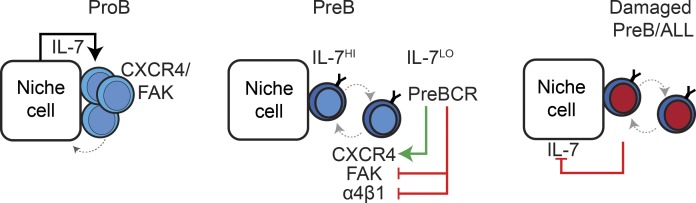
B cell development is characterized by well-defined transitions. Fistonich et al. demonstrate that two distinct cell circuits formed between proB, preB, and IL-7+ cells regulate the size and quality of B cell progenitors and control B cell development.
Abstract
B cell progenitors require paracrine signals such as interleukin-7 (IL-7) provided by bone marrow stromal cells for proliferation and survival. Yet, how B cells regulate access to these signals in vivo remains unclear. Here we show that proB and IL-7+ cells form a cell circuit wired by IL-7R signaling, which controls CXCR4 and focal adhesion kinase (FAK) expression and restricts proB cell movement due to increased adhesion to IL-7+CXCL12Hi cells. PreBCR signaling breaks this circuit by switching the preB cell behavior into a fast-moving and lower-adhesion state via increased CXCR4 and reduced FAK/α4β1 expression. This behavioral change reduces preB cell exposure to IL-7, thereby attenuating IL-7R signaling in vivo. Remarkably, IL-7 production is downregulated by signals provided by preB cells with unrepaired double-stranded DNA breaks and by preB acute lymphoblastic leukemic cells. Combined, these studies revealed that distinct cell circuits control the quality and homeostasis of B cell progenitors.
Graphical Abstract
Introduction
B cell development is highly dependent on IL-7 receptor signaling mediated by IL-7 (Grabstein et al., 1993; Peschon et al., 1994; von Freeden-Jeffry et al., 1995; Puel et al., 1998; Carvalho et al., 2001). At the proB cell stage, IL-7R signals mostly through the JAK1/3 and STAT5a/b pathway to promote survival and proliferation. However, at the preB cell stage, IL-7R signaling plays both positive and negative roles in B cell development. It is still required for preB cell proliferation and anti-apoptotic gene expression (e.g., Bcl2, Bcl2l1, and Mcl1). Yet, it also inhibits Igκ germline transcription through STAT5 binding of Eκi and recruitment of Polycomb repressive complex 2, which results in histone H3 lysine 27 trimethylation and inaccessibility of Jκ and Cκ regions to the RAG proteins (Mandal et al., 2011). Furthermore, IL-7R–induced cyclin D3 expression negatively regulates Vκ transcription, resulting in inhibited RAG protein accessibility to the Vκ genes (Powers et al., 2012). IL-7R signaling in preB cells has also been proposed to repress Rag1 and Rag2 transcription (Johnson et al., 2008) through phosphatidylinositol-3-OH kinase (PI3K) activation, AKT phosphorylation, and Foxo1 inactivation (Ochiai et al., 2012). Thus, IL-7R signaling simultaneously promotes preB cell survival and proliferation while also profoundly inhibiting IgL chain gene rearrangement and developmental progression into the immature B cell stage. How can preB cells then balance positive and negative effects of IL-7R signaling to allow developmental progression in vivo?
Some evidence suggests that preB cell receptor (preBCR) signaling activates IRF4 expression (Thompson et al., 2007). In addition to enabling IgL gene rearrangement, IRF4 promotes CXCR4 expression and increases preB cell migration toward CXCL12 (Johnson et al., 2008). BCR signaling at the immature B cell stage also promotes CXCR4 upregulation and increases B-lineage cell migration to CXCL12 in vitro and in vivo (Beck et al., 2014). Initial attempts to identify bone marrow (BM) stromal cell subsets that express IL-7 and CXCL12, using an anti–IL-7 antibody and Cxcl12GFP/+ reporter mice, concluded that nonhematopoietic CXCL12-GFPHi stromal cells did not express IL-7. Reciprocally, cells detected with an anti–IL-7 antibody were not marked by Cxcl12-driven GFP expression. It was therefore concluded that two distinct stromal cell subsets existed in the BM of adult mice that expressed either IL-7 or CXCL12, but not both (Tokoyoda et al., 2004). In light of these findings, Singh and colleagues proposed that preBCR-mediated upregulation of CXCR4 localizes preB cells near CXCL12-producing cells and, in this manner, lowers IL-7R signaling by positioning preB cells in IL-7low microenvironments (Johnson et al., 2008). This model gained traction because it provided a reasonable explanation for how preB cells coordinate positive and negative roles of IL-7R signaling in vivo (Clark et al., 2014). However, blinded analyses of IL-7–producing cells in tissue sections of wild-type and IL-7–deficient mice stained with an anti–IL-7 antibody failed to reliably identify IL-7+ cells, and, since then, multiple IL-7 reporter mice have been generated (Mazzucchelli et al., 2009). Using Il7GFP/+ knock-in mice and Il7-CFP transgenic mice in combination with Cxcl12dsRed/+ reporter mice, we identified a nonhematopoietic Lepr+ cell subset with mesenchymal progenitor potential that not only expressed IL-7 but also expressed the highest amount of CXCL12 of all BM cells (Cordeiro Gomes et al., 2016). Furthermore, hematopoietic multipotent progenitor cells and common lymphoid progenitor cells are strictly dependent on CXCR4 for optimal IL-7R signaling and consequently for lymphoid lineage development. However, these findings raise the possibility that preBCR signaling would actually promote preB cell localization near CXCL12Hi cells where IL-7–production is highest. Thus, how preB cells regulate this juxtaposition between preBCR and IL-7R signaling to successfully progress into the immature and mature B cell stages still remains enigmatic (Lim et al., 2017).
The behavior of proB and preB cells in vivo is presumably physiologically relevant also in extreme situations such as when B cell precursors cannot repair double-stranded DNA breaks (DSBs). Indeed, unrepaired RAG-mediated DSBs result in the activation of NF-κB and SpiC-controlled gene expression programs that downregulate preBCR signaling components and upregulate the expression of genes involved in lymphocyte migration and adhesion (Bredemeyer et al., 2008; Bednarski et al., 2016). Furthermore, Ikaros-deficient proB cells and both mouse and human Ikaros-deficient BCR-ABL–expressing preB acute lymphoblastic leukemic (preB-ALL) cells are aberrantly adherent to stromal cells (Joshi et al., 2014; Schwickert et al., 2014; Schjerven et al., 2017). Thus, understanding how normal and DSB-damaged proB and preB cells behave in vivo and interact with BM niches can lead to novel insights into B cell development and homeostasis.
Here we show that proB cells are essentially nonmotile and intimately associated with CXCL12+ IL-7+ mesenchymal cells. CXCL12 guides proB cells toward IL-7Hi niches that in turn activate IL-7R signaling to promote CXCR4 and focal adhesion kinase (FAK; encoded by Ptk2) expression. This enforces proB cell/IL-7+ cell interactions and favors IL-7–mediated proB cell expansion. PreBCR signaling then breaks this circuit by further increasing CXCR4 expression while substantially lowering α4β1/FAK-mediated adhesion to VCAM-1. Consequently, preB cells spend little time in direct contact with CXCL12- and IL-7–producing cells, attenuating IL-7R signaling in vivo. Remarkably, IL-7+ niche cells modulate IL-7 production in response to the quality of neighboring preB cells: preB cells with unrepaired DSBs and BCR-ABL+ preB-ALLs instruct IL-7+ niche cells to decrease IL-7 and CXCL12 production, which significantly reduces endogenous B lymphopoiesis. These findings provide evidence supporting a model in which cell circuits operate in the BM to control the number and quality of B cell precursors.
Results
Overlapping BM niches for proB and preB cells
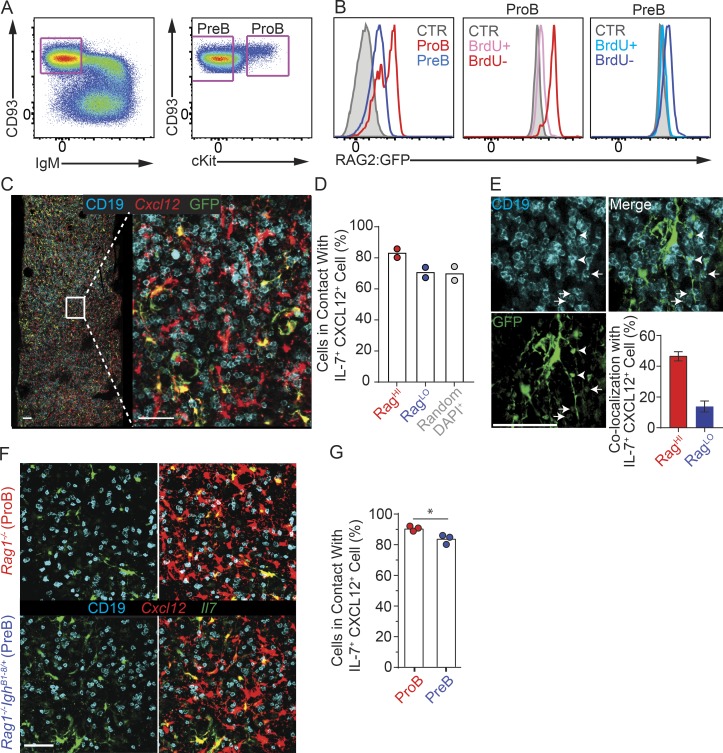
Although proB and preB cells can be readily distinguished by flow cytometry using a variety of cell surface markers (Fig. 1 A), these phenotypic differences were insufficient for their reliable visualization in situ (not shown). To overcome this limitation, we took advantage of the fact that proB cells express significantly higher amounts of Rag1 and Rag2 than preB cells and used RAG2:GFP knock-in mice (Monroe et al., 1999) to examine the distribution of proB and preB cells in vivo. As expected, cKit+ CD19+ CD93Hi IgM− proB cells expressed bimodal and notoriously high amounts of RAG2:GFP, whereas cKit− CD19+ CD93Hi IgM− preB cells expressed detectable but significantly lower amounts of RAG2:GFP (Fig. 1 B). A short, 20-min pulse in vivo with the thymidine analogue BrdU confirmed that RAG2:GFP is predominantly abundant in BrdU-negative proB and preB cells (Fig. 1 B), in agreement with studies showing active RAG2 phosphorylation, ubiquitination, and degradation during the S phase of the cell cycle (Desiderio, 2010). To examine the distribution of proB and preB cells in relation to CXCL12 and IL-7+ mesenchymal progenitor cells (MPCs), we reconstituted lethally irradiated Il7GFP/+ Cxcl12dsred/+ mice with BM cells isolated from RAG2:GFP knock-in mice. Analyses of femur whole mounts by confocal microscopy revealed that RAG2:GFPHi B-lineage cells were slightly closer to CXCL12+ IL-7+ MPCs than RAG2:GFPLo B cells, whereas the distribution of RAG2:GFPLo B cells was similar to that of randomly picked CD19− cells marked by DAPI+ nuclei (Fig. 1, C and D). Furthermore, ∼50% of RAG2:GFPHi proB cells (29 out of 62) were found in direct contact with a CXCL12+ IL-7+ cell that also directly interacted with a RAG2:GFPLo preB cell, although only ∼15% of RAG2:GFPLo preB cells (40 out of 284) in contact with a CXCL12+ IL-7+ cell had a neighboring RAG2:GFPHi proB cell (Fig. 1 E).
