Abstract
Eosinophil-associated diseases (EADs) are rare, heterogeneous disorders characterized by the presence of eosinophils in tissues and/or peripheral blood resulting in immunopathology. The heterogeneity of tissue involvement, lack of sufficient animal models, technical challenges in working with eosinophils, and lack of standardized histopathologic approaches have hampered progress in basic research. Additionally, clinical trials and drug development for rare EADs are limited by the lack of primary and surrogate endpoints, biomarkers, and validated patient-reported outcomes. Researchers with expertise in eosinophil biology and eosinophil-related diseases reviewed the state of current eosinophil research, resources, progress, and unmet needs in the field since the 2012 meeting of the NIH Taskforce on the Research of Eosinophil-Associated Diseases (TREAD). RE-TREAD focused on gaps in basic science, translational, and clinical research on eosinophils and eosinophil-related pathogenesis. Improved recapitulation of human eosinophil biology and pathogenesis in murine models was felt to be of importance. Characterization of eosinophil phenotypes, the role of eosinophil subsets in tissues, identification of biomarkers of eosinophil activation and tissue load, and a better understanding of the role of eosinophils in human disease were prioritized. Finally, an unmet need for tools for use in clinical trials was emphasized. Histopathologic scoring, patient- and clinician-reported outcomes, and appropriate coding were deemed of paramount importance for research collaborations, drug development, and approval by regulatory agencies. Further exploration of the eosinophil genome, epigenome, and proteome was also encouraged. Although progress has been made since 2012, unmet needs in eosinophil research remain a priority.
Keywords: biomarkers, eosinophil-related disorders, eosinophilia, hypereosinophilic syndromes, murine models, translational research
1 ∣. INTRODUCTION
In June 2012, a meeting of the National Institute of Health (NIH) Taskforce on the Research of Eosinophil-Associated Diseases (TREAD) was convened with the aim of defining, clarifying, and prioritizing the unmet research and supportive needs of EADs, defined as rare conditions presenting with eosinophils in the peripheral blood and/or tissues with associated pathology. Although the number of citations with “eosinophil” in the subject or as a keyword has increased in the past 5 years (Fig. 1) and the clinical development and approval of therapeutic agents targeting eosinophils in more common eosinophil-related diseases, such as asthma, has brought new focus to the eosinophil, unmet needs remain in research of basic eosinophil biology and rare EADs. The Taskforce consisted of nationally and internationally recognized, basic, and clinical eosinophil researchers and patient advocacy groups (PAGs). The following 7 common unmet needs across the spectrum of EADs were identified: (1) creation of diagnostic codes to identify and define patient subsets; (2) in depth analysis of biological samples (to help standardize markers of eosinophilic tissue involvement and better understand disease pathogenesis); (3) development and validation of reliable testing for diagnosis and assessment of exacerbation of eosinophilic disease; (4) mechanistic understanding of the role of eosinophils in various EADs; (5) establishment of patient registries; (6) improved guidance and success of relevant clinical trials in rare disease patient populations; and (7) expansion of the number of useful therapeutics for treating EADs.1 In response to this publication, the National Institute of Allergy and Infectious Diseases (NIAID) issued two funding announcements, which were co-sponsored by the National Heart Lung and Blood Institute, the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the National Institute of Diabetes and Digestive and Kidney Diseases. These announcements covered both exploratory (R21) and more established (R01) investigator-initiated basic and clinical/translational projects.
FIGURE 1.
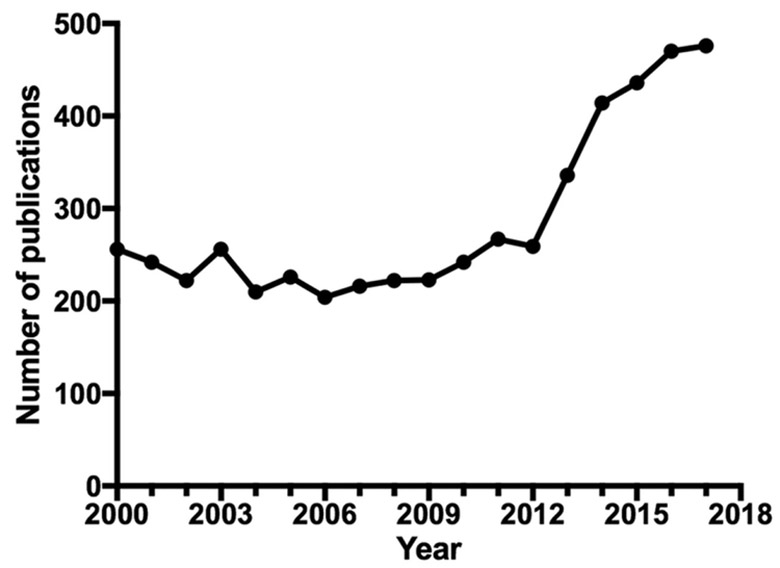
Publications for “eosinophil”, “eosinophilia”, or “eosinophils” as the subject or keyword from 2000 until 2017 (Pubmed)
In 2017, an updated taskforce, “RE-TREAD”, was organized during a meeting of the International Eosinophil Society to bring together a multidisciplinary group of clinicians and scientists with expertise in rare eosinophil-related disorders to discuss issues related to patient care and clinical trial design. A second meeting goal was to continue to foster collaborative relationships between multidisciplinary clinical and basic science researchers with the aim of improving diagnosis and treatment of EADs. This publication summarizes the conclusions of RE-TREAD and the state of eosinophil research in 2017. It also creates a road map for clinicians, PAGs, and other entities concerned with the health and treatment of patients with rare EADs.
2 ∣. METHODS
RE-TREAD was convened during the biennial meeting of the International Eosinophil Society in Göthenburg, Sweden in July 2017. Presentations covered the current status of eosinophil clinical research and disease categorization including diagnostic codes, use of registries, tissue markers of eosinophil pathogenesis including granule proteins, “omics”, histopathology, basic mechanisms in humans and animal models, biomarkers of eosinophilia and eosinophil-associated clinical outcome metrics, clinical trials, and novel therapeutics. Focus groups discussed prioritization of unmet needs and identification of action items for clinicians, researchers, and stakeholders involved in eosinophil-related research.
3 ∣. RESULTS
In contrast to TREAD, which took an organ and disease-specific approach to define unmet needs in individual EADs, RE-TREAD focused on the following 3 main areas important to EADs as a whole: (1) The role of eosinophils and eosinophil activation in the pathogenesis of EADs, (2) clinical and translational studies in EADs, and (3) development and standardization of novel techniques for the study of eosinophils and eosinophil-mediated pathogenesis.
3.1 ∣. The role of eosinophils in the pathogenesis of EADs
Despite the presence of eosinophils and evidence of eosinophil activation in the blood and tissues of patients with EADs, the precise role of eosinophils in disease pathogenesis remains unclear. The reasons for this are multifactorial and include the heterogeneity and complexity of these disorders, the lack of standardized methods to evaluate and target eosinophil-associated pathology, limitations in the recapitulation of human EADs in animal models, and gaps in our understanding of the basic biology of human eosinophils. Recent progress in these areas in the context of novel therapeutics targeting eosinophils, as well as recommendations for future research, are summarized in the next section.
3.1.1 ∣. Eosinophil histopathology
Standardization of methods for evaluation of eosinophil involvement in tissue pathology was identified by TREAD as an unmet need common to all EADs. Specific suggestions for eosinophilic esophagitis (EoE) included counting of eosinophils in biopsies as cells per high power field (/(HPF)) or per unit area, use of mean versus peak counts, consideration of pathologic features other than eosinophil numbers, and detection and validation of eosinophil involvement using immunohistochemical staining for eosinophil granule proteins. Despite significant progress, a number of issues remain. For example, although interobserver agreement among pathologists trained in the counting of eosinophils in esophageal biopsies (per unit area and per HPF) was high,2 re-review of slides reported to have peak eosinophil counts 1-14/HPF yielded higher counts in >20% of the cases.3 To diminish over-reliance on a single pathologic feature, an EoE histology scoring system (EoEHSS) was developed. The EoEHSS scores 8 features (eosinophil inflammation, basal zone hyperplasia, eosinophil abscess, eosinophil surface layering, dilated intercellular spaces, surface epithelial alteration, dysker-atotic epithelial cells, and lamina propria fibrosis) for both severity (grade) and extent (stage) of disease and was shown to outperform peak eosinophil count in differentiating treated from untreated EoE patients.4 Using a different approach, immunohistochemical staining for eosinophil peroxidase, EoE could be differentiated from gastroesophageal reflux disease (GERD),5 although the sensitivity and specificity of this assay has still not been directly compared to EoEHSS.
Standardized methods to evaluate eosinophils, or their products, outside of the esophagus, were recognized as an unmet need by TREAD, but still do not exist. Comprehensive evaluations of eosinophilic gastritis (EG) and eosinophilic colitis (EC) biopsies by a panel of pathologists are underway in the Consortium of Eosinophilic Gastrointestinal Researchers (CEGIR (Table 1)). It is anticipated that data generated from this consortium will help standardize methods to evaluate gastrointestinal biopsies across multiple centers and identify the relationship, if any, between eosinophilic gastrointestinal disorders (EGID) and other inflammatory bowel diseases.
TABLE 1.
