Abstract
Lipids have been analyzed in applications including drug discovery, disease etiology elucidation, and natural products. The chemical and structural diversity of lipids requires a tailored lipidomics workflow for each sample type. Therefore, every protocol in the lipidomics workflow, especially those involving sample preparation, should be optimized to avoid the introduction of bias. The coupling of ultra-high-performance liquid chromatography (UHPLC) with high-resolution mass spectrometry (HRMS) allows for the separation and identification of lipids based on class and fatty acid acyl chain. This work provides a comprehensive untargeted lipidomics workflow that was optimized for various sample types (mammalian cells, plasma, and tissue) to balance extensive lipid coverage and specificity with high sample throughput. For identification purposes, both data-dependent and data-independent tandem mass spectrometric approaches were incorporated, providing more extensive lipid coverage. Popular open-s ource feature detection, data processing, and identification software are also outlined.
Keywords: Lipidomics, Ultra-high performance liquid chromatography (UHPLC), High-resolution mass spectrometry, Sample preparation, Biomarker discovery
1. Introduction
Lipids are analyzed in numerous settings including synthetic and natural products industries [1–6] and medical fields [7–10]. The diverse biological functions and ubiquitous occurrence of lipids highlight their potential as clinical biomarkers and as indicators of pathways perturbed by disease or environmental exposure. Lipid diversity in function is enabled by lipid diversity in structure, with over 180,000 possible lipid species at the level of fatty acid constituents [11]. Lipid structural diversity, amphiphilic nature, and concentrations range several million-fold [12], all pose unique analytical challenges in the field of lipidomics.
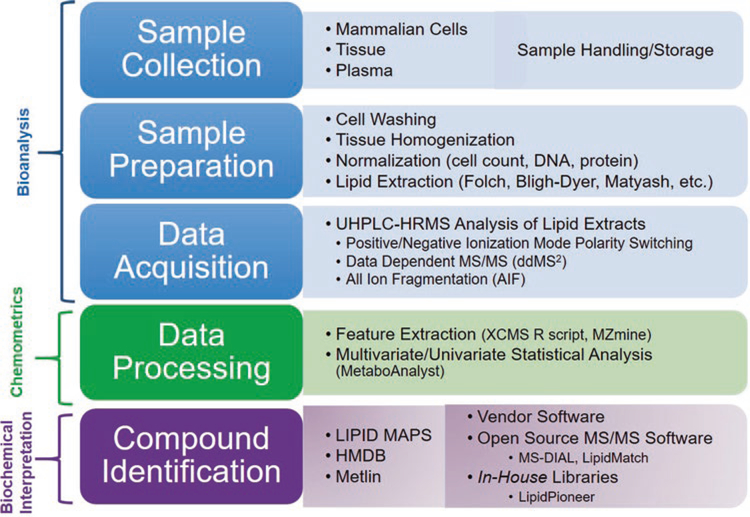
The most comprehensive lipidomics workflows in terms of the number and accuracy of lipid species analyzed include sample homogenization and extraction, separation by liquid chromatography, ionization and detection by electrospray ionization high- resolution tandem mass spectrometry (ESI-HR MS/MS), feature detection and identification, and statistical analysis (Fig. 1). Because lipids encompass a broad range of chemical and physical properties, lipidomics sample preparation workflows have not been standardized in the literature. In addition, every step of the sample preparation workflow (e.g., storage, sample handling, homogenization, and lipid extraction) may bias the results. Therefore, the lipidomics sample preparation workflow should be tailored to the sample type. The amphiphilic lipids are amenable to reverse phase (RP) chromatography, where research has shown these lipids under RP conditions to separate based on class, fatty acid constituents, and even by sn1 and sn2 positional isomers and double bond positions [13]. Chromatographic separation enhances specificity, reduces ion suppression in ESI-HRMS/MS, and aids in the identification of lipid species. High-resolution tandem mass spectra of precursor ions and fragment ions containing information on lipid class and fatty acid acyl chain information can be used to confidently identify lipid structures.
Fig. 1.
Lipidomics workflow including the components for bioanalysis, chemometrics with open-source software, and biochemical interpretation of lipid extracts
Here, we describe a workflow that is optimized for various sample types (mammalian cells, plasma, and tissue), which balances extensive lipid coverage and specificity with high sample throughput. In addition, quality control procedures are included to characterize any introduction of non-biological variability. The workflow provides semi-quantitative lipid concentrations using available lipid class-representative standards, allowing comparison of lipid data between different techniques and labs. For identification purposes, both data-dependent and data-independent tandem mass spectrometric approaches are used, providing more extensive lipid coverage. Popular open-source feature detection and identification software are described in the notes with suggested parameters. Data-processing and interpretation strategies are outlined to reduce the chance of false positives and false negatives, providing as much information on up- and down-regulated lipids and lipid signatures as possible. In addition, multivariate statistical techniques and open-source software for predicting and categorizing biological perturbations based on lipid profiles are described.
