Abstract
The fission yeast clade, which has a distinct life history from other yeasts, can provide important clues about evolutionary changes. To reveal these changes the large S. cryophilus supercontigs were assembled into chromosomes using synteny relationships and the conserved pericentromeric, subtelomeric genes. Togetherness of the supercontigs was confirmed by PCR. Investigation of the gene order revealed localisation of the rDNA arrays, more than 300 new conserved orthologues and proved that S. cryophilus supercontigs were mosaics of collinear blocks. PFGE analysis showed that size of the S. cryophilus chromosomes differ from the S. pombe chromosomes. Comparative genomic analyses of the newly assembled chromosomes confirmed that the closest relative of S. cryophilus was S. octosporus not just in sequence similarity but also in a structural way, and revealed that preservation of the conserved regions did not arise from the lower number of chromosomal rearrangements. Translocations were more typical in the closely related species, while the number of inversions increased with the phylogenetic distances. Our data suggested that sites of the chromosomal rearrangements were not random and often associated with repetitive sequences, structural- and nucleotide evolution might correlate. Chromosomal rearrangements of the fission yeasts compared to other lineages were also discussed.
Introduction
Although sequencing processes are becoming more and more accurate and fast, assembly of draft genomic sequences remains a serious challenge in many cases, in turn they are necessary to perform extensive and thorough comparative evolutionary studies. These large-scale comparative studies of the ever-increasing numbers of sequences enable us to discover similarities and differences between the genomes, gain insight into genome structures and learn how genomes function and evolve.
Thus, investigation of Haemophilus influenzae and Escherichia coli sequences revealed an important role of the gene shuffling in bacterial evolution1, while comparison of Saccharomyces cerevisiae and Candida albicans genome sequences shed light on the fact that small inversions could be common forms of the chromosomal rearrangements2. Other analyses helped us to identify rapidly evolving genes3,4, while comparison of Hemiascomycetes whole genome sequences drew our attention to numerous interesting features, like mechanisms of a single gene-, segmental- and whole genome duplications or showed that nucleotide and structural evolution depend on two different molecular clocks reviewed in5. Moreover, a novel form of evolution (mesosynteny) was also identified by studying filamentous fungi genomes6. A genome and proteome sequence comparison of the Schizosaccharomyces pombe and S. cereviaise provided insight into the functional similarities and differences between the budding and fission yeasts7, while a study of Schizosaccharomyces species showed that the fission yeasts could have unusually stable genome structures8.
As Schizosaccharomyces species (Schizosaccharomyces pombe, S. japonicus, S. octosporus and S. cryophilus) have haploid genomes and distinct life history from other yeasts, this clade can provide an attractive model for the genome evolution studies, which was hindered by lack of the assembled S. cryophilus chromosomes. In order to expand our knowledge obtained from previous analyses, the aim of this project was to assemble the S. cryophilus large supercontigs into chromosomes based on the data available and use these chromosomes for comparative genomic studies. Accordingly, we suggest here a hypothetical genome assembly based on synteny relationships and validated by molecular experiments. Finally, the newly assembled S. cryophilus genome was used for comparative analyses, which revealed important features of the Schizosaccharomyces genomes.
Results
Assembly of the S. cryophilus supercontigs based on synteny and the conserved pericentromeric- and subtelomeric genes revealed that S. cryophilus supercontigs were mosaics of collinear blocks belonging to the different chromosomes of its related species
Since the S. cryophilus database (Broad) contained only supercontigs (Scs) and no chromosomes, which would be necessary to perform extensive comparative evolutionary studies with the S. cryophilus genome, we decided to assemble those Scs. Earlier results showed that gene order and gene content were remarkably conserved in genomes of the related fission yeast species8, thus we supposed that identification of locally collinear blocks (LCBs)(conserved regions of the chromosomes) could allow us to set order of the S. cryophilus largest Scs. To identify the LCBs, sequence alignments were carried out by Mauve program using the closely related S. octosporus and S. pombe DNA sequences as reference genomes. In the first alignments, the S. cryophilus Scs were in default order (1–9) and orientation (Supplementary Fig. S1a,b), while later we reordered the Scs with Mauve by using the synteny relationships (Supplementary Fig. S1c,d). These alignments revealed that the S. cryophilus Scs are mosaics of LCBs belonging to different chromosomes of the related species (Fig. S1). At the same time these alignments showed that “automatic” reordering of the Scs based on global synteny could not provide a reliable order. Namely, completely different Sc orders were obtained depending on the reference genomes (Sc order: Sc5,3,2,6,8,9,1,7,4 - with S. pombe reference genome and Sc4,9,5,8,7,2,6,3,1-with S. octosporus reference genome) (Supplementary Fig. S1c,d). Consequently, we tried to reveal the true order and orientation of the Scs by manual identification of the pericentromeric and subtelomeric genes based on earlier results that higher degree of conservation were typical in these regions8–12. Thus, the translated sequences of 70 S. pombe and S. octosporus pericentromeric- and 180 subtelomeric genes were collected and their putative orthologues in S. cryophilus genome were identified by BLASTp program. Our results showed that pericentromeric- and subtelomeric gene orders were highly conserved also in the S. cryophilus genome (Supplementary Tables S1 and S2).
Although we failed to find the corresponding orthologues of every subtelomeric genes (Supplementary Table S2), the successfully identified S. cryophilus genes enabled us to find those Scs which contained subtelomeric genes. Based on these findings, we concluded that Sc3, Sc4, Sc6 and Sc7 could have subtelomeric ends, while the pericentromeric genes of S. cryophilus could be found on the Sc2, Sc3, Sc4, Sc5, Sc7, Sc9 (Supplementary Table S1). tRNA genes, which tend to be located close to the centromeres13,8 were also found on these supercontigs.
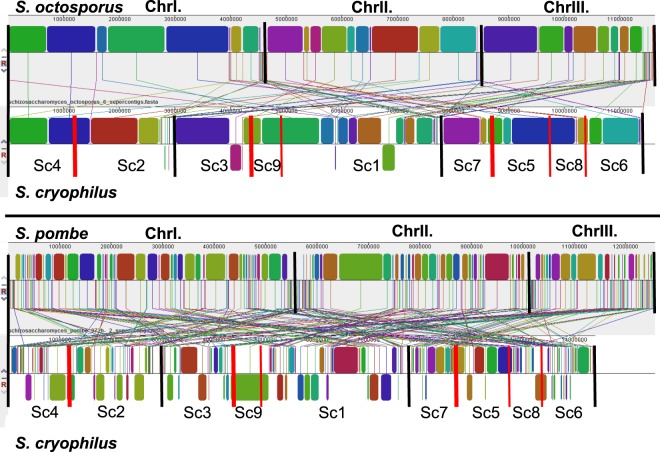
Considering these data, additional Mauve alignments were carried out which suggested the following Sc order: Sc4-Sc2; Sc3-Sc9-Sc1 and Sc7-Sc5-Sc8-Sc6 (Fig. 1). This state of the hypothetic assembly seemed to be reliable and also shed light on further neighbouring contigs, such as Sc9-Sc1; Sc5-Sc8, Sc8-Sc6 (Fig. 1).
