Conventional kinesin is essential for Drosophila melanogaster posterior determination by localizing oskar mRNA at the oocyte posterior pole. In this study, we show that two essential kinesin functions, cargo transport and cytoplasmic streaming, together with a myosin V–mediated cortical anchorage ensure correct posterior determination.
Abstract
The posterior determination of the Drosophila melanogaster embryo is defined by the posterior localization of oskar (osk) mRNA in the oocyte. Defects of its localization result in a lack of germ cells and failure of abdomen specification. A microtubule motor kinesin-1 is essential for osk mRNA posterior localization. Because kinesin-1 is required for two essential functions in the oocyte—transport along microtubules and cytoplasmic streaming—it is unclear how individual kinesin-1 activities contribute to the posterior determination. We examined Staufen, an RNA-binding protein that is colocalized with osk mRNA, as a proxy of posterior determination, and we used mutants that either inhibit kinesin-driven transport along microtubules or cytoplasmic streaming. We demonstrated that late-stage streaming is partially redundant with early-stage transport along microtubules for Staufen posterior localization. Additionally, an actin motor, myosin V, is required for the Staufen anchoring to the actin cortex. We propose a model whereby initial kinesin-driven transport, subsequent kinesin-driven streaming, and myosin V–based cortical retention cooperate in posterior determination.
Introduction
Cell polarization and compartmentalization are the cornerstones of differentiation and development of multicellular organisms. The Drosophila melanogaster oocyte is a classical model of cell polarization. The anterior–posterior body axis is defined by bicoid (bcd) mRNA and oskar (osk) mRNA localization at anterior and posterior parts of the oocyte, respectively (Roth and Lynch, 2009). Mutations in osk lead to the failure of pole cell (germ cells in the future gonads) specification along with abdomen development and therefore embryonic lethality (Lehmann and Nüsslein-Volhard, 1986, 1991). A key component of posterior localization of osk mRNA is Staufen, an evolutionarily conserved RNA-binding protein that forms RNP particles with osk mRNA and colocalizes with it throughout most of the oogenesis (hereafter referred to as osk/Staufen; St Johnston et al., 1991; Palacios and St Johnston, 2002). In stau mutants, osk mRNA fails to localize correctly at the posterior pole of the oocyte (Ephrussi et al., 1991). Staufen contains five double-stranded RNA (dsRNA)-binding domains that are required for proper localization and translation of osk mRNA (Micklem et al., 2000).
Forward and reverse genetics have identified multiple factors involved in posterior determination in Drosophila (Newmark and Boswell, 1994; Erdélyi et al., 1995; Tetzlaff et al., 1996; Micklem et al., 1997; Hachet and Ephrussi, 2001; Mohr et al., 2001; van Eeden et al., 2001) including components required for microtubule-dependent transport during mid-oogenesis (Glotzer et al., 1997; Cha et al., 2002; Palacios and St Johnston, 2002; Serbus et al., 2005). It has been shown that a microtubule motor kinesin-1 (conventional kinesin) is required for posterior determination (Brendza et al., 2000; Palacios and St Johnston, 2002; Krauss et al., 2009). In the absence of kinesin-1 heavy chain (KHC), osk/Staufen completely loses its posterior localization. These data were interpreted as evidence that transport of osk/Staufen along microtubules by kinesin-1 drives its posterior localization. osk/Staufen is initially localized at the posterior pole at stage 9, before the onset of fast streaming. The stage 9 oocyte displays an anterior–posterior gradient of microtubules, with more microtubule minus ends anchored at the anterior and lateral cortex and much less at the posterior cortex (Theurkauf et al., 1992; Clark et al., 1997; Shulman et al., 2000; Nashchekin et al., 2016). An in vivo imaging study has shown that osk mRNA displays a biased movement toward the posterior pole on the polarized microtubule network (Zimyanin et al., 2008), and a recent modeling study indicated that a simple gradient of cortical microtubules could be sufficient to localize osk mRNA at the posterior pole of stage 9 oocytes (Khuc Trong et al., 2015). Therefore, it is very likely that kinesin-driven directed transport is the driving force for the initial localization of osk/Staufen.
However, during rapid oocyte growth and massive “dumping” of the cytoplasm from the nurse cells, the oocyte cytoskeleton reorganizes dramatically (Theurkauf et al., 1992; Dahlgaard et al., 2007), and the oocyte initiates fast microtubule-based and kinesin-driven cytoplasmic streaming that efficiently mixes the cytoplasmic contents in the oocytes (Gutzeit and Koppa, 1982; Glotzer et al., 1997; Palacios and St Johnston, 2002; Serbus et al., 2005; Quinlan, 2016). Within that period, newly synthesized mRNA and proteins including osk mRNA and Staufen continue to be deposited to the oocyte (St Johnston et al., 1991; Mische et al., 2007). Furthermore, the posterior localization of RNPs is dynamic, and RNPs can escape from the posterior cap (Sinsimer et al., 2013). Thus, mechanisms defining posterior localization have to be in place not only during initial localization but also during ooplasmic streaming (Sinsimer et al., 2011). It has been proposed that cytoplasmic flow contributes to the long-range transport of osk RNPs as anteriorly injected osk mRNA can be found at the posterior pole (Glotzer et al., 1997). Depolymerization of microtubules prevented the proper localization of the anterior-injected osk mRNA (Glotzer et al., 1997), demonstrating that this localization is microtubule dependent. Depolymerization of microtubules blocked ooplasmic streaming, resulting in a less compact posterior localization of the core components of germ plasm, nanos mRNA, and Vasa protein, implying that streaming translocates posterior determinants from the nurse cell dumping sites (Forrest and Gavis, 2003). However, depolymerization of microtubules also eliminates microtubule-dependent directed transport, which makes it impossible to determine the contribution of ooplasmic streaming to the posterior localization of these RNPs.
This “translocating by streaming” hypothesis was further supported by a more recent study showing that ectopic expression of a constitutively active form of Cappuccino (CapuΔN; Drosophila Formin) caused overstabilization of cytoplasmic actin mesh and delayed the onset of fast streaming, which in turn delayed localization of posterior determinants (Bor et al., 2015). Microtubules remained intact in these oocytes, but reorganization of actin filaments could potentially explain the delay as the osk RNPs were not localized as precisely as in the WT even before the fast streaming onset (Bor et al., 2015). Thus, it is still unclear whether the delayed posterior determination is due to defects in streaming or anchorage.
As ooplasmic streaming, like cargo transport along microtubules, requires kinesin-1 (Palacios and St Johnston, 2002; Serbus et al., 2005), it is difficult to determine to what extent these two kinesin-1 activities contribute to osk/Staufen posterior localization. Therefore, although the role of streaming in posterior determination was initially proposed two decades ago, it has not been fully tested (Quinlan, 2016).
In this study, we studied the contribution of ooplasmic streaming in delivery of posterior RNPs. Recently, we showed that kinesin-driven microtubule sliding is required for streaming and created mutant kinesins that are deficient in either directed transport or in the ability to generate microtubule sliding/streaming (Lu et al., 2016; Winding et al., 2016). In this study, we used these mutants to dissociate these functions of kinesin-1. Using Staufen as a proxy of posterior determinants, we showed that kinesin-driven ooplasmic streaming is essential for precise posterior localization. Remarkably, streaming alone without the contribution of directed transport can deliver Staufen to the correct site. Thus, streaming and directed transport are partially redundant, together providing a robust mechanism of posterior determination. Finally, we showed that the myosin V (MyoV) motor traps Staufen particles at the actin cortex and thus is essential for the posterior accumulation of Staufen. Together, three distinct functions of two motors generate precise posterior determination and ensure the correct body planning for future embryonic development.
Results
The use of SunTagged Staufen for live imaging of posterior determinants
To examine the behavior of Staufen particles in fast-streaming oocytes, we initially used a published functional construct of GFP-tagged Staufen (mat atub-GFP-Staufen; Schuldt et al., 1998; Palacios and St Johnston, 2002) to visualize Staufen in Drosophila oocytes. We found that in addition to a tight posterior crescent, a small amount of GFP-Staufen clusters circulated in streaming oocytes (Video 1). However, these clusters were dim, which made it difficult to analyze their movement. To improve Staufen signal, we tagged the C terminus of Staufen with a recently developed bright fluorescent probe, SunTag (Tanenbaum et al., 2014), generating Staufen-SunTag (Fig. 1 A). SunTag is a two-component labeling system that allows Staufen to recruit up to 24 copies of superfolder GFP (sfGFP), making Staufen particles suitable for long-term live imaging. Furthermore, the SunTag system minimizes the fluorescent background as the sfGFP component of the system contains the nuclear localization signal, so all unbound sfGFP is sequestered in the nucleus (Tanenbaum et al., 2014). We generated a transgenic line carrying UASp-Staufen-SunTag and expressed Staufen-SunTag by a germline-specific Gal4, nos-Gal4[VP16] (Van Doren et al., 1998). We first tested whether Staufen-SunTag is functional by putting the construct into a stau loss-of-function mutant background (stauD3/staury9; St Johnston et al., 1991). We found that Staufen-SunTag formed a tight crescent in the absence of endogenous Staufen protein (Fig. S1, A–A′′) and could fully rescue the maternal defects of stau loss-of-function mutant including posterior Vasa localization (Fig. S1, A–D; St Johnston et al., 1991; Breitwieser et al., 1996) and female fertility (Table 1; Lehmann and Nüsslein-Volhard, 1991; St Johnston et al., 1991).
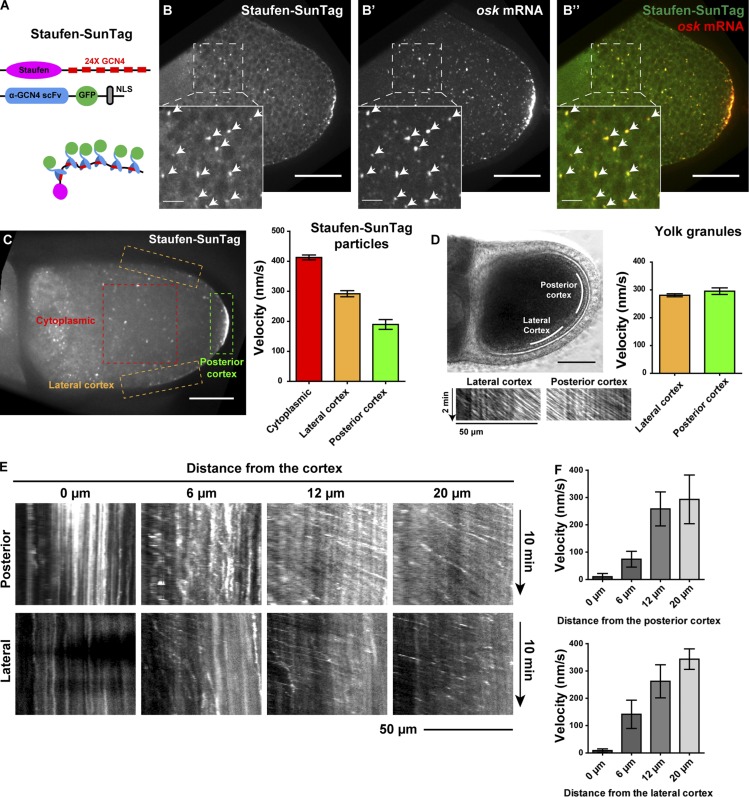
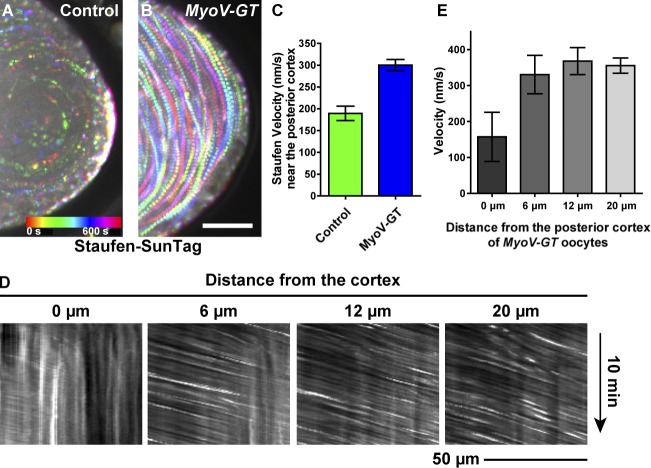
Figure 1.
