Abstract
Skeletal muscle fitness and plasticity is an important determinant of human health and disease. Mitochondria are essential for maintaining skeletal muscle energy homeostasis by adaptive re-programming to meet the demands imposed by a myriad of physiologic or pathophysiological stresses. Skeletal muscle mitochondrial dysfunction has been implicated in the pathogenesis of many diseases, including muscular dystrophy, atrophy, type 2 diabetes, and aging-related sarcopenia. Notably, exercise counteracts the effects of many chronic diseases on skeletal muscle mitochondrial function. Recent studies have revealed a finely tuned regulatory network that orchestrates skeletal muscle mitochondrial biogenesis and function in response to exercise and in disease states. In addition, increasing evidence suggests that mitochondria also serve to “communicate” with the nucleus and mediate adaptive genomic re-programming. Here we review the current state of knowledge relevant to the dynamic remodeling of skeletal muscle mitochondria in response to exercise and in disease states.
The specialized skeletal muscle mitochondrial system
Skeletal muscle structural and functional plasticity is a key feature that allows for adaptation to myriad physiological stimuli. Physiological conditions are often dictated by acute and chronic changes in physical activity and workload of the muscle. As such, muscle responds through changes in contractile machinery, calcium handling, and energy metabolic capacity. In particular, mitochondrial ATP production must match demands imposed by changes in physical activity and exercise. Accordingly, the skeletal muscle mitochondrial system has evolved for this purpose.
Based on subcellular location, skeletal muscle mitochondria can be generally classified into subsarcolemmal (SS) and intermyofibrillar (IMF). SS mitochondria are characterized by a large, lamellar shape located underneath the sarcomere, while the IMF mitochondria are relatively smaller, more compact and located between the contractile filaments. Based on recent three-dimensional electron microscopy findings, skeletal muscle mitochondria can be further classified into paravascular mitochondria (PVM), I-band mitochondria (IBM), fiber parallel mitochondria (FPM), and cross fiber connection mitochondria (CFCM).1 All four different mitochondrial populations are highly connected with each other to form a complex network that can rapidly distribute energy throughout myofibers.1
Skeletal muscle mitochondria are highly dynamic organelles, undergoing remarkable remodeling in response to a variety of physiological and pathophysiological stresses to meet energy and contraction demands of the muscle. Changes in energy demands of the muscle are most often associated with changes in physical activity. For example, increases in mitochondrial content and mass (biogenesis) accompany endurance exercise training (reviewed in ref.2). Likewise, decreases in mitochondrial content and oxidative energy metabolism are associated with obesity, diabetes and aging. It should be noted, however, that solely increasing mitochondrial biogenesis is not sufficient to support high oxidative respiration. A highly regulated system exists as a quality control mechanism to replace damaged mitochondria through orchestrated autophagy and mitophagy in muscle. Finally, mitochondrial biogenesis and mitophagy most likely do not exist independently as there is increasing evidence that at least in certain instances, these pathways are coordinately regulated. In this review, we focus on the mitochondrial remodeling that takes place in response to exercise and disease. Skeletal muscle possesses a highly integrated system that senses and coordinates changes in physical activity or extracellular cues with the control of mitochondrial biogenesis. In addition, we discuss quality control of muscle mitochondria through autophagic and mitophagic mechanisms. Finally, we review the relationship between muscle mitochondrial remodeling and disease and aging.
Integration of physical activity with muscle mitochondrial biogenesis
A nuclear receptor network controls mitochondrial fuel selection and respiratory function in muscle
Nuclear receptors constitute a large family of DNA-binding transcription factors. Several of these factors are critical regulators of muscle fuel metabolism and mitochondrial biogenesis and function. The peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors was originally identified as a regulator of peroxisome biogenesis.3 The prototypical member of this family, PPARα, plays a central role in the regulation of muscle fuel preferences. Specifically, PPARα drives mitochondrial fatty acid oxidation (FAO) enzyme gene expression in skeletal muscle and other oxidative tissues such as heart and liver.4–7 PPARα expression also parallels the oxidative capacity of specific muscle fibers, exhibiting the highest expression in slow, oxidative type I fibers.8 PPARα, as well as the other two members of this family PPARβ (also known as PPARδ) and PPARγ, binds to their cognate DNA elements as an obligate heterodimer with the retinoid X receptor (RXR). The PPAR factors also have large hydrophobic ligand binding domains that bind a variety of endogenous fatty acid or fatty acid-derived ligands.9–11 Although the precise endogenous ligands for PPARα, and the more closely related PPARδ, have remained elusive, long-chain unsaturated fatty acids have been shown to bind and activate these receptors.12 In this way, PPARα serves as a cellular sensor for increased delivery of fatty acids into muscle and subsequent upregulation of the enzymatic machinery needed for mitochondrial FAO and ATP production. Supporting this notion, overexpression of PPARα in mouse skeletal muscle results in a robust increase of the expression of genes involved in fatty acid import, binding, and oxidation as well as increased FAO rates.13 Interestingly, this comes at the expense of glucose utilization and is accompanied by a decrease in glycolytic enzyme expression, glucose intolerance and reduced exercise capacity.13,14 Somewhat surprisingly, FAO rates in skeletal muscle of PPARα knockouts (KO) are lower than wild-type controls in the fasted but not fed state.15 This most likely reflects compensation by PPARδ in muscle. Although not identical, there is a large degree of overlap in the gene targets of PPARα and PPARδ.16,17 Remarkably, activation of PPARδ synergizes with exercise to promote endurance capacity.17 In addition to direct effects on FAO, increases in the exercise capacity by PPARδ may be a result of glucose sparing.18
The estrogen-related receptor (ERR) is also a critical regulator of mitochondrial biogenesis in muscle. The ERR family is composed of 3 members (α, β, and γ) and characterized as orphan receptors as no endogenous ligand has been identified.19 The ERR factors have been shown to activate almost all aspects of mitochondrial energy metabolism including FAO, the TCA cycle, and electron transport chain and oxidative phosphorylation (ETC/OXPHOS) in many cell types.20 Multiple lines of evidence support a critical role for ERR signaling in maintenance of skeletal muscle energy metabolism and function. ERRα-deficient mice display lower activity and impaired exercise performance concomitant with lower expression of muscle mitochondrial energy metabolic genes.21 ERRα KO mice also have diminished muscle mass suggesting a possible crosstalk between energy metabolism and the control of muscle growth.21 ERRα also activates PPARα in skeletal muscle providing a feed forward loop to activate oxidative metabolism.22 ERRα is also linked to muscle repair and regeneration. ERRα KO mice have impaired regeneration following muscle injury.23 In addition to ERRα, overexpression of ERRγ in muscle results in marked activation of oxidative metabolic pathways, mitochondrial biogenesis and a corresponding increase in slow type I fibers.24,25 The switch to type I fibers by ERRγ is mediated by direct activation of the slow myosin heavy chain 7 and 7b (Myh7, Myh7b) genes and expression of the miR-499/208b pathway that reinforces the slow fiber phenotype.26,27 ERRγ is also correlated with type I fiber %, ATPmax, and VO2max in human skeletal muscle.26 As such, this ERR-triggered circuit provides a mechanism to coordinately regulate slow myosin heavy chain expression with high rates of mitochondrial ATP production.
Integration of upstream signaling pathways with mitochondrial biogenesis in muscle
The increased metabolic demands of exercise require a system to match increased energy requirements with enhanced oxidative capacity to generate ATP. A key discovery in this regard was the identification of PPARγ coactivator-1α (PGC-1α) as a cold-inducible transcriptional coactivator in brown adipose tissue.28 Subsequent studies demonstrated that PGC-1α directly interacts and coactivates multiple nuclear receptors, often in a ligand-dependent manner, to increase oxidative metabolism and mitochondrial biogenesis in multiple cell types.29–31 A key link between exercise and control of mitochondrial biogenesis came with the discovery that expression of PGC-1α is induced in skeletal muscle following an acute bout of exercise.32–34 Forced expression of PGC-1α in muscle in transgenic mice results in a robust mitochondrial biogenic response, an increase of type I and oxidative type IIa fibers and improved fatigue resistance.35 Conversely, loss of PGC-1α and the closely related PGC-1β in muscle leads to markedly reduced mitochondrial respiration, expression of ETC/OXPHOS genes and exercise performance.36,37 However, loss of PGC-1 does not alter fiber-type composition in muscle.36,37 Furthermore, loss of PGC-1α does not affect training-induced increases in OXPHOS/ETC gene expression.38–40 These results suggest that PGC-1 signaling in muscle is required for developmental mitochondrial biogenesis and to respond to acute dynamic physiological stresses but not necessarily chronic metabolic adaptations in response to exercise.