Figure 1.
Pairwise whole genome alignments of the species created by Mauve49 aligner, where the newly established supercontig (Sc) order of S. cryophilus was used. The large overlapping locally collinear blocks (LCBs) between the adjacent Scs provide support for the current Sc order. The alignments also indicate that S. cryophilus closest relative is S. octosporus. Colourful rectangles represent LCBs. Sizes of the rectangles are proportional to the genomic extensions of LCBs. LCBs below a genome’s centre line are in inverted orientation relative to the reference genome. Vertical black lines indicate the chromosome boundaries, vertical red lines show the inner supercontig boundaries of S. cryophilus. Wider red lines indicate positions of the centromeric regions in the case of S. cryophilus.
Validation of the neighbouring supercontigs by PCR
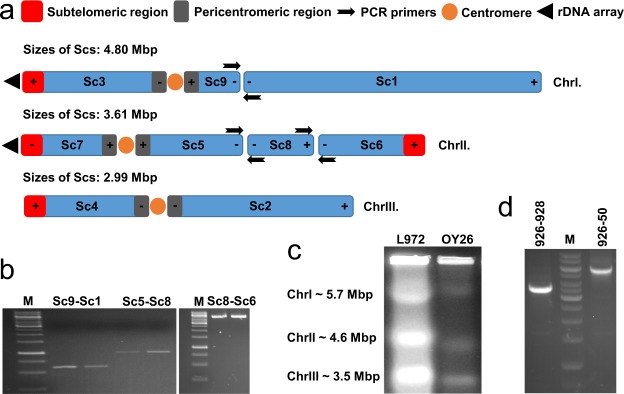
To validate adjacency of the neighbouring Scs (Sc9-Sc1; Sc5-Sc8, Sc8-Sc6) suggested by the last Mauve alignment (Fig. 1), PCR amplifications were carried out with sequence specific primers (Table 1). Primers were designed to hybridize to the corresponding contig ends of the concerning Scs (Fig. 2a). After optimisation of the PCR parameters, we managed to amplify those PCR products which confirmed togetherness of the Sc9-Sc1, Sc5-Sc8 and Sc8-Sc6 (Fig. 2b). PCR fragments were also approved by sequencing (Genbank accessions: MH605091- MH605096, Supplementary File S1). Concatenated chromosome sequences of S. cryophilus in fasta format are available in Supplementary Files S2–S4.
Table 1.
List of strains and primers used in this study. Sc: supercontig.
| Species | Collection number | Description |
|---|---|---|
| S. pombe | 0–1 | Wild-type strain L972h- |
| S. cryophilus | 6–21; CBS11777 | Wild-type strain |
| Primers | Sequence (5′-3′) | Description |
| For proving relation of supercontigs | ||
| Sc9 Forw_B | tagtttatggccgccacagt | Sc9 and Sc1 |
| Sc1 Rev_B | ccgtctgctttctcagtttg | Sc9 and Sc1 |
| Sc5 Forw_C | gcttcaagctgccacatttt | Sc5 and Sc8 |
| Sc8 Rew_C | gcgatctctttagcatttcca | Sc5 and Sc8 |
| Sc6endF | ggaaataccttttggcgact | Sc6 and Sc8 |
| Sc8startR | ggtctaagggggcagattta | Sc6 and Sc8 |
| For detecting localization of | ||
| NL455 | ggtccgtgtttcaagacgg | ribosomal DNA |
| 18S rDNA1 | tcattacggcggtcctagaa | ribosomal DNA |
| SPOG_04999 | tgttggtgttgatgagcagc | ribosomal DNA |
Figure 2.
(a) Hypothetic chromosomes of S. cryophilus and positions of the primers (black arrows). “+” and “−” indicate the original orientation of the supercontigs available in the corresponding databases. In the case of (+−) the orientation coincides with the default orientation, (−+) refers to inverted orientations. (b) PCR validation of togetherness of Sc9-Sc1 (lane: 1., 2.; primers: 666–667), Sc5-Sc8 (lane: 3.,4.; primers: 668–669) and Sc8-Sc6 (lane: 5.,6.; primers: 794–795) supercontig pairs. M: 1 Kb DNA ladder. (c) Pulsed-field gel electrophoresis of the chromosomal DNA of S. pombe (L972) and S. cryophilus (OY26). (d) PCR validation of localisation of the rDNA arrays on the S. cryophilus chromosomes. M: 1 Kb DNA ladder; 926, 928, 50: PCR primers. Gel photos of (b–d) are cropped, full-length gels are presented in Supplementary Figs S7–S10.
Determination of the sizes of S. cryophilus chromosomes and localisation of the rDNA arrays
To investigate total sizes of the S. cryophilus chromosomes, we carried out a karyotypic analysis which confimed that S. cryophilus had three chromosomes similarly to the related S. pombe (Fig. 2c) (https://www.pombase.org/)8, and revealed that S. cryophilus chromosomes differ in size from the S. pombe chromosomes (Fig. 2c). Since S. cryophilus Scs were mosaics of collinear blocks belonging to the different chromosomes of its closely related species, we classified its chromosomes depending on their sizes (Figs 1 and S1). Thus hereinafter the largest chromosome was designated as ChrI, while the smallest one as ChrIII, similarly to S. pombe chromosomes (Fig. 2c). Consequently, S. cryophilus ChrI seemed to be larger in the karyotypic analysis than S. pombe ChrI (5.7Mbp). S. cryophilus ChrII and ChrIII were smaller than 4.6 Mbp and 3.5 Mbp, respectively (Fig. 2c). At the same time, we have to mention that differences between calculated length of the coherent Scs (Fig. 2a) and real sizes of the chromosomes found in the karyotypic analysis (Fig. 2c) could arise from the lack of certain chromosomal regions, such as unplaced small contigs (~0,2Mbp overall), centromeres, telomeres, or the unknown localisation of rDNA arrays.
