Abstract
Vitamin B12 or cobalamin is an essential metabolite for humans, which makes it an interesting compound for many research groups that focus in different producer-strains synthesis pathways. In this work, we report the influence of key intermediaries for cobalamin synthesis added to the culture medium in two Lactobacillus (L.) strains, L. reuteri CRL 1098 and L. coryniformis CRL 1001. Here, we report that addition of Co2+ and 5,6-dimethylbenzimidazole increased the corrinoid compounds production in both strains while addition of L-threonine increased only the corrinoid compounds production by CRL 1001 strain. Then, we purified and characterized by LC-MS the corrinoid compounds obtained. Physiological studies besides in silico analysis revealed that L. reuteri CRL 1098 and L. coryniformis CRL 1001 follow different pathways for the last steps of the corrinoid compounds synthesis.
Keywords: lactic acid bacteria, Lactobacillus, corrinoid synthesis, cobalamin gene cluster, biosynthetic intermediaries
Introduction
Vitamin B12 belongs to group B vitamins and is the most complex water-soluble vitamin. Many analogous of vitamin B12 have been described; all of them are porphyrin compounds with a common structure: a corrinoid ring contracted with a chelated cobalt ion at the center of the macrocycle. In this vitamin, the cobalt ion is covalently bound to an upper or β-ligand, and is coordinated with a lower or α-ligand (Rucker et al., 2001). In nature, the vitamin B12 analogs present either an adenosyl group or a methyl group as β-ligand. The best-studied vitamin B12 is cobalamin, a cobamide where 5,6-dimethylbenzimidazole (DMB) is the α-ligand aglycon (Johnson and Escalante-Semerena, 1992). In the vitamin B12 synthetic preparations, the cyano group is present as β-ligand in contrast to naturally occurring analogs. Furthermore, changes in the α-ligand result in different vitamin B12 analogs. Several vitamin B12 analogs containing benzimidazoles, purines, and phenolic compounds as bases of the α-ligand have been described (Chan et al., 2015).
De novo biosynthesis of all these corrinoid compounds can be divided into three main steps: (i) the Uroporphyrinogen III synthesis; (ii) the corrinoid ring synthesis, and (iii) the adenosylation, the amino-propanol arm attachment and the nucleotide loop bridging assembly of the lower ligand to the cobalt of the corrinoid ring core (Martens et al., 2002). Corrinoid ring synthesis follows two different pathways according to oxygen requirements. In the aerobic pathway, the cobalt chelation occurs as one of the final steps of the corrinoid ring formation while in the anaerobic pathway this reaction takes place in the first step of the synthesis (Warren et al., 2002).
Animals cannot synthesize vitamin B12, which is involved in many important enzymatic reactions, thus making it an essential metabolite. These reactions are the conversion of homocysteine to methionine and the interconversion of (2R)-methylmalonyl-CoA to succinyl-CoA. Certain bacteria strains and archaea are able to synthetize vitamin B12 by de novo biosynthetic pathway (Roth et al., 1996). For this reason, many research studies are focused on the production of vitamins by bacteria in food and on improving the synthesis in large-scale production as well.
Currently, there are several Lactobacillus (L.) strains described as vitamin B12 producers, such as L. reuteri CRL 1098, L. rossiae DSM 15814, L. coryniformis CRL 1001 and L. plantarum BCF 20, BHM 10 and LZ 95 (Taranto et al., 2003; De Angelis et al., 2014; Torres et al., 2016; Bhushan et al., 2017) whose genomes have been partially sequenced.
Previous genome analysis of L. reuteri CRL 1098 and L. coryniformis CRL 1001 showed that both strains contain all necessary genes for de novo corrinoid compound biosynthesis. In this work, we put in evidence key differences in the last steps of the biosynthetic pathway of these strains. Besides, an increased corrinoid compounds synthesis was observed when adding certain intermediaries to the culture medium. These results were confirmed by HPLC quantification, and compounds were characterized by mass spectrometry. The expression profile of key genes of the biosynthetic pathway was also studied. Finally, we demonstrated the existence of two different biosynthetic pathways of cobalamin-type corrinoid compounds in Lactobacillus strains.
Materials and Methods
Strains, Media, and Culture Conditions
Lactobacillus reuteri CRL 1098 and L. coryniformis CRL 1001 were previously described as cobalamin producer strains (Taranto et al., 2003; Torres et al., 2016). These strains belong to CERELA-CONICET culture collection. The strains CRL 1098 and CRL 1001 were grown overnight without shaking in Man-Rogosa-Sharpe (MRS) broth and in Vitamin B12 Assay Medium (Merck, Germany), at 37°C.
Salmonella (S.) Typhimurium AR 2680 was used as indicator strain in the bioassays for cobalamin determination in minimal A medium (NaCl, 0.5 g/l, Na2HPO4, 6 g/l; KH2PO4, 3 g/l, NH4Cl 1 g/l; glucose, 4 g/l; MgSO4, 2 mM, CaCl2 0.1 mM). This strain has two mutations in metE and cbiB genes. The AR 2680 strain was grown with aeration in Luria-Bertani (LB) broth at 37°C. As negative control, L. plantarum ATCC 8014 strain was used.
