Lele et al. review sources of cellular forces on the nucleus and the structural contributors to its mechanical response.
Abstract
Positioning and shaping the nucleus represents a mechanical challenge for the migrating cell because of its large size and resistance to deformation. Cells shape and position the nucleus by transmitting forces from the cytoskeleton onto the nuclear surface. This force transfer can occur through specialized linkages between the nuclear envelope and the cytoskeleton. In response, the nucleus can deform and/or it can move. Nuclear movement will occur when there is a net differential in mechanical force across the nucleus, while nuclear deformation will occur when mechanical forces overcome the mechanical resistance of the various structures that comprise the nucleus. In this perspective, we review current literature on the sources and magnitude of cellular forces exerted on the nucleus, the nuclear envelope proteins involved in transferring cellular forces, and the contribution of different nuclear structural components to the mechanical response of the nucleus to these forces.
Introduction
As the cell migrates, it must position the nucleus to maintain polarity and deform the nucleus to pass through narrow spaces (Wolf et al., 2013). Positioning and shaping the nucleus represents particular challenges because of its large size and resistance to deformation and therefore requires transferring active forces generated in (or transmitted through) the cytoskeleton onto the nucleus. These forces are generated by actin or microtubule polymerization, actomyosin contraction, and/or microtubule motor activity to compress, shear, or pull on the nucleus (Gundersen and Worman, 2013), and in some cases, they can cause nuclear membrane rupture (Denais et al., 2016; Raab et al., 2016). Cytoskeletal forces can act directly on the nucleus or be transmitted to the nucleus by molecular linkages with cytoskeletal elements. In the vast majority of the cases, the linker of nucleoskeleton and cytoskeleton (LINC) complex establishes the linkage and transmits mechanical force from the cytoskeleton to the nucleus (Luxton and Starr, 2014; Lee and Burke, 2017; Uhler and Shivashankar, 2017; Kirby and Lammerding, 2018). The LINC complex is composed of outer nuclear membrane KASH proteins (or nesprins in vertebrates) and inner nuclear membrane SUN proteins, which are anchored by an interaction with the nuclear lamina (principally lamins A and C; Starr and Fridolfsson, 2010; Chang et al., 2015b).
Cytoskeletal forces exerted on the nucleus can broadly elicit two responses: the nucleus can deform, and/or it can move. Forces can also move intranuclear structures, which we do not consider in this perspective (reviewed by Hiraoka and Dernburg, 2009; Starr, 2009; Tajik et al., 2016; Katsumata et al., 2017; Burke, 2018). Nuclear movement will occur when there is a net differential in mechanical force across the nucleus. Understanding how the nucleus moves requires identifying the sources and magnitudes of the competing forces that are components of the nuclear force balance as well as how these forces change dynamically during processes like cell migration.
The nuclear response to cytoskeletal forces is determined by the mechanical properties of structures in the nucleus, which include the nuclear lamina, chromatin, the nuclear matrix, nuclear bodies, RNA, and proteins. In this perspective, we discuss the contribution of different nuclear structural components to the mechanical response of the nucleus to mechanical force as well as the sources of cellular forces exerted on the nucleus.
Mechanical deformation of the nucleus in response to force
Mechanical measurements of isolated nuclei
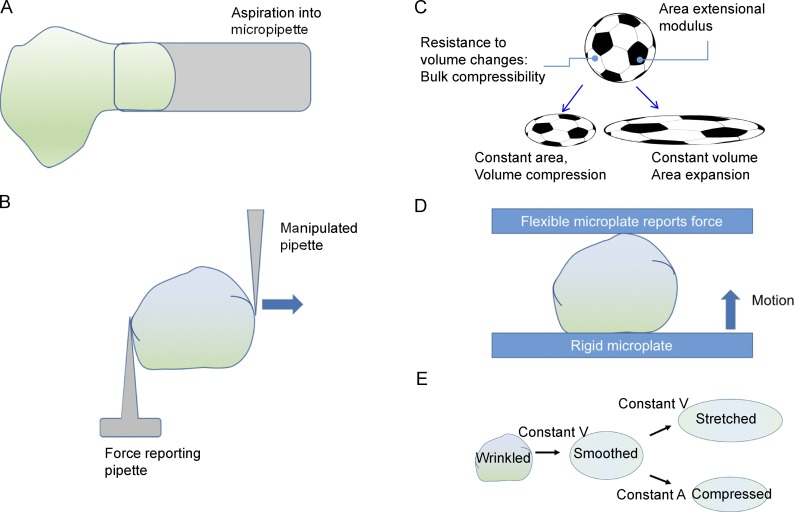
The mechanical properties of the nucleus were first measured in isolated nuclei aspirated into micropipettes (Fig. 1 A), which revealed that the length of an aspirated chondrocyte nucleus displayed asymptotic behavior with time (Guilak et al., 2000). Under force, a purely elastic solid will instantly reach a new, deformed shape, while a purely viscous fluid will continuously deform without reaching a steady state. The asymptotic behavior of the chondrocyte nucleus suggested that the nucleus behaves like a viscoelastic solid, with a steady-state strain reached on the time scale of tens of seconds. Related experiments revealed that nuclear deformation under force can have two contributions: one from elastic deformation (Dahl et al., 2004), which is reversible (i.e., the nucleus relaxes back to its unstressed shape upon removal of the force), and the other from plastic deformation, which reflects nonelastic changes in nuclear structure under force (Pajerowski et al., 2007).
Figure 1.
Methods to measure nuclear mechanics and key mechanical parameters important for describing nuclear shaping. (A) An isolated nucleus is aspirated into a micropipette. The outer boundary of the nucleus represents the nuclear membrane, while the inner boundary represents the nuclear lamina. The wrinkles represent folds in the nuclear envelope and lamina. (B) The micromanipulation technique in which a pipette is attached to one end of the isolated nucleus, and a force-measuring pipette is attached to the other end. The manipulating pipette is translated away (indicated by arrow), and force versus nuclear extension is quantified. (C) A sphere with minimum surface area to volume ratio must increase in its area or decrease in its volume (or a combination of both) during flattening. The resistance to volume changes and to area expansion are natural mechanical parameters relevant in nuclear shaping. (D) Schematic of one type of a nuclear compression measurement in which a rigid microplate is translated toward a flexible microplate, and the flexible microplate reports force. (E) During cell spreading, the nucleus flattens at constant volume and constant area until excess area in the lamina is smoothed out. After that, the nucleus can flatten only if the area (A) is stretched or if the volume (V) is compressed; the resistance to both changes is high, and forces in a typical fibroblast cell are not large enough to cause such changes. As a result, the nucleus reaches a steady-state shape.
While providing early understanding of the mechanical properties of the nucleus, micropipette aspiration can impose potentially nonphysiological large (100–500%; Stephens et al., 2017a) and spatially inhomogeneous strains (see text box; Vaziri et al., 2007) on nuclei. Smaller, physiologically relevant, and homogeneous nuclear strains have been achieved by stretching an isolated nucleus by moving a pipette attached to one side while the opposite side was attached to a pipette reporting force (Fig. 1 B). Under such strains, the nucleus deformed linearly with force and purely elastically such that removal of force resulted in complete relaxation of the nucleus to the original shape (Stephens et al., 2017a). Thus, at physiologically relevant strains, the nucleus does not appear to undergo plastic deformations reported with micropipette aspiration methods. Differences in these techniques may also have resulted in differing conclusions about the contribution of chromatin and the nuclear lamina to the mechanical behavior of the nucleus, as will be discussed.
•Force on the nucleus can pull, push, or shear the nuclear surface. Nuclear stress is equal to the local force per unit area of the nuclear surface.
•Nuclear deformation refers to a change in shape or size of the nucleus due to a force that acts on it.
•When the nuclear contents flow under applied force, this dissipates mechanical energy. If subsequent removal of force does not cause a relaxation of the nucleus to its original shape, this is called plastic deformation. If the nucleus recovers its initial shape, this is an elastic deformation.
•Nuclear strain is a dimensionless measure of nuclear deformation (sometimes expressed as percent strain). Generally, it is calculated as the change in nuclear length divided by the original nuclear length.
•The nuclear spring constant is defined as the applied force divided by the resulting change in length of the deformed nucleus.
The nuclear lamina and chromatin determine nuclear resistance to an extensional force
A familiar instance of nuclear deformation is the flattening of the nucleus after suspended cells land on a surface and spread. Two key mechanical parameters of this process can be understood by analogy to a spherical object such as a soccer ball (Fig. 1 C). A sphere has the minimum possible surface area to volume ratio, and when flattened against a surface, two possibilities (or combinations thereof) exist. The sphere can be flattened by reducing its volume at constant surface area (e.g., by letting air out of a soccer ball), extending its surface area while keeping its volume constant (e.g., by stretching the soccer ball), or a combination of both processes. Thus, the resistance of the nuclear surface (more specifically the nuclear lamina) to extension and the interior nucleoplasm (chromatin and other subnuclear structures) to changes in volume are likely important in determining the mechanical behavior of the nucleus during changes in its shape.