2. Materials
Prepare all solutions using LC-MS grade solvents. Refrigerate solvents used for sample preparation and store all samples in temperatures at −80 °C or below. For the purpose of this study, analytical grade solvents (formic acid, chloroform, and methanol) were purchased from Fisher-Scientific (Fairlawn, NJ). Mobile phase solvents were Fisher Optima LC/MS grade (acetonitrile, isopropanol, and water). Triacylglyceride lipid standards (TG 15:0/15:0/15:0 and TG 17:0/17:0/17:0) were purchased from Sigma-Aldrich (St. Louis, MO). Exogenous lysophosphatidylcholine (LPC 17:0 and LPC 19:0), phosphatidylcholine (PC 17:0/17:0 and PC 19:0/19:0), phosphatidylethanolamine (PE 15:0/15:0 and PE 17:0/17:0), phosphatidylserine (PS 14:0/14:0 and PS 17:0/17:0), and phosphatidylglycerol (PG 14:0/14:0 and PG 17:0/17:0) lipid standards were purchased from Avanti Polar Lipids (Alabaster, AL) (see Note 1). All lipid standards were diluted prior to analysis in 1:2 (v/v) chloroform:methanol (CHCl3:MeOH) and a working standard mix was then prepared by diluting the stock solution with the same solvent mixture. Butylated hydroxytoluene (BHT) was purchased from Sigma Aldrich. All protein and DNA assay measurements were obtained using a Qubit 3.0 Fluorometer purchased from Thermo Fisher Scientific. Lipid extracts were dried down using a MultiVap 118 nitrogen dryer set at 30 °C (Organomation Associates, Inc.).
2.1. Sample Preparation
Mammalian cells: minimum of 106 cells, cell rinsing solution: deionized water with ammoniated cell washing buffer (see Note 2).
Tissue: liquid nitrogen-pooled mortar and ceramic pestle, liquid nitrogen, homogenization beads (zirconium oxide or ceramic for soft tissue, and stainless steel for muscle and harder tissues).
Plasma collected using EDTA anticoagulant. Stored for long term at −80 °C in polypropylene Eppendorf tubes.
Centrifuge (see Note 3).
Polypropylene Eppendorf tubes (1.5 mL and/or 2 mL), polypropylene conical tubes (5 mL and/or 15 mL) (see Note 4).
2.2. Sample- Dependent Folch Lipid Extraction
Lipid internal standard mix: create a stock solution [1:2 (v/v) chloroform: methanol] of the following lipids: PC 17:0/17:0, PC 19:0/19:0, PE 15:0/15:0, PE 17:0/17:0, PS 14:0/14:0, PS 17:0/17:0, PG 14:0/14:0, PG 17:0/17:0, TG 15:0/ 15:0/15:0, and TG 17:0/17:0/17:0 (see Note 1).
Folch lipid extraction solvents: methanol with 1 mM butylated hydroxytoluene (BHT), chloroform, water (see Note 5).
Re-extraction solvent: 2:1 (v/v) chloroform/methanol (see Note 6).
Vortex.
Nitrogen dryer.
2.3. UHPLC-HRMS Data Acquisition
UHPLC C18 column (see Note 7).
Reconstitution solvent: 100% isopropanol.
Solvent A: acetonitrile:water (60:40, v/v) with 10 mM ammonium formate and 0.1% formic acid (see Note 8).
Solvent B: isopropanol:acetonitrile:water (90:8:2, v/v) with 10 mM ammonium formate and 0.1% formic acid (see Note 8).
UHPLC system coupled to a high-resolution mass spectrometer capable of employing positive and negative ionization with a heated electrospray probe (see Note 9).
Glass vials with 200 μL inserts.
3. Methods
All samples and solvents during the Folch lipid extraction should be kept on ice. Avoid exposing samples to room temperature for more than 5 min.
3.1. Sample Preparation
Cell preparation: Pellet mammalian cells in a 15 mL conical tube at 311 × g for 5 min at 4 °C. Wash cell pellet 2–3 times by adding 1 mL of the cell rinsing solution. During the last washing step, reconstitute the cells in the rinsing solution and obtain a 5 μL aliquot for each assay (protein and/or DNA measurement). Store the cell pellet at −80 °C or perform the lipid extraction (see Note 10).
Plasma preparation: Thaw plasma samples on ice before pipetting 40 μL into a 2.0 mL centrifuge tube prior to the Folch extraction. Maintain samples on ice for the remainder of extraction (see Note 11).
Tissue Pulverization/Homogenization: Flash freeze tissues in liquid nitrogen quickly after collection and store at −80 °C. Further pulverize individual tissue samples in a liquid nitrogen- cooled mortar with a ceramic pestle. Weigh the fine powder into a homogenization tube using balance tared with homogenization beads (ceramic or zirconium oxide for soft tissues, and stainless steel for muscle or harder tissues). Aim for 50 mg of tissue. Record the weight of the tissue powder as further extraction volumes will be adjusted based on weight of tissue. Add internal standard to the tissue (125 μL of 160 ppm) and homogenize tissues for lipid extraction in the Folch solvents [chloroform:methanol, (2:1, v:v)] at a volume (μL) at 20 times the weight (mg) (see Note 12).
