Abstract
Axons need to be properly guided to their targets to form synaptic connections, and this requires interactions between highly conserved extracellular and transmembrane ligands and their cell surface receptors. The majority of studies on axon guidance signaling pathways have focused on the role of these pathways in rearranging the local cytoskeleton and plasma membrane in growth cones and axons. However, a smaller body of work has demonstrated that axon guidance signaling pathways also control gene expression via local translation and transcription. Recent studies on axon guidance ligands and receptors have begun to uncover the requirements for these alternative mechanisms in processes required for neural circuit formation: axon guidance, synaptogenesis and cell migration. Understanding the mechanisms by which axon guidance signaling regulates local translation and transcription will create a more complete picture of neural circuit formation, and may be applied more broadly to other tissues where axon guidance ligands and receptors are required for morphogenesis.
Keywords: Axon Guidance, Neural Development, Slit, Robo, Netrin, Dcc, Ephrin, Semaphorin, Plexin, Local translation, transcription factor
Introduction
The precise establishment of neural circuits during development is essential for coordinated animal behavior. Cell migration, axon guidance, and synaptogenesis are all processes required for proper neural circuit formation, and axon guidance ligands and receptors regulate these processes. At the tip of the axon is the highly motile growth cone, which encounters a variety of diverse cues, mainly attractants and repellants, as it navigates through its environment. Extracellular cues interact with receptors expressed on growth cones to mediate axon outgrowth, growth cone collapse, and turning. The following axon guidance cues and receptors will be the focus of this review: 1) semaphorins and their neuropilin and plexin receptors, 2) Slits and their roundabout (Robo) receptors, 3) netrins and their deleted in colorectal carcinoma (Dcc), Frazzled (Fra, in Drosophila), Unc40 (in C. elegans), neogenin, and Unc5 receptors, and 4) ephrins and their Eph receptors (Hou et al., 2008). Sonic hedgehog, Wnt, bone morphogenetic protein and other signaling pathways (Yam and Charron, 2013) have also been shown to play roles in axon guidance, and we refer the reader to previous reviews that discuss these pathways (Bovolenta, 2005; Charron and Tessier-Lavigne, 2007; Sanchez-Camacho and Bovolenta, 2009). We will not cover these pathways in this review, as their involvement in gene regulation is already well studied and reviewed.
Most axon guidance receptors impinge on cytoplasmic proteins to regulate Rho family small GTPases, which in turn modulate cytoskeletal and membrane dynamics through diverse downstream effectors. Thus, Rho family GTPases can integrate signals from multiple cues to direct growth cone dynamics (Luo, 2002; O'Donnell et al., 2009). Recent reports implicate the SCAR/WAVE complex in axon guidance, and suggest that SCAR/WAVE may interact directly with axon guidance receptors through the conserved WIRS motif to regulate Arp2/3-dependent actin polymerization (Zallen et al., 2002; Lin et al., 2009; Bernadskaya et al., 2012; Chen et al., 2014). For comprehensive reviews on actin and microtubule dynamics in navigating growth cones and axons, we refer the reader to reviews that explore this topic (Krause et al., 2003; Lowery and Van Vactor, 2009; Dent et al., 2011; Vitriol and Zheng, 2012; Gomez and Letourneau, 2014; Spillane and Gallo, 2014; Stankiewicz and LinSeman, 2014).
The majority of studies on axon guidance receptor signaling have been focused on how axon guidance receptors signal locally to regulate the cytoskeleton and growth cone plasma membrane. In contrast, a smaller body of work has demonstrated that axon guidance cues and receptors also act non-canonically to control cell proliferation, cell migration, and axon guidance by regulating gene expression through translational or transcriptional mechanisms. In this review we aim to synthesize the studies that investigate these mechanisms in an attempt to demonstrate that axon guidance ligands and receptors broadly function to regulate gene expression across a range of neuron subtypes, developmental processes, and organisms.
Part 1: Local translation
Local translation is required for axon guidance in vitro
Axons continue to grow and respond to guidance cues even after being severed from their cell bodies (Harris et al., 1987), indicating that all of the required signaling components to mediate these responses are present in growth cones. The observation that growth cones also contain messenger RNAs (mRNAs), translation machinery, and molecules involved in protein degradation (Tennyson, 1970; Bassell et al., 1998; Campbell and Holt, 2001), led to the suggestion that protein synthesis and degradation may occur locally in growth cones. Indeed, vertebrate neurons translate proteins in their growth cones and dendrites (Davis et al., 1992; Crino and Eberwine, 1996). In vitro, specific axon guidance cues can rapidly induce local protein synthesis in growth cones and axons to affect axon turning and collapse, and preventing protein synthesis blocks these responses (Farrar and Spencer, 2008; Lin and Holt, 2008). Thus, local translation in growth cones and axons is clearly necessary in order for some axon guidance cues to modulate growth cone behavior. For example, the axon guidance cues Sema3A, Slit2, and netrin1 can all induce local protein translation, and this is required to steer axons in both intact neurons and severed axons in vitro (Campbell and Holt, 2001; Wu et al., 2005; Leung et al., 2006; Piper et al., 2006; Lin and Holt, 2007).
The requirement for local translation depends on cell type and the concentration of guidance cues
Despite the fact that several independent studies demonstrated a role for local translation in guidance responses in vitro, the limited in vivo evidence and conflicting results from in vitro experiments caused significant skepticism in the field as to the importance of local translation in axon guidance. The majority of experiments were initially done with Xenopus laevis retinal ganglion cell (RGC) axons, but later reports tested the requirement for local translation in axon guidance in other organisms and neuronal subtypes.
In one report, which contrasted substantially from earlier work, Letourneau and colleagues (2009) found that Sema3A-mediated growth cone collapse in cultured chick dorsal root ganglion (DRG) neurons could still occur in the presence of protein translation inhibitors, strongly suggesting that growth cone responses to Sema3A do not strictly depend on protein synthesis. To account for the differences seen in the requirement for local translation in axon guidance, the authors speculated that different neuronal populations might respond differently to guidance cues, as a result of both their intrinsic properties as well as the extrinsic cues the neurons encounter (Roche et al., 2009).
More recently, this apparent conflict has been revisited, leading to the discovery that different concentrations of a ligand that growth cones encounter can result in significant differences in the requirement for local translation (Manns et al., 2012; Nedelec et al., 2012). In chick DRG neurons and mouse and human embryonic stem cell-derived spinal motor neurons (ES-MNs), growth cone collapse in response to treatment with low Sema3A concentrations (<100 ng/ml) requires local protein synthesis (Manns et al., 2012; Nedelec et al., 2012). In contrast, when neurons are treated with high Sema3A concentrations (>625 ng/ml), growth cone collapse still occurs even when protein synthesis is blocked (Manns et al., 2012; Nedelec et al., 2012). Human ES-MNs and mouse brachiothoracic motor neurons show the same bimodal concentration-dependent responses to both Sema3A and Sema3F, suggesting that multiple semaphorins induce local translation. Strikingly, one of the neuronal subtypes analyzed, cervical ES-MNs, lacks the local protein synthesis-dependent response to low Sema3A concentrations. This is thought to be due to lack of local protein synthesis machinery in the growth cones of these neurons (Nedelec et al., 2012).
