Abstract
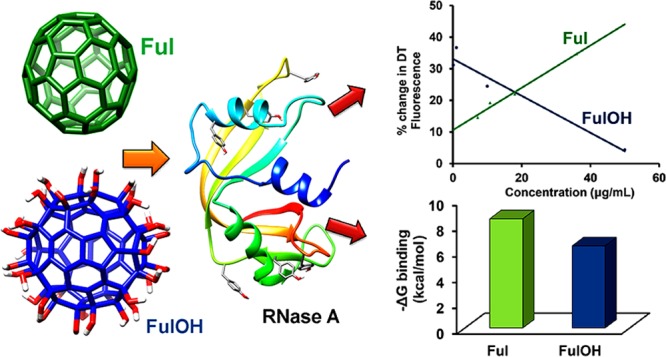
Fullerene–protein interaction studies have been a key topic of investigation in recent times, but the lower water solubility of fullerene somewhat limits its application in the biological system. In this work, we have compared the activities of fullerene and its water-soluble hydrated form, that is fullerenol, on ribonuclease A (RNase A) under physiological conditions (pH 7.4). The interaction studies of fullerene and fullerenol with protein suggest that the binding depends on the hydrophobic interactions between the protein and the ligand. In addition, fullerene and fullerenol slow down the ribonucleolytic activity of RNase A through noncompetitive and mixed types of inhibition, respectively. This precisely gives the idea about the ligand-binding sites in RNase A, which has further been explored using docking studies. Both these nanoparticles show a reduction in dityrosine formation in RNase A caused due to oxidative stress and also prevent RNase A dimer formation to different extents depending on their concentration.
1. Introduction
Nanoparticles have found widespread applications in the field of polymer science,1 green synthesis,2 electronics,3 and also absorption of species, which would otherwise be harmful to health.4,5 Their contribution in the field of biology and medicinal chemistry has highlighted their ever-increasing necessity as the nanometer-level dimensions render them possible for use in monitoring cellular activities at a minute scale.6 They find applications in the field of fluorescent labeling,7 purification of biomolecules,8 drug delivery,9 probing proteins and DNA,10,11 treatment of tumors,12 etc.
Fullerene (Ful) is a carbon allotrope that can take the forms of hollow sphere, ellipsoid, tube, and other geometries, of which spherical fullerenes are buckyballs. The smallest member of the buckyball family is C20, and the most common member is C60 (buckminsterfullerene). The cagelike structure of fullerenes has led to much focus on their properties (Figure 1). The production of fullerene-specific antibodies have shown that they are capable of specific interactions with proteins.13 The widespread applications of fullerene in biology are mainly due to the presence of significant interactions with proteins in aqueous solution. Fullerenes and their derivatives have been extensively explored for biomedical applications. Examples of fullerene bioactivity include antibacterial activity,14 neuroprotection,15 DNA cleavage,16 apoptosis,17 ion channel inhibition,18 and inhibition of amyloid formation.19 Due to the high degree of unsaturation, the molecule is able to effectively trap free radicals, thereby making it a radical sponge.20 In vivo studies on rats have shown that at moderate concentrations fullerene increases the life span significantly without any toxic effects.21 It shows nanomolar-level affinity toward peptides, leading to the formation of fullerene-based tetrameric helix bundles.22 Apart from this, it shows fluorescence quenching in human serum albumin (HSA) as well as significant perturbation in the chemical shift in the presence of lysozyme.23,24 As a result, fullerene and its derivatives have been utilized as inhibitors for certain proteins like neuronal nitric oxide synthase,25 human immunodeficiency virus protease,26 glutathione reductase,27 and tyrosine phosphatases.28 Studies on fullerene are, however, limited due to its poor solubility in aqueous medium. In aqueous solution, it has a solubility of less than 10–9 mg/L with an octanol–water partition coefficient log KOW of 6.67 and readily forms aggregates of nC60, thereby making it sparingly soluble in water.23
Figure 1.
Structure of (a) fullerene and (b) fullerenol.
Several techniques are employed to make nanoparticles soluble in aqueous medium to exploit their biological properties. Generally, carbohydrate-based compounds like chitosan and cyclodextrin have been found to increase the solubility of nanoparticles for their use in drug delivery and other biological applications.29,30 This laboratory has also reported methods to prepare protein-based nanoparticles as well as poly(lactic-co-glycolic acid)-loaded nanoparticles to improve the solubility of bioactive compounds.31,32 In this article, the focus is on the development of water-soluble fullerene derivatives to investigate their inhibitory and antioxidant abilities. Hydroxylated fullerenes, generally known as fullerenols (FulOH), are commonly used water-soluble fullerene derivatives. The solubility of these nanoparticles depends on the number of OH (hydroxyl) groups attached to the fullerene unit and is denoted C60(OH)x [x = number of hydroxyl groups attached]. Chiang et al. have shown that fullerenols with 12 OH groups are not as soluble in water33 as C60(OH)22–2634 under neutral conditions. On the other hand, higher fullerenol compounds prepared using H2O2 in the absence and presence of a phase-transfer catalyst showed much improved solubility in water to about 17.5 mg/mL [C60(OH)36] and 64.9 mg/mL [C60(OH)44], respectively.35,36 The higher solubility of fullerenol renders it possible to be used in biological systems with almost negligible toxicity.37 The unsaturated fullerene unit along with the large number of attached −OH groups also makes fullerenol a better radical scavenger. Polyphenolic compounds having several −OH groups are known to possess high antioxidant activity as estimated using the 2,2-diphenyl-1-picrylhydrazyl assay.38 However, encapsulation of such polyphenols using β-cyclodextrin has shown to increase the overall solubility and prevent oligomerization of proteins caused due to oxidative stress.39 Research on the antioxidant properties of fullerenol showed its ability to quench the superoxide radical generated in the presence of xanthine and xanthine oxidase,40 which further led to the scope for in vivo studies. Fullerenol has the ability to increase the survival rates of mice by quenching reactive oxygen species41 and can also prevent the damage of A549 cells in the presence of H2O2 with much higher efficiency.42 The interaction of fullerenols with proteins has been studied by Yang et al. with lysozyme using both experimental and theoretical methods, where a high association constant was observed followed by tryptophan (Trp) quenching.24 The protein binding ability of fullerenol was further explored for studying the enzymatic inhibition properties where it showed significant reduction in enzymatic activity at high fullerenol concentrations. Detailed studies on the enzyme (E) monooxygenase showed that fullerenol acts as a noncompetitive and mixed-type inhibitor depending on the nature of the substrate with Ki values at the micromolar level.43 Wu et al. used docking studies to explore the interaction of fullerene and fullerenol with different proteins. The results highlighted that the extent of binding interactions between the protein and ligand depends on the size of fullerene unit and number of OH groups attached.44 However, the study was restricted to theoretical calculations only.
In the present work, fullerenol has been prepared and the effect of both fullerene and fullerenol on ribonuclease A (RNase A) has been investigated experimentally using several spectroscopic studies. Ribonucleases (RNases) are a group of enzymes that catalyze the degradation of RNA into smaller components. RNases catalytically cleave RNA to maintain an optimal level of balance in protein synthesis during the transcription and translation processes. One of the most widely studied proteins of this family is bovine pancreatic ribonuclease A45,46 (RNase A). Higher levels of RNases can lead to cytotoxic effects by entering into the cell cytosol and preventing protein synthesis by breaking down RNA, thereby causing cell death.47 Earlier reports from our laboratory have shown that polyphenols like rutin and quercetin as well as green tea polyphenols like (−)-epicatechin gallate (ECG) and (−)-epigallocatechin gallate have inhibitory effects on RNase A.48,49
Because RNase A plays a crucial role in protein synthesis, a proper balance of this protein within our body is highly essential. Deficiency of ribonucleases can lead to brain infection; thus, retention of enzyme activity is equally important.50 Oxidative stress is one of the main causes for protein damage caused due to reactive free radical species.51 Out of several modifications in amino acid residues occurring due to oxidative stress, dityrosine (DT) formation is one such modification often found in patients suffering from Alzheimer’s disease, cataracts, etc.52 Dityrosine (DT) formation can result in intermolecular covalent bond formation between two Tyr residues of different monomers, leading to a protein dimer.53 Because both fullerene and fullerenol have antioxidant properties, these molecules have been used to study the changes in DT formation caused due to oxidation of RNase A in the presence of peroxide. The extent of dimer formation has been monitored using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF). The role of fullerene and fullerenol as an inhibitor as well as an antioxidant molecule has also been explored. The number of binding sites and Tyr fluorescence quenching using these two compounds have been monitored to further understand protein–nanoparticle interactions. In the present work, we investigated the interaction of fullerene and fullerenol with RNase A and compared the extent of inhibition of the ribonucleolytic activity of the protein.
2. Results and Discussion
Fullerene and fullerenol have been characterized using Fourier transform infrared (FTIR), dynamic light scattering (DLS), and MALDI-TOF along with microscopic techniques field emission scanning electron microscopy (FESEM) and atomic force microscopy (AFM). The nanoparticles were initially used for interaction studies with RNase A, after which their effects on the enzymatic action of the protein were investigated. Due to their high radical quenching ability, both fullerene and fullerenol were used to prevent the dityrosine (DT) formation in RNase A due to oxidative stress.
