein et al. (2003) adopted a non-parametric Bayesian approach to analysing incomplete randomised trial data, and argued that sensitivity parameters are more plausibly a priori independent of other parameters of interest in a selection model than in a pattern-mixture model. Scharfstein et al. (2014) proposed a fully parametric approach based on a selection model. On the other hand, Daniels and Hogan (2000) advocated a pattern-mixture framework as “a convenient and intuitive framework for conducting sensitivity analyses”.
We use the pattern-mixture model in this paper because its sensitivity parameters are usually more easily interpreted (White et al., 2007, 2008). For a binary outcome, a convenient sensitivity parameter is the informative missing odds ratio (IMOR), defined, conditional on covariates, as the odds of positive outcome in missing values divided by the odds of positive outcome in observed values (Higgins et al., 2008; Kaciroti et al., 2009). For a continuous outcome, a convenient sensitivity parameter is the covariate-adjusted mean difference between missing and observed outcomes (Mavridis et al., 2015).
Most estimation procedures described for general pattern-mixture models are likelihood-based (Little, 1993, 1994; Little and Yau, 1996; Hedeker and Gibbons, 2006), while the National Research Council (2010) describes point estimation using sensitivity parameters with bootstrap standard errors. In this paper we propose instead using the mean score method, a computationally convenient method which was originally proposed under a MAR assumption for incomplete outcome data (Pepe et al., 1994) and for incomplete covariates (Reilly and Pepe, 1995). The method is particularly useful to allow for auxiliary variables, so that outcomes can be assumed MAR given model covariates and auxiliary variables but not necessarily MAR given model covariates alone (Pepe et al., 1994). We are not aware of the mean score method having been used for sensitivity analysis.
The aim of this paper is to propose methods for principled sensitivity analysis that are fast, non-stochastic, available in statistical software, and agree exactly with standard methods in the special cases where standard methods are appropriate. We focus on randomised trials with outcome measured at a single time, allowing for continuous or binary outcomes, or indeed any generalised linear model, and for covariate adjustment.
The paper is organised as follows. Section 2 describes our proposed method. Section 3 proposes small-sample corrections which yield exact equivalence to standard procedures in special cases. Section 4 illustrates our method in QUATRO, a mental health trial with outcome measured at a single time. Section 5 describes a simulation study. Section 6 discusses the implementation of our method, possible alternatives, limitations and extensions.
2. Mean score approach
Assume that for the ith individual (i = 1 to n) in an individually randomised trial, there is an outcome variable yi, and let ri be an indicator of yi being observed. Let nobs and nmis = n − nobs be the numbers of observed and missing values of y respectively. Let xi be a vector of covariates including the pS-dimensional fully-observed covariates xSi in the substantive model, comprising an intercept, an indicator zi for the randomised group, and (optionally) baseline covariates. xi may also include fully-observed auxiliary covariates xAi that are not in the substantive model but that help to predict yi, and/or covariates xRi that are only observed in individuals with missing yi and describe the nature of the missing data: for example, the reason for missingness.
The aim of the analysis is to estimate the effect of randomised group, adjusting for the baseline covariates. We assume the substantive model is a generalised linear model (GLM) with canonical link,
| (1) |
where h(.) is the inverse link function. We are interested in estimating βSz, the component of the pS-dimensional vector βS corresponding to z.
If we had complete data, we would estimate βS by solving the estimating equation where and
| (2) |
The mean score approach (Pepe et al., 1994; Reilly and Pepe, 1995) handles missing data by replacing with US(βS), its expectation over the distribution of the missing data given the observed data. We write US(βS) = ∑i USi(βS) and Then by the repeated expectation rule, since xi includes xSi, so US(βS) = 0 is an unbiased estimating equation if is.
To compute USi(βS), we need only E [yi|xi, ri = 0], because (2) is linear in yi. We estimate this using the pattern-mixture model
| (3) |
where xPi = (xSi, xAi) of dimension pP; the subscript P distinguishes the parameters βP and covariates xPi of the pattern-mixture model from the parameters βS and covariates xSi of the substantive model. Models (1) and (3) are typically not both correctly specified: we return to this issue in the simulation study.
In equation (3), Δ(xi) is a user-specified departure from MAR for individual i. MAR in this setting means that p(ri = 1|xi, yi) = p(ri = 1|xi), which implies E [yi|xi, ri] = E [yi|xi] and hence Δ(xi) = 0 for all i. A simple choice of Δ(xi) that expresses MNAR is Δ(xi) = δ, where the departure from MAR is the same for all individuals. Differences in departure from MAR between randomised groups are often plausible and can have strong impact on the estimated treatment effect (White et al., 2007), so an alternative choice is Δ(xi) = δzi . The departure Δ(xi) could also depend on reasons for missingness coded in xRi: for example, it could be 0 for individuals lost to follow-up (if MAR seemed plausible for them), and δzi for individuals who refused follow-up.
Putting it all together, the mean score method solves
| (4) |
where ỹi(βP) is defined as yi if ri = 1 and if ri = 0.
2.1. Estimation using full sandwich variance
The parameter βP in (3) is estimated by regressing yi on xPi in the complete cases (ri = 1). Once βP is estimated, we calculate the ỹi(β̂P) using the known values Δ(xi) and solve (4) for βS. The whole procedure amounts to solving the set of estimating equations U(β) = 0 where U(β) = (US(β)T, UP(βP)T)T, US(β) = ∑i USi(β), UP(βP) = ∑i UPi(βP),
| (5) |
Pepe et al. (1994) derived a variance expression for βS assuming categorical xSi. To accommodate any form of xSi, we instead obtain standard errors by the sandwich method, based on both estimating equations. The sandwich estimator of var (β̂) is
| (6) |
where B = −dU/dβ evaluated at β = β̂, C = ∑iUi(β̂)Ui(β̂)T and Ui(β) = (USi(β)T, UPi(βP)T)T. B and C are given in Section A of the Supplementary Materials.