The movement of Staufen-SunTag in streaming oocytes. (A) A cartoon of SunTag labeling of Staufen. The Staufen-SunTag contains two components: (1) 24 copies of GCN4 epitope fused to the C terminus of Staufen and (2) an anti-GCN4 single-chain variable fragment (scFv) tagged with sfGFP and a nuclear localization signal (NLS). SunTagging attaches 24 copies of GFP to the Staufen C-terminal tail; free GFP-fused antibody is sequestered in the nucleus. (B–B’’) Staufen-SunTag labeling particles all contains osk mRNA. Staufen-SunTag and osk mRNA were colocalized both at the posterior crescent and in individual cytoplasmic particles (arrowheads). (C) Average velocities (mean ± 95% confidence intervals) of Staufen-SunTag particles in the cytoplasmic area, lateral cortex, and posterior cortex (outlined boxes). Unpaired t test with Welch's correction between the following groups: cytoplasmic area and lateral cortex, P < 0.0001; cytoplasmic area and posterior cortex, P < 0.0001; lateral cortex and posterior cortex, P < 0.0001. (D) Kymographs and velocities (mean ± 95% confidence intervals) of yolk granules at lateral cortex and posterior cortex of control oocytes. (E and F) Kymographs and average velocity (mean ± 95% confidence intervals) of Staufen-SunTag particles at different distances from the posterior cortex and the lateral cortex. Bars: 50 µm (main images); 10 µm (insets).
Table 1. Embryonic hatching rate from different maternal genetic backgrounds.
| Maternal genotype | Embryonic hatching rate | Maternal genotype |
|---|---|---|
| Control (w/+; nos-Gal4VP16/+) | 92.4% | n = 463 |
| KhcmutA/KhcmutA | 48.8% | n = 172 |
| nos>Klc-RNAi + Btz-RNAi | 19.1% | n = 591 |
| KhcmutA/KhcmutA; nos>Klc-RNAi + btz-RNAi | 0% | n = 388 |
| nos>Khc-RNAi | 0% | n = 458 |
| nos>Khc-RNAi + Unc104-KTail | 0.8% | n = 358 |
| stau[D3]/stau[ry9] | 0% | n = 205 |
| stau[D3]/stau[ry9]; nos>Staufen-SunTag | 82.2% | n = 118 |
Single inhibition of either streaming (by KhcmutA) or directed transport along microtubules (by Klc and btz double RNAi) maternally led to a reduced embryonic hatching rate. The inhibition of directed transport had a stronger effect on embryo hatching rates than the inhibition of streaming. Simultaneous inhibition of streaming and directed transport caused complete failure of embryogenesis comparable with knockdown of KHC (Khc-RNAi). Partial rescue of streaming by the expression of a chimeric motor (Unc104-KTail) in Khc knockdown background did not rescue the embryonic lethality. Lack of Staufen (stau[D3]/stau[ry9]) caused embryonic lethality, while ectopic expression of Staufen-SunTag maternally significantly suppressed the embryonic lethality resulting from the absence of endogenous Staufen.
To further validate the functionality of the Staufen-SunTag probe, we visualized osk mRNA by single-molecule FISH (smFISH) in the Staufen-SunTag oocytes. We found that all Staufen-SunTag particles contain osk mRNA (Fig. 1, B–B′′, arrowheads). We also stained the Staufen-SunTag oocytes with anti-Staufen antibody and confirmed that SunTag particles indeed contain Staufen protein (not just unbound sfGFP escaped from the nucleus; Fig. S1, E–G, arrowheads). The results of these experiments show that Staufen-SunTag is fully functional, faithfully reports behavior of the endogenous protein, and can be used as a proxy of posterior determinants that is convenient for live imaging and tracking.
Circulation and anchorage of Staufen particles in streaming oocytes
Live imaging of Staufen-SunTag particles during ooplasmic streaming showed that although most of them carried with the flow of the cytoplasm, particles close to the cortex slowed down. This slowdown was especially pronounced near the posterior pole. Some of the particles got trapped there and became associated with the posterior crescent (Videos 1 and 2). Quantitative analysis of Staufen-SunTag showed that the particles moved at the highest rate (>400 nm/s) in the cytoplasmic area, while particles near the posterior cortex had much lower velocities (<200 nm/s; Figs. 1 C and S1 H). The particles clearly decelerated in a close proximity to the cortex (within ∼10 µm) and became almost immobilized at the cortex (Fig. 1, E and F; and Fig. S1 I). The deceleration was more pronounced near the posterior cortex (Fig. 1, E and F). Interestingly, unlike Staufen, the yolk granules did not display any decrease in velocity at the posterior cortex as compared with the lateral cortex (Fig. 1 D), indicating that streaming itself did not slow down in the posterior pole vicinity. Thus, posterior pole deceleration is specific to Staufen along with probably other posterior determinants. Together, our data support the hypothesis that flow circulation delivers Staufen to the posterior cortex for anchorage.
Inhibition of microtubule sliding suppresses Staufen circulation and causes localization defects
If circulation of Staufen by streaming contributes to its posterior localization, we would expect to see that streaming inhibition results in posterior localization defects. Recently, we have demonstrated that kinesin-driven microtubule sliding is required for ooplasmic streaming, and we created a Khc mutant, KhcmutA, that is defective in sliding but not in microtubule-dependent transport (Lu et al., 2016; Winding et al., 2016). KhcmutA replaces four conserved residues in the ATP-independent microtubule binding site at the KHC C-terminal tail, reducing the ability of kinesin to slide microtubules (Winding et al., 2016). As a result, KhcmutA flies have dramatically reduced ooplasmic streaming (Lu et al., 2016), allowing us to separate the roles of kinesin-driven streaming and transport.
We first wanted to determine whether Staufen transport along microtubules is affected by KhcmutA. Initially, we examined Staufen transport in Drosophila S2 cells. Treatment of S2 cells with low concentrations of cytochalasin D (CytoD) results in formation of processes filled by uniformly polarized microtubules (Ling et al., 2004; Kim et al., 2007; Ally et al., 2009; Barlan et al., 2013; Lu et al., 2013a). GFP-Staufen expressed in Drosophila S2 cells (Fig. S2 A) formed particles that moved bidirectionally in the processes (Video 3). A pulldown assay showed that GFP-Staufen interacts in cells with endogenous Staufen as well as KHC (Fig. S2, B–D). Most importantly, these GFP-Staufen complexes contained osk mRNA (Fig. S2 E), consistent with the data showing that Staufen is a component of osk RNPs (Veeranan-Karmegam et al., 2016; Gáspár et al., 2017). Therefore, we concluded that GFP-Staufen forms RNP particles in S2 cells. GFP-Staufen particles robustly moved in processes of S2 cells (Figs. S2 F and S3, A and B); knockdown of KHC by dsRNA against Khc 3′-UTR abolished GFP-Staufen movement (Figs. S2 F and S3 B), consistent with previous studies on the role of kinesin-1 in Staufen transport (Palacios and St Johnston, 2002; Serbus et al., 2005). We then transfected the S2 cells either with the construct containing just the coding region of WT Khc (KhcWT) or KhcmutA while suppressing the endogenous KHC expression with Khc 3′-UTR dsRNA. We found that both KhcWT and KHCmutA efficiently rescued movement of GFP-Staufen (Fig. S3 B), demonstrating that KHCmutA can efficiently transport Staufen particles in S2 cells.
Recently, nonmuscle Tropomyosin 1 (Tm1) has been shown as an adaptor recruiting kinesin-1 to the osk RNPs (Veeranan-Karmegam et al., 2016; Gáspár et al., 2017). Tm1C binds to the C terminus of KHC (residues 824–975; Veeranan-Karmegam et al., 2016), containing the ATP-independent microtubule binding site where four conserved residues were mutated in KhcmutA (Lu et al., 2016; Winding et al., 2016). Therefore, it raised the question of whether Tm1C can efficiently recruit KHCmutA to osk/Staufen. We examined the interaction between Tm1C and KHCWT or KHCmutA tail in a pulldown experiment and found that Tm1C interacts with both proteins at a comparable level, suggesting that KHCmutA can be efficiently recruited to Staufen particles through Tm1C (Fig. S3 C).
Next, we examined the Staufen-SunTag movement in the cytoplasm of stage 9 oocytes before the onset of fast uniform streaming. Stage 9 oocytes display slow uncoordinated streaming (Gutzeit and Koppa, 1982; Palacios and St Johnston, 2002; Serbus et al., 2005; Dahlgaard et al., 2007; Quinlan, 2016; Drechsler et al., 2017). We quantified the motility events in stage 9 oocytes using two different cutoff values. Quantification of all motility events demonstrated that Staufen-SunTag velocities are comparable between control and KhcmutA (Fig. 2, A–D), but this does not allow us to distinguish between the effect of slow streaming (that can carry Staufen particles) and rapid transport along microtubules. We therefore next quantified only the fast-moving (>0.1 µm/s) particles as the slow stage 9 streaming does not exceed this rate (Zimyanin et al., 2008). We found that (1) the percentages of fast-moving Staufen particles in control and KhcmutA are similar (15.2% and 12.8%, respectively) and (2) that the velocities of fast-moving Staufen particles in KhcmutA are comparable with the control (Fig. 2 D, inset). Together, we conclude that as in S2 cells, KhcmutA efficiently transports Staufen particles along microtubules in the oocytes.
