Abstract
Viruses are the most abundant component of the human microbiota. Recent evidence has uncovered a rich diversity of viruses within the female bladder, including both bacteriophages and eukaryotic viruses. We conducted whole-genome sequencing of the bladder microbiome of 30 women: 10 asymptomatic ‘healthy’ women and 20 women with an overactive bladder. These metagenomes include sequences representative of human, bacterial and viral DNA. This analysis, however, focused specifically on viral sequences. Using the bioinformatic tool virMine, we discovered sequence fragments, as well as complete genomes, of bacteriophages and the eukaryotic virus JC polyomavirus. The method employed here is a critical proof of concept: the genomes of viral populations within the low-biomass bladder microbiota can be reconstructed through whole-genome sequencing of the entire microbial community.
Keywords: urinary microbiome, urinary virome, bacteriophage, JC polyomavirus
Full-Text
The old paradigm that the bladder is sterile results from the use of standard urine culture-dependent methods that are optimized for Escherichia coli [1, 2]. However, there is definitive evidence that communities of bacteria exist within the bladder [3–6], as well as for associations between these bladder microbiota and urinary symptom levels, treatment response and urinary tract infection (UTI) risk [7–15]. Furthermore, the bladder microbiota of individuals both with and without urinary symptoms include viral species. The viruses isolated from urine include several that infect human cells [16–22], as well as those that infect bacteria [bacteriophages (phages)] [23–25]. Metagenomic sequencing of the urinary virome, which detects eukaryotic viruses and phages in the lytic cycle, revealed an abundance of phages [26, 27].
Because the bladder microbiota exist at a substantially lower biomass [1, 5, 6] than many other human niches (e.g. the gut [28]), sequencing the bladder’s virome presents unique technical difficulties. From the gut, the viral biomass can be separated and the extracted DNA can be sequenced directly [29, 30]. In contrast, previous urine virome metagenomic studies have relied on DNA amplification prior to sequencing to increase DNA concentrations [26]. These amplification methods, however, have well-documented biases [31]. As such, the complete diversity of the virome may not be captured. Alternatively, we hypothesized that the challenges of sequencing the bladder virome could be overcome bioinformatically. Bioinformatic approaches have successfully identified complete viral genomes from bacterial metagenomes (e.g. [32]). Moreover, complete viral genomes have been reconstructed from viral metagenomes containing significant quantities of non-viral (bacterial and eukaryotic) DNA (e.g. [33]). Thus, we conducted whole-genome sequencing of the bladder microbiota and examined the sequence data specifically for viral sequences. This approach has the potential to capture both lytic and lysogenic phage sequences present in the community.
In a previously published study [10], urine was collected aseptically via transurethral catheter from 10 women without urinary symptoms (control) and 20 women with reported overactive bladder symptoms (OAB) and stored with the DNA preservative AssayAssure (Sierra Molecular) at −80 °C. In the current study, 5 ml of each urine sample was thawed and the DNA was extracted, as described previously [10, 34]. Briefly, the urine was incubated in a lysis solution containing mutanolysin and lysozyme and the DNA extracted from the sample using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. The Illumina Nextera kit was used for whole-genome library preparation with fragment sizes of 200–300 bp. Sequencing was conducted on the Illumina HiSeq 2500 platform, producing paired-end 100 bp ×2 reads. Human contaminating reads were filtered out by mapping to the human reference genome (hg19) with Bowtie2 [35]. Table S1 (available in the online version of this article) lists the number of raw reads and filtered reads for each sequenced patient sample. Most of the reads produced represent bacterial and viral species; on average, only 5.3 % of the reads mapped to the human reference genome sequence. Raw sequencing data are available from the NCBI’s Sequence Read Archive (SRA) database, BioProject accession number PRJEB8104. The accession numbers for each sample are listed in Table S1.
Fig. S1 outlines the analytical process. Each individual metagenome dataset was assembled separately. The raw reads were first trimmed for quality using the tool Sickle [36] and then assembled by SPAdes (v3.10.1) with the ‘meta’ (metagenomic) option [37]. There was only a weak correlation between the number of reads produced for a given sample and the number of contigs assembled from those reads (r=0.23). Next, the virMine [38] tool was used to classify the contigs produced. Briefly, virMine first filtered out contigs that were less than 1000 bp in length; this length is a user-defined parameter and was selected to eliminate partial gene sequences and repetitive elements from downstream analyses. For the remaining contigs, open reading frames were predicted, translated and compared to virMine’s bacterial and viral protein sequence databases (RefSeq protein sequences). These comparisons enabled us to classify each contig as bacterial, viral, or unknown (exhibiting no similarity to bacterial or viral contigs). The genome assembly and virMine statistics are listed in Table S1. The microbiomes were dominated by bacterial contigs (90 % on average). The contigs classified as ‘unknown’ were queried against the NCBI nr/nt database via megablast, and we found that the overwhelming majority were human in origin (results not shown). Thus, here we will focus on the 252 contigs from the 30 metagenomes that were predicted to be viral.