Create a pooled sample group quality control (see Note 13) and/or use a standard reference material (SRM) (e.g., Red Cross Blood Plasma or National Institute of Standards and Technology (NIST) SRM) as a quality control (see Note 14).
3.2. Sample- Dependent Folch Lipid Extraction
Spike in an aliquot of the lipid internal standard mix into plasma or mammalian cells and an empty Eppendorf/conical tube as an extraction blank (see Note 15).
Add ice-cold methanol with 1 mM BHT and chloroform (1:2, v/v) directly to the sample.
Incubate on ice for 30 min and vortex occasionally.
Add ice-cold water to a final ratio of chloroform/methanol/water (8:4:3, v/v/v) and incubate on ice for an additional 10 min.
Centrifuge the sample at 311 × g for 5 min at 4 °C to separate the aqueous and organic layer.
Pipette through aqueous layer (upper phase) and transfer the organic layer (lower phase) to a separate Eppendorf/conical tube without contaminating the organic phase with the protein layer.
Re-extract on the remaining aqueous layer by adding the re- extraction solvent, vortexing, and centrifuging for 5 min at 4 °C.
Dry down the organic layer under nitrogen at 30 °C (see Note 16).
Reconstitute the dried lipid extract with 100% isopropanol (see Note 17).
Transfer lipid extract to an LC vial with a 200 μL glass insert.
3.3. UHPLC-HRMS Data Acquisition
Create an instrument sequence that begins with solvent and extraction blanks to give time for column and instrument equilibration (see Note 18).
Equilibrate the UHPLC C18 column at 50 °C with starting percentages of Solvent A and B as mobile phases (see Note 19).
Apply the following LC gradient: 32% B at 0 min, 40% B at 1 min, a hold at 40% B until 1.5 min, 45% B at 4 min, 50% B at 5 min, 60% B at 8 min, 70% B at 11 min, and 80% B at 14 min at a flow rate of 0.5 mL/min (see Note 20).
Maintain the autosampler at 5 °C.
The following heated electrospray ionization (HESI) parameters were used in positive ion mode: spray voltage at 3.3 kV, sheath gas and auxiliary nitrogen pressure at 30 and 5 arbitrary units, respectively, and capillary and heater temperatures at 300 °C and 350 °C, respectively. HESI parameters that differed in negative ion mode were sheath gas and auxiliary gas at 25 and 15 arbitrary units, respectively, and a capillary temperature of 250 °C.
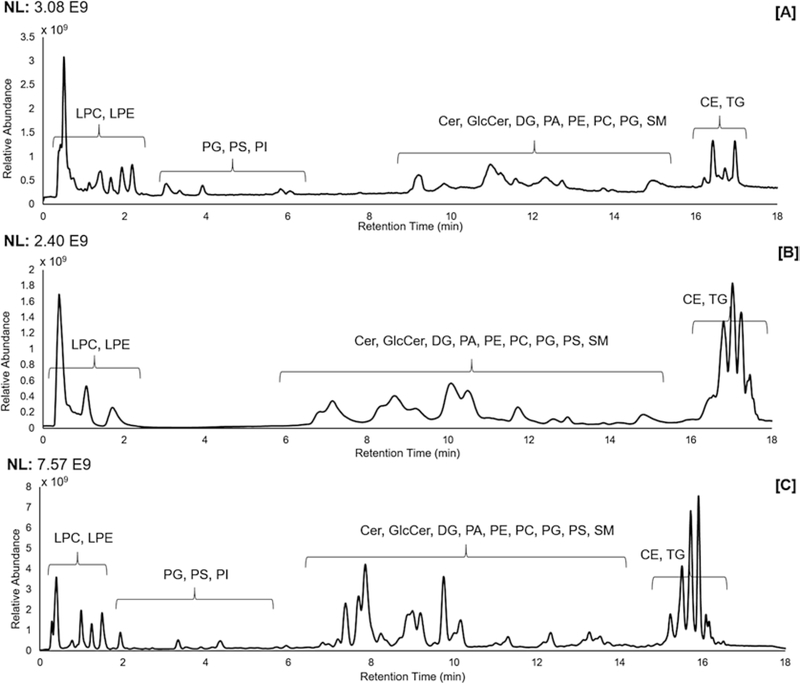
The following full-scan MS conditions were used with polarity switching (see Note 21) following calibration of the instrument (see Note 22): S-lens RF level at 35 V, a resolution of 70,000 with an automatic gain control of 5 × 106 ions, and maximum injection time of 256 ms, scanning from m/z 200– 1500 (see Note 23). All data were acquired in profile mode. See Fig. 2 for the TIC of lipid extracts from mammalian cells, tissue, and plasma collected in positive ion mode.