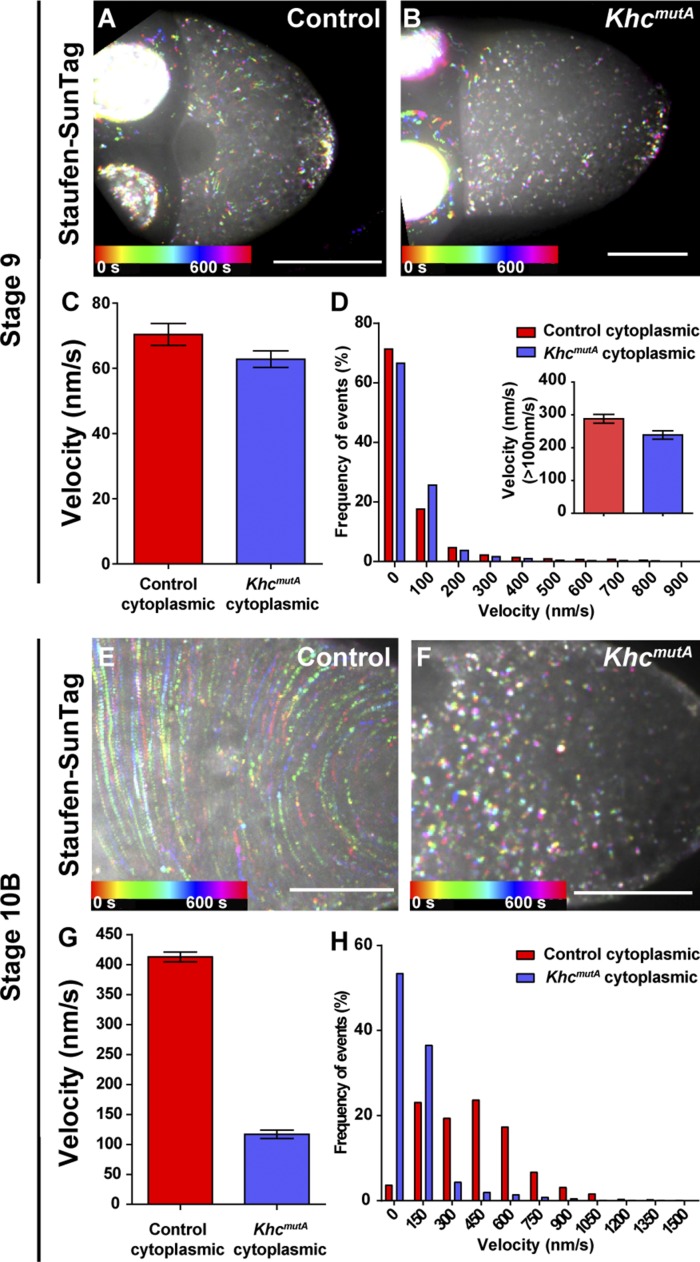
Figure 2.
Effects of KhcmutA on Staufen movement. (A–D) Replacement of endogenous Khc with KhcmutA does not affect overall Staufen movement in stage 9 oocytes. (A and B) Temporal color-coded images showing Staufen-SunTag particle movement in cytoplasmic areas of a stage 9 control oocyte (A) and a stage 9 KhcmutA oocyte (B). (C and D) Quantification of Staufen-SunTag movement in cytoplasm of stage 9 oocytes. (C) Average velocities (mean ± 95% confidence interval) of Staufen-SunTag in cytoplasm of control oocytes and of KhcmutA oocytes. (D) Histograms of frequency distribution (percentages) of Staufen-SunTag velocity in cytoplasm of control and of KhcmutA oocytes. The inset shows the average velocity of fast-moving Staufen-SunTag particles (>0.1 µm/s) in cytoplasm of control and of KhcmutA oocytes. (E–H) Replacement of endogenous Khc with KhcmutA leads to slower movement of Staufen particles in cytoplasm of stage 10B oocytes. (E and F) Temporal color-coded images showing Staufen-SunTag particle movement in cytoplasmic areas of a stage 10B control oocyte (E) and a stage 10B KhcmutA oocyte (F). (G and H) Quantification of Staufen-SunTag movement in cytoplasm of stage 10B oocytes. (G) Average velocities (mean ± 95% confidence interval) of Staufen-SunTag in cytoplasm of control oocytes (the same set of data as Fig. 1 C) and of KhcmutA oocytes. (H) Histograms of frequency distribution (percentages) of Staufen-SunTag velocity in cytoplasm of control and of KhcmutA oocytes. Bars, 50 µm.
We then examined Staufen-SunTag movement in stage 10B KhcmutA oocytes. As we expected, the circulation speed of Staufen-SunTag was sharply reduced, especially in the cytoplasmic area (Fig. 2, E and F). Tracking of Staufen-SunTag movement in the KhcmutA cytoplasmic area showed an approximately fourfold decrease in velocity (Fig. 2, G and H). Staufen staining showed more diffuse posterior localization in KhcmutA oocytes (Figs. 3 and S4 R), consistent with our previous study (Lu et al., 2016). Thus, circulation of Staufen particles with the ooplasm is necessary to maintain the posterior localization of Staufen in fast-growing oocytes.
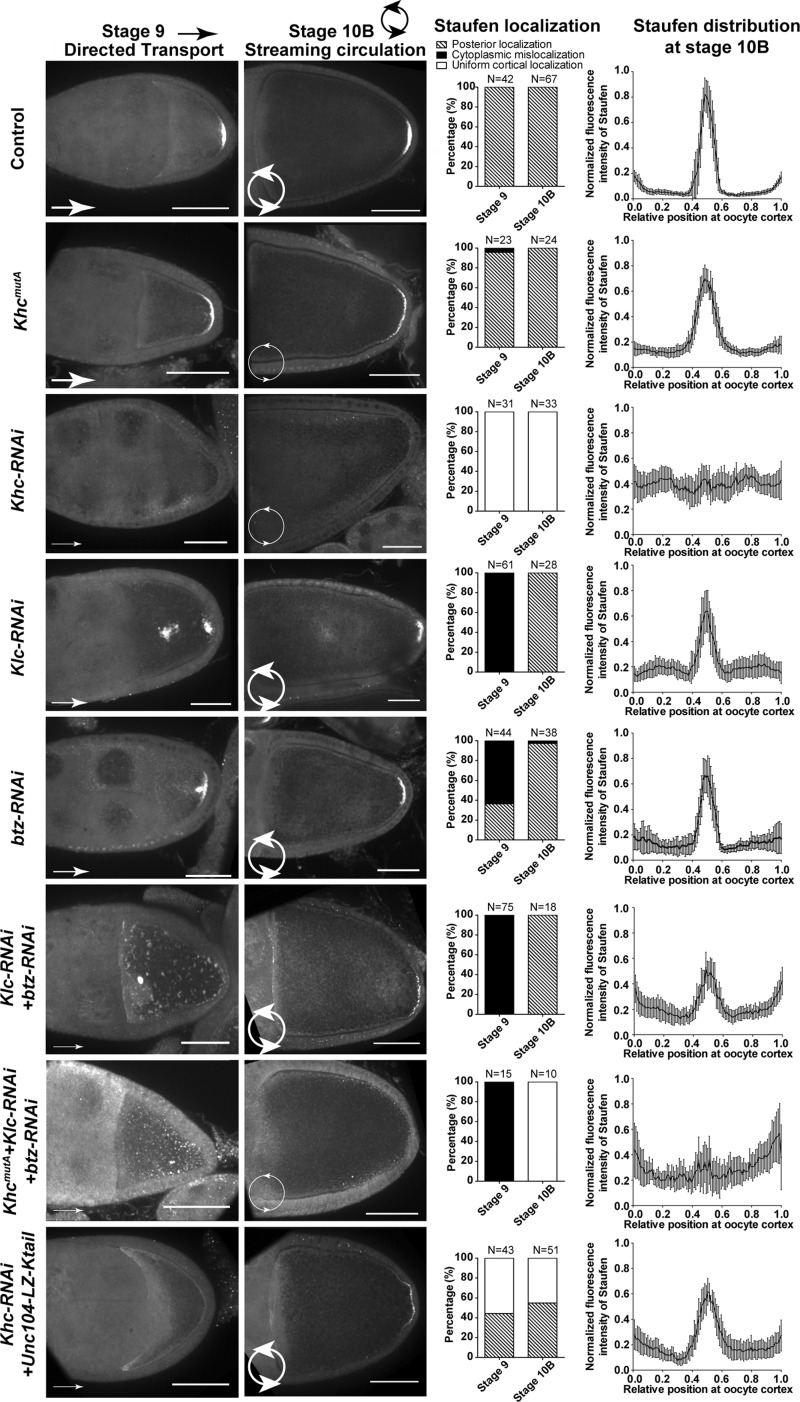
Figure 3.
Directed transport and streaming cooperate for robust Staufen posterior localization. Staufen staining and summary of Staufen localization patterns at stage 9 and stage 10B and Staufen distribution (normalized fluorescence intensity; mean ± 95% confidence interval) at stage 10B oocyte cortex were shown in following genotypes: control, KhcmutA, Khc-RNAi (nos>Khc-TRiP RNAi-GL00330), Klc-RNAi (nos>Klc-TRiP RNAi-GL00535), btz-RNAi (nos>btz- TRiP.RNAi-GLC01869), Klc-RNAi + btz-RNAi, KhcmutA +Klc-RNAi + btz-RNAi, and Khc-RNAi + Unc104-Ktail. To note: only the rescued Khc-RNAi +Unc104-Ktail stage 10B oocytes were analyzed in Staufen distribution on the oocyte cortex. Bars, 50 µm. Arrows and circular arrows stand for directed transport and streaming circulation, respectively. The widths of the arrow lines represent the level of transport or streaming.
Defects in Staufen transport are partially rescued by streaming
Having shown streaming requirement for the enrichment of Staufen at the posterior cortex, we next asked whether it is also sufficient for it. As kinesin-1 is required for both directed transport and streaming, knockdown of KHC completely abolished the Staufen localization at the posterior cortex (Figs. 3 and S4 R), in agreement with previously published data (Brendza et al., 2000; Palacios and St Johnston, 2002; Krauss et al., 2009). To test whether streaming is sufficient, we wanted to inhibit transport without affecting streaming. Thus, we searched for the proteins required for Staufen transport along microtubules but not for streaming. Initially, we used S2 cells for these experiments. Knockdown of kinesin-1 light chain (KLC), a typical adaptor for cargo transport, resulted in an ∼70% reduction in Staufen movement (Fig. S2 F), suggesting involvement of additional factor(s). Barentsz (Btz) is required for osk mRNA localization and has been proposed to link Staufen to kinesin-1 (van Eeden et al., 2001; Macchi et al., 2003; Wilhelm et al., 2003). Knockdown of Btz resulted in an ∼60% reduction in Staufen movement (Fig. S2 F). When both KLC and Btz were knocked down, Staufen movements dropped to a level comparable with KHC knockdown (Fig. S2 F), indicating that both KLC and Btz are required for KHC-driven Staufen transport.
As KLC knockdown only causes a slight reduction in streaming rate (∼20% reduction; Palacios and St Johnston, 2002; Ganguly et al., 2012) while Btz is not required for streaming (Palacios and St Johnston, 2002), we knocked down KLC or Btz to inhibit Staufen transport in the oocytes. Consistent with S2 data, we found partial mislocalization of Staufen in Klc and btz single knockdown at stage 9 (Figs. 3 and S4, A–I). Double knockdown of KLC and Btz caused complete Staufen mislocalization in stage 9 oocytes (Fig. 3), consistent with transport inhibition results in S2 cells (Fig. S2 F).
As anterior–posterior gradient of microtubule density is required for posterior localization of osk/Staufen (Zimyanin et al., 2008; Parton et al., 2011; Khuc Trong et al., 2015; Nashchekin et al., 2016), we tested whether the microtubule gradient is disrupted by Klc-RNAi and btz-RNAi. Microtubule distribution in stage 9 oocytes was examined by either tubulin antibody staining or by expression of a GFP-tagged minus end marker, Patronin (Goodwin and Vale, 2010; Khanal et al., 2016). Both techniques showed that knockdown of KLC, Btz, or both together did not affect overall anterior–posterior microtubule gradient (Fig. S4, J–Q). This suggests that the mislocalization of Staufen after knockdown of these kinesin adaptors is not caused by the defect of microtubule organization.
Next, we examined the effect of knockdown of these adaptors on Staufen localization in fast-streaming stage 10B oocytes. We found that in these RNAi knockdown oocytes, Staufen posterior localization was partially restored (Figs. 3 and S4, A–I and R), implying that a later-stage mechanism rescues the early localization defects. Together, it indicates that posterior localization of Staufen at stage 9 relies on transport along microtubules (Zimyanin et al., 2008; Parton et al., 2011; Khuc Trong et al., 2015; Nashchekin et al., 2016), but streaming can circulate mislocalized Staufen and partially rescue the posterior localization at stage 10B.