Twenty-seven of the 30 bladder metagenomes examined included contigs predicted to be viral. To further evaluate these contigs, each was queried against the nr/nt database via the NCBI web interface using the megablast algorithm (Table S2). In comparing the contigs to this database, eight samples were identified as containing sequences of human origin. The virMine software characterized these contigs as viral, as they did not resemble bacterial sequences and had moderate sequence similarity to a sequence in the viral database. The contigs within another seven samples were uniformly short (~1 kbp) and only exhibited sequence similarity to annotated transposases. Transposases, along with integrases, can be encoded by a phage to allow that phage to enter its lysogenic (latent) life cycle by inserting itself into the bacterial genome (the inserted phage genome is now called the ‘prophage’) [39]. Thus, while these contigs suggest the presence of lysogenic phages within the bladder microbiota, they do not provide an insight into the phage species. The remaining 12 metagenomes, however, had recognizable phage and/or eukaryotic virus sequences.
Two patient samples – OAB045 and OAB052 – contained numerous contigs with homologies to annotated phage genes, including genes annotated as encoding tail proteins, phage tail tape measure proteins, phage DNA packaging proteins, phage portal proteins, terminases and capsid proteins. Furthermore, these contigs represented phage genome fragments, including several coding regions. For instance, in the OAB052 sample, a 4898 bp contig was identified, containing annotated regions for a phage terminase, phage portal protein, endopeptidase Clp, major capsid protein, phage DNA packaging protein and two hypothetical proteins. This contig is homologous to a region within the 18.3 kbp putative prophage (determined via phast [40]) in the Gardnerella vaginalis HMP9231 genome. As such, it is unlikely that the contig identified here represents a complete, intact phage genome. Nevertheless, it may represent a Gardnerella prophage, which we previously showed to be prevalent within Gardnerella strains of the bladder [41]. We next examined the contigs that were classified as bacterial by the virMine tool. blast queries found significant homology (e-score=0) between the larger contigs within the OAB052 metagenome and G. vaginalis genome records in GenBank. Thus, we hypothesize that the larger viral contigs detected within the OAB052 patient sample represent lysogenic phages. While here we have presented the analysis of just one of these contigs, similar observations were made for the other contigs from these samples: viral sequences exhibited homologies to annotated prophages within bacterial species that were also found within the sample’s metagenome.
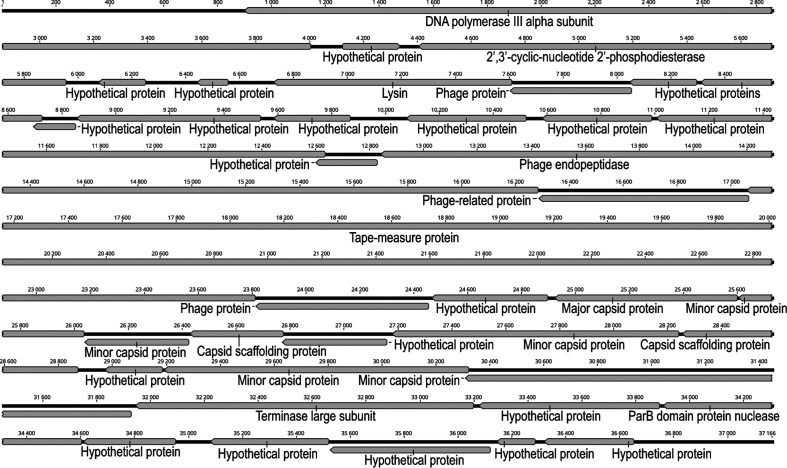
Larger phage sequences were identified in three patient samples – OAB010, OAB018 and OAB039. Table 1 lists the contigs identified in each of these samples. While many of these larger phage sequences include novel genic content (i.e. low or no sequence homology to records in GenBank), each exhibited some homology to recognized prophage sequences within bacterial genomes (per phast [40]). The most similar phage species are listed in Table 1. Based upon the size of the assembled genome and the presence of ‘hallmark’ viral genes [42], we were able to confidently predict the completeness of several of these assembled sequences. The phage sequences listed in Table 1 were then annotated using the RAST server [43] (Table S3). The genome map for the putative complete phage genome sequence within the OAB018 patient sample is shown in Fig. 1 (generated using