Figure 1.
Localization of proB and preB cells in relationship to IL-7– and CXCL12-producing cells. (A) Gating strategy for proB and preB cells. Left, BM cells gated on live CD19+ cells. Right, cKit expression distinguishes proB from preB cells. (B) Left, RAG2:GFP fusion protein expression in proB (red) and preB (blue) cells isolated from RAG2:GFP mice. Filled histograms show background fluorescence in wild-type preB cells. RAG2:GFP expression in proB (middle panel) and preB (right panel) in S phase (BrdU+) and in non–S phase (BrdU–). (C) Longitudinal 30-µm-thick femur section from lethally irradiated Il7GFP/+ Cxcl12dsRed/+ mouse reconstituted with RAG2:GFP BM cells stained to detect GFP (green), CD19 (cyan), and nuclei (DAPI, not shown). Cxcl12-dsRed fluorescent protein (red) was directly visualized. Data were pooled from two independent experiments (n = 2). (D) Quantification of proB (RAG2:GFPHi) and preB (RAG2:GFPLo) cells in contact with IL-7– and CXCL12-producing cells (see Materials and methods for details). Bars indicate average; circles indicate individual mice. (E) Quantification of colocalized RAG2:GFPHi (proB) and RAG2:GFPLo (preB) in contact with individual IL-7+ CXCL12+ cells (bottom right). Example of a RAG2:GFPHi (arrowheads) and a RAG2:GFPLo (arrows) cell in contact with the same IL-7+ cell. The same datasets were analyzed as in D. Error bars indicate SD. (F) Longitudinal 30-µm-thick section of a femur from a lethally irradiated Il7GFP/+ Cxcl12dsRed/+ mouse reconstituted with Rag1−/− (top panels) or Rag1−/− IghB1-8/+ (bottom panels) BM cells, stained to detect GFP (green) and CD19 (cyan). Cxcl12-dsRed was directly visualized (red). An entire femur/mouse was acquired; data were pooled from three independent experiments (n = 3). (G) Frequency of proB (Rag1−/−) and preB (Rag1−/− IghB1-8/+) cells in contact with IL-7+ CXCL12+ cells. Bars indicate average; circles depict individual mice. *, P < 0.05 by unpaired Student’s t test. Scale bars, 50 µm. Data in all panels are representative of two to five independent experiments.
Next, we analyzed mice in which B cell development is blocked at the proB (Rag1−/−) and at the preB (Rag1−/− IghB1-8/+) cell stage. As before, we generated BM chimeras by transplanting BM cells from either mice into lethally irradiated Il7GFP/+ Cxcl12dsred/+ mice and analyzed proB and preB cell distribution in femur whole mounts by confocal microscopy. ProB cells were found in direct contact with IL-7+ CXCL12+ stromal cells at slightly but significantly higher frequencies than preB cells (Fig. 1, F and G). In summary, while the large majority of proB and preB cells occupy IL-7 niches, a small fraction of preB cells is more frequently found in niches farther away from IL-7+ cells.
ProB and preB cell dynamic behavior in vivo
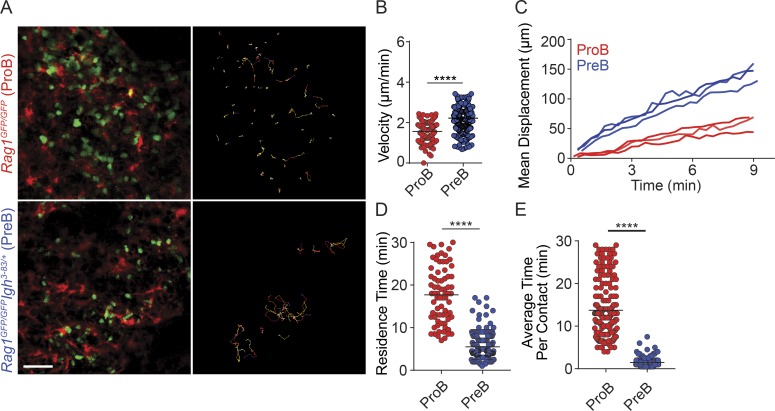
Next, we sought to understand the mechanisms by which preB cells position away from IL-7–expressing cells, with the broader goal of understanding whether positional differences in B cell precursors are physiologically important. To gain insight into how proB and preB cells localize in the BM, we visualized their behavior in vivo by intravital two-photon microscopy, as previously described (Pereira et al., 2009). We generated mice in which proB cells are labeled genetically with Rag1GFP/GFP (B cell development is arrested at the proB cell stage) and mice in which preB cells are similarly labeled but development is allowed to proceed until the preB cell stage due to transgenic expression of a recombined heavy chain locus (Igh3-83/+). While proB cells were predominantly static and in direct contact with CXCL12+ cells, with a few proB cells migrating short distances, nearly all preB cells were motile (Fig. 2 A and Video 1). PreB cells displayed increased median velocity (Fig. 2 B) and mean displacement (Fig. 2 C) when compared with proB cells. This drastic difference in dynamic behavior was also reflected in the prolonged time individual proB cells spent in contact with CXCL12+ cells throughout a 30-min imaging period (Fig. 2 D), as well as in an increased average time individual proB cells spent in contact with individual CXCL12+ cells when compared with preB cells (Fig. 2 E). These observations showed that the small differences found in proB and preB cell static positioning in situ (Fig. 1) were caused by dramatically different dynamic behaviors in vivo, and suggest that they might be physiologically relevant.
Figure 2.
ProB and preB cell migratory behavior in vivo. (A) Distribution of GFP+ proB cells (green, top left) and of preB cells (green, bottom left) in BM calvaria of lethally irradiated Cxcl12dsRed/+ mice reconstituted with Rag1GFP/GFP (top panels) or with Rag1GFP/GFP Igh3-83/+ (bottom panels) BM cells. Movement of GFP+ B cells tracked for 30 min in vivo by IVM. Colored lines (right panels) represent cell trajectories. Scale bar, 50 µm. (B) Median velocity (micrometers per minute). Each circle represents a single cell analyzed. (C) Mean motility coefficient of proB and preB cells. Cell displacement from starting coordinates is plotted against time (minutes). Lines represent the mean motility coefficient from three separate videos per mouse. (D) Time (minutes) spent in direct contact with CXCL12+ cells over 30 min. (E) Average time (minutes) proB and preB cells spent in contact with individual CXCL12+ cells. Data are from Video 1. Circles represent single cells analyzed. ****, P < 0.00005 by unpaired Student’s t test. Data in all panels are representative of three independent experiments.
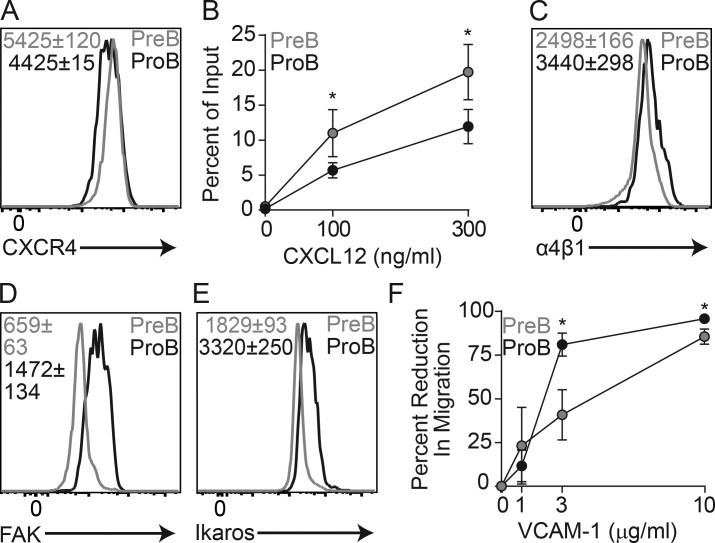
As developing B cell migration in vivo is controlled by CXCR4 (Beck et al., 2014), we measured CXCR4 expression in proB and preB cells and migration in vitro using a transwell assay. Indeed, CXCR4 expression is enhanced in preB cells (Fig. 3 A), and preB cells migrated toward a gradient of CXCL12 significantly more than did proB cells (Fig. 3 B). Nevertheless, this relatively small difference in migration toward CXCL12 in vitro contrasted with a remarkably different dynamic behavior in vivo, suggesting that additional mechanisms might be involved in vivo. Other than CXCR4, B-lineage cells required α4β1 integrin-mediated adhesion to VCAM-1 for retention and migration within the BM (Pereira et al., 2009; Beck et al., 2014), and proB cells expressed higher amounts of α4β1 than preB cells (Fig. 3 C). Previous studies showed that the transcription factor Ikaros acts as a transcriptional repressor of multiple genes involved in cell migration and adhesion (Schjerven et al., 2013; Joshi et al., 2014; Schwickert et al., 2014), of which FAK (encoded by Ptk2) played a major role in supporting aberrant proB cell adhesion to stromal cells and VCAM-1 in vitro (Schwickert et al., 2014). FAK is also required for the retention of Ikaros-sufficient proB cells in the BM (Park et al., 2013). Interestingly, proB cells expressed significantly higher amounts of FAK (and Ikaros) than preB cells (Fig. 3, D and E), suggesting that proB cells might be more adherent to VCAM-1 than preB cells. To test this hypothesis, we coated transwells with a range of VCAM-1 concentrations and quantified proB and preB cell migration toward a CXCL12 gradient in vitro. As expected, VCAM-1–mediated adhesion prevented proB cell migration significantly more than in preB cells (Fig. 3 F). Combined, these data strongly suggest that the reduced migratory behavior of proB cells is due to a small reduction in CXCR4 expression in combination with increased FAK/α4β1 integrin-mediated adhesion to VCAM-1.