Since extensions of rDNA arrays can significantly influence sizes of the chromosomes, we wanted to establish the possible locations of 18S-5,8S-28S rDNA genes on the S. cryophilus chromosomes. Based on synteny between S. octosporus and S. cryophilus we managed to find orthologues of the S. octosporus rDNA genes in the subtelomeric region of Sc7. This data could also be confirmed by PCR (Fig. 2d), as PCR fragments were successfully amplified with primers 928–926 and 926–50 (928 binds to the 18 S subunit, 50 binds to the 28 S subunit (D1/D2 domain), while 926 binds to the gene SPOG04999 which is the closest one to the rDNA array according to the sequence file). A further rDNA array was supposed to be on the Sc3 by synteny investigations of the S. octosporus rDNA close genes (Supplementary Table S2, Supplementary Fig. S2), which might correlate with the large size of S. cryophilus ChrI observed in the PFGE analysis (Fig. 2c).
Nucleotide sequence similarity and gene order comparisons confirmed that S. cryophilus closest relative was S. octosporus
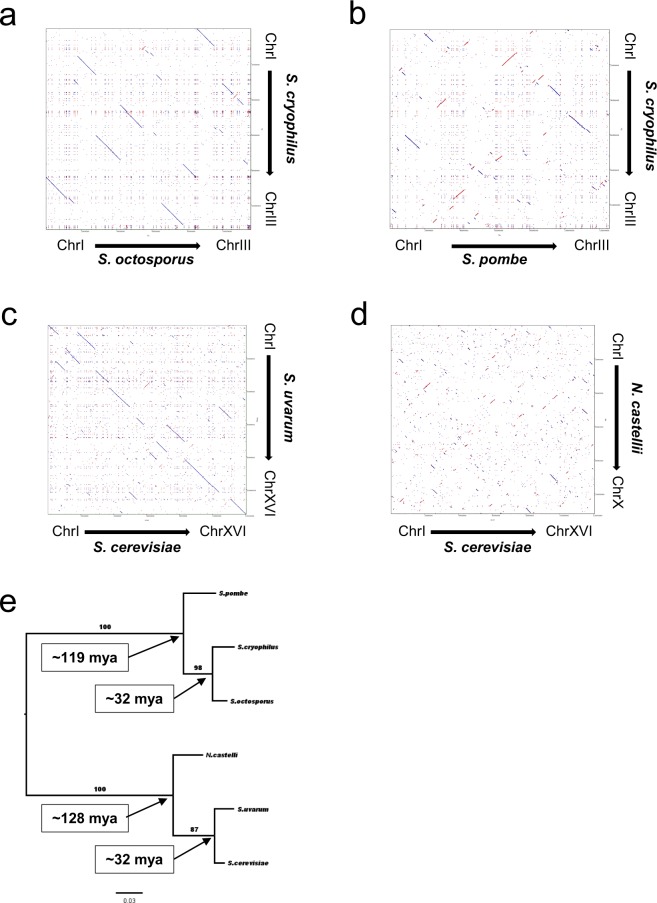
To gain more information on the relation of the species, the common putative orthologues shared by S. pombe, S. octosporus and S. cryophilus were manually identified (Supplementary Table S3). As a result, we managed to find 4580 1:1:1 putative orthologous genes (89–92% of the whole gene content of the three species), in contrast to 4218 genes shared by the four fission yeast species8. This was in a good agreement with the phylogenetic distances of the species. Synteny analyses of these common orthologous proteins confirmed the close relation of S. cryophilus and S. octosporus8,14 (Supplementary Fig. S3), which was also supported by additional DNA level studies. Namely, we created whole genome dot-plots using the concatenated DNA sequences of the species and the newly established Sc order of the S. cryophilus genome with different parameters (E = 0; E < 1.0E-30; alignment size >1000 nt) (Supplementary Table S4). These dot-plot alignments revealed more consecutive homologous DNA sequences between S. cryophilus and S. octosporus (Fig. 3a, Supplementary Fig. S4a) than between S. cryophilus and S. pombe (Fig. 3b, Supplementary Fig. S4b) regardless of the level of strictness. Statistical analyses of the pairwise alignments also proved that S. cryophilus closest relative was S. octosporus (Fig. 4, Supplementary Table S4).
Figure 3.
Dot-plot alignments created with YASS50 (E < 1.0E-30) using the concatenated whole genome sequences of the species. The alignments clearly show that interchromosomal translocations were more frequent than inversions in short evolutionary terms. Blue lines represent homologous segments in the same orientation, while red lines represent inverted segments. (a) S. octosporus and S. cryophilus alignment; (b) S. pombe and S. cryophilus alignment; (c) S. cerevisiae and S. uvarum alignment; (d) S. cerevisiae and N. castelli alignment. First species located to the horizontal axis and the latter located to the vertical axis. High resolution pictures of these alignments are available in Supplementary Fig. 4(a–d). (e) Phylogenetic relations of the concerning species. Values at the branches are statistical supports come from 100 bootstrap replicates. Numbers in the boxes indicate the dates of divergences8. Mya: million years ago.
Figure 4.
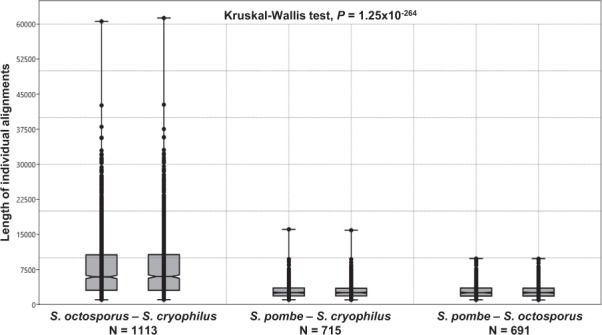
Length distributions of individual DNA alignments created by YASS50 (E = 0). Box plots indicate the 25–75 percent quartiles. Horizontal lines within the boxes show the medians of the samples, notches indicate the 95 percent confidence intervals for the medians. Minimal and maximal values are depicted by the whiskers, plots on the whiskers show individual values. N: sample size. Alignments of S. octosporus-S. cryophilus are significantly different from the other alignments (Kruskal-Wallis test, P = 1.25 × 10−264). Pairwise statistics (Mann-Whitney U tests) are presented in Supplementary Table 3.
Study of chromosomal rearrangements showed that interchromosomal translocations were more frequent than inversions in the closely related species, while the number of inverted segments became higher with the increasing phylogenetic distances
Hereinafter, we were interested in the chromosomal rearrangements of S. cryophilus. To obtain information about their number and types, we examined the created whole genome dot-plots and compared them with certain Saccharomycotina genomes. These dot-plot alignments shed light on that the frequency of translocations (that are mainly interchromosomal translocations) was higher compared to the frequency of inversions in the closest relatives than in the distantly related species. While number of the inverted sequences became higher with increasing phylogenetic distances (Fig. 3a,b). We observed the same tendency in the alignments of S. pombe and S. octosporus (Supplementary Fig. S6a,b). This latter observation was also supported by GRIMM analysis15, which could estimate the minimal number of changes in the lineages. As we were interested only in gross chromosomal changes, we used manually selected LCBs larger than 20.000 nucleotides extracted from Mauve alignments. According to optimal rearrangement scenarios provided by GRIMM, 7 translocations and 4 inversions could occur between the closely related S. octosporus and S. cryophilus, 9 and 54 between the distantly related S. pombe and S. cryophilus, and 14 and 53 between S. pombe-S. octosporus, respectively. Interestingly, the same tendency was emerged in those Saccharomycotina species, whose dates of divergences approximately matched with the Schizosaccharomyces species investigated8 (Fig. 4c–e).