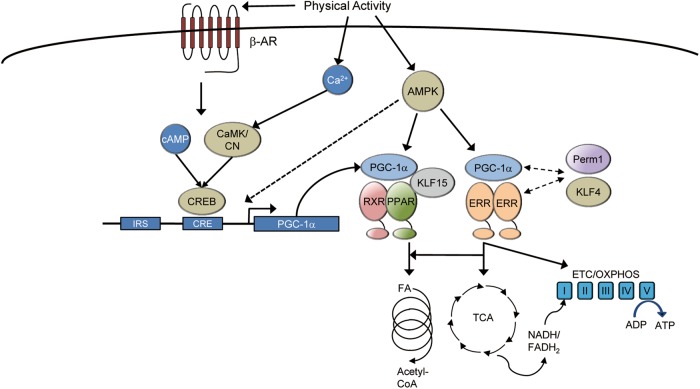
PGC-1α is a highly inducible transcriptional coregulator providing a mechanism to integrate extracellular and intracellular signals with the control of mitochondrial energy metabolism and quality control. Multiple signaling pathways appear to converge on regulation of PGC-1α (Fig. 1). Exercise-induced activation of PGC-1α expression is mediated, at least in part, by β-adrenergic signaling.41 PGC-1α expression is highly responsive to cAMP and activation of the cAMP response element binding protein (CREB).42 This almost certainly is a key mechanism downstream of β-adrenergic signaling in muscle. Exercise-induced activation of PGC-1α can be also mediated by the AMP-activated protein kinase (AMPK). AMPK is activated by depletion of ATP levels following exercise in muscle. AMPK was demonstrated to activate PGC-1α either via direct phosphorylation of PGC-1α or by promoting Sirt1-mediated PGC-1α activation.43,44 In skeletal muscle, AMPK activation is sufficient to drive the PGC-1α regulatory circuit and mitochondrial function.17 Signaling through the calcium-dependent protein phosphatase, calcineurin, and kinase, calmodulin-dependent kinase, has also been shown to activate PGC-1α following exercise.45 Induction of PGC-1α expression by calcium is also mediated by the MEF2 transcription factor which is repressed by histone deacetylase (HDAC) and activated by de-repression.46 The identification of insulin response sequences (IRS) in the promoter of PGC-1α also confirms insulin signaling as a key regulator of PGC-1α expression.47 Increases in PGC-1α in heart in response to exercise are also dependent on the insulin receptor substrate 1 and 2 (IRS1 and IRS2) molecules that act as key mediators of insulin signaling.48 Finally, considerable evidence demonstrates that skeletal muscle PGC-1α expression is diminished in diabetics.49,50 Although a direct causal relationship between diminished PGC-1α signaling and insulin resistance has not been established, a clear correlation between them exists.
Fig. 1.
Integration of upstream signaling pathways with mitochondrial biogenesis in the muscle. Multiple signaling pathways in the skeletal muscle serve to transmit changes in physical activity or other extracellular signals to mitochondrial biogenesis. Many of these pathways directly activate the peroxisome-proliferator activated receptor γ (PPARγ) coactivator-1α (PGC-1α). PGC-1α interacts directly with its effector nuclear receptors such as the estrogen-related receptor (ERR) and the peroxisome-proliferator activated receptor (PPAR) to regulate genes involved in virtually all aspects of mitochondrial energy metabolism. CaMK calmodulin-dependent kinase, CN calcineurin, AMPK AMP-dependent kinase, β-AR beta adrenergic receptor, IRS insulin response sequence, CRE cAMP response element, CREB cAMP response element binding protein, RXR retinoid X receptor, KLF Krüppel-like factor, FA fatty acid, ETC electron transport chain, OXPHOS oxidative phosphorylation
Other factors also converge on PGC-1/nuclear receptor signaling providing additional layers of regulation. For instance, the PGC-1 and ERR-induced regulator in muscle 1 (PERM1) is activated in muscle by exercise and required for maximal activity of PGC-1α in muscle.51 Although its precise function is not clear, ectopic expression of PERM1 in skeletal muscle also increases ERRα and PGC-1α expression, mitochondrial content and respiratory capacity.51 The Krüppel-like factors (KLF) are zinc finger transcription factors that also regulate skeletal muscle metabolism and mitochondrial function. Early work demonstrated that one KLF, KLF15, positively regulated the insulin-responsive glucose transporter 4 (GLUT4) in muscle.52 It was subsequently shown that KLF15 is induced in muscle acutely in human exercise and regulates a broad array of lipid and fatty acid metabolic genes in both muscle and heart.53,54 Further, KLF15 knockout mice have reduced endurance capacity and an increased reliance on carbohydrates during exercise.53 There is also evidence that actions of KLFs are regulated in coordination with nuclear receptors. For instance, KLF15 directly interacts with PPARα to regulate mitochondrial FAO enzyme gene expression.55 Likewise, KLF5 may be a direct negative regulator of PPARδ in skeletal muscle and inhibit activation of genes involved in mitochondrial fatty acid oxidation and energy uncoupling.56 In heart, KLF4 cooperates with ERR and PGC-1α to regulate mitochondrial biogenesis and is necessary for maximal mitochondrial function.57 A similar role of KLF4 in muscle has not yet been elucidated. Other transcription factors in skeletal muscle also cooperate with PGC-1α. Yin Yang 1 (YY1) was identified through a computational approach to be a common target interacting with both PGC-1α and mTOR, perhaps linking nutrient sensing with mitochondrial biogenesis.58 Muscle-specific YY1 knockout mice display profound defects in mitochondrial structure and energy production.59 Additional factors also act as negative regulators of PGC-1 signaling. For instance, loss of the nuclear receptor corepressor RIP140 in muscle results in increased FAO and ETC/OXPHOS gene expression.60 These effects are meditated through RIP140 action on multiple nuclear receptors including PPAR and ERR.61 In addition, knockout of the gene encoding folliculin (Flcn) or its partner folliculin interacting partner 1 (Fnip1), has also been shown to induce the PGC-1α cascade and mitochondrial function in muscle cells.62–64 The inhibitory effects of the FLCN/FNIP1 complex on PGC-1α signaling may be mediated through suppression of AMPK signaling.64
Orchestration of muscle mitochondrial quality control and biogenesis
Mitochondrial dynamics
Mitochondria are highly adaptable organelles that require continuous surveillance to maintain their function integrity. First, mitochondria constantly undergo fission and fusion. Two mitochondria can form a single, tubular like mitochondria by fusing their outer and inner membranes. Conversely, mitochondrial fission fragments the tubular mitochondria network into small organelles. The sophisticated regulation of mitochondrial fission and fusion has been an area of intense interest and research. The membrane bound GTPases mitofusin1 (MFN1) and mitofusin 2 (MFN2) initiate the outer membrane (OM) fusion of two mitochondria, followed by the fusion of the inner membranes (IM) mediated by the optic atrophy protein 1 (OPA1). Loss-of-function studies have demonstrated that MFN1, MFN2 and OPA1 are essential for mitochondrial fusion. Cells lacking both MFN1 and MFN2 are unable to undergo mitochondrial fusion, whereas cells lacking OPA1 do undergo mitochondrial OM fusion, but fail to complete the IM fusion.65 Mitochondrial morphology can be rapidly affected by mitochondrial fission and fusion, thus, these morphologic changes of mitochondria can reflect the relative rates of fusion and fission. Surprisingly, different muscle fiber types display distinct patterns of mitochondrial dynamics. In fast-twitch muscle fibers, small block-like mitochondria are arranged in rows and columns, while in fibers from slow-twitch muscle, mitochondria are more elongated and interconnected to each other from multiple sarcomeres.66 Furthermore, switching from glycolytic to oxidative muscle fiber promotes mitochondria elongation in parallel.66 Given the distinct energy metabolic profiles of slow and fast fiber types, it is tempting to speculate that changes in mitochondrial dynamics adapt to match a change in fuel substrate preference.
