Abstract
Non-invasive human neuroimaging has yielded many exciting discoveries about the brain. Numerous methodological advances have also occurred, though inertia has slowed their adoption. This paper presents an integrated approach to data acquisition, analysis, and sharing that builds upon recent advances, particularly from the Human Connectome Project (HCP). The “HCP-style” paradigm has seven core tenets: (1) collect multimodal imaging data from many subjects; (2) acquire data at high spatial and temporal resolution; (3) preprocess data to minimize distortions, blurring, and temporal artifacts; (4) represent data using the natural geometry of cortical and subcortical structures; (5) accurately align corresponding brain areas across subjects and studies; (6) analyze data using neurobiologically accurate brain parcellations; and (7) share published data via user-friendly databases. We illustrate the HCP-style paradigm using existing HCP datasets and provide guidance for future research. Widespread adoption of this paradigm should accelerate progress in understanding the brain in health and disease.
Introduction
The Human Connectome Project (HCP) began in 2010 with ~$40M awarded by the National Institutes of Health (NIH) to two consortia to develop improved neuroimaging methods and to acquire a dataset of unprecedented size and quality for mapping the normal human macro-scale connectome1,2 that is, the long-distance connections between all of the brain’s areas. Better maps of the brain’s areas and their connections will deepen our understanding of healthy brain function and may improve our ability to understand and treat neurological and psychiatric disorders. The “WU-Minn-Ox” HCP consortium, centered at Washington University, University of Minnesota, and Oxford University, spent its first two years developing the state of the art methods discussed here, and then proceeded to acquire, analyze, and share high quality multimodal neuroimaging, behavioral, and genotype data from 1200 healthy young adult twins and non-twin siblings3,4. The HCP focused primarily on MRI-based imaging modalities to measure brain architecture, function, connectivity, and the regular, topographic, organization of some brain areas; however, magnetoencephalography (MEG) data was also acquired in some subjects (see Box 2). As the HCP’s core activities draw to a close in 2016, its contributions include (i) a set of broadly useful MRI acquisition protocols, consisting of thoroughly tested and optimized pulse sequences and image reconstruction algorithms; (ii) an unprecedented collection of exceptionally high quality, freely shared neuroimaging data; (iii) numerous publicly available neuroimaging software and informatics tools; (iv) a growing number of intriguing discoveries emerging from analyses of HCP data; (v) the emergence of an “HCP-style” paradigm for neuroimaging data acquisition, analysis, and sharing; and (vi) a growing number of HCP-style projects that will study different age ranges and brain disorders.
BOX 2— Accessing and using HCP data, software, and protocols.
Overview of the HCP’s Data Sharing. ConnectomeDB (http://db.humanconnectome.org) is the primary repository of HCP imaging, behavioral, and demographic data. 3T structural, functional, and diffusion imaging data for ~1100 subjects is shared in unprocessed, minimally preprocessed (MPP), and more extensively processed forms and staged over 4 major releases (Q1, Q1–Q3, S500, S900) with the final S1100 release slated for summer, 2016. 7T data acquired on 180 subjects has been released in part with the rest to follow shortly. MEG data acquired from 95 subjects gives a window on fast temporal dynamics of the brain and was released in the fall of 2015114. Importantly, the S900 release and the forthcoming S1100 release include the improved intersubject registration provided by “MSMAll”. The HCP_MMP1.0 parcellation is available in BALSA (http://balsa.wustl.edu), and individual subject parcellations for each HCP subject will be released on ConnectomeDB in the future (anticipated fall, 2016).
HCP Data Is Much Larger than that of Past Studies. With the high spatial and temporal resolution of HCP data, the dataset is at least an order of magnitude larger than widely used open access neuroimaging data sets like ADNI115. For a single subject, the compressed NIFTI-formatted unprocessed data is around 10 GB, and the preprocessed data is nearly 30GB.
HCP Data Is Widely Used via Multiple Modes of Data Access. To date, >5,200 investigators have agreed to HCP Open Access Data Use Terms (~520 to Restricted Access terms for accessing family structure and other sensitive data). Users can (i) access ConnectomeDB directly to download ‘packages’ for individual subjects, user-selected subject groups, or HCP-specified groups; (ii) purchase “Connectome-in-a-Box” for the cost of the hard drives, which can be shared by investigators at a given institution; and (iii) access data via the Amazon cloud for processing on the cloud (or for download). Data downloaded directly from ConnectomeDB exceeds 5400 terabytes (TB), with an additional 2000 TB transferred via hard drives (Connectome-in-a-Box) and the Amazon cloud.
Data documentation. The richness and complexity of the HCP data requires extensive documentation for users to understand what is available, how the datasets are organized, and how they were processed, including quality control measures. Available resources include (i) a Reference Manual associated with each data release (http://humanconnectome.org/documentation); (ii) HCP course materials (lectures, tutorials, and associated data) available at http://humanconnectome.org/courses/); (iii) publications, e.g. for database organization90; and (iv) the HCP public wiki (https://wiki.humanconnectome.org/display/PublicData/Home), which provides additional documentation, FAQs, and updates (including known issues and planned fixes).
Software Sharing. Also important to the replicability of neuroimaging studies is the sharing of the software used for analysis. Scripts for HCP pipelines are available on GitHub (https://github.com/Washington-University/Pipelines/releases), and their usage is described in the HCP course materials. This includes pipelines for MEG as well as MRI data. Connectome Workbench is available as binaries and source code (http://www.humanconnectome.org/software/get-connectome-workbench.html, on GitHub, and in http://neuro.debian.net) along with tutorials. Additionally many of the labs that make up the HCP consortia share their software on either their own websites or the HCP website.
MRI protocols. The MRI protocols used for the main HCP 3T and 7T scans are available at http://protocols.humanconnectome.org/HCP/; HCP-style pulse sequences have been widely distributed (http://www.cmrr.umn.edu/multiband/index.shtml). The HCP-Short protocols used for the HCP Lifespan (see below) are at http://protocols.humanconnectome.org/lifespan/
This article is both a position paper and a primer, promoting awareness and adoption of the integrated HCP-style paradigm along with guidance on its use. We provide an accessible conceptual overview of seven core tenets of the paradigm and summarize several recent discoveries made possible through its application. Additional technical information and discussion is provided in the cited publications and in 18 sections of the Supplementary Information (SI) that expand on specific topics raised in the main text. Investigators interested in practical aspects of implementing the HCP paradigm in their own lab can reference the MRI protocols, HCP software, and HCP Course materials listed below.
The seven tenets of the HCP-style paradigm span the domains of acquisition, analysis, and sharing of MRI-based data: (1) acquiring large amounts of high quality, multi-modal data on as many subjects as feasible to measure architectural, functional, connectional, and topographical information4 (2) acquiring this MRI data with high spatial, temporal, and angular resolution for structural, functional, and diffusion MRI using cutting-edge accelerated acquisition protocols5;(3) minimizing blurring introduced by preprocessing and removing distortions, noise, and temporal artifacts as selectively and completely as possible6,7; (4) representing cortical data (surface vertices) and subcortical data (volume voxels) in a common geometrical framework (“CIFTI grayordinates”) that is optimal for each6; (5) accurately aligning corresponding brain areas across subjects and studies using “areal features” related to connectivity and architecture8,9; (6) using structurally and functionally relevant brain parcellations (preferably based on multiple modalities) to provide a strong neuroanatomical framework for condensing complex neuroimaging data and enhancing statistical sensitivity and power without blurring across areal boundaries8; and (7) routine sharing of extensively analyzed results such as statistical maps10 (plus raw and preprocessed data when feasible11) together with the code used for the analysis, so that other neuroscientists can make precise comparisons across studies, along with replicating and extending findings.
The HCP-style paradigm builds on recent advances in data acquisition, analysis, and sharing from many neuroimaging labs worldwide, both within and outside the HCP consortia. It benefitted greatly from the NIH’s emphasis on methodological development prior to high-throughput data acquisition. The HCP-style paradigm differs from common practices in the human neuroimaging community that emerged during the early years of PET and fMRI (the ‘traditional paradigm’), which often involve (i) voxel-based (‘volumetric’) analysis of data acquired at relatively coarse spatial and temporal resolutions on small groups of subjects; (ii) extensive unconstrained spatial smoothing that attempts to compensate for imperfect crosssubject alignment, improve signal-to-noise ratio (SNR), and satisfy oversimplified statistical assumptions12; (iii) not using parcellations at all or using parcellations derived from Brodmann’s 2D schematic drawing; and (iv) reporting summary results using 3D volumetric coordinates while keeping the extensively analyzed data private. Relative to the new paradigm, the traditional paradigm has limitations in spatial fidelity, neuroanatomical fundamentals, and the robustness of cross-study comparisons, along lines also noted by Turner13,14.
Though most neuroimaging studies still use the traditional paradigm, important exceptions dating as far back as the 1990’s include numerous “retinotopy” studies that use functional MRI (fMRI) data to map the arrangement of visual areas on cortical surface meshes of individual subjects15,16. Thanks in part to this emphasis on surface-based analyses, the visual system is one of the better understood functional systems in the human brain. We anticipate accelerated progress in understanding other brain regions as the new paradigm is applied to them more routinely.
A solid conceptual understanding of MRI acquisition, preprocessing, and analysis (tenets 1–5) is important for applying the HCP-style paradigm to one’s own studies, and for understanding the methodological choices made by the HCP and its successors (see below). Many of these choices were driven by fundamental properties of human brain anatomy, MR physics, and human physiology.
Tenets 1 and 2: Multi-modal Data Acquisition Informed by Brain Anatomy and Physiology.
Comprehensive non-invasive brain mapping of many subjects using multi-modal MRI.