Geneious, Auckland, NZ). The phage sequences identified here are not necessarily unique to the microbiota of the urinary tract (Table S2). For instance, the sequence of contig 28 from the OAB010 sample is 99 % identical to a prophage found within a Streptococcus agalactiae strain isolated from a patient’s blood sample [44], as well as from a strain isolated from a diseased tilapia (GenBank record CP016501). These larger sequences are informative about both the bioinformatic approach employed here and the samples themselves. First, complete (or near-complete) phage genomes can be reconstructed by sequencing bladder microbiome samples. Second, because we sequenced the bacterial and viral fractions together, it is possible to associate phages and their bacterial host. Last, we found evidence of related phages being present in the bladder microbiota of different patients. For instance, the OAB018 and OAB039 patient samples both contain phage sequences that are similar to those of the Lactobacillus-infecting phages PLE2 and phi adh. These phages were first detected as prophages within the genomes of the probiotic strains Lactobacillus casei BL23 [45] and Lactobacillus gasseri ADH, respectively. Further sequencing of the bladder microbiota is necessary to ascertain whether these phage families are common constituents of the bladder virome.
Table 1. Putative complete/near-complete phage genomes identified within bladder microbiome samples.
The most similar phage sequences were determined using phast [40].
| Sample | Contig # | Length (kbp) | Coverage | Bacterial blast homology (sequence ID/query coverage) | Most similar phage (length) |
|---|---|---|---|---|---|
| OAB010 | 28 | 17.5 | 16.56 | S. agalactiae (99 %/100 %) | phiCT453B (36.7 kbp) |
| 31 | 8.1 | 11.61 | S. agalactiae (95 %/99 %) | phiCT453B (36.7 kbp) | |
| 39 | 3.4 | 14.21 | S. agalactiae (100 %/100 %) | phiARI0923 (33.5 kbp) | |
| OAB018 | 28 | 37.1 | 9.54 | L. helveticus (87 %/71 %) | phig1e (42.3 kbp) |
| 49 | 26.8 | 10.89 | L. helveticus (85 %/15 %) | phig1e (42.3 kbp) | |
| 66 | 17.8 | 7.30 | L. allii (72 %/3 %) | PLE2 (35.1 kbp) | |
| 148 | 7.6 | 6.96 | L. helveticus (76 %/25 %) | phi adh (43.8 kbp) | |
| OAB039 | 55 | 13.6 | 18.08 | L. allii (72 %/4 %) | PLE2 (35.1 kbp) |
| 79 | 8.5 | 23.09 | L. gasseri (67 %/57 %) | phi adh (43.8 kbp) |
Fig. 1.
Genome map for the 37.1 kbp contig 28 from the OAB018 patient sample.
Five patient samples, OAB021, OAB026, OAB032, OAB042 and OAB045, contained recognizable complete genomes for the human polyomavirus JC (JCV). Furthermore, a partial genome sequence, 1023 bp, was retrieved from patient sample OAB025. JCV is a circular double-stranded DNA virus (~5130 bp) and occurs naturally in the urine of healthy individuals. A previous study found that up to 80 % of adults excreted JCV in their urine [46]. Furthermore, JCV quasispecies have been detected in healthy individuals [47]. JCV, however, was not detected within any of the 10 asymptomatic ‘healthy’ individuals (controls) included in this study. While JCV infection has been associated with progressive multifocal leukoencephalopathy, a fatal neurological disorder [48], JCV within individuals with overactive bladder has yet to be studied. The prevalence of JCV within these five samples varied. Raw reads were mapped to the RefSeq for the species (GenBank accession: NC_001699) using Bowtie2 (v 2.2.6) [35], revealing coverage of the JCV genome ranging from 12x to 726.9x. Coverage correlated with the % reads in the sample corresponding to the JCV genome (r2=0.9570). JCV was most abundant in patient samples OAB042 and OAB045, in which 4.4 and 3.2 %, respectively, of the total reads generated were classified as JCV.
