Abstract
The atomic structure of the infectious, protease-resistant, β-sheet-rich and fibrillar mammalian prion remains unknown. Through the cryo-EM method, MicroED, we reveal the sub-1Å resolution structure of a protofibril formed by a wild-type segment from the β2-α2 loop of the bank vole prion protein. The structure of this protofibril reveals a stabilizing network of hydrogen bonds that link polar zippers within a sheet, producing motifs we name ‘polar clasps’.
Keywords: cryo-EM, electron diffraction, prion, amyloid, protofibril
Summary sentence:
Ultrahigh-resolution cryo-EM structure reveals a prion protofibril stabilized by a dense three-dimensional network of hydrogen bonds.
Main text
Micro electron diffraction (MicroED) is a cryo-electron microscopy (cryo-EM) method that facilitates the determination of atomic structures from sub-micrometer thin protein nanocrystals and fragmented crystallites1–3. Several high-resolution amyloid structures have been determined by MicroED. These include: two structures from the toxic core of the Parkinson’s associated protein, α-synuclein4, two from the type-2 diabetes associated protein, IAPP5, and five ab initio structures 2,6. These add to structures of the fibril cores of α-synuclein7, amyloid-beta8, tau9, and a fungal prion10 recently determined by complementary methods. However, an atomic structure of the infectious, scrapie form of the mammalian prion protein (PrPSc) remains unknown11. PrPSc shares some structural hallmarks of amyloid – appearing as ropelike filaments or rods made of tightly mating beta sheets12,13. However, PrPSc also differs from other amyloids: it resists proteolysis and denaturation, is infectious, and can spread within and between species to cause disease11,14,15.
To evaluate the source of PrPSc stability, we investigated segments in mammalian prion proteins (PrP) that might form the core of PrPSc fibrils. Informed by structure-based prediction of amyloid-prone sequences16 we identified 168QYNNQNNFV176, a segment of the β2-α2 loop of bank vole (Myodes glareolus) PrP (Figure 1), a universal prion acceptor17,18. This segment lies within the predicted cross-beta core of PrP fibrils (Supplementary Fig. 1), shows high conservation in rodents and other mammals (Supplementary Fig. 1), and is rich in asparagines19,20, which may stabilize prion fibrils21. At sub-millimolar concentrations this segment produces highly ordered aggregates (Supplementary Fig. 2) that, when aligned and illuminated by x-rays, produce cross-β diffraction (Supplementary Fig. 2). Like PrPSc, aggregates formed by this segment are resistant to high concentrations of urea, guanidine, and a range of pH, but are sensitive to sodium hydroxide (Supplementary Fig. 3). Given its shared biophysical properties with PrPSc, we labeled our segment proto-PrPSc and set out to uncover the structural basis for its stability.
Figure 1.
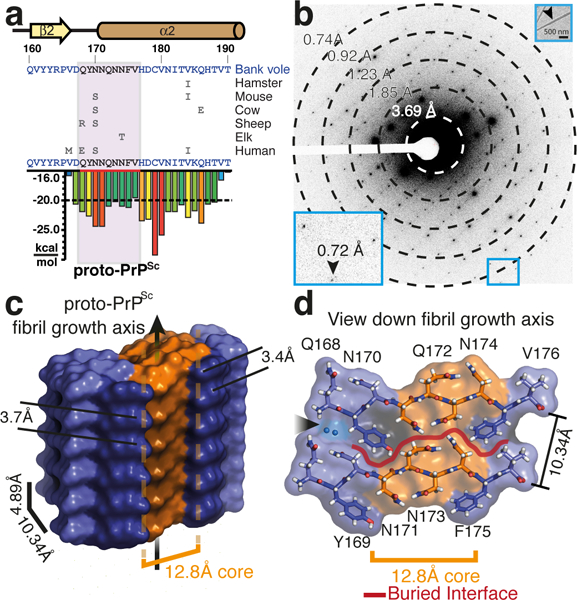
Structure of proto-PrPSc from sub-Å diffraction of nano-scale crystals by MicroED. a, Sequence of the bank vole PrP β2-α2 loop, highlighting local secondary structure and variation in sequence compared to hamster, mouse, cow, sheep, elk, and human. The sequence of proto-PrPSc is highlighted in a plot that indicates propensity for amyloid formation (scored in kcal/mol). The region encoding for proto-PrPSc is shown in magenta. b, Single frame of MicroED data showing strong diffraction to 0.72Å (inset). A second inset shows a proto-PrPSc crystal diffracted by MicroED (arrow). Scale bar 500nm. c, Side and d, top view of an isosurface model of proto-PrPSc. Stacked aromatics are highlighted in dark purple, a highly constrained core region in orange, two waters are noted with a black arrow, and a red line outlines the interface between sheets in panel d.
