Abstract
Allelic diversity of the KIR2DL receptors drive differential expression and ligand-binding affinities that impact natural killer cell function and patient outcomes for diverse cancers. We have developed a global intermediate resolution amplification-refractory mutation system (ARMS) PCR-SSP method for distinguishing functionally relevant subgroups of the KIR2DL receptors, as defined by phylogenetic study of the protein sequences. Use of the ARMS design makes the method reliable and usable as a kit, with all reactions utilizing the same conditions. Six reactions define six subgroups of KIR2DL1; four reactions define three subgroups of KIR2DL2; and five reactions define four subgroups of KIR2DL3. Using KIR allele data from a cohort of 426 European-Americans, we identified the most common KIR2DL subtypes and developed the high-throughput PCR-based methodology, which was validated on a separate cohort of 260 healthy donors. Linkage disequilibrium analysis between the different KIR2DL alleles revealed that seven allelic combinations represent more than 95% of the observed population genotypes for KIR2DL1/L2/L3. In summary, our findings enable rapid typing of the most common KIR2DL receptor subtypes, allowing more accurate prediction of co-inheritance and providing a useful tool for the discrimination of observed differences in surface expression and effector function among NK cells exhibiting disparate KIR2DL allotypes.
Subject terms: Immunogenetics, Innate lymphoid cells
Introduction
As key members of the innate immune response, natural killer (NK) cells survey surrounding cells, discriminating damaged or infected cells from healthy cells, in part via receptor recognition of altered self-MHC on damaged cells1. This process, termed “education” or “licensing” is enabled through interactions between inhibitory receptors on NK cells with “self” MHC, that permit cytotoxic granule release for target cell killing, but also inhibition of the NK cell upon binding to cognate MHC. In humans, the principal receptors mediating education are the polygenic, polymorphic inhibitory killer cell immunoglobulin-like receptors (KIR), which recognize antigens presented by HLA-A, -B, and –C molecules2.
The KIR2DL receptors exclusively recognize HLA-C molecules: KIR2DL1 recognizes HLA-C allotypes characterized by Lys80 (collectively referred to as HLA-C group 2); KIR2DL3 recognizes predominantly HLA-C allotypes characterized by Asn80 (HLA-C group 1); while KIR2DL2 recognizes members of both HLA-C groups 1 and 22. Between the KIR2DL receptors and their specificities, nearly all HLA-C allotypes have a cognate inhibitory KIR. Additional inhibitory KIR molecules include KIR3DL1, which recognizes the Bw4 epitope exhibited by some HLA-A and HLA-B allotypes, and KIR3DL2, which recognizes the HLA-A3, HLA-A11, and HLA-B27 proteins2,3.
Significant diversity exists from individual to individual both at the KIR gene content and allele level. Some patterns of genetic combination are well-recognized and have led to the designation of the canonical KIR haplotype-A, characterized by gene content as presence of the centromeric KIR2DL3 and KIR2DL1 and the telomeric KIR3DL1, in the absence of all activating KIR, with the exception of the telomeric KIR2DS4. The remaining haplotypes collectively comprise the KIR B-haplotypes, exhibiting differing numbers and types of activating KIR in the centromeric or telomeric portions4–6. Clinical consequences of KIR diversity, even at the level of gene content, have provided some clues to the importance of differentially educated NK cells in control of viral infection, such as hepatitis C7, HIV8 and hematologic malignancy9,10.
Allelic polymorphism further diversifies the educational breadth of the NK repertoire. It is increasingly clear that different alleles of the same receptor, KIR3DL1, exhibit different surface expression properties and affinities for the same HLA ligand11–13, leading to substantial variations in NK education and sensitivity to inhibition11,12. These findings have enabled a more intricate understanding of NK education, again with important implications in viral control11,14 and malignancy15. Whether allele subtype variation for KIR2DL similarly impacts NK cell function and disease outcomes has not been extensively studied. However, it is known that the KIR2DL receptors are highly polymorphic, and that allelic variation may influence cell surface expression16,17, as well as avidity and specificity for HLA-C ligands18,19, potentially leading to benefits in infectious disease7,20.
While the clinical ramifications of allele-driven KIR diversity continue to emerge, a lack of straightforward technology to discriminate KIR content at the allele level has hampered large-scale clinical studies. Next-generation sequencing technology for KIR allele typing remains investigational21 or out of practical reach for research laboratories. We previously reported an accessible multiplex PCR assay for the cost-effective discrimination of KIR3DL1 alleles, and we have employed this assay in a large retrospective analysis of hematopoietic cell transplantation patients to demonstrate the clinical impact of functional KIR subtyping15,22. We now present a similar approach for the centromeric inhibitory KIR genes KIR2DL1, KIR2DL2, and KIR2DL3, identifying the nucleotide sites potentially important for functional discrimination among receptor alleles and devising an amplification-refractory mutation system (ARMS) PCR-SSP typing methodology. We anticipate that functional classification of the centromeric inhibitory KIR, as has been done from the telomeric KIR3DL1/S1, will broaden our understanding of how these alleles influence human health and disease.
