Abstract
Respiration is one of the most basic features of living organisms, and the electron transport chain complexes are probably the most complicated protein system in mitochondria. Complex-IV is the terminal enzyme of the electron transport chain, existing either as randomly scattered complexes or as a component of supercomplexes. NDUFA4 was previously assumed as a subunit of complex-I, but recent biochemical data suggested it may be a subunit of complex-IV. However, no structural evidence supporting this notion was available till now. Here we obtained the 3.3 Å resolution structure of complex-IV derived from the human supercomplex I1III2IV1 and assigned the NDUFA4 subunit into complex-IV. Intriguingly, NDUFA4 lies exactly at the dimeric interface observed in previously reported crystal structures of complex-IV homodimer which would preclude complex-IV dimerization. Combining previous structural and biochemical data shown by us and other groups, we propose that the intact complex-IV is a monomer containing 14 subunits.
Introduction
Mitochondria are critical to many cellular activities. Respiration is the central function of mitochondria, and is exquisitely regulated in multiple ways in response to varying cell conditions.1–3 The assembly of respiratory chain complexes, Complex I–IV (CI, NADH:ubiquinone oxidoreductase; CII, succinate:ubiquinone oxidoreductase; CIII, cytochrome bc1 complex; and CIV, cytochrome c oxidase) into supercomplexes (I1III2IV1–2, III2IV1–2) or even a megacomplex (I2III2IV2) is a critical form of respiration regulation.3–10 Many lines of evidence show that the formation of higher-order complexes can structurally stabilize the individual complexes,11–15 raise the efficiency of electron transfer,16–22 and significantly reduce the generation of reactive oxygen species (ROS).23–25 Functional studies in vivo also confirmed the validity of these higher-order assemblies in living cells. Several recently acquired structures of supercomplex I1III2IV1 (SCI1III2IV1) and megacomplex I2III2IV2 provided exciting insights into the function and mechanism of these huge machines and led to several interesting hypotheses.4,6,9,10 All these structures, especially that of the megacomplex,10 indicated the existence of Q-pool and the close spatial proximity of the binding sites of cytochrome c in CIII and CIV, although no protein subunits were shown to directly form a physical channel that can assist CoQ and cytochrome c transport.
CIV is the terminal oxidase of the electron transport chain in mitochondria.26–29 It accepts electrons from cytochrome c to reduce the oxygen to water and meanwhile pumps two protons from the matrix side to the intermembrane space (IMS).30–32 Previous studies suggested that the mammalian CIV consists of 13 distinct subunits, along with three copper II ions, one magnesium ion, one zinc ion, two heme A molecules, and several phospholipid molecules.33–35 Current opinions point out that CIV exists in two states under physiological conditions, either being assembled into supercomplexes or freely scattered on mitochondrial inner membrane.36 NDUFA4 was originally considered as a subunit of Complex-I37 but was proposed to belong to Complex-IV recently.38 However, the precise location of NDUFA4 in CIV remains unknown, and thus this notion still lacks structural support. In the present study, with the 3.3 Å density map of human CIV, for the first time, we obtained the entire CIV structure containing 14 subunits, which includes the extra subunit NDUFA4. In all the previous crystal structures, CIV is assembled to form a homodimer with each protomer containing 13 subunits, while in all supercomplex structures CIV exists as a monomer. Here, we found NDUFA4 is incorporated into CIV and binds exactly to the dimeric interface in the crystal structures of CIV dimer, indicating that NDUFA4 actually hampered the formation of CIV dimer. Structural analyses indicated that the CIV homodimer with each protomer containing 13 subunits was obtained because of the usage of strong detergent cholic acid for crystallization which expelled NDUFA4 from the supercomplexes. Therefore, CIV should be a 14-subunit monomer instead of a homodimer with each protomer containing 13 subunits which is described by the previous 37 X-ray crystal structures (Supplementary information, Table S1).
Results and discussion
Protein purification and structure determination
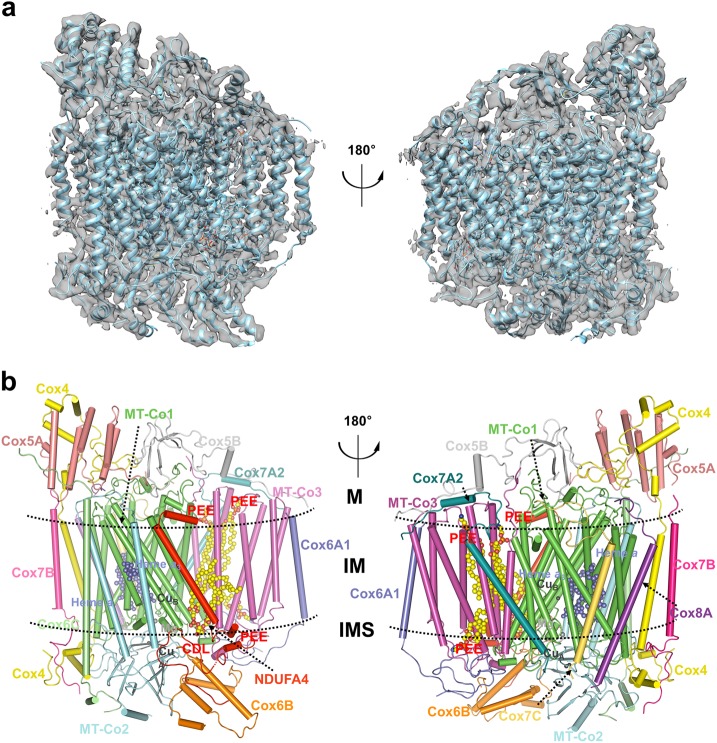
In this study, the human mitochondrial electron transport chain supercomplex I1III2IV1 (SCI1III2IV1) was purified from cultured HEK293F cells as previously described.10 Super-resolution images were collected on a Titan Krios microscope operated at a voltage of 300 kV with a K2 direct electron detector instead of the Falcon-II detector that we used before.10 The process of 3D reconstruction was described in Supplementary information, Figure S1. Further refinement with the mask encircling CIV results in a 3.3 Å resolution map of CIV (Supplementary information, Figures S1 and S2). Using a combination of structure docking and de novo modeling, we obtained the atomic structure of the CIV. The well-resolved density maps allowed us to build the structure models of almost all the residues with side chains in COOT39 (Fig. 1; Supplementary information, Figure S3 and Tables S2 and S3). The “gold standard” Fourier shell correlation (FSC) criteria between the models and maps have no significant differences, indicating the absence of overfitting (Supplementary information, Figure S3g). In the final CIV structure, we could accurately place 14 subunits, including NDUFA4, as well as a multitude of cofactors, including 4 phospholipids, 2 heme molecules, 2 copper centers, and 1 magnesium ion (Fig. 1b). Among 1888 residues of the mature human CIV, 1,857 (98.36%) residues were modeled in our structure and 1824 (96.61%) residues were assigned with side chains (Supplementary information, Figure S2).