Clinical, scientific, and translational resources for eosinophil researchers
| Tool | Description | Resource |
|---|---|---|
| Rare Diseases Research Tools | ||
| RDCRN program | Resource with support for clinical studies, consortia, collaborations, data sharing and management in Rare Diseases |
http://rarediseasesnetwork.epi.usf.edu/ Example Consortia: CEGIR: https://www.rarediseasesnetwork.org/cms/cegir/About-Us/CEGIR VCRC: https://www.rarediseasesnetwork.org/cms/vcrc |
| Tissue Chip Program | Funded projects for development of 3-D platforms (Chips) to evaluate safety of new drug candidates | https://ncats.nih.gov/tissuechip/about |
| TRND | Program supports pre-clinical development of therapeutic candidates for diseases with unmet needs | https://ncats.nih.gov/trnd/about |
| NCATS New Therapeutic Uses program | Resource for repurposing therapeutic agents for new indications | https://ncats.nih.gov/ntu/about |
| Omics Databases | ||
| NCBI GEO Profiles and Datasets | Searchable database of individual gene expression profiles from curated DataSets in the Gene Expression Omnibus (GEO) repository | https://www.ncbi.nlm.nih.gov/gds |
| Haemosphere | Searchable gene expression profiles of hematopoietic cell types | haemosphere.org125 |
| Strain | Description | Reference |
|---|---|---|
| Mouse modelsa | ||
| iPHIL | Eosinophil depletion via inducible diphtheria toxin receptor knockin (EPX promoter driven DTR expression) | 12 |
| eoCre | Removal of floxed gene of interest specifically in eosinophils upon crossing (EPX promoter driven Cre) | 24 |
| PHIL | Congenital eosinophil deficiency mediated EPX-promoter driven expression of Diphtheria Toxin A chain |
126 |
| EPX−/− and MBP-1−/− | Mice with knockout of EPX, MBP-1 or both | 127-129 |
| NJ.1638/hE2 or NJ.1726 | Mice with chronic severe asthma, overexpression of IL-5 (Cd3δ promoter) and eotaxin-2 (CC10 promoter) or IL-5 from CC10 promoter | 15,130,131 |
| Reagents | ||
| IHC/IFa | Antibodies to MBP-1 (mouse) and to EPX (Human/mouse) available | 5,132 |
| EPX ELISAa | EPX specific ELISA (mouse and human) patient samples | 84,85,133,134 |
| MBP-1 and CLC/Gal-10 ELISAsb | MBP-1 and CLC/Galectin-10 specific ELISAs (human patient samples) | 9 |
| Eosinophil Granule protein Multiplexc | Simultaneous measurement of MBP, ECP, EDN and EPO in a multiplex suspension array system. | 135 |
CEGIR, Consortium for Eosinophilic Gastrointestinal Disease Researchers; CLC, Charcot-Leyden crystal; ELISA, enzyme-linked immunosorbent assay; EPX, eosinophil protein X; GEO, Gene Expression Omnibus; IF, immunofluorescence; IHC, immunohistochemistry; MBP, major basic protein; RDCRN, Rare Diseases Clinical Research Network; TRND, Treatment for Rare and Neglected Diseases; VCRC, Vasculitis Clinical Research Consortium.
May be performed in collaboration or obtained by contacting Jacobsen.elizabeth@mayo.edu;
May be performed or obtained by contacting sackerma@uic.edu;
May be performed in collaboration with aklion@niaid.nih.gov.
TREAD also recommended the standardization and validation of methods to evaluate tissues to improve the differential diagnosis and treatment for organ-restricted EAD. Some progress has been made. For example, using stains to identify Th1 (T-bet and CD4) or Th2 (GATA and CD4), a significantly lower Th1/Th2 cell ratio was identified in morphea compared to eosinophilic fasciitis, facilitating the distinction between these two disorders that share multiple histopathologic features (dermal inflammation including eosinophil infiltration and fibrosis extending into fascia).6 Similarly, in the esophagus, T-bet (Th1) and GATA-3 (Th2) expression may help distinguish between EoE and gastroesophageal reflux disease (GERD). Finally, additional noninvasive approaches have been developed since TREAD, including the Cytosponge7,8 and esophageal string test (EST).9 Assessment of eosinophil granule protein levels using either of these methods has been shown in preliminary studies to correlate with eosinophil counts in matched biopsies.
Future efforts should aim to (i) continue to standardize and validate general methods for evaluating tissue eosinophilia and eosinophil-mediated pathology; (ii) develop additional noninvasive approaches for use as histopathologic surrogates in eosinophil-related diseases; (iii) standardize and validate methods for evaluation of eosinophils in organ-specific conditions (in particular EG, EC), but also in other organs, including skin; and (iv) examine the relationships between EADs and other overlapping inflammatory conditions with associated eosinophilia (e.g., EGID and inflammatory bowel disease, or hypereosinophilic syndromes (HES), EGPA, and IgG4-related disease).
3.1.2 ∣. Animal models of eosinophilic disease
At the time of TREAD, experts felt that existing murine models did not accurately recapitulate EADs, although they were felt to be important in understanding the basic biologic functions of eosinophils. TREAD recommended utilization of “humanized” murine models to better model human disease but recognized that species differences in soluble mediators and tissue proteins needed to be addressed. Since 2012, significant progress has been made in both the development of new murine models and their use in elucidating the complex role of eosinophils in allergic diseases. Further, these models have facilitated the recognition of beneficial functions of eosinophils, including their role in metabolism, tissue regeneration, immune homeostasis, and putative functions for eosinophils in non-eosinophilic diseases (reviewed in Ref. 10).
Because murine models can be manipulated in ways that are not currently possible in humans, they are critical to studies of eosinophil function.11 Murine models have provided significant insight into allergic inflammation,12 including knowledge of eosinophil structure and function.11 Importantly, mice deficient in IL-5,13 overexpressing IL-5,14,15 or injected with IL-516 have provided information relevant to development of biologics targeting the IL-5 axis. Eosinophils in mice contribute to maintenance of long-lived plasma cells in the bone marrow,17 IgA production in the intestine,17,18 modulation of the intestinal microbiome,19 the metabolic activity of alternatively activated macrophages in adipose tissue,20 and tissue regeneration in skeletal muscle21 and liver.22 Murine models (as well as some data in humans) suggest that recruited, IL-5 dependent, inflammatory eosinophils in allergic inflammation are distinct from IL-5 independent, resident lung eosinophils.23 Since the last TREAD meeting, novel strains of mice, including iPHIL12 and eoCre,24 have been developed to manipulate the kinetics or gene-specific expression within eosinophils (Table 1). Together, these models and earlier strains (for review Ref. 25,26) highlight underappreciated roles for mouse eosinophils, both in disease and in the maintenance of normal tissue homeostasis.
Eosinophils have multifaceted interactions with the immune system, which have been studied extensively in mice. Eosinophils have been shown to exhibit phenotypic changes in response to signals from the extracellular matrix in murine asthma models.27,28 They can recruit Th2 CD4+ T cells,29 activate dendritic cells,30 polarize M2 macrophages31 cause nerve cell branching,32 recruit and activate CD8 T cells,33,34 and secrete nerve growth factor or eosinophil peroxidase which activate mast cells through TrkA35 or Mas-related gene X2 (MrgX2)36 receptors, respectively. The interactions between eosinophils and other cells are just starting to be explored in humans.
In a mouse model of severe asthma, EPX, but not eosinophil granule major basic protein-1 (eMBP-1) induced goblet cell metaplasia,37 suggesting that the pattern of eosinophil granule protein release may be an important driver of a particular disease pathology. As such, granule specific assays, such as the EPX ELISA or EPX immunohistochemistry (Table 1, Supplementary Table S1), will help clarify these relationships and are likely to play a significant role in future assessments of eosinophil function in human eosinophilic and non-eosinophilic disorders.
Despite expanded availability of murine models (Table 1, Supplementary Table S2 and S3) for the study of eosinophil biology, participants recognized the need for better recapitulation of human pathology. For example, although murine models have provided unique insight into roles of individual granule proteins, murine eosinophils are relatively refractory to degranulation in mostmodels. Additional issues that complicate extrapolation of murine findings to humans include differential activation of Th1/Th2/Th17 pathways between the species, altered expression patterns and function of murine and human surface receptor paralogues and orthologues (e.g., Siglec-F and Siglec-8, EGF-like module-containing mucin-like hormone receptor-like 1 (EMR-1) and F4/80), and the absence of murine genes encoding soluble mediators, such as eotaxin-3, EoE gene products, such as calpain-14,38 and Charcot-Leyden Crystal (CLC) protein/Galectin-1039 for which intracellular and extracellular roles, for example, in granulogenesis during human eosinophil development, secretion of granule cationic proteins, and glycan-containing ligands, remain to be determined. Finally, no murine models exist that create “humanized” eosinophil mice as has been done, for example, with mast cells,40 and current data from newly derived murine models (e.g., non-irradiated NBSGW mice engrafted with hematopoietic cells) have not been specifically evaluated for engraftment of eosinophils using available eosinophil cell surface markers.41 None of the currently available models adequately replicate the various subtypes of HES or EGPA.
Future efforts should aim to further refine and recapitulate eosinophil biology in murine models including (i) improvement of murine models of eosinophil degranulation in tissues; (ii) further assessment of unique human targets (e.g., eotaxin-3, calpain-14, Charcot-Leyden Crystal Protein (CLC)/Galectin-10, and ECP); (iii) recapitulation of EADs in murine models; (iv) investigations of hematopoietic transfer models in mice to evaluate eosinophil engraftment with the hope that humanized eosinophil mice might allow better investigation of the development, maintenance, and inflammatory responses of human eosinophils; and (v) further assessment of the role of eosinophils, both beneficial and harmful, in homeostasis and non-EADs.
3.1.3 ∣. Eosinophils in human disease
Delineating the basic biology of human eosinophils remains an important goal. Many critical insights into the basic biology of human eosinophils or other cells that interact with eosinophils have been gained since the TREAD meeting in 2012, including the recognition that eosinophils may: (1) interact with and activate innate type 2 lymphoid cells (ILC2s),42 (2) serve as important sources of cytokines and other mediators, (3) possess the ability to secrete functional, cell-free, tissue-deposited granules, (4) exhibit differential phenotypes (e.g., resident and inflammatory subsets), and (5) exhibit crosstalk with other cells, such as mast cells,43 and other non-hematopoietic cells (though a discussion of all these interactions is beyond the scope of this review [reviewed in Ref. 44-46], and 6) may be beneficial in some non-EADs (e.g., in some cancers [reviewed in Ref. 47]. Despite this progress, there is still much to learn regarding the interaction of eosinophils and their response to tissue components, such as matrix proteins,48 fibroblasts,49 epithelium, and other tissue-resident cells.50 It is known that the engagement and expression of integrins can engage matrix proteins, such as fibronectin and laminin, and that these interactions alter eosinophil biology including degranulation and survival.51,52 Whether other changes in the tissue microenvironment (e.g., during morphogenesis, aging, wound development, and repair) influence the biology of resident and recruited eosinophils is less clear. Cell surface lectin receptors, such as Siglec-8, can engage specific glycan ligands on tissue and may selectively influence eosinophils.53,54 This bidirectional response and understanding of tissue microenvironments will be an important concept for future study in EADs.