From optimized microcrystals (Supplementary Fig. 4), we determined a micro-focus x-ray diffraction structure of proto-PrPSc to 1.1Å resolution by molecular replacement (Supplementary Fig. 4 and Supplementary Table 1). Conditions in which we observed microcrystals of proto-PrPSc also produced showers of nanocrystals, evident in electron micrographs (Figure 1). These nanocrystals diffracted to 0.72Å by MicroED (Figure 1). Merging diffraction from multiple crystals we achieved a high completeness, 0.75Å resolution dataset (Supplementary Table 1). From these data, we obtained an ab initio solution that was overall similar to our microfocal x-ray diffraction structure of proto-PrPSc (Supplementary Fig. 4) and suitable for atomic refinement (Supplementary Fig. 5). This ultrahigh resolution MicroED structure of proto-PrPSc shows features invisible in the x-ray structure and critical to our understanding of its stability.
The structure of proto-PrPSc reveals a prion protofibril with amyloid like features: beta strands arranged parallel and in register as a class two steric zipper12 in which sheets pair front to back (Figure 2). Two tightly mating curved sheets make up the proto-PrPSc fibril (Figure 2), although side chains in these sheets interdigitate less in proto-PrPSc than is observed in conventional amyloid structures12. Although sheets stack in a parallel, face to back configuration, the convex face of one sheet nestles against the concave face of its neighbor, approximately 10.3Å away, with a high degree of surface complementarity (Sc, 0.807) (Figure 2). The interface between these sheets is large, concealing 204.5Å2 per strand, and is entirely devoid of waters at its core (Figure 1). Atoms in the MicroED structure of proto-PrPSc are extremely well ordered, with an average B-factor of 6.0Å2 overall and 2.8Å2 within its core (Supplementary Fig. 6 and Supplementary Table 1). These values are less than half the overall B-factor in our x-ray diffraction structure of proto-PrPSc (Supplementary Table 1), confirming a greater degree of order in our nanocrystallites compared to larger microcrystals of the same segment.
Figure 2.
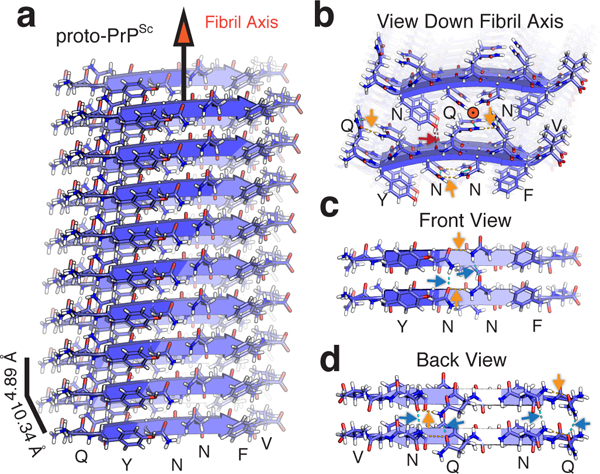
Filament structure of proto-PrPSc and its stabilizing network of hydrogen bonds. a, Structure of proto-PrPSc determined by MicroED shown as an assembly of ten chains. Distances between strands and sheets, and the fibril growth axis are labeled. b, A view down the fibril axis highlighting the interface between adjacent sheets. An orange circle denotes the direction of the fibril axis. c, Front and d, back views of the fibril with two strands representing a sheet. Polar contacts are labeled as orange arrows for intra-chain and blue arrows inter-chain.