Fig. 1.
Overall structure of human CIV monomer. a Cryo-EM density map of human CIV monomer along with the cartoon-presented models after soft mask subregion refinement being aligned to the 3.3 Å map are shown in two differently rotated views along the membrane. b The overall structure of human CIV. Subunits are colored and labeled with text in the same colors, respectively. The cofactors and phospholipids are shown in spheres. CDL, cardiolipin; PEE, phosphatidylethanolamine. The transmembrane region is indicated by two dashed lines. M, matrix; IM, inner membrane; IMS, intermembrane space
Overall structure of 14-subunit human CIV
Although numerous structures of CIV from different species have been determined in the past two decades, none identified the position of the NDUFA4 subunit (Supplementary information, Table S1). NDUFA4 was originally considered as an accessory subunit of CI,37 but further studies indicated that this protein might belong to CIV.38 In all previous crystal structures, CIV alone forms a homodimer, however, in all supercomplex structures, CIV exists as a monomer.4–6,9 In the present study, we got the 3.3 Å density map of CIV using cryo-EM single particle reconstruction method to unequivocally assign NDUFA4 as the 14th subunit of human CIV (Figs. 1 and 2). Our structural and biochemical data confirmed that the CIV either from a supercomplex or at a monomer state contains the NDUFA4 subunit (Fig. 3). Thus, we have determined the first structure of the intact human CIV containing 14 distinct subunits.
Fig. 2.
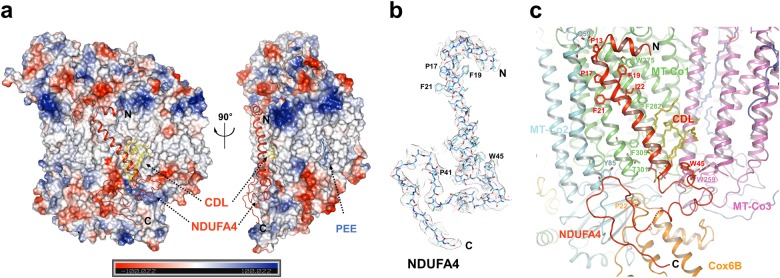
Interactions between NDUFA4 and other subunits of CIV. a Charge-smoothed potential map of CIV generated by PyMOL, except that NDUFA4 is shown in cartoon. Shown in two differently rotated views along the mitochondrial inner membrane. CDL and PEE are shown in stick and colored yellow and blue, respectively. The N- and C-terminals are indicated. b Cryo-EM densities of the NDUFA4 in human CIV monomer, superimposed with atomic model. The N and C terminals are indicated. c Interactions between NDUFA4 and other subunits of CIV. Residues and subunits are labeled and shown in stick and cartoon, respectively. CDL is colored yellow and labeled red. d NDUFA4 interact with the cardiolipin molecule in CIV monomer. Cardiolipin is shown in yellow sticks. Residues and subunits directly involved in cardiolipin binding are labeled and shown in stick and cartoon, respectively. Hydrogen bonds are shown in dashed lines
Fig. 3.