Cultures and Cell-Extracts
The biosynthetic intermediaries added to the Vitamin B12 Assay Medium were: 5-aminolevulinic acid [ALA; final concentration (FC): 25 ng/ml]; Cobalt Chloride (CoCl2, FC: 250 ng/ml), 5,6-dimethylbenzimidazole (DMB; FC: 200 ng/ml); Porphobilinogen (PBG, FC: 250 ng/ml); L-threonine (L-Thr, FC: 50 μg/ml) and Uroporphyrinogen III (UIII, FC: 250 ng/ml). The corrinoids produced in the presence of different intermediaries were extracted as described by Torres et al. (2016).
Cobalamin Detection
The production of corrinoid compounds in the cell extract (CE) obtained from each strain grown for 16 h was analyzed with the bioassay using S. Typhimurium AR 2680 as indicator strain. L. plantarum ATCC 8014 and a commercial cyanocobalamin solution (0.5 μg/ml) were used as negative and positive control, respectively. In addition, the concentration of vitamin B12 was determined by commercial enzyme immunoassay (RIDASCREEN-FAST Vitamin B12. R-Biopharm, Rhone Ltd., Glasgow, Scotland). The competitive immunoassay for the vitamin B12 determination was performed following the protocol described by the manufacturer.
Purification, Characterization, and Quantification of the Corrinoid Produced by Lactobacillus Strains
Based on previous results of our research group, key intermediaries (alone or in combination) were added to the growth culture media in order to increase the corrinoid concentration produced. The following conditions were used for L. coryniformis CRL 1001 and L. reuteri CRL 1098: (i) CoCl2, (ii) CoCl2 plus DMB, and (iii) CoCl2 plus L-Thr only for CRL 1001 strain.
Then, the cells were broken and the corrinoid in the intracellular fraction (cell extract -CE-) was evinced by bioassay. The corrinoid produced was purified and quantified by RP-HPLC. The corrinoid concentration was calculated from the peaks areas and a commercial CN-Cbl standard curve made with 0.5, 1, and 2 μg/ml. The peaks of the chromatogram were collected for the mass spectrometry analysis. RP-HPLC and mass spectrometry were carried out as described by Torres et al. (2016). Experiments were performed three times in duplicate and results were expressed as means ± SD. Statistical analysis was conducted using MINITAB software (version 15 for Windows). Tukey’s post hoc test was used to test for differences between the mean values. Significance was set at P < 0.05.
Relative Expression of Key Genes of Cobalamin Synthesis in Lactobacillus Strains
The primers for relative gene expression analysis (RT-qPCR) were designed on the basis of the corresponding gene sequences of L. coryniformis CRL 1001 (NZ_LNUL00000000.1) and L. reuteri CRL 1098 (NZ_LYWI00000000.1) using Primer Design tool (Bio-Rad) (Table 1). As normalizing reporter, 16S rRNA gene was used.
Table 1.
Primer sequences of key genes of de novo biosynthesis of cobalamin-type corrinoid compounds.
| Strain | Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon length (bp) |
|---|---|---|---|---|
| CRL 1098 | 16S rRNA | ACGTGCTACAATGGACGGTA | ACTAGCGATTCCGACTTCGT | 120 |
| cbiF | TGCGTTCCTGGTGTTAGTTC | GTCCTGCCATACGCGTAATAA | 106 | |
| hemB | CGCGAAGTAGCTAGTGATGAA | CAACTAACGGCAGCAAAGTATG | 115 | |
| cobS | AAGCACATGGCGAATTATTG | TTCGTGACCTTTCGTCGAT | 132 | |
| cobT | AAACAGCAGCAGAAGTTGTTGGGC | CATTGCTCCAGCCATTGCACCTAA | 182 | |
| CRL 1001 | 16S rRNA | GACGAAAGTCTGATGGAGCA | TTCTGGTTGGATACCGTCAA | 122 |
| cbiF | AGTGTTTCTGGGACTTTGGC | CGGTATTCATGGGTGAAATG | 146 | |
| hemB | GATGGTATCGTGCAACAAGC | ACGACACCACAATGACCAGT | 113 | |
| cobS | TCTTCACGAACACCCGATAA | TTGCAATGGTGTTACCTGCT | 141 | |
| pduX | GTACGGCAGATCTAGTGGCA | GCATCGATCACAGTCAATCC | 148 | |
| cblS | TCAGTCGGTGTAACCGTGAT | GTCGCACTTCTTGGTAAGCA | 148 | |
| cblT | CAATTATGCATCAAGCCGTT | TGGTAAGGCCACGTAGACAA | 98 |
Total RNA from L. coryniformis CRL 1001 and L. reuteri CRL 1098 grown in presence of the intermediaries were extracted at different growth phases (lag, exponential, and late exponential). Briefly, 10 mL of the cell pellet was suspended in 500 μL TE buffer (Tris–HCl 10 mM pH 8 – EDTA 1 nM pH 8) and added to 170 μL macaloid 2%, 500 μL TE buffer saturated with phenol-chloroform-isoamylic solution (chloroform-isoamylic 24:1, phenol-chloroform-isoamylic 1:1), 50 μL SDS 10% and 0,6 g glass beads (Sigma-Aldrich, Buenos Aires, Argentina). This suspension was subjected to Mini-BeadBeater-8 cell disrupter (BioSpec Products Inc.) at maximum speed in 10 cycles of 1 min each cycle, with intervals of 1 min on ice, to disrupt the cells. Subsequently, samples were centrifuged (5,000 ×g, 15 min, 4°C). The upper phase was separated and added phenol-chloroform-isoamylic solution (chloroform-isoamylic 24:1, phenol-chloroform-isoamylic 1:1). Again, the samples were centrifuged (5,000 ×g, 15 min, 4°C). After separating the upper phase, RNA precipitation was done with absolute ethanol and 3 M sodium acetate at -70°C overnight. Next day, samples were centrifuged (10,000 ×g, 15 min, 4°C), washed with ethanol 70°, and dried at room temperature. RNA samples were suspended in 20 μL miliQ water. DNase treatment was performed adding 10 μL of DNase (Turbo Dnase, Ambion, Thermo Fisher Scientific, Buenos Aires, Argentina) followed by incubation at 37°C for 180 min. DNase was inactivated by adding 1 μl of RQ1 DNase Stop Solution (Ambion) to the reaction mixture and heating at 65°C for 10 min. After verifying the absence of DNA by conventional PCR using the purified RNA as template, the samples were quantified with a Qubit®2.0 fluorometer (InvitrogenTM, Life Technologies Co., Carlsbad, CA, United States) using Qubit®HS RNA Assay Kit (Molecular ProbesTM, Life Technologies Co.). All RNA samples were stored at -70°C until use.