A better understanding of the Sema3A signaling pathway may provide insight into these concentration-dependent responses. Sema3A treatment leads to the activation of glycogen synthase kinase 3 beta (Gsk3b), which appears to act downstream of Sema3A regardless of the concentration, and Gsk-3beta activation is necessary for Sema3A-mediated growth cone collapse (Manns et al., 2012). At low concentrations, Sema3A also signals through the mechanistic target of rapamycin (Mtor), to activate local protein synthesis of Rhoa (Wu et al., 2005; Manns et al., 2012). Inhibiting Gsk-3beta activation results in an increase in protein synthesis, as demonstrated by the increased fluorescence of phosphorylated Eif4ebp1 (4EBP1), a marker for translation. This observation suggests that activated Gsk3b may antagonize Mtor. Therefore, high concentrations of Sema3A may lead to a significant increase in Gsk3b activity, which can overcome the need for local protein synthesis in Sema3A-mediated growth cone collapse by inhibiting Mtor and thus protein synthesis. It is unclear how these guidance cue concentrations might relate to the in vivo concentrations of cues encountered by growth cones, but it is likely that differential concentration-dependent signaling outputs may serve to diversify axonal responses to a limited set of cues.
Local translation of specific proteins are induced by guidance cues
It is clear that diverse guidance cues can induce local translation and that this activity is important to affect downstream signaling and axon responsiveness. We turn now to the consideration of the proteins that are specifically translated in response to different cues, and, how in turn these proteins contribute to distinct axon guidance responses. In recent studies of cue-induced local translation, a number of distinct mechanisms that control how specific mRNAs are translated locally have begun to emerge (Table 1).
Table 1. The targets and mechanisms for cue-dependent local translation.
Sema3, Slit2, and netrin1 all induce local translation of specific mRNAs. Thus far, two mechanisms have begun to be elucidated: Slit2 indirectly induces local translation of cofilin1 by antagonizing miR-182, and netrin1 causes Dcc to directly release translation machinery, allowing local translation to occur.
| Ligand | Receptor | Translation target | Mechanism |
|---|---|---|---|
| Sema3A | Nrp1 | Rhoa, nfpc | ? |
| Slit2 | Robo2/3 | cofilin1 | Indirect- Slit2 signaling antagonizes the microRNA miR-124, resulting in the release of cofilin1 mRNA |
| netrin1 | Dcc? | Beta-actin, Dscam, Sensorin | ? |
| netrin1 | Dcc | ? | Direct-interaction between Dcc and translational machinery |
Sema3A induces the local translation of RhoA and NF-protocadherin
Sema3A has been reported to induce the local translation of two specific proteins, Rhoa and NF-protocadherin (Nfpc). In DRGs, Sema3A-mediated growth cone collapse depends on the Rhoa effector Rock1 (rho-associated protein kinase), which acts downstream of axon guidance receptors to regulate cytoskeletal dynamics (Dontchev and Letourneau, 2002). Unsurprisingly then, Rhoa activation is required for Sema3A-mediated growth cone collapse (Wu et al., 2005). Interestingly, Rhoa transcripts are found in axons at higher levels than other transcripts and are localized in puncta throughout the axon (Wu et al., 2005). Sema3A treatment increases the fluorescence intensity of Rhoa protein, while growth associated protein 43 (Gap43), which is expressed at high levels in neurons during development, is not affected, suggesting that Sema3A specifically induces local translation of Rhoa. In addition, a translation reporter for Rhoa reveals that Rhoa mRNA is translated in growth cones following Sema3A treatment, and translation inhibitors block this effect (Wu et al., 2005). These experiments indicate that Sema3A induces local translation of Rhoa in DRG axons and growth cones.
In X. laevis RGCs, Sema3A also induces the local translation of the cell adhesion molecule Nfpc in vitro (Leung et al., 2013). Nfpc is necessary in RGC axons to maintain the correct levels of adhesion with the optic tract and helps guide RGC axons to their targets. In vivo imaging demonstrates the Sema3A-dependent local translation of an nfpc translational reporter in the growth cone, and the observation that a function-blocking antibody for the neuropilin 1 (Nrp1) receptor prevents this effect, reveals a partial requirement for Nrp1 in this process (Leung et al., 2013). In summary, Sema3A induces the local translation of specific mRNAs, Rhoa and nfpc, in vitro, and in vivo imaging data strongly supports the conclusion that this regulated translation is likely to contribute to axon guidance.
Slit2 induces the local translation of cofilin1
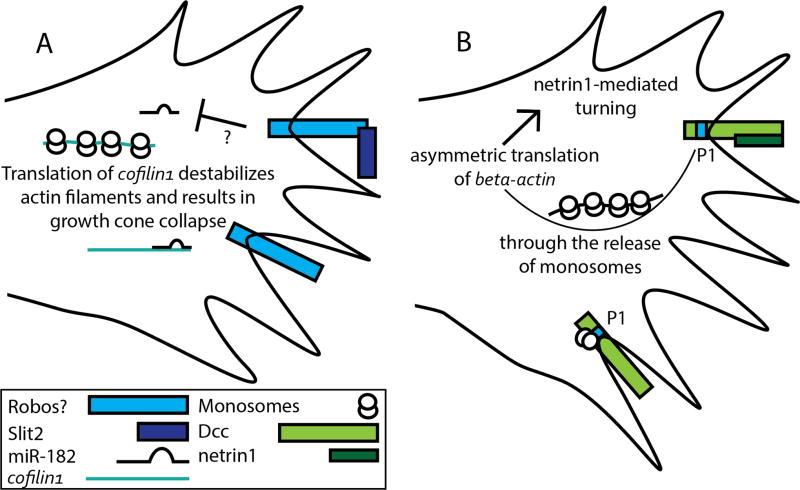
In X. laevis RGC axons, there is considerable evidence that slit2 can induce the translation of cofilin1, which destabilizes Filamentous actin and may act downstream of Slit2-Robo signaling to cause axon retraction and collapse (Figure 1A). Cofilin1 mRNA interacts with Vg1RBP, an RNA-binding protein implicated in the localization of specific mRNAs to growth cones. Inhibitors of protein synthesis block Slit2-induced cofilin1 translation, and prevent growth cone collapse (Piper et al., 2006). In addition, a cofilin1 translation reporter, where the 3’ UTR of cofilin1 mRNA is fused to a photo-convertible Kaede protein (Leung and Holt, 2008) is translated in response to slit2 (Bellon et al., 2017). Thus, Slit2 treatment induces local translation of cofilin1 in RGC growth cones in vitro. One method for controlling the specificity of mRNAs translated in response to axon guidance cues could be a relationship between miRNAs with specific targets and axon guidance pathways. miR-182 is the most highly expressed miRNA in X. laevis RGC axons. In slit morphants, X. laevis RGC axons exhibit targeting defects in vivo, where RGC axons target a wider area than in wild-type animals, and the loss of miR-182 in RGCs results in defects that resemble slit morphant phenotypes (Bellon et al., 2017). An algorithm to identify potential targets of miR-182 found cofilin1 mRNA as a top target (Zivraj et al., 2010), suggesting a link between miR-182 and Slit-cofilin1 growth cone collapse. The loss of miR-182 causes an increase in cofilin1 immunostaining intensity in RGC axons similarly to the fluorescence intensity visualized in control RGC axons treated with slit2, suggesting miR-182 can block cofilin1 translation (Bellon et al., 2017). Unexpectedly, despite having increased cofilin1 present in the miR-182 morphant RGCs, their axons fail to turn away from Slit2 (Bellon et al., 2017). Perhaps a tighter regulation of where cofilin1 is translated is required for Slit2-mediated growth cone repulsion, and this is lost when miR-182 is knocked down throughout the entire growth cone. While these observations suggest that the effect of Slit2 on local translation is important in vivo, it is important to point out that the effects observed upon miR-182 manipulation cannot be directly attributed to a role in Slit-dependent local translation. Nevertheless, these findings are among the strongest evidence for the in vivo importance for local translation in axon guidance. The ability of Slit2 to regulate miRNAs provides an intriguing mechanism to explain how specific mRNAs are selected for local translation.