2.2. Estimation using two linear regressions
A special case arises if there are no auxiliary variables, so xPi = xSi for all i, and h(.) is the identity function, as in linear regression. Then we can rearrange (5) to give
Thus (βS − βP) may be estimated by linear regression of (1 − ri)Δ(xi) on xSi. This estimate is uncorrelated with β̂P because cov (USi(β) − UPi(βP), UPi(βP)) = 0. This gives a direct way to estimate βS, and its variance var (β̂P) + var (β̂S − β̂P), from standard linear regressions.
In particular, consider a two-arm trial with no covariates, and write the coefficients of zi in (1) and (3) as βSz and βPz. Then βSz − βPz is estimated as the difference between arms in the mean of (1 − ri)Δ(xi), which is a1δ1 − a0δ0 where aj is the proportion of missing data in arm j = 0, 1 and δj is the average of Δ(xi) over individuals with missing data in arm j = 0, 1. Therefore the estimated parameter of interest is β̂Sz = β̂Pz + a1δ1 − a0δ0 as in White et al. (2007); the same result can be derived in other ways.
3. Equivalence to standard procedures
We now consider two special cases which can be fitted by standard procedures: (1) when MAR is assumed and there are no auxiliary variables, so incomplete cases contribute no information and the standard procedure is an analysis of complete cases, and (2) when ‘missing = failure’ is assumed for a binary outcome, so the standard procedure is to replace missing values with failures. Our aim is that point estimates, standard errors and confidence intervals produced by the mean score procedure should agree exactly with those produced by the standard procedures in these cases.
Equality of point estimates is easy to see. In case (1), we have Δ(xi) = 0 and xPi = xSi for all i, so if βP solves UP(βP) = 0 then β = (βP, βP) solves US(β) = 0. In case (2), ‘missing = failure’ can be expressed as Δ(xi) = −∞ for all i, so the mean score procedure gives whenever ri = 0, and solving US(βS, βP) = 0 gives the same point estimate as replacing missing values with failures.
Exact equality of variances between mean score and standard procedures depends on which finite sample corrections (if any) are applied. Many such corrections have been proposed to reduce the small-sample bias of the sandwich variance estimator and to improve confidence interval coverage (Kauermann and Carroll, 2001; Lu et al., 2007). Here, we assume that the standard procedures use the commonly used small-sample correction factor for the sandwich variance where f = n/(n − p*), n is the sample size and p* is the number of regression parameters (in linear regression) or 1 (in other GLMs): this is for example the default in Stata (StataCorp, 2011).
Exact equality of confidence intervals between mean score and standard procedures additionally depends on the distributional assumptions used to construct confidence intervals. Here, we assume that standard procedures for linear regression construct confidence intervals from the t distribution with n − p* degrees of freedom, and that standard procedures for other GLMs construct confidence intervals from the Normal distribution.
With missing data, we propose using the same small-sample correction factor, distributional assumptions and degrees of freedom, but replacing n by an effective sample size neff as shown below. Thus we propose forming confidence intervals for linear regression by assuming and for other GLMs by assuming
3.1. Full sandwich method
For the full sandwich method of Section 2.1, we propose computing neff as nobs + (Imis/Imis*)nmis where Imis is the influence of the individuals with missing values, and Imis* is the same individuals’ influence if the missing values had been observed. The comparison of the same individuals is crucial, because missing individuals, if observed, would have different influence from observed individuals.
To determine Imis, we consider weighted estimating equations ∑i wiUi(β) = 0 with solution βw. Differentiating with respect to w = (w1, . . . , wn)T at w = 1 gives where U is a n × (pS + pP) matrix with ith row Ui(β). Since also we get
| (7) |
We now define the influence of observation i as
where the S subscript denotes the elements corresponding to βS. Hence we define the influence of the individuals with missing values as Imis = ∑i(1 − Ri)Imis,i.
To determine Imis* , we let be the (unknown) parameter estimate that would be obtained if the complete data had been observed, following the pattern-mixture model (3). In this case the influence would be
We define the “full-data influence” as the expectation of over the distribution of the complete data given the observed data, under the pattern-mixture model (3). From (7) we get and so that
| (8) |
and is evaluated as the squared residual plus the residual variance from model (1) for individual i. Finally, In case (1), Imis = 0 so neff = nobs, as in standard analysis. In case (2), Imis* = Imis so neff = n, again as in standard analysis.
3.2. Two linear regressions method
For the two linear regressions method of Section 2.2, the small-sample correction to the variance is naturally applied separately to each variance in say. We can derive the corresponding variances without small-sample correction as say. To compute neff, we use the heuristic and hence we estimate neff by solving In case (1), so neff = nobs, as in standard analysis. Case (2) does not apply to linear regression.
4. Example: QUATRO trial
The QUATRO trial (Gray et al., 2006) was a randomised controlled trial in people with schizophrenia, to evaluate the effectiveness of a patient-centred intervention to improved adherence to prescribed antipsychotic medications. The trial included 409 participants in four European centres. The primary outcome, measured at baseline and 1 year, was participants’ quality of life, expressed as the mental health component score (MCS) of the SF-36 (Ware, 1993). The MCS is designed to have mean 50 and standard deviation 10 in a standard population, and a higher MCS score implies a better quality of life. The data are summarised in Table 1.
Table 1.