A three-dimensional network of hydrogen bonds stabilizes proto-PrPSc (Supplementary Table 2): hydrogens at its core participate in intra-residue C5 bonds22 (Figure 3), and asparagine and glutamine residues stack along its fibril axis (Figure 2). Glutamines and asparagines in proto-PrPSc form networks of hydrogen bonds reminiscent of proton wires23, polar ladders24, and the polar zippers first proposed by Max Perutz25,26 (Supplementary Fig. 7). Neighboring polar ladders in proto-PrPSc are additionally linked by hydrogen bonds within a strand (Figure 3), a motif we refer to as a ‘polar clasp’ (Supplementary Fig. 7). Stacks of phenylalanine and tyrosine residues shield clasps at the core of proto-PrPSc (Figures 1 and 2). The importance of this aromatic embrace is underscored by a lack of clasps in structures of shorter segments from this region of PrP that lack tyrosine 16912,27. On this evidence, we hypothesize that polar clasps and stacked aromatic residues act in concert to stabilize proto-PrPSc, as they might for PrPSc.
Figure 3.
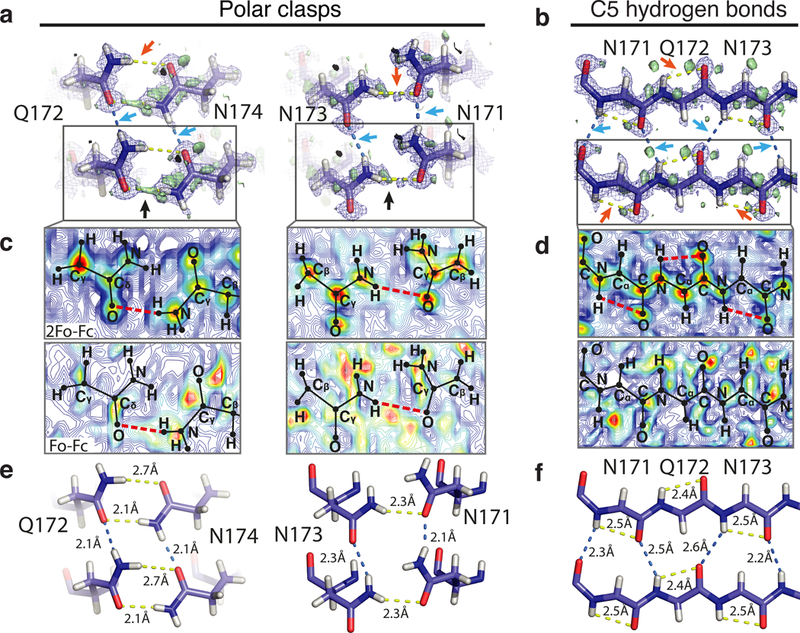
Sub-Å cryo-EM maps show stabilizing hydrogen bonds in proto-PrPSc. a, Polar clasps formed by hydrogen bonds that link Q172 to N174, and N171 to N173, and b, C5 hydrogen bonds (orange arrows) in the context of backbone hydrogen bonds at the core of proto-PrPSc. Bridging density can be seen in both the 2Fo-Fc and Fo-Fc maps (blue mesh and green isosurface respectively) for a subset of the hydrogen bonds (black arrows). Contour maps show 2Fo-Fc or Fo-Fc density for c, polar clasps or d, C5 bonds. Contours are averaged over a ~1.5Å thick volume with high contour values are shown in red, low values in blue, and hydrogen bonds as dashed red lines. Distances for both intra (yellow lines) and inter (blue lines) chain hydrogen bonds are highlighted in e, polar clasps or f, C5 bonds.
Our hypothesis of proto-PrPSc stability relies on the locations of hydrogen atoms throughout the structure. Hydrogens in the MicroED structure of proto-PrPSc are unambiguously assigned, informed by pronounced difference density in ultrahigh resolution maps (Supplementary Figs. 5, 8, and 9 and Supplementary Table 4). Hydrogens at the core of proto-PrPSc are as evident as those seen in structures of small organic compounds determined by electron diffraction28, including our own structure of carbamazepine (Supplementary Fig. 10 and Supplementary Table 3). Density in ultrahigh resolution maps of proto-PrPSc suggests hydrogen may occupy positions that deviate from idealized geometry (Supplementary Fig. 5). Improved hydrogen positions indicated by ultrahigh resolution maps in MicroED could bolster the accuracy of calculations based on observed hydrogen bond networks.