For identification (see Note 24), pooled samples from each sample group were analyzed in both polarities separately using alternate full scans and all ion fragmentation (AIF) scans with AIF parameters as follows: a resolution of 70,000 with an automatic gain control of 5 × 106 ions and maximum injection time of 256 ms, scanning from m/z 100–1500 with a stepped normalized collision energy (NCE) of 15, 20, and 25 (see Note 25).
Fragmentation of ions obtained in each polarity separately in pooled samples was acquired using data-dependent top10 (ddMS2-top10) analysis as well. Ions were isolated using a 1 amu window, and isolation was triggered using an intensity threshold of 5 × 104 (setting the underfill ratio to reach this desired target), an apex trigger of 10–20 s, isotope exclusion on, and a dynamic exclusion of 4 s. Ions were fragmented by HCD using NCEs of 15, 20, and 25, and fragments were measured with a resolution of 35,000 with an automatic gain control of 5 × 106 ions and maximum injection time of 175 ms.
Fig. 2.
Total ion chromatogram of the reverse-phase endogenous lipid elution profile for extracted (a) Jurkat T lymphocyte cells, (b) skin tissue, (c) plasma in positive ion mode using a Supelco Analytical Titan C18 column
3.4. HRMS Lipidomic Data Processing
Convert .raw files to an mzXML output using software such as ProteoWizard MSConvert (see Note 26).
Process the mzXML files using a feature detection and alignment software (see Note 27).
Export the peak-picked data as a .csv file that can be imported for data analysis and interpretation.
3.5. Data Analysis and Interpretation
Apply a univariate and/or multivariate statistical analysis software to the peak-picked data (see Note 28).
Match features that significantly differ between predefined sample groups from feature detection and alignment software to a database or in-house library for annotation. Confidence in annotation increases with information including m/z, adduct, retention time, and fragmentation pattern (see Notes 24 and 29).
4. Notes
Other lipid internal standards that represent common lipid classes that can be purchased from Avanti Polar lipids include diacylglyceride (DG 14:0/14:0), sphingomyelin SM (d18:1/17:0), and ceramide Cer (d18:1/17:0).
A minimum of 106 suspension and/or adherent cells are needed for lipidomic studies to ensure a sufficient instrument signal (107–108 peak intensity). Cells should be washed with an ammoniated cell washing buffer as these rinsing solvents are compatible with ESI-LC-MS conditions [14].
For mammalian cell lipidomics studies, a centrifuge is needed that can support 5 or 15 mL conical tubes. For plasma and tissue lipidomics studies, a centrifuge is needed that can support 1.5 or 2 mL Eppendorf tubes.
Polypropylene tubes should be used when working with chloroform.
The methanol contains 1 mM BHT to minimize oxidation of lipid species [15].
Because 1 mM BHT was added during the initial steps of the extraction, it is not needed here.
Options for a UHPLC C18 columns include: Supelco Analytical Titan C18 (2.1 × 75 mm, 1.9 μm) and Waters Acquity™ BEH C18 column (2.1 × 50 mm, 1.7 μm). Polar lipid species can be separated on a HILIC column such as the Agilent HILIC Plus RRHD column (2.1 × 150 mm, 1.8 μm).
Use glass pipettes to aliquot formic acid into mobile phases. A 1 mL ampule of formic acid for each liter of mobile phase can be used to avoid contamination.
This work incorporated a Dionex Ultimate 3000 UHPLC system coupled to a Q Exactive Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA).
If possible, normalize all samples using the cell count before performing sample preparation. If the cell count information is unavailable, data can be normalized to DNA or protein concentrations post-data acquisition [16].
Typically, a consistent volume used for plasma is adequate; however, protein content may be assessed to provide an alternative method of normalization. Another extraction method that works well for lipids from plasma is the Matyash MTBE method [17]. Notes comparing the two methods were published by Patterson, et al. [18].
Maintain tissue weight:homogenization solvent ratio at 1 mg:20 μL to avoid tissue weight normalization post-data acquisition and differences in extraction efficiencies based on lipid concentrations. For example, 50 mg of tissue will require 1000 μL of Folch solvent. Alternatively, tissue can be homogenized in an aqueous buffer (ammonium formate, etc.) and then aliquoted like plasma; however, we have found most efficient homogenization for lipid analysis occurs with organic solvents during homogenization. If the tissue is to be homogenized in the Folch solvents, continue with Subheading 3.2, step 3.