QUATRO trial: data summary.
| Intervention (n=204) |
Control (n=205) |
||
|---|---|---|---|
| Centre | Amsterdam (%) | 50 (25%) | 50 (24%) |
| Leipzig (%) | 49 (24%) | 48 (23%) | |
| London (%) | 45 (22%) | 47 (23%) | |
| Verona (%) | 60 (29%) | 60 (29%) | |
| MCS at baseline | Mean (SD) | 38.4 (11.2) | 40.1 (12.1) |
| Missing (%) | 13 (6%) | 10 (5%) | |
| MCS at 1 year | Mean (SD) | 40.2 (12.0) | 41.3 (11.5) |
| > 40 (%) | 99 (57%) | 104 (54%) | |
| Missing (%) | 29 (14%) | 13 (6%) | |
We first estimate the intervention effect on MCS, adjusted for baseline MCS and centre. Thus in the substantive model (1), h(.) is the identity link, yi is MCS at 1 year for participant i, and xSi is a vector containing 1, randomised group zi (1 for the intervention group and 0 for the control group), baseline MCS and dummy variables for three centres. We have no xAi or xRi.
We also estimate the intervention effect on a binary variable, MCS dichotomised at an arbitrary value of 40, where for illustration we use baseline MCS and dummy variables for centre as auxiliary variables xAi. Thus in the substantive model (1), h(η) = 1/{1 + exp (−η)} is the inverse of the logit link, yi is dichotomised MCS at 1 year for participant i, and xSi is a vector containing 1 and zi.
In both analyses, we replace the 23 missing values of baseline MCS with the mean baseline MCS: while such mean imputation is not valid in general, it is appropriate and efficient in the specific case of estimating intervention effects with missing baseline covariates in randomised trials (White and Thompson, 2005; Groenwold et al., 2012).
As expected (results not shown), the point estimate, standard error and confidence interval from the mean score method agree exactly with standard methods under MAR and under missing=failure, using the small-sample corrections of Section 3.
We consider three sets of sensitivity analyses using the mean score method around a MAR assumption, with departures from MAR (1) in the intervention arm only (Δ(xi) = δzi), (2) in both arms (Δ(xi) = δ), or (3) in the control arm only (Δ(xi) = δ(1 − zi)) (White et al., 2012). For the quantitative outcome, the investigators suggested that the mean of the missing data could plausibly be lower than the mean of the observed data by up to 10 units (equal to nearly one standard deviation of the observed data), so we allow δ to range from 0 to -10 (Jackson et al., 2010). The investigators were not asked about missing values of the dichotomised outcome, so for illustrative purposes we allow δ to range from 0 to -6, which is close to “missing=failure”.
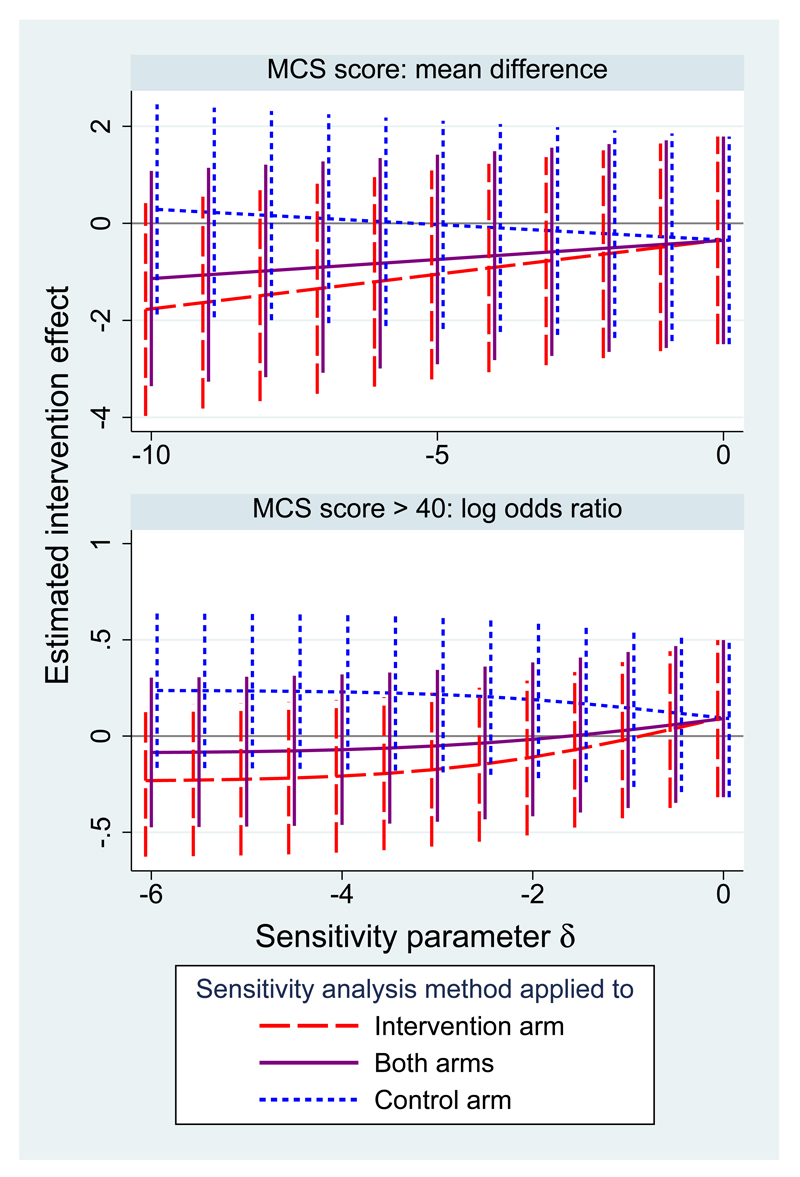
Figure 1 shows the results of these sensitivity analysis using the two linear regressions method for the quantitative outcome (upper panel) and using the full sandwich method for the dichotomised outcome (lower panel). Results are more sensitive to departures from MAR in the intervention arm because there are more missing data in this arm. However, the finding of a non-significant intervention effect is unchanged over these ranges of sensitivity analyses. This means that the main results of the trial are robust even to quite strong departures from MAR.