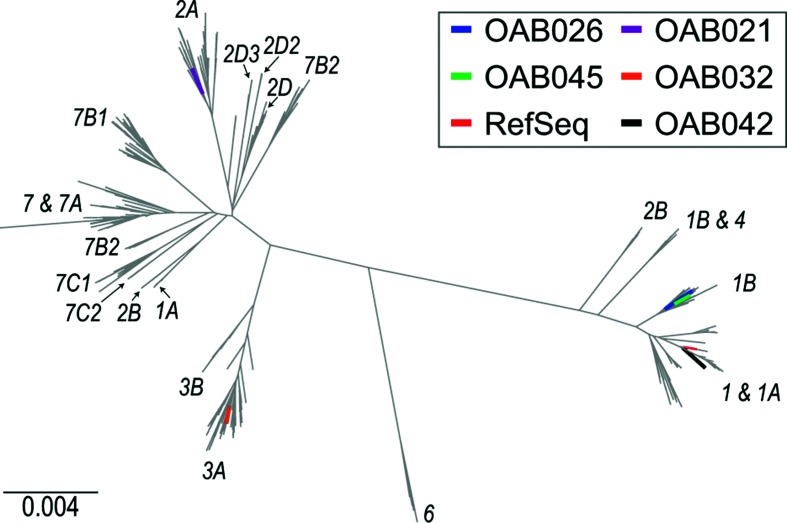
Previous research has identified subtypes of JCV and found that these subtypes can correspond to different human population groups [49]. Thus, we next determined the subtypes of the five JCV complete genomes from the bladder microbiome samples by comparing their genomes to 605 publicly available genomes representative of the diversity of the species (Table S2). The sequences were aligned using muscle through Geneious; the alignments were trimmed, removing the tandem repeats (as their placement at the 5′ or 3′ end of the genome sequence varied among the genome sequence records), and a phylogenetic tree was inferred using FastTree [50] (Fig. 2). Clades were labelled according to their documented genotype, determined from the literature [49] and from GenBank records. Genotype classifications rely on coding sequence variation, most notably the VP1 capsid coding sequence [51]. This tree aids in gaining greater insight into the JCV genomes detected within the patient samples. The JCV strains identified in patient samples OAB026 and OAB045 were representative of subtype 1, genotype 1B (exhibiting the greatest sequence similarity to isolates from individuals of German heritage [49]). The JCV virus from patient sample OAB042 was also categorized as subtype 1 (genotype 1A) via sequence homology [50]. Subtype 1 is relatively common in the United States and Europe [52] and these three patients self-reported as ‘white/non- Hispanic’. The JCV strains identified in patient samples OAB032 and OAB021 were classified as belonging to genotypes 3A (prevalent in Africa and southwestern Asia) and 2A (prevalent amongst individuals of Japanese and Native American decent), respectively, based upon their nearest neighbours and placement within the phylogenetic tree (Fig. 2) [49, 53]. However, the self-reported ethnicities of these patients are incongruent with the ethnicities typically associated with these subtypes; patient OAB032 self-reported as ‘white/Hispanic’ and patient OAB021 self-reported as ‘black/non-Hispanic.’ As the majority of sequencing and genotyping studies of JCV have largely been restricted to individuals with or without neurological diseases, our findings here should prompt further investigation of the presence and genotypes of JCV in individuals with and without lower urinary tract symptoms to ascertain whether JCV plays any role in urinary tract symptoms or disease.
Fig. 2.
Phylogenetic tree for 610 complete genomes of JCV, including strains isolated in this study (tree branches shown in black and labelled) and the reference sequence (NC_001699) for the species (shown in red).
Here, we have shown that challenges in isolating viral species from the low-biomass bladder microbiome can be circumvented via bioinformatic classification tools; whole-genome, as well as partial-genome, sequences can be reconstructed from complex samples. While the sheer size of bacterial genomes lends to greater representation in whole-genome sequencing data, viral genomes were detected without amplification within 27 of the 30 urinary samples examined here. This further supports prior estimates of the abundance of viruses within the bladder microbiota [25, 26]. Moreover, as our results show, our strategy can detect both lysogenic and lytic phages, as well as eukaryotic viruses.
Funding information
This work was supported by the NIH (R01 DK104718 to A. J. W) and NSF (ABI 1149387 and 1661357 to C. P.). A. G. is supported by the Carbon Research Fellowship at Loyola University Chicago and the CREU research fellowship (CRA).