Ultrahigh resolution maps of proto-PrPSc mirror features that appear in electron density maps of the highest resolution x-ray structures and show similar residual density, which in the latter is assigned to valence electrons29 (Supplementary Fig. 9). In addition, in polar clasps at the core of proto-PrPSc, density is seen surrounding donor hydrogens and oxygen acceptors (Figure 3 and Supplementary Figs. 5 and 9). This density is not explicitly accounted for by our atomic model during refinement, and is not evident in maps of other small molecules determined by MicroED or in lower resolution maps of proto-PrPSc (Supplementary Figs. 8 and 10, and Supplementary Table 4). However, this density is preserved across maps produced by refinement programs with different scattering factor libraries (Supplementary Fig. 11). A full account of the complex features present in ultrahigh resolution cryo-EM maps may require advances in refinement that incorporate molecular vibrations, crystal bending, non-kinematic scattering and chemical bonding29–33.
Through MicroED, cryo-EM now surpasses the one ångstrom barrier and yields the structure of proto-PrPSc, a denaturant-resistant prion protofibril. Cryo-EM maps of proto-PrPSc at 0.75Å resolution reveal a three-dimensional network of stabilizing hydrogen bonds that link residues between and within its beta strands through polar clasps.
Materials and Methods
Phylogenetic and Sequence Analysis of bank vole PrP 27–30
The top 130 most similar wild type sequences to bank vole PrP 90–231 were generated using NCBI BLAST. Of these 130 sequences, all sequences containing the proto-PrPsc peptide were selected and aligned.
Characterization of proto-PrPSc peptide
The synthetic peptide representing sequence 168QYNNQNNFV176 of the bank vole prion protein was purchased from GenScript (Piscataway, NJ). The peptide used for our experiments was received at greater than 98% purity achieved by reverse phase HPLC. The peptide was qualified on a Bruker (ultrafleXtreme) MALDI-TOF/TOF mass spectrometer, reported as m/z (intensity, arbitrary units). The spectrum has a mass list that includes a [M+H]+ peak at 1141.3 Da (expected 1140.5 Da), a [M+Na]+ peak at 1163.4 Da (expected 1162.49 Da), and a [M+K]+ peak at 1179.3 Da (1178.46 Da).
Characterization of carbamazepine
Lyophilized powder of carbamazepine (5H-dibenzo[b,f]azepine-5-carboxamide, C15H12N2O) with purity > 99% was purchased from Sigma-Aldrich (St Louis, MO) and crystallized without further purification.
Aggregation of proto-PrPSc
Proto-PrPSc peptide was solubilized in ultrapure water at 0.22–1.75 mM. 50 μL of each sample was added to a 96-well clear flat bottom plate in triplicate and evaluated for aggregate formation by reading absorbance at 350 nm on an Infinite M1000 Pro plate reader (Tecan). Readings were measured immediately after solubilization, after three hours, and after six hours of shaking at 900 rpm at 37o C. Wells were imaged after six hours using a Leica M205 C light microscope (Leica Microsystems) and after three and six hours by electron microscopy as described below. Electron microscope images are representative of more than five images taken at each concentration and time point.
Fibril diffraction from proto-PrPSc aggregates
Solutions containing aggregates of proto-PrPSc were clarified by centrifugation. Pelleted aggregates were resuspended in a concentrated volume in water, applied between two pulled capillaries and left to dry overnight. Oriented aggregates formed between the capillary ends, were resupplied with additional solution containing aggregates, and left to dry again. This was repeated several times to grow the bulk of aligned aggregates on a capillary. Aligned aggregates were then diffracted using 5 minute exposures to a FRE+ rotating anode generator with VARIMAX HR confocal optics producing Cu K-α radiation (Rigaku, Tokyo, Japan) and detected using a RIGAKU R-AXIS HTC imaging plate detector at a distance of 156 mm from the source.