Figure 3.
CXCR4-mediated migration and VLA4/FAK-mediated adhesion to VCAM-1 in proB and preB cells. (A) CXCR4 expression in proB and preB cells. (B) In vitro transwell (5-µm pore) migration assay of C57BL/6 BM cells toward CXCL12 gradients. Y axis indicates frequency of migrated cells; x axis shows CXCL12 concentration (nanograms per milliliter). (C) α4 integrin expression in proB and preB cells. (D) FAK expression (encoded by Ptk2) in proB and preB cells. (E) Ikaros (encoded by Ikzf1) expression in proB and preB cells. (F) ProB and preB cell migration across VCAM-1–coated transwells (5 µm) toward CXCL12 gradient (300 ng/ml). Y axis indicates percent reduction in migratory cells; x axis shows VCAM-1 concentration (micrograms per milliliter) used for coating transwells. (B and F) Error bars indicate SD of three individual experiments. Numbers inside all histograms indicate geometric mean fluorescence intensity ± SD (n = 3 mice in each group). *, P < 0.05 by unpaired Student’s t test. Data in all panels are representative of three to five independent experiments.
ProB cell/IL-7+ cell circuits control IL-7R signaling
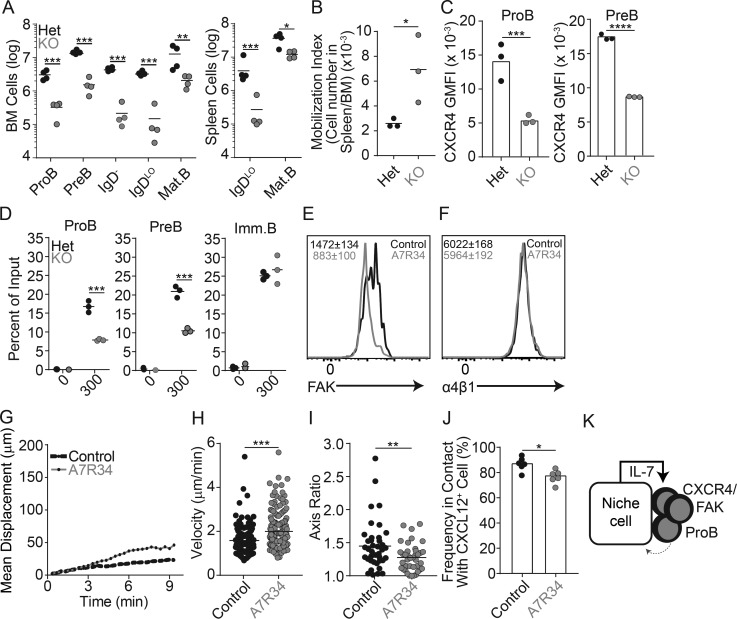
The observation that proB cells are predominantly nonmotile and stably adherent to BM IL-7– and CXCL12-producing MPCs suggests that IL-7R signaling is constitutively high in proB cells, which led us to hypothesize that IL-7R signaling might, in turn, regulate the expression of genes involved in proB cell localization and dynamic behavior. To test this hypothesis, we conditionally deleted IL-7Ra in proB cells by crossing Il7rafl/fl mice with Mb1cre/+ mice. B cell development was severely impaired at the proB and preB cell stages, which led to an overall reduction in total B cell numbers (Fig. 4 A). We confirmed by flow cytometry that IL-7R protein expression was not detected on the few proB and preB cells that could be found in the BM of Il7rafl/fl Mb1cre/+ mice (data not shown). Interestingly, we noted that the proportion of IL-7Ra–deficient proB and preB cells in the spleen was higher than that seen in the spleen of littermate control mice (Fig. 4 B), suggesting that IL-7Ra–deficient B-lineage precursors might be less retained in the BM than wild-type counterparts. The few IL-7Ra–deficient B-lineage cells that nevertheless developed expressed significantly reduced amounts of CXCR4 in proB and preB cells (Fig. 4 C and data not shown, respectively). Furthermore, IL-7Ra–deficient proB and preB cells migrated toward CXCL12 in vitro with lower efficiency than IL-7Ra–sufficient proB and preB cells, while IL-7Ra deficiency had no impact on migration to CXCL12 (or CXCR4 expression) in immature B cells that no longer expressed IL-7Ra (Fig. 4 D and data not shown, respectively). To further validate these findings, we measured the impact of IL-7Ra signaling on motility and adhesion mechanisms of wild-type proB and preB cells developing in vivo. Indeed, IL-7Ra signaling actively controlled the proB adhesion program as measured by significantly reduced FAK expression after treatment with an anti–IL-7Ra blocking antibody (clone A7R34) for 36 h (Fig. 4 E), whereas α4β1 expression was unchanged (Fig. 4 F). Similar changes were observed in IL-7Ra–deficient proB cells (data not shown). To test if IL-7Ra signaling also controls proB cell behavior in vivo, we analyzed proB cell motility in Rag1GFP/GFP mice treated with IL-7Ra blocking antibodies for 36 h by intravital two-photon microscopy. Although the majority of proB cells remained largely sessile, we could detect a subtle but significant increase in the mean displacement and median cell velocity of proB cells treated with anti–IL-7R antibody (Fig. 4, G and H; and Video 2) that did not seem to be caused by active, amoeboid cell motility, given that proB cells became more rounded (Fig. 4 I) and presumably less adherent to the extracellular matrix. In agreement with this possibility, proB cells became slightly less associated with CXCL12+ cell processes (Fig. 4 J). Loosely adherent proB cells likely become more motile as a consequence of shear stress generated by intrasinusoidal turbulent blood flow and plasma transudation into BM parenchyma (Beck et al., 2014). We propose that CXCR4 and integrin signaling (α4β1/FAK/VCAM-1) locks proB cells in contact with IL-7–producing cells in BM. This tight interaction likely drives constitutive IL-7Ra signaling that further promotes CXCR4 and FAK/α4β1 integrin-mediated adhesion to IL-7–producing cells (Fig. 4 K) in a feed-forward loop manner that resembles a two-cell circuit system (Zhou et al., 2018).
Figure 4.
Relationship between IL-7R signaling and CXCR4-mediated proB cell motility. (A) Total number of developing B cell subsets from femur and tibia (left) and from spleen (right) of mice conditionally deficient in IL-7Ra in B-lineage cells: Mb1Cre/+ Il7raFl/+ (black) and Mb1Cre/+ Il7raFl/Fl (gray) mice. (B) Mobilization index: ratio of proB and preB cell number in the spleen to that in the BM of Mb1Cre/+ Il7raFl/+ (Het, black) and Mb1Cre/+ Il7raFl/Fl (KO, gray) mice. Circles depict individual mice. (C) CXCR4 geometric mean fluorescence intensity (GMFI) in proB cells (left) and preB cells (right) gated as in A in Mb1Cre/+ Il7raFl/+ (Het, black) and Mb1Cre/+ Il7raFl/Fl (KO, gray) mice. (D) In vitro transwell (5 µm) migration assay of Mb1Cre/+ Il7raFl/+ (Het, black) and Mb1Cre/+ Il7raFl/Fl (KO, gray) BM cells toward CXCL12 gradients. Y axis indicates frequency of migrated cells; x axis shows CXCL12 concentration (nanograms per milliliter). (A–D) Lines indicate mean; circles depict individual mice (n = 3). (E) FAK expression in proB cells isolated from C57BL/6 mice untreated (littermate controls, black) or treated with anti–IL-7Ra blocking antibodies (gray) for 36 h. (F) α4 integrin expression in proB cells isolated from C57BL/6 mice treated with isotype control (black) or anti–IL-7Ra blocking antibody (gray) for 36 h. (E and F) Numbers inside histograms indicate geometric mean fluorescence intensity ± SD (n = 3 mice in each group). (G) Mean motility coefficient of proB cells in mice untreated (black) or treated with IL-7Ra blocking antibodies (gray) for 36 h. GFP+ proB cells were visualized in calvaria of lethally irradiated Cxcl12dsRed/+ mice reconstituted with BM cells from Rag1GFP/GFP mice. ProB cell movement was tracked for 30 min in vivo by IVM. Lines represent the mean motility coefficient from three separate videos per mouse. (H) Median velocity. Circles, individual cells. (I) Measurement of cell axis ratio of GFP+ proB cells before (black) and after (gray) treatment with anti–IL-7Ra blocking antibody. Each point represents an individual cell analyzed. (J) Frequency of GFP+ proB cells found in contact with Cxcl12-dsRed cells in mice treated with IL-7Ra blocking antibody (gray) or untreated (black). Circles, average from individual fields of view. (K) Model of the proB/IL-7+ cell circuit. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005 by unpaired Student’s t test. Data in all panels are representative of two to four independent experiments.