The cDNA was synthetized using 1 μg of total RNA and qScriptTM cDNA SuperMix kit (Quanta BiosciencesTM) according to the manufacturer’s instructions in a T100TM Thermal Cycler (Bio-Rad). A conventional PCR was performed to confirm cDNA synthesis. The cDNA was stored at -70°C until use.
Quantitative PCR assays was performed using an iQTM5 Multicolor Real-Time PCR Detection System iCycler (Bio-Rad Laboratories Inc.). Amplicons were detected with PerfeCtaTM SYBER® Green SuperMix for iQTM (Quanta BiosciencesTM). Each reaction contained 1X SYBER® Green SuperMix, 300 nM of each primer and 50 ng of total cDNA, genomic DNA as positive control and no template as negative control. All reactions were done in duplicate. The amplification program consisted of 1 cycle of 94°C for 5 min and 40 cycles of amplification (94°C for 1 min, 55°C for 1 min, and 72°C for 30 s) followed by a melting curve (81 cycles of 10 s at 60°C). The relative expression of the cbiF, hemB, cobS, pduX, cblS, and cblT genes for L. coryniformis CRL 1001 and cbiF, hemB, cobS, and cobT for L. reuteri CRL 1098 with the addition of different synthesis intermediaries at 3, 6, and 9 h was estimated according to the 2-ΔΔCT method (Livak and Schmittgen, 2001). The condition with the addition of CoCl2 at 3, 6, and 9 h was used as control for each point of the growth curve for each condition. The reported values are the changes in the gene expression of the strain grown with the intermediaries and without (control, given value = 1) and normalized against 16S rRNA gene expression.
Results
In silico Comparative Genomic Analysis of Cobalamin Biosynthetic Gene Cluster
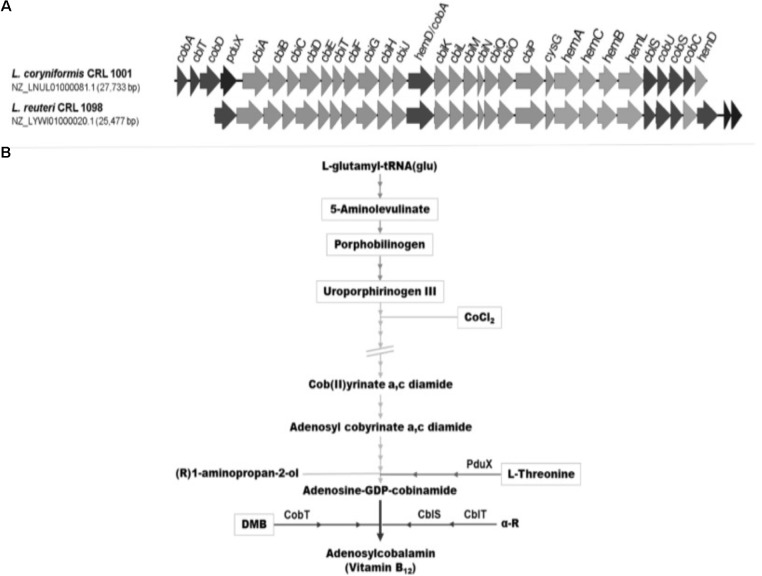
Lactobacillus reuteri CRL 1098 and L. coryniformis CRL 1001 are well-known cobalamin producer strains and they have gained attention for novel functional foods development. Previous in silico analyses of genome sequences of both strains revealed the presence of all the necessary genes for the de novo synthesis of cobalamin-type compounds although some differences in the cobalamin biosynthetic gene cluster were evidenced (Figure 1A). The genes hemALBCD encoding proteins involved in the first steps, the synthesis of uroporphyrinogen III from L-glutamyl-tRNA(glu), are present in both genomes. In addition, both strains produce the corrinoid ring by the anaerobic pathway and they bear all the cbi genes necessary for the complete ring formation. However, the last biosynthetic steps showed genes involved in different pathways for the aminopropanol arm formation and the assembly of the lower ligand.
