Abstract
What is the core of the human brain is a fundamental question that has been mainly addressed by studying the anatomical connections between differently specialized areas, thus neglecting the possible contributions from their functional interactions. While many methods are available to identify the core of a network when connections between nodes are all of the same type, a principled approach to define the core when multiple types of connectivity are allowed is still lacking. Here, we introduce a general framework to define and extract the core–periphery structure of multi-layer networks by explicitly taking into account the connectivity patterns at each layer. We first validate our algorithm on synthetic networks of different size and density, and with tunable overlap between the cores at different layers. We then use our method to merge information from structural and functional brain networks, obtaining in this way an integrated description of the core of the human connectome. Results confirm the role of the main known cortical and subcortical hubs, but also suggest the presence of new areas in the sensori-motor cortex that are crucial for intrinsic brain functioning. Taken together these findings provide fresh evidence on a fundamental question in modern neuroscience and offer new opportunities to explore the mesoscale properties of multimodal brain networks.
Keywords: complex networks, multilayer networks, rich-club, brain connectivity, multimodal integration
1. Introduction
Complex networks are characterized by the existence of non-random structures at different topological scales [1–3]. A peculiar structure is the so-called core–periphery organization [4], where the network exhibits a group of tightly connected nodes (i.e. the core), and a group made by the remaining weakly connected nodes (i.e. the periphery).
Core–periphery organization has been recognized as a fundamental property of complex networks to support integration of information [5–12]. A related concept is that of rich-club behaviour, where the tightly connected nodes are the network hubs, i.e. the nodes with a large number of links [13,14]. A rich-club organization has been observed in various real-world systems, such as social, technological and biological networks [13–16], including the brain [17–20]. More recently, a refined version of the rich-club analysis, based not only on the number of connections of the hubs, but also on their capability to bridge different communities, has been shown to be relevant to support the integrative properties of the nervous system [21].
In the human brain, rich-club and rich-core organization, associated with the efficiency in communication and distribution of information, have been mainly reported in anatomical, or structural, connectivity networks obtained experimentally from diffusion tensor imaging (DTI) data. It has been conjectured that rich cores, rather than shortest paths, may actually be responsible for the efficient integration of information between remote brain areas [17], which is a crucial prerequisite for normal cognitive performance [22,23]. Current evidence suggests that posterior medial and parietal cortical regions mainly constitute the core of the human connectome [17,24], while the role of other areas, such as the medial prefrontal cortex (mPFC) and the sensori-motor system [25], is yet to be clarified. Because brain regions are also characterized by functional interactions inferred from neuroimaging data, such as functional magnetic resonance imaging (fMRI) [26,27], we hypothesize that integrating information from both structural and functional networks can give a more accurate estimate of the regions that eventually constitute the core of the human cortex.
Instead of aggregating the two different types of connectivity or analysing them separately, we adopt a multiplex network approach that preserves and exploits the original information on how brain regions are structurally and functionally interconnected. In a multiplex network, different connectivity types are mathematically represented as networks at different layers. Notably, in a multiplex—a particular case of multilayer network—there is a one-to-one correspondence between the nodes at different layers [28–32]. Multiplex network theory has been recently used to successfully extract higher-order properties of multimodal [33] and multifrequency brain networks that cannot be retrieved by standard approaches [34,35].
Interestingly, the detection of core–periphery organization in multiplex networks has been poorly explored, with the exception of approaches based on k-core decomposition [36,37]. To address this gap, we introduce a criterion to define and detect core–periphery organization in multiplex networks. Our method works for any number of layers and is scalable to large networks, being non-parametric and based on local node information [16]. In the following, we first introduce the general framework and then we validate it on synthetic multiplex networks with tunable core similarity.
We finally apply our method to integrate information from structural and functional brain networks and extract the multiplex core–periphery organization of the human brain. The obtained results confirm the main core areas in the posterior medial and parietal cortex, but also highlights the central role played by the regions of the sensori-motor system, which has been surprisingly neglected by previous studies on core–periphery organization, despite being considered a fundamental component of the default-mode network [25].
Our research sheds new light on the emergence of core regions in the human connectome, and we hope it will spur further work towards a better understanding of the complex relationships in the nervous system.
2. Results
2.1. Extracting the rich core of a multiplex network
Let us consider a multiplex network described by a vector of adjacency matrices 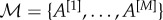 , where all interactions of type α,
, where all interactions of type α, 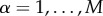 , are encoded in a different layer described by a binary adjacency matrix
, are encoded in a different layer described by a binary adjacency matrix 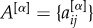 . To detect the core–periphery structure of a multiplex network, we first compute the multiplex degree vector
. To detect the core–periphery structure of a multiplex network, we first compute the multiplex degree vector 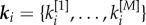 of each node i [31], where
of each node i [31], where 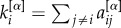 . From now on, we refer to
. From now on, we refer to 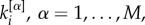 as the richness of node i at layer α. Notice that this is the simplest way to define the richness of a node, and different measures of richness, such as other measures of node centrality, can be as well used.
as the richness of node i at layer α. Notice that this is the simplest way to define the richness of a node, and different measures of richness, such as other measures of node centrality, can be as well used.
For each layer α, we then divide the links of a node i in two groups, those towards nodes with lower richness and those towards nodes with higher richness. Hence, in our case, we can specifically decompose the degree of node i at layer α as 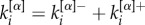 . Finally, the multiplex richness μi of node i is obtained by aggregating single-layer information:
. Finally, the multiplex richness μi of node i is obtained by aggregating single-layer information:
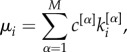 |
2.1 |
where the coefficients c[α] modulate the relative relevance of each layer and can, for instance, be determined by exogenous information. In analogy to the single-layer case, we define the multiplex richness of a node towards richer nodes as:
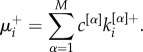 |
2.2 |
In the most simple set-up, we can assume 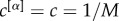
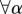 . More general functional forms to aggregate the contributions from different layers, giving rise to alternative measures of μi and μi+, are presented in the Methods section.
. More general functional forms to aggregate the contributions from different layers, giving rise to alternative measures of μi and μi+, are presented in the Methods section.
The nodes of the multiplex are ranked according to their richness μ, so that the node i with the best rank, i.e. ranki = 1, is the node with the largest value of μ, the node ranked 2 is the one with the second largest value of μ, and so on. We then compute for each node i the value of μi+ as a function of ranki. The value of the rank corresponding to the maximum of μi+ finally determines the core–periphery structure. All nodes with rank lower than such a value are assigned to the multiplex core, whereas the remaining ones become part of the periphery. Nodes in the multiplex core are not necessarily part of the core of each layer, but are topologically the most valuable ones when all types of connectivity are considered. Moreover, we notice that also in the simplest case, when 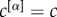
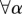 , the multiplex core–periphery partition cannot be obtained by simply combining the cores of the different layers, or by applying the single-layer algorithm on the corresponding aggregated network.
, the multiplex core–periphery partition cannot be obtained by simply combining the cores of the different layers, or by applying the single-layer algorithm on the corresponding aggregated network.
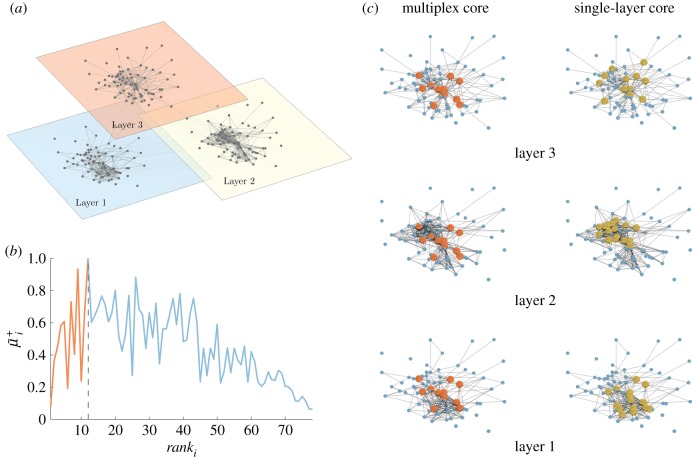
As an illustrative example, we report in figure 1 the curve μi+ as a function of ranki obtained in the case of the Top Noordin Terrorist network, a multiplex network of N = 78 individuals with three layers (encoding information about mutual trust, common operations and exchanged communication between terrorists), which has been used as a benchmark to test measures and models of multiplex networks [31].