Results
KIR2DL1 allele typing
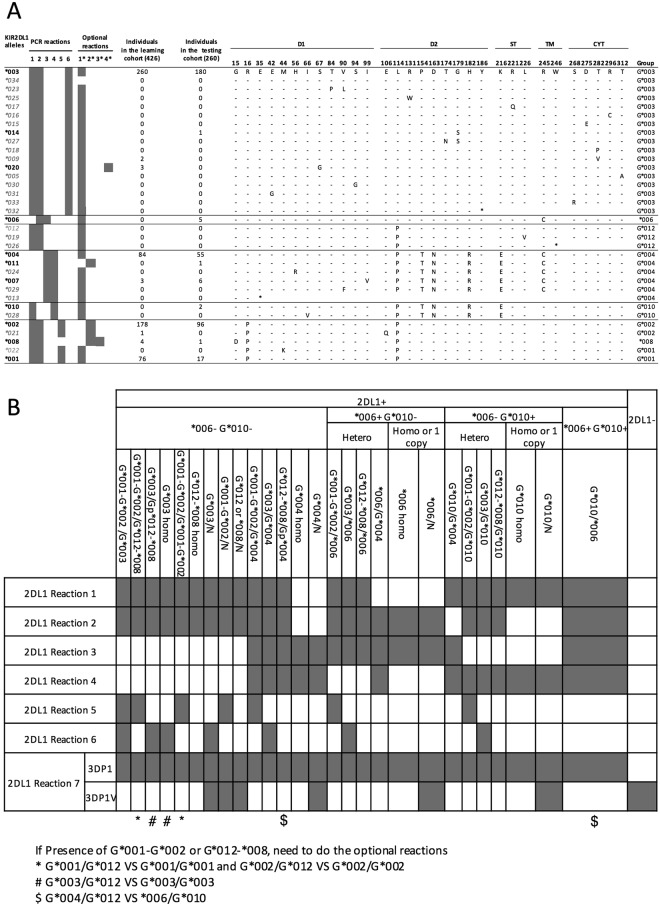
We examined the KIR alleles previously identified by sequence-based typing in a cohort of 426 healthy individual donors23. Of the 34 known KIR2DL1 alleles (EMBL-EBI IPD KIR), four (*001, *002, *003, *004) occurred frequently, with a presence for each allele in 18% or more of the individuals in the cohort (Fig. 1A). Five other alleles (*007, *008, *009, *020, *021) were found in fewer than 1% of all individuals. The remaining alleles were not identified in the cohort. Allelic distribution and phylogenetic analysis (Supplemental Fig. 1A) identified six non-overlapping groups. Six distinct ARMS PCR reactions specific for these six groups and four supplemental reactions to further discriminate alleles present within the group (Table 1) were optimized using DNA from 178 of the original 426 donors. We designed an additional reaction to identify the pseudogene KIR3DP1 and its variants (KIR3DP1V), the latter characterized by the presence of an exon 2. The presence of KIR3DP1V on the chromosome 19 is associated with the absence of KIR2DL1 on the same haplotype5,24,25. Detection of KIR3DP1 and KIR3DP1V can therefore be used to estimate KIR2DL1 copy number.
Figure 1.
KIR2DL1 allele typing method. (A) Alignment of the amino acid sequences of the 34 known KIR2DL1 allelic variants. A dash indicates identity with the consensus KIR2DL1*003, and an (*) indicates a stop codon. Structural domains are indicated: Ig-like domains (D1 and D2), stem domain (ST), transmembrane domain (TM), and cytoplasmic domain (CYT). Six PCR reactions separate the six subgroups identified by phylogenetic analysis. Four additional PCR reactions separate alleles within subgroups. Frequencies of the alleles present in the learning cohort of 426 individuals and in the testing cohort of 260 individuals are indicated. The group identification number for each allele is indicated. Alleles identified by PCR are in bold black font, and alleles that were not tested are in gray italics. (B) KIR2DL1 PCR interpretation guide. PCR profiles marked by *, #, or $ are similar and require a higher resolution of genotyping using supplemental reactions.
Table 1.
KIR2DL PCR primers and target site positions.