The HCP acquired ~4 hours of structural, functional, and diffusion images (along with auxiliary ‘field map’ images to help with image preprocessing). The structural images were T1-weighted (T1w) and T2-weighted (T2w) images (see Supplementary Topic #1 for a discussion of the relationship between these terms and MRI contrast). These images allow non-invasive measurement of architectural properties like myelin content within the cerebral cortex and cortical thickness that match well with corresponding invasive measures17–19, see also Supplementary Topic #1. The structural images also generate geometrical models of brain anatomy (cortical surface meshes and subcortical segmentations17,20–27) that are critical for all other analyses. The functional images included task-based fMRI, where the subjects carried out behavioral tasks while in the scanner28, and resting-state scans, where subjects fixated on a crosshair. These functional images enable measurement of functional activation within brain areas, and ‘functional connectivity’ based on correlations of the fMRI signal between brain areas in the absence of a task or controlled external stimuli. They can also map the internal topographic organization within brain areas, such as the ordered representation of visual space within visual areas using task driven15,16,29 or resting-state-based methods (visuotopic organization)8. The similarity of these detailed maps across species30 and in comparison with invasive studies in non-human primates,31,32 confirms that fMRI can accurately capture properties of brain organization. Diffusion MRI uses directionally dependent diffusion of water within white matter to encode average axonal fiber orientations, which can be connected by streamlining tractography algorithms to estimate structural brain connectivity, yielding results that are modestly correlated with invasively measured structural connectivity33,34. Diffusion MRI also allows estimates of microscopic structural properties of brain tissue, such as the integrity and orderliness of axons (e.g.35–37. Thus, the HCP MRI data enables brain architecture, function, connectivity, and topography to be estimated non-invasively. The large number of subjects (~1100) imaged by the HCP enables statistically precise estimation of these measures in the healthy young adult population and investigation of the nature and extent of their individual variability, while the focus on twins (and their non-twin siblings) enables assessment of their heritability.
How brain anatomy drives MRI spatial resolution.
The cerebral cortex, the seat of human cognition, is a folded sheet, 1.6 – 4mm thick (mean ~2.6 mm), with a surface area of ~1,000 cm2/hemisphere18,38. High quality structural images (both T1w and T2w) are vital for generating accurate geometrical models upon which all other analyses are based (surface meshes for the cerebral cortex and grey matter ROIs for the subcortical structures6. For the main HCP, we acquired these images at an isotropic spatial resolution of 0.7mm (voxel cubes 0.7mm on each side), enabling accurate individual subject maps of cortical myelin content and thickness together with precise models of the inner (white matter) and outer (pial) surfaces. In general, we recommend acquiring T1w and T2w images at 0.8mm isotropic or better (half the minimum thickness of the cortex), in contrast to the single 1 mm T1w image traditionally acquired. Such high-resolution images benefit from tightly fitting multi-channel MR receive head coils, such as the Siemens 32-channel head coil used by the HCP.
For fMRI, it is important to acquire images of sufficient spatial resolution to distinguish between cortical gray matter, cerebrospinal fluid (CSF), and white matter, and between opposite banks of cortical folds. The HCP acquired high-resolution fMRI data at 2 mm isotropic (with high resolution fMRI being defined as having voxels smaller than the mean cortical thickness of 2.6 mm, see Supplementary Fig. 1) on its customized 3-tesla (3T) scanner (and at 1.6 mm isotropic at 7T). High-resolution fMRI enables more precise localization of functional signals 6 in contrast to resolutions as low as 4 mm that have been traditionally used in fMRI.
Diffusion MRI (dMRI) aims to address a fundamentally resolution-starved problem: mapping connections through brain white matter whose individual axons are mostly <1 micron diameter 39 with MRI voxels that exceed 1 mm isotropic, contain hundreds of thousands of axons 40, and typically include multiple populations of fibers crossing in various directions. Both of the HCP consortia used customized MRI scanners and customized MRI pulse sequences to acquire in vivo human diffusion data of unprecedented resolution5,41–46. The 3T WU-Minn HCP scanner uses very strong magnetic field gradients (100 mT/m), approximately double that which commercial scanners could achieve when the project began in 2010. Along with new and optimized dMRI pulse sequences, this enabled 1.25mm isotropic resolution in human dMRI in vivo (see Supplementary Topic #2 and Supplementary Figs. 2 and 3 for more details) and 1.05mm isotropic at 7T45,47, in contrast to traditional dMRI with 2–2.5 mm resolution. The recently introduced Siemens Prisma MRI scanner (80 mT/m) developed in part because of the HCP enables the community to acquire similar dMRI data.
Faster, longer imaging for cleaner fMRI data and more robust white matter fiber orientation estimates.
Most MRI-based functional imaging relies on the Blood Oxygen Level Dependent (BOLD) contrast mechanism in which increased neural activity indirectly alters the regional deoxygenated hemoglobin content over a time course (‘hemodynamic response function’, or HRF) of many seconds48. Accordingly, one might think that it is sufficient to acquire fMRI images at intervals modestly faster than the HRF (2–3 s Repetition Time, TR, has been typical for whole brain imaging). However, fMRI images are corrupted by many sources of time-dependent (temporal) artifacts, including subject movement, physiology, and scanner instability. Many artifacts occur more rapidly than the typical fMRI volume TR and thus are aliased into the signal. High temporal resolution fMRI data (i.e., TR < 1 s) facilitates more selective and effective artifact removal7, and improves statistical efficiency, particularly for multi-variate statistics49. Recent data also suggest that there may be faster components to the BOLD response50,51, which would be better captured with faster scans. The HCP acquired fMRI data at TR = 0.72 s for 3T and 1 s for 7T using MRI pulse sequences that acquire many brain slices at a time (‘multi-band’ - also known as ‘simultaneous multi slice’ - sequences, see Supplementary Topic #3 5,43,52–54 and Supplementary Fig. 4). For diffusion imaging, using these sequences to reduce the TR allows for more images to be acquired with diffusion weightings at different strengths and along a larger number of unique spatial directions in the same amount of time. The resulting higher angular resolution and effective SNR enables multiple fiber populations to be more robustly identified in each imaging voxel, helping to resolve crossing fiber pathways and improve tractography5,44.
The HCP used 1 hour of scan time for each of task fMRI, resting state fMRI, and diffusion imaging. This enabled seven fMRI tasks to be acquired covering multiple cognitive domains and involving most of the brain28 http://protocols.humanconnectome.org/HCP/3T/task-fMRI-protocol-details.html, along with four 15-min resting state fMRI runs (helping to provide more stable connectivity estimates55), and 270 different diffusion measures (90 unique diffusion directions for each of three different diffusion weighting strengths (i.e. “shells”), enabling better fiber orientation and tissue microstructure modeling at high resolution56,57. See Supplementary Topic #4 for particulars on various “HCP-Short” protocols optimized for subjects who are less tolerant of long scans than healthy young adults.
Tenets 3 and 4: Optimizing the Precision of Data Analysis
Aligning all MRI modalities within the subject’s physical space.
A key early task of preprocessing is to align all MRI modalities so that every image volume is in the subject’s undistorted physical space. Head movements occur in the scanner, both between acquisition of each image and during acquisition. Because head movement during T1w and T2w images causes uncorrectable blurring, the HCP acquired at least two pairs of T1w and T2w images, and used all scans that were rated good or excellent in overall quality (11; see Supplementary Topic #5 for further discussion of this issue and potential solutions). Spatial effects of movement during fMRI or dMRI acquisitions can be corrected after the fact using image registration to realign the images; however, movement also induces temporal variations of image intensities that must be corrected through other means described below. Movement between images of different modalities is corrected with advanced registration algorithms that use knowledge of brain tissue boundaries and the expected image intensity gradients across those boundaries to align images58.
Ideally, image registration within each subject would entail simple translations or rotations, but MRI images have many forms of spatial distortion that must be unwarped before an image faithfully represents the individual brain’s real physical dimensions and thus can be precisely aligned. These distortions all derive from imperfections in the scanner’s encoding of physical space using perturbations (linear gradients) of the main magnetic field. All MRI images are slightly distorted by small nonlinearities in the space-encoding gradients that are correctible using information measured and specified by the manufacturer (gradient distortion, see Supplementary Topic #6). In addition, placement of the subject’s head in the scanner induces idiosyncratic variations in the magnetic field (field inhomogeneities), particularly near the air-filled spaces in the head (sinuses and ear canals), which can be measured using the auxiliary ‘field map’ scan mentioned above. Correcting these distortions with a field map is critical for accurately aligning data across modalities within each subject, though traditionally this scan is often omitted, leading to poor alignment in some brain regions. Scanner vendors provide a standard field map scan (based on gradient echoes), but the HCP developed an alternative ‘spin echo EPI field map’, that is faster, at least as accurate, and also enables correction of fMRI image intensity biases (see Supplementary Topic #7). These field map scans should exactly match the geometry and distortion properties of the functional imaging scans. Diffusion imaging scans have additional ‘eddy current’ distortions caused by switching on and off the strong magnetic field gradients used to generate diffusion weighting. The HCP developed a new algorithm for correcting eddy current distortions that outperforms previous methods and allows quality control and outlier detection (see Supplementary Topic #8;59–61). With field maps, advanced distortion correction methods, and anatomically informed registration algorithms, the HCP’s spatial preprocessing pipelines6 precisely align the T1w, T2w, fMRI, and diffusion data to the subject’s physical space while minimizing interpolation-induced blurring by combining all spatial transformations in a single spline interpolation.
Cleaning time-dependent artifacts from MRI timeseries data.