Figure 1. Slit2 and netrin1: Different mechanisms to regulate local translation?
A) A model for indirect regulation of local translation by the axon guidance cue Slit2. Slit2 causes the miRNA miR-182 to release cofilin1 mRNA, potentiating cofilin1 local translation and resulting in growth cone collapse. B) A model for direct regulation of local translation by netrin1-Dcc signaling. Dcc interacts with translational machinery through the conserved P1 motif indicated. Interaction between netrin1 and Dcc induces the release of monosomes from Dcc, allowing them to form polysomes and translate mRNAs locally. Netrin1 mediates the asymmetric translation of beta-actin, resulting in attractive turning. The induction of beta-actin translation by netrin could be due to the direct release of translational machinery from Dcc, or through an alternative mechanism via cytoplasmic proteins that link netrin signaling with translational machinery.
To determine the receptor that Slit2 signals through, truncated Robo2 and Robo3 receptors that lack their cytoplasmic domains were expressed in RGC growth cones, causing elevated activity of miR-182. This observation suggests that Slit2 may require the Robo2 and Robo3 receptors in this process (Bellon et al., 2017). However, the use of these ‘dominant negative’ receptors does raise the question of whether Robo2 and Robo3 3 are acting cell-autonomously in this context, as well as whether the dominant negative receptors are sequestering Slits away from other receptors or specifically blocking Robo2/3 activity. The use of morpholinos or RNAi to knockdown robo2 and robo3 in X. laevis RGCs, would be useful to further confirm that Robo2 and Robo3 are the receptors involved in Slit2-dependent cofilin1 translation.
netrin1 induces the local translation of beta-actin and Dscam
Similar to Sema3A and Slit2, netrin1 has also been found to induce the local translation of proteins already implicated in axon guidance, beta-actin and the cell adhesion molecule Dscam. Beta-actin protein is highly expressed in growth cones and filopodia, and Beta-actin mRNA co-localizes with translational machinery in granules detected in neurites, axons, and growth cones (Bassell et al., 1998). The 3’ UTR of Beta-actin mRNA contains a short sequence, called a zipcode, that is required for the localization of Beta-actin mRNA to the plasma membrane (Condeelis and Singer, 2005), and two members of the VICKZ (Vg1 RBP/Vera, IMP-1,2,3, CRD-BP, KOC, ZBP-1) family of RNA-binding proteins, Vg1rbp and Zbp1, interact with Beta-actin mRNA via the zipcode sequence to regulate its localization (Zhang et al., 2001; Yisraeli, 2005; Leung et al., 2006; Yao et al., 2006; Welshhans and Bassell, 2011). In X. laevis RGC growth cones treated with netrin1 in vitro, granules containing the RNA trafficking protein Vg1rbp move into filopodia that are closer to the source of netrin1, and beta-actin mRNA is asymmetrically translated, with higher levels of beta-actin protein present on the side of the growth cone encountering higher levels of netrin1 (Leung et al., 2006). Netrin1 can induce the local translation of beta-actin mRNA in vitro in both X. laevis RGCs, and mammalian cortical neurons (Leung et al., 2006; Welshhans and Bassell, 2011). In mammalian cortical neurons cultured from mice lacking the RNA-binding protein Zbp1, netrin1 no longer induces axon attraction in a turning assay and does not increase local translation of a beta-actin translational reporter to the levels seen in wild-type neurons (Welshhans and Bassell, 2011). These observations indicate that Zbp1 is required for netrin1-mediated local translation of Beta-actin mRNA in mammalian cortical neurons in vitro.
Recently, Strohl et al. (2017) developed an imaging technique to visualize translation of single molecules in an in vitro culture system. Using this system, the authors determined that Beta-actin mRNA is locally translated at multiple sites within growth cones treated with netrin1, and that, remarkably, translation of Beta-actin mRNA is induced within 20 seconds of applying netrin1 to neurons in culture (Strohl et al., 2017). It would be interesting to determine whether sites of rapidly induced actin translation co-localize with the Dcc receptor. In addition to inducing the translation of Beta-actin mRNA, there is also some evidence that suggests netrin1 can induce the local translation of Dscam. Dscam mRNA is detected throughout the soma, axon, and growth cone of mouse hippocampal neurons, and blocking translation prevents an increase in the expression of the cell adhesion protein Dscam in response to netrin1 (Jain and Welshhans, 2016).
Several salient points have risen from studies on local translation in axon guidance, including: the local translation of specific mRNAs by guidance cues, and asymmetric translation of certain mRNAs, which are both often required for downstream receptor signaling to regulate axon guidance. Still, several aspects of how guidance cues regulate translation at the growth cone are still unknown. In particular, our current understanding of how receptors interact with and signal to translational machinery is limited. Indeed, the only axon guidance receptor currently known to directly interact with translational machinery is Dcc (Tcherkezian et al., 2010) (Figure 1B).
Dcc directly associates with translational machinery
In the previous mechanisms discussed here, axon guidance receptors might regulate local translation by recruiting cytoplasmic signaling proteins, or receptors could directly interact with translation machinery to regulate local translation. Indeed, the axon guidance receptor Dcc has been shown in vitro to directly interact with translation machinery, including eukaryotic initiation factors, ribosomal proteins, small and large ribosomal subunits, and monosomes. Both electron microscopy and immunofluorescence analysis show that Dcc co-localizes with both translation machinery and with newly synthesized protein in axons and dendrites (Tcherkezian et al., 2010). The interaction between Dcc and translation machinery is dependent on netrin1, which causes Dcc to release ribosomal subunits and monosomes, allowing for polysomes to form and translation to occur (Tcherkezian et al., 2010). Removal of the extracellular domain of Dcc inhibits translation in response to netrin1 (Tcherkezian et al., 2010). The conserved P1 motif within the cytoplasmic domain of Dcc is required for Dcc to interact with translation machinery (Figure 1B). While the in vitro biochemical links between Dcc and translation machinery is quite compelling, the in vivo significance of these observations for axon guidance is less clear. In vivo evidence linking Dcc-dependent translational regulation to axon guidance is limited to a single experiment where a Dcc receptor lacking the P1 motif (DccΔP1) is mis-expressed in chick commissural neurons in the developing spinal cord. Neurons expressing DccΔP1 are less likely to extend their axons to the midline in comparison to wild type axons (Tcherkezian et al., 2010). However, the axon guidance defects resulting from over-expressing a dominant negative DccΔP1 cannot be solely attributed to a loss of interaction between Dcc and translational machinery without further analysis. A homolog of Dcc has not been found in the chick, although a homolog of neogenin, a closely related family member that can substitute for Dcc, contains the conserved P1 motif (Phan et al., 2011). Still, the defects resulting from the expression of DccΔP1 could result from blocking netrin1 interactions with neogenin, or alternatively they could be due to an unknown factor that binds to the P1 motif of Dcc. In the Drosophila embryo, rescue experiments show that FraΔP1, where Fra is the invertebrate orthologue of Dcc, is able to rescue the midline crossing of a subset of commissural axons in fra mutants comparably to the full length Fra receptor, suggesting the P1 motif is not required for commissural axon guidance (Garbe et al., 2007). However, these experiments were performed with receptors that were expressed at higher than endogenous expression levels, potentially overcoming a requirement for the P1 motif. A more precise analysis to elucidate the function of the P1 motif in axon guidance is necessary. Dcc directly interacting with translational machinery is an exciting finding, and future studies should determine if this interaction is required for Dcc-mediated axon guidance, both in vitro and in vivo. For example, it would be interesting to determine if the netrin1-induced local translation of Beta-actin mRNA requires Dcc, and if Dcc interacts directly with translational machinery to mediate local translation of either Beta-actin or Dscam. An interesting alternative possibility is that Dcc control of local translation is important for other neuronal functions of Dcc, such as the regulation of synapse formation or function.