Figure 1.
QUATRO trial: sensitivity analysis for the estimated intervention effect on the MCS (with 95% confidence interval) over a range of departures from MAR.
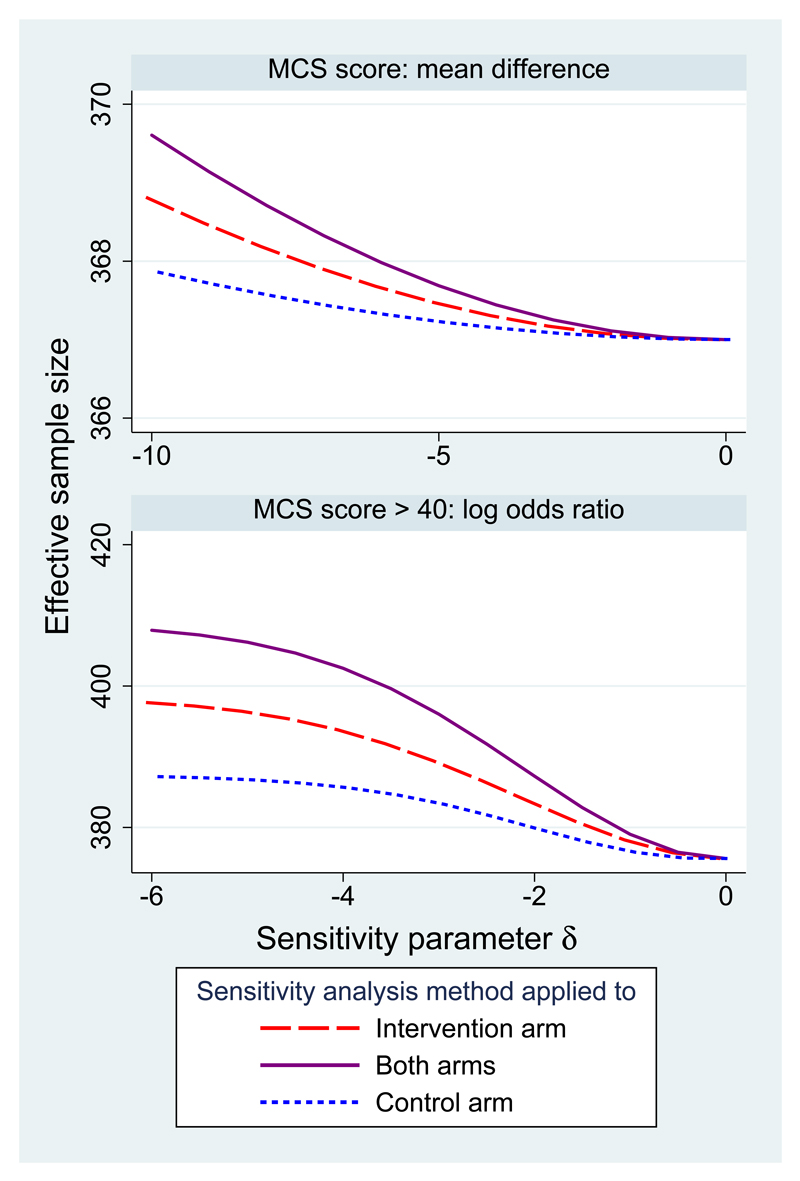
Figure 2 shows the effective sample size for these two analyses. Effective sample size increases from 367 at MAR for both analyses. For the dichotomised outcome it is near the total sample size of 409 when δ = −6 in both arms. For the quantitative outcome it does not pass 370 because the range of δ is more moderate.
Figure 2.
QUATRO data: effective sample size in sensitivity analysis.
5. Simulation study
We report a simulation study aiming (i) to evaluate the performance of the mean score method when it is correctly specified, (ii) to compare the mean score method with alternatives, and (iii) to explore the impact of the incompatibility of models (1) and (3). We assume the sensitivity parameters Δ(xi) are correctly specified.
5.1. Data generating models
We generate data under four data generating models (DGMs), each with four choices of parameters. We focus on the case of a binary outcome. In DGMs 1-3, we generate data under a pattern-mixture model. In DGM 1, there are no baseline covariates, so xi = xSi = xPi = (1, zi). We generate a treatment indicator zi ∼ Bern(0.5); a missingness indicator ri with logit P(ri|zi) = α1 + αzzi; and a binary outcome yi following model (3) with logit P(yi|zi, ri) = βP1 + βPzzi + βPr(1 − ri). The substantive model (1) is then logit P(yi|zi) = βS1 + βSzzi. Because this substantive model contains only a single binary covariate, it is saturated and cannot be mis-specified. Therefore both substantive model and pattern-mixture model are correctly specified
DGM 2 extends DGM 1 by including xi ∼ N(0, 1) as a single baseline covariate independent of zi, so xi = (1, xi, zi). The missingness indicator follows logit P(ri|xi, zi) = α1 + αxxi + αzzi and the binary outcome follows model (3) with logit P(yi|xi, zi, ri) = βP1 + βPxxi + βPzzi + βPr(1 − ri). The substantive model is as in DGM 1 with xSi = (1, zi), and xAi = (xi) is an auxiliary variable in the analysis. Thus the substantive model and pattern-mixture model are again both correctly specified.
DGM 3 is identical to DGM 2, but now xi is included in the substantive model, which is therefore logit P(yi|xi, zi) = βS1 + βSxxi + βSzzi with xSi = xPi = (1, xi, zi). Now the substantive model is incorrectly specified while the pattern-mixture model remains correctly specified.