FIGURE 1.
(A) cbi-cob-hem gene clusters of L. coryniformis CRL 1001 and L. reuteri CRL 1098. (B) Simplified cobalamin anaerobic synthesis pathway. Addition of different pathway intermediaries are highlighted with a box.
The genome of L. reuteri CRL 1098 possesses cobT gene encoding for a nicotinate mononucleotide (NaMN): base phosphoribosyltransferase, which activates the lower ligand base. In contrast, L. coryniformis CRL 1001 does not own cobT gene but possesses the cblTS genes encoding for an α-ribasol transporter and a kinase protein, respectively, as described for other Firmicutes strains. The α-ribazole salvaging and α-ribazole-P synthesis were reported previously and presented as an alternative pathway of lower ligand activation (Gray and Escalante-Semerena, 2010). It is important to note that only L. coryniformis CRL 1001 harbors the pduX gene that encodes a kinase able to phosphorylate L-Thr. This compound is a precursor of the aminopropanol arm that binds the lower ligand to the corrinoid (Figure 2). The in silico studies point out differences in the set of genes involved in the cobalamin-type compounds synthesis. On this basis, the production of these compounds by the strains under study in the presence of different intermediaries was analyzed.
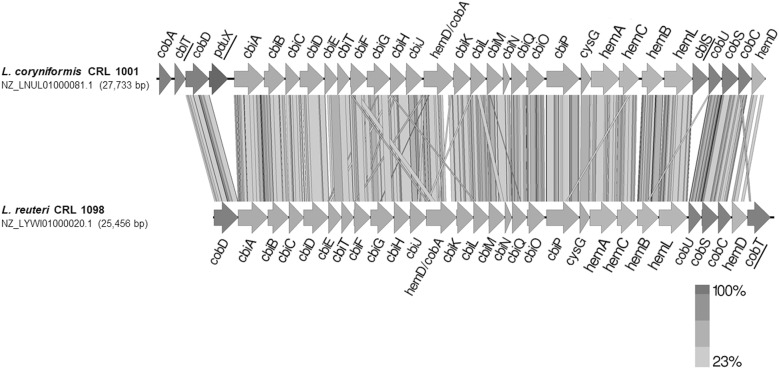
FIGURE 2.
cbi-cob-hem gene clusters comparison between L. coryniformis CRL 1001 and L. reuteri CRL 1098. Arrows indicate the transcription direction depict genes. Orthologous conserved genes are depicted as gray and black bars.
Effect of Biosynthetic Pathway Intermediaries on Corrinoid Compounds Production
Lactobacillus coryniformis CRL 1001 and L. reuteri CRL 1098 were grown in vitamin B12-free culture medium in the presence of the following biosynthetic pathway intermediaries: ALA, PBG, UIII, CoCl2, DMB plus CoCl2, L-Thr plus CoCl2. The sequential addition and roles of intermediaries have been shown in Figure 1B. The effect of DMB and L-Thr was evaluated in the presence of supplementary CoCl2 considering that the addition of this ion is an essential condition for the synthesis of corrinoids in stages subsequent to the insertion of this element. The Figure 3 show the relative quantification using a commercial enzyme immunoassay of total corrinoids in the presence of different intermediaries respect to the condition without intermediaries. The corrinoid production by L. coryniformis CRL 1001 was greater with the CoCl2, DMB + CoCl2 and L-Thr + CoCl2 compared with the condition without intermediaries; ALA, PBG, and UIII did not improve the corrinoid synthesis in this strain. In L. reuteri CRL 1098, only DMB + CoCl2 and CoCl2 increased vitamin B12 production while the other intermediaries evaluated (L-Thr, ALA, PBG, and UIII) had no positive effect.
FIGURE 3.
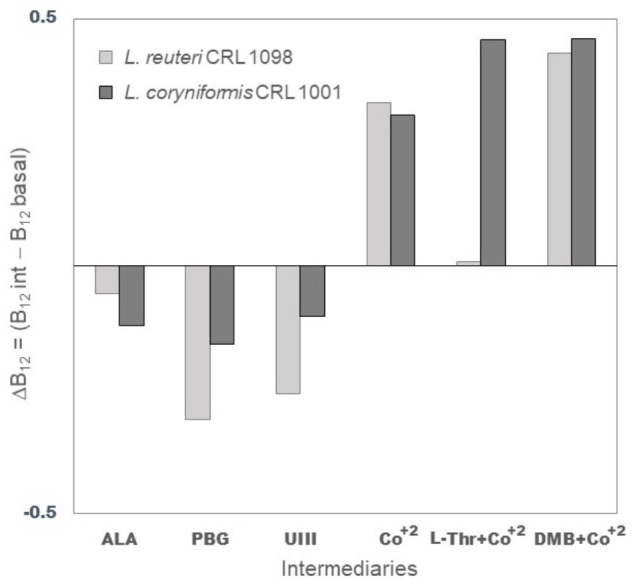
Relative quantification of corrinoid compounds produced by L. reuteri CRL 1098 and L. coryniformis CRL 1001 with addition of different intermediaries. Results are presented as ΔB12 (net difference of the corrinoid quantification with [B12 int.] and without intermediary [B12basal]). ALA (5-aminolevulinic acid), PBG (Porphobilinogen), UIII (Uroporphyrinogen III), CoCl2 (Cobalt, Co+), L-Thr (L-threonine) + CoCl2, DMB (5,6-dimethylbenzimidazole) + CoCl2.