Multiple studies support a critical role for mitochondrial fusion in skeletal muscle quality control. Skeletal muscle Mfn2 KO mice display reduced mitochondrial respiration function, impaired muscle insulin signaling, and features of age-related sarcopenia.67,68 Although the exact role of Mfn2 in ER-mitochondrial interactions is under debate,69–73 notably, loss of MFN2 has been shown to activate an ER stress signaling in skeletal muscle.67 Remarkably, mice with targeted ablation of both Mfn1 and Mfn2 genes show a profound muscle atrophy phenotype.74 Combined deficiency of Mfn1 and Mfn2 in skeletal muscle causes significant mitochondrial dysfunction, compensatory mitochondrial proliferation and mtDNA depletion as well. As expected, the muscle-specific Mfn1/2 double-KO mice are exercise intolerant and have a higher post-exercise blood lactate level.74 Transgenic and gene knockout approaches in mice have also been used to define the physiological roles of OPA1 in skeletal muscle. Two independent studies have shown that overexpression of OPA1 can improve muscle mitochondrial respiration function.75,76 OPA1 transgenic mice are protected against denervation-induced muscle atrophy and the transgene expression rescues muscle loss in a model of myopathy caused by COX15 deletion in muscle.75,76 In contrast to the gain-of-function models, muscle-specific KO of Opa1 results in a severe detrimental phenotype including mitochondrial dysfunction, muscle loss and premature death.77,78 Specifically, muscle-specific Opa1 deletion in newborns is lethal, while acute tamoxifen-induced deletion of Opa1 in 5-month-old mice leads to a precocious aging phenotype including metabolic derangement in multiple organs, a systemic inflammatory response and precocious epithelial senescence.77 Interestingly, mice with a skeletal muscle-specific, tamoxifen-induced Opa1 deficiency show an improved glucose tolerance and insulin sensitivity at basal level and in response to a HFD challenge despite reduce muscle mass, and the favorable metabolic phenotype is linked to muscle-derived FGF21.77,78 The importance of mitochondrial fission in skeletal muscle has also been studied. Overexpression of dynamin related protein 1 (Drp1) in skeletal muscle induces muscle mass loss and decreased exercise performance.79 Interestingly, similar to the Mfn2 KO mice, manipulation of OPA1 or Drp1 in skeletal muscle also activates mitochondria-stress signals, as discussed below.77–79 Taken together, these data clearly demonstrate that proper maintenance of mitochondria dynamics, i.e. the balance between fission and fusion, is necessary for optimal muscle mitochondrial function and fitness.
Mitochondrial proteolysis
Mitochondria adopt two main mechanisms for quality control, proteolysis and mitophagy, that respond to damage caused by oxidative stress, misfolded or damaged proteins, or defects in the electron transport chain. Mitochondrial proteases serve as the first line of quality-control mechanism against low degree of mitochondrial stress. Both ATP-dependent and ATP-independent mitochondrial proteases have been identified for selective clearance of the excess non-assembled, misfolded, or damaged proteins within mitochondria. The two ATP-independent proteases are mitochondrial inner membrane metalloprotease Atp23 homolog (ATP23) and serine protease HTRA2. There are four main ATP-dependent proteases comprising the ATPases associated with diverse cellular activities (AAA+) superfamily. The intermembrane AAA (i-AAA) member such as YME1L1, has a protease domain facing the intermembrane space. The membrane-bound AAA (m-AAA) protease faces the mitochondrial matrix and is comprised of homo- or heterohexamers of the AFG3L2 and SPG7 gene products. The other AAA+ proteases are the Lon protease homolog (LONP, encoded by the LONP1 gene), and Clp protease proteolytic subunit (CLPP).80 Approximately two thirds of the 1200 mitochondrial proteins reside in the matrix, thus the main matrix AAA proteases such as LONP and CLPP, serve as the major protein-folding homeostasis control mechanism in mitochondria.
Mutations in the main mitochondrial matrix proteases LONP and CLPP have been implicated in genetic diseases in human. For example, mutations in the human LONP1 gene result in CODAS syndrome, a multisystem developmental disorder characterized by cerebral, ocular, dental, auricular, and skeletal anomalies.81 Mutations in the human CLPP gene cause Perrault syndrome, a heterogeneous autosomal recessive disease characterized by sensorineural hearing loss and ovarian failure.82 Both in vitro and in vivo studies have demonstrated that LONP and CLPP are key regulators of mitochondrial proteostasis.83,84 Several proteins such as aconitase (ACO), cytochrome c oxidase subunit 4 isoform 1 (Cox4i1), steroidogenic acute regulatory protein (STAR), and transcription factor A (TFAM) have been identified as targets for degradation by LONP under basal or stress conditions.80 Most interestingly, as discussed below, mitochondrial matrix proteases LONP and CLPP have been implicated in the mitochondrial unfolded protein response (UPRmt) and their expression is induced in response to UPRmt in mammalian cells.85,86
LONP is also essential for survival given the embryonic lethality of mouse with deletion of the Lonp1 gene.87 However, unlike the extensive studies of mitochondrial dynamics in skeletal muscle, there is no animal model of conditional tissue-specific LONP knockout. Therefore little is known about the contribution of mitochondrial matrix proteases to mitochondrial function and tissue homeostasis in skeletal muscle in vivo. Similarly, the function of CLPP in muscle is also not entirely clear. Homozygous deletion of the gene encoding CLPP in mice causes infertility, hearing loss, and growth retardation,88 thus limiting the study of the role for CLPP in skeletal muscle. Taken together, although mitochondrial proteolysis has been implicated to be critical for maintenance of mitochondrial function, the causal link between the derangement of muscle mitochondrial proteostasis and physiological outcomes under normal and disease states remains unclear.
Skeletal muscle mitochondrial quality control through mitophagy
Emerging evidence demonstrates that mitophagy serves as a critical quality-control mechanism for selective targeting and removal of damaged or dysfunctional mitochondria.89–92 Both ubiquitin-dependent and receptor-dependent mitophagy pathways such as those involving PINK1/Parkin, NIX/BNIP3, FUNDC1, BCL2L13, and PBH2 have been identified in mammals.91–95 Elegant studies in cultured cells have demonstrated that these pathways engage the core autophagy machinery to activate mitochondrial clearance in response to various stress stimuli. Among those mitophagy mediators, the role of PINK1-Parkin signaling has been well studied. PINK1 mainly serves as a sensor for the mitochondrial polarization state. Under normal conditions, PINK1 is transported into the mitochondria for degradation by the serine protease presenilins-associated rhomboid-like protein (PARL). Upon acute mitochondrial stress, mitochondrial depolarization stabilizes PINK1, which phosphorylates MFN2 and subsequently recruits the E3 ubiquitin ligase Parkin onto mitochondria.96 Parkin acts downstream of PINK1 to trigger selective autophagy of depolarized mitochondria.91 Recent results demonstrate that Parkin is required for maintenance of baseline mitochondrial function in skeletal muscle and for exercise-induced increases in mitophagy flux.97 However, in different stress conditions, mitophagy receptors such as NIX/BNIP3, FUNDC1, BCL2L13, and PBH2 have been shown to mediate the degradation of damaged mitochondrial independently of PINK1/Parkin, by interacting directly with LC3. For example, in response to hypoxia, the mitophagy receptor FUNDC1 is dephosphorylated to enhance its interaction with LC3 through a LC3-binding motif, thereby triggering mitophagy.92,94,98 Loss of FUNDC1 in muscle leads to significantly reduced mitophagy flux, mitochondrial energetics and exercise performance.99 Interestingly, however, muscle FUNDC1 deficiency alleviates HFD-induced obesity and improves systemic glucose homeostasis.99 NIX and BNIP3 expression are also induced during hypoxia, likely promoting the clearance of unwanted mitochondria complementary to the FUNDC1 pathway.90,92
Although several pieces of evidence have suggested an important role for general autophagy or mitophagy in regulating muscle mass and metabolism, our understanding of the in vivo physiological relevance and mechanisms of mitophagy in skeletal muscle remains largely incomplete. Notably, previous studies suggested that finely tuned regulation of autophagy flux is necessary for maintaining muscle metabolic homeostasis and quality. Genetic manipulations that inhibit muscle autophagy/mitophagy lead to conflicting results when examining HFD-induced glucose intolerance.99–101 Whether and how the various mitophagy pathways contribute to the impact of autophagy on muscle metabolism is unclear. An important future goal is to establish the metabolic impact of the divergent mitophagy pathways in skeletal muscle under physiological and pathological conditions.
Integration of mitochondrial quality control and biogenesis
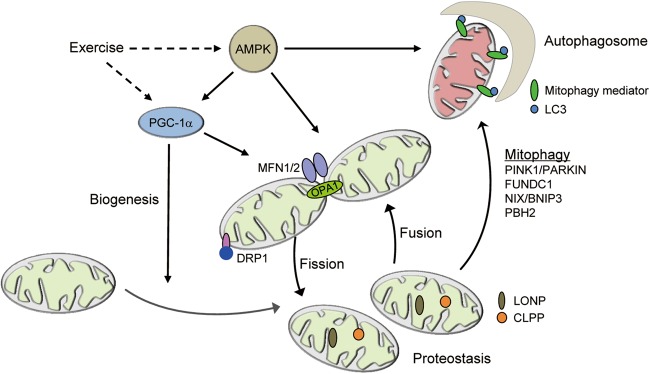
Accumulating evidence suggests that there is an integrated signaling mechanism that coordinately regulates mitochondrial biogenesis, mitochondrial dynamics, and mitochondrial quality control (Fig. 2). An interesting interconnection between muscle mitochondrial biogenesis and dynamics was discovered in PGC-1α/β double KO mice. As described above, combined deficiency of PGC-1α/β in skeletal muscle results in a dramatic exercise performance deficit related to a severely impaired mitochondrial function.37 Mitochondria from PGC-1α/β double KO muscle show significant heterogeneity in size, including many small, fragmented mitochondria juxtaposed to elongated, thin mitochondria, indicative of a defect in mitochondrial dynamics as well as biogenesis.37 The levels of several proteins critical for mitochondrial fusion and fission were also downregulated in the PGC-1α/β-deficient muscle, including MFN1, MFN2, and Drp1.37 Mechanistically, PGC-1α regulates MFN1 and MFN2 expression through ERRα.102,103 These results support a direct link between the transcriptional regulation of mitochondrial biogenesis and mitochondrial dynamics. PGC-1α has also been recently shown to regulate mitophagy in a denervation-induced muscle atrophy model as well as acute exercise.104,105 Given that nuclear receptor PPARα is found to bind to a broad array of autophagy gene promoters in liver,106 it is possible that PGC-1α signaling may also regulate expression of autophagy genes in skeletal muscle by co-activating nuclear receptors. Taken together, these results demonstrate a PGC-1α−mediated mechanism for the coordination of mitochondrial biogenesis and quality control in skeletal muscle.