Sample pools can be created before or after extraction depend-ing on the sample type. If pooled before extraction, an even aliquot from all samples in the experiment or group should be pooled. For plasma, 10 μL (for example) can be pooled from each sample and combined into one Eppendorf tube. Once mixed well, that pooled sample can be prepared alongside the other samples in the experiment. For tissue, equal weights of pulverized powder can be combined in a separate tube. Once mixed well, this pooled powder can be weighed out and prepared alongside other samples in the experiment. For cells, a reconstituted aliquot of cells (in an ammoniated buffer) can be pooled for each sample group. The advantage of pooling before extraction is that the sample can be maintained as a QC for future experiments using the same samples. If samples are pooled after extraction and reconstitution, an even volume from each sample can be pooled in to a separate LC vial prior to analysis. A dilution series of the pooled sample (e.g., 1:1, 1:2, 1:4, 1:8) analyzed at the end of a sequence can enable discrimination between artifacts and features of biological origin [19]. These samples are made post reconstitution and each contains an aliquot of the pooled QC with increasing amounts of isopropanol.
The standard reference material (SRM) can be used in addition to the pooled sample group QCs. The SRM can be prepared alongside samples every day of sample preparation. This SRM provides injections for inter- and intra-day variability comparisons and consistency of instrument and column quality. If pooled QCs are not made, representative samples from each sample group can be used to obtain lipid identification using MS/MS, see Subheading 3.3, steps 7 and 8.
The amount of internal standard spiked in should be compa-rable to the intensity level of endogenous lipids in the sample. The concentration of the internal standard required varies with the sample type but should be close to 10–15 ppm. The purpose of the extraction blank is to monitor the sample preparation reproducibility and to identify background interferences due to the sample preparation process. The volume of the Folch extraction solvents is dependent on the sample type. However, the final ratios of the solvents should be chloroform/methanol/water (8:4:3, v/v/v).
If the samples cannot be processed on the instrument on the same day, store the dried lipid extract at −80 °C until analysis.
Lipid extracts reconstituted with 100% IPA should undergo data acquisition on the same day to avoid sample degradation and lipid oxidation. Mammalian cell extracts containing 106– 107 cells should be reconstituted in 50 μL of IPA. Plasma extracts (from 40 μL of plasma) should be reconstituted in 100 μL IPA. Tissue extracts (from 50 mg of tissue) should be reconstituted in 500 μL IPA. Do not use inserts in vials in this case. For HILIC analysis, the lipid extract should be reconstituted in no less than 80% acetonitrile.
Following initial blanks, two solvent blanks followed by QCs should be run and incremented every 10–15 sample injections. Sample injections must be randomized throughout the sequence. The sequence ends with the dilution series of the pooled sample, if available.
The column temperature was increased to 50 °C to reduce the column back pressure when the gradient increases to 100% IPA.
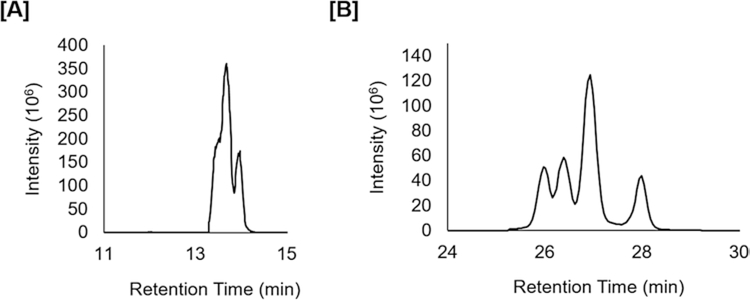
In order to increase throughput for large-scale lipidomic studies, we recommend shorter chromatographic runs. Be aware that lipids with different fatty acid constituents, but the same total number of carbons and degrees of unsaturation are isomeric and will often coelute. This is especially problematic for triacylglycerides where over ten species identified by MS/MS for a given feature have been determined in our previous work (unpublished). Fig. 3 shows an example for TG(50:6) where increasing the time for chromatographic separation shows multiple peaks (Fig. 3b) which were indistinguishable in shorter chromatographic runs (Fig. 3a). Therefore, changes in concentrations across sample groups for one feature might represent changes in an average of various lipid species. Even at longer (e.g., 70 min) chromatographic runs, TGs molecular species will often overlap.
Polarity switching reduces the number of scans across a peak significantly as it takes 1 s to switch between polarities for which time nearly four full scans can be acquired. In the case that more scans are desired across a peak (e.g., to improve deconvolution of closely eluting peaks), samples can be run in both positive and negative polarity separately, although this nearly doubles data acquisition time. In addition, lipid ions in negative mode are often at least an order of magnitude lower in signal than respective lipid ions in positive mode. In the case where the negative ion mode signal must be increased without increasing positive ion mode signal (due to saturation effects at high concentrations), separate negative mode analysis of more concentrated samples or separate analysis with higher injection volumes for negative mode can be performed. Note that higher injection volumes should not exceed 10 μL, as this could shift retention times.