Figure 1.
An illustrative example of the multiplex rich core analysis. In panel (a), we show a multiplex social network obtained from the Top Noordin Terrorists’ contacts, with N = 78 nodes, M = 3 layers and 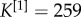 ,
, 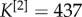 and
and 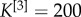 , for the three layers respectively. Panel (b) shows the curve
, for the three layers respectively. Panel (b) shows the curve 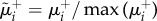 as a function of ranki. All nodes from rank equal to 1 up to the node with maximum
as a function of ranki. All nodes from rank equal to 1 up to the node with maximum 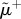 are part of the core of the multiplex, which is shown in red colour in panel (c), first column. The cores obtained at each layer by the standard single-layer analysis are reported in yellow for the sake of comparison in the second column. The percentages of core nodes in the single layers that are in the multiplex core are 83.3% for layer 1, 66.7% for layer 2, and 58.3% for layer 3.
are part of the core of the multiplex, which is shown in red colour in panel (c), first column. The cores obtained at each layer by the standard single-layer analysis are reported in yellow for the sake of comparison in the second column. The percentages of core nodes in the single layers that are in the multiplex core are 83.3% for layer 1, 66.7% for layer 2, and 58.3% for layer 3.
Coefficients 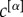 were chosen, in this case, to be inversely proportional to
were chosen, in this case, to be inversely proportional to 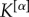 to compensate for the different densities of the three layers. The resulting multiplex rich core integrates information from all the layers and looks different from the rich cores obtained at each of the three layers by a standard single-layer rich core analysis. More details about the results of this analysis are reported in electronic supplementary material, table S1.
to compensate for the different densities of the three layers. The resulting multiplex rich core integrates information from all the layers and looks different from the rich cores obtained at each of the three layers by a standard single-layer rich core analysis. More details about the results of this analysis are reported in electronic supplementary material, table S1.
2.2. Testing the method on multiplex networks with tunable core similarity
A network with a well-defined core–periphery structure has a high density of links among core nodes. With a suitable labelling of the nodes, the adjacency matrix of the network can be decomposed into four different blocks: a dense diagonal block encoding information on core–core links, a sparser diagonal block describing links among peripheral nodes and two off-diagonal blocks encoding core–periphery edges. The key feature of this block structure is that 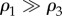 , i.e. the density ρ1 of the core–core block is much higher than that of the periphery–periphery block, ρ3. As first noted by Borgatti & Everett [4], the density ρ2 of the off-diagonal blocks is typically not a crucial factor to characterize a core–periphery structure.
, i.e. the density ρ1 of the core–core block is much higher than that of the periphery–periphery block, ρ3. As first noted by Borgatti & Everett [4], the density ρ2 of the off-diagonal blocks is typically not a crucial factor to characterize a core–periphery structure.
To test how our method works on multiplex networks with different structures, we have introduced a model to produce synthetic multiplex networks with tunable core similarity. In particular, we have constructed multiplexes where each of the M = 2 layers contains N = 250 nodes, only Nc = 50 of them belonging to the core. Each layer has the same average node degree 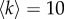 , and the same set of parameters
, and the same set of parameters 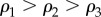 to describe its core–periphery structure. Our model enables control of the number of nodes that are both in the core of layer 1 and 2. (see Methods for more details).
to describe its core–periphery structure. Our model enables control of the number of nodes that are both in the core of layer 1 and 2. (see Methods for more details).
To quantify the similarity among cores at different layers, we introduce the core similarity 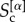 of layer α with respect to the other layers as:
of layer α with respect to the other layers as:
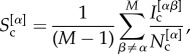 |
2.3 |
where 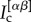 is the number of nodes in the core of both layer α and layer β, whereas
is the number of nodes in the core of both layer α and layer β, whereas 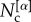 is the size of the core at layer α. The core similarity
is the size of the core at layer α. The core similarity 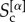 ranges in
ranges in 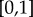 . When layer α does not share core nodes with any other layers we have
. When layer α does not share core nodes with any other layers we have 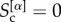 , when all its core nodes also belong to the cores of the other layers
, when all its core nodes also belong to the cores of the other layers 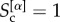 , and when on average only half of them are part of the cores on each other level
, and when on average only half of them are part of the cores on each other level 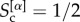 . The average core similarity of the multiplex can then be computed as
. The average core similarity of the multiplex can then be computed as 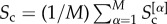 .
.
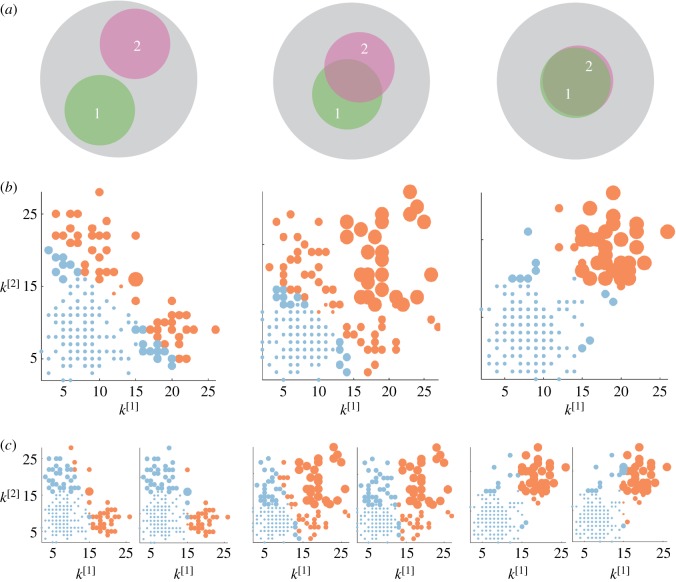
In figure 2, we show the results for three multiplex networks with different core similarity. In the left column of figure 2, we consider a multiplex with 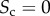 . The cores of the two layers are not overlapping, as shown in panel (a). As a consequence, many nodes with high degree in one layer have low degree in the other one. When
. The cores of the two layers are not overlapping, as shown in panel (a). As a consequence, many nodes with high degree in one layer have low degree in the other one. When 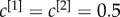 , the multiplex core of the system is formed by those nodes with sufficiently high multiplex richness, as shown in panel (b). In panel (c), we show the changes in the multiplex core when we partially (
, the multiplex core of the system is formed by those nodes with sufficiently high multiplex richness, as shown in panel (b). In panel (c), we show the changes in the multiplex core when we partially (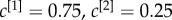 , left subplot) or completely (
, left subplot) or completely (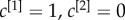 , right subplot) bias the algorithm towards the first layer.
, right subplot) bias the algorithm towards the first layer.
Figure 2.
Core–periphery structure in synthetic multiplex networks with different core similarity. In panel (a), we sketch multiplex networks with M = 2 layers, N = 250 nodes and different levels of core similarity, namely 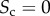 (left column),
(left column), 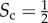 (central column) and
(central column) and 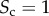 (right column). In panel (b), the nodes are placed in a two-dimensional plane according to their degree at each layer. The size of each dot is proportional to the multiplex richness μi of the node (unbiased case, c[1] = c[2] = c = 0.5). Nodes belonging to the multiplex cores are usually placed in the right-top corner of the plots and are coloured in orange, while the multiplex periphery is in blue. In panel (c), we report results obtained for two cases with
(right column). In panel (b), the nodes are placed in a two-dimensional plane according to their degree at each layer. The size of each dot is proportional to the multiplex richness μi of the node (unbiased case, c[1] = c[2] = c = 0.5). Nodes belonging to the multiplex cores are usually placed in the right-top corner of the plots and are coloured in orange, while the multiplex periphery is in blue. In panel (c), we report results obtained for two cases with 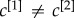 , namely:
, namely: 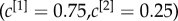 where the core is biased towards the important nodes of the first layer (left), and
where the core is biased towards the important nodes of the first layer (left), and 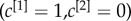 , where the core corresponds to the core of the first layer (right).