Purification, Quantification, and Characterization of the Corrinoid Compounds Produced by Lactobacillus Strains
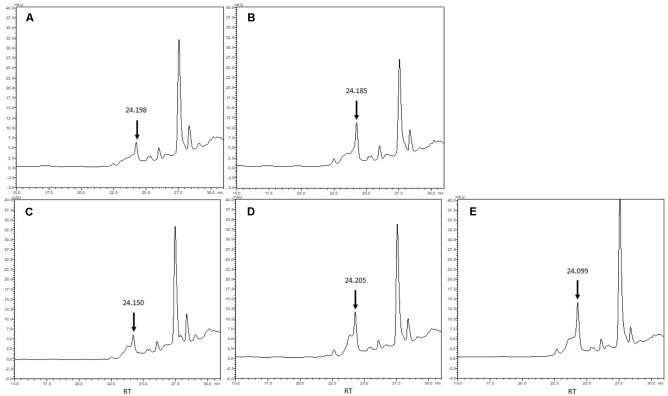
Bioassay and immunoassay demonstrated that both CoCl2 and DMB + CoCl2 addition increased the production of corrinoid compound by L. coryniformis CRL 1001 and L. reuteri CRL 1098; while the presence of L-Thr + CoCl2 induced the corrinoid production only in CRL 1001 strain. L-Thr was not evaluated in further assays for CRL 1098 strain as it had no effect on the corrinoid formation. To evaluate the optimum cobalamin production of CRL 1001 strain, a set of different growth conditions were evaluated (i) CoCl2, (ii) CoCl2 + DMB, and (iii) CoCl2 + L-Thr. In subsequent experiments, the corrinoid production by L. reuteri CRL 1098 was evaluated only in the presence of CoCl2 and CoCl2 + DMB. In order to purify the corrinoid compounds, the peaks with retention time (RT) close to cyanocobalamin (CN-Cbl) RT (24.98 min) were collected (Figure 4) and analyzed for cobalamin activity by bioassay using S. Typhimurium AR 2680 as indicator strain. All the collected peaks showed the same B12 complementation ability than CN-Cbl standard (data not shown). Furthermore, higher corrinoid levels with DMB + CoCl2 addition compared with the basal condition (CoCl2 alone) were obtained. The corrinoid production in the presence of DMB was twofold and 2.2-fold greater for CRL 1098 and CRL 1001 strains, respectively, compared to the basal condition. The L-Thr + CoCl2 addition to the medium increased by 2.6-fold the corrinoid synthesis in L. coryniformis CRL 1001 respect to the basal condition (Table 2).
FIGURE 4.
RP-HPLC chromatogram of the cell extract (CE) from L. reuteri CRL 1098 (A CoCl2 B DMB + CoCl2) and L. coryniformis CRL 1001 (C- CoCl2 D- DMB + CoCl2 E- L-Thr + CoCl2). An arrow indicates peaks corresponding to the corrinoid compound.
Table 2.
Corrinoid compounds quantification by HPLC.
| L. reuteri CRL 1098 | Intermediaries | Corrinoid (μg/mL)# |
| CoCl2 | 1.28 ± 0.26∗ | |
| DMB + CoCl2 | 2.52 ± 0.20 | |
| L. coryniformis CRL 1001 | Intermediaries | Corrinoid (μg/mL) |
| CoCl2 | 1.34 ± 0.18 | |
| DMB + CoCl2 | 2.98 ± 0.43 | |
| L-Thr + CoCl2 | 3.45 ± 0.48 |
The corrinoid concentration in CEs was calculated with a commercial CN-Cbl standard curve and the active peaks areas. #Corrinoid concentration in CEs was calculated with a commercial CN-Cbl standard curve and the active peaks areas. ∗Standard deviation.