2.1. Characterization of Aqueous Suspension of Fullerene and Fullerenol Prepared
2.1.1. UV–Vis Studies
The UV–vis spectra for both fullerene and fullerenol were measured in the range of 200–600 nm. Fullerene gives two characteristic peaks, one at around 340 nm and the other at around 260 nm, which changes slightly depending on the degree of association of fullerene in water.54 In our present work, the concentration of fullerene has been determined by taking the absorbance value at 347 nm (ε347 = 18 900 M–1 cm–1).55 Fullerenol, on the other hand, being a clear and colorless solution, does not exhibit any characteristic peaks like fullerene (Figure S1 in Supporting Information).56
2.1.2. FTIR Studies
Fullerene shows a distinct peak at around 1647 cm–1, which is due to the C=C stretching, whereas the other peaks around at 1400, 1153, 580, and 522 cm–1 are due to the IR-active F1u modes of fullerene C60.57 In the case of fullerenol, a sharp peak of very high intensity at around 1107 cm–1 (corresponding to C–O stretching) confirms the linking of hydroxyl groups to the fullerene system along with C=C peaks at around 1618 cm–1 (Figure 2). The broad region 3415 cm–1 corresponds to the −OH stretching in addition to −OH bending vibrations at around 1446 cm–1 (in plane) and 987 cm–1 (out of plane).58,59
Figure 2.
FTIR spectra of Fullerene and Fullerenol.
2.1.3. MALDI-TOF
Mass spectrometric analysis (MALDI-TOF) of the fullerene sample gave a sharp, highly intense peak at around 720 Da m/z, which is the molecular weight for C60 fullerene (Figure S2 in Supporting Information).60 This suggests that the aqueous suspension of fullerene is highly pure. MALDI-TOF analysis of the fullerenol sample gave a distribution of peaks ranging from 1.3 to 1.4 kDa m/z (Figure 3), indicating that about 30–40 hydroxyl groups are present in the prepared sample of fullerenol, thus contributing to its high solubility in aqueous medium.61,62
Figure 3.
MALDI-TOF spectra of fullerenol showing a series of peaks within 1300–1400 m/z.
2.1.4. Morphology, Size, and ζ Potential Measurements
The morphology and size range of fullerene and fullerenol were initially measured from field emission scanning electron microscopy (FESEM) and atomic force microscopy (AFM). Both these microscopy images showed that both fullerene and fullerenol particles form aggregates in aqueous solution having somewhat spherical shapes (Figure S3 in Supporting Information). The FESEM images show that the fullerene and fullerenol nanoparticles are almost spherical in shape and have smooth surfaces. The average size distribution of the nanoparticles in FESEM for fullerene is around 140–160 nm, and it increases to 230–250 nm for fullerenol. In both the cases, larger sizes of the nanoparticles can be seen, which results from aggregation of smaller particles.63 AFM also shows the presence of spherical nanoparticles in the system, and the size distribution gives the sizes of the nanoparticles present within the system. The sizes of fullerene nanoparticles from AFM are around 150 nm and just over 250 nm for fullerenol nanoparticles. The greater size of fullerenol may be due to the increase in size of the pristine C60 due to addition of −OH groups. However, in AFM, it can be seen that the fullerene nanoparticles are well distributed and are separated from one another; however, the nanoparticles are closely packed in the case of fullerenol. This may be due to the fact that the −OH groups in fullerenol are involved in intermolecular hydrogen bonding with neighboring fullerenol molecules, leading to a closely associated structure. This may be manifested in the larger fullerenol aggregates, which are closely packed, as seen in AFM images. The size of the nanoparticles was also determined using dynamic light scattering experiments. It was observed that the average size of the fullerene nanoparticles was 158 ± 27.71 nm (Figure 4), which is close to that obtained from AFM. Upon hydration, the average size of the fullerenol nanoparticles increases by ∼50% to 237.5 ± 24.7 nm. The ζ potential of fullerene solution in water at pH 7.4 was found to be −7.34 ± 2.80 mV, and it decreases to −14.45 ± 1.90 mV for fullerenol solution in water. The presence of −OH groups in fullerenol may increase the negative charge density as compared to that in fullerene, accounting for the decrease in the ζ potential value.
Figure 4.

DLS measurements of fullerene and fullerenol.
2.2. Interaction Studies of Fullerene and Fullerenol with RNase A
The interaction studies were carried out by studying the Tyr fluorescence quenching in RNase A in the presence of fullerene and fullerenol. RNase A exhibits fluorescence emission at 304 nm on excitation at 278 nm due its tyrosine residues.64 The emission peak intensity decreases with an increase in both fullerene and fullerenol concentrations in the solution (Figure 5). However, the decrease in intensity is much more significant in the case of fullerene compared to that in fullerenol. From Table 1, it can be seen that the Ksv values for fullerene are ∼5 times higher than those for fullerenol. The lower Ksv value for fullerenol suggests that fullerene is able to quench the Tyr residue more effectively than fullerenol. Although both nanoparticles bind to the protein in a 1:1 ratio, the binding constant (Kb) values indicate that the binding of fullerenol decreases by ∼34% in comparison to that of fullerene. It may be concluded from the tyrosine quenching experiment that although fullerene and fullerenol both bind to RNase A the binding sites are different and fullerenol binds at a site further away from the Tyr residue compared with fullerene.
Figure 5.
Fluorescence quenching of RNase A in the presence of fullerene (0–10 μg/mL) and fullerenol (0–2 μg/mL).
Table 1. Binding Parameters for Fullerene and Fullerenol with RNase A.
| Ksv, (g/mL)−1 × 104 | Kb, (g/mL)−1 × 104 | n | |
|---|---|---|---|
| fullerene | 8.2 ± 0.65 | 5.3 ± 0.42 | 1.19 |
| fullerenol | 1.7 ± 0.11 | 3.5 ± 0.13 | 1.09 |
In the presence of proteins, the sizes of the fullerene nanoparticles increase to a significant extent as seen from DLS results (Figure S4 in Supporting Information). The average size of fullerene increases by ∼42% (275.15 ± 28.5 nm) and of fullerenol by ∼35% (369.05 ± 38.2 nm) in the presence of RNase A at pH 7.4. This may be due to the fact that the protein gets bound to the surface of the nanoparticles, which leads to the increase in their size by a significant amount. Moreover, on addition of RNase A to the nanoparticle solution, the overall charge on the nanoparticles becomes almost neutral with the fullerene–protein complex having a slightly higher positive charge (0.16 ± 0.01 mV) than that on fullerenol/protein complex (0.06 ± 0.01 mV) at pH 7.4. RNase A is positively charged at pH 7.4 (pI 9.6) and attaches to the surface of the nanoparticles (as seen from DLS studies), thereby reducing the negative charge on the nanoparticles. Both these study signify that in the presence of protein, fullerene and fullerenol bind with RNase A, which increases their size in solution, and also, due to the higher pI of the protein (pI 9.6), the overall negative charge on the nanoparticles decreases. Moreover, the increase in size for protein–fullerene is higher as compared to that for protein–fullerenol. This can be explained from the interaction studies, which show that the binding affinity of fullerene is higher than that of fullerenol, which indicates that in the system more number of RNase A can bind to the fullerene nanoparticle than with fullerenol.
2.3. Inhibition Studies
2.3.1. Agarose Gel-Based Assay
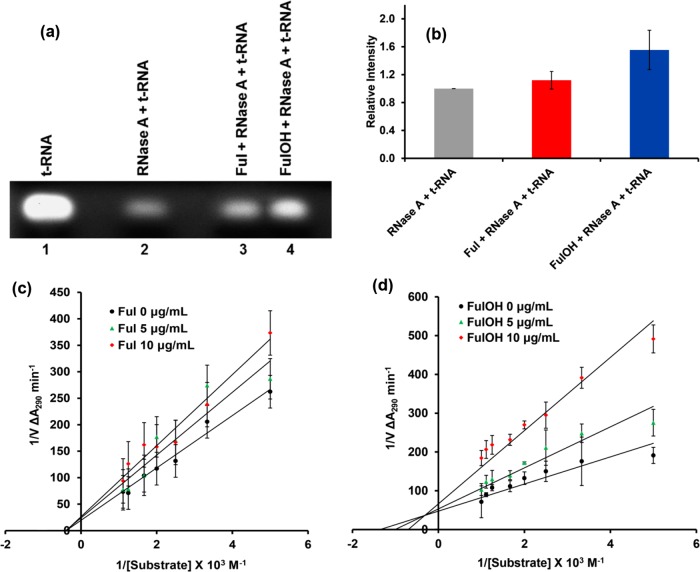
The effect of fullerene and fullerenol on RNase A was assayed qualitatively by the degradation of t-RNA in an agarose gel-based assay. The extent of degradation of t-RNA was estimated by observing the intensity of stained ethidium bromide, as shown in Figure 6a. The ability of the ligand to affect the enzyme activity is measured in terms of the extent of t-RNA degradation from the differential intensity observed. Lane 2 displays degraded t-RNA in the presence of RNase A, with intensity significantly decreased with respect to that in lane 1 (only t-RNA). Lane 3 displays slightly enhanced intensity compared with lane 2, indicating reduced degradation of t-RNA in the presence of fullerene (Figure 6b). The intensity of lane 4 is higher than that of lane 3, indicating that the extent of degradation is the least in the presence of fullerenol. This suggests that both fullerene and fullerenol inhibit the RNase A activity with fullerenol showing somewhat greater inhibition compared with fullerene.