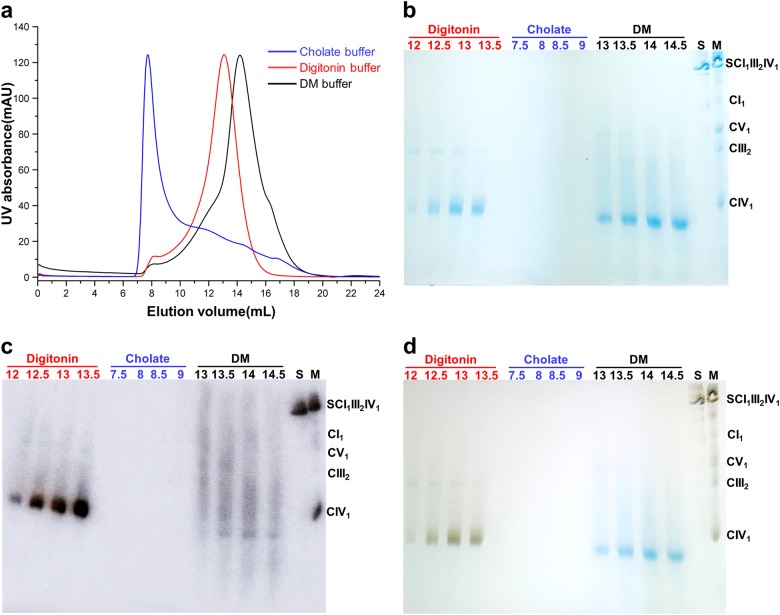
Biochemical characterization of monomeric porcine CIV in different detergents. a Representative trace of size-exclusion chromatography by the superdex 200 (10/300 GL) column of monomeric CIV with different detergents. b Fractions of different elution volumes were analyzed by BN-PAGE. Samples with different detergents were labeled and colored. S, SCI1III2IV1; M, maker. c Western blot analyses were used to probe for the composition of the monomeric CIV complex with antibody against the NDUFA4 subunit. d In-gel enzyme activity staining of CIV by DAB/cytochrome c
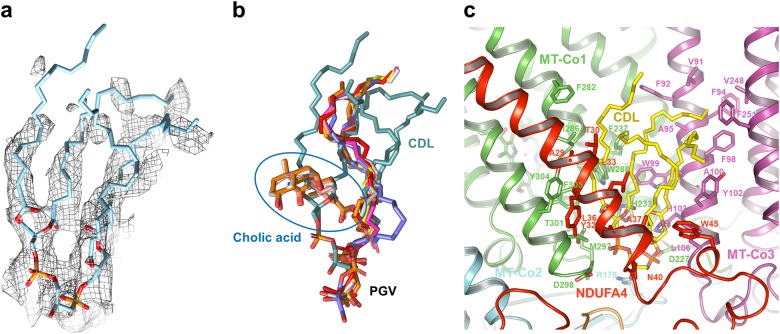
The 13 traditional subunits of human CIV adopt similar arrangements as in other mammalian CIV structures reported previously.33 Intriguingly, NDUFA4 is identified in our final structure. The N- and C-terminals of the NDUFA4 are located at the matrix side and the IMS, respectively. The middle region of NDUFA4 forms a transmembrane helix (TMH) that parallels the main TMHs of MT-CO1 (mitochondrially encoded cytochrome c oxidase I) and is adjacent to the TMH2 of MT-CO2 (Figs. 1b and 2). The head of a well-defined CDL (cardiolipin) molecule inserts into the deep pocket formed by the main TMHs of MT-CO1, MT-CO3, the C-terminal region and the TMH of NDUFA4, and Cox6B1 at the IMS side (Figs. 1b and 4). This CDL molecule might be important for stabilizing NDUFA4 in CIV, as this position is occupied by a cholic acid and a phosphatidylglycerol (PGV) molecule in most crystal structures of the CIV dimer (Fig. 4b). By contrast, the other three phosphatidylethanolamine (PEE) molecules bind at the same sites in both our structure and the previous crystal structures (Supplementary information, Figure S4).
Fig. 4.
The CDL molecule is important for the NDUFA4 binding. a Cryo-EM density of the CDL molecule in human CIV monomer. b Comparison of the CDL molecule in human CIV monomer with molecules in former CIV crystal structures that clash with CDL. All the molecules are shown in stick. Blue circle indicates the position of cholic acid, CDL is colored cyan, and the position of PGV is indicated, respectively. Molecules from PBD ID: 1V54, 2DYR, 2EIJ, 2Y69, 2ZXW, 5XDQ, and 5B1A are color red, magenta, yellow, salmon, gray, slate, and orange, respectively. c CDL interacts with NDUFA4. CDL is shown in yellow sticks. Residues and subunits directly involved in CDL binding are labeled and shown in stick and cartoon, respectively
Three different subunits of human CIV
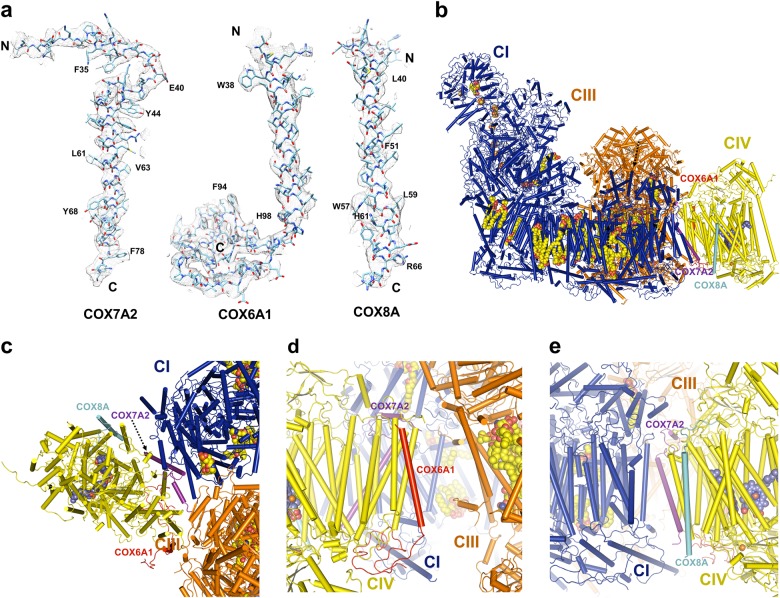
It is clear that Cox7A1, Cox6A2 and Cox8B in the 13-subunit bovine homodimeric CIV crystal structures were replaced by the human Cox7A2, Cox6A1, and Cox8A, respectively, in the 14-subunit human monomeric CIV structure (Fig. 5a; Supplementary information, Table S3). This might be because of tissue specificity,40 since the previous CIV samples were derived from bovine heart while the current CIV samples were purified from human epithelial kidney cells. Genetic analyses suggest that humans only have two Cox8 isoforms, Cox8A and Cox8C, while the rodents have all three Cox8 isoforms, Cox8A, Cox8B, and Cox8C.41
Fig. 5.