, where the core corresponds to the core of the first layer (right).
In the central column of figure 2, we consider a multiplex with 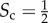 . Half of the core nodes are common to both layers while half are typical of each layer. The block structure of the two layers is partially overlapping, and the nodes are spread uniformly over the
. Half of the core nodes are common to both layers while half are typical of each layer. The block structure of the two layers is partially overlapping, and the nodes are spread uniformly over the 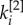 versus
versus 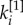 plane. In the unbiased case, the multiplex core of the system is formed by nodes which are part of the core on both layers, but also by nodes scoring extremely high in one layer, despite being in the periphery in the other one (panel b). When
plane. In the unbiased case, the multiplex core of the system is formed by nodes which are part of the core on both layers, but also by nodes scoring extremely high in one layer, despite being in the periphery in the other one (panel b). When 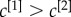 , this is particularly true for nodes which have high richness in the first layer and low richness in the second, while the opposite is much more unlikely (panel c).
, this is particularly true for nodes which have high richness in the first layer and low richness in the second, while the opposite is much more unlikely (panel c).
In the right column of figure 2, we consider a multiplex with 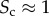 . The block structure of the two layers is now almost identical; the node degrees
. The block structure of the two layers is now almost identical; the node degrees 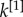 and
and 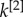 are correlated and most of the nodes belonging to each core are in the multiplex core (panel b). As the core structure at the two layers are extremely similar, the biased cases do not differ significantly from the unbiased one (panel c).
are correlated and most of the nodes belonging to each core are in the multiplex core (panel b). As the core structure at the two layers are extremely similar, the biased cases do not differ significantly from the unbiased one (panel c).
2.3. Merging structure and function to extract the connectome’s core
We have applied our method to investigate the human connectome by considering, at the same time, structural and functional information. We have therefore constructed a multiplex brain network formed by one structural layer and one functional layer. The two layers were obtained by first averaging brain connectivity matrices estimated, respectively, from DTI and fMRI data in 171 healthy individuals. Each of the two layers is then thresholded by fixing the average node degree 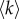 . We have focused our analysis on 158 regions of interest (ROIs) of the cortex (see Methods for more details).
. We have focused our analysis on 158 regions of interest (ROIs) of the cortex (see Methods for more details).
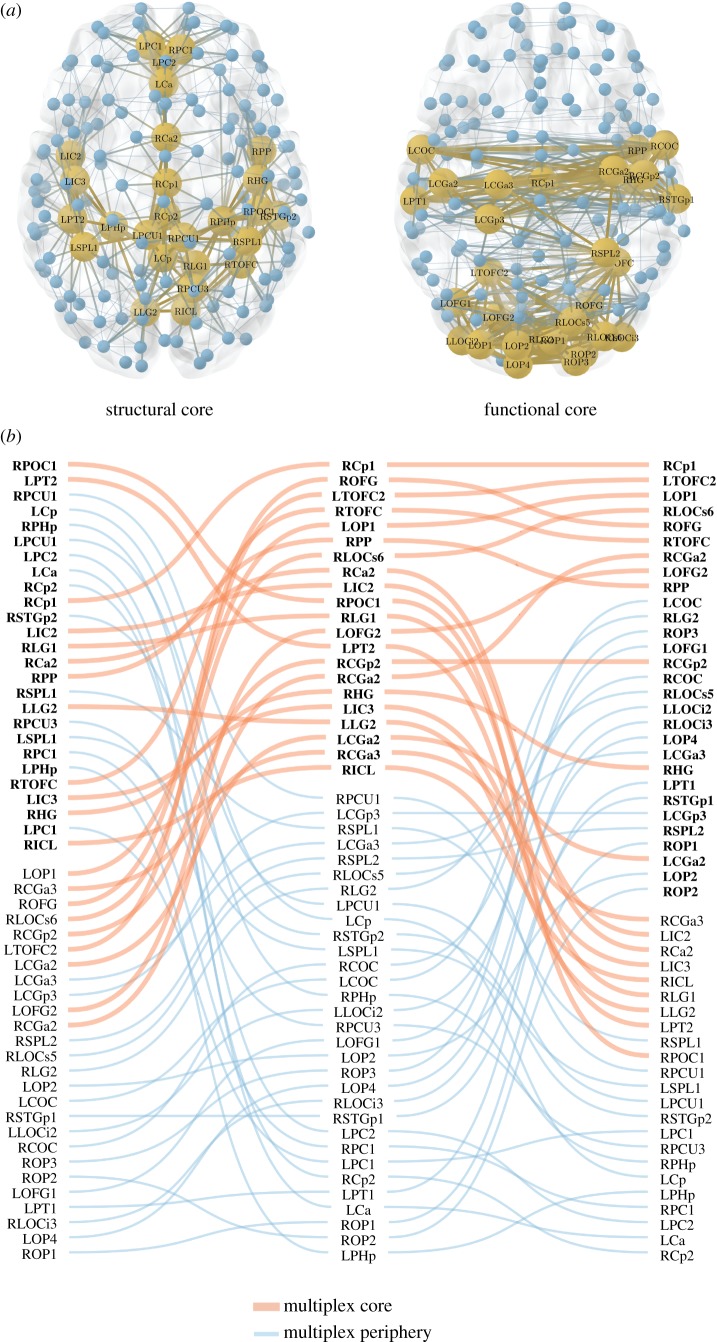
In figure 3, we report the cores found by analysing the two layers separately, as well as the multiplex core obtained with our method. The figure refers to the case of a representative threshold corresponding to an average node degree 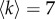 . We notice that the cores of the structural and functional layers are only partially overlapping, with a value of core similarity of
. We notice that the cores of the structural and functional layers are only partially overlapping, with a value of core similarity of 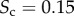 . For the sake of completeness, we also report the Sc values for the entire threshold range (electronic supplementary material, figure S1). A detailed analysis on the robustness of the multiplex core detection in the presence of random fluctuations is reported in the electronic supplementary material, text S1.
. For the sake of completeness, we also report the Sc values for the entire threshold range (electronic supplementary material, figure S1). A detailed analysis on the robustness of the multiplex core detection in the presence of random fluctuations is reported in the electronic supplementary material, text S1.
Figure 3.
Extracting the multiplex core of the human brain from structural and functional information. (a) The structural and functional brain networks filtered with an average node degree 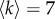 are shown, respectively, on the left and right side. They are represented from above with the frontal lobe pointing upward. The position of the nodes corresponds to the actual location of the brain ROIs (electronic supplementary material, table S2). Yellow and large nodes represent the brain regions belonging to the core according to the standard single-layer method. Blue and small nodes code for the ROIs in the periphery. Links are yellow and thick if they connect two ROIs in the core, while they are blue and thin if they connect two peripheral nodes. (b) ROIs are ranked from top to bottom according to their richness in the structural (left column), functional (right column) and multiplex network (central column). In each column, the labels in bold/normal font stand for the ROIs that are in the core/periphery. For the sake of simplicity, only ROIs that are at least in one core (structural, functional or multiplex) are listed in the three columns. Red/blue and thick/thin lines identify ROIs that go into the core/periphery according to the multiplex approach.
are shown, respectively, on the left and right side. They are represented from above with the frontal lobe pointing upward. The position of the nodes corresponds to the actual location of the brain ROIs (electronic supplementary material, table S2). Yellow and large nodes represent the brain regions belonging to the core according to the standard single-layer method. Blue and small nodes code for the ROIs in the periphery. Links are yellow and thick if they connect two ROIs in the core, while they are blue and thin if they connect two peripheral nodes. (b) ROIs are ranked from top to bottom according to their richness in the structural (left column), functional (right column) and multiplex network (central column). In each column, the labels in bold/normal font stand for the ROIs that are in the core/periphery. For the sake of simplicity, only ROIs that are at least in one core (structural, functional or multiplex) are listed in the three columns. Red/blue and thick/thin lines identify ROIs that go into the core/periphery according to the multiplex approach.