| Reaction | Primers name | Nucleotide targeted | Sequence Primers | Size amplicon (bp) |
|---|---|---|---|---|
| Control | ControlF | NA | CCAAGCCCAACCTTAAGAAGAAAATTGGAG | 813 |
| ControlR | NA | CCAAACCCACGGTACGCATGGGAACACTGC | ||
| 2DL1 Reaction 1 | 2DL1R1F | 3680 | AGAGATAAGACACCAGGAAGGGGAAGCCCG | 388 |
| 2DL1R1R | 4011 | TGTCCAGAGGGTCACTGGGAGCTGACTC | ||
| 2DL1 Reaction 2 | 2DL1R2F | 5499 | GAGAGAGAGAGAGAGAGAGCATTAGGTCATAGTA | 383 |
| 2DL1R2R | 5820 | TGACTTTGACCACTCGTATGGAGAGTCTT | ||
| 2DL1 Reaction 3 | 2DL1R3F | 13420 | ATCCTCTTCATCCTCCTCTTCTTTCTCCTTCACT | 252 |
| 2DL1R3R | 13609 | CAGTTCAGAATCAGGCAACGGTCTGTGAAT | ||
| 2DL1 Reaction 4 | 2DL1R4F | 5499 | GAGAGAGAGAGAGAGAGAGCATTAGGTCATAGGA | 297 |
| 2DL1R4R | 5735 | TGGCCTGGAATGTTCCGTTGACCTTGCT | ||
| 2DL1 Reaction 5 | 2DL1R5F | 3790 | AACCTTCCCTCCTGGCCCACCCAGGTAC | 278 |
| 2DL1R5R | 4011 | GATGTCCAGAGGGTCACTGGGAGCTGACGC | ||
| 2DL1 Reaction 6 | 2DL1R6F | 5616 | ATATGAGAAACCTTCTCTCTCAGCCCAGTT | 202 |
| 2DL1R6R | 5761 | GTGGGTGGCAGGGCCCAGAGGAAAGTAA | ||
| 2DL1 Reaction 7 | 3DP1F | NA | ACGTGTTGTGAGTTGGTCATAGTGA | 649 |
| 3DP1VF | NA | AAGTGGAAATGGGAGAATCTTCTGAC | 382 | |
| 3DP1R | NA | GCCCTCTGACCTGTGACCATGATC | ||
| 2DL1 Optional 1 | 2DL1O1F | 71 | GTTGGTCATAGTGAAGGACACTAGGTGTCAAATTCTATC | 274 |
| 2DL1O1R | 281 | TCACCAACACACGCCATGCTGACGTC | ||
| 2DL1 Optional 2 | 2DL1O2F | 281 | CTCCGGCAGCACCATGTCGCTCTTAT | 390 |
| 2DL1O2R | 620 | CCGTAACTCCACCTCCAGGCCCATTA | ||
| 2DL1 Optional 3 | 2DL1O3F | 3787 | AAACCTTCCCTCCTGGCCCACCCAAA | 376 |
| 2DL1O3R | 4110 | CTTCCTTACAGCCACCTGGGTCTCCAGT | ||
| 2DL1 Optional 4 | 2DL1O4F | 3942 | GGGTCTCCAAGGCCAACTTCTCCATGG | 222 |
| 2DL1O4R | 4110 | CTTCCTTACAGCCACCTGGGTCTCCACT | ||
| 2DL2 Reaction 1 | 2DL2R1F | 5663 | TATCCAGGGAGGGGGAGGCCCATGATT | 211 |
| 2DL2R1R | 5820 | TGAGACAGATATGGGGTTTCCTCACCAG | ||
| 2DL2 Reaction 2 | 2DL2R2F | 5663 | TATCCAGGGAGGGGGAGGCCCATGATT | 210 |
| 2DL2R2R | 5820 | GAGACAGATATGGGGTTTCCTCACCCA | ||
| 2DL2 Reaction 3 | 2DL2R3F | 13995 | ACAGATGCTGCGGTAATGGACCAAGATT | 309 |
| 2DL2R3R | 14249 | ATCTGGACTCAGCATTTGGAAGTTCCCC | ||
| 2DL2 Reaction 4 | 2DL2R4F | 11984 | CTACTTCCAATCACCTGTGGAGATTCATG | 2322 |
| 2DL2R4R | 14249 | ATCTGGACTCAGCATTTGGAAGTTCCTT | ||
| 2DL2 Optional 1 | 2DL2O1F | 3754 | AACCTTCCCTCCTGGCCCACCCAGGTTC | 191 |
| 2DL2O1R | 3890 | CATCATGGGACCGATGGAGAAGTTGGTT | ||
| 2DL2 Optional 2 | 2DL2O2F | 3754 | AACCTTCCCTCCTGGCCCACCCAGGTAG | 191 |
| 2DL2O2R | 3890 | CATCATGGGACCGATGGAGAAGTTGGGT | ||
| 2DL3 Reaction 1 | 2DL3R1F | 13892 | ATGAAATGAGGGCCCAGAAGTGCCCTGT | 314 |
| 2DL3R1R | 14154 | GGTGTCTTGGGCCTCTGAGAAGGAC | ||
| 2DL3 Reaction 2 | 2DL3R2F | 3825 | CACAGAGAAGGGAAGTTTAAGGACACTTTGTG | 399 |
| 2DL3R2R | 4168 | TGTATGGCCCCTGTGTCTGTCCTTT | ||
| 2DL3 Reaction 3 | 2DL3R3F | 9063 | CTGTCTCATGTTCTAGGAAACCCTTCAAATAGTTGGGT | 319 |
| 2DL3R3R | 9303 | GAAGGATGTCAGATTGGCAATCATTCTTCTAGCTTGTAGGAAA | ||
| 2DL3 Reaction 4 | 2DL3R4F | 13973 | GCCTGCAGGGAACAGAACAGTGAACAAG | 233 |
| 2DL3R4R | 14154 | GGTGTCTTGGGCCTCTGAGAAGGCT | ||