Fig. 2.
Integration of mitochondrial quality control and biogenesis. Muscle mitochondria undergo extensive turnover and remodeling in response to a variety of physiological stimuli. PGC-1α activates mitochondrial biogenesis. Mitochondrial fission is mediated by DRP1 and its receptors. Mitochondrial fusion is mediated by MFN1/2 for the fusion of the outer membrane and OPA1 for the inner membranes. The main mitochondria matrix proteases such as LONP and CLPP maintain mitochondrial proteostasis and regulate mitochondrial function. The damaged or dysfunctional mitochondria are cleared via mitophagy. Mitophagy is mediated by mitophagy mediators such as PINK1/PARKIN, FUNDC1, NIX/BNIP3, and PBH2 in mammals. Exercise coordinately regulates mitochondrial biogenesis, mitochondrial dynamics, and mitochondrial quality-control program via AMPK and the PGC-1α. Mfn1/2 mitofusin 1/2, Drp1 dynamin-related protein 1, OPA1 optic atrophy 1, LONP Lon peptidase 1, CLPP caseinolytic mitochondrial matrix peptidase proteolytic subunit, PINK1 PTEN-induced putative kinase 1, FUNDC1 FUN14 domain containing 1, Bnip3 BCL2 interacting protein 3
There is also evidence to suggest a close link between mitochondrial dynamics and mitophagy. Fusion between healthy and damaged mitochondria is known to dilute the damaged organelles into the healthy mitochondrial network, thus maintaining overall mitochondrial fitness.107 In contrast, mitochondrial fragmentation allows the segregation of damaged components from the mitochondrial network, triggering the mitophagy process.108 Consistently, defective mitochondrial fusion in Mfn2 KO skeletal muscle cells leads to activation of mitophagy.68
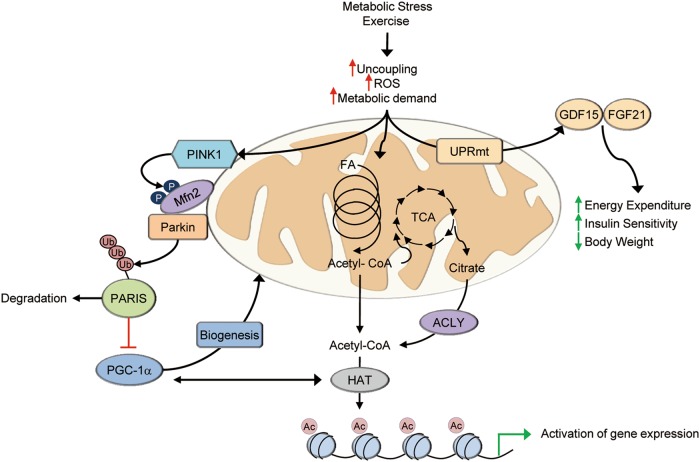
Direct links between mitophagy and mitochondrial biogenesis may also exist. Expression of a MFN2 mutant lacking PINK1 phosphorylation sites prevents induction of PGC-1α and mitochondrial biogenesis in the postnatal heart.109 One potential mechanism linking mitophagy to mitochondrial biogenesis involves the Parkin-interacting substrate (PARIS).110 PARIS regulates mitochondrial biogenesis by transcriptionally repressing PGC-1α, likely through binding of the IRS within the PGC-1α promoter region. Phosphorylation by PINK1 primes PARIS for ubiquitination by Parkin resulting in the degradation of PARIS and subsequent de-repression and activation of PGC-1α expression (Fig. 3).111 In skeletal muscle, recent results demonstrate that loss of Parkin results in an increase in nuclear-localized PARIS and a subsequent decrease in PGC-1α following exercise.112 Although these regulating systems are likely active in skeletal muscle, further investigation is needed to determine the role of these systems in the regulation of mitochondrial quality control and biogenesis.
Fig. 3.
Skeletal muscle mitochondria serve as cellular sensors of metabolic demand and stress. Various forms of metabolic stress including exercise triggers responses in muscle mitochondria that result in signals to other cellular compartments or tissues. Increased mitophagy is integrated with biogenesis through activation of PGC-1α, either directly or through mechanisms such as PARIS. Activation of gene expression by PGC-1a also occurs in cooperation with histone acetyltransferases (HAT). Acetyl-CoA generated by increased mitochondrial respiration can donate acetyl groups for histone modifications. Paracrine or endocrine signals can also be generated, at least in part, by activation of the mitochondrial unfolded protein response (UPRmt) that leads to increases in GDF15 and FGF21 production and secretion. PARIS Parkin interacting substrate, ACLY ATP citrate lyase, HAT histone acetyltransferase, Ac acetyl, Ub ubiquitin, FA fatty acid, UPRmt mitochondrial unfolded protein response
Extensive studies have supported an important role of AMPK in orchestrating mitochondrial biogenesis, dynamics and mitochondrial quality control. Firstly, as described above, AMPK can activate mitochondrial biogenesis through a PGC-1α dependent mechanism.43 AMPK activation in skeletal muscle is sufficient to drive fatty acid oxidation and mitochondrial biogenesis. In addition, AMPK has been shown to promote mitochondrial fission by direct phosphorylation of mitochondrial fission factor (MFF), a receptor for Drp1.113 Moreover, AMPK is able to directly phosphorylate and activate the unc-51 like autophagy activating kinase 1 (ULK1),114 triggering the autophagic cascade and mitophagy. More recently, it has been shown that exercise-induced mitophagy in skeletal muscle depends on AMPK-ULK1 signaling.115 Together, in this fashion, AMPK serves as a critical regulator of muscle mitochondria function through coordinate regulation of mitochondrial biogenesis and mitophagy. Consistent with the central role of AMPK in maintenance of muscle mitochondrial function, muscle-specific AMPK β1β2 double-KO in mice results in significantly impaired exercise performance related to reduced mitochondrial content and function.116
Exercise training is well known to enhance muscle mitochondrial function, leading to improvements in whole-body metabolic homeostasis.2 Exercise activates signaling networks that coordinately control mitochondrial remodeling, including mitochondrial biogenesis, dynamics, and mitophagy. Given that AMPK and PGC-1α are induced by exercise training, it is tempting to speculate that AMPK/PGC-1α signaling may orchestrate the activity of multiple pathways in response to exercise to control mitochondrial biogenesis, dynamics and quality control in skeletal muscle. The actions of AMPK and PGC-1α on mitochondrial remodeling could lead to a net effect of replacing existing unhealthy mitochondria with new functional mitochondria. This may also be necessary for remodeling of mitochondria to a more oxidative phenotype that is known to occur following exercise. However, future studies are needed to fully understand the coordinate control of mitochondrial biogenesis and quality that define muscle fitness.