Most mechanical models for the nucleus assume that the chromatin and the nuclear lamina determine nuclear resistance to deformation under an external force; however, there is disagreement on the roles of these components. A micropipette aspiration study of large Xenopus laevis oocyte nuclei (Dahl et al., 2004) found no difference in the mechanical resistance of the nuclear envelope and associated lamina to deformation between osmotically swollen and unswollen nuclei. As chromatin and most of the nucleoplasm separate from the nuclear envelope during swelling, this suggests that the nuclear lamina determines elastic resistance of the nucleus to an extensional force. Assuming a 2D nuclear lamina, the extensional resistance of the Xenopus oocyte nucleus is then equal to the resistance of the nuclear lamina to an area expansion (Dahl et al., 2004). Micropipette aspiration experiments in mouse embryonic fibroblasts (MEFs) and HeLa cells similarly showed that the nuclear lamina primarily resists initial deformation, and nucleoplasmic contents resist only very large deformations (Rowat et al., 2006).
In contrast, the micromanipulation technique shows that for isolated nuclei from HeLa cells, human BJ5-ta cells, and vimentin-null MEFs, chromatin, rather than the lamina, elastically resists an extensional force below ∼30% strain (Stephens et al., 2017a). After ∼30% strain, the flexible nuclear lamina becomes taut and resists further strain, causing an abrupt increase in the nuclear resistance to force such that the nucleus becomes “stiff” to deformation. Experiments with isolated endothelial nuclei or cells, or C2C12 mouse cells, compressed between glass microplates (Fig. 1 D) also support that the force-compression curves are nonlinear, with the nucleus initially soft and then stiffening to compression with increasing deformation (Caille et al., 2002; Peeters et al., 2005). Nuclear rigidity in micromanipulation experiments also depends on the state of chromatin histone modifications (Stephens et al., 2017b). In contrast with the elastic behavior of chromatin in micromanipulation experiments, micropipette aspiration of embryonic stem cell nuclei, which lack the principal lamins A/C, caused chromatin to flow, thereby causing plastic deformations of the nucleus (Pajerowski et al., 2007).
The nuclear lamina is predominantly composed of four lamins: lamin A, lamin C lamin B1, and lamin B2. The A-type (lamins A and lamin C) and B-type lamins form distinct, nonoverlapping, filamentous networks under the inner nuclear membrane (Shimi et al., 2015; Xie et al., 2016; Turgay et al., 2017). There is consensus among experimental methods that lamin A/C is a major contributor to resistance. The nucleus softens greatly in the absence of lamin A/C as observed in micropipette aspiration experiments (Pajerowski et al., 2007; Swift et al., 2013; Davidson et al., 2014), in micromanipulation experiments (Stephens et al., 2017a), in direct forcing of nuclei in intact cells with micropipettes (Neelam et al., 2015), in stretching of cells attached to flexible membranes (Lammerding et al., 2004), in atomic force microscopy measurements (Schäpe et al., 2009), and in cell compression experiments (Broers et al., 2004). As lamin A expression scales with stiffness of tissues (Raab et al., 2012; Swift et al., 2013), it is likely that nuclei in cells from stiff tissues such as muscle and bone will have stiffer nuclei than those in softer tissues such as adipose or brain.
In contrast with A-type lamins, B-type lamins do not appear to play a major role in nuclear stiffness (Lammerding et al., 2006; Stephens et al., 2017a). Rather, nuclear stiffness increased upon knockdown of lamin B1 in erythroid precursors and in U251 glioma cells and correlated with the ratio of lamin A to B (Shin et al., 2013). Lamin B1 may contribute to stiffness in cells that have very low levels of lamin A/C. The different lamins may act in mechanically distinct ways to resist deformation, with lamin A providing viscous resistance and lamin B1 providing elastic resistance (Shin et al., 2013; Swift et al., 2013).
Disease-related mutations in lamin proteins may affect nuclear stiffness. Hutchinson-Gilford progeria syndrome is caused by a point mutation in LMNA that encodes lamin A/C, resulting in the production of abnormally processed lamin A that accumulates on the inner nuclear membrane and causes the nucleus to become stiffer (Goldman et al., 2004; Dahl et al., 2006; Verstraeten et al., 2008). Other mutations in lamin A/C produce nuclear phenotypes consistent with softer nuclei, although there is no consistent relationship between nuclear stiffness and type of disease. How different mutations in lamins result in differences in nuclear mechanical properties is unclear.
An important caveat to consider when interpreting studies that measure the mechanical properties of the nucleus with physical means is that they assume that the physical properties of the nucleus do not change in response to the applied force. However, isolated nuclei from HeLa cells, MRC5 fibroblasts, and human umbilical cord endothelial cells (HUVECs) rapidly stiffen (tens of seconds) in response to forces applied to nesprins on the outer nuclear surface (Guilluy et al., 2014). Relaxation of tension on the nucleus through myosin inhibition increases lamin A phosphorylation and softens the nucleus (Buxboim et al., 2014). Therefore, how the nucleus responds to physical forces through molecular adaptations is worth considering in studies of the physical properties of nuclei.
Compressibility of the nucleus
In addition to extensional stiffness of the nucleus or extensional modulus of the nuclear lamina, the resistance to compression of the nuclear volume is another important mechanical parameter. It is likely that osmotic stresses contribute to the resistance to nuclear volume expansion or compression in the cell.
Osmotic properties of the nucleus have been extensively studied (Finan et al., 2009, 2011; Finan and Guilak, 2010). In isolated articular chondrocyte nuclei, only large macromolecules that cannot passively leave or enter the nucleus contribute to osmotic stresses (Finan et al., 2009). Solutes smaller than the nuclear pore opening pass freely through the pore and therefore do not exert an osmotic stress. Thus, the size exclusion behavior of the nuclear membrane and nuclear pores, rather than semipermeability to water, may determine osmotic stresses in the nucleus. When the cell volume is osmotically changed by varying solute concentrations in the media, the nuclear volume changes proportionately to cell volume (Finan and Guilak, 2010; Guo et al., 2017); this is consistent with free flow of water and small solutes across the nuclear pore complexes while equilibrating the osmotic stress generated by larger impermeable molecules. These findings are supported by parallel changes in cell and nuclear volume after trypsinization of MEFs (Kim et al., 2016) and a correlation of nuclear size with cell size in yeast (Jorgensen et al., 2007; Neumann and Nurse, 2007). Overall, nuclear volume changes may be secondary effects of cell volume regulation and not necessarily due to changes in cytoskeletal forces directly exerted on the nuclear surface. At mitotic exit, however, the transient polymerization of nuclear F-actin can increase the volume of the nucleus throughout the early G1 phase of the cell cycle, resulting in local nuclear protrusions associated with nuclear F-actin (Baarlink et al., 2017). Inhibitors of nuclear actin prevented the change in nuclear volume, suggesting that at least for this stage of the cell cycle, nuclear F-actin polymerization may contribute to nuclear volume changes.
The mechanical behavior of the nucleus during nuclear shaping in cells
There are at least two competing models to explain the mechanical behavior of the nucleus in cultured cells based on different assumptions about how the nucleus stores elastic energy. In the elastic deformation model, mechanical energy is stored in the shape of the nucleus, hence deformed steady-state nuclear shapes require sustained mechanical forces. By analogy, force is required to flatten a rubber ball against a surface, but its spherical shape is restored once the force is removed. In contrast, the nonelastic deformation model posits that mechanical energy due to shape deformations dissipates quickly, and the ultimate nuclear shape is the culmination of incremental nonelastic shape changes. An appropriate analogy is the deformation of a honey-filled wrinkled bag suspended in water. If the bag has excess surface area relative to a sphere of the same volume, the bag can be deformed into many different shapes as long as the deformation is not so extreme as to require stretching of the bag or compression of the contents. A small force is required only to overcome the viscous resistance to deformations, but no force is required to sustain the deformed shape, since the shape changes are not elastic. Below, we consider evidence for these two models.
The elastic deformation hypothesis
One might intuit elastic behavior of the nucleus from the observations that nuclear shape mimics the shape of the cell (Chen et al., 1999) and that upon cell trypsinization, the nucleus rounds up in shape (Wang et al., 1993). Early work modeled the nucleus as an object storing elastic energy and mechanically integrated with tensile cytoskeletal structures (Sims et al., 1992). Furthermore, micropipette aspiration experiments with isolated nuclei (Dahl et al., 2005; Pajerowski et al., 2007; Stephens et al., 2017a) show reversible deformation of the nucleus upon relaxing the applied force, suggesting the expectation that nuclei must similarly be elastically deformed inside cells.
To understand the coordination of nuclear shape with cell shape, endothelial cells were micropatterned into elongated shapes, which resulted in correspondingly elongated nuclei (Versaevel et al., 2012). They proposed a mechanical model in which actomyosin stress fibers orient parallel to the elongated cell compress and elongate the nucleus. In this model, the nuclear shape is fully determined, given the instantaneous distribution of stresses from actomyosin stress fibers. Furthermore, the deformed nucleus balances this stress by storing elastic energy in its elongated shape. Others have modeled nuclear flattening in spread cells as a result of compressive stresses from the actomyosin-tensed cortex above the nucleus such that the deformed nuclear shape stores elastic energy (Vishavkarma et al., 2014). Similar models have been proposed by others (Khatau et al., 2009; Li et al., 2014). Models of the nucleus behaving as an elastic shell containing plastic chromatin or some variation thereof have been proposed in the literature (e.g., Cao et al., 2016). Such models predict that a removal of mechanical stresses on the nuclear surface should cause a relaxation of the elongated nuclear shape to its original undeformed shape. However, these studies stopped short of a direct validation of the key assumption that the nucleus stores elastic energy in the living cell, which recent experiments argue against.