Acknowledgements
For prior patient recruitment, we want to acknowledge the Loyola Urinary Education and Research Collaborative (LUEREC), specifically Mary Tulke RN and Drs Linda Brubaker, Elizabeth Mueller, Cynthia Brincat, Susanne Taege and Tanaka Dune, and the patients who provided the samples for this study.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
The GenBank SRA accession numbers for the 30 metagenomes produced here are as follows: ERR926109 through ERR926123 and ERR926139 through ERR926153, under the BioProject Accession #PRJEB8104.
Three supplementary tables and one supplementary figure are available with the online version of this article.
Abbreviations: JCV, human polyomavirus JC; OAB, overactive bladder.
References
- 1.Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54:1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass EH. Pyelonephritis and bacteriuria. A major problem in preventive medicine. Ann Intern Med. 1962;56:46–53. doi: 10.7326/0003-4819-56-1-46. [DOI] [PubMed] [Google Scholar]
- 3.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50:1376–1383. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012;10:174. doi: 10.1186/1479-5876-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol. 2013;51:2054–2062. doi: 10.1128/JCM.03314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas-White KJ, Kliethermes S, Rickey L, Lukacz ES, Richter HE, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol. 2017;216:55.e1–55.e16. doi: 10.1016/j.ajog.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brubaker L, Nager CW, Richter HE, Visco A, Nygaard I, et al. Urinary bacteria in adult women with urgency urinary incontinence. Int Urogynecol J. 2014;25:1179–1184. doi: 10.1007/s00192-013-2325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. 2015;213:347.e1–347.e11. doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5:e01283-14. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J. 2016;27:723–733. doi: 10.1007/s00192-015-2847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nienhouse V, Gao X, Dong Q, Nelson DE, Toh E, et al. Interplay between bladder microbiota and urinary antimicrobial peptides: mechanisms for human urinary tract infection risk and symptom severity. PLoS One. 2014;9:e114185. doi: 10.1371/journal.pone.0114185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karstens L, Asquith M, Davin S, Stauffer P, Fair D, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol. 2016;6:78. doi: 10.3389/fcimb.2016.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas-White KJ, Lin H, Gao X, Fok C, Mueller ER, et al. Urinary symptoms and associated urinary microbes in urogynecologic surgical patients. 2018. [DOI] [PMC free article] [PubMed]
- 15.Thomas-White KJ, Gao X, Lin H, Fok C, Ghanayem K, et al. Urinary microbes and post-operative urinary tract infection risk in urogynecologic surgical patients. 2018. [DOI] [PMC free article] [PubMed]
- 16.Iwasawa A, Kumamoto Y, Maruta H, Fukushima M, Tsukamoto T, et al. Presence of human papillomavirus 6/11 DNA in condyloma acuminatum of the urinary bladder. Urol Int. 1992;48:235–238. doi: 10.1159/000282342. [DOI] [PubMed] [Google Scholar]
- 17.Echavarria M, Forman M, Ticehurst J, Dumler JS, Charache P. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol. 1998;36:3323–3326. doi: 10.1128/jcm.36.11.3323-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karim RZ, Rose BR, Brammah S, Scolyer RA. Condylomata acuminata of the urinary bladder with HPV 11. Pathology. 2005;37:176–178. doi: 10.1080/00313020500058615. [DOI] [PubMed] [Google Scholar]
- 19.Burián Z, Szabó H, Székely G, Gyurkovits K, Pankovics P, et al. Detection and follow-up of torque teno midi virus ("small anelloviruses") in nasopharyngeal aspirates and three other human body fluids in children. Arch Virol. 2011;156:1537–1541. doi: 10.1007/s00705-011-1021-0. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch HH, Kardas P, Kranz D, Leboeuf C. The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS. 2013;121:685–727. doi: 10.1111/apm.12128. [DOI] [PubMed] [Google Scholar]
- 21.Rinaldo CH, Tylden GD, Sharma BN. The human polyomavirus BK (BKPyV): virological background and clinical implications. APMIS. 2013;121:728–745. doi: 10.1111/apm.12134. [DOI] [PubMed] [Google Scholar]
- 22.Assetta B, Atwood WJ. The biology of JC polyomavirus. Biol Chem. 2017;398:839–855. doi: 10.1515/hsz-2016-0345. [DOI] [PubMed] [Google Scholar]