Muscle mitochondria as a metabolic sensor hub in response to physiological stimuli
Mitochondria-to-nucleus communication (retrograde response)
Increasing evidence suggests that mitochondria are not only the powerhouse of the cell that generates ATP but also function as a central node for cellular signaling. Evidence is emerging that mitochondria can communicate to the nucleus (retrograde) to alter the transcription of nuclear genes in response to various cellular stresses.117 Among these mitochondrial retrograde responses, the mitochondrial unfolded protein response (UPRmt) has been the best studied. The UPRmt is provoked when there is an accumulation of unassembled mitochondrial respiratory proteins triggering an unknown mechanism to communicate with the nucleus, leading to the induction of genes involved in maintaining mitochondrial protein-folding homeostasis.85,118,119 Although originally identified in mammalian cells, UPRmt signaling has been extensively described in Caenorhabditis elegans. The transcription factor ATFS-1 is responsible for the UPRmt response in C. elegans. Under basal conditions, the ATFS-1 is imported into the mitochondrial matrix for degradation by the Lon protease. Multiple forms of mitochondrial stresses including accumulation of unfolded proteins, OXPHOS defects, accumulation of ROS, or mitochondrial–nuclear protein imbalance can lead to reduced mitochondrial import of ATFS-1. This results in accumulation of ATFS-1 in the cytosol and its translocation to the nucleus with consequent activation of a broad array of stress response genes to maintain mitochondrial function.120 Activation of the UPRmt response by various forms of mitochondrial stress has also been validated in mammalian cells. Interestingly, the mitochondrial matrix proteases LONP and CLPP have been linked to the mammalian UPRmt and their expression is induced in response to unfolded protein stress.85,121 Recent studies conducted in C. elegans have also demonstrated that UPRmt can trigger a non-cell-autonomous mitochondrial regulatory pathway that has profound effects on aging and metabolism.122 In addition to UPRmt, recent studies have also suggested that intermediary metabolites derived from mitochondrial metabolism signal to the nucleus resulting in epigenetic modifications to regulate gene expression (reviewed in ref.123). For example, acetyl-CoA can be generated from citrate, a product of the mitochondrial TCA cycle, through the action of ATP-citrate lyase (ACLY), as well as from mitochondrial FAO, providing a source of acetyl groups for histone acetylation (Fig. 3).124,125 Interestingly, increases in histone acetylation by fatty acids were associated with activation of genes involved in fatty acid utilization suggesting a specificity to this pathway.125 Similarly, S-adenosylmethionine is an essential substrate for histone or DNA methylation.126
Muscle mitohormesis
Skeletal muscle comprises ~40% of total body mass and is the largest consumer of glucose and fatty acids, via mitochondrial oxidation. As such, mitochondria function in skeletal muscle is a critical determinant of systemic metabolic homeostasis. Numerous studies in mice, and in humans, have shown a close association between derangements in muscle mitochondrial function and the development of insulin resistance.127,128 As discussed further below, evidence suggests that mitochondrial dysfunction in skeletal muscle results in a reduction of FAO, leading to the accumulation of various lipid species and derangements in insulin signaling.128–130 However, other studies have shown that mice with defective mitochondrial function in skeletal muscle are more insulin sensitive and protected against diet-induced obesity. For example, muscle-specific deletion of Aifm1 (apoptosis inducing factor mitochondria associated 1) results in impaired mitochondrial oxidative activity but protection against HFD-induced obesity and insulin resistance.131 Likewise, mice with targeted ablation of Tfam in skeletal muscle also exhibit enhanced glucose tolerance, despite reduced mitochondrial DNA content and function in skeletal muscle.132 In addition, defective autophagy in muscle-specific Atg7 KO mice leads to impaired muscle mitochondrial function but resistance to HFD-induced obesity with improved systemic insulin sensitivity and glucose tolerance.101 Mice lacking FUNDC1 in muscle show mitophagy defect but are protected against chronic HFD-induced obesity and insulin resistance despite reduced muscle mitochondrial energetics.99 Acute muscle Opa1 deficiency in 4-week-old mice also causes mitochondrial defects but protection against HFD-induced obesity and insulin resistance.78 Therefore, whereas mitochondrial dysfunction may contribute to insulin resistance, it is not a pre-requisite and depending on the defect, may actually lead to insulin sensitivity. Some data suggest that the seemingly contradictory phenotypes may be related to a non-cell-autonomous role of the muscle mitochondrial stress response.78,99,101
The observation that perturbations in muscle mitochondrial function and quality control influence systemic metabolism suggests the intriguing possibility that this response may involve extracellular signals. This notion is consistent with the concept of mitohormesis, an adaptive response triggered by mild mitochondrial stress that was originally postulated based on observations in C. elegans.133,134 The concept of “mitokines” (soluble factors that can be released from cells with stressed mitochondria) was put forth to explain systemic metabolic benefits in the context of mitochondrial stress in skeletal muscle (Fig. 3). In mammals, the hormones FGF21 and GDF15, and mitochondria-derived peptides have been described as putative mitokines (Fig. 3).99,101,135,136 FGF21 has been shown to be a direct ATF4 target downstream of UPRmt,99,101 while GDF15 secretion is induced in a CHOP-dependent manner during UPRmt.135 The identification of FGF21 as a mitokine is of interest given its well known favorable effect on systemic metabolic control.137–139 In multiple mouse models of mitochondrial defects and in human patients, skeletal muscle secretes FGF21 into circulation to exert an endocrine effect.78,99,101,140,141 GDF15 has also been shown to be induced and released from skeletal muscle upon multiple forms of mitochondrial stress.121,135 Similar to FGF21, pharmacological administration of recombinant GDF15 leads to many effects including decreased food intake and reduced body weight, improved insulin sensitivity, and increased energy expenditure in obese mice.135 Other potential mitokines have been identified including a short peptide encoded by the mitochondrial open-reading-frame of the 12S rRNA-c (MOTS-c).136 MOTS-c also exerts beneficial effects on obesity and insulin resistance.
These data taken together provide significant evidence that muscle mitochondria are not merely the “powerhouse of the cell” but act as cellular sensors of increased energy demand or other cellular stresses and subsequently signal to other cellular compartments such as the nucleus or other organs in the body (Fig. 3). These signals influence gene expression and epigenomic status of the cell in addition to direct effects on the mitochondria, e.g. mitophagy. These responses can also generate a mitohormesis signal that results in beneficial effects for metabolic diseases. It is also tempting to speculate that muscle mitochondria may function as an early checkpoint prior to the propagation of energy metabolic stress. This raises several interesting questions: (1) Are there other mitokines that may mediate muscle mitochondrial communications? (2) Although a few mitokines have been identified, what are the mechanism by which the muscle UPRmt or other pathways trigger the expression of such factors? (3) Will it be possible to exploit such muscle mitochondrial signals as treatment strategies?
Muscle mitochondrial remodeling in disease states
Muscular dystrophy
Duchenne muscular dystrophy (DMD), caused by mutations in the X-linked gene dystrophin, is the most common and severe form of muscular dystrophy and remains incurable to date.142–144 Individuals with DMD display muscle weakness and increased muscle exhaustion. Interestingly, fast glycolytic myofibers are more sensitive to the dystrophic pathology in DMD patients as well as in the preclinical mdx mouse model of DMD.145 Previous studies have shown mitochondrial dysfunction in DMD muscle,146,147 contributing to a vicious cycle and further promoting the progression of muscular dystrophy. Conversely, numerous studies have demonstrated that enhancing the muscle mitochondrial functional capacity in the mdx mouse model of DMD may prevent or ameliorate dystrophic pathology. For example, overexpression of PGC-1α specifically in skeletal muscle was shown to improve muscular dystrophy.148,149 In addition, activation of PGC-1α through upstream signaling pathways such as AMPK or Sirt1 also prevents the hallmarks of muscular dystrophy in mdx mice.150–152 Recent work has also demonstrated that promoting autophagy may have therapeutic benefits in muscular dystrophy.153,154 It is certainly possible that muscle function could be enhanced by promoting the clearance of defective mitochondria, contributing to a favorable impact on the muscular dystrophy phenotype. Interestingly, direct manipulation of miR-499 is able to restore the oxidative muscle mitochondrial program and prevent the hallmarks of DMD, including serum CK release and diminished exercise capacity in mdx mice.64 In addition, Fnip1 knockout mice on an mdx genetic background have also been shown to be protected against muscular dystrophy.63 Notably, activation of miR-499 directly inhibits FNIP1, leading to activation of PGC-1α signaling in skeletal muscle.64 As many of the routes to muscular dystrophy rescue in mdx mice converge on the activation of PGC-1α, this provides intriguing evidence that PGC-1α and mitochondrial energy metabolism are central to the pathogenesis of muscular dystrophy and suggests a therapeutic approach for DMD.