Whereas a better understanding of basic eosinophil function is clearly a priority, well-designed translational studies are required for the application of basic science advances to human disease. These, in turn, depend on the development of clear diagnostic criteria, consistent nomenclature for standardization of acquisition, analyses, and histopathologic evaluation of biopsies and establishment of validated biomarkers of disease activity and treatment outcomes in clinical trials and daily practice. Although blood biomarkers tend to be favored for simplicity and ease of access, biomarkers based on other biological specimens, such as urine or nasal secretions, and organ-specific biomarkers and scoring systems are also needed.
3.1.4. ∣. Eosinophil granule proteins
Eosinophils store a diverse array of cytokines and chemokines preformed within intracellular granules. Tissue eosinophils respond to external stimuli with rapid and differential secretion of eosinophil granule-derived cytokines through a vesicle-mediated process called piecemeal degranulation, ultrastructural evidence of which is readily observed in tissue eosinophils across all EADs. In addition, tissues and secretions from multiple EADs exhibit cell-free, membrane-bound eosinophil granules (reviewed in Ref. 10) that express ligand-reactive receptors on their outer membranes and remain competent to undergo stimulus-dependent, differential secretion.55 Therefore, there is evolving recognition that eosinophil-derived organelles (i.e., vesicles and free granules) are deposited within tissues as a consequence of cytolytic modes of eosinophil cell death and function as instruments of local tissue damage and/or immunoregulation even in the absence of intact eosinophils. Since the 2012 TREAD report, several studies have begun to identify mechanisms that regulate the expulsion of eosinophil cell-free granules in tissues, both independent from and in association with the extrusion of DNA “traps”.56-60 A more comprehensive understanding of regulated cell death pathways contributing to this process, and the physiological consequences of tissue-deposited eosinophil cell-free granules is needed. Moreover, efficacies of new and existing proposed therapies (further described in the section on Expansion of new Therapeutic Areas in EADs) must be measured by criteria not only based on depletion of intact eosinophils, but also with careful attention paid to the presence of residual free granules within tissues.
3.1.5 ∣. Tissue eosinophils and eosinophil subtypes
Eosinophils are produced in the bone marrow from CD34+ eosinophil lineage-committed progenitors (EoP) that can mobilize from the bone marrow and accumulate at sites of allergic inflammation.61 EoPs may undergo in situ differentiation (i.e., extramedullary hematopoiesis [EMH])62,63 into mature cells and can contribute to tissue inflammation through production of locally produced cytokines and promotion of EMH with resultant tissue eosinophilia. Better understanding of the role of locally produced cytokines that promote EoP differentiation, EoP accumulation, and blockade of this process is needed to help clarify the processes underlying maintenance and resolution of tissue eosinophilia in EADs.
Although it has long been recognized that eosinophils can have an activated phenotype upon stimulation with IL-5, IL-3, IL-33, or GM-CSF, recent data23 suggest that there may be differential eosinophil activation by cytokines,64 and perhaps different subtypes in the human lung. Resident lung eosinophils from non-asthmatic lungs have distinct surface marker expression (Siglec-8+CD62L+IL-3Rlo) in comparison to inflammatory eosinophils in the sputum of subjects with asthma (Siglec-8+CD62LloIL-3Rhi), and different eosinophil subtypes may exist in human skin under pathological conditions.65 The functional consequences of these differences in surface marker expression have not been determined definitively in humans, although corresponding murine data suggest that resident eosinophils have a regulatory phenotype. Recent and prior studies have noted differential gene expression patterns between tissue-resident eosinophils,23,66 suggesting that the local environment promotes an eosinophil phenotype with tissue-specific functions. Colonic eosinophils have been shown to be protective in Clostridium difficile infections in both mice and humans.67 Notably, eosinophils promoted intestinal barrier integrity and survival in mice that was independent of eosinophil-derived IL-4 or bactericidal activity. The specific protective mechanism remains unknown, highlighting the need for a deeper understanding of the factors regulating the development of functional eosinophil subtypes, their responses to environmental signals, and the beneficial and harmful consequences of their activities in eosinophil homeostasis, pathogenesis, and trafficking. Although the mechanisms of trafficking have begun to be delineated68-70 in asthma, additional focus on mechanisms of cell trafficking in specific EADs would be timely. Utilization of single cell analyses, including single cell RNAseq and CyTOF mass spectroscopy, to tease out clusters in heterogeneous populations will likely be helpful in this regard.
Future efforts should aim to (i) delineate mechanisms that control cardinal eosinophil functions (e.g., migration and survival), that both elicit and shape the differential secretion of functionally diverse preformed mediators; (ii) identify subtypes and functions of eosinophils unique to different tissues and mechanisms of eosinophil recruitment to specific organs; (iii) delineate the contribution of local eosinophilopoiesis in EAD pathogenesis via identification of EoP-specific targeting strategies in affected tissues; (iv) better characterize the normal homeostatic, protective, and pathogenic functions of eosinophils in EADs and non-EADs; (v) better characterize cell death pathways of eosinophils, particularly the mechanisms and triggers for, and outcomes of, deposition of cell-free granules in humans; and (vi) continue to explore the relationships between eosinophils and other cells, including lymphocytes, mast cells, ILC2s, fibroblasts, adipocytes, mesenchymal stem cells, and epithelial cells, and their role in EADs.
3.2 ∣. Clinical and translational studies in EADs
Since the TREAD meeting, there has been a dramatic increase in the development and approval of novel therapies for treatment of both common and rare EADs. While this is exciting, some rare EADs remain understudied due to (1) difficulties in recruitment of rare patients and collecting of prospective samples during multicenter studies from which to perform mechanistic research, (2) adequately powered studies from which to draw evidence-based conclusions, and (3) logistical challenges that arise from attempts to create cohesive research plans with uniform collection of biological samples, data-coordination, and analysis. Investigators studying rare EADs would benefit from the creation of research networks and registries for collaboration and pooling of resources. In addition, a comprehensive guide to the resources (Table 1) and organizations that provide funding (Table 3) to researchers in rare diseases, specifically in EADs, would help foster increased engagement of new investigators in this area. Recommendations for overcoming these challenges are summarized in the next section.
TABLE 3.
Patient advocacy groups,funding organizations,and other groups relevant to EAD research
| Organization | Website |
|---|---|
| AAAAIa | http://www.aaaai.org/professional-education-and-training/grants-awards |
| APFEDa | http://apfed.org/research/ |
| AUSEEa | http://www.ausee.org/medicalresearchgrants.htm |
| Burroughs Wellcome Fund |
https://www.bwfund.org/feature-research-rare-diseases |
| CUREDa | https://curedfoundation.org/ |
| EFC | http://eoscoalition.org/ |
| FARE | https://www.foodallergy.org/ |
| NORDa | https://rarediseases.org/for-clinicians-and-researchers/research-opportunities/research-grant-program/ |
| Vasculitis Foundationa |
http://www.vasculitisfoundation.org/research/ |
Abbreviations: APFED, American Partnership for Eosinophilic Disorders; CURED, Campaign Urging Research for Eosinophilic Disease; EFC-Eosinophilic Family Coalition; FARE-Food Allergy Research & Education; NORD-National Organization for Rare Disorders.
Organization or Patient Advocacy Group that has provided funding for rare disease or EAD research.
3.2.1 ∣. Registries
The Rare Disease Clinical Research Network (RDCRN) program allows multidisciplinary teams to partner across several institutions to facilitate patient recruitment, accelerate young investigator training, and engage patient support through partnership with PAGs. RDCRN has collaborated with PAGs and the pharmaceutical industry on government-sponsored clinical research studies to be completed in a timely fashion and to bring novel therapeutics to market. An EAD-associated registry was borne out of RDCRN through the CEGIR (U54 AI117804), an initiative of the Office of Rare Disease Research (ORDR) that is jointly funded by NCATS, NIAID, and NIDDK.71 This registry (part of a longitudinal observational study) is designed to understand the natural history of EGID and will help standardize histopathologic evaluation and assessment of these rare disorders. Although some EADs are included in larger registries (e.g., EGPA is included in the Vasculitis Clinical Research Consortium (VCRC)), the only network to date that attempts to capture all of the varied forms of HES is the French network, Centre de Référence National des Syndromes Hyper-eosinophiliques. This network has recruited more than 800 patients to date and has resulted in the publication of several large clinical series.72,73 Replication of these efforts in other countries would allow confirmation of the French findings in different epidemiologic settings, identify pools of patients for potential participation in clinical studies, and provide patient data to address important issues in the diagnosis and treatment of EADs.
3.2.2 ∣. Patient and clinician reported outcome measures
The development and validation of disease-specific quality of life (QOL) and disease control indices are needed to measure the patient’s perspective of symptoms and disease impacts, and to evaluate the efficacy of treatment and/or other interventions in EADs. These measures, whether patient reported outcomes (PRO) or clinician-reported outcomes, are often required by regulatory authorities for use in clinical licensing trials. Currently validated metrics exist for only a handful of EADs and focus only on symptoms related to the affected organ system. In asthma, previously validated endpoints (such as AQLQ/ACQ)74,75 and exacerbation rates have been used for trials of eosinophilic asthma; whereas, newly developed PROs and QOLs had to be created for eosinophilic esophagitis (PedsQL (Pediatric Quality of Life Inventory),76 EoE-QOL-A (EoE Quality of Life-Adults),77 and PEESS v2.0 (Pediatric Eosinophilic Esophagitis Symptom Severity)).78 The lack of similar metrics in other EADs, including EGID involving the stomach, small bowel, and colon, are due, in large part, to the heterogeneity of symptoms when multiple organs are affected. The confounding effects of medication-related side effects can also hinder the development of PROs. Because of the limited opportunities for validation of instruments in rare diseases, careful consideration of the design and choice of instruments across centers of excellence in EADs are important so that results from different studies can be compared. In a recent double-blind, placebo controlled trial evaluating mepolizumab in patients with EGPA,79 a clinician reported outcome measure (EGPA-CAS) specific for EGPA was developed and is being compared to the Birmingham Vasculitis Activity Scale(BVAS) that was developed for use in vasculitis in general. Results from this study, when available, may provide additional tools for study of eosinophil-related symptoms.