For external calibration, less than a 3 ppm error in mass accuracy should be obtained in positive and negative polarity, with our lab consistently obtaining less than a 1 ppm error in mass accuracy. Lock masses greatly increase the accuracy of mass measurements. Lock masses used for positive ion mode were diisooctyl phthalate (m/z 391.2842) and polysiloxanes (m/z 371.1012 and 445.1200). No known background ions with stable signal across the entire chromatographic region are available to be used as lock masses in negative ion mode.
The m/z scan window was increased to m/z 1500 in order to include the [M−H]− adduct of the cardiolipin species.
When identifying features, assumptions should be clearly indicated in assigning a specific level of structural resolution. Note that not all lipid identification software annotates lipid structures based on MS/MS data properly, with the danger of over reporting structural resolution. Exact mass alone only gives tentative identification of lipid class and total carbons and double bonds, with class-specific fragments giving more confidence in identifications. If fragments containing fatty acid constituents occur, care should be taken in identifying lipids at this level. Using nominal mass isolation, fatty acid fragments could come from multiple coeluting species, including those without the same exact mass and from different lipid classes. A table of currently accepted notation for lipid assignment with the structural resolution determined by information obtained using mass spectrometry is provided for reference (Table 1).
The injection volume will change depending on the sample type. Inject at least 5 μL for the mammalian cell lipid extracts. Because of the lower ion intensity observed in negative ionization mode, a larger injection volume may be required in which case, polarity switching cannot be used (see Note 21).
Files should be converted to an mzXML format to centroid the data and reduce the file size. The mzXML files can be directly uploaded to peak picking/detection software.
(1) XCMS [20] has an online and R script workflow that can be used for data processing. The built-in workflow was developed for filtering, peak picking/integration, peak matching, RT alignment, and gap filling. (2) The MZmine 2 [21] workflow used in our lab consists of creating a mass peak list, building chromatograms, smoothing chromatograms, peak deconvolution and integration (using a local minimum algorithm), deisotoping peaks, peak list alignment, gap filling, and filtering. A batch-processing file for MZmine containing optimized parameters for column-dependent lipidomic analysis can be created and applied to all samples. (3) MS-DIAL [22] can also be used with the same steps above as in MZmine for feature detection. In our lab MS-DIAL often crashed or outputted erroneous peak picking results when samples sizes greater than eight were used. The advantage of MS-DIAL is that it also incorporates the identification of the lipids using LipidBlast [23] libraries and either data-dependent fragmentation spectra or data-independent fragmentation spectra. While originally designed for SWATH, a text file containing AIF parameters can be used in conjunction with MS-DIAL to identify lipids from AIF data. Both MS-DIAL and MZmine provide user interfaces to view peaks and the resulting peak integration, while XCMS does not. Manual quality control of feature detection is an important step for insuring the correct parameters are used and data quality is sufficient for research goals.
MetaboAnalyst is an online processing and bioinformatics tool used to analyze and visualize statistical differences in complex lipidomic datasets [24, 25]. Options for univariate statistical analysis in MetaboAnalyst include fold change, t-test, volcano plots, ANOVA, correlations, and significance analysis of microarrays (SAM). Multivariate analysis platforms available include principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA). Clustering/classification options include, but are not limited to, random forest, K-means clustering, and hierarchical clustering. Additional data analysis/identification pro-grams include the web-based MeltDB [26], commercial SIMCA-P software [27], and commercial SAS software.
Examples of lipidomic databases used for lipid identification include LIPID MAPS [28], the Human Metabolome Database (HMDB) [29], Metlin [30], vendor software, and in- house libraries. Software for annotating lipid structure based on frag-mentation for LC-MS/MS data (see Note 22) includes LipidSearch [31, 32], MS-DIAL [22], and LipidMatch [33] or an in-house fragmentation library/software. The lipid identification software, LipidMatch [33], provides additional unique features in comparison to MS-Dial, including the ability to incorporate MZmine and XCMS outputs, to obtain structurally defined lipid assignments, and to mine simulated MS/MS libraries for oxidized lipids. Table 2 provides a list of 31 of the 56 lipid types with in silico fragmentation libraries in LipidMatch for identification of various lipid species. It is common to have less than 50% of features match annotations in databases, depending on thresholds used in peak detection and depth of libraries.
Fig. 3.
Reconstructed ion chromatograms of ammoniated TG(50:6) ions using an 18 min chromatographic run (a) and a 45 min chromatographic run (b) on a Waters™ Acquity BEH C18 column. Short chromatographic runs exhibit features that often represent not one, but multiple lipid species
Table 1.
Lipid assignments using tandem mass spectrometry should take into account the structural resolution known. One problem in the field of lipidomics is over reporting structural resolution
| Structural resolution | Example |
|---|---|
| Carbons and double bonds | PC(34:2) |
| Fatty acid constituents | PC(16:0_18:2) |
| Positional isomers | PC(16:0/18:2) |
| Double bond position | PC(16:0/18:2(9,12)) |
| Double bond cis (Z) or trans (E) | PC(16:0/18:2(9Z, 12E)) |
Table 2.