Netrin-mediated local translation at the synapse
In addition to its role in axon guidance, netrin1 is also required for synaptogenesis in C. elegans and mammals (Colon-Ramos et al., 2007; Park et al., 2011; Stavoe and Colon-Ramos, 2012; Stavoe et al., 2012; Goldman et al., 2013). For example, C. elegans netrin (Unc6) induces synaptogenesis through the Dcc (Unc40) receptor (Colon-Ramos et al., 2007), and this requires Unc40 to interact with CED5/Dock180 (a rac GEF) and activate CED10/Rac1 to mediate local cytoskeletal rearrangements (Stavoe and Colon-Ramos, 2012). In mammalian cortical neurons, netrin1 also promotes synaptogenesis (Goldman et al., 2013), but the requirement for Dcc as the receptor in this context has not been tested. In Aplysia sensory and motor neuron co-cultures in vitro, bath application of netrin1 stimulates local translation of the sensory neuron-specific neuropeptide sensorin at synapses. In response to netrin1 application, a translation-dependent increase in Sensorin protein is observed in sensory neurons (Kim and Martin, 2015). Notably, while treatment with netrin1 does not convert non-synaptic sites to synaptic sites, it does result in an increase in amplitude of the excitatory post-synaptic potential (EPSP) in sensory neurons, as well as an increase in sites of synaptic connections, suggesting netrin1 increases synaptic strength between Aplysia sensory neurons and motor neurons (Kim and Martin, 2015). The over-expression of Aplysia netrin1 in motor neurons is sufficient to induce increases in sensorin protein in the sensory neurons with which they are co-cultured (Kim and Martin, 2015), suggesting that netrin1 can act in trans to induce local translation in the sensory neurons. The authors demonstrate that Dcc is required for netrin-mediated induction of Sensorin translation by using a function-blocking antibody against Dcc. These experiments imply that Dcc is the receptor that netrin interacts with to increase synaptic strength, and that this is controlled by netrin-Dcc induction of local protein translation. However, the ability of netrin1 to increase synaptic strength has not been tested in a Dcc-deficient or local translation-blocking assay, which would more definitively demonstrate that Dcc and/or local translation, respectively, are required. Additionally, it remains to be seen whether netrin induction of local translation is required in vivo for synaptogenesis or synaptic plasticity.
The control of local translation in axons and growth cones by extracellular cues provides an enticing model for how axon guidance and synaptogenesis can be precisely tuned. The specific expression of proteins in certain compartments may increase the spatial and temporal control provided by axon guidance cues. Still, further investigation of the in vivo role for local translation in axon guidance and synaptogenesis is needed to fill in the gaps in our fragmentary knowledge of how receptors signal to translation machinery, and how specific mRNAs are selected for translation.
Part 2: Transcriptional Regulation
The ability of axon guidance signaling pathways to control protein synthesis presents an intriguing mechanism to regulate protein expression in specific areas of the cell. In a similar vein, axon guidance receptors and their ligands have also been implicated in controlling gene expression at the level of transcription in several contexts. There had been hints that axon guidance receptors might regulate transcription similarly to the way that notch controls transcription. However, the evidence was primarily from in vitro systems, or only demonstrated a correlational relationship between axon guidance receptors and altered expression of specific genes. In this section of the review, we will discuss recent findings that indicate that guidance receptors can signal to regulate gene transcription, in some cases in surprisingly direct ways.
Axon guidance receptors are transcriptional activators
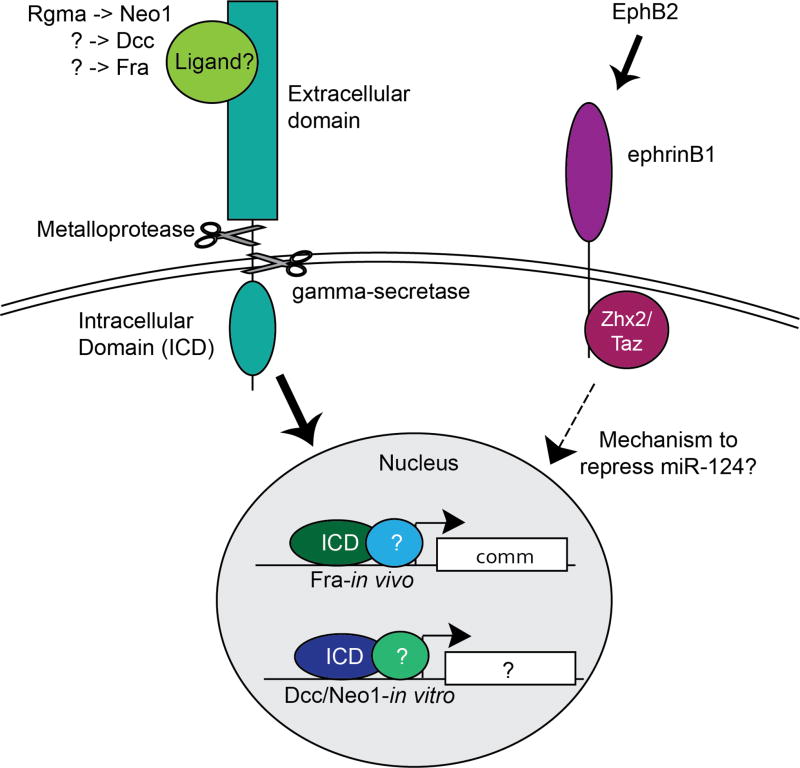
It is now clear that Dcc, neogenin (Neo1), and Frazzled (Fra, the Drosophila orthologue of Dcc) are able to function as transcriptional activators (Figure 2) (Taniguchi et al., 2003; Goldschneider et al., 2008; Neuhaus-Follini and Bashaw, 2015). While early work suggests Dcc and Neo1 could act as transcriptional activators in in vitro assays, recent reports demonstrate an in vivo role for Fra as a transcriptional activator in Drosophila. Preliminary evidence in vertebrates demonstrated that Dcc is cleaved by gamma-secretase, a protease that cleaves single-pass transmembrane proteins in their transmembrane domain, to release the intracellular domain (ICD) of the protein. Cleavage of Dcc by gamma-secretase is necessary for Dcc to activate a transcriptional reporter in cell culture (Taniguchi et al., 2003).