As shown in figure 3, ventral brain areas tend in general to form the structural core, while more dorsal regions appear in the functional core. Notably, brain ROIs (electronic supplementary material, table S2) that are in the core of both structural and functional layers also tend to be in the core of the multiplex. Instead, ROIs being in the periphery of both layers tend to be excluded from the multiplex core. However, exceptions may exist depending on the multiplex richness of the nodes. For example, the posterior part of the right precentral gyrus (RCGa3), which is in the periphery of both the structural and functional layer, is eventually assigned to the multiplex core, because of its relatively high rank score in the two layers. The situation appears even less predictable for ROIs that are in the core of one layer and in the periphery of the other layer. Only occasionally these will belong to the multiplex core. This is the case, for example, of the anterior part of right precentral gyrus (RCGa2) which exhibits a relatively low structural richness but high functional richness, i.e. ranked seventh in the functional core, or of the anterior part of the right parietal operculum (RPOC1), which has the highest structural richness but a low functional richness.
2.4. Revealing new core regions of the human brain
We have extracted the multiplex core–periphery structure of the human brain for the full range of available thresholds 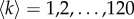 (see Methods for more details). In this way, we have been able to calculate the coreness
Ci of each node i, defined as the normalized number of thresholds at which the corresponding ROI is present in the rich core. This allows us to rank ROIs according to their likelihood to be part of the multiplex core and to compare these to the rankings obtained separately for structural and functional layers. We note that the same approach of investigating the persistence across a set of different filtering thresholds can be applied to any node property. This can turn useful for statistical validation in the case no threshold is universally accepted, as often happens for brain networks [38–40].
(see Methods for more details). In this way, we have been able to calculate the coreness
Ci of each node i, defined as the normalized number of thresholds at which the corresponding ROI is present in the rich core. This allows us to rank ROIs according to their likelihood to be part of the multiplex core and to compare these to the rankings obtained separately for structural and functional layers. We note that the same approach of investigating the persistence across a set of different filtering thresholds can be applied to any node property. This can turn useful for statistical validation in the case no threshold is universally accepted, as often happens for brain networks [38–40].
Parietal (pre/cuneus PCU/LOC, superior parietal lobe SPL), cingulate (anterior Ca, posterior Cp), temporal (superior temporal gyrus), insular (insular cortex IC), as well as frontal ROIs (paracingulate PC) mainly constitute the structural core, as shown in electronic supplementary material, figure S2. While some overlap exists between the structural and the functional cores, the latter rather tends instead to include occipital (occipital fusiform gyrus OFG, temporo-occipital fusiform cortex TOFC) and central (pre/post central gyrus CGa/CGp) ROIs and, notably, to exclude regions in the frontal lobe (top 25% ROIs, electronic supplementary material, figure S3).
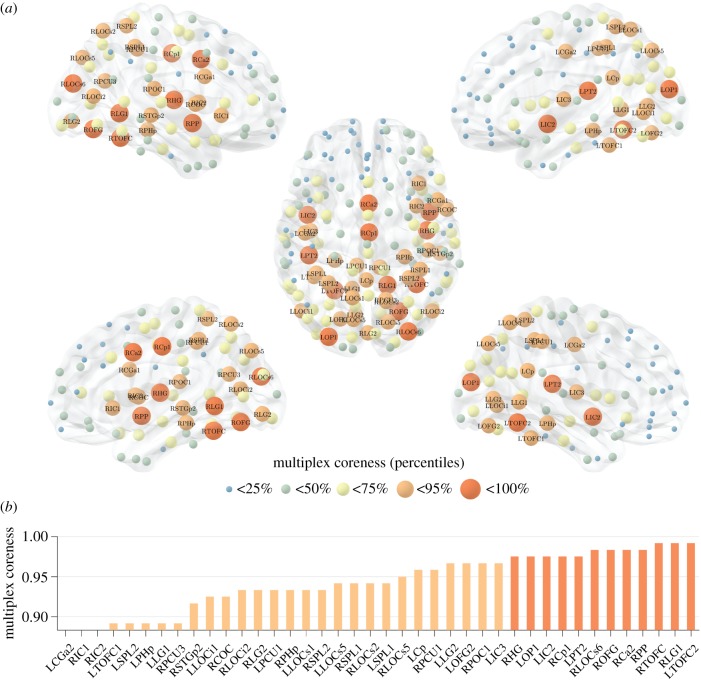
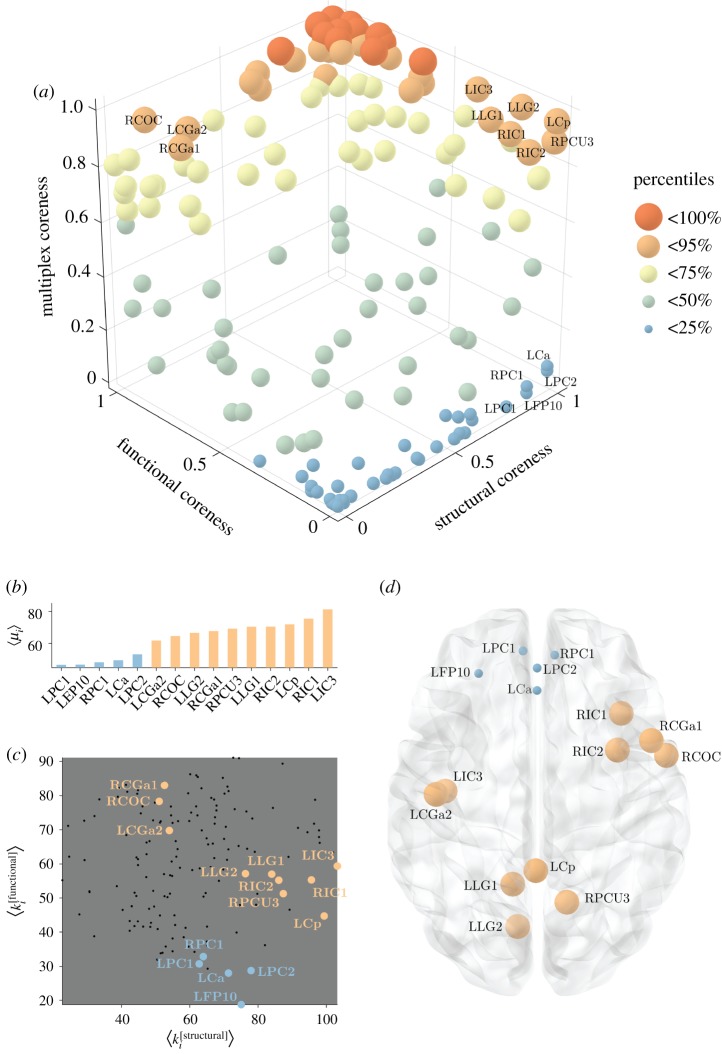
Figure 4 shows the coreness of the multiplex network. As expected, ROIs that are peripheral (i.e. low coreness) in both layers are also peripheral in the multiplex, while ROIs with both a high structural and high functional coreness are typically observed in the multiplex core (e.g. TOFC, OFG, Ca, Cp). Interesting behaviours emerge for those regions typically characterized by high coreness in one layer and low coreness in the other layer. In fact, some of these ROIs are part of the multiplex core, while others are usually found in the multiplex periphery, as shown in figure 5a. For areas with a different assignment in the two layers, we note that the main contribution to the multiplex richness μi comes from the richness in the layer where node i is identified as core. Interestingly, not only is the average richness of the node in the core layer higher than the one in the peripheral layer, but also its fluctuations around the mean.