DGM 4 is a selection model. Here zi and xi are generated as in DGM 2, then yi is generated following the substantive model logit P(yi|xi, zi) = βS1 + βSxxi + βSzzi and ri is generated following logit P(ri|xi, zi, yi) = α1 + αxxi + αzzi + αyyi. Here the substantive model is correctly specified while the pattern mixture model is mis-specified.
For the parameter values, we consider scenarios a-d for each DGM. In scenario a, the sample size is nobs = 500; the missingness model has αx = αz = αy = 1 and we choose α1 to fix πobs = P(r = 1) = 0.75; and the pattern mixture model has βP1 = 0, βPx = βPz = 1, βPr = −1. (αx and βPx are ignored in DGM 1, αy is ignored in DGM 1-3 and βPr is ignored in DGM 4.) Scenarios b-d vary scenario a by setting nobs = 2000, πobs = 0.5, and βPr = −2 respectively. 1000 data sets were simulated in each case. Table 2 summarises the simulation design.
Table 2.
Simulation study: data generating models.
| DGM | Type | PM correct? | SM correct? | Auxiliary variables? |
|---|---|---|---|---|
| 1 | Pattern-mixture | Yes | Yes | No |
| 2 | Pattern-mixture | Yes | Yes | Yes |
| 3 | Pattern-mixture | Yes | No | No |
| 4 | Selection | No | Yes | No |
| Parameters | Description | |||
| a | Base case | |||
| b | Larger sample size | |||
| c | More missing data | |||
| d | Larger departure from MAR | |||
5.2. Analysis methods
The mean score (MS) method is implemented as described in sections 2 and 3, with logit link. xi is used as an auxiliary in DGM 2. In DGM 1-3, Δ(xi) is taken to equal the known value −βPr for all individuals; in DGM 4, Δ(xi) is not known but (for the purposes of the simulation study) is estimated by fitting the pattern-mixture model to a data set of size 1,000,000 before data deletion.
The MS method is compared with analysis of data before data deletion (Full); analysis of complete cases (CC), which wrongly assumes MAR; and two alternative methods that allow for MNAR, multiple imputation (MI) and selection model with inverse probability weighting (SM).
In the MI approach (Rubin, 1987; White et al., 2011b), the imputation model is equation (3), and data are imputed with an offset Δ(xi) in the imputation model. The number of imputations is fixed at 30.
In the SM approach, we use the response model logit p(ri = 1|yi, xi) = αTxPi+Δ*(xi)yi where the sensitivity parameter Δ*(xi) expresses departure from MAR as the log odds ratio for response per 1-unit change in yi. In DGM 4, Δ*(xi) is taken to equal the known value αy; in DGM 1-3, Δ*(xi) is not known but (for the purposes of the simulation study) is estimated by fitting the selection model to a data set of size 1,000,000 before data deletion. The parameters α cannot be estimated by standard methods, since some yi are missing, so we use a weighted estimating equation which does not involve the missing yi’s (Rotnitzky et al., 1998; Dufouil et al., 2004; National Research Council, 2010):
The substantive model is then fitted to the complete cases with stabilised weights where is estimated by the same procedure as but with no Δ*(xi)yi or xAi terms (Robins et al., 2000). Variances are computed by the sandwich variance formula, ignoring uncertainty in α̂.
5.3. Estimand
The estimand of interest is the coefficient βSz in the substantive model. It is computed by fitting the substantive model to the data set of size 1,000,000 before data deletion. We explore bias, empirical and model-based standard errors, and coverage of estimates β̂Sz.
5.4. Results
Results are shown in Table 3. CC is always biased, often inefficient, and poorly covering. Small bias (at most 3% of the true value) is observed in the “Full” analysis (i.e. before data deletion) in some settings: this is a small-sample effect (Nemes et al., 2009), since as noted above, the true value and “Full” are calculated in the same way with large and small samples respectively. Taking “Full” as a gold standard, MS, MI and SM methods all have minimal bias (at most 2% of true values). Precisions of MS and MI are similar, with SM slightly inferior in DGM 2. Coverages are near 95%, with some over-coverage for SM in some settings, as a consequence of slightly overestimated standard errors (results not shown) due to ignoring uncertainty in α̂ (Lunceford and Davidian, 2004). The performance of MS is not appreciably worse when the selection model is mis-specified (DGM 3) or when the pattern-mixture model is mis-specified (DGM 4). Computation times for MI are 15-18 times longer than for MS, which is 10-30% longer than SM.
Table 3.
Simulation results: bias, empirical standard error and coverage of nominal 95% confidence interval for methods Full (before data deletion), CC (complete cases analysis), MS (mean score), MI (multiple imputation), SM (selection model + IPW). Error denotes maximum Monte Carlo error.