| 2DL3 Reaction 5 | 2DL3R5F | 3853 | CCTCATTGGAGAGCACCATGATGGGGCT | 430 |
| 2DL3R5R | 4222 | CCTCTCTCTGGGACATGTCTGTCTGTCTGTCTGT | ||
| 2DL3 Optional 1 | 2DL3O1F | 3708 | TAGGAGTCCACAGAAAACCTTCCCTCGG | 323 |
| 2DL3O1R | 3976 | GAATGTCCGGACACTCTCACCTGTGACG | ||
| 2DL3 Optional 2 | 2DL3O2F | 16795 | CCCTCCATCTGGGTGCTTGTCCTAAAGGCG | 213 |
| 2DL3O2R | 16949 | GCGATGAAGGAGAAAGAAGAGGAGGAGGTC | ||
| 2DL3 Optional 3 | 2DL3O3F | 17646 | TGAACAAGACCCTCAGGAGGTGACATTT | 169 |
| 2DL3O3R | 17761 | TCATGGGCAGGAGACAACTTTGGATAT | ||
| 2DL3 Optional 4 | 2DL3O4F | 7315 | TCCTGCAATGTTGGTCAGATGTCAGGTTCG | 643 |
| 2DL3O4R | 7903 | AGGCCACAGGGCCCAACTCAGGTCGT | ||
| 2DL3 Optional 5 | 2DL3O5F | 13892 | ATGAAATGAGGGCCCAGAAGTGCCCTGT | 278 |
| 2DL3O5R | 14111 | CTCTGTGTGAAAACGCAGTGATTCAACTGTTT | ||
| 2DL3 Optional 6 | 2DL3O6F | 13892 | ATGAAATGAGGGCCCAGAAGTGCCCTGT | 278 |
| 2DL3O6R | 14111 | CTCTGTGTGAAAACGCAGTGATTCAACTGTTC |
Altogether, by using all eleven PCR reactions, we were able to separate ten different individual alleles or groups of alleles (KIR2DL1-G*001, -G*002, -G*003, -G*004, -G*012, *006, *008, *010, *011, *020) exhibited individually or in combination (Supplemental Fig. 2A,B). A few combinations of alleles, involving KIR2DL1-G*012 in particular, cannot be resolved with our method (Fig. 1B). It should be noted that these alleles are rare, and none of them was found in the 426 sequenced individuals.
KIR2DL2 allele typing
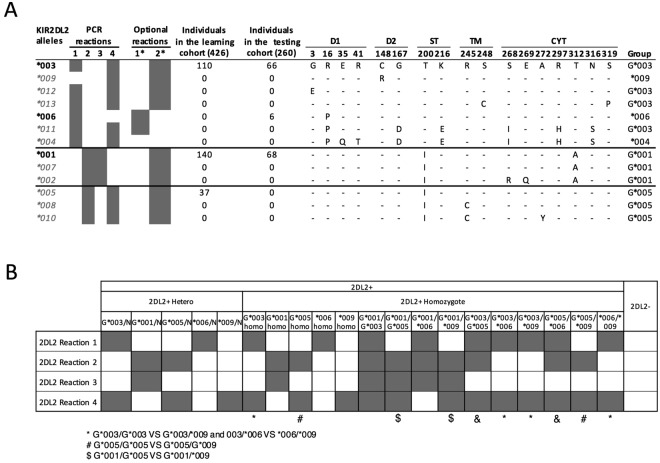
Among the 13 KIR2DL2 alleles published in EMBL-EBI IPD KIR database, only three alleles (*001, *003, *005) occur in our cohort of 426 healthy individuals (Fig. 2A). Phylogenetic analysis suggests that these three alleles typify separate functional groups (Supplemental Fig. 1B). Four ARMS PCR reactions define the three groups, and two supplemental reactions increase the resolution of the method (Table 1). With this approach, we can separate six different individual alleles or groups of alleles (KIR2DL2-G*001, -G*003, -G*005, *004, *006, *009) (Supplemental Fig. 2C,D).
Figure 2.
KIR2DL2 allele typing method. (A) Alignment of the amino acid sequences of the 13 known KIR2DL2 allelic variants. A dash indicates identity with the consensus KIR2DL2*003. Structural domains are indicated: Ig-like domains (D1 and D2), stem domain (ST), transmembrane domain (TM), and cytoplasmic domain (CYT). Four PCR reactions separate the three subgroups identified by phylogenetic analysis. Two additional PCR reactions separate alleles within subgroups. Frequencies of the alleles present in the learning cohort of 426 individuals and in the testing cohort of 260 individuals are indicated. The group identification of each allele is indicated. The alleles tested by PCR are in bold black font, and non-tested alleles are in gray italics. (B) KIR2DL2 PCR interpretation guide. PCR profiles marked by *, #, $ or & are similar and require a higher resolution of genotyping using supplemental reactions.