Furthermore, we tested the effect of simultaneous inhibition of streaming and transport by introducing KhcmutA into Klc and btz double RNAi. In this triple mutant, we found complete mislocalization of Staufen at stage 9 and no posterior accumulation at stage 10B (Figs. 3 and S4 R). To confirm that Staufen staining truly represents osk mRNA localization, we examined the osk mRNA by smFISH and found that its pattern is similar to Staufen staining in all tested genotypes (Fig. S4, S–Z). Together, the data demonstrated that kinesin-dependent transport and streaming are both important and partially redundant for posterior determination.
To independently verify the role of streaming in Staufen localization, kinesin-1 was replaced with a chimeric motor that has been shown to drive ooplasmic streaming in a Khc-RNAi background but is unable to move cargoes (Lu et al., 2016; Winding et al., 2016). This chimeric motor contains an Unc104 (Caenorhabditis elegans kinesin-3) motor domain dimerized by a leucine zipper followed by a short C-terminal fragment of KHC tail (Unc104-Ktail residues 905–975) that contains the ATP-independent microtubule-binding site. The chimera cannot move normal kinesin cargoes but is active in microtubule sliding (Winding et al., 2016). It partially rescues streaming in KHC-depleted oocytes (Lu et al., 2016).
As this chimeric motor carries a part of the Tm1 binding domain at the KHC-tail and Tm1 recruits kinesin-1 to osk/Staufen RNPs (Veeranan-Karmegam et al., 2016; Gáspár et al., 2017), we first tested whether the chimeric motor is able to directly transport Staufen. Unlike full-length KHC, Unc104-Ktail failed to rescue Staufen movement in S2 cells (Fig. S3 D). We further tested whether this part of the KHC tail (residues 905–975) used in the chimeric motor can interact with Tm1. Pulldown experiments showed that unlike the residues 345–975 fragment of KHC, this smaller fragment does not interact Tm1 (Fig. S3 E). Then, we expressed the Unc104-Ktail in Khc-RNAi oocytes and found that it partially rescued Staufen posterior localization (Figs. 3 and S4 R), indicating that streaming alone is sufficient to localize Staufen to the posterior pole. Intriguingly, we also found partial rescue of Staufen localization in a fraction of stage 9 oocytes (Fig. 3). We examined the Staufen-SunTag particle movement in stage 9 oocytes of Unc104-Ktail + Khc-RNAi oocytes. We found that instead of rescuing Staufen transport along microtubules, chimeric motor occasionally rescued slow streaming (Fig. S3, F–H). We speculate that constitutively active sliding-only chimeric motor can slide free microtubules and drive slow streaming in stage 9 oocytes. Because of the small size of stage 9 oocytes, slow streaming by the chimera can deliver Staufen to its posterior anchorage place. This is consistent with computational simulation showing that slow streaming contributes to the accumulation of osk mRNA (Ganguly et al., 2012).
Together, these data strongly indicate that transport and streaming are important contributors to posterior localization of osk/Staufen. These two processes are partially redundant to ensure the robust and error-free posterior determination.
Two Staufen localization processes are required for robust posterior determination
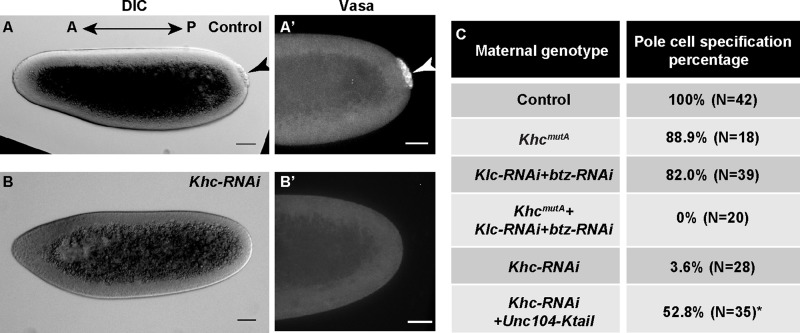
To understand the biological implications of imprecise Staufen localization, we examined a biological readout of posterior determination, pole cell specification (Lehmann and Nüsslein-Volhard, 1986, 1991; Ephrussi et al., 1991). In WT embryos, pole cells could be easily identified at the posterior end of blastoderm embryos as a cluster of cells excluded from the syncytial somatic layers (Fig. 4 A, black arrowhead) and by positive Vasa staining (Fig. 4 A′, white arrowhead; Hay et al., 1988, 1990; Lu et al., 2012). Depletion of KHC maternally resulted in >95% of embryos lacking pole cells (Fig. 4, B and C). Simultaneous inhibition of transport and streaming by Klc and btz double RNAi and KhcmutA led to the same result (Fig. 4 C). In contrast, inhibition of only one of the two KHC activities resulted in only ∼10–20% of embryos without pole cell specification (Fig. 4 C). These results further demonstrate that transport and streaming have partially overlapping functions in posterior determination. When streaming was (partially) recovered by ectopic expression of the Unc104-Ktail chimera in Khc-RNAi background, ∼50% of embryos that reached syncytial blastoderm stage displayed pole cell specification (compared with only 4% in Khc-RNAi; Fig. 4 C).
Figure 4.
Genetic redundancy between streaming and transport ensures the patterning for future embryonic development. (A and A’) Pole cell specification at the posterior side of a syncytial blastoderm embryo from a WT mother shown as a cluster of cells segregated from the somatic cells (A, arrowhead) and positive for anti-Vasa staining (A’, arrowhead). (B and B’) No pole cell specification in a syncytial blastoderm embryo from a Khc-RNAi mother. Bars, 50 µm. DIC, differential interference contrast. (C) Summary of pole cell specification percentages from the listed maternal genotypes. Asterisk marks a percentage that may not be an accurate estimate of pole cell specification percentage as some embryos showed apoptotic/necrotic blebbing and did not reach the syncytial blastoderm stage.
We further quantified the embryo hatching rate in all the genotypes mentioned above. Consistent with previously published results on the Khc protein-null allele (Brendza et al., 2000), maternal knockdown of KHC by RNAi resulted in 100% embryonic lethality (Table 1). Similar to Khc-RNAi, maternal inhibition of both functions of kinesin-1 led to the complete failure of hatching (Table 1). Maternal down-regulation of streaming by KhcmutA caused ∼50% embryonic lethality as we previously reported (Table 1; Winding et al., 2016). Maternal inhibition of KLC and Btz by RNAi caused a higher lethality than KhcmutA (∼80% embryonic lethality; Table 1), suggesting that these two adaptors have additional functions during embryogenesis. In agreement with this idea, 99.2% embryos from Khc-RNAi + chimeric Unc104-Ktail mothers failed to complete embryogenesis (Table 1) despite ∼50% of the oocytes having partial Staufen posterior localization and ∼50% embryos showing pole cell specification (Figs. 3, 4 C, and S4 R). Not surprisingly, kinesin-driven cargo transport is vital for overall embryogenesis, while streaming is more essential for early development (Fig. 4 C and Table 1).
MyoV functions as a local anchor of Staufen particles
Accumulation of Staufen at the posterior cortex involves not only transport and circulation but also anchoring in order to counteract diffusion and shear force from streaming. Loss of KHC or microtubule depolymerization results in uniform cortical localization of osk/Staufen; this uniform cortical localization is significantly reduced when F-actin is fragmented by CytoD (Cha et al., 2002). These data strongly suggest that osk/Staufen cortical localization is dependent on the actin cytoskeleton but is microtubule independent. Previous work demonstrated that MyoV (or didum), is involved in the Staufen cortical localization (Krauss et al., 2009).
Therefore, we directly tested the ability of MyoV to anchor Staufen to actin using S2 cells. S2 cells have a dense network of actin filaments at lamellipodia (Rogers et al., 2003; Lu et al., 2013a). The retrograde flow of actin in lamella pushes back the microtubules. Thus, we stopped the retrograde flow of actin with an Arp2/3 inhibitor, CK-666 (Nolen et al., 2009), which allowed microtubules to penetrate into the actin-rich lamella (Fig. S5 A). Movement of GFP-Staufen along microtubules was mostly suppressed under these conditions. Knockdown of MyoV resulted in a significant increase in Staufen motility (Fig. S5, B–D). Furthermore, we were able to detect MyoV on the GFP-Staufen particles in a BirA*-dependent proximity biotinylation (BioID) assay (Fig. S5 E; see the BioID pulldown assay section in Materials and methods for more details). These results strongly suggest that in S2 cells, MyoV is a component of Staufen particles and functions as an actin-dependent anchor. Intriguingly, we were not able to detect MyoV in a regular pulldown assay on GFP-Staufen particles, indicating that MyoV interaction with the Staufen particles is highly dynamic and transient.
To test the role of MyoV in oocytes, we used a MyoV dominant-negative mutant, MyoV-GT (Krauss et al., 2009). Ectopic expression of this mutant caused aggregation and mislocalization of Staufen in both stage 9 and stage 10B oocytes (Fig. S5, F–I), consistent with data published by Krauss et al. (2009). We then combined the MyoV mutant with the Staufen-SunTag probe and imaged the movements of Staufen particles. Remarkably, MyoV inhibition eliminated the deceleration of Staufen particles at the posterior cortex (Figs. 5 and S1 H and Video 4). This implies that MyoV functions as one of the components anchoring posterior determinants in the streaming oocytes (Fig. 6).
Figure 5.
MyoV functions to anchor Staufen at the posterior cortex of streaming oocytes. (A and B) Temporal color-coded images showing Staufen-SunTag movement near the posterior cortex in a control oocyte (A) and a MyoV-GT mutant oocyte (B). Bar, 25 µm. (C) Quantification of Staufen-SunTag velocities near the posterior cortex in MyoV-GT mutant and control oocytes (the same set of data as Fig. 1 C). Unpaired t test with Welch's correction between control posterior cortex and MyoV-GT posterior cortex. P < 0.0001. (D) Kymographs of Staufen-SunTag particles at different distances from the posterior cortex of the MyoV-GT mutant oocyte (B). (E) Average velocities (mean ± 95% confidence intervals) of Staufen-SunTag particles at different distances from the posterior cortex of the MyoV-GT oocyte.
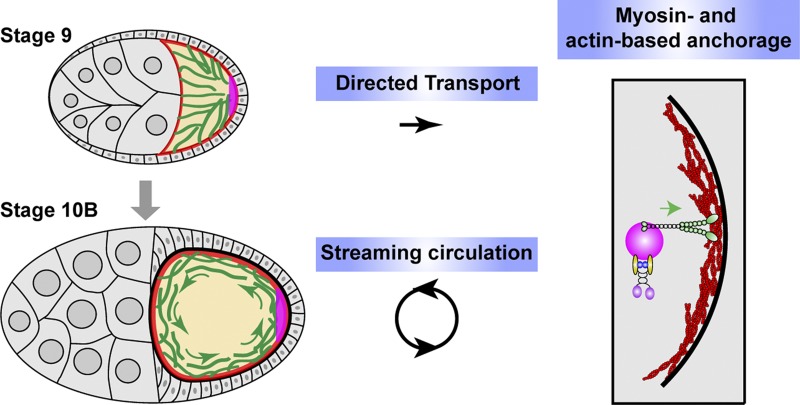
Figure 6.
Two cytoskeletal systems, two motor proteins, and three processes contribute to posterior determination of Drosophila oocytes. Two kinesin-dependent processes, directed transport and streaming circulation, deliver osk/Staufen RNPs from the anterior side of the oocyte to the posterior pole at stage 9 and stage 10B, respectively. At the posterior cortex, MyoV-based anchorage on actin cortex anchors and accumulates osk/Staufen particles locally.