Three different subunits in human CIV. a Cryo-EM densities of three different subunits in human CIV monomer, superimposed with respective atomic models. The N- and C-terminals are indicated. b Position of three different subunits of CIV in supercomplex I1III2IV1. Protein complexes and subunits are in the same colors with the corresponding labels, respectively. Proteins are shown in cartoon and the cofactors are shown in spheres. c Top view from the matrix side of the position of three different subunits of CIV in supercomplex I1III2IV1. d Position of COX6A1 and COX7A2 in supercomplex I1III2IV1. View from CIII and CIV side of supercomplex I1III2IV1. e Position of COX8A and COX7A2 in supercomplex I1III2IV1. View from CI and CIV side of supercomplex I1III2IV1. c–e are shown in the same way as in (b)
Previous studies demonstrated that the cytochrome c oxidase subunit VIIa polypeptide 2-like protein (Cox7A2L) was required for the CIII-CIV interaction.20,42–44 Indeed, we show that Cox7A2 in our CIV structure replaces Cox7A1 in the structure of bovine CIV dimer (Fig. 5a; Supplementary information, Table S3). Our result is also consistent with the finding that mutation of the Cox7A2L isoform in the mouse strain C57BL/6 impairs the formation of the supercomplex III2IV1,44 since our CIV structure is derived from the SCI1III2IV1. We also find that human Cox6A1 and Cox8A replace Cox6A2 and Cox8B in the bovine CIV structures, respectively (Fig. 5a; Supplementary information, Table S3). In the structure of supercomplex I1III2IV1, the Cox8A is close to ND5 of CI while the Cox6A1 is close to CIV (Fig. 5b, e). It is also known that these subunits have the capacity to exchange for one another and the different isoforms of CIV indeed render this enzyme more plastic than others. Whether Cox6A1 and Cox8A, which are functionally similar to Cox7A2L, are required for the CIII-CIV interaction or supercomplex formation needs further biological, biochemical and structural studies. Especially, the high resolution structures of CIV in the supercomplex I1III2IV1 derived from different bovine organs may help to explain why these three subunits are replaced by different homologs in our human CIV.
NDUFA4 is a subunit of human CIV
A previous study showed that CIV purified from bovine heart mitochondria was a monomer with a native molecular mass of about 200 kDa on the blue-native page in the presence of either n-dodecyl-D-maltopyranoside (DDM) or digitonin as detergents,45 and same results were observed in different experiments and species.8,14,19 Our data also show that human CIV in digitonin is a 200 kDa monomer, rather than a dimer (Fig. 3; Supplementary information, Figure S5). Recently, a monomeric bovine CIV model was proposed according to the data observed in 2D crystals in lipid bilayer,46 which suggested that CIV is a monomer in membranes. Consistent with a previous study suggesting that the purified CIV alone contains NDUFA4 and the evolutionary evidence given,38 we propose that NDUFA4 is an inherent subunit of CIV. It possibly dissociated from the complex during the multistep detergent-based purification and crystallization, producing an artificial dimerization of two 13-subunit protomers that lack NDUFA4. Pitceathly and colleagues confirmed that NDUFA4 dissociated from CIV when the concentration of DDM exceeded 0.08%.47 We treated the isolated CIV monomer purified from porcine hearts with different detergents including digitonin, cholate and n-Decyl-D-maltopyranoside (DM). The size-exclusion chromatography result and BN-PAGE analysis clearly showed that CIV dissociated in cholate buffer, and the molecular weight of CIV in DM buffer was slightly smaller than that in the digitonin buffer, indicating that certain subunits or detergents might be dissociating (Fig. 3a, b). Further western blot analysis confirmed that NDUFA4 dissociated from CIV in cholate and DM buffer (Fig. 3c), which was consistent with previous biochemical results.47 Finally we tested the in-gel enzyme activity of CIV in different buffer (Fig. 3d): CIV in digitonin buffer had enzyme activity while the CIV lacking NDUFA4 in DM buffer lost enzyme activity. A recent opinion claimed that CIV monomer only contains 13 subunits and NDUFA4 is an assembly factor guiding CIV into SCI1III2IV1.48 Since our CIV is derived from SCI1III2IV1, we conclude that NDUFA4 is not an assembly factor, otherwise it should dissociate from the supercomplex when the CIV assembly is finished. More interestingly, the binding site of NDUFA4 is on the opposite side of the interacting interface between CI and CIV in SCI1III2IV1 (Figs. 1 and 2), a position that is unlikely for an assembly factor to bind. Consistent with a recent review,40 we conclude that the NDUFA4 is a bona fide COX subunit.