Figure 4.
The multiplex core of the human connectome. Panel (a) shows the human brain, where ROIs are highlighted based on their multiplex coreness. The colour and size of the nodes are associated with the percentiles of multiplex coreness in each brain region, so that core nodes are larger in size and coloured in red. The left side shows the lateral view of the left hemisphere (top, dorsal; bottom, ventral). The right side shows the lateral view of the right hemisphere (top, dorsal; bottom, ventral). In the middle, the brain is shown from above, with the frontal lobe pointing upward. In panel (b), we report the ROIs corresponding to the 25% highest values of multiplex coreness. The colour follows the same legend as in panel (a).
Figure 5.
Emergent non-trivial core regions in the multiplex brain. Panel (a) shows the scatter plot of the structural, functional and multiplex coreness of the ROIs in the brain. The colour and size of the nodes are associated with the percentile of multiplex coreness across the set of brain regions, as in figure 4. Panel (b) reports the average value of multiplex richness 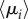 across the different thresholds for the ROIs with the strongest differences in structural and functional coreness. The colour follows the same legend as in panel (a). Panel (c) illustrates the distribution of the ROIs (black points) as a function of their averaged structural and functional degree across all the thresholds. Only the ROIs listed in panel (b) are highlighted according to the same colour legend as in panel (a).
across the different thresholds for the ROIs with the strongest differences in structural and functional coreness. The colour follows the same legend as in panel (a). Panel (c) illustrates the distribution of the ROIs (black points) as a function of their averaged structural and functional degree across all the thresholds. Only the ROIs listed in panel (b) are highlighted according to the same colour legend as in panel (a).
As a consequence, among regions that are core in the structural layer but peripheral in the functional one, those with relatively higher structural richness (degree), such as precuneus PCU, insular cortex IC and posterior cingulate Cp, finally tend to join the multiplex core no matter the exact value of their functional richness (upper right corner of figure 5a). Conversely, ROIs with relatively lower structural degree are usually peripheral in the multiplex, and typically located in the pre-frontal cortex PC and frontal lobe FP (lower right corner of figure 5a), as illustrated in figure 5b,c. Similarly, among areas in the functional core, those with relatively higher functional degree, such as precentral gyrus CGa and central operculum COC, tend to join the multiplex core (upper left corner of figure 5a). By contrast, ROIs with relatively lower functional degree, are mostly peripheral in the multiplex, and are located in the parietal operculum POC and superior frontal gyrus SFG (lower left corner of figure 5a).
In a separate analysis, we have extracted the multiplex brain coreness from each individual and we show that, despite a normal inter-subject variability, the average multiplex brain coreness is very similar to the multiplex coreness of the group-averaged brain networks (electronic supplementary material, figure S4). Finally, we have evaluated the robustness of the results when also including subcortical ROIs in the brain networks. We report that thalamus, putamen and hippocampus are among the regions with highest coreness and therefore become part of the multiplex core (electronic supplementary material, figure S5). Interestingly, their presence does not significantly alter the coreness of the other ROIs (electronic supplementary material, figure S6), suggesting an assortative structure where highly connected subcortical regions preferentially get connected with core regions in the cortex.
3. Discussion
The existence of a network core in the brain is a prerequisite for neural functioning and cognition, and damage to the core have been associated with several neurological or psychiatric diseases [19,41,42]. Finding the router regions that ensure integration between the different brain modules and communication in the system is therefore a fundamental question in neuroscience. Previous studies have mainly considered the structural connectivity of the brain through disparate techniques, such as k-core decomposition, centrality measures and rich-club analysis [17,24]. While the results obtained agree on the implication of posterior medial and parietal cortical regions—as well as subcortical thalamus, putamen and hippocampus—in the network core [17,24], they neglect the possible role of other areas that are crucial from a functional perspective, such as those in the default-mode network (DMN) [25].
To integrate information from both structural and functional brain connectivity at the network level, we introduce a general criterion to define and extract the core when nodes are connected through links which can vary in meaning and nature, and the whole system can be described as a network with multiple layers [28–32]. Compared to standard approaches, this method has the theoretical advantage of providing a more robust solution, taking into account the relative importance of the nodes at each layer, rather than simply considering the union or intersection of the cores across layers, or extracting the core from the aggregated network.
The results obtained shed new light on the role of the regions characterizing the intrinsic brain function to eventually form the core of the human brain. First, we show that mPFC (e.g. PC and FP), exhibiting a high structural but low functional coreness, is eventually assigned to the periphery (figure 5a, lower-right corner). This outcome can be predicted by the lower multiplex richness and relatively low structural degree, and not solely by the attitude of frontal areas to be peripheral in the functional brain network (figure 5b,c). The exclusion of the mPFC from the rich core supports the hypothesis that default-mode network activity may be mainly driven from highly coupled areas of the posterior medial and parietal cortex, which in turn link to other highly connected regions, such as the medial orbitofrontal cortex [24].
Second, while frontal ROIs are excluded, new regions gain importance and become part of the core because of their higher multiplex richness (see figure 5a, upper left corner). Among them, we report areas of the central gyrus (CGa, CGp to a minor extent), which are characterized by a low structural but relatively high functional degree, as shown in figure 5b,c. These regions are part of the primary sensori-motor cortex, which has been shown to be the most extensive of the resting-state components, or networks (out of eight [43]), covering 27% of the total grey matter in the brain [44]. The primary sensori-motor component has a high degree of integration (overlap and activity coupling) with all other resting-state networks (e.g. DMN), which is consistent with the increased synchronization of neural activity in cortical regions during sensory processing [45]. Notably, ongoing functional connectivity in the primary sensori-motor network, originally revealed by seed-based analysis [46,47], has been extensively verified by ICA and clustering methods [48,49].
Our method provides an effective tool to integrate mesoscale topological information in brain networks derived from multimodal neuroimaging data. Multimodal integration of brain networks is gaining more and more interest [50–53] due, on the one hand, to the increasing availability of large heterogeneous datasets (e.g. HCP http://www.humanconnectomeproject.org, ADNI http://adni.loni.usc.edu) and, on the other hand, to the need of principled ways to characterize multiscale neural mechanisms (e.g. cross-frequency coupling) and to provide predictive diagnostics for multifactor brain diseases, such as Alzheimer's disease.
It is important to note, that our analysis of the human connectome relies on the assumption that each layer contributes with the same intensity to the definition of the multiplex core. In general, however, the contribution of a layer α can be weighted differently through a choice of the parameter 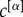 , and this can be used to enhance or reduce the importance of the different types of connectivity. A larger value of
, and this can be used to enhance or reduce the importance of the different types of connectivity. A larger value of 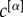 increases the relevance of the corresponding layer until when, in the limit in which
increases the relevance of the corresponding layer until when, in the limit in which  and the coefficients of all the other layers go to zero, the multiplex core is no longer defined by the topology of all the M layers, but coincides with the core at layer α. For instance, setting
and the coefficients of all the other layers go to zero, the multiplex core is no longer defined by the topology of all the M layers, but coincides with the core at layer α. For instance, setting 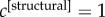 and
and 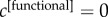 returns a core based on the anatomical information only, and in agreement with most of the previous literature on such topic (see electronic supplementary material, figure S2). As an unbiased way to characterize the multiplex core of the human brain, we have focused our analysis on the simplest and symmetric case,
returns a core based on the anatomical information only, and in agreement with most of the previous literature on such topic (see electronic supplementary material, figure S2). As an unbiased way to characterize the multiplex core of the human brain, we have focused our analysis on the simplest and symmetric case, 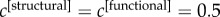 . We show in electronic supplementary material, figure S7 that the results are relatively stable for small perturbations around this unbiased condition. However, other combinations are in general possible and should be adopted if supported by a plausible rationale. For example, in the case of multifrequency brain networks one could assign different weights to the network layers taking into account the frequency scaling of the brain activity’s power spectra [54,55].