| DGM | Bias | Empirical SE | Coverage | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full | CC | MS | MI | SM | Full | CC | MS | MI | SM | Full | CC | MS | MI | SM | |
| 1a | 0.007 | -0.128 | 0.010 | 0.010 | 0.009 | 0.191 | 0.223 | 0.218 | 0.219 | 0.218 | 95.0 | 90.6 | 95.1 | 95.3 | 95.8 |
| 1b | 0.004 | -0.130 | 0.007 | 0.006 | 0.006 | 0.095 | 0.113 | 0.111 | 0.112 | 0.111 | 94.3 | 76.7 | 93.8 | 93.4 | 94.6 |
| 1c | 0.007 | -0.167 | 0.018 | 0.018 | 0.018 | 0.183 | 0.267 | 0.258 | 0.259 | 0.258 | 96.5 | 91.9 | 95.9 | 95.4 | 96.6 |
| 1d | 0.000 | -0.177 | 0.003 | 0.004 | 0.004 | 0.187 | 0.223 | 0.203 | 0.204 | 0.203 | 95.1 | 88.3 | 95.6 | 95.5 | 97.1 |
| 2a | 0.009 | -0.151 | 0.010 | 0.010 | 0.005 | 0.178 | 0.219 | 0.203 | 0.203 | 0.209 | 96.3 | 90.1 | 95.4 | 95.4 | 97.0 |
| 2b | 0.004 | -0.157 | 0.003 | 0.003 | -0.001 | 0.092 | 0.109 | 0.101 | 0.101 | 0.104 | 94.6 | 71.5 | 95.1 | 95.3 | 96.9 |
| 2c | 0.005 | -0.206 | 0.002 | -0.001 | -0.004 | 0.185 | 0.296 | 0.256 | 0.257 | 0.290 | 94.9 | 88.1 | 95.0 | 94.7 | 97.2 |
| 2d | 0.011 | -0.175 | 0.011 | 0.010 | 0.007 | 0.186 | 0.227 | 0.200 | 0.200 | 0.202 | 95.0 | 86.1 | 94.5 | 95.2 | 98.2 |
| 3a | 0.016 | -0.110 | 0.021 | 0.020 | 0.015 | 0.208 | 0.249 | 0.245 | 0.246 | 0.249 | 96.5 | 92.7 | 95.4 | 94.7 | 95.3 |
| 3b | 0.003 | -0.127 | 0.003 | 0.003 | -0.003 | 0.106 | 0.122 | 0.120 | 0.120 | 0.121 | 94.8 | 81.8 | 95.3 | 95.2 | 95.3 |
| 3c | 0.012 | -0.160 | 0.015 | 0.012 | 0.018 | 0.217 | 0.327 | 0.316 | 0.318 | 0.323 | 95.1 | 90.3 | 95.0 | 95.1 | 95.4 |
| 3d | 0.019 | -0.163 | 0.020 | 0.020 | 0.020 | 0.216 | 0.254 | 0.238 | 0.239 | 0.249 | 94.2 | 88.6 | 94.5 | 94.6 | 95.3 |
| 4a | 0.005 | -0.148 | 0.014 | 0.012 | 0.006 | 0.218 | 0.255 | 0.251 | 0.252 | 0.253 | 94.2 | 90.0 | 94.8 | 94.5 | 95.0 |
| 4b | 0.006 | -0.147 | 0.015 | 0.015 | 0.008 | 0.107 | 0.124 | 0.122 | 0.122 | 0.123 | 94.0 | 77.1 | 94.0 | 94.1 | 94.6 |
| 4c | 0.008 | -0.203 | 0.004 | 0.002 | 0.011 | 0.213 | 0.345 | 0.336 | 0.337 | 0.343 | 94.2 | 90.8 | 94.2 | 94.3 | 95.0 |
| 4d | 0.026 | -0.118 | 0.045 | 0.045 | 0.036 | 0.211 | 0.250 | 0.246 | 0.248 | 0.249 | 94.2 | 91.2 | 94.3 | 94.5 | 94.8 |
| Error | 0.007 | 0.011 | 0.011 | 0.011 | 0.011 | 0.005 | 0.008 | 0.008 | 0.008 | 0.008 | 0.8 | 1.4 | 0.8 | 0.8 | 0.7 |
6. Discussion
We have proposed a mean score method which works well when the sensitivity parameters are known. In practice, of course, the sensitivity parameters are unknown, and a range of values will be used in a sensitivity analysis.
The main practical difficulty in implementing any principled sensitivity analysis is choosing the value(s) of the sensitivity parameters. This is a subjective process requiring subject-matter knowledge and is best done by discussion between statisticians and suitable ‘experts’, typically the trial investigators. By using the pattern-mixture model, we use a sensitivity parameter Δ(xi) that is easier to communicate with ‘experts’ than the corresponding parameters in selection models or shared parameter models. The procedure has been successfully applied in several trials (White, 2015). Special attention is needed to the possibility that Δ(xi) varies between randomised groups, because estimated treatment effects are highly sensitive to such variation (White et al., 2007). As with all aspects of trial analysis, plausible ranges of the Δ(xi) parameters should be defined before the data are collected or before any analysis. An alternative approach would report the “tipping point”, the value of Δ(xi) for which the main results are substantively affected, leaving the reader to make the subjective decisions about the plausibility of more extreme values (Liublinska and Rubin, 2014). Presenting this information could be complex without subjective decisions about the difference in Δ(xi) between randomised groups. Effective methods are therefore needed for eliciting sensitivity parameters.
Our method does not incorporate data on discontinuation of treatment, unless this can be included as an auxiliary variable. Our method would however be a suitable adjunct to estimation of effectiveness in a trial with good follow-up after discontinuation of treatment. Further work is needed to combine our sensitivity analysis with models for outcome before and after discontinuation of treatment (Little and Yau, 1996). In a drug trial in which follow-up ends on discontinuation of treatment, we see our method as estimating efficacy or a de jure estimand; if effectiveness or a de facto estimand is required then post-discontinuation missing data in each arm may be imputed by the methods of Carpenter et al. (2013). Further work is needed to perform a full sensitivity analysis in this setting.
Our use of parametric models makes our results susceptible to model mis-specification, and indeed in many cases models (1) and (3) cannot both be correctly specified except under MAR. However, the simulation study shows that the impact of such model inconsistency is small relative to the impact of assumptions about the missing data and the difficulty of knowing the values of the sensitivity parameters.
We compared the mean score method with multiple imputation and inverse probability weighting. In the case of a quantitative outcome, MI can be simplified by imputing under MAR and then adding the offset to the imputed data before fitting the substantive model and applying Rubin’s rules. We could also impute under MAR and then use a weighted version of Rubin’s rules to allow for MNAR (Carpenter et al., 2007). Both MI methods are subject to Monte Carlo error and so seem inferior to the mean score method. A full likelihood-based analysis of the selection model would also be possible, and a Bayesian analysis could directly allow for uncertainty about Δ(xi) in a single analysis (Mason et al., 2012). These alternative approaches are both more computationally complex.