Dysregulation of mitochondrial energetics during disuse atrophy
Skeletal muscle mass is controlled by a fine balance between protein synthesis and degradation pathways (reviewed in ref.155). This balance is sensitive to changes in physical activity levels. For instance, decreased activity as a result of bed rest, chronic illness or even microgravity experienced during space flight results in a loss of muscle mass. Prolonged disuse can produce a loss of approximately 0.5–0.6% of skeletal mass per day.156 Muscle strength and the intrinsic contractile capacity of the myofiber are also decreased with disuse or immobility.157,158 Elderly individuals are particularly susceptible to disuse atrophy. Studies have shown that older individuals do not fully recover muscle function following disuse.159,160 This can lead to a vicious cycle of muscle atrophy followed by incomplete recovery and subsequent debilitating events or illnesses that exacerbate immobility and reduce independence. Recent studies have shown that adverse mitochondrial remodeling occurs in muscle during disuse atrophy. For example, 1 week of strict bed rest in healthy young adults results in diminished OXPHOS protein expression and citrate synthase activity.161 Multiple preclinical and clinical studies have now demonstrated that a reduction in PGC-1α expression accompanies these changes in mitochondria in various forms of muscle atrophy.162–165 In addition, a protective role for preserved mitochondrial function during muscle atrophy is supported by numerous preclinical studies. Overexpression of PGC-1α or PGC-1β prevents or reduces muscle atrophy in various experimental conditions such as hindlimb unloading, denervation and heart failure.165–169 Although the precise mechanism involved in this cross-talk is not fully understood, studies have shown that PGC-1 inhibits FoxO3-mediated induction of the ubiquitin ligases, muscle ring-finger 1 (MuRF1) and atrogin-1, without affecting protein synthesis pathways.165,168
A decline in mitochondrial function in aging and sarcopenia
Sarcopenia is defined as the steady decline in muscle mass and function that occurs with aging. Numerous factors contribute to the decline in muscle structure and function including reduced rates of protein synthesis and increased proteolysis, insulin resistance, changes in the neuromuscular junction, etc. A number of studies have shown a decline in oxidative capacity and mitochondrial content in aging muscle.170–172 Not surprisingly, both PGC-1α mRNA and protein levels are reduced in aged muscles.172–175 More recently, changes in mitochondrial function are recognized as a contributing factor in muscle performance in sarcopenia. For instance, maximal mitochondrial ATP production (ATPmax), as determined by measurements of phosphocreatine recovery using 31P magnetic resonance spectroscopy (MRS), is negatively correlated with fatigability in a walking test in older adults.176 ATPmax and mitochondrial respiration rates measured in permeabilized muscle fibers also correlate with walking speed in older adults.177,178 Walking speed and function is a particularly relevant readout as this is directly related to functional independence in the elderly.
Animal studies have also implicated dysregulation of mitochondrial dynamics in the progression of sarcopenia during aging. During aging, increased reactive oxygen species (ROS) production causes damage to mitochondrial proteins.179 The expression levels of many autophagy and mitophagy components such as Atg7, p62, beclin1, Bnip3, Parkin, and LAMP2 are known to decline with age.180–182 Interestingly, muscle-specific Atg7 knockout mice display many features of aging including muscle loss and weakness suggesting a possible link between impaired autophagy and mitophagy with sarcopenia.183
Targeting mitochondria as a therapeutic approach for muscle disease
The abundant data demonstrating deficits in mitochondrial function and content in a number of muscle diseases make therapeutic targeting of these pathways attractive. These discoveries have popularized the somewhat fanciful notion of “exercise in a pill” for muscle and metabolic diseases. Regardless of whether this is actually achievable or not, certain aspects of this concept are attractive as therapeutic strategies and certain molecules have shown promise in preclinical proof-of-concept studies. Several approaches have been shown to increase mitochondrial function and oxidative capacity in muscle. For instance, PPARδ activating ligands were shown to synergize with exercise and improve running endurance.17 PPARδ ligands are now progressing into the clinic for treating DMD. Recently, there has also been intense interest in activation of the sirtuin pathway as a mechanism for boosting mitochondrial function and oxidative metabolism. The primary means to achieve this has been the use of NAD+ analogs and precursors such as nicotinamide riboside (NR) to activate sirtuin 1. NR supplementation in mice was recently shown to increase muscle NAD+ levels and activate expression of mitochondrial energy metabolic genes.184 Other studies have also shown that NR enhances muscle energetics and function in the mdx model of DMD.185 An increase in mitochondrial respiration has been demonstrated in permeabilized skeletal muscle fibers from human subjects following 2 weeks of supplementation with acipimox, another NAD+ precursor.186 Although a corresponding increase in muscle NAD+ levels was not reported, this provides further evidence for the promise of targeting this pathway to boost muscle mitochondrial oxidative metabolism. These data warrant further investigation of these agents in conditions of impaired muscle function including DMD or other muscular dystrophies, disuse atrophy, and sarcopenia.
Conclusions and perspectives
Mitochondria are highly dynamic organelles that require continuous surveillance to maintain their functional integrity. A complex regulatory network is required to orchestrate skeletal muscle mitochondrial biogenesis, respiratory function, and maintenance in response to exercise and in disease states. It is clear that these mitochondrial quality control mechanisms are essential for maintaining skeletal muscle metabolic homeostasis. This is particularly important to maintaining muscle function in response to a myriad of physiological stressors such as changes in physical activity, altered fuel substrate availability, and even aging. Mechanisms have evolved to regulate and coordinate mitochondrial biogenesis, proteolysis, and mitophagy to maintain mitochondrial health and function. Although several pathways of proteolysis and mitophagy have been identified, it will be important to further understand how these quality control pathways contribute to the maintenance of muscle mitochondrial function under physiological and pathological conditions. Importantly, mitochondrial quality control pathways do not exist independently. Mitochondrial biogenesis, dynamics, proteostasis, and mitophagy are highly interconnected and regulated by a tightly linked regulatory network. In this way, the skeletal myocyte coordinates biogenesis of healthy mitochondria with replacement of damaged mitochondria to maintain optimum muscle function.
Mitochondria also communicate with the endoplasmic reticulum, lysosomes, and nucleus to relay information regarding energy metabolic status and stress. These communications may occur in the form of direct contact as in the case of mitophagy or through the generation of certain metabolites that signal to other organelles such as the nucleus to influence gene expression. Finally, mitochondria have developed non-cell-autonomous mechanisms to communicate on an organism-wide basis. Messengers for this type of communication include secreted myokines or mitokines.
The progress made in understanding the role of mitochondria in skeletal muscle health has created a desire to develop therapeutics directed at mitochondria. Some of these proposed therapies could be touted as “exercise mimetics”; however, one should treat with caution the notion that a single therapeutic is capable of mimicking the multitude of effects produced by exercise. Further, increasing mitochondrial function or energy metabolic capacity through a single focused target or without a corresponding increase in demand may produce untoward and unintended consequences. Nonetheless, despite these caveats, the importance of mitochondrial quality to muscle and cardiovascular/metabolic fitness justifies pursuit of therapies aimed at mitochondria for many skeletal muscle or metabolic diseases.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 31471110, 31771291); the Ministry of Science and Technology of China (973 Program 2015CB856300); Natural Science Foundation of Jiangsu Province (BK20170014); and Fundamental Research Funds for the Central Universities (090314380023) to Z.G. and NIH grants R01 DK045416, R01 HL058493, and R01 HL128349 to D.P.K.
Competing interests
The authors declare no competing interests.
Contributor Information
Zhenji Gan, Phone: +86-25-58641546, Email: ganzj@nju.edu.cn.