NDUFA4 is required for the formation of CIV
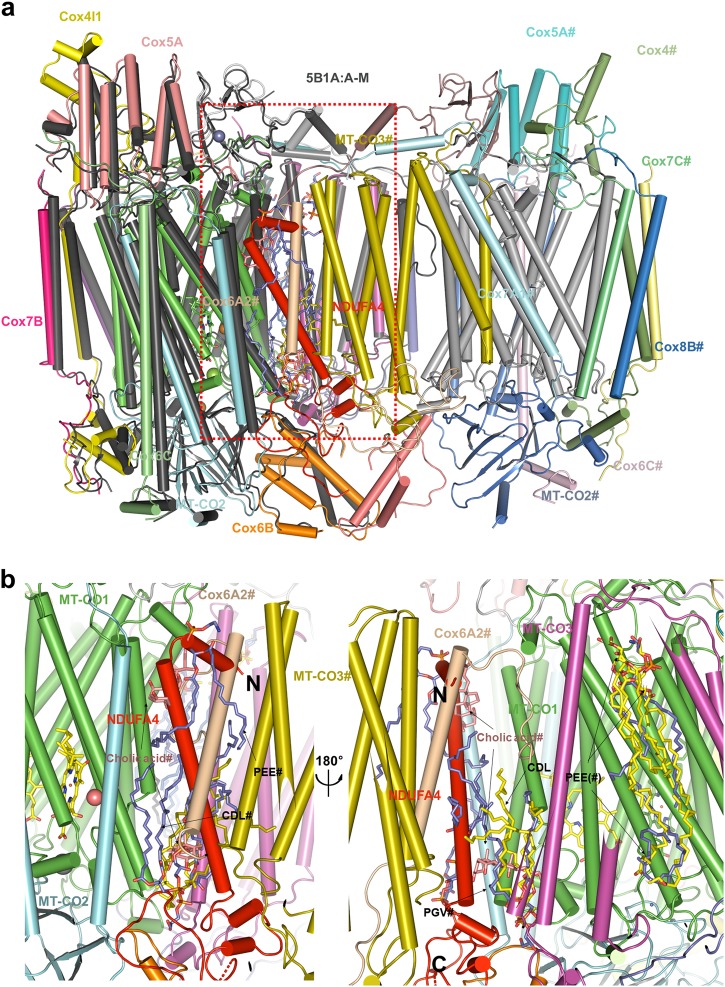
All crystal structures of CIV solved to date show CIV as a dimer formed by two 13-subunit protomers, but CIV exists as a monomer in the supercomplexes (Supplementary information, Table S3). When we superpose one of the 13-subunit protomer49 (PDB: 5B1A) onto our structure, the N-terminal region and the middle section of the TMH of NDUFA4 clash with the TMH of Cox6A2 in another protomer of the CIV dimer (Fig. 6a). Moreover, other parts of the NDUFA4-binding site are occupied by a cholic acid, a PEE and a CDL molecule (Figs. 4 and 6b). Interestingly, the well positioned CDL molecule in our CIV structure was replaced by a PGV molecule in the crystal structures (Figs. 4b and 6b). Our 14-subunit CIV structure is clearly incompatible with all the published crystal structures of the 13-subunit CIV dimer. Coincidentally, the newly identified NDUFA4 subunit binds exactly at the dimeric interface of the previous crystal structures, which indicates that NDUFA4 actually hampers the formation of the CIV dimer. Interestingly, previous studies demonstrated that NDUFA4 was important for the stability of CIV in vivo.38 CIV levels were reduced upon NDUFA4 knockdown, and were restored when exogenous NDUFA4 was overexpressed, indicating that NDUFA4 was required for the formation of CIV. More importantly, no dimeric CIV was detected when NDUFA4 was depleted,38 which suggested that the 13-subunit CIV is unstable and does not have the ability to form a dimer in vivo. Together with our biochemical analyses, we have determined the structure of the intact CIV with 14 distinct subunits as a monomer, instead of a homodimer with each protomer containing 13 subunits which is revealed by previous X-ray crystal structures (Supplementary information, Table S1).
Fig. 6.
Structure comparison of the CIV monomer and dimer. a Structure comparison of the human CIV monomer and the bovine dimeric CIV (PDB:5B1A). Different subunits of CIV are labeled and shown in different colors. Subunits from 5B1A are distinguished by “#”. b The NDUFA4 hampers the dimerization of CIV. Shown in two differently rotated views along the mitochondrial inner membrane. Cholic acids are shown in salmon sticks. Phospholipids from human CIV structure and bovine CIV structure are shown in yellow and blue, respectively. CDL, cardiolipin; PEE, phosphatidylethanolamine; PGV, phosphatidylglycerol
Conclusion
In conclusion, we purified SCI1III2IV1 from cultured human cells, collected cryo-EM data and performed 3D reconstruction to get the high-quality 3.3 Å CIV density map with sub-region refinement. Assignment of the 14th subunit, NDUFA4, in the monomeric CIV not only confirms that NDUFA4 is a bona fide subunit of CIV, but also provides a straightforward explanation for why CIV is a monomer. Unconventionally, we assigned the NDUFA4 subunit into the CIV structure for the first time, and discovered that the binding site of NDUFA4 lies exactly at the dimeric interface of the previously reported CIV crystal structure. After comparing previous structural data with our CIV monomer structure, we found the stronger cholic acid salt used in crystallization buffer replaced the CDL molecule, which might be important for NDUFA4 stabilization, in our structure. Although we cannot fully exclude the possibility that the CIV could form a dimer in vivo, our data clearly demonstrate that the dimerization state in the crystal structures is suspectable. Our 14-subunit CIV monomer is consistent with the previous biochemical data showing the CIV monomer45 and 2D-crystal studies in lipid bilayer.46 The previous functional studies of CIV that based on the crystal structures of CIV homodimer may have to be revisited. Since our CIV structure is a part of SCI1III2IV1, which is in a more native state, we propose that the intact monomeric complex-IV contains 14 subunits including NDUFA4.