3.2.3 ∣. Academia-industry collaborations
The importance of leveraging clinical trials to collect data to enhance understanding of underlying disease mechanisms in EADs was uniformly endorsed by the RE-TREAD participants. One example of this is the EGPA study mentioned above that was co-sponsored by the NIH and GlaxoSmithKline (GSK), and included extramural NIH-funding to support mechanistic studies at 5 US academic sites.79 Standard operating procedures were implemented for tracking blood, sputum, and urine samples through a data coordinating center in order to collect blood samples for quantification of cytokines and other mediators for analysis of treatment response and evaluation of exacerbations, and to collect blood and tissue for transcriptome profiling for exploration of mechanisms of disease. During the conduct of the study, special attention was paid to operating procedures for internal governance and transparency requirements, especially in consideration of FDAsubmission of this pivotal phase 3 study. This collaboration serves as a model for future design of clinical studies in EADs with incorporation of a priori defined research goals, and pooling of resources, expertise, and rare-disease patients across multiple institutions.
3.2.4 ∣. Creation of more specific diagnostic codes for EADs
The United States applies a clinical modifier (ICD10-CM) to the International Classification of Diseases (ICD) codes approved by the World Health Organization. These are then approved by major organizations, including the American Hospital Association and Centers for Medicare and Medicaid Services, and govern classification of disease and reasons for visits in most health care settings. ICD-CM codes are important for payors and providers of healthcare services, but, more importantly, availability of specific ICD codes allows clinicians to identify and accurately diagnose rare diseases. Unique ICD codes can improve disease management and treatment choices, and may result in improved resource allocation and understanding of unmet needs in rare EADs. From a research perspective, utilization of more specific codes is important for better understanding of prevalence, treatment, and healthcare utilization associated with these diseases as well as the identification of subjects for potential recruitment into clinical trials. Once codes are available, dissemination of this key change will be important for incorporation by clinicians into workflows.
In the 2012 TREAD, the lack of unique codes for EADs was identified as an unmet need since most EADs, with the exception of Löffler’s syndrome and Pulmonary Eosinophilia, would have been classified under the umbrella ICD9 code of “Eosinophilia”. Fortunately, since 2012, more specific codes for EGIDs (eosinophilic esophagitis, eosinophilic gastritis or gastroenteritis, and eosinophilic colitis) have been incorporated into ICD-10-CM. Unfortunately, little progress had been made for other EADs as of the 2017 RE-TREAD meeting. A clinical workgroup was convened to prioritize additional specific codes. Within a few months of the meeting, 11 EADs (Table 2) were suggested to APFED (American Partnership for Eosinophilic Disorders) in light of their prior success of this organization in advancing codes for specific EGIDs.
TABLE 2.
2017 IES clinical workgroup proposed list of ICD-CM codes
| 1. Idiopathic Hypereosinophilic Syndrome |
| 2. Lymphocytic Variant Hypereosinophilic Syndrome |
| 3. Myeloid Hypereosinophilic Syndrome with mutations in PDGFRA, PDGFRB, FGFR1, or JAK2a |
| 4. Myeloid Hypereosinophilic Syndrome without a known molecular abnormality |
| 5. Hypereosinophilic Syndrome NOS |
| 6. Eosinophilic Hepatitis |
| 7. Eosinophilic Fasciitis |
| 8. Acute Eosinophilic Pneumonia |
| 9. Chronic Eosinophilic Pneumonia |
| 10. Eosinophilic Granulomatosis with Polyangiitis (EGPA) |
| 11. Drug Rash with Eosinophilia and Systemic Symptoms (DRESS) |
Abbreviations: HES, hypereosinophilic syndrome, PDGFR, platelet derived growth factor, FGFR, fibroblast growth factor receptor, JAK, janus associated kinase, NOS, not otherwise specified.
with consideration of modifiers for specific mutations.
3.2.5 ∣. Biomarkers
Although peripheral absolute eosinophil counts (AEC) have traditionally been used for diagnosis and monitoring of treatment in EADs, their use as biomarkers in the setting of clinical trials has not been validated. Nevertheless, placebo-controlled studies of mepolizumab for HES80 and EGPA79 that used a central laboratory to standardize counts across centers demonstrated a reduction in AEC associated with disease improvement and control, suggesting that reduction of the AEC is a useful biomarker of disease activity in certain EADs. Although intuitively it would seem that tissue eosinophilia should be a superior biomarker of eosinophil-mediated pathology, technical issues, including the lack of standardization across affected tissues and variability between readers (see Histopathology section above), as well the requirement for biopsy samples, have precluded the widespread use of tissue eosinophilia as a biomarker in clinical trials. Where this has been attempted, the results have been mixed. For example, clinical trials of mepolizumab and reslizumab in EoE did not meet their primary endpoints despite reduction in tissue eosinophilia in esophageal biopsies.81-83 Whether this was due to lack of sufficient tissue depletion, irreversible structural changes in the esophagus, a lack of direct eosinophil contribution to disease pathogenesis, or other factors is unclear.
Markers of eosinophil activation, including eosinophil granule proteins levels in blood, tissue, and body fluids, may be more easily quantifiable markers of disease activity and have been used as biomarkers in some studies, including a multicenter, double-blind, placebo-controlled trial of mepolizumab in HES,80 asthma,84,85 and EoE.9 However, differences in the specific granule proteins measured, and methodologies used between studies, currently make it difficult to draw generalizable conclusions.
A major advancement since the TREAD meeting is the development of noninvasive techniques to assess eosinophil granule deposition. These include the use of body fluids, such as saliva, stool, and urine, as surrogates for tissue biopsies and the esophageal string test.9 To capture a standardized metric of blood eosinophilia and activation states, a gene expression metric was derived for blood samples collected in PAXgene RNA tubes as part of a clinical trial of anti-IL-13 antibody in asthma.37 This eosinophil-related gene signature metric is neither age dependent nor lab dependent and may capture activation states as well as proportions of eosinophils in a peripheral blood sample. The applicability of this metric to other EADs remains to be seen.
Several biomarkers have been developed to characterize type 2/eosinophilic inflammation in asthma, including fractional exhaled nitric oxide (FeNO), and serum or plasma periostin protein levels.86 These biomarkers correlate with eosinophilic airway inflammation in asthma, but have not been explored more broadly in EADs. Additional examples of biomarkers with applicability restricted to a single or selected group of EADs include TARC/CCL17,87 or quantification of the clonal population88 in lymphoid HES, functional or structural markers of disease in EoE (e.g., EndoFLIP to measure esophageal distensibility), and tyrosine kinase mutations in myeloid HES. Further demonstration of the validity of these biomarkers is needed.
3.2.6 ∣. Expansion into new therapeutic areas in EADs
Since the TREAD meeting in 2012, 3 biologics that specifically target eosinophils by binding IL-5 (mepolizumab, reslizumab) or its receptor (benralizumab) have been approved for the treatment of eosinophilic asthma.89-91 Despite this exciting advance in therapeutic options for asthma and the recent FDA approval of mepolizumab for adult patients with EGPA (at a dose of 300 mg sc monthly), progress in the clinical development of targeted therapies for other rare EADs has been slow. The safety and efficacy of mepolizumab (Nucala®, GlaxoSmithKline) as a steroid-sparing agent in HES was demonstrated in a multicenter, double-blind, placebo-controlled phase 2 trial nearly 10 years ago;80 yet, mepolizumab at the dose used in this study is currently available only off-label or in the setting of a compassionate use protocol for HES. Both mepolizumab and reslizumab (Cinqair®; Teva Pharmaceuticals) have been studied in eosinophilic esophagitis, although results have been equivocal (see above). Benralizumab (Fasenra™, AstraZeneca), a mAb against the IL-5 receptor that causes antibody-dependent cell-mediated cytotoxicity, has been shown to deplete blood and tissue eosinophils in patients with asthma.89,92 A phase 2 trial was recently completed in patients with treatment-refractory HES (NCT02130882), and pilot studies are currently underway in EGPA and EGID (Clinicaltrials.gov NCT03010436).
Unrelated to the IL-5 axis, a new small molecule agent, dex-pramipexole (Knopp Biosciences), was noted to have coincidental eosinophil-lowering activity during its initial clinical development in amyotrophic lateral sclerosis.93 In an open-label phase 2 study in patients with chronic rhinosinusitis with nasal polyps (CRSwNP), dexpramipexole lowered blood eosinophils by 94% from baseline (P < 0.001) and polyp eosinophils by 97% (P = 0.001).94 Promising results of a recently completed phase 2 trial in HES should be available soon.95 Clinical trials of other agents that target eosinophils, including antibodies to Siglec-8, are planned or ongoing for non-EADs (NCT02734849; NCT02808793; NCT03379311) and are likely to provide additional insights into EADs in future planned trials.
Finally, a number of agents that target pathways important in eosinophilic inflammation, rather than eosinophils themselves, are being studied or have been approved for a variety of common disorders where eosinophils may play a role. These include anti-IL-13 (lebrikizumab, tralokinumab, RCP4046), anti-IL-4Rα (dupilumab), anti-TSLP (tezepelumab),96 and anti-IL-31 (nemolizumab), various of which have demonstrated efficacy in eosinophilic asthma,97 atopic dermatitis (NCT0252509498,99), nasal polyposis,100 and EoE (NCT02379052101).
An important secondary benefit of clinical trials of targeted agents in these disorders is the mechanistic insights provided regarding the role of eosinophils in disease pathogenesis. For example, the efficacy of mepolizumab, reslizumab, and benralizumab in eosinophilic asthma confirmed a role for eosinophils in asthma exacerbations, but also helped to promote the concept of asthma endotypes.102 In contrast, the demonstration that antibodies to IL-13 and IL-4Rα are also effective in reducing exacerbations and FeNO levels in eosinophilic asthma, despite a transient increase in circulating eosinophils,34 highlights the differences between blood and tissue eosinophils and their roles in disease. As the numbers of targeting therapies continue to expand, it will become increasingly important to determine the EADs, and perhaps even subtypes of EADs, that are most likely to benefit from available agents. Future studies should address the positive and negative consequences of targeting multiple pathways in EADs.