Discussion
Generation and maintenance of cell polarity is one of the fundamental processes during development. In Drosophila oocytes, posterior determination is achieved by posterior localization of osk mRNA, which is colocalized with the RNA-binding protein Staufen. Posterior localization of osk/Staufen is initiated in mid-oogenesis and maintained in late oogenesis. osk/Staufen localization requires kinesin-1 (Brendza et al., 2000; Cha et al., 2002). Kinesin-1 has two essential roles in development: transport of cargoes along microtubules and sliding cytoplasmic microtubules against each other (Jolly et al., 2010; Lu et al., 2013b, 2015; del Castillo et al., 2015; Lu and Gelfand, 2017). Because microtubule sliding by kinesin-1 had not been previously known, kinesin-dependent transport was considered by default as the mechanism for the posterior determination. Recently, we generated kinesin mutants that allowed us to dissociate these two kinesin functions (Winding et al., 2016), and we demonstrated that microtubule sliding is essential for ooplasmic streaming (Lu et al., 2016).
In this study, we used these mutants to determine the contributions of streaming and transport to posterior determination. Although the role of streaming in osk/Staufen localization has been proposed, our data demonstrate for the first time that it indeed contributes significantly to osk/Staufen localization. Furthermore, we showed that MyoV functions as a cortical anchor for posterior determinants. Thus, osk/Staufen posterior localization requires three processes: transport by kinesin-1 along microtubules, circulation by streaming, and retention by MyoV in the actin-rich posterior cortex (Fig. 6). Spatial and temporal coordination of these activities ensures correct posterior determination.
Directed transport on microtubules by kinesin-1
Multiple factors required for localization of the osk/Staufen have been identified in Drosophila oocytes including kinesin-1. In the absence of KHC, osk/Staufen becomes uniformly localized throughout the oocyte cortex (Brendza et al., 2000; Cha et al., 2002). As kinesin-1 motor domain fused to β-galactosidase (Kin-βGal) is accumulated at the posterior pole (Clark et al., 1994; Brendza et al., 2000), it was first suggested that microtubules are highly polarized along the anterior–posterior axis with plus ends pointing toward the posterior pole, and kinesin-1 transports osk/Staufen RNPs from anterior to posterior side along these polarized microtubules (Brendza et al., 2000; Palacios and St Johnston, 2002). However, in vivo live imaging showed that the movement of osk/Staufen RNPs in the oocyte was less organized and displayed only a slight posterior bias (Zimyanin et al., 2008; Parton et al., 2011). This bias is probably due to the asymmetry of cortical microtubules in the oocyte, with more cortical microtubules attached by their minus ends to the anterior and lateral than to the posterior cortex (Shulman et al., 2000; Parton et al., 2011; Nashchekin et al., 2016). Computational simulation suggested that this bias is sufficient for posterior accumulation of osk/Staufen (Khuc Trong et al., 2015). Recently, it has been shown that localized dynactin protects growing microtubules from catastrophe and allows them to extend further into the posterior region, thus delivering osk/Staufen to posterior pole (Nieuwburg et al., 2017).
An important question is what adaptor(s) recruit kinesin-1 to osk/Staufen. A general adaptor for kinesin-driven cargo transport is KLC (Karcher et al., 2002; Dodding et al., 2011). KLC was first considered to be dispensable for Staufen localization in oocytes. However, in Klc8ex94 germline clones used in this study, Staufen staining appeared less compact and occasionally was found in a dot near the posterior pole (Palacios and St Johnston, 2002). Later, it was reported that Staufen was mislocalized in ∼20% of klc8ex94 stage 9 oocytes (Loiseau et al., 2010). Staufen mislocalization was more pronounced in double mutant of Klc and a KLC-like protein, pat1 (Loiseau et al., 2010), suggesting that KLC is involved in osk/Staufen transport. In this study, we knocked down KLC by three RNAi lines that target either its coding region or 3′UTR region and found that all three RNAi resulted in Staufen partial mislocalization in the cytoplasm of the stage 9 oocytes (Figs. 3 and S4, A–F). The discrepancy between the Klc null and Klc-RNAi phenotypes can be explained by stability of its mRNA. Because we used RNAi to knock down KLC (including the existing Klc mRNA), it had an earlier and stronger effect on Staufen localization than the Klc8ex94 germline clones that still contained residual Klc mRNA transcribed before the clone induction. Therefore, we conclude that KLC contributes to kinesin-driven Staufen transport, although to a lesser extent than typical kinesin cargoes.
Another important component of osk/Staufen transport is Btz. Btz was first identified in a mutant screen for defects in osk/Staufen localization in oocytes (van Eeden et al., 2001; Ghosh et al., 2012). Interestingly, Btz is only transiently localized at the posterior pole at stage 9 but disappears from the site at stage 10B (van Eeden et al., 2001), implying that this Btz is only required for the early accumulation. In btz null mutant (btz2), osk mRNA posterior localization is completely disrupted; but the embryos laid by btz2 mutant females can still have an abdomen, which is much milder than osk and stau mutants (van Eeden et al., 2001). This implies that another mechanism partially restores osk mRNA (or Osk protein) at the posterior pole at later stages, in a good agreement with our data showing that streaming can partially rescue osk/Staufen localization.
Two recent studies showed that nonmuscle Tm1 could serve as an adaptor recruiting kinesin-1 to osk/Staufen. osk mRNA fails to localize at the posterior pole in Tm1 mutants (Erdélyi et al., 1995; Tetzlaff et al., 1996). Veeranan-Karmegam et al. (2016) demonstrated that Tm1C, a specific isoform of Tm1, binds to KHC tail and functions to localize osk mRNA. They proposed that Tm1C links kinesin-1 to osk/Staufen, facilitating its posterior transport. Gáspár et al. (2017) showed that Tm1I/C directly binds to osk mRNA and is required for recruiting kinesin-1 upon the nuclear export in nurse cells.
Interestingly, Tm1C binds to the C terminus of KHC (amino acids 824–975; Veeranan-Karmegam et al., 2016), the region containing the microtubule binding site required for ooplasmic streaming (Lu et al., 2016; Winding et al., 2016). Although our KhcmutA carries four amino acid changes in this region, it does not affect its interaction with Tm1C, as KhcmutA efficiently transports Staufen in both Drosophila S2 cells as well as in oocytes and KHCmutA can still bind Tm1C (Figs. 2 and S3).
Cytoplasmic streaming as a delivery mechanism
Cytoplasmic streaming circulates cytoplasmic contents in the rapidly growing oocytes (Gutzeit and Koppa, 1982). The Ephrussi and Gavis groups demonstrated that posterior determinants are translocated by streaming and enriched at the posterior cortex (Glotzer et al., 1997; Forrest and Gavis, 2003; Sinsimer et al., 2011, 2013; Little et al., 2015). Recently, mitochondria have been shown to be churned with ooplasmic streaming and are selectively accumulated at the posterior cortex in an actin-dependent manner (Hurd et al., 2016). In our study, we used streaming-deficient and streaming-only kinesin mutants to demonstrate that streaming contributes to osk/Staufen localization by delivering them to the posterior cortex for anchorage by MyoV (Figs. 1, 3, and 5). Therefore, streaming functions as a critical step to position the cytoplasmic components that are delivered from nurse cells to the late-stage oocytes.
MyoV as an anchor of osk/Staufen RNPs
To achieve the posterior accumulation of osk/Staufen RNPs, a local anchorage is required in addition to delivery. Depolymerization of microtubules inhibited long-range movement of anterior-injected osk RNA but did not prevent its cortical localization (Glotzer et al., 1997; Cha et al., 2002). Depolymerization of F-actin, in contrast, caused a partial dissociation of osk mRNA from the posterior cortex (Cha et al., 2002). Moreover, in the Khc mutant, osk/Staufen became uniformly cortical (Brendza et al., 2000; Cha et al., 2002). These data indicate that osk/Staufen anchorage is an actin-based mechanism. The actin motor MyoV has been implicated in the cortical anchorage of osk/Staufen (Krauss et al., 2009). Dominant-negative inhibition of MyoV results in delocalization of Staufen from the posterior pole (Fig. S5; Krauss et al., 2009), and inhibition of MyoV causes faster movement of Staufen particles in S2 cells (Fig. S5). More importantly, we found that in WT oocytes, Staufen particles significantly slowed down when they got close to the cortex (Fig. 1), while in MyoV mutant oocytes, this deceleration was abolished (Fig. 5). Together, it strongly supports the idea that MyoV functions as an actin-based anchor of Staufen (Fig. 6).
The function of MyoV as a cortical anchor is evolutionarily conserved. For example, MyoV functions in anchoring melanosomes (Wu et al., 1998), peroxisomes (Kapitein et al., 2013), and mitochondria (Pathak et al., 2010), and it tethers organelles at F-actin–enriched areas (Rodionov et al., 1998; Rogers and Gelfand, 1998). A clear advantage of using a motor protein rather than a regular cross-linker for this anchorage process is that reversible binding and release characteristic for a motor protein allows for refinement of localization and removal of mislocalized components.
In addition to MyoV, Long Osk protein is essential for Staufen anchorage in late-stage fast-streaming oocytes but not for initial posterior localization (Vanzo and Ephrussi, 2002). Osk protein is translated when the osk mRNA reaches the posterior cortex. Osk protein interacts with Staufen (Breitwieser et al., 1996) and with actin filaments (Hurd et al., 2016), enhancing the initial posterior anchoring by MyoV. Thus, Osk protein anchorage forms a positive feedback loop: (1) osk mRNA at the posterior pole translates to Osk protein, and then (2) Osk protein binds to actin filaments and interacts with Staufen, bringing more osk/Staufen RNPs to the posterior pole. Furthermore, Long Osk protein induces endocytosis for cortical F-actin remodeling and forms long F-actin projections through actin regulators, which facilitates the anchorage of pole plasm components for pole plasm assembly (Tanaka et al., 2011; Tanaka and Nakamura, 2011). However, this positive feedback loop requires an initial accumulation of osk/Staufen RNPs. We propose that the kinesin-driven transport and myosin-mediated anchorage provides the initial accumulation of osk/Staufen RNPs at posterior pole of the stage 9 oocytes. This is consistent with the data that MyoV is required for correct Staufen localization initially (Fig. S5; Krauss et al., 2009), while Osk protein becomes essential at the later fast-streaming stages (Vanzo and Ephrussi, 2002). Together, MyoV and Osk protein ensure precise anchorage in the oocytes and thus correct posterior determination for the future embryos.
Oocytes are the biggest cells in animals, and correct patterning of oocytes/early embryos is essential for development. Even after the initial polarization, new osk/Staufen is being delivered from interconnected nurse cells to the rapidly growing oocyte. It has to travel >100 µm to get to the posterior pole and then get deposited there in an error-free manner. Our work revealed that two functionally overlapping kinesin-driven processes are required to achieve perfect posterior determination and ensure the robust patterning of future embryos. Furthermore, our data support an evolutionarily conserved myosin-dependent local anchorage at the F-actin–rich cortex. Together, we propose a multistep model for generation of robust polarity in oocytes (Fig. 6).