Mauve alignments and GRIMM analyses suggested that a higher number of gross chromosomal rearrangements occurred in the fission yeast genomes than in the budding yeast genomes
As reported by Rhind8, conservation of the gene content is significantly higher in Schizosaccharomyces than within Saccharomyces or Kluyveromyces, both of which have much lower amino acid divergence. We assumed that the observed conservation of gene content/order might be in relation to the fact that lower number of rearrangements could occur in the genomes of fission yeast. Thus, to compare dynamics of the genome evolutionary changes of the fission yeasts to the sampled Saccharomycotina species we created Mauve alignments (Supplementary Fig. S4e,f) and performed GRIMM rearrangement analyses with all of the extracted LCBs regardless of their size (Table 2).
Table 2.
Comparisons of multi chromosomal distances (MCDs) among the species.
| Number of Chrs | Multi Chromosomal Distance (MCD) | Gross changes/all changes percentages | ||||||
|---|---|---|---|---|---|---|---|---|
| per whole genomes | per chromosomes | per Megabases | ||||||
| all changes | gross changes | all changes | gross changes | all changes | gross changes | |||
| So - Scry | 3 -3 | 46 | 11 | 15.33 | 3.67 | 3.97 | 0.95 | 24% |
| Sp - Scry | 3-3 | 150 | 63 | 50.00 | 21.00 | 12.50 | 5.25 | 42% |
| Su - Scer | 16-16 | 72 | 5 | 4.50 | 0.31 | 6.10 | 0.42 | 7% |
| Nc - Scer | 10–16 | 607 | 102 | 37.94 | 6.38 | 51.88 | 8.72 | 17% |
The data indicate that the chromosomes of fission yeasts bore more gross rearrangements than the chromosomes of budding yeasts. Values were estimated by GRIMM15 using the data of LCBs extracted from the pairwise whole genome alignments created by Mauve aligner49. Since GRIMM estimates optimal rearrangement scenarios by transforming one genome to another via rearrangement events, the given values in the table correspond to one genome. For example MCD values per chromosomes in the case of So – Scry correspond to 3 chromosomes not 6. So: S. octosporus; Scry: S. cryophilus; Sp: S. pombe; Su: S. uvarum; Scer: S. cerevisiae; Nc: N. castelli. Chrs: chromosomes. All changes mean that we considered every rearrangement events regardless of the sizes and positions of the concerning LCBs. While in the case of gross changes we excluded the subtelomeric regions because these are inclined to undergo rearrangements and just the LCBs > 20 000 nucleotides were considered.
Based on optimal rearrangement scenarios, the multichromosomal distances (MCDs) (by means of the number of changes that possibly occurred) were proved to be higher in the Saccharomycotina species than in the fission yeasts when we considered all the changes possibly occurred (Table 2). These findings might suggest that fewer chromosomal rearrangements occurred in the genomes of the fission yeasts than in the budding yeast. However, according to an alternative analysis, where we excluded the subtelomeric regions, because these segments are inclined to undergo rearrangements and used only LCBs larger than 20000 nucleotides showed different result. That is, considering only the gross changes we could find less chromosomal rearrangements in all pairs of species and our data coincided with the findings of Fischer in the case of S. cerevisiae and S. uvarum16,17 (Table 2). Later, numbers of the gross changes were compared to the chromosome numbers and sizes of the genomes, and these ratios clearly suggest that more gross rearrangements happened per chromosome (or per megabase) in the fission yeasts genomes (Table 2). That is, individual chromosomes of the fission yeasts bore many more large scale translocations and inversions than chromosomes of the budding yeasts (Table 2).
Breakpoint analyses suggested that sites of chromosomal rearrangements could not be random
In order to obtain information on the sites of chromosomal rearrangements, we identified the chromosomal breakpoints between large LCBs (>20 000 nts) in the YASS and Mauve alignments. To ensure that breakpoints were correctly revealed, we examined both S. octosporus – S. cryophilus and S. cryophilus – S. octosporus alignments. The analyses revealed 19 breakpoints. In the next step, the genes located to the edges of the LCBs were identified and mainly 5S rDNA genes were found (5S rDNAs were associated with 12 breakage sites from the 19) (Supplementary Fig. S5). Thus, we assumed that rearrangements could happen along these repeated sequences. A similar data was found earlier, where inversion endpoints were correlated with repeated sequences18,19. In other cases, the rearrangements occurred in large intergenic regions (>1000 nts).
The random breakage model of chromosomal evolution considers distribution of lengths between breakpoints and supposes that lengths of LCBs between rearrangements should be exponentially distributed20–22. To reveal trends in chromosomal evolution of the fission yeasts we also computed distribution of the lengths of LCBs between rearrangements and observed in the S. octosporus – S. cryophilus analysis that the concerning values did not show agreement with the random breakage model (Fig. 5), in contrast to some Aspergillus species23. However, values of the more distantly related S. pombe – S. cryophilus and of S. pombe – S. octosporus did not diverge largely from the model prediction (Fig. 5).
Figure 5.