Daniel P. Kelly, Phone: +(215) 898-0768, Email: dankelly@pennmedicine.upenn.edu
Rick B. Vega, Phone: +407-303-1387, Email: richard.vega@flhosp.org
References
- 1.Glancy B, et al. Mitochondrial reticulum for cellular energy distribution in muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 4.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 1998;273:23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 6.van der Meer DL, et al. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010;38:2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMullen PD, et al. A map of the PPARalpha transcription regulatory network for primary human hepatocytes. Chem. Biol. Interact. 2014;209:14–24. doi: 10.1016/j.cbi.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Cresci S, Wright LD, Spratt JA, Briggs FN, Kelly DP. Activation of a novel metabolic gene regulatory pathway by chronic stimulation of skeletal muscle. Am. J. Physiol. 1996;270:C1413–C1420. doi: 10.1152/ajpcell.1996.270.5.C1413. [DOI] [PubMed] [Google Scholar]
- 9.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kliewer SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krey G, et al. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarthy MV, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finck BN, et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 2005;1:133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Gan Z, et al. The nuclear receptor PPARbeta/delta programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 2011;25:2619–2630. doi: 10.1101/gad.178434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muoio DM, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J. Biol. Chem. 2002;277:26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- 16.Burkart EM, et al. Nuclear receptors PPARbeta/delta and PPARalpha direct distinct metabolic regulatory programs in the mouse heart. J. Clin. Invest. 2007;117:3930–3939. doi: 10.1172/JCI32578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narkar VA, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan W, et al. PPARdelta promotes running endurance by preserving glucose. Cell Metab. 2017;25:1186–1193.e1184. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 20.Dufour CR, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Perry MC, Dufour CR, Tam IS, B’Chir W, Giguere V. Estrogen-related receptor-alpha coordinates transcriptional programs essential for exercise tolerance and muscle fitness. Mol. Endocrinol. 2014;28:2060–2071. doi: 10.1210/me.2014-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaBarge S, McDonald M, Smith-Powell L, Auwerx J, Huss JM. Estrogen-related receptor-alpha (ERRalpha) deficiency in skeletal muscle impairs regeneration in response to injury. FASEB J. 2014;28:1082–1097. doi: 10.1096/fj.13-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narkar VA, et al. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metab. 2011;13:283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangwala SM, et al. Estrogen-related receptor gamma is a key regulator of muscle mitochondrial activity and oxidative capacity. J. Biol. Chem. 2010;285:22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan Z, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Invest. 2013;123:2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 29.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 30.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 32.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J. Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baar K, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 34.Terada S, Kawanaka K, Goto M, Shimokawa T, Tabata I. Effects of high-intensity intermittent swimming on PGC-1alpha protein expression in rat skeletal muscle. Acta Physiol. Scand. 2005;184:59–65. doi: 10.1111/j.1365-201X.2005.01423.x. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 36.Rowe GC, et al. Disconnecting mitochondrial content from respiratory chain capacity in PGC-1-deficient skeletal muscle. Cell Rep. 2013;3:1449–1456. doi: 10.1016/j.celrep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zechner C, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leick L, et al. PGC-1alpha is not mandatory for exercise- and training-induced adaptive gene responses in mouse skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008;294:E463–E474. doi: 10.1152/ajpendo.00666.2007. [DOI] [PubMed] [Google Scholar]
- 39.Ballmann C, Tang Y, Bush Z, Rowe GC. Adult expression of PGC-1alpha and -1beta in skeletal muscle is not required for endurance exercise-induced enhancement of exercise capacity. Am. J. Physiol. Endocrinol. Metab. 2016;311:E928–e938. doi: 10.1152/ajpendo.00209.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowe GC, El-Khoury R, Patten IS, Rustin P, Arany Z. PGC-1alpha is dispensable for exercise-induced mitochondrial biogenesis in skeletal muscle. PLoS ONE. 2012;7:e41817. doi: 10.1371/journal.pone.0041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miura S, et al. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology. 2007;148:3441–3448. doi: 10.1210/en.2006-1646. [DOI] [PubMed] [Google Scholar]
- 42.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 43.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. USA. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 48.Riehle C, et al. Insulin receptor substrates are essential for the bioenergetic and hypertrophic response of the heart to exercise training. Mol. Cell. Biol. 2014;34:3450–3460. doi: 10.1128/MCB.00426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 51.Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J. Biol. Chem. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray S, et al. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 53.Haldar SM, et al. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc. Natl. Acad. Sci. USA. 2012;109:6739–6744. doi: 10.1073/pnas.1121060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prosdocimo DA, et al. Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J. Biol. Chem. 2014;289:5914–5924. doi: 10.1074/jbc.M113.531384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prosdocimo DA, et al. KLF15 and PPARalpha cooperate to regulate cardiomyocyte lipid gene expression and oxidation. PPAR Res. 2015;2015:201625. doi: 10.1155/2015/201625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oishi Y, et al. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nat. Med. 2008;14:656–666. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 57.Liao X, et al. Kruppel-like factor 4 is critical for transcriptional control of cardiac mitochondrial homeostasis. J. Clin. Invest. 2015;125:3461–3476. doi: 10.1172/JCI79964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 59.Blattler SM, et al. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol. Cell. Biol. 2012;32:3333–3346. doi: 10.1128/MCB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seth A, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 2007;6:236–245. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, et al. Roles of transcriptional corepressor RIP140 and coactivator PGC-1alpha in energy state of chronically infarcted rat hearts and mitochondrial function of cardiomyocytes. Mol. Cell. Endocrinol. 2012;362:11–18. doi: 10.1016/j.mce.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 62.Hasumi H, et al. Regulation of mitochondrial oxidative metabolism by tumor suppressor FLCN. J. Natl. Cancer Inst. 2012;104:1750–1764. doi: 10.1093/jnci/djs418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reyes NL, et al. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2015;112:424–429. doi: 10.1073/pnas.1413021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 2016;8:1212–1228. doi: 10.15252/emmm.201606372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schrepfer E, Scorrano L. Mitofusins, from mitochondria to metabolism. Mol. Cell. 2016;61:683–694. doi: 10.1016/j.molcel.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 66.Mishra P, Varuzhanyan G, Pham AH, Chan DC. Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell Metab. 2015;22:1033–1044. doi: 10.1016/j.cmet.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebastian D, et al. Mitofusin 2 (Mfn2) links mitochondrial and endoplasmic reticulum function with insulin signaling and is essential for normal glucose homeostasis. Proc. Natl. Acad. Sci. USA. 2012;109:5523–5528. doi: 10.1073/pnas.1108220109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sebastian D, et al. Mfn2 deficiency links age-related sarcopenia and impaired autophagy to activation of an adaptive mitophagy pathway. EMBO J. 2016;35:1677–1693. doi: 10.15252/embj.201593084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 70.Filadi R, et al. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc. Natl. Acad. Sci. USA. 2015;112:E2174–E2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Filadi R, et al. On the role of Mitofusin 2 in endoplasmic reticulum-mitochondria tethering. Proc. Natl. Acad. Sci. USA. 2017;114:E2266–e2267. doi: 10.1073/pnas.1616040114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naon D, et al. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc. Natl. Acad. Sci. USA. 2016;113:11249–11254. doi: 10.1073/pnas.1606786113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naon D, et al. Reply to Filadi et al.: Does Mitofusin 2 tether or separate endoplasmic reticulum and mitochondria? Proc. Natl. Acad. Sci. USA. 2017;114:E2268–e2269. doi: 10.1073/pnas.1618610114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varanita T, et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 2015;21:834–844. doi: 10.1016/j.cmet.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Civiletto G, et al. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metab. 2015;21:845–854. doi: 10.1016/j.cmet.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tezze C, et al. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab. 2017;25:1374–1389.e1376. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira RO, et al. OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 2017;36:2126–2145. doi: 10.15252/embj.201696179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Touvier T, et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015;6:e1663. doi: 10.1038/cddis.2014.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 81.Strauss KA, et al. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Am. J. Hum. Genet. 2015;96:121–135. doi: 10.1016/j.ajhg.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenkinson EM, et al. Perrault syndrome is caused by recessive mutations in CLPP, encoding a mitochondrial ATP-dependent chambered protease. Am. J. Hum. Genet. 2013;92:605–613. doi: 10.1016/j.ajhg.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 84.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 85.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quiros PM, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 88.Gispert S, et al. Loss of mitochondrial peptidase ClpP leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 2013;22:4871–4887. doi: 10.1093/hmg/ddt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J. Cell Biol. 2014;205:435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu L, Sakakibara K, Chen Q, Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 95.Wei Y, Chiang WC, Sumpter R, Jr., Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–238.e210. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen CCW, Erlich AT, Hood DA. Role of Parkin and endurance training on mitochondrial turnover in skeletal muscle. Skelet. Muscle. 2018;8:10. doi: 10.1186/s13395-018-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen G, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 99.Fu T, et al. Mitophagy directs muscle-adipose crosstalk to alleviate dietary obesity. Cell Rep. 2018;23:1357–1372. doi: 10.1016/j.celrep.2018.03.127. [DOI] [PubMed] [Google Scholar]