Future efforts in clinical and translational research should aim to (i) discover biomarkers of tissue specificity and scoring systems for assessment of disease activity and treatment outcomes for use in clinical trials and daily practice; (ii) delineate epidemiology, diagnostic criteria, organ-specific biomarkers, and endotypes of EADs so that a personalized medicine approach can be used; (iii) identify novel biomarkers of eosinophil involvement, activation status, and disease activity in prospectively collected and archived human samples; (iv) design adequately powered clinical studies to identify biomarkers for disease management and for discovery of potential novel therapeutic approaches, for example, by inhibition of additional key players in eosinophil biology such as eosinophil chemoattractants, as well as IL-33 and TSLP; (v) facilitate interactions between academia and industry early in clinical trial development for biomarker discovery. Of particular importance is the comparison of the efficacy of conventional and novel therapies for specific EADs and the creation of optimized treatment algorithms and personalized approaches to patient selection for novel therapeutics; (vi) advance new codes for EADs for incorporation and use in ICD10-CM and ICD11-CM; (vii) promote funding guidelines that include support for eosinophil mechanistic studies not strictly aligned to a particular EAD entity; and (vii) perform clinical trials for comparison of the efficacy of conventional and novel therapies for specific EADs and for creation of optimized treatment algorithms.
3.3 ∣. New biological techniques applied to EADs
Major advances in biomedical technology, bioinformatics, and their application since 2012 have led to novel approaches to study EADs. A significant reduction in the cost of genome sequencing and a corresponding ability to aggregate and analyze large datasets has made the application of these approaches to the “eosinophilome” possible. The potential of these developments to deliver precision and personalized care103 in EADs cannot be overemphasized.
3.3.1 ∣. “Omics”
Notable advances include application of “omic” approaches, particularly proteomics and transcriptomics to human eosinophil immunobiology. An in-depth assessment of the eosinophil proteome with demonstration of upregulation of phosphoproteins with IL-5 stimulation was described.104 The proteome and phosphoproteome were also noted to change after stimulation, including responses to IL-5, IL-3, and GM-CSF.105 Other studies show the value of applying transcriptomics to human eosinophil biology, with evolution from gene array studies to RNAseq methodology. Gene array analysis showed upregulated transcripts in bronchoalveolar lavage eosinophils after segmental allergen challenge,106 and TGF-β target genes in peripheral blood eosinophils were shown to be expressed by RNAseq.107 Transcriptome analysis of esophageal tissue specimens was shown to be effective in making molecular diagnoses of EoE and differentiating EoE from reflux disease,108 and glycomics analyses are being used to identify glycoproteins in mucins and other endogenous tissue structures capable of directly interacting with eosinophil lectin receptors109 further paving the way for personalized approaches to diagnosis of EADs. Identified future challenges include mapping the interaction between the microbiome and eosinophil functional responses and characterization of eosinophil-specific and eosinophil subset-specific signatures in healthy and diseased tissues using single cell sequencing and other novel techniques.
The use of laser capture dissection and single cell sequencing,110 multiplex florescence in-situ hybridization analysis,111 and omics112 have already proven useful in the study of new targets in murine models. Gene editing and clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated (Cas) technology provide faster generation of floxed mice to genes of interest in eosinophils, and in cell lines CRISPR/Cas has been used to study the FIP1L1-PDGFRA fusion gene to further understand mechanisms in myeloid HES.113 Conceivably, CRISPR may be used with humanized mice in the future as a tool to manipulate transferred human eosinophils.
Outstanding questions in the field of EADs include the lack of predictors for the development of EADs, as well as optimal treatments for tissue specific manifestations, such as in EGIDs. Using an omics approach, it is hopeful that phenotypic and genomic data, can be analyzed as a function of environment, epigenome, transcriptome, proteome, metabolome, glycome, and microbiome. Indeed, genomic data has already identified regions that confer susceptibility to eosinophilia.114 The key transcriptional programs responsible for eosinophil development are emerging.115 The eosinophil proteome has been preliminarily mapped out,104 and eosinophils have been shown to regulate the intestinal microbiome.18 De novo mutations and genome wide methylation patterns responsible for HES have been initiated.116 The EoE transcriptome has been uncovered, providing new insight into disease pathogenesis and providing the framework for a diagnostic PCR-based diagnostic panel.108,117 Such an approach has facilitated new drug development such as anti-Type 2 cytokine therapy.118
Future efforts in the area of technology, biomedical advances, and “big data” should aim to (i) expand and further analyze the eosinophil epigenome, including eosinophil subtypes; (ii) characterize the metabolome of eosinophils, including eosinophil subtypes; (iii) define the environmental factors that may interact with eosinophilomes; (iv) integrate the eosinophil genome, transcriptome, epigenome, proteome, glycome, and metabolome and share these data as a resource to eosinophil researchers; (v) apply the eosinophilome to EADs to improve fundamental knowledge of mechanisms of disease onset, prognosis, and response to treatment; and (vi) explore the role of paradigm-shifts in research as they relate to the diagnosis of EADs and incorporation of these practices into clinical practice (e.g., EoE Diagnostic Panel108).
4 ∣. DISCUSSION
The goals of both the prior TREAD meeting1 and the most recent RE-TREAD meeting were to convene leading international experts in eosinophil biology and EADs to work together to help move the field toward improved understanding, diagnosis, and treatment of EADs. The topics considered ranged from basic eosinophil biology to clinical trials. While progress has been made during these past 5 years, significant gaps remain in our understanding of EAD initiation, development, evolution, progression, organ specificity, and remission. With the unique exception of the highly effective and perhaps “curative” use of tyrosine kinase inhibitors for PDGFR-associated myeloid neoplasms,119,120 treatment paradigms for HES remain mostly empiric.121 Fortunately, the last 5 years have witnessed the sequential FDA approval of 3 new-in-class anti-eosinophil biologics that target IL-5 or its receptor.122 Such drugs are not only a welcome addition to the arsenal of therapies for EADs but are highly targeted and specific pharmacologic tools for dissecting the contribution of eosinophils to disease, adding timeliness to RE-TREAD. A prime example is the successful use of mepolizumab to treat EGPA,79 which provided some mechanistic insight into the question of whether eosinophils truly play a role in the pathogenesis of a complex multisystem vasculitic disorder. At a minimum, withdrawal of corticosteroids did not result in worsening of vasculitis as has occurred with various other steroidsparing medications used for asthma.123 Although not yet approved for use in eosinophilic disorders other than asthma and EGPA, there is strong evidence that mepolizumab (and other biologics targeting the IL-5 axis) have clinically beneficial activity in HES, lymphoid HES, and perhaps EGID.122,124 Since their cost and the paucity of available data, including uncertainty on optimal dosing, are likely to make off-label use in these disorders difficult, the importance of well-designed clinical trials, especially in rare EAD, remains a priority. Moreover, as the number of approved targeted agents increases, direct comparisons between the different agents and the safety and efficacy of combination therapies will be essential to create optimized outcomes based recommendations to help guide physicians.
It is hoped that this RE-TREAD report will once again help to keep the field of basic eosinophil immunobiology and translational efforts aimed at EADs focused on common goals that will benefit patients. Outlined in this document are specific recommendations addressing a wide range of research issues that can directly or indirectly impact the development of new therapeutic modalities and their implementation for the diagnosis and treatment of patients with EADs. Although great strides have been made since the 2012 TREAD report, significant unmet needs remain.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the International Eosinophil Society, Kris Heijnen, and Kate Filipiak for organizing the workshop, the workshop attendees, and prior TREAD participant Dr. Wendy Book for her thoughtful discussions.
FUNDING STATEMENT
This work was funded in part by the Division of Intramural Research, NIAID, NIH (P.K., A.D.K.); NIH K08 HL116429 and UG1 HL139117 (P.A.); in part by grants from the FDA (R01FD004086), NIH (R21HL118588), APFED (HOPE Grant program), and University of Illinois (Chancellors Innovation Fund-Proof of Concept Award) (S.J.A.); R01 AI072265, P01 HL107151, and R01 AI105839 (B.S.B.); NIH grant R01AI130033 (P.C.F.); NIH P01 HL088594, NIH R21 AI122103 (S.K.M.); NIH U19 AI070235, NIH R01 AI124355, R37 A1045898, NIH K24DK100303, NIAID-U-01 AI097073, the Campaign Urging Research for Eosinophilic Disease (CURED) Foundation, the Buckeye Foundation, and the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning. The study is also funded by U54 AI117804, which is part of the Rare Disease Clinical Research Network (RDCRN), an initiative of the ORDR, NCATS, and is funded through collaboration between NCATS, NIAID, and NIDDK, as well as the patient advocacy groups American Partnership for Eosinophilic Disorders (APFED), CURED, and the Eosinophilic Family Coalition (EFC), which have collectively resulted in the CEGIR (M.E.R.); AI132840-01A1, HL065228, RAR061567A, HL124165(EAJ); the Belgian National Fund for Scientific Research (grant F 5/4/150/4)(F.R.); Israel Science Foundation grant (ISF 472/15) (F.L.S.); AI121186 (L.A.S.); the NIHR Leicester Biomedical Research Centre, and the views expressed are those of the author and not necessarily those of the National Institute for Health Research or the Department of Health (A.J.W.); NIAID-U-01 AI097073 (M.E.W.); NIAID, R37020241 (P.F.W.).