All KIR2DL2 reactions were optimized using DNA from 178 of the 426 healthy donors. Previously reported analysis of these same DNA samples could not completely resolve KIR2DL2*005 from KIR2DL2*00123. With our method, we did not identify the presence of KIR2DL2*005 and instead found that 21 of the 21 samples with ambiguous typing exhibited KIR2DL2*001. The presence of KIR2DL2*001 and the absence of KIR2DL2*005 in these samples were confirmed by sequencing by an independent laboratory (data not shown). Figure 2A displays the amino acid alignment of KIR2DL2 alleles and segregation of alleles by the typing methodology. Ambiguity in interpretation for certain combinations of alleles occurs, as indicated (Fig. 2B).
KIR2DL3 allele typing
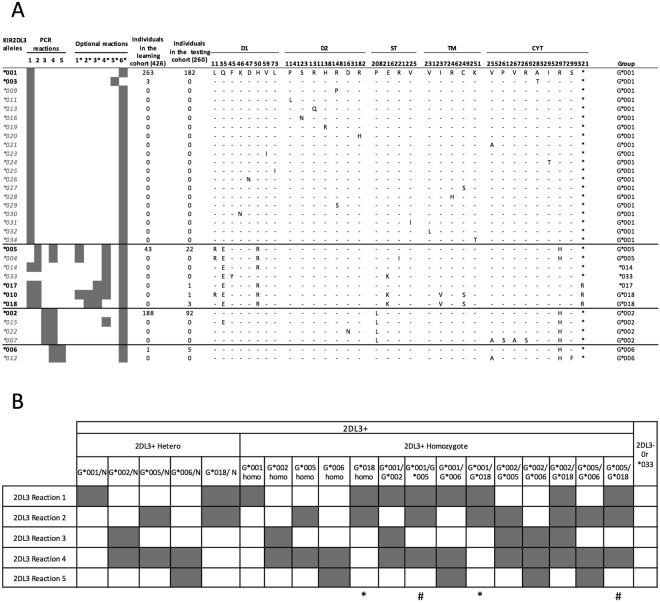
Of 34 KIR2DL3 alleles published in an EMBL-EBI IPD KIR database, only five (*001, *002, *003, *005, *006) were identified in the cohort of 426 healthy individuals (Fig. 3A). Phylogenetic analysis (Supplemental Fig. 1C) suggested four separate groups, which could be segregated by five distinct main ARMS PCR reactions (Fig. 3B), complemented by six supplemental reactions to further optimize resolution. With these eleven PCR reactions (Table 1), we could separate eleven different alleles or groups of alleles (KIR2DL3-G*001, -G*002, -G*005, *003, *006, *009, *010, *014, *015, *017, *018) (Supplemental Fig. 1E,F).
Figure 3.
KIR2DL3 allele typing method. (A) Alignment of the amino acid sequences of the 34 known KIR2DL3 allelic variants. A dash indicates identity with the consensus KIR2DL3*001, an (*) indicates a stop codon. Structural domains are indicated: Ig-like domains (D1 and D2), stem domain (ST), transmembrane domain (TM), and cytoplasmic domain (CYT). Five PCR reactions separate the four subgroups identified by phylogenetic analysis. Six additional PCR reactions separate alleles within subgroups. Frequencies of the alleles present in the learning cohort of 426 individuals and in the testing cohort of 260 individuals are indicated. The group identification for each allele is indicated. The alleles tested by PCR are in bold black font, and the non-tested alleles are in gray italics. (C) KIR2DL3 PCR interpretation guide. PCR profiles marked by * or # are similar and require a higher resolution of genotyping using supplemental reactions.
We confirmed accuracy of the typing methods for KIR2DL1, KIR2DL2, and KIR2DL3 by comparing results using this methodology for 200 donors of the validation cohorts to results generated by another laboratory by a sequence-based typing methodology, finding 99.6% concordance26,27.
Linkage disequilibrium between KIR2DL alleles
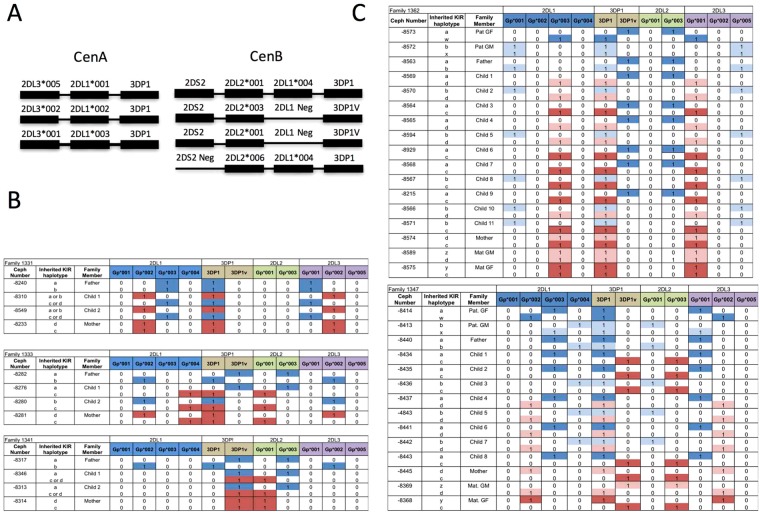
We calculated LD between alleles of KIR2DL1, KIR2DL2, KIR2DL3, KIR3DP1, and the presence or absence of gene KIR2DS2 in 260 individuals (Fig. 4A). Seven allele combinations, three belonging to the canonical centromeric haplotype-A (KIR2DL1-KIR2DL3-KIR3DP1)23,28 and four belonging to the centromeric haplotype-B, were represented in more than 95% of the donors. Of interest, the centromeric haplotype-B allele combination comprised of KIR2DL1*004, KIR2DL2*006 and the absence of KIR2DS2 was found in 2.3% of individuals typed.