Chemical denaturation of proto-PrPSc aggregates
Proto-PrPSc was solubilized at approximately 3.5 mM and allowed to form aggregates. This solution was diluted to approximately 1:4 before treatment with either 0.5 – 6.0 M urea, 0.5–4.5 M guanidinium-HCl, 0.75 M HCl, 0.1 M citrate pH 2 or 4, 0.1M Sodium/potassium phosphate pH 6 or 8, 0.1 M CAPS pH 10, or 0.75 M NaOH. Aggregate content was measured by absorbance compared to a control solution consisting of 0.01% (w/v) 1um latex spheres in water. Spectra across the visible range (250–700 nm) were collected using a Nanodrop One (Thermo). A single point of reference, OD at 600 nm, was used for comparison across sampled conditions.
Growth of proto-PrPSc crystals
Peptide was weighed out as powder and dissolved in ultrapure water at near maximum solubility, 3.5 mM. Crystals were grown at room temperature by the hanging-drop method in a 96-well Wizard screen. Crystals appeared in various conditions, and were further optimized in 24-well hanging-drop vapor diffusion experiments. The best crystals of proto-PrPSc grew within 24 hours at a peptide concentration of 1.75 mM in the presence of 0.1 M 2-(N-morpholino) ethanesulfonic acid (MES), pH 6.0 and either 10% ethanol or 10% 2-Methyl-2,4-pentanediol (MPD).
Transmission electron microscopy
Approximately two microliters of aggregated proto-PrPSc were applied to 300-mesh formvar-carbon coated grids (Ted Pella Inc., Redding, Ca.) for two minutes before excess liquid was removed and grids left to dry. Grids were imaged either on a Tecnai T12 or F20 electron microscope (Thermo-Fisher; formerly FEI). Samples were imaged at a magnification of 2100× with a dose rate of less than 30 e-/Å2.
Microfocus x-ray data collection
Crystals grown in 0.1M MES pH 6.0 and 10% ethanol and mixed with 100% glycerol as cryoprotectant were harvested from 24-well hanging drops using MiTeGen loops and flash frozen in liquid nitrogen. Seventy-three diffraction images were collected, each spanning an 3o wedge, from a single crystal at a temperature of 100K at the advanced photon source (APS) beamline 24-ID-E, equipped with an ADSC Q315 CCD detector, using a 10 μm beam with a 0.98 Å wavelength.
Microfocus x-ray data processing and structure determination
Diffraction images collected from a single crystal of proto-PrPSc were indexed and integrated in Denzo, yielding a dataset with 80.75% overall completeness at 1.1 Å resolution in space group P1. A suitable molecular replacement solution was obtained from this data using the PHASER program and an idealized beta strand nonapeptide alanine model as a probe. The model was refined using REFMAC, against the measured data to a final Rwork/Rfree of 0.14/0.16.
MicroED sample preparation
Nano-scale needle crystals of proto-PrPSc were grown in batch in 0.1 M MES, pH 6.0 and 10% ethanol. Crystals were diluted in mother liquor and fragmented by force of pipetting to create an approximately monodisperse solution of crystals. Carbamazepine was crystallized in batch by dilution into neat isopropanol at 100 μg/mL. Two microliters of either solution were placed on a holey carbon grid (1/4, 2/2, 2/4, #300 copper; Ted Pella Inc.) prior to plunge freezing into liquid ethane and transferring into liquid nitrogen for storage. Grids were held by a liquid nitrogen cooled Gatan 626 cryo-holder for transfer into and manipulation within the electron microscope.