Figure 6.
(a) Agarose gel assay for fullerene and fullerenol; (b) relative intensity in agarose gel; Lineweaver–Burk plot for (c) fullerene (Ful) and (d) fullerenol (FulOH).
2.3.2. Inhibition Kinetics
The mode of inhibition in RNase A by fullerene is noncompetitive, whereas for fullerenol, it is of a mixed type, as shown in Figure 6c,d. The inhibition constant (Ki) value for fullerene is 26.49 ± 2.17 μg/mL, and being a noncompetitive inhibitor, it binds to a region away from the active site generally referred to as the allosteric site (Table 2). In the case of fullerenol, it can bind to the active site as well as the allosteric site with a Ki value of 4.66 ± 1.04 μg/mL. Both the nanoparticles significantly lower the enzymatic activity of RNase A, with fullerenol being slightly more effective compared with fullerene.
Table 2. Inhibition Constant Values (Ki) and Type of Inhibition for Fullerene and Fullerenol against Ribonucleolytic Activity of RNase A.
| inhibitor | Ki (μg/mL) | type of Inhibition |
|---|---|---|
| fullerene | 26.49 ± 2.17 | noncompetitive |
| fullerenol | 4.66 ± 1.04 | mixed type |
2.4. Docking Studies
Both fullerene and fullerenol were optimized in Avogadro software. MALDI-TOF suggested that the number of hydroxyl groups present in fullerenol ranges from 30 to 40. In our case, fullerenol was prepared by placing OH groups in the equatorial position, which is considered to be the most stable,65 and 35 −OH groups were placed accordingly and used for docking studies after optimization.
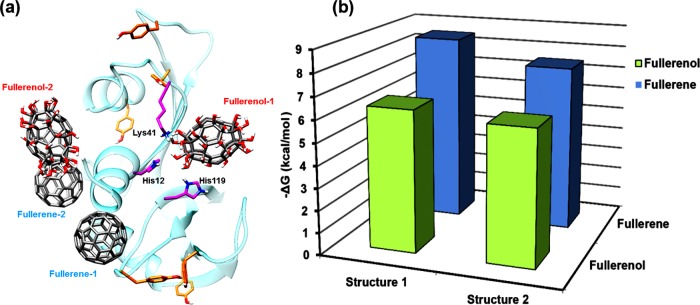
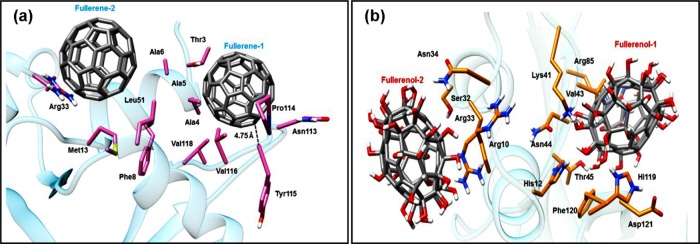
Autodock Vina gives two probable positions for the attachment of both fullerene and fullerenol with a slight difference in their −ΔG values. The two different modes of attachment of fullerene and fullerenol are represented by fullerene-1/fullerene-2 and fullerenol-1/fullerenol-2, respectively. It can be seen that Tyr115 lies within a distance of 0.5 nm from fullerene-1, whereas no Tyr residues are present near the binding sites of both the fullerenols (Figure 7a). The results obtained show that −ΔG (free energy of binding) is higher in the case of fullerene as compared to that in fullerenol despite the attachment of hydroxyl groups to the latter (Figure 7b). This explains the higher Tyr quenching ability of fullerene over fullerenol. The residues located close to fullerene-1 and fullerene-2 are mostly nonpolar (Ala, Leu, Val) and aromatic (Phe, Tyr), thereby implying that the protein–ligand binding is stabilized by hydrophobic interactions (Figure 8a). On the other hand, in Figure 8b, both the fullerenols are surrounded mostly by polar residues (Lys, Arg), which might prevent any hydrophobic interaction, thus reducing the extent of binding between the protein and the ligand. The ∼25% decrease in free energy can be attributed to the lowering in hydrophobic interactions, which has been studied earlier using quantum mechanical calculations.66 The lower interaction value for fullerenol can be correlated with the binding energy obtained from Tyr fluorescence quenching studies, which also showed a similar trend.
Figure 7.
(a) Probable positions for binding of fullerene and fullerenol in RNase A as obtained from AutoDock Vina. (b) Free energy of binding for fullerene and fullerenol.
Figure 8.
RNase A residues located within 0.5 nm distance from (a) fullerene and (b) fullerenol.
Fullerenol-1 binds almost directly with the active site residues (His12, Lys41, His119), making it an effective inhibitor as compared with fullerene, which lies slightly away from the active site. This supports the experimentally obtained results for inhibition constant values for fullerene and fullerenol, where Ki (fullerenol) < Ki (fullerene). Figure 9 gives a comparative idea about the distance of fullerene-1 and fullerenol-1 from the Cα backbone atom of three active site residues of RNase A. It has been previously observed that the Lys41 residue in RNase A plays a vital role during RNA cleavage by stabilizing the transition state through hydrogen bond formation with the substrate.67,68 From Figure 8b, the proximity of fullerenol-1 to Lys41 clearly indicates that the hydroxyl groups of fullerenol may get associated with hydrogen bonding with Nζ of Lys41, thereby preventing the amino acid from stabilizing the transition state and finally reducing the enzyme activity. In addition, both fullerene-2 and fullerenol-2 occupy a common binding site, which is away from the active site of RNase A. Previous reports from this laboratory have shown that noncompetitive inhibitors of RNase A (like (−)-epicatechin, ECG) bind near residues like Arg10, Asn34, Leu35, and Lys37.49 From Figure 9, it can be seen that the nearby residues of fullerene-2 and fullerenol-2 are Arg10, Arg33, and Asn34 and thus it can be stated that this region may be the allosteric site for RNase A.
Figure 9.
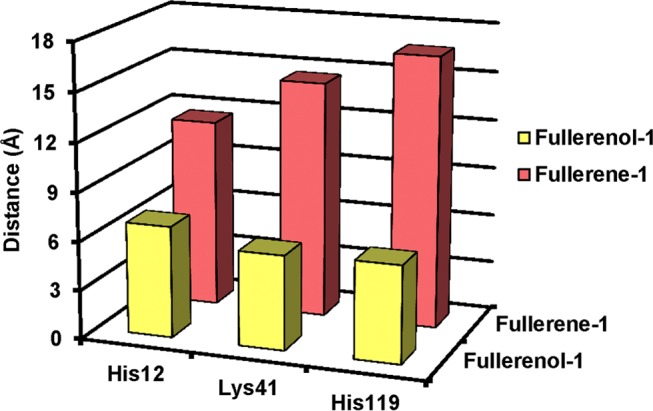
Distance of active site residues of RNase A in the docked structure.
2.5. Prevention of Dityrosine (DT) Cross-Linkage in RNase A
The RNase A dimer obtained by treating pure RNase A with K2S2O8 and Co(OAc)2, can be detected using dityrosine fluorescence, SDS-PAGE and MALDI-TOF. RNase A upon treatment with oxidant shows an enhanced fluorescence peak around 405 nm upon excitation at 317 nm which is absent in native RNase A. The sulfate free radical produced abstracts one proton from the tyrosine residue and forms a tyrosyl free radical which has been reported earlier by using a radiolysis study.69 As a result two such tyrosyl free radicals formed from two different protein molecules come close to one another leading to the formation of a DT bond.
2.5.1. Inhibition of DT Fluorescence
To check how fullerene and fullerenol affect the DT fluorescence, RNase A samples were treated with fullerene and fullerenol prior to the addition of K2S2O8 and Co(OAc)2. A significant decrease in the DT fluorescence intensity was observed with an increase in the concentration of fullerene along with a gradual blue shift from 403 to 400 nm (Figure 10a). The decrease in DT intensity suggests that fullerene reduces the formation of dityrosine cross-linkage, thereby acting as a potential inhibitor against oxidative stress. The shift of around ∼3 nm in the peak intensity in the lower-wavelength region may be due to the fact that with an increase in the concentration of the nanoparticles a significant population of fullerene molecules comes close to the DT bond by replacing the water molecules around it. Fullerene itself being highly hydrophobic decreases the polarity around the DT bond, thereby causing the blue shift.70 In the case of fullerenol, the DT fluorescence intensity decreases initially with an initial increase in concentration. However, the DT intensity increases at a high concentration range of fullerenol (10 μg/mL), indicating that fullerenol could have a tendency to enhance oxidation and subsequent dimerization at high concentrations (Figure 10b).71 In the case of fullerenol, no shift in emission maximum is noticed with change in concentration. This difference from fullerene can be explained on the basis of the extent of interaction of these nanoparticles with Tyr residues. Interaction studies highlighted that fullerene is able to quench the Tyr fluorescence more than fullerenol, thus suggesting that the former is likely to be in a closer proximity to the Tyr, as observed from docking studies. Thus, the population of fullerenol near modified Tyr residues (DT bond) is expected to be lower than fullerene due to which no such significant change in the wavelength shift is observed for fullerenol.