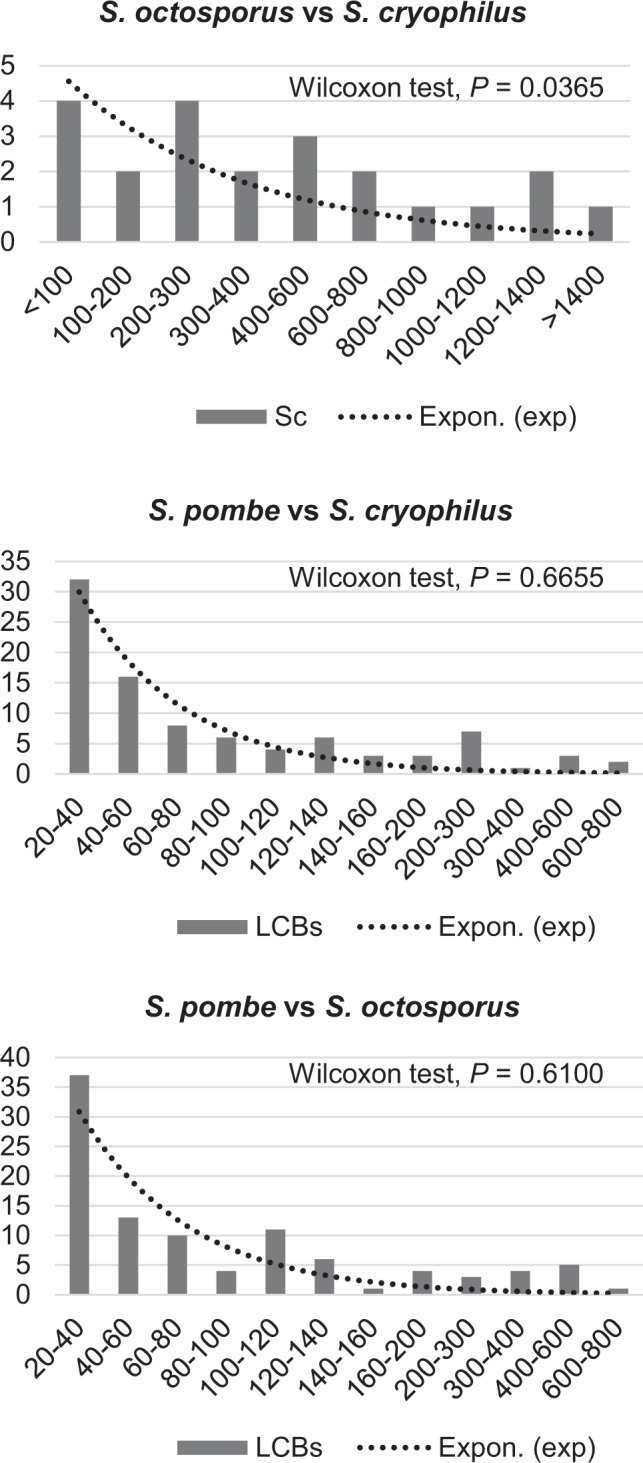
Comparison of histograms of LCBs’ lengths to random breakage model of chromosomal evolution (dashed lines). According to random breakage model distances between breakpoints should follow an exponential distribution of the form f(x) = 1/L e − x/L, where L is the average size of all syntenic segments. Numbers on the horizontal axes indicate the distribution of the lengths of LCBs in Kb. Values of the vertical axes show the frequency of LCBs at a given length. Analysis of S. octosporus – S. cryophilus proved that the concerning values did not show agreement with the random breakage model. However, values of S. pombe – S. cryophilus and of S. pombe – S. octosporus did not diverge largely from the model prediction.
Structural and sequence evolution in the fission yeast genomes might be correlated
Later, structural and sequence evolution in the fission yeasts was investigated in two considered scenarios. In the first case - depending on the Mauve alignments - we found 224 LCBs in S. cryophilus (Fig. 1) and 226 LCBs in S. octosporus using S. pombe as reference genome (Supplementary Fig. S6), while estimation of MCDs established by GRIMM showed 150 for S. cryophilus and 156 for S. octosporus (Table 3). Based on these findings, the overall rate of genome reorganization seemed to be almost the same in the two different lineages.
Table 3.
Comparison of sequence and structural evolution of the fission yeasts.
| Amino acid identity8 | Mauve analysis | Manual analysis | |||
|---|---|---|---|---|---|
| Mauve LCBs | MCD | Manual LCBs | MCD | ||
| S. cryophilus - S. octosporus | 85.00% | 59 | 46 | 26 | 11 |
| S. pombe - S. cryophilus | 66.40% | 224 | 150 | 111 | 63 |
| S. pombe - S. octosporus | 65.60% | 226 | 156 | 112 | 67 |
The results suggested that structural differences might correlate (Pearson’s r = 0.99; P = 0.0176) with the established amino acid divergence (1-identity). Values of amino acid identity originated from8. Mauve LCBs: locally collinear blocks (conserved regions of the chromosomes) established by Mauve49. Manual LCBs: manually selected LCBs larger than 20 000 nucleotides. MCD: multi-chromosomal distance (by means of number of changes occurred) estimated by GRIMM15.
Later we examined the previous results of GRIMM analysis (submission of the LCBs larger than 20 000 nucleotides). The overall MCDs were 63 in the S. cryophilus and 67 in the S. octosporus lineage (Table 3). These results suggested that structural differences might correlate (Pearson’s r = 0.99; P = 0.0176) with the established amino acid divergence8 (Table 3), similarly to certain vertebrates, nematodes and arthropods24–26 and differently from the Aspergillus species23. However, genome evolution (either sequence or structural) of S. octosporus seems to be somewhat faster than genome evolution of S. cryophilus (Table 3).
Discussion
As genome sequencing has become less expensive, thousands of genome projects have been launched recently. However, performing a genome assembly with good quality is still a serious challenge27. Consequently, many genomes remained in the state of draft genomes, which could be sufficient for certain experimental studies, but not for the analysis of large scale genomic changes28,29.
Since fission yeasts have a distinct life history from other yeasts30,31, share important biological features, such as chromosome structure and metabolism, G2/M cell cycle control, cytokinesis, or the spliceosome components with metazoans32,33, reviewed in34, and they have haploid chromosome sets, these species (S. pombe, S. octosporus, S. japonicus, S. cryophilus) can provide an attractive model for genome evolution studies.
In order to introduce the genome of the recently described species S. cryophilus14 into comparative genomic analyses of the fission yeasts and study of the chromosomal changes, we decided to assemble the S. cryophilus large Scs into chromosomes. Thus, genome sequence alignments, BLAST searches and investigation of the synteny relationships were carried out with the S. cryophilus Scs. Their results revealed that S. octosporus is the closest relative of S. cryophilus (Figs 1, 3 and 4, Supplementary Figs S1, S3 and S4), which was in good agreement with earlier data obtained from protein sequences alignments and investigation of rRNA genes8,14.
These results also revealed that S. cryophilus Scs were mosaics of collinear blocks belonging to the different chromosomes of its related species (Supplementary Fig. S1, Fig. 1) and the subtelomeric- and pericentomeric genes were conserved between S. cryophilus and S. octosporus (Supplementary Tables S1 and S2, Supplementary Fig. S2), similarly to its relatives and several Saccharomyces species8,9. These conserved genes and the following Mauve alignments allowed us to determine order of the Scs. They suggested the following order: Sc3-9-1 (ChrI), Sc7-5-8-6 (ChrII) and Sc4-2 (ChrIII) (Fig. 1). Togetherness of Sc9-Sc1, Sc5-Sc8-Sc6 was also proved by PCR reactions (Fig. 2a,b) and confirmed by the re-sequencing of the S. cryophilus genome35. At the same time, sequencing of the S. cryophilus centromers suggested an exchange in the localisation of Sc7 and Sc435.