MicroED data collection
MicroED data collection from nine sub-micron thick needle crystals and a single sub-micron carbamazepine crystal was performed as previously described34. Briefly, crystals of either proto-PrPSc or carbamazepine lying in a frozen-hydrated state on holey carbon grids were inspected visually in overfocused diffraction mode on either a cryo-cooled FEI Tecnai F20 microscope operated at 200 kV (Janelia Research Campus), or a Titan environmental TEM operated at 300 kV (Environmental Molecular Sciences Lab, PNNL). Diffraction patterns used for structure determination were collected on a TVIPS TemCam-F416 CMOS detector in rolling-shutter mode with three second exposures while proto-PrPSc crystals were unidirectionally rotated at a constant rate of 0.27o s−1 over angular wedges ranging from −55o to +72o. A single carbamazepine crystal was rotated at a speed of 0.2o s−1 over an angular wedge ranging between −45 o to +45 o with 5 second exposures. Beam intensity was held constant, with an average dose rate of 0.003–0.005 e- Å −1 sec−1, corresponding to a total dose of ~1–3 e- Å−2 per data set. We used a camera length of 520 mm, the equivalent of a sample to detector distance of 950 mm in a corresponding lensless system. All diffraction was performed using a circular selected area aperture of ~1 μm2 in projection.
MicroED data processing
Diffraction movies were converted to the SMV file format using TVIPS tools as previously described35. Indexing and integration were performed in XDS36. Integrated diffraction intensities from partial data sets of nine different proto-PrPSc crystals were sorted and merged in XSCALE36. Merged intensities were converted to amplitudes at various resolution cutoffs to produce separate 1.1Å, 1.0Å, 0.9Å, 0.8Å, and 0.75Å data sets. Ab initio structure determination was performed on each of these data sets using SHELXD37. Phases obtained from the atomic assembly generated by direct methods were used to produce maps of sufficient quality for subsequent model building in Coot38 and refinement in Phenix39 using electron scattering form factors to produce a final structure in space group P 1 with a final Rwork/Rfree of 24/25. Refinement in REFMAC was carried out in parallel to a final Rwork/Rfree of 23/25. The structure refined in Phenix (PDB ID 6AXZ) was used in all subsequent analysis and is shown in figures. Ab initio structure determination for carbamazepine was performed in SHELXT37 in which a solution was found in space group P 21m with no errors in chemical assignment or atom positions for all carbon, nitrogen, and oxygen atoms. This solution was refined in SHELXL37 using electron form factors to an R value of 21.8%. A structure with hydrogen positions refined to best match difference density in the map lowered the R value to 19.8%.
Analysis of buried surface area (Sa) and surface complementarity (Sc) for proto-PrPSc
The structure of proto-PrPSc was used to calculate both Sa and Sc from an assembly consisting of two sheets generated by translational symmetry, each consisting of ten stacked beta strands. Sa was computed as an average of the buried surface area per chain in our assembly, calculated as the difference between the sum of the solvent accessible surface area of the two sheets and the solvent accessible surface area of the entire complex, divided by the total number of strands in both sheets. Sc was calculated using the CCP4 suite for all points at the interface between the two aforementioned sheets.
Comparison of x-ray and MicroED structures
Sequence alignment and structural superposition of both an individual chain and an assembly of two sheets from x-ray and MicroED structures of proto-PrPSc were performed in PyMOL40. This alignment produced an all atom R.M.S.D. of 0.162Å on which structural similarity was assessed.
Analysis of aromatic residues in proto-PrPSc
Distances were measured between stacked aromatic residues that flank the core of proto-PrPSc. Aromatics stacked between strands along planes separated by 3.7 or 3.4Å for Y169 or F175, respectively, in a parallel-displaced configuration 41. We measured the inter-sheet hydrogen bond created by Y169 to the backbone carbonyl of N171 on an opposing strand. Both Y169 and F175 formed aromatic ladders that channel polar residues into a 12.8Å long region at the core.
Analysis of hydrogen bond networks
Informed by the locations of hydrogens, visible in our maps at a 0.7–2.5 σ range, we evaluated hydrogen bond networks determined by the program HBplus (v.3.06)42 and by manual inspection using PyMOL. We measured asparagine ladders between residues N170, N171, N173, and N174 and the corresponding asparagine residues on the strands above and below. We also measured glutamine ladders between residues Q168 and Q172 and their corresponding residues on strands above and below. We analyzed distances associated with intra-strand hydrogen bonds formed by three pairs of residues: Q168-N170, N171-N173 and Q172-N174. Polar contacts were also measured between Q168-N170, with a 3.1 Å donor-acceptor (D-A) distance and 2.3Å H-O distance, Q172-N174, with a 3.0 Å D-A distance and 2.1Å H-O distance and N171-N173, with a 3.1Å D-A distance and 2.3Å H-O distance. In each case, the linked residues faced the same side of the beta strand and bridged residues at positions [N/Q]i and [N]i+2 within the strand. While the HBplus program does not identify C5 hydrogen bonds in our structure, we measured these bonds in residues N171, Q172 and N173, based on the criteria that carbonyl oxygens bond with intra-residue amide protons if their geometry permits, with H-O distances shorter than 2.5Å22.