Number of simulated fragments for selected lipid classes used by the LipidMatch software [33]
| Class | Adducts | Species |
|---|---|---|
| Ac(2,3,4)PIM(1,2) | [M−H]− | 22,621 |
| AcCa | [M+H]+ | 53 |
| (L)PC | [M+H]+, [M+Na]+, [M+HCO2]− | 779 |
| LPC(O−34:2) | [M+H]+, [M+HCO2]− | 114 |
| LPC(P−36:3) | [M+H]+, [M+HCO2]− | 228 |
| (L)PE | [M+H]+, [M+Na]+, [M−H]− | 779 |
| (L)PE(O− | [M+H]+, [M−H]− | 114 |
| (L)PE(P− | [M+H]+, [M−H]− | 228 |
| (L)PI | [M−H]−, [M+H]+, [M+NH4]+ | 779 |
| CE | [M+NH4]+ | 38 |
| Cer | [M+H]+ | 1445 |
| CerG | [M+H]+ | 30 |
| CerP | [M+H]+, [M−H]− | 169 |
| CL | [M-2H]− | 3065 |
| CoQ | [M+NH4]+ | 5 |
| DG | [M+NH4]+ | 741 |
| DGDG | [M+HCO2]− | 780 |
| Ganglioside | [M−H]− | 1353 |
| LipidA_PP | [M−H]− | |
| MMPE | [M−H]− | |
| DMPE | [M−H]− | |
| MG | [M+NH4]+ | 50 |
| MGDG | [M+NH4]+, [M+H]+, [M+NH4−CO]+ | 2305 |
| Ox(L)PC | [M+H]+, [M+Na]+, [M+HCO2]− | 31,326 |
| Ox(L)PE | [M+H]+, [M−H]−, [M+Na]+ | 31,326 |
| OxTG | [M+NH4]+ | 8557 |
| PA | [M−H]−, [M+H]+ | 741 |
| PG | [M−H]−, [M+NH4]+, [M+Na]+ | 741 |
| PS | [M−H]−, [M+H]+, [M+Na]+ | 741 |
| PS(O- | [M+H]+, [M−H]− | 114 |
| PS(P- | [M+H]+, [M−H]− | 228 |
| SM | [M+H]+ | 1445 |
| So | [M+H]+ | 13 |
| Sulfatide | [M−H]− | 169 |
| TG | [M+NH4]+ | 115,520 |
Acknowledgments
This work was supported by the Southeast Center for Integrated Metabolomics (SECIM)—NIH Grant #U24 DK097209.
References
- 1.Anonymous (2015) Major fats and oils industry overview In: Chemical economics handbook. IHS Markit, London [Google Scholar]
- 2.Batenburg JJ (1992) Surfactant phospholipids: synthesis and storage. Am J Phys 262(4 Pt 1):L367–L385 [DOI] [PubMed] [Google Scholar]
- 3.Bhuyan S, Sundararajan S, Yao L, Hammond EG, Wang T (2006) Boundary lubrication properties of lipid-based compounds evaluated using microtribological methods. Tribol Lett 22(2):167–172. doi: 10.1007/s11249-006-9076-x [DOI] [Google Scholar]
- 4.Brennan L, Owende P (2010) Biofuels from microalgae—a review of technologies for production, processing, and extractions of biofuels and co-products. Renew Sust Energ Rev 14(2):557–577. doi: 10.1016/j.rser.2009.10.009 [DOI] [Google Scholar]
- 5.Piazza GJ, Foglia TA (2001) Rapeseed oil for oleochemical usage. Eur J Lipid Sci Technol 103(7):450–454. doi: [DOI] [Google Scholar]
- 6.Uner M, Wissing SA, Yener G, Muller RH (2005) Skin moisturizing effect and skin penetration of ascorbyl palmitate entrapped in solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC) incorporated into hydrogel. Pharmazie 60(10):751–755 [PubMed] [Google Scholar]
- 7.Fernandis AZ, Wenk MR (2009) Lipid-based biomarkers for cancer. J Chromatogr B Anal Technol Biomed Life Sci 877(26):2830–2835. doi: 10.1016/j.jchromb.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 8.Kwan BCH, Kronenberg F, Beddhu S, Cheung AK (2007) Lipoprotein metabolism and lipid management in chronic kidney disease. J Am Soc Nephrol 18(4):1246–1261. doi: 10.1681/asn.2006091006 [DOI] [PubMed] [Google Scholar]
- 9.Lukyanov AN, Torchilin VP (2004) Micelles from lipid derivatives of water-soluble polymers as delivery systems for poorly soluble drugs. Adv Drug Deliv Rev 56(9):1273–1289. doi: 10.1016/j.addr.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 10.Sutphen R, Xu Y, Wilbanks GD, Fiorica J, Grendys EC Jr, LaPolla JP, Arango H, Hoffman MS, Martino M, Wakeley K, Griffin D, Blanco RW, Cantor AB, Xiao YJ, Krischer JP (2004) Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol Biomarkers Prev 13(7):1185–1191 [PubMed] [Google Scholar]
- 11.Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M (2008) Informatics and computational strategies for the study of lipids. Mol BioSyst 4(2):121–127. doi: 10.1039/b715468b [DOI] [PubMed] [Google Scholar]
- 12.Lintonen TP, Baker PR, Suoniemi M, Ubhi BK, Koistinen KM, Duchoslav E, Campbell JL, Ekroos K (2014) Differential mobility spectrometry- driven shotgun lipidomics. Anal Chem 86(19):9662–9669. doi: 10.1021/ac5021744 [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Snyder L, McKeon TA (1998) Prediction of relative retention times of triacylglycerols in non-aqueous reversed-phase high-performance liquid chromatography. J Chromatogr 808(1):43–49. doi: 10.1016/S0021-9673(98)00134-4 [DOI] [Google Scholar]