Figure 4.
Linkage disequilibrium between KIR2DL alleles. (A) Linkage disequilibrium analysis identifies seven common combinations of centromeric KIR2DL alleles in a cohort of 260 individuals. (B) Allelic segregation of the KIR2DL and KIR3DP1 alleles in CEPH families. Paternal KIR2DL alleles are shown in blue; maternal alleles in red. (C) KIR2DL allele typing from three generations of CEPH family individuals demonstrates Mendelian inheritance of allele combinations established by the LD study.
To confirm the LD observed by calculation in our cohort, we also genotyped immortalized B cell lines from the Centre d’Etude Polymorphisme Humaine (CEPH). We selected 5 families and performed KIR2DL allele typing using our methodology (Fig. 4B,C). The typing results demonstrated a Mendelian inheritance of allele combinations established by the LD study.
Discussion
We have established a comprehensive genotyping method to distinguish alleles and allele groups for the KIRL2DL1, KIR2DL2 and KIR2DL3 genes. We validated the methodology using 178 donors from a learning cohort and 260 samples from a validation cohort, further confirming its robustness by sequence-based typing. Designed as a typing kit for the centromeric region of the KIR haplotype, our methodology provides a reliable, cost-effective alternative to sequencing methods that can be employed using basic laboratory equipment.
The KIR2DL1 typing method identifies the four most common KIR2DL1 alleles, in addition to the less common KIR2DL1*006 and KIR2DL1*010 alleles found in three individuals. The learning cohort of 178 individuals was mostly comprised of Caucasian individuals, while the testing cohort was more ethnically diverse. For the 136 individuals in the testing cohort for whom we could obtain ethnicity, 59.6% were Caucasian,17.6% Asian, 16.2% African-American, 3.7% Hispanic, and 2.9% mixed ethnicity. Consistent with this population diversity, our typing revealed in several individuals the presence of KIR2DL1*006, an allele previously reported to be relatively well-represented (7%) in an African-American cohort29. With our KIR2DL2 typing method, we can now faithfully resolve ambiguities between KIR2DL2*005 and KIR2DL2*001 reported with previous methodologies23. A third typing methodology validated our results26,27.
Copy number for the KIR2DL1 alleles can be estimated using typing for the framework pseudogene KIR3DP1, where the KIR3DP1V (*001, *002, *004, *007, *009, *011, *012) alleles are associated with the absence of KIR2DL1. Concerning KIR2DL2 and KIR2DL3, copy number estimation was based on the mutual non-co-expression due to the allelic relationship of the two genes for the same locus. Our method has its limitations and is accurate for content only with copy number and haplotype inferred by LD. It has previously been reported that an additional KIR2DL1, KIR2DL2 or KIR2DL3 allele occurs in 1 to 2% of the population30. To identify these individuals, a quantitative PCR assay would be informative in addition to our typing method to calculate more accurately copy number of each KIR allele31.
The LD analysis identified the seven predominant combinations of KIR2DL receptors, representing more than 95% of the 260 donors studied. Genotyping of the CEPH family further validated the utility of our method and confirmed the Mendelian inheritance of the allele combinations established by the LD study.
This typing methodology will facilitate future studies aimed at determining if functional differences exist between alleles, as suggested by phylogenetic segregation. As has previously been demonstrated for the KIR3DL1 alleles, diversity in cell surface expression, ligand affinity, and effector function for the KIR2DL alleles may combine to modulate NK education and influence innate immune response to viral pathogens and malignancy.