In functional imaging, many processes cause time-dependent intensity fluctuations other than the BOLD-related fluctuations of interest. This structured temporal noise is especially problematic for resting-state functional connectivity because it uses correlations of noisy signals in different parts of the brain, rather than correlation of a predefined noiseless task design with noisy data. Temporal noise can either increase or reduce functional connectivity artifactually and lead to incorrect conclusions62-64. Major sources of structured temporal noise include subject movement, subject physiology, and the physics of MRI. It is useful to consider the fMRI timeseries as containing fluctuations arising from 1) the BOLD signals of interest, 2) structured temporal noise, 3) and unstructured random ‘Gaussian’ temporal noise11. Temporal cleanup aims to remove temporal noise as selectively as possible while minimally impacting the BOLD signals of interest. The HCP devised a method based on Independent Components Analysis (ICA,65, which splits the fMRI timeseries into structured components and unstructured noise, representing all of the timeseries fluctuations. The structured components are then categorized by a machine learning classifier (FIX) into signal and noise7, and the noise components are removed by regressing them out of the data. For HCP data, the ICA+FIX approach removes spatially specific (i.e. affecting only part of the brain in a particular way) structured noise components and retains spatially specific BOLD signal components (i.e. resting state networks, RSNs) with better than 99% accuracy7. In contrast, traditional approaches that include bandpass filtering, WM and CSF regression, and censoring of high-motion frames63,64) are not as selectively at targeting structured noise while preserving BOLD signal. However, a controversy remains about “global” fluctuations in fMRI (see Box 1 and also Supplementary Topic #9 and Supplementary Figs. 5 and 6), as the spatial ICA+FIX approach is not designed to separate these ‘global’ fluctuations in the average fMRI signal into signal and noise. For dMRI, subject motion and physiology can cause signal loss. Such ‘outliers’ can be detected using a new approach that builds a model of the diffusion signal from all of the data, allowing artifactual measurements to be identified and replaced with estimates of what the signal should have been (see Supplementary Topic #8, 59–61).
Box 1: Global fMRI Temporal Fluctuations.
A hotly debated topic is how best to handle ‘global’ signals and artifacts present in fMRI data62,63,105–107. The core phenomenon involves slow and spatially widespread fluctuations in the fMRI timeseries. The average timeseries across the brain includes both neural signal (e.g. the average of all the resting state networks) and structured noise (e.g. physiological noise), and the debate centers on whether the average timeseries should be regressed out of the data to remove the effects of global noise62 or whether it should be preserved to avoid distorting neural signals by removing their average. Additive global noise will by definition artifactually increase ‘full’ correlations across the brain (especially problematic if it differs across groups), but removal of a global neural signal will artifactually decrease correlations across the brain and, more problematically, distort the pattern of correlations for particularly large/strong networks62,108,109.
What is needed to get past the current impasse is a method that selectively separates global signal and global noise. Though no such method is yet available, we offer several observations about global fMRI fluctuations. 1) In HCP data, the global signal (after ICA+FIX cleanup) is primarily a gray matter signal, with a near zero amplitude in white matter voxels and low-level anti-correlation with CSF (Supplementary Fig. 5). 2) Within grey matter, the mean grey time course has much higher amplitude in sensory regions (e.g. visual, auditory, vestibular, and somatosensory cortex, plus the lateral geniculate nucleus and other thalamic nuclei) and is lower in the cerebellum and many neocortical cognitive regions (Supplementary Fig. 6). Thus, the global signal is not uniform across the brain, or even across grey matter, and these observations constrain the possible sources of the global fluctuations:
-
(i)
Physiological effects. Substantial fMRI timeseries variance is attributable to physiological variables, including rate and depth of breathing, heart rate, and end tidal pCO2110–112). Indeed, any physiological process that affects global perfusion pressure of the brain (i.e. the above three, or even yawning) may transiently alter the amount of deoxyhemoglobin in the brain and contribute to global, physiologically artifactual fluctuations of the BOLD signal. Different brain areas and tissue types may be differentially sensitive to such changes in perfusion pressure because of differing metabolic rates, which may explain regional differences in global signal amplitude.
-
(ii)
Head movements (direct, biophysical). Transient head movements are sometimes followed by global fluctuations in the fMRI signal, leading to suggestions that head movements could directly cause these fluctuations62,64,106. However, head movements are unlikely to cause global fluctuations directly, given the lack of a plausible biophysical mechanism for how head movement might induce tissue-specific and brain-area-specific effects on the fMRI timeseries, let alone globally uniform changes in signal intensity short of complete head coil exit and reentry (See Supplementary Topic #9).
-
(iii)
Head movements (indirect, neural). Head movements may induce neural responses in somatosensory, vestibular, and/or visual systems that may in turn modulate wider brain activity in a pattern very consistent with that mentioned above.
-
(iv)
Widespread/High amplitude RSNs, average of the RSNs. As discussed above, a portion of the global fluctuations will represent the average of the RSNs with particular weighting to more widespread or stronger networks.
It will be important to separate global physiological effects from spatially widespread neural effects (using either extrinsic physiological monitoring or intrinsic fMRI-data-driven means yet to be developed). Short of a complete solution to the problem of global fluctuations, we note that some approaches to analysis (such as the use of partial correlation when estimating functional connectivity, and multi-variate measures in general) are much more immune to the effects of global fluctuations than others (e.g., full correlation measures and uni-variate measures in general)113.
The traditional approach to reducing unstructured Gaussian noise involves spatial and temporal smoothing, which unfortunately affect both signal and noise unselectively. A common problem in traditional neuroimaging is that smoothing mixes the grey matter functional signals of a given brain area with signals from white matter, CSF, and nearby brain areas, markedly reducing the spatial fidelity of neuroimaging data. Parcellation (tenet 6) enables averaging of data without mixing signal across different tissue types or between different brain areas in so far as the parcellation is neurobiologically accurate, and acts as a neuroanatomically informed method of nonlinear smoothing. Another option is to selectively smooth unstructured noise more than structured BOLD signal (see Supplementary Topic #10). This enables reduction of certain artifacts (see Supplementary Fig. 7) and enhancement of SNR without concomitant spatial blurring and without invoking a brain parcellation.
Preserving precision from voxels to CIFTI “grayordinates.”
It is vital that the preprocessing and analysis techniques used on high resolution HCP-style data maintain this hard-won spatial resolution. Besides removing distortions and accurately aligning data across modalities within each subject, the data must be put into an analysis framework that enables accurate crosssubject and cross-study comparisons (tenet 5 below). The traditional neuroimaging paradigm represents spatial data in a regular 3D array of voxels (an image volume) that reflects the output of the MRI image reconstruction process. Because the cerebral cortex has a sheet-like folded geometry, it is advantageous to visualize and analyze cortical data on surface mesh vertices, instead of volume voxels15,25,26,38. However, subcortical grey matter structures are mostly globular in shape and remain best represented using volume voxels. To represent the inherently dual geometry of the brain, the HCP developed the CIFTI file format6, which integrates surface vertices and subcortical voxels into a single file and represents each major brain gray matter structure using the appropriate geometry (see Figure 1). CIFTI gray matter vertices and voxels are called ‘grayordinates.’ Analyses focusing only on grayordinates (e.g., fMRI) dramatically reduce file sizes and computational burdens (see Figure 1 legend). For dMRI, surface representations allow more accurate anatomical constraints in tractography (e.g., the white surface can be used as the destination for tractography). The final common outputs of the HCP preprocessing and analysis pipelines are datasets in the CIFTI standard space6. CIFTI is natively supported by Connectome Workbench, and other tools are currently implementing CIFTI compatibility (e.g., FSL, FreeSurfer, FieldTrip, nibabel). In particular, FSL’s PALM software enables permutation-based statistical inference on CIFTI datasets (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/PALM 66). The CIFTI grayordinates analysis framework allows each brain structure to be registered using the approach best suited to it (see Tenet 5).
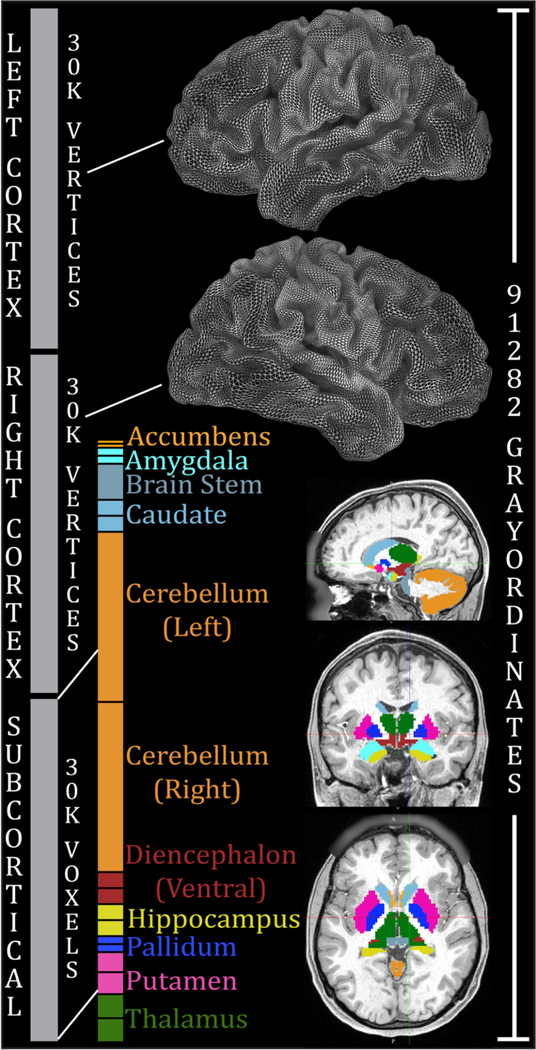
Figure 1.