The Scs investigated belonged to three chromosomes, as the Pulsed Field Gel Electrophoresis proved it. However the S. cryophilus chromosomes had different sizes compared to S. pombe chromosomes (Fig. 2c). Since calculated length of the coherent Scs differed from the real size of the chromosomes (Fig. 2a,c), we assumed that there were unassembled and unidentified regions of the S. cryophilus chromosomes. To reduce the missing regions, we tried to find positions of the rDNA arrays, which often located in subtelomeric regions and their extensions could exceed 1Mbp32,36,37. According to synteny analysis of the sequences deposited on Broad, one rDNA array could be found on Sc7 (ChrIII35)(Fig. 2a), which was confirmed also by PCR reactions (Fig. 2d). While study of the gene content and order in the regions located next to the S. octosporus rDNA arrays shed light on a further S. cryophilus rDNA array on the Sc3 (ChrI) (Supplementary Table S2, Supplementary Fig. S2, Fig. 2a). These localisations are in good agreement with the re-sequencing data35, which also has revealed a third array on the ChrII and further atypical centromere-proximal rDNA repeats35.
Later, the genome conservation and chromosomal rearrangements were investigated using the newly established S. cryophilus chromosomes. Since the reordering of genetic elements could occur by different mechanisms, we primarily wanted to learn what kind of rearrangements formed the current S. cryophilus chromosome structure. YASS and Mauve analyses revealed a high number of chromosomal rearrangements, which were mainly interchromosomal translocations in the closely related species (Figs 1 and 3a, Supplementary Fig. S1). At the same time, the whole genome alignments also showed that the number of inversions increased with phylogenetic distance, which was also supported by GRIMM analysis (Fig. 3b). Interestingly, these data were not fission yeast-specific, since similar tendency was obtained from those Saccharomycotina species, whose dates of divergences approximately matched the Schizosaccharomyces species investigated (Fig. 3c,d,e). These data arose the question, whether the interchromosomal translocations could be more sustainable than inversions in short evolutionary terms? Since effects of both rearrangements types can be extensive, as they can change the gene expression pattern of a genome38, consequently they can lead to elevation of the fitness in certain environments38,39 or even reproductive isolation40,41, this possibility does not seem probable. Instead, we suppose that the underlying mechanisms generating rearrangements were responsible for the greater frequency of translocations. Accordingly, a decreasing number of translocations between the distantly related species do not necessarily originate from less translocation events; rather we suppose that the greater number of inversions tend to blur the traces of interchromosomal translocation events2.
Study of genomes of the related species8 and our earlier sequence alignments shed light on the highly conserved gene orders of the fission yeasts (Fig. 1, Supplementary Figs S2 and S3). Thus, we could suppose that number of chromosomal rearrangements were low, which could preserve the large LCBs. In contrast, our analyses suggested that more gross chromosomal changes could occur in the genomes of S. cryophilus and S. octosporus than in S. cerevisiae and S. uvarum (Table 2). Moreover, the breakpoint analysis pointed out that 5S rDNAs were often associated with breakage sites (Supplementary Fig. S5). These latter results resembled those data where rearrangement endpoints were correlated with repeated sequences18,19,42. The random breakage model prediction also supported that the chromosomal rearrangements in these two fission yeasts species probably did not occur randomly (Fig. 5). At the same time we have to take note that this trend seemed to be less obvious with increasing phylogenetic distances (Fig. 5). Consequently, we assume that inner regions of the large LCBs could contain a lower number of those sequences which predisposing to DNA breakage or recombination. This idea might be supported by data of other studies, where these large structural variations mainly occurred in positions of the low gene density regions43. Furthermore, repetitive sequences, which are inclined to attract insertions of the transposons that can also cause changes in the genome, seemed to be situated rather in the centromere or telomere regions35. That is, LCBs can be under stronger selection pressure. Moreover, the 3D architecture of the genomes could also contribute to the highly conserved gene order reviewed in44.
Besides this, we proved that the structural and sequence evolution in the fission yeast genomes might be correlated with the previously established amino acid divergences8 (Table 3), similarly to certain vertebrates, nematodes and arthropods24–26 and differently from the Aspergillus species23. However, a slight difference was discernible between S. octosporus and S. cryophilus compared to S. pombe, since genome evolution of the former might be faster (Table 3).
Taken together, we propose here a hypothetical assembly of the S. cryophilus Scs, whose comparative genomic analyses provided insights into genome evolution of the haploid Schizosaccharomyces species.
Materials and Methods
Yeast strains and media
The strains used in this study are listed in Table 1. Compositions of the rich culture media were the following: YPL: 2% glucose, 1% Scharlau casein tryptic peptone, 1% yeast extract, pH 6.7–6.9. YPA: YPL + 2% agar. YEL: 1% yeast extract (Scharlau, 07-079-500), 3% glucose (VWR). YEA: YEL + 2% agar. S. pombe cells were cultured at 30 °C, while S. cryophilus was incubated at 25 °C.
DNA isolation and PCR amplification
Genomic DNA was isolated from exponential-phase yeast cultures grown either in YEL or in YPL with the glass bead method45. These genomic DNA and the primers listed in Table 1 were used in the PCR reactions. Since certain Scs contained overlapping sequences at their ends, the PCR primers were designed to hybridize outside these overlapping regions. Parameters for PCR reactions were optimised individually for each reaction. Parameters of Sc adjacency validation and rDNA amplification: 95 °C-3 min; 95 °C-30 sec; 54 °C-30 sec; 72 °C-3-5 min (steps 2–4 were repeated 30X); 72 °C-10 min. Gel electrophoresis was carried out in 1% agarose gel in 1xTBE buffer. Gels were stained with ethidium bromide and photos were taken by UV-Transilluminator (UVP Bio-Doc-It Imaging System). Gel photos were cropped in Microsoft Office PowerPoint 2013.
Pulsed-field electrophoresis of the chromosomal DNA
Chromosomal preparations were obtained as described previously46. The samples (chromosomes in 1.5% LM agarose) were placed into the wells of 1% agarose gel. 0.5 × TBE cooled to 14 °C was used as a buffer. Electrophoresis was carried out on the CHEF-DR III apparatus (Bio-Rad) at 50 V in the following mode: (1) 48 h 2400 sec; (2) 70 h 3000 sec; and (3) 24 h 3300 sec41. After electrophoresis, the gel was stained with ethidium bromide, washed in distilled water and photographed with Olympus C-4000 Zoom digital camera under UV light. Background and contrast of gel photo was adjusted in and was cropped in Microsoft Office PowerPoint 2013.