Materials and methods
Plasmid constructs
EGFP and Staufen coding DNA sequence were amplified by PCR with KpnI/EcoRI sites and NotI/AgeI sites, respectively, and cloned into pAc5.1/V5-HisA vector (Invitrogen) to generate pAC-EGFP-Staufen construct. Staufen coding DNA sequence and 24× GCN4 (SunTagV1; Tanenbaum et al., 2014; Lu et al., 2016) were amplified by PCR with SpeI/NotI sites and NotI/XbaI sites, respectively, and cloned into pUASp vector (Rørth, 1998) to generate pUASp-Staufen-SunTagV1 construct for making transgenic flies. Tm1C coding region was amplified by PCR with KpnI/BamHI sites from the pAGW-Tm1C (GFP-Tm1) from Veeranan-Karmegam et al. (2016) and was inserted into pEGFP-C1 vector (Takara Bio Inc.). TagRFP-tagged KhcWT-tail (residues 349–975 with 910–975 RKRYQ), KhcmutA-tail (residues 349–975 with 910–975 AAAYA), and leucine-zipper (VKQLEDKVEELASKNYHLENEVARLKKLV)-Ktail (residues 905–975; Winding et al., 2016) were subcloned into pcDNA3.1+ by KpnI/XbaI sites.
Drosophila genetics
Fly stocks and crosses were kept on standard cornmeal food supplemented with dry active yeast at room temperature (∼24–25°C). The following fly stocks were used in this study: UASp-scFv (second-chromosome and third-chromosome insertions; Lu et al., 2016), UASp-Staufen-SunTagV1 (second-chromosome and third-chromosome insertions; generated in this study), nos-Gal4-VP16 (III; Van Doren et al., 1998; Lu et al., 2012), mat αtub-Gal4-VP16[V37] (7063; Bloomington Drosophila Stock Center), KhcmutA (II; Lu et al., 2016; Winding et al., 2016), Khc-TRiP RNAi-GL00330 (attP2; III; 35409; Bloomington Drosophila Stock Center), Klc-TRiP RNAi-GL00535 (attP40; II; 36795; Bloomington Drosophila Stock Center), Klc-TRiP RNAi-HMS00883 (attP2; III; 33934; Bloomington Drosophila Stock Center), Klc-TRiP RNAi- HMS02429 (attP40; II; 42597; Bloomington Drosophila Stock Center), btz-TRiP RNAi-GLC01869 (attP2; III; 61165; Bloomington Drosophila Stock Center), btz-TRiP RNAi-HMS04337 (attP40; II; 58353; Bloomington Drosophila Stock Center); UASp-MyoV-GT (III; from A. Ephrussi; Krauss et al., 2009); stauD3 (St Johnston et al., 1991), mat atub-GFP-Staufen (from D. St Johnson; Schuldt et al., 1998; Palacios and St Johnston, 2002), staury9 (10742; Bloomington Drosophila Stock Center; St Johnston et al., 1991), and UASp-GFP-Patronin (II; a generous gift from U. Abdu, Ben Gurion University of the Negev, Beer-Sheva, Israel). Standard double balancing and homologous recombination were used to combine UASp-Staufen-SunTagV1 and UASp-scFv on second and third chromosomes (named as UASp-Staufen-SunTag) and to combine KhcmutA and Klc-TRiP RNAi-GL00535 on second chromosomes.
Live imaging and analysis of Staufen-SunTag movement in streaming oocytes
Young mated female adults expressing nos>Staufen-SunTag in various genetic backgrounds were fattened on dry active yeast for ∼24 h and dissected in Halocarbon oil 700 (Sigma-Aldrich) as previously described (Pokrywka, 2013; Lu et al., 2016). Stage 10B–11 egg chambers were imaged within 1 h after dissection on a Nikon Eclipse U2000 inverted stand with a 20× 0.75 NA lens and a Yokogawa CSU10 spinning-disk confocal head using an Evolve electron-multiplying charge-coupled device (EMCCD; Photometrics) controlled by Nikon Elements software. Images were acquired every 5 s for 10–20 min.
Staufen-SunTag tracking
Staufen-SunTag samples were analyzed in ImageJ (National Institutes of Health) and tracked by DiaTrack 3.04 (Lu et al., 2013a; Vallotton and Olivier, 2013). The near cortex area was defined as a <20-µm distance from the cortex (where the fluorescent signal ends as the Staufen-SunTag was only expressed in the oocyte). The posterior cortex was defined as the ∼70-µm posteriormost cortical region, while lateral cortex was defined as the ∼70–80-µm cortical region flanking the posterior cortex zone. The cytoplasmic region was defined in the middle of the oocyte, >20 µm from the cortex. The maximum velocity of Staufen particles was set as 1.5 µm/s, and the minimal lifetime of Staufen particles was set as 15 s. For kymographs and manual tracking, a line of 70 µm (∼2.7 µm width) was drawn along the cortex and positioned at different distances from the cortex. Kymographs were generated using an ImageJ plugin, Multi Kymograph (https://imagej.net/Multi_Kymograph). Velocity of Staufen particles at different distances from the cortex was measured in the kymographs (Fig. 1) or tracked by Nikon Elements (Fig. S1 I), respectively. Velocities were plotted, and statistical analysis was performed in Prism 5 (GraphPad Software).
Immunostaining of Drosophila oocytes
For Staufen and Vasa staining, a standard fixation and staining protocol was used (Lu et al., 2012, 2016): Ovaries were dissected from young mated females fed with dry active yeast for 18–24 h (∼10–15 females + 5 males in a vial) in 1× PBS and fixed in 4% EM-grade methanol-free formaldehyde diluted in PBT (1× PBS and 0.1% Triton X-100), washed with PBTB (1× PBS, 0.1% Triton X-100, and 0.2% BSA) five times for 10 min each, and blocked in 5% (vol/vol) normal goat serum–containing PBTB for 1 h. Samples were then incubated with primary antibody at 4°C overnight, washed with PBTB five times for 10 min each time, incubated with secondary antibody at room temperature (∼24–25°C) for 4 h, and washed with PBTB five times for 10 min each before mounting. The following primary and secondary antibodies were used in this study: mouse anti-Staufen (1:50; a gift from C.Q. Doe, University of Oregon, Eugene, OR), rat anti-Vasa (1:500; a gift from E.L. Ferguson, the University of Chicago, Chicago, IL; Lu et al., 2012), goat anti-mouse FITC and TRITC, and donkey anti-rat TRITC (1:100; Jackson ImmunoResearch Laboratories, Inc.). Samples were imaged on a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning-disk confocal head and a 40× 1.30 NA lens using an Evolve EMCCD controlled by Nikon Elements software. Images were acquired every 0.5 µm/step in z stacks.
Immunostaining of microtubules in Drosophila oocytes
The tubulin staining protocol of Drosophila oocytes was previously described (Legent et al., 2015). A mouse monoclonal antibody against α-tubulin (DM1α) and goat anti–mouse FITC secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) were used (both at 1:500). Images of tubulin staining were acquired on a Nikon A1-plus microscope at Northwestern University’s Nikon Imaging Center with an S Fluor 40× 1.30 NA oil lens using Galvano scanning and a DU4 Detector DU4 GaAsP CH2/3 controlled by Nikon Elements software. Single-frame images were then processed with Log function in ImageJ (ImageJ>Process>Math>Log) to avoid the overexposure of the bright signal at the follicle cells.
Quantification of Staufen staining in the oocytes
A 5-µm maximum intensity z projection was generated in each sample. The plot profile was either generated along a 3.6-µm-wide line delineating the oocyte cortex (starting and ending at the area where the oocyte meets the nurse cells; Figs. 3 and S4 R) or an 8-µm-wide line along the anterior–posterior axis (starting the boundary between nurse cells and oocytes and ending at the posteriormost follicle cells; Fig. S4 R). We normalized the distance in the plot profiles using a custom MatLab program. We also normalized fluorescence intensity of Staufen (maximum and minimum) in Fig. 3.
Quantification of GFP-Patronin in the oocytes
A 5-µm maximum-intensity z projection was generated in each sample. A line (4.4 µm wide) was drawn along the oocyte cortex (starting and ending at the area where the oocyte meets the nurse cells), and a plot profile of fluorescence intensity of GFP-Patronin was generated along this line at the cortex. We normalized fluorescence intensity (maximum and minimum) and the distance in the plot profiles using a custom MatLab program.
smFISH of Drosophila oocytes
smFISH was performed to visualize osk mRNA in certain genotypes (Little et al., 2013, 2015). The smFISH protocol was previously described (Veeranan-Karmegam et al., 2016). The Quasar570-conjugated Stellaris DNA oligonucleotide probes against osk mRNA (LGC Biosearch Technologies) was a generous gift from G. Gonsalvez (Augusta University, Augusta, GA) and used at 1:100 dilution.
Drosophila cell culture and stable cell line selection
Drosophila S2 cells were maintained in Insect-XPRESS protein-free insect cell medium with L-glutamine (Lonza). To select stable cell line expressing transgenes, S2 cells were cotransfected with pAC-EGFP-Staufen and pCoHygro (Invitrogen) in 20:1 molar ratio using Cellfectin (Invitrogen). Hygromycin was added to normal growth medium 48 h after transfection to a final concentration of 300 ng/µl. Expression of the transgene was confirmed by fluorescence microscopy and immunoblotting.
RNAi treatment
RNAi treatment for S2 cells was as described previously (Ling et al., 2004; Kim et al., 2007; Ally et al., 2009; Barlan et al., 2013; del Castillo et al., 2015; Winding et al., 2016). 106 cells in 35-mm dishes were incubated with 30 µg dsRNA for 3 d. Cells were split 1:3 on the third day and incubated with a fresh aliquot of 30 µg dsRNA. On day 5, cells were plated onto Concanavalin A–coated coverslips with 5 µM CytoD added to induce process outgrowth. Images were taken on the fifth day after the initial RNAi treatment.
GFP-Staufen image acquisition, tracking, and image analysis
Images of live cells expressing GFP-Staufen were acquired using an inverted microscope (Nikon Eclipse U2000) with a Plan Apochromat 60× 1.4 NA lens. A 100-W halogen light source was used for epifluorescence illumination to minimize phototoxicity. Images were captured using an Orca II-ER cooled charge-coupled device camera (Hamamatsu Photonics) run by MetaMorph software every 2 s for a total length of 2 min. The coordinates of the cell body and each particle were recorded, and the behavior of the particles was tracked using DiaTrack software. Particles that traveled away from the cell body were defined as moving in the anterograde direction, whereas particles that traveled toward the cell body were defined as moving in the retrograde direction. At least two to three independent experiments were recorded for each condition, and 10 cells from each experiment were randomly chosen for analysis.
Immunoprecipitation (IP) and RT-PCR
Approximately 1–1.5 × 108 S2 cells were used per IP assay. Cells were pelleted and washed twice with HL3 buffer (70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 5 mM Trehalose, 115 mM sucrose, and 5 mM Hepes, pH 7.2). Cell pellets were resuspended in homogenization buffer (50 mM Hepes, pH 7.4, 50 mM KCl, 2 mM MgCl2, 2 mM DTT, 1 mM PMSF, 50 mM ribonucleoside vanadyl complex [New England BioLabs, Inc.], 10 µg/ml each chymostatin, leupeptin, and pepstatin, and 40 U/ml RNasin [Promega]) and homogenized using a ball-bearing homogenizer. Post-mitochondrial supernatant was generated by centrifuging at 10,000 g for 5 min and precleared with rabbit IgG attached to protein A beads (Bio-Rad Laboratories) for 1 h at 4°C. GFP antibodies were prebound to protein A or protein A/G beads and were incubated with precleared extract for another 1 h at 4°C. The beads were washed four times with IPP-50 buffer (25 mM Tris, 50 mM KCl, 2 mM MgCl2, 1 mM DTT, and 0.01% Triton X-100) and either solubilized directly with SDS-PAGE sample buffer or with TriReagent (Molecular Research Center, Inc.) for RNA isolation. cDNA was synthesized using OligodT primers and Moloney murine leukemia virus (Ambion). Selective mRNA was then amplified by PCR using Taq polymerase.