. We show in electronic supplementary material, figure S7 that the results are relatively stable for small perturbations around this unbiased condition. However, other combinations are in general possible and should be adopted if supported by a plausible rationale. For example, in the case of multifrequency brain networks one could assign different weights to the network layers taking into account the frequency scaling of the brain activity’s power spectra [54,55].
In practice, the proposed method to detect the core–periphery organization of multiplex networks has two clear advantages: (i) it is fast and scalable, since it works using only local information; (ii) it is non-parametric, e.g. no need to input a priori information such as the core size. Moreover, it can be generalized in a straightforward way to the case of directed networks. A drawback of the method is that it focuses on highly connected rich nodes, and neglects the possible important role of the so-called connectors, i.e. central nodes with low degree [56]. We note that alternative core–periphery structures which include connectors can be detected by more computationally demanding methods such as those based on stochastic block models, which have been recently proposed to extract the mesoscale structure of time-varying and multilayer networks [57]. We hope that our work can trigger further developments in the exploration of core–periphery structure of real-world large-scale multiplex networks.
To conclude, our method to investigate multiplex core–periphery organization in complex networks suggests that the core of the human cortex is made up of known cortical and subcortical hubs, as well as of areas in the sensori-motor system that were previously overlooked by standard approaches, but that are crucial for the brain functioning. Our findings offer an augmented definition of the rich core of the human brain, which takes into account not only the anatomical structure but also its function.
We hope that our work will contribute to advance our understanding of the mesoscale connectivity mechanisms in multiplex brain networks, in an effort to better integrate the one-to-many relationships that exist between structure and function in the human brain [26].
4. Methods
4.1. Multiplex stochastic block model with tunable core similarity
Stochastic block models for multiplex networks have been recently introduced by Peixoto [57]. Here, we introduce a stochastic block model that enables sampling of multiplex networks with an assigned value of core similarity SC (see equation (3)). Suppose we have N nodes and we want to construct a multiplex network having a core–periphery structure at each layer 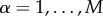 , with
, with 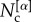 nodes in the core of layer α.
nodes in the core of layer α.
In particular, we set M = 2, N = 250, 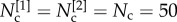 , and we create at each layer a core–periphery structure with the same set of densities:
, and we create at each layer a core–periphery structure with the same set of densities: 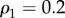 ,
, 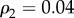 and
and 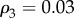 . Namely, for each of the two layers, we connect with a probability ρ1 two nodes both in the core, with probability ρ2 a node in the core and a node in the periphery, and finally with probability ρ3 two peripheral nodes. The values of the three parameters were chosen in a way that
. Namely, for each of the two layers, we connect with a probability ρ1 two nodes both in the core, with probability ρ2 a node in the core and a node in the periphery, and finally with probability ρ3 two peripheral nodes. The values of the three parameters were chosen in a way that 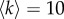 on both layers, and the core–periphery structure of each layer is sufficiently strong to be detected with good accuracy, as discussed in the electronic supplementary material, text S2.
on both layers, and the core–periphery structure of each layer is sufficiently strong to be detected with good accuracy, as discussed in the electronic supplementary material, text S2.
Different levels of core similarity are achieved by varying the overlap between core nodes at the two layers. When the two sets of core nodes are completely overlapping, 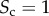 , whereas when the two sets are disjoint
, whereas when the two sets are disjoint 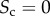 . Despite other related formulations of Sc are possible, our definition reflects the intuition that when two layers with equal core size share half of the core nodes, then
. Despite other related formulations of Sc are possible, our definition reflects the intuition that when two layers with equal core size share half of the core nodes, then 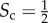 .
.
4.2. Multiplex richness μi and μi+
The multiplex richness μi and μi+ introduced in equations (1) and (2) are obtained by means of a simple aggregation of information based on the single layers. In the simplest set-up 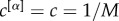 for
for 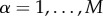 , and the multiplex richness μi of a node i is simply proportional to its overlapping degree oi [31]. A layer with higher density weighs more in the computation of the multiplex core of a network.
, and the multiplex richness μi of a node i is simply proportional to its overlapping degree oi [31]. A layer with higher density weighs more in the computation of the multiplex core of a network.
In general, coefficients 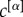 can be used to modulate the relevance of the layers of the network in order to extract its core. If one wants to have equal contributions to μi and μi+ from all the layers but their number of links
can be used to modulate the relevance of the layers of the network in order to extract its core. If one wants to have equal contributions to μi and μi+ from all the layers but their number of links 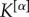 is different—for instance, because in some layers it might be easier to establish or measure a connection than in others—a natural choice is to set
is different—for instance, because in some layers it might be easier to establish or measure a connection than in others—a natural choice is to set 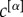 to be proportional to
to be proportional to 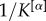 . In other cases, independently from their density, it might be reasonable to assign different importance to different layers, because of exogenous information. Once again this can be achieved by assigning different values of the coefficients
. In other cases, independently from their density, it might be reasonable to assign different importance to different layers, because of exogenous information. Once again this can be achieved by assigning different values of the coefficients 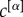 .
.
Our method inherits many advantageous properties of the original algorithm proposed for single-layer networks [16]. First, it can be easily extended to directed layers by replacing 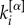 with
with 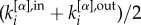 in equation (1), where
in equation (1), where 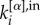 and
and 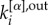 correspond, respectively, to the in-degree and out-degree of node i at layer α, and by substituting
correspond, respectively, to the in-degree and out-degree of node i at layer α, and by substituting 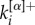 with
with 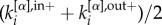 in equation (2). Second, for weighted networks μi and μi+ can be obtained by replacing the adjacency matrix binary entries
in equation (2). Second, for weighted networks μi and μi+ can be obtained by replacing the adjacency matrix binary entries 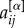 with their weighted counterparts
with their weighted counterparts 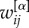 , and by substituting the node degree with the strength
, and by substituting the node degree with the strength 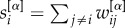 . Third, the core size is relatively stable with respect to randomly chosen different rankings of nodes with equal degree.
. Third, the core size is relatively stable with respect to randomly chosen different rankings of nodes with equal degree.
We finally notice that equation (1) is a particular choice of a more general scenario, where the multiplex richness μi is a generic function f of the degree of a node at the different layers:
| 4.1 |
and μi+ is a generic function g:
| 4.2 |
4.3. Multimodal brain networks
We have considered 171 healthy human subjects from the NKI Rockland dataset http://fcon_1000.projects.nitrc.org/indi/pro/nki.html. We have used diffusion weighted magnetic resonance imaging (dwMRI) and fMRI to derive, respectively, structural and functional brain networks in each subject.
We have gathered the corresponding connectivity matrices from the USC Multimodal Connectivity Database (http://umcd.humanconnectomeproject.org) [58].
In particular, structural connectivity has been obtained using anatomical fibre assignment through the continuous tracking (FACT) algorithm [59]. Functional connectivity has been computed by means of the Pearson’s correlation coefficient between fMRI signals recorded during a 10 min resting state (RS). RS-based functional connectivity measures the amount of interaction—or temporal dependence—between different brain areas during spontaneous brain activity [27]. More details about the processing steps can be found here [60]. A total number of N = 188 ROIs are available for both structural and functional brain networks, thus resulting in connectivity matrices of size N × N, spatially matched with the MNI152 template [61].
Because we are mainly interested in cortical networks, we focused our analysis on the network obtained by removing all subcortical ROIs and obtained connectivity matrices of size 158 × 158. The full names and acronyms for all the ROIs can be found in electronic supplementary material, table S1. We have then averaged the resulting connectivity matrices (after Fisher transformation) across subjects in order to have a population-level representation. At the end, we obtained a structural weighted connectivity matrix 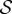 , whose entry
, whose entry 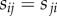 contain the group-average number of axonal fibres between ROIs i and j, and a functional weighted connectivity matrix
contain the group-average number of axonal fibres between ROIs i and j, and a functional weighted connectivity matrix 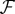 , whose entry
, whose entry 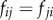 correspond to the group-average correlation coefficient between the fMRI signals of ROIs i and j.
correspond to the group-average correlation coefficient between the fMRI signals of ROIs i and j.