The proposed mean score method can be extended in various ways. We have illustrated the method for departures from a MAR assumption, but it can equally be used if the primary analysis with a binary outcome assumed “missing = failure”, by varying Δ(xi) from −∞ rather than from 0. The method is also appropriate in observational studies, except that mean imputation for missing covariates is not appropriate in this context. The method can be applied to a cluster-randomised trial as described in Section B of the Supplementary Materials. Further work could allow the imputation and substantive models to have different link functions, non-canonical links to be used with suitable modification to US, and extension to trials with more than two arms.
Sensitivity analysis should be more widely used to assess the importance of departures from assumptions about missing data. The proposed mean score approach provides data analysts with a fast and fully theoretically justified way to perform the sensitivity analyses. It is implemented in a Stata module rctmiss available from Statistical Software Components (SSC) at https://ideas.repec.org/s/boc/bocode.html.
Supplementary Materials
The supplementary materials give details of the sandwich variance in equation (6) and sketch an extension to clustered data.
Acknowledgements
IRW was funded by the Medical Research Council [Unit Programme number U105260558]. NJH was funded by NIH grant 5R01MH054693-12. JC was funded by ESRC Research Fellowship RES-063-27-0257. We thank the QUATRO Trial Team for access to the study data.
References
- Carpenter JR, Kenward MG, White IR. Sensitivity analysis after multiple imputation under missing at random: a weighting approach. Statistical Methods in Medical Research. 2007;16(3):259–275. doi: 10.1177/0962280206075303. [DOI] [PubMed] [Google Scholar]
- Carpenter JR, Roger JH, Kenward MG. Analysis of longitudinal trials with protocol deviation: a framework for relevant, accessible assumptions, and inference via multiple imputation. Journal of Biopharmaceutical Statistics. 2013;23(3):1352–71. doi: 10.1080/10543406.2013.834911. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Hogan JW. Reparameterizing the Pattern Mixture Model for Sensitivity Analyses Under Informative Dropout. Biometrics. 2000;56(4):1241–1248. doi: 10.1111/j.0006-341x.2000.01241.x. [DOI] [PubMed] [Google Scholar]
- Diggle P, Kenward MG. Informative drop-out in longitudinal data analysis. Applied Statistics. 1994;43(1):49–93. [Google Scholar]
- Dufouil C, Brayne C, Clayton D. Analysis of longitudinal studies with death and drop-out: a case study. Statistics in Medicine. 2004;23(14):2215–2226. doi: 10.1002/sim.1821. [DOI] [PubMed] [Google Scholar]
- Follmann D, Wu M. An approximate generalized linear model with random effects for informative missing data. Biometrics. 1995;51:151–168. [PubMed] [Google Scholar]
- Gray R, Leese M, Bindman J, Becker T, Burti L, David A, Gournay K, Kikkert M, Koeter M, Puschner B, Schene A, et al. Adherence therapy for people with schizophrenia: European multicentre randomised controlled trial. The British Journal of Psychiatry. 2006;189(6):508–514. doi: 10.1192/bjp.bp.105.019489. [DOI] [PubMed] [Google Scholar]
- Groenwold RHH, White IR, Donders ART, Carpenter JR, Altman DG, Moons KGM. Missing covariate data in clinical research: when and when not to use the missing-indicator method for analysis. Canadian Medical Association Journal. 2012;184(11):1265–1269. doi: 10.1503/cmaj.110977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedeker DR, Gibbons RD. Longitudinal data analysis. John Wiley and Sons; 2006. [Google Scholar]
- Higgins JP, White IR, Wood AM. Imputation methods for missing outcome data in meta-analysis of clinical trials. Clinical Trials. 2008;5(3):225–239. doi: 10.1177/1740774508091600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, White IR, Leese M. How much can we learn about missing data?: an exploration of a clinical trial in psychiatry. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2010;173(3):593–612. doi: 10.1111/j.1467-985X.2009.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaciroti NA, Schork MA, Raghunathan T, Julius S. A Bayesian sensitivity model for intention-to-treat analysis on binary outcomes with dropouts. Statistics in Medicine. 2009;28(4):572–585. doi: 10.1002/sim.3494. [DOI] [PubMed] [Google Scholar]
- Kauermann G, Carroll RJ. A note on the efficiency of sandwich covariance matrix estimation. Journal of the American Statistical Association. 2001;96(456):1387–1396. [Google Scholar]
- Kenward M, Goetghebeur E, Molenberghs G. Sensitivity analysis for incomplete categorical data. Statistical Modelling. 2001;1(1):31–48. [Google Scholar]
- Kenward MG. Selection models for repeated measurements with non-random dropout: an illustration of sensitivity. Statistics in Medicine. 1998;17(23):2723–2732. doi: 10.1002/(sici)1097-0258(19981215)17:23<2723::aid-sim38>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Little R, Yau L. Intent-to-treat analysis for longitudinal studies with drop-outs. Biometrics. 1996;52(4):1324–1333. [PubMed] [Google Scholar]
- Little RJA. Pattern-mixture models for multivariate incomplete data. Journal of the American Statistical Association. 1993;88(421):125–134. [Google Scholar]
- Little RJA. A class of pattern-mixture models for normal incomplete data. Biometrika. 1994;81(3):471–483. [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. second edition Hoboken: N. J.: Wiley; 2002. [Google Scholar]