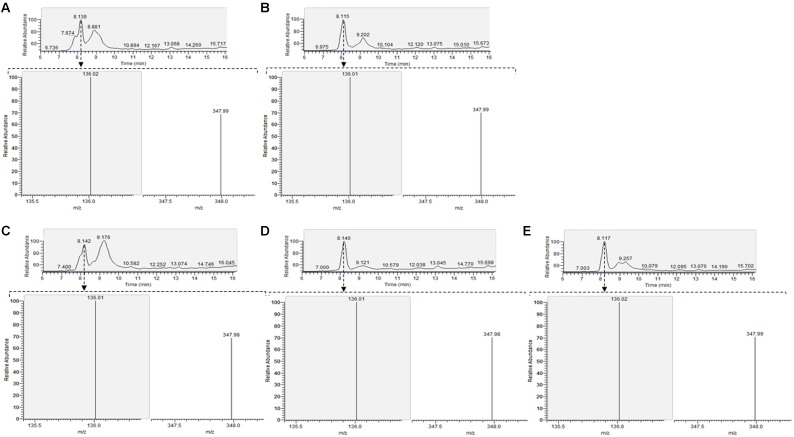
To characterize the corrinoid compounds, liquid chromatography–electrospray ionization/tandem mass spectrometry (LC/ESI–MS/MS) to the collected peaks was performed. Transitions were sought in the MS and MS/MS spectra of the peaks with cobalamin activity. The transitions 678.3 (m/z) [M + 2H+]++ to 358.7 (m/z) and 678.3 (m/z) [M + 2H+]++ to 146.9 (m/z) were examined for corrinoid compounds where DMB is the aglycon attached to ribofuranose 3-phosphate. For corrinoid compounds where adenine is the aglycon attached to ribofuranose 3-phosphate, transitions 672.5 (m/z) [M + 2H+]++ to 347.8 (m/z) and 672.5 (m/z) [M + 2H+]++ to 135.9 (m/z) were sought. For all active peaks analyzed, the MS spectra indicated that a double charged ion with an approximately m/z of 673 [M + 2H+]++ was prominent. The MS/MS spectrum showed that the dominant ions with a value approximate m/z 347.8 [M + 2H+]++ were attributable to Coα-[α-(7-adenyl)]-Coβ-cyanocobamide (Pseudo B12). In this compound type, adenine is the aglycon attached to ribofuranose-3-phosphate in the lower ligand (Figure 5).
FIGURE 5.
Liquid chromatography–electrospray ionization/tandem-mass spectrometry (LC/ESI–MS/MS) chromatograms of peaks with cobalamin activity. The transitions are shown together (SRMs, 672.5 m/z – > 136.0 m/z calculated for adenine y 672.5 m/z – > 348.0 m/z correspond to the lower ligand in which adenine is the aglycon attached to ribofuranose 3-phosphate). Total ion chromatogram (TIC) of the active peaks of L. reuteri CRL 1098 (A- CoCl2 B- DMB + CoCl2) and L. coryniformis CRL 1001 (C- CoCl2 D- DMB + CoCl2 E- L-Thr + CoCl2).
Expression of Key Genes of the Corrinoid Biosynthesis Pathway
Since the addition of biosynthetic intermediaries improved cobalamin production in both strains, the relative expression of key genes of the biosynthetic pathway in cells grown during 3 h (lag phase), 6 h (exponential phase), and 9 h (late-exponential phase) in B12 free medium in the presence of different intermediaries was tested by qPCR. The time of incubation (3, 6, and 9 h) in the B12 free medium with CoCl2 was the condition used as reference, to which the arbitrary value of 1 was assigned for both strains. Basal level of expression of key genes in the vitamin B12 free medium with different intermediaries was observed for both strains at all growth phases. The relative expression levels of the key genes with DMB and L-Thr addition remained unchanged in the strains under study after 3, 6, and 9 h of incubation with respect to the basal condition (data not shown). These data demonstrate that the key genes studied for L. reuteri CRL 1098 and L. coryniformis CRL 1001 may be regulated at a post-transcriptional or translational level.
Discussion
We have previously reported that L. reuteri CRL 1098 supplementation efficiently correct the nutritional vitamin B12 deficiency in an in vivo model (Molina et al., 2008, 2009). In this work, cobalamin biosynthesis intermediaries were added to the vitamin B12 free medium to analyse their effect on the corrinoid production by two different Lactobacillus strains.
The common precursor of all tetrapyrrole molecules (cobalamin, heme, and chlorophyll) is ALA. The result of two condensed ALA molecules is PBG. Finally, four PBG molecules are polymerized and cyclized to form UIII, the last common intermediary (Kang et al., 2012). Mohammed et al. (2014) reported an increase in vitamin B12 production by Bacillus (B.) megaterium with addition of ALA to the culture medium (Mohammed et al., 2014). Different results were obtained for both L. reuteri CRL 1098 and L. coryniformis CRL 1001 strains, in which the decreased corrinoid compound production with ALA, PBG, and UIII addition may be due to a negative regulation by a classical feedback control (Robin et al., 1991).
Regarding CoCl2 and DMB addition, an increased cobalamin production is described for Propionibacterium and Bacillus strains (Mohammed et al., 2014; Wang et al., 2015). Similar results are reported for both Lactobacillus strains, CRL1001 and CRL1098, in the present study.
In the anaerobic pathway of the cobalamin biosynthesis, precorrin-2 is chelated with cobalt a reaction that is catalyzed by CbiK enzyme. Cobalt requirements for cobalamin-type corrinoids synthesis is proposed as screening criteria for detecting producer strains of Propionibacterium and Lactobacillus genera (Seidametova et al., 2004; Bhushan et al., 2016). In this work, we report that cobalt addition as chloride salt allowed the growth of both CRL 1098 and CRL 1001 Lactobacillus strain. Moreover, the cobalt addition also increased the corrinoid production in both strains compared with our previous data without cobalt addition (Taranto et al., 2003; Torres et al., 2016).