Amylome profiling and informatic analysis of polar clasps
A subset of the predicted amylome was analyzed16, consisting of six residue segments found to score favorably when threaded onto a template based on the structure of the yeast prion NNQQNY. We chose all segments that scored two standard deviations better than the mean score for all peptides (Z-score >2). This subset of six residue segments represents 95,381 out of 7,900,599 total segments, or 1.2% of all possible segments of this size in the human proteome. For each segment, we searched for a [Y/F] X [N/Q] X [N/Q] X [Y/F] motif across the region of the protein to which the segment belonged; a 10-resiue window including two residues upstream and downstream of a profiled segment. Protein identifiers were obtained from the RefSeq database for each of the 35 catalogued segments. Where a segment appeared in more than one protein, each was counted independently.
Calculation of contour maps
2Fo-Fc and Fo-Fc density maps were calculated from the final refined MTZ file using the FFT tool in CCP4. Maps were converted to MRC format in Chimera and imported into MATLAB. Contour plots were calculated such that the number of contours spanned the minimum to the maximum values of the maps with intervals of one standard deviation (σ) between contour levels.
Data Availability
Atomic coordinates and structure factors for proto-PrPSc have been deposited in the Electron Microscopy and Protein Data Banks as EMD-7017, 6AXZ respectively. Source data for all figures and files is available from the authors upon reasonable request, please see author contributions for specific datasets. A life sciences reporting summary for this article is available.
Supplementary Material
Acknowledgments:
We thank Colin Ophus (Molecular Foundry-NCEM), John Miao (UCLA) and Hosea Nelson (UCLA) for helpful discussions. This work is supported by DOE Grant DE-FC02–02ER63421, the EICN in the CNSI at UCLA, the Janelia Research Visitor Program, and the NE-CAT beamline 24-ID-E, funded by NIH-NIGMS P41 GM103403. This work was also supported by STROBE: A National Science Foundation Science and Technology Center under Grant No. DMR 1548924. G.F.H. is supported by BEC.AR program, Fundación Bunge y Born, Williams, and René Barón, and is a member of CONICET. M.G.J is supported by a QCB Collaboratory Postdoctoral Fellowship. J.A.R. is supported as a Searle Scholar and a Beckman Young Investigator. T.G. and D.S.E are supported by the Howard Hughes Medical Institute (HHMI); JEE and IVN by the DOE-BER Molecules to Mesoscale Bioimaging Project FWP #66382.
Footnotes
Competing interests: The authors declare no competing interests.