Transfection of HEK293 cells and pulldown by GFP binder
Cells at 60–70% confluency have been transfected with four different combinations of DNAs (for Fig. S3 C): (1) pEGFP-C1 + pcDNA3.1+-TagRFP-KHCWT tail, (2) pEGFP-C1 + pcDNA3.1+-TagRFP-KHCmutA tail, (3) pEGFP-Tm1C + pcDNA3.1+-TagRFP-KHCWT tail, and (4) pEGFP-Tm1C+pcDNA3.1+-TagRFP-KHCmutA tail as well as two combinations of DNAs (for Fig. S3 E): (1) pEGFP-Tm1C + pcDNA3.1+-TagRFP-KHCWT tail and (2) pEGFP-Tm1C + pcDNA3.1+-TagRFP-LZ-Ktail using the calcium phosphate method. Cells were harvested 48 h after transfection into 500 µl of a 10-mM Tris buffer, pH 7.4, containing 150 mM NaCl, 0.5 mM MgCl2, and protease inhibitor cocktail. Cells were homogenized, and 1% Triton X-100 was added. After centrifugation at 5,000 g at 4°C, soluble fractions were incubated for 4 h at 4°C with 30 µl Sepharose beads conjugated with single-chain GFP antibody (GFP binder; GFP-Trap-M; ChromoTek). Beads were washed with the same buffer without Triton X-100 and resuspended in 30–50 µl Laemmli buffer. Samples were boiled for 5 min and analyzed by Western blotting. Rabbit polyclonal anti-tRFP antibody (AB233; Sapphire North America) and rabbit polyclonal anti-GFP antibody (produced in our laboratory) were both used at 1:1,000 dilution.
Drosophila embryo Vasa staining and imaging
Embryos of certain genotypes were first collected within 2 h after egg laying from apple juice agar plates into a 50-µm cell strainer and rinsed with water containing 0.4% NaCl and 0.03% Triton X-100. Embryos were then dechorionated in 50% bleach and 50% EtOH for 5 min and then were rinsed several times in 1× PBS + 0.1% Triton X-100 to remove the bleach. Embryos were fixed with 50% heptane/50% PEM-formaldehyde solution (0.1 M Pipes, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde) and vigorously vortexed for 30 min at room temperature. The PEM-formaldehyde layer was replaced with 100% MeOH, and embryos with heptane/MeOH were vortexed vigorously for 1 min. The devitellinized embryos on the bottom were collected and rehydrated in the following steps: 100% MeOH for 10 min, 50% MeOH and 50% PBS + 0.1% Triton X-100 for 10 min, and PBS + 0.1% Triton X-100 for 10 min. Embryos were then blocked in PBS + 0.1% Triton X-100 + 5% normal goat serum for 1 h and stained with rat anti-Vasa antibody (1:500) at 4°C overnight; washed with 1× PBS + 0.1% Triton X-100 five times for 10 min each time, incubated with secondary TRITC-conjugated anti-rat antibody (1:100) at 4°C overnight, and washed with 1× PBS + 0.1% Triton X-100 five times for 10 min each time before mounting in 90% glycerol/10% PBS. Embryos were imaged on a Nikon Eclipse U2000 inverted stand with a Yokogawa CSU10 spinning-disk confocal head and a 20× 0.75 NA lens using an Evolve EMCCD for fluorescent images and CoolSnap charge-coupled device (Roger Scientific) for differential interference contrast images, respectively, controlled by Nikon Elements software. Fluorescent images were acquired every 1 µm/step in z stacks.
BioID pulldown assay
A promiscuous biotin ligase fusion protein BirA* was amplified from the original construct, pcDNA3.1 MCS-BirA(R118G)-HA (36047; Addgene; Roux et al., 2012), and inserted into a pMT-EGFP-Staufen construct (EGFP-GFP was subcloned from pAC-EGFP-GFP into pMT/V5-His vector by KpnI/AgeI sites to generate pMT-EGFP-Staufen construct) to the C terminus of Staufen by AgeI site. A Drosophila S2 stable cell carrying pMT-EGFP-Stau-BirA* construct was made and induced with CuSO4 and 50 µM biotin for 24 h. Streptavidin bead pulldown was performed as described previously (Roux et al., 2013) with an extra wash with wash buffer 4 (2 M urea and 10 mM Tris-HCl, pH 8) for 8 min after wash buffer 3. A rabbit polyclonal anti-Drosophila MyoV antibody was raised against MyoV coiled-coil domain (ProteinTech). Rabbit α-MyoV antibody was used 1:2,000 for Western blotting.
Online supplemental material
Fig. S1 shows how Staufen-SunTag particles are functional and move at different velocities in different areas of the oocyte. Fig. S2 shows how GFP-Staufen forms an RNP complex in S2 cells and is transported by kinesin-1. Fig. S3 shows how KHCmutA does not affect Staufen transport along microtubules and how chimeric Unc104-Ktail motor cannot transport Staufen particles. Fig. S4 shows how inhibition of transport only causes osk/Staufen mislocalization in stage 9 oocytes, while simultaneous inhibition of transport and streaming complete abolishes osk/Staufen posterior localization in stage 9 and stage 10B oocytes. Fig. S5 shows how MyoV functions as a local anchorage for Staufen. Video 1 shows Staufen localization and movement in streaming oocytes. Video 2 shows how streaming circulates Staufen particles to the anchorage site. Video 3 shows how Staufen particles move on microtubules in S2 cells. Video 4 shows how inhibition of MyoV abolishes the deceleration of Staufen particles near the posterior cortex.
Supplementary Material
Acknowledgments
We thank Dr. Chris Q. Doe for anti-Staufen antibody, Dr. Edwin L. Ferguson for anti-Vasa antibody, Dr. Ronald Vale (University of California, San Francisco, San Francisco, CA) and Dr. Marvin Tanenbaum (Hubrecht Institute, Utrecht, Netherlands) for SunTag constructs, Dr. Graydon Gonsalvez for smFISH protocol, osk DNA probe, and Tm1C DNA construct, Dr. Anne Ephrussi (EMBL Heidelberg, Heidelberg, Germany) for MyoV mutant fly stock, Dr. Daniel St Johnston (University of Cambridge, Cambridge, UK) for Staufen mutant and GFP-Staufen flies, the Bloomington Drosophila Stock Center (supported by National Institutes of Health grant P40OD018537) for fly stocks, and Dr. Joshua Z. Rappoport for expert help in imaging. We also thank the Gelfand laboratory members for support, discussion, and suggestions. S.-C. Ling dedicates this work to Sheue-Houy Tyan, and A.S. Serpinskaya and V.I. Gelfand to memory of our teacher J. M. Vasiliev.
Research reported in this study was supported by the National Institute of General Medical Sciences grants R01GM052111 and R01GM124029 to V.I. Gelfand.
The authors declare no competing financial interests.
Author contributions: W. Lu, S.-C. Ling, and V. I. Gelfand conceived the project and designed the experiments; W. Lu, M. Lakonishok, A.S. Serpinskaya, and S.-C. Ling performed the experiments; W. Lu, A.S. Serpinskaya, D. Kirchenbüechler, and S.-C. Ling analyzed the data; W. Lu, and V.I. Gelfand wrote the manuscript.
References
- Ally S., Larson A.G., Barlan K., Rice S.E., and Gelfand V.I.. 2009. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol. 187:1071–1082. 10.1083/jcb.200908075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K., Lu W., and Gelfand V.I.. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23:317–322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor B., Bois J.S., and Quinlan M.E.. 2015. Regulation of the formin Cappuccino is critical for polarity of Drosophila oocytes. Cytoskeleton (Hoboken). 72:1–15. 10.1002/cm.21205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitwieser W., Markussen F.H., Horstmann H., and Ephrussi A.. 1996. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 10:2179–2188. 10.1101/gad.10.17.2179 [DOI] [PubMed] [Google Scholar]
- Brendza R.P., Serbus L.R., Duffy J.B., and Saxton W.M.. 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 289:2120–2122. 10.1126/science.289.5487.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha B.J., Serbus L.R., Koppetsch B.S., and Theurkauf W.E.. 2002. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4:592–598. 10.1038/ncb832 [DOI] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L.Y., and Jan Y.N.. 1994. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4:289–300. 10.1016/S0960-9822(00)00068-3 [DOI] [PubMed] [Google Scholar]
- Clark I.E., Jan L.Y., and Jan Y.N.. 1997. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 124:461–470. [DOI] [PubMed] [Google Scholar]
- Dahlgaard K., Raposo A.A., Niccoli T., and St Johnston D.. 2007. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell. 13:539–553. 10.1016/j.devcel.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo U., Lu W., Winding M., Lakonishok M., and Gelfand V.I.. 2015. Pavarotti/MKLP1 regulates microtubule sliding and neurite outgrowth in Drosophila neurons. Curr. Biol. 25:200–205. 10.1016/j.cub.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding M.P., Mitter R., Humphries A.C., and Way M.. 2011. A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. EMBO J. 30:4523–4538. 10.1038/emboj.2011.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler M., Giavazzi F., Cerbino R., and Palacios I.M.. 2017. Active diffusion and advection in Drosophila oocytes result from the interplay of actin and microtubules. Nat. Commun. 8:1520 10.1038/s41467-017-01414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L.K., and Lehmann R.. 1991. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 66:37–50. 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]
- Erdélyi M., Michon A.M., Guichet A., Glotzer J.B., and Ephrussi A.. 1995. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 377:524–527. 10.1038/377524a0 [DOI] [PubMed] [Google Scholar]
- Forrest K.M., and Gavis E.R.. 2003. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13:1159–1168. 10.1016/S0960-9822(03)00451-2 [DOI] [PubMed] [Google Scholar]
- Ganguly S., Williams L.S., Palacios I.M., and Goldstein R.E.. 2012. Cytoplasmic streaming in Drosophila oocytes varies with kinesin activity and correlates with the microtubule cytoskeleton architecture. Proc. Natl. Acad. Sci. USA. 109:15109–15114. 10.1073/pnas.1203575109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáspár I., Sysoev V., Komissarov A., and Ephrussi A.. 2017. An RNA-binding atypical tropomyosin recruits kinesin-1 dynamically to oskar mRNPs. EMBO J. 36:319–333. 10.15252/embj.201696038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Marchand V., Gáspár I., and Ephrussi A.. 2012. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 19:441–449. 10.1038/nsmb.2257 [DOI] [PubMed] [Google Scholar]
- Glotzer J.B., Saffrich R., Glotzer M., and Ephrussi A.. 1997. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr. Biol. 7:326–337. 10.1016/S0960-9822(06)00156-4 [DOI] [PubMed] [Google Scholar]
- Goodwin S.S., and Vale R.D.. 2010. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 143:263–274. 10.1016/j.cell.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit H.O., and Koppa R.. 1982. Time-Lapse Film Analysis of Cytoplasmic Streaming during Late Oogenesis of Drosophila. J. Embryol. Exp. Morphol. 67:101–111. [Google Scholar]
- Hachet O., and Ephrussi A.. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11:1666–1674. 10.1016/S0960-9822(01)00508-5 [DOI] [PubMed] [Google Scholar]
- Hay B., Jan L.Y., and Jan Y.N.. 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 55:577–587. 10.1016/0092-8674(88)90216-4 [DOI] [PubMed] [Google Scholar]
- Hay B., Jan L.Y., and Jan Y.N.. 1990. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 109:425–433. [DOI] [PubMed] [Google Scholar]
- Hurd T.R., Herrmann B., Sauerwald J., Sanny J., Grosch M., and Lehmann R.. 2016. Long Oskar Controls Mitochondrial Inheritance in Drosophila melanogaster. Dev. Cell. 39:560–571. 10.1016/j.devcel.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly A.L., Kim H., Srinivasan D., Lakonishok M., Larson A.G., and Gelfand V.I.. 2010. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc. Natl. Acad. Sci. USA. 107:12151–12156. 10.1073/pnas.1004736107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L.C., van Bergeijk P., Lipka J., Keijzer N., Wulf P.S., Katrukha E.A., Akhmanova A., and Hoogenraad C.C.. 2013. Myosin-V opposes microtubule-based cargo transport and drives directional motility on cortical actin. Curr. Biol. 23:828–834. 10.1016/j.cub.2013.03.068 [DOI] [PubMed] [Google Scholar]