Materials and Methods
Reagents
The following reagents were used: Trizma base (T4661, Sigma), D-Sorbitol (S1876, Sigma), KCl (746436, Sigma-Aldrich), EGTA (E4378, Sigma-Aldrich), BSA (0332, AMRESCO), Percoll(17-0891-09, GE Healthcare), Digitonin (D141. Sigma), Glycine (G7126, Sigma), 4-nitro blue tetrazolium chloride (NBT; N6876, Sigma-Aldrich), β-nicotinamide adenine dinucleotide, reduced (β-NADH; N8129, Sigma), 6-aminohexanoic acid (07260, Sigma-Aldrich), Coomassie brilliant blue G-250 (0615, AMRESCO), glycerol (G6279, Sigma-Aldrich), Ponceau S (P3504, Sigma-Aldrich), acrylamide (V900845, Vetec), N,N′-methylenebis(acrylamide) (V900301, Vetec), Tricine (T0377, Sigma), imidazole (I202, Aldrich), ammonium persulfate (A3678, Sigma), TEMED (T22500, Sigma-Aldrich), SMM 293-TI medium (Sino Biological Inc.), Penicillin/Streptomycin (P1400, Solarbio), and RNase (AP-MN-P-250G-RA, Axygen).
Cell culture
HEK293F cells (R790–07, Invitrogen) were cultured in SMM 293-TI medium supplemented with 1× penicillin/streptomycin at 37 °C with 8% CO2 and seeded every day.
Human mitochondria preparation
All procedures are carried out at 4 °C. HEK293F cells were harvested before cell density reached 2.0 × 106 cells/mL and suspended by buffer-A (50 mM Tris pH 7.4, 225 mM sorbitol, 75 mM sucrose, 1 mM EGTA and 0.1% BSA). The suspension was homogenized by a soft blender for 150 s and the homogenate was centrifuged at 3,000 × g for 10 min. Supernatant was further centrifuged at 20,000 × g for 30 min to obtain the crude mitochondria. The pellet was suspended in buffer-B (10 mM Tris pH 7.4, 250 mM sucrose, 40% Percoll and 1 mM EGTA) and centrifuged at 66,000 × g for 50 min. The clear mitochondria layer was extracted carefully and diluted with buffer-C (10 mM Tris, pH 7.4, 75 mM sorbitol, 25 mM sucrose, 0.05 mM EGTA). The highly pure mitochondria were collected by centrifugation for 30 min at 20,000 × g.
Supercomplex purification
Pure mitochondria were homogenized and extracted by 0.5% (w/v) digitonin for 4 h with slow stirring at 4 °C in buffer-C, RNase was added by a final concentration of 0.1 mg/mL to digest the mitochondrial ribosome. The extraction was centrifuged at 150,000 × g for 30 min at 4 °C and the supernatant was concentrated to 1 mL by 100 kDa cutoff centrifugal filter (Millipore). Concentrated sample was loaded and centrifuged on 0.3–1.3 M sucrose gradients in buffer-D (10 mM HEPES, pH 7.4, 20 mM KCl, 0.1% digitonin) at 150,000 × g for 21 h at 4 °C with a SW41 rotor (Beckman). Gradients were fractionated and investigated by 3%–10% BN-PAGE. The supercomplexes were concentrated and finally purified by Superose 6 increase 10/300 GL column (GE Healthcare) in buffer-D. The peak fractions were collected for EM sample preparation, and the purity of supercomplexes was verified by BN-PAGE and NBT staining as described previously9.
The porcine heart monomeric CIV purification and the NDUFA4 subunit identification
After the sucrose density gradient centrifugation mentioned in the last step, gradient fractions containing pure porcine heart monomeric complex IV were concentrated and purified by Superdex 200 10/300 GL column (GE Healthcare) in buffer-D. The peak fractions were collected and diluted 10-fold in buffer-E (40 mM sodium phosphate, pH 6.8, 0.5% sodium cholate). After incubation for 30 min, the diluted fractions were concentrated and dissolved in buffer-E by Superdex 200 10/300 GL column (GE Healthcare). Finally, the buffer dissolving the monomeric complex IV was changed to buffer-F (40 mM sodium phosphate, pH 6.8, 0.2% DM) by Superdex 200 10/300 GL column (GE Healthcare) in the same way as the previous step.
Fractions of porcine heart monomeric CIV size-exclusion chromatography in buffers containing different detergents were applied to 3%–13% BN-PAGE. After electrophoresis, the gel was incubated in buffer-G (40 mM sodium phosphate, pH 7.4, 2.5 mM 3,3′-diaminobenzidine, 0.1 mM cytochrome c) for 40 min to detect the in-gel catalytic activity of CIV. At the same time, the monomeric CIV protein was electroblotted onto Immobilon-P transfer membranes (Millipore) and sequentially probed with specific antibody against NDUFA4 subunit.