Combined representation of cortical surface vertices and subcortical voxels in the CIFTI grayordinates standard space. Left and right cerebral cortices contribute about 30k surface vertices each (fewer than the 32k vertex standard meshes for each hemisphere, because the non-cortical medial wall is not included). Additionally, 19 subcortical grey matter structures combine to contribute about 30k volume voxels. In total, there are 91,282 grayordinates corresponding to all of the grey matter sampled at a 2 mm average vertex spacing on the surface and as 2 mm voxels subcortically. The HCP’s minimal preprocessing pipelines ensure that each subject has 91,282 aligned grayordinates, thereby facilitating cross-subject comparisons of data within this coordinate system. This entails more than a two-fold reduction in file size relative to the >200,000 voxels needed for an equivalent 2 mm isotropic volume representation. For the 1.6mm 7T HCP data, we have developed a 1.6mm standard grayordinates space with 170,494 grayordinates using “59k” surface meshes. Reproduced with permission from Reference 6.
Tenet 5: Accurately A ligning Brain Areas Across Subjects and Studies.
Cross-subject comparisons.
Analyses that combine neuroimaging data obtained from different individuals benefit greatly from accurately aligning corresponding brain areas, so that like is compared with like14, Alignment of subcortical and cortical structures is best achieved through differing means. Subcortical structures are less variable in size, shape, and position across subjects, and reasonable alignment is achieved by nonlinear 3D registration to a standard template, as in the traditional paradigm. The HCP currently uses FSL’s FNIRT for nonlinear registration of T1w images to MNI space, but registrations that also incorporate dMRI fiber orientations in the white matter may improve alignment of subcortical white matter fiber tracts (See Supplementary Topic #1167,68).
For human cerebral cortex, the complexity and variability of folding patterns and of areas relative to folds38,69–71 poses a profound challenge that is an ill-posed problem for nonlinear 3D registration. Though alternative approaches such as hyperalignment (where alignment is not spatially constrained) are useful in some contexts, many neuroscientific questions are best addressed by aligning subjects to a common space. Registering cortical surfaces rather than brain volumes enables better alignment because accurate cortical segmentation of each subject reduces the dimensionality of the registration problem from 3D to 2D—alignment need only occur tangential to the cortical sheet as opposed to a 3D alignment of the folded sheet itself26,38,72–76.
Though surface-based registration of the cerebral cortex to an atlas is not a part of the traditional paradigm, it has been adopted by a growing number of investigators. Most use surface registration based on cortical folding patterns26,38,72–76, which achieves ‘geographic correspondence’ in regions where folding patterns are consistent across individuals and ‘functional correspondence’ in regions where areal boundaries are consistently correlated with folding. Because cortical folding patterns are highly variable in many regions of cortex and their relationships to cortical areas are similarly variable, folding patterns alone are not enough to achieve alignment of (functional) cortical areas across most of cerebral cortex. Registration needs instead to be driven by features more closely tied to cortical areas, such as architecture, function, connectivity, and topography77. Such areal-feature-based cortical surface registration can dramatically improve the sharpness of group average maps and enhance group statistics, as it largely compensates for differences across subjects in cortical areal size, shape, and position. Indeed, alignment based on T1w/T2w myelin maps, resting state network maps, and visuotopic connectivity maps yields sharper task fMRI contrast maps with higher statistics, even though the task fMRI data were not used in the registration (Figure 2)8,9. Areal-feature-based aligned data was recently released by the HCP (dubbed ‘MSMAll’ because it uses the Multimodal Surface Matching algorithm,9 and all of the useful modalities listed above). We believe that many studies will benefit from using areal-feature-based surface registration as a starting point, even if they choose to make final comparisons across subjects using other means (e.g. hyperalignment78, or individual subject parcellation (see below and ref.8).
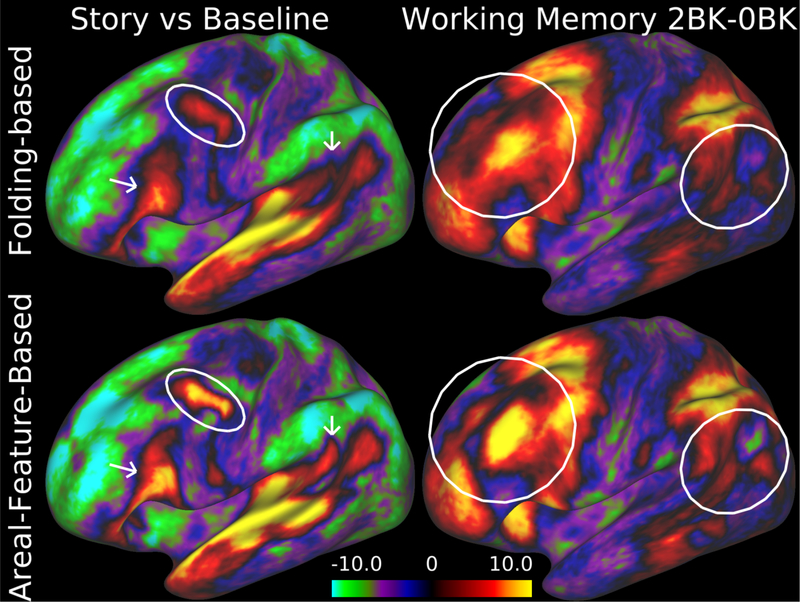
Figure 2.
Improved intersubject registration using information based on areal features in addition to cortical folding. The top row shows group task-fMRI z-stat maps (“Story vs Baseline” contrast from the language task on the left and the two-back vs zero-back contrast in the working memory task on the right) from 120 Q1 and Q2 HCP subjects after intersubject registration using the Multimodal Surface Matching (MSM) method constrained only by folding (FreeSurfer’s ‘sulc’ maps). Bottom row shows sharper group task-fMRI maps and higher z-statistics when using resting-state networks (RSN) along with myelin maps to constrain the registration. For the data in this figure and figure 6, subject recruitment procedures and informed consent forms, including consent to share de-identified data, were approved by the Washington University institutional review board. Data at http://balsa.wustl.edu/97V4.
Cross-study comparisons.
Traditionally, cross-study comparisons based on 3D stereotaxic coordinates were assumed to align if everyone used a common standard volume space (e.g., MNI). These 3D coordinates were typically derived from thresholded statistical maps of the ‘significant’ activations (or connectivity seed locations). Though such standard space coordinates allow for gross comparisons across studies at the gyral level (e.g., ref.79, questions of whether a particular cortical area was activated in particular task contrast typically reveal clouds of points whose distribution may be suggestive, but often inconclusive (e.g., ref.19). Stereotaxic coordinates have high spatial uncertainty (relatively small changes in the thresholded statistical maps used to produce them can have large effects on the resulting coordinate locations, see Supplementary Topic #12 and Supplementary Fig. 8), and they do not contain information about the spatial extent or boundaries of activation. Conclusive cross-study comparisons require assessment of the overlap of activated regions and whether their boundaries align or not. Cross-study comparisons also rely on the absence of any “drift” in alignment between the studies (see Supplementary Topic #13 and Supplementary Fig. 9). When drift is eliminated and boundaries are compared, direct and conclusive assessments of cross-study agreement are possible (see Supplementary Fig. 10, also Figure 6).
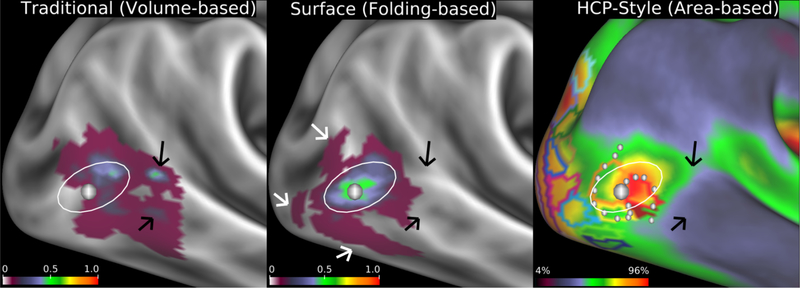
Figure 6.
Fidelity of localizing area MT+ (architectonic area hOc5) when mapped to the cortical surface by different methods. Left: volume-based mapping of probabilistic cytoarchitectonic area hOc5103 from 10 postmortem subjects mapped to a cortical atlas surface. Black arrows point to locations where the volumebased mapping spreads across gyral and sulcal folds. Center: Surface-based registration of hOc5 from the same 10 subjects mapped to individual surface reconstructions, then to a surface-based atlas using FreeSurfer’s folding-based surface registration method73. White arrows identify ‘outlier’ hOc5 from individual subjects that are not well aligned to the FreeSurfer group average owing to imperfect correspondence between areal boundaries and sulcal folds. Left and center panels adapted from ref. 38. The white oval is in the same location across all panels showing how the volume-based alignment drifts away from the surfacebased alignments, in addition to having substantially lower cross-subject overlap. Right: Group average cortical myelin map (from 196 HCP subjects) with a yellow/orange/red hotspot indicating the MT+ complex and retinotopic areal Maximum Probability Maps (MPMs) (from 12 non-HCP subjects) both registered using arealfeature-based surface registration and ‘de-drifted’ (see Supplementary Topic #13) (adapted from ref.104). As shown in Supplementary Fig. 10, the white dots represent a contour along which functional connectivity rapidly changes that aligns with the border between retinotopic areas MT and pMST from a separate study. This is an example of the more conclusive cross-study, cross-modal boundary comparisons made possible by the HCP-style analysis paradigm. Data at http://balsa.wustl.edu/kNpD.
Tenet 6: Neuroanatomically Accurate Maps of Brain Areas.