- 100.He C, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim KH, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 102.Soriano FX, et al. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 2006;55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 103.Martin OJ, et al. A role for peroxisome proliferator-activated receptor gamma coactivator-1 in the control of mitochondrial dynamics during postnatal cardiac growth. Circ. Res. 2014;114:626–636. doi: 10.1161/CIRCRESAHA.114.302562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vainshtein A, Desjardins EM, Armani A, Sandri M, Hood DA. PGC-1alpha modulates denervation-induced mitophagy in skeletal muscle. Skelet. Muscle. 2015;5:9. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vainshtein A, Tryon LD, Pauly M, Hood DA. Role of PGC-1alpha during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am. J. Physiol. Cell Physiol. 2015;308:C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JM, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J. Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- 109.Gong G, et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shin JH, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee Y, et al. PINK1 primes Parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 2017;18:918–932. doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen Chris Chin Wah, Erlich Avigail T., Crilly Matthew J., Hood David A. Parkin is required for exercise-induced mitophagy in muscle: impact of aging. American Journal of Physiology-Endocrinology and Metabolism. 2018;315(3):E404–E415. doi: 10.1152/ajpendo.00391.2017. [DOI] [PubMed] [Google Scholar]
- 113.Toyama EQ, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laker RC, et al. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat. Commun. 2017;8:548. doi: 10.1038/s41467-017-00520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O’Neill HM, et al. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc. Natl. Acad. Sci. USA. 2011;108:16092–16097. doi: 10.1073/pnas.1105062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 118.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J. Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 119.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 120.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shpilka T, Haynes CM. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018;19:109–120. doi: 10.1038/nrm.2017.110. [DOI] [PubMed] [Google Scholar]
- 122.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Matilainen O, Quiros PM, Auwerx J. Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol. 2017;27:453–463. doi: 10.1016/j.tcb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 124.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McDonnell E, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chiang PK, et al. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- 127.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 128.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2011;8:92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 129.Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15:595–605. doi: 10.1016/j.cmet.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pospisilik JA, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 132.Wredenberg A, et al. Respiratory chain dysfunction in skeletal muscle does not cause insulin resistance. Biochem. Biophys. Res. Commun. 2006;350:202–207. doi: 10.1016/j.bbrc.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 133.Rattan SI. Hormesis in aging. Ageing Res. Rev. 2008;7:63–78. doi: 10.1016/j.arr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 134.Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chung HK, et al. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 2017;216:149–165. doi: 10.1083/jcb.201607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee C, et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kharitonenkov A, et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 139.Owen BM, et al. FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab. 2014;20:670–677. doi: 10.1016/j.cmet.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suomalainen A, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tyynismaa H, et al. Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 142.Jay V, Vajsar J. The dystrophy of Duchenne. Lancet. 2001;357:550–552. doi: 10.1016/S0140-6736(00)04052-6. [DOI] [PubMed] [Google Scholar]
- 143.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Briguet A, Courdier-Fruh I, Foster M, Meier T, Magyar JP. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 2004;14:675–682. doi: 10.1016/j.nmd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 145.Webster C, Silberstein L, Hays AP, Blau HM. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503–513. doi: 10.1016/0092-8674(88)90463-1. [DOI] [PubMed] [Google Scholar]
- 146.Kuznetsov AV, et al. Impaired mitochondrial oxidative phosphorylation in skeletal muscle of the dystrophin-deficient mdx mouse. Mol. Cell. Biochem. 1998;183:87–96. doi: 10.1023/a:1006868130002. [DOI] [PubMed] [Google Scholar]
- 147.Timmons JA, et al. Human muscle gene expression responses to endurance training provide a novel perspective on Duchenne muscular dystrophy. FASEB J. 2005;19:750–760. doi: 10.1096/fj.04-1980com. [DOI] [PubMed] [Google Scholar]
- 148.Handschin C, et al. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Selsby JT, Morine KJ, Pendrak K, Barton ER, Sweeney HL. Rescue of dystrophic skeletal muscle by PGC-1alpha involves a fast to slow fiber type shift in the mdx mouse. PLoS ONE. 2012;7:e30063. doi: 10.1371/journal.pone.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ljubicic V, Burt M, Lunde JA, Jasmin BJ. Resveratrol induces expression of the slow, oxidative phenotype in mdx mouse muscle together with enhanced activity of the SIRT1-PGC-1alpha axis. Am. J. Physiol. Cell Physiol. 2014;307:C66–C82. doi: 10.1152/ajpcell.00357.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ljubicic V, et al. Chronic AMPK activation evokes the slow, oxidative myogenic program and triggers beneficial adaptations in mdx mouse skeletal muscle. Hum. Mol. Genet. 2011;20:3478–3493. doi: 10.1093/hmg/ddr265. [DOI] [PubMed] [Google Scholar]
- 152.Chalkiadaki A, Igarashi M, Nasamu AS, Knezevic J, Guarente L. Muscle-specific SIRT1 gain-of-function increases slow-twitch fibers and ameliorates pathophysiology in a mouse model of Duchenne muscular dystrophy. PLoS Genet. 2014;10:e1004490. doi: 10.1371/journal.pgen.1004490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pauly M, et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am. J. Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 154.De Palma C, et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. Biol. 2014;49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wall BT, Dirks ML, van Loon LJ. Skeletal muscle atrophy during short-term disuse: implications for age-related sarcopenia. Ageing Res. Rev. 2013;12:898–906. doi: 10.1016/j.arr.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 157.Hvid LG, et al. Four days of muscle disuse impairs single fiber contractile function in young and old healthy men. Exp. Gerontol. 2013;48:154–161. doi: 10.1016/j.exger.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 158.Paddon-Jones D, et al. Essential amino acid and carbohydrate supplementation ameliorates muscle protein loss in humans during 28 days bedrest. J. Clin. Endocrinol. Metab. 2004;89:4351–4358. doi: 10.1210/jc.2003-032159. [DOI] [PubMed] [Google Scholar]
- 159.Suetta C, et al. Effects of aging on human skeletal muscle after immobilization and retraining. J. Appl. Physiol. (1985) 2009;107:1172–1180. doi: 10.1152/japplphysiol.00290.2009. [DOI] [PubMed] [Google Scholar]
- 160.Hvid LG, et al. Aging impairs the recovery in mechanical muscle function following 4 days of disuse. Exp. Gerontol. 2014;52:1–8. doi: 10.1016/j.exger.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 161.Dirks ML, et al. One week of bed rest leads to substantial muscle atrophy and induces whole-body insulin resistance in the absence of skeletal muscle lipid accumulation. Diabetes. 2016;65:2862–2875. doi: 10.2337/db15-1661. [DOI] [PubMed] [Google Scholar]
- 162.Suetta C, et al. Aging affects the transcriptional regulation of human skeletal muscle disuse atrophy. PLoS ONE. 2012;7:e51238. doi: 10.1371/journal.pone.0051238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brocca L, et al. The time course of the adaptations of human muscle proteome to bed rest and the underlying mechanisms. J. Physiol. 2012;590:5211–5230. doi: 10.1113/jphysiol.2012.240267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Feng HZ, Chen X, Malek MH, Jin JP. Slow recovery of the impaired fatigue resistance in postunloading mouse soleus muscle corresponding to decreased mitochondrial function and a compensatory increase in type I slow fibers. Am. J. Physiol. Cell Physiol. 2016;310:C27–C40. doi: 10.1152/ajpcell.00173.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sandri M, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Geng T, Li P, Yin X, Yan Z. PGC-1alpha promotes nitric oxide antioxidant defenses and inhibits FOXO signaling against cardiac cachexia in mice. Am. J. Pathol. 2011;178:1738–1748. doi: 10.1016/j.ajpath.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cannavino J, Brocca L, Sandri M, Bottinelli R, Pellegrino MA. PGC1-alpha over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. J. Physiol. 2014;592:4575–4589. doi: 10.1113/jphysiol.2014.275545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J. Biol. Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kang C, Goodman CA, Hornberger TA, Ji LL. PGC-1alpha overexpression by in vivo transfection attenuates mitochondrial deterioration of skeletal muscle caused by immobilization. FASEB J. 2015;29:4092–4106. doi: 10.1096/fj.14-266619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tonkonogi M, et al. Reduced oxidative power but unchanged antioxidative capacity in skeletal muscle from aged humans. Pflugers Arch. 2003;446:261–269. doi: 10.1007/s00424-003-1044-9. [DOI] [PubMed] [Google Scholar]
- 171.Conley KE, et al. Ageing, muscle properties and maximal O(2) uptake rate in humans. J. Physiol. 2000;526(Pt 1):211–217. doi: 10.1111/j.1469-7793.2000.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Short KR, et al. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA. 2005;102:5618–5623. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chabi B, et al. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell. 2008;7:2–12. doi: 10.1111/j.1474-9726.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 174.Safdar A, et al. Aberrant mitochondrial homeostasis in the skeletal muscle of sedentary older adults. PLoS ONE. 2010;5:e10778. doi: 10.1371/journal.pone.0010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Joseph AM, et al. The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell. 2012;11:801–809. doi: 10.1111/j.1474-9726.2012.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Santanasto AJ, et al. Skeletal muscle mitochondrial function and fatigability in older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70:1379–1385. doi: 10.1093/gerona/glu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Coen PM, et al. Skeletal muscle mitochondrial energetics are associated with maximal aerobic capacity and walking speed in older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2013;68:447–455. doi: 10.1093/gerona/gls196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Santanasto AJ, et al. The relationship between mitochondrial function and walking performance in older adults with a wide range of physical function. Exp. Gerontol. 2016;81:1–7. doi: 10.1016/j.exger.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lourenco dos Santos S, et al. Oxidative proteome alterations during skeletal muscle ageing. Redox Biol. 2015;5:267–274. doi: 10.1016/j.redox.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Joseph AM, et al. Dysregulation of mitochondrial quality control processes contribute to sarcopenia in a mouse model of premature aging. PLoS ONE. 2013;8:e69327. doi: 10.1371/journal.pone.0069327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Carnio S, et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 2014;8:1509–1521. doi: 10.1016/j.celrep.2014.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gouspillou G, et al. Increased sensitivity to mitochondrial permeability transition and myonuclear translocation of endonuclease G in atrophied muscle of physically active older humans. FASEB J. 2014;28:1621–1633. doi: 10.1096/fj.13-242750. [DOI] [PubMed] [Google Scholar]
- 183.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 184.Canto C, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ryu D, et al. NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med. 2016;8:361ra139. doi: 10.1126/scitranslmed.aaf5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.van de Weijer T, et al. Evidence for a direct effect of the NAD+ precursor acipimox on muscle mitochondrial function in humans. Diabetes. 2015;64:1193–1201. doi: 10.2337/db14-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]