Abstract
It has become clear that early life (including in utero exposures), is a key window of vulnerability where environmental exposures can alter developmental trajectories and initiate allergic disease development. However, recent evidence suggests that there may be additional windows of vulnerability to environmental exposures in the parental generation before conception, or even in previous generations. There is evidence suggesting that information of prior exposures can be transferred across generations, and experimental animal models suggest that such transmission may be conveyed through epigenetic mechanisms. While the molecular mechanisms of inter- and trans-generation epigenetic transmission have yet to be determined, the realisation that environment before conception may alter risks of allergic diseases, has profound implications for the development of public health interventions to prevent disease. Future research in both experimental models and in multigenerational human cohorts is needed to better understand the role of inter- and trans-generational effects in asthma and allergic disease. This will provide the knowledge basis for a new approach to efficient intervention strategies aimed at reducing the major public health challenge of these conditions.
Keywords: transgenerational, intergenerational, multigenerational, parental, epigenetics, DNA methylation, asthma, allergy, environment
INTRODUCTION
Asthma and allergies have increased exponentially over recent decades of industrialization and urbanization. The impact and severity of these multifactorial diseases are still rising in many low and lower-middle income countries, particularly among younger age groups (1–4), causing a substantial burden of disease from early childhood years. Despite major initiatives for prevention, no strategies have so far succeeded in substantially decreasing morbidity. Asthma and allergy now constitute major common chronic inflammatory diseases worldwide, and are recognized as a global public health concern (5).
Extensive literature has addressed a large number of factors shown to be associated with asthma and allergic disease (6, 7). The more traditional risk factors include environmental toxicants (810), indoor mould and dampness (11), outdoor air pollution (12, 13), occupation (14, 15), and dietary factors (16–18). Women’s hormonal/metabolic status (19, 20), climate factors (21, 22), tuberculosis (23), parasitic worms (24), and overall loss of protective factors such as reduced exposure to infectious agents and symbiotic microorganisms (25) are also of interest. Epidemiological research has increasingly acknowledged the importance of developmental origins, with early environmental exposures, being key determinants for later onset of allergic diseases (26–28). In particular, early life biodiversity (29–31) is believed to play a role in the causality of allergies. This focus on early life development has driven a search for new approaches starting during pregnancy and early childhood to prevent allergies. However, to date, no intervention has proved effective to substantially reduce or prevent asthma and allergies.
An emerging understanding of the pathophysiological mechanisms involved in development and persistence of allergic diseases, reveals complex gene-environment interactions, with many genes have been identified in which genetic variants are associated with allergic phenotype (32–35), and interact with multiple environmental factors. However it is clear that the inherited sequence variation associated with allergic disease across the genome identified to date only explains a part of the heritability of allergic disease (36).
The epigenome refers to the information in the genome, that lies “above” the DNA sequence, controls the expression of genes by mechnisms such as DNA methylation and histone modifications. Importantly, the epigenome is in part heritable through cell division (mitosis) and is fundamental to control tissue differentiation and cellular responsiveness. The epigenome of a cell or tissue is determined by both DNA sequence and cellular or organismal environmental exposures, as well as by stochasticity. Partially stable in the course of mitosis, epigenetic information establishes a memory (or signature) of past exposures particularly in developmental transitions. Thus, the epigenome integrates influences of the genome, development and environmental exposures, and is increasingly being recognised to play a key role in the pathophysiology of disease (37).
Epigenetics has been defined by Ptashne in 2007 by three criteria: (I) a change in the activity of a gene that does not involve a mutation, (II) that is initiated by a signal, and (III) that can result in altered disease risk in the absence of the signal that initiated its change (38). Classically, four epigenetic mechanisms have been identified: (a) DNA methylation, (b) histone modification, (c) chromatin remodeling, and (d) small (21- to 26-nt) non-coding RNAs. There is ample evidence that DNA methylation (DNAm) fulfills all three criteria required to be considered as an epigenetic mechanism (39–41). Histone modifications fulfill the criteria as they have the potential to result from exogenous signals such as cigarette smoke, alter gene activity, and are maintained through mitosis (42–44). However, meiotic inheritance of histone modification has only been demonstrated in C. elegans (45). DNAm usually works hand in hand with histone modifications to activate or silence genes by influencing chromatin structure and it’s accessibility by transcription factors (46). MicroRNAs (miRNAs) are also controlled by exogenous factors and alter gene activity by either inhibiting translation or degrading messenger RNAs (mRNA) (47, 48). For instance, in humans, miRNAs have been demonstrated to be differentially expressed in current and never smokers, and to be related to particulate matter exposure (42, 49). Currently there is little evidence that environmentally induced miRNAs expression patterns can be inherited (50). However, since miRNAs are part of the genetic code, it is possible that DNAm may affect the activity of miRNAs and thus facilitate inheritance.
The role of epigenetic regulation in the aetiology of asthma and allergy is becoming increasingly evident (51–56). Further, elucidating the epigenetic mechanisms involved in inflammation and the immune response to allergens will provide better understanding of the pathophysiology of allergic disease and a mechanistic understanding of how genes and environment interact to determine disease susceptibility. While the majority of studies of the epigenetics of allergic disease have focused on identifying epigenetic marks that are present before disease development (e.g. in cord blood) or in individuals with disease, this approach cannot explain the missing heritability (the problem where single genetic variations are unable to explain for much of the heritability in diseases) in allergic disease described above. However, the recognition that epigenetic information may be transmitted across generations (i.e. through meiosis) provides a mechanism whereby epigenetics could contribute to heritability of disease, and explain observations of trans-generational effects of environmental exposure on risk of allergic disease (57). This review aims to summarize the evidence for trans- and inter-generational inheritance of allergic disease, and the role of epimutations and epigenetic inheritance in allergic disease.
Transgenerational versus intergenerational inheritance
It is important to note that while early-life, including in utero, exposure to environmental factors has been shown to represent a key susceptibility window for allergic disease (58), however, this does not represent true transgenerational inheritance where epigenetic information is passed between generations. As discussed by Arshad at al. (57), there are a number of ways in which cross-generational effects may be transmitted and result in apparent transmission of disease risk between generations. Genetic inheritance across generations can explain familial resemblance in phenotypes, but cannot account for alterations in disease risk as a result of environmental exposures of prior generations in the absence of continued exposure. Shared familial environment or other cultural effects can also result in similarity of disease phenotypes between generations. In addition, there is the possibility of epigenetically mediated effects to explain transmission of disease or the effect of environmental factors across generations. With regard to epigenetic effects it is important to distinguish between intergenerational and transgenerational inheritance (Figure 1). Intergenerational effects occur when maternal environmental exposures (F0) have direct effects on the germ cells, or developing fetus (including the germ line of the fetus, leading to altered phenotype of the child (F1) and possibly grandchild (F2). On the paternal line environmental exposures of the father can have direct effects on the germ cells that will form child (F1). A true transgenerational effect, where epigenetic information is transmitted across generations, can only be proven if the effect of exposure is transmitted to the F2 (on the paternal line, or in a maternal line where exposure occurred only prior to conception), or F3 (on the maternal line when exposure occurs during pregnancy), and possibly future generations, in the absence of further environmental exposure or germline mutations (Figure 1).