The nonelastic deformation hypothesis
To test whether nuclear shapes store mechanical energy, the cell was excised from around the nucleus of elongated fibroblasts or breast cancer cells with highly irregular nuclei by micromanipulation (Tocco et al., 2018). The elongated or irregular nuclear shape was maintained for 5–10 min after isolation, far longer than the seconds for shape relaxations of an elastic nucleus (Neelam et al., 2015). Furthermore, elongated shapes of nuclei persist after isolation of nuclei by differential centrifugation (Deguchi et al., 2005), and laser ablation of stress fibers along elongated nuclei causes no relaxation in elongated nuclear shapes (Alam et al., 2015). These results suggest that the nucleus in cells does not store elastic energy.
How could a nucleus deform nonelastically despite the large changes in its shape? A computational model suggests that the flattening of a round nucleus during cell spreading does not require the nucleus to store elastic energy, rather that the nucleus in fibroblasts should reach a steady-state “flat” shape during cell spreading as the wrinkled lamina in the rounded nucleus becomes fully taut during flattening (Li et al., 2015). Once the lamina is taut, further flattening of the nucleus would require compression of the nuclear volume or extension of the lamina area (Fig. 1 E), events unlikely to be achieved by the cellular forces exerted during cell spreading given the measured bulk compressibility of the nucleus and the extensional modulus of the nuclear lamina. Nuclear volume remains virtually constant as nuclei flatten during fibroblast spreading (Li et al., 2015) as well as during nuclear deformations in WT and lamin A/C–deficient fibroblasts as they squeeze through narrow pores (Davidson et al., 2015). Folds in the nuclear lamina in MCF10A cells are observed to decrease during the process of cell spreading and establishment of nuclear shape (Neelam et al., 2016). The isolated nucleus becomes stiffer after an initial threshold strain, where the lamina is predicted to become taut (Banigan et al., 2017; Stephens et al., 2017a). All this evidence is supportive of a model where the excess lamina surface area relative to that of a sphere of the same volume allows the nucleus in the cell to undergo dramatic changes in shape at constant nuclear area and volume. As a result, the nucleus does not store elastic energy in its shape.
A mechanical model of the nucleus consisting of a stiff lamina with excess surface area and resistance to volume compression/expansion but little resistance to shape changes on the time scale of cell migration also helps interpret observations of nuclear shape changes and mechanical resistance of the nucleus to cell migration through narrow spaces (Wolf et al., 2013; Davidson et al., 2014; Thomas et al., 2015). The nucleus can deform freely until extreme changes in shape make the lamina taut, at which point further deformation requires a reduction in nuclear volume and/or an increase in the lamina area and disruption of the nuclear lamina potentially accompanied by nuclear membrane rupture (Denais et al., 2016; Hatch and Hetzer, 2016; Raab et al., 2016). This might explain why cells lacking lamin A/C more easily migrate through narrow pores than WT cells (Rowat et al., 2013; Davidson et al., 2014; Harada et al., 2014): the resistance of the lamina to extension in some cells might be too large for cellular forces to overcome. A good example of this is dendritic cells, which have a level of lamin A expression intermediate between fibroblasts and neutrophils. Dendritic cells are able to pass through narrow constrictions by partially disrupting the lamina and facilitating passage of the cells through narrow constrictions (Thiam et al., 2016). Circumferential nuclear buckling is observed in MEFs migrating in 3D environments (Davidson et al., 2014). Such results suggest that the lamina is not uniformly under tension during extreme nuclear deformations but rather behaves as a 2D solid that may be anisotropic in its deformation under tension.
Changes in the condensation state of the chromatin can affect nuclear volume (Tamada et al., 2006; Mazumder et al., 2008; Chan et al., 2017). Consequently, mechanical perturbations that affect chromatin condensation may change the mechanical responses of nuclei. For example, embryonic stem cells in a defined transition state between naive pluripotency and lineage restriction exhibit an abnormal auxetic mechanical response, i.e., a cross-sectional expansion to compression and a cross-sectional contraction to stretching, caused (at least in part) by decondensation of chromatin and volume expansion of the strained nuclei (Pagliara et al., 2014).
Dynamic changes in cell shape are required for changes to nuclear shape
Extreme nuclear deformations are observed when cells migrate through tight spaces. Fibrosarcoma cells that enter capillaries of mice show strikingly elongated nuclear shapes (Yamauchi et al., 2005), as do neutrophils extravasating through endothelia. There are limits on how much the cell can deform the nucleus, which in turn limits the ability of the cell to squeeze through 3D matrices. Different cell types including tumor cells, T cells, and neutrophils are unable to squeeze through pore sizes of the ECM at ≤10% of the nuclear cross section (Wolf et al., 2013). Such arrested cells send out cytoplasmic protrusions through the porous environment, but they are unable to deform their nuclei enough to squeeze through the narrow pores. A number of papers have reported a similar requirement for nuclear deformation for the cells to squeeze through narrow constrictions (Davidson et al., 2014; Harada et al., 2014; Thomas et al., 2015; Thiam et al., 2016).
Nuclear shape changes accompany cell shape changes when cells migrate from 1D fibronectin lines to 2D fibronectin patterns (Tocco et al., 2018), consistent with the observation that the nucleus deforms reversibly in response to local proximal protrusions (Alam et al., 2015) and the concept that dynamic changes in cell shape can generate compressive or tensile dissipative stresses on the nuclear surface to shape it (Li et al., 2015). Such stresses are likely generated through friction between structures such as F-actin filaments that are present between the nucleus and the moving cell boundary (Li et al., 2015). Thus, nuclear shape in the cell at any instant may be a cumulative result of incremental changes to its shape in the past, making the history of cell shape changes crucially important. The concept that local assembly of the cytoskeletal network can exert stresses on the nuclear surface is supported by observations of a perinuclear Arp2/3-dependent F-actin network that assembles around the nucleus as dendritic cells migrate through narrow constrictions (Thiam et al., 2016). Consistent with the observations that myosin activity is not required for nuclear flattening during cell spreading (Li et al., 2015; Neelam et al., 2016), myosin activity is not required for shaping dendritic cell nuclei during their passage through narrow constrictions or for translocating the cell through the constrictions (Thiam et al., 2016).
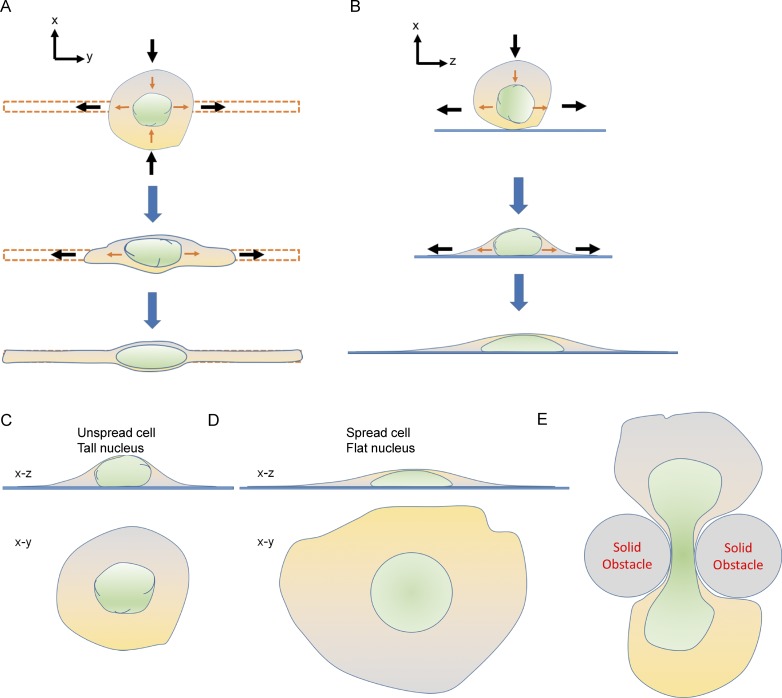
The model for nuclear shaping proposed by Li et al. (2015) explains a number of observations. First, it explains the relationship between the shape of the cell and the shape of the nucleus. For example, cells spread and elongate only in one direction on a 1D fibronectin line; therefore, the nuclear boundary, which follows the direction of motion of the cell boundary, adopts a 1D elongated nuclear shape (Fig. 2 A). The nuclear shape can continuously change in response to changes in cell shape provided the lamina has excess area stored in its folds or wrinkles. After it becomes taut, the nucleus will reach a steady-state shape (Fig. 2 A). The x–z cross-sectional shape of the nucleus would similarly follow changes to the x–z cross section of the cell (Fig. 2 B). By a similar mechanism, if the cell stops spreading, the nucleus will stop flattening (Fig. 2, C and D). Thus, the model explains the observed inverse relationship between the degree of nuclear flattening and the degree of cell spreading (Li et al., 2015). It can also explain how the nucleus takes on complex shapes such as an hourglass shaped in migration through small pores (Fig. 2 E; Wolf et al., 2013; Denais et al., 2016) because the cell itself adopts such a shape through motion of its boundaries. Similarly, elongated nuclear shapes in fibroblasts observed during migration in 3D ECM (Petrie et al., 2014) are likely due to directional motion of the cell boundaries.