Figure 2. Axon guidance receptors are cleaved and enter the nucleus to regulate transcription.
A schematic depicting a general mechanism for axon guidance factors to enter the nucleus and regulate transcription. On the left, a ligand interacts with the extracellular domain of the receptor, triggering ectodomain-shedding by a metalloprotease, and subsequent cleavage in the transmembrane domain by the single-pass transmembrane protease gamma-secretase. The resulting intracellular domain product then enters the nucleus and interacts with nuclear proteins to regulate transcription. It should be noted that while gamma-secretase cleavage and transcriptional activation has been demonstrated to be required for the Drosophila protein Fra, and that in vivo, and in vitro experiments with vertebrate proteins Dcc and Neo1 also support this model, the experiments linking the transcriptional regulation downstream of EphB2-ephrinB1 signaling to this model is substantially weaker. EphB2-ephrinB1 signaling does repress the expression of miR-124, and this is mediated by the transcriptional repressor Zhx2.
Neo1 is cleaved by a metalloprotease, potentially Tace/Adam17 (Okamura et al., 2011), which is followed by gamma-secretase cleavage, and the Neo1 intracellular domain (ICD) can subsequently enter the nucleus (Goldschneider et al., 2008). In the nucleus, the Neo1 ICD activates transcription of a reporter in cells, and ChIP on cells reveals several different loci where the Neo1 ICD interacts with chromatin near specific genes (Goldschneider et al., 2008). Several proteins that were found to interact with the N-terminal domain of the Neo1 ICD in a yeast two-hybrid screen are implicated in transcriptional regulation, including the histone acetyltransferase Tip60/Kat5. In vitro, Neo1 also interacts with the LIM domain only 4 (Lmo4) transcription factor in human neurons and in embryonic rat cortical neurons. Neo1 may also regulate gene expression indirectly, as Neo1 releases Lmo4 in response to repulsive guidance molecule family member A (Rgma), which allows Lmo4 to translocate to the nucleus (Goldschneider et al., 2008; Schaffar et al., 2008). Chick RGCs cultured in vitro on Rgma have short axons, but a miRNA designed to target Lmo4 causes these RGC axons to appear longer, indicating that Lmo4 has a role in Neo1-mediated growth cone repulsion (Banerjee et al., 2016). Interestingly, in chick RGC explant cultures, overexpression of the Neo1 ICD inhibits outgrowth of neurites, yet the Neo1 ICD with its nuclear localization signal removed only partially inhibits neurite outgrowth (Banerjee et al., 2016). This observation suggests that the Neo1 ICD has a nuclear function that can affect neurite outgrowth inhibition in vitro. Additional experiments are required to examine whether and how the Neo1 ICD regulates transcription in vivo, and to determine what the transcriptional targets are that the Neo1 ICD regulates. The cleavage of Neo1 by Tace/Adam17, as well as the ability of the Neo1 ICD to interact with chromatin is dependent on Rgma in vitro (Goldschneider et al., 2008; Okamura et al., 2011). However, Rgma does not interact with Dcc, which leaves open the question of what regulates the transcriptional function of Dcc.
In Drosophila, transcriptional activation by Fra is independent of its canonical ligand Netrin (Yang et al., 2009). Fra has also been shown to be cleaved by gamma-secretase (Neuhaus-Follini and Bashaw, 2015), and this cleavage is necessary in vivo for Fra to activate transcription of commissureless (comm), whose protein product antagonizes repulsive Slit-Robo1 signaling in Drosophila (Neuhaus-Follini and Bashaw, 2015). Both in vitro and in vivo experiments show that the Fra ICD moves in and out of the nucleus, and the conserved P3 motif is the activation domain required for the Fra ICD to activate transcription (Neuhaus-Follini and Bashaw, 2015). The in vivo requirement for the Fra ICD to activate transcription was demonstrated in rescue experiments in fra null mutants. A Fra full-length receptor with a point mutation, which abolishes transcriptional activity while leaving other known Fra signaling activities intact, fails to rescue comm expression in vivo. However, the same receptor with a VP16 activation domain fused to the c-terminus is able to rescue, demonstrating that Fra needs an intact activation domain to regulate comm expression (Neuhaus-Follini and Bashaw, 2015).
Fra regulates the transcription of one known gene in Drosophila, the Neo1 ICD interacts with several promoters in cells, and there are likely more genes that Dcc, Neo1, and Fra regulate to control axon guidance or other Dcc-, Neo1- and Fra-dependent processes. The sole gene currently known to be regulated by Fra is the endosomal sorting protein Comm (Yang et al., 2009; Neuhaus-Follini and Bashaw, 2015), which does not have an orthologue in vertebrates. It is also unclear whether the Fra ICD activates transcription of comm directly by binding to the comm promoter, or indirectly by regulating the transcription of other genes.
Control of progenitor dynamics: axon guidance receptors controlling transcription?
Unlike Dcc, Neo1, and Fra, the Robo receptors have not been implicated in regulating transcription directly. Still, both Drosophila Robo1 and Human ROBO1 receptors are cleaved by the metalloprotease Kuzbanian/ADAM10, and this cleavage is necessary for Slit-Robo1 signaling in Drosophila (Coleman et al., 2010). In addition, human ROBO1 has been shown to undergo a subsequent cleavage by gamma-secretase, which allows the ROBO1 ICD to enter the nucleus in cancer cell lines (Seki et al., 2010). These observations suggest ROBO1 has the potential to enter the nucleus and act as a transcription factor; however, there is no in vivo evidence supporting this idea.
Slit-Robo signaling is required in mammalian cortical neurogenesis, and some evidence suggests Robo receptors may regulate transcription in this context; however, whether Robo receptors regulate transcription directly or indirectly is unclear. Furthermore, reports in the field have often produced conflicting results that complicate our understanding of how Robo receptors might regulate cortical neurogenesis. In the developing mammalian cortex, progenitor cells must strike a balance between dividing for self-renewal, and generating post-mitotic neurons, such as excitatory pyramidal neurons (Noctor et al., 2007). Apical (radial glial cells) and basal (intermediate progenitors) progenitor populations can divide to produce pyramidal neurons. Radial glia typically divide symmetrically to self-renew, and asymmetrically to give rise to either pyramidal neurons, or, more likely, intermediate progenitors (Noctor et al., 2004). Intermediate progenitors always divide symmetrically, either to self-renew or to produce two pyramidal neurons (Miyata et al., 2004; Noctor et al., 2004).
Robo receptors had already been implicated in the regulation of cortical interneuron proliferation (Andrews et al 2006, Hernandez-Miranda 2011), and the expression of Robo1, Robo2, and Slit in the ventricular and subventricular zones (VZ and SVZ) of the cortex suggested Slit-Robo signaling may also have a role in proliferation of pyramidal neurons (Borrell et al., 2012; Yeh et al., 2014). Here we focus on two recent reports that provide some evidence for Robo receptors regulating transcription, yet they directly contradict each other in several key aspects (Borrell et al., 2012; Yeh et al., 2014). Despite some conflicting observations, both studies support the idea that Slit-Robo signaling plays important roles in regulating progenitor dynamics in the developing mammalian cortex (Borrell et al., 2012; Yeh et al., 2014).