Methods
Genomic analyses and Primer design
All allele-coding sequences of KIR2DL1, KIR2DL2 and KIR2DL3 from the EMBL-EBI IPD KIR database sequences (http://www.ebi.ac.uk/ipd/kir/alleles.html) were included in our alignment analyses. We performed gene alignments and phylogenetic analyses using MacVector software version 13.5.5. Protein sequences of alleles for KIR2DL1 (Supplemental Fig. 1A), KIR2DL2 (Supplemental Fig. 1B) and KIR2DL3 (Supplemental Fig. 1C) were aligned and analyzed by tree building methods: neighbor joining (Uncorrected method, Best Tree) with MacVector. Genomic sequencing in a cohort of 426 European-American healthy donors previously identified nine KIR2DL1, three KIR2DL2 and five KIR2DL3 alleles respectively23. Among KIR2DL1, KIR2DL2, and KIR2DL3 alleles, four, three, and three alleles were found with >1% frequency respectively. Sequence homology was then used in conjunction with the phylogenetic analyses to categorize alleles into KIR allele subgroups, for which PCR primer combinations were then designed. Low frequency alleles were assigned to subgroups based on sequence homology in the exon coding regions. Primer pairs targeting SNPs present in each subtype group were identified and their specificity for KIR2DL1, KIR2DL2 or KIR2DL3 was confirmed using NCBI primer blast. To provide an internal control for DNA quality, an 813 bp control band derived from a conserved region of the APC gene was multiplexed into each reaction. Specific primer sequences and PCR conditions are shown in Table 1. The position of the SNP targeted is based on the following genomic sequences: for KIR2DL1 primers KIR2DL1*00303 (IPD Acc No: KIR00005), for KIR2DL2 primers KIR2DL2*0030101 (IPD Acc No: KIR00012), for KIR2DL3 primers KIR2DL3*0010101 (IPD Acc No: KIR00014).
For KIR2DL1, we designed six PCR reactions to delineate six distinct allele groups based on the coding sequences and four supplemental reactions to identify additional subgroups or individual alleles represented in the n = 426 cohort (Fig. 1A). We designed one additional reaction for KIR3DP1-3DP1V to determine KIR2DL1 copy number5. For KIR2DL2 alleles, we designed four PCR reactions to separate three distinct groups with two supplemental reactions to identify subgroups (Fig. 2A). For KIR2DL3 alleles, five PCR reactions separate alleles into four distinct groups, with six supplemental reactions to identify some subgroups or individual alleles (Fig. 3A). The design of the primers was optimized using the software AmplifX (V1.7.0, http://crn2m.univ-mrs.fr/pub/recherche/equipe-t-brue/jullien-nicolas/programmation/amplifx/), following the principles of amplification refractory mutation system (ARMS)-PCR32. All primers are ARMS-PCR primers, with the exceptions of the control primers and KIR2DL1 allele primer pair #7, and were designed for an annealing temperature of 63 °C (Table 1). We used a testing cohort of 260 healthy individuals whose KIR genotypes had been identified by sequence-based typing to verify the specificity of the primers, as well as 178 DNA from the European-American healthy donors (Figs 1A–3A).
PCR Reactions
The ProFlex PCR system (Life Technologies) was used to optimize and validate the PCR reaction conditions. Each 20 µL reaction included 50–100 ng of DNA and was prepared with Taq polymerase (0.25 µL), dNTP (0.5 µL) and PCR buffer (2 µL) (Roche). Each primer was used at a final concentration of 0.5 µM. All reactions used the following PCR template: 95 °C 5 min, (95 °C 15 s, 63 °C 20 s, 72 °C 1 min) X 40 cycles, 72 °C 7 min, with the exception of 2DL2 PCR reaction #4, which utilized the following conditions: 95 °C 5 min, (95 °C 15 s, 63 °C 20 s, 72 °C 2.5 min) X 40 cycles, 72 °C 7 min. (Control primers were designed to amplify a fragment of the APC gene. All reactions utilize reaction-specific primers and the control primers, except for KIR2DL1 PCR reaction 7 and KIR2DL2 PCR reaction 4, which do not include control primers. We analyzed all PCR products using electrophoresis on 1.5% agarose gels for 40 min at 125 V. Control bands (813 bp) confirmed DNA quality. Specific product sizes ranged from 0.2–2.3 kb (Table 1)
PCR interpretation
The KIR2DL1, KIR2DL2 and KIR2DL3 PCR profiles are displayed respectively in Figs 1B–3B. In cases where the observed results prompt multiple interpretations, supplemental reactions can be used to increase resolution. A few very rare allelic combinations cannot be discerned from each other and are indicated as ambiguous combinations in the table footnotes. Gel electrophoresis and interpretation of the basic and supplemental reactions for KIR2DL1, KIR2DL2, and KIR2DL3 are shown in Supplemental Fig. 2.
Cells, DNA Sources and Preparation
Genomic DNA was extracted from cell lines, frozen peripheral blood mononuclear cells (PBMC) and whole blood using blood mini kits according to the manufacturer’s instructions (Qiagen). EBV-immortalized cell lines derived from multi-generational families were produced by the Centre d’Etude Polymorphisme Humaine (CEPH) (http://www.cephb.fr/en/familles_CEPH.php-presentation). Samples were anonymized by the CEPH. DNA samples from unrelated hematopoietic stem cell donors were collected under National Marrow Donor Program (NMDP) Institutional Review Board-informed research consent and provided by the NMDP Research Repository. We collected PBMC from consenting healthy human donors at MKSCC and the New York Blood Center, following approval from the MSKCC Institutional Review Board. Additional PBMC were isolated from buffy coats obtained from healthy volunteer donors via the New York Blood Center (http://nybloodcenter.org/). The MSKCC IRB waived the need for additional research consent for anonymous NYBC samples.