Figure 2.
Nuclear shape during cell spreading is a result of incremental changes in nuclear shape caused by incremental changes in cell shape. (A and B) x–y and x–z cross sections of cells and nuclei based on data from spreading fibroblasts on 1D fibronectin lines (red; Tocco et al., 2018). The direction of motion of the cell boundary is shown by black arrows, while orange arrows show the direction of motion of the nuclear boundary. The nuclear boundaries follow the motion of the cell boundaries, which results in nuclear shape mimicking cell shape. This correlation between the nuclear and cell shape is observed both in the x–y plane (A) and in the x–z plane (B). Coordination between nuclear and cell shape. (C and D) Illustration of observations by Li et al. (2015) that nuclear flattening is inversely correlated with cell spreading. In less-spread cells, the nucleus remains round, while in well-spread cells, it flattens out and reaches a steady-state shape after the lamina becomes taut. (E) An hourglass-type shape of the nucleus that can be observed during cell migration through confined spaces. These types of observations (C–E) can be explained by positing a mechanical stress on the nucleus caused by motion of cell boundaries that drives motion of nuclear boundaries, which naturally results in a coordination between nuclear and cell shape.
Rounding of cell shape caused by disruption of actin filaments, inhibition of nonmuscle myosin II with blebbistatin (Buxboim et al., 2014, 2017; Driscoll et al., 2015), or trypsinization (Ingber, 2003) will result in rounded nuclear shapes (following a dynamic path that is the reverse of the sequence in Fig. 2) because inward motion of cell boundaries will exert a compressive stress on the nuclear surface. That the nucleus rounds up in these experiments may not represent the relaxation of stored elastic energy in the nucleus, but rather the requirement of a change in cell shape for changing nuclear shape. Similarly, persistent elongated nuclear shapes in cells migrating through pores were interpreted as evidence for plastic nuclear deformation (Harada et al., 2014), but given the coupling between cell and nuclear shapes, it is also possible that cell shapes remain elongated for long times after passing through tiny pores, which causes elongated nuclear shapes to persist.
Although myosin activity may not be necessary for overall shaping of the nucleus, it does influence nuclei in a number of ways. Once overall nuclear shape is established, actomyosin stress fibers may indent the apical surface of the nucleus to shape it locally (Cramer et al., 1997; Li et al., 2014; Versaevel et al., 2014). A nesprin–actin tension sensor has directly revealed actomyosin forces on the apical and equatorial surfaces of the nucleus (Arsenovic et al., 2016). Retrogradely flowing apical actomyosin bundles interact with the nucleus and position it in wounded fibroblasts (Luxton et al., 2010); perinuclear actin fibers can reorient the long axis of the nucleus along the long axis of the cell (Maninová and Vomastek, 2016); and myosin activity is required for directional nuclear motion toward lamellipodia formed by Rac photoactivation (Wu et al., 2014). A requirement for myosin activity in propelling the nucleus for invasion by MDA–MB-231 cells through 3D collagen gels (Thomas et al., 2015) and for moving the nucleus forward to generate pressure in fibroblasts and fibrosarcoma cells for lobopodial migration in dense 3D matrices has been reported (Petrie et al., 2014, 2017).
The centrosome can also locally affect nuclear shape as reflected in the commonly observed indentation of the nucleus near the centrosome (Schermelleh et al., 2008). The basis for this is unknown but presumably reflects the maintenance of the centrosome near the nucleus by dynein and/or the LINC complex (Malone et al., 2003; Semenova et al., 2008; Wu et al., 2011a).
The mechanics of nuclear positioning
Conceptual model: Nuclear position is a result of a balance of forces
The position of nuclei in cells is typical of cell type and activity. For example, in fibroblasts and most cultured cells, the nucleus is positioned near the cell center. Yet upon migration stimuli, nuclei become repositioned toward the cell rear in fibroblasts migrating in 2D (Gomes et al., 2005; Luxton et al., 2010) and are actively moved forward during single fibroblast migration in 2D (Wu et al., 2014; Alam et al., 2015) and lobopodia migration in 3D (Petrie et al., 2014). In epithelia, the nucleus is positioned preferentially at the base, apex, or center depending on the epithelial type (Gundersen and Worman, 2013). In multinucleated muscle fibers, a small number of nuclei cluster under the neuromuscular junction, whereas the others are spaced equidistantly at the cell periphery (Metzger et al., 2012; Wilson and Holzbaur, 2015). As nuclear mispositioning is associated with several diseases including muscular dystrophies and lissencephaly (Méjat and Misteli, 2010; Folker and Baylies, 2013; Gundersen and Worman, 2013), understanding the mechanisms and forces responsible for nuclear positioning has taken on added importance (Fig. 3).
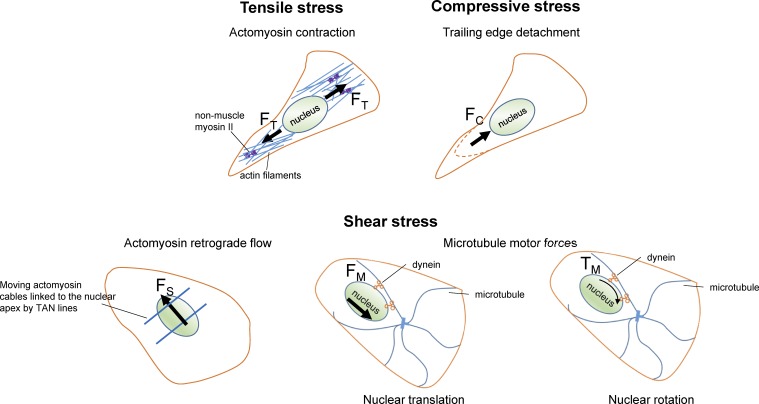
Figure 3.
Examples of mechanical stresses that can position the nucleus. These include tensile stress (corresponding to a net force FT), compressive stress (FC), or shear stress (FS). Depending on the cell context, different stresses can be dominant. For example, tensile actomyosin forces may position the nucleus during 2D cell crawling, and actomyosin retrograde flow can position the nucleus away from the leading edge during the initial phase of wound healing. Compressive stresses generated, for example, by trailing edge detachment can translate the nucleus toward the leading edge. Microtubule motors can translate the nucleus by shearing it (FS) or rotate it by exerting a torque on it (TM). Kinesin motor action is not shown in the figure for clarity. Dissipative nuclear stresses generated due to moving boundaries may also contribute to nuclear positioning (not shown). A balance between a subset of or all of these forces determines nuclear position. TAN lines, transmembrane actin-associated nuclear lines.
A stationary position of the nucleus implies that either there are no forces acting on it or that the sum of forces acting on it from all directions is zero. As more cases are examined, it is becoming clear that the nucleus is almost always under force even when it is stationary (Wu et al., 2014; Neelam et al., 2015; Arsenovic et al., 2016; Zhu et al., 2017a). Centering of the nucleus is possible when the magnitude of radially directed “pushing” or “pulling” cytoskeletal forces depends on distance from the cell periphery in such a way that forces become balanced when the nucleus is centered (Wu et al., 2014; Zhu et al., 2017a). When forces on the nucleus are unbalanced, the nucleus will move directionally until restoring forces on the nucleus increase and a new balance of forces is established, at which point the nucleus will become stationary and stably positioned. Such a model is consistent with dynamic positioning of the nucleus to specific positions in different cell types (Gomes et al., 2005; Alam et al., 2015; Almonacid et al., 2015) and tissues like developing muscle cells (Metzger et al., 2012; Wilson and Holzbaur, 2012).
The nucleus can also move without reaching an apparent steady position. This is particularly evident in interkinetic nuclear migration in neural epithelial cells (Tsai et al., 2010), where the nucleus migrates from one end of the cell to the other and back, upon which the cell divides. The migration of the nucleus is directional with little diffusive behavior (Baye and Link, 2007) except when it stops before changing the direction of its motion. The mechanics of the motion again is suggestive of a balance of forces on the nucleus. Changes in the direction of nuclear motion, driven by microtubule motors, have similarly been observed in developing Caenorhabditis elegans embryos (Fridolfsson and Starr, 2010) and during meiotic prophase in Schizosaccharomyces pombe (Vogel et al., 2009). In these motions, the nucleus is likely acted upon continually by mechanical forces, with a stoppage and then a change in the direction of the motion reflecting a balance of forces and then a change in direction of the net force on the nucleus.
Quantitative measurements of cytoskeletal forces on the nucleus in cells
The force balance on the nucleus is complex in migrating cells because the cell shape deforms continually and cytoskeletal structures turn over regularly (Lauffenburger and Horwitz, 1996). However, the nucleus maintains a relatively stable position rear of the cell centroid for many types of cell migration (Luxton and Gundersen, 2011; Gundersen and Worman, 2013). From a physical standpoint, understanding nuclear positioning requires identifying the molecular source of mechanical forces on the nucleus, the magnitude of these forces, and the magnitude of passive resistance to nuclear motion. Quantitative measurements of nuclear forces in migrating cells are rare. By traction force microscopy, the maximum tension across the nucleus in fibroblasts migrating in 2D was estimated to be of the order of 100 nN (Alam et al., 2015). This force is the combined tension of 10–100 stress fibers (tension in a stress fiber is of the order of 1–10 nN; Kumar et al., 2006) or the maximum force generated by 30,000 nonmuscle myosin IIA motor molecules (one motor generates 3.4 pN; Hundt et al., 2016). This tension decreased significantly upon disrupting the LINC complex but without changing overall cell traction (Alam et al., 2015). LINC disruption has been variously reported to increase cell traction (Chancellor et al., 2010), decrease traction (Graham et al., 2018), or have no effect (Alam et al., 2015; Elosegui-Artola et al., 2017).