We have used density-based thresholding to derive structural and functional brain networks by removing the lowest values from the connectivity matrices and binarizing the remaining ones [27]. We have considered a full range of density thresholds, corresponding to an increasing average node degree 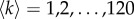 . The last value was given by the maximal
. The last value was given by the maximal 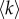 observed in the native structural connectivity matrices, which are originally not fully connected. After filtering, for each threshold we have combined the resulting structural and functional brain networks into a multiplex network
observed in the native structural connectivity matrices, which are originally not fully connected. After filtering, for each threshold we have combined the resulting structural and functional brain networks into a multiplex network 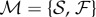 .
.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Data accessibility
All the experimental data used in this work can be downloaded from the USC Multimodal Connectivity Database http://umcd.humanconnectomeproject.org. The Matlab code used to compute the core–periphery structure of multiplex networks is made available at https://github.com/brain-network/bnt.
Authors' contributions
F.B. carried out the theoretical work, participated in data analysis, participated in the design of the study and drafted the manuscript; J.G. participated in data analysis and drafted the manuscript; M.C. conceived and designed the study, and helped draft the manuscript. V.L. conceived the study and drafted the manuscript; F.D.V.F. coordinated the study, participated in the design of the study, participated in data analysis and drafted the manuscript. All authors gave final approval for publication.
Competing interests
We have no competing interests.
Funding
F.D.V.F. and M.C. acknowledge support by the ANR French programme through the contracts ANR-10-IAIHU-06 and ANR-15-NEUC-0006-02. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Reference
- 1.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298, 824–827. ( 10.1126/science.298.5594.824) [DOI] [PubMed] [Google Scholar]
- 2.Girvan M, Newman MEJ. 2002. Community structure in social and biological networks. Proc. Natl Acad. Sci. USA 99, 7821–7826. ( 10.1073/pnas.122653799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortunato S. 2010. Community detection in graphs. Phys. Rep. 486, 75–174. ( 10.1016/j.physrep.2009.11.002) [DOI] [Google Scholar]
- 4.Borgatti SP, Everett MG. 2000. Models of core/periphery structures. Soc. Networks 21, 375–395. ( 10.1016/S0378-8733(99)00019-2) [DOI] [Google Scholar]
- 5.Csermely P, London A, Wu L-Y, Uzzi B. 2013. Structure and dynamics of core/periphery networks. J. Complex Networks 1, 93–123. ( 10.1093/comnet/cnt016) [DOI] [Google Scholar]
- 6.Rombach M, Porter M, Fowler J, Mucha P. 2014. Core–periphery structure in networks. SIAM J. Appl. Math. 74, 167–190. ( 10.1137/120881683) [DOI] [Google Scholar]
- 7.Boyd JP, Fitzgerald WJ, Mahutga MC, Smith DA. 2010. Computing continuous core/periphery structures for social relations data with MINRES/SVD. Soc. Networks 32, 125–137. ( 10.1016/j.socnet.2009.09.003) [DOI] [Google Scholar]
- 8.Zhang X, Martin T, Newman MEJ. 2015. Identification of core–periphery structure in networks. Phys. Rev. E 91, 032803 ( 10.1103/PhysRevE.91.032803) [DOI] [PubMed] [Google Scholar]
- 9.Luo F, Li B, Wan X-F, Scheuermann RH. 2009. Core and periphery structures in protein interaction networks. BMC Bioinformatics 10, S8 ( 10.1186/1471-2105-10-S4-S8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barucca P, Lillo F. 2016. Disentangling bipartite and core–periphery structure in financial networks. Chaos Solitons Fractals 88, 244–253. ( 10.1016/j.chaos.2016.02.004) [DOI] [Google Scholar]
- 11.Verma T, Russmann F, Araújo NaM, Nagler J, Herrmann HJ. 2016. Emergence of core–peripheries in networks. Nat. Commun. 7, 10441 ( 10.1038/ncomms10441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagiolo G, Reyes J, Schiavo S. 2010. The evolution of the world trade web: a weighted-network analysis. J. Evol. Econ. 20, 479–514. ( 10.1007/s00191-009-0160-x) [DOI] [Google Scholar]
- 13.Colizza V, Flammini A, Serrano MA, Vespignani A. 2006. Detecting rich-club ordering in complex networks. Nat. Phys. 2, 110–115. ( 10.1038/nphys209) [DOI] [Google Scholar]
- 14.Zhou S, Mondragon RJ. 2004. The rich-club phenomenon in the Internet topology. IEEE Commun. Lett. 8, 180–182. ( 10.1109/LCOMM.2004.823426) [DOI] [Google Scholar]
- 15.Vaquero LM, Cebrian M. 2013. The rich club phenomenon in the classroom. Sci. Rep. 3, 1174 ( 10.1038/srep01174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma A, Mondragón RJ. 2015. Rich-cores in networks. PLoS ONE 10, e0119678 ( 10.1371/journal.pone.0119678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heuvel MPvd, Sporns O. 2011. Rich-club organization of the human connectome. J. Neurosci. 31, 15 775–15 786. ( 10.1523/JNEUROSCI.3539-11.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harriger L, Heuvel MPvd, Sporns O. 2012. Rich club organization of macaque cerebral cortex and its role in network communication. PLoS ONE 7, e46497 ( 10.1371/journal.pone.0046497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Heuvel MP, Sporns O, Collin G, Scheewe T, Mandl RCW, Cahn W, Goñi J, Hulshoff Pol HE, Kahn RS. 2013. Abnormal rich club organization and functional brain dynamics in schizophrenia. JAMA Psychiatry 70, 783–792. ( 10.1001/jamapsychiatry.2013.1328) [DOI] [PubMed] [Google Scholar]
- 20.Ball G. et al. 2014. Rich-club organization of the newborn human brain. Proc. Natl Acad. Sci. USA 111, 7456–7461. ( 10.1073/pnas.1324118111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertolero MA, Yeo BTT, D’Esposito M. 2017. The diverse club. Nat. Commun. 8, 1277 ( 10.1038/s41467-017-01189-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198. ( 10.1038/nrn2575) [DOI] [PubMed] [Google Scholar]
- 23.Stam CJ. 2014. Modern network science of neurological disorders. Nat. Rev. Neurosci. 15, 683–695. ( 10.1038/nrn3801) [DOI] [PubMed] [Google Scholar]
- 24.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159 ( 10.1371/journal.pbio.0060159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckner RL, Andrews-Hanna JR, Schacter DL. 2008. The brain’s default network. Ann. NY Acad. Sci. 1124, 1–38. ( 10.1196/annals.1440.011) [DOI] [PubMed] [Google Scholar]
- 26.Friston KJ. 2011. Functional and effective connectivity: a review. Brain Connect 1, 13–36. ( 10.1089/brain.2011.0008) [DOI] [PubMed] [Google Scholar]
- 27.De Vico Fallani F, Richiardi J, Chavez M, Achard S. 2014. Graph analysis of functional brain networks: practical issues in translational neuroscience. Phil. Trans. R. Soc. B 369, 20130521 ( 10.1098/rstb.2013.0521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Domenico M, Solé-Ribalta A, Cozzo E, Kivelä M, Moreno Y, Porter MA, Gómez S, Arenas A. 2013. Mathematical formulation of multilayer networks. Phys. Rev. X 3, 041022 ( 10.1103/PhysRevX.3.041022) [DOI] [Google Scholar]
- 29.Kivelä M, Arenas A, Barthelemy M, Gleeson JP, Moreno Y, Porter MA. 2014. Multilayer networks. J. Complex Networks 2, 203–271. ( 10.1093/comnet/cnu016) [DOI] [Google Scholar]