Accurately subdividing the brain into its constituent parts (areas or parcels) has been a holy grail for brain cartographers and other neuroanatomists for more than a century80–82. The goal is to reliably identify parcels that are distinct from one another in architecture, function, connectivity, and/or topographic organization and are consistently identifiable in most or all individuals. For neuroimaging, parcellation serves to: (i) clarify ‘where we are’ in the brain when describing results14; (ii) facilitate comparisons across individuals and studies; and (iii) reduce the dimensionality of complex and noisy datasets while respecting neuroanatomically distinct boundaries. Parcellation can dramatically improve statistical sensitivity and power by averaging across neuroanatomically similar units of the brain, enhancing SNR and reducing the vast number of statistical comparisons often carried out in neuroimaging (circumventing problematic corrections for multiple comparisons that have affected a substantial fraction of the neuroimaging literature 12). As noted above, parcellation is an attractive alternative to traditional spatial smoothing for analyses that are interested in effects at the areal level (in practice, most studies that provide a standard space coordinate table).
A cortical areal parcellation should ideally have four key qualities: (i) It should be based on many (hundreds of) well aligned subjects so that it represents the typical areal arrangement in the studied population (e.g., healthy young adults for the HCP). (ii) It should reflect complementary and converging evidence from multiple modalities across the whole cerebral neocortex for completeness and for higher confidence in boundaries. Most parcellation efforts to date have relied instead on information from a single modality, such as architecture70,83, retinotopy16,29, or resting-state fMRI84–86, and often do not cover the whole hemisphere. (iii) It should reflect existing terminology for areas previously reported in the neuroanatomical literature and a rational terminology for newly defined areas. (iv) It should be possible to automatically replicate the parcellation in individual subjects’ datasets based on multi-modal areal fingerprints - even in subjects with atypical areal arrangements or those from future studies. The recently reported “HCP_MMP1.0” (HCP MultiModal Parcellation, version 1.0) meets these criteria8, but we expect this will not be the final word on cortical parcellation. Also subcortical structures and the cerebellum need analogous multi-modal parcellations.
The HCP_MMP1.0 parcellation (Figure 3) contains 180 distinct areas per hemisphere, symmetrically arranged across the two hemispheres. 83 areas corresponded reasonably well with areas defined in one or more prior studies and were assigned the same names, while 97 were assigned new names. The group average parcellation was used to train a machine-learning classifier (a multi-layer perceptron87) to enable a fully automated process for identifying areas in individual subjects based on each area’s multi-modal fingerprint, even in individuals having atypical topological arrangements of areas (Supplementary Fig. 11).
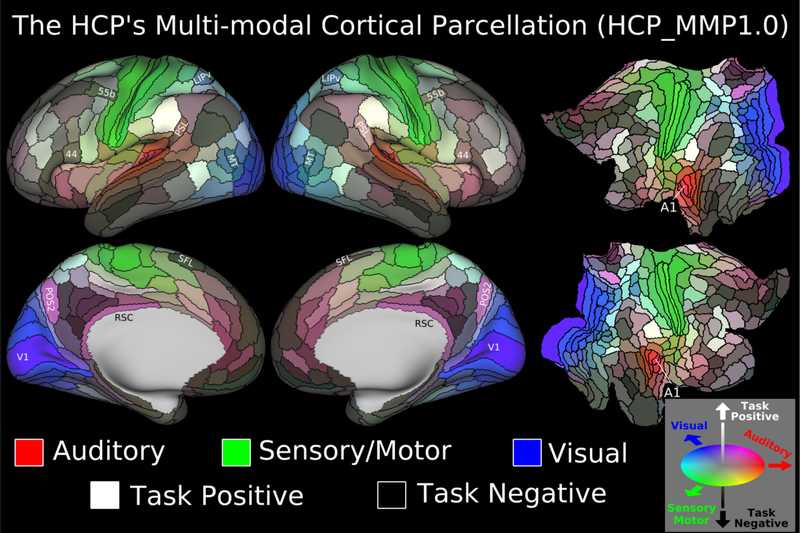
Figure 3.
The HCP_MMP1.0 (HCP Multi-Modal Parcellation, v 1.0). Each panel shows 180 cortical areas delineated and identified in the left or right hemisphere, displayed on an inflated or flattened cortical surface. Black outlines indicate areal borders. Colors indicate to what extent the areas are associated in the resting state to auditory (red), somatosensory (green), visual (blue), task positive (towards white), or task negative (towards black) systems. The legend on the bottom right illustrates the 3D color space used in the figure. Reprinted from ref8. Data at http://balsa.wustl.edu/WN56.
More generally, the classifier can reliably detect cortical areas in individual subjects without using the HCP’s task fMRI data (using only myelin maps, thickness maps, resting state network maps, and resting state visuotopic maps), enabling generalization of the approach to many non-HCP neuroimaging datasets. Individual investigators wishing to apply the HCP Pipelines, areal-feature-based registration, and trained areal classifier to their own individual subjects, need only acquire T1w, T2w, field map, and fMRI data as described above, as even non-resting state fMRI data include similar patterns of connectivity88. The population-based parcellation can also be applied directly to registered individual subjects, though it will not be accurate where individual subjects deviate from the typical layout of areas. The HCP’s multimodal ‘hard’ parcellation v1.0 provides a neuroanatomical foundation for future neuroimaging analyses, though other ‘weighted’ approaches provide valuable complementary information as well (see Supplementary Topic #14). It’s worth noting that FreeSurfer uses an algorithm to automatically label gyri and sulci in individual subjects based on manually generated training labels that is similar in spirit to our areal classifier89.
Tenet 7: Routine Data Sharing and Advanced Informatics for Data Management
Data sharing for individual laboratories.
Data may be publicly shared at many levels, including extensively processed, preprocessed, and unprocessed stages together with the code used for analysis. Data sharing is increasingly mandated by journals and funding agencies. Although the HCP has shared its data using an advanced informatics infrastructure described below, most individual investigators only publish their results in journal articles, as the additional time and informatics skills required for more expansive data sharing have made it impractical. Generating and editing manuscript figures is typically a tedious process involving multiple software applications that neither work efficiently together nor aid in preparing data for post-publication sharing. Routine sharing of extensively processed data would be expedited by a streamlined process for generating publication-ready figures that is integrated with a service for easy postpublication sharing of the associated data.
An integrated approach to data visualization, figure generation, and data sharing is now available. Connectome Workbench (a freely available general-purpose neuroimaging visualization platform) provides a flexible interface for displaying rich multi-modal neuroimaging datasets. “Workbench” enables visualization of various data types overlaid on surfaces or volumes (NIFTI, GIFTI, or CIFTI files). Multi-panel montage figures can be generated by combining web browser-like tabs that can also be annotated (e.g., text, symbols, and imported image panels) directly in Workbench. For example, Figure 4 is one of 42 published figures in one study 8 exported from Workbench without further editing. Importantly, Workbench allows such complex displays to be saved as “scenes” that can be quickly re-opened, edited, re-saved, and exported as publication-ready images. Data from such figures can be shared by uploading the scene file to the BALSA database (http://balsa.wustl.edu;10 directly from Workbench, together with basic information about the publication. Scene-specific URLs enable one-click linking from a published figure to the corresponding page in BALSA (see Figure legends). Adoption of similar strategies by other neuroimaging software and databases would accelerate sharing of extensively analyzed data and facilitate more accurate and diverse comparisons across studies.
Figure 4.
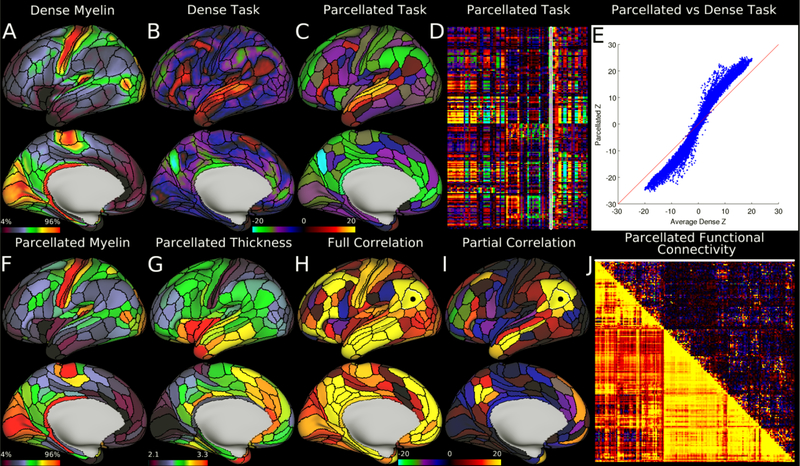
Example parcellated analyses run using HCP data and the HCP_MMP1.0 cortical parcellation. Panels A and F, showing dense and parcellated myelin maps respectively, are very similar despite a dramatic dimensionality reduction. Panel G shows a map of parcellated cortical thickness (in millimeters; corrected for folding by regressing out mean surface curvature). Panels B and C show example dense and parcellated task fMRI analysis (LANGUAGE Story vs Baseline). Panel D shows the entire HCP task fMRI battery’s Z statistics (86 contrasts; 47 distinct and 39 sign-reversed versions) analyzed in parcellated form and displayed as a matrix (rows are parcels, columns are contrasts, white outline indicates the map displayed in Panel C). Panel E demonstrates a major improvement in Z statistics from fitting a task design on parcellated data instead of fitting it on dense data, and then parcellating afterwards. Panels H and I show parcellated functional connectivity maps on the brain (seeded from area PGi, black dot). In both cases, the task negative (default mode) network is apparent. Panel J shows a parcellated connectome matrix view with the full correlation connectome below the diagonal and the partial correlation connectome above the diagonal (white line shows the displayed partial correlation brain map). The figure matches the Connectome Workbench scene available on the BALSA database (http://balsa.wustl.edu/RG0x). Notably, Panel E was generated in matlab and saved as a PNG and then loaded in to Workbench as its own tab. Reprinted from ref8.