Figure 10.
Change in dityrosine (DT) fluorescence of RNase A with different concentrations of (a) fullerene and (b) fullerenol. Effect of (c) fullerene and (d) fullerenol on dimer formation of RNase A upon oxidation. Oxi: K2S2O8 + Co(OAc)2.
2.5.2. SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed to investigate the effect of fullerene and fullerenol on dimer formation. Figure 10c shows that with an increase in the concentration of fullerene the dimer band intensity around 25 kDa decreases. A similar observation was made for fullerenol (Figure 10d) except that at very high concentration (10 μg/mL) the band intensity at 25 kDa did not show much decrease and thus reinforcing the conclusions derived from the dityrosine (DT) fluorescence experiment.
2.5.3. MALDI-TOF
The observations in MALDI-TOF analysis showed a peak at around 27.6 kDa for the RNase A dimer (monomer peak at around 13.6 kDa) upon oxidation (Figure 11). On addition of fullerene (36 μg/mL) and fullerenol (1.0 μg/mL), the dimer peak intensity decreases significantly. This shows that both fullerene and fullerenol are effective in preventing oxidation and subsequent dimerization of RNase A and have potential antioxidant properties.
Figure 11.
Change in dimer peak intensity of RNase A upon addition of fullerene and fullerenol. Oxi: K2S2O8 + Co(OAc)2.
2.6. Relevance of the Study
Both fullerene and fullerenol show significant interaction with RNase A under physiological conditions (pH 7.4) but with a notable difference in the binding constant values. Fullerene showed greater quenching in Tyr fluorescence compared with fullerenol, which is highlighted from the Stern–Volmer constant (Ksv) values. Previous studies showed that attachment of organophosphate groups to fullerene decreases the Ksv value compared to that for pristine fullerene in the case of HSA, which suggests that the extent of quenching depends on the functional group attached to the fullerene.23,72 Moreover, the extent of quenching for C60 is higher in the case of Tyr (RNase A) than Trp (HSA), which shows that the mode of interaction also depends on the protein residue.23
The binding constant for fullerene and fullerenol is comparable to that for previously reported polyphenolic compounds like rutin and quercetin with RNase A as well as their Cu complexes (on the order 104 M–1).48 On the other hand, carboxy-functionalized fullerene showed enhanced Kb values for HSA, which is about 103 times higher than that for fullerene or its hydrated form for RNase A.73 Because the residues present in the binding pocket differ from one protein to another, the above comparison suggests that the extent of binding depends more on the nature of the protein pocket than the interacting ligand. In the present case, fullerene shows 1:1 binding with RNase A, whereas attachment of carboxy group to fullerene increases the binding to 3:1 in the case of bovine β-lactoglobulin.74 On the other hand, fullerenol shows 1:1 binding in the case of lysozyme, which is similar to our result for RNase A.24
The Ki value shows that fullerenol acts as a better inhibitor than fullerene for RNase A. This difference can be attributed to the fact that Ki calculation takes into consideration the attachment of the ligand with enzyme (E) as well as the enzyme substrate complex, whereas in the case of Kd, the substrate concentration is not involved. The presence of negatively charged oxygen atoms in fullerenol assists the approach to the active site, thereby showing greater inhibition. The nonpolar fullerene molecule has a lower tendency to approach close to the active site and thus binds to the allosteric site (away from the active site), thereby showing lower inhibition.
The enzymatic activity of the dityrosine (DT) cross-linked RNase A dimer was compared to that of the monomer using the agarose gel-based assay. It was found that the intensity of the dimer peak is almost 2 times that of the monomer, indicating the presence of a higher amount of active t-RNA in the dimer system as compared with the monomer (Figure S5a,b). This shows that the activity of dimer is somewhat lower than that of the monomer. The kinetics data (using cytidine 2′,3′-cyclic monophosphate (2′,3′-cCMP) as substrate) (Figure S5c) showed that the maximum rate of substrate degradation decreases by 16% (from Vmax values) in the case of the dimer, which is also reflected from the Km values, which increases by almost 31%, indicating the reduced activity of the dimer in comparison to that of the monomer (Table S1). This was further quantified using the Kcat value that is the turn-over number of the enzyme, which was reduced by ∼14% for the DT cross-linked dimer as compared with the native protein. A previous study on the enzymatic activity of the RNase A dimer showed that the photoirradiated DT cross-linked dimer has lower substrate affinity than the RNase A monomer.53
Fullerene acts as a potential antioxidant, thereby suppressing oxidative stress in proteins. The introduction of OH groups into fullerene is expected to increase the antioxidant power of fullerenol compared to that of the parent molecule. The antioxidant properties of these two species were studied in the presence of RNase A by measuring the DT fluorescence intensity. Incubation of the protein with oxidant (Co2+ and S2O82–) in the presence of these nanoparticles showed a decrease in DT intensity in a concentration-dependent manner. It has been observed that the antioxidant activity increases with the increase in fullerene concentration, as seen from the gradual decrease in DT spectra. The case is somewhat different for fullerenol, which shows antioxidant activity at very low concentrations that increases at elevated concentrations. A comparative analysis of the change in DT fluorescence intensity is shown in Figure 12. The antioxidant effect of fullerenol for RNase A is almost twice that of fullerene at concentrations less than 10 μg/mL. There is a slight increment in the change in DT fluorescence with an increase in concentration for fullerenol at very low concentration, after which it shows a regular decrease. On the other hand, the antioxidant activity of fullerene is proportional to its concentration. Fullerene being a radical scavenger may have the tendency to react with the sulfate ion present, thereby preventing its effect on the Tyr residue. This antioxidant effect is reversed for fullerenol at higher concentrations. Although previous reports highlighted the aggregation of fullerenol at higher concentration as a key factor for promoting oxidation,71 the exact mechanism is still to be understood. Both fullerene and fullerenol prevent oxidative stress in protein, and their presence in the system may inhibit the enzymatic activity. In the present study, the concentrations of fullerene and fullerenol both are kept above and below the Ki value while studying their effect during oxidative stress. However, for fullerene, significant antioxidant activity can be observed for concentrations 7.2 and 21.6 μg/mL, which are lower than the inhibition constant value (26.49 μg/mL) of fullerene. On the other hand, fullerenol prevents DT formations at 0.1 and 1.0 μg/mL, which are also lower than its Ki value (4.66 μg/mL). Thus, both fullerene and fullerenol decrease DT formation and prevent oxidative stress in RNase A much at a much lower concentration than the inhibition constant value. This suggests both fullerene and fullerenol can be employed as an antioxidant without affecting the enzymatic activity of the protein if the concentrations of these components are kept below their respective Ki values.
Figure 12.

Percentage change in DT fluorescence peak intensity at various concentrations of fullerene and fullerenol.
3. Conclusions
Our present work investigates the interaction of fullerene and fullerenol with RNase A with the possible prevention of oxidative stress in the protein. Fullerene being highly nonpolar is sparingly soluble in water, and to overcome this, we have prepared highly water soluble fullerenol having ∼30–40 −OH groups within a span of 1–2 h without any catalyst with certain modification of previously used techniques. Although both fullerene and fullerenol bind to the protein in a 1:1 ratio, the extent of binding is much higher in the case of fullerene compared to that in fullerenol. This has been further verified using molecular docking studies, which showed that the ΔG value is more favorable in the case of fullerene compared to that of fullerenol. The interaction is governed mainly by hydrophobic as well as π–π stacking interactions with the aromatic protein residues. The unsaturated fullerene prevents oxidative stress in RNase A with an increase in concentration, whereas for fullerenol, this effect is more pronounced at lower concentrations. The nanoparticles also show significant inhibition of the ribonucleolytic activity of RNase A with fullerenol showing a lower Ki value than fullerene. Fullerene shows noncompetitive inhibition by binding to the allosteric site, whereas fullerenol shows a mixed-type inhibition by binding to both the allosteric site and the active site. The experiments show that fullerene and fullerenol have the ability to control the damage of the protein caused due to oxidative stress and can also prevent significant cleavage of RNA by inhibiting RNase A at a comparatively lower concentration. This shows that hydroxylation of fullerene to fullerenol not only affects the extent of binding with RNase A but also has a considerable effect on DT formation caused due to oxidative stress.
4. Materials and Methods
4.1. Materials
RNase A, yeast (t-RNA), and cytidine 2′,3′-cyclic monophosphate (2′,3′-cCMP) were purchased from Sigma Chemical Co. (St. Louis). Fullerene (C60) was purchased from Alfa Aesar. Cobalt acetate [Co(OAc)2], potassium persulfate (K2S2O8), and other analytical-grade reagents were obtained from SRL India.