PreBCR signaling breaks the proB/IL-7+ cell circuit to attenuate IL-7R signaling at the preB cell stage
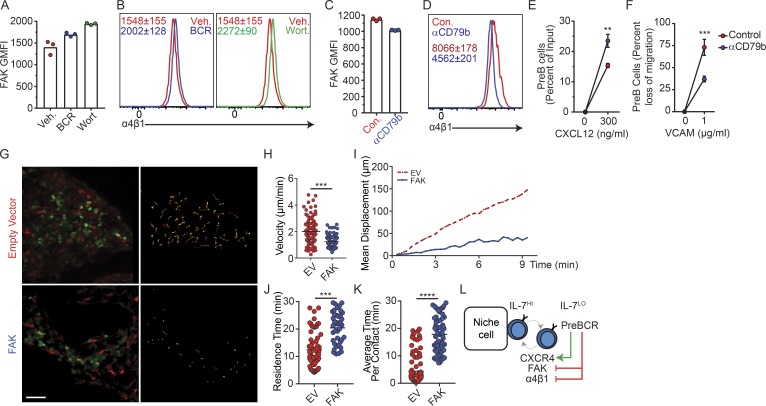
In addition to signaling from the preBCR, the transition from a constitutively high to an intermittent and low IL-7Ra signaling from proB to preB cells has been proposed to control RAG gene expression and Ig light chain gene accessibility (Johnson et al., 2008; Ochiai et al., 2012; Clark et al., 2014). We reasoned that the rapid movement and reduced time spent in contact with CXCL12 and IL-7+ MPCs might enable preB cells to attenuate IL-7R signaling in vivo. Furthermore, as preBCR signaling induces IRF4 expression and IRF4 drives CXCR4 upregulation (Muljo and Schlissel, 2003; Thompson et al., 2007; Johnson et al., 2008), we hypothesized that preBCR signaling is also responsible for the switch from higher to lower FAK/α4β1 integrin-mediated adhesion to VCAM-1. Indeed, preB cells treated with a cocktail of SYK and BTK inhibitors (fostamatinib and ibrutinib, respectively) or the PI3K inhibitor wortmannin promoted α4β1 and FAK expression (Fig. 5, A and B), whereas stimulatory anti-CD79b antibody treatment reduced α4β1 and FAK expression (Fig. 5, C and D), increased CXCR4 expression in preB cells (data not shown), and increased preB cell migration toward CXCL12 (Fig. 5 E). The changes induced by anti-CD79b stimulation, albeit also small, were significant and sufficient for causing significant differences in α4β1-mediated preB cell adhesion to VCAM-1 in vitro (Fig. 5 F). These findings suggested that preB cells acquire rapid motility through the combined small but significant and physiologically relevant changes in CXCR4, α4β1, and FAK expression in vivo. To test this possibility, we took advantage of the fact that Philadelphia chromosome–positive BCR-ABL preB-ALL cells are developmentally arrested at the preB cell stage (Schjerven et al., 2017) to measure the effects of increased FAK signaling in preB cell behavior in vivo. Indeed, preB-ALL cells retrovirally transduced with a Ptk2 expression vector significantly reduced their migratory behavior in vivo at day 7 after transplantation (Fig. 5, G–I; and Video 3) and increased their time in contact with CXCL12Hi stromal cells (Fig. 5, J and K). Combined, these findings suggest that productive V(D)J rearrangements in proB cells and signaling through the newly assembled preBCR acts as a proB/IL-7+ cell circuit breaker through increased CXCR4 expression and decreased integrin-mediated adhesion, which promotes rapid movement between IL-7Hi and IL-7Lo BM microenvironments (Fig. 5 L). These results are consistent with studies showing differential adhesion of B cell precursors to VCAM-1 in vitro (Glodek et al., 2003).
Figure 5.
Relationship between preBCR signaling and CXCR4- and integrin-mediated preB cell motility. (A) FAK geometric mean fluorescence intensity (GMFI) in preB cells from C57BL/6 mice treated with BCR signaling inhibitors (blue, fostamatinib and ibrutinib) or vehicle (red), or with PI3K inhibitor wortmannin (green) or vehicle (red) for 6 h in vitro. (B) α4 integrin expression in preB cells of C57BL/6 mice treated as in A. Numbers indicate geometric mean fluorescence intensity ± SD (n = 3 mice per condition). (C and D) FAK geometric mean fluorescence intensity (C) and α4 integrin expression (D) in preB cells treated with stimulatory CD79b antibody for 6 h in vitro. Con., control. (A and C) Bars indicate mean; circles depict individual mice/assays. (D) Numbers inside histogram indicate geometric mean fluorescence intensity ± SD (n = 3 individual mice/assays). (E) Transwell migration toward CXCL12 of preB cells treated with stimulatory CD79b antibody for 6 h in vitro. Y axis indicates frequency of migrated cells; x axis indicates CXCL12 concentration (nanograms per milliliter). (F) PreB cell migration across VCAM-1–coated transwells (5 µm) toward CXCL12 gradient (300 ng/ml). Y axis indicates percent reduction in migrated cells; x axis indicates VCAM-1 concentration (micrograms per milliliter) used for coating transwells. (E and F) Circles represent average of three independent experiments. Error bars indicate SD. (G) Distribution of EV-transduced GFP+ preB-ALL cells (green, top left) and of FAK-transduced GFP+ preB-ALL cells (green, bottom left) in BM calvaria of Cxcl12dsRed/+ mice at day 14 after transplantation. Movement of GFP+ B cells tracked for 30 min by IVM. Colored lines represent cell trajectories (right panels). Scale bar, 50 µm. (H) PreB-ALL cell velocity (micrometers per minute). Each circle represents a single cell analyzed. (I) Mean motility coefficient of EV- and FAK-transduced preB-ALL cells. Cell displacement from starting coordinates is plotted against time (minutes). Lines depict the mean motility coefficient from three videos imaged/mouse. (J) Time (minutes) spent in direct contact with CXCL12+ cells over a 30-min period. (K) Average time (minutes) that EV- and FAK-transduced preB-ALL cells spent in direct contact with individual CXCL12+ cells. (J and K) Circles represent single cells. Data in panels G–K are from Video 3. (L) Model of the preB/IL-7+ cell circuit. **, P < 0.005; ***, P< 0.0005; ****, P < 0.00005 by unpaired Student’s t test. Data in all panels are representative of two to four independent experiments.
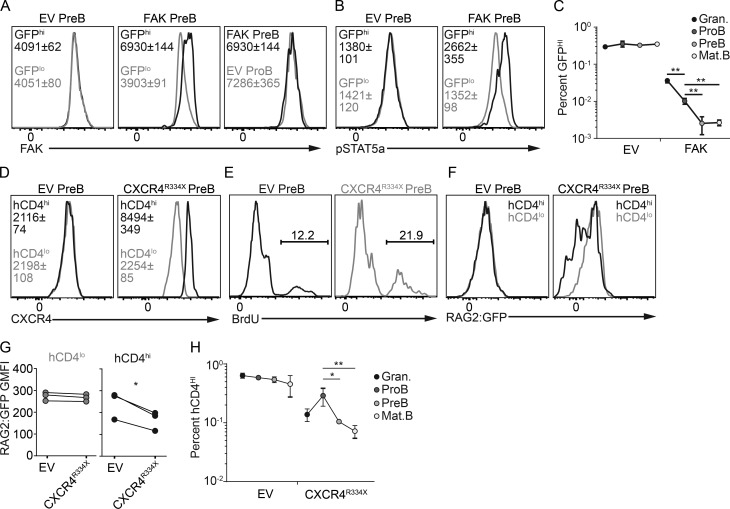
Such tight control of preB cell dynamic behavior in vivo lends support to the long proposed but never tested hypothesis that CXCR4-regulated positioning attenuates IL-7Ra signaling required for Rag gene reexpression and developmental progression into the immature B cell stage (Johnson et al., 2008; Clark et al., 2014). To experimentally test this model, we antagonized the FAK downregulation that normally occurs at the preB cell stage by overexpressing Ptk2 (reported by GFP expression) in hematopoietic stem cell (HSC)–enriched BM cells from C57BL/6 mice and analyzed B cell developmental subsets in FAKHi versus control transduced cells. Lethally irradiated recipient mice were reconstituted with empty vector (EV)– or Ptk2-transduced HSCs for a period of 4–6 wk. Hematopoietic progenitor cells overexpressing FAK showed a strong inability to differentiate into blood cell types in vivo for unknown reasons, but a few FAKHi lymphoid and myeloid cells could nevertheless be detected in the BM. Approximately twofold overexpression of FAK was achieved in Ptk2-transduced preB cells, which made it equivalent to that detected in EV-transduced proB cells (Fig. 6 A). Furthermore, FAKHi preB cells showed significantly higher amounts of phosphorylated STAT5a than EV-transduced preB cells (Fig. 6 B), suggesting that FAKHi preB cells may have been more exposed to IL-7 in vivo. Interestingly, the frequency of FAKHi cells within the preB cell and subsequent B cell subsets was fourfold lower than FAKHi cells within the proB stage (Fig. 6 C). We cannot rule out the possibility that increased FAK signaling prevents preB cell proliferation. However, these findings suggest that antagonism of the normal FAK downregulation that occurs at the preB cell stage blocks the transition between the proB and preB cell stages, which significantly reduces mature B cell production in vivo. To validate these findings and to gain further insight, we asked if proximity to IL-7 alters the preB cell cycle and perturbs RAG2 protein stability in vivo. To address this question, we overexpressed the R334X desensitization mutant form of CXCR4 that causes WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome (Gorlin et al., 2000; Hernandez et al., 2003; Balabanian et al., 2005) in HSCs isolated from RAG2:GFP mice and reconstituted lethally irradiated recipient mice with EV- or CXCR4R334X-transduced HSCs for a period of 4–6 wk. CXCR4R334X-transduced preB cells expressed higher amounts of CXCR4 than EV-transduced preB cells (Fig. 6 D). Interestingly, overexpression of the CXCR4R334X mutant increased the fraction of preB cells that was in the S phase of the cell cycle, as measured by a 20-min pulse of BrdU incorporation in vivo (Fig. 6 E). As expected, it also reduced the fraction of RAG2:GFPHi preB cells (Fig. 6, F and G). These findings suggest that enforcement of preB cell interactions with IL-7+ cells is sufficient for reducing the time preB cells spend in the G1 phase of the cell cycle and hence to perturb RAG2 protein stability. These changes were reflected in a significant expansion of CXCR4R334X-Hi proB cells when compared with granulocytes (Fig. 6 H), a result that is consistent with increased IL-7R signaling due to increased exposure to IL-7 (Carvalho et al., 2001; Rother et al., 2016). In contrast, CXCR4R334X-Hi preB cells were reduced by threefold, presumably due to reduced RAG2 protein half-life and time spent in the G1 phase of the cell cycle. This led to a greater than threefold reduction in the generation of CXCR4R334X-Hi mature B cells (Fig. 6 H). Reduced granulocyte production from CXCR4R334X-Hi progenitors is likely a consequence of reduced differentiation capacity of HSCs that express CXCR4 desensitization mutant (McDermott et al., 2015; Freitas et al., 2017).