Statistics
Linkage disequilibrium (LD) between pairs of KIR alleles was calculated according to Mattiuz et al.33, and the significance of LD values was assessed by χ2 analysis.
Statement of Informed consent
All methods were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Acknowledgements
This work was supported by funding from NIH U01 AI069197 and P01 CA23766 to K.C.H., the Francois Wallace Monahan Fellowship to J.B.L., and the Cancer Center Core grant (NIH P03 CA008748) to MSKCC. We thank DKMS Life Science Lab for KIR allele sequencing of our cohort.
Author Contributions
J.B.L. designed the method and primers. J.B.L. and A.K. performed the experiments. J.B.L. and J.E.B. created the sample banks. J.B.L. and K.C.H. wrote the manuscript. All authors discussed the results and contributed to the final manuscript. K.C.H. supervised the project.
Competing Interests
The authors have the following interests: The method of KIR2DL alleles typing described in this manuscript was submitted by Drs Jean-Benoît LE LUDUEC and Katharine C. Hsu (Methods and Kits for typing KIR2DL alleles, U.S. Provisional Patent Application No. PCT/US2017/054172). The authors confirm that this does not alter their adherence to Scientific Reports policies on materials and data sharing.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/18/2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33135-1.
References
- 1.Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55:221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 2.Parham P, Guethlein LA. Genetics of Natural Killer Cells in Human Health, Disease, and Survival. Annu Rev Immunol. 2018;36:519–548. doi: 10.1146/annurev-immunol-042617-053149. [DOI] [PubMed] [Google Scholar]
- 3.Wong-Baeza I, et al. KIR3DL2 binds to HLA-B27 dimers and free H chains more strongly than other HLA class I and promotes the expansion of T cells in ankylosing spondylitis. J Immunol. 2013;190:3216–3224. doi: 10.4049/jimmunol.1202926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065X.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 5.Hsu KC, et al. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 6.Nakimuli A, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci USA. 2015;112:845–850. doi: 10.1073/pnas.1413453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khakoo SI, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 8.Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol. 2018;50:102–111. doi: 10.1016/j.coi.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giebel S, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 10.Hsu KC, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. 2005;105:4878–4884. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J Immunol. 2016;196:3398–3410. doi: 10.4049/jimmunol.1502469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders PM, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med. 2016;213:791–807. doi: 10.1084/jem.20152023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yawata M, et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boudreau JE, et al. KIR3DL1/HL A-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation. J Clin Oncol. 2017;35:2268–2278. doi: 10.1200/JCO.2016.70.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bari R, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. 2006;177:5347–5357. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- 18.Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. 2013;190:6198–6208. doi: 10.4049/jimmunol.1300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilton HG, et al. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol. 2015;195:3160–3170. doi: 10.4049/jimmunol.1501358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendzekhadze K, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci USA. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norman PJ, et al. Defining KIR and HLA Class I Genotypes at Highest Resolution via High-Throughput Sequencing. Am J Hum Genet. 2016;99:375–391. doi: 10.1016/j.ajhg.2016.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudreau JE, Le Luduec JB, Hsu KC. Development of a novel multiplex PCR assay to detect functional subtypes of KIR3DL1 alleles. PLoS One. 2014;9:e99543. doi: 10.1371/journal.pone.0099543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vierra-Green C, et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS One. 2012;7:e47491. doi: 10.1371/journal.pone.0047491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin MP, Single RM, Wilson MJ, Trowsdale J, Carrington M. KIR haplotypes defined by segregation analysis in 59 Centre d’Etude Polymorphisme Humain (CEPH) families. Immunogenetics. 2008;60:767–774. doi: 10.1007/s00251-008-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nature Reviews Immunology. 2013;13:133–144. doi: 10.1038/nri3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruschke, J. et al. An updated neXtype algorithm for high-throughput KIR genotyping delivering allele-level resolution. In KIR Workshop, Cambridge, UK (2017).
- 27.Wagner, I. et al. Allele-level KIR genotyping of almost 200,000 registry samples – towards a new standard profile for potential blood stem-cell donors. In KIR Workshop, Cambridge, UK (2017).
- 28.Shilling HG, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 29.Hou L, et al. In contrast to other stimulatory natural killer cell immunoglobulin-like receptor loci, several KIR2DS5 alleles predominate in African Americans. Hum Immunol. 2009;70:733–737. doi: 10.1016/j.humimm.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beziat V, et al. Influence of KIR gene copy number on natural killer cell education. Blood. 2013;121:4703–4707. doi: 10.1182/blood-2012-10-461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little, S. Amplification-refractory mutation system (ARMS) analysis of point mutations. Current protocols in human genetics Chapter 9, Unit 98, 10.1002/0471142905.hg0908s07 (2001). [DOI] [PubMed]
- 33.Mattiuz, P. L., Piazza, I. D. Ceppelini, A.& Bodmer, R. WF. In Terasaki P, ed. Histocompatibility Testing1970, 193–205 (1971).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.