4.2. Methods
4.2.1. Preparation of Aqueous Suspension of Fullerene and Fullerenol
An aqueous suspension of fullerene was prepared using the general protocol, which includes dissolution of solid fullerene in toluene followed by a solvent-exchange method using toluene as the organic phase and water as the aqueous phase.75,76 In brief, solid fullerene was initially dissolved in toluene and stirred for 30 min. Water was added to the solution in a 2:1 v/v ratio, and the entire mixture was subjected to ultrasonication till complete evaporation of the organic solvent. The aqueous residue was filtered using a 0.22 μm ultrafiltration membrane. The clear yellow solution thus obtained was stored at room temperature away from light.
Several methods of fullerenol preparation have been reported, which include oxidation of the double bond using peroxide and NaOH. Earlier methods involved the initial acidification under drastic conditions with the reaction being carried out at a high temperature of around 85–95 °C.77 Wang et al. devised a unique method for the preparation of fullerenol using solid-state organic reactions, which gave ∼16 −OH groups attached to fullerene with very high yield.78 More recently, Kokubo et al. showed that the number of −OH groups increased to more than 40 when peroxides were used in the presence of a phase-transfer catalyst (tetrabutylammonium hydroxide) at 60 °C.36 We have employed the acidification technique followed by addition of peroxide at room temperature. For preparation of fullerenol, 2 mL of the water suspension of fullerene (having concentration 1 mg/mL) was acidified by conc. H2SO4 and stirred for ∼25 min. H2O2 was added dropwise to a glass vial containing NaOH pellets followed by simultaneous addition of the acidified fullerene solution. The mixture was stirred for another 1–2 h during which the yellow color fades to give a colorless solution. This colorless solution was taken in a clean, dried separating funnel, and ∼5 mL of ethyl acetate was added to it. Around 2 mL of double-distilled water was added to the separating funnel, and the solvents were mixed thoroughly. This led to the separation of two colorless solvent phases, which were collected in two test tubes separately. Fullerenol having a relatively more hydrophobic surface prefers the organic phase and remains in the ethyl acetate part, whereas the salts are dissolved exclusively in the aqueous part. The organic phase was again added to the separating funnel followed by double-distilled water, and the same process was repeated 2–3 times. The organic part was collected and vacuum-dried, and solid colorless fullerenol was obtained by evaporating the solvent. An aqueous solution of fullerenol was used for further studies. The pictorial representation of the preparation of fullerene and fullerenol is shown in Scheme 1.
Scheme 1. Preparation of Water-Soluble Fullerenol from Fullerene.
4.2.2. Characterization of Fullerene and Fullerenol from UV–Vis Studies
UV–vis spectroscopy was used to characterize the fullerene suspension and fullerenol solution. Spectral measurements were carried out using a Shimadzu UV-1800 UV–vis spectrophotometer over a wavelength range 200–600 nm at 25 °C.
4.2.3. Morphology, Size, and ζ Potential Measurements
Field emission scanning electron microscopy (FESEM) was used to determine the morphology of the fullerene and fullerenol aggregates. A drop of sample was mounted on a glass slide, air-dried, and then scanned in Nova NanoSEM 450 operating at 10 kV. The morphology of the aggregates was also monitored by atomic force microscopy (AFM) using Agilent Technologies, model 5500. A drop of sample was deposited on a freshly cleaved mica foil. The mica foil was then allowed to dry in air and then scanned. The images were taken in tapping mode using a silicon probe cantilever of 215–235 μm length, at a resonance frequency of 146–236 kHz, and a force constant of 21–98 N/m.
The sizes of fullerene and fullerenol particles were analyzed by dynamic light scattering (DLS). The particles were initially sonicated, and sizes were determined using a Malvern Nano ZS instrument, which operates using a 4 mW He–Ne laser (λ = 632 nm) and a scattering angle of 173°. ζ potential measurements were carried out using the same instrument with very dilute solutions of the sample. The reported value is the measurement of 100 runs and is the average of three independent readings.
4.2.4. Fourier Transform Infrared Spectroscopy
The characteristic vibrational bands for fullerene and fullerenol were identified using Shimadzu IRTracer-100. The aqueous solutions of the nanoparticles were dried over a KBr pellet before being subjected to IR radiation.
4.2.5. Fluorescence Quenching of Tyrosine by Fullerene and Fullerenol
Fluorescence measurements were performed using a Jobin Yvon Spex Fluorolog-3 spectrofluorimeter in a 1 cm quartz cell using an excitation wavelength of 278 nm.64 It may be mentioned here that RNase A lacks tryptophan (Trp). The excitation and emission bandwidths were 5 nm. The emission spectra were recorded from 298 to 350 nm. A quantitative analysis of the interaction with RNase A was performed by a fluorimetric titration of a solution of RNase A in 10 mM phosphate buffer (2 μM) with addition of fullerene (0–10 μg/mL) and fullerenol (0–2 μg/mL), total volume of the solution being 600 μL. The binding parameters for fullerene/fullerenol have been calculated from fluorescence quenching data using the following equations79
| 1 |
| 2 |
where ΔF = F0 – F; F0 and F are the fluorescence intensities of protein in the absence and presence of fullerene/fullerenol, respectively, Ksv is the Stern–Volmer constant, [Q] is the concentration of the quencher, n is the number of binding sites, and Kb is the equilibrium binding constant.
4.2.6. Inhibition Studies
4.2.6.1. Agarose Gel-Based Assay
RNase A was mixed with Tris–acetic acid buffer (pH 8) and fullerene/fullerenol. Aliquots (20 μL) were mixed with 20 μL of t-RNA (16.6 mg/mL, freshly dissolved in RNase A-free buffer) and incubated for 30 min. The sample buffer (10 μL) with 10% glycerol and 0.025% bromophenol blue was added to quench the reaction. Aliquots (10 μL) were loaded onto a 1.1% agarose gel, and the undegraded t-RNA was visualized by ethidium bromide staining.
4.2.6.2. Inhibition Kinetics
The effect of fullerene and fullerenol on the ribonucleolytic activity of RNase A was checked following the method described by Anderson et al.80 The assay was carried out in oligovinylsulfonic acid-free 0.1 M N-morholinoethane sulfonic acid–NaOH buffer, pH 6.0, containing 0.1 M NaCl and 2′,3′-cCMP as the substrate. The substrate concentration was varied from 0.2 to 0.9 mM and the inhibitor concentration from 0 to 10 μg/mL for both fullerene and fullerenol. The inhibition constant and the type of binding enzyme were established from Lineweaver–Burk plots.81
4.2.7. Docking Studies
The Protein Data Bank (PDB) file of the crystal structure of the wild-type native bovine pancreatic RNase A (PDB ID: 1FS3)82 was downloaded from RCSB (http://www.rcsb.org) and used for further study. The structures of fullerene and fullerenol (30 −OH attached) were prepared and optimized in Avogadro software.83 The binding region of fullerene and fullerenol in RNase A was identified from docking studies using AutoDock Vina with the center positioned at the following coordinates: x = 20.489, y = 18.046, and z = 25.639 and grid size 32 Å × 28 Å × 44 Å.84 The structures were further analyzed using Chimera software.85
4.2.8. Oxidation of RNase A
To 200 μM Co(OAc)2 and 2 mM K2S2O8 solution in 10 mM phosphate buffer (pH 7.4), the RNase A solution was added so as to make the final concentration of protein 100 μM. For studying the antioxidant property, different concentrations of fullerene (5–40 μg/mL) and fullerenol (0.1–10 μg/mL) were added to the oxidizing medium prior to the addition of protein. The resultant solutions were incubated at 37 °C for 24 h.
4.2.9. Dityrosine (DT) Fluorescence
The formation of DT in RNase A during oxidative stress was confirmed by measuring the DT fluorescence emission intensity around 405 nm.86 The samples were excited at 317 nm, and the emission scan range was set at 330–450 nm. Both the excitation and emission slits were set at 5 nm with an integration time of 0.3 s.
4.2.10. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
The oxidized and fullerene-/fullerenol-treated protein samples were analyzed by SDS polyacrylamide gel electrophoresis. The samples were mixed with buffer containing SDS and β-mercaptoethanol and heated for 5 min prior to loading onto a 15% SDS polyacrylamide gel. Staining with Coomassie Brilliant Blue (SRL, India) was followed by destaining using an aqueous solution with 8% acetic acid and 6% methanol. The molecular mass of the fractions was compared with that of a standard-molecular-weight marker (10–250 kDa).
4.2.11. Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF)
To ensure the formation of an aqueous solution of fullerene and estimate the probable molecular weight of fullerenol, MALDI-TOF analyses were performed. This method was also used to confirm the formation of the RNase A dimer during oxidation. The matrix was prepared using a saturated solution of sinapinic acid (20 mg/mL) in 1:1 (v/v) CH3CN/H2O solution containing 1% trifluoroacetic acid. Equal volumes of the fullerene and fullerenol samples and the prepared matrix were mixed for analysis. A Bruker ultrafleXtreme mass spectrometer was used for the measurement with a 19 kV acceleration voltage along with pulsed ion extraction. A reflection mode of 20 kV was used for the detection of the positive ions. Laser desorption was done using a smartbeam-II Laser system, and 500 laser shots were summed.