- 30.Boccaletti S, Bianconi G, Criado R, del Genio CI, Gómez-Gardeñes J, Romance M, Sendiña-Nadal I, Wang Z, Zanin M. 2014. The structure and dynamics of multilayer networks. Phys. Rep. 544, 1–122. ( 10.1016/j.physrep.2014.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battiston F, Nicosia V, Latora V. 2014. Structural measures for multiplex networks. Phys. Rev. E 89, 032804 ( 10.1103/PhysRevE.89.032804) [DOI] [PubMed] [Google Scholar]
- 32.Battiston F, Nicosia V, Latora V. 2017. The new challenges of multiplex networks: measures and models. Eur. Phys. J. Spec. Top. 226, 401–416. ( 10.1140/epjst/e2016-60274-8) [DOI] [Google Scholar]
- 33.Battiston F, Nicosia V, Chavez M, Latora V. 2017. Multilayer motif analysis of brain networks. Chaos 27, 047404 ( 10.1063/1.4979282) [DOI] [PubMed] [Google Scholar]
- 34.De Domenico M, Sasai S, Arenas A. 2016. Mapping multiplex hubs in human functional brain networks. Front. Neurosci. 10, 326 ( 10.3389/fnins.2016.00326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillon J, Attal Y, Colliot O, Corte VL, Dubois B, Schwartz D, Chavez M, De Vico Fallani F. 2017. Loss of brain inter-frequency hubs in Alzheimer’s disease. Sci. Rep. 7, 10879 ( 10.1038/s41598-017-07846-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azimi-Tafreshi N, Gómez-Gardeñes J, Dorogovtsev SN. 2014. K-core percolation on multiplex networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 90, 032816 ( 10.1103/PhysRevE.90.032816) [DOI] [PubMed] [Google Scholar]
- 37.Corominas-Murtra B, Thurner S. 2016. The weak core and the structure of elites in social multiplex networks. In Interconnected networks, understanding complex systems (ed. Garas A.), pp. 165–177. Cham, Switzerland: Springer. [Google Scholar]
- 38.van Wijk BCM, Stam CJ, Daffertshofer A. 2010. Comparing brain networks of different size and connectivity density using graph theory. PLoS ONE 5, e13701 ( 10.1371/journal.pone.0013701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornito A, Zalesky A, Breakspear M. 2013. Graph analysis of the human connectome: promise, progress, and pitfalls. NeuroImage 80, 426–444. ( 10.1016/j.neuroimage.2013.04.087) [DOI] [PubMed] [Google Scholar]
- 40.De Vico Fallani F, Latora V, Chavez M. 2017. A topological criterion for filtering information in complex brain networks. PLoS Comput. Biol. 13, e1005305 ( 10.1371/journal.pcbi.1005305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gollo LL, Zalesky A, Hutchison RM, van den Heuvel M, Breakspear M. 2015. Dwelling quietly in the rich club: brain network determinants of slow cortical fluctuations. Phil. Trans. R. Soc. B 370, 20140165 ( 10.1098/rstb.2014.0165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daianu M, Jahanshad N, Nir TM, Jack CR, Weiner MW, Bernstein MA, Thompson PM. 2015. Alzheimer’s disease neuroimaging initiative. Rich club analysis in the Alzheimer’s disease connectome reveals a relatively undisturbed structural core network. Hum. Brain Mapp. 36, 3087–3103. ( 10.1002/hbm.22830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heuvel MPvd, Pol HEH. 2010. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 20, 519–534. ( 10.1016/j.euroneuro.2010.03.008) [DOI] [PubMed] [Google Scholar]
- 44.Tomasi D, Volkow ND. 2011. Association between functional connectivity hubs and brain networks. Cereb. Cortex 21, 2003–2013. ( 10.1093/cercor/bhq268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivasan R, Russell DP, Edelman GM, Tononi G. 1999. Increased synchronization of neuromagnetic responses during conscious perception. J. Neurosci. 19, 5435–5448. ( 10.1523/jneurosci.19-13-05435.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541. ( 10.1002/mrm.1910340409) [DOI] [PubMed] [Google Scholar]
- 47.Xiong J, Parsons LM, Gao JH, Fox PT. 1999. Interregional connectivity to primary motor cortex revealed using MRI resting state images. Hum. Brain Mapp. 8, 151–156. ( 10.1002/(sici)1097-0193(1999)8:2/3%3C151::aid-hbm13%3E3.0.co;2-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvador R, Suckling J, Coleman M, Pickard J, Menon D, Bullmore E. 2005. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb. Cortex 15, 1332–2342. ( 10.1093/cercor/bhi016) [DOI] [PubMed] [Google Scholar]
- 49.Damoiseaux J, Rombouts S, Barkhof F, Scheltens P, Stam C, Smith S, Beckmann C. 2006. Consistent resting-state networks across healthy subjects. Proc. Natl Acad. Sci. USA 103, 13 848–13 853. ( 10.1073/pnas.0601417103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rykhlevskaia E, Gratton G, Fabiani M. 2008. Combining structural and functional neuroimaging data for studying brain connectivity: a review. Psychophysiology 45, 173–187. ( 10.1111/j.1469-8986.2007.00621.x) [DOI] [PubMed] [Google Scholar]
- 51.Lei X, Ostwald D, Hu J, Qiu C, Porcaro C, Bagshaw AP, Yao D. 2011. Multimodal functional network connectivity: an EEG-fMRI fusion in network space. PLoS ONE 6, e24642 ( 10.1371/journal.pone.0024642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simas T, Chavez M, Rodriguez PR, Diaz-Guilera A. 2015. An algebraic topological method for multimodal brain networks comparisons. Front. Psychol. 6, 904 ( 10.3389/fpsyg.2015.00904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amico E, Goñi J.2017. Mapping hybrid functional-structural connectivity traits in the human connectome, [q-bio]. arXiv (http://arxiv.org/abs/1710.02199. )
- 54.Bédard C, Kröger H, Destexhe A. 2006. Does the 1/f frequency scaling of brain signals reflect self-organized critical states? Phys. Rev. Lett. 97, 118102 ( 10.1103/PhysRevLett.97.118102) [DOI] [PubMed] [Google Scholar]
- 55.Dehghani N, Bédard C, Cash SS, Halgren E, Destexhe A. 2010. Comparative power spectral analysis of simultaneous elecroencephalographic and magnetoencephalographic recordings in humans suggests non-resistive extracellular media. J. Comput. Neurosci. 29, 405–421. ( 10.1007/s10827-010-0263-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corominas-Murtra B, Fuchs B, Thurner S. 2014. Detection of the elite structure in a virtual multiplex social system by means of a generalised K-core. PLoS ONE 9, e112606 ( 10.1371/journal.pone.0112606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peixoto TP. 2015. Inferring the mesoscale structure of layered, edge-valued, and time-varying networks. Phys. Rev. E 92, 042807 ( 10.1103/PhysRevE.92.042807) [DOI] [PubMed] [Google Scholar]
- 58.Brown JA, Van Horn JD. 2016. Connected brains and minds—the UMCD repository for brain connectivity matrices. NeuroImage 124, 1238–1241. ( 10.1016/j.neuroimage.2015.08.043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mori S, van Zijl PCM. 2002. Fiber tracking: principles and strategies—a technical review. NMR Biomed. 15, 468–480. ( 10.1002/nbm.781) [DOI] [PubMed] [Google Scholar]
- 60.Brown JA, Rudie JD, Bandrowski A, Van Horn JD, Bookheimer SY. 2012. The UCLA multimodal connectivity database: a web-based platform for brain connectivity matrix sharing and analysis. Front. Neuroinform. 6, 28 ( 10.3389/fninf.2012.00028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Craddock RC, James G, Holtzheimer PE, Hu XP, Mayberg HS. 2012. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 33, 1914–1928. ( 10.1002/hbm.21333) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the experimental data used in this work can be downloaded from the USC Multimodal Connectivity Database http://umcd.humanconnectomeproject.org. The Matlab code used to compute the core–periphery structure of multiplex networks is made available at https://github.com/brain-network/bnt.