Bioinformatics
Genome sequence data
The nucleotid sequences of Schizosaccharomyces pombe (L972 h−), S. octosporus (yFS286) and S. cryophilus (OY26) were downloaded from the database of Broad Institute (http://www.broadinstitute.org/annotation/genome/schizosaccharomyces_group/MultiDownloads.html), whose data were relocated in the meantime to the FungiDB (http://fungidb.org/fungidb/). Individual chromosome sequences with annotations were downloaded from NCBI with the following accession numbers: CU329670, CU329671 and CU329672 for S. pombe, KE503206, KE503207 and KE503208 for S. octosporus, KE546988, KE546989, KE546990, KE546991, KE546992, KE546993, KE546994, KE546995 and KE546996 for the contigs of S. cryophilus. The annotated files were imported to the SnapGene Viewer software (http://www.snapgene.com/products/snapgene_viewer). Chromosome sequences of Saccharomyces cerevisiae (S288C) were downloaded from Saccharomyces Genome Database (http://www.yeastgenome.org/download-data/sequence), S. bayanus var. uvarum (CBS7001) sequences were obtained from (http://www.saccharomycessensustricto.org)47. Individual chromosome sequences of Naumovozyma castelli (CBS 4309) were downloaded from GenBank with the following accessions: HE576752-HE576761.
BLAST analyses and sequence comparison
BLASTp search was performed in the website of Broad Institute (http://www.broadinstitute.org/annotation/genome/schizosaccharomyces_group/MultiHome.html) with the following parameters: E value: 1e-3, matrix: BLOSUM62 and BLOSUM45 and default parameters were used for the others. After the retirement of the Schizosaccharomyces website at Broad Institute, NCBI BLASTp search (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) and BLASTp search at PomBase (http://genomebrowser.pombase.org/Multi/Tools/Blast?db=core) were performed with standard parameters. We used the sequences of S. pombe as reference to identify the putative orthologues of S. octosporus and S. cryophilus. To make sure the results are reliable, reciprocal BLAST analyses were also carried out. Beside the sequence similarities, genes in the neighbourhood and predicted protein domains were also considered in the orthology inference. We ignored the single genes, only orthologues within synteny blocks were considered. To perform pairwise alignment a Needleman-Wunsch algorithm was used at the website of EMBL-EBI (http://www.ebi.ac.uk/Tools/psa/emboss_needle/nucleotide.html)48.
Whole genome alignments and rearrangement analyses
Whole genome alignments were generated with Mauve aligner using the progressive Mauve algorithm with standard parameters except minimum LCB weight, which was adjusted to 4049. Whole genome dot-plots were created with YASS (http://bioinfo.lifl.fr/yass/yass.php)50 with the following parameters: E value: 1.0E-30; X-drop: 50; window range: 100–200000; window incr.: 2X; hit criterion: double and default parameters were used for the others. For the nucleotide comparison we extracted the individual alignments in tabular form from all three pairwise alignments (S. octosporus – S. cryophilus, S. pombe – S. cryophilus, S. pombe – S. octosporus), but we considered only the statistically most significant (E value: 0) alignments and filtered out the non-syntenic repetitive regions like 5S RNAs, tRNAs and high copy number genes. The number of large scale inversions and translocations between the compared genomes were estimated with GRIMM v2.01 (http://grimm.ucsd.edu/cgi-bin/grimm.cgi)15.
Synteny analyses
Shared synteny of the subtelomeric genes were presented with the online tool Genome Synteny Viewer GSV (http://cas-bioinfo.cas.unt.edu/gsv/homepage.php)51 using the manually curated list of putative orthologues. Visualizations of whole genome syntenic relationships were displayed using the OrthoClusterDB online platform with the following parameters: order and strandedness: -r -s, synteny block size lower bound: 2, upper bound: 2000 and default parameters were used for the others (http://genome.sfu.ca/cgi-bin/orthoclusterdb/runortho.cgi)52.
Breakpoint analyses and breakage model
Chromosomal breakpoints were determined in the whole genome alignments (either in Mauve or in YASS) and inspected manually. Breakpoint associated sequences were extracted from the generated dot-plots and were identified in the corresponding annotated sequence files using SnapGene Viewer. To determine whether conserved LCBs in fission yeasts follow random breakage, the distribution of lengths of syntenic regions between large rearrangements were analysed. According to the random breakage model distances between breakpoints should follow an exponential distribution of the form f(x) = 1/L e−x/L, where L is the average size of all syntenic segments20,23.
Phylogenetic tree construction
Phylogenetic tree was created at the website of Phylogeny.fr (http://www.phylogeny.fr/)53 using the concatenation of 3 evolutionarily conserved protein sequences of the concerning species (Supplementary Table S5). The sequences were submitted to a manually adjusted workflow consisting of MUSCLE for alignment, GBLOCKS for curation of the alignment and PhyML with WAG substitution model for phylogeny. The number of substitution rate category was adjusted to 4, gamma distribution parameter and proportions of invariable sites were both estimated. Branch support was estimated from bootstrap analysis (100 replicates). The tree was displayed with FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analyses
Normal distributions of the data were tested by Shapiro-Wilk and Anderson-Darling tests. Since most of our datasets proved not to be normally distributed, Kruskal-Wallis test was used for multiple comparison followed by pairwise Mann-Whitney U as post-hoc test. Wilcoxon signed rank test was used in the case of related pairwise datasets. Correlation of the data was tested by linear Pearson correlation test. P values were considered significant below the alpha level 0.05. All statistical analyses were performed in PAST v.3.20 software (https://folk.uio.no/ohammer/past/)54 and in Microsoft Office Excel 2013.
Electronic supplementary material
Acknowledgements
We thank Anita Kovács for technical assistance and Edina Karanyicz for the help in PFGE. This work was partly supported by the European Union and the European Social Fund through project EFOP-3.6.1-16-2016-00022 and Higher Education Institutional Excellence Programme of the Ministry of Human Capacities in Hungary, within the framework of the Biotechnology thematic programme of the University of Debrecen.
Author Contributions
Conceived and designed the study: L.Á.Sz., S.M. and I.M. Performed the bioinformatics analyses: L.Á.Sz. Implemented PCR analysis: L.A.P. Implemented PFGE analysis: Z.A. Analysed the data: L.Á.Sz. and I.M. Contributed reagents and materials: S.M. Wrote the paper: L.Á.Sz. and I.M. All authors read and approved the manuscript.
Data Availability
The sequences generated during the current study are available in the GenBank repository with the following accession numbers: MH605091- MH605096. Other data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32525-9.