Acknowledgments
S.D. is grateful to Department of Biotechnology, Ministry of Science and Technology, New Delhi (Project No.: BT/PR6189/BRB/10/1150/2012). The authors are thankful to Central Research Facility (CRF), IIT Kharagpur, for instrumental facilities. The authors would like to acknowledge Prof. Sunando DasGupta (Department of Chemical Engineering, IIT Kharagpur) and his group for providing DLS facilities. The authors are grateful to Professor Nilmoni Sarkar (Department of Chemistry, IIT Kharagpur) and his group for providing ζ potential facilities. P.R. and S.B. acknowledge IIT Kharagpur, and CSIR, New Delhi, respectively for financial support.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01584.
UV–vis spectra of fullerene and fullerenol, MALDI-TOF spectrum of fullerene in water, microscopic studies of fullerene and fullerenol using field emission scanning electron microscopy (FESEM) and atomic force microscopy (AFM), dynamic light scattering (DLS) studies of fullerene and fullerenol in the presence of RNase A, enzyme activity of DT cross-linked RNase A dimer and monomer (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Dirix Y.; Bastiaansen C.; Caseri W.; Smith P. Oriented Pearl-Necklace Arrays of Metallic Nanoparticles in Polymers: A New Route toward Polarization-Dependent Color Filters. Adv. Mater. 1999, 11, 223–227. 10.1002/(SICI)1521-4095(199903)11:33.0.CO;2-J. [DOI] [Google Scholar]
- Saha S.; Pal A.; Kundu S.; Basu S.; Pal T. Photochemical Green Synthesis of Calcium-Alginate-Stabilized Ag and Au Nanoparticles and Their Catalytic Application to 4-Nitrophenol Reduction. Langmuir 2010, 26, 2885–2893. 10.1021/la902950x. [DOI] [PubMed] [Google Scholar]
- Maekawa K.; Yamasaki K.; Niizeki T.; Mita M.; Matsuba Y.; Terada N.; Saito H. Drop-on-Demand Laser Sintering with Silver Nanoparticles for Electronics Packaging. IEEE Trans. Compon., Packag., Manuf. Technol. 2012, 2, 868–877. 10.1109/TCPMT.2011.2178606. [DOI] [Google Scholar]
- Wadhawan A.; Garrett D.; Perez J. M. Nanoparticle-Assisted Microwave Absorption by Single-Wall Carbon Nanotubes. Appl. Phys. Lett. 2003, 83, 2683–2685. 10.1063/1.1615679. [DOI] [Google Scholar]
- Zhang W.-x. Nanoscale Iron Particles for Environmental Remediation—An Overview. J. Nanopart. Res. 2003, 5, 323–332. 10.1023/A:1025520116015. [DOI] [Google Scholar]
- Salata O. Applications of Nanoparticles in Biology and Medicine. J. Nanobiotechnol. 2004, 2, 3. 10.1186/1477-3155-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. C.; Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- Molday R. S.; Mackenzie D. Immunospecific Ferromagnetic Iron-Dextran Reagents for the Labeling and Magnetic Separation of Cells. J. Immunol. Methods 1982, 52, 353–367. 10.1016/0022-1759(82)90007-2. [DOI] [PubMed] [Google Scholar]
- De Jong W. H.; Borm P. J. A. Drug Delivery and Nanoparticles: Applications and Hazards. Int. J. Nanomed. 2008, 3, 133–149. 10.2147/IJN.S596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzold C.; Lisdat F. Fast Protein Detection Using Absorption Properties of Gold Nanoparticles. Analyst 2012, 137, 2821–2826. 10.1039/c2an35054h. [DOI] [PubMed] [Google Scholar]
- Mahtab R.; Rogers J. P.; Murphy C. J. Protein-Sized Quantum Dot Luminescence Can Distinguish between “Straight”, “Bent”, and “Kinked” Oligonucleotides. J. Am. Chem. Soc. 1995, 117, 9099–9100. 10.1021/ja00140a040. [DOI] [Google Scholar]
- Yanase M.; Shinkai M.; Honda H.; Wakabayashi T.; Yoshida J.; Kobayashi T. Intracellular Hyperthermia for Cancer Using Magnetite Cationic Liposomes: An in Vivo Study. Jpn. J. Cancer Res. 1998, 89, 463–470. 10.1111/j.1349-7006.1998.tb00586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. X.; Wilson S. R.; Das M.; Coughlin D. J.; Erlanger B. F. Antigenicity of Fullerenes: Antibodies Specific for Fullerenes and Their Characteristics. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 10809–10813. 10.1073/pnas.95.18.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon D. Y.; Adams L. K.; Falkner J. C.; Alvarez P. J. J. Antibacterial Activity of Fullerene Water Suspensions: Effects of Preparation Method and Particle Size. Environ. Sci. Technol. 2006, 40, 4360–4366. 10.1021/es0603655. [DOI] [PubMed] [Google Scholar]
- Dugan L. L.; Turetsky D. M.; Du C.; Lobner D.; Wheeler M.; Almli C. R.; Shen C. K.; Luh T. Y.; Choi D. W.; Lin T. S. Carboxyfullerenes as Neuroprotective Agents. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 9434–9439. 10.1073/pnas.94.17.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama H.; Yamago S.; Nakamura E.; Shiraki T.; Sugiura Y. Photoinduced Biochemical Activity of Fullerene Carboxylic Acid. J. Am. Chem. Soc. 1993, 115, 7918–7919. 10.1021/ja00070a064. [DOI] [Google Scholar]
- Huang Y.-L.; Shen C. K.-F.; Luh T.-Y.; Yang H. C.; Hwang K. C.; Chou C.-K. Blockage of Apoptotic Signaling of Transforming Growth Factor-Beta in Human Hepatoma Cells by Carboxyfullerene. Eur. J. Biochem. 1998, 254, 38–43. 10.1046/j.1432-1327.1998.2540038.x. [DOI] [PubMed] [Google Scholar]
- Park K. H.; Chhowalla M.; Iqbal Z.; Sesti F. Single-Walled Carbon Nanotubes Are a New Class of Ion Channel Blockers. J. Biol. Chem. 2003, 278, 50212–50216. 10.1074/jbc.M310216200. [DOI] [PubMed] [Google Scholar]
- Xie L.; Luo Y.; Lin D.; Xi W.; Yang X.; Wei G. The Molecular Mechanism of Fullerene-Inhibited Aggregation of Alzheimer’s β-Amyloid Peptide Fragment. Nanoscale 2014, 6, 9752–9762. 10.1039/C4NR01005A. [DOI] [PubMed] [Google Scholar]
- Gharbi N.; Pressac M.; Hadchouel M.; Szwarc H.; Wilson S. R. [60]Fullerene is a Powerful Antioxidant in Vivo with No Acute or Subacute Toxicity. Nano Lett. 2005, 5, 2578–2585. 10.1021/nl051866b. [DOI] [PubMed] [Google Scholar]
- Baati T.; Bourasset F.; Gharbi N.; Njim L.; Abderrabba M.; Kerkeni A.; Szwarc H.; Moussa F. The Prolongation of the Lifespan of Rats by Repeated Oral Administration of [60]Fullerene. Biomaterials 2012, 33, 4936–4946. 10.1016/j.biomaterials.2012.03.036. [DOI] [PubMed] [Google Scholar]
- Kim K.-H.; Ko D.-K.; Kim Y.-T.; Kim N. H.; Paul J.; Zhang S.-Q.; Murray C. B.; Acharya R.; DeGrado W. F.; Kim Y. H.; et al. Protein-Directed Self-Assembly of a Fullerene Crystal. Nat. Commun. 2016, 7, 11429 10.1038/ncomms11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.; Liu S.; Yin J.; Wang H. Interaction of Human Serum Album and C60 Aggregates in Solution. Int. J. Mol. Sci. 2011, 12, 4964–4974. 10.3390/ijms12084964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-T.; Wang H.; Guo L.; Gao Y.; Liu Y.; Cao A. Interaction of Fullerenol with Lysozyme Investigated by Experimental and Computational Approaches. Nanotechnology 2008, 19, 395101 10.1088/0957-4484/19/39/395101. [DOI] [PubMed] [Google Scholar]
- Wolff D. J.; Barbieri C. M.; Richardson C. F.; Schuster D. I.; Wilson S. R. Trisamine C60-Fullerene Adducts Inhibit Neuronal Nitric Oxide Synthase by Acting as Highly Potent Calmodulin Antagonists. Arch. Biochem. Biophys. 2002, 399, 130–141. 10.1006/abbi.2002.2766. [DOI] [PubMed] [Google Scholar]