In fibroblasts migrating in dense 3D matrices, there is a pressure differential in the front versus back of the cell (Petrie et al., 2014), and these pressure measurements can be used to estimate a nuclear force. The pressure difference between the front and the back is reported to be ∼1,500 Pa, which, multiplied by the projected area of the cell cross section (assumed to be circular with a radius of 5 µm; see Fig. 2 C in Petrie et al., 2014), yields a force of 100 nN, which is similar to the force estimated for fibroblasts in 2D (Alam et al., 2015).
Application of a centrifugal force to the nucleus in fibroblast monolayers for 30 min caused nuclei to displace from the center, but the nuclei recovered their central position when the force was removed, suggesting the presence of a restoring force that recenters the nucleus (Zhu et al., 2017a). Applying a centrifugal acceleration on the order of 2,000–5,000 g caused noticeable nuclear displacements; therefore, for an assumed nuclear mass of ∼100 pg (Schürmann et al., 2016), this amounts to a force of 20–50 nN, which is of the order of the estimates by traction force microscopy (Alam et al., 2015). Overall, such measurements suggest that a force on the order of tens of nanonewtons is required for nuclear displacement in cells.
In Drosophila melanogaster oocytes, growing microtubules may position the nucleus by pushing and indenting the nucleus with much lower forces of several piconewtons (Zhao et al., 2012). Yet, whether (larger) dynein-mediated forces that could pull on microtubules associated with the centrosome in order to center the centrosomal array of microtubules (Wu et al., 2011b), and hence position the associated nucleus, has not been ruled out.
Sources of cytoskeletal forces and their linkage to the nucleus
Forces that move the nucleus in cells are of different origins and vary depending on cell type and context (Gundersen and Worman, 2013). Furthermore, there is considerable variability in which LINC complex components are harnessed to transmit nuclear mechanical forces. Of the four different nesprin genes implicated in nuclear positioning, three are widely expressed (SYNE1, 2, and 3), and one (SYNE4) is more highly expressed in secretory epithelia (Starr and Fridolfsson, 2010; Lee and Burke, 2017). SYNE1 and 2 encode related giant proteins: nesprin-1G and nesprin-2G, respectively. These spectrin repeat–containing proteins have paired calponin homology domains at their N termini that directly bind actin and sites near their C-terminal transmembrane domain that bind kinesin and dynein microtubule motor proteins. Through alternative start sites and splicing, a number of smaller versions of nesprin-1 and nesprin-2 are produced, and these tend to lack the actin-binding domain of the giant forms but retain the regions that bind to microtubule motors (Rajgor et al., 2012; Duong et al., 2014). Nesprin-3α, one of the two splice variants encoded by SYNE3, binds intermediate filaments through plectin (Wilhelmsen et al., 2005; Roux et al., 2009). Nesprin-4, encoded by SYNE4, is a short nesprin that interacts with kinesin-1. All four nesprins can bind to either of the widely expressed SUN1 and SUN2 proteins (Stewart-Hutchinson et al., 2008; Ostlund et al., 2009). There is some evidence that binding of a single nesprin to SUN1 versus SUN2 may alter its specificity of interaction with the cytoskeleton. For example, SUN1 is required for microtubule-dependent nuclear positioning, whereas SUN2 supports actin-dependent nuclear positioning of nesprin-2G after centrifugal displacement (Zhu et al., 2017a). Below, we consider examples of cases where specific nesprins and SUNs have been implicated in nuclear positioning.
Wounded monolayers of cells have been a useful model system to study cell polarization and nuclear movement as it relates to cell migration (Luxton and Gundersen, 2011). In wounded monolayers of fibroblasts or myoblasts, serum or the serum factor lysophosphatidic acid stimulates a Cdc42 signaling pathway that activates rearward nuclear movement and results in the positioning of centrosome anterior to the nucleus (so called “centrosome orientation”; Gomes et al., 2005; Luxton et al., 2010; Chang et al., 2015a; Saunders et al., 2017). The rearward motion of the nucleus is caused by retrograde flow of actomyosin, which is coupled to the nucleus through a LINC complex composed of nesprin-2G and SUN2 reinforced by several actin-bundling proteins that interact with nesprin-2G (Luxton et al., 2010; Kutscheidt et al., 2014; Jayo et al., 2016). The rearward motion reflects an unbalanced force on the (previously centered) nucleus. Upon rearward motion, the nucleus reaches a new stable position. However, how the nucleus is moved forward as the cells at the wound edge crawl to close the wound is not as clear.
The nucleus in fibroblasts is displaced from the center toward the direction of the lamellipodia formed by Rac1 photoactivation in a manner dependent on myosin II activity (Wu et al., 2014). This is consistent with results obtained by traction force microscopy measurements of migrating fibroblasts that showed that the protruding (or retracting) cell boundary imposes a tensile (or compressive) stress on the nucleus to position it near the point of maximum tension in the cell (Alam et al., 2015). Alternative explanations for the forward motion of the nucleus involve apical actin fibers (or the so-called actin cap; Kim et al., 2014) and dynein (see below). However, apical actin fibers typically are stationary and parallel to the direction of motion of the moving nucleus in a migrating fibroblast (Wu et al., 2014). It is unclear how such fibers could generate a forward force on the nucleus. One study suggests that zippering of apical actin stress fibers at the rear end of the nucleus could push the nucleus forward, although a mechanical model for how this could happen was not proposed (Shiu et al., 2018).
Fibroblasts, fibrosarcoma cells, and breast carcinoma cells migrating in dense 3D matrices move by a unique lobopodial form of migration in which myosin contraction anterior to the nucleus moves the nucleus forward to pressurize the front of the cells (Petrie et al., 2014, 2017). Nesprin-3, which attaches the nucleus to intermediate filaments through plectin, is required for the movement of the nucleus and the generation of anterior compartment pressure (Petrie et al., 2014), leading to a model in which actomyosin contraction pulls on the nucleus through intermediate filaments, though the mechanism remains to be determined. Depending on the density and cross-linking of the matrix, fibroblasts can also use lamellipodia-based migration, but how the nucleus is positioned in this case is not yet clear (Zhu et al., 2017b).
Uniaxial lobopodial motility, where cell protrusions occur predominately in one direction, is a highly efficient mode of translocation through 3D matrices. In contrast, cells that extend protrusions in multiple directions tend to be immobile. In the piston model (Petrie et al., 2014), the nucleus physically partitions the cell into front and back compartments. Actomyosin tension creates a higher hydrostatic pressure in the front compartment, and this pressure helps generate bleb-like membrane protrusions at the leading edge for motility. If a front–back differential in hydrostatic pressure is an important mechanism for sustained unidirectional lobopodial motility, then the lack of a nucleus would hinder 3D motility, as observed by Graham et al. (2018).
Microtubule motor forces are also important for determining nuclear position. In migrating cortical neurons, the centrosome advances into the thin leading process followed by the movement of the nucleus toward the centrosome (Schaar and McConnell, 2005; Tsai et al., 2010; Bertipaglia et al., 2017). This movement of the nucleus requires dynein, which is likely coupled to the nucleus through nesprin-1 and/or nesprin-2 (Shu et al., 2004; Tsai et al., 2007; Zhang et al., 2009). Myosin II is localized behind the nucleus and may contribute to its forward movement during neuronal migration (Solecki et al., 2009). Microtubules and dynein may also be important for moving the nucleus forward in migrating fibroblasts (Levy and Holzbaur, 2008).
Nuclei undergo a series of movements during muscle development, culminating in the positioning of a small cluster of nuclei under the neuromuscular junction and the equal spacing of the remaining nuclei at the periphery of the syncytial muscle fiber (Folker and Baylies, 2013; Roman and Gomes, 2017). These movements include initial clustering after myoblast fusion, alignment and spreading along the muscle fiber, and finally, movement to the muscle periphery. Most of these movements involve microtubules, the microtubule motors dynein and kinesin, and specific LINC complexes (Cadot et al., 2012; Metzger et al., 2012; Folker et al., 2014; Wilson and Holzbaur, 2015; Gache et al., 2017; Gimpel et al., 2017). The centripetal movement of the nucleus to the myofiber periphery involves an unusual mechanism in which the zippering of the sarcomeres drives nuclear movement (Roman et al., 2017).
Overexpressing nesprin-4, which recruits kinesin-1 to the nuclear envelope, displaces the nucleus toward the edge of epithelial cells (Roux et al., 2009). In hair cells of the inner ear, nesprin-4 is required to maintain the basal position of nuclei, and loss of nesprin-4 from nuclei results in deafness in mice and humans (Horn et al., 2013). Compared with other systems, relatively little is understood of the nuclear positioning mechanism and forces that are responsible for the specific positioning of nuclei in their different locations in epithelial cells.