In another step of the biosynthesis, DMB is the nucleotide attached to the aminopropanol arm as lower ligand in the cobalamin molecule (Hazra et al., 2015). The cobalamin-type corrinoid compounds produced by CRL 1001 and CRL 1098 strains increased with DMB addition. Nevertheless, LC-MS results were different as expected since the molecules synthesized corresponded to pseudo-B12 despite the positive regulation on the synthesis. This compound, with adenine instead of DMB as lower ligand base is synthesized in most bacteria able to produce cobalamin-type corrinoid compounds via the anaerobic pathway (Taga and Walker, 2008). Hazra et al. (2013) proposed that CobT enzyme of L. reuteri CRL 1098 expressed in E. coli activates DMB rather than other molecules as base of the lower ligand (Hazra et al., 2013). According to our results, CRL 1098 strain is not able to use DMB instead of adenine for vitamin B12 synthesis. As previously published, L. coryniformis CRL 1001 does not have the cobT gene but the genes encoding CblS (a kinase enzyme) and CblT (a transporter protein) (Torres et al., 2016). Mattes and Escalante-Semerena (2017) observed that cblST genes of Geobacillus (G.) kaustophilus expressed in S. enterica ΔcobT that CblT transports DMB into the cell and CblS activate preferably ribazole over adenine (Mattes and Escalante-Semerena, 2017). Our results are not in agreement with this statement since pseudo-B12 is obtained, thus suggesting that CblS is unable to phosphorylate DMB despite entering the cell. Further studies are oncoming in both strains for a better understanding of the changes taking place in the lower ligand nucleotide of the molecule.
In a different way to the previously described, the L-Thr addition to the vitamin B12 free medium showed different results in L. reuteri CRL 1098 and L. coryniformis CRL 1001 strains. The increased production of cobalamin-type corrinoid compounds in CRL 1001 strain could be ascribed to the presence of pduX gene in the genome (Torres et al., 2016). The pduX encodes a protein kinase able to phosphorylate free L-Thr prior to the formation of lower ligand aminopropanol arm (Fan and Bobik, 2008). For this reason, we propose that L-Thr may exert some positive regulation in the synthesis of this type of compounds.
The study of the expression profile of key genes encoding proteins involved in cobalamin-type corrinoid compound synthesis showed no expression change despite the increased cobalamin production in both strains. Data obtained by different work groups showed that cobalamin synthesis is regulated by a riboswitch mechanism (Nahvi et al., 2004). Briefly, binding of cobalamin to the cob mRNA inhibits the initiation of translation and stabilizes the complex formed between them (Polaski et al., 2016). For these reasons, we suggest that the regulation is at the post-transcriptional or translational level. Our results are in agreement with previous reports since no changes in regulation of key synthesis genes of the mRNA transcription phase were evidenced (Nahvi et al., 2004).
In this study, we demonstrate that addition of key intermediaries to the vitamin B12 free medium increases the corrinoid production by both CRL1001 and CRL 1098 lactobacilli strains. In addition, the existence of two different cobalamin-type corrinoid compound biosynthetic pathways in two close phylogenetic strains was confirmed. The results obtained in this study would improve the production of these cobalamin-type corrinoid compounds by the food grade L. coryniformis CRL 1001 and L. reuteri CRL 1098 strains.
Author Contributions
AT carried out biochemical and molecular genetic studies and participated in the drafting the manuscript. VV participated in physiological studies. GF participated in the discussion of the study. LS participated in the design, discussion, and coordination of this study and drafting the manuscript. MT carried out the coordination of this study and participated in the design, discussion, and drafting the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Gastón Pourrieux (Centro de Referencia para Lactobacilos, CERELA-CONICET, Argentina) for the technical support with HPLC and spectrometry studies.
Footnotes
Funding. This study was carried out with the financial support from CONICET (PIP0406/12) and MinCyT (PICT2015 N∘1705) from Argentina.
References
- Bhushan B., Tomar S. K., Chauhan A. (2017). Techno-functional differentiation of two vitamin B12 producing Lactobacillus plantarum strains: an elucidation for diverse future use. Appl. Microbiol. Biotechnol. 101 697–709. 10.1007/s00253-016-7903-z [DOI] [PubMed] [Google Scholar]
- Bhushan B., Tomar S. K., Mandal S. (2016). Phenotypic and genotypic screening of human-originated lactobacilli for vitamin B12 production potential: process validation by micro-assay and UFLC. Appl. Microbiol. Biotechnol. 100 6791–6803. 10.1007/s00253-016-7639-9 [DOI] [PubMed] [Google Scholar]
- Chan C. H., Newmister S. A., Keenan T., Klaas K. R., Rayment I., Escalante-Semerena J. C. (2015). Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica. Biochim. Biophys. Acta 1 464–475. 10.1016/j.bbagen.2013.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M., Bottacini F., Fosso B., Kelleher P., Calasso M., Di Cagno R., et al. (2014). Lactobacillus rossiae, a vitamin B12 producer, represents a metabolically versatile species within the Genus Lactobacillus. PLoS One 9:e107232. 10.1371/journal.pone.0107232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Bobik T. A. (2008). The PduX enzyme of Salmonella enterica is an L-threonine kinase used for coenzyme B12 synthesis. J. Biol. Chem. 283 11322–11329. 10.1074/jbc.M800287200 [DOI] [PubMed] [Google Scholar]