References
- 1.Tatusov RL, et al. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with. Escherichia coli. Curr Biol. 1996;6(3):279–91. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 2.Seoighe C. Prevalence of small inversions in yeast gene order evolution. Proc Natl Acad Sci USA. 2000;97(26):14433–14437. doi: 10.1073/pnas.240462997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maguire SL, et al. Comparative Genome Analysis and Gene Finding in Candida Species Using CGOB. Mol Biol Evol. 2013;30(6):1281–1291. doi: 10.1093/molbev/mst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haerty W, et al. Evolution in the Fast Lane: Rapidly Evolving Sex-Related Genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dujon B. Yeast evolutionary genomics. Nature Reviews Genetics. 2010;11:512–24. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 6.Hane JK, et al. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 2011;12:R45. doi: 10.1186/gb-2011-12-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood, V. Schizosaccharomyces pombe comparative genomics; from sequence to systems. In Topics in Current Genetics 15 (eds) Sunnerhagen, P. P. & Piskur, J. Springer Verlag, 10.1007/4735_97 (2006).
- 8.Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabre E, et al. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing and subtelomeres. Mol Biol Evol. 2005;22(4):856–873. doi: 10.1093/molbev/msi070. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germline. Nature reviews, Molecular Cell Biology. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res. 2014;24(10):1650–64. doi: 10.1101/gr.172122.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellermeier C, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn RM, Clarke L, Carbon J. Clustered tRNA genes in Schizosaccharomyces pombe centromeric DNA sequence repeats. Proc Natl Acad Sci USA. 1991;88(4):1306–1310. doi: 10.1073/pnas.88.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helston RM, Box JA, Tang W, Baumann P. Schizosaccharomyces cryophilus sp. nov., a new species of fission yeast. FEMS Yeast Res. 2010;10:779–786. doi: 10.1111/j.1567-1364.2010.00657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- 16.Fischer G, James SA, Roberts IN, Oliver SG, Louis EJ. Chromosomal evolution in Saccharomyces. Nature. 2000;405:451–454. doi: 10.1038/35013058. [DOI] [PubMed] [Google Scholar]
- 17.Fischer G, Neuvéglise C, Durrens P, Gaillardin C, Dujon B. Evolution of gene order in the genomes of two related yeast species. Genome Res. 2001;11:2009–2019. doi: 10.1101/gr.212701. [DOI] [PubMed] [Google Scholar]
- 18.Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15(1):1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe CJ. The endpoints of an inversion in wheat chloroplast DNA are associated with short repeated sequences containing homology to att-lambda. Curr Genet. 1985;10(2):139–145. doi: 10.1007/BF00636479. [DOI] [PubMed] [Google Scholar]
- 20.Nadeau J, Taylor B. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci USA. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q, Pevzner PA, Tesler G. The fragile breakage versus random breakage models of chromosome evolution. PLoS Comput Biol. 2006;2:e14. doi: 10.1371/journal.pcbi.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker TS, Lenhard B. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genom. 2007;278:487–491. doi: 10.1007/s00438-007-0287-0. [DOI] [PubMed] [Google Scholar]
- 23.Galagan JE, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 24.Burt DW, et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- 25.Coghlan A, Wolfe KH. Fourfold faster rate of genome rearrangement in nematodes than in. Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharakhov IV, et al. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- 27.Bradnam KR, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Giga Science. 2013;2(1):10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics. 2005;21:4320–4321. doi: 10.1093/bioinformatics/bti769. [DOI] [PubMed] [Google Scholar]
- 29.Sims GE, Jun SR, Wu GA, Kim SH. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proc Natl Acad Sci USA. 2009;106:2677–2682. doi: 10.1073/pnas.0813249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sipiczki Matthias. Genome Biology. 2000;1(2):reviews1011.1. doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forsburg SL. The best yeast? Trends Genet. 1999;15:340–344. doi: 10.1016/S0168-9525(99)01798-9. [DOI] [PubMed] [Google Scholar]
- 32.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z. Big data mining powers fungal research: recent advances in fission yeast systems biology approaches. Curr Genet. 2017;63(3):427–433. doi: 10.1007/s00294-016-0657-4. [DOI] [PubMed] [Google Scholar]
- 34.Olsson I, Bjerling P. Advancing our understanding of functional genome organisation through studies in the fission yeast. Curr Genet. 2011;57:1–12. doi: 10.1007/s00294-010-0327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong P. et al. Inter-species conservation of organisation and function between non-homologous regional centromeres. bioRxiv 10.1101/309815.
- 36.Pasero P, Marilley M. Size variation of rDNA clusters in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol Gen Genet. 1993;236:448–452. doi: 10.1007/BF00277147. [DOI] [PubMed] [Google Scholar]
- 37.Brown WRA, et al. A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 (Bethesda) 2011;1:615–626. doi: 10.1534/g3.111.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avelar AT, Perfeito L, Gordo I, Ferreira MG. Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nat. Commun. 2013;4:2235. doi: 10.1038/ncomms3235. [DOI] [PubMed] [Google Scholar]
- 39.Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanders SE, et al. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife. 2014;3:e02630. doi: 10.7554/eLife.02630.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naumov GI, Kondratieva VI, Naumova ES. Hybrid sterility of the yeast Schizosaccharomyces pombe: genetic genus and many species in statu nascendi? Microbiology (Moscow) 2015;84:159–169. doi: 10.1134/S0026261715010099. [DOI] [PubMed] [Google Scholar]
- 42.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeffares DC, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nature Communications. 2016;8:14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature reviews. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 45.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 46.Naumova ES, Sukhotina NN, Naumov GI. Molecular markers for differentiation between the closely related dairy yeast Kluyveromyces lactis var. lactis and wild Kluyveromyces lactis strains from the European “krassilnikovii” population, Microbiology (Moscow) 2005;74(3):329–335. [PubMed] [Google Scholar]
- 47.Scannell DR, et al. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for Saccharomyces sensus stricto genus. G3 (Bethesda) 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 49.Darling AE, Mau B, Perna NT. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noe L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Research. 2005;33(2):W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Revanna KV, Chiu CC, Bierschank E, Dong Q. GSV: A web-based genome synteny viewer for customized data. BMC Bioinformatics. 2011;12:316. doi: 10.1186/1471-2105-12-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng MP, et al. OrthoClusterDB: an online platform for synteny blocks. BMC Bioinformatics. 2009;10:192. doi: 10.1186/1471-2105-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36(Web Server):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1):9. [Google Scholar]
- 55.O’Donnell, K. Fusarium and its near relatives. In: Reynolds DR, Taylor JW, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International, Wallingford, 225–233 (1993).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences generated during the current study are available in the GenBank repository with the following accession numbers: MH605091- MH605096. Other data generated or analysed during this study are included in this published article (and its Supplementary Information files).