Abbreviations:
- AEC
absolute eosinophil count
- CEGIR
Consortium of Eosinophilic Gastrointestinal Researchers
- CRISPR
clustered regularly interspaced short palindromic repeat
- EAD
eosinophil-associated diseases
- EC
eosinophilic colitis
- EG
eosinophilic gastritis
- EGID
eosinophilic gastrointestinal disorders
- EGPA
eosinophilic granulomatosis with polyangiitis
- EMH
extramedullary hematopoiesis
- EMR-1
EGF-like module-containing mucin-like hormone receptor-like 1
- EoE
eosinophilic esophagitis
- EoP
eosinophil progenitor
- HES
hypereosinophilic syndromes
- HPF
high power field
- NCATS
National Center for Advancing Translational Science
- NIAID
National Institute of Allergic and Infectious Diseases
- NIH
National Institute of Health
- ORDR
Office of Rare Diseases Research
- PAG
Patient Advocacy Group
- PRO
patient reported outcome
- QOL
quality of life
- RDCRN
Rare Disease Clinical Research Network
- TREAD
Taskforce on the Research of Eosinophil-Associated Diseases
- TRND
therapeutics for rare and neglected diseases
Footnotes
DISCLOSURE
P.A. has received honoraria for advisory boards for G.S.K. and AstraZeneca, receives royalties from UpToDate™, and has consulted for Ambrx and Advance Medical. J.R.A. is an employee of Genentech, Inc., holds stock/stock options in Roche, and is a named inventor on patent applications related to biomarkers of eosinophilic disorders. S.J.A. is a co-inventor of the Esophageal String Test (EST) for EoE and co-founder of EnteroTrack, LLC that is developing the EST for clinical use, and is the CSO and a paid consultant for EnteroTrack. He has had recent consulting agreements with and received honoraria from Receptos (Celgene) and Knopp Biosciences, and is a member of the APFED Medical Advisory Panel. B.S.B. receives research funding from the National Institutes of Health and Acerta Pharma. He has current or recent consulting or scientific advisory board arrangements with, or has received honoraria from Sanofi-Aventis, TEVA, GSK, AstraZeneca, and Allakos, Inc., and owns stock in Allakos. He receives publication-related royalty payments from Elsevier and Wolters Kluwer, and is a co-inventor on existing Siglec-8-related patents and thus may be entitled to a share of royalties received by Johns Hopkins University on the potential sales of such products. B.S.B. is also a co-founder of Allakos, Inc. which makes him subject to certain restrictions under University policy. The terms of this arrangement are being managed by the Johns Hopkins University and Northwestern University in accordance with their conflict of interest policies. MHC is a consultant for Shire, Regeneron, Receptos; has received research funding from Shire, Regeneron, Receptos; is Chair for a Medical Advisory Panel, American Partnership for Eosinophilic Disorders (APFED); and is Member of The International Gastrointestinal Eosinophil Researchers (TIGERS). P.C.F. has received grants from the National Institutes of Health, has served as a consultant for Genentech, Inc., and has received research funding from Knopp Biosciences, LLC. J.E.K. has received honoraria for advisory boards from GSK. G.J.G. and K.M.L. (spouses) receive royalties or have royalty sharing agreements with Ception, Cephalon, Teva, Mayo Foundation, and UpToDate™, have consulted for GSK and Knopp, have received grants from AstraZeneca, GSK, and NIH, and serve on the board of APFED and the Beiersdorf Dermatology Advisory Panel. S.K.M. has served on advisory boards for Astra-Zeneca. C.P. is an employee of Knopp Biosciences and has equity interest in the company.
M.E.R. is a consultant for Pulm One, Spoon Guru, Celgene, Shire, Astra Zeneca, GSK, Allakos, Adare, Regeneron, and Novartis, and has an equity interest in the first 3 listed and immune pharmaceuticals, and royalties from Reslizumab (Teva Pharmaceuticals). M.E.R. is an inventor of patents, owned by Cincinnati Children’s. F.R. has received honoraria for serving on advisory boards from GSK and Knopp. H.U.S. has received honorarium for serving on a scientific advisory board for Knopp. J.S. is a full-time employee and shareholder of GSK. A.J.W. has received honorariums for advisory boards from GSK, Astra Zeneca, TEVA, and Knopp. M.E.W. has received honoraria from Astra-Zeneca, Teva, GSK, Boehringer Ingelheim, Novartis, Boston Scientific, Sanofi, and Regeneron. P.F.W. has received honorariums for advisory boards from GSK and Knopp.
P.K., A.D.K., E.A.J., M.M., K.S., L.S., F.L.S., and R.G.S. have no conflicts of interest to disclose.
AUTHORSHIP
P.K., P.A., and A.K. wrote and edited the manuscript. All listed authors were either participants, speakers, or key experts in the development of this manuscript and have critically evaluated, edited (B.S.B., S.J.A.), and/or written parts of this review.
SUPPORTING INFORMATION
Additional information may be found online in the supporting information tab for this article.
REFERENCES
- 1.Bochner BS, Book W, Busse WW, et al. Workshop report from the National Institutes of Health Taskforce on the Research Needs of Eosinophil-Associated Diseases (TREAD). J Allergy Clin Immunol. 2012;130:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dellon ES, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Inter-and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia. Dig Dis Sci. 2010;55:1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stucke EM, Clarridge KE, Collins MH, Henderson CJ, Martin LJ, Rothenberg ME. Value of an additional review for eosinophil quantification in esophageal biopsies. J Pediatr Gastroenterol Nutr. 2015;61:65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins MH, Martin LJ, Alexander ES, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Protheroe C, Woodruff SA, de Petris G, et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:749–755.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moy AP, Maryamchik E, Nikolskaia OV, Nazarian RM. Th1-and Th17-polarized immune infiltrates in eosinophilic fasciitis—a potential marker for histopathologic distinction from morphea. J Cutan Pathol. 2017;44:548–552. [DOI] [PubMed] [Google Scholar]
- 7.Katzka DA, Geno DM, Ravi A, et al. Accuracy, safety, and tolerability of tissue collection by cytosponge vs endoscopy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:77–83.e2. [DOI] [PubMed] [Google Scholar]
- 8.Katzka DA, Smyrk TC, Alexander JA, et al. Accuracy and safety of the cytosponge for assessing histologic activity in eosinophilic esophagitis: a two-center study. Am J Gastroenterol. 2017;112:1538–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuta GT, Kagalwalla AF, Lee JJ, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62:1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller PF, Spencer LA. Functions of tissue-resident eosinophils. Nat Rev Immunol. 2017;17:746–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JJ, Jacobsen EA, Ochkur SI, et al. Human versus mouse eosinophils: “that which we call an eosinophil, by any other name would stain as red”. J Allergy Clin Immunol. 2012;130:572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen EA, Lesuer WE, Willetts L, et al. Eosinophil activities modulate the immune/inflammatory character of allergic respiratory responses in mice. Allergy. 2014;69:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopf M, Brombacher F, Hodgkin PD, et al. IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 1996;4:15–24. [DOI] [PubMed] [Google Scholar]
- 14.Borchers MT, Crosby J, Justice P, et al. Intrinsic AHR in IL-5 transgenic mice is dependent on CD4(+) cells and CD49d-mediated signaling. Am J Physiol Lung Cell Mol Physiol. 2001;281:L653–L659. [DOI] [PubMed] [Google Scholar]
- 15.Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol. 1997;158:1332–1344. [PubMed] [Google Scholar]
- 16.Hitoshi Y, Yamaguchi N, Korenaga M, Mita S, Tominaga A, Takatsu K. In vivo administration of antibody to murine IL-5 receptor inhibits eosinophilia of IL-5 transgenic mice. Int Immunol. 1991;3:135–139. [DOI] [PubMed] [Google Scholar]
- 17.Chu VT, Fröhlich A, Steinhauser G, et al. Eosinophils are required for the maintenance of plasma cells in the bone marrow. Nat Immunol. 2011;12:151–159. [DOI] [PubMed] [Google Scholar]
- 18.Jung Y, Wen T, Mingler MK, et al. IL-1β in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal Immunol. 2015;8:930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg HF, Masterson JC, Furuta GT. Eosinophils, probiotics, and the microbiome. J Leukoc Biol. 2016;100:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Molofsky AB, Liang H-E, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heredia JE, Mukundan L, Chen FM, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goh YPS, Henderson NC, Heredia JE, et al. Eosinophils secrete IL-4 to facilitate liver regeneration. Proc Natl Acad Sci USA. 2013;110: 9914–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mesnil C, Raulier S, Paulissen G, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126:3279–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle AD, Jacobsen EA, Ochkur SI, et al. Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J Leukoc Biol. 2013;94:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen EA, Lee NA, Lee JJ. Re-defining the unique roles for eosinophils in allergic respiratory inflammation. Clin Exp Allergy. 2014;44:1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdala Valencia H, Loffredo LF, Misharin AV, Berdnikovs S. Phenotypic plasticity and targeting of Siglec-F(high) CD11c(low) eosinophils to the airway in a murine model of asthma. Allergy. 2016;71:267–271. [DOI] [PubMed] [Google Scholar]
- 28.Johansson MW. Eosinophil activation status in separate compartments and association with asthma. Front Med (Lausanne). 2017;4:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen EA, Ochkur SI, Pero RS, et al. Allergic pulmonary inflammation in mice is dependent on eosinophil-induced recruitment of effector T cells. J Exp Med. 2008;205:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen EA, Zellner KR, Colbert D, Lee NA, Lee JJ. Eosinophils regulate dendritic cells and Th2 pulmonary immune responses following allergen provocation. J Immunol. 2011;187:6059–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolarski B, Kurowska-Stolarska M, Kewin P, Xu D, Liew FY. IL-33 exacerbates eosinophil-mediated airway inflammation. J Immunol. 2010;185:3472–3480. [DOI] [PubMed] [Google Scholar]