- Liublinska V, Rubin DB. Sensitivity analysis for a partially missing binary outcome in a two-arm randomized clinical trial. Statistics in Medicine. 2014;33(24):4170–4185. doi: 10.1002/sim.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Preisser JS, Qaqish BF, Suchindran C, Bangdiwala SI, Wolfson M. A comparison of two bias-corrected covariance estimators for generalized estimating equations. Biometrics. 2007;63(3):935–941. doi: 10.1111/j.1541-0420.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Lunceford JK, Davidian M. Stratification and weighting via the propensity score in estimation of causal treatment effects: a comparative study. Statistics in Medicine. 2004;23:2937–2960. doi: 10.1002/sim.1903. [DOI] [PubMed] [Google Scholar]
- Mason A, Richardson S, Plewis I, Best N. Strategy for modelling nonrandom missing data mechanisms in observational studies using Bayesian methods. Journal of Official Statistics. 2012;28(2):279–302. [Google Scholar]
- Mavridis D, White IR, Higgins JPT, Cipriani A, Salanti G. Allowing for uncertainty due to missing continuous outcome data in pair-wise and network meta-analysis. Statistics in Medicine. 2015;34:721–741. doi: 10.1002/sim.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. The Prevention and Treatment of Missing Data in Clinical Trials. Washington, DC: Panel on Handling Missing Data in Clinical Trials. Committee on National Statistics, Division of Behavioral and Social Sciences and Education. The National Academies Press; 2010. [Google Scholar]
- Nemes S, Jonasson JM, Genell A, Steineck G. Bias in odds ratios by logistic regression modelling and sample size. BMC Medical Research Methodology. 2009;9(1):56. doi: 10.1186/1471-2288-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Reilly M, Fleming TR. Auxiliary outcome data and the mean score method. Journal of Statistical Planning and Inference. 1994;42:137–160. [Google Scholar]
- Reilly M, Pepe MS. A mean score method for missing and auxiliary covariate data in regression models. Biometrika. 1995;82(2):299–314. [Google Scholar]
- Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;11:550–560. doi: 10.1097/00001648-200009000-00011. [DOI] [PubMed] [Google Scholar]
- Rogers WH. Regression standard errors in clustered samples. Stata Technical Bulletin. 1993;13:19–23. [Google Scholar]
- Rotnitzky A, Robins JM, Scharfstein DO. Semiparametric regression for repeated outcomes with nonignorable nonresponse. Journal of the American Statistical Association. 1998;93(444):1321–1339. [Google Scholar]
- Roy J. Modeling longitudinal data with nonignorable dropouts using a latent dropout class model. Biometrics. 2003;59(4):829–836. doi: 10.1111/j.0006-341x.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. New York: John Wiley and Sons; 1987. [Google Scholar]
- Scharfstein D, McDermott A, Olson W, Wiegand F. Global sensitivity analysis for repeated measures studies with informative dropout: a fully parametric approach. Statistics in Biopharmaceutical Research. 2014;6(4):338–348. [Google Scholar]
- Scharfstein DO, Daniels MJ, Robins JM. Incorporating prior beliefs about selection bias into the analysis of randomized trials with missing outcomes. Biostatistics. 2003;4:495–512. doi: 10.1093/biostatistics/4.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. Stata Statistical Software: Release 12. College Station, TX: StataCorp LP; 2011. [Google Scholar]
- Ware JE. SF-36 Health Survey: Manual and Interpretation Guide. Boston: The Health Institute, New England Medical Center; 1993. [Google Scholar]
- Ware JH. Interpreting incomplete data in studies of diet and weight loss. New England Journal of Medicine. 2003;348(21):2136–2137. doi: 10.1056/NEJMe030054. [DOI] [PubMed] [Google Scholar]
- West R, Hajek P, Stead L, Stapleton J. Outcome criteria in smoking cessation trials: proposal for a common standard. Addiction. 2005;100:299–303. doi: 10.1111/j.1360-0443.2004.00995.x. [DOI] [PubMed] [Google Scholar]
- White IR. Sensitivity analysis: The elicitation and use of expert opinion. In: Molenberghs G, Fitzmaurice G, Kenward MG, Tsiatis A, Verbeke G, editors. Handbook of Missing Data Methodology. Chapman and Hall; 2015. [Google Scholar]
- White IR, Thompson SG. Adjusting for partially missing baseline measurements in randomized trials. Statistics in Medicine. 2005;24(7):993–1007. doi: 10.1002/sim.1981. [DOI] [PubMed] [Google Scholar]
- White IR, Carpenter J, Evans S, Schroter S. Eliciting and using expert opinions about dropout bias in randomized controlled trials. Clinical Trials. 2007;4(2):125–139. doi: 10.1177/1740774507077849. [DOI] [PubMed] [Google Scholar]
- White IR, Higgins JPT, Wood AM. Allowing for uncertainty due to missing data in meta-analysis - Part 1: Two-stage methods. Statistics in Medicine. 2008;27(5):711–727. doi: 10.1002/sim.3008. [DOI] [PubMed] [Google Scholar]
- White IR, Horton NJ, Carpenter J, Pocock SJ. Strategy for intention to treat analysis in randomised trials with missing outcome data. British Medical Journal. 2011a;342:d40. doi: 10.1136/bmj.d40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White IR, Wood A, Royston P. Tutorial in biostatistics: Multiple imputation using chained equations: issues and guidance for practice. Stat Med. 2011b;30:377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- White IR, Carpenter J, Horton NJ. Including all individuals is not enough: lessons for intention-to-treat analysis. Clinical Trials. 2012;9:396–407. doi: 10.1177/1740774512450098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, White IR, Thompson SG. Are missing outcome data adequately handled? A review of published randomized controlled trials in major medical journals. Clinical Trials. 2004;1(4):368–376. doi: 10.1191/1740774504cn032oa. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.