Large-Scale Data Sharing and an Advanced Informatics Infrastructure.
Well designed data management tools and processes are vital for the successful collection and analysis of high quality HCP-Style data on a large scale. ConnectomeDB, the primary database for HCP data, was designed for user-friendly sharing of unprocessed and preprocessed data from a large number of individual subjects, which can be selectively downloaded using a variety of flexible criteria https://db.humanconnectome.org 11,90. ConnectomeDB is based on the XNAT informatics platform (http://www.xnat.org 91, which manages imaging data through a structured workflow from acquisition through quality control and automated processing90. It also provides data entry and import tools for task and biometric data collected during image acquisition as well as behavioral and other non-imaging data. ConnectomeDB also serves as the data management platform for the Lifespan Connectome projects, has been deployed by some Connectomes Related to Human Disease projects (see below), and can benefit other groups carrying out large-scale HCP-style studies (source code at http://www.xnat.org/download/). Given the complexity of the HCP project and the massive size of the HCP dataset, documentation and distribution required a multi-faceted approach (Box 2).
HCP-Style Follow Up Studies.
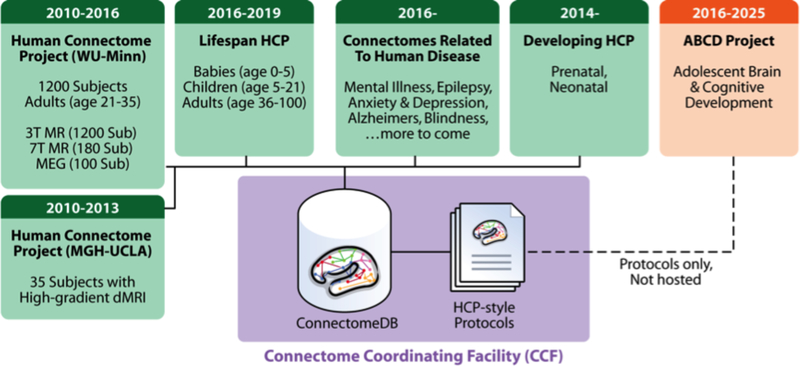
The original (healthy young adult) HCP will be succeeded by several large-scale imaging projects that use “HCP-style” data acquisition, analysis, and sharing to study different populations in health and disease. Data sharing for many of these projects will be managed by the Connectome Coordination Facility (www.humanconnectome.org/ccf), an NIH-funded resource devoted to supporting data sharing from the Connectomes Related to Human Disease (CRHD) projects (11 currently funded, with more anticipated in 2017) plus three Lifespan HCP projects (see Figure 5). The CCF is an extension of the ConnectomeDB infrastructure and will provide a common interface for searching within and across projects. In addition, it is developing common acquisition protocols and maintaining the HCP’s processing pipelines to maximize cross-study comparisons and analysis, and partnering with other connectomics studies such as the Adolescent Brain Cognitive Development (ABCD) study and UK Developing HCP (dHCP http://www.developingconnectome.org/) to encourage harmonization of acquisition and analysis methods. Versions of the HCP-Short Protocol (see Supplementary Topic #4) have been adapted for a variety of vendors and scanner models, with the goal of producing images of similar spatial and temporal resolution.
Figure 5.
Current and future projects that use HCP-style data acquisition, analysis, and sharing. Projects shown in green will use the Connectome Coordination Facility (CCF, in purple) as their primary mode of data sharing.
Concluding comments
The NIH had two interrelated core goals in launching the HCP: 1) to generate a publicly shared, widely used repository of high quality multi-modal neuroimaging data suitable for mapping the human connectome, and 2) to advance the methods used in neuroimaging research.
More than 140 studies have acknowledged the use of HCP data, and for half of these the authors were largely or entirely outside the HCP consortium (https://wiki.humanconnectome.org/x/YQDJAw). These studies address a wide range of questions using various imaging modalities. Five brief examples highlight some of this progress. Smith et al.,92 identified a ‘positive-negative’ phenotypic axis that covaries with differences in the strength of functional connectivity in a subset of brain regions that are generally implicated in higher cognitive functions. Yeo et al93 investigated the overlap and segregation of the brain’s functional networks, finding that association regions tend to overlap with at least two networks whereas somatosensory and visual regions are more isolated. Wang et al.94 investigated functional network parcellations at the individual level using HCP data. Hawrylycz et al.95 demonstrated correlations between gene expression patterns in postmortem human cortex and HCP-based in vivo functional connectivity. Tavor et al.96 showed that a model relating task-independent (rfMRI) measurements to task activity can accurately predict task activation maps for unseen subjects, suggesting a coupling between brain connectivity and function at the level of individual subjects. After release of the final “S1100” imaging and genotyping data (summer, 2016) and the individual-subject cortical parcellations (projected fall, 2016), we anticipate that a growing fraction of studies using HCP data will use the full set of available subjects, better-aligned data, multimodal parcellations of group average and individual subjects, and family structure (e.g., twins). Capitalizing on the full richness of the HCP data offers the best prospects for robust and neurobiologically grounded findings. Even with large amounts of high quality data, it remains extremely important to use caution in interpreting complex neuroimaging datasets and the indirect inferences related to brain connectivity available from neuroimaging. Neither functional connectivity based on rfMRI nor structural connectivity based on dMRI tractography provides good quantitative estimates of the actual strength of direct anatomical connectivity between areas or regions. This reflects a variety of methodological biases and other limitations that have become better understood in recent years 2,34,97 see Supplementary Topic #15), but remain serious limitations even with the advances provided by the HCP-style paradigm. Research continues to further validate non-invasive connectivity methods and maximize their accuracy.
The HCP-style paradigm brings together a wide range of methodological advances, many provided by investigators or projects outside the HCP, and others reflecting direct HCP innovations. New MRI scanner technology (Siemens Prisma) and pulse sequence technology (multi-band) were developed in part through the HCP, enabling imaging of finer details faster. Many improvements in image distortion correction, registration, and data-driven image denoising enable selective removal of artifacts while preserving the precision of the signals of interest. The CIFTI grayordinates analysis framework together with areal-feature-based surface registration largely compensates for individual variability across subjects without causing or resorting to blurring of the data. A multi-modal cortical parcellation provides a new neuroanatomical foundation for studies of the human cerebral cortex and is a valuable prerequisite for generating area-to-area connectomes in individuals and group averages. Intuitive, user-friendly data visualization software together with integrated neuroimaging databases and software pipelines enable easier data processing and sharing.
The overarching objectives of the HCP-style paradigm are (i) to acquire and analyze neuroimaging data as accurately as possible in each individual from their original brain imaging voxels to their individually mapped brain areas; (ii) to use precise inter-subject alignment while avoiding smoothing to minimize blurring across brain areal boundaries when comparing results across individuals, groups, and studies; and (iii) to routinely share neuroimaging data in a way that facilitates conclusive cross-study comparisons. Unlike traditional volume-based approaches that often result in statistically significant ‘blobs’ or 3D coordinates of unclear neuroanatomical identity, the HCP-style paradigm aims to remain as faithful as possible to the underlying neuroanatomy, even in the face of the remarkable individual variability of human brains. Indeed, the gains in spatial sharpness and clarity provided by the HCP-style paradigm (see Figure 6) are qualitatively analogous to those made in astronomy after the introduction of adaptive optics and space telescopes to overcome the atmospheric blurring that plagues conventional ground-based telescopes98. We provide concrete, generally applicable guidelines for individual labs wishing to embark on a new study using the HCP-style paradigm, though of course not all recommendations will fit every study, and methods will continue to improve (Supplementary Topic #16).
We propose that the diverse technological and conceptual advances in imaging integrated by (and developed within) the HCP meet the criteria of a new paradigm for neuroimaging research99. The “HCP-style” paradigm has a differing set of assumptions (e.g., blurriness in brain images is not a ‘fact of life’ but an avoidable artifact of traditional methods and that neuroanatomical localization is critical to understanding the brain). It invokes new terminology, and provides substantial new capabilities for further research. It is also disruptive to ‘business as usual’ as practiced by the majority of neuroimaging investigators, and legacy data may not be easily comparable with data from the HCP-style paradigm while maintaining the new paradigm’s improved standards of spatial fidelity and neuroanatomical localization (Supplementary Topic #17, 18). Thus, moving to the new paradigm should not be undertaken lightly. That said, we believe the benefits decidedly outweigh the costs, as the new paradigm opens up a variety of novel analysis strategies that will likely accelerate our understanding of human brain structure and function and may even prove useful in a clinical setting. Instead of relying on rough evidence of spatial proximity as in the old paradigm, we aim in the new paradigm to precisely compare the overlap and boundaries of results from differing studies to see if they likely refer to the same brain areas or not. Additionally, we are able to separate differences in brain function or connectivity much more precisely from differences in brain areal size, shape, and position. We can investigate the significance of variations in individual subject areal topologies from the population’s typical topology. Finally, the HCP-style paradigm is better positioned to exploit further improvements in brain image acquisition that will enable even finer grained study of the brain at the level of cortical layers or columns, as well as finer subcortical organization100–102. Nonetheless, the challenges to neuroimaging remain daunting, and we hope that new technologies and ideas will lead to further paradigmatic advances in the future.
Supplementary Material
Acknowledgments.