In mouse oocytes, which lack centrosomes, the nucleus is centered by a mechanism independent of centrosomal microtubules (Almonacid et al., 2015). Oocytes lacking formins, which are involved in the polymerization of actin filaments, are incapable of centering their nuclei. However, following microinjection of formin-2, nuclei centered in ∼200 min. Myosin Vb activity was required as well, leading Almonacid et al. (2015) to propose a statistical mechanical model whereby nuclear centering is driven by a pressure gradient created by myosin Vb–driven fluctuations. However, the combined requirement of formin and myosin Vb is also consistent with nuclear centering by centripetal F-actin network flow from the cell cortex since myosin V transports formin-containing vesicles along actin-filament tracks to the plasma membrane (Schuh, 2011). Hence, greater formin-mediated F-actin assembly at the cortex and the resulting centripetal flow should be expected with myosin Vb activity. Consistent with the transport function of myosin Vb, actin-rich vesicles near the cell periphery preferentially moved toward the cortex in a myosin Vb–dependent fashion (Almonacid et al., 2015). Thus, forces due to F-actin centripetal flow is a feasible alternative explanation for nuclear centering in oocytes in this study.
Conclusions
In this perspective, we have described studies that have revealed mechanical properties and cytoskeletal forces that contribute to nuclear shape and position. While it has been proposed that the nucleus stores elastic energy, an alternative is that the nucleus behaves as a nonelastic object inside cells. Energy is dissipated in stretching of the folds in the lamina until the lamin becomes taut, after which it resists further deformation; in extreme cases, the nucleus may rupture. For positioning the nucleus, unbalanced forces must be generated by the actin and/or microtubule cytoskeletons and transmitted to the nucleus to move it from its set position. This is usually accomplished by attaching the cytoskeleton to the nucleus through specific LINC complexes.
Nevertheless, there is still much to be learned. Outstanding questions in the field include how the forces on the nucleus stimulate structural and mechanical responses that stiffen or soften the nucleus and/or lead to changes in its connections to the cytoskeleton and how nuclear positioning influences forces exerted on other mechanically responsive elements in the cell such as focal adhesions and cell–cell adhesions. Most of our knowledge of nuclear shape and positioning comes from a relatively small number of cultured cell lines, and it will be important to extend studies to other cell types and tissues where nuclear behavior may be atypical. The field of nuclear mechanics will benefit from the continued development of new technologies such as nesprin tension sensors to measure nuclear mechanical forces in living cells and tissues and to probe and mechanically manipulate nuclei in vitro and in vivo. High-resolution imaging techniques to map the effect of mechanical force on chromatin structure will also help determine how nuclear forces could impact gene expression. Nuclear shape, structure, and positioning are frequently altered in human pathologies like cancer (Zink et al., 2004; Denais and Lammerding, 2014; Uhler and Shivashankar, 2018) and muscular dystrophies (Gundersen and Worman, 2013; Davidson and Lammerding, 2014). A grand future goal is to determine how abnormal mechanical force transfers to the nucleus, how changes in nuclear mechanical properties and positioning might contribute to the development of pathologies like cancer, and how the extent to which reverting these properties can mitigate disease.
Acknowledgments
T.P. Lele acknowledges support from National Institutes of Health (R01 EB014869). R.B. Dickinson’s contribution is based upon work supported by (and while serving at) the National Science Foundation. G.G. Gundersen acknowledges National Institutes of Health grants R01 GM099481 and R01 AR068636.
The authors declare no competing financial interests.
References
- Alam S.G., Lovett D., Kim D.I., Roux K.J., Dickinson R.B., and Lele T.P.. 2015. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J. Cell Sci. 128:1901–1911. 10.1242/jcs.161703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M., Ahmed W.W., Bussonnier M., Mailly P., Betz T., Voituriez R., Gov N.S., and Verlhac M.H.. 2015. Active diffusion positions the nucleus in mouse oocytes. Nat. Cell Biol. 17:470–479. 10.1038/ncb3131 [DOI] [PubMed] [Google Scholar]
- Arsenovic P.T., Ramachandran I., Bathula K., Zhu R., Narang J.D., Noll N.A., Lemmon C.A., Gundersen G.G., and Conway D.E.. 2016. Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension. Biophys. J. 110:34–43. 10.1016/j.bpj.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarlink C., Plessner M., Sherrard A., Morita K., Misu S., Virant D., Kleinschnitz E.M., Harniman R., Alibhai D., Baumeister S., et al. . 2017. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 19:1389–1399. 10.1038/ncb3641 [DOI] [PubMed] [Google Scholar]
- Banigan E.J., Stephens A.D., and Marko J.F.. 2017. Mechanics and Buckling of Biopolymeric Shells and Cell Nuclei. Biophys. J. 113:1654–1663. 10.1016/j.bpj.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye L.M., and Link B.A.. 2007. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci. 27:10143–10152. 10.1523/JNEUROSCI.2754-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertipaglia C., Gonçalves J.C., and Vallee R.B.. 2017. Nuclear migration in mammalian brain development. Semin. Cell Dev. Biol.. 10.1016/j.semcdb.2017.11.033 [DOI] [PubMed] [Google Scholar]
- Broers J.L., Peeters E.A., Kuijpers H.J., Endert J., Bouten C.V., Oomens C.W., Baaijens F.P., and Ramaekers F.C.. 2004. Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 13:2567–2580. 10.1093/hmg/ddh295 [DOI] [PubMed] [Google Scholar]
- Burke B. 2018. LINC complexes as regulators of meiosis. Curr. Opin. Cell Biol. 52:22–29. 10.1016/j.ceb.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Buxboim A., Swift J., Irianto J., Spinler K.R., Dingal P.C., Athirasala A., Kao Y.R., Cho S., Harada T., Shin J.W., and Discher D.E.. 2014. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 24:1909–1917. 10.1016/j.cub.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A., Irianto J., Swift J., Athirasala A., Shin J.W., Rehfeldt F., and Discher D.E.. 2017. Coordinated increase of nuclear tension and lamin-A with matrix stiffness outcompetes lamin-B receptor that favors soft tissue phenotypes. Mol. Biol. Cell. 28:3333–3348. 10.1091/mbc.e17-06-0393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B., Gache V., Vasyutina E., Falcone S., Birchmeier C., and Gomes E.R.. 2012. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 13:741–749. 10.1038/embor.2012.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caille N., Thoumine O., Tardy Y., and Meister J.J.. 2002. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 35:177–187. 10.1016/S0021-9290(01)00201-9 [DOI] [PubMed] [Google Scholar]
- Cao X., Moeendarbary E., Isermann P., Davidson P.M., Wang X., Chen M.B., Burkart A.K., Lammerding J., Kamm R.D., and Shenoy V.B.. 2016. A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration. Biophys. J. 111:1541–1552. 10.1016/j.bpj.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.J., Li W., Cojoc G., and Guck J.. 2017. Volume Transitions of Isolated Cell Nuclei Induced by Rapid Temperature Increase. Biophys. J. 112:1063–1076. 10.1016/j.bpj.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor T.J., Lee J., Thodeti C.K., and Lele T.. 2010. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 99:115–123. 10.1016/j.bpj.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Antoku S., Östlund C., Worman H.J., and Gundersen G.G.. 2015a Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus. 6:77–88. 10.1080/19491034.2015.1004947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Worman H.J., and Gundersen G.G.. 2015b Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.S., Brangwynne C., and Ingber D.E.. 1999. Squaring up to the cell-shape debate. Trends Cell Biol. 9:283 10.1016/S0962-8924(99)01551-2 [DOI] [PubMed] [Google Scholar]
- Cramer L.P., Siebert M., and Mitchison T.J.. 1997. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: implications for the generation of motile force. J. Cell Biol. 136:1287–1305. 10.1083/jcb.136.6.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Kahn S.M., Wilson K.L., and Discher D.E.. 2004. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 117:4779–4786. 10.1242/jcs.01357 [DOI] [PubMed] [Google Scholar]
- Dahl K.N., Engler A.J., Pajerowski J.D., and Discher D.E.. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89:2855–2864. 10.1529/biophysj.105.062554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L., and Misteli T.. 2006. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 103:10271–10276. 10.1073/pnas.0601058103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., and Lammerding J.. 2014. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol. 24:247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., Denais C., Bakshi M.C., and Lammerding J.. 2014. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 7:293–306. 10.1007/s12195-014-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., Sliz J., Isermann P., Denais C., and Lammerding J.. 2015. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 7:1534–1546. 10.1039/C5IB00200A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi S., Maeda K., Ohashi T., and Sato M.. 2005. Flow-induced hardening of endothelial nucleus as an intracellular stress-bearing organelle. J. Biomech. 38:1751–1759. 10.1016/j.jbiomech.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Denais C., and Lammerding J.. 2014. Nuclear mechanics in cancer. Adv. Exp. Med. Biol. 773:435–470. 10.1007/978-1-4899-8032-8_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll T.P., Cosgrove B.D., Heo S.J., Shurden Z.E., and Mauck R.L.. 2015. Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophys. J. 108:2783–2793. 10.1016/j.bpj.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong N.T., Morris G.E., Lam L.T., Zhang Q., Sewry C.A., Shanahan C.M., and Holt I.. 2014. Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS One. 9:e94380 10.1371/journal.pone.0094380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola A., Andreu I., Beedle A.E.M., Lezamiz A., Uroz M., Kosmalska A.J., Oria R., Kechagia J.Z., Rico-Lastres P., Le Roux A.L., et al. . 2017. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell. 171:1397–1410. 10.1016/j.cell.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Finan J.D., and Guilak F.. 2010. The effects of osmotic stress on the structure and function of the cell nucleus. J. Cell. Biochem. 