- 32.Foster EL, Simpson EL, Fredrikson LJ, et al. Eosinophils increase neuron branching in human and murine skin and in vitro. PLoS One. 2011;6:e22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carretero R, Sektioglu IM, Garbi N, Salgado OC, Beckhove P, Häm-merling GJ. Eosinophils orchestrate cancer rejection by normalizing tumor vessels and enhancing infiltration of CD8(+) T cells. Nat Immunol. 2015;16:609–617. [DOI] [PubMed] [Google Scholar]
- 34.Samarasinghe AE, Melo RCN, Duan S, et al. Eosinophils promote antiviral immunity in mice infected with influenza A virus. J Immunol. 2017;198:3214–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kritas SK, Saggini A, Cerulli G, et al. Neuropeptide NGF mediates neuro-immune response and inflammation through mast cell activation. J Biol Regul Homeost Agents. 2014;28:177–181. [PubMed] [Google Scholar]
- 36.Fujisawa D, Kashiwakura J-I, Kita H, et al. Expression of Mas-related gene X2 on mast cells is upregulated in the skin of patients with severe chronic urticaria. J Allergy Clin Immunol. 2014;134:622–633.e9. [DOI] [PubMed] [Google Scholar]
- 37.Jacobsen EA, Ochkur SI, Doyle AD, et al. Lung pathologies in a chronic inflammation mouse model are independent of eosinophil degranulation. Am J Respir Crit Care Med. 2017;195:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kottyan LC, Davis BP, Sherrill JD, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya KR, Ackerman SJ. Eosinophil granule proteins: form and function. J Biol Chem. 2014;289:17406–17415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bryce PJ, Falahati R, Kenney LL, et al. Humanized mouse model of mast cell-mediated passive cutaneous anaphylaxis and passive systemic anaphylaxis. J Allergy Clin Immunol. 2016;138:769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntosh BE, Brown ME, Duffin BM, et al. Nonirradiated NOD,B6.SCID Il2rγ−/− Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Rep. 2015;4:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bal SM, Bernink JH, Nagasawa M, et al. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol. 2016;17:636–645. [DOI] [PubMed] [Google Scholar]
- 43.Elishmereni M, Bachelet I, Nissim Ben-Efraim AH, Mankuta D, Levi-Schaffer F. Interacting mast cells and eosinophils acquire an enhanced activation state in vitro. Allergy. 2013;68:171–179. [DOI] [PubMed] [Google Scholar]
- 44.Jacobsen EA, Taranova AG, Lee NA, Lee JJ. Eosinophils: singularly destructive effector cells or purveyors of immunoregulation. J Allergy Clin Immunol. 2007;119:1313–1320. [DOI] [PubMed] [Google Scholar]
- 45.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy. 2010;40:563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akuthota P, Wang HB, Spencer LA, Weller PF. Immunoregulatory roles of eosinophils: a new look at a familiar cell. Clin Exp Allergy. 2008;38:1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BP, Rothenberg ME. Eosinophils and cancer. Cancer Immunol Res. 2014;2:1–8. [DOI] [PubMed] [Google Scholar]
- 48.Abdala-Valencia H, Coden ME, Chiarella SE, et al. Shaping eosinophil identity in the tissue contexts of development, homeostasis, and disease. J Leukoc Biol. 2018;104:95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landolina N, Gangwar RS, Levi-Schaffer F. Mast cells’ integrated actions with eosinophils and fibroblasts in allergic inflammation: implications for therapy. Adv Immunol. 2015;125:41–85. [DOI] [PubMed] [Google Scholar]
- 50.Bochner BS. Eosinophil interactions with extracellular matrix proteins: effects on eosinophil function and cytokine production Adhesion Molecules in Allergic Disease. 1st ed. Boca Raton, FL: CRC Press; 1997:187–200. [Google Scholar]
- 51.Kita H, Horie S, Gleich GJ. Extracellular matrix proteins attenuate activation and degranulation of stimulated eosinophils. J Immunol. 1996;156:1174–1181. [PubMed] [Google Scholar]
- 52.Seminario MC, Bochner BS. Expression and function of beta 1 integrins on human eosinophils. Mem Inst Oswaldo Cruz. 1997;92:157–164. [DOI] [PubMed] [Google Scholar]
- 53.Kiwamoto T, Katoh T, Evans CM, et al. Endogenous airway mucins carry glycans that bind Siglec-F and induce eosinophil apoptosis. J Allergy Clin Immunol. 2015;135:1329–1340.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll DJ, O’Sullivan JA, Nix DB, Cao Y, Tiemeyer M, Bochner BS. Sialic acid-binding immunoglobulin-like lectin 8 (Siglec-8) is an activating receptor mediating β2-integrin-dependent function in human eosinophils. J Allergy Clin Immunol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neves JS, Perez SAC, Spencer LA, et al. Eosinophil granules function extracellularly as receptor-mediated secretory organelles. Proc Natl Acad Sci USA. 2008;105:18478–18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radonjic-Hoesli S, Wang X, de Graauw E, et al. Adhesion-induced eosinophil cytolysis requires the receptor-interacting protein kinase 3 (RIPK3)-mixed lineage kinase-like (MLKL) signaling pathway, which is counterregulated by autophagy. J Allergy Clin Immunol. 2017;140:1632–1642. [DOI] [PubMed] [Google Scholar]
- 57.ZhuX Hogan SP, Molkentin JD, Zimmermann N. Cyclophilin D regulates necrosis, but not apoptosis, of murine eosinophils. Am J Physiol Gastrointest Liver Physiol. 2016;310:G609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueki S, Melo RCN, Ghiran I, Spencer LA, Dvorak AM, Weller PF. Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood. 2013;121:2074–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Germic N, Stojkov D, Oberson K, Yousefi S, Simon H-U. Neither eosinophils nor neutrophils require ATG5-dependent autophagy for extracellular DNA trap formation. Immunology. 2017;152:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yousefi S, Gold JA, Andina N, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. [DOI] [PubMed] [Google Scholar]
- 61.Sehmi R, Smith SG, Kjarsgaard M, et al. Role of local eosinophilopoietic processes in the development of airway eosinophilia in prednisone-dependent severe asthma. Clin Exp Allergy. 2016;46:793–802. [DOI] [PubMed] [Google Scholar]
- 62.Hui CCK, McNagny KM, Denburg JA, Siracusa MC. In situ hematopoiesis: a regulator of TH2 cytokine-mediated immunity and inflammation at mucosal surfaces. Mucosal Immunol. 2015;8:701–711. [DOI] [PubMed] [Google Scholar]
- 63.Salter BM, Sehmi R. Hematopoietic processes in eosinophilic asthma. Chest. 2017;152:410–416. [DOI] [PubMed] [Google Scholar]
- 64.Esnault S, Kelly EA. Essential mechanisms of differential activation of eosinophils by IL-3 compared to GM-CSF and IL-5. Crit Rev Immunol. 2016;36:429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roth N, Städler S, Lemann M, Hösli S, Simon HU, Simon D. Distinct eosinophil cytokine expression patterns in skin diseases the possible existence of functionally different eosinophil subpopulations. Allergy. 2011;66:1477–1486. [DOI] [PubMed] [Google Scholar]
- 66.Wen T, Mingler MK, Blanchard C, Wahl B, Pabst O, Rothenberg ME. The pan-B cell marker CD22 is expressed on gastrointestinal eosinophils and negatively regulates tissue eosinophilia. J Immunol. 2012;188:1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buonomo EL, Cowardin CA, Wilson MG, Saleh MM, Pramoonjago P, Petri WA. Microbiota-regulated IL-25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Rep. 2016;16:432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farahi N, Loutsios C, Peters AM, Condliffe AM, Chilvers ER. Use of technetium-99m-labeled eosinophils to detect active eosinophilic inflammation in humans. Am J Respir Crit Care Med. 2013;188:880–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farahi N, Loutsios C, Simmonds RP, et al. Measurement of eosinophil kinetics in healthy volunteers. Methods Mol Biol. 2014;1178:165–176. [DOI] [PubMed] [Google Scholar]
- 70.Loutsios C, Farahi N, Simmonds R, et al. Clinical application of autologous technetium-99m-labelled eosinophils to detect focal eosinophilic inflammation in the lung. Thorax. 2015;70:1085–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng K, Gupta SK, Kantor S, et al. Creating a multi-center rare disease consortium—the Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). Transl Sci Rare Dis. 2017;2:141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lefèvre G, Copin M-C, Roumier C, et al. , French Eosinophil Network. CD3-CD4+ lymphoid variant of hypereosinophilic syndrome: nodal and extranodal histopathological and immunophenotypic features of a peripheral indolent clonal T-cell lymphoproliferative disorder. Haematologica. 2015;100:1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Legrand F, Renneville A, Macintyre E, et al. , on behalf of the French Eosinophil Network. The spectrum of FIP1L1-PDGFRA-associated chronic eosinophilic leukemia: new insights based on a survey of 44 cases. Medicine. 2013;92 DOI: 10.1097/MD.0b013e3182a71eba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juniper EF, O’Byrne PM, Guyatt GH, Ferrie PJ, King DR. Development and validation of a questionnaire to measure asthma control. Eur Respir J. 1999;14:902–907. [DOI] [PubMed] [Google Scholar]
- 75.Juniper EF, Guyatt GH, Willan A, Griffith LE. Determining a minimal important change in a disease-specific quality of life questionnaire. J Clin Epidemiol. 1994;47:81–87. [DOI] [PubMed] [Google Scholar]
- 76.Franciosi JP, Hommel KA, Bendo CB, et al. PedsQL eosinophilic esophagitis module: feasibility, reliability, and validity. J Pediatr Gastroenterol Nutr. 2013;57:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Safroneeva E, Coslovsky M, Kuehni CE, et al. , International EEsAI Study Group. Eosinophilic oesophagitis: relationship of quality of life with clinical, endoscopic and histological activity. Aliment Pharmacol Ther. 2015;42:1000–1010. [DOI] [PubMed] [Google Scholar]
- 78.Martin LJ, Franciosi JP, Collins MH, et al. Pediatric eosinophilic esophagitis symptom scores (PEESS v2.0) identify histologic and molecular correlates of the key clinical features of disease. J Allergy Clin Immunol. 2015;135:1519–1528.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wechsler ME, Akuthota P, Jayne D, et al. , EGPA Mepolizumab Study Team. Mepolizumab or placebo for eosinophilic granulomatosis with polyangiitis. N Engl J Med. 2017;376:1921–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rothenberg ME, Klion AD, Roufosse FE, et al. , Mepolizumab HES Study Group. Treatment of patients with the hypereosinophilic syndrome with mepolizumab. N Engl J Med. 2008;358:1215–1228. [DOI] [PubMed] [Google Scholar]
- 81.Straumann A, Conus S, Grzonka P, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59:21–30. [DOI] [PubMed] [Google Scholar]