Figure 6.
Relationship between enforced positioning within IL-7+ niches, RAG2 protein stability, and preB cell developmental progression. (A) Overexpression of FAK (Ptk2) or EV reported by GFP in retrovirally transduced BM cells isolated from C57BL/6 mice. FAK expression in preB cells transduced with EV (left) or FAK (middle). Representative histogram of preB cells transduced with FAK compared with proB cells transduced with EV (right). (B) pSTAT5a in EV- or FAK-transduced preB cells. (C) Frequency of EV- (left) or FAK-transduced (right) GFPHi BM B-lineage cells and granulocytes from femur and tibia. (D) Overexpression of CXCR4 R334X (WHIM mutation) in retrovirally transduced BM cells isolated from RAG2:GFP knock-in mice. Transduced cell subsets reported by human CD4. CXCR4 expression in EV- (left) or CXCR4R334X-transduced (right) preB cells gated on hCD4Lo and hCD4Hi cells. (E) BrdU incorporation in EV- (left) or CXCR4R334X-transduced preB cells. Numbers indicate the frequency of BrdU+ cells. (F) RAG2:GFP expression in gated hCD4Hi and hCD4Lo preB cells (EV, left; CXCR4R334X, right). (G) Quantification of RAG2:GFP fusion protein geometric mean fluorescence intensity (GMFI). Circles depict individual mice. (H) Frequency of hCD4Hi B-lineage cells and granulocytes in femur and tibia BM. Circles indicate mean ± SD (n = 3). (C and H) Error bars indicate SD of three independent experiments. Numbers inside histograms (A, B, and D) indicate geometric mean fluorescence intensity ± SD (n = 3 mice in each group). Data in all panels were pooled from three independent experiments. *, P < 0.05; **, P < 0.01 by unpaired Student’s t test (C and H). *, P < 0.05 by paired Student’s t test (G).
Preleukemic preB and BCR-ABL preB cells regulate IL-7 production in vivo
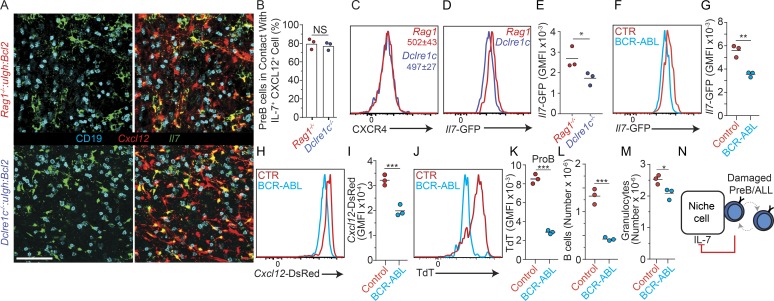
Previous studies suggested that RAG-mediated DSBs elicit an NF-κB–driven and Spic-driven gene expression program in preB cells that downregulates the expression of preBCR signaling components and upregulates the expression of genes involved in cell migration (Bredemeyer et al., 2008; Bednarski et al., 2016). Thus, we reasoned that the accumulation of unrepaired DSBs that occurs in Artemis (encoded by Dclre1c)-deficient preB cells might alter their distribution in relationship to IL-7+ cells in vivo. To address this question, we compared the distribution of Igh and Bcl2 transgenic Rag1−/− preB cells (no RAG-mediated DSBs) and of Igh and Bcl2 transgenic Dclre1c−/− preB cells (accumulated unrepaired DSBs) relative to IL-7+ and CXCL12+ cells. However, we did not detect significant differences in Artemis-deficient and -sufficient preB cells in vivo (Fig. 7, A and B), which was consistent with no measurable differences in CXCR4 expression (Fig. 7 C). Unexpectedly, we noted that Il7 expression was lower in MPCs isolated from the BM of mice in which preB cells were deficient in Artemis (Fig. 7, D and E), suggesting that IL-7+ cells sense the quality of neighboring preB cells through signals provided by Artemis-deficient preB cells. As Artemis deficiency is a risk factor for leukemogenesis, we finally asked if bona fide preB-ALLs also regulate IL-7 production in BM. We adoptively transferred 105 BCR-ABL preB-ALL cells into nonirradiated Cxcl12dsRed/+ Il7GFP/+ mice and measured Il7-GFP and Cxcl12-dsRed expression in MPCs 2 wk after transfer. Remarkably, Il7-GFP and Cxcl12-dsRed expression was significantly reduced in mice recipient of BCR-ABL preB-ALLs (Fig. 7, F–I). Furthermore, terminal deoxynucleotidyl transferase expression was significantly reduced in nonleukemic proB cells (Fig. 7, J and K), consistent with reduced IL-7R signaling in these cells (Wei et al., 2000; Carvalho et al., 2001; Rother et al., 2016). These changes resulted in a threefold reduction in endogenous nonleukemic B cell production (Fig. 7 L), while granulocytes were only reduced by ∼20% (Fig. 7 M). Combined, these results suggest that preB/IL-7+ cell circuits evolved to not only regulate preB cell transition into the immature B cell compartment, but also to facilitate the removal of dangerous preleukemic preB cells (Fig. 7 N).
Figure 7.
DNA repair deficiency in preB cells and control of IL-7 expression in vivo. (A) Longitudinal 30-µm-thick femur section of lethally irradiated Il7GFP/+ Cxcl12dsRed/+ mice reconstituted with Rag1−/− VH147Igh Eμ-Bcl2 (top panels) or Dclrec1−/− VH147Igh Eμ-Bcl2 BM stained to detect GFP (green), CD19 (cyan), and cell nuclei (DAPI, not shown). Cxcl12-dsRed fluorescent protein (red) was directly visualized. Scale bar, 50 µm. Images are representative of three individual mice. (B) Quantification of Artemis-deficient (blue) and -sufficient (red) preB cells in contact with IL-7– and CXCL12-producing cells. Bars indicate mean; circles depict individual mice (n = 3). (C) CXCR4 expression in Artemis-deficient (blue) and -sufficient (red) preB cells in vivo. Numbers indicate geometric mean fluorescence intensity (GMFI) ± SD (n = 3). (D) Il7-GFP expression in nonhematopoietic Lepr+ BM cells of lethally irradiated Il7GFP/+ mice reconstituted with Rag1−/− VH147Igh Eμ-Bcl2 or Dclrec1−/− VH147Igh Eμ-Bcl2 BM cells. (E) Quantification of Il7-GFP expression from D. (F) Geometric mean fluorescence intensity of Il7-GFP expression in nonhematopoietic Lepr+ BM cells of Il7GFP/+ mice 2 wk after transfer of 105 BCR-ABL transduced preB cells. (G) Quantification of Il7-GFP expression from F. (H) Geometric mean fluorescence intensity of Cxcl12-dsRed expression in nonhematopoietic Lepr+ BM cells of Cxcl12dsred/+ Il7GFP/+ mice 2 wk after transfer of 105 BCR-ABL preB cells. (I) Quantification of Cxcl12-dsRed expression from H. Lines indicate mean; circles depict individual mice analyzed (n = 3). (J) Terminal deoxynucleotidyl transferase (Tdt) expression in nonleukemic proB cells isolated from mice described in H. (K) Quantification of terminal deoxynucleotidyl transferase expression from J. (L and M) Total CD19+ host B lymphocytes (L) and granulocytes (M) 2 wk after transfer of 105 BCR-ABL transduced preB cells. (E, G, and K–M) Lines indicate mean; circles depict individual mice analyzed (n = 3). (N) Model of damaged preB or preB-ALL cell/IL-7+ cell circuit. Data in all panels are representative of two to four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
B lymphocyte progenitors have been proposed to occupy distinct microenvironments in the BM according to their developmental stage (Dorshkind, 1990; Jacobsen and Osmond, 1990; Pereira et al., 2009; Mourcin et al., 2011). ProB cells are highly dependent on extrinsic growth factors (mostly IL-7, stem cell factor, and possibly FLT3 ligand) for survival and proliferation (Grabstein et al., 1993; Peschon et al., 1994; von Freeden-Jeffry et al., 1995; Waskow et al., 2002; Sitnicka et al., 2003). Most, if not all, of these factors are produced by Leptin receptor (Lepr)–positive nonhematopoietic cells that express the highest amounts of CXCL12 in the BM (Ding et al., 2012; Cordeiro Gomes et al., 2016). In later stages, preB cell survival and proliferation continue to depend on IL-7, albeit to a lower extent than proB cells due to pro-survival and pro-proliferative signals mediated by the preBCR (Clark et al., 2014). However, IL-7R signaling also prevents preB cell developmental progression by inhibiting Rag gene expression and Ig light chain gene accessibility (Ochiai et al., 2012). Therefore, it was unclear how preB cells achieve an appropriate level of IL-7R signaling that allows cells to developmentally progress in vivo. Here we show that proB and preB cells display remarkably different dynamic behaviors in vivo. ProB cells are predominantly nonmotile and intimately associated with IL-7+ cells. IL-7R signaling controls this stable cell–cell interaction by promoting high CXCR4 and FAK expression. This type of feed-forward regulation resembles an unstable two-cell circuit (Zhou et al., 2018) in that it would predict uncontrolled proB cell growth under normal homeostatic conditions. Instead, our data suggest that cell circuit stability is partly ensured by preBCR signaling, which acts as a circuit breaker by altering the behavior of preB cells in vivo. Importantly, this is achieved through a relatively subtle increase in CXCR4 expression and reduction in α4β1/FAK expression. These combined changes in chemoattractant receptor expression and integrin signaling presumably promote a switch from a nonmotile and highly adherent to a highly motile and less adherent state. This change in behavior reduces the time preB cells spend in contact with IL-7+ cells in the BM, thereby attenuating IL-7R signaling. Indeed, IL-7R signaling could be significantly increased in preB cells by simply overexpressing FAK or a desensitization mutant CXCR4 (R334X), which reduced RAG2 protein half-life and inhibited both preB cell and subsequent B cell developmental stages by more than threefold. Given that proB cells do not undergo uncontrolled cell growth in Rag-deficient mice, other negative feedback mechanisms independent of preBCR signaling likely operate to ensure cell circuit stability.