References and Notes:
- 1.Shi D, Nannenga BL, Iadanza MG & Gonen T Three-dimensional electron crystallography of protein microcrystals. eLife 2, e01345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Cruz MJ et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat. Methods 14, 399–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez JA, Eisenberg DS & Gonen T Taking the measure of MicroED. Curr. Opin. Struct. Biol. 46, 79–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez JA et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krotee P et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. eLife 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya MR et al. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc. Natl. Acad. Sci. 201606287 (2016). doi: 10.1073/pnas.1606287113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuttle MD et al. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat. Struct. Mol. Biol. 23, 409–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt M et al. Peptide dimer structure in an Aβ(1–42) fibril visualized with cryo-EM. Proc. Natl. Acad. Sci. 112, 11858–11863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick AWP et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature (2017). doi:10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasmer C et al. Amyloid Fibrils of the HET-s(218–289) Prion Form a β Solenoid with a Triangular Hydrophobic Core. Science 319, 1523–1526 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez JA, Jiang L & Eisenberg DS Toward the Atomic Structure of PrPSc. Cold Spring Harb. Perspect. Biol. a031336 (2017). doi:10.1101/cshperspect.a031336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawaya MR et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg D & Jucker M The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKinley MP, Bolton DC & Prusiner SB A protease-resistant protein is a structural component of the Scrapie prion. Cell 35, 57–62 (1983). [DOI] [PubMed] [Google Scholar]
- 15.Kurt TD & Sigurdson CJ Cross-species transmission of CWD prions. Prion 10, 83–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldschmidt L, Teng PK, Riek R & Eisenberg D Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. 107, 3487–3492 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts JC et al. Evidence That Bank Vole PrP Is a Universal Acceptor for Prions. PLOS Pathog 10, e1003990 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurt T et al. The molecular basis for cross-species prion transmission. FASEB J. 30, 814.7–814.7 (2016). [Google Scholar]
- 19.Halfmann R et al. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol. Cell 43, 72–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zambrano R et al. PrionW: a server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res. 43, W331–W337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurt TD et al. Asparagine and glutamine ladders promote cross-species prion conversion. J. Biol. Chem. jbc.M117.794107 (2017). doi:10.1074/jbc.M117.794107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newberry RW & Raines RT A prevalent intraresidue hydrogen bond stabilizes proteins. Nat. Chem. Biol. 12, 1084–1088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagle JF & Morowitz HJ Molecular mechanisms for proton transport in membranes. Proc. Natl. Acad. Sci. U. S. A. 75, 298–302 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoder MD, Lietzke SE & Jurnak F Unusual structural features in the parallel β-helix in pectate lyases. Structure 1, 241–251 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Perutz MF, Staden R, Moens L & De Baere I Polar zippers. Curr. Biol. CB 3, 249–253 (1993). [DOI] [PubMed] [Google Scholar]
- 26.Perutz MF, Johnson T, Suzuki M & Finch JT Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. U. S. A 91, 5355–5358 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiltzius JJW et al. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palatinus L et al. Hydrogen positions in single nanocrystals revealed by electron diffraction. Science 355, 166–169 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Hirano Y, Takeda K & Miki K Charge-density analysis of an iron-sulfur protein at an ultra-high resolution of 0.48 Å. Nature 534, 281–284 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Glaeser RM & Downing KH High-resolution electron crystallography of protein molecules. Ultramicroscopy 52, 478–486 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Zhong S, Dadarlat VM, Glaeser RM, Head-Gordon T & Downing KH Modeling chemical bonding effects for protein electron crystallography: the transferable fragmental electrostatic potential (TFESP) method. Acta Crystallogr. A 58, 162–170 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Jelsch C et al. Accurate protein crystallography at ultra-high resolution: Valence electron distribution in crambin. Proc. Natl. Acad. Sci. 97, 3171–3176 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuo JM, Kim M, O’Keeffe M & Spence JCH Direct observation of d-orbital holes and Cu–Cu bonding in Cu2O. Nature 401, 49–52 (1999). [Google Scholar]
- 34.Shi D et al. The collection of MicroED data for macromolecular crystallography. Nat. Protoc. 11, 895–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hattne J, Shi D, de la Cruz MJ, Reyes FE & Gonen T Modeling truncated pixel values of faint reflections in MicroED images. J. Appl. Crystallogr. 49, 1029–1034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W XDS. Acta Crystallogr . D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheldrick GM A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD et al. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delano W The PyMOL Molecular Graphics System. (Schrödinger LLC; ). [Google Scholar]
- 41.McGaughey GB, Gagné M & Rappé AK π-Stacking Interactions ALIVE AND WELL IN PROTEINS. J. Biol. Chem. 273, 15458–15463 (1998). [DOI] [PubMed] [Google Scholar]
- 42.McDonald IK & Thornton JM Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for proto-PrPSc have been deposited in the Electron Microscopy and Protein Data Banks as EMD-7017, 6AXZ respectively. Source data for all figures and files is available from the authors upon reasonable request, please see author contributions for specific datasets. A life sciences reporting summary for this article is available.