- Karcher R.L., Deacon S.W., and Gelfand V.I.. 2002. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12:21–27. 10.1016/S0962-8924(01)02184-5 [DOI] [PubMed] [Google Scholar]
- Khanal I., Elbediwy A., Diaz de la Loza M.C., Fletcher G.C., and Thompson B.J.. 2016. Shot and Patronin polarise microtubules to direct membrane traffic and biogenesis of microvilli in epithelia. J. Cell Sci. 129:2651–2659. 10.1242/jcs.189076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuc Trong P., Doerflinger H., Dunkel J., St Johnston D., and Goldstein R.E.. 2015. Cortical microtubule nucleation can organise the cytoskeleton of Drosophila oocytes to define the anteroposterior axis. eLife. 4:e06088 10.7554/eLife.06088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., and Gelfand V.I.. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651. 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss J., López de Quinto S., Nüsslein-Volhard C., and Ephrussi A.. 2009. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr. Biol. 19:1058–1063. 10.1016/j.cub.2009.04.062 [DOI] [PubMed] [Google Scholar]
- Legent K., Tissot N., and Guichet A.. 2015. Visualizing Microtubule Networks During Drosophila Oogenesis Using Fixed and Live Imaging. Methods Mol. Biol. 1328:99–112. 10.1007/978-1-4939-2851-4_7 [DOI] [PubMed] [Google Scholar]
- Lehmann R., and Nüsslein-Volhard C.. 1986. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 47:141–152. 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Lehmann R., and Nüsslein-Volhard C.. 1991. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 112:679–691. [DOI] [PubMed] [Google Scholar]
- Ling S.C., Fahrner P.S., Greenough W.T., and Gelfand V.I.. 2004. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 101:17428–17433. 10.1073/pnas.0408114101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.C., Tikhonov M., and Gregor T.. 2013. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 154:789–800. 10.1016/j.cell.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F., and Gavis E.R.. 2015. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17:558–568. 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau P., Davies T., Williams L.S., Mishima M., and Palacios I.M.. 2010. Drosophila PAT1 is required for Kinesin-1 to transport cargo and to maximize its motility. Development. 137:2763–2772. 10.1242/dev.048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., and Gelfand V.I.. 2017. Moonlighting Motors: Kinesin, Dynein, and Cell Polarity. Trends Cell Biol. 27:505–514. 10.1016/j.tcb.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Casanueva M.O., Mahowald A.P., Kato M., Lauterbach D., and Ferguson E.L.. 2012. Niche-associated activation of rac promotes the asymmetric division of Drosophila female germline stem cells. PLoS Biol. 10:e1001357 10.1371/journal.pbio.1001357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Del Castillo U., and Gelfand V.I.. 2013a Organelle transport in cultured Drosophila cells: S2 cell line and primary neurons. J. Vis. Exp. (81):e50838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Fox P., Lakonishok M., Davidson M.W., and Gelfand V.I.. 2013b Initial neurite outgrowth in Drosophila neurons is driven by kinesin-powered microtubule sliding. Curr. Biol. 23:1018–1023. 10.1016/j.cub.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Lakonishok M., and Gelfand V.I.. 2015. Kinesin-1-powered microtubule sliding initiates axonal regeneration in Drosophila cultured neurons. Mol. Biol. Cell. 26:1296–1307. 10.1091/mbc.e14-10-1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Winding M., Lakonishok M., Wildonger J., and Gelfand V.I.. 2016. Microtubule-microtubule sliding by kinesin-1 is essential for normal cytoplasmic streaming in Drosophila oocytes. Proc. Natl. Acad. Sci. USA. 113:E4995–E5004. 10.1073/pnas.1522424113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P., Kroening S., Palacios I.M., Baldassa S., Grunewald B., Ambrosino C., Goetze B., Lupas A., St. Johnston D., and Kiebler M.. 2003. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 23:5778–5788. 10.1523/JNEUROSCI.23-13-05778.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem D.R., Dasgupta R., Elliott H., Gergely F., Davidson C., Brand A., González-Reyes A., and St Johnston D.. 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7:468–478. 10.1016/S0960-9822(06)00218-1 [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Adams J., Grünert S., and St Johnston D.. 2000. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 19:1366–1377. 10.1093/emboj/19.6.1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., Li M., Serr M., and Hays T.S.. 2007. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary. Mol. Biol. Cell. 18:2254–2263. 10.1091/mbc.e06-10-0959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S.E., Dillon S.T., and Boswell R.E.. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15:2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashchekin D., Fernandes A.R., and St Johnston D.. 2016. Patronin/Shot Cortical Foci Assemble the Noncentrosomal Microtubule Array that Specifies the Drosophila Anterior-Posterior Axis. Dev. Cell. 38:61–72. 10.1016/j.devcel.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P.A., and Boswell R.E.. 1994. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development. 120:1303–1313. [DOI] [PubMed] [Google Scholar]
- Nieuwburg R., Nashchekin D., Jakobs M., Carter A.P., Khuc Trong P., Goldstein R.E., and St Johnston D.. 2017. Localised dynactin protects growing microtubules to deliver oskar mRNA to the posterior cortex of the Drosophila oocyte. eLife. 6:e27237 10.7554/eLife.27237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I.M., and St Johnston D.. 2002. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 129:5473–5485. 10.1242/dev.00119 [DOI] [PubMed] [Google Scholar]
- Parton R.M., Hamilton R.S., Ball G., Yang L., Cullen C.F., Lu W., Ohkura H., and Davis I.. 2011. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194:121–135. 10.1083/jcb.201103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak D., Sepp K.J., and Hollenbeck P.J.. 2010. Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci. 30:8984–8992. 10.1523/JNEUROSCI.1621-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrywka N.J. 2013. Live imaging of GFP-labeled proteins in Drosophila oocytes. J. Vis. Exp. (73). 10.3791/50044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan M.E. 2016. Cytoplasmic Streaming in the Drosophila Oocyte. Annu. Rev. Cell Dev. Biol. 32:173–195. 10.1146/annurev-cellbio-111315-125416 [DOI] [PubMed] [Google Scholar]
- Rodionov V.I., Hope A.J., Svitkina T.M., and Borisy G.G.. 1998. Functional coordination of microtubule-based and actin-based motility in melanophores. Curr. Biol. 8:165–168. 10.1016/S0960-9822(98)70064-8 [DOI] [PubMed] [Google Scholar]
- Rogers S.L., and Gelfand V.I.. 1998. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr. Biol. 8:161–164. 10.1016/S0960-9822(98)70063-6 [DOI] [PubMed] [Google Scholar]
- Rogers S.L., Wiedemann U., Stuurman N., and Vale R.D.. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78:113–118. 10.1016/S0925-4773(98)00157-9 [DOI] [PubMed] [Google Scholar]
- Roth S., and Lynch J.A.. 2009. Symmetry breaking during Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 1:a001891 10.1101/cshperspect.a001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., and Burke B.. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196:801–810. 10.1083/jcb.201112098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., and Burke B.. 2013. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 74 10.1002/0471140864.ps1923s74 [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams J.H., Davidson C.M., Micklem D.R., Haseloff J., St Johnston D., and Brand A.H.. 1998. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 12:1847–1857. 10.1101/gad.12.12.1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L.R., Cha B.J., Theurkauf W.E., and Saxton W.M.. 2005. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development. 132:3743–3752. 10.1242/dev.01956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman J.M., Benton R., and St Johnston D.. 2000. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 101:377–388. 10.1016/S0092-8674(00)80848-X [DOI] [PubMed] [Google Scholar]
- Sinsimer K.S., Jain R.A., Chatterjee S., and Gavis E.R.. 2011. A late phase of germ plasm accumulation during Drosophila oogenesis requires lost and rumpelstiltskin. Development. 138:3431–3440. 10.1242/dev.065029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsimer K.S., Lee J.J., Thiberge S.Y., and Gavis E.R.. 2013. Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Reports. 5:1169–1177. 10.1016/j.celrep.2013.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Beuchle D., and Nüsslein-Volhard C.. 1991. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 66:51–63. 10.1016/0092-8674(91)90138-O [DOI] [PubMed] [Google Scholar]
- Tanaka T., and Nakamura A.. 2011. Oskar-induced endocytic activation and actin remodeling for anchorage of the Drosophila germ plasm. Bioarchitecture. 1:122–126. 10.4161/bioa.1.3.17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kato Y., Matsuda K., Hanyu-Nakamura K., and Nakamura A.. 2011. Drosophila Mon2 couples Oskar-induced endocytosis with actin remodeling for cortical anchorage of the germ plasm. Development. 138:2523–2532. 10.1242/dev.062208 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Gilbert L.A., Qi L.S., Weissman J.S., and Vale R.D.. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159:635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff M.T., Jäckle H., and Pankratz M.J.. 1996. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 15:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., Smiley S., Wong M.L., and Alberts B.M.. 1992. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 115:923–936. [DOI] [PubMed] [Google Scholar]
- Vallotton P., and Olivier S.. 2013. Tri-track: free software for large-scale particle tracking. Microsc. Microanal. 19:451–460. 10.1017/S1431927612014328 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A.L., and Lehmann R.. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8:243–246. 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- van Eeden F.J., Palacios I.M., Petronczki M., Weston M.J., and St Johnston D.. 2001. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J. Cell Biol. 154:511–523. 10.1083/jcb.200105056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N.F., and Ephrussi A.. 2002. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 129:3705–3714. [DOI] [PubMed] [Google Scholar]
- Veeranan-Karmegam R., Boggupalli D.P., Liu G., and Gonsalvez G.B.. 2016. A new isoform of Drosophila non-muscle Tropomyosin 1 interacts with Kinesin-1 and functions in oskar mRNA localization. J. Cell Sci. 129:4252–4264. 10.1242/jcs.194332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm J.E., Hilton M., Amos Q., and Henzel W.J.. 2003. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 163:1197–1204. 10.1083/jcb.200309088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winding M., Kelliher M.T., Lu W., Wildonger J., and Gelfand V.I.. 2016. Role of kinesin-1-based microtubule sliding in Drosophila nervous system development. Proc. Natl. Acad. Sci. USA. 113:E4985–E4994. 10.1073/pnas.1522416113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bowers B., Rao K., Wei Q., and Hammer JA III. 1998. Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J. Cell Biol. 143:1899–1918. 10.1083/jcb.143.7.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V.L., Belaya K., Pecreaux J., Gilchrist M.J., Clark A., Davis I., and St Johnston D.. 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 134:843–853. 10.1016/j.cell.2008.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.