Cryo-EM data acquisition and processing
Cryo-EM samples were prepared using 400-mesh Quantifoil R1.2/1.3 grids (Quantifoil, Micro Tools GmbH, Germany). The grids were coated with a home-made continuous thin layer of carbon. Aliquots of 4 μL of digitonin-solubilized SC at a concentration of 0.2 mg/mL were applied to the grids pre-glow discharged for 30 s. The grids were blotted for 2 s and plunged into liquid ethane cooled by liquid nitrogen using FEI Mark IV Vitrobot operated at 4 °C and 100% humidity. Grids were then transferred into and kept in the liquid nitrogen until being used for data collection. Micrographs were collected on a Titan Krios microscope operated at a voltage of 300 kV with a Gatan K2 Summit direct electron detector (Gatan Company) instead of the Falcon II direct electron detector we previously used. Automated single-particle data acquisition was performed with AutoEMation (written by Jianlin Lei) with a nominal magnification of ×105,000 using super-resolution mode. The defocus range were set from −1.3 to −2.3 μm. The total dose rate on the detector was about 50 e/Å2 with a total exposure time of 5.44 s. Each micrograph stack contains 32 frames. Four batches of data were collected with six cryo-samples, obtaining 24,593 micrographs in total.
Image processing
Correction of electron beam-induced sample motion was first done by MotionCorr.50 The frames were binned by 2-fold, resulting in a final pixel size of 1.091 Å/pixel. Patch-based motion detection combined with dose weighting was then performed with MotionCor2.51 Micrographs' screening, particle picking, and normalization were processed with EMAN2.152 and RELION2.1,53 and the details were the same as described before.10 Program of Gctf54 was used to estimate the contrast transfer function (CTF) parameters of each single particle. The 2D and 3D averaging classification and 3D auto-refinement were performed with RELION2.1. 1.47 million particles extracted from 24,953 micrographs (15,217 from first three batches of data) were subjected to a cascade of 2D and 3D classification. Two rounds of 2D classification and one round of 3D classification were performed on the first three batches of data separately to select the best particles, and 316,000 particles from six classes were finally selected in total. The 3.9 Å model of homo SC which we reported before10 was low-pass filtered to 60 Å to be the initial reference. 3D auto-refinement was performed on these 316,000 particles, resulting in an overall resolution of 3.6 Å within the region defined by a soft mask. We used the RELION software to generate soft masks, and the threshold was set to 0.0024. The initial binary mask was extended for 3 pixels in all directions, and is further extended with a raised-cosine soft edge of a width of 5 pixels. A second round of refinement was performed by applying the same soft mask used for post-processing of the SC, improving the resolution to 3.4 Å. The sub-region refinement method was performed to improve the local resolution of our density map.
To improve the quality of CIV density map, we classify the 316,000 particles into 6 classes with a soft mask of CIV generated from the summed refined map of SC applied without performing particle aligning. The fourth batch of data was processed independently by the same procedure as described above. Finally, a total of 156,000 homogeneous particles was preserved to reconstruct the structure of CIV, resulting in the reported resolution of CIV to be 3.8 Å within the region defined by the soft mask. A second round of 3D auto-refinement was performed with a tighter mask generated from the summed refined map of CIV applied to improve the relatively low translational and rotational accuracies. The final resolution of CIV was 3.3 Å within the region defined by the soft mask. All reported resolutions are based on the gold-standard FSC = 0.143 criteria,55 and the final FSC curve were corrected for the effect of a soft mask using high-resolution noise substitution.56 Final density maps were sharpened by B-factors calculated with the RELION post-processing program. Local resolution map was calculated using ResMap.57
Model building and refinement
To build atomic models of Homo sapiens CIV, we used 1.5 Å bovine CIV structure (PDB code: 5B1A)49as the initial model. With 3.3 Å CIV density map, we were able to model side chains and local geometry to high accuracy. CIV models were adjusted by COOT58 and real-space refined using PHENIX.59 All ligand and phospholipid models were generated using elbow60 module by their geometric constraints. The ligands and phospholipids were docked into densities and refined in COOT. All the figures were created in PyMOL (www.pymol.org), COOT, and UCSF Chimera.61
For cross-validation against overfitting, we random displaced the atom positions of the final model by up to a maximum of 0.5 Å,62 and refined against the half map 1 generated by RELION 3D auto-refine procedure, resulting in a model named Test. Then we calculated the FSC curve of both half map against the model Test, and compared with the FSC curve of final model against the summed map generated by RELION 3D auto-refine procedure (Supplementary information, Figure S3).
All reported resolutions are based on the gold-standard FSC = 0.143 criteria,55 and the final FSC curve was corrected for the effect of a soft mask using high-resolution noise substitution.56 Final density maps were sharpened by B-factor and calculated by RELION post-process procedure. Local resolution map was calculated using ResMap57 (Supplementary information, Figure S2). The models with the ligands and phospholipids were subjected to global refinement and minimization in real space refinement using PHENIX. Refinement and validation statistics were calculated by MolProbity63 and EMRinger64 module in PHENIX (Supplementary information, Table S2).
Accession codes
The atomic coordinates of human CIV has been deposited in the Worldwide Protein Data Bank with the accession code 5Z62. The corresponding maps have been deposited in the Electron Microscopy Data Bank with the accession code EMD-6896.
Electronic supplementary material
Acknowledgements
We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the facility support. The computation was completed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the National Key R&D Program of China (2017YFA0504600 and 2016YFA0501100), the National Science Fund for Distinguished Young Scholars (31625008), and the National Natural Science Foundation of China (21532004 and 31570733).
Author contributions
M.Y. conceived, designed, and supervised the project; built the model; analyzed the data; and wrote the manuscript. J.G., S.Z., T.L. and R.G. did the protein purification and detergent screening. M.W., S.Z., and T.L. performed EM sample preparation, data collection, and structural determination. All authors discussed the data of the manuscript.
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0071-1.