- 82.Spergel JM, Rothenberg ME, Collins MH,et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a doubleblind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2012;129:456–463,463.e1. [DOI] [PubMed] [Google Scholar]
- 83.Assa’ad AH, Gupta SK, Collins MH, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141:1593–1604. [DOI] [PubMed] [Google Scholar]
- 84.Ochkur SI, Kim JD, Protheroe CA, et al. A sensitive high throughput ELISA for human eosinophil peroxidase: a specific assay to quantify eosinophil degranulation from patient-derived sources. J Immunol Methods. 2012;384:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nair P, Ochkur SI, Protheroe C, et al. Eosinophil peroxidase in sputum represents a unique biomarker of airway eosinophilia. Allergy. 2013;68:1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doran E, Cai F, Holweg CTJ, Wong K, Brumm J, Arron JR. Interleukin-13 in asthma and other eosinophilic disorders. Front Med (Lausanne). 2017;4:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Lavareille A, Roufosse F,Schmid-Grendelmeier P,et al. High serum thymus and activation-regulated chemokine levels in the lymphocytic variant of the hypereosinophilic syndrome. J Allergy Clin Immunol. 2002;110:476–479. [DOI] [PubMed] [Google Scholar]
- 88.Khoury P, Makiya M, Klion AD. Clinical and biological markers in hypereosinophilic syndromes. Front Med (Lausanne). 2017;4:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bleecker ER, FitzGerald JM, Chanez P, et al. , SIROCCO study investigators. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high-dosage inhaled corticosteroids and long-acting β2-agonists (SIROCCO): a randomised, multicentre, placebo-controlled phase 3 trial. The Lancet. 2016;388:2115–2127. [DOI] [PubMed] [Google Scholar]
- 90.Park H-S, Kim M-K, Imai N, et al. , Asian Benralizumab Study Group. A phase 2a study of benralizumab for patients with eosinophilic asthma in South Korea and Japan. Int Arch Allergy Immunol. 2016;169:135–145. [DOI] [PubMed] [Google Scholar]
- 91.Pavord ID, Korn S, Howarth P, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. 2012;380:651–659. [DOI] [PubMed] [Google Scholar]
- 92.Nair P, Wenzel S, Rabe KF, et al. , ZONDA Trial Investigators. Oral glucocorticoid-sparing effect of benralizumab in severe asthma. N Engl J Med. 2017;376:2448–2458. [DOI] [PubMed] [Google Scholar]
- 93.Dworetzky SI, Hebrank GT, Archibald DG, Reynolds IJ, Farwell W, Bozik ME. The targeted eosinophil-lowering effects of dexpramipexole in clinical studies. Blood Cells Mol Dis. 2017;63:62–65. [DOI] [PubMed] [Google Scholar]
- 94.Prussin C, Laidlaw TM, Panettieri RA, et al. Dexpramipexole effectively lowers blood and tissue eosinophils in subjects with chronic rhinosinusitis with nasal polyps. J Allergy Cli Immunol. 2017;139. [Google Scholar]
- 95.Panch SR, Bozik M, Brown T, et al. Dexpramipexole as a steroidsparing agent in hypereosinophilic syndromes (HES): an open-label proof-of-concept study. Blood. 2016;128:1327. [Google Scholar]
- 96.Corren J, Parnes JR, Wang L, et al. Tezepelumab in adults with uncontrolled asthma. N Engl J Med. 2017;377:936–946. [DOI] [PubMed] [Google Scholar]
- 97.Wenzel S, Ford L, Pearlman D, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368:2455–2466. [DOI] [PubMed] [Google Scholar]
- 98.Beck LA, Thaçi D, Hamilton JD, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014;371:130–139. [DOI] [PubMed] [Google Scholar]
- 99.Ruzicka T, Hanifin JM, Furue M, et al. , XCIMA Study Group. Anti-Interleukin-31 receptor A antibody for atopic dermatitis. N Engl J Med. 2017;376:826–835. [DOI] [PubMed] [Google Scholar]
- 100.Bachert C, Mannent L, Naclerio RM, et al. Effect of subcutaneous dupilumab on nasal polyp burden in patients with chronic sinusitis and nasal polyposis: a randomized clinical trial. JAMA. 2016;315:469–479. [DOI] [PubMed] [Google Scholar]
- 101.Hirano I, Dellon ES, Hamilton JD, et al. Dupilumab efficacy and safety in adult patients with active eosinophilic esophagitis: a randomized double-blind placebo-controlled phase 2 trial. Am J Gastroenterol. 2017;112(1). AB 20 (ACG 2017). [Google Scholar]
- 102.Ray A, Oriss TB, Wenzel SE. Emerging molecular phenotypes of asthma. Am J Physiol Lung Cell Mol Physiol. 2015;308:L130–L140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kottyan LC, Weirauch MT, Rothenberg ME. Making it big in allergy. J Allergy Clin Immunol. 2015;135:43–45. [DOI] [PubMed] [Google Scholar]
- 104.Wilkerson EM, Johansson MW, Hebert AS, et al. The peripheral blood eosinophil proteome. J Proteome Res. 2016;15:1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soman KV, Stafford SJ, Pazdrak K, et al. Activation of human peripheral blood eosinophils by cytokines in a comparative time-course proteomic/phosphoproteomic study. J Proteome Res. 2017;16:2663–2679. [DOI] [PubMed] [Google Scholar]
- 106.Esnault S, Kelly EA, Schwantes EA, et al. Identification of genes expressed by human airway eosinophils after an in vivo allergen challenge. PLoS One. 2013;8:e67560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shen Z-J, Hu J, Esnault S, Dozmorov I, Malter JS. RNA Seq profiling reveals a novel expression pattern of TGF-β target genes in human blood eosinophils. Immunol Lett. 2015;167:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.O’Sullivan JA, Carroll DJ, Bochner BS. Glycobiology of eosinophilic inflammation: contributions of Siglecs, glycans, and other glycan-binding proteins. Front Med (Lausanne). 2017;4:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li G, Dzilic E, Flores N,Shieh A, Wu SM. Strategies for the acquisition of transcriptional and epigenetic information in single cells. J Thorac Dis. 2017;9:S9–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiao L, Guo J. Multiplexed single-cell in situ RNA analysis by reiterative hybridization. Anal Methods. 2015;7:7290–7295. [Google Scholar]
- 112.Breschi A, Gingeras TR, Guigó R. Comparative transcriptomics in human and mouse. Nat Rev Genet. 2017;18:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vanden Bempt M, Demeyer S, Mentens N, et al. Generation of the Fip1l1-Pdgfra fusion gene using CRISPR/Cas genome editing. Leukemia. 2016;30:1913–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. [DOI] [PubMed] [Google Scholar]
- 115.Bouffi C, Kartashov AV, Schollaert KL, et al. Transcription factor repertoire of homeostatic eosinophilopoiesis. J Immunol. 2015;195:2683–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andersen CL, Nielsen HM, Kristensen LS, et al. Whole-exome sequencing and genome-wide methylation analyses identify novel disease associated mutations and methylation patterns in idiopathic hypereosinophilic syndrome. Oncotarget. 2015;6:40588–40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rothenberg ME, Wen T, Greenberg A, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135:500–507. [DOI] [PubMed] [Google Scholar]
- 119.Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–1214. [DOI] [PubMed] [Google Scholar]
- 120.Khoury P, Desmond R, Pabon A, et al. Clinical features predict respon siveness to imatinib in platelet-derived growth factor receptor-alpha-negative hypereosinophilic syndrome. Allergy. 2016;71:803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ogbogu PU, Bochner BS, Butterfield JH, et al. Hypereosinophilic syndrome: a multicenter, retrospective analysis of clinical characteristics and response to therapy. J Allergy Clin Immunol. 2009;124:1319–1325.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bochner BS. Novel therapies for eosinophilic disorders. Immunol Allergy Clin North Am. 2015;35:577–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lilly CM, Churg A, Lazarovich M, et al. Asthma therapies and Churg-Strauss syndrome. J Allergy Clin Immunol. 2002;109:S1–19. [DOI] [PubMed] [Google Scholar]
- 124.Kuang FL, Klion AD. Biologic agents for the treatment of hypereosinophilic syndromes. J Allergy Clin Immunol Pract. 2017;5:1502–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.de Graaf CA, Choi J, Baldwin TM, et al. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Reports. 2016;7:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee JJ, Dimina D, Macias MP, et al. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004;305:1773–1776. [DOI] [PubMed] [Google Scholar]
- 127.Denzler KL, Borchers MT, Crosby JR, et al. Extensive eosinophil degranulation and peroxidase-mediated oxidation of airway proteins do not occur in a mouse ovalbumin-challenge model of pulmonary inflammation. J Immunol. 2001;167:1672–1682. [DOI] [PubMed] [Google Scholar]
- 128.Denzler KL, Farmer SC, Crosby JR, et al. Eosinophil major basic protein-1 does not contribute to allergen-induced airway pathologies in mouse models of asthma. J Immunol. 2000;165:5509–5517. [DOI] [PubMed] [Google Scholar]
- 129.Doyle AD, Jacobsen EA, Ochkur SI, et al. Expression of the secondary granule proteins major basic protein 1 (MBP-1) and eosinophil peroxidase (EPX) is required for eosinophilopoiesis in mice. Blood. 2013;122:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee JJ, McGarry MP, Farmer SC, et al. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med. 1997;185:2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ochkur SI, Jacobsen EA, Protheroe CA, et al. Coexpression of IL-5 and eotaxin-2 in mice creates an eosinophil-dependent model of respiratory inflammation with characteristics of severe asthma. J Immunol. 2007;178:7879–7889. [DOI] [PubMed] [Google Scholar]
- 132.Willetts L, Parker K, Wesselius LJ, et al. Immunodetection of occult eosinophils in lung tissue biopsies may help predict survival in acute lung injury. Respir Res. 2011;12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rank MA, Ochkur SI, Lewis JC, et al. Nasal and pharyngeal eosinophil peroxidase levels in adults with poorly controlled asthma correlate with sputum eosinophilia. Allergy. 2016;71:567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ochkur SI, Kim JD, Protheroe CA, et al. The development of a sensitive and specific ELISA for mouse eosinophil peroxidase: assessment of eosinophil degranulation ex vivo and in models of human disease. J Immunol Methods. 2012;375:138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Makiya MA, Herrick JA, Khoury P, Prussin CP, Nutman TB, Klion AD. Development of a suspension array assay in multiplex for the simultaneous measurement of serum levels of four eosinophil granule proteins. J Immunol Methods. 2014;411:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