- Gray M. J., Escalante-Semerena J. C. (2010). A new pathway for the synthesis of alpha-ribazole-phosphate in Listeria innocua. Mol. Microbiol. 77 1429–1438. 10.1111/j.1365-2958.2010.07294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A. B., Han A. W., Mehta A. P., Mok K. C., Osadchiy V., Begley T. P., et al. (2015). Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc. Natl. Acad. Sci. U.S.A. 112 10792–10797. 10.1073/pnas.1509132112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra A. B., Tran J. L., Crofts T. S., Taga M. E. (2013). Analysis of substrate specificity in CobT homologs reveals widespread preference for DMB, the lower axial ligand of vitamin B12. Chem. Biol. 20 1275–1285. 10.1016/j.chembiol.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Johnson M. G., Escalante-Semerena J. C. (1992). Identification of 5,6-dimethylbenzimidazole as the Co alpha ligand of the cobamide synthesized by Salmonella Typhimurium. Nutritional characterization of mutants defective in biosynthesis of the imidazole ring. J. Biol. Chem. 267 13302–13305. [PubMed] [Google Scholar]
- Kang Z., Zhang J., Zhou J., Qi Q., Du G., Chen J. (2012). Recent advances in microbial production of delta-aminolevulinic acid and vitamin B12. Biotechnol. Adv. 30 1533–1542. 10.1016/j.biotechadv.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Martens J. H., Barg H., Warren M. J., Jahn D. (2002). Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58 275–285. 10.1007/s00253-001-0902-7 [DOI] [PubMed] [Google Scholar]
- Mattes T. A., Escalante-Semerena J. C. (2017). Salmonella enterica synthesizes 5,6-dimethylbenzimidazolyl-(DMB)-alpha-riboside. Why some Firmicutes do not require the canonical DMB activation system to synthesize adenosylcobalamin. Mol. Microbiol. 103 269–281. 10.1111/mmi.13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed Y., Lee B., Kang Z., Du G. (2014). Development of a two-step cultivation strategy for the production of vitamin B12 by Bacillus megaterium. Microb. Cell Fact. 13:102. 10.1186/s12934-014-0102-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina V., Médici M., Taranto M. P., Font de Valdez G. (2008). Effects of maternal vitamin B12 deficiency from end of gestation to weaning on the growth and haematological and immunological parameters in mouse dams and offspring. Arch. Anim. Nutr. 62 162–168. 10.1080/17450390801892567 [DOI] [PubMed] [Google Scholar]
- Molina V. C., Médici M., Taranto M. P., Font de Valdez G. (2009). Lactobacillus reuteri CRL 1098 prevents side effects produced by a nutritional vitamin B deficiency. J. Appl. Microbiol. 106 467–473. 10.1111/j.1365-2672.2008.04014.x [DOI] [PubMed] [Google Scholar]
- Nahvi A., Barrick J. E., Breaker R. R. (2004). Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32 143–150. 10.1093/nar/gkh167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaski J. T., Holmstrom E. D., Nesbitt D. J., Batey R. T. (2016). Mechanistic insights into cofactor-dependent coupling of RNA folding and mRNA Transcription/Translation by a Cobalamin riboswitch. Cell Rep. 15 1100–1110. 10.1016/j.celrep.2016.03.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin C., Blanche F., Cauchois L., Cameron B., Couder M., Crouzet J. (1991). Primary structure, expression in Escherichia coli, and properties of S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase from Bacillus megaterium. J. Bacteriol. 173 4893–4896. 10.1128/jb.173.15.4893-4896.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Lawrence J. G., Bobik T. A. (1996). Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50 137–181. 10.1146/annurev.micro.50.1.137 [DOI] [PubMed] [Google Scholar]
- Rucker R. B., Suttie J. W., McCormick D. B. (eds) (2001). Handbook of Vitamins, 3rd Edn. Abingdon: Taylor & Francis. [Google Scholar]
- Seidametova E. A., Shakirzianova M. R., Ruzieva D. M., Guliamova T. G. (2004). [Isolation of cobalt-resistant strains of propionic acid bacteria, potent producers of vitamin B12]. Prikl. Biokhim. Mikrobiol. 40 645–648. 10.1023/B:ABIM.0000046990.49021.07 [DOI] [PubMed] [Google Scholar]
- Taga M. E., Walker G. C. (2008). Pseudo-B12 joins the cofactor family. J. Bacteriol. 190 1157–1159. 10.1128/JB.01892-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranto M. P., Vera J. L., Hugenholtz J., De Valdez G. F., Sesma F. (2003). Lactobacillus reuteri CRL1098 produces cobalamin. J. Bacteriol. 185 5643–5647. 10.1128/JB.185.18.5643-5647.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. C., Vannini V., Bonacina J., Font G., Saavedra L., Taranto M. P. (2016). Cobalamin production by Lactobacillus coryniformis: biochemical identification of the synthetized corrinoid and genomic analysis of the biosynthetic cluster. BMC Microbiol. 16:240. 10.1186/s12866-016-0854-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang Z., Jiao Y., Liu S., Wang Y. (2015). Improved propionic acid and 5,6-dimethylbenzimidazole control strategy for vitamin B12 fermentation by Propionibacterium freudenreichii. J. Biotechnol. 193 123–129. 10.1016/j.jbiotec.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Warren M. J., Raux E., Schubert H. L., Escalante-Semerena J. C. (2002). The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19 390–412. 10.1039/b108967f [DOI] [PubMed] [Google Scholar]