Borrell and colleagues show that although Robo1 and Robo2 are both detected in the VZ of the cortex, Robo2 appears to be much more highly expressed (Borrell et al., 2012). Accordingly, while both Robo1 and Robo2 single mutants have an increase in basal progenitors (albeit less severe than the double mutant), Robo2 mutants have a more severe phenotype than the Robo1 mutants, suggesting Robo2 has a larger role in regulating progenitor populations in the developing cortex. Similarly, single mutants of Slit1 and Slit2 had no significant effect on the progenitor populations in the cortex, yet the Slit1/2 double mutant resulted in an increase in basal progenitors (Borrell et al., 2012). In direct contrast to these observations, Yeh and colleagues show that Robo1 is expressed in the proliferative zones of the cortex, while Robo2 is undetectable (Yeh et al., 2014). Furthermore, Robo2 single mutants did not have any defects in the progenitor populations in the cortex, while Robo1 mutants resulted in an increase in both the apical and basal progenitor populations (Yeh et al., 2014). While the role for Robo receptors reported by the two groups are clearly at odds, there is agreement that Slit1 and 2 are necessary for proper regulation of progenitor populations in the cortex. Notably, the two groups used different Robo1 and Robo2 single mutants, raising the possibility that differences in genetic background may explain some of the phenotypic differences that were reported; however, in both cases the mutants used are null mutants, and both groups used the same Robo1/2 double mutants. While both reports find that Slit-Robo signaling is involved in controlling progenitor dynamics, the mechanism each proposes differs greatly. Borrell and colleagues report that there is no difference in apoptosis, and the cell cycle of the basal progenitors is found to be disrupted in Robo1/2 mutants: basal progenitors divide less frequently, their cell cycle length is significantly longer, and progenitors fail to separate from the ventricular surface (Borrell et al., 2012). Progenitors that stay attached to the ventricular surface are known to have decreased proliferation (Cappello et al., 2006), suggesting this may be a cause for the slow and less frequent divisions of basal progenitors. Yeh and colleagues, however, find that fewer progenitors undergo apoptosis, progenitors are proliferative for an increased amount of time, but their cell cycle appears otherwise normal, and the Robo1 mutants have a small but significant decrease in microglia (Yeh et al., 2014), which are reported to cause an increase in progenitor pools (Cunningham et al., 2013). Analyzing conditional knockouts for Robo1/2 single and double mutants may help to clarify the discrepancies observed in regards to the Robo receptor required for proper cortical neurogenesis, and the mechanism required for proper progenitor dynamics.
Robo receptor signaling and the control of neural progenitor dynamics
How might Robo receptors signal downstream to regulate progenitor dynamics? Interestingly, expression of the Notch1 effector Hes1 is significantly reduced in the cortex in Robo1/2 double mutants, and over-expression of Hes1 in Robo1/2 double mutants recues the progenitor defect (Borrell et al., 2012). In addition, RNAi knock down of Hes1 leads to a reciprocal effect and increases the number of progenitors. These observations suggest that Robo receptors may regulate Hes1 expression to mediate progenitor dynamics in the developing cortex. The effect of Robo2 on Hes1 expression was further tested using an in vitro primary culture system, where a myristolated Robo2 construct was found to activate the Hes1 reporter (Hes-luc). Robo2 was able to activate transcription of the Hes-luc reporter independently of Notch1, although co-expression of Notch1 and Robo2 led to a synergistic effect on reporter expression (Borrell et al., 2012). These findings suggest Robo2 may regulate progenitor dynamics in the cortex through the regulation of transcription. Additional evidence pointing to a potential role for Robo receptors in regulating transcription comes from microarray analysis on tissue from the developing cortex, where it was found that over 300 genes are either up- or down-regulated in Robo1 mutants compared to wildtype controls (Yeh et al., 2014). Thus, in the context of progenitor proliferation in the developing mammalian cortex, the Robo receptors may regulate the transcription of genes involved in neurogenesis.
Robo receptors and progenitor dynamics in intestinal stem cells
In the adult Drosophila midgut epithelium Robo2 plays a role in maintaining progenitor dynamics. In the midgut, intestinal stem cells (ISCs) give rise to both enteroblast progenitor cells and secretory enteroendocrine (EE) cells (Zeng and Hou, 2015). Robo2 RNAi and robo2 homozygous clones generated using MARCM (Lee and Luo, 2001), and the specific knockdown of robo2 in only ISCs, all result in an increase in EE cells. These observations suggest that Robo2 normally functions in ISCs to control progenitor dynamics and restrict the differentiation of EEs (Biteau and Jasper, 2014). The transcription factor Prospero (Pros) is necessary but not sufficient to specify EE cell fate (Zeng and Hou, 2015), and genetic interactions with Robo2 suggest Robo2 and Pros might act in the same process (Biteau and Jasper, 2014). While the relationship between Pros and Robo2 in the Drosophila midgut remains unclear, one intriguing idea is that Robo2 may regulate transcription of pros in this system.
ephrin-Eph signaling and the regulation of neurogenesis
In contrast to the uncertainty over whether Robo receptors can control transcriptional regulation to mediate progenitor dynamics, there is stronger evidence that Eph-ephrin signaling regulates transcription during neurogenesis, as reviewed in Laussu et al. (2014). The transmembrane ephrinBs are cleaved by gamma-secretase (Georgakopoulos et al., 2006; Tomita et al., 2006), and the ephrinB1 ICD can interact with zinc fingers and homeoboxes 2 (Zhx2), a transcriptional repressor that is expressed in cortical neural progenitors and inhibits neuronal differentiation (Wu et al., 2009). One transcriptional target of ephrinB1 signaling in neural progenitors is the pro-neurogenic miRNA miR-124 (Arvanitis et al., 2010). EphrinB1 mutant neural progenitor cells have an increase in miR-124 RNA, and cortical sections from EphrinB1 mutant mice have increased levels of miR-124 RNA (Arvanitis et al., 2010). Interestingly, miR-124 in turn represses expression of EphrinB1 along with other genes (Arvanitis et al., 2010). While ephrinB1 signaling is implicated in repressing transcription, the evidence that ephrinB1 regulates transcription directly is weak. While there are reports that ephrinBs are cleaved by gamma-secretase, it has not been shown that gamma-secretase cleavage or the translocation of ephrinB1 ICD are required for ephrinB1 to repress transcription of miR-124. Indeed, ephrinB1 ICD interacts with transcriptional coactivator with PDZ-binding motif (Taz), and phosphorylation of the ephrinB1 ICD results in translocation of Taz to the nucleus in bone marrow stromal cells (Xing et al., 2010). However, whether transcriptional regulation also requires ephrinB1 to translocate to the nucleus remains unknown.
Discussion and Future Directions
Axon guidance pathways regulate axon guidance, synaptogenesis, progenitor dynamics, and cell migration using a variety of mechanisms. Originally found to control local cytoskeletal rearrangements, axon guidance pathways may also regulate gene expression to control these complex developmental processes. Mounting evidence demonstrates that axon guidance ligands have the ability to induce local translation, and that this is often a requirement for growth cones to respond to axon guidance cues in vitro. Axon guidance cues also induce the local translation of specific proteins that are required for the growth cone to respond to the cue. This presents an interesting model where guidance cues induce translation of specific proteins at local sites in the growth cone to mediate growth cone steering, axon branching, and synaptogenesis. However, further research is necessary to demonstrate that local protein synthesis is required in vivo for specific axon guidance pathways. In addition, it is not clear how the axon guidance receptors required for local translation signal to translation machinery. Thus far, the only receptor shown to directly interact with translational machinery is Dcc, and this interaction has yet to be shown to be required for netrin1-Dcc in vivo functional outputs. A more thorough understanding of the receptor signaling mechanisms that converge on translational machinery might allow for the design of more specific receptor manipulations that would directly test their in vivo requirement in local translation. Since it is clear that multiple guidance cues regulate translation, at least in vitro, how broad of a role does local translation play in vivo in axon guidance? A recent report describing the transcripts that were linked with ribosomes in the axons from both embryonic mice as well as postnatal mice shows an enrichment for transcripts with axon-specific functions (Shigeoka et al., 2016), suggesting that local translation of these mRNAs may play a role in axon guidance and synaptogenesis.