We thank the other investigators and staff members of the Human Connectome Project consortium for invaluable contributions to data acquisition, analysis, and sharing. Additionally, we thank the many colleagues outside the HCP upon whose methodological contributions the paradigm espoused in this paper are also based. We thank S. Danker for assistance in manuscript preparation. Supported in part by the Human Connectome Project, WU-Minn Consortium (1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; the McDonnell Center for Systems Neuroscience at Washington University, NIH F30 MH097312 (MFG), RO1 MH-60974 (DCVE), P41 EB015894 (NIBIB; KU), Wellcome Trust 098369/Z/12/Z (SMS, JA, TEJB, MJ, ECR, SNS), and 5R01EB009352 (DSM), 5P30NS048056 (DSM), and 5R24MH108315 (DSM).
Footnotes
The authors declare no competing financial interests.
References
- 1.Behrens TE & Sporns O Human connectomics. Current opinion in neurobiology 22, 144–153, doi: 10.1016/j.conb.2011.08.005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jbabdi S, Sotiropoulos SN, Haber SN, Van Essen DC & Behrens TE Measuring macroscopic brain connections in vivo. Nature neuroscience 18, 1546–1555, doi: 10.1038/nn.4134 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Smith SM Introduction to the Neuroimage Special Issue “Mapping the Connectome”. Neuroimage 80, 1 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Van Essen DC et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79, doi: 10.1016/j.neuroimage.2013.05.041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ugurbil K et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage 80, 80–104, doi: 10.1016/j.neuroimage.2013.05.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glasser MF et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124, doi: 10.1016/j.neuroimage.2013.04.127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffanti L et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage 95, 232–247, doi: 10.1016/j.neuroimage.2014.03.034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glasser MF et al. A multi-modal parcellation of human cerebral cortex. Nature (in press) doi 10.1038/nature18933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson EC et al. MSM: A new flxible framework for multimodal surface matching. Neuroimage 100, 414–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Essen D et al. The brain analysis of spatial maps and atlases (BALSA) databas. Neuroimage (in press) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcus DS et al. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage 80, 202–219, doi: 10.1016/j.neuroimage.2013.05.077 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eklund A, Nichols TE & Knutsson H Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. Proceedings of the National Academy of Sciences, 201602413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner R Where matters: New approaches to brain analysis In: Microstructural Parcellation of the Human Cerebral Cortex Springer; Berlin Heidelberg, pp. 179–196 (2013). [Google Scholar]
- 14.Turner R & Geyer S Comparing like with like: the power of knowing where you are. Brain connectivity 4, 547–557, doi: 10.1089/brain.2014.0261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sereno MI et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science 268, 889–893 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Mruczek RE, Arcaro MJ & Kastner S Probabilistic Maps of Visual Topography in Human Cortex. Cereb Cortex 25, 3911–3931, doi: 10.1093/cercor/bhu277 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischl B & Dale AM Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America 97, 11050–11055, doi: 10.1073/pnas.200033797 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasser MF, Goyal MS, Preuss TM, Raichle ME & Van Essen DC Trends and properties of human cerebral cortex: correlations with cortical myelin content. NeuroImage 93 Pt 2, 165–175, doi: 10.1016/j.neuroimage.2013.03.060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasser MF & Van Essen DC Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 11597–11616, doi: 10.1523/JNEUR0SCI.2180-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dale AM, Fischl B & Sereno MI Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 9, 179–194 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Dale AM & Sereno MI Improved localizadon of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. Journal of cognitive neuroscience 5, 162–176 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Fischl B, Liu A & Dale AM Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE transactions on medical imaging 20, 70–80 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Fischl B et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Fischl B et al. Sequence-independent segmentation of magnetic resonance images.Neuroimage 23, S69–S84 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Fischl B, Sereno MI & Dale AM Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 9, 195–207, doi: 10.1006/nimg.1998.0396 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Fischl B, Sereno MI, Tootell RB & Dale AM High-resolution intersubject averaging and a coordinate system for the cortical surface. Human brain mapping 8, 272–284 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ségonne F, Pacheco J & Fischl B Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE transactions on medical imaging 26, 518–529 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Barch DM et al. Function in the human connectome: task-fMRI and individual differences in behavior. NeuroImage 80, 169–189, doi: 10.1016/j.neuroimage.2013.05.033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wandell BA & Winawer J Imaging retinotopic maps in the human brain. Vision Res 51, 718–737, doi: 10.1016/j.visres.2010.08.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orban GA, Zhu Q & Vanduffel W The transition in the ventral stream from feature to real-world entity representations. Front Psychol 5, 695, doi: 10.3389/fpsyg.2014.00695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brewer AA, Press WA, Logothetis NK & Wandell BA Visual areas in macaque cortex measured using functional magnetic resonance imaging. The Journal of neuroscience: the official journal of the Society for Neuroscience 22, 10416–10426 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Essen DC, Felleman DJ, DeYoe EA, Olavarria J & Knierim J Modular and hierarchical organization of extrastriate visual cortex in the macaque monkey. Cold Spring Harbor symposia on quantitative biology 55, 679–696 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Calabrese E, Badea A, Cofer G, Qi Y & Johnson GA A Diffusion MRI Tractography Connectome of the Mouse Brain and Comparison with Neuronal Tracer Data. Cereb Cortex 25, 4628–4637, doi: 10.1093/cercor/bhv121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donahue C et al. Using diffusion tractography to predict cortical connection strength and distance: A quantitative comparison with tracers in the monkey. Neuroimage. 2016. pii: S1053–8119(16)30035–0. doi: 10.1016/j.neuroimage.2016.04.002. [Epub ahead of prin t] (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basser PJ, Mattiello J & LeBihan D MR diffusion tensor spectroscopy and imaging. Biophysical journal 66, 259–267, doi: 10.1016/S0006-3495(94)80775-1 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fieremans E, Jensen JH & Helpern JA White matter characterization with diffusional kurtosis imaging. Neuroimage 58, 177–188, doi: 10.1016/j.neuroimage.2011.06.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Schneider T, Wheeler-Kingshott CA & Alexander DC NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016, doi: 10.1016/j.neuroimage.2012.03.072 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Van Essen DC, Glasser MF, Dierker DL, Harwell J & Coalson T Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb Cortex 22, 2241–2262, doi: 10.1093/cercor/bhr291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liewald D, Miller R, Logothetis N, Wagner HJ & Schuz A Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque. Biological cybernetics 108, 541–557, doi: 10.1007/s00422-014-0626-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Essen DC & Ugurbil K The future of the human connectome. Neuroimage 62, 1299–1310, doi: 10.1016/j.neuroimage.2012.01.032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang SY et al. The impact of gradient strength on in vivo diffusion MRI estimates of axon diameter. Neuroimage 106, 464–472, doi: 10.1016/j.neuroimage.2014.12.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNab JA et al. The Human Connectome Project and beyond: initial applications of 300 mT/m gradients. Neuroimage 80, 234–245, doi: 10.1016/j.neuroimage.2013.05.074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Setsompop K et al. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magnetic resonance in medicine 67, 1210–1224, doi: 10.1002/mrm.23097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Setsompop K et al. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage 80, 220–233, doi: 10.1016/j.neuroimage.2013.05.078 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sotiropoulos SN et al. Fusion in diffusion MRI for improved fibre orientation estimation: An application to the 3T and 7T data of the human connectome project. Neuroimage 134, 396–409, doi: 10.1016/j.neuroimage.2016.04.014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sotiropoulos SN et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 80, 125–143, doi: 10.1016/j.neuroimage.2013.05.057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vu AT et al. High resolution whole brain diffusion imaging at 7T for the Human Connectome Project. Neuroimage 122, 318–331, doi: 10.1016/j.neuroimage.2015.08.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hillman EM Coupling mechanism and significance of the BOLD signal: a status report. Annual review of neuroscience 37, 161–181, doi: 10.1146/annurev-neuro-071013-014111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feinberg DA et al. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One 5, e15710, doi: 10.1371/journal.pone.0015710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JE & Glover GH BOLD fractional contribution to resting-state functional connectivity above 0.1 Hz. Neuroimage 107, 207–218, doi: 10.1016/j.neuroimage.2014.12.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niazy RK, Xie J, M iller K, Beckmann CF & Smith SM Spectral characteristics of resting state networks. Prog Brain Res 193, 259–276, doi: 10.1016/B978-0-444-53839-0.00017-X (2011). [DOI] [PubMed] [Google Scholar]
- 52.Auerbach EJ, Xu J, Yacoub E, Moeller S & Ugurbil K Multiband accelerated spin-echo echo planar imaging with reduced peak RF pow er using time-shifted RF pulses. Magnetic resonance in medicine 69, 1261–1267, doi: 10.1002/mrm.24719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moeller S et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magnetic resonance in medicine 63, 1144–1153, doi: 10.1002/mrm.22361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J et al. Evaluation of slice accelerations using multiband echo planar imaging at 3 T. Neuroimage 83, 991–1001, doi: 10.1016/j.neuroimage.2013.07.055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laumann TO et al. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670, doi: 10.1016/j.neuron.2015.06.037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jbabdi S, Sotiropoulos SN, Savio AM, Grana M & Behrens TE Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magnetic resonance in medicine 68, 1846–1855, doi: 10.1002/mrm.24204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeurissen B, Tournier JD, Dhollander T, Connelly A & Sijbers J Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411–426, doi: 10.1016/j.neuroimage.2014.07.061 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Greve DN & Fischl B Accurate and robust brain image alignment using boundary-based registration. NeuroImage 48, 63–72, doi: 10.1016/j.neuroimage.2009.06.060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersson JL & Sotiropoulos SN Non-parametric representation and prediction of single- and multi-shell diffusion-weighted MRI data using Gaussian processes. Neuroimage 122, 166–176, doi: 10.1016/j.neuroimage.2015.07.067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson JL & Sotiropoulos SN An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078, doi: 10.1016/j.neuroimage.2015.10.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersson JLR, Graham MS, Zsoldos E & Sotiropoulos SN Incorporating outlie rdetection and replacement into a non-parameteric framework for movement and distortion correction of diffusion MR images. Neuroimage (in review) (2016). [DOI] [PubMed] [Google Scholar]