A surprising observation from our studies was the finding that IL-7 production is downregulated by signals provided by preB cells bearing unrepaired DSBs. It is possible that normal preB cells undergoing transient DSB responses also downregulate IL-7 production and that Artemis deficiency allows DSBs to persist long enough for changes in IL-7 expression to be detected. Nevertheless, these observations revealed an unsuspected layer of regulation of preB/IL-7+ cell circuits that may have evolved to prevent the survival and proliferation of preleukemic preB cells. However, the fact that BCR-ABL+ preB-ALLs could also induce IL-7 (and CXCL12) downregulation suggests that leukemic cells exploit certain cell circuit pathways to reduce the production of nonleukemic B-lineage cells. This would suggest that normal and leukemic preB cells compete for similar factors, with nutrients fueling anabolic metabolism being likely candidates. Future studies addressing nutrient requirements of normal and leukemic progenitor B cells will be of high value.
The fact that DSB-damaged preB cells and normal preB cells express similar amounts of CXCR4 and are similarly distributed in the BM suggests that DSB-damaged and normal B cell progenitors interact with the same niche. It remains unclear if the presence of a single DSB-damaged preB cell is sufficient for downregulating IL-7 expression in individual IL-7+ cells that harbor nondamaged B cell progenitors, or if a threshold of damaged preB cells is needed. Nevertheless, these observations raise the interesting possibility that individual IL-7+ cells are regulated independently, and also that individual cells may return back to expressing normal amounts of IL-7. In this case, individual IL-7+ cells would be expected to act as single niche units of lymphopoiesis.
In conclusion, our findings provide support for a novel conceptual model of B cell development where cell circuits between proB, preB, and IL-7+ cells are wired to control the number and developmental progression of B cell progenitors, while being equipped with negative feedback loops that may disfavor the survival of preleukemic cells. Our findings also suggest that BCR-ABL+ preB-ALLs exploit IL-7 downregulation to favor leukemic cell growth at the expense of nonleukemic B cells. It will be important to unravel the molecular and cellular mechanism(s) controlling IL-7 production in BM niches.
Materials and methods
Mice
C57BL/6 (CD45.2), Boy/J (CD45.1), and Il7raF/F mice were obtained from the Jackson Laboratory. Il7raF/F mice were crossed to Mb1cre/+ mice (Pelanda et al., 2002). Rag1GFP/+ mice (Kuwata et al., 1999) were crossed to each other to generate Rag1GFP/GFP mice. Rag1−/−, IghB1-8/+, and Rag2:GFP mice were kindly provided by Dr. David Schatz (Yale University, New Haven, CT). Dclrec1−/−:uIgH:BCL2 and Rag1−/−:uIgH:BCL2 mice were kindly provided by Dr. Jeffrey Bednarski (Washington University in St. Louis, St. Louis, MO). Cxcl12DsRed/+ and Il7GFP/+ mice were from our internal colony. A detailed list of all mouse strains and other materials and reagents used is provided (Table 1). Mice were maintained under specific pathogen–free conditions and used according to protocols approved by the Yale University Institutional Animal Care and Use Committee.
Table 1. List of reagents, DNA constructs and mouse strains, sources, and catalog numbers.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| APC BrdU Flow Kit | BD Biosciences | Cat#557892 |
| Donkey polyclonal anti-goat biotin | Jackson ImmunoResearch | Cat#705-065-003 |
| Donkey polyclonal anti-rabbit Alexa488 | Life Technologies | Cat#A21206 |
| Donkey polyclonal anti-rat Alexa488 | Jackson ImmunoResearch | Cat#112-545-003 |
| Donkey polyclonal anti-rat biotin | Jackson ImmunoResearch | Cat#112-065-003 |
| FITC BrdU Flow Kit | BD Biosciences | Cat#559619 |
| Goat polyclonal anti-GFP | Rockland | Cat#600-101-215 |
| Mouse monoclonal anti-human CD4 biotin | BD Biosciences | Clone RPA-T4; Cat#555345 |
| Mouse monoclonal anti-human CD4 PE | BD Biosciences | Clone L200; Cat#550630 |
| Mouse monoclonal anti-mouse FAK | Invitrogen | Clone ZF002; Cat#39-6500 |
| Rat monoclonal anti-CXCR4 biotin | eBioscience | Clone 2B11; Cat#13-9991-82 |
| Rat monoclonal anti-mouse CD19 BV711 | BioLegend | Clone 6D5; Cat#115555 |
| Rat monoclonal anti-mouse CD19 PE | BioLegend | Clone 6D5; Cat#115508 |
| Rat monoclonal anti-mouse CD93 APC | BioLegend | Clone AA4.1; Cat#136510 |
| Rat monoclonal anti-mouse cKIT BV421 | BioLegend | Clone ACK2; Cat#135124 |
| Rat monoclonal anti-mouse cKIT BV711 | BioLegend | Clone 2B8; Cat#105835 |
| Rat monoclonal anti-mouse IgD BV711 | BioLegend | Clone 11-26c.2A; Cat#405731 |
| Rat monoclonal anti-mouse IgD PE | BioLegend | Clone 11-26c.2A; Cat#405706 |
| Rat monoclonal anti-mouse IgG1 PE | BD Biosciences | Clone A85-1; Cat#550083 |
| Rat monoclonal anti-mouse IgM PECy7 | eBioscience | Clone II/41; Cat#25-5790-81 |
| Rat monoclonal anti-mouse Ikaros PE | BioLegend | Clone 2A9; Cat#653304 |
| Rat monoclonal anti-mouse Integrin α4 PE | Southern Biotech | Clone PS/2; Cat#1520-09L |
| Rat monoclonal anti-mouse pSTAT5 (Y694) BV421 | BD Biosciences | Clone 47/Stat5; Cat#562077 |
| Streptavidin Alexa700 | Life Technologies | Cat#S21383 |
| Streptavidin BV605 | BioLegend | Cat#405229 |
| Streptavidin BV711 | BioLegend | Cat#405241 |
| Bacterial and virus strains | ||
| “Subcloning efficiency” DH5α competent cells (F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ−) | Invitrogen | Cat#18265017 |
| Biological samples | ||
| N/A | ||
| Chemicals, peptides, and recombinant proteins | ||
| DMSO | American Bioanalytical | CAS:67-68-5 |
| Ibrutinib (PCI-32765) | Selleck Chemicals | Cat#S2680 |
| Fostamatinib (R788) | Selleck Chemicals | Cat#S2625 |
| Critical commercial assays | ||
| N/A | ||
| Deposited data | ||
| N/A | ||
| Experimental models: Cell lines | ||
| HEK293T | Gift from Dr. David Schatz, Yale University | |
| BCR-ABL | Schjerven et al., 2017 | Gift from Dr. Hilde Schjerven, University of California, San Francisco |
| Experimental models: Organisms/strains | ||
| Mus musculus, strain Mb1cre | Pelanda et al., 2002 | Gift from J.G. Cyster, University of California, San Francisco |
| Mus musculus, strain Il7rtm1.1Asin/J (Il7rFlox) | McCaughtry et al., 2012 | The Jackson Laboratory, Cat#022143 |
| Mus musculus, strain RAG2:GFP | Monroe et al., 1999 | Gift from Dr. David Schatz, Yale University |
| Mus musculus, strain Rag1−/− | Mombaerts et al., 1992 | Gift from Dr. David Schatz, Yale University |
| Mus musculus, strain Rag1GFP | Kuwata et al., 1999 | Our internal colony |
| Mus musculus, strain IghB1-8 (B6.129P2(C)-Ightm2Cgn/J) | Sonoda et al., 1997 | Gift from Dr. David Schatz, Yale University |
| Mus musculus, strain Igh3-83 (C.129P2(B6)-Igktm1Rsky Ightm2Rsky/PldJ) | Pelanda et al., 1997 | The Jackson Laboratory, Cat#011074 |
| Mus musculus, strain Il7GFP | Miller et al., 2013 | Gift from Dr. Susan Kaech, Yale University |
| Mus musculus, strain Cxcl12DsRed | Ding and Morrison, 2013 | Gift from Dr. Sean Morrison, University of Texas Southwestern Medical Center |
| Mus musculus, strain C57BL/6J | The Jackson Laboratory | Stock no. #000664 |
| Oligonucleotides | ||
| N/A | ||
| Recombinant DNA | ||
| pMXs-puro-EGFP-FAK | Hara et al., 2008 | Addgene plasmid #38194 |
| pMSCV CXCR4R334X IRES GFP | Pereira laboratory | N/A |
| pMSCV CXCR4R334X IRES hCD4 | This paper | N/A |
| Software and algorithms | ||
| FlowJo version 9.6.4 | Tree Star | http://www.flowjo.com |
| Imaris version 7.2 | Bitplane | http://www.bitplane.com/imaris/imaris |
| Other | ||
| N/A | ||
BM chimeras
Il7GFP/+Cxcl12DsRed/+ mice were lethally irradiated with two doses of 550 rads separated by 3 h and reconstituted with 3 × 106 whole BM cells of either Rag1−/−, Rag1−/− IghB1-8/+, Rag1GFP/GFP, Rag1GFP/GFP Igh3-83/+, Dclrec1−/−:VH147Igh:BCL2, or Rag1−/−:VH147Igh:BCL2. Chimeric mice were analyzed 4 wk after reconstitution. Boy/J mice were lethally irradiated in the same way and reconstituted with CXCR4R334X or EV-transduced RAG2:GFP BM.