- 23.Brown-Jaque M, Muniesa M, Navarro F. Bacteriophages in clinical samples can interfere with microbiological diagnostic tools. Sci Rep. 2016;6:33000. doi: 10.1038/srep33000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malki K, Sible E, Cooper A, Garretto A, Bruder K, et al. Seven bacteriophages isolated from the female urinary microbiota. Genome Announc. 2016;4:e01003-16. doi: 10.1128/genomeA.01003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller-Ensminger T, Garretto A, Brenner J, Thomas-White K, Zambom A, et al. Bacteriophages of the urinary microbiome. J Bacteriol. 2018;200:e00738-17. doi: 10.1128/JB.00738-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santiago-Rodriguez TM, Ly M, Bonilla N, Pride DT. The human urine virome in association with urinary tract infections. Front Microbiol. 2015;6:14. doi: 10.3389/fmicb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rani A, Ranjan R, McGee HS, Metwally A, Hajjiri Z, et al. A diverse virome in kidney transplant patients contains multiple viral subtypes with distinct polymorphisms. Sci Rep. 2016;6:33327. doi: 10.1038/srep33327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, et al. Healthy human gut phageome. Proc Natl Acad Sci USA. 2016;113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yilmaz S, Allgaier M, Hugenholtz P. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods. 2010;7:943–944. doi: 10.1038/nmeth1210-943. [DOI] [PubMed] [Google Scholar]
- 32.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mokili JL, Dutilh BE, Lim YW, Schneider BS, Taylor T, et al. Identification of a novel human papillomavirus by metagenomic analysis of samples from patients with febrile respiratory illness. PLoS One. 2013;8:e58404. doi: 10.1371/journal.pone.0058404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi NA, Fass JN. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. 2011 https://github.com/najoshi/sickle Available at.
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garretto A, Hatzopoulos T, Kalesinskas L, Putonti C. virMine: Automated detection and annotation of complete viral genomes from complex metagenomic samples. 2017 doi: 10.7717/peerj.6695. https://github.com/thatzopoulos/virMine Available at. [DOI] [PMC free article] [PubMed]
- 39.Krupovic M, Forterre P. Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann N Y Acad Sci. 2015;1341:41–53. doi: 10.1111/nyas.12675. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malki K, Shapiro JW, Price TK, Hilt EE, Thomas-White K, et al. Genomes of Gardnerella strains reveal an abundance of prophages within the bladder microbiome. PLoS One. 2016;11:e0166757. doi: 10.1371/journal.pone.0166757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koonin EV, Senkevich TG, Dolja VV. The ancient virus world and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teatero S, McGeer A, Li A, Gomes J, Seah C, et al. Population structure and antimicrobial resistance of invasive serotype IV group B Streptococcus, Toronto, Ontario, Canada. Emerg Infect Dis. 2015;21:585–591. doi: 10.3201/eid2014.140759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dieterle ME, Fina Martin J, Durán R, Nemirovsky SI, Sanchez Rivas C, et al. Characterization of prophages containing "evolved" Dit/Tal modules in the genome of Lactobacillus casei BL23. Appl Microbiol Biotechnol. 2016;100:9201–9215. doi: 10.1007/s00253-016-7727-x. [DOI] [PubMed] [Google Scholar]
- 46.Chang H, Wang M, Tsai RT, Lin HS, Huan JS, et al. High incidence of JC viruria in JC-seropositive older individuals. J Neurovirol. 2002;8:447–451. doi: 10.1080/13550280260422758. [DOI] [PubMed] [Google Scholar]
- 47.van Loy T, Thys K, Tritsmans L, Stuyver LJ. Quasispecies analysis of JC virus DNA present in urine of healthy subjects. PLoS One. 2013;8:e70950. doi: 10.1371/journal.pone.0070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khalili K, Gordon J, White MK. The polyomavirus, JCV and its involvement in human disease. Adv Exp Med Biol. 2006;577:274–287. doi: 10.1007/0-387-32957-9_20. [DOI] [PubMed] [Google Scholar]
- 49.Shackelton LA, Rambaut A, Pybus OG, Holmes EC. JC virus evolution and its association with human populations. J Virol. 2006;80:9928–9933. doi: 10.1128/JVI.00441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agostini HT, Ryschkewitsch CF, Stoner GL. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J Clin Microbiol. 1996;34:159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid CE, Li H, Sur G, Carmillo P, Bushnell S, et al. Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J Infect Dis. 2011;204:237–244. doi: 10.1093/infdis/jir256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saruwatari L, Sugimoto C, Kitamura T, Ohno N, Sakai E, et al. Asian domains of four major genotypes of JC virus, Af2, B1-b, CY and SC. Arch Virol. 2002;147:1–10. doi: 10.1007/s705-002-8299-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.