The axon guidance receptors Fra, Neo1, and Dcc can act as transcription factors, and Robo receptors and ephrins have the potential to at least interact with transcription factors to regulate transcription indirectly. Fra, Neo1, and Dcc activate transcription in vitro, and Fra also has one characterized transcriptional target in vivo in Drosophila. It remains to be determined whether Fra activates transcription of comm directly, or through the transcriptional regulation of other genes. Fra, Neo1, and Dcc are all sequentially cleaved, and their ICDs can enter the nucleus. Future studies should determine the mechanism through which they activate transcription, and whether they have multiple targets. Axon guidance receptors are also expressed in other tissues besides the nervous system, and determining whether they function as transcription factors in other tissues will provide insight into general non-canonical mechanisms, and a better understanding of developmental processes. The evidence that ephrinB1 acts as a transcription factor is promising, but definitive evidence that ephrinB1 has a nuclear function is still lacking. The Robo receptors have a clear role in progenitor dynamics, and they have been tied to alterations in gene expression in mammalian neurogenesis and the Drosophila midgut. Whether Robo receptors can directly regulate transcription in these tissues to control progenitor dynamics remains to be determined.
Continuing research into the mechanisms by which axon guidance signaling pathways regulate transcription and local translation will provide a more thorough understanding of axon guidance, synaptogenesis, and ultimately neural circuit formation. Clearly, precise regulation of axon guidance requires more than cytoskeletal rearrangements, and a better understanding of how axon guidance cues and receptors regulate gene expression will be informative for elucidating these processes. Axon guidance cues and receptors are also expressed in tissues outside of the nervous system in normal development, and in cancer cells. Understanding how axon guidance pathways signal to control gene expression will also more broadly provide insight into developmental processes, disease states, and may suggest new therapeutic strategies.
Acknowledgments
Supported by:
National Institute of Neurological Disorders and Stroke
R01NS-046333
R01NS-054739
R35NS-097340
National Institutes of Health
T32-HD083185
References
- Arvanitis DN, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol Cell Biol. 2010;30:2508–2517. doi: 10.1128/MCB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P, Harada H, Tassew NG, Charish J, Goldschneider D, Wallace VA, Sugita S, Mehlen P, Monnier PP. Upsilon-secretase and LARG mediate distinct RGMa activities to control appropriate layer targeting within the optic tectum. Cell Death Differ. 2016;23:442–453. doi: 10.1038/cdd.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon A, Iyer A, Bridi S, Lee FC, Ovando-Vazquez C, Corradi E, Longhi S, Roccuzzo M, Strohbuecker S, Naik S, Sarkies P, Miska E, Abreu-Goodger C, Holt CE, Baudet ML. miR-182 Regulates Slit2-Mediated Axon Guidance by Modulating the Local Translation of a Specific mRNA. Cell Rep. 2017;18:1171–1186. doi: 10.1016/j.celrep.2016.12.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernadskaya YY, Wallace A, Nguyen J, Mohler WA, Soto MC. UNC-40/Dcc, SAX-3/Robo, and VAB-1/Eph polarize F-actin during embryonic morphogenesis by regulating the WAVE/SCAR actin nucleation complex. PLoS Genet. 2012;8:e1002863. doi: 10.1371/journal.pgen.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7:1867–1875. doi: 10.1016/j.celrep.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, Cardenas A, Ciceri G, Galceran J, Flames N, Pla R, Nobrega-Pereira S, Garcia-Frigola C, Peregrin S, Zhao Z, Ma L, Tessier-Lavigne M, Marin O. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovolenta P. Morphogen signaling at the vertebrate growth cone: a few cases or a general strategy? J Neurobiol. 2005;64:405–416. doi: 10.1002/neu.20161. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, Brakebusch C, Gotz M. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Charron F, Tessier-Lavigne M. The Hedgehog, TGF-beta/BMP and Wnt families of morphogens in axon guidance. Adv Exp Med Biol. 2007;621:116–133. doi: 10.1007/978-0-387-76715-4_9. [DOI] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HA, Labrador JP, Chance RK, Bashaw GJ. The Adam family metalloprotease Kuzbanian regulates the cleavage of the roundabout receptor to control axon repulsion at the midline. Development. 2010;137:2417–2426. doi: 10.1242/dev.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Ramos DA, Margeta MA, Shen K. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science. 2007;318:103–106. doi: 10.1126/science.1143762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Dou P, DeWit M, Kater SB. Protein synthesis within neuronal growth cones. J Neurosci. 1992;12:4867–4877. doi: 10.1523/JNEUROSCI.12-12-04867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontchev VD, Letourneau PC. Nerve growth factor and semaphorin 3A signaling pathways interact in regulating sensory neuronal growth cone motility. J Neurosci. 2002;22:6659–6669. doi: 10.1523/JNEUROSCI.22-15-06659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar NR, Spencer GE. Pursuing a 'turning point' in growth cone research. Dev Biol. 2008;318:102–111. doi: 10.1016/j.ydbio.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Garbe DS, O'Donnell M, Bashaw GJ. Cytoplasmic domain requirements for Frazzled-mediated attractive axon turning at the Drosophila midline. Development. 2007;134:4325–4334. doi: 10.1242/dev.012872. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos A, Litterst C, Ghersi E, Baki L, Xu C, Serban G, Robakis NK. Metalloproteinase/Presenilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. Embo j. 2006;25:1242–1252. doi: 10.1038/sj.emboj.7601031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JS, Ashour MA, Magdesian MH, Tritsch NX, Harris SN, Christofi N, Chemali R, Stern YE, Thompson-Steckel G, Gris P, Glasgow SD, Grutter P, Bouchard JF, Ruthazer ES, Stellwagen D, Kennedy TE. Netrin-1 promotes excitatory synaptogenesis between cortical neurons by initiating synapse assembly. J Neurosci. 2013;33:17278–17289. doi: 10.1523/JNEUROSCI.1085-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschneider D, Rama N, Guix C, Mehlen P. The neogenin intracellular domain regulates gene transcription via nuclear translocation. Mol Cell Biol. 2008;28:4068–4079. doi: 10.1128/MCB.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. J Neurochem. 2014;129:221–234. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WA, Holt CE, Bonhoeffer F. Retinal axons with and without their somata, growing to and arborizing in the tectum of Xenopus embryos: a time-lapse video study of single fibres in vivo. Development. 1987;101:123–133. doi: 10.1242/dev.101.1.123. [DOI] [PubMed] [Google Scholar]
- Hou ST, Jiang SX, Smith RA. Permissive and repulsive cues and signalling pathways of axonal outgrowth and regeneration. Int Rev Cell Mol Biol. 2008;267:125–181. doi: 10.1016/S1937-6448(08)00603-5. [DOI] [PubMed] [Google Scholar]