Tissue preparation, cell enumeration, and flow cytometry
BM cells were flushed from both femurs and tibias in DMEM containing 10% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 1 mM L-glutamine, and 10 mM Hepes. Spleens were mashed through 70-µm strainers containing 5 ml of the same media. Cells were counted with a Beckman Coulter Counter.
Flow cytometry analyses and cell gating were performed as previously described (Nevius et al., 2015; Cordeiro Gomes et al., 2016). Briefly, proB cells were gated as CD19+IgM−CD93+cKit+, preB cells as CD19+IgM−IgD−CD93+cKit−, immature IgD− as CD19+IgM+IgD−CD93+, immature IgDlo as CD19+IgM+IgDloCD93+, and Mat.B as CD19+IgM+IgDhiCD93−. Dead cell dyes (DAPI) were used for all freshly stained (nonpermeabilized) samples.
Two-photon intravital imaging
Mice were anesthetized with a mixture of ketamine and xylazine before immobilization on a custom-made stereotactic apparatus. Laser-scanning microscopy images were collected using an Olympus BX61 fluorescence microscope with a 20× 0.95 NA water-immersion objective and dedicated single-beam TriM Scope II (LaVision BioTec). The image acquisition software used was Imspector. Stacks of 18–21 optical sections of 3-µm z-spacing were acquired every 30 s for 30 min. The laser was tuned to a wavelength of 880 nm. Videos with excessive levels of tissue drifting were excluded from analysis.
Retroviral transductions of hematopoietic stem and progenitor cells and BCR-ABL preB-ALLs
The R334X CXCR4 cytoplasmic tail mutant plasmid generation has been described (Beck et al., 2014). pMXs-puro-EGFP-FAK was acquired from Addgene (plasmid #38194). Donor Rag2:GFP or C57BL/6 mice were injected intraperitoneally with 300 µl of 10-mg/ml 5-fluorouracil (5-FU). 5 d after 5-FU injection, BM was harvested and spin infected with harvested retrovirus for 2 h at 2,450 rpm at room temperature. Recipient mice were then reconstituted with the transduced BM for 4 wk before analysis.
BCR-ABL+ preB-ALL adoptive transfer
BCR-ABL cells were prepared as previously described (Schjerven et al., 2017). For imaging experiments, 105 BCR-ABL cells transduced with a retroviral vector containing GFP alone or Ptk2-IRES-GFP were adoptively transferred into Cxcl12dsRed/+ mice via intravenous tail vein injection. 14 d later, BM calvaria were imaged by two-photon intravital microscopy (IVM). For Il7-GFP evaluation experiments (Fig. 7), 105 BCR-ABL cells transduced with a retroviral vector containing GFP were adoptively transferred into Il7GFP/+ mice via intravenous tail vein injection and compared with mock injection. 14 d later, mice were euthanized and BM IL-7+ mesenchymal cells were analyzed by flow cytometry.
In vivo IL-7R antibody blockade
Mice were intravenously injected with PBS or 200 µg antibody (clone A7R34). In all cases, treatments lasted for 36 h before mice were analyzed.
Transwell migration and adhesion assay
Chemotaxis assays were performed with 106 BM cells or 2.5 × 105 BCR-ABL cells. Cells were incubated for 30 min in RPMI containing 0.5% fatty acid–free BSA, 1% penicillin/streptomycin, 1% L-glutamine, and 1% Hepes. Cells were then deposited onto the top of a transwell containing 5-µm pores and allowed to migrate for 3 h toward varying concentrations of CXCL12 in the bottom chamber at 37°C. Cells were then harvested, stained, and resuspended in 40 µl of staining buffer before analysis for 40 s by flow cytometry. For adhesion assays, transwells were precoated with 1, 3, or 10 µg/ml mouse VCAM-1 at 4°C overnight before performing the assay as otherwise described.
In vitro stimulations and inhibitions
Approximately 106 whole BM cells were cultured in IMDM containing 20% FBS, 1% penicillin/streptomycin, 1% L-glutamine, 1% Hepes, and 50 µM β-mercaptoethanol. For all stimulations, anti-CD79b (clone HM79b) was given to cells at a concentration of 10 µg/ml for 8 h. Stimulations were confirmed by observing IgM internalization of mature B lymphocytes by flow cytometry. For inhibitions, cells were given a combination of fostamatinib (1 µM) and ibrutinib (1 µM) or wortmannin (1 µM) for 6 h and were compared with vehicle (DMSO) control.
BM immunofluorescence microscopy
The distinction of proB and preB cells based on the intensity of RAG2:GFP expression was determined as follows: a histogram distribution of voxels for RAG2:GFP in individual CD19+ cells was generated from individual images captured from longitudinal femur sections (z = 30 µm). The top 10% of CD19+ cells with the highest GFP expression (voxels) was classified as proB (GFPHi), and the remaining cells were classified as preB (GFPLo). In Fig. 1, D, E, and G, and Fig. 7 B, each circle represents the average frequency of proB or preB cells in contact with CXCL12+ cells obtained in one mouse. The average frequency was calculated from >200–300 proB and preB cells analyzed from 10 images obtained from longitudinal femur sections imaged by confocal microscopy.
Femurs and tibias were fixed in 4% paraformaldehyde in PBS for 5 h followed by washing with PBS three times. Bones were submerged in 30% sucrose overnight before being snap frozen in optimal cutting temperature media (Tissue-Tek) in a bath of hexane surrounded by dry ice. Bone was shaved off using a cryostat (Leica) until the surface of BM was visible. The bone was then placed in a bath of room temperature PBS to melt remaining optimum cutting temperature compound, followed by permeabilization in 0.1% Triton X-100 for 2 h. Bones were stained with primary antibodies for 3 d at 4°C, followed by secondary antibodies for 1 d at 4°C and tertiary antibodies for 1 d at 4°C. Images were acquired on a Leica SP8 confocal microscope using a 20× oil-immersion objective (0.75 NA). For random positioning analysis, spots placed on DAPI+ and CD19− cells in Imaris software were given a random identifier in Microsoft Excel using the RANDBETWEEN() function, and randomly assigned spots were analyzed.
BrdU incorporation in vivo
Rag2:GFP mice were injected with 1 mg BrdU for 20 min before immediate euthanasia. BM cells were harvested, cell surface markers were stained, and then cells were fixed, and BrdU was detected using BD PharMingen BrdU Flow Kits per the manufacturer’s specification.
Statistics
Student’s two-tailed t test or χ2 tests were performed using PRISM 6 (Graphpad Software) or Excel (Microsoft). P values <0.05 were considered significant.
Online supplemental material
Video 1 shows proB and preB cell movement within calvaria BM of live mice. Video 2 shows proB cell movement in calvaria BM of live mice treated with PBS or with 200 µg of anti–IL-7Ra antibody (clone A7R34). Video 3 shows the in vivo behavior of BCR-ABL+ preB cells retrovirally transduced to overexpress FAK or EV.
Supplementary Material
Acknowledgments
We thank Dr. Alt (Harvard Medical School) for RAG2:GFP mice and Dr. Schatz (Yale University) for RAG2:GFP and Rag1–/– IghB1–8/+ mice. We thank E.F. Ayongaba (University of California, San Francisco) for providing BCR-ABL+ preB cells. We are grateful to L. White (Washington University in St. Louis), and N. Ewing-Crystal and M. Creamer (Yale University) for technical assistance. We also thank A. Cumano (Institut Pasteur, Paris) for critical review of the manuscript.
C. Fistonich was funded by the National Institutes of Health (T32 AI007019); S. Zehentmeier was funded by the German Research Foundation (DFG ZE1060/1-1). This work was funded by the National Institutes of Health (RO1AI113040 to J.P. Pereira).
The authors declare no competing financial interests.
Author contributions: C. Fistonich designed and performed experiments and revised the paper. S. Zehentmeier, J.J. Bednarski, and R. Miao contributed to experiments and revised the paper. H. Schjerven and B.P. Sleckman provided preB-ALL cells and Artemis-deficient BM cells, respectively, and revised the paper. J.P. Pereira designed and performed experiments and wrote the paper.