Figure 1:
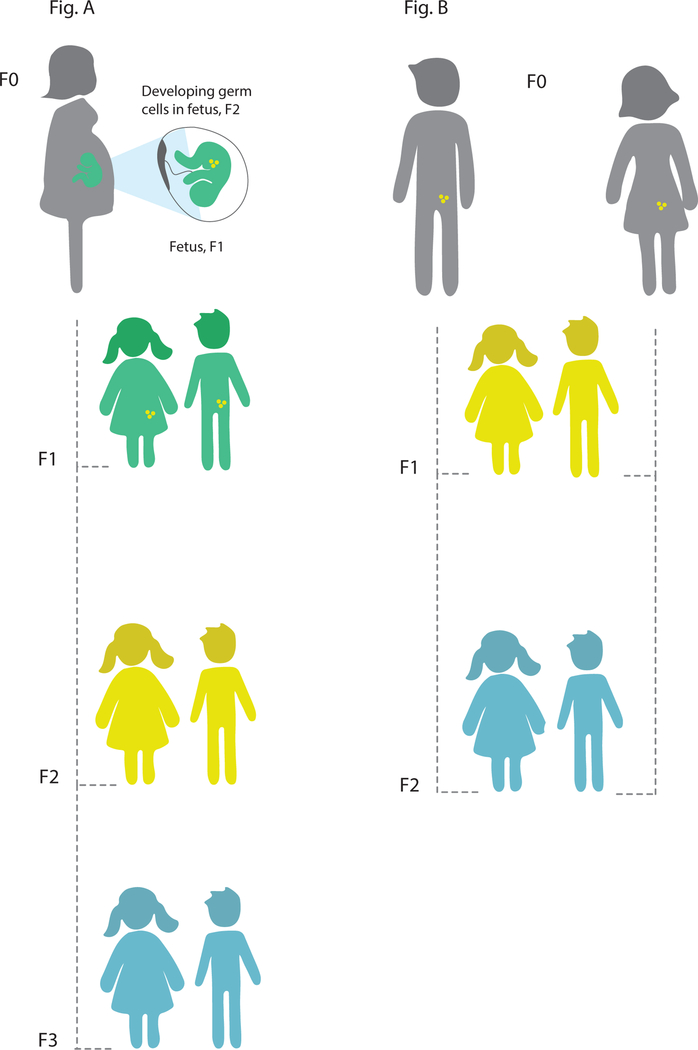
Principles of inter- and transgenerational epigenetic inheritance. (A) If a pregnant woman (F0) is exposed to an environmental stressor, her son/daughter (F1, green) and his/her germ cells that will form F2 (yellow) are also directly exposed and this may result in intergenerational effects. The third generation (F3, blue) is the 1st generation that could represent transgenerational epigenetic inheritance. (B) If a man or a woman (F0) and their germ cells to F1 (yellow) is directly exposed to an enironmental stressor, the F2 offspring (blue) is the 1st generation that could represent transgenerational epigenetic inheritance.
Others have suggested that transgenerational similarity in DNAm is attributable to genetic effects by methylation quantitative trait loci (methQTL) (59–61), i.e., single nucleotide polymorphisms (SNPs) that increase the susceptibility for the methylation of specific CpGs, such as those observed at the 17q21 asthma susceptibilty locus where there is strong association between SNPs and CpG sites related to gene expression, illustrating the complex relationship between sequence variation, CpG methylation and gene expression (62, 63). Another mechanism whereby genetic effects can cause transgenerational similarity in the epigenome is Metastable epialleles. These are alleles that are variably expressed in genetically identical individuals due to epigenetic modifications established during early development and are thought to be particularly vulnerable to environmental influences (64), such as the Agouti locus in mice (65). A genetic contribution is also supported by findings that methylation and gene expression differences were smaller in monozygotic compared to dizygotic twins (66, 67). Investigation of monozygotic twins have been considered to offer a human analog of inbred animal studies (68).
Evidence for inter- and trans-generational inheritance
A number of studies have shown that environmental exposures can lead to transgenerational inheritance of phenotypes in animal models. For example, in Drosophila, maternal high sugar caloric intake has been found to affect body composition and metabolism of at least two generations (69). In another study, exposures of mothers in early life (the larval period) to a transient high caloric diet was found to result in significant difference in offspring development and metabolism, and this also extended to the next generation (70). In C. elegans, it has been found that the manipulation of H3K4me3 chromatin modifiers can induce an epigenetic memory of longevity in subsequent generations (45) and the effect of starvation-induced developmental arrest can inherited through at least three generations (71).
Evidence for transgenerational effects of environmental exposures have also been found in vertebrate models. For example, exposure of zebrafish embryos to the environmental toxin benzo[a]pyrene has been found leading to neurobehavioral and physiological deficits in the F2 generation (72). In mammals, it has been demonstrated that early life traumatic stress in the paternal line resulted in altered microRNA (miRNA) expression, and behavioural and metabolic responses in the progeny (73).
Exploring potential trans- and intergenerational epigenetic inheritance in multi-generational human studies is difficult due to the long life-cycle of humans, lack of data accuracy (often using participant recall of their own and previous generations’ exposures and outcomes), difficulty in controlling for confounding factors, and ethical issues (74). None-the-less, observational studies have suggested that transgenerational effects may exist that cannot easily be attributed to cultural and/or genetic inheritance (75). For example, a study of the Överkalix population in northern Sweden suggested paternal transgenerational effects in humans. In these studies, longevity and specific causes of death were linked to detailed historical records of harvests and food supply experienced by previous generations in early life (76, 77). Studies of the Dutch famine of 194445 have also revealed that offspring born during the famine were smaller compared to those born the year before the famine, and that they had increased risk of metabolic and cardiovascular disease in adulthood. Although differences in DNA methylation have been found in adult female offspring exposed to the famine in utero, and that these offspring effects persist for two generations, it is not established that these differences are present in germ cells and are truly reflecting an epigenetic transgenerational inheritance (78).