- 62.Power JD, Schlaggar BL & Petersen SE Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage 105, 536–551, doi: 10.1016/j.neuroimage.2014.10.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Power JD, Barnes KA, Snyder AZ, Schlaggar BL & Petersen SE Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59, 2142–2154, doi: 10.1016/j.neuroimage.2011.10.018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Satterthwaite TD et al. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. NeuroImage 60, 623–632, doi: 10.1016/j.neuroimage.2011.12.063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beckmann CF & Smith SM Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging 23, 137–152, doi: 10.1109/TMI.2003.822821 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Winkler AM, Webster MA, Vidaurre D, Nichols TE & Smith SM Multi-level block permutation. Neuroimage 123, 253–268, doi: 10.1016/j.neuroimage.2015.05.092 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H et al. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE Trans Med Imaging 26, 1585–1597, doi: 10.1109/TMI.2007.906784 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Yushkevich PA, Alexander DC & Gee JC Deformable registration of diffusion tensor MR images with explicit orientation optimization. Medical image analysis 10, 764–785, doi: 10.1016/j.media.2006.06.004 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Amunts K, Malikovic A, Mohlberg H, Schormann T & Zilles K Brodmann’s areas 17 and 18 brought into stereotaxic space-where and how variable? NeuroImage 11, 66–84, doi: 10.1006/nimg.1999.0516 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Eickhoff SB et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage 25, 1325–1335, doi: 10.1016/j.neuroimage.2004.12.034 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Van Essen DC A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. NeuroImage 28, 635–662, doi: 10.1016/j.neuroimage.2005.06.058 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Anticevic A et al. Comparing surface-based and volume-based analyses of functional neuroimaging data in patients with schizophrenia. NeuroImage 41, 835848, doi: 10.1016/j.neuroimage.2008.02.052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischl B et al. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex 18, 1973–1980, doi: 10.1093/cercor/bhm225 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frost MA & Goebel R Measuring structural-functional correspondence: spatial variability of specialised brain regions after macro-anatomical alignment. NeuroImage 59, 1369–1381, doi: 10.1016/j.neuroimage.2011.08.035 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Smith SM et al. Resting-state fMRI in the Human Connectome Project. NeuroImage 80, 144–168, doi: 10.1016/j.neuroimage.2013.05.039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tucholka A, Fritsch V, Poline JB & Thirion B An empirical comparison of surface-based and volume-based group studies in neuroimaging. NeuroImage 63, 1443–1453, doi: 10.1016/j.neuroimage.2012.06.019 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Felleman DJ & Van Essen DC Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex 1, 1–47 (1991). [DOI] [PubMed] [Google Scholar]
- 78.Haxby JV et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72, 404–416, doi: 10.1016/j.neuron.2011.08.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Glasser MF & Rilling JK DTI tractography of the human brain’s language pathways. Cereb Cortex 18, 2471–2482, doi: 10.1093/cercor/bhn011 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Brodmann K Vergleichende Lokalisationslehre der G rosshirnrinde. Leipzig:. (1909). [Google Scholar]
- 81.Nieuwenhuys R The myeloarchitectonic studies on the human cerebral cortex of the Vogt-Vogt school, and their significance for the interpretation of functional neuroimaging data. Brain structure & function 218, 303–352, doi: 10.1007/s00429-012-0460-z (2013). [DOI] [PubMed] [Google Scholar]
- 82.Vogt C & Vogt O Allgemeinere ergebnisse unswerer hirnforschung. J Psychol Neurol 25, 279–468 (1919). [Google Scholar]
- 83.Nieuwenhuys R, Broere CA & Cerliani L A new myeloarchitectonic map of the human neocortex based on data from the Vogt-Vogt school. Brain structure & function 220, 2551–2573, doi: 10.1007/s00429-014-0806-9 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Gordon EM et al. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex 26, 288–303, doi: 10.1093/cercor/bhu239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Power JD et al. Functional network organization of the human brain. Neuron 72, 665–678, doi: 10.1016/j.neuron.2011.09.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo BT et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106, 1125–1165, doi: 10.1152/jn.00338.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hacker CD et al. Resting state network estimation in individual subjects. NeuroImage 82, 616–633, doi: 10.1016/j.neuroimage.2013.05.108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cole MW, Bassett DS, Power JD, Braver TS & Petersen SE Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251, doi: 10.1016/j.neuron.2014.05.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fischl B et al. Automatically parcellating the human cerebral cortex. Cerebral cortex 14, 11–22 (2004). [DOI] [PubMed] [Google Scholar]
- 90.Hodge MR et al. Connectom eDB-Sharing human brain connectivity data. Neuroimage 124, 1102–1107, doi: 10.1016/j.neuroimage.2015.04.046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marcus DS, Olsen TR, Ramaratnam M & Buckner RL The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroim aging data. Neuroinformatics 5, 11–34 (2007). [DOI] [PubMed] [Google Scholar]
- 92.Smith SM et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nature neuroscience 18, 1565–1567, doi: 10.1038/nn.4125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeo BT, Krienen FM, Chee MW & Buckner RL Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. Neuroimage 88C, 212–227, doi: 10.1016/j.neuroimage.2013.10.046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang D et al. Parcellating cortical functional networks in individuals. Nature neuroscience 18, 1853–1860, doi: 10.1038/nn.4164 (2015b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hawrylycz M et al. Canonical genetic signatures of the adult human brain. Nature neuroscience 18, 1832–1844, doi: 10.1038/nn.4171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tavor I et al. Task-free MRI predicts in dividual differences in brain a ctivity during task performance. Science 352, 216–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reveley C et al. Superficial white matter fiber systems impede detection of long- range cortical connections in diffusion MR tractography. Proceedings of the National Academy of Sciences of the United States of America 112, E2820–2828, doi: 10.1073/pnas.1418198112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dalcanton JJ 18 years of science with the Hubble Space Telescope. Nature 457, 4150, doi: 10.1038/nature07621 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Kuhn T The Structure of Scientific Revolutions. Chicago: The University of Chicago Press; (1962). [Google Scholar]
- 100.De Martino F et al. Frequency preference and attention effects across cortical depths in the human primary auditory cortex. Proceedings of the National Academy of Sciences of the United States of America 112, 16036–16041, doi: 10.1073/pnas.1507552112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.De Martino F et al. Spatial organization of frequency preference and selectivity in the human in ferior colliculus. Nature communications 4, 1386, doi: 10.1038/ncomms2379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Muckli L et al. Contextual Feedback to Superficial Layers of V1. Current biology: CB 25, 2690–2695, doi: 10.1016/j.cub.2015.08.057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.M alikovic A et al. Cytoarchitectonic analysis of the human extrastriate cortex in the region of V5/MT+: a probabilistic, stereotaxic map of area hOc5. Cereb Cortex 17, 562–574, doi: 10.1093/cercor/bhj181 (2007). [DOI] [PubMed] [Google Scholar]
- 104.Abdollahi RO et al. Correspondences between retinotopic areas and myelin maps in human visual cortex. NeuroImage 99, 509–524, doi: 10.1016/j.neuroimage.2014.06.042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Murphy K, Birn RM, Handwerker DA, Jones TB & Bandettini PA The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? NeuroImage 44, 893–905, doi: 10.1016/j.neuroimage.2008.09.036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Power JD et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage 84, 320–341, doi: 10.1016/j.neuroimage.2013.08.048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saad ZS et al. Correcting brain-wide correlation differences in resting-state FMRI. Brain connectivity 3, 339–352, doi: 10.1089/brain.2013.0156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gotts SJ et al. The perils of global signal regression for group comparisons: a case study of Autism Spectrum Disorders. Frontiers in human neuroscience 7, 356, doi: 10.3389/fnhum.2013.00356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saad ZS et al. Trouble at rest: how correlation patterns and group differences become distorted after global signal regression. Brain Connect 2, 25–32, doi: 10.1089/brain.2012.0080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Birn RM The role of physiological noise in resting-state functional connectivity. NeuroImage 62, 864–870, doi: 10.1016/j.neuroimage.2012.01.016 (2012). [DOI] [PubMed] [Google Scholar]
- 111.Chang C & Glover GH Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage 47, 1381–1393, doi: 10.1016/j.neuroimage.2009.04.048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Golestani AM, Chang C, Kwinta JB, Khatamian YB & Jean Chen J Mapping the end-tidal CO2 response function in the resting-state BOLD fMRI signal: spatial specificity, test-retest reliability and effect of fMRI sam pling rate. NeuroImage 104, 266–277, doi: 10.1016/j.neuroimage.2014.10.031 (2015). [DOI] [PubMed] [Google Scholar]
- 113.Smith SM et al. Network modelling methods for FMRI. NeuroImage 54, 875–891, doi: 10.1016/j.neuroimage.2010.08.063 (2011). [DOI] [PubMed] [Google Scholar]
- 114.Larson-Prior LJ et al. Adding dynamics to the Human Connectome Project with MEG. NeuroImage 80, 190–201, doi: 10.1016/j.neuroimage.2013.05.056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jack CR Jr. et al. The Alzheimer’s Disease Neuroim aging Initiative (ADNI): MRI methods. Journal of magnetic resonance imaging : JMRI 27, 685–691, doi: 10.1002/jmri.21049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.