References
- Balabanian K., Lagane B., Pablos J.L., Laurent L., Planchenault T., Verola O., Lebbe C., Kerob D., Dupuy A., Hermine O., et al. 2005. WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood. 105:2449–2457. 10.1182/blood-2004-06-2289 [DOI] [PubMed] [Google Scholar]
- Beck T.C., Gomes A.C., Cyster J.G., and Pereira J.P.. 2014. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J. Exp. Med. 211:2567–2581. 10.1084/jem.20140457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarski J.J., Pandey R., Schulte E., White L.S., Chen B.R., Sandoval G.J., Kohyama M., Haldar M., Nickless A., Trott A., et al. 2016. RAG-mediated DNA double-strand breaks activate a cell type-specific checkpoint to inhibit pre-B cell receptor signals. J. Exp. Med. 213:209–223. 10.1084/jem.20151048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. 2008. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 456:819–823. 10.1038/nature07392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T.L., Mota-Santos T., Cumano A., Demengeot J., and Vieira P.. 2001. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(-/)- mice. J. Exp. Med. 194:1141–1150. 10.1084/jem.194.8.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.R., Mandal M., Ochiai K., and Singh H.. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14:69–80. 10.1038/nri3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro Gomes A., Hara T., Lim V.Y., Herndler-Brandstetter D., Nevius E., Sugiyama T., Tani-Ichi S., Schlenner S., Richie E., Rodewald H.R., et al. 2016. Hematopoietic Stem Cell Niches Produce Lineage-Instructive Signals to Control Multipotent Progenitor Differentiation. Immunity. 45:1219–1231. 10.1016/j.immuni.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S. 2010. Temporal and spatial regulatory functions of the V(D)J recombinase. Semin. Immunol. 22:362–369. 10.1016/j.smim.2010.09.001 [DOI] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., and Morrison S.J.. 2012. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 481:457–462. 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshkind K. 1990. Regulation of hemopoiesis by bone marrow stromal cells and their products. Annu. Rev. Immunol. 8:111–137. 10.1146/annurev.iy.08.040190.000551 [DOI] [PubMed] [Google Scholar]
- Freitas C., Wittner M., Nguyen J., Rondeau V., Biajoux V., Aknin M.L., Gaudin F., Beaussant-Cohen S., Bertrand Y., Bellanné-Chantelot C., et al. 2017. Lymphoid differentiation of hematopoietic stem cells requires efficient Cxcr4 desensitization. J. Exp. Med. 214:2023–2040. 10.1084/jem.20160806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glodek A.M., Honczarenko M., Le Y., Campbell J.J., and Silberstein L.E.. 2003. Sustained activation of cell adhesion is a differentially regulated process in B lymphopoiesis. J. Exp. Med. 197:461–473. 10.1084/jem.20021477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin R.J., Gelb B., Diaz G.A., Lofsness K.G., Pittelkow M.R., and Fenyk J.R. Jr. 2000. WHIM syndrome, an autosomal dominant disorder: clinical, hematological, and molecular studies. Am. J. Med. Genet. 91:368–376. [DOI] [PubMed] [Google Scholar]
- Grabstein K.H., Waldschmidt T.J., Finkelman F.D., Hess B.W., Alpert A.R., Boiani N.E., Namen A.E., and Morrissey P.J.. 1993. Inhibition of murine B and T lymphopoiesis in vivo by an anti-interleukin 7 monoclonal antibody. J. Exp. Med. 178:257–264. 10.1084/jem.178.1.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P.A., Gorlin R.J., Lukens J.N., Taniuchi S., Bohinjec J., Francois F., Klotman M.E., and Diaz G.A.. 2003. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat. Genet. 34:70–74. 10.1038/ng1149 [DOI] [PubMed] [Google Scholar]
- Jacobsen K., and Osmond D.G.. 1990. Microenvironmental organization and stromal cell associations of B lymphocyte precursor cells in mouse bone marrow. Eur. J. Immunol. 20:2395–2404. 10.1002/eji.1830201106 [DOI] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., and Singh H.. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345. 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Joshi I., Yoshida T., Jena N., Qi X., Zhang J., Van Etten R.A., and Georgopoulos K.. 2014. Loss of Ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat. Immunol. 15:294–304. 10.1038/ni.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata N., Igarashi H., Ohmura T., Aizawa S., and Sakaguchi N.. 1999. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J. Immunol. 163:6355–6359. [PubMed] [Google Scholar]
- Lim V.Y., Zehentmeier S., Fistonich C., and Pereira J.P.. 2017. A Chemoattractant-Guided Walk Through Lymphopoiesis: From Hematopoietic Stem Cells to Mature B Lymphocytes. Adv. Immunol. 134:47–88. 10.1016/bs.ai.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., and Clark M.R.. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220. 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucchelli R.I., Warming S., Lawrence S.M., Ishii M., Abshari M., Washington A.V., Feigenbaum L., Warner A.C., Sims D.J., Li W.Q., et al. 2009. Visualization and identification of IL-7 producing cells in reporter mice. PLoS One. 4:e7637 10.1371/journal.pone.0007637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughtry T.M., Etzensperger R., Alag A., Tai X., Kurtulus S., Park J.H., Grinberg A., Love P., Feigenbaum L., Erman B., and Singer A.. 2012. Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus. J. Exp. Med. 209:2263–2276. 10.1084/jem.20121505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott D.H., Gao J.L., Liu Q., Siwicki M., Martens C., Jacobs P., Velez D., Yim E., Bryke C.R., Hsu N., et al. 2015. Chromothriptic cure of WHIM syndrome. Cell. 160:686–699. 10.1016/j.cell.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.N., Hartigan-O’Connor D.J., Lee M.S., Laidlaw G., Cornelissen I.P., Matloubian M., Coughlin S.R., McDonald D.M., and McCune J.M.. 2013. IL-7 production in murine lymphatic endothelial cells and induction in the setting of peripheral lymphopenia. Int. Immunol. 25:471–483. 10.1093/intimm/dxt012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., and Papaioannou V.E.. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. 10.1016/0092-8674(92)90030-G [DOI] [PubMed] [Google Scholar]
- Monroe R.J., Seidl K.J., Gaertner F., Han S., Chen F., Sekiguchi J., Wang J., Ferrini R., Davidson L., Kelsoe G., and Alt F.W.. 1999. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 11:201–212. 10.1016/S1074-7613(00)80095-3 [DOI] [PubMed] [Google Scholar]
- Mourcin F., Breton C., Tellier J., Narang P., Chasson L., Jorquera A., Coles M., Schiff C., and Mancini S.J.. 2011. Galectin-1-expressing stromal cells constitute a specific niche for pre-BII cell development in mouse bone marrow. Blood. 117:6552–6561. 10.1182/blood-2010-12-323113 [DOI] [PubMed] [Google Scholar]
- Muljo S.A., and Schlissel M.S.. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37. 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Nevius E., Pinho F., Dhodapkar M., Jin H., Nadrah K., Horowitz M.C., Kikuta J., Ishii M., and Pereira J.P.. 2015. Oxysterols and EBI2 promote osteoclast precursor migration to bone surfaces and regulate bone mass homeostasis. J. Exp. Med. 212:1931–1946. 10.1084/jem.20150088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai K., Maienschein-Cline M., Mandal M., Triggs J.R., Bertolino E., Sciammas R., Dinner A.R., Clark M.R., and Singh H.. 2012. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 13:300–307. 10.1038/ni.2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Wolfram P., Canty K., Harley B., Nombela-Arrieta C., Pivarnik G., Manis J., Beggs H.E., and Silberstein L.E.. 2013. Focal adhesion kinase regulates the localization and retention of pro-B cells in bone marrow microenvironments. J. Immunol. 190:1094–1102. 10.4049/jimmunol.1202639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelanda R., Schwers S., Sonoda E., Torres R.M., Nemazee D., and Rajewsky K.. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 7:765–775. 10.1016/S1074-7613(00)80395-7 [DOI] [PubMed] [Google Scholar]
- Pelanda R., Hobeika E., Kurokawa T., Zhang Y., Kuppig S., and Reth M.. 2002. Cre recombinase-controlled expression of the mb-1 allele. Genesis. 32:154–157. 10.1002/gene.10070 [DOI] [PubMed] [Google Scholar]
- Pereira J.P., An J., Xu Y., Huang Y., and Cyster J.G.. 2009. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat. Immunol. 10:403–411. 10.1038/ni.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon J.J., Morrissey P.J., Grabstein K.H., Ramsdell F.J., Maraskovsky E., Gliniak B.C., Park L.S., Ziegler S.F., Williams D.E., Ware C.B., et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180:1955–1960. 10.1084/jem.180.5.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S.E., Mandal M., Matsuda S., Miletic A.V., Cato M.H., Tanaka A., Rickert R.C., Koyasu S., and Clark M.R.. 2012. Subnuclear cyclin D3 compartments and the coordinated regulation of proliferation and immunoglobulin variable gene repression. J. Exp. Med. 209:2199–2213. 10.1084/jem.20120800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A., Ziegler S.F., Buckley R.H., and Leonard W.J.. 1998. Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat. Genet. 20:394–397. 10.1038/3877 [DOI] [PubMed] [Google Scholar]
- Rother M.B., Jensen K., van der Burg M., van de Bovenkamp F.S., Kroek R., van IJcken W.F., van der Velden V.H., Cupedo T., Olstad O.K., van Dongen J.J., and van Zelm M.C.. 2016. Decreased IL7Rα and TdT expression underlie the skewed immunoglobulin repertoire of human B-cell precursors from fetal origin. Sci. Rep. 6:33924 10.1038/srep33924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerven H., McLaughlin J., Arenzana T.L., Frietze S., Cheng D., Wadsworth S.E., Lawson G.W., Bensinger S.J., Farnham P.J., Witte O.N., and Smale S.T.. 2013. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 14:1073–1083. 10.1038/ni.2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerven H., Ayongaba E.F., Aghajanirefah A., McLaughlin J., Cheng D., Geng H., Boyd J.R., Eggesbø L.M., Lindeman I., Heath J.L., et al. 2017. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J. Exp. Med. 214:793–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert T.A., Tagoh H., Gültekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 15:283–293. 10.1038/ni.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E., Brakebusch C., Martensson I.L., Svensson M., Agace W.W., Sigvardsson M., Buza-Vidas N., Bryder D., Cilio C.M., Ahlenius H., et al. 2003. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J. Exp. Med. 198:1495–1506. 10.1084/jem.20031152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Pewzner-Jung Y., Schwers S., Taki S., Jung S., Eilat D., and Rajewsky K.. 1997. B cell development under the condition of allelic inclusion. Immunity. 6:225–233. 10.1016/S1074-7613(00)80325-8 [DOI] [PubMed] [Google Scholar]
- Thompson E.C., Cobb B.S., Sabbattini P., Meixlsperger S., Parelho V., Liberg D., Taylor B., Dillon N., Georgopoulos K., Jumaa H., et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 26:335–344. 10.1016/j.immuni.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Tokoyoda K., Egawa T., Sugiyama T., Choi B.I., and Nagasawa T.. 2004. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 20:707–718. 10.1016/j.immuni.2004.05.001 [DOI] [PubMed] [Google Scholar]
- von Freeden-Jeffry U., Vieira P., Lucian L.A., McNeil T., Burdach S.E., and Murray R.. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181:1519–1526. 10.1084/jem.181.4.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskow C., Paul S., Haller C., Gassmann M., and Rodewald H.R.. 2002. Viable c-Kit(W/W) mutants reveal pivotal role for c-kit in the maintenance of lymphopoiesis. Immunity. 17:277–288. 10.1016/S1074-7613(02)00386-2 [DOI] [PubMed] [Google Scholar]
- Wei C., Zeff R., and Goldschneider I.. 2000. Murine pro-B cells require IL-7 and its receptor complex to up-regulate IL-7R alpha, terminal deoxynucleotidyltransferase, and c mu expression. J. Immunol. 164:1961–1970. 10.4049/jimmunol.164.4.1961 [DOI] [PubMed] [Google Scholar]
- Zhou X., Franklin R.A., Adler M., Jacox J.B., Bailis W., Shyer J.A., Flavell R.A., Mayo A., Alon U., and Medzhitov R.. 2018. Circuit Design Features of a Stable Two-Cell System. Cell. 172:744–757.e17. 10.1016/j.cell.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.