- Jain S, Welshhans K. Netrin-1 induces local translation of down syndrome cell adhesion molecule in axonal growth cones. Dev Neurobiol. 2016;76:799–816. doi: 10.1002/dneu.22360. [DOI] [PubMed] [Google Scholar]
- Kim S, Martin KC. Neuron-wide RNA transport combines with netrin-mediated local translation to spatially regulate the synaptic proteome. Elife. 2015;4 doi: 10.7554/eLife.04158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Laussu J, Khuong A, Gautrais J, Davy A. Beyond boundaries--Eph:ephrin signaling in neurogenesis. Cell Adh Migr. 2014;8:349–359. doi: 10.4161/19336918.2014.969990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung LC, Urbancic V, Baudet ML, Dwivedy A, Bayley TG, Lee AC, Harris WA, Holt CE. Coupling of NF-protocadherin signaling to axon guidance by cue-induced translation. Nat Neurosci. 2013;16:166–173. doi: 10.1038/nn.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. Embo j. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Huang CH, Kao HH, Liou GG, Yeh SR, Cheng CM, Chen MH, Pan RL, Juang JL. Abi plays an opposing role to Abl in Drosophila axonogenesis and synaptogenesis. Development. 2009;136:3099–3107. doi: 10.1242/dev.033324. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Manns RP, Cook GM, Holt CE, Keynes RJ. Differing semaphorin 3A concentrations trigger distinct signaling mechanisms in growth cone collapse. J Neurosci. 2012;32:8554–8559. doi: 10.1523/JNEUROSCI.5964-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Nedelec S, Peljto M, Shi P, Amoroso MW, Kam LC, Wichterle H. Concentration-dependent requirement for local protein synthesis in motor neuron subtype-specific response to axon guidance cues. J Neurosci. 2012;32:1496–1506. doi: 10.1523/JNEUROSCI.4176-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus-Follini A, Bashaw GJ. The Intracellular Domain of the Frazzled/Dcc Receptor Is a Transcription Factor Required for Commissural Axon Guidance. Neuron. 2015;87:751–763. doi: 10.1016/j.neuron.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Neural stem and progenitor cells in cortical development. Novartis Found Symp. 2007;288:59–73. discussion 73-58, 96-58. [PubMed] [Google Scholar]
- O'Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32:383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Kohmura E, Yamashita T. TACE cleaves neogenin to desensitize cortical neurons to the repulsive guidance molecule. Neurosci Res. 2011;71:63–70. doi: 10.1016/j.neures.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Park J, Knezevich PL, Wung W, O'Hanlon SN, Goyal A, Benedetti KL, Barsi-Rhyne BJ, Raman M, Mock N, Bremer M, Vanhoven MK. A conserved juxtacrine signal regulates synaptic partner recognition in Caenorhabditis elegans. Neural Dev. 2011;6:28. doi: 10.1186/1749-8104-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan KD, Croteau LP, Kam JW, Kania A, Cloutier JF, Butler SJ. Neogenin may functionally substitute for Dcc in chicken. PLoS One. 2011;6:e22072. doi: 10.1371/journal.pone.0022072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling Mechanisms Underlying Slit2-Induced Collapse of Xenopus Retinal Growth Cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Camacho C, Bovolenta P. Emerging mechanisms in morphogen-mediated axon guidance. Bioessays. 2009;31:1013–1025. doi: 10.1002/bies.200900063. [DOI] [PubMed] [Google Scholar]
- Schaffar G, Taniguchi J, Brodbeck T, Meyer AH, Schmidt M, Yamashita T, Mueller BK. LIM-only protein 4 interacts directly with the repulsive guidance molecule A receptor Neogenin. J Neurochem. 2008;107:418–431. doi: 10.1111/j.1471-4159.2008.05621.x. [DOI] [PubMed] [Google Scholar]
- Seki M, Watanabe A, Enomoto S, Kawamura T, Ito H, Kodama T, Hamakubo T, Aburatani H. Human Robo1 is cleaved by metalloproteinases and gamma-secretase and migrates to the nucleus in cancer cells. FEBS Lett. 2010;584:2909–2915. doi: 10.1016/j.febslet.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE. Dynamic Axonal Translation in Developing and Mature Visual Circuits. Cell. 2016;166:181–192. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane M, Gallo G. Involvement of Rho-family GTPases in axon branching. Small GTPases. 2014;5:e27974. doi: 10.4161/sgtp.27974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz TR, LinSeman DA. Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front Cell Neurosci. 2014;8:314. doi: 10.3389/fncel.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AK, Colon-Ramos DA. Netrin instructs synaptic vesicle clustering through Rac GTPase, MIG-10, and the actin cytoskeleton. J Cell Biol. 2012;197:75–88. doi: 10.1083/jcb.201110127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AK, Nelson JC, Martinez-Velazquez LA, Klein M, Samuel AD, Colon-Ramos DA. Synaptic vesicle clustering requires a distinct MIG-10/Lamellipodin isoform and ABI-1 downstream from Netrin. Genes Dev. 2012;26:2206–2221. doi: 10.1101/gad.193409.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl F, Lin JQ, Laine RF, Wong HH, Urbancic V, Cagnetta R, Holt CE, Kaminski CF. Single Molecule Translation Imaging Visualizes the Dynamics of Local beta-Actin Synthesis in Retinal Axons. Sci Rep. 2017;7:709. doi: 10.1038/s41598-017-00695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Kim SH, Sisodia SS. Presenilin-dependent "gamma-secretase" processing of deleted in colorectal cancer (Dcc) J Biol Chem. 2003;278:30425–30428. doi: 10.1074/jbc.C300239200. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane Receptor Dcc Associates with Protein Synthesis Machinery and Regulates Translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J Cell Biol. 1970;44:62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita T, Tanaka S, Morohashi Y, Iwatsubo T. Presenilin-dependent intramembrane cleavage of ephrin-B1. Mol Neurodegener. 2006;1:2. doi: 10.1186/1750-1326-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Qiu R, Wang J, Zhang H, Murai K, Lu Q. ZHX2 Interacts with Ephrin-B and regulates neural progenitor maintenance in the developing cerebral cortex. J Neurosci. 2009;29:7404–7412. doi: 10.1523/JNEUROSCI.5841-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Kim J, Wergedal J, Chen ST, Mohan S. Ephrin B1 regulates bone marrow stromal cell differentiation and bone formation by influencing TAZ transactivation via complex formation with NHERF1. Mol Cell Biol. 2010;30:711–721. doi: 10.1128/MCB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam PT, Charron F. Signaling mechanisms of non-conventional axon guidance cues: the Shh, BMP and Wnt morphogens. Curr Opin Neurobiol. 2013;23:965–973. doi: 10.1016/j.conb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Yang L, Garbe DS, Bashaw GJ. A frazzled/Dcc-dependent transcriptional switch regulates midline axon guidance. Science. 2009;324:944–947. doi: 10.1126/science.1171320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yeh ML, Gonda Y, Mommersteeg MT, Barber M, Ypsilanti AR, Hanashima C, Parnavelas JG, Andrews WD. Robo1 modulates proliferation and neurogenesis in the developing neocortex. J Neurosci. 2014;34:5717–5731. doi: 10.1523/JNEUROSCI.4256-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Hou SX. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142:644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]