References
- 1.Daum B, Walter A, Horst A, Osiewacz HD, Kuhlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl. Acad. Sci. USA. 2013;110:15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enriquez JA. Supramolecular organization of respiratory complexes. Annu. Rev. Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 4.Gu J, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 5.Guo R, Gu J, Wu M, Yang M. Amazing structure of respirasome: unveiling the secrets of cell respiration. Protein Cell. 2016;7:854–865. doi: 10.1007/s13238-016-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 7.Melber A, Winge DR. Inner secrets of the respirasome. Cell. 2016;167:1450–1452. doi: 10.1016/j.cell.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sousa JS, Mills DJ, Vonck J, Kuhlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. eLife. 2016;5:e21290. doi: 10.7554/eLife.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609 e1510. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Guo R, Zong S, Wu M, Gu J, Yang M. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170:1247–1257 e1212. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- 11.Acin-Perez R, et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/S1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamantea E, et al. A novel nonsense mutation (Q352X) in the mitochondrial cytochrome b gene associated with a combined deficiency of complexes I and III. Neuromuscul. Disord. 2002;12:49–52. doi: 10.1016/S0960-8966(01)00244-9. [DOI] [PubMed] [Google Scholar]
- 14.Moreno-Lastres D, et al. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vempati UD, Han XL, Moraes CT. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 2009;284:4383–4391. doi: 10.1074/jbc.M805972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benard G, et al. Functional dynamic compartmentalization of respiratory chain intermediate substrates: Implications for the control of energy production and mitochondrial diseases. Int. J. Biochem. Cell Biol. 2008;40:1543–1554. doi: 10.1016/j.biocel.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 17.Moreno-Loshuertos R, Enriquez JA. Respiratory supercomplexes and the functional segmentation of the CoQ pool. Free Radic. Biol. Med. 2016;100:5–13. doi: 10.1016/j.freeradbiomed.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Schafer E, et al. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 19.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Cogliati S, et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- 21.Cogliati S, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapuente-Brun E, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 23.Genova ML. Electron transport in the mitochondrial respiratory chain. Adv. Photosynth. Resp. 2014;39:401–417. doi: 10.1007/978-94-017-8742-0_21. [DOI] [Google Scholar]
- 24.Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan CL, Perevoshchikova IV, Hey-Mogensen M, Orr AL, Brand MD. Sites of reactive oxygen species generation by mitochondria oxidizing different substrates. Redox Biol. 2013;1:304–312. doi: 10.1016/j.redox.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostermeier C, Iwata S, Michel H. Cytochrome c oxidase. Curr. Opin. Struct. Biol. 1996;6:460–466. doi: 10.1016/S0959-440X(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 27.Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 28.Brown GC. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta. 2001;1504:46–57. doi: 10.1016/S0005-2728(00)00238-3. [DOI] [PubMed] [Google Scholar]
- 29.Malatesta F, Antonini G, Sarti P, Brunori M. Structure and function of a molecular machine: cytochrome c oxidase. Biophys. Chem. 1995;54:1–33. doi: 10.1016/0301-4622(94)00117-3. [DOI] [PubMed] [Google Scholar]
- 30.Verkhovsky MI, Jasaitis A, Verkhovskaya ML, Morgan JE, Wikstrom M. Proton translocation by cytochrome c oxidase. Nature. 1999;400:480–483. doi: 10.1038/22813. [DOI] [PubMed] [Google Scholar]
- 31.Capaldi RA. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- 32.Ostermeier C, Harrenga A, Ermler U, Michel H. Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc. Natl. Acad. Sci. USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 34.Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 35.Yoshikawa S, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 36.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta. 2003;1604:135–150. doi: 10.1016/S0005-2728(03)00059-8. [DOI] [PubMed] [Google Scholar]
- 38.Balsa E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Pitceathly Robert D.S., Taanman Jan-Willem. NDUFA4 (Renamed COXFA4) Is a Cytochrome -c Oxidase Subunit. Trends in Endocrinology & Metabolism. 2018;29(7):452–454. doi: 10.1016/j.tem.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Sinkler CA, et al. Tissue- and condition-specific isoforms of mammalian cytochrome c oxidase subunits: from function to human disease. Oxid. Med. Cell. Longev. 2017;2017:1534056. doi: 10.1155/2017/1534056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda K, Shiba S, Horie-Inoue K, Shimokata K, Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat. Commun. 2013;4:2147. doi: 10.1038/ncomms3147. [DOI] [PubMed] [Google Scholar]
- 43.Williams EG, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mourier A, Matic S, Ruzzenente B, Larsson NG, Milenkovic D. The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab. 2014;20:1069–1075. doi: 10.1016/j.cmet.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 46.Osuda Y, et al. Two-dimensional crystallization of monomeric bovine cytochrome c oxidase with bound cytochrome c in reconstituted lipid membranes. Microscopy (Oxf.) 2016;65:263–267. doi: 10.1093/jmicro/dfv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitceathly RDS, et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 2013;3:1795–1805. doi: 10.1016/j.celrep.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kadenbach B. Regulation of mammalian 13-subunit cytochrome c oxidase and binding of other proteins: role of NDUFA4. Trends Endocrinol. Metab. 2017;28:761–770. doi: 10.1016/j.tem.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Yano N, et al. The Mg2+-containing water cluster of mammalian cytochrome c oxidase collects four pumping proton equivalents in each catalytic cycle. J. Biol. Chem. 2016;291:23882–23894. doi: 10.1074/jbc.M115.711770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1108. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.