- Nakamura E.; Tokuyama H.; Yamago S.; Shiraki T.; Sugiura Y. Biological Activity of Water-Soluble Fullerenes. Structural Dependence of DNA Cleavage, Cytotoxicity, and Enzyme Inhibitory Activities Including HIV-Protease Inhibition. Bull. Chem. Soc. Jpn. 1996, 69, 2143–2151. 10.1246/bcsj.69.2143. [DOI] [Google Scholar]
- Mashino T.; Okuda K.; Hirota T.; Hirobe M.; Nagano T.; Mochizuki M. Inhibitory Effect of Fullerene Derivatives on Glutathione Reductase. Fullerene Sci. Technol. 2001, 9, 191–196. 10.1081/FST-100102966. [DOI] [Google Scholar]
- Kobzar O. L.; Trush V. V.; Tanchuk V. Y.; Zhilenkov A. V.; Troshin Pa; Vovk A. I. Fullerene Derivatives as a New Class of Inhibitors of Protein Tyrosine Phosphatases. Bioorg. Med. Chem. Lett. 2014, 24, 3175–3179. 10.1016/j.bmcl.2014.04.110. [DOI] [PubMed] [Google Scholar]
- Singh K.; Mishra A. Water Soluble Chitosan Nanoparticle for the Effective Delivery of Lipophilic Drugs: A Review. Int. J. Appl. Pharm. 2013, 5, 1–6. [Google Scholar]
- Shi Y.; Goodisman J.; Dabrowiak J. C. Cyclodextrin Capped Gold Nanoparticles as a Delivery Vehicle for a Prodrug of Cisplatin. Inorg. Chem. 2013, 52, 9418–9426. 10.1021/ic400989v. [DOI] [PubMed] [Google Scholar]
- Ghosh P.; Bag S.; Roy A. S.; Subramani E.; Chaudhury K.; Dasgupta S. Solubility Enhancement of Morin and Epicatechin through Encapsulation in an Albumin Based Nanoparticulate System and Their Anticancer Activity against the MDA-MB-468 Breast Cancer Cell Line. RSC Adv. 2016, 6, 101415 10.1039/C6RA20441D. [DOI] [Google Scholar]
- Ghosh P.; Patwari J.; Dasgupta S. Complexation With Human Serum Albumin Facilitates Sustained Release of Morin From Polylactic-Co-Glycolic Acid Nanoparticles. J. Phys. Chem. B 2017, 121, 1758–1770. 10.1021/acs.jpcb.6b08559. [DOI] [PubMed] [Google Scholar]
- Chiang L. Y.; Wang L.-Y. Y.; Swirczewski J. W.; Soled S.; Cameron S. Efficient Synthesis of Polyhydroxylated Fullerene Derivatives Via Hydrolysis of Polycyclosulfated Precursors. J. Org. Chem. 1994, 59, 3960–3968. 10.1021/jo00093a030. [DOI] [Google Scholar]
- Li J.; Takeuchi A.; Ozawa M.; Li X.; Saigo K.; Kitazawa K. C60 Fullerol Formation Catalysed by Quaternary Ammonium Hydroxides. J. Chem. Soc., Chem. Commun. 1993, 1784. 10.1039/c39930001784. [DOI] [Google Scholar]
- Kokubo K.; Matsubayashi K.; Tategaki H.; Takada H.; Oshima T. Facile Synthesis of Highly Water-Soluble Fullerenes More Than Half-Covered By. ACS Nano 2008, 2, 327–333. 10.1021/nn700151z. [DOI] [PubMed] [Google Scholar]
- Kokubo K.; Shirakawa S.; Kobayashi N.; Aoshima H.; Oshima T. Facile and Scalable Synthesis of a Highly Hydroxylated Water-Soluble Fullerenol as a Single Nanoparticle. Nano Res. 2011, 4, 204–215. 10.1007/s12274-010-0071-z. [DOI] [Google Scholar]
- Oberdörster E. Manufactured Nanomaterials (Fullerenes, C60) Induce Oxidative Stress in the Brain of Juvenile Largemouth Bass. Environ. Health Perspect. 2004, 112, 1058–1062. 10.1289/ehp.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaño D.; Fernández-Pachón M. S.; Moyá M. L.; Troncoso A. M.; García-Parrilla M. C. Radical Scavenging Ability of Polyphenolic Compounds towards DPPH Free Radical. Talanta 2007, 71, 230–235. 10.1016/j.talanta.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Roy P.; Dinda A. K.; Chaudhury S.; Dasgupta S. β-Cyclodextrin Encapsulated Polyphenols as Effective Antioxidants. Biopolymers 2018, 109, e23084 10.1002/bip.23084. [DOI] [PubMed] [Google Scholar]
- Chiang L. Y.; Lu F.-J.; Lin J.-T. Free Radical Scavenging Activity of Water-Soluble Fullerenols. J. Chem. Soc., Chem. Commun. 1995, 1283–12884. 10.1039/c39950001283. [DOI] [Google Scholar]
- Andrievsky G. V.; Bruskov V. I.; Tykhomyrov A. A.; Gudkov S. V. Peculiarities of the Antioxidant and Radioprotective Effects of Hydrated C60 Fullerene Nanostuctures in Vitro and in Vivo. Free Radic. Biol. Med. 2009, 47, 786–793. 10.1016/j.freeradbiomed.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Yin J. J.; Lao F.; Fu P. P.; Wamer W. G.; Zhao Y.; Wang P. C.; Qiu Y.; Sun B.; Xing G.; Dong J.; et al. The Scavenging of Reactive Oxygen Species and the Potential for Cell Protection by Functionalized Fullerene Materials. Biomaterials 2009, 30, 611–621. 10.1016/j.biomaterials.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueng T. H.; Kang J. J.; Wang H. W.; Cheng Y. W.; Chiang L. Y. Suppression of Microsomal Cytochrome P450-Dependent Monooxygenases and Mitochondrial Oxidative Phosphorylation by Fullerenol, a Polyhydroxylated Fullerene C60. Toxicol. Lett. 1997, 93, 29–37. 10.1016/S0378-4274(97)00071-4. [DOI] [PubMed] [Google Scholar]
- Wu X.; Yang S.-T.; Wang H.; Wang L.; Hu W.; Cao A.; Liu Y. Influences of the Size and Hydroxyl Number of Fullerenes/Fullerenols on Their Interactions with Proteins. J. Nanosci. Nanotechnol. 2010, 10, 6298–6304. 10.1166/jnn.2010.2623. [DOI] [PubMed] [Google Scholar]
- Raines R. T. Ribonuclease A. Chem. Rev. 1998, 98, 1045–1065. 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Marshall G. R.; Feng J. A.; Kuster D. J. Back to the Future: Ribonuclease A. Biopolymers 2008, 90, 259–277. 10.1002/bip.20845. [DOI] [PubMed] [Google Scholar]
- Ledoux L. Action of Ribonuclease on Two Solid Tumours in Vivo. Nature 1955, 176, 36–37. 10.1038/176036a0. [DOI] [PubMed] [Google Scholar]
- Tripathy D. R.; Roy A. S.; Dasgupta S. Complex Formation of Rutin and Quercetin with Copper Alters the Mode of Inhibition of Ribonuclease A. FEBS Lett. 2011, 585, 3270–3276. 10.1016/j.febslet.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Ghosh K. S.; Maiti T. K.; Debnath J.; Dasgupta S. Inhibition of Ribonuclease A by Polyphenols Present in Green Tea. Proteins: Struct., Funct., Bioinf. 2007, 69, 566–580. 10.1002/prot.21484. [DOI] [PubMed] [Google Scholar]
- Henneke M.; Diekmann S.; Ohlenbusch A.; Kaiser J.; Engelbrecht V.; Kohlschütter A.; Krätzner R.; Madruga-Garrido M.; Mayer M.; Opitz L.; et al. RNASET2-Deficient Cystic Leukoencephalopathy Resembles Congenital Cytomegalovirus Brain Infection. Supplementary Information. Nat. Genet. 2009, 41, 773–775. 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- Berlett B. S.; Stadtman E. R. Protein Oxidation in Aging, Disease, and Oxidative Stress. J. Biol. Chem. 1997, 272, 20313–20316. 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Shacter E. Quantification and Significance of Protein Oxidation in Biological Samples. Drug Metab. Rev. 2000, 32, 307–326. 10.1081/DMR-100102336. [DOI] [PubMed] [Google Scholar]
- Dinda A. K.; Tripathy D. R.; Das A.; Dasgupta S. Comparison of the Ribonucleolytic Activity of the Dityrosine Cross-Linked Ribonuclease A Dimer with Its Monomer in the Presence of Inhibitors. Int. J. Biol. Macromol. 2014, 63, 107–113. 10.1016/j.ijbiomac.2013.10.033. [DOI] [PubMed] [Google Scholar]
- Hare J. P.; Kroto H. W.; Taylor R. Reprint of: Preparation and UV/Visible Spectra of Fullerenes C60 and C70. Chem. Phys. Lett. 2013, 589, 57–60. 10.1016/j.cplett.2013.08.068. [DOI] [Google Scholar]
- Chen Z.; Westerhoff P.; Herckes P. Quantification of C60 Fullerene Concentrations in Water. Environ. Toxicol. Chem. 2008, 27, 1852–1859. 10.1897/07-560.1. [DOI] [PubMed] [Google Scholar]