- 14.Ulmer CZ, Yost RA, Chen J, Mathews CE, Garrett TJ (2015) Liquid c hromatography- mass spectrometry metabolic and lipidomic sample preparation workflow for suspension-c ultured mammalian cells using Jurkat t lymphocyte cells. J Proteomics Bioinform 8(6):126–132. doi: 10.4172/jpb.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis A, Rudnitskaya A, Blackburn GJ, Mohd Fauzi N, Pitt AR, Spickett CM (2013) A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J Lipid Res 54(7):1812–1824. doi: 10.1194/jlr.M034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva LP, Lorenzi PL, Purwaha P, Yong V, Hawke DH, Weinstein JN (2013) Measurement of DNA concentration as a normalization strategy for metabolomic data from adherent cell lines. Anal Chem 85(20):9536–9542. doi: 10.1021/ac401559v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D (2008) Lipid extraction by methyl-tert-butyl ether for high- throughput lipidomics. J Lipid Res 49(5):1137–1146. doi: 10.1194/jlr.D700041-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson RE, Ducrocq AJ, McDougall DJ, Garrett TJ, Yost RA (2015) Comparison of blood plasma sample preparation methods for combined LC-MS lipidomics and metabolomics. J Chromatogr B Anal Technol Biomed Life Sci 1002:260–266. doi: 10.1016/j.jchromb.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorkas PA, Isaac G, Anwar MA, Davies AH, Want EJ, Nicholson JK, Holmes E (2015) Untargeted UPLC-MS profiling pipeline to expand tissue metabolome coverage: application to cardiovascular disease. Anal Chem 87(8):4184–4193. doi: 10.1021/ac503775m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78(3):779–787. doi: 10.1021/ac051437y [DOI] [PubMed] [Google Scholar]
- 21.Pluskal T, Castillo S, Villar-Briones A, Orešič M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11(1):1–11. doi: 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M (2015) MS-DIAL: data- independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12(6):523–526. doi: 10.1038/nmeth.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O (2013) LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 10(8):755–758. doi: 10.1038/nmeth.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia J, Wishart DS (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6(6):743–760. doi: 10.1038/nprot.2011.319 [DOI] [PubMed] [Google Scholar]
- 25.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS (2012) MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40(Web Server issue):W127–W133. doi: 10.1093/nar/gks374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler N, Neuweger H, Bonte A, Langenkamper G, Niehaus K, Nattkemper TW, Goesmann A (2013) MeltDB 2.0-advances of the metabolomics software system. Bioinformatics 29(19):2452–2459. doi: 10.1093/bioinformatics/btt414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Li D, Meng J, Wang H (2010) Introduction to SIMCA-P and its application In: Esposito V, Chin W, Henseler J, Wang H (eds) Handbook of partial least squares: concepts methods and applications. Springer, Berlin, Heidelberg, pp 757–774 [Google Scholar]
- 28.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA (2009) Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res 50(Suppl):S9–14. doi: 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L (2007) HMDB: the human metabolome database. Nucleic Acids Res 35(Database issue):D521–D526. doi: 10.1093/nar/gkl923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27(6):747–751 [DOI] [PubMed] [Google Scholar]
- 31.Taguchi R, Nishijima M, Shimizu T (2007) Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods Enzymol 432:185–211. doi: 10.1016/s0076-6879(07)32008-9 [DOI] [PubMed] [Google Scholar]
- 32.Taguchi R, Ishikawa M (2010) Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine lipid search. J Chromatogr A 1217(25):4229–4239. doi: 10.1016/j.chroma.2010.04.034 [DOI] [PubMed] [Google Scholar]
- 33.Koelmel JP, Kroeger NM, Gill EL et al. (2017) Expanding lipidome coverage using LC-MS/MS data-dependent acquisition with automated exclusion list generation. J Am Soc Mass Spectrom 1–10. doi: 10.1007/s13361-017-1608-0 [DOI] [PMC free article] [PubMed] [Google Scholar]