109:460–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan J.D., Chalut K.J., Wax A., and Guilak F.. 2009. Nonlinear osmotic properties of the cell nucleus. Ann. Biomed. Eng. 37:477–491. 10.1007/s10439-008-9618-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan J.D., Leddy H.A., and Guilak F.. 2011. Osmotic stress alters chromatin condensation and nucleocytoplasmic transport. Biochem. Biophys. Res. Commun. 408:230–235. 10.1016/j.bbrc.2011.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., and Baylies M.K.. 2013. Nuclear positioning in muscle development and disease. Front. Physiol. 4:363 10.3389/fphys.2013.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E.S., Schulman V.K., and Baylies M.K.. 2014. Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development. 141:355–366. 10.1242/dev.095612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H.N., and Starr D.A.. 2010. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191:115–128. 10.1083/jcb.201004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gache V., Gomes E.R., and Cadot B.. 2017. Microtubule motors involved in nuclear movement during skeletal muscle differentiation. Mol. Biol. Cell. 28:865–874. 10.1091/mbc.e16-06-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel P., Lee Y.L., Sobota R.M., Calvi A., Koullourou V., Patel R., Mamchaoui K., Nédélec F., Shackleton S., Schmoranzer J., et al. . 2017. Nesprin-1α-Dependent Microtubule Nucleation from the Nuclear Envelope via Akap450 Is Necessary for Nuclear Positioning in Muscle Cells. Curr. Biol. 27:2999–3009. 10.1016/j.cub.2017.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., and Collins F.S.. 2004. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 101:8963–8968. 10.1073/pnas.0402943101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Graham D.M., Andersen T., Sharek L., Uzer G., Rothenberg K., Hoffman B.D., Rubin J., Balland M., Bear J.E., and Burridge K.. 2018. Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J. Cell Biol. 217:895–914. 10.1083/jcb.201706097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F., Tedrow J.R., and Burgkart R.. 2000. Viscoelastic properties of the cell nucleus. Biochem. Biophys. Res. Commun. 269:781–786. 10.1006/bbrc.2000.2360 [DOI] [PubMed] [Google Scholar]
- Guilluy C., Osborne L.D., Van Landeghem L., Sharek L., Superfine R., Garcia-Mata R., and Burridge K.. 2014. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 16:376–381. 10.1038/ncb2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G.G., and Worman H.J.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Pegoraro A.F., Mao A., Zhou E.H., Arany P.R., Han Y., Burnette D.T., Jensen M.H., Kasza K.E., Moore J.R., et al. . 2017. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. USA. 114:E8618–E8627. 10.1073/pnas.1705179114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C., Ivanovska I.L., and Discher D.E.. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204:669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., and Hetzer M.W.. 2016. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 215:27–36. 10.1083/jcb.201603053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., and Dernburg A.F.. 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 17:598–605. 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Horn H.F., Brownstein Z., Lenz D.R., Shivatzki S., Dror A.A., Dagan-Rosenfeld O., Friedman L.M., Roux K.J., Kozlov S., Jeang K.T., et al. . 2013. The LINC complex is essential for hearing. J. Clin. Invest. 123:740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt N., Steffen W., Pathan-Chhatbar S., Taft M.H., and Manstein D.J.. 2016. Load-dependent modulation of non-muscle myosin-2A function by tropomyosin 4.2. Sci. Rep. 6:20554 10.1038/srep20554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D.E. 2003. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 116:1157–1173. 10.1242/jcs.00359 [DOI] [PubMed] [Google Scholar]
- Jayo A., Malboubi M., Antoku S., Chang W., Ortiz-Zapater E., Groen C., Pfisterer K., Tootle T., Charras G., Gundersen G.G., and Parsons M.. 2016. Fascin Regulates Nuclear Movement and Deformation in Migrating Cells. Dev. Cell. 38:371–383. 10.1016/j.devcel.2016.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., and Futcher B.. 2007. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 18:3523–3532. 10.1091/mbc.e06-10-0973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K., Nishi E., Afrin S., Narusawa K., and Yamamoto A.. 2017. Position matters: multiple functions of LINC-dependent chromosome positioning during meiosis. Curr. Genet. 63:1037–1052. 10.1007/s00294-017-0699-2 [DOI] [PubMed] [Google Scholar]
- Khatau S.B., Hale C.M., Stewart-Hutchinson P.J., Patel M.S., Stewart C.L., Searson P.C., Hodzic D., and Wirtz D.. 2009. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 106:19017–19022. 10.1073/pnas.0908686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Cho S., and Wirtz D.. 2014. Tight coupling between nucleus and cell migration through the perinuclear actin cap. J. Cell Sci. 127:2528–2541. 10.1242/jcs.144345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Li B., Si F., Phillip J.M., Wirtz D., and Sun S.X.. 2016. Volume regulation and shape bifurcation in the cell nucleus. J. Cell Sci. 129:457 10.1242/jcs.185173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T.J., and Lammerding J.. 2018. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 20:373–381. 10.1038/s41556-018-0038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Maxwell I.Z., Heisterkamp A., Polte T.R., Lele T.P., Salanga M., Mazur E., and Ingber D.E.. 2006. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J. 90:3762–3773. 10.1529/biophysj.105.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutscheidt S., Zhu R., Antoku S., Luxton G.W., Stagljar I., Fackler O.T., and Gundersen G.G.. 2014. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat. Cell Biol. 16:708–715. 10.1038/ncb2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., and Lee R.T.. 2004. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113:370–378. 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., and Lee R.T.. 2006. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281:25768–25780. 10.1074/jbc.M513511200 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D.A., and Horwitz A.F.. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Lee Y.L., and Burke B.. 2017. LINC complexes and nuclear positioning. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.11.008 [DOI] [PubMed] [Google Scholar]
- Levy J.R., and Holzbaur E.L.. 2008. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121:3187–3195. 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Kumar A., Makhija E., and Shivashankar G.V.. 2014. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 35:961–969. 10.1016/j.biomaterials.2013.10.037 [DOI] [PubMed] [Google Scholar]
- Li Y., Lovett D., Zhang Q., Neelam S., Kuchibhotla R.A., Zhu R., Gundersen G.G., Lele T.P., and Dickinson R.B.. 2015. Moving cell boundaries drive nuclear shaping during cell spreading. Biophys. J. 109:670–686. 10.1016/j.bpj.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., and Gundersen G.G.. 2011. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr. Opin. Cell Biol. 23:579–588. 10.1016/j.ceb.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., and Starr D.A.. 2014. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr. Opin. Cell Biol. 28:69–75. 10.1016/j.ceb.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Vintinner E., and Gundersen G.G.. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., and White J.G.. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836. 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Maninová M., and Vomastek T.. 2016. Dorsal stress fibers, transverse actin arcs, and perinuclear actin fibers form an interconnected network that induces nuclear movement in polarizing fibroblasts. FEBS J. 283:3676–3693. 10.1111/febs.13836 [DOI] [PubMed] [Google Scholar]
- Mazumder A., Roopa T., Basu A., Mahadevan L., and Shivashankar G.V.. 2008. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys. J. 95:3028–3035. 10.1529/biophysj.108.132274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjat A., and Misteli T.. 2010. LINC complexes in health and disease. Nucleus. 1:40–52. 10.4161/nucl.1.1.10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., and Baylies M.K.. 2012. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 484:120–124. 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Chancellor T.J., Li Y., Nickerson J.A., Roux K.J., Dickinson R.B., and Lele T.P.. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl. Acad. Sci. USA. 112:5720–5725. 10.1073/pnas.1502111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam S., Hayes P.R., Zhang Q., Dickinson R.B., and Lele T.P.. 2016. Vertical uniformity of cells and nuclei in epithelial monolayers. Sci. Rep. 6:19689 10.1038/srep19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F.R., and Nurse P.. 2007. Nuclear size control in fission yeast. J. Cell Biol. 179:593–600. 10.1083/jcb.200708054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C., Folker E.S., Choi J.C., Gomes E.R., Gundersen G.G., and Worman H.J.. 2009. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 122:4099–4108. 10.1242/jcs.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliara S., Franze K., McClain C.R., Wylde G., Fisher C.L., Franklin R.J.M., Kabla A.J., Keyser U.F., and Chalut K.J.. 2014. Auxetic nuclei in embryonic stem cells exiting pluripotency. Nat. Mater. 13:638–644. 10.1038/nmat3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., and Discher D.E.. 2007. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA. 104:15619–15624. 10.1073/pnas.0702576104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters E.A., Oomens C.W., Bouten C.V., Bader D.L., and Baaijens F.P.. 2005. Mechanical and failure properties of single attached cells under compression. J. Biomech. 38:1685–1693. 10.1016/j.jbiomech.2004.07.018 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Koo H., and Yamada K.M.. 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 345:1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Harlin H.M., Korsak L.I., and Yamada K.M.. 2017. Activating the nuclear piston mechanism of 3D migration in tumor cells. J. Cell Biol. 216:93–100. 10.1083/jcb.201605097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Swift J., Dingal P.C.D.P., Shah P., Shin J.-W., and Discher D.E.. 2012. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 199:669–683. 10.1083/jcb.201205056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Rajgor D., Mellad J.A., Autore F., Zhang Q., and Shanahan C.M.. 2012. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 7:e40098 10.1371/journal.pone.0040098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W., and Gomes E.R.. 2017. Nuclear positioning in skeletal muscle. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Roman W., Martins J.P., Carvalho F.A., Voituriez R., Abella J.V.G., Santos N.C., Cadot B., Way M., and Gomes E.R.. 2017. Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat. Cell Biol. 19:1189–1201. 10.1038/ncb3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., and Burke B.. 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 106:2194–2199. 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat A.C., Lammerding J., and Ipsen J.H.. 2006. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J. 91:4649–4664. 10.1529/biophysj.106.086454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D.E., Olins A.L., Herrmann H., Weitz D.A., and Lammerding J.. 2013. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 288:8610–8618. 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.A., Harris N.J., Willey P.T., Woolums B.M., Wang Y., McQuown A.J., Schoenhofen A., Worman H.J., Dauer W.T., Gundersen G.G., and Luxton G.W.. 2017. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol. 216:657–674. 10.1083/jcb.201507113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., and McConnell S.K.. 2005. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA. 102:13652–13657. 10.1073/pnas.0506008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäpe J., Prausse S., Radmacher M., and Stick R.. 2009. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys. J. 96:4319–4325. 10.1016/j.bpj.2009.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G., et al. . 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M. 2011. An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13:1431–1436. 10.1038/ncb2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann M., Scholze J., Müller P., Guck J., and Chan C.J.. 2016. Cell nuclei have lower refractive index and mass density than cytoplasm. J. Biophotonics. 9:1068–1076. 10.1002/jbio.201500273 [DOI] [PubMed] [Google Scholar]
- Semenova I., Burakov A., Berardone N., Zaliapin I., Slepchenko B., Svitkina T., Kashina A., and Rodionov V.. 2008. Actin dynamics is essential for myosin-based transport of membrane organelles. Curr. Biol. 18:1581–1586. 10.1016/j.cub.2008.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Kittisopikul M., Tran J., Goldman A.E., Adam S.A., Zheng Y., Jaqaman K., and Goldman R.D.. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell. 26:4075–4086. 10.1091/mbc.e15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.W., Spinler K.R., Swift J., Chasis J.A., Mohandas N., and Discher D.E.. 2013. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 110:18892–18897. 10.1073/pnas.1304996110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu J.Y., Aires L., Lin Z., and Vogel V.. 2018. Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat. Cell Biol. 20:262–271. 10.1038/s41556-017-0030-y [DOI] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M.D., Xie Z., Gleeson J.G., and Tsai L.H.. 2004. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron. 44:263–277. 10.1016/j.neuron.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Sims J.R., Karp S., and Ingber D.E.. 1992. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J. Cell Sci. 103:1215–1222. [DOI] [PubMed] [Google Scholar]
- Solecki D.J., Trivedi N., Govek E.E., Kerekes R.A., Gleason S.S., and Hatten M.E.. 2009. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 63:63–80. 10.1016/j.neuron.2009.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A. 2009. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 122:577–586. 10.1242/jcs.037622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., and Fridolfsson H.N.. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Banigan E.J., Adam S.A., Goldman R.D., and Marko J.F.. 2017a Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell. 28:1984–1996. 10.1091/mbc.e16-09-0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Liu P.Z., Banigan E.J., Almassalha L.M., Backman V., Adam S.A., Goldman R.D., and Marko J.F.. 2017b Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell. 29:220–233. 10.1091/mbc.E17-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson P.J., Hale C.M., Wirtz D., and Hodzic D.. 2008. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 314:1892–1905. 10.1016/j.yexcr.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.W., Tewari M., et al. . 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341:1240104 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A.S., and Wang N.. 2016. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15:1287–1296. 10.1038/nmat4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada H., Van Thuan N., Reed P., Nelson D., Katoku-Kikyo N., Wudel J., Wakayama T., and Kikyo N.. 2006. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell. Biol. 26:1259–1271. 10.1128/MCB.26.4.1259-1271.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam H.R., Vargas P., Carpi N., Crespo C.L., Raab M., Terriac E., King M.C., Jacobelli J., Alberts A.S., Stradal T., et al. . 2016. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 7:10997 10.1038/ncomms10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D.G., Yenepalli A., Denais C.M., Rape A., Beach J.R., Wang Y.L., Schiemann W.P., Baskaran H., Lammerding J., and Egelhoff T.T.. 2015. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 210:583–594. 10.1083/jcb.201502039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocco V.J., Li Y., Christopher K.G., Matthews J.H., Aggarwal V., Paschall L., Luesch H., Licht J.D., Dickinson R.B., and Lele T.P.. 2018. The nucleus is irreversibly shaped by motion of cell boundaries in cancer and non-cancer cells. J. Cell. Physiol. 233:1446–1454. 10.1002/jcp.26031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.W., Bremner K.H., and Vallee R.B.. 2007. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10:970–979. 10.1038/nn1934 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Lian W.N., Kemal S., Kriegstein A.R., and Vallee R.B.. 2010. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 13:1463–1471. 10.1038/nn.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y., Eibauer M., Goldman A.E., Shimi T., Khayat M., Ben-Harush K., Dubrovsky-Gaupp A., Sapra K.T., Goldman R.D., and Medalia O.. 2017. The molecular architecture of lamins in somatic cells. Nature. 543:261–264. 10.1038/nature21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler C., and Shivashankar G.V.. 2017. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 18:717–727. 10.1038/nrm.2017.101 [DOI] [PubMed] [Google Scholar]
- Uhler C., and Shivashankar G.V.. 2018. Nuclear Mechanopathology and Cancer Diagnosis. Trends Cancer. 4:320–331. 10.1016/j.trecan.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Vaziri A., Gopinath A., and Deshpande V.. 2007. Continuum-based computational models for cell and nuclear mechanics. J. Mech. Mater. Struct. 2:1169–1191. 10.2140/jomms.2007.2.1169 [DOI] [Google Scholar]
- Versaevel M., Grevesse T., and Gabriele S.. 2012. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 3:671 10.1038/ncomms1668 [DOI] [PubMed] [Google Scholar]
- Versaevel M., Braquenier J.B., Riaz M., Grevesse T., Lantoine J., and Gabriele S.. 2014. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep. 4:7362 10.1038/srep07362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten V.L., Ji J.Y., Cummings K.S., Lee R.T., and Lammerding J.. 2008. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 7:383–393. 10.1111/j.1474-9726.2008.00382.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishavkarma R., Raghavan S., Kuyyamudi C., Majumder A., Dhawan J., and Pullarkat P.A.. 2014. Role of actin filaments in correlating nuclear shape and cell spreading. PLoS One. 9:e107895 10.1371/journal.pone.0107895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S.K., Pavin N., Maghelli N., Jülicher F., and Tolić-Nørrelykke I.M.. 2009. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 7:e1000087 10.1371/journal.pbio.1000087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Butler J.P., and Ingber D.E.. 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 260:1124–1127. 10.1126/science.7684161 [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., and Sonnenberg A.. 2005. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171:799–810. 10.1083/jcb.200506083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., and Holzbaur E.L.. 2012. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci. 125:4158–4169. 10.1242/jcs.108688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.H., and Holzbaur E.L.. 2015. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development. 142:218–228. 10.1242/dev.114769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., and Friedl P.. 2013. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Lee K.C., Dickinson R.B., and Lele T.P.. 2011a How dynein and microtubules rotate the nucleus. J. Cell. Physiol. 226:2666–2674. 10.1002/jcp.22616 [DOI] [PubMed] [Google Scholar]
- Wu J., Misra G., Russell R.J., Ladd A.J., Lele T.P., and Dickinson R.B.. 2011b Effects of dynein on microtubule mechanics and centrosome positioning. Mol. Biol. Cell. 22:4834–4841. 10.1091/mbc.e11-07-0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kent I.A., Shekhar N., Chancellor T.J., Mendonca A., Dickinson R.B., and Lele T.P.. 2014. Actomyosin pulls to advance the nucleus in a migrating tissue cell. Biophys. J. 106:7–15. 10.1016/j.bpj.2013.11.4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Chojnowski A., Boudier T., Lim J.S., Ahmed S., Ser Z., Stewart C., and Burke B.. 2016. A-type Lamins Form Distinct Filamentous Networks with Differential Nuclear Pore Complex Associations. Curr. Biol. 26:2651–2658. 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- Yamauchi K., Yang M., Jiang P., Yamamoto N., Xu M., Amoh Y., Tsuji K., Bouvet M., Tsuchiya H., Tomita K., et al. . 2005. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 65:4246–4252. 10.1158/0008-5472.CAN-05-0069 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., and Han M.. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187. 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Graham O.S., Raposo A., and St Johnston D.. 2012. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 336:999–1003. 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R., Antoku S., and Gundersen G.G.. 2017a Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr. Biol. 27:3097–3110. 10.1016/j.cub.2017.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R., Liu C., and Gundersen G.G.. 2017b Nuclear positioning in migrating fibroblasts. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D., Fischer A.H., and Nickerson J.A.. 2004. Nuclear structure in cancer cells. Nat. Rev. Cancer. 4:677–687. 10.1038/nrc1430 [DOI] [PubMed] [Google Scholar]