- Singh R.; Goswami T. Highly Luminescent Multifunctional Hemi-Ortho Ester Derivatives of Fullerenol. Synth. Met. 2007, 157, 951–955. 10.1016/j.synthmet.2007.09.006. [DOI] [Google Scholar]
- Song P.; Liu H.; Shen Y.; Du B.; Fang Z.; Wu Y. Fabrication of Dendrimer-like Fullerene (C60)-Decorated Oligomeric Intumescent Flame Retardant for Reducing the Thermal Oxidation and Flammability of Polypropylene Nanocomposites. J. Mater. Chem. 2009, 1305–1313. 10.1039/b815610g. [DOI] [Google Scholar]
- Yang S.; Mulet X.; Gengenbach T.; Waddington L.; Seeber A.; Zhen M.; Wang C.; Muir B. W.; Such G. K.; Hao X. Limitations with Solvent Exchange Methods for Synthesis of Colloidal Fullerenes. Colloids Surf., A 2017, 514, 21–31. 10.1016/j.colsurfa.2016.11.021. [DOI] [Google Scholar]
- Alves G. C.; Ladeira L. O.; Righi A.; Krambrock K.; Calado H. D.; de Freitas Gil R. P.; Pinheiro M. V. B. Synthesis of C60(OH)18-20 in aqueous alkaline solution under O2-atmosphere. J. Braz. Chem. Soc. 2006, 17, 1186–1190. 10.1590/s0103-50532006000600017. [DOI] [Google Scholar]
- Levi N.; Hantgan R. R.; Lively M. O.; Carroll D. L.; Prasad G. L. C60-Fullerenes: Detection of Intracellular Photoluminescence and Lack of Cytotoxic Effects. J. Nanobiotechnol. 2006, 4, 14. 10.1186/1477-3155-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjević A.; Ajdinović B.; Dopudja M.; Trajković S.; Milovanović Z.; Maksin T.; Nešković O.; Bogdanović V.; Trpkov D.; Cvetićanin J. Scintigraphy of the Domestic Dog Using [99mTc(CO)3(H2O)3]-C60(OH)22-24. Dig. J. Nanomater. Biostruct. 2010, 6, 99–106. [Google Scholar]
- Silion M.; Dascalu A.; Pinteala M.; Simionescu B. C.; Ungurenasu C. A Study on Electrospray Mass Spectrometry of Fullerenol C60(OH)24. Beilstein J. Org. Chem. 2013, 9, 1285–1295. 10.3762/bjoc.9.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Zhang H.; Ruan L.; Chen L.; Li H.; Chang X.-L.; Zhang X.; Yang S.-T. Bioaccumulation of 13C-Fullerenol Nanomaterials in Wheat. Environ. Sci. Nano 2016, 3, 799–805. 10.1039/C5EN00276A. [DOI] [Google Scholar]
- Pundak S.; Roche R. S. Tyrosine and Tyrosinate Fluorescence of Bovine Testes Calmodulin: Calcium and PH Dependence. Biochemistry 1984, 23, 1549–1555. 10.1021/bi00302a032. [DOI] [PubMed] [Google Scholar]
- He H.; Zheng L.; Jin P.; Yang M. The Structural Stability of Polyhydroxylated C60(OH)24: Density Functional Theory Characterizations. Comput. Theor. Chem. 2011, 974, 16–20. 10.1016/j.comptc.2011.07.005. [DOI] [Google Scholar]
- Rivelino R.; Malaspina T.; Fileti E. E. Structure, Stability, Depolarized Light Scattering, and Vibrational Spectra of Fullerenols from All-Electron Density-Functional-Theory Calculations. Phys. Rev. A: At., Mol., Opt. Phys. 2009, 79, 013201 10.1103/PhysRevA.79.013201. [DOI] [Google Scholar]
- Messmore J. M.; Fuchs D. N.; Raines R. T. Ribonuclease A: Revealing Structure—Function Relationships with Semisynthesis. J. Am. Chem. Soc. 1995, 117, 8057–8060. 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt E. S.; Wittmayer P. K.; Templer B. M.; Raines R. T. Contribution of a Tyrosine Side Chain to Ribonuclease A Catalysis and Stability. Protein Sci. 1996, 5, 1697–1703. 10.1002/pro.5560050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath J. L.; Willson R. L. Chain Reactions and Radiosensitization: Model Enzyme Studies. Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med. 1975, 27, 389–398. 10.1080/09553007514550361. [DOI] [PubMed] [Google Scholar]
- Mahmoud S. F.; Bialkowski S. E. Laser-Excited Fluorescence of Dityrosine. Appl. Spectrosc. 1995, 49, 1669–1676. 10.1366/0003702953965678. [DOI] [Google Scholar]
- Zha Y.-y.; Yang B.; Tang M.-l.; Guo Q.-c.; Chen J.-t.; Chen L.-p.; Wang M. Concentration-Dependent Effects of Fullerenol on Cultured Hippocampal Neuron Viability. Int. J. Nanomed. 2012, 3099–3109. 10.2147/IJN.S30934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-f.; Shu C.-y.; Xie L.; Wang C.-r.; Zhang Y.-z.; Xiang J.-f.; Li L.; Tang Y.-l. Protein Conformation Changes Induced by a Novel Organophosphate-Containing Water-Soluble Derivative of a C60Fullerene Nanoparticle. J. Phys. Chem. C 2007, 14327–14333. 10.1021/jp073267au. [DOI] [Google Scholar]
- Belgorodsky B.; Fadeev L.; Ittah V.; Benyamini H.; Zelner S.; Huppert D.; Kotlyar A. B.; Gozin M. Formation and Characterization of Stable Human Serum Albumin–Tris-Malonic Acid [C60]Fullerene Complex. Bioconjugate Chem. 2005, 16, 1058–1062. 10.1021/bc050103c. [DOI] [PubMed] [Google Scholar]
- Belgorodsky B.; Fadeev L.; Kolsenik J.; Gozin M. Biodelivery of a Fullerene Derivative. Bioconjugate Chem. 2007, 18, 1095–1100. 10.1021/bc060363+. [DOI] [PubMed] [Google Scholar]
- Andrievsky G. V.; Kosevich M. V.; Vovk O. M.; Shelkovsky V. S.; Vashchenko L. A. On the Production of an Aqueous Colloidal Solution of Fullerenes. J. Chem. Soc., Chem. Commun. 1995, 98, 1281. 10.1039/c39950001281. [DOI] [Google Scholar]
- Torres V. M.; Posa M.; Srdjenovic B.; Simplício A. L. Solubilization of Fullerene C60 in Micellar Solutions of Different Solubilizers. Colloids Surf., B 2011, 82, 46–53. 10.1016/j.colsurfb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Chiang L. Y.; Upasani R. B.; Swirczewski J. W.; Soled S. Evidence of Hemiketals Incorporated in the Structure of Fullerols Derived from Aqueous Acid Chemistry. J. Am. Chem. Soc. 1993, 115, 5453–5457. 10.1021/ja00066a014. [DOI] [Google Scholar]
- Wang S.; He P.; Zhang J.-M.; Jiang H.; Zhu S.-Z. Novel and Efficient Synthesis of Water-Soluble [60]Fullerenol by Solvent-Free Reaction. Synth. Commun. 2005, 35, 1803–1808. 10.1081/SCC-200063958. [DOI] [Google Scholar]
- Lakowicz J. R. In Principles of Fluorescence Spectroscopy, 3rd ed.; Lakowicz J. R., Ed.; Springer: New York, 2006; pp 1–923. [Google Scholar]
- Anderson D. G.; Hammes G. G.; Walz F. G. Binding of Phosphate Ligands to Ribonuclease A. Biochemistry 1968, 7, 1637–1645. 10.1021/bi00845a004. [DOI] [PubMed] [Google Scholar]
- Lineweaver H.; Burk D. The Determination of Enzyme Dissociation Constants. J. Am. Chem. Soc. 1934, 56, 658–666. 10.1021/ja01318a036. [DOI] [Google Scholar]
- Chatani E.; Hayashi R.; Moriyama H.; Ueki T. Conformational Strictness Required for Maximum Activity and Stability of Bovine Pancreatic Ribonuclease A as Revealed by Crystallographic Study of Three Phe120 Mutants at 1.4 A Resolution. Protein Sci. 2002, 11, 72–81. 10.1110/ps.31102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanwell M. D.; Curtis D. E.; Lonie D. C.; Vandermeersch T.; Zurek E.; Hutchison G. R. Avogadro: An Advanced Semantic Chemical Editor, Visualisation, and Analysis Platform. J. Cheminf. 2012, 4, 1–17. 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization and Multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Heineckes J. W.; Li W.; Daehnke H. L. 3rd; Goldstein J. A. Dityrosine, a specific marker of oxidation, is synthesized by the myeloperoxidase-hydrogen peroxide system of human neutrophils and macro phages. J. Biol. Chem. 1993, 268, 4069–4077. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.