Molecular mechanisms of inter- and trans-generational inheritance
The germ cells undergo extensive epigenetic reprogramming, from their earliest presence in the embryo until the mature reproductive cells, and the best described reprogramming phases occur in early embryonic development and in the pre-puberty period (79). The germ cells are believed to be more susceptible to environmental influences during these reprogramming phases. However the precise molecular mechanisms underlying transgenerational inheritance still remain unclear. It is hypothesized that transmission of information occurs through epigenetic variation in sperm, oocytes, or both sets of gametes. There are several mechanisms, such as DNAm, histone modification, or changes in non-coding RNA (ncRNA) that could play an important role in transmitting epigenetic information from one generation to the next (79–81). Due to it’s stability in stored DNA samples and comparative ease of measurement, DNA methylation has been the most studied epigenetic mechanism in human studies of inter- and 225 transgenerational effects. However, DNAm undergoes two rounds of erasure, in the formation of gametes and shortly after fertilisation, and it is unclear whether, or how, memory of CpG site methylation is maintained through meiosis. None-the-less, it has been found that the sperm epigenome may be altered by chemical compounds, such as the endocrine disruptor vinclozolin, and result in transgenerational inheritance via DNAm of induced adult-onset disease to the F3 generation (82). In Agouti mice, methyl donor supplementation during pregnancy altered the trajectory of obesity across generations due to altered expression of the agouti gene resulting from changes in DNAm in the offspring (83). Histone modification is another potential route for transgenerational inheritance. C. elegans, though they do not exhibit DNAm like mammals, can impart heritable epigenetic changes, generated from histone modification, to subsequent generations (45). Another possible mechanism for conveying epigenetic information between generations is ncRNAs, such as microRNA (miRNA), small interfering RNA (siRNA), and piwi-interacting RNA (piRNA), which can potentially act as mediators of environmentally induced transgenerational inheritance. These ncRNAs show enhancer-like function and can control chromatin structure. Gapp et al. demonstrated that traumatic stress in early life altered mouse miRNA expression, and behavioural and metabolic responses in the progeny. The phenotype of the progeny could be recapitulated by injection of sperm miRNAs into fertilised oocytes (73).
Epigenetic transmission across generations in allergic disease
Evidence for transmission across generations in allergic disease in animal models
Several intergenerational murine models provide evidence that preconception allergen sensitization impacts on the development of antigen-specific (T and B cell) immune responses in offspring, predisposing to development of asthma and atopy (84–86). Mechanisms involved in regulation of allergic response have been associated with epigenetic changes of the IL-4 gene promoter (86) as well as altered DNAm in dendritic cells (87).
A number of studies have demonstrated adverse effects of maternal smoking and nicotine exposure on offspring pulmonary function. In utero smoking has been demonstrated to affect lung growth and maturation (88), causing alveolarization defects and decreased expression of retinoic acid signalling pathway elements (89), as well as induced airway remodelling and lung structure changes in mice offspring (90). Prenatal nicotine exposure has been shown to decrease forced expiratory flow rates mediated through α7 nicotinic acetylcholine receptors (nAChRs) (91), and to affect global lung methylation levels and down-regulate PPARy expression in the progeny (92).
Maternal particle exposure has also been linked to adverse effects on offspring’s lung health. Murine models have found associations between diesel exhaust particles (DEP), and increased asthma susceptibility in F1 pups, with distinct methylation changes located to promoter regions of genes related to lung development, interleukin (IL)-4 and interferon (IFN)-y signaling (93–95), as well as an activation of aryl hydrocarbon receptor (AhR) and oxidative stress-regulated genes (96). Maternal exposure to specific phthalates (mono-n-butyl phthalate, a metabolite of butyl benzyl phthalate (BBP)), has been shown to increase the risk for persistent airway inflammation in offspring and to induce aberrant DNAm in genes involved in Th2 differentiation (97).
Murine models have demonstrated that maternal exposure to microbial components and supplementation of probiotic bacteria can modulate the immune response in the offspring by suppressing allergic sensitization and airway inflammation in the F1 generation (98–100). It has also been shown that maternal glucocorticoid-induced stress during pregnancy can increase airway inflammation and susceptibility to allergy in the offspring (101).
Multigenerational murine models are emerging, and effects of phthalate exposures through enhanced eosinophilic airway inflammation have been reported to persist in the F2 generation (97). It has been shown that exposure to fungi of the F0 generation was associated with decreased immunoglobulin E and airway eosinophilia as well as altered methylation in genes regulating T helper cells in third-generation (F2) mice (102). In a recent study by Gregory et al., elevated asthma risk following intrauterine exposure to particulate air pollution was identified up to the F3 generation (93). This model suggests a transgenerational effect on asthma susceptibility from exposure to environmental particles. The transgenerational murine model developed by Rehan et al. shows that nicotine exposure of pregnant rats is associated with increased airway resistance in F3 offspring when challenged with metacholine (103).
Evidence for transmission across generations in allergic disease in humans
The long life-cycle of humans makes investigating epigenetic transmission across generations in human a challenge. However, recently several studies with various solutions as to obtaining multi-generation data have been published (Table 1). In different cohorts, higher asthma risk in persons whose maternal grandmother smoked has been found, even if the mother did not smoke (104–109). In the North European RHINE study, higher asthma risk was found in persons whose paternal grandmother smoked (110). Further, this study found that father’s smoking before age 15 years was associated with particularly high asthma risk in future offspring. This finding was replicated in an analysis of two generations in the RHINESSA cohort, using advanced statistical modelling and also accounting for unmeasured confounders. Ongoing analyses of RHINESSA give supportive evidence for a role of father’s early puberty exposure in offspring health; showing lower lung function in offspring whose father smoked before age 15 (111), differential DNAm related to father’s smoking (112), and higher asthma risk in offspring of fathers that became overweight before voice break (113). In an analysis of the ECRHS (European Community Respiratory Health Survey) cohort, in which asthmatic/allergic disease status was measured in the parent generation at three time points over twenty years and offspring allergies reported by the parents at the third study wave, the authors found stronger associations of offspring allergies with parental asthmatic and allergic disease activity as measured before conception as compared to parental status after birth (114). This indicates that disease activity might induce changes that are transmissible to the next generation, rather than a role of shared environment, this has been termed “induced epigenetic transmission” (57). Finally, a study of helminths and allergies in two generations in Norway found that fathers’ Toxocara exposure was associated with daughters’ allergies, and mother’s Toxocara with sons’ allergies (24). While parental exposure was not measured preconception, the sex-specific pattern might indicate a role for epigenetic transmission given parent of origin effects are seen for both genetic variation and epigenetic variation (115, 116), and risk of asthma in offspring from parental asthma has also been shown to be related to the sex of the affected parent (117).
Table 1:
Evidence for inter- and trasngeneration inhertiance of allergic disease in humans
| Reference | Key findings/Study cohort | Exposure across generations |
|---|---|---|
| Accordini S et al. A three-generation study on the association of tobacco smoking with asthma. International journal of epidemiology. 2018. |
Increased asthma risk in F2 generation due to grandmaternal smoking (F0) and paternal smoking (F1) prior to conception The European Community of Respiratory Health Study (ECRHS) |
Intergenerational: F0-F1- F2 |
| Li YF et al. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest. 2005;127(4):1232–41 |
Increased asthma risk in F2 generation due to maternal (F1) and grandmaternal (F0) smoking during pregnancy The Children’s Health Study in southern California (CHS) |
Intergenerational: F0-F1- F2 |
| Miller LL et al. Do grandmaternal smoking patterns influence the etiology of childhood asthma? Chest. 2014;145(6):1213–8. |
Increased asthma risk in F2 generation (female offspring) due to paternal grandmother (F0) smoking during pregnancy The Avon Longitudinal Study of Parents and children (ALSPAC) |
Intergenerational: F0-F2 |
| Magnus MC et al. Grandmother’s smoking when pregnant with the mother and asthma in the grandchild: the Norwegian Mother and Child Cohort Study. Thorax. 2015;70(3):237–43. |
Increased asthma risk in F2 generation due to grandmaternal (F0) smoking during pregnancy, independent of the mother’s smoking status The Norwegian Mother and Child Cohort Study (MoBa) |
Intergenerational: F0-F2 |
| Braback L et al. Childhood asthma and smoking exposures before conception - a three-generational cohort study. Pediatr Allergy Immunol. 2018. |
Increased asthma risk in F2 generation due to paternal grandmother smoking (F0) The Respiratory Health In Northern Europe study (RHINE) |
Intergenerational: F0-F2 |
| Lodge CJ et al. Grandmaternal smoking increases asthma risk in grandchildren: A nationwide Swedish cohort. Clin Exp Allergy. 2018;48(2):167–74. |
Increased asthma risk in F2 generation due to paternal grandmother smoking (F0) The Respiratory Health In Northern Europe study (RHINESSA) |
Intergenerational: F0-F2 |
| Svanes C et al. Father’s environment before conception and asthma risk in his children: a multi-generation analysis of the Respiratory Health In Northern Europe study. International journal of epidemiology. 2017;46((1):235–45. |
Increased asthma risk in F1 generation due to paternal smoking (F0) prior to conception (RHINE study) |
Intergenerational: F0-F1 |
| Accordini S et al. Three-generation effects of tobacco smoking on lung function within the paternal line. European Respiratory Journal. 2017;50((suppl 61)):PA1178. |
Lower lung function in F1 generation due to paternal smoking (F0) prior to conception (RHINESSA study) |
Intergenerational: F0-F1 |
| Bertelsen RJ et al. Clinical markers of asthma and IgE assessed in parents before conception predict asthma and hayfever in the offspring. Clin Exp Allergy. 2017;47(5):627–38. |
Stronger associations of offspring (F1) allergies with parental (F0) asthmatic and allergic disease activity measured prior to conception as compared to parenta status after birth (ECRHS study) |
Intergenerational: F0-F1 |
While maternal diet is increasingly recognised as a risk factor for offspring asthma and atopy (118), there is no current evidence to suggest that inter- or transgenerational effects occur in allergic disease. However maternal dietary factors such as Vitamin D and Fatty Acids that have been associated with asthma risk have also been shown to be associated with DNA Methylation changes at birth in offspring (119, 120). Further research is needed to understand whether these methylation changes lie on the casual pathway between maternal diet and offspring allergic phenotype.
Methodology for studying epigenetic transmission across generations in allergic disease
Several approaches have been undertaken to explore transgenerational epigenetic inheritance in multi-generational human studies, including recruiting the offspring of birth cohort participants who are now reaching reproductive age, recruiting offspring / grandoffspring of adult cohorts, and use of offspring recall and/or registry data to determine phenotype and/or exposures in parental generations. As mentioned before, all these approaches come with advantages and disadvantages, with compromises between prospective data collection and ease / length of cohort recruitment required. However there are a number of multigenerational cohorts available that are already beginning to allow the assessment of inter- and transgenerational effects in allergic disease (57). While most studies have used regression models to assess the effects of prior exposure on outcome, other approaches such as logistic regression analyses with generalized estimating equations and multilevel mediation models within a hierarchical framework (104) are being ustilised to account for familial clustering.
Several statistical approaches have been used to evaluate epigenetic inheritance of methylation in multigenerational cohorts. Correlation is one of the most used methods (121, 122). Strong positive correlation between parent-offspring pairs indicate a higher level of similarity of DNAm between generations. Some studies choose weighted correlation instead of Pearson correlation to minimize the variance of the correlation estimate (123). However, observed similarity of DNAm could also be due to the fact that parent-offspring share the same environmental factors. To distinguish environmental factors from inheritance, narrow sense heritability is defined as , where Var(A) is the variance due to the average effects of inheritance and Var(P) is the total variance. Two major approaches, path analysis model (PAM) and variance of component model (VOM), are generally used to estimate heritability (124). The component of variance can be obtained by ANOVA or fitting linear mixed models (123, 125). The linear mixed model is more flexible in adjusting for covariates, accounting different types of study designs, and explicity addressing environmental variation (123, 126). In addition to studying epigenetic inheritance at level of individual CpGs, transgenerational inheritance can also be evaluated for groups of CpGs that share similar pattern of DNAm transmission (127). This approach, which incorporates unsupervised cluster into beta regression, was recently developed by Han et al. (127), and was able to identify sets of CpGs that have same/different inheritance patterns between mother-offspring and father-offspring.
Conclusions
In conclusion, there is inceasing evidence from both invertebrate and vertebrate expirmental models that transmission of epigenetic information across generations occurs. Furthermore, experimental animal models also suggest this can lead to altered lung and immune development in response to environmental exposures in previous generations. In humans, studies based on historical data suggest a role for transgenerational inheritance in general, and analyses of human multi-generation data suggest intergenerational environmental effects in asthma and allergies. Unmeasured confounding is a matter of concern in non-experimental studies in which the exposure is not randomized (128). The only human study addressing unmeasured confounding in this context found that this error was very small (104), still human studies will need to be informed and complemented by careful studies in experimental models where duration of exposures can be tightly controlled to determine precise windows of vulnerability and randomised to avoid confounding.
Careful study design will be needed to show that the changes to the epigenome induced by environmental effects actually are passed across generations in humans, and the underlying epigenetic mechanisms determined. Multi-generational cohort studies based on national and international collaboration should be established to prospectively and with a clear time order address the question on whether inter- and transgenerational inheritance are contributing to the risk of allergic diseases, and maximium use should be made of registry data, which can provide retrospective validated information for some generations, shortening the time frame necessary to study effects over multiple decades.
Another important area for future research is the issue of tissue specificity of DNA methylation. In epigenetic studies, unlike studies of DNA sequence variation, the cellular source of DNA samples is an essential consideration in study design given the extent of tissue specific methylation (129). The majority of studies of the epigenetics of allergic disease have utilized peripheral blood leukocytes due to ease of sampling and availability of stored samples from historical cohorts, though both nasal brushings (130, 131) and saliva (132) have also been used. Recently a comparison of blood, buccal, nasal and bronchial epithelial tissue methylation profiles has demonstrated that nasal epithelium represents the best proxy for bronchial epithelial cells (133). However, with respect to inter- and transgenerational effects, it is likely that the effects on the epigenome of exposures to the developing embryo, or transmitted through meiosis, may manifest in multiple tissues, though remains to be established.
If it is firmly established that inter- and transgenerational effects are of importance in asthma and allergic disease, the potential practical consequences for public health policies are considerable. What are the time windows in which health promotion would be most efficient? A perspective on asthma and allergies might provide the knowledge basis for a new approach to efficient intervention strategies aimed at reducing the major public health challenge of asthma and allergies.
Acknowledgments:
The authors want to thank the members of the Isle of Wight, ALEC and RHINESSA research teams who have contributed to the study of multigenerational responses in allergy and asthma. JWH and CS are members of inVIVO Planetary Health, a Group of the Worldwide Universities Network (WUN)
W. Karmaus receives grant support from the University of Memphis and NIH. J. W. Holloway receives grant support from the MRC UK, NIH, and European Union. C. Svanes receives grant support from the Western Norway Health Authorities and European Union.
Funding: This study was supported by the National Institutes of Health, grants R01AI091905 and R01HL132321 (PI: Karmaus), and R01AI121226 (MPI: Zhang and Holloway), the Western Norway Health Authorities grant 912011 and the European Union (Horizon 2020, GA-633212) Ageing Lungs in European Cohorts study.
Abbreviations:
- ncRNA
(non-coding RNA)
- miRNA
(microRNA)
- siRNA
(small interfering RNA)
- piRNA
(piwi-interacting RNA)
- DNAm
(DNA methylation)
- RHINESSA
(Respiratory Health In Northern Europe, Spain and Australia generation study)
- ECRHS
(European Community Respiratory Health Survey)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest:
REFERENCES
- 1.Pawankar R Allergic diseases and asthma: a global public health concern and a call to action. World Allergy Organization Journal 2014;7(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjorksten B, Clayton T, Ellwood P, Stewart A, Strachan D. Worldwide time trends for symptoms of rhinitis and conjunctivitis: Phase III of the International Study of Asthma and Allergies in Childhood. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 2008;19(2):110–24. [DOI] [PubMed] [Google Scholar]
- 3.Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2007;62(9):758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asher MI, Montefort S, Bjorksten B, Lai CK, Strachan DP, Weiland SK, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet (London, England) 2006;368(9537):733–43. [DOI] [PubMed] [Google Scholar]
- 5.Beaglehole R, Bonita R, Alleyne G, Horton R, Li L, Lincoln P, et al. UN High-Level Meeting on Non-Communicable Diseases: addressing four questions. Lancet (London, England) 2011;378(9789):449–55. [DOI] [PubMed] [Google Scholar]
- 6.Beasley R, Semprini A, Mitchell EA. Risk factors for asthma: is prevention possible? Lancet 2015;386(9998):1075–85. [DOI] [PubMed] [Google Scholar]
- 7.Burbank AJ, Sood AK, Kesic MJ, Peden DB, Hernandez ML. Environmental determinants of allergy and asthma in early life. J Allergy Clin Immunol 2017;140(1):1–12. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertelsen RJ, Longnecker MP, Lovik M, Calafat AM, Carlsen KH, London SJ, et al. Triclosan exposure and allergic sensitization in Norwegian children. Allergy 2013;68(1):84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton EM, Todd M, Dowd JB, Aiello AE. The impact of bisphenol A and triclosan on immune parameters in the U.S. population, NHANES 2003–2006. Environmental health perspectives 2011;119(3):390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell MJ, Macher JM, Kumagai K. Measured moisture in buildings and adverse health effects: a review. Indoor air 2018. [DOI] [PubMed]
- 11.Norback D, Zock JP, Plana E, Heinrich J, Svanes C, Sunyer J, et al. Mould and dampness in dwelling places, and onset of asthma: the population-based cohort ECRHS. Occupational and environmental medicine 2013;70(5):325–31. [DOI] [PubMed] [Google Scholar]
- 12.Peterson B, Saxon A. Global increases in allergic respiratory disease: the possible role of diesel exhaust particles. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology 1996;77(4):263–8; quiz 9–70. [DOI] [PubMed] [Google Scholar]
- 13.Thurston GD, Kipen H, Annesi-Maesano I, Balmes J, Brook RD, Cromar K, et al. A joint ERS/ATS policy statement: what constitutes an adverse health effect of air pollution? An analytical framework. Eur Respir J 2017;49(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svanes O, Skorge TD, Johannessen A, Bertelsen RJ, Bratveit M, Forsberg B, et al. Respiratory Health in Cleaners in Northern Europe: Is Susceptibility Established in Early Life? PloS one 2015;10(7):e0131959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid PA, Reid PT. Occupational lung disease. J R Coll Physicians Edinb 2013;43(1):44–8. [DOI] [PubMed] [Google Scholar]
- 16.Patel S, Murray CS, Woodcock A, Simpson A, Custovic A. Dietary antioxidant intake, allergic sensitization and allergic diseases in young children. Allergy 2009;64(12):1766–72. [DOI] [PubMed] [Google Scholar]
- 17.Weiland SK, von Mutius E, Husing A, Asher MI. Intake of trans fatty acids and prevalence of childhood asthma and allergies in Europe. ISAAC Steering Committee. Lancet (London, England) 1999;353(9169):2040–1. [DOI] [PubMed] [Google Scholar]
- 18.Ellwood P, Asher MI, Bjorksten B, Burr M, Pearce N, Robertson CF. Diet and asthma, allergic rhinoconjunctivitis and atopic eczema symptom prevalence: an ecological analysis of the International Study of Asthma and Allergies in Childhood (ISAAC) data. ISAAC Phase One Study Group. The European respiratory journal 2001;17(3):436–43. [DOI] [PubMed] [Google Scholar]
- 19.Macsali F, Real FG, Plana E, Sunyer J, Anto J, Dratva J, et al. Early age at menarche, lung function, and adult asthma. Am J Respir Crit Care Med 2011;183(1):8–14. [DOI] [PubMed] [Google Scholar]
- 20.Triebner K, Johannessen A, Puggini L, Benediktsdottir B, Bertelsen RJ, Bifulco E, et al. Menopause as a predictor of new-onset asthma: A longitudinal Northern European population study. J Allergy Clin Immunol 2016;137(1):50–7 e6. [DOI] [PubMed] [Google Scholar]
- 21.Beck I, Jochner S, Gilles S, McIntyre M, Buters JT, Schmidt-Weber C, et al. High environmental ozone levels lead to enhanced allergenicity of birch pollen. PloS one 2013;8(11):e80147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiland SK, Husing A, Strachan DP, Rzehak P, Pearce N. Climate and the prevalence of symptoms of asthma, allergic rhinitis, and atopic eczema in children. Occupational and environmental medicine 2004;61(7):609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Mutius E, Pearce N, Beasley R, Cheng S, von Ehrenstein O, Bjorksten B, et al. International patterns of tuberculosis and the prevalence of symptoms of asthma, rhinitis, and eczema. Thorax 2000;55(6):449–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jogi NO, Svanes C, Siiak SP, Logan E, Holloway JW, Igland J, et al. Zoonotic helminth exposure and risk of allergic diseases: A study of two generations in Norway. Clin Exp Allergy 2018;48(1):66–77. [DOI] [PubMed] [Google Scholar]
- 25.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clinical and experimental immunology 2010;160(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svanes C, Omenaas E, Jarvis D, Chinn S, Gulsvik A, Burney P. Parental smoking in childhood and adult obstructive lung disease: results from the European Community Respiratory Health Survey. Thorax 2004;59(4):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duijts L Growing large and fast: is infant growth relevant for the early origins of childhood asthma? Thorax 2016;71(12):1071–2. [DOI] [PubMed] [Google Scholar]
- 28.Svanes C, Sunyer J, Plana E, Dharmage S, Heinrich J, Jarvis D, et al. Early life origins of chronic obstructive pulmonary disease. Thorax 2010;65(1):14–20. [DOI] [PubMed] [Google Scholar]
- 29.von Mutius E The microbial environment and its influence on asthma prevention in early life. The Journal of allergy and clinical immunology 2016;137(3):680–9. [DOI] [PubMed] [Google Scholar]
- 30.Haahtela T, Holgate S, Pawankar R, Akdis CA, Benjaponpitak S, Caraballo L, et al. The biodiversity hypothesis and allergic disease: world allergy organization position statement. The World Allergy Organization journal 2013;6(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proceedings of the National Academy of Sciences of the United States of America 2012;109(21):8334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirvonen A Gene-environment interactions in chronic pulmonary diseases. Mutation research 2009;667(1–2):132–41. [DOI] [PubMed] [Google Scholar]
- 33.Douwes J, Pearce N. Asthma and the westernization ‘package’. International journal of epidemiology 2002;31(6):1098–102. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz DA. Gene-environment interactions and airway disease in children. Pediatrics 2009;123 Suppl 3:S151–9. [DOI] [PubMed] [Google Scholar]
- 35.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet 2011;27(3):107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. The Lancet Respiratory medicine 2014;2(5):405–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feinberg AP. The Key Role of Epigenetics in Human Disease Prevention and Mitigation. N Engl J Med 2018;378(14):1323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ptashne M On the use of the word ‘epigenetic’. Curr Biol 2007;17:R233–R6 [DOI] [PubMed] [Google Scholar]
- 39.Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature reviews 2012;13(3):153–62. [DOI] [PubMed] [Google Scholar]
- 40.Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics 2011;3(3):267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13(7):484–92. [DOI] [PubMed] [Google Scholar]
- 42.Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep 2012;12(3):211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol 2012;189(2):819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Royce SG, Karagiannis TC. Histone deacetylases and their role in asthma. J Asthma 2012;49(2):121–8. [DOI] [PubMed] [Google Scholar]
- 45.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 2011;479(7373):365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet 2006;15 Spec No 1:R95–101. [DOI] [PubMed] [Google Scholar]
- 47.Angulo M, Lecuona E, Sznajder JI. Role of MicroRNAs in Lung Disease. Arch Bronconeumol 2012;48(9):325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su WY, Xiong H, Fang JY. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem Biophys Res Commun 2010;396(2):177–81. [DOI] [PubMed] [Google Scholar]
- 49.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med 2011;183(10):1295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012; ;489(7416):447–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. The Journal of allergy and clinical immunology 2015;136(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu CJ, Soderhall C, Bustamante M, Baiz N, Gruzieva O, Gehring U, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. The Lancet Respiratory medicine 2018; 6(5):379–388. [DOI] [PubMed] [Google Scholar]
- 53.Everson TM, Lyons G, Zhang H, Soto-Ramirez N, Lockett GA, Patil VK, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med 2015;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang L, Willis-Owen SAG, Laprise C, Wong KCC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature 2015;520(7549):670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang IV, Lozupone CA, Schwartz DA. The environment, epigenome, and asthma. J Allergy Clin Immunol 2017;140(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harb H, Renz H. Update on epigenetics in allergic disease. J Allergy Clin Immunol 2015;135(1):15–24. [DOI] [PubMed] [Google Scholar]
- 57.Arshad SH, Karmaus W, Zhang H, Holloway JW. Multigenerational cohorts in patients with asthma and allergy. J Allergy Clin Immunol 2017;139(2):415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lockett GA, Huoman J, Holloway JW. Does Allergy Begin in Utero? Pediatr Allergy Immunol 2015;In Press. [DOI] [PubMed]
- 59.McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome biology 2014;15(5):R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet 2011;7(8):e1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 2010;20(7):883–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acevedo N, Reinius LE, Greco D, Gref A, Orsmark-Pietras C, Persson H, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet 2015;24(3):875–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kothari PH, Qiu W, Croteau-Chonka DC, Martinez FD, Liu AH, Lemanske RF Jr., et al. Role of local CpG DNA methylation in mediating the 17q21 asthma susceptibility gasdermin B (GSDMB)/ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) expression quantitative trait locus. J Allergy Clin Immunol 2018;141(6):2282–6 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res 2007;61(5 Pt 2):30R–7R. [DOI] [PubMed] [Google Scholar]
- 65.Blewitt M, Whitelaw E. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol 2013;5(11):a017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loke YJ, Novakovic B, Ollikainen M, Wallace EM, Umstad MP, Permezel M, et al. The Peri/postnatal Epigenetic Twins Study (PETS). Twin Res Hum Genet 2013;16(1):13–20. [DOI] [PubMed] [Google Scholar]
- 67.Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 2010;19(21):417688. [DOI] [PubMed] [Google Scholar]
- 68.Van Baak TE, Coarfa C, Dugue PA, Fiorito G, Laritsky E, Baker MS, et al. Epigenetic supersimilarity of monozygotic twin pairs. Genome Biol 2018;19(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buescher JL, Musselman LP, Wilson CA, Lang T, Keleher M, Baranski TJ, et al. Evidence for transgenerational metabolic programming in Drosophila. Dis Model Mech 2013;6(5):1123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matzkin LM, Johnson S, Paight C, Markow TA. Preadult parental diet affects offspring development and metabolism in Drosophila melanogaster. PLoS One 2013;8(3):e59530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, et al. Starvationinduced transgenerational inheritance of small RNAs in C. elegans. Cell 2014;158(2):277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knecht AL, Truong L, Marvel SW, Reif DM, Garcia A, Lu C, et al. Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol Appl Pharmacol 2017;329:148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014;17(5):667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rothstein MA, Cai Y, Marchant GE. The ghost in our genes: legal and ethical implications of epigenetics. Health Matrix Clevel 2009;19(1):1–62. [PMC free article] [PubMed] [Google Scholar]
- 75.Pembrey M, Saffery R, Bygren LO, Network in Epigenetic E, Network in Epigenetic E. Human transgenerational responses to early-life experience: potential impact on development, health and biomedical research. J Med Genet 2014;51(9):563–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, et al. Sexspecific, male-line transgenerational responses in humans. Eur J Hum Genet 2006;14(2):159–66. [DOI] [PubMed] [Google Scholar]
- 77.Kaati G, Bygren LO, Pembrey M, Sjostrom M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet 2007;15(7):784–90. [DOI] [PubMed] [Google Scholar]
- 78.Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, et al. Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine. BJOG : an international journal of obstetrics and gynaecology 2013;120(5):548–53. [DOI] [PubMed] [Google Scholar]
- 79.Wu H, Hauser R, Krawetz SA, Pilsner JR. Environmental Susceptibility of the Sperm Epigenome During Windows of Male Germ Cell Development. Curr Environ Health Rep 2015;2(4):356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 2014;157(1):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sales VM, Ferguson-Smith AC, Patti ME. Epigenetic Mechanisms of Transmission of Metabolic Disease across Generations. Cell Metab 2017;25(3):559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guerrero-Bosagna C, Covert TR, Haque MM, Settles M, Nilsson EE, Anway MD, et al. Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod Toxicol 2012;34(4):694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 1998;12(11):949–57. [PubMed] [Google Scholar]
- 84.Victor JR, Muniz BP, Fusaro AE, de Brito CA, Taniguchi EF, Duarte AJ, et al. Maternal immunization with ovalbumin prevents neonatal allergy development and up-regulates inhibitory receptor Fc gamma RIIB expression on B cells. BMC immunology 2010;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herz U, Joachim R, Ahrens B, Scheffold A, Radbruch A, Renz H. Prenatal sensitization in a mouse model. American journal of respiratory and critical care medicine 2000;162(3 Pt 2):S62–5. [DOI] [PubMed] [Google Scholar]
- 86.Song Y, Liu C, Hui Y, Srivastava K, Zhou Z, Chen J, et al. Maternal allergy increases susceptibility to offspring allergy in association with TH2-biased epigenetic alterations in a mouse model of peanut allergy. J Allergy Clin Immunol 2014;134(6):1339–45 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fedulov AV, Kobzik L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol 2011;44(3):285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larcombe AN, Foong RE, Berry LJ, Zosky GR, Sly PD. In utero cigarette smoke exposure impairs somatic and lung growth in BALB/c mice. The European respiratory journal 2011;38(4):932–8. [DOI] [PubMed] [Google Scholar]
- 89.Manoli SE, Smith LA, Vyhlidal CA, An CH, Porrata Y, Cardoso WV, et al. Maternal smoking and the retinoid pathway in the developing lung. Respiratory research 2012;13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blacquiere MJ, Timens W, Melgert BN, Geerlings M, Postma DS, Hylkema MN. Maternal smoking during pregnancy induces airway remodelling in mice offspring. Eur Respir J 2009;33(5):1133–40. [DOI] [PubMed] [Google Scholar]
- 91.Wongtrakool C, Wang N, Hyde DM, Roman J, Spindel ER. Prenatal nicotine exposure alters lung function and airway geometry through alpha7 nicotinic receptors. American journal of respiratory cell and molecular biology 2012;46(5):695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rehan VK, Liu J, Naeem E, Tian J, Sakurai R, Kwong K, et al. Perinatal nicotine exposure induces asthma in second generation offspring. BMC Med 2012;10:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gregory DJ, Kobzik L, Yang Z, McGuire CC, Fedulov AV. Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy. American journal of physiology Lung cellular and molecular physiology 2017:ajplung.00035.2017. [DOI] [PMC free article] [PubMed]
- 94.Fedulov AV, Leme A, Yang Z, Dahl M, Lim R, Mariani TJ, et al. Pulmonary exposure to particles during pregnancy causes increased neonatal asthma susceptibility. American journal of respiratory cell and molecular biology 2008;38(1):57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicological sciences : an official journal of the Society of Toxicology 2008;102(1):7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manners S, Alam R, Schwartz DA, Gorska MM. A mouse model links asthma susceptibility to prenatal exposure to diesel exhaust. The Journal of allergy and clinical immunology 2014;134(1):63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jahreis S, Trump S, Bauer M, Bauer T, Thurmann L, Feltens R, et al. Maternal phthalate exposure promotes allergic airway inflammation over 2 generations through epigenetic modifications. The Journal of allergy and clinical immunology 2017. [DOI] [PubMed]
- 98.Blumer N, Herz U, Wegmann M, Renz H. Prenatal lipopolysaccharide-exposure prevents allergic sensitization and airway inflammation, but not airway responsiveness in a murine model of experimental asthma. Clinical and experimental allergy 2005;35(3):397–402. [DOI] [PubMed] [Google Scholar]
- 99.Blumer N, Sel S, Virna S, Patrascan CC, Zimmermann S, Herz U, et al. Perinatal maternal application of Lactobacillus rhamnosus GG suppresses allergic airway inflammation in mouse offspring. Clinical and experimental allergy 2007;37(3):348–57. [DOI] [PubMed] [Google Scholar]
- 100.Brand S, Teich R, Dicke T, Harb H, Yildirim AO, Tost J, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. The Journal of allergy and clinical immunology 2011;128(3):618–25.e1–7. [DOI] [PubMed] [Google Scholar]
- 101.Lim R, Fedulov AV, Kobzik L. Maternal stress during pregnancy increases neonatal allergy susceptibility: role of glucocorticoids. American journal of physiology Lung cellular and molecular physiology 2014;307(2):L141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, et al. Prenatal exposure to allergen, DNA methylation, and allergy in grandoffspring mice. Allergy 2012;67(7):904–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rehan VK, Liu J, Sakurai R, Torday JS. Perinatal nicotine-induced transgenerational asthma. American journal of physiology Lung cellular and molecular physiology 2013;305(7):L501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Accordini S, Calciano L, Johannessen A, Portas L, Benediktsdottir B, Bertelsen RJ, et al. A three-generation study on the association of tobacco smoking with asthma. International journal of epidemiology 2018. March 9. doi: 10.1093/ije/dyy031. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li YF, Langholz B, Salam MT, Gilliland FD. Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 2005;127(4):1232–41. [DOI] [PubMed] [Google Scholar]
- 106.Miller LL, Henderson J, Northstone K, Pembrey M, Golding J. Do grandmaternal smoking patterns influence the etiology of childhood asthma? Chest 2014;145(6):1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Magnus MC, Haberg SE, Karlstad O, Nafstad P, London SJ, Nystad W. Grandmother’s smoking when pregnant with the mother and asthma in the grandchild: the Norwegian Mother and Child Cohort Study. Thorax 2015;70(3):237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Braback L, Lodge CJ, Lowe AJ, Dharmage SC, Olsson D, Forsberg B. Childhood asthma and smoking exposures before conception - a three-generational cohort study. Pediatr Allergy Immunol 2018. March 9. doi: 10.1093/ije/dyy031. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 109.Lodge CJ, Braback L, Lowe AJ, Dharmage SC, Olsson D, Forsberg B. Grandmaternal smoking increases asthma risk in grandchildren: A nationwide Swedish cohort. Clin Exp Allergy 2018;48(2):167–74. [DOI] [PubMed] [Google Scholar]
- 110.Svanes C, Koplin J, Skulstad SM, Johannessen A, Bertelsen RJ, Benediktsdottir B, et al. Father’s environment before conception and asthma risk in his children: a multi-generation analysis of the Respiratory Health In Northern Europe study. International journal of epidemiology 2017;46((1):235–45. [DOI] [PubMed] [Google Scholar]
- 111.Accordini S, Johannessen A, Calciano L, Jogi R, Martinez-Moratalla Rovira J, Benediktsdottir B, et al. Three-generation effects of tobacco smoking on lung function within the paternal line. European Respiratory Journal 2017;50((suppl 61)):PA1178. [Google Scholar]
- 112.Mørkve Knudsen T, Rezwan F, Skulstad SM, Bertelsen RJ, Gomez Real F, ProbstHensch N, et al. Late Breaking Abstract - Epigenome-wide association of father’s smoking on offspring DNA methylation. European Respiratory Journal 2017;50(Suppl. 61):OA2948. [Google Scholar]
- 113.Johannessen A, Calciano L, Lonnebotn M, Bertelsen RJ, Braback L, Holm M, et al. Late Breaking Abstract - Fathers’ overweight and offspring asthma – an intergenerational perspective. European Respiratory Journal 2017;50(suppl 61):PA2615. [Google Scholar]
- 114.Bertelsen RJ, Rava M, Carsin AE, Accordini S, Benediktsdottir B, Dratva J, et al. Clinical markers of asthma and IgE assessed in parents before conception predict asthma and hayfever in the offspring. Clin Exp Allergy 2017;47(5):627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martino D, Prescott S. Epigenetics and prenatal influences on asthma and allergic airways disease. Chest 2011;139(3):640–7. [DOI] [PubMed] [Google Scholar]
- 116.Moffatt MF, Cookson WO. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy 1998;28 Suppl 1:56–61 [DOI] [PubMed] [Google Scholar]
- 117.Arshad SH, Karmaus W, Raza A, Kurukulaaratchy RJ, Matthews SM, Holloway JW, et al. The effect of parental allergy on childhood allergic diseases depends on the sex of the child. J Allergy Clin Immunol 2012. [DOI] [PMC free article] [PubMed]
- 118.Lee-Sarwar K, Litonjua AA. As You Eat It: Effects of Prenatal Nutrition on Asthma. J Allergy Clin Immunol Pract 2018;6(3):711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suderman M, Stene LC, Bohlin J, Page CM, Holvik K, Parr CL, et al. 25Hydroxyvitamin D in pregnancy and genome wide cord blood DNA methylation in two pregnancy cohorts (MoBa and ALSPAC). J Steroid Biochem Mol Biol 2016;159:102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Dijk SJ, Zhou J, Peters TJ, Buckley M, Sutcliffe B, Oytam Y, et al. Effect of prenatal DHA supplementation on the infant epigenome: results from a randomized controlled trial. Clin Epigenetics 2016;8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kile ML, Baccarelli A, Tarantini L, Hoffman E, Wright RO, Christiani DC. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PLoS One 2010;5(10):e13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cullati S, Courvoisier DS, Gayet-Ageron A, Haller G, Irion O, Agoritsas T, et al. Patient enrollment and logistical problems top the list of difficulties in clinical research: a crosssectional survey. BMC Med Res Methodol 2016;16:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Day K, Waite LL, Alonso A, Irvin MR, Zhi D, Thibeault KS, et al. Heritable DNA Methylation in CD4+ Cells among Complex Families Displays Genetic and Non-Genetic Effects. PLoS One 2016;11(10):e0165488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McArdle JJ, Prescott CA. Mixed-effects variance components models for biometric family analyses. Behav Genet 2005;35(5):631–52. [DOI] [PubMed] [Google Scholar]
- 125.Rowlatt A, Hernandez-Suarez G, Sanabria-Salas MC, Serrano-Lopez M, Rawlik K, Hernandez-Illan E, et al. The heritability and patterns of DNA methylation in normal human colorectum. Hum Mol Genet 2016;25(12):2600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Heckerman D, Gurdasani D, Kadie C, Pomilla C, Carstensen T, Martin H, et al. Linear mixed model for heritability estimation that explicitly addresses environmental variation. Proc Natl Acad Sci U S A 2016;113(27):7377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Han S, Zhang H, Lockett GA, Mukherjee N, Holloway JW, Karmaus W. Identifying heterogeneous transgenerational DNA methylation sites via clustering in beta regression. Ann Appl Stat 2015;9(4):2052–72. [Google Scholar]
- 128.Uddin MJ, Groenwold RH, Ali MS, de Boer A, Roes KC, Chowdhury MA, et al. Methods to control for unmeasured confounding in pharmacoepidemiology: an overview. Int J Clin Pharm 2016;38(3):714–23. [DOI] [PubMed] [Google Scholar]
- 129.Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individualspecific DNA methylation patterns. Hum Mol Genet 2009;18(24):4808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Somineni HK, Zhang X, Biagini Myers JM, Kovacic MB, Ulm A, Jurcak N, et al. Teneleven translocation 1 (TET1) methylation is associated with childhood asthma and trafficrelated air pollution. J Allergy Clin Immunol 2016;137(3):797–805 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol 2017;139(5):1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Langie SAS, Moisse M, Szarc Vel Szic K, Van Der Plas E, Koppen G, De Prins S, et al. GLI2 promoter hypermethylation in saliva of children with a respiratory allergy. Clinical epigenetics 2018;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brugha R, Lowe R, Henderson AJ, Holloway JW, Rakyan V, Wozniak E, et al. DNA methylation profiles between airway epithelium and proxy tissues in children. Acta Paediatr 2017;106(12):2011–6. [DOI